Abstract
Caves are special environments that harbour an incredible diversity of life, including fungal species. Brazilian caves have been demonstrated to be biodiversity hotspots for known and unknown fungal species. We investigated the richness of culturable fungi in a tropical cave in Brazil by isolating these microorganisms from the sediment and air. The fungal abundance of colony-forming units (CFUs) was 3 178 in sediment and 526 in air. We used morphological features and phylogenetic analyses of actin (actA), calmodulin (cmdA), internal transcribed spacer regions and intervening 5.8S rRNA (ITS), large subunit (LSU) rDNA, RNA polymerase II second largest subunit (rpb2), translation elongation factor 1-alpha (tef1), and β-tubulin (tub2) genes to identify these isolates. Forty-one species belonging to 17 genera of Ascomycota and two of Basidiomycota were identified, and the genus Aspergillus was most commonly observed in the cave (13 taxa). Twenty-four species were found in sediment (16 exclusives) and 25 species were found in air (17 exclusives). In this study, we introduced a new genus (Pseudolecanicillium gen. nov.) in the family Cordycipitaceae and six new species (14 % of the total taxa identified) of fungal isolates obtained from sediment and air: Aspergillus lebretii sp. nov., Malbranchea cavernosa sp. nov., Pseudohumicola cecavii sp. nov., Pseudolecanicillium caatingaense sp. nov., Talaromyces cavernicola sp. nov., and Tritirachium brasiliense sp. nov. In addition, we built a checklist of the fungal taxa reported from Brazilian caves. Our results highlight the contribution of Brazilian caves to the estimation of national and global fungal diversity.
Citation: Alves VCS, Lira RA, Lima JMS, Barbosa RN, Bento DM, Barbier E, Bernard E, Souza-Motta CM, Bezerra JDP (2022). Unravelling the fungal darkness in a tropical cave: richness and the description of one new genus and six new species. Fungal Systematics and Evolution 10: 139–167. doi: 10.3114/fuse.2022.10.06
Keywords: Aspergillus, Caatinga dry forest, cave environment, fungal taxonomy, novel taxa
INTRODUCTION
Fungi are cosmopolitan organisms that inhabit aquatic and terrestrial environments (Peay et al. 2016). Fungal species also inhabit extreme environments (Tiquia-Arashiro & Grube 2019, Coleine et al. 2022), and caves could be one of these extreme environments in which fungal species play key ecological roles (Ferreira et al. 2000, Nieves-Rivera 2003, Nováková 2009). Caves are natural subterranean cavities that may have a different lithological origin (e.g. carbonate and ferruginous caves, Travassos 2019) and harbour a special subterranean biota, with several species of fauna and flora in the speleological environment (Ogórek et al. 2013, Piló & Auler 2013). In this subterranean environment, various species have been reported and described as troglobites, troglophiles, and trogloxenes (Zhang et al. 2017, 2021). Troglobites are those living only in the subterranean environments (called hypogeal), the troglophiles live in the epigeal (external) and hypogeal environment, completing their life cycle inside or outside the caves, and trogloxenes depend on the epigean as part of their life cycle (Holsinger & Culver 1988, Pinto-da-Rocha 1993, Bento et al. 2021). Caves have a typical environment with low variations in abiotic conditions such as light, temperature, air current, and relative humidity (Lobo & Boggiani 2013).
Cave fungal diversity has been reported over the years (Vanderwolf et al. 2013) with an emphasis on airborne fungi, a group whose spores are disseminated by the air (Flores & Onofre 2010). Cave environments have higher chances to harbour special fauna, flora, and microorganisms (including fungi) that have a high capacity to tolerate abiotic and biotic factors (Nováková 2009, Bastian et al. 2010, Ogórek et al. 2013). Several studies have been conducted in caves to determine their fungal diversity (Vanderwolf et al. 2013, Cunha et al. 2020, Zhang et al. 2021), and recently, this hypogeal diversity was reported to have originated in the surface environment (Zhang et al. 2018).
The Brazilian territory has approximately 22 809 known caves, but this is estimated to be 10 % of the total number of caves in Brazil, emphasising the importance of speleological heritage in the country (Jansen et al. 2012, ICMBio/CECAV 2022). The North-eastern region of Brazil currently has approximately 4 303 known caves distributed in nine states, most of which are in Bahia (1 694) and Rio Grande do Norte (1 365) (ICMBio/CECAV 2022). According to Auler & Zogbi (2005), the North-eastern region of Brazil has high potential for new cave discoveries. This region is mainly characterised by the Caatinga domain, a part of the dry diagonal of South America, and is recognised as one of the most diverse dry forests in the world (Silva et al. 2017). Bat caves, that is, caves harbouring exceptionally high bat populations (some with more than 100 000 bats), are known in the North-eastern region of Brazil (Otálora-Ardila et al. 2019), and their mycobiome has recently been unravelled (Cunha et al. 2020, Pereira et al. 2022). Due to the presence of thousands of bats in these caves, there is a large deposition of guano, which is a special substrate for the growth of fungal species (Ogórek 2016).
The cave environment frequently harbours rare species of fungi, or even species not yet described, mainly in association with minimally explored substrates, which have great potential for the description of new species (Taylor et al. 2013, Cunha et al. 2020). Nearly 2 000 species of fungi are currently known from caves (see Zhang et al. 2021), but only 180 fungal taxa have been reported from Brazilian caves (Ferreira et al. 2000, Pedro & Bononi 2007, Taylor et al. 2013; Taylor et al. 2014, Paula et al. 2016, Hornick 2017, Crous et al. 2018, Fonseca et al. 2019, Cunha et al. 2020, Carvalho et al. 2022, Pereira et al. 2022). Less than 12 caves in Brazil have had their mycobiome studied, indicating a large knowledge gap on cave fungal diversity in such a megadiverse country. The most recent study on fungi from a Brazilian cave (Cunha et al. 2020) showed 59 taxa belonging to 39 genera obtained from the air, guano, and body of bats from a bat cave in the Caatinga. Pereira et al. (2022) surveyed another bat cave in the same region and reported eight Cladosporium spp., including the description of two new species. Carvalho et al. (2022) studied culturable fungi from bat ectoparasites of a cave in the Caatinga and described new species in Allophoma and Pyrenochaetopsis. Such studies highlight the great potential for new fungal discoveries in the country, and a potentially higher contribution to global fungal diversity. Moreover, these studies emphasized the importance of the mycobiome for cave environments and the conservation of subterranean environments (Cunha et al. 2020).
In this study, we investigated the cultivable fungal richness in sediment and air in a tropical cave in the Caatinga dry forest in Brazil. Based on our data and a literature review, we built a checklist of fungal taxa reported from Brazilian caves since the first known study in 1963. Our data may also help Brazilian environmental authorities plan conservation policies for the tourist trade.
MATERIALS AND METHODS
Study area
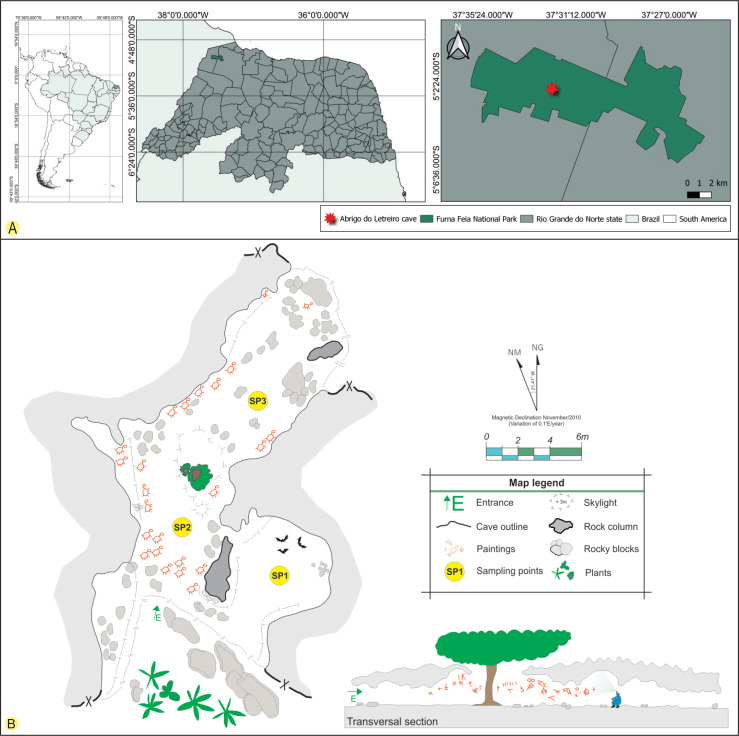
The Abrigo do Letreiro cave is located in Furna Feia National Park (5°4’14.88’’S and 37°32’1.51’’W), between the municipalities of Baraúna and Mossoró in the state of Rio Grande do Norte, North-eastern Brazil (Fig. 1). The national park was created in 2012 to protect the conservation area of a speleological complex and the Caatinga dry forest. The park has an area of approximately 8 494 ha and 25 322 ha of a buffer zone, totalling 33 816 hectares (Brasil 2012). The region’s climate is BWh’ (arid and hot), according to the Köppen’s classification (Dubreuil et al. 2018), and is characterised by strong insolation with high temperatures throughout the year (average annual temperature: 27.5 °C), relative humidity of approximately 70 %, and annual precipitation around 673.9 mm (ICMBio 2020).
Fig. 1.
A. The geographical location of the Abrigo do Letreiro cave in the Furna Feia National Park, Brazil. B. Cave sketch showing the sampling points (SP1, SP2 and SP3). The cave sketch was adapted from one drawn by the CECAV/ICMBio-MMA, Brazil.
The cave has a limestone lithology and is formed by a single chamber of approximately 64 m of horizontal projection and an altitude of approximately 154 m, with two entrances at its ends and a skylight near the centre (Sobrinho et al. 2016). The morphology of the Abrigo do Letreiro cave facilitates the flow of air from the external environment, which indicates that it is a cavity with a high level of energy (Heaton 1986). The Abrigo do Letreiro cave has a very common tree in areas of the Caatinga dry forest, Erythrina velutina (Fabaceae), at the centre of the cave in a portion where there is a skylight. In addition to its ecological and speleological importance, it has anthropological significance because several cave paintings with geometric traditions and symbolist styles are distributed along the walls and ceiling of the cave (Bento 2013) (Fig. 2).
Fig. 2.

Abrigo do Letreiro cave in the Furna Feia National Park, Brazil. A. Outside skylight view and the tree Erythrina velutina (Fabaceae). B. Inside skylight view and the tree E. velutina. C. Sampling point 2 and cave paintings with geometric tradition and symbolist style distributed along the walls and ceiling of the cave. D. A small colony of bats (Peropteryx macrotis) at the sampling point 1. E. Petri dishes used to sample airborne fungi at the sampling point 2. Photos were taken by D.M. Bento.
The collection was authorised by the Ministério do Meio Ambiente (MMA)/Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) (SISBIO number 54274).
Sampling points
The cave was treated as a single chamber and three collection points were chosen from the cave’s main entrance: the first was 5.7 m (point 1) from the entrance, followed by the second 5.2 m (point 2), and the third 14 m (point 3). The points were chosen based on easy access to the cave environment and the abiotic conditions observed during the collection. Point 1 had a small colony of bats (Peropteryx macrotis), point 2 was located near the skylight, and point 3 was located deep within the cave (Fig. 1).
Isolation of airborne fungi
Airborne fungi were collected, using the sedimentation methodology, on a culture medium contained in Petri dishes, as described by Cunha et al. (2020). At each sampling point, three 90 mm Petri dishes containing Sabouraud dextrose (SAB) agar supplemented with chloramphenicol (100 mg/L) were used for fungal isolation. The plates were placed equidistant from each other, 1 m from the cave floor, and opened for 20 min. After exposure, the plates were closed, identified, packaged, and transported to the laboratory. Plates were incubated at 28 °C for up to 14 d in the dark. Fungal colonies were observed daily and counted. Selected colonies were isolated, purified, and preserved for further identification.
Isolation of fungi from the sediment of the cave
Approximately 10 g of cave superficial sediment was collected at each sampling point using sterilised 50 mL centrifuge tubes. The sediment at Point 1 was composed of soil, bat guano, and rodent faeces (Kerodon rupestris), Point 2 contained soil and rodent faeces (K. rupestris), and Point 3 was composed of soil and bat guano. In the laboratory 1 g of each sampled sediment was placed in a 250 mL Erlenmeyer flask containing 9 mL of distilled and sterilised water plus chloramphenicol (0.1 mg/L). The flasks were manually shaken and used to make dilutions of 10−2, 10−3, and 10−4, from which 1 mL was transferred to Petri dishes with Brain and Heart Infusion (BHI) agar and SAB media plus chloramphenicol (100 mg/L). Petri dishes were incubated at 28 °C for at least 7 d in the dark, and representative isolates of the total fungal colonies grown were chosen and purified using SAB plus chloramphenicol (100 mg/L) (Cunha et al. 2020).
Fungal isolates
Ex-type cultures are deposited in the University Recife Mycology (URM) culture collection (Micoteca URM Profa. Maria Auxiliadora Cavalcanti, WDCM 604), and holotypes (as permanent slide preparations) in the URM fungarium (Herbário URM Pe. Camille Torrend) at the Universidade Federal de Pernambuco (UFPE), Recife, Brazil (Barbosa et al. 2020). In addition, all isolates are deposited in the working collection of the Laboratório de Taxonomia e Biotecnologia Utilizando Fungos housed at the Micoteca URM/UFPE and ex-type strains and representative isolates in the working collection FCCUFG of Jadson D.P. Bezerra housed at the Laboratório de Micologia of the Instituto de Patologia Tropical e Saúde Pública, Universidade Federal de Goiás, Goiânia, Brazil.
Fungal identification
Morphology
Strains of the new genus and species were grown on various culture media. For Aspergillus and Talaromyces species, the isolates were cultivated on malt extract agar (MEA), Czapek agar (CZ), Czapek yeast extract agar (CYA), CYA supplemented with 5 % NaCl (CYAS), dichloran 18 % glycerol agar (DG18), oatmeal agar (OA), yeast extract sucrose agar (YES) and creatine sucrose agar (CREA) (Samson et al. 2010). All agar media were incubated at 25 °C for 7 d in the dark. Additional CYA and MEA plates were incubated at 10, 30, and 37 °C and the colony diams were measured after 7 d of incubation in the dark. Microscopic observations were made of colonies grown on MEA (Samson et al. 2010). For Malbranchea species and the new genus, isolates were subcultured on potato dextrose agar (PDA), OA, and synthetic nutrient-deficient agar (SNA) and incubated in the dark at 25 °C for 14 or 28 d (Zhang et al. 2021). The isolates of Pseudohumicola were cultivated on PDA, OA, SNA, and MEA and incubated at 25 °C for 14 d in the dark (Zhang et al. 2017, Wang et al. 2019). For Tritirachium species, the isolate was cultivated on CYA, MEA, OA, PDA, SNA, and YES and incubated at 25 °C for 7 d in the dark (Bezerra et al. 2020). Colours of colonies were evaluated using the colour charts of Rayner (1970). Lactic acid (60-80 %) was used as the mounting fluid, and 96 % ethanol was used to remove excess conidia. For microscopic analysis, at least 15–30 reproductive structures were analysed to describe each new species. The Nikon Eclipse Ni microscope, equipped with Nikon DS-Fi2 camera, was used to capture images using NIS-Elements AR v. 4.20 software and photos were later edited.
DNA extraction, PCR amplification, and sequencing
Total genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA), according to the manufacturer’s protocol. Actin (actA), calmodulin (cmdA), internal transcribed spacer regions and intervening 5.8S rRNA (ITS), large subunit (LSU) rDNA, RNA polymerase II second largest subunit (rpb2), translation elongation factor 1-alpha (tef1), and β-tubulin (tub2) genes were amplified using the primer pairs ACT-512F and ACT-783R (Carbone & Kohn 1999), CMD5 and CMD6 (Hong et al. 2006), ITS1 and ITS4 (White et al. 1990), LR0R and LR5 (Vilgalys & Hester 1990), rpb2-5F2 and frpb2-7cR (Liu et al. 1999, Sung et al. 2007), 983F and 2218R (Rehner & Buckley 2005), EF-728F and EF-986R (Carbone & Kohn 1999), and Bt2a and Bt2b (Glass & Donaldson 1995), respectively. PCR amplification, sequencing, and sequence analysis were performed as described by Bezerra et al. (2017a, b) and Barbosa et al. (2020). The DNA sequences generated in this study have been deposited in GenBank (Supplementary Table S1).
Phylogenetic analyses
Phylogenetic analyses were performed using our sequences with reference sequences retrieved from the GenBank database, following recently published papers (e.g. Aspergillus – Houbraken et al. 2020, Cordycipitaceae – Mongkolsamrit et al. 2020 and Wang et al. 2020, Malbranchea – Rodríguez-Andrade et al. 2021, Pseudohumicola – Wang et al. 2022, Talaromyces – Zhang et al. 2021 and Wang & Zhuang 2022, Tritirachium – Bezerra et al. 2020). Sequences were aligned using the online tool MAFFT v. 7 (Katoh et al. 2013) and manually edited using MEGA v.7 (Kumar et al. 2016). Initially, each alignment, along with the combined dataset, was analysed based on maximum likelihood (ML) analysis using RAxML-HPC BlackBox v. 8.2.12 (Stamatakis et al. 2008) at the CIPRES Science Gateway (Miller et al. 2010). Later, the combined datasets were analysed based on Bayesian inference (BI) analysis conducted using MrBayes v. 3.2.7a (Ronquist et al. 2012) with XSEDE at the CIPRES Science Gateway. BI analysis was conducted with 1 × 106 generations and a burning value of 25 %, with chains sampled every 1 000 generations, and ML analysis with 1 000 bootstrap replicates. The best nucleotide model for BI analysis was estimated using MrModelTest v. 2.3 software (Nylander 2004), and the GTR + I + G model was used for all ML analyses. Phylogenetic trees were visualised using FigTree software (Rambaut 2010). Values ≥ 0.95 BI posterior probability (BPP) and 70 % ML bootstrap support (ML-BS) are shown near the nodes. The alignments were deposited in TreeBASE (study ID 29511).
Data analyses
The number of colonies (colony-forming units, CFU) was considered to be the fungal abundance, and the values were analysed based on ANOVA using the F test with a probability level of 5 %, and Tukey’s test was applied for significant results (Cunha et al. 2020). Airborne fungi and those obtained from the sediment were evaluated as the total number of CFUs and richness of fungal species (Cunha et al. 2020).
Checklist of fungal species in Brazilian caves
Based on the study by Vanderwolf et al. (2013), we conducted an active search for papers reporting mycological studies in Brazilian caves using the keywords “Brazilian cave” and “cave fungi in Brazil” in scientific and popular platforms (e.g. Web of Science, Scopus, SciELO, and Portal de Periódicos da CAPES). The obtained results were treated, and fungal taxa names were extracted from these papers to build a checklist of mycospeleological studies in Brazil. In this checklist, we observed the Brazilian state/region where the study was conducted, years of publication, and fungal names that were taxonomically checked using the Index Fungorum and MycoBank databases.
RESULTS
General fungal abundance and richness
The abundance of cultivable fungi in Abrigo do Letreiro cave was determined based on the number of colony forming-units (CFUs) at each sampling point (Supplementary Table S2). The fungal abundance was 526 CFU in the air, with point 3 having the highest number of CFU (200); however, no significant difference was observed among the numbers of CFU at each point (F = 0.6145). The number of 3 178 CFU were found in the sediment, with point 1 having the highest number (1 254 CFU); however, no significant difference was observed among the numbers of CFU at each point (F = 2.0075). Seventy-seven isolates (40 from air and 37 from sediment) were selected among UFC observed and identified based on morphology (e.g., genus level) and DNA sequence analyses. These isolates were identified as belonging to 19 genera of Ascomycota and Basidiomycota, with the phylum Ascomycota having the greatest number of taxa (39); only two taxa were Basidiomycota. In Ascomycota (17 genera), Aspergillus had the largest number of taxa (13), followed by Cladosporium (4) and Penicillium (5). Fourteen other ascomycetous genera were represented by at least one or two taxa (e.g. Edenia and Humicola). In Basidiomycota, the identified isolates were Sympodiomycopsis (from the air) and Tritirachium (from the sediment) (Table 1).
Table 1.
Fungal taxa richness. List of fungal taxa isolated from air and sediment in the Abrigo do Letreiro cave, Furna Feia National Park, Rio Grande do Norte, North-eastern region, Brazil. P = fungal taxon present (observed) and – = fungal taxon absent (not observed).
| Taxon | Recorda | Air | Sediment | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Collection points | Point 1 | Point 2 | Point 3 | Point 1 | Point 2 | Point 3 | |
| Ascomycota | |||||||
| Alternaria jacinthicola | S | – | – | P | – | – | – |
| Amesia sp. | – | – | – | – | P | – | |
| Aspergillus alboluteus | – | – | – | P | – | – | |
| Aspergillus brunneoviolaceus | – | – | P | – | – | – | |
| Aspergillus cf. niger | – | P | – | – | – | – | |
| Aspergillus dimorphicus | S | – | P | – | – | P | – |
| Aspergillus eburneocremeus | S | – | – | – | – | P | – |
| Aspergillus germanicus | – | P | P | – | – | – | |
| Aspergillus lebretii sp. nov. | S | – | – | – | – | – | – |
| Aspergillus neoniger | P | – | – | – | – | – | |
| Aspergillus sp. | – | – | – | – | P | – | |
| Aspergillus sp. section Aspergillus | – | – | – | P | – | – | |
| Aspergillus stellatus | P | – | – | – | P | – | |
| Aspergillus sydowii | P | – | – | P | – | P | |
| Aspergillus tubingensis | – | P | – | P | – | – | |
| Candida sp. | – | P | – | – | – | – | |
| Cercospora vignigena | S | P | – | – | – | – | – |
| Cercospora cf. canescens | S | P | – | – | – | – | – |
| Cladosporium oxysporum | P | – | – | – | P | P | |
| Cladosporium subuliforme | – | – | – | – | – | P | |
| Cladosporium tenuissimum | – | – | P | – | P | – | |
| Cladosporium xanthocromaticum | S | P | – | – | – | – | – |
| Edenia gomezpompae | G, S | – | – | P | – | – | – |
| Gymnascella hyalinospora | – | – | – | P | – | – | |
| Humicola lutea | S | – | – | – | P | – | – |
| Humicola sp. | – | – | – | – | P | – | |
| Lasiodiplodia iranensis | G, S | – | P | P | – | – | – |
| Leptosphaerulina sp. | – | P | – | – | – | – | |
| Malbranchea cavernosa sp. nov. | S | P | – | – | P | – | – |
| Neocosmospora sp. | – | – | – | P | – | – | |
| Ovatospora sp. | G | – | – | – | P | – | – |
| Pseudohumicola cecavii sp. nov. | G, S | P | – | – | P | – | – |
| Pseudolecanicillium caatingaense gen. et sp. nov. | G, S | P | – | – | – | – | – |
| Penicillium citrinum | – | – | – | – | P | – | |
| Penicillium copticola | – | – | – | P | – | – | |
| Penicillium sp. section Fasciculata | – | – | – | – | P | – | |
| Penicillium sp. section Paradoxa | – | – | – | – | P | – | |
| Penicillium guanacastense | – | P | – | – | – | – | |
| Talaromyces cavernicola sp. nov. | S | P | – | – | – | – | – |
| Basidiomycota | |||||||
| Sympodiomycopsis paphiopedili | G, S | – | P | – | – | – | – |
| Tritirachium brasiliense sp. nov. | S | – | – | – | P | – | – |
a G = genus and S = species first record in a cave environment.
Airborne fungi
Morphological and phylogenetic analyses of the DNA sequences identified 40 isolates of airborne fungi belonging to 25 species (Table 1). Considering the sampling points defined from the main entrance of the cave, point 1 had the highest richness (12 taxa), followed by points 2 (eight) and 3 (six). Aspergillus was the most representative genus in Ascomycota with nine taxa, being present at all sampling points. Other genera commonly reported as airborne in several environments (including caves), such as Cladosporium, Penicillium, and Talaromyces, were also identified in our study. Interestingly, at point 1, we identified isolates of the new genus described below in the family Cordycipitaceae (a family with entomopathogenic species) and a keratinophilic fungus (Malbranchea). At points 2 and 3, we found isolates reported as phytopathogenic fungi (Cercospora and Lasidiodiplodia), and at point 2, a representative of Candida. At point 3, we found a representative of the genus Edenia which was first isolated from plants. Nine airborne fungal isolates are described below as belonging to one new genus and five species, representing 20 % of the identified taxa.
Fungi from sediment
Thirty-seven fungal isolates obtained from cave sediment were identified as twenty-four species and taxa belonging to 10 genera (Table 1). Nine of the genera were Ascomycota, with Aspergillus being the most commonly reported (eight taxa), and Tritirachium was the only basidiomycete genus found in the sediment. The sediment from point 1 had the greatest number of species (12), followed by points 2 (11) and 3 (three). Only Aspergillus sydowii (points 1 and 3) and Cladosporium oxysporum (points 2 and 3) were found at more than one collection point. Nine isolates from the sediments were identified as eight putative new species (33 % of the identified taxa).
Brazilian checklist of mycospeleological studies
We identified 13 mycospeleological studies in Brazil published between 1963 and 2022 (Supplementary Table S3). Among the five macro regions of Brazil, one study was conducted in the Amazon (North), seven in the South-east region, five in the North-east region, and no studies were found in the South and Central West regions. Among the fungal taxa, the largest number were Ascomycota, with the genera Aspergillus, Candida, Cladosporium, and Penicillium the most commonly reported. A few taxa were included in the Basidiomycota and Mucoromycota phyla. In these studies, only three new species were introduced based on fungal isolates obtained from the cave environment. Our study enriches this list with a new genus and six new fungal species found in Brazilian caves.
Phylogeny of new species
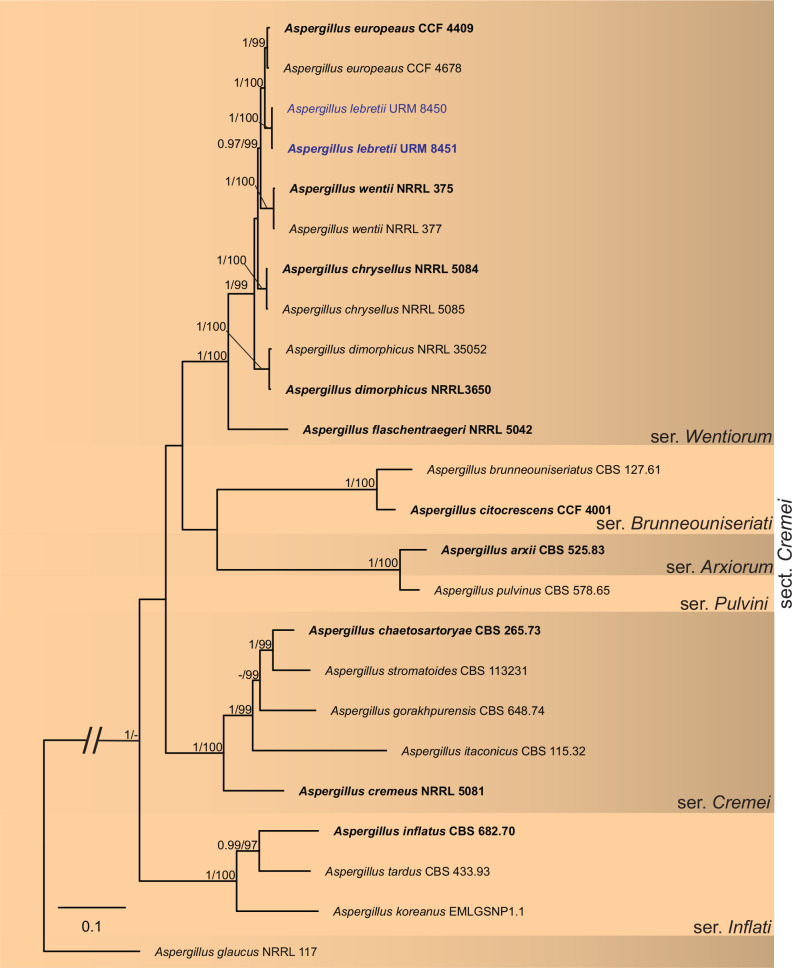
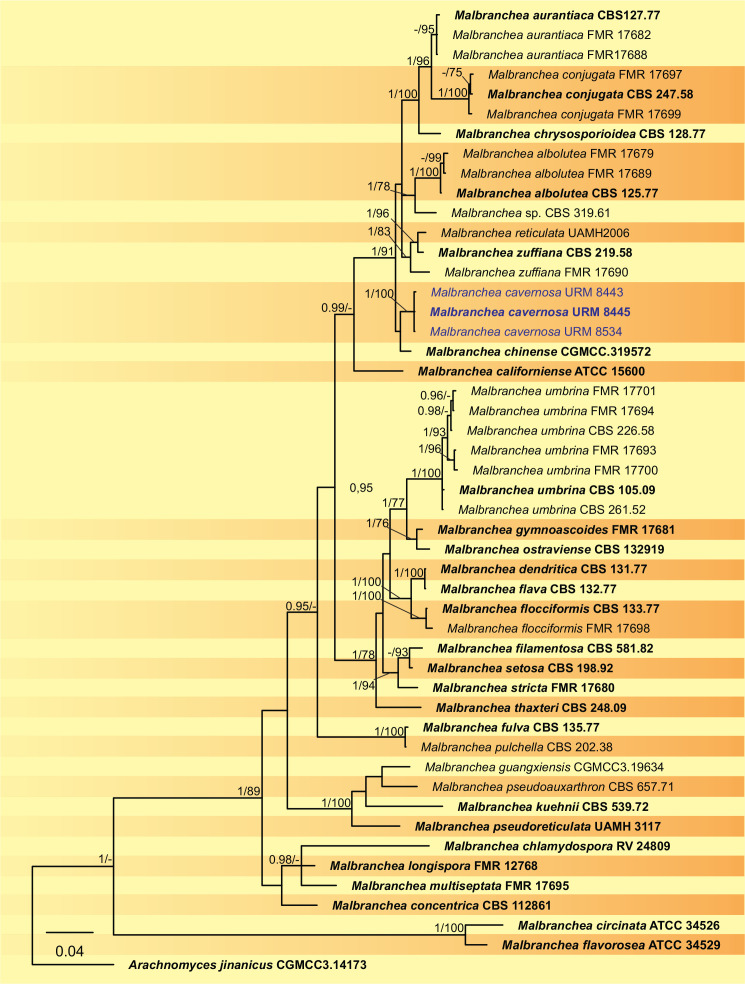
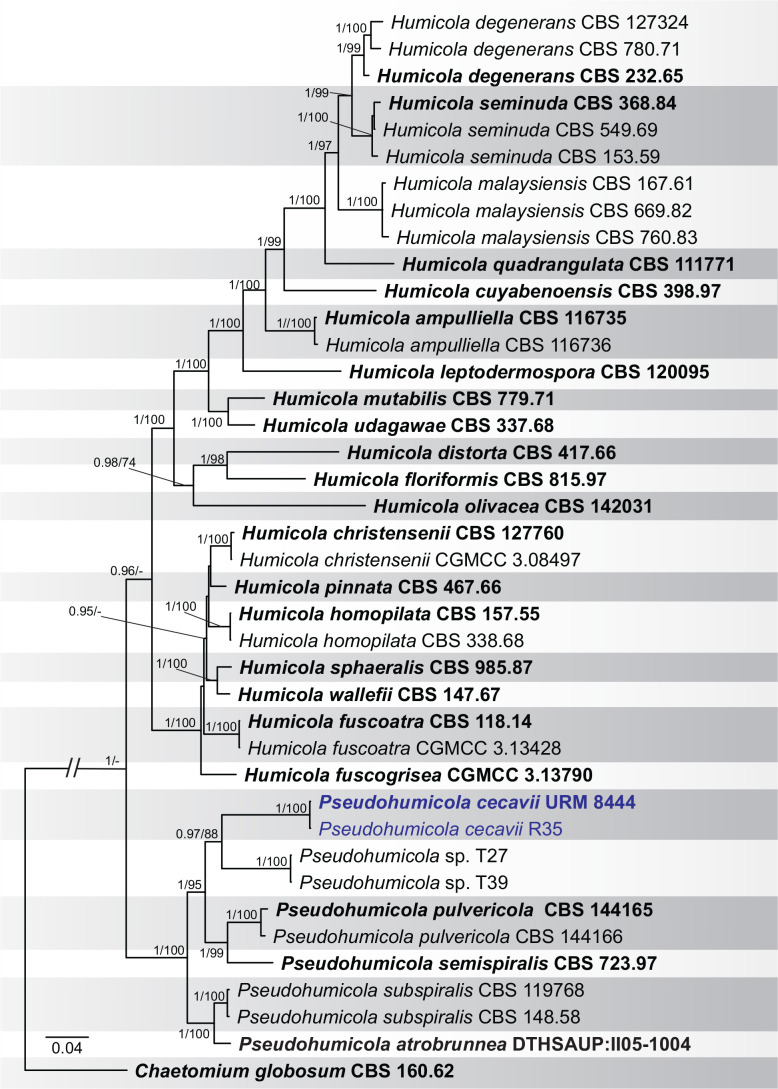
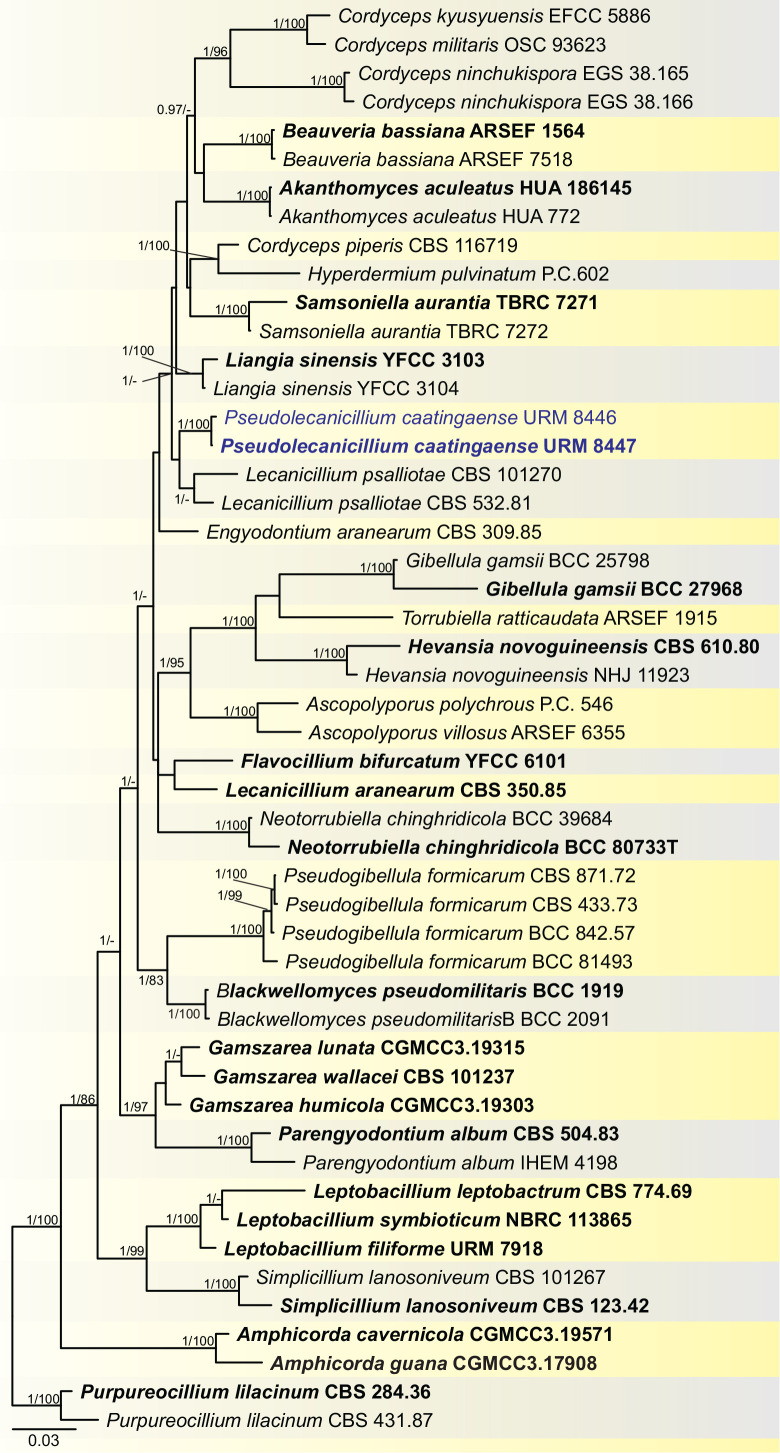
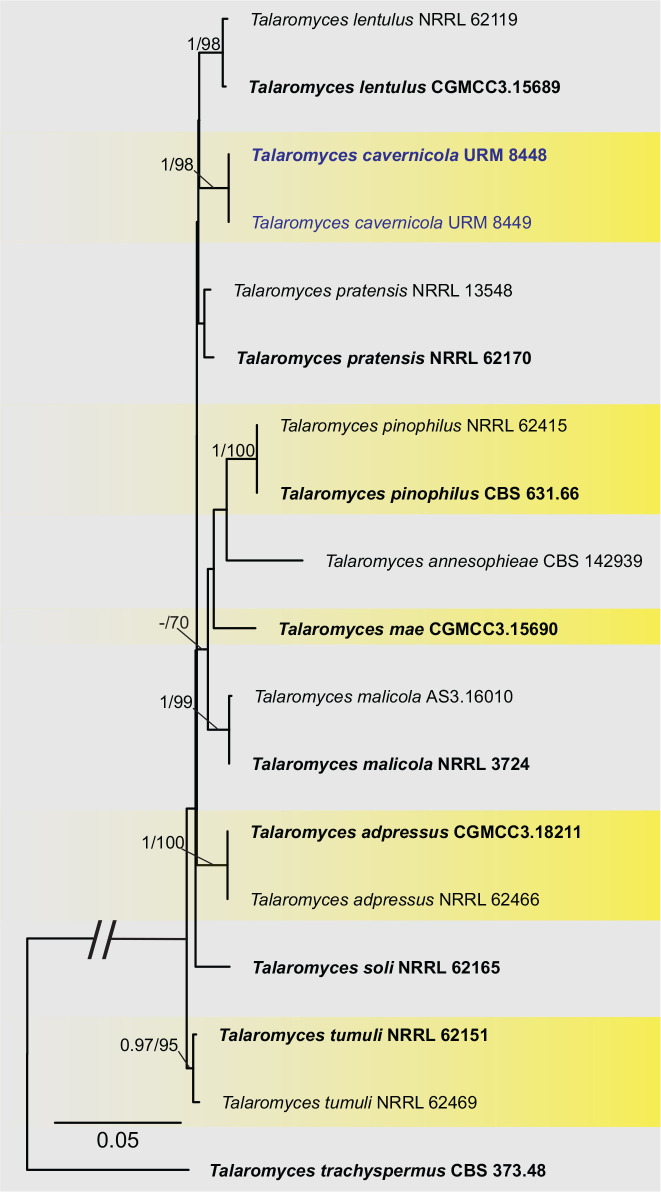
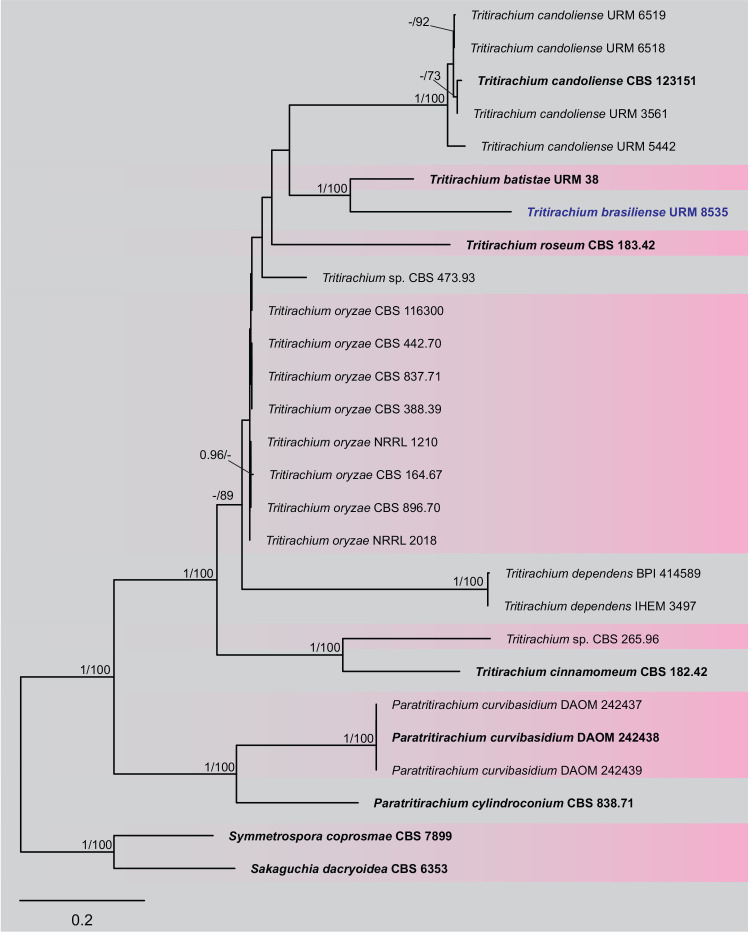
BLASTn searches using sequences of the 12 isolates, which are introduced below as new species, showed the relationship among our DNA sequences and others deposited in GenBank, which helped us to build specific alignments for each genus having a new species and based on previous studies. Details of the combined datasets (number of species/sequences and length of the datasets (bp)) and the best nucleotide models for BI analysis are shown in Supplementary Table S4. In the phylogenetic analysis, using a combined matrix of cmdA-tub2-rpb2 sequences, Aspergillus isolates (URM 8450 and URM 8451 from the air) were grouped as an independent lineage (BPP = 1 and BS-ML = 100) with A. europaeus as the closest species in the series Wentiorum of Aspergillus section Cremei (Fig. 3). Three Malbranchea isolates (URM 8445 and URM 8443 from the air; isolate R23 from the sediment) are introduced here as new species based on ITS-LSU phylogenetic inference that placed them as a well-supported clade (BPP = 1 and BS-ML = 100) and related to M. chinense (Fig. 4). Two Pseudohumicola isolates (URM 8444 from the air and isolate R35 from the sediment) were placed in the ITS-tub2-rpb2 tree as a unique lineage (BPP = 1 and BS-ML = 100), with P. pulvericola and P. semispiralis as related species (Fig. 5). Two other isolates (URM 8446 and URM 8447) were obtained from the air and placed in the ITS-LSU-rpb2-tef1 tree as an unknown and well-defined lineage (BPP = 1 and BS-ML = 100) in the family Cordycipitaceae, and are introduced below as a new genus and species (Fig. 6). The fifth new species was introduced based on two Talaromyces isolates (URM 8448 and URM 8449) found in the air, and phylogenetically clustered as an independent lineage (ITS-cmdA-tub2: BPP = 1 and BS-ML = 100) in the T. pinophilus species complex of Talaromyces section Talaromyces (Fig. 7 and Supplementary Figs S1 and S2). Sequences from one isolate of Tritirachium URM 8535, obtained from the sediment, grouped as an independent lineage in the ITS alignment (BPP = 1 and BS-ML = 100) having Tritirachium batistae as related species (Fig. 8).
Fig. 3.
Bayesian phylogenetic tree using sequences of cmdA-tub2-rpb2 of species included in Aspergillus section Cremei. The new species described in this study (Aspergiilus lebretii) is highlighted in blue. Ex-type strains are in bold. Values for ML-BS ≥70 % and BPP ≥0.95 are included near nodes. The tree was rooted to Aspergillus glaucus NRRL 117.
Fig. 4.
Bayesian phylogenetic tree using sequences of ITS-LSU of species included in Malbranchea. The new species described in this study (Malbranchea cavernosa) is highlighted in blue. Ex-type strains are in bold. Values for ML-BS ≥70 % and BPP ≥0.95 are included near nodes. The tree was rooted to Arachnomyces jinanicus CGMCC3.14173.
Fig. 5.
Bayesian phylogenetic tree using sequences of ITS-rpb2-tub2 of species included in Humicola and Pseudohumicola. The new species described in this study (Pseudohumicola cecavii) is highlighted in blue. Ex-type strains are in bold. Values for ML-BS ≥70 % and BPP ≥0.95 are included near nodes. The tree was rooted to Chaetomium globosum CBS 160.62.
Fig. 6.
Bayesian phylogenetic tree using sequences of ITS-LSU-rpb2-tef1 of species included in the family Cordycipitaceae. The new genus and species described in this study (Pseudolecanicillium caatingaense) are highlighted in blue. Ex-type strains are in bold. Values for ML-BS ≥70 % and BPP ≥0.95 are included near nodes. The tree was rooted to Purpureocillium lilacinus CBS 284.36 and CBS 431.87.
Fig. 7.
Maximum likelihood tree using sequences of cmdA-tub2-rpb2 of species included in Talaromyces section Talaromyces. The new species described in this study (Talaromyces cavernicola) is highlighted in blue. Ex-type strains are in bold. Values for ML-BS ≥70 % and BPP ≥0.95 are included near nodes. The tree was rooted to Talaromyces trachyspermus CBS 373.48.
Fig. 8.
Maximum likelihood tree using sequences of ITS of species included in Tritirachium and related genera. The new species described in this study (Tritirachium brasiliense) is highlighted in blue. Ex-type strains are in bold. Values for ML-BS ≥70 % and BPP ≥0.95 are included near nodes. The tree was rooted to Sakaguchia dacryoidea CBS 6353 and Symmetrospora coprosmae CBS 7899.
TAXONOMY
Aspergillus lebretii V.C.S. Alves, J.D.P. Bezerra & R.N. Barbosa, sp. nov. MycoBank MB 846119. Fig. 9.
Fig. 9.

Aspergillus lebretii URM 8451. A. Colonies from left to right (top row) MEA, CYA, CZ, YES, CYAS and OA; (bottom row) MEA, CYA, CZ, YES, CYAS reverse and CREA, after 7 d at 25 °C in the dark. B–G. Conidiophore and conidia. H. Conidia. I. Schematic line drawing of Aspergillus lebretii. Scale bars: B = 100 μm; C–E = 50 μm; F, G = 25 μm; H = 10 μm.
Etymology: lebretii reflects the name of Michel Le Bret, founder of the Brazilian Society of Speleology.
Suborder classification: Eurotiales, Aspergillaceae.
Infrageneric classification: subgenus Cremei, section Cremei, series Wentiorum.
Typus: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento (holotype URM 95150, culture ex-type URM 8451 = FCCUFG 09 = isolate F32).
Culture characteristics (25 °C, 7 d, in the dark): MEA: 26–27 mm diam; mycelium white to buff (45); margins entire, centrally raised, non-sulcate, colony texture cottony, sclerotia absent, poor sporulation, conidial colour en masse indeterminate, exudate and pigment soluble absent; reverse saffron (10). CYA: 27–28 mm diam; mycelium white to buff (45), centre slightly elevated, radially sulcate, colony texture cottony, entire to regular margins, sclerotia absent, poor sporulation, conidial colour en masse indeterminate, exudate and pigment soluble absent; reverse primrose (66). CZ: 21–23 mm diam; mycelium white to buff (45); margins entire, centrally raised, non-sulcate, colony texture cottony, entire to regular margins, sclerotia absent, poor sporulation, conidial colour en masse indeterminate, exudate and pigment soluble absent; reverse primrose (66). YES: 39–41 mm diam; mycelium white, margins entire, centrally crateriform, radially sulcate, colony texture cottony, entire to regular margins, sclerotia absent, sparse sporulation, conidial colour en masse cinnamon (62), exudate and soluble pigments absent, reverse ochreous (44). OA: 14–15 mm diam; centrally raised (convex), non-sulcate, colony texture cottony, entire to regular margins, white to buff (45) coloured mycelium, sclerotia absent, poor sporulation, conidia in undetermined mass, no exudate and soluble pigments; reverse primrose (66). CYAS: 39–40 mm diam; mycelium white, elevated colonies (convex), non-sulcate, colony texture cottony, entire to regular margins, sclerotia absent, sparse sporulation, conidial colour en masse buff (45), exudate and soluble pigments absent; reverse saffron (10). CREA: slightly positive for acid production. Colonies at 30 °C, 7 d in the dark: MEA: 20–21 mm diam; CYA: 22–23 mm diam; no growth was observed at 37 °C and 10 °C.
Conidiophores biseriate, stipes, hyaline, smooth-walled, aseptate and occasionally septate, 558–604.5(–930) × 8–13.5 μm, occasionally longer. Vesicle globose and occasionally subglobose, 16–40 μm. Metulae wedge-shaped, occasionally septate, 13.5–16 × 5–5.5 μm. Phialides ampulliform, (7.5–)8–8.5 × (4.5–)5–5.5 μm. Conidia globose, yellow to yellowish-brown, smooth to slightly roughened, 2.5–4.5 μm. Hülle cells and sclerotia were absent. Sexual morph not observed.
Additional material examined: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento, URM 8450 = FCCUFG 10 = isolate F23.
Notes: BLASTn searches using ITS sequences showed that Aspergillus lebretii has a high identity (99–100 %) to species in the series Wentiorum of Aspergillus section Cremei. The tub2 sequences of A. lebretii showed an identity of 98.08 % to 98.17 % with the ex-type strain of A. europaeus CCF 4409 and 95.91 % to 96.15 % with the ex-type strains of A. chrysellus NRRL5084 and A. dimorphicus NRRL 3650. The cmdA sequences of A. lebretii showed an identity of 96.55 %–97.5 % with the ex-type strain of A. europaeus CCF 4409, and 91.54 % to 96 % identity with the ex-type strains of A. chrysellus NRRL5084 and A. dimorphicus NRRL 3650. The rpb2 sequences of A. lebretii showed an identity of 98.31 % with the ex-type strain of A. europaeus CCF 4409 and 97.40 % with A. dimorphicus NRRL 3650. Phylogenetic analyses based on a combined dataset tub2-cmdA-rpb2 indicated that A. lebretii forms a unique, well supported lineage related to A. europaeus CCF 4409 (Fig. 3). Based on a nucleotide pairwise comparison of tub2-cmdA-rpb2, A. lebretii differed from the ex-type culture of A. europaeus CCF 4409 by 1/535 bp of ITS, 6/421 of tub2, 10/618 bp of cmdA and 19/884 bp of rpb2. Macromorphologically, A. lebretii differs from A. europaeus by having colonies of cottony texture, poor sporulation, reverse of colonies saffron colour, and absence of soluble pigments in MEA, whereas in A. europaeus, the colonies have a granular texture, sporulation occurs on almost the entire colony surface, and the reverse of colonies is yellow to bright yellow and there is the production of yellow soluble pigments. On CYA, the colonies of A. lebretii showed low growth (27–28 mm diam) and cottony texture, whereas A. europaeus colonies were larger (32–45 mm diam) and velvety to floccose textures. In addition, A. lebretii shows weak production of acidic compounds in CREA, whereas A. europeaus shows intensive production (Hubka et al. 2016). Micromorphologically, A. lebretii commonly has long stipes [558–604.5(–930) × 8–13.5 μm, occasionally longer] than A. europeaus (300–750 × 7–15 μm); vesicles in A. lebretii are globose and occasionally subglobose (16–40 μm) and A. europeaus has vesicles piriform or globose (11–44 μm); conidia in A. lebretii are globose, pigmented and slightly smaller (2.5–4.5 μm) and A. europaeus has conidia variable in shape, colourless when young and pigmented with age, and slightly larger (3.5–5 × 3–4.5 μm) (Hubka et al. 2016).
Malbranchea cavernosa V.C.S. Alves, R.A. Lira, Souza-Motta & J.D.P. Bezerra, sp. nov. MycoBank MB 846121. Fig. 10.
Fig. 10.
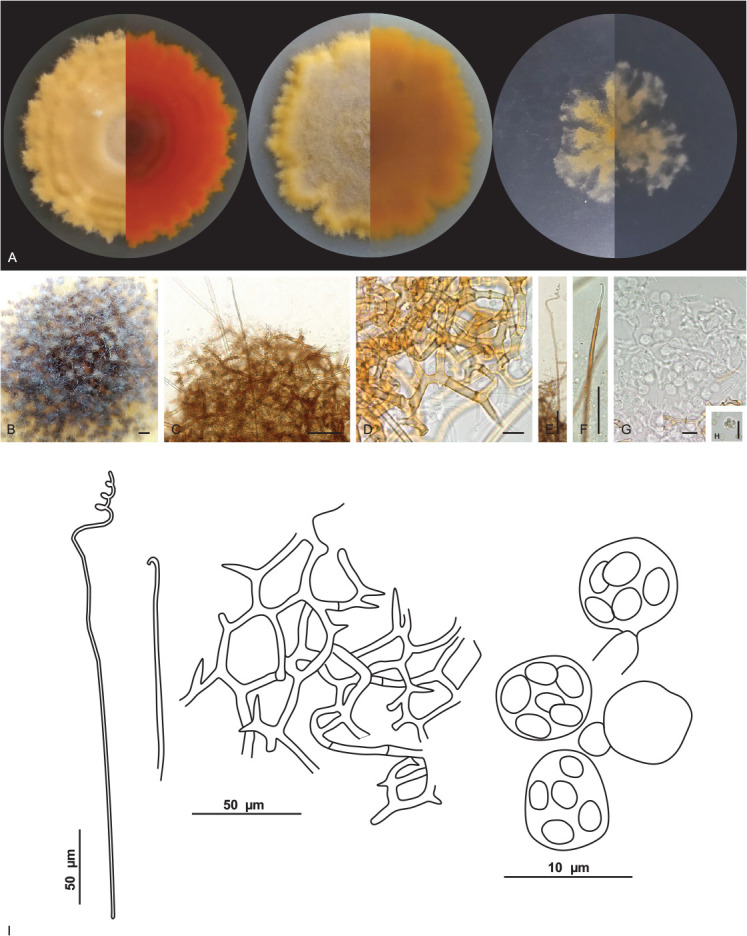
Malbranchea cavernosa URM 8445. A. Colonies on PDA, OA and SNA after 4 wk at 25 °C in the dark. B. Ascoma. C. Ascoma gymnothecium. D. Reticuloperidium terminated by spine-like projection. E, F. Appendages. G. Asci. H. Ascospores. I. Schematic line drawing of Malbranchea cavernosa. Scale bars: B = 300 μm; C, E, F = 50 μm; D, G, H = 10 μm.
Etymology: cavernosa reflects the location, a cave, where the species was first isolated.
Suborder classification: Onygenales, Malbrancheaceae.
Typus: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento (holotype URM 95151, culture ex-type URM 8445 = FCCUFG 13 = isolate E27).
Culture characteristics (25 °C, 4 wk, in the dark): PDA: 65–67 mm diam, centrally raised, lobed and irregular margins, velutinous, non-sulcate, ochreous (44); exudates or soluble pigments were not observed; reverse scarlet (5) to orange (7). OA: 63–65 mm diam, flat, lobed, and irregular margins, velutinous, non-sulcate, ochreous (44) to apricot (42), centre fawn (87); exudate hyaline and abundant and soluble pigments are absent; reverse sienna (8) to orange (7). SNA: 29–30 mm diam, flat, lobed margins, cottony, non-sulcate, ochreous (44) to saffron (10); exudates or soluble pigments were not observed; reverse saffron (10) to ochreous (44). Sporulation at 10–14 d.
Hyphae hyaline, septate, 1–1.5(–2) μm wide. Sexual morph: Ascomata abundant, solitary or in clusters, globose, white at first and becoming orange to brown when mature, (232–) 251–325(–335) μm diam. Peridial hyphae rough, thick walled, septate, pale brown, branched forming a reticuloperidium, terminated by spine-like projection, 2–6.5 μm wide. Appendages with a curved tip, smooth, aseptate, 456–576(–688) × 3–4.5 μm. Asci 8-spored, globose, hyaline, 5.5–6.5 μm. Ascospores were globose to subglobose, smooth-walled to roughened, 2.5–3.5 × 2–2.5 μm. Asexual morph not observed.
Additional material examined: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento, URM 8443 = FCCUFG 14 = isolate E13; Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, isolated from sediment, 5 Mar. 2020; R.A. Lira & D.M. Bento, URM 8534 = isolate R23.
Notes: Based on BLASTn searches, the ITS sequences of Malbranchea cavernosa have a low identity of 96.10 % with the ex-type strain of M. chinense CGMCC 3.19572 and with Auxarthron thaxteri CBS 297.63 (a species synonymized with Malbranchea umbrina, Rodríguez-Andrade et al. 2021). The LSU sequences of M. cavernosa showed high identity (99.23 %) with the ex-type strain of M. chinense CGMCC 3.19572. The tub2 sequences of M. cavernosa showed low identity (92.94 %) with the sequences of M. chinense CGMCC 3.19572. The tef1 sequences of M. cavernosa showed low identity (95.94 %) with the sequences of M. chinense CGMCC 3.19572 and M. guangxiensis LC12464 (94.64 %.) Phylogenetic analyses based on a combined dataset of ITS-LSU rDNA sequences, showed that M. cavernosa, is a unique and well supported lineage related to M. chinense CGMCC 3.19572 (Fig. 4). Based on a pairwise nucleotide comparison of ITS-LSU, M. cavernosa differs from that of M. chinense with 19/712 bp of ITS and 4/833 bp of LSU. Macromorphologically, colonies of M. cavernosa differ from that of M. chinense by fast growth on all culture media (PDA: 18–23 mm diam; OA: 18–23 mm diam; SNA: 21–25 mm diam in M. chinense) (Zhang et al. 2021). Micromorphologically, M. cavernosa differs from M. chinense, which only has an asexual morph, by having only a sexual morph. Malbranchea cavernosa morphologically resembles Malbranchea ostraviensis, which differs from the size of the ascoma (300–400 μm in M. ostraviensis) and by peridial hyphae that are sometimes dichotomously branched in M. ostraviensis. The appendages of M. cavernosa are slightly larger and wider than those of M. ostraviensis (350–600 × 2.8–3.2 μm); asci in M. cavernosa are globose (5.5–6.5 μm) and smaller than those of M. ostraviensis, which are globose, subglobose or pyriform (7.4–8.7 × 6.6–7.7 μm); ascospores of M. cavernosa are globose to subglobose and in M. ostraviensis are oblate (2.5–3 μm) (Hubka et al. 2013) and they are not phylogenetically related.
Pseudohumicola cecavii V.C.S. Alves, R.A. Lira, Souza-Motta & J.D.P. Bezerra, sp. nov. MycoBank MB 846122. Figs 11, 12.
Fig. 11.
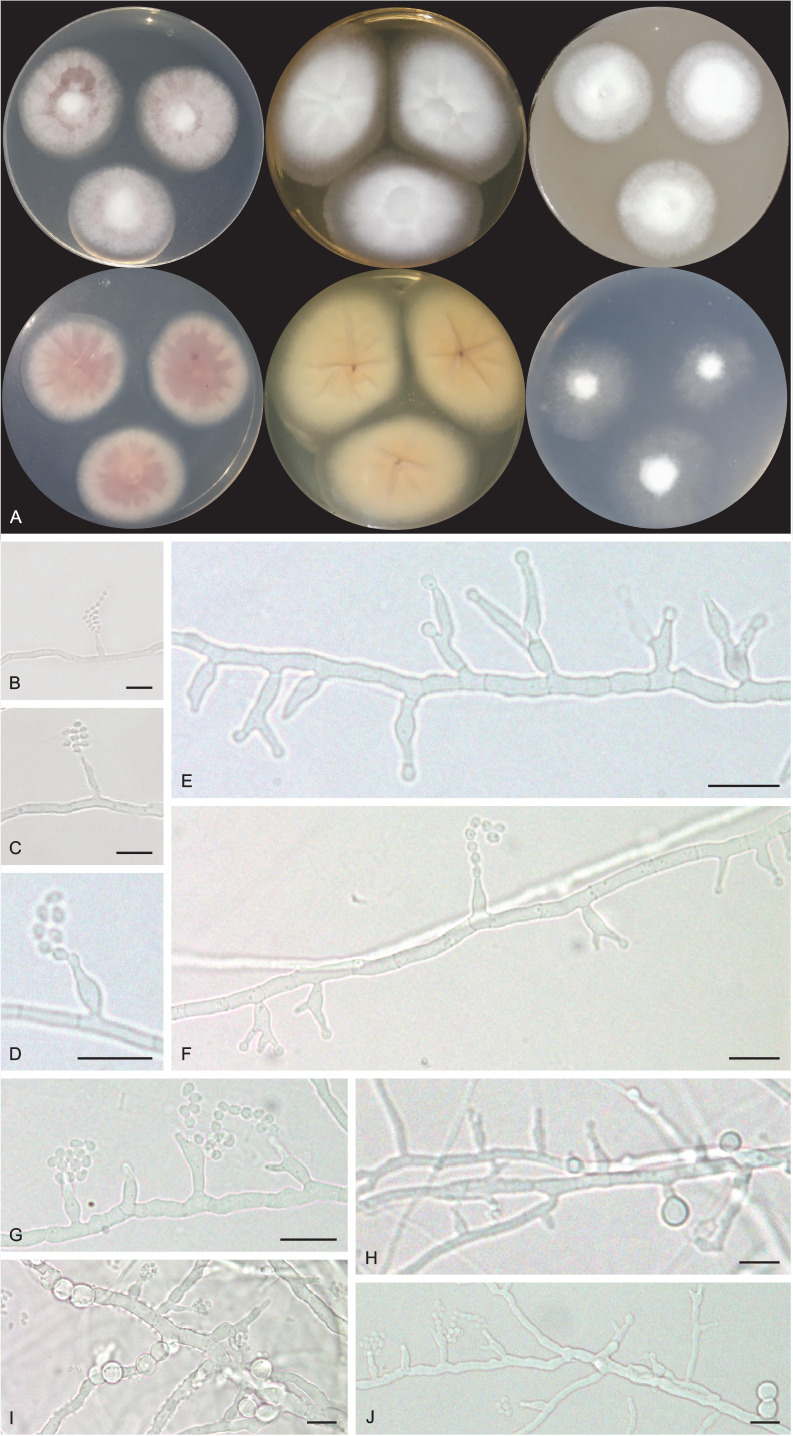
Pseudohumicola cecavii URM 8444. A. Colonies from left to right (top row) PDA, MEA and OA; (bottom row) PDA reverse, MEA reverse and SNA after 7 d at 25 °C in the dark. B–G. Conidiophore, conidiogenous cells and conidia. H–J. Conidiophore, conidiogenous cells, conidia and chlamydospores. Scale bars = 10 μm.
Fig. 12.
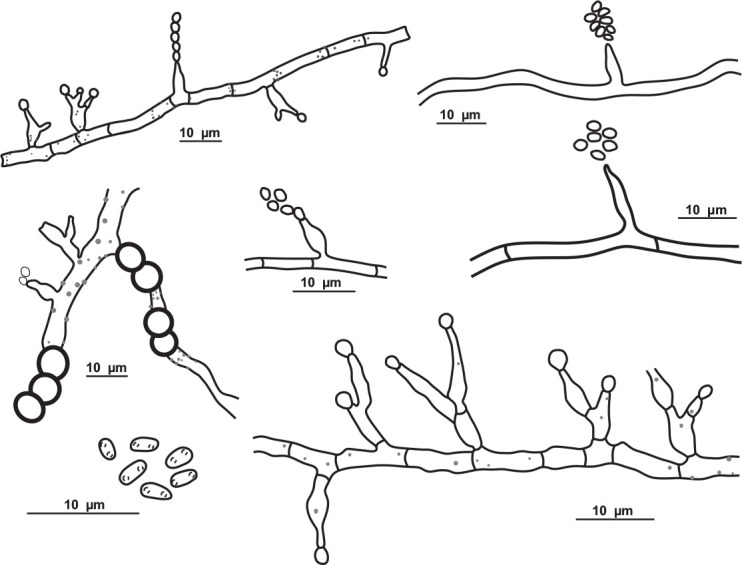
Schematic line drawing of Pseudohumicola cecavii URM 8444.
Etymology: cecavii reflects the name of the Brazilian federal institution devoted to the study and conservation of caves, the Centro Nacional de Pesquisa e Conservação de Cavernas, whose acronym is CECAV.
Suborder classification: Sordariales, Chaetomiaceae.
Typus: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento (holotype URM 95154, culture ex-type URM 8444 = FCCUFG 17 = isolate E21).
Culture characteristics (25 °C, 2 wk, in the dark): PDA: 34–35 mm diam, white to rosy vinaceous (58), cottony, centrally raised, entire to regular margin, radially sulcate, exudate, and soluble pigments absent; reverse rosy vinaceous (58) to buff (45). MEA: 30–35 mm diam, white, cottony, centrally raised, entire to regular margin, radially sulcate, exudate and soluble pigments absent; reverse ochreous (44). OA: 29–30 mm diam, white mycelium, cottony, centrally crateriform, entire to regular margin, non-sulcate, exudate and soluble pigments absent; reverse saffron (10). SNA: (25–)30–32 mm diam, white mycelium, cottony, centrally raised, filiform margin, non-sulcate, hyaline exudate, and soluble pigments absent; reverse buff (45).
Hyphae hyaline, septate, branched and tortuous, 1–3 μm wide. Conidiogenous cells phialidic, cylindrical to ampuliform, 4.5–7.5 × 1.5–3 μm. Conidia globose to subglobose, chains or slime heads, guttulate, 2–3 × 1–1.5(–2) μm. Chlamydospores globose, occasionally subglobose, intercalary and terminal, occasionally in chains, 5–7 μm. Sexual morph not observed.
Additional material examined: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88′ S, 37°32’1.51′ W, isolated from sediment, 5 Mar. 2020, R.A. Lira & D.M. Bento, isolate R35.
Notes: Based on BLASTn searches of ITS sequences, Pseudohumicola cecavii has a high identity (99.79 %) to a sequence deposited as “Humicola sp. 1002-FVD3 1” which was obtained from the frugivorous guano of a Brazilian bat (Cunha et al. 2020), amongst other sequences of the recently introduced genus Pseudohumicola (Wang et al. 2022). The rpb2 sequence had low identity (93.72 %) with P. atrobrunnea HSAUP II05-1004 isolated from soil, and the tub2 sequence also had low identity (89.24 %, P. subspiralis CBS 148.58 isolated from leaf fragments in soil) (Wang et al. 2019). Phylogenetic analyses of a combined ITS-rpb2-tub2 alignment placed our sequences as a unique lineage in a clade encompassing P. pulvericola and P. semispiralis and related to other Pseudohumicola isolates (T27 and T39) obtained during this study (Fig. 5). However, we were not able to obtain new cultures during the subculturing process of these two isolates which also appeared as a putative new phylogenetic species. Macromorphologically, P. cecavii differs from P. pulvericola which has faster colony growth on OA (51–57 mm diam) and MEA (46–52 mm diam), and from P. semispiralis (OA: 36–42 mm diam, producing a soluble pigment ochreous to umber; MEA: 39–45 mm diam, producing a soluble pigment) (Wang et al. 2019). Micromorphologically, P. cecavii differs from P. pulvericola which has thick-walled and larger conidia (2.5–4 × 1–2(–2.5) μm) and larger conidiophores/conidiogenous cells (13–30 × 1–3 μm). Pseudohumicola cecavii mainly differs from P. semispiralis by its only known sexual morph and production of conidia usually lateral on the hyphae (Wang et al. 2019).
Pseudolecanicillium V.C.S. Alves, Souza-Motta & J.D.P. Bezerra, gen. nov. MycoBank MB 846123.
Etymology: Reflects the phylogenetic relationship with species previously described in Lecanicillium and related genera.
Suborder classification: Hypocreales, Cordycipitaceae.
Hyphae septate, hyaline, smooth. Conidiophores long, lecanicillium-like, wider at the base, tapering towards the tip, reduced to conidiogenous cells, and semimacronematous or somewhat differentiated with conidiogenous cells arranged solitary or in 2–4 verticills. Conidiogenous cells phialidic, discrete, wider at the base and tapering gradually towards the tip, straight to slightly flexuous, terminally and laterally in the conidiophores, mono or polyphialidic, proliferating sympodially, and forming a rachis-like structure in the upper region, with multiple denticules, producing solitary or short chains of conidia. Conidia dry, solitary or in short chains, ellipsoid to slightly fusiform, apices acute, smooth, aseptate, hyaline. Chlamydospores were not observed. Sexual morph not observed.
Type species: Pseudolecanicillium caatingaense V.C.S. Alves, Souza-Motta & J.D.P. Bezerra
Pseudolecanicillium caatingaense V.C.S. Alves, Souza-Motta & J.D.P. Bezerra, sp. nov. MycoBank MB 846124. Figs 13, 14.
Fig. 13.
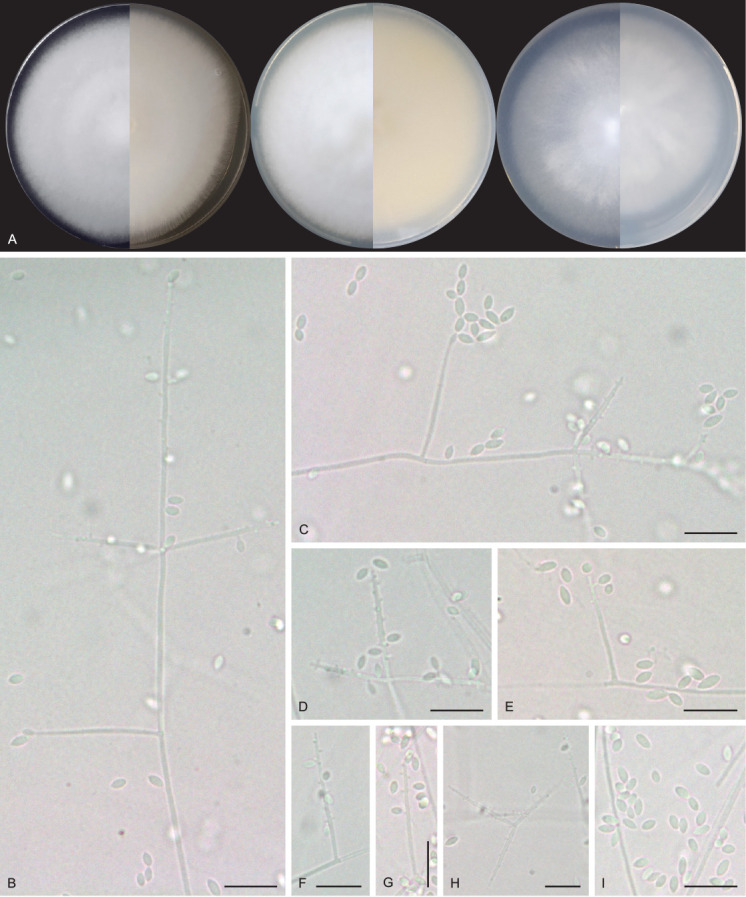
Pseudolecanicillium caatingaense URM 8447. A. Colonies on PDA, OA and SNA after 2 wk at 25 °C in the dark. B, C. Conidiophores, conidiogenous cells and conidia. D–H. Details of conidiogenous cells and conidia. I. Conidia. Scale bars = 10 μm.
Fig. 14.
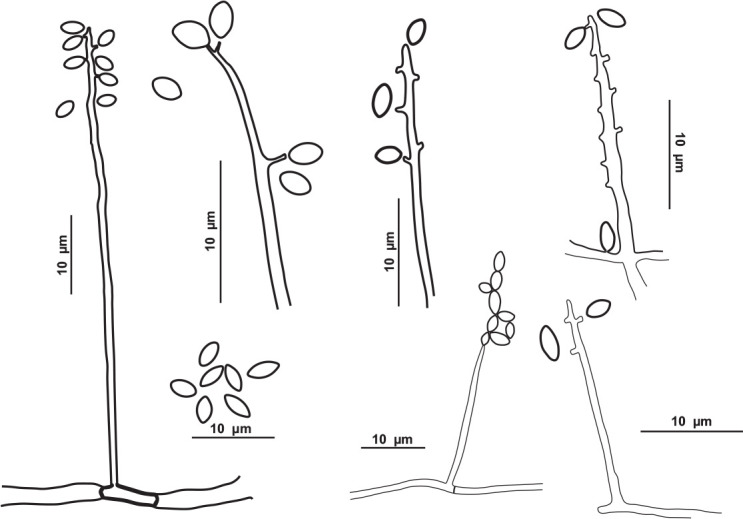
Schematic line drawing of Pseudolecanicillium caatingaense URM 8447.
Etymology: caatingaense reflects the location, the Caatinga tropical dry forest, where the species was first isolated in Brazil.
Typus: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento (holotype URM 95152, culture ex-type URM 8447 = FCCUFG 15 = isolate E34).
Culture characteristics (25 °C, 2 wk, in the dark): PDA: 74–76 mm diam, white, cottony, centrally raised, entire to regular margin, non-sulcate, exudate, and soluble pigments absent; reverse white to creamish. OA: 70–72 mm diam, white, cottony, centrally raised, entire to regular margin, non-sulcate, exudate, and soluble pigments absent; reverse creamish to straw (46). SNA: 74–76 mm diam, white, cottony, centrally raised, entire to regular margin, non-sulcate, exudate, and soluble pigments absent; reverse white to creamish. Sporulation within 7–10 d.
Hyphae 1–2 μm wide, septate, hyaline, and smooth. Conidiophores long, straight to flexuous, lecanicillium-like, wider at the base, tapering towards the tip, reduced to conidiogenous cells, and semimacronematous or somewhat differentiated with conidiogenous cells arranged solitary or in 2–4 verticills. Conidiogenous cells phialidic, discrete, wider at the base and tapering gradually towards the tip, straight to slightly flexuous, terminally and laterally in the conidiophores, mono or polyphialidic, proliferating sympodially, and forming a rachis-like structure in the upper region, with multiple denticules, producing solitary or short chains of conidia, 10–18.5 × 1–1.5 μm. Conidia dry, solitary or in short chains, ellipsoid to slightly fusiform, apices acute, smooth, aseptate, hyaline, 2–4.5 × 1.5–2 μm. Chlamydospores were not observed. Sexual morph not observed.
Additional material examined: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento, URM 8446 = FCCUFG 16 = isolate E31.
Notes: BLASTn searches using ITS sequences of Pseudolecanicillium caatingaense demonstrated low identity (95.03 %) to a sequence deposited as Blackwellomyces pseudomilitaris TBRC 3662 isolated from Lepidoptera (larvae) (Mongkolsamrit et al. 2020), amongst other sequences of Cordyceps and Lecanicillium. The tef1 sequence had low identity (95.36 %) to a sequence deposited as L. psalliotae CBS 532.81, isolated from soil (Sung et al. 2007). The LSU sequences showed high identity (99 %) with sequences deposited as Lecanicillium species (e.g. Lecanicillium fusisporum CBS 164.70), tub2 had low identity (89.67 %) with L. tenuipes CBS 658.80, and the rpb2 sequences also had low identity (83.95 %) to the ex-type strain of L. fungicola var. aleophilum CBS 992.69. Phylogenetic analysis of the ITS-LSU-rpb2-tef1 matrix identified sequences of the new genus and species as a unique and well supported lineage in the family Cordycipitaceae (Fig. 6) and related to sequences deposited as L. psalliotae (CBS 101270 and CBS 532.81). Cordycipitoid fungi have been widely studied over the years, and the systematic position of some taxa in the family Cordycipitaceae have also been resolved based on multigene phylogenetic analyses (Wang et al. 2020). Recently, new genera were introduced in this family (e.g. Flavocillium, Gamszarea, Jenniferia, Liangia, and Polystromomyces) (Wang et al. 2020; Zhang et al. 2021; Mongkolsamrit et al. 2022). Pseudolecanicillium caatingaense mainly differs from the related genera by having phialides proliferating sympodially and forming a rachis-like structure in the upper region and with multiple denticules; and by having conidia ellipsoid to slightly fusoid and apices acute.
Talaromyces cavernicola V.C.S. Alves, J.D.P. Bezerra & R.N. Barbosa, sp. nov. MycoBank MB 846125. Fig. 15.
Fig. 15.
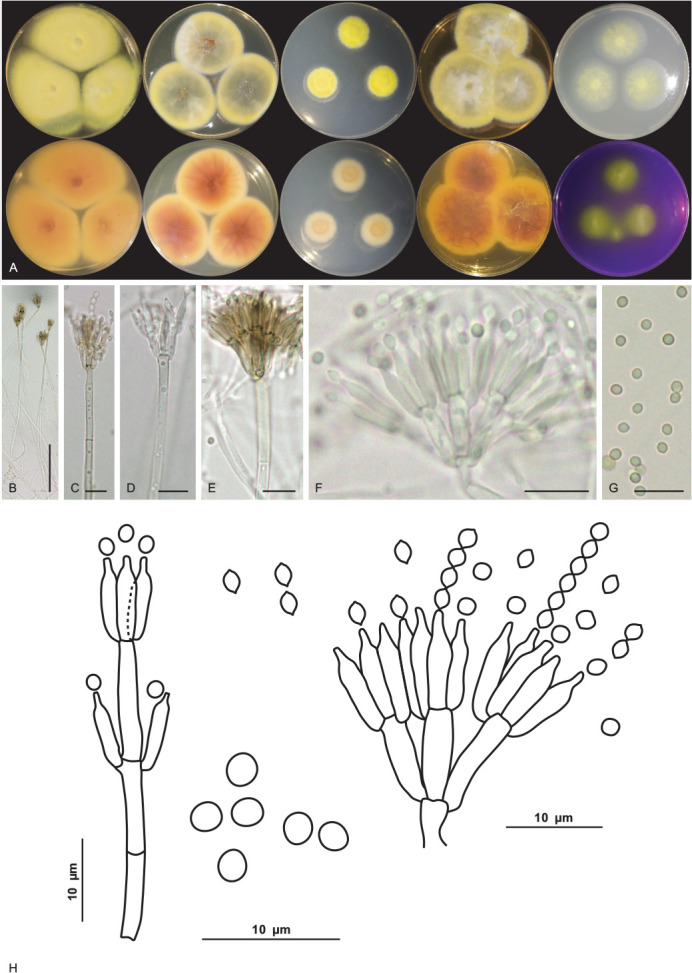
Talaromyces cavernicola URM 8448. A. Colonies from left to right (top row) MEA, CYA, CZ, YES and OA; (bottom row) MEA, CYA, CZ, YES reverse and CREA after 7 d at 25 °C in the dark. B–F. Conidiophore and conidia. G. Conidia. H. Schematic line drawing of Talaromyces cavernicola. Scale bars: B = 100 μm; C–G = 10 μm.
Etymology: cavernicola reflects the location, a cave, where the species was first isolated.
Suborder classification: Eurotiales, Trichocomaceae.
Infrageneric classification: section Talaromyces.
Typus: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento (holotype URM 95155, culture ex-type URM 8448 = FCCUFG 11 = isolate E35A).
Culture characteristics (25 °C, 7 d, in the dark): MEA: 35–41 mm diam, mycelium pure yellow (14) to greenish yellow (16), entire to regular margins colonies slightly elevated at the centre, non-sulcate, colony texture velutinous, sclerotia absent, sparse sporulation, conidial colour en masse greyish green (50), colourless exudates in small droplets, soluble pigments absent; reverse umber (9) to ochreous (44). CYA: 31–35 mm diam, mycelium pure yellow (14) to yellow green (71), colony plane, radially sulcate, regular to entire margins, sclerotia absent, sparse sporulation, conidial colour en masse yellow green (71), exudates in small to large droplets of hyaline to cinnamon (62), soluble pigments absent; reverse sienna (8) to ochreous (44). CZ: 18–19 mm diam, mycelium pure yellow (14), margins entire to regular umbonate, radially sulcate, colony texture velutinous, sclerotia absent, poor sporulation, conidial colour en masse indeterminate, colourless exudates in small droplets, soluble pigments absent; reverse saffron (10) to salmon (41). YES: (40–) 45–47 mm diam, mycelium pure yellow (14) to yellow-green (71), colonies slightly elevated at the centre, non-sulcate, colony texture cottony, entire to regular margins, coloured mycelium, sclerotia absent, sparse sporulation, conidial colour en masse yellow-green (71), soluble pigments and exudate absent; reverse sienna (8) to ochreous (44). OA: 24–28 mm diam, flat, non-sulcate, colony texture cottony, entire to regular margins, citrine green (67) to yellow green (71) mycelium, sclerotia absent, poor sporulation, conidia in undetermined mass, exudates in small droplets of yellowish colour, soluble pigments absent; reverse primrose (66). DG18: 4–7 mm diam, mycelium white, colonies umbonate, non-sulcate, colony texture cottony, entire to regular margins, sclerotia absent, poor sporulation, conidial colour en masse indeterminate, soluble pigments and exudate absent; reverse saffron (10) to umber (9). CREA: low acid production. Colonies at 30 °C, 7 days, in the dark: MEA: 49–50 mm diam; CYA: 34–35 mm diam; colonies at 37 °C: MEA: 27–28 mm diam; CYA: 34–38 mm diam; no growth was observed at 10 °C.
Conidiophores biverticillate, stipes septate, hyaline to pigmented, smooth to roughened, commonly long, 21–420 × 3–4 μm. Metulae 3–5, 8–10 × 2.5–3.5 μm. Phialides 2–4, acerose, 8–9 × 2–4 μm. Conidia globose to subglobose, greenish, ornamented and occasionally smooth, 2 × 1.5–2 μm. Sclerotia absent. Sexual morph not observed.
Additional material examined: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S and 37°32’1.51’’W, as an airborne fungus, 5 Mar. 2020, V.C.S. Alves & D.M. Bento, URM 8449 = FCCUFG 12 = isolate E35B.
Notes: Based on BLASTn searches, the ITS sequences of Talaromyces cavernicola have a high identity (99.62–99.81 %) with species of Talaromyces section Talaromyces. The tub2 of T. cavernicola showed 98.26 % identity with the ex-type strain of T. pratensis and 98 % to 98.5 % identity with the sequences of T. stollii NRRL62122, T. lentulus NRRL62143, T. tumuli NRRL62151, and T. sayulitensis NRRL62203. The cmdA sequence of T. cavernicola showed low identity (96.39%) with the ex-type strain of T. pinophilus CBS 631.66. The rpb2 sequences of T. cavernicola showed an identity of 98.75% with the ex-type strain of T. pinophilus CBS 631.66. Phylogenetic analyses based on a combined dataset tub2-cmdA-rpb2 showed that T. cavernicola is a unique, well supported lineage related to T. soli NRRL62165 (Fig. 7 and Supplementary Figs S1 and S2). Based on a nucleotide pairwise comparison of tub2-cmdA-rpb2, T. cavernicola differs from T. soli by 6/512 bp for tub2, 16/533 bp for cmdA and 19/753 bp for rpb2; no differences were observed in ITS. Macro morphologically, colonies of T. cavernicola differ from those of T. soli by having small growth on DG18 (4–7 mm diam in T. cavernicola and 7–14 days in T. soli). On OA, colonies of T. cavernicola were smaller (24–28 mm diam) than that of T. soli (23–43 mm diam). On CREA, T. cavernicola showed low production of acidic compounds, whereas T. soli showed moderate production of acidic compounds (Peterson & Jurjević 2019). Micromorphologically, T. cavernicola has longer conidiophores (21–420 × 3–4 μm) than T. soli [(5–)30–200(–320, rare) × (2.5–)3–4(–4.5) μm]; metulae in T. cavernicola are slightly smaller [(8–10 × 2.5–3.5 μm) and in verticils of 3–5], while in T. soli metulae are 8–12(–14) × 2.5–4 μm and in verticils of (3–)5–9(–11); phialides in T. cavernicola are smaller (8–9 × 2–4 μm and in verticils of 2–4) than in T. soli (9–11(–20) × 2.5–3(–3.5) μm and in verticils of (3–)5–9(–11)); and conidia smaller (2 × 1.5–2 μm) in T. cavernicola than T. soli [2.5–3.5(–5.5) × 2.5–3.5(–4.5) μm] (Peterson & Jurjević 2019).
Tritirachium brasiliense R.A. Lira, V.C.S. Alves, Souza-Motta & J.D.P. Bezerra, sp. nov. MycoBank MB 846126. Figs 16, 17.
Fig. 16.
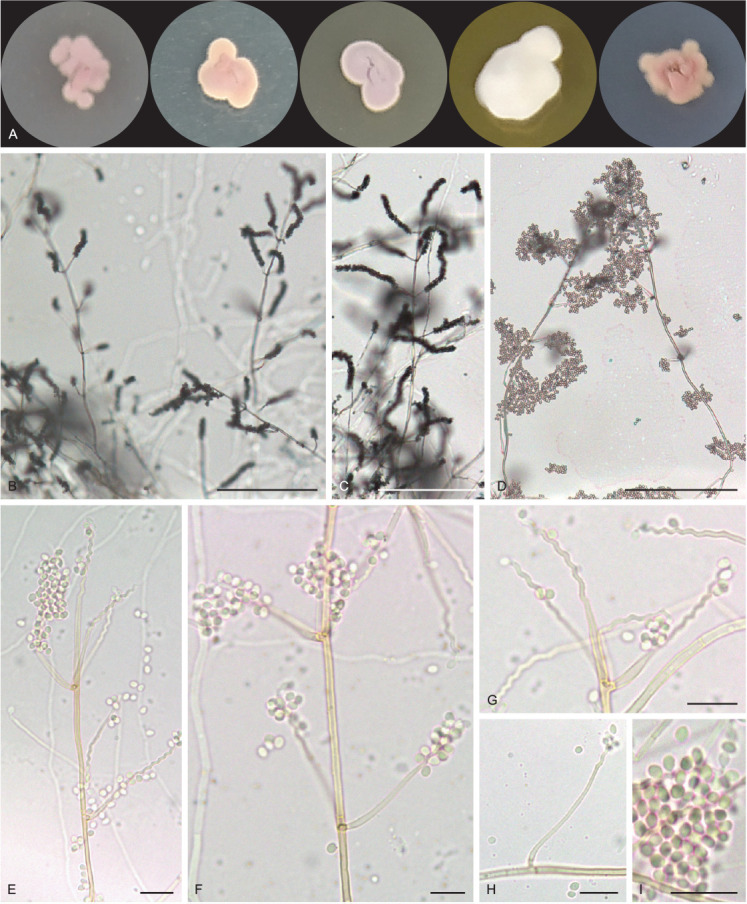
Tritirachium brasiliense URM 8535. A. Colonies from left to right OA, PDA, CYA, YES and SNA after 7 d at 25 °C in the dark. B–F. Conidiophores, conidiogenous cells and conidia. G, H. Details of conidiogenous cells. I. Conidia. Scale bars: B–D = 100 μm; E–I = 10 μm.
Fig. 17.
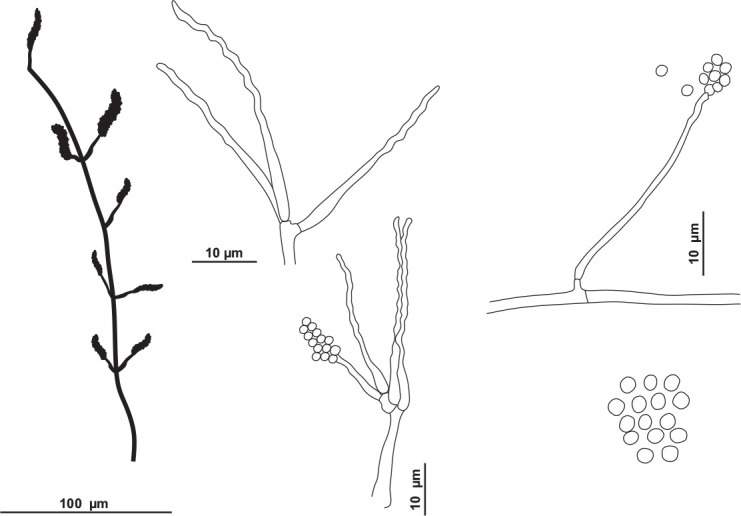
Schematic line drawing of Tritirachium brasiliense URM 8535.
Etymology: brasiliense reflects the name of the country, Brazil, where the fungus was firstly isolated from the sediment of a cave.
Suborder classification: Tritirachiales, Tritirachiaceae.
Typus: Brazil, Rio Grande do Norte state, Furna Feia National Park, Abrigo do Letreiro cave, 5°4’14.88’’S, 37°32’1.51’’W, isolated from sediment, 5 Mar. 2020, R.A. Lira & D.M. Bento (holotype URM 95153, culture ex-type URM 8535 = FCCUFG 18 = isolate R11).
Culture characteristics (25 °C, 7 d, in the dark): PDA: 5–7 mm diam, mycelium rosy buff (61), wavy margin, slightly raised, without furrows, colony texture velvety, exudate and soluble pigments absent; reverse hazel (88). MEA: 4–6 mm diam, mycelium pale vinaceous (85), wavy margin, slightly raised colony, without furrows, colony texture velvety, exudate and soluble pigments absent; reverse dark brick (60) to fawn (87). OA: 5–9 mm diam, mycelium pale vinaceous (85), irregular margins, flat, not furrowed, colony texture velvety, exudate and soluble pigments absent; reverse buff (61). SNA: 6–9 mm diam, mycelium rosy vinaceous (58), irregular margins, flat, not furrowed, colony texture velvety, exudate and soluble pigments absent; reverse cinnamon (62) to park brick (60). YES: 7–11 mm diam, white, entire margins slightly wavy, convex, without furrows, cottony texture, exudate and soluble pigments absent; reverse cinnamon (62) and the centre isabelline (65). CYA: 6–8 mm de diam, mycelium pale purplish grey (127), irregular margins, raised, without furrows, colony texture velvety, exudate and soluble pigments absent; reverse hazel (88) and the centre isabelline (65).
Hyphae pale brownish, smooth walled, 1–2 μm wide. Conidiophores reddish brown, lighter towards the base, smooth and somewhat thick-walled, 100–140 × 1.5–2 μm, slightly tapering towards the tip, bearing mainly on the upper part 3–8 whorls conidiogenous cells and occasionally a side branch which bears (1–)3–4 conidiogenous cells. Conidiogenous cells consisting of an elongate basal part, slightly swollen below the middle, tapering towards the tip, 9–20 × 1–1.5, and a well-developed geniculate part. Conidia reddish brown, smooth, thin-walled, globose to subglobose, rarely with a slightly apiculate base, 2–3 × 1–3 μm. Chlamydospores not observed. Sexual morph not observed.
Notes: Based on BLASTn searches of ITS sequence, Tritirachium brasiliense had a low identity (90.70 %) with Tritirachium oryzae TO-BRK2. The LSU sequence showed low identity with the sequences of Tritirachium batistae URM 38 (96.56 %) and T. oryzae TO-BRK2 (95.62 %). The rpb2 sequences also showed low identity (85.12 %) with sequences of T. batistae URM 38. Phylogenetic analyses of ITS sequences placed the new species as a unique lineage related to T. batistae URM 38 (Fig. 8). Tritirachium brasiliense differs from T. batistae by differences in colony colour on the PDA, MEA, CYA, YES, and OA culture media; in T. brasiliense the predominant colour is pale vinaceous to rosy buff and in T. batistae the predominant colour is white to greyish. The size of colonies also differs between T. brasiliense and T. batistae which has larger colonies sizes. The texture of colonies in T. brasilense is predominantly velvety, except on YES, while in T. batistae the cottony texture was predominantly observed. Conidiophores of T. brasiliense are slightly reddish and larger (100–140 μm) than those of T. batistae that are brownish with maturity and smaller (9–23.5 μm). The conidia of T. brasiliense are reddish brown and in T. batistae they are hyaline (Bezerra et al. 2020).
DISCUSSION
Studies on fungi in caves have been conducted to investigate different substrates and hosts (Zhang et al. 2021). Our study observed high fungal abundance in the sediment and air of the Abrigo do Letreiro cave in the Caatinga drylands of North-east Brazil at all sampling points. Morphological features and phylogenetic analyses indicated 41 species belonging to 17 genera of Ascomycota and two genera of Basidiomycota, with Aspergillus being the most commonly observed genus in the cave (13 taxa). One new genus and six species were described in our study. Despite the growing quest to unravel the mycobiota of the cave environment, there is still a knowledge gap regarding the fungi living in these environments, especially when associated with special caves, such as the caves of the Caatinga dry forest in Brazil (Cunha et al. 2020, Pereira et al. 2022).
Differences in the spatial distribution of fungi: the role of biotic and abiotic factors
In our study, the abundance of airborne fungi was spatially heterogeneous, being the region furthest from the cave entrance the most representative of the number of CFUs. On the other hand, the highest fungal abundance in sediment was at the point 1 (near the main cave entrance). The point 1 had the highest species richness for both airborne and sediment fungi. Taylor et al. (2013) investigated a cave in Minas Gerais state (Brazil) and obtained a different result for the abundance of airborne fungi, which was higher near the entrance; however, the studied cave was highly visited as a tourist attraction. Similar to our results, Cunha et al. (2020) demonstrated that the collection points distant from the bat cave entrance had the highest abundance of airborne fungi, which was attributed to the large presence and movement of bats at this sampling point. In contrast, the abundance of fungi in the sediment was higher at a collection point closer to the main entrance (being the closest point of the Abrigo do Letreiro cave), and the deposition of guano at this point of the cave was higher due to a small colony of bats roosting at this point. In a study of fungi from the terrestrial Gruta do Catão (Bahia state, Brazil), similar results were observed regarding the abundance of sediment fungi, which was higher in the closed areas of the cave (Paula et al. 2016). In an iron-ore cave in Brazil, Taylor et al. (2014) showed that the highest abundance of fungi from sediment was obtained in the twilight zone in the cave, where there was a transition from the entrance to the darkest zone. In addition, Zhang et al. (2018) suggested that fungi from caves may have their origin in the surface environment, as the reported fungal species are also known in outside environments, and because the speleogenesis seems to be short for fungal speciation.
Forty-nine fungal taxa were identified in Abrigo do Letreiro cave, with the collection point located near the main entrance and the highest richness of fungi (22 taxa, almost half of the total species identified in our study). The Abrigo do Letreiro cave has a special morphology, that is, a skylight in the middle of the cave where we found a tree species (Erythrina velutina, Fabaceae) of the Caatinga dry forest (Sobrinho et al. 2016); that skylight allows a connection between the hypogeal and the external environments, resembling the same conditions found at the main cave entrance and also has a similar fungal richness shared between these two collection points. The air current, the direct connection with the land surface, and the presence of bats at this sampling point may influence this result because the fungal community can be influenced by factors that are responsible for transporting fungal propagules and consequently causing their dispersion (Vanderwolt et al. 2013, Vanderwolf et al. 2013b, Johnson et al. 2013, Holz et al. 2018, Cunha et al. 2020).
The mycobiota in caves has a distinct distribution pattern that can be influenced by biotic and abiotic factors, such as the entrances that caves may have, air currents, the presence of bats, and consequently, the deposition of guano (Cunha et al. 2020). This fact can be verified with fungal findings that are normally also found in the external environment in association with plants or with insects and other animals, and this was also verified in the cave environment (Zhang et al. 2017, Cunha et al. 2020). Places with greater deposition of guano and organic material are where greater fungal development can be observed (Taylor et al. 2013, Cunha et al. 2020). The origin of fungi in caves was demonstrated to be from the external environment (Zhang et al. 2018).
Richness and noteworthy records
The fungal isolates were grouped into 19 genera, with Aspergillus being the most common, followed by Penicillium and Cladosporium. These fungi are commonly the most reported taxa in caves around the world, in tropical and subtropical caves, and European countries (Varderwolf et al. 2013, Zhang et al. 2017, Cunha et al. 2020, Zhang et al. 2021). Similar to our results, Zhang et al. (2017) reported that these genera are most frequently isolated from the air in karst caves in China; similar results were obtained by Kokurewicz et al. (2016) in Poland. In Brazil, Aspergillus and Penicillium species were also more abundant in air samples, bat guano, soil, sediment, and other substrates (see Supplementary Table S3; Castrillón et al. 1976, Taylor et al. 2013, Paula et al. 2016, Cunha et al. 2020). Studying a bat cave in the Caatinga, Cunha et al. (2020) obtained the largest number of taxa (12) of Aspergillus, five of Penicillium and three of Cladosporium from different substrates/hosts of the Meu Rei cave (Catimbau National Park, Pernambuco state, Brazil); similar to our results, the genus Aspergillus was the most representative in the air of the Meu Rei cave. Recently, Pereira et al. (2022) reported the richness of eight Cladosporium species, including two new species, from the air in Furna do Morcego cave located in the Caatinga forest in Brazil and Carvalho et al. (2022) reported the isolation of 13 fungal species from bat ectoparasites of a cave in the Caatinga with the description of two new species. Taylor et al. (2014) and Paula et al. (2016) studied the sediments of Brazilian caves and reported Aspergillus and Penicillium as the most commonly found genera. These genera of fungi can be found widely in environments other than caves, and are very common in the isolation of fungi from various substrates that are considered ubiquitous fungi and adaptable to different environments (Crous et al. 2014, Barbosa et al. 2016, Diao et al. 2018).
We also isolated fungi in the Abrigo do Letreiro cave, which are commonly associated with plants and have been reported as entomopathogens. Cunha et al. (2020) also reported isolates of Diaporthe, Aplosporella (mainly found in plants), and Beauveria bassiana (mainly reported as an entomopathogen) in their study of a tropical cave in Brazil. Other fungi are known to have species treated as opportunistic pathogens (e.g. Candida and Malbranchea) were also found in our study. We did not observe the presence of the pathogenic fungus Histoplasma capsulatum, probably because of the methods used to isolate this fungal species; however, human infection by this fungus has already been reported in caves of Brazil (Vicentini et al. 2012) and it has been reported in cavities in other countries (Varderwolf et al. 2013).
New taxa discovered
One new genus and six new species were described for the fungi obtained in our study, accounting for 14 % of the total taxa identified. Aspergillus lebretii sp. nov. has been described in the series Wentiorum of the section Cremei and other species in this section have been related to caves (Vanderwolf et al. 2013, Hubka et al. 2016) and are commonly found in soil and as food spoilage (Hubka et al. 2016); some species are also used in the production of enzymes and organic acids and have been reported as mycotoxin producers and contaminants of cereals (Flannigan 1986, Roehr et al. 1992). Interesting, A. europeaus, the species mostly related to A. lebretii, were collected from soils near European caves, and other isolates were also obtained from the soil in the Czech Republic (Nováková et al. 2012, 2014b, Hubka et al. 2015) and a new species A. citoscrescens was isolated from cave sediment in Spanish Castañar de Ibor cave (Crous et al. 2015); Aspergillus wentii, the most known species of section Cremei, has also been reported in several caves around the world (Vanderwolf et al. 2013). Similar to Aspergillus, species of Talaromyces section Talaromyces, where T. cavernicola sp. nov. is placed, are commonly found in soil, have biotechnological potential to produce pigments and enzymes, and are reported as opportunistic pathogens in humans (Guevara-Suarez et al. 2017, Morales-Oyervides et al. 2020).
The new species Malbranchea cavernosa was introduced in a genus mainly reported to have keratinophilic potential and has been reported as an opportunistic etiological agent in humans and other animals (Hubka et al. 2013). In addition, species of Malbranchea are mainly isolated from the soil and excrement of animals and have been previously reported in caves (as Auxarthron species, Zhang et al. 2021). Pseudolecanicillium caatingaense gen. et sp. nov. has been described in the family Cordycipitaceae, which includes species mainly found as entomopathogenic fungi of several insects and has great potential for use in biological control (Goettel et al. 2008, Mongkolsamrit et al. 2020, Wang et al. 2020). Some fungi belonging to the family Cordycipitaceae have also been reported in caves (Varderwolf et al. 2013, Cunha et al. 2020, Zhang et al. 2021). The fifth new species introduced here, Pseudohumicola cecavii, was found in air and sediment with guano from insectivorous bats and excrements of small mammals (Kerodon rupestris), and other species in this genus have been reported from different substrates (e.g. dung of horse, leaves, paper, and soil) (Wang et al. 2019). Species of Pseudohumicola, a recently introduced genus (Wang et al. 2022), and the related genus Humicola have been isolated from caves in several countries (Vanderwolf et al. 2013, Zhang et al. 2017) and in Brazil, they have also been reported from the guano of frugivore bats (Cunha et al. 2020). In addition, some Humicola species have the potential as bio-organic fertilisers or biological control organisms for plant diseases (Lang et al. 2012). The new species of Tritirachium, T. brasiliense, was isolated from sediment; species of this genus are reported in different substrates and habitats, such as decaying organic matter and plants (see Bezerra et al. 2020) and they have also been reported as human opportunistic pathogens (Martínez-Herrera et al. 2015).
As it is an environment little explored in terms of mycobiota studies, the Abrigo do Letreiro cave presented a high richness of airborne and sediment fungi when compared to other caves in Brazil (see Supplementary Table S3), considering that 14 % of the obtained species were described as novel. Caves in the Caatinga dry forest in Brazil have great potential for the discovery of known and unknown fungal species (see Cunha et al. 2020, Pereira et al. 2022, Carvalho et al. 2022 and Supplementary Table S3) which need to be treated as mycodiversity hotspots in the country.
Acknowledgments
We would like to thank the team of the Laboratório de Taxonomia e Biotecnologia Utilizando Fungos/UFPE, the team/collaborators of the ICMBio/CECAV for their logistical support, Mateus Cruz (MSc) and Daniel Monte (MSc) for their help with the confection of the map and fungal drawings, Tiago Cavalcante (MSc) for his help in the organization of data and statistical analysis, and the staff of the Micoteca URM and Herbário URM for their help depositing the cultures and permanent slides preparations. We also thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Finance Code 001; CAPES-PRInt process number 88887.311891/2018-00), the Financiadora de Estudos e Projetos (FINEP, process number 01.19.0171.00), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, process number 310298/2018-0), and the Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE, process number APQ-0350-2.12/19). V.C.S. Alves was supported by a master scholarship from CNPq (process 132009/2020-0), and E. Barbier was supported by a postdoctoral grant from CAPES and FACEPE (process #88887.353052/2019-00). C.M. Souza-Motta and E. Bernard have fellowships from CNPq.
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
Supplementary Material: http://fuse-journal.org/
Maximum likelihood tree using sequences of tub2-cmdA-rpb2 of species included in Talaromyces section Talaromyces.
Maximum likelihood tree using an independent dataset of ITS, tub2, cmdA, and rpb2 of species included in Talaromyces section Talaromyces.
GenBank accession numbers of sequences obtained in this study (in bold) and sequences from other studies are ordered according to the phylogenetic analyses of the new species described here.
Number of fungal colonies (CFU) from air and sediment.
Checklist of mycoespeleological studies in Brazil.
Details on the combined datasets (number of taxa, sequences, and length of dataset (bp)) and the best–fit models for each partition proposed by MrModelTest.
REFERENCES
- Auler A, Zogbi L. (2005). Espeleologia: noções básicas. 1st edn. Redespeleo Brasil, Brasil. [Google Scholar]
- Barbosa RN, Bezerra JDP, Costa PMO, et al. (2016). Aspergillus and Penicillium (Eurotiales: Trichocomaceae) in soils of the Brazilian tropical dry forest: diversity in an area of environmental preservation. Revista de Biología Tropical 64: 45–53. [DOI] [PubMed] [Google Scholar]
- Barbosa RN, Bezerra JDP, Santos ACS, et al. (2020). Brazilian tropical dry forest (Caatinga) in the spotlight: an overview of species of Aspergillus, Penicillium and Talaromyces (Eurotiales) and the description of P. vascosobrinhous sp. nov. Acta Botanica Brasilica 34: 409–429. [Google Scholar]
- Bastian F, Jurado V, Nováková A, et al. (2010). The microbiology of Lascaux Cave. Microbiology 3: 644–652. [DOI] [PubMed] [Google Scholar]
- Bento DM, Cruz JB, Santos DJ, et al. (2013). Parque Nacional da Furna Feia – o parque nacional com a maior quantidade de cavernas do Brasil. 32th Congresso Brasileiro de Espeleologia, Barreiras-BA. SBE Campinas: 31–43. [Google Scholar]
- Bento DM, Souza-Silva M, Vasconscellos A, et al. (2021). Subterranean “oasis” in the Brazilian semiarid region: neglected sources of biodiversity. Biodiversity and Conservation 30: 3837–3857. [Google Scholar]
- Bezerra JDP, Oliveira RJV, Paiva LM, et al. (2017a). Bezerromycetales and Wiesneriomycetales ord. nov. (class Dothideomycetes), with two novel genera to accommodate endophytic fungi from Brazilian cactus. Mycological Progress 16: 297–309. [Google Scholar]
- Bezerra JDP, Sandoval-Denis M, Paiva LM, et al. (2017b). New endophytic Toxicocladosporium species from cacti in Brazil, and description of Neocladosporium gen. nov. IMA Fungus 8: 77–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra JDP, Felipe MTC, Paiva LM, et al. (2020). Phylogenetic placement of Tritirachium strains from the URM culture collection originally founded by Augusto Chaves Batista (1916-1967) in Brazil, and the description of T. batistae sp. nov. Acta Botanica Brasilica 34: 290–300. [Google Scholar]
- Brasil (2012). Decreto de 5 de junho de 2012. Dispõe sobre a criação do Parque Nacional da Furna Feia, nos municípios de Baraúna e Mossoró, Estado do Rio Grande do Norte. Diário Oficial da República Federativa do Brasil, Brasília, DF. [Google Scholar]
- Carbone I, Kohn LM. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Carvalho JLVR, Lima JMS, Barbier E, et al. (2022) Ticket to ride: fungi from bat ectoparasites in a tropical cave and the description of two new species. Brazilian Journal of Microbiology: 10.1007/s42770-022-00841-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillón AL, Moraes MAP, Furtado MSS. (1976). Isolamento de Microsporum amazonicum do solo do estado do Amazonas, Brasil. Acta Amazônica 6: 487–490. [Google Scholar]
- Coleine C, Stajich JE, Selbmann L. (2022). Fungi are key players in extreme ecosystems. Trends in Ecology & Evolution 37: 517–518. [DOI] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. (2014). Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ. et al. (2015). Fungal planet description sheets: 371–399. Persoonia 35: 264–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Luangsa-Ard JJ, Wingfield MJ, et al. (2018). Fungal Planet description sheets: 785-867. Persoonia 41: 238–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha AOB, Bezerra JDP, Oliveira TGL, et al. (2020). Living in the dark: Bat caves as hotspots of fungal diversity. PLoS ONE 15: e0243494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao YZ, Chen Q, Jiang XZ, et al. (2018). Penicillium section Lanata-divaricata from acidic soil. Cladistics 35: 514–549. [DOI] [PubMed] [Google Scholar]
- Dubreuil V, Fante KP, Planchon O. (2018). The types of annual climates in Brazil: an application of the classification of Koppen from 1961 to 2015. Confins Revue Franco-bresilienne de Geographie 37: 23. [Google Scholar]
- Ferreira RL, Nonaka E, Rosa CA. (2000). Riqueza e abundância de fungos associados ao guano de morcegos hematófagos na Gruta da Lavoura. Carste 12: 46–51. [Google Scholar]
- Flannigan B. (1986). Aspergillus clavatus - an allergenic, toxigenic deteriorgen of cereals and cereal products. International Biodeterioration 22: 79–89. [Google Scholar]
- Flores LH, Onofre SB. (2010). Determinação da presença de fungos anemófilos e leveduras em unidade de saúde da cidade de São Francisco Beltrão–PR. Revista Saúde e Biologia 9: 50–55. [Google Scholar]
- Fonseca RMP, Paula CCP, Bichuette ME, et al. (2019). First record of Amphoromorpha/Basidiobolus fungus on centipedes (Geophilomorpha, Geophilidae) from Brazilian caves. Subterranean Biology 32: 61–67. [Google Scholar]
- Glass NL, Donaldson GC. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goettel MS, Koike M, Kim JJ, et al. (2008). Potential of Lecanicillium spp. for management of insects, nematodes and plant diseases. Journal of Invertebrate Pathology 98: 256–261. [DOI] [PubMed] [Google Scholar]
- Guevara-Suarez M, Sutton DA, Gene J, et al. (2017). Four new species of Talaromyces from clinical sources. Mycoses 60: 651–662. [DOI] [PubMed] [Google Scholar]
- Heaton T. (1986). Caves: a tremendous range in energy environments on earth. National Speleological Society News 44: 301–4. [Google Scholar]
- Holsinger JR, Culver DC. (1988). The invertebrate cave fauna of Virginia and a part of eastern Tennessee: zoogeography and ecology. Brimleyana 14: 1–162. [Google Scholar]
- Holz PH, Lumsden LF, Marenda MS, et al. (2018). Two subspecies of bent-winged bats (Miniopterus orianae bassanii and oceanensis) in southern Australia have diverse fungal skin flora but not Pseudogymnoascus destructans. PLoS ONE 13: e0204282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Cho HS, Shin HD, et al. (2006). Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology 56: 477–485. [DOI] [PubMed] [Google Scholar]
- Hornick BL. (2017). Fungal iron oxidation in Brazilian iron caves. Honors Research Projects: 432. [Google Scholar]
- Houbraken J, Kocsubé S, Visagie CM, et al. (2020). Classification of Aspergillus, Penicillium and Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Dobiasova S, Lyskova P, et al. (2013). Auxarthron ostraviense sp. nov., and A. umbrinum associated with non-dermatophytic onychomycosis. Medical Mycology 51: 614–624. [DOI] [PubMed] [Google Scholar]
- Hubka V, Nováková A, Kolarik M, et al. (2015) Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani sect. nov. Mycologia 107: 169–208. [DOI] [PubMed] [Google Scholar]
- Hubka V, Nováková A, Samsom RA, et al. (2016). Aspergillus europaeus sp. nov., a widely distributed soil-borne species related to A. wentii (section Cremei). Plant Systematics and Evolution 302: 641–650. [Google Scholar]
- Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) (2022). Anuário estatístico do patrimônio espeleológico brasileiro 2022. Centro Nacional de Pesquisa e Conservação de Cavernas (ICMBio/CECAV), Brasília – DF. [Google Scholar]
- Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) (2020). Plano de manejo do Parque Nacional da Furna Feia. Brasília – DF. [Google Scholar]
- Jansen DC, Cavalcanti LF, Lamblém HS. (2012). Mapa de potencialidade de ocorrência de cavernas no Brasil, na escala 1:2.500.000. Revista Brasileira de Espeleologia 2: 42–57. [Google Scholar]
- Johnson LJAN, Miller AN, McCleery RA, et al. (2013). Psychrophilic and psychrotolerant fungi on bats and the presence of Geomyces spp. on bat wings prior to the arrival of white nose syndrome. Applied and Environmental Microbiology 79: 5465–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokurewicz T, Ogórek R, Pusz W, et al. (2016). Bats increase the number of cultivable airborne fungi in the “Nietoperek” bat reserve in Western Poland. Microbial Ecology 72: 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JJ, Hu J, Ran W, et al. (2012). Control of cotton Verticillium with and fungal diversity of rhizosphere soils by bio-organic fertilizer. Biology and Fertilly of Soils 48: 191–203. [Google Scholar]
- Liu YJ, Whelen S, Hall B. (1999). Phylogenetic relationship among Ascomycetes: Evidence from an RNA Polymerase II Subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lobo HAS, Boggiani PC. (2013). Cavernas como patrimônio geológico. Boletim Paranaense de Geociências 70: 190–199. [Google Scholar]
- Martínez-Herrera EO, Arroyo-Camarena S, Tejada-García DL, et al. (2015). Onychomycosis due to opportunistic molds. Anais Brasileiros de Dermatologia 90: 334–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010). Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Gateway Computing Environments Workshop (GCE): 1–8. [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Tasanathai K, et al. (2020). Molecular phylogeny and morphology reveal cryptic species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycological Progress 19: 957–983. [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Tasanathai K, et al. (2022) Comprehensive treatise of Hevansia and three new genera Jenniferia, Parahevansia and Polystromomyces on spiders in Cordycipitaceae from Thailand. MycoKeys 91: 113–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Oyervides L, Ruiz-Sanchez JP, Oliveira JC, et al. (2020). Biotechnological approaches for the production of natural colorants by Talaromyces/Penicillium: A review. Biotechnology Advances 43: 107601. [DOI] [PubMed] [Google Scholar]
- Nieves-Rivera AM. (2003). Mycological survey of Río Camuy Caves Park, Puerto Rico. Journal of Cave and Karst Studies 65: 23–28. [Google Scholar]
- Nováková A. (2009). Microscopic fungi isolated from the Domica Caves system (Slovak Karst National Park, Slovakia). A review. International Journal of Speleology 38: 71–82. [Google Scholar]
- Nováková A, Hubka V, Saiz-Jimenez C, et al. (2012). Aspergillus baeticus sp. nov. and Aspergillus thesauricus sp. nov., two species in section Usti from Spanish caves. International Journal of Systematic and Evolutionary Microbiology 62: 2778–2785. [DOI] [PubMed] [Google Scholar]
- Nováková A, Hubka V, Saíz-Jiménez C. (2014). Microscopic fungi isolated from cave air and sediments in the Nerja Cave - preliminary results. In: The Conservation of Subterranean Cultural Heritage (Saíz-Jiménez C, ed). CRC Press, London: 239–246. [Google Scholar]
- Nylander JAA. (2004). MrModeltest v. 2. Program distributed by the author. [Google Scholar]
- Ogórek R, Lejman A, Matkowski K. (2013). The fungi isolated from the NiedŸwiedzia Cave in Kletno (Lower Silesia, Poland). International Journal of Speleolgy 42: 161–166. [Google Scholar]
- Ogórek R, Visòovská Z, Tanèinová D. (2016). Mycobiota of underground habitats: case study of Harmanecká cave in Slovakia. Microbiology Ecology 71: 87–99. [DOI] [PubMed] [Google Scholar]
- Otálora-Ardila A, Torres JM, Barbier E, et al. (2019). Thermally-assisted monitoring of bat abundance in an exceptional cave in Brazil’s Caatinga drylands. Acta Chiropterologica 21: 411–423. [Google Scholar]
- Paula CCP, Montoya QV, Rodrigues A, et al. 2016. Terrestrial filamentous fungi from Gruta do Catão (São Desidério, Bahia, Northeastern Brazil) show high levels of cellulose degradation. Journal of Cave and Karst Studies 78: 208–217. [Google Scholar]
- Peay K, Kennedy P, Talbot J. (2016). Dimensions of biodiversity in the Earth mycobiome. Nature Reviews Microbiology 14: 434–447. [DOI] [PubMed] [Google Scholar]
- Pedro EG, Bononi VLR. (2007). Cave fungi of the karst region of the State Touristic Park of the Upper Ribeira Valley (PETAR) in the State of São Paulo in Brazil. Focus 5: 65–78 [Google Scholar]
- Pereira MLS, Carvalho JLVR, Lima JMS, et al. (2022). Richness of Cladosporium in a tropical bat cave with the description of two new species. Mycological Progress 21: 345–357. [Google Scholar]
- Piló LB, Auler A. (2013). Introdução à Espeleologia. In: Curso de Espeleologia e Licenciamento Ambiental. ICMBio, Brasília: 7–23. [Google Scholar]
- Pinto-da-Rocha R. (1993). Invertebrados cavernícolas da porção meridional da província espeleológica do Vale do Ribeira, Sul do Brasil. Revista Brasileira de Zoologia 10: 229–255. [Google Scholar]
- Rambaut A. (2010). FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. [Google Scholar]
- Rayner RW. (1970). A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society. Kew, Surrey, UK. [Google Scholar]
- Rehner S, Buckley E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Andrade E, Cano-Lira JF, Wiederhold N, et al. (2021). A revision of malbranchea-like fungi from clinical specimens in the United States of America reveals unexpected novelty. IMA Fungus 12: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehr M, Kubicek CP, Kominek J. (1992). Industrial acids and other small molecules. In: Aspergillus: Biology and industrial applications (Bennett JW, Klich MA, eds) Butterworth-Heinemann, Boston: 91–131. [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systems Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Houbraken J, Thrane U, et al. (2010). Food and indoor fungi. 2nd edn. CBS Laboratory Manual Series 2, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. [Google Scholar]
- Silva JMC, Leal IR, Tabarelli M. (2017). Caatinga: the largest tropical dry forest region in South America. 1st ed. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- Sobrinho JE, Silva STA, Ribeiro GB, et al. (2016). Monitoramento do microclima e análise da contribuição de visitações sobre seus parâmetros, em cavernas do Parque Nacional da Furna Feia. Centro de Engenharias/Centro de Ciências Agrárias, Universidade Federal Rural do Semi-Árido, Brasil. [Google Scholar]
- Stamatakis A, Rougemont JPH. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Bioinformatics 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Sung GH, Sung JM, Hywel-Jones NL, et al. (2007). A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution 44: 1204–1223. [DOI] [PubMed] [Google Scholar]
- Taylor ELS, Resende-Stoianoff MA, Ferreira RL. (2013). Mycological study for a management plan of a Neotropical show cave (Brazil). International Journal of Speleology 42: 267–277. [Google Scholar]
- Taylor ELS, Ferreira RL, Cardoso PG, et al. (2014). Cave entrance dependent spore dispersion of filamentous fungi isolated from various sediments of iron ore cave in Brazil: a colloquy on human threats while caving. Ambient Science 1: 16–28. [Google Scholar]
- Tiquia-Arashiro SM, Grube M. (2019) Fungi in Extreme Environments: Ecological Role and Biotechnological Significance. (Tiquia-Arashiro SM, Grube M, eds). Springer International Publishing, Berlin/Heidelberg, Germany: 626. [Google Scholar]
- Travassos LEP. (2019). Geomorfologia cárstica subterrânea. In: Princípios de Carstologia e Geomorfologia Cárstica. ICMBio, Brasília: 133–161. [Google Scholar]
- Vanderwolf KJ, Malloch D, Mcalpine DF, et al. (2013). World review of fungi, yeasts, and slime molds in caves. International Journal of Speleology 42: 77–96. [Google Scholar]
- Vanderwolf KJ, McAlpine DF, Malloch D, et al. (2013). Ectomycota associated with hibernating bats in Eastern Canadian caves prior to the emergence of white-nose syndrome. Northeastern Naturalist 20: 115–130. [Google Scholar]
- Vicentini AP, Passos AN, Silva DF, et al. (2012). Histoplasmose: um risco ocupacional entre pesquisadores que realizam trabalho de campo? Revista Instituto Adolfo Lutz 71: 747–752. [Google Scholar]
- Vilgalys R, Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4239–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Yang FY, Meijer M, et al. (2019). Redefining Humicola sensu stricto and related genera in the Chaetomiaceae. Studies in Mycology 63: 65–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YB, Wang Y, Fan Q, et al. (2020). Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Diversity 103: 1–46. [Google Scholar]
- Wang XC, Zhuang WY. (2022). New species of Talaromyces (Trichocomaceae, Eurotiales) from Southwestern China. Journal of Fungi 8: 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Han PJ, Bai FY, et al. (2022). Taxonomy, phylogeny and identification of Chaetomiaceae with emphasis on thermophilic species. Studies in Mycology 101: 121–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylognetics. In: A Guide to Molecular Methods and 76 Applications (Innis MA, Gelfand DH, Sninsky JJ, White JW, eds). Academic Press: 315–322. [Google Scholar]
- Zhang ZF, Liu F, Zhou X, et al. (2017). Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia 39: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZF, Peng Z, Cai L. (2018). Origin of Cave Fungi. Frontiers in Microbiology 9: 1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZF, Zhou SY, Eurwilaichitr L, et al. (2021). Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Diversity 106: 29–136. [Google Scholar]
- Zhang ZK, Wang XC, Zhuang WY, et al. (2021). New Species of Talaromyces (Fungi) isolated from soil in Southwestern China. Biology 10: 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood tree using sequences of tub2-cmdA-rpb2 of species included in Talaromyces section Talaromyces.
Maximum likelihood tree using an independent dataset of ITS, tub2, cmdA, and rpb2 of species included in Talaromyces section Talaromyces.
GenBank accession numbers of sequences obtained in this study (in bold) and sequences from other studies are ordered according to the phylogenetic analyses of the new species described here.
Number of fungal colonies (CFU) from air and sediment.
Checklist of mycoespeleological studies in Brazil.
Details on the combined datasets (number of taxa, sequences, and length of dataset (bp)) and the best–fit models for each partition proposed by MrModelTest.