Abstract
Sorghum production is seriously threatened by the root parasitic weeds (RPWs) Striga hermonthica and Striga asiatica in sub-Saharan Africa. Research has shown that Striga control depends on eliminating its seed reserves in soil. Several species of the genus Fusarium (Nectriaceae, Hypocreales), which have been isolated from diseased Striga plants have proven to be highly pathogenic to all developmental stages of these RPWs. In the present study 439 isolates of Fusarium spp. were found associated with soils from Sorghum growing fields, Sorghum rhizosphere, or as endophytes with Sorghum roots and seeds, or as endophytes of Striga stems and seeds. Based on multi-locus phylogenies of combinations of CaM, tef1, rpb1 and rpb2 alignments, and morphological characteristics, 42 species were identified, including three species that are newly described, namely F. extenuatum and F. tangerinum from Sorghum soils, and F. pentaseptatum from seed of Striga hermonthica. Using a previously published AFLP-derived marker that is specific to detect isolates of F. oxysporum f.sp. strigae, an effective soil-borne biocontrol agent against Striga, we also detected the gene in several other Fusarium species. As these isolates were all associated with the Striga/Sorghum pathosystem, the possibility of horizontal gene transfer among these fusaria will be of interest to further investigate in future.
Citation: Lombard L, van Doorn R, Groenewald JZ, Tessema T, Kuramae EE, Etolo DW, Raaijmakers JM, Crous PW (2022). Fusarium diversity associated with the Sorghum-Striga interaction in Ethiopia. Fungal Systematics and Evolution 10: 177–215. doi: 10.3114/fuse.2022.10.08
Keywords: biological control agent, molecular phylogeny, systematics, three novel taxa
INTRODUCTION
Root parasitic weeds (RPWs) of the genera Orobanche, Phelipanche and Striga are major yield-limiting factors of a wide range of cereal crops such as maize, rice, millet, sorghum and the legume cowpea, particularly in Sub-Saharan Africa, India and Southeast Asia (Masteling et al. 2019). In sub-Saharan Africa, Sorghum production is seriously threatened by Striga hermonthica and Striga asiatica. Striga species are obligate hemiparasitic root parasites which penetrate the roots of host plants and absorb nutrients using a specialised organ known as haustoria. These RPWs have been estimated to infest some 64 % of the total cereal production area in West Africa (Gressel et al. 2004, Ejeta 2007, Parker 2012), and this figure is still increasing. For Striga hermonthica it has been estimated that 50–300 M ha (approximately equivalent to the size of France and India) of field soils in Africa are currently infested (Vurro et al. 2019). Crop infections can lead to grain yield losses of 20–80 % in Africa, but total crop losses have also been reported (Gurney et al. 2002).
Seed germination of obligate RPWs reply on host-derived signals released by the roots (e.g. strigolactones). Seed germination is followed by haustorium formation, leading to root infection. Other stages of the life cycle that are possible targets for disease control include the Striga soil seed bank, and seed production. Present control strategies include resistance breeding, hand weeding, alternative cropping practices, and chemical control (Eteja 2007). Because these strategies are only effective if applied in combination, an integrated systems approach is needed to provide effective and sustainable control of RWPs (Masteling et al. 2019).
Striga control depends on eliminating its seed reserves in soil (Daffalla et al. 2014). Species of the common soil-borne genus Fusarium, which have been isolated from diseased Striga hermonthica have proven to be highly pathogenic to all developmental stages of the parasite, including seeds. Some species are highly host-specific and non-pathogenic to a wide range of crops tested, thus rendering them as candidates to be used as mycoherbicides (Kroschel et al. 1999). Anteyi et al. (2022) reported that an isolate of F. venenatum produced the exometabolite diacetoxyscirpenol (DAS), which consistently inhibited seed germination of diverse S. hermonthica seedlots. Furthermore, surveys for fungal pathogens of Striga spp. revealed that Fusarium species (especially F. oxysporum) were the most prominent pathogens associated with diseased Striga spp. (Sauerborn et al. 2007), with several species showing significant disease development in this host when tested under controlled and/or field conditions (Abbasher & Sauerborn 1992, Ciotola et al. 1995, 2000, Abbasher et al. 1996, Kroschel et al. 1996, Sauerborn et al. 1996a, 2007, Hess et al. 2002, Marley et al. 2004). In such trials the application of isolates of F. nygamai and F. oxysporum caused more than 90 % reduction of S. hermonthica emergence (Abbasher & Sauerborn, 1992, Ciotola et al. 1995). Furthermore, isolates of F. oxysporum f.sp. strigae (Abbasher et al. 1995, Marley et al. 1999) resulted in a positive result when it was used as a biological control agent against Striga (Abbasher et al. 1995, Ciotola et al. 1995, Kroschel et al. 1996, Marley et al. 1999). It has also been shown that F. incarnatum greatly reduced the germination, survival and attachment of S. asiatica on maize roots (Abbasher et al. 1996).
Based on these findings, the aim of the present study was to determine which Fusarium species are commonly associated with Sorghum and rhizosphere soils, or as endophytes with Sorghum roots and seeds, or Striga stems and seeds.
MATERIALS AND METHODS
Isolates
Soil
Soil samples with varying levels of Striga infestation were collected in Ethiopia (samples E1–E47). To collect sorghum rhizophere-associated fungi, cultivar Teshale was grown in pots containing the 47 soils under greenhouse conditions and the rhizosphere suspension was collected from 5-wk-old plants. Fungal isolations followed the methods of Groenewald et al. (2018) and Giraldo et al. (2019). Colonies were sub-cultured on 2 % potato dextrose agar (PDA), oatmeal agar (OA), malt extract agar (MEA) (Crous et al. 2019), synthetic nutrient-poor agar (SNA; Nirenberg 1976), carnation leaf agar (CLA; Fisher et al. 1982), and incubated at 25 °C under continuous near-ultraviolet light to promote sporulation. Reference isolates and specimens of the studied Fusarium spp. are maintained in the Ethiopian Biodiversity Institute (EBI), Addis Ababa, Ethiopia (EMCC-F) and in the working collection of Lorenzo Lombard, housed at the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands (LLC).
Endophytes
Fungal endophytes were isolated from 53 Striga plant stems, 107 Sorghum roots and root collars, from 20 seeds from each of 59 different Sorghum genotypes, and from seeds of Striga asiatica collected in Derashe and from S. hermonthica collected in Abergelle, Asosa, Feddis, Humera, Kobo, with 150 seeds per sample.
Plant stems and roots were washed with running tap water, and 5-mm segments were cut with a sterilised scalpel, and immersed into the following series of solutions: sterile distilled H2O for 60 s, 70 % ethanol for 60 s, 2.5 % sodium hypochlorite for 4 min, 70 % ethanol for 30 s, and a final rinse in sterile distilled H2O. Striga and Sorghum seeds were treated in a similar fashion, except that they were rinsed for longer in 2.5 % sodium hypochlorite (5 min), and afterwards the seedcoat was broken with a pincette. All tissues were plated onto malt extract agar with antibiotics (Penicillin-Streptomycin), to inhibit bacterial growth and incubated on a laboratory bench (21 °C). Plates were checked daily for fungal growth, and emerging colonies were hyphal tipped, re-inoculated on fresh MEA plates, and colonised agar plugs maintained at -80 °C in 10 % (v/v) glycerol (Crous et al. 2019).
DNA extraction, amplification (PCR), phylogeny and AFLP-based marker testing
Protocols for genomic DNA isolation, PCR amplification of partial calmodulin (CaM) gene, DNA-directed RNA polymerase II largest (rpb1) and second largest subunit (rpb2) genes, and translation elongation factor 1-alpha (tef1) gene, and sequencing of the novel isolates (Supplementary Table S1) followed Crous et al. (2021b). Sequences derived in this study were deposited in GenBank (Supplementary Table S1), the alignments and phylogenetic trees in figshare (doi: 10.6084/m9.figshare.21080776).
Initial identifications to species complex level were made using megablast searches (Zhang et al. 2000) of the obtained sequences against NCBIs GenBank nucleotide database and the Fusarioid-ID database (www.fusarium.org; Crous et al. 2021b). Reference sequences (Supplementary Table S2) and based on megablast searches were then used to construct single-gene and multi-gene alignments for the different Fusarium species complexes. Phylogenetic analyses using RAxML v. 8.0.0 (Stamatakis 2014), IQ-TREE v. 2.1.3 (Nguyen et al. 2015, Minh et al. 2020) and MrBayes v. 3.2.7 (Ronquist & Huelsenbeck 2003) followed Crous et al. (2021b), with the exception that trees were saved every 10 or 100 generations (Supplementary Table S3). All resulting trees were printed with Geneious Prime v. 2022.1.1 (Biomatters Ltd, Auckland, New Zealand) and the layout of the trees was done in Adobe Illustrator v. CC 2021.
The absence or presence of an explicit AFLP-based marker associated with Fusarium oxysporum f.sp. strigae (Fos) was also tested for all novel isolates (Supplementary Table S1). DNA amplification conditions and primers follow Zimmermann et al. (2015). Sanger sequencing was performed on all positive amplicons and compared to the reference sequence provided by Zimmermann et al. (2015) to confirm that the correct gene fragment was obtained.
Morphology
Slide preparations were mounted in water, from colonies sporulating on CLA, following the protocols described by Crous et al. (2021b). Observations were made with a Nikon SMZ25 dissection microscope, and with a Zeiss Axio Imager 2 light microscope using differential interference contrast (DIC) illumination and images recorded on a Nikon DS-Ri2 camera with associated software. Colony characters and pigment production were noted after 7 d of growth on MEA, PDA and OA incubated at 25 °C. Colony colours (surface and reverse) were scored using the colour charts of Rayner (1970). Taxonomic novelties were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
RESULTS
Isolates
In the present study, 439 isolates representing 42 species (including two as “Fusarium sp.” and three novel species) from eight species complexes were obtained from the abovementioned substrates (Supplementary Table S1). These are treated below in the Taxonomy section.
Phylogeny
Four multigene alignments were generated in the present study and subjected to the three phylogenetic analyses described above. Statistical values for the alignments and phylogenetic trees are summarised in Supplementary Table S3.
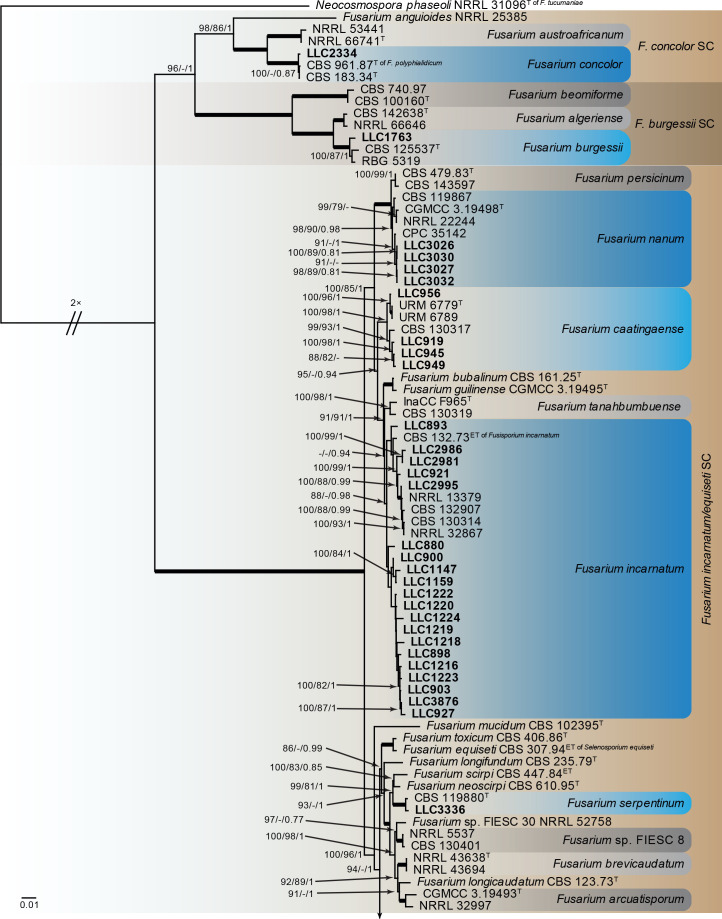
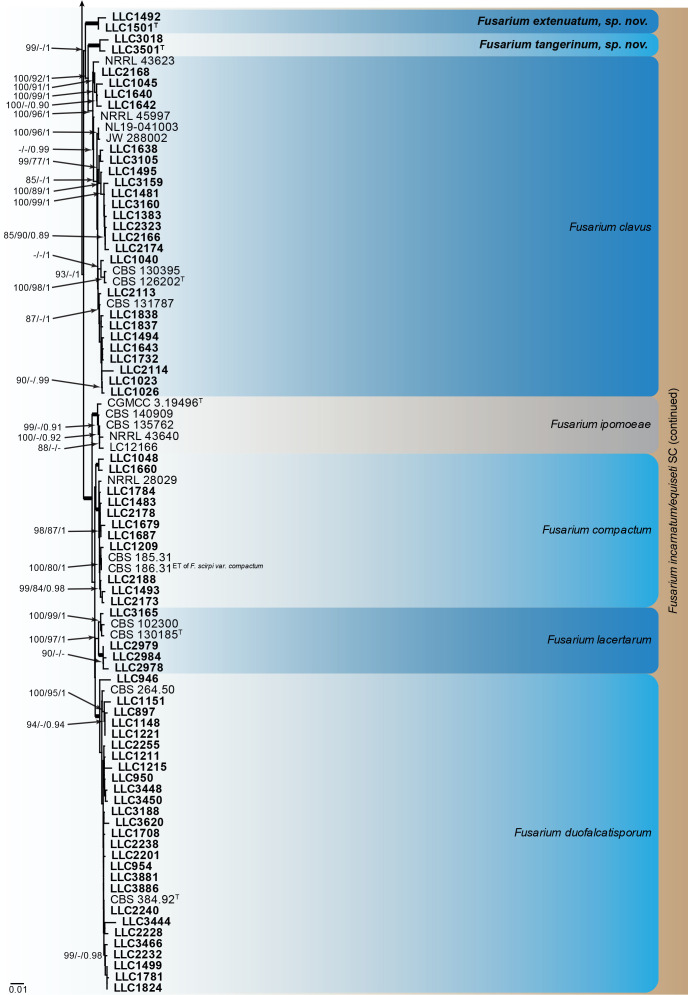
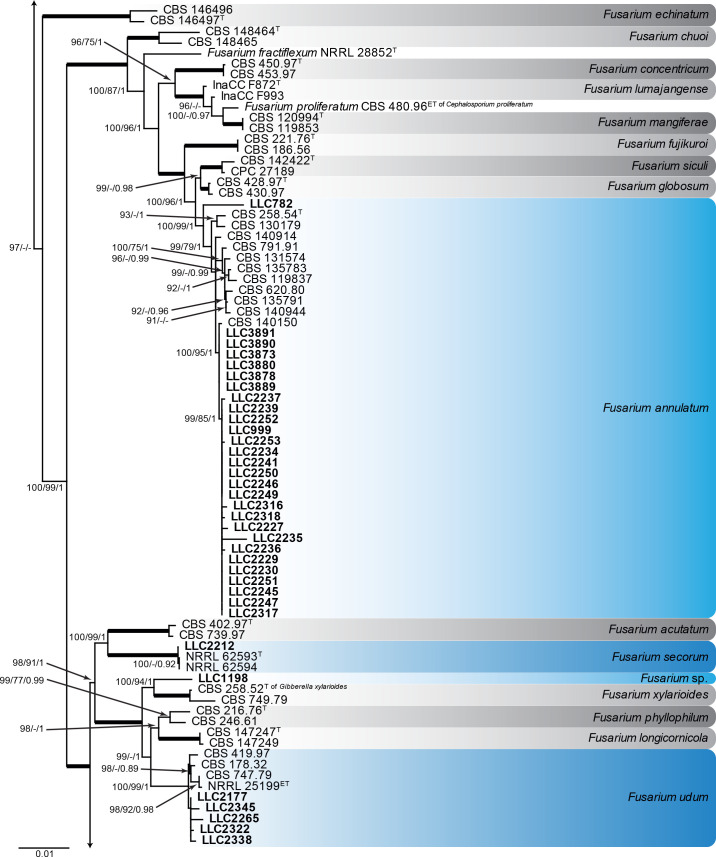
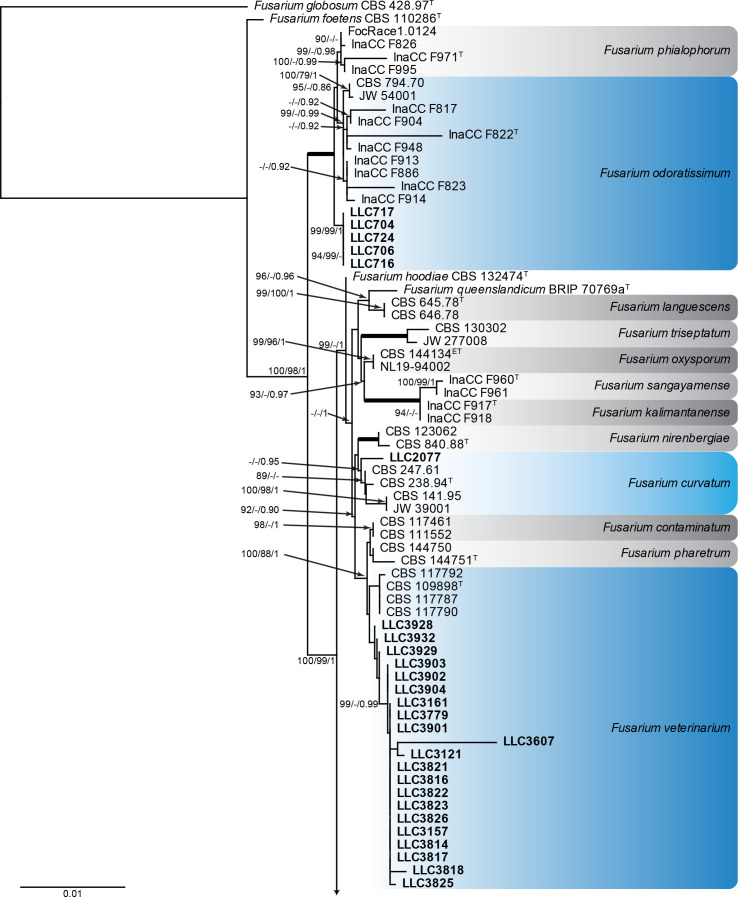
Fusarium burgessii, F. concolor and F. incarnatum species complexes (Fig. 1): Isolates clustered with 10 known species, and two novel lineages which are formally named below. Known species are F. concolor (FCOSC), F. burgessii (FburSC) and F. caatingaense, F. clavus, F. compactum, F. duofalcatisporum, F. incarnatum, F. lacertarum, F. nanum and F. serpentinum (FIESC). The two novel species, F. extenuatum and F. tangerinum, belong to the FIESC. The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) overall displayed the same species clades and mainly differed with regards to the backbone relationships between species clades/lineages (data not shown, support and posterior probability values are superimposed on the presented figure).
Fig. 1.
The IQ-TREE maximum likelihood consensus tree inferred from the combined CaM, rpb1, rpb2 and tef1 sequence alignment. Thickened lines indicate nodes with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other nodes indicated at the branches (IQ-TREE > 84 % / RAxML > 74 % / PP > 0.74). The tree is rooted to Neocosmospora phaseoli (NRRL 31096, ex-type culture). The scale bar indicates the number of expected changes per site. Species complexes are indicated on the right and highlighted with coloured blocks (brown tints). Species clades containing the novel isolates (in bold) are highlighted with coloured blocks (blue tints). Additional species clades are shown in coloured blocks with grey tints. Taxonomic novelties recognised in this study are shown in bold text.
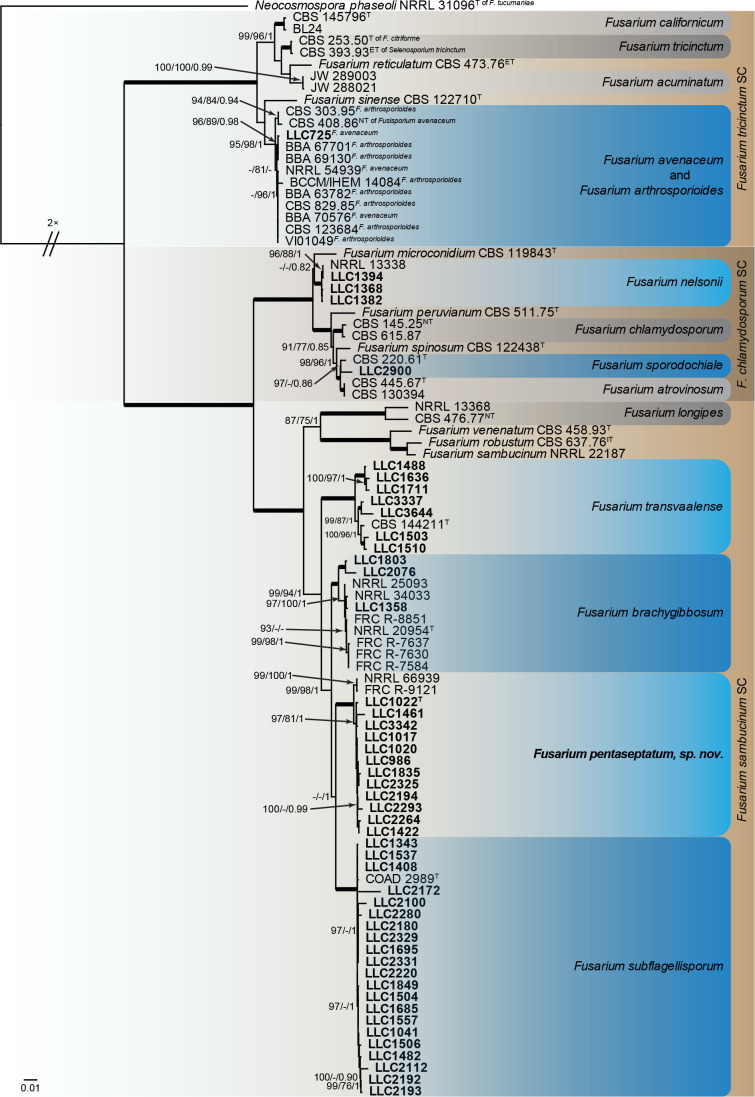
Fusarium chlamydosporum, F. sambucinum and F. tricinctum species complexes (Fig. 2): Novel isolates clustered with six known species, and one novel lineage (previously called Fusarium sp. FSAMSC 28) which are formally named below. Known species are F. avenaceum (FTSC), F. nelsonii and F. sporodochiale (FCSC) and F. brachygibbosum, F. subflagellisporum and F. transvaalense (FSAMSC). The novel species, F. pentaseptatum, belongs to the FSAMSC. The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) overall displayed the same species clades and mainly differed with regards to the backbone relationships between species clades/lineages (data not shown, support and posterior probability values are superimposed on the presented figure).
Fig. 2.
The IQ-TREE maximum likelihood consensus tree inferred from the combined CaM, rpb1, rpb2 and tef1 sequence alignment. Thickened lines indicate nodes with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other nodes indicated at the branches (IQ-TREE > 84 % / RAxML > 74 % / PP > 0.74). The tree is rooted to Neocosmospora phaseoli (NRRL 31096, ex-type culture). The scale bar indicates the number of expected changes per site. Species complexes are indicated on the right and highlighted with coloured blocks (brown tints). Species clades containing the novel isolates (in bold) are highlighted with coloured blocks (blue tints). Additional species clades are shown in coloured blocks with grey tints. The taxonomic novelty recognised in this study is shown in bold text.
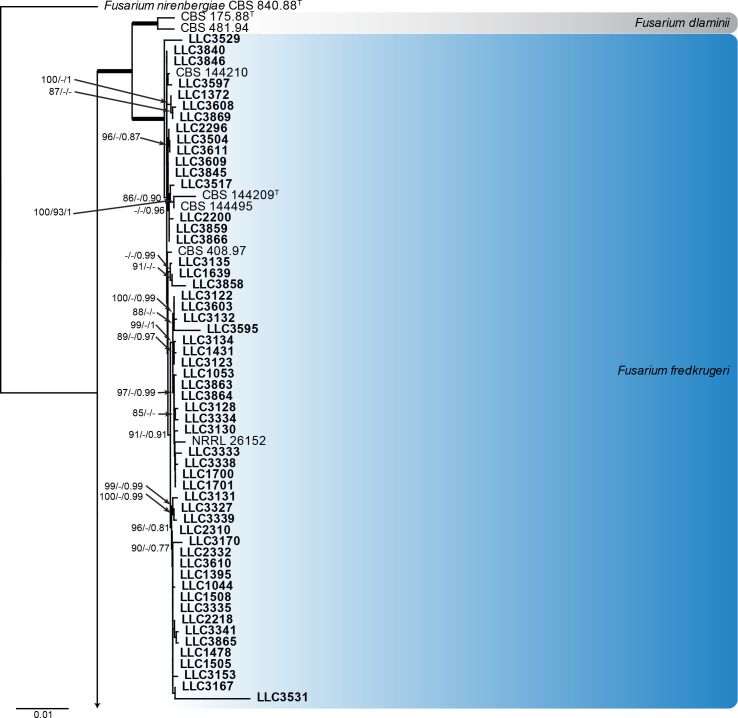
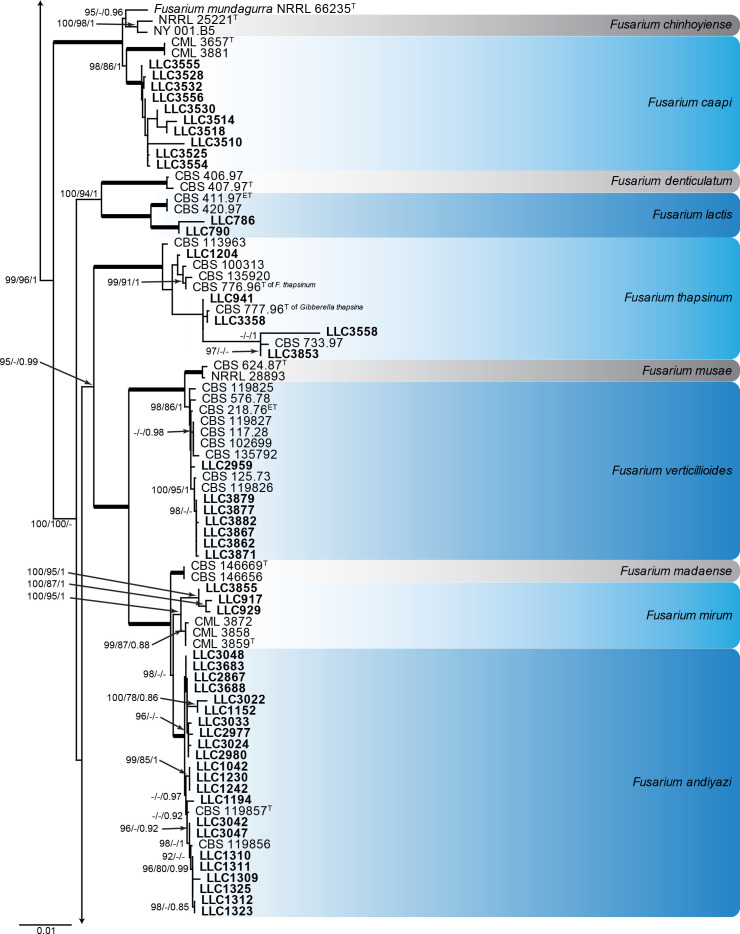
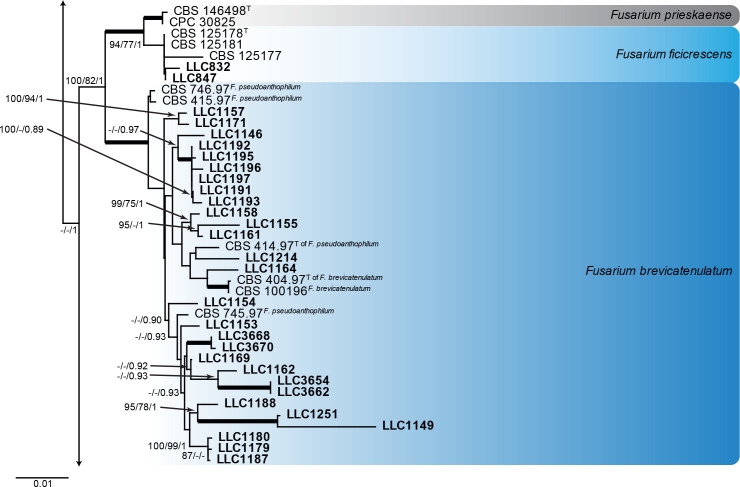
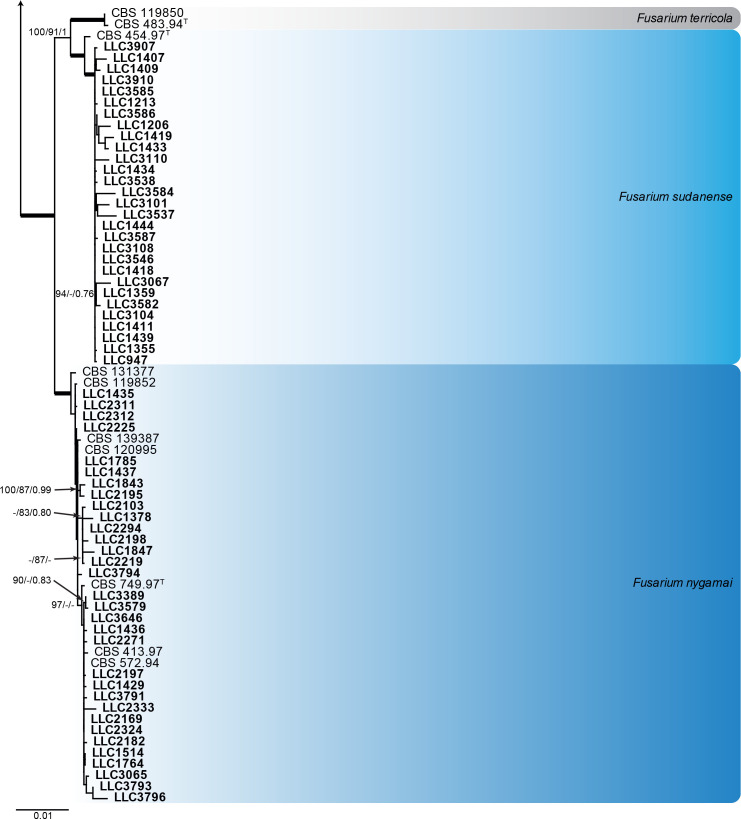
Fusarium fujikuroi species complex (Fig. 3): Novel isolates represent one undescribed species and clustered with 14 known species, namely F. andiyazi, F. annulatum, F. brevicatenulatum, F. caapi, F. ficicrescens, F. fredkrugeri, F. lactis, F. mirum, F. nygamai, F. secorum, F. sudanense, F. thapsinum, F. udum and F. verticillioides. The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) overall displayed the same species clades and mainly differed with regards to the backbone relationships between species clades/lineages (data not shown, support and posterior probability values are superimposed on the presented figure).
Fig. 3.
The IQ-TREE maximum likelihood consensus tree inferred from the combined CaM, rpb1, rpb2 and tef1 sequence alignment. Thickened lines indicate nodes with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other nodes indicated at the branches (IQ-TREE > 84 % / RAxML > 74 % / PP > 0.74). The tree is rooted to Fusarium nirenbergiae (CBS 840.88, ex-type culture). The scale bar indicates the number of expected changes per site. Species clades containing the novel isolates (in bold) are highlighted with coloured blocks (blue tints). Additional species clades are shown in coloured blocks with grey tints.
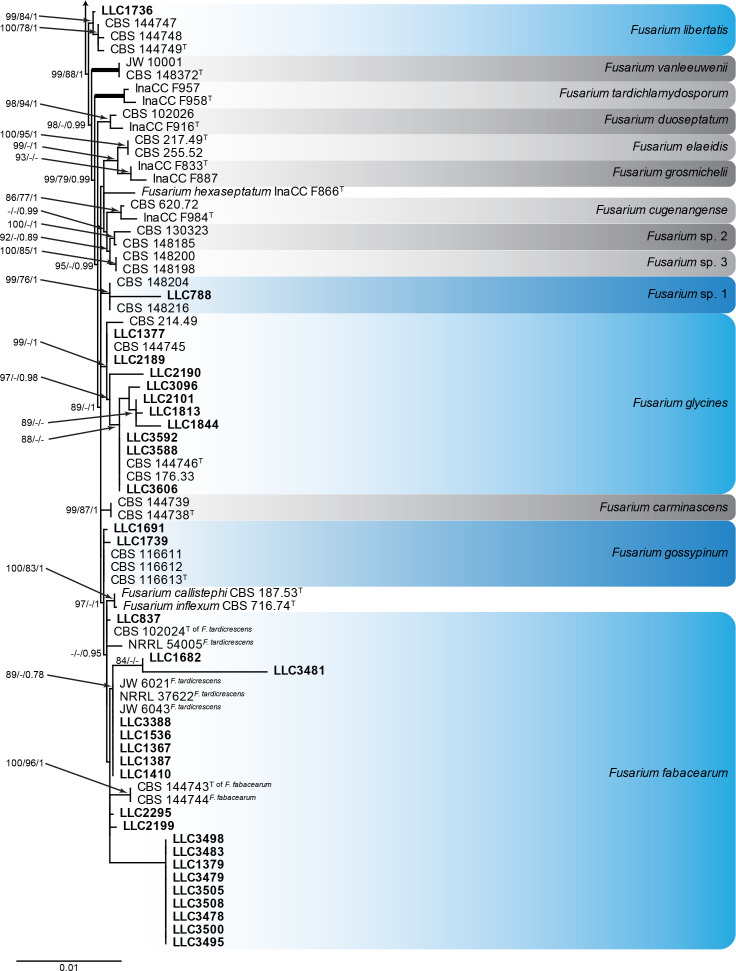
Fusarium oxysporum species complex (Fig. 4): Novel isolates clustered with seven known species, namely F. curvatum, F. fabacearum, F. glycines, F. gossypinum, F. libertatis, F. odoratissimum and F. veterinarium, as well as one species clade which was labelled as “Fusarium sp. 1” by Crous et al. (2021a). The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) overall displayed the same species clades and mainly differed with regard to the backbone relationships between species clades/lineages (data not shown, support and posterior probability values are superimposed on the presented figure).
Fig. 4.
The IQ-TREE maximum likelihood consensus tree inferred from the combined CaM, rpb1, rpb2 and tef1 sequence alignment. Thickened lines indicate nodes with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other nodes indicated at the branches (IQ-TREE > 84 % / RAxML > 74 % / PP > 0.74). The tree is rooted to Fusarium globosum (CBS 428.97, ex-type culture). The scale bar indicates the number of expected changes per site. Species clades containing the novel isolates (in bold) are highlighted with coloured blocks (blue tints). Additional species clades are shown in coloured blocks with grey tints.
Based on these phylogenetic trees, several taxonomic decisions were made, and the individual and combined trees are discussed under the Notes in the Taxonomy section below, where applicable.
AFLP-based marker for Fos
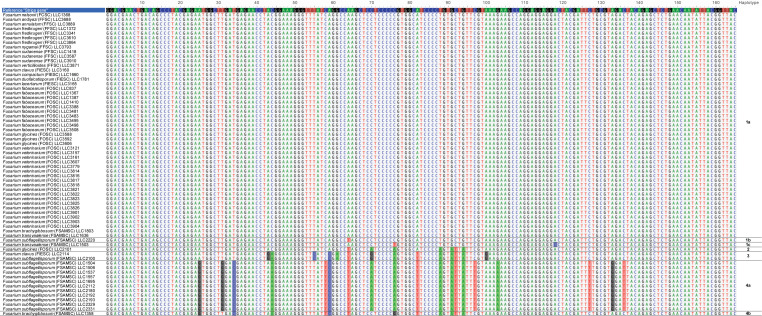
DNA of all 439 isolates obtained in this study were subjected to a PCR amplification of the AFLP-based marker for Fos, resulting in 66 amplicons (Supplementary Table S1). Sequence analyses of these amplicons revealed that most of the sequences (49 isolates) match the reference sequence from Zimmermann et al. (2015) (labelled here as haplotype 1a). However, six other sequence haplotypes are also observed (Fig. 5). Two of these differ by one and two substitutions from the reference sequence, respectively (haplotypes 1b and 1c; one isolate each). Although haplotype 2 (one isolate) differs with 7 substitutions from the reference sequence. These differences could be derived from haplotype 4a, and could be indicative of recombination events between haplotypes 1a and 4a. Haplotype 3 (two isolates) also appears to be an intermediate form between haplotypes 1a and 4a but has five unique substitutions not present in any of the other haplotypes. There are 19 substitutions between haplotypes 1a and 4a (11 isolates), with haplotype 4b (one isolate) appearing to be an additional intermediate form between haplotypes 1a and 4a.
Fig. 5.
Sequence alignment of sequences obtained for the Striga AFLP-derived marker amplicons with nucleotide substitutions compared to the reference sequence highlighted and haplotypes indicated to the right of the figure. Sequences in this figure were deposited in GenBank under accession numbers OP600320–OP600385.
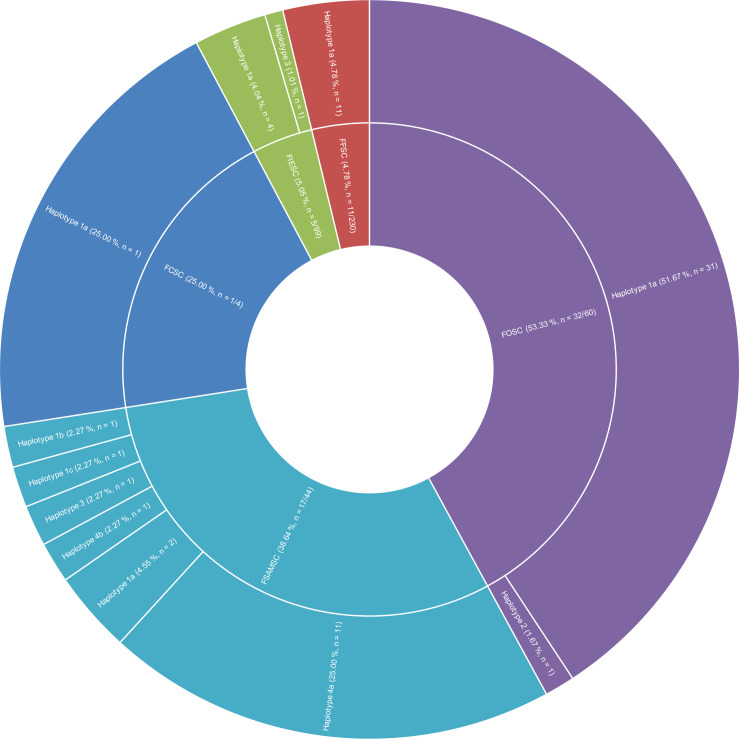
The AFLP-based marker for Fos is quite prevalent in the Fusarium oxysporum species complex (FOSC) (32 amplicons out of 60 isolates tested) and FSAMSC (17 amplicons out of 44 isolates tested). A single isolate out of the four Fusarium chlamydosporum species complex (FCSC) isolates is positive but given the low number of isolates tested, this value should not be considered significant until more isolates are tested. The two remaining species complexes, Fusarium fujikuroi species complex (FFSC) and Fusarium incarnatum/equiseti species complex (FIESC), only have 11 and 5 amplicons out of 230 and 99 isolates tested, respectively (Fig. 6). Haplotype 1a is present and the dominant haplotype in all of the species complexes, except for the Fusarium sambucinum species complex (FSAMSC) complex where haplotype 4a is the dominant haplotype (Fig. 6). None of the isolates from Fusarium burgessii species complex (FburSC) or Fusarium concolor species complex (FconSC) tested positive (Fig. 7).
Fig. 6.
Graph showing the distribution of haplotypes per species complex. The percentage is calculated per species complex from the number of positive tests divided by the total number of tests performed in the species complex. The pie part for the FCSC complex is artificially large due to being represented by only four isolates. FCSC: Fusarium chlamydosporum species complex; FFSC: Fusarium fujikuroi species complex; FIESC: Fusarium incarnatum/equiseti species complex; FOSC: Fusarium oxysporum species complex; FSAMSC: Fusarium sambucinum species complex.
Fig. 7.
Graph showing the distribution of haplotypes and negative PCR reactions for the striga marker across the tested species. FCSC: Fusarium chlamydosporum species complex; FFSC: Fusarium fujikuroi species complex; FIESC: Fusarium incarnatum/equiseti species complex; FOSC: Fusarium oxysporum species complex; FSAMSC: Fusarium sambucinum species complex.
Haplotype 1a is present in all species that amplified for the AFLP-based marker for Fos, except for F. subflagellisporum (FSAMSC) which mainly contains haplotype 4a and haplotypes 1b and 3 to a much lesser extent (Fig. 7). Fusarium fabacearum and F. veterinarium (both FOSC) contain the largest number of amplicons of haplotype 1a, with the haplotype being present in almost all F. veterinarium isolates tested and in roughly half of the F. fabacearum isolates tested (Fig. 7). The AFLP-based marker for Fos amplified in more than half of the F. subflagellisporum (FSAMSC) isolates, but sequence analyses of these amplicons revealed the dominant haplotype to be haplotype 4a (Fig. 7).
Taxonomy
Based on the results obtained, 42 Fusarium species were identified, including three species which are newly described below. Collection details of the material examined can be found in Supplementary Table S1.
Fusarium burgessii species complex (FburSC)
Fusarium burgessii M.H. Laurence et al., Fungal Diversity 49: 109. 2011.
Materials examined: Supplementary Table S1.
Notes: Isolate LLC1763 clusters with the ex-type and additional isolate of F. burgessii with full support (Fig. 1, part 1).
Fusarium chlamydosporum species complex (FCSC)
Fusarium sporodochiale L. Lombard & Crous, Fungal Syst. Evol. 4: 196. 2019.
Materials examined: Supplementary Table S1.
Notes: Isolate LLC2900 (Fig. 2) is a sister lineage to F. sporodochiale with low support (IQ-TREE bootstrap = 97 % / RAxML bootstrap = <74 % / PP = 0.86). The CaM sequence of isolate LLC2900 is 98.87 % (524/530 nt) identical to the ex-type isolate of F. sporodochiale (CBS 220.61), the rpb1 sequence 98.57 % (827/839 nt), the rpb2 sequence 99.89 % (873/874 nt), and the tef1 sequence 98.84 % (681/689 nt).
Fusarium nelsonii Marasas & Logrieco, Mycologia 90: 508. 1998.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster with F. nelsonii isolate NRRL 13338 with full support (Fig. 2).
Fusarium concolor species complex (FCOSC)
Fusarium concolor Reinking, Centbl. Bakt. ParasitKde, Abt. II 89: 512. 1934.
Synonym: Fusarium polyphialidicum Marasas et al., Mycologia 78: 678. 1986.
Materials examined: Supplementary Table S1.
Note: Isolate LLC2334 clusters with the ex-types of F. concolor and F. polyphialidicum with full support (Fig. 1, part 1).
Fusarium fujikuroi species complex (FFSC)
Fusarium andiyazi Marasas et al., Mycologia 93: 1205. 2001.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster with the ex-type of F. andiyazi with full support (Fig. 3, part 3). Although there is some genetic variation, the internal structure of the species is poorly supported.
Fusarium annulatum Bugnic., Rev. Gén. Bot. 59: 17. 1952. Fig. 8.
Fig. 8.
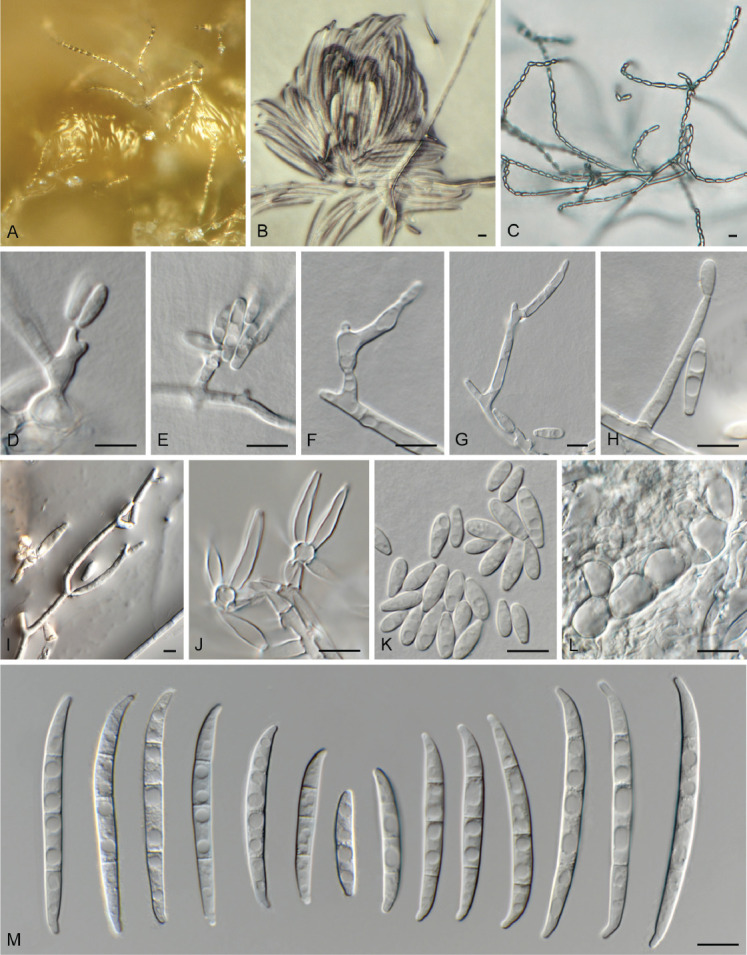
Fusarium annulatum (LLC 782). A, B. Sporodochia on SNA. C. Chains of microconidia. D–I. Mono- and polyphialides giving rise to microconidia. J. Sporodochial conidiophores. K. Microconidia. L. Chlamydospores. M. Sporodochial macroconidia. Scale bars = 10 μm.
Materials examined: Supplementary Table S1.
Notes: Fusarium annulatum LLC782 (Fig. 3, part 2) clusters basal to the F. annulatum clade. The F. annulatum s. str. subclade containing the ex-type isolate is moderately supported (IQ-TREE bootstrap = 99 % / RAxML bootstrap = 79 % / PP = 1) whereas the association between LLC782 and F. annulatum s.str. was highly supported (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 99 % / PP = 1). No CaM or rpb1 sequence of isolate LLC782 was available for comparison. The rpb2 sequence of isolate LLC782 is 98.86 % (954/965 nt) identical to the ex-type isolate of F. annulatum (CBS 258.54), and the tef1 sequence 98.52 % (665/675 nt). As we were unable to generate CaM and rpb1 sequence data for this isolate, the sequence similarity to F. annulatum is quite high, and F. annulatum is already genetically quite variable, we refrain from introducing a novel species for this this isolate.
Fusarium brevicatenulatum Nirenberg et al., Mycologia 90: 460. 1998.
Synonym: Fusarium pseudoanthophilum Nirenberg et al., Mycologia 90: 461. 1998.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster with the ex-types of F. brevicatenulatum and F. pseudoanthophilum with full support (Fig. 3, part 4). Although there is quite a bit of genetic variation in the species clade, the internal structure of the species is poorly supported. The present study expands the sampling for these two species published by Yilmaz et al. (2021; names from that publication shown in superscript). Based on this broader sampling and genetic diversity, we follow the suspicion of Leslie & Summerell (2006), and synonymise F. pseudoanthophilum under F. brevicatenulatum.
Fusarium caapi M.M. Costa et al., Mycol. Progr. 20: 67. 2021. Fig. 9.
Fig. 9.
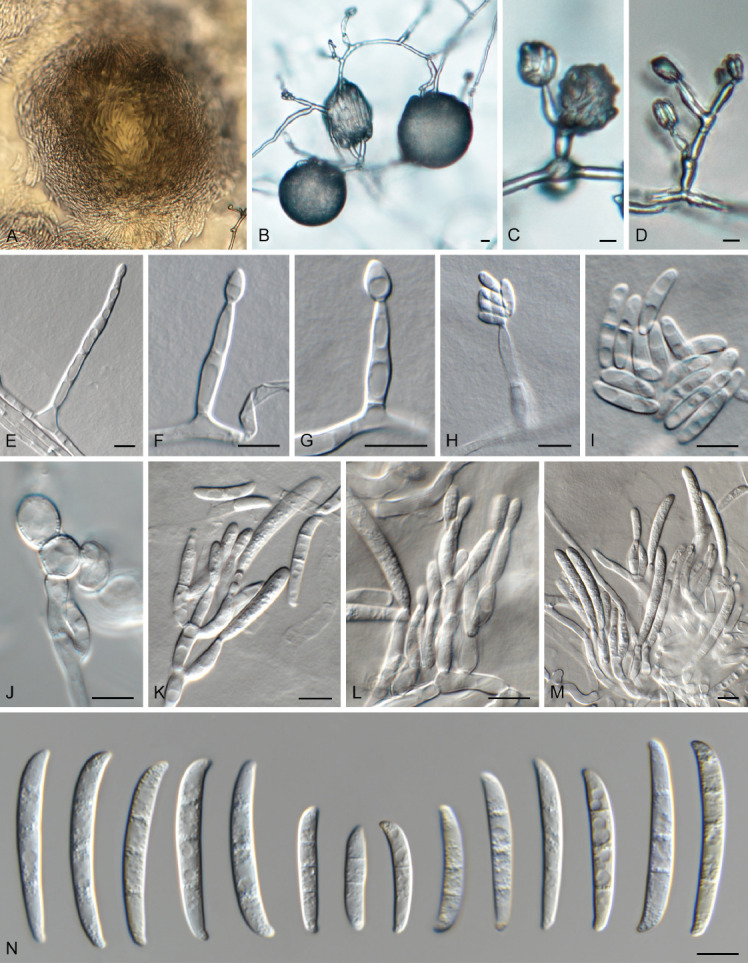
Fusarium caapi (LLC 3528). A, B. Sporodochia on SNA. C–H. Phialides giving rise to microconidia. I. Microconidia. J. Chlamydospores. K–M. Sporodochial conidiophores. N. Sporodochial macroconidia. Scale bars = 10 μm.
Materials examined: Supplementary Table S1.
Notes: The CaM sequence of isolate LLC3528 is 99.67% (608/610 nt) and 99.02% (606/612 nt) identical to the ex-type isolates of F. chinhoyiense (NRRL 25221) and F. mundagurra (NRRL 66235), respectively (Fig. 3, part 3). No CaM sequence is available for F. caapi (CML 3657). The rpb1 sequence of isolate LLC3528 is 99.46 % (741/745 nt) and 99.72 % (723/725 nt) identical to the ex-type isolates of F. chinhoyiense (NRRL 25221) and F. mundagurra (NRRL 66235), respectively. No rpb1 sequence is available for F. caapi (CML 3657). The rpb2 sequence of isolate LLC3528 is 98.56 % (821/833 nt), 99.53 % (853/857 nt) and 99.14 % (806/813 nt) identical to the ex-type isolates of F. caapi (CML 3657), F. chinhoyiense (NRRL 25221) and F. mundagurra (NRRL 66235), respectively. The tef1 sequence of isolate LLC3528 is 98.86 % (435/440 nt), 98.02 % (644/657 nt) and 98.35 % (537/546 nt) identical to the ex-type isolates of F. caapi (CML 3657), F. chinhoyiense (NRRL 25221) and F. mundagurra (NRRL 66235), respectively.
Fusarium ficicrescens Al-Hatmi et al., Fungal Biol. 120: 274. 2015.
Materials examined: Supplementary Table S1.
Notes: The two isolates cluster with the ex-type of F. ficicrescens with some to moderate support (IQ-TREE bootstrap = 94 % / RAxML bootstrap = 77 % / PP = 1; Fig. 3, part 4).
Fusarium fredkrugeri Sand.-Den. et al., MycoKeys 34: 79. 2018.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster with the ex-type of F. fredkrugeri with full support (Fig. 3, part 1). Although there is some genetic variation in the species clade, the internal structure of the species is poorly supported.
Fusarium lactis Pirotta, Arch. Labor. Bot. Critt. Univ. Pavia 2 & 3: 316. 1879. Fig. 10.
Fig. 10.
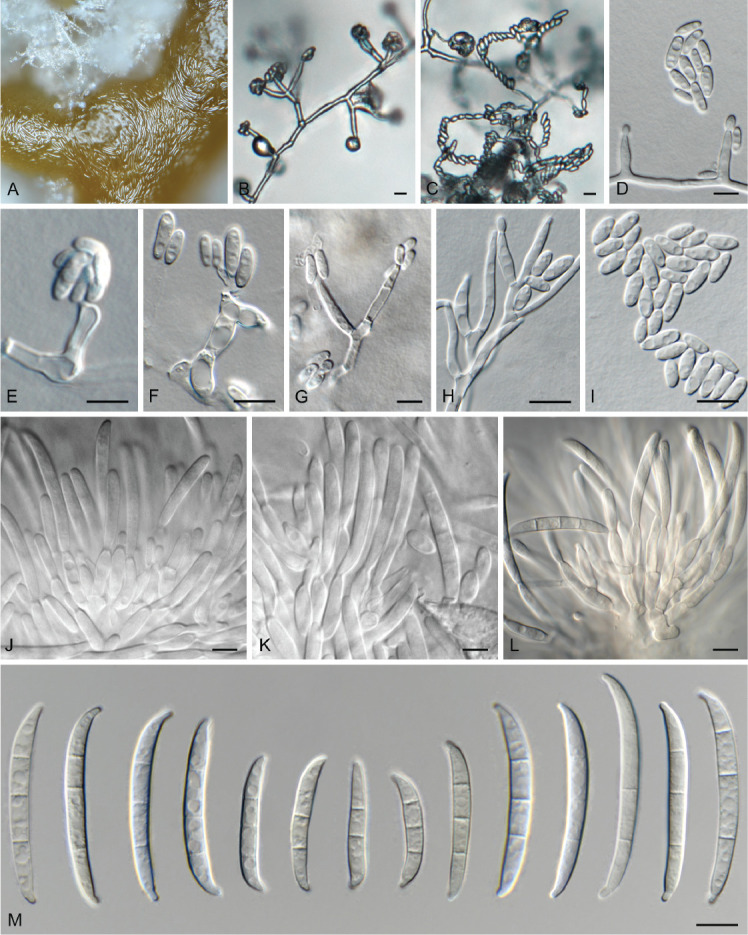
Fusarium lactis (LLC 790). A. Sporodochium on SNA. B–H. Phialides giving rise to microconidia. I. Microconidia. J–L. Sporodochial conidiophores. M. Sporodochial macroconidia. Scale bars = 10 μm.
Materials examined: Supplementary Table S1.
Notes: The CaM sequence of isolate LLC790 is 99.18 % (608/613 nt) identical to the ex-epitype isolate of F. lactis (CBS 411.97), the rpb1 sequence 99.61 % (1 520/1 526 nt), the rpb2 sequence 99.75 % (797/799 nt), and the tef1 sequence 97.53 % (633/649 nt).
Fusarium mirum M.M. Costa et al., Fungal Biol. 126: 262. 2022. Fig. 11.
Fig. 11.
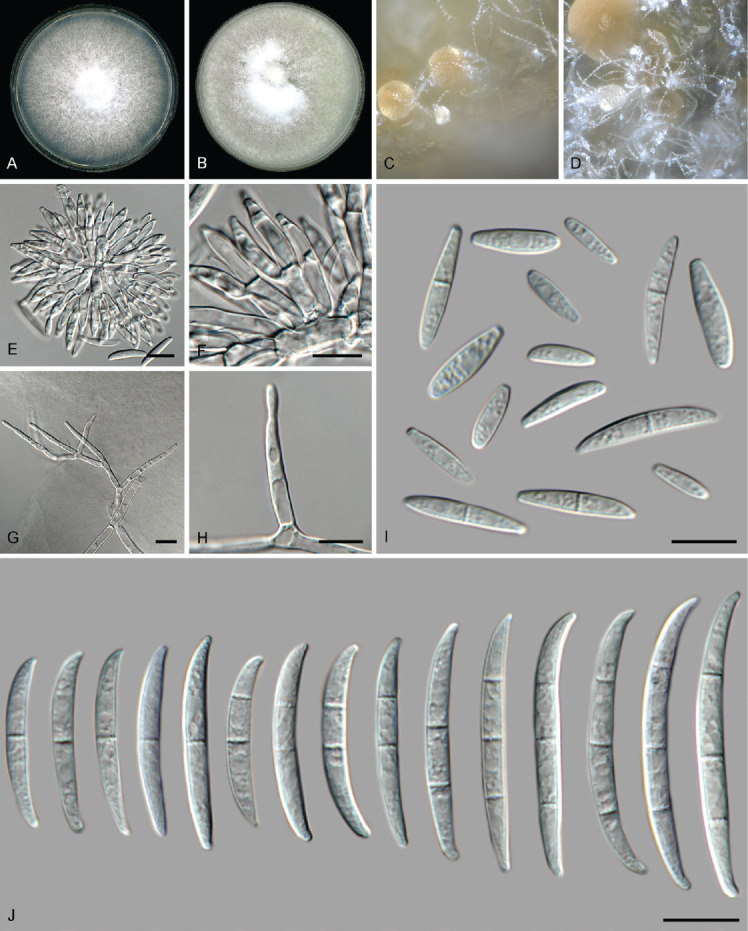
Fusarium mirum (LLC 917). A. Colony on PDA. B. Colony on OA. C, D. Sporodochia. E, F. Sporodochial conidiophores. G, H. Monophialides on aerial mycelium. I. Aerial microconidia. J. Sporodochial macroconidia. Scale bars = 10 μm.
Materials examined: Supplementary Table S1.
Notes: The F. mirum s.str. subclade (Fig. 3, part 3) containing the ex-type isolate is highly supported (IQ-TREE bootstrap = 99 % / RAxML bootstrap = 87 % / PP = 0.88) as is the association between LLC917 and F. mirum s.str. clades (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 95 % / PP = 1). No CaM and rpb1 sequences were available for comparison to the ex-type isolate of F. mirum (CML 3859). The rpb2 sequence of isolate LLC917 is 99.89 % (897/898 nt) identical to the ex-type isolate of F. mirum (CML 3859), and the tef1 sequence 99.53 % (641/644 nt). As no CaM and rpb1 sequences are available of F. mirum and rpb2 and tef1 are highly identical to the ex-type isolate of F. mirum, we refrain from introducing a new species for this clade at this point.
Fusarium nygamai L.W. Burgess & Trimboli, Mycologia 78: 223. 1986.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster with the ex-type of F. nygamai with full support (Fig. 3, part 5). Although there is some genetic variation in the species clade, the internal structure of the species is poorly supported.
Fusarium secorum Secor et al., Fungal Biol. 118: 767. 2014.
Materials examined: Supplementary Table S1.
Notes: The isolate clusters with the ex-type of F. secorum with full support (Fig. 3, part 2).
Fusarium sp. (LLC1198)
Materials examined: Supplementary Table S1.
Notes: Fusarium sp. LLC1198 (Fig. 3, part 2) is a single isolate basal to the F. xylarioides clade. The F. xylarioides s.str. subclade containing the ex-type isolate is fully supported (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 100 % / PP = 1) whereas the association between the LLC1198 and F. xylarioides s.str. clades was also highly supported (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 94 % / PP = 1). The CaM sequence of isolate LLC1198 is 96.98 % (643/663 nt) identical to the ex-type isolate of F. xylarioides (CBS 258.52), the rpb1 sequence (1 525/1 525 nt), the rpb2 sequence 96.78 % (873/902 nt), and the tef1 sequence 97.07 % (629/648 nt). Isolate LLC1198 forms part of another study, and will be described elsewhere.
Fusarium sudanense S.A. Ahmed et al., Antonie van Leeuwenhoek 110: 826. 2017.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster as a fully-supported clade sister to the ex-type of F. sudanense (Fig. 3, part 5). Although there is some genetic variation in the species clade, the internal structure of the species is poorly supported. The CaM sequence of isolate LLC1434 is 99.84 % (612/613 nt) identical to the ex-type isolate of F. sudanense (CBS 454.97), the rpb1 sequence is identical (803/803 nt), the rpb2 sequence 99.89 % (875/876 nt), and the tef1 sequence 99.64 % (546/548 nt). Given the high similarity on all four loci with the ex-type isolate of F. sudanense we treat this subclade as belonging to that species rather than introducing a new species here.
Fusarium thapsinum Klittich et al., Mycologia 89: 644. 1997.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster interspersed with isolates of F. thapsinum (including the ex-type) in a fully-supported clade (Fig. 3, part 3). Although there is quite some genetic variation in the species clade, the internal structure of the species is not to poorly supported.
Fusarium udum E.J. Butler, Mem. Dept. Agric. India, Bot. Ser. 2: 54. 1910.
Synonyms: see www.fusarium.org
Materials examined: Supplementary Table S1.
Notes: The isolates cluster sister to isolates of F. udum (including the ex-type) in an almost fully-supported clade (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 99 % / PP = 1; Fig. 3, part 3). Although there is some genetic variation in the species clade, the internal structure of the species is not to poorly supported.
Fusarium verticillioides (Sacc.) Nirenberg, Mitt. Biol. Bundesanst. Land- Forstw. Berlin-Dahlem 169: 26. 1976.
Synonyms: see www.fusarium.org
Materials examined: Supplementary Table S1.
Notes: The isolates cluster sister to isolates of F. verticillioides (including the ex-type) in a highly-supported clade (IQ-TREE bootstrap = 98 % / RAxML bootstrap = 86 % / PP = 1; Fig. 3, part 3). Although there is some genetic variation in the species clade, the internal structure of the species is not to poorly supported.
Fusarium incarnatum/equiseti species complex (FIESC)
Fusarium caatingaense A.C.S. Santos et al., Mycologia 111: 248. 2019.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster interspersed with isolates of F. caatingaense (including the ex-type) in an almost fully-supported clade (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 98 % / PP = 1; Fig. 1, part 1). Although there is some genetic variation in the species clade, the internal structure of the species is partly to highly supported but with short branches.
Fusarium clavus J.W. Xia et al., Persoonia 43: 199. 2019.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster interspersed with isolates of F. clavus (including the ex-type) in an almost fully-supported clade (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 96 % / PP = 1; Fig. 1, part 2). Although there is some genetic variation in the species clade, the internal structure of the species is partly to highly supported but with short branches.
Fusarium compactum (Wollenw.) Raillo, Fungi of the Genus Fusarium: 180. 1950. Fig. 12.
Fig. 12.
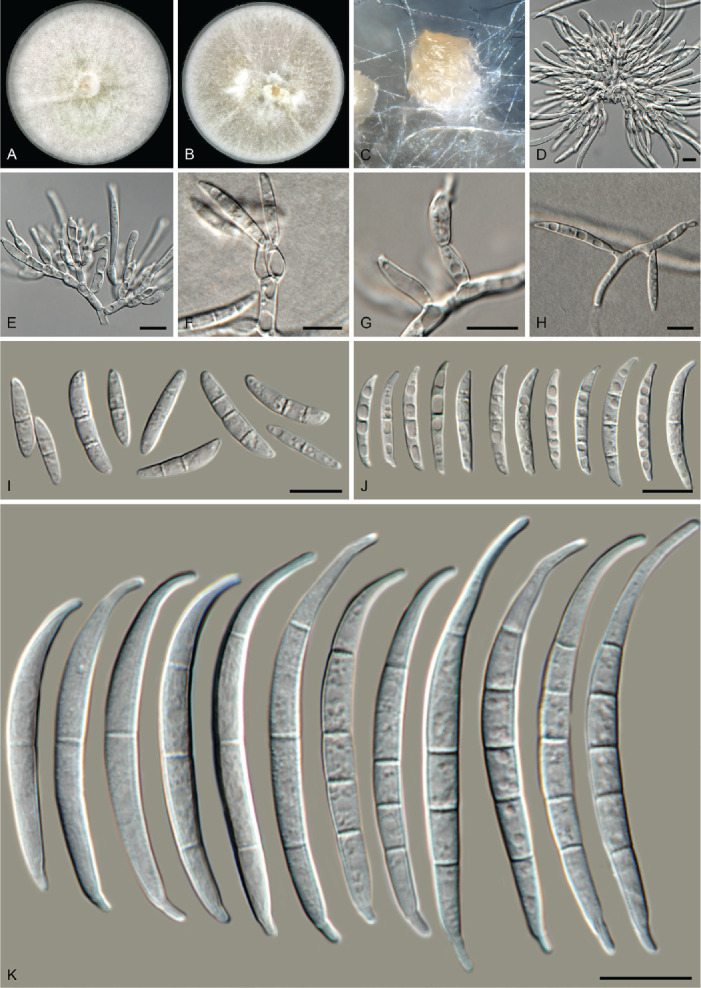
Fusarium compactum (LLC 1660). A. Colony on PDA. B. Colony on OA. C. Sporodochium on carnation leaves. D, E. Sporodochia and sporodochial conidiophores. F–H. Phialides on aerial mycelium. I, J. Aerial macroconidia. K. Sporodochial macroconidia. Scale bars = 10 μm.
Basionym: Fusarium scirpi var. compactum Wollenw., Fusaria Autographica Delineata 3: no. 924. 1930. MB 124046.
Synonyms: see www.fusarium.org
Materials examined: Supplementary Table S1.
Notes: The F. compactum s.str. subclade containing the ex-epitype isolate is fully supported whereas the association with isolates LLC1048/LLC1660 and F. compactum s.str. clades was not supported (all support values are below the threshold values for display on the tree) (Fig. 1, part 2).
In the phylogenetic tree (Fig. 1, part 2), the F. compactum clade consists of two main individually highly supported subclades. The CaM sequence of isolate LLC1048 is 98.44 % (567/576 nt) identical to the ex-epitype isolate of F. compactum (CBS 186.31). The rpb1 sequence of isolate LLC1048 is 99.42 % (1 549/1 558 nt) identical to F. compactum isolate NRRL 28029 (GenBank HM347150); no rpb1 sequence is available for the ex-epitype isolate of F. compactum (CBS 186.31). The rpb2 sequence of isolate LLC1048 is 99.89 % (878/879 nt) identical to the ex-epitype isolate of F. compactum (CBS 186.31), and the tef1 sequence 97.02 % (652/672 nt). However, as this subclade is highly similar to F. compactum based on three of the four loci used here and only tef1 is proving to be genetically the most diverse, we choose not to introduce a new species for this subclade pending a further definition of the species boundaries of F. compactum.
Fusarium duofalcatisporum J.W. Xia et al., Persoonia 43: 201. 2019.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster interspersed with isolates of F. duofalcatisporum (including the ex-type) in a fully-supported clade (Fig. 1, part 2). Although there is some genetic variation in the species clade, the internal structure of the species is partly to highly supported but with short branches.
Fusarium extenuatum L. Lombard, sp. nov. — MycoBank MB 846717. Fig. 13.
Fig. 13.
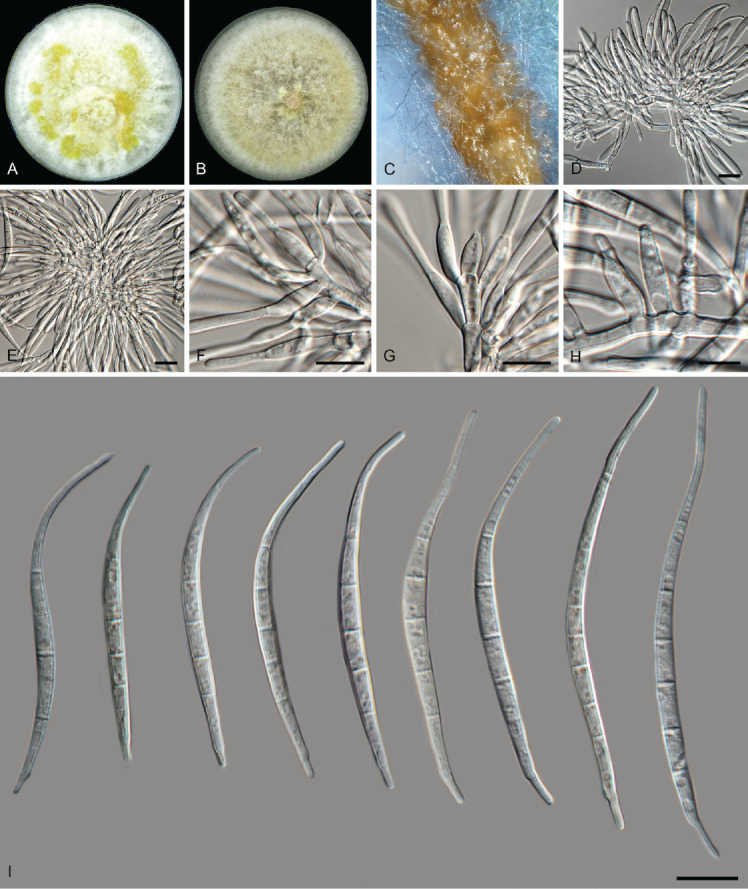
Fusarium extenuatum (LLC 1501). A. Colony on PDA. B. Colony on OA. C. Sporodochia on carnation leaves. D, E. Sporodochia. F–H. Sporodochial conidiophores. I. Sporodochial macroconidia. Scale bars = 10 μm.
Etymology: Name refers to the conidiophores that are reduced to lateral phialides on the aerial mycelium.
Typus: Ethiopia, Tigray Region, Central (Meakelewi) Zone, Tanqua Abergele District, Yechela locality (Kebele), from soil collected in a sorghum field, 2017, D.W. Etolo & L. Lombard (holotype EMCC-F333, preserved as metabolic inactive culture, culture ex-type LLC1501 = EMCC-F333).
Conidiophores reduced to solitary conidiogenous cells borne laterally on hyphae; aerial conidiogenous cells monophialides, subulate to subcylindrical, smooth- and thin-walled, 14–22 × 4 μm, periclinal thickening and collarettes often inconspicuous. Aerial macroconidia similar to sporodochial conidia. Sporodochial conidiophores 22–38 μm tall, irregularly branched, bearing terminal solitary or whorls of 2–3 phialides. Sporodochial conidiogenous cells monophialidic, doliiform, subulate to subcylindrical, smooth- and thin-walled, (10–)11–13(–15) × (3–) 4–5 μm. Sporodochial conidia straight to moderately curved and slender, tapering towards the basal part, apical cell elongated (papillate) to whip-like; basal cell well-developed, foot-shaped, mostly papillate, (3–)5(–6)-septate, hyaline, thin- and smooth-walled: 3-septate conidia 32–46(–61) × (3–)4–5 μm (av. 38 × 5 μm) ; 4-septate conidia (34–)36–40(–42) × 4–5 μm (av. 38 × 5 μm); 5-septate conidia (49–)47–69(–73)× (3–)4–5 μm (av. 63 × 4 μm); 6-septate conidia (75–)77–87(–90) × (4–)5–6 μm (av. 82 × 5 μm; n = 7). Chlamydospores not observed.
Culture characteristics: Colonies on PDA reaching 85–90 mm diam at 25 °C after 7 d. Surface white with luteus to amber flames, raised, woolly to cottony with abundant aerial mycelium, margin regular and filiform. Reverse pale luteus. On OA, white to amber, raised, woolly to cottony with abundant aerial mycelium, margin regular and filiform. Reverse pale luteus.
Additional materials examined: Supplementary Table S1.
Notes: Fusarium extenuatum represents a unique fully-supported clade in the Equiseti Clade (Xia et al. 2019) in the FIESC. Although morphologically similar to F. longifundum, this species did not produce any chlamydospores in culture. In addition, these two species are not closely related in the phylogenetic tree (Fig. 1, part 1 vs part 2).
In the phylogenetic tree (Fig. 1, part 2), the F. clavus s.l. clade consists of three main highly supported subclades. The F. clavus clade at its most basal position is highly supported in two of the three analyses (IQ-TREE bootstrap = 99 % / RAxML bootstrap < 75 % / PP = 1); the third subclade contains the species’ ex-type isolate and is highly supported (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 96 % / PP = 1). The first subclade is described here as F. extenuatum sp. nov.; see F. tangerinum below for a discussion on the second subclade. The CaM sequence of isolate LLC1501 from this subclade is 98.69 % similar (527/534 nt) to that of F. ipomoeae isolate LC12163 and 98.0 % similar (539/550 nt) to the ex-type isolate of F. clavus (CBS 126202). The rpb1 sequence of isolate LLC1501 is most identical to F. equiseti isolate NL19-97009 (835/841 nt = 99.29 %). No rpb1 sequence is available for the ex-type isolate of F. clavus, but a comparison to the rpb1 sequence of F. clavus isolate JW 288002 reveals a similarity of 98.26 % (789/803 nt). The rpb2 sequence of isolate LLC1501 is 97.74 % similar (820/839 nt) to that of the ex-type isolate of F. clavus (CBS 126202). The tef1 sequence of isolate LLC1501 is 94.81 % identical (566/597 nt) to that of the ex-type isolate of F. clavus (CBS 126202).
Fusarium incarnatum (Desm.) Sacc., Syll. Fung. (Abellini) 4: 712. 1886. Fig. 14.
Fig. 14.
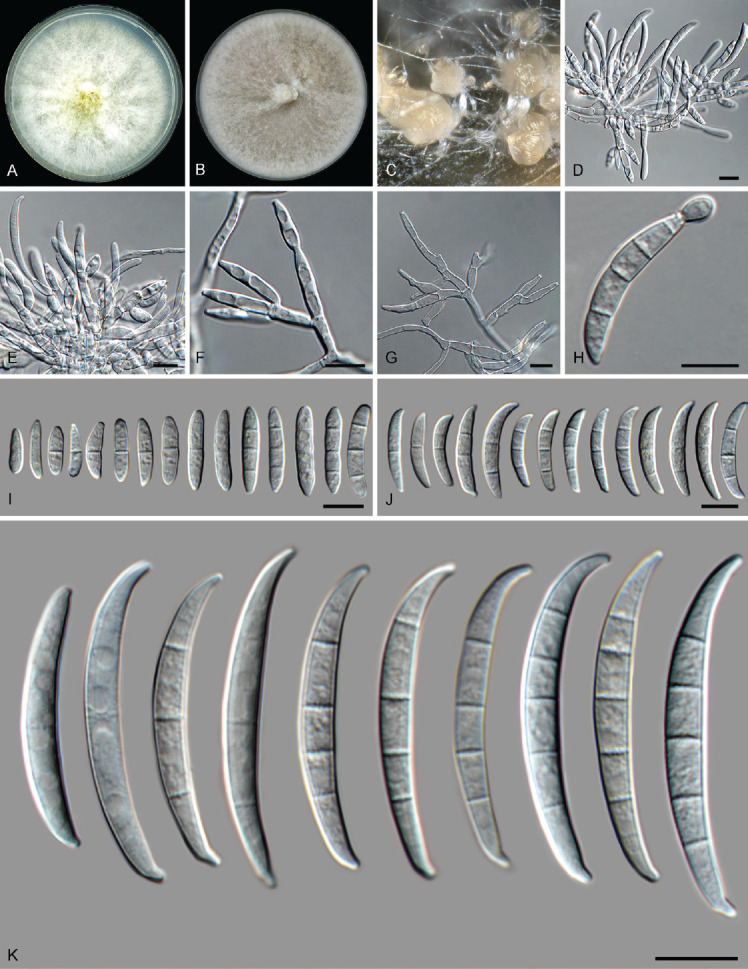
Fusarium incarnatum (LLC 1220). A. Colony on PDA. B. Colony on OA. C. Sporodochia on carnation leaves. D, E. Sporodochia and sporodochial conidiophores. F, G. Conidiophores with mono- and polyphialides on aerial mycelium. H. Microcyclic conidiogenesis. I. Aerial microconidia. J. Aerial macroconidia. K. Sporodochial macroconidia. Scale bars = 10 μm.
Materials examined: Supplementary Table S1.
Notes: In the phylogenetic tree (Fig. 1, part 1), the F. incarnatum clade consists of two subclades not supported in any of phylogenetic analyses. The F. incarnatum clade itself is moderately supported in the IQ-TREE analysis (88 %), not supported in the RAxML analysis (< 75 %) and highly supported in the Bayesian analysis (PP = 0.98). The CaM sequence of isolate LLC1220 from the second subclade is identical (549/549 nt) to that of the ex-epitype isolate of F. incarnatum (CBS 132.73) in the first subclade whereas the rpb1 sequence is more identical to F. guilinense isolate FRC R-8480 (793/798 nt = 99.37 %). No rpb1 sequence is available for the ex-epitype of F. incarnatum but a comparison to the rpb1 sequence of F. incarnatum isolate NRRL 32866 reveals a similarity of 98.75 % (788/798 nt). The rpb2 sequence of isolate LLC1220 differs 7 nt from several species such as F. caatingaense, F. nanum and F. hainanense while it is 98.81 % identical (829/839 nt) to that of the ex-epitype isolate of F. incarnatum (CBS 132.73). The tef1 sequence of isolate LLC1220 is 99.83 % identical (599/600 nt) to that of the ex-epitype isolate of F. incarnatum (CBS 132.73). Given that both intron-rich genes CaM and tef1 are identical to almost identical to F. incarnatum, and rpb1 and rpb2 only have up to 10 nt differences, we refrain for now from introducing a new species for the second clade and treat it as F. incarnatum.
Fusarium lacertarum Subrahm. [as ‘laceratum’], Mykosen 26: 478. 1983.
Materials examined: Supplementary Table S1.
Notes: The F. lacertarum s.str. subclade containing the ex-type isolate is highly supported (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 99 % / PP = 1) and the association between the clade containing isolate LLC2984 and the F. lacertarum s.str. clade is also highly supported (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 97 % / PP = 1).
In the phylogenetic tree (Fig. 1, part 2), the F. lacertarum clade consists of two main highly supported subclades. The CaM sequence of isolate LLC2984 is 99.09 % (546/551 nt) identical to the ex-type isolate of F. lacertarum (CBS 130185). No rpb1 sequences of isolates belonging to LLC2984 are available for comparison to the ex-type isolate of F. lacertarum (CBS 130185). The rpb2 sequence of isolate LLC2984 is 98.98 % (870/879 nt) identical to the ex-type isolate of F. lacertarum (CBS 130185). The tef1 sequence of isolate LLC2984 is 98.80 % (661/669 nt, including a single indel of 3 nt) identical to the ex-type isolate of F. lacertarum (CBS 130185). However, as this subclade is highly similar to F. lacertarum based on three available loci used here, we choose not to introduce a new species for this subclade pending a further definition of the species boundaries of F. lacertarum.
Fusarium nanum M.M. Wang et al., Persoonia 43: 85. 2019.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster interspersed with isolates of F. nanum (including the ex-type) in a partly-supported clade (IQ-TREE bootstrap = 98 % / RAxML bootstrap = 90 % / PP = 0.98; Fig. 1, part 1). Although there is some genetic variation in the species clade, the internal structure of the species is partly supported but with short branches.
Fusarium serpentinum J.W. Xia et al., Persoonia 43: 217. 2019.
Materials examined: Supplementary Table S1.
Note: The isolate clusters with the ex-type isolate of F. serpentinum in a fully-supported clade (Fig. 1, part 1).
Fusarium tangerinum Crous, Sand.-Den. & M.M. Costa, sp. nov. MycoBank MB 846718. Fig. 15, 16.
Fig. 15.
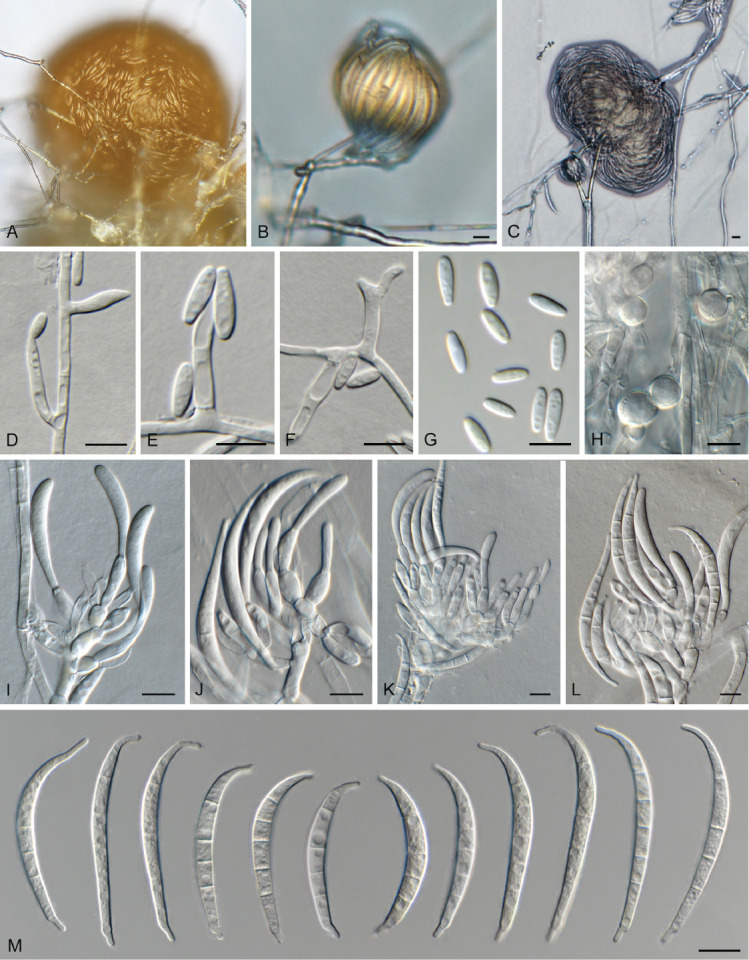
Fusarium tangerinum (LLC 3501). A–C. Sporodochia on SNA. D–F. Phialides giving rise to microconidia. G. Microconidia. H. Chlamydospores. I–L. Sporodochial conidiophores. M. Sporodochial macroconidia. Scale bars = 10 μm.
Fig. 16.
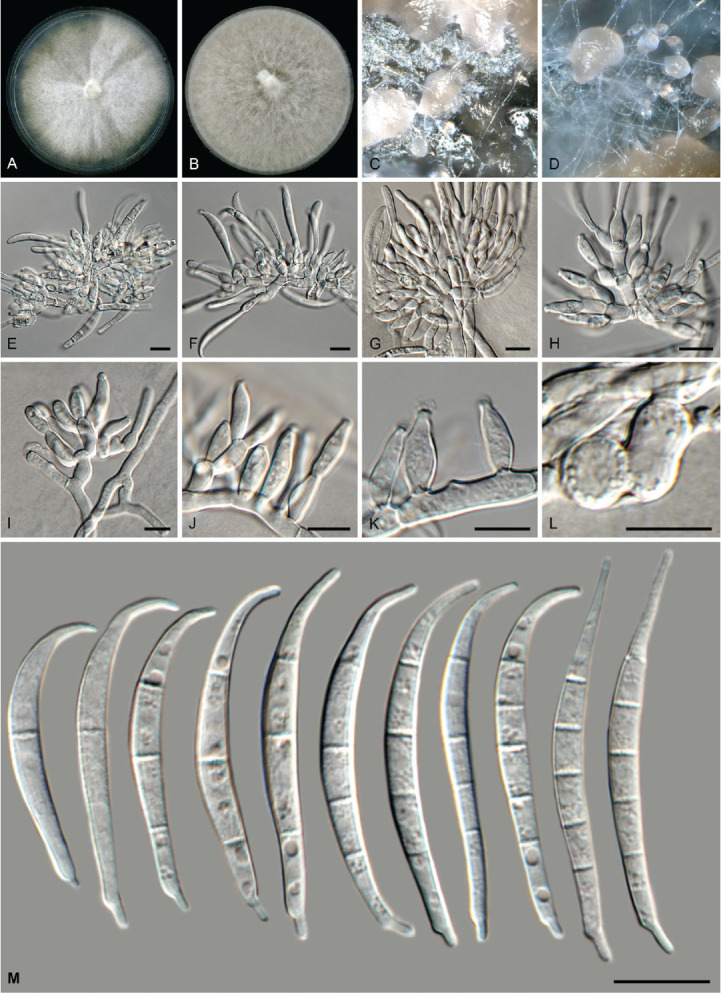
Fusarium tangerinum (LLC 3018). A. Colony on PDA. B. Colony on OA. C, D. Sporodochia. E–H. Sporodochial conidiophores. I–K. Monophialides on aerial mycelium. L. Chlamydospores. M. Sporodochial macroconidia. Scale bars = 10 μm.
Etymology: Name refers to its abundant, orange sporodochia.
Typus: Ethiopia, Amhara Region, Oromia Zone, Artuma Fursi District, Hula Tukuye locality (Kebele), from Sorghum cv. Teshale rhizosphere soil field, 2019, D.W. Etolo & L. Lombard (holotype EMCC-F377, preserved as metabolically inactive culture, culture ex-type LLC3501 = EMCC-F377).
Aerial conidiophores sparingly branched, 15–40 μm tall, bearing terminal, rarely lateral monophialides (polyphialides rarely observed), mostly reduced to conidiogenous cells on hyphae; aerial conidiogenous cells monophialidic, subulate to subcylindrical, thin-walled, 10–25 × 2.5–3 μm, with flared collarette and minute periclinal thickening. Aerial conidia aggregating in false heads, fusoid-ellipsoid, aseptate, apex subobtuse, base truncate, (6–)8–9(–11) × (2–)2.5(–3) μm. Sporodochia orange, abundant on CLA. Sporodochial conidiophores densely aggregated, branched, consisting of a stipe bearing whorls or 2–4 monophialides; sporodochial conidiogenous cells monophialidic, subulate to subcylindrical, 8–20 × 3.5–5 μm, smooth- and thin-walled with periclinal thickening and minute collarette. Sporodochial conidia falcate, curved dorsiventrally, tapering from middle towards both ends; apical cell elongated, hooked, whip-like with parallel sides and subobtuse apex; basal cell foot-shaped, notch well developed, (3–)5-septate, hyaline, smooth-walled, guttulate; 3-septate conidia (23–)30–40(–44) × (3.5–)4(–5) μm, 5-septate conidia (40–)45–50(–55) × (4–)4.5(–5) μm. Chlamydospores on SNA after 1 wk, solitary or in groups of 2–3, terminal or intercalary, subglobose, 8–12 μm diam.
Culture characteristics: Colonies erumpent, spreading, covering dish after 7 d at 25 °C. On PDA surface and reverse saffron; on OA surface saffron, reverse saffron to dark vinaceous.
Additional material examined: Supplementary Table S1.
Notes: Fusarium tangerinum is proposed here for the second fully supported clade in the F. clavus s.l. clade; also see F. extenuatum sp. nov. above.
In the phylogenetic tree (Fig. 1, part 2), the F. clavus clade consists of three main highly supported subclades. The CaM sequence of isolate LLC3018 from the second subclade is 98.06 % (555/566 nt) and 99.27 % (546/550 nt) identical to the ex-type isolates of F. extenuatum (LLC1501) and F. clavus (CBS 126202), respectively. No rpb1 sequence is available for isolate LLC3018 and therefore the sequence of isolate LLC3501 was used for comparison. The rpb1 sequence of isolate LLC3501 is 96.42 % (727/754 nt) identical to the ex-type isolate of F. extenuatum (LLC1501). No rpb1 sequence is available for the ex-type isolate of F. clavus (CBS 126202), but a comparison to the rpb1 sequence of F. clavus isolate JW 288002 reveals a similarity of 96.55 % (728/754 nt). The rpb2 sequence of isolate LLC3018 is 97.67 % (713/730 nt) and 96.99 % (708/730 nt) identical to the ex-type isolates of F. extenuatum (LLC1501) and F. clavus (CBS 126202), respectively. The tef1 sequence of isolate LLC3018 is 94.09 % (637/677 nt) and 96.64 % (575/595 nt) identical to the ex-type isolates of F. extenuatum (LLC1501) and F. clavus (CBS 126202), respectively.
Fusarium oxysporum species complex (FOSC)
Fusarium curvatum L. Lombard & Crous, Persoonia 41: 21. 2018.
Materials examined: Supplementary Table S1.
Notes: The F. curvatum s.str. subclade containing the ex-type isolate is poorly supported (IQ-TREE bootstrap = 89 % / RAxML bootstrap = < 75 % / PP = <0.74; Fig. 4, part 1) and the node including LLC2077 only receives support from the Bayesian analysis (PP = 0.95). The CaM sequence of isolate LLC2077 is identical (559/559 nt) to the ex-type isolate of F. curvatum (CBS 238.94), the rpb1 sequence 99.88 % (1 665/1 667 nt), the rpb2 sequence is identical (877/877 nt), and the tef1 sequence 99.31 % (577/581 nt). Due to the absent to minor differences between LLC2077 and the ex-type isolate of F. curvatum (CBS 238.94), we treat this isolate as F. curvatum.
Fusarium fabacearum L. Lombard et al., Persoonia 43: 24. 2019. Fig. 17.
Fig. 17.
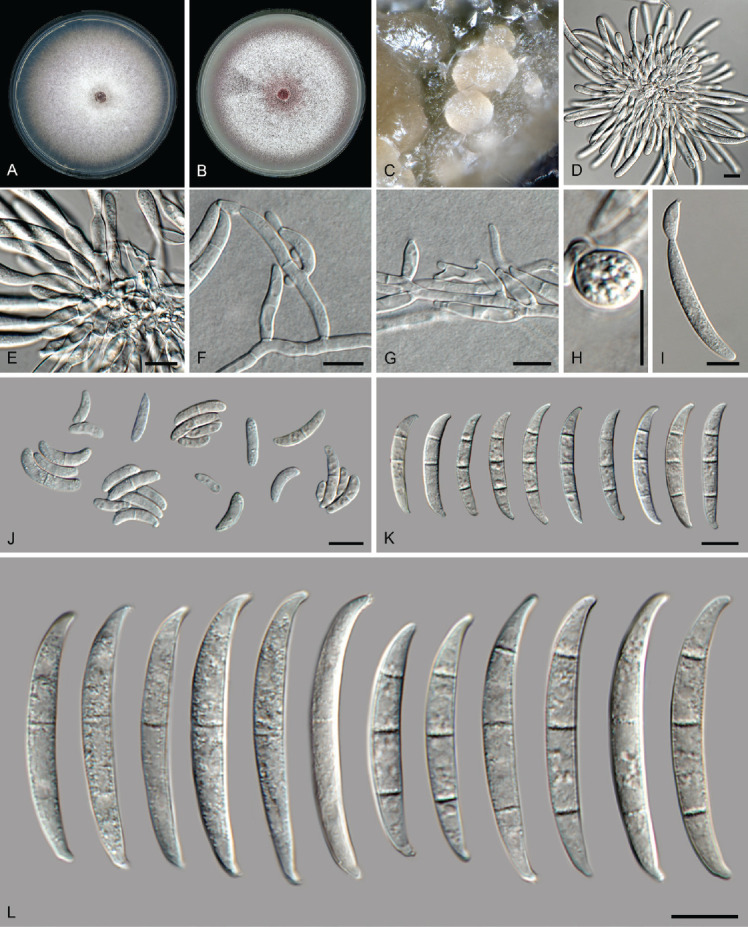
Fusarium fabacearum (LLC 2199). A. Colony on PDA. B. Colony on OA. C. Sporodochia on carnation leaves. D, E. Sporodochia and sporodochial conidiophores. F, G. Conidiophores with mono- and polyphialides on aerial mycelium. H. Chlamydospore. I. Microcyclic conidiogenesis. J. Aerial microconidia. K. Aerial macroconidia. L. Sporodochial macroconidia. Scale bars = 10 μm.
New synonym: Fusarium tardicrescens Maryani et al., Persoonia 43: 69. 2019.
Materials examined: Supplementary Table S1.
Notes: The isolates are interspersed with isolates previously treated as F. fabacearum and F. tardicrescens (including the ex-types of both species) in an unresolved clade (Fig. 4, part 2). As no CaM sequence is available for the ex-type isolate of F. tardicrescens (CBS 102024), the CaM sequence of isolate LLC2199 was compared to that of F. tardicrescens isolate JW 6021 and a 99.66 % similarity (590/592 nt) was found. The CaM sequence of isolate LLC2199 is 99.32 % (588/592 nt) identical to the ex-type isolate of F. fabacearum (CBS 144743). The rpb1 sequence of isolate LLC2199 is 99.66 % (879/882 nt) and 99.87 % (791/792 nt) identical to the ex-type isolates of F. fabacearum (CBS 144743) and F. tardicrescens (CBS 102024), respectively. The rpb2 sequence of isolate LLC2199 is identical (877/877 and 835/835 nt) to the ex-type isolates of F. fabacearum (CBS 144743) and F. tardicrescens (CBS 102024), respectively. The tef1 sequence of isolate LLC2199 is 99.67 % (613/615 nt) and 100 % (574/574 nt) identical to the ex-type isolates of F. fabacearum (CBS 144743) and F. tardicrescens (CBS 102024), respectively. In the present study, we treat all newly sequenced isolates under the earlier validly described name F. fabacearum (Lombard et al. 2019; published online 18 Dec. 2018) rather than the later possible synonym F. tardicrescens (Maryani et al. 2019a; invalidly published online 5 Jul. 2018, validated by Maryani et al. 2019b on 14 Mar. 2019).
Fusarium glycines L. Lombard et al., Persoonia 43: 25. 2018.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster interspersed with isolates of F. glycines (including the ex-type) in a partly-supported clade (IQ-TREE bootstrap = 99 % / RAxML bootstrap = < 75 % / PP = 1; Fig. 4, part 2). Although there is some genetic variation in the species clade, the internal structure of the species is only partly supported.
Fusarium gossypinum L. Lombard & Crous, Persoonia 43: 26. 2018.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster with isolates of F. gossypinum (including the ex-type) in an unresolved clade (Fig. 4, part 2). Although there is some genetic variation, there is no internal structure.
Fusarium libertatis L. Lombard & Crous, Persoonia 41: 29. 2018.
Materials examined: Supplementary Table S1.
Notes: The F. libertatis s.str. subclade containing the ex-type isolate is well-supported (IQ-TREE bootstrap = 100 % / RAxML bootstrap = 78 % / PP = 1) and the node including LLC1736 also receives high support (IQ-TREE bootstrap = 99 % / RAxML bootstrap = 84 % / PP = 1). The CaM sequence of isolate LLC1736 is identical (602/602 nt) to the ex-type isolate of F. libertatis (CBS 144749). No rpb1 sequence are available for isolates of F. libertatis and therefore comparisons were not possible. The rpb2 sequence of isolate LLC1736 is 99.89 % (876/877 nt) identical to the ex-type isolate of F. libertatis (CBS 144749), and the tef1 sequence 99.31 % (577/581 nt). Due to the absent to minor differences between LLC2077 and the ex-type isolate of F. libertatis (CBS 144749), we treat this isolate as F. libertatis.
Fusarium odoratissimum Maryani et al., Stud. Mycol. 92: 159. 2018. Fig. 18.
Fig. 18.
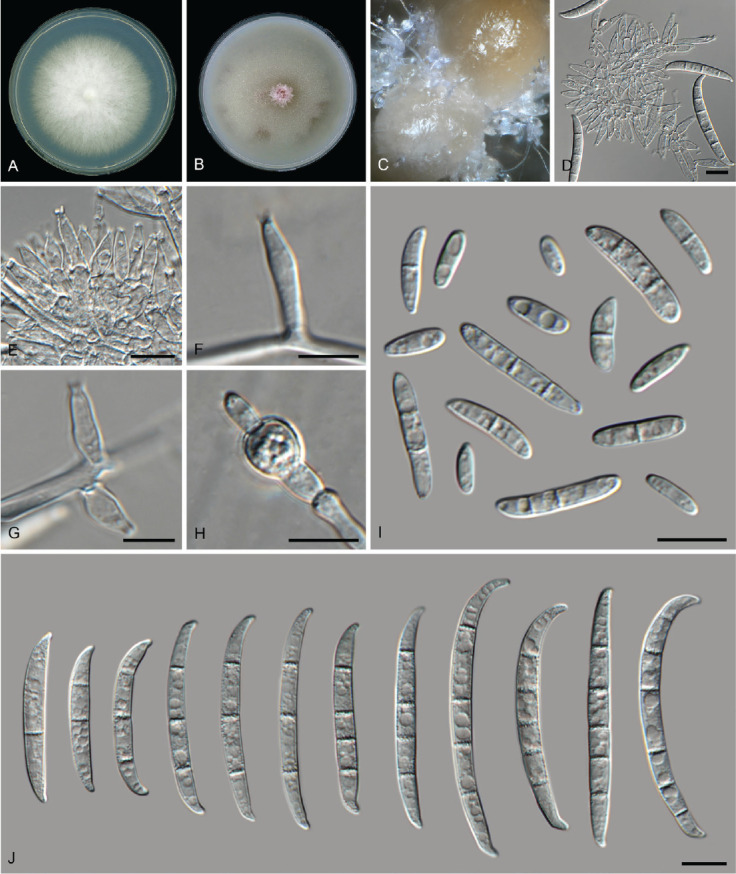
Fusarium odoratissimum (LLC 706). A. Colony on PDA. B. Colony on OA. C. Sporodochia on carnation leaves. D, E. Sporodochia and sporodochial conidiophores. F, G. Lateral conidiogenous cells on aerial mycelium. H. Chlamydospore. I. Aerial microconidia. J. Sporodochial macroconidia. Scale bars = 10 μm.
Materials examined: Supplementary Table S1.
Notes: Isolate LLC706 clustered in a highly-supported clade, closely related to F. odoratissimum and F. phialophorum. In the phylogenetic tree (Fig. 4, part 1), the clade containing LLC706 is almost fully supported (IQ-TREE bootstrap = 99 % / RAxML bootstrap = 99 % / PP = 1). As no CaM sequences are available for the ex-type isolate of F. odoratissimum (InaCC F822) and for none of the isolates of F. phialophorum, the CaM sequence of isolate LLC706 was only compared to F. odoratissimum (isolates JW 54001 and CBS 102030), and a 100 % similarity (574/574 and 570/570 nt), was found. The rpb1 sequence of isolate LLC706 is 98.38 % (1 338/1 360 nt) and 99.10 % (1 431/1 444 nt) identical to the ex-type isolates of F. odoratissimum (InaCC F822) and F. phialophorum (InaCC F971), respectively. The rpb2 sequence of isolate LLC706 is 99.77 % (858/860 nt) and 99.77 % (858/860 nt) identical to the ex-type isolates of F. odoratissimum (InaCC F822) and F. phialophorum (InaCC F971), respectively. The tef1 sequence of isolate LLC706 is 99.48 % (569/572 nt) and 99.65 % (570/572 nt) identical to the ex-type isolates of F. odoratissimum (InaCC F822) and F. phialophorum (InaCC F971), respectively. However, as this subclade is highly similar to both F. odoratissimum and F. phialophorum based on the loci used here and only rpb1 is proving to be genetically the most diverse, we choose not to introduce a new species for this subclade pending a further definition of the species boundaries of F. odoratissimum and F. phialophorum.
Fusarium sp. 1
Materials examined: Supplementary Table S1.
Notes: The isolate clusters with two isolates of Fusarium sp. 1 sensu Crous et al. (2021a) in a moderately to highly supported clade (IQ-TREE bootstrap = 99 % / RAxML bootstrap = 76 % / PP = 1; Fig. 4, part 2). The species was treated by Crous et al. (2021a), but not formally described as none of the loci used could individually discriminate it as unique. The long branch for this isolate is due to the missing rpb2 sequence.
Fusarium veterinarium L. Lombard & Crous, Persoonia 41: 35. 2018. Figs 19.
Fig. 19.
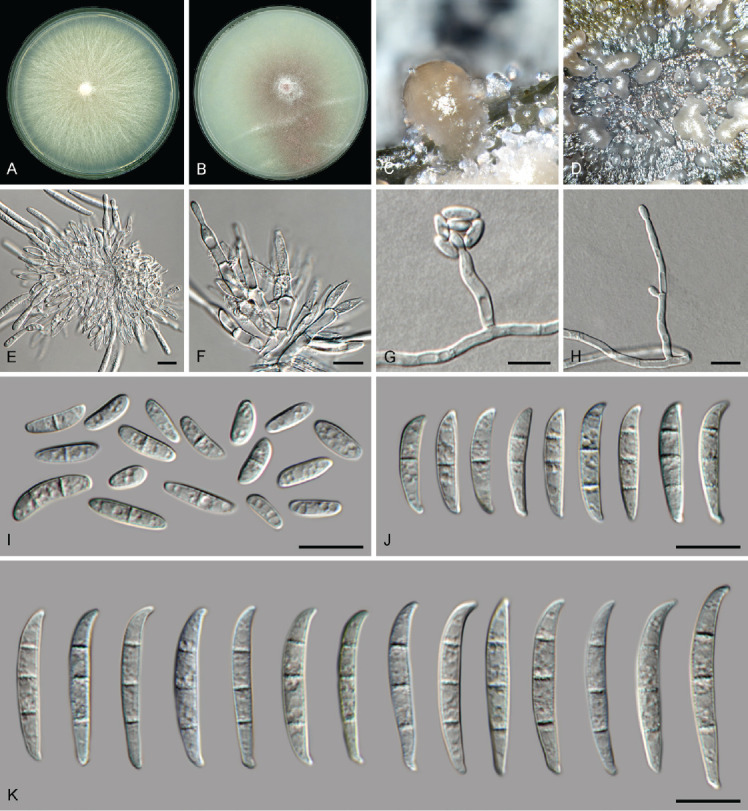
Fusarium veterinarium (LLC 3929). A. Colony on PDA. B. Colony on OA. C, D. Sporodochia on carnation leaves. E, F. Sporodochia and sporodochial conidiophores. G. False head carried on a phialide on aerial mycelium. H. Conidiophore and phialides on aerial mycelium. I. Aerial microconidia. J. Aerial macroconidia. K. Sporodochial macroconidia. Scale bars = 10 μm.
Materials examined: Supplementary Table S1.
Notes: In the phylogenetic tree (Fig. 4, part 1), the F. veterinarium s.l. clade is not supported above the display threshold values (IQ-TREE bootstrap = >84 % / RAxML bootstrap = >74 % / PP = >0.74) and none of the internal “subclades” receive any significant support. No CaM and rpb2 sequences are available for LLC3779, therefore comparisons were made using the sequences of LLC3901. The CaM sequence of isolate LLC3901 is 99.83 % (601/602 nt) identical to the ex-type isolate of F. veterinarium (CBS 109898). No rpb1 sequences of F. veterinarium isolates were available for comparison. The rpb2 sequence of isolate LLC3901 is 99.89 % (876/877 nt) identical to the ex-type isolate of F. veterinarium (CBS 109898), and the tef1 sequence 99.51 % (613/616 nt). The CaM sequence of isolate LLC3929 is identical (580/580 nt) to the ex-type isolate of F. veterinarium (CBS 109898). No rpb1 sequences of F. veterinarium isolates were available for comparison. The rpb2 sequence of isolate LLC3929 is identical (877/877 nt) to the ex-type isolate of F. veterinarium (CBS 109898). The tef1 sequence of isolate LLC3929 is identical (423/423 nt) to the ex-type isolate of F. veterinarium (CBS 109898). The observed tree topology seems to be an artifact of the missing F. veterinarium rpb1 sequences and the isolates are therefore treated as belonging to F. veterinarium.
Fusarium sambucinum species complex (FSAMSC)
Fusarium brachygibbosum Padwick, Mycol. Pap. 12: 11. 1945. Fig. 20.
Fig. 20.
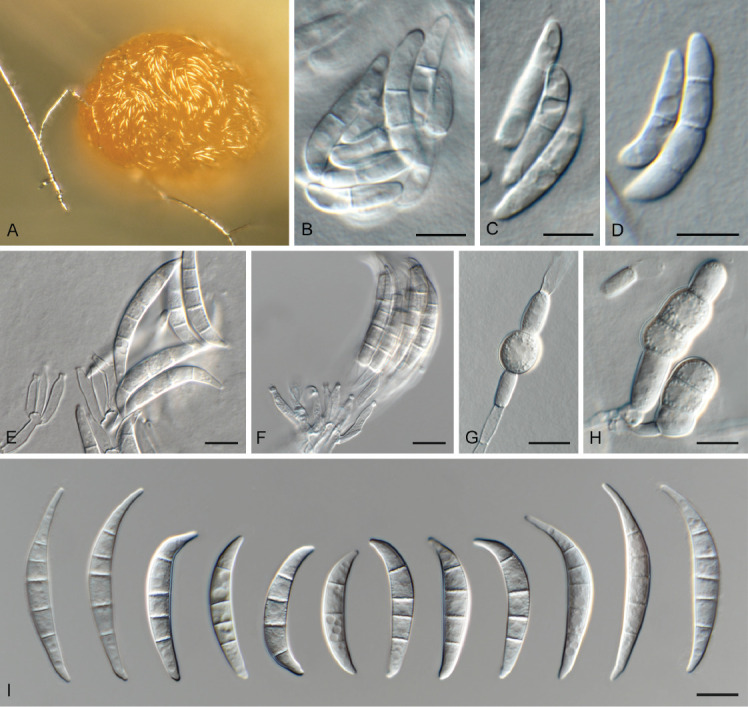
Fusarium brachygibbosum (LLC 1803). A. Sporodochium on SNA. B–D. Macroconidia submerged in SNA. E, F. Sporodochial conidiophores. G, H. Chlamydospores. I. Sporodochial macroconidia. Scale bars = 10 μm.
Materials examined: Supplementary Table S1.
Notes: In the phylogenetic tree (Fig. 2), the F. brachygibbosum s.l. clade is fully supported and the two internal subclades were fully supported and highly supported (IQ-TREE bootstrap = 97 % / RAxML bootstrap = 100 % / PP = 1), respectively. No CaM sequence is available for the ex-type isolate of F. brachygibbosum (NRRL 20954) and therefore a comparison was made with the sequence of isolate NRRL 34033. The CaM sequence of isolate LLC1803 is 99.29 % (559/563 nt) identical to F. brachygibbosum isolate NRRL 34033, the rpb1 sequence 99.44 % (881/886 nt), the rpb2 sequence 98.45 % (888/902 nt), and the tef1 sequence 97.74 % (605/619 nt).
Fusarium pentaseptatum Crous, Sand.-Den. & M.M. Costa, sp. nov. MycoBank MB 846719. Fig. 21.
Fig. 21.
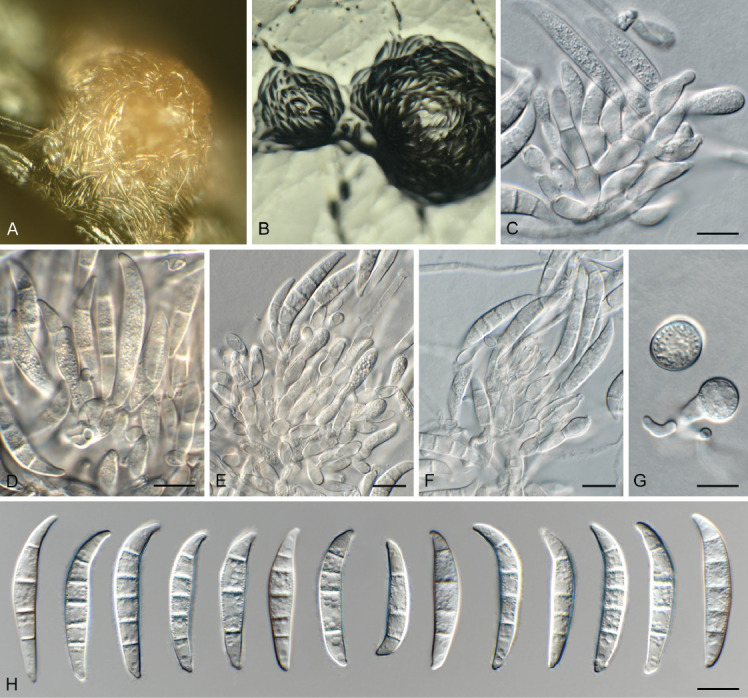
Fusarium pentaseptatum (LLC 1022). A, B. Sporodochia on SNA. C–F. Sporodochial conidiophores. G. Chlamydospores. H. Sporodochial macroconidia. Scale bars = 10 μm.
Etymology: Name refers to its predominantly 5-septate macroconidia.
Typus: Ethiopia, Tigray Region, Western (Mirab) Zone, Tahtay Adiyabo District, Kushet locality (Kebele), endophytic from seed of Striga hermonthica, 2017, T. Tessema & L. Lombard (holotype EMCC-F321, preserved as metabolically inactive culture, culture ex-type LLC1022 = EMCC-F321).
Aerial conidiophores not observed. Sporodochia pale luteous, abundant on CLA. Sporodochial conidiophores densely aggregated, branched, consisting of a stipe bearing whorls or 2–4 monophialides; sporodochial conidiogenous cells monophialidic, doliiform to subcylindrical, 8–13 × 4–5 μm, smooth- and thin-walled with periclinal thickening at apex. Sporodochial conidia falcate, curved dorsiventrally, widest in second or third cell from apex; apical cell curved, pointed; basal cell foot-shaped, notch poorly developed, (3–)5-septate, hyaline, smooth-walled, guttulate; 4-septate conidia (32–)34–36(–38) × 6–7 μm, 5-septate conidia (33–)37–38(–45) × 6–7 μm. Chlamydospores on SNA after 1 wk, solitary or in pairs, terminal or intercalary, subglobose, 12–15 μm diam.
Culture characteristics: Colonies erumpent, spreading, covering dish after 7 d at 25 °C. On PDA surface rosy vinaceous to vinaceous, reverse scarlet to violet; on OA surface and reverse pale luteous.
Additional materials examined: Supplementary Table S1.
Notes: Fusarium pentaseptatum FSAMSC 28 (F. sambucinum SC) (Fig. 2) is fully supported in all three analyses and a sister clade to F. subflagellisporum; the node joining the two clades is only fully supported in the Bayesian analysis. This lineage was introduced by Laraba et al. (2021) as “F. sp. nov.-28” for two isolates from China, associated with soil and soybean roots, respectively. The authors did not formally name this clade and therefore a name is introduced in the present study. The phylogenetic tree shows two potential subclades; however, these differences are minor (comparison of NRRL 66939 with LLC1020; CaM: no sequences for NRRL 66939 & FRC R-9121 to compare; rpb1: 883/886 nt; rpb2: 898/902 nt; tef1: 609/612 nt). No CaM and rpb1 sequences are available for the ex-type isolate of F. subflagellisporum (COAD 2989). The rpb2 sequence of isolate LLC1020 is 95.23 % (859/902 nt) identical to the ex-type isolate of F. subflagellisporum (COAD 2989). The tef1 sequence of isolate LLC1020 is 97.03 % (653/673 nt) identical to the ex-type isolate of F. subflagellisporum (COAD 2989). A comparison was also made against the ex-type of F. brachygibbosum (NRRL 20954): CaM: no sequences for NRRL 20954 to compare; rpb1: 868/886 nt; rpb2: 877/900 nt; tef1: 593/617 nt.
Fusarium subflagellisporum T.F. Nóbrega & R.W. Barreto, Persoonia 47: 313. 2021.
Materials examined: Supplementary Table S1.
Notes: The isolates cluster with the ex-type of F. subflagellisporum in a fully-supported clade (Fig. 2). Fusarium subflagellisporum is listed as FSAMSC 27 in Laraba et al. (2021).
Fusarium transvaalense Sand.-Den. et al., MycoKeys 34: 82. 2018.
Materials examined: Supplementary Table S1.
Notes: In the phylogenetic tree (Fig. 2), the F. transvaalense s.l. clade is fully supported and the two internal subclades were fully and highly supported (IQ-TREE bootstrap = 99 % / RAxML bootstrap = 87 % / PP = 1), respectively. The rpb1 sequence of isolate LLC1488 is 99.24 % (131/132 nt) identical to the ex-type isolate of F. transvaalense (CBS 144211) – only partial overlap is available for comparison. The rpb2 sequence of isolate LLC1488 is 98.56 % (889/902 nt) identical to the ex-type isolate of F. transvaalense (CBS 144211), and the tef1 sequence 96.75 % (655/677 nt).
Fusarium tricinctum species complex (FTSC)
Fusarium avenaceum (Fr.) Sacc., Syll. Fung. 4: 713. 1886.
Synonyms: see www.fusarium.org
Materials examined: Supplementary Table S1.
Notes: Isolate LLC725 clusters with isolates labelled as F. avenaceum and F. arthrosporioides (including the ex-neotype of F. avenaceum) in the phylogenetic tree (Fig. 2). We choose to place this isolate in F. avenaceum for now as there are currently no (ex-)type sequences available for F. arthrosporioides.
DISCUSSION
In the present study we investigated which Fusarium spp. were associated with soil from Sorghum fields in Ethiopia, or dominate the rhizosphere of Sorghum when grown in these soils or occurred as endophytes in Sorghum roots and seeds, or Striga stems and seeds. A total of 42 Fusarium species distributed over eight different Fusarium Species Complexes were identified, including three species which we believe are new to science and two undescribed species.
Several species complexes were poorly represented, namely FburSC (F. burgessii, 1 isolate), FCSC (F. nelsonii, 3 isolates and F. sporodochiale, 1 isolate), FCOSC (F. concolor, 1 isolate), and FTSC (F. avenaceum, 1 isolate). Fusarium burgessii was initially described from soils associated with native vegetation in Australia (Laurence et al. 2011). Fusarium nelsonii was isolated from plant debris in wheat soil, the roots of Medicago and from sorghum malt and corn kernels (Marasas et al. 1998) and F. sporodochiale from soil collected in South Africa (Lombard et al. 2019). Fusarium concolor (described from Hordeum vulgare in Uruguay; Reinking 1934) was recently reported from various soils associated with Hordeum, Triticum and Zea in Africa (South Africa, Zimbabwe), Australia, Europe (Italy, Spain), South America (Uruguay), and North America (Hawaii, Georgia) (Jacobs-Venter et al. 2018). Lastly, F. avenaceum (neotype from Hordeum vulgare in Denmark; Crous et al. 2021b) is a common soilborne fungus, and can cause stem and root diseases in various pasture legumes in temperate regions of the world (Leslie & Summerell 2006). Although soilborne, none of these four species were well represented in the present study, and thus probably play a minor role in the Striga/Sorghum pathosystem.
Four complexes that were well represented however, include the FFSC, FIESC, FOSC and FSAMSC. Fifteen species were delineated in the FFSC. Although F. proliferatum has been extensively studied in the past, most isolates were representative of F. annulatum (Yilmaz et al. 2021). Fusarium annulatum is common in tropical and temperate zones, and has been reported from a wide host range (Domsch et al. 2007), occurring in various soil samples in the present study. Fusarium caapi was recently described from Brazil, where it was isolated from seed of Brachiaria brizantha (Costa et al. 2021), and this is the first record from Africa, where it was found in Sorghum rhizospere soil samples. Fusarium lactis was originally identified from clotted milk in Europe, and later shown to be associated with endosepsis of figs in California (Leslie & Summerell 2006), but was here found associated with Sorghum root collors in a Dutch experimental greenhouse. Fusarium mirum was recently described from Sorghum bicolor in Cameroon and Egypt (Costa et al. 2022), and here isolated from Sorghum soil and Striga seed in Ethiopia. Fusarium brevicatenulatum was originally described from symptomatic plants of Striga asiatica collected in Madagascar (Nirenberg et al. 1998), and shown here to be synonymous with F. pseudoanthophilum (Zea mays, Zimbabwe), a synonymy already suspected by Leslie & Summerell (2006), and confirmed via mating studies by Amata et al. (2010). The Ethiopian isolates studied here were all recovered from Striga seed, again underlining the strong association of F. brevicatenulatum with Striga. Fusarium sudanense was described from Striga hermonthica in Sudan (Moussa et al. 2017), and later also reported to cause seedling blight and seed rot of wheat in Argentina (Larran et al. 2020). The isolates recovered in the present study originated from either Striga seed, or Sorghum field or rhizosphere soil in Ethiopia. Fusarium fredkrugeri was described from soil samples and the rhizosphere of Melhania acuminata collected in South Africa, and is also known from Striga hermonthica in Madagascar (Sandoval-Denis et al. 2018), and isolated from Striga seed and Sorghum field and rhizosphere soil in Ethiopia in the present study. Fusarium secorum, a sugar beet pathogen from the USA (Secor et al. 2014), is reported here as a single isolate from Sorghum field soil in Ethiopia. Fusarium udum causes a wilt disease of Cajanus cajan and Crotalaria spp. in tropical regions (Pfenning et al. 2019), and is reported here from Sorghum field soil in Ethiopia. Fusarium thapsinum is widely distributed, causing a stalk rot and grain mould of Sorghum (Leslie & Summerell 2006), and was also isolated from Striga seed and Sorghum rhizosphere soil in Ethiopia in this study. Fusarium verticillioides is globally widely distributed, causing a stalk and cob rot of maize (Leslie & Summerell 2006), and was isolated from Sorghum seed and rhizosphere soil in Ethiopia in the present study. Fusarium andiyazi is pathogenic to Sorghum, and may be the most dominant Fusarium species on this host (Marasas et al. 2001). It has also been reported from Zea mays in Portugal (Simões et al. 2022), and Saccharum officinarum in China (Bao et al. 2020). It was isolated from Striga seed and stems, and Sorghum seed in Ethiopia in the present study. Fusarium ficicrescens, isolated from figs in Iran (Al-Hatmi et al. 2016), is reported here from Striga asiatica seed collected in Ethiopia. Fusarium nygamai is widely distributed and has been associated with root rot of Asparagus, Gossypium, Oryza, Pennisetum, Sorghum, Vicia faba and Zea, and may cause systemic infections in humans (Leslie & Summerell 2006). It was isolated from Sorghum field and rhizosphere soil in Ethiopia in the present study. The last species in this SC remains unnamed for now (Fusarium sp. isolate LLC1198) pending the collection of more isolates.
Ten species were associated with the FIESC, including F. extenuatum and F. tangerinum, which were described here as new species isolated from Ethiopian Sorghum field soil and Sorghum seed and rhizosphere, respectively. Fusarium compactum is commonly isolated from grasslands and dry dessert soils in hot climates (Australia, Middle East, Africa; Leslie & Summerell 2006), and is generally regarded as a saprobe. In the present study it was isolated from Striga hermonthica seed and Sorghum field soil in Ethiopia. Fusarium lacertarum is known as a pathogen of Vigna unguiculata and Nopalea cochenillifera in Brazil (do Amaral et al. 2022), and Sorghum bicolor in the USA (Beacorn & Thiessen 2021). It the present study it was isolated as endophyte from Sorghum seed and from Sorghum rhizosphere soil in Ethiopia. Fusarium nanum was described from Musa nana in China (Wang et al. 2019), and has since been found to cause decay of Cucumis melo in China (Zhang et al. 2022), and Glycine max in the USA (Okello et al. 2020). It was isolated as an endophyte from Ethiopian Sorghum seed in the present study. Fusarium caatingaense, which was described from insects in Brazil (Santos et al. 2019), was isolated as an endophyte from Ethiopian Striga hermonthica seed in the present study. Fusarium serpentinum was described from an unknown substrate by Xia et al. (2019), and isolated in the present study from Sorghum rhizosphere soil in Ethiopia. Fusarium clavus was described from environmental, plant and human samples originating from Africa, Asia, Europe and North America (Xia et al. 2019). It proved to be a dominant taxon in the present study, being isolated as endophyte from Striga hermonthica seed as well as Sorghum field and rhizosphere soils in Ethiopia. Fusarium duofalcatisporum appears to be restricted to North- and South-eastern Africa (O’Donnell et al. 2009), and is here shown to be a dominant taxon in the Striga/Sorghum pathosystem, associated as endophyte with Striga hermonthica seed, as well as Sorghum field and rhizosphere soil in Ethiopia. Fusarium incarnatum has a complicated taxonomic history with numerous synonyms (Xia et al. 2019, Crous et al. 2021b) and was here isolated from Striga and Sorghum seeds as well as Sorghum rhizosphere soil from Ethiopia.
Eight species are associated with the FOSC. Fusarium odoratissimum is the well-known causal agent of Panama disease of banana (Maryani et al. 2019a). Surprisingly, an isolate from this pathogen was recently collected during a Citizen Science project sampling garden soils in the Netherlands (Crous et al. 2021a), and was again isolated from the roots of Sorghum plants growing in Dutch greenhouse soils during the present study. Fusarium fabacearum was described from Glycine max and Zea mays collected in South Africa (Lombard et al. 2019), and was isolated from seed of Striga asiatica and Sorghum field and rhizosphere soils in Ethiopia in the present study. Fusarium veterinarium was reported from diverse, mainly veterinary samples collected in Europe and the USA (Lombard et al. 2019), and shown to be common in Ethiopian as well as Dutch Sorghum rhizosphere soil in the present study. Fusarium curvatum is known from Beaucarnia sp., Hedera helix and Matthiola incana collected in Europe, and reported here as a single isolate from Sorghum field soil in Ethiopia. Fusarium libertatis was described from the rock surfaces in the stone quarry on Robben Island (Van Riebeeck’s Quarry), where Nelson Mandela and other political prisoners were forced to undergo manual hard labour. This species is here also reported as a single isolate from Sorghum field soil in Ethiopia. Fusarium glycines is known from Glycine max collected in South Africa and Linum usitatissium from an unknown location (Lombard et al. 2019), while F. gossypinum is known from wilted Gossypium hirsutum collected in the Ivory coast; both species were isolated here from Sorghum field and rhizosphere soil in Ethiopia. Fusarium sp. 1 is an unnamed species in the FOSC from garden soil in the Netherlands (Crous et al. 2021a) and was also isolated from Sorghum roots in a Dutch greenhouse.
Four species are associated with FSAMSC in this study. Fusarium brachygibbosum was originally described from diseased Sorghum vulgare plants collected in India (Crous et al. 2021b), and is now regarded as a a potential European Union quarantine pest (EFSA PLH Panel 2021). In the present study it was isolated from Sorghum field soil in Ethiopia. Fusarium pentaseptatum (=FSAMSC 28; Laraba et al. 2021) is described here as endophyte from Striga hermonthica seed and Sorghum field and rhizosphere soils in Ethiopia, and is known from soybeans and soil samples in China (Laraba et al. 2021). Fusarium subflagellisporum (=FSAMSC 27; Laraba et al. 2021) was described based on hypertrophied floral and vegetative branches of Mangifera indica trees in Brazil, but has been reported from Arachis hypogaea and Zea mays soil in the USA, Sorghum debris in Puerto Rico, from Nigeria, and Pennisetum soils in Zimbabwe (Crous et al. 2021b). In the present study it is reported from Ethiopian Striga hermonthica seed and Sorghum field soil. Lastly, Fusarium transvaalense is known from the rhizosphere of Sida cordifolia, Kyphocarpa angustifolia and Melhania acuminata in South Africa (Sandoval-Denis et al. 2018), and is reported here from Ethiopian Sorghum field and rhizosphere soil.
Zimmermann et al. (2015) developed a specific AFLP-based marker to identify isolates of F. oxysporum f.sp. strigae (Fos) among other Fusarium isolates isolated from Striga hermonthica collected in Africa (Burkina Faso, Ghana, Kenya, Mali, Nigeria and Niger). Barring one isolate of F. oxysporum f.sp. melonis from Israel, the test proved highly effective in detecting this DNA fragment in isolates of Fos, suggesting it an effective marker to use in the detection of Fos in African soils. In screening for this marker DNA fragment among the isolates obtained in the present study, however, it was detected in several species in diverse species complexes, namely (FCSC) F. nelsonii; (FFSC) F. andiyazi, F. annulatum, F. fredkrugeri, F. nygamai, F. sudanense, F. verticillioides; (FIESC) F. clavus, F. compactum, F. duofalcatisporum, F. lacertarum; (FOSC) F. fabacearum, F. glycines, F. veterinarium and (FSAMSC) F. brachygibbosum, F. subflagellisporum and F. transvaalense. Interestingly, many isolates from the FSAMSC contain a positive amplicon with a sequence which is not completely identical to the original reference sequence (19 substitutions over the length of 165 nucleotides). Although it is to be expected that the AFLP-based marker DNA fragment could be transferred among isolates in the FOSC via the parasexual cycle (Ma et al. 2010), it is rather surprising to find it in other Fusarium species complexes, suggesting that the parasexual cycle (or horizontal chromosome transfer) could be more prominent in Fusarium than expected. Further studies are now underway to determine if the presence of this DNA fragment also correlates with effective biocontrol of Striga, as it does in the case of Fos. Further research is therefore required to establish the potential role that these various Fusarium spp. could play in inhabiting Striga.
Acknowledgments
The Bill and Melinda Gates Foundation supported this research work through grant number OPP1082853: PROMISE ‘Promoting Root Microbes for Integrated Striga Eradication’ project via Ethiopian Institute of Agricultural Research and Netherlands Institute of Ecology. We thank Marjan Vermaas for assistance with the photographic plates.
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
Supplementary Material: http://fuse-journal.org/
Collection details, striga gene haplotype and GenBank accession numbers of novel strains used in the phylogenetic trees.
Collection details and GenBank accession numbers of strains used in the phylogenetic trees.
Summary of phylogenetic information for the different analyses in this study.
REFERENCES
- Abbasher AA, Kroschel J, Sauerborn J. (1995). Micro-organisms of Striga hermonthica in northern Ghana with potential as biocontrol agents. Biocontrol Science and Technology 5: 157–161. [Google Scholar]
- Abbasher AA, Sauerborn J. (1992). Fusarium nygamai a potential bioherbicide for Striga hermonthica control in sorghum. Biological Control 2: 291–296. [Google Scholar]
- Abbasher AA, Sauerborn J, Kroschel J, et al. (1996). Evaluation of Fusarium semitectum var. majus for biological control of Striga hermonthica. In: Ninth International Symposium on Biological Control of Weeds (Moran VC, Hoffmann JH, eds). University of Cape Town, South Africa: 115–120. [Google Scholar]
- Al-Hatmi AMS, Mirabolfathy M, Hagen F, et al. (2016). DNA barcoding, MALDI-TOF, and AFLP data support Fusarium ficicrescens as a distinct species within the Fusarium fujikuroi species complex. Fungal Biology 120: 265–278. [DOI] [PubMed] [Google Scholar]
- Amata RL, Burgess LW, Summerell BA, et al. (2010). An emended description of Fusarium brevicatenulatum and F. pseudoanthophilum based on isolates recovered from millet in Kenya. Fungal Diversity 43: 11–25. [Google Scholar]
- Anteyi WO, Klaiber I, Rasche F. (2022). Diacetoxyscirpenol, a Fusarium exometabolite, prevents efficiently the incidence of the parasitic weed Striga hermonthica. BMC Plant Biology 22: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Huang Z, Li T, et al. (2020). First report of Fusarium andiyazi causing sugarcane Pokkah Boeng Disease in China. Plant Disease 104: 286–286. [Google Scholar]
- Beacorn JA, Thiessen LD. (2021). First report of Fusarium lacertarum causing Fusarium Head Blight on Sorghum in North Carolina. Plant Disease 105: 699. [Google Scholar]
- Ciotola M, DiTommaso A, Watson AK. (2000). Chlamydospore production, inoculation methods and pathogenicity of Fusarium oxysporum M12-4A, a biocontrol for Striga hermonthica. Biocontrol Science and Technology 10: 129–145. [Google Scholar]
- Ciotola M, Watson AK, Hallett SG. (1995). Discovery of an isolate of Fusarium oxysporum with potential to control Striga hermonthica in Africa. Weed Research 35: 303–309. [Google Scholar]
- Costa MM, Melo MP, Sandin FC, et al. (2021). Fusarium species from tropical grasses and description of two new taxa. Mycological Progress 20: 61–72. [Google Scholar]
- Costa MM, Saleh AA, Melo MP, et al. (2022). Fusarium mirum sp. nov., intertwining Fusarium madaense and Fusarium andiyazi, pathogens of tropical grasses. Fungal Biology 126: 250–266. [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Hernández-Restrepo M, van Iperen AL, et al. (2021a). Citizen science project reveals novel fusarioid fungi (Nectriaceae, Sordariomycetes) from urban soils. Fungal Systematics and Evolution 8: 101–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Lombard L, Sandoval-Denis M, et al. (2021b). Fusarium: more than a node or a foot-shaped basal cell. Studies in Mycology 98: 100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkleij GJM, Groenewald JZ, et al. (2019). Fungal Biodiversity. [Westerdijk Laboratory Manual Series No. 1]. Westerdijk Fungal Biodiversity Institute publishing, Utrecht, Netherlands. [Google Scholar]
- Daffalla HM, Hassan MM, Osman MG, et al. (2014). Effect of seed priming on early development of Sorghum (Sorghum bicolor L. Moench) and Striga hermonthica (Del.) Benth. International Scholarly Research Notices 2014: 134931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Amaral ACT, Koroiva R, da Costa AF, et al. (2022). First report of Fusarium lacertarum as the causal agent of wilt in Vigna unguiculata. Journal of Plant Patholology 104: 1173. [Google Scholar]
- Domsch K, Gams W, Anderson T-H. (2007). Compendium of Soil Fungi 2 edn. IHW-Verlag, Eching. [Google Scholar]
- EFSA PLH Panel (EFSA Panel on Plant Health), Bragard C, Di Serio F, Gonthier P, et al. (2021). Scientific Opinion on the pest categorisation of Fusarium brachygibbosum. EFSA Journal 19: 6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejeta G. (2007). Breeding for Striga resistance in sorghum: exploitation of an intricate host–parasite biology. Crop Science 47 (Suppl. 3): S216–S227. [Google Scholar]
- Fisher NL, Burgess LW, Toussoun TA, et al. (1982). Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151–153. [Google Scholar]
- Giraldo A, Hernández-Restrepo M, Crous PW. (2019). New plectosphaerellaceous species from Dutch garden soil. Mycological Progress 18: 1135–1154. [Google Scholar]
- Gressel J, Hanafi A, Head G, et al. (2004). Major heretofore intractable biotic conisolatets to African food security that may be amenable to novel biotechnological solutions. Crop Protection 23: 661–689. [Google Scholar]
- Groenewald M, Lombard L, de Vries M, et al. (2018). Diversity of yeast species from Dutch garden soil and the description of six novel Ascomycetes. Federation of European Microbiological Societies Yeast Research 18: foy076. [DOI] [PubMed] [Google Scholar]
- Gurney AL, Press MC, Scholes JD. (2002). Can wild relatives of sorghum provide new sources of resistance or tolerance against Striga species? Weed Research 42: 317–324. [Google Scholar]
- Hess DE, Kroschel J, Traoré D, et al. (2002). Striga: biological control strategies for a new millennium. In: Sorghum and Millet Diseases 2000 (Leslie JF, ed.), Iowa State Press, Ames, Iowa, USA: 165–170. [Google Scholar]
- Jacobs-Venter A, Laraba I, Geiser DM, et al. (2018). Molecular systematics of two sister clades, the Fusarium concolor and F. babinda species complexes, and the discovery of a novel microcycle macroconidium–producing species from South Africa. Mycologia 110: 1189–1204. [DOI] [PubMed] [Google Scholar]
- Kroschel J, Hundt A, Abbasher A, et al. (1996). Pathogenicity of fungi collected in northern Ghana to Striga hermonthica. Weed Research 36: 515–520. [Google Scholar]
- Kroschel J, Jost A, Sauerborn J. (1999). Insects for Striga control – possibilities and conisolatets. In: Advances in Parasitic Weed Control at On-farm Level. Vol. I. Joint Action to Control Striga in Africa (Kroschel J, Mercer-Quarshie H, Sauerborn J, eds.), Margraf Verlag, Weikersheim, Germany: 117–132. [Google Scholar]
- Laraba I, McCormick SP, Vaughan MM, et al. (2021). Phylogenetic diversity, trichothecene potential, and pathogenicity within Fusarium sambucinum species complex. PLoS ONE 16: e0245037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larran S, Siurana MPS, Caselles JR, et al. (2020). Fusarium sudanense, endophytic fungus causing typical symptoms of seedling blight and seed rot on wheat. Journal of King Saud University – Science 32: 468–474. [Google Scholar]
- Laurence MH, Summerell BA, Burgess LW, et al. (2011). Fusarium burgessii sp. nov. representing a novel lineage in the genus Fusarium. Fungal Diversity 49: 101–112. [Google Scholar]
- Leslie JF, Summerell BA. (2006). The Fusarium laboratory manual. Blackwell Publishing Professional, USA. [Google Scholar]
- Lombard L, Sandoval-Denis M, Lamprecht SC, et al. (2019). Epitypification of Fusarium oxysporum – clearing the taxonomic chaos. Persoonia 43: 1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, van der Does HC, Borkovich KA, et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasas WFO, Rheeder JP, Lamprecht SC, et al. (2001). Fusarium andiyazi sp. nov., a new species from sorghum. Mycologia 93: 1203–1210. [Google Scholar]
- Marasas WFO, Rheeder JP, Logrieco A, et al. (1998). Fusarium nelsonii and F. musarum: two new species in Section Arthrosporiella related to F. camptoceras. Mycologia 90: 505–513. [Google Scholar]
- Marley PS, Aba DA, Shebayan JAY, et al. (2004). Integrated management of Striga hermonthica in sorghum using a mycoherbicide and host plant resistance in the Nigerian Sudano-Sahelian savanna. Weed Research 44: 157–162. [Google Scholar]
- Marley PS, Ahmed SM, Shebayan JAY, et al. (1999). Isolation of Fusarium oxysporum with potential for biocontrol of the witchweed (Striga hermonthica) in the Nigerian Savanna. Biocontrol Science and Technology 9: 159–163. [Google Scholar]
- Maryani N, Lombard L, Poerba YS, et al. (2019a). Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Studies in Mycology 92: 155–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryani N, Sandoval-Denis M, Lombard L, et al. (2019b). New endemic Fusarium species hitch-hiking with pathogenic Fusarium isolates causing Panama disease in small-holder banana plots in Indonesia. Persoonia 43: 48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masteling R, Lombard L, de Boer W, et al. (2019). Harnessing the microbiome to control plant parasitic weeds. Current Opinion in Microbiology 49: 26-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh Q, Schmidt HA, Chernomor O, et al. (2020). IQ-TREE2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa TAA, Al-Zharani HS, Kadasa NMS, et al. (2017). Two new species of the Fusarium fujikuroi species complex isolated from the natural environment. Antonie van Leeuwenhoek 110: 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, et al. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg H, O’Donnell K, Kroschel J, et al. (1998). Two new species of Fusarium: Fusarium brevicatenulatum from the noxious weed Striga asiatica in Madagascar and Fusarium pseudoanthophilum from Zea mays in Zimbabwe. Mycologia 90: 459–463. [Google Scholar]
- Nirenberg HI. (1976). Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. (2009). Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-equiseti and F. chlamydosporum species complexes within the United States. Journal of Clinical Microbiology 47: 3851–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okello PN, Petrovic K, Singh AK, et al. (2020). Characterization of species of Fusarium causing root rot of Soybean (Glycine max L.) in South Dakota, USA, Canadian Journal of Plant Pathology 42: 560–571. [Google Scholar]
- Parker C. (2012). Parasitic Weeds: A world Challenge. Weed Science 60: 269–276. [Google Scholar]
- Pfenning LH, De Melo MP, Costa MM, et al. (2019). Fusarium udum revisited: a common, but poorly understood member of the Fusarium fujikuroi species complex. Mycological Progress 18: 107–117. [Google Scholar]
- Rayner RW. (1970). A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, UK. [Google Scholar]
- Reinking OA. (1934). Interesting new Fusaria. Zentralblatt für Bakteriologie und Parasitenkunde, Abteilung 2. 89: 509–514. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sandoval-Denis M, Swart WJ, Crous PW. (2018). New Fusarium species from the Kruger National Park, South Africa. MycoKeys 34: 63–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos ACDS, Trindade JVC, Lima CS, et al. (2019). Morphology, phylogeny, and sexual stage of Fusarium caatingaense and Fusarium pernambucanum, new species of the Fusarium incarnatum-equiseti species complex associated with insects in Brazil. Mycologia 111: 244–259. [DOI] [PubMed] [Google Scholar]
- Sauerborn J, Abbasher AA, Kroschel J, et al. (1996). Biological control of Striga hermonthica with Fusarium nygamai in maize. In: Ninth International Symposium on Biological Control of Weeds (Moran VC, Hoffmann JH, eds), University of Cape Town, South Africa: 461–466. [Google Scholar]
- Sauerborn J, Müller-Stöver D, Hershenhorn J. (2007). The role of biological control in managing parasitic weeds. Crop Protection 26: 246–254. [Google Scholar]
- Secor GA, Rivera-Varas V, Christ DS, et al. (2014). Characterization of Fusarium secorum, a new species causing Fusarium yellowing decline of sugar beet in north central USA. Fungal Biology 118: 764–75. [DOI] [PubMed] [Google Scholar]
- Simões D, Diogo E, de Andrade E. (2022). First report of Fusarium andiyazi presence in Portuguese maize kernels. Agriculture 12: 336. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurro M, Boari A, Thiombiano B, et al. (2019). Strigolactones and parasitic plants. In: Strigolactones - Biology and Applications (Koltai H, Prandi C, eds). Springer International Publishing: 89–120. [Google Scholar]
- Wang MM, Chen Q, Diao YZ, et al. (2019). Fusarium incarnatum-equiseti complex from China. Persoonia 43: 70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia JW, Sandoval-Denis M, Crous PW, et al. (2019). Numbers to names – restyling the Fusarium incarnatum-equiseti species complex. Persoonia 43: 186–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N, Sandoval-Denis M, Lombard L, et al. (2021). Redefining species limits in the Fusarium fujikuroi species complex. Persoonia 46: 129–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Dang QQ, Cao XD, et al. (2022). First report of muskmelon fruit rot caused by Fusarium nanum in China. Plant Disease 106: 1763. [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, et al. (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational Biology 7: 203–214. [DOI] [PubMed] [Google Scholar]
- Zimmermann J, de Klerk M, Musyoki MK, et al. (2015). An explicit AFLP-based marker for monitoring Fusarium oxysporum f.sp. strigae in tropical soils. Biological Control 89: 42–52. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Collection details, striga gene haplotype and GenBank accession numbers of novel strains used in the phylogenetic trees.
Collection details and GenBank accession numbers of strains used in the phylogenetic trees.
Summary of phylogenetic information for the different analyses in this study.