Abstract
CRISPR-based assays can be adopted as ultrasensitive molecular diagnostics in resource-limited settings, but point-of-care applications must address additional requirements. Here, we discuss the major obstacles for developing these assays and offer insights into how to surmount them.
Subject terms: Assay systems, Biochemical assays
Rapid and accurate diagnosis is essential for effective treatment, prognostic evaluation and containment of many diseases. However, almost half of the global population has limited access to molecular diagnostics1. Assays based on the activity of clustered regularly interspaced short palindromic repeats (CRISPR) could address the long-standing problem of diagnosis in poor, rural and marginalized communities globally owing to their low cost, versatility and ease-of-use.
CRISPR–Cas and guide RNA (gRNA) complexes can recognize and cleave exogenous nucleic acid sequences to attenuate virus replication2. The specificity of these complexes can be readily adjusted by a gRNA substitution, which has led to their rapid adoption in gene-editing applications and later in high-specificity ultrasensitive nucleic acid detection. Despite being robust and able to detect scarce targets in complex biological samples3, most current CRISPR nucleic acid detection methods do not meet the World Health Organization’s ‘ASSURED’ criteria4 for point-of-care (POC) testing, which require assays to be affordable, deliverable and usable in remote or resource-limited areas.
Refinements for CRISPR POC assays
Most CRISPR assays involve nucleic acid extraction and target amplification steps that require training and expensive material and equipment. Refinements can simplify assay workflows and instrumentation but must be balanced against assay performance. For example, extraction-free nucleic acid isolation protocols that use reagents and/or heat to release nucleic acid from cells, viruses or nucleic acid complexes from clinical specimens are preferred because CRISPR reactions tolerate factors present in biospecimens5. Streamlined affinity matrix procedures can also be used to concentrate scarce nucleic acid targets and purify them from inhibitory factors to address sensitivity issues. Similarly, methods that constrain CRISPR reactions in small volumes can intensify signal generation. This strategy can be particularly useful for in situ analysis of nucleic acid targets in extracellular vesicles captured from liquid biopsies, which can be induced to fuse with reagent-loaded liposomes6.
Separate nucleic acid amplification steps increase assay time and complexity and introduce a cross-contamination risk during sample transfer to the CRISPR reaction. Alternatively, nucleic acid amplification and CRISPR reactions can be performed successively or concurrently without fluid transfer, using microfluidic devices or single-well reactions with optimized reaction parameters, respectively. Eliminating the target nucleic acid amplification step can reduce the assay time, cost and contamination risk, but requires an ultrasensitive signal readout system to ensure acceptable assay performance. For example, multiplex assays that use distinct CRISPR–Cas and gRNA complexes to recognize different sites in the same nucleic acid target can improve the performance of amplification-free CRISPR assays, but at the same time increase assay costs and reduce specificity in proportion to the number of added targets7.
Therefore, instead of simplifying individual steps, refinements for POC applications should focus on systematically integrating CRISPR assay procedures into user-friendly devices (Fig. 1), such as microfluidic chips8 or lateral flow assays9. This approach can also be used to develop wearable devices that could function as multiplex tests for disease diagnosis or environmental exposure9.
Fig. 1. CRISPR assay requirements for use in resource-limited settings.
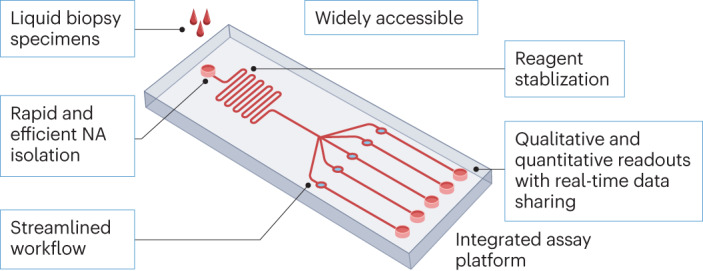
New assays intended for broad use should analyse minimally invasive liquid biopsy specimens (such as blood, saliva or urine) on inexpensive assay cassettes that automate assay workflows (such as isolation, amplification and detection of nucleic acids (NA)) and are pre-loaded with stabilized assay reagents that reduce cold-chain concerns. Such assays should not require technical expertise or expensive equipment, and their cassette-based workflows should improve accuracy by reducing sources of user error. Finally, these assays should be readable by eye (qualitative or semi-quantitative) or by a smart terminal (quantitative) reporting real-time results to a central system to facilitate disease control and telemedicine efforts.
Readouts, analyses and data transfer
Sensitive and quantitative CRISPR assay readouts currently rely primarily on fluorescent or electrical signal changes detected by sophisticated laboratory equipment not usable in resource-limited or remote settings. Nanomaterial-based colorimetric sensors can, however, be easily integrated into CRISPR assays to replace fluorescence signal outputs3,5. Colour changes that reflect target nucleic acid abundance in these assays can be read by the naked eye but do not provide quantitative data. However, standard dilution control dipsticks, or their equivalents, could be incorporated into these assays to provide semi-quantitative or quantitative results, when read by the naked eye or common, portable and inexpensive devices, including smartphones7,9. Notably, the network connection of these devices allows real-time encrypted data sharing with national agencies or central hospitals to facilitate epidemiological investigations, disease control decisions and enable telemedicine programs that could improve patient care in remote, resource-limited or otherwise underserved areas (Fig. 1).
Clinical validation studies
Reports of new CRISPR assays often lack the rigorous clinical validation data necessary to accurately evaluate their diagnostic performance and clinical utility. Most of these studies used small retrospective sample cohorts to report diagnostic sensitivity and specificity, which can introduce data artefacts, particularly if the analysed samples were not collected or handled in an appropriate manner. Studies intended to validate CRISPR assay diagnostic performance require appropriate reference standards because high-sensitivity CRISPR assays can exceed the diagnostic performance of reference clinical assays and thus require comparisons against composite reference standards that integrate results from several clinical tests or findings. The development and adoption of standard validated approaches for CRISPR data analysis are also urgently needed to allow uniform data reporting and comparison of results from different CRISPR assays.
Global access
Transport and storage aspects need to be accounted for when considering resource-limited settings. Lyophilized CRISPR reagents can be stored at 4 °C for over 5 months and at room temperature for up to 30 days without substantial performance losses5,10. However, less is known about reagent stability under more variable ambient temperature conditions, and development and adoption of standard reagent stabilization protocols should greatly benefit the development of POC CRISPR assays. Early in their development, new POC tests should, as a minimum requirement, evaluate and optimize assay performance after reagent lyophilization and rehydration to extend their shelf life in the absence of robust cold-chain management (Fig. 1).
Finally, streamlined patent licensing and market authorization procedures need to be established for CRISPR reagents and CRISPR assays to standardize and scale-up production, reduce assay development and production costs and barriers to market entry, and to diversify and stabilize supply chains. Given the potential for CRISPR diagnostics, it is advisable to streamline these processes, as was done for SARS-CoV-2 PCR diagnostics early in the COVID-19 pandemic when governmental authorities in multiple countries used emergency use authorization policies to expand testing capacity.
Acknowledgements
The work was primarily supported by research funding provided by the National Institutes of Health (NIH): U01CA252965, R01HD090927, R01AI144168, R01HD103511 and R21NS130542, and W8IXWH1910926 from the US Department of Defense.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Fleming KA, et al. The Lancet Commission on diagnostics: transforming access to diagnostics. Lancet. 2021;398:1997–2050. doi: 10.1016/S0140-6736(21)00673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in Bacteria and Archaea. Ann. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 3.Gootenberg JS, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Land KJ, Boeras DI, Chen X-S, Ramsay AR, Peeling RW. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019;4:46–54. doi: 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arizti-Sanz J, et al. Simplified Cas13-based assays for the fast identification of SARS-CoV-2 and its variants. Nat. Biomed. Eng. 2022;6:932–943. doi: 10.1038/s41551-022-00889-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ning B, et al. Liposome-mediated detection of SARS-CoV-2 RNA-positive extracellular vesicles in plasma. Nat. Nanotech. 2021;16:1039–1044. doi: 10.1038/s41565-021-00939-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fozouni P, et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333.e329. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrasekaran SS, et al. Rapid detection of SARS-CoV-2 RNA in saliva via Cas13. Nat. Biomed. Eng. 2022;6:944–956. doi: 10.1038/s41551-022-00917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen PQ, et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat. Biotech. 2021;39:1366–1374. doi: 10.1038/s41587-021-00950-3. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen LT, Rananaware SR, Pizzano BLM, Stone BT, Jain PK. Clinical validation of engineered CRISPR/Cas12a for rapid SARS-CoV-2 detection. Commun. Med. 2022;2:7. doi: 10.1038/s43856-021-00066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


