Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)-based gene editing has been widely used in various cell types and organisms. To make genome editing with Clustered regularly interspaced short palindromic repeats far more precise and practical, we must concentrate on the design of optimal gRNA and the selection of appropriate Cas enzymes. Numerous computational tools have been created in recent years to help researchers design the best gRNA for Clustered regularly interspaced short palindromic repeats researches. There are two approaches for designing an appropriate gRNA sequence (which targets our desired sites with high precision): experimental and predicting-based approaches. It is essential to reduce off-target sites when designing an optimal gRNA. Here we review both traditional and machine learning-based approaches for designing an appropriate gRNA sequence and predicting off-target sites. In this review, we summarize the key characteristics of all available tools (as far as possible) and compare them together. Machine learning-based tools and web servers are believed to become the most effective and reliable methods for predicting on-target and off-target activities of Clustered regularly interspaced short palindromic repeats in the future. However, these predictions are not so precise now and the performance of these algorithms -especially deep learning one’s-depends on the amount of data used during training phase. So, as more features are discovered and incorporated into these models, predictions become more in line with experimental observations. We must concentrate on the creation of ideal gRNA and the choice of suitable Cas enzymes in order to make genome editing with Clustered regularly interspaced short palindromic repeats far more accurate and feasible.
Keywords: CRiSPR/Cas, gRNA design, on-target, off-target, computational approach, machine learning
1 Introduction
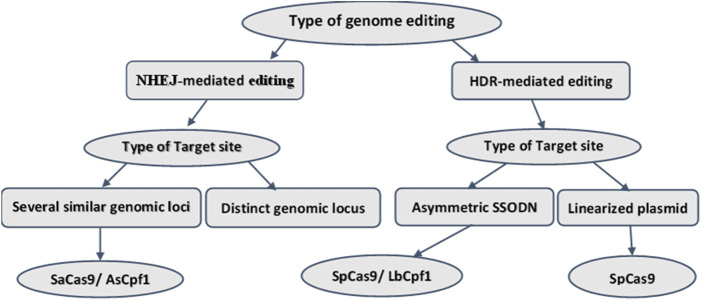
Over the last decade, the Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system has become the dominant tool for genome editing due to its simplicity, high performance, accuracy, and programmability (Gaj et al., 2013; Jacquin et al., 2019; Afzal et al., 2020). In addition, other influential factors such as ease of use, low cost, high speed, multiplex potential, and higher specific DNA targeting ability have increased the success and popularity of CRISPR across the global scientific community (Mali et al., 2013). The unique characteristics of this technology have made it one of the broad topics in molecular biology, synthetic biology, and genetic engineering (Jinek et al., 2012). Gene activation (CRISPRa), gene repression, CRISPR interference (CRISPRi), and epigenome editing are popular tasks in genome engineering using CRISPER. The basic overflow of the CRISPR systems is illustrated in Figure 1.
FIGURE1.
Basic overflow of CRISPR systems.
As shown in Figure 2, CRISPR systems have three main components. The first one is a short synthetic guide RNA sequence (gRNA) necessary for Cas binding. The gRNA targets the Cas9 endonuclease (a protein which can cleave the DNA sequences) to define DNA. The gRNA can be supplied as a two-part system consisting of crRNA and tracrRNA, or as a single guide RNA (sgRNA), where the crRNA and tracrRNA are connected by a linker. The target’s recognition is facilitated by the protospacer-adjacent motif (PAM). Cleavage occurs on both strands 3 bp upstream of the PAM.
FIGURE 2.
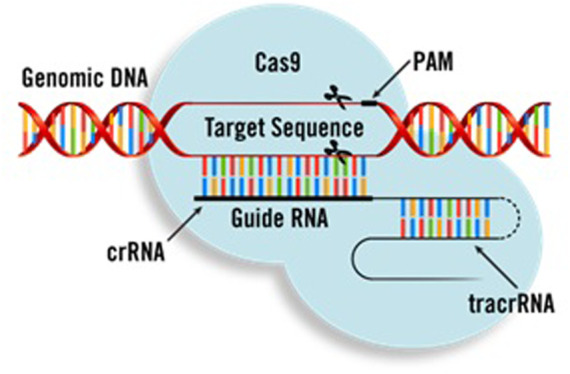
Main components of CRISPR (Duan et al., 2021).
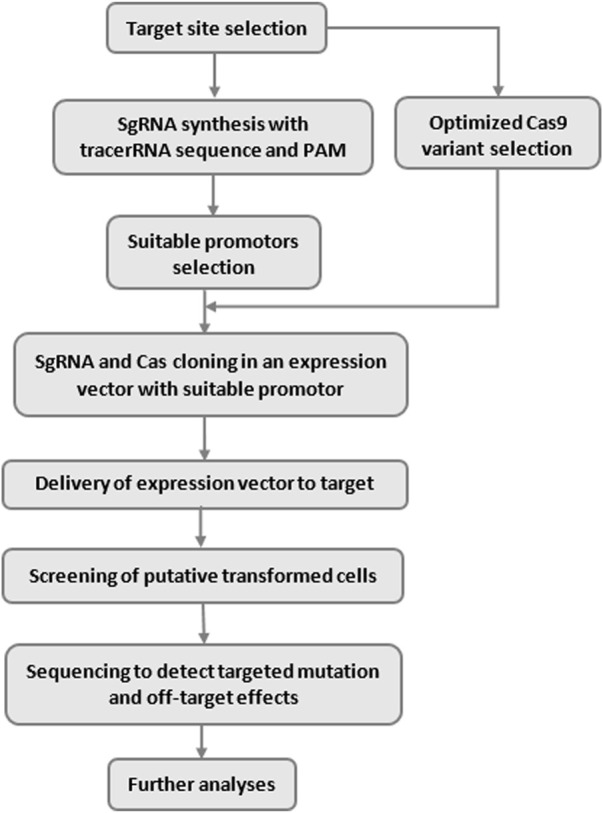
To use CRISPR for genome engineering, we need to select two components: Cas9 and gRNA (Gasiunas et al., 2012; Cox et al., 2015). Once a genome modification is decided, the first step is to identify the best site/sites for targeting Cas-induced DSBs (Jinek et al., 2014). The second step is to design the appropriative gRNA (Cui et al., 2018).
After designing gRNA, the only requirement for cleaving a CRISPR target site is finding a 3-base pair (3 bp) PAM. The form of PAM varies depending on the bacterial species of the Cas9 gene. For example, the most commonly used Cas9 nuclease, derived from S.pyogenes, recognizes a PAM sequence of NGG (Rabinowitz et al., 2020). Using the frequency of “GG” = 5.21% in the reference human genome, there would be an expected 161,284,793 NGG PAM sites in the human genome, or roughly one “GG” dinucleotide every 42 bases. So, cleaving unwanted sites, called off-target sites, is very common (Duan et al., 2021). Therefore, CRISPR target sites should be selected in such a way that minimizes potential off-target cleavage (Herai, 2019; Rabinowitz et al., 2020). But this is not always straightforward as it is not guaranteed that the desired cleaves will appear on just the selected site. Unfortunately, the existence of these unwanted cleaves is possible in every experiment. Therefore, activity (on-target) and specificity (off-target) are two critical factors considered when designing a genomic edition with CRISPR (Herai, 2019).
According to research, the accuracy of CRISPR-based genomic edition depends on two issues: 1) the choice of Cas enzyme with suitable cutting power, 2) the choice of the appropriate cutting site, which relies on the performance of the gRNA. To achieve this, in the first step, we must select the optimal gRNAs contains high on-target activity and low (no) off-target efficiency (Moreno-Mateos et al., 2015; Luo et al., 2019; Manibalan et al., 2020). We will discuss this issue later. In the second step, we must select a suitable Cas enzyme [15]. In recent years, different variants of the Cas enzyme have been discovered. We can proceed according to Figure 3 to choose the proper Cas, depending on the type of editing. The choice of the Cas enzyme is effective on the PAM and the gRNA design.
FIGURE 3.
Selection of Cas enzyme.
In recent years, researchers have taken two main approaches for designing gRNAs, including experimental and machine learning-based methods (ML) (Lin and Luo, 2019). ML-based methods utilize the results of computational algorithms trained with real data to predict the effects of gRNAs instead of designing an actual experiment. Experimental methods are very costly and time-consuming (Chuai et al., 2017; Lin and Luo, 2019). In contrast, ML models are inexpensive and manageable. However, in terms of accuracy, they are still very different from experimental methods (Höijer et al., 2020). The accuracy of ML methods is highly dependent on the training process and the availability of adequate training data. Recent advances in the genome-wide analyses help researchers to discover all off-target sites, while the detection methods like Polymerase Chain Reaction (PCR) based methods, cannot find all of these sites. Using new sequencing technology, such as next-generation sequencing (NGS), and third generation sequencing which based on long-reads, can help us to detect more off-target sites. Mainly, single-molecule real-time sequencing (SMRT), has shown promising performance in genome sequencing. Researchers use these techniques to find more accurate information about off-target sites and use them in training their computational models (Lin and Wong, 2018; Höijer et al., 2020). Also, there are some repetitive, low complexity, AT/GC-rich regions, known as dark, in which ML-based tools cannot predict on-target and off-target sites in these areas. But amplification-free long-read sequencing technology helps to reveal Cas9 target sites even in these dark regions (Höijer et al., 2020). As the number of available features about on-target and off-target sites and the creation of large databases in this field increases, the predictions of ML-based methods become closer to experimental observations (Jiang et al., 2016; Abadi et al., 2017).
Some recent research has shown that ML-based methods can determine the extent of effective interactions and side-effects (changing unwanted sites) of each gRNA precisely (Abadi et al., 2017; Lin and Wong, 2018). Such a process can significantly accelerate the process of gRNA design for any part of human DNA, thus allowing us to edit anywhere in DNA (Jiang et al., 2016). However, existing models still have challenging issues, such as data imbalance, data heterogeneity, insufficient training data, generalizability, and cross-species inefficiency (Chuai et al., 2017).
We described the basic concepts of CRISPR systems and introduced activity and specificity as two main challenges in this area (Moreno-Mateos et al., 2015; Herai, 2019). In the rest of the paper, we provide an overview of computational approaches, especially machine and deep learning (MDL) algorithms, which we believe are the most effective and reliable methods for predicting gRNAs effects. The summary of our review is presented in Tables 1–Tables3, only for tools with active access link. Table 1 illustrates computational tools and software packages related to CRISPR systems; Table 2 summarizes tools and software packages related to finding off-target sites; Table 3 shows those related to gRNA design; and finally, Table 4 reports MDL-based tools and software packages related to CRISPR systems.
TABLE 1.
Tools and software packages related to CRISPR systems.
| Name | Main functionality | Input | Cell type | Interface | Year | Source |
|---|---|---|---|---|---|---|
| CRISPRidentify Mitrofanov et al. (2021)* | It detects All possible CRISPR arrays | Genome sequences | Bacteria and archaeal | Standalone application | 2021 | https://github.com/BackofenLab/CRISPRidentify |
| CRISPRloci Alkhnbashi et al. (2021)* | Definition of CRISPR leaders for each locus; Prediction of all CRISPR arrays in the correct orientation; annotation of Cas genes and associated information, include the Cas subtypes | Protein, genomic DNA, CRISPR repeats or viral sequences are accepted | Bacteria, archaeal and viral | Webserver and standalone versions (Python, Perl and Java) | 2021 | Webserver: https://rna.informatik.unifreiburg.deCRISPRloci |
| Standalone version: https://github.com/BackofenLab/CRISPRloci | ||||||
| ANNOgesic Yu et al. (2018) | It can detect several genomic features, including genes, CDSs, tRNAs, rRNAs, TSSs, PSs, transcripts, terminators, UTRs, sRNAs, sORFs, circular RNAs, CRISPR-related RNAs, riboswitches, and RNA-thermometers | RNA-seg | Bacterial and archaeal genome | Command-line (Python) | 2018 | The software: https://pypi.org/project/ANNOge/ https://hub.docker.com/r/silasysh/annogesic/Documentation: http://annogesic.readthed.ocs.io/ |
| CRISPR-DAV Wang et al. (2017) | A pipeline to analyze the CRISPR NGS data in a high-throughput manner. Output: read counts in various stages; read depths and indel frequencies in amplicon; counts and percentages of indel reads; frequencies of allele, SNP and HDR. | Files that describe software paths, parameters, mplicon, CRISPR sites, and FASTQ sources | Any selected genome | Command line Interface (Perl and R) | 2017 | https://github.com/pinetree1/crispr-dav.git and https://hub.docker.com/r/pinetree1/crispr-dav |
| Cas-analyzer Park et al. (2017)* | It is an NGS data analyzer. It categorizes and sorts the results. The position and size of insertions or deletions are depicted as interactive graphs | Deep sequencing data | Any selected genome | Web user interface (JavaScript) | 2017 | http://www.rgenome.net/cas-analyzer/ |
| CRISPRAnalyzeR Winter et al. (2017)* | An application to analyze, document, and explore pooled CRISR/Cas9 screens. Reagent phenotypes such as efficiency scores and predicted genomic binding sites are displayed | An sgRNA library or screening data | Any selected genome | Open-source web or standalone application | 2017 | http://www.crispranalyzer.org |
| source code at: http://www.github.com/boutroslab/CRISPRAnalyzeR | ||||||
| CRISPRcloud Jeong et al. (2017) | An application to extract, cluster, and analyze raw next-generation sequencing files derived from pooled screening experiments | sgRNA read counts data | Human and mouse | Cloud-based web application | 2017 | http://crispr.nrihub.org |
| CRISPRdigger Ge et al. (2016) | can Discover Direct Repeats (DRs) for CRISPRs and achieve a higher accuracy for a query genome | A genome sequence | Any selected genome | Command line application | 2016 | http://www.healthinformaticslab.org/supp/ |
| BATCH-GE Boel et al. (2016) | It detects and reports indel mutations and other precise genome editing events and calculates the corresponding mutagenesis efficiencies | NGS-derived sequencing data, DNA of interest | Any selected genome | Command line application | 2016 | https://github.com/WouterSteyaert/BATCH-GE.git |
| CRISPRleader O’Brien and BaileyGT-Scan. (2014) | It detects leader sequences and shows full annotation of the CRISPR array and its strand orientation as well as conserved core leader boundaries | Genome sequence | Archaea and bacteria | Command line application (HTML pages) | 2016 | http://www.bioinf.unifreiburg.de/Software/CRISPRleader/ |
| CRISPRDetect Biswas et al. (2016)* | It enables accurate identification of CRISPR arrays in genomes and their direction, repeat spacer boundaries, substitutions, insertions or deletions in repeats and spacers. It lists Cas genes that are annotated in the genome | Four inputs: genomic sequence, word size, min of word repeat, and max gap between repeats | Archaea and bacteria | Web application and command line (PERL) | 2016 | http://bioanalysis.otago.ac.nz/CRISPRDetect/ |
| CRISPR-GA Güell et al. (2014) | It estimates the HR, NHEJ, and a complete report of the location and characteristics of the indels | The genomic region | Any selected genome | Web user interface (implemented in R) | 2014 | http://crispr-ga.net. Documentation at: http://crispr-ga.net/documentation.html |
| Crass Skennerton et al. (2013) | It identifies and reconstructs CRISPR loci from raw metagenomic data without the need for assembly or prior knowledge of CRISPR in the data set. | Raw file in FASTA or FASTq format | All genome | Command line interface | 2013 | http://bioinformatics.ninja/crass |
TABLE 3.
Tools and software packages related to gRNA design.
| Name | Main functionality | Input | Cell type | Interface | Year | Source |
|---|---|---|---|---|---|---|
| SNP-CRISPR Chen et al. (2020) | It designs gRNAs for non-reference genomes to support allelic targeting. SNP-CRISPR calculates the gRNA efficiency score for the variant and the References sequences | Target genome, variant information including the genome coordinates and sequence changes | Human, Mouse, Zebrafish, Fly | Web application | 2020 | https://www.flyrnai.org/tools/snp_crispr/ |
| AlleleAnalyzer Keough et al. (2019) | It designs allele-specific dual gRNAs. It incorporates single-nucleotide variants and short insertions and deletions to design sgRNAs for precisely editing one or multiple haplotypes of a sequenced genome, currently supporting 11 Cas proteins | Target genome (with genetic variant information) | Human | Application | 2019 | https://github.com/keoughkath/AlleleAnalyzer |
| CRISPR-Local Sun et al. (2019) | It designs sgRNAs in plants and other organisms that factor in genetic variation and is optimized to generate genome-wide sgRNAs | whole-genome sequencing, mRNA sequencing or known variants for specific transgenic receptor lines | Plants | Application | 2018 | http://crispr.hzau.edu.cn/CRISPR-Local/ |
| CRISPR-P Liu et al. (2017)* | It helps to design of gRNA. It output: all targetable sites; the details and GC content of each gRNA; the restriction enzyme site in the targeting region; and synthetic DNA oligos; as well as the microhomology score and the secondary structure of sgRNA. | The gene locus tag, genomic position, or sequence | 49 plant genomes | Web user interface | 2017 | http://cbi.hzau.edu.cn/crispr2/ |
| CRISPR FOCUS Cao et al. (2017)* | It retrieves all possible gRNA and prioritize them. It also provides a rational and high-throughput approach for sgRNA library design | Gene symbols or RefSeq IDs | Human or mouse genome | Web application | 2017 | http://cistrome.org/crispr-focus/ |
| Guide Picker Hough et al. (2017)* | It provides rapid guide RNA generation and selection. It retrieves guide sequences with on and off-target sites | The genome and the gene name | Mouse or human gene | Web application (JavaScript) | 2017 | https://www.deskgen.com/guide-picker/ |
| SgTiler Ahmed and He. (2017)* | It generates graphical representation for distribution of sgRNA. It shows four outputs: i) all candidate sgRNAs; ii) list of filtered sgRNAs; iii) list of sgRNA details; and iv) a summary report with important statistics | Three input files: i) FASTA file; ii) A file with exon coordinates; and iii) a file of regulatory regions | Any selected genome | Command line application (Python) | 2017 | https://github.com/HansenHeLab/sgTiler |
| CRISPOR Concordet and Haeussler. (2018) | It finds guide RNAs in an input sequence and ranks them according to different scores. It evaluates potential off-targets in the genome of interest and predicts on-target activity | A sequence (typically an exon), a genome, and the type of CRISPR nuclease | More than 150 genomes | Web and standalone command line application | 2016 | http://crispor.org |
| CRISPR-DO Ma et al. (2016) | It retrieves information about target sequences, overlaps with exons, putative regulatory sequences and SNPs in the spCas9 CRISPR system | sgRNA | Human, mouse, zebrafish, fly and worm | Web application | 2016 | http://cistrome.org/crispr/ |
| Breaking-Cas Oliveros et al. (2016)* | It retrieves all sequences, coordinates, scores, and annotation details of every gRNA and off-targets | The name of the References organism, the characteristics of the Cas-like nuclease, and the sequence(s) of the intended target genomic | All eukaryotic genomes | Web application | 2016 | http://bioinfogp.cnb.csic.es/tools/breakingcas |
| CT-Finder Zhu et al. (2016) | It helps users to design gRNAs optimized for specificity and shows Graphic visualization of on and off-target sites in Cas9n and RFNs | DNA sequence, a References genome, the on and off-target PAM sequences, and length of gRNA and seed region | Human, mouse, Arabidops | Web application | 2016 | http://bioinfolab.miamioh.edu/ct-finder |
| CRISPETa Pulido-Quetglas et al. (2017) | It helps to design sgRNAs | One or more target regions | Human, mouse, zebrafish, Drosophila, melanogaster and Caenorhabditis elegans | Command-line and web application | 2016 | Server: http://crispeta.crg.eu/manual Source code: https://github.com/guigolab/CRISPETA |
| CLD Heigwer et al. (2016)* | It helps to design sgRNAs | Three files: i) the genome sequence, ii) a parameter (Hwang and Bae, 2021) file, and iii) a gene list | All organisms | Command line application | 2016 | htts://github.com/ |
| CRISPy-web Blin et al. (2016)* | It scans for gRNAs and potential off-targets | Target sequence or gene | Any microbial genome | Web application | 2016 | http://crispy.secondarymetabolites.org |
| EuPaGDT Peng and Tarleton. (2015) | It finds all gRNAs. It also scores, and ranks them. Additionally, it assists users in designing single-stranded oligonucleotides for homology-directed repair | Sequence or gene | Eukaryotic organisms | Web application | 2015 | http://grna.ctegd.uga.edu |
| Spacer Scoring for CRISPR(SSC) Xu et al. (2015)* | It predicts SgRNA efficiency | DNA sequence | Any selected genome | Web application | 2015 | http://crispr.dfci.harvard.edu/SSC/ |
| Cas-Designer Park et al. (2015)* | It aids researchers in choosing appropriate target sites in a gene of interest. It outputs a list of all possible gRNAs and their potential off-target sites, including bulge-type sites, and also an out-of-frame score for each | DNA sequence | Most of genomes (Wang et al. (2019a) | Command line interface | 2015 | http://rgenome.net/cas-designer/ |
| CRISPR multitargeter Prykhozhij et al. (2015) | It searches input sequences for single-sgRNA and two-sgRNA/Cas9 nickase targeting | sgRNA, GC% | 12 genomes like zebrafish | Web application | 2015 | http://www.multicrispr.net/ |
| CRISPR-ERA Liu et al. (2015)* | It designs gRNA. It outputs sgRNAs, on and off target location, and details of them with their E- and S-scores etc. | Target gene or genomic site | 9 common prokaryotic and eukaryotic organisms | Web application | 2015 | http://crisprera.stanford.edu/InitAction.action |
| CCTop Stemmer et al. (2015)* | It identifies and ranks all candidate sgRNA target sites according to their off-target quality and displays full documentation | Target genome site | 15 common prokaryotic and eukaryotic organisms | application (python) | 2015 | http://crispr.cos.uniheidelberg.de/ |
| CRISPRseek Zhu et al. (2014)* | It identifies gRNAs and also scores and ranks them to minimize off-target cleavage | Any sequence | Any selected genome | Command line application ® | 2014 | http://www.bioconductor.org |
*Means the tools are free of charge to access.
TABLE 2.
Tools and software packages related to finding off-target sites.
| Name | Main functionality | Input | Cell type | Interface | Year | Source |
|---|---|---|---|---|---|---|
| CALITAS Fennell et al. (2021) | CALITAS is a CRISPR-Cas-aware aligner and integrated off-target search algorithm. It supports an unlimited number of mismatches and gaps and allows PAM mismatches or PAM-less searches | gRNA, one or more local regions of a target sequence | Human | Standalone application | 2021 | https://github.com/editasmedicine/calitas |
| CRISPR-SE Li et al. (2021) | It is an accurate and fast search engine using a brute force approach to find all off-target sites | gRNA | Human and mouse genomes | Web user interface | 2021 | The webserver: http://renlab.sdsc.edu/CRISPRSE/ |
| The source code: https://github.com/bil022/CRISPR-SE | ||||||
| CRISPRitz Cancellieri et al. (2020)* | It enumerates and annotates putative off-target sequences and assesses their potential impact on the functional genome. It has three outputs: i) all off-target sites; ii) an overall mismatch and bulge profile for each guide; iii) motif matrices | PAM sequence, a list of guides, References genome (required) and genomic annotations and number of mismatches (optional) | All genome | Standalone application | 2019 | https://github.com/pinellolab/CRISPRitz |
| https://github.com/InfOmics/CRISPRitz | ||||||
| CHOPCHOP v3.0 Labun et al. (2019) | It Identifies sgRNA targets. Five outputs: i) the number of off-targets, ii) whether the off-targets contain mismatches or are perfect hits, and iii) where the target site lies within the gene iv) the results are ranked by GC-content | Four inputs: i) the target; ii) species; iii) CRISPR effector and iv) the purpose of the experiment | 200 genomes | Command-line program and web user interface | 2019 | Server: https://chopchop.cbu.uib.no |
| The local installation: https://bitbucket.org/valenlab/chopchop | ||||||
| CRSeek Dampier et al. (2018) | It finds all on-target and off-target sites | Interested sequence | All genome | Command line interface (Python) | 2018 | https://github.com/DamLabResources/crseek) |
| CRISPR-RT Zhu et al. (2018)* | It retrieves all the protentional targets and relevant information for gRNAs in CRISPR-C2c2 system | An RNA/DNA sequence | 10 genomes include human | Web application | 2017 | http://bioinfolab.miamioh.edu/CRISPR-RT |
| PhytoCRISPEX Rastogi et al. (2016) | It finds potential targets and shows the gene name with start, stop, and sequence of the sgRNA targets. It also shows the results of checks at level one and two | DNA sequences | 13 algae (diatoms, haptophytes, etc.), or any user defined genome | Web interface and UNIX-based standalone application | 2016 | http://www.phytocrispex.biologie.ens.fr/CRISPEx/crispexdownloads/ |
| CRISPResso Pinello et al. (2015)* | It finds potential on and off-targets | Two files for paired-end reads or a single file for single-end reads, and the References amplicon sequence | Any selected genome | Command line interface or web server | 2015 | http://github.com/lucapinello/CRISPResso. Web application www.crispresso.rocks |
| Cas-OFFinder Bae et al. (2014)* | It searches for potential off-target sites and shows their locations, position, direction, and number of mismatches | Genome sequence | Any selected genome | Command line program (written in OpenCL) and website | 2014 | http://www.rgenome.net/cas-offinder |
| CasOT Xiao et al. (2014) | It finds potential off-target sites in any given genome with user-specified types of PAMs, and number of mismatches | target sites or genome and a genome annotation file (optional) | Any selected genome | Command-line program (a Perl script) | 2014 | http://eendb.zfgenetics.org/casot/ |
| COSMID Cradick et al. (2014)* | It retrieves all off-target sites matching the user-supplied criteria in comparison to the guide strand with chromosomal location | The guide sequence, type of PAM, allowed number of mismatches, insertions and deletions, genome of interest, and primer design parameters | 7 genomes including human and mouse | web user interface | 2014 | http://crispr.bme.gatech.edu |
| CRISPRdirect Naito et al. (2015)* | It outputs a list of on and off-target sites with details (target position, target sequence, the number of target sites in the genome, GC content, and calculated melting temperature) | Two inputs: i) an accession number, and ii) a genome coordinate or an arbitrary nucleotide sequence up to 10 kbp | 9 genomes including human and mouse, rat etc. | Web user interface | 2014 | http://crispr.dbcls.jp |
| E-CRISP Heigwer et al. (2014) | It retrieves positions of CRISPR targets | Gene Id or gene sequence | More than 40 genomes | Web user interface | 2014 | http://www.e-crisp.org/E-CRISP |
| GT-Scan O’Brien and BaileyGT-Scan. (2014) | It ranks all potential on and off-targets | Genomic region and target rule (target length, constrained positions and positions with high-, low- or no-target and off-target specificity) | More than 25 genomes | Web user interface | 2014 | http://gt-scan.braembl.org.au |
| sgRNAcas9 Xie et al. (2014)* | It predicts all single or paired CRISPR target sequences and the corresponding information for each target site (such as start and end values, sequence pattern, GC content, sgRNA offset, etc.) | Sequences of target position | All genome | Command line interface (Perl script) | 2014 | www.biootools.com |
| SSFinder Upadhyay and Sharma. (2014)* | It identifies potential off-target sites and classifies them | File name and directory of input sequences | All genome | Command line interface (python) | 2014 | https://code.google.com/p/ssfinder/ |
| CRISPRTarget Biswas et al. (2013) | It predicts the most likely targets of gRNAs. Targets can be displayed and scored for flanking sequences and PAMs | Spacers | Any selected genome | Web application | 2013 | http://bioanalysis.otago.ac.nz/CRISPRTarget |
*Means the tools are free of charge to access.
TABLE 4.
MDL-based tools and software packages related to CRISPR systems.
| Name | Main functionality | Input | Cell type | Interface | Model | Year | Source |
|---|---|---|---|---|---|---|---|
| C-RNNCrispr Zhang et al. (2020) | It predicts sgRNA on-target activity. It is a transfer learning approach by using small-sized datasets to fine-tune | Datasets to fine-tune | 4 cell line | Standalone software | CNN and BGRU | 2020 | https://github.com/Peppags/C_RNNCrispr |
| CRISPRpred Muhammad Rafid et al. (2020) | It predicts sgRNA on-target activity | Position independent and position specific features | Human | Standalone software | SVM and random forest | 2020 | https://github.com/Rafid013/CRISPRpredSEQ |
| DeepCpf1 Kwon et al. (2019) | It predicts the activity of AsCpf1 (location of all targetable sequences and efficiency of each; information on GC contents, positions, strands, and DeepCpf1 scores.) | Cell line types, information on the sequences of a target and its surroundings, and References sequences | All genome | Web tool | CNN | 2019 | http://deepcrispr.info/ |
| DeepHF Wang et al. (2019a) | It predicts SpCas9 activity for each gRNA (all targetable sequences, restriction sites, strands, and predicted efficiency) | Various types of SpCas9 nucleases, DNA sequences | All genomes | Web tool | CNN | 2019 | http://www.DeepHF.com/ |
| CINDEL Iyombe. (2019) | It predicts the indel frequencies of CRISPR/Cas12 with TTTV PAM sequence (targetable sequences, positions, strands, GC contents, and INDEL scores) | References sequences | Web tool | - | 2019 | http://big.hanyang.ac.kr/cindel | |
| DeepSpCas9 Kim et al. (2019) | It predicts SpCas9 activity for each gRNA (positions, GC content, and DeepSpCas9 scores) | Target sequence information with its surroundings, and gene symbols | Human | Web tool | CNN | 2019 | http://deepcrispr.info/DeepSpCas9 |
| Microhomology-Predictor Hwang et al. (2021) | It predicts the deletion patterns by calculating the scores of possible deletion patterns produced by a MMEJ pathway following DNA cleavage by ZFNs, TALENs, or Cas9. All possible deletion patterns and the pattern scores can be checked | Target sites with high out-of-frame scores | All genome | Web tool | - | 2019 | http://www.rgenome.net/mich-calculator |
| inDelphi Cloney. (2019) | It predicts the spectrum of cut-site, possible sgRNA sequences, predicted mutation patterns, possible frameshift codons, and their frequencies | Sequences of both sides of cleavage in various cell types | Human and mouse | Standalone software | - | 2019 | https://indelphi.giffordlab.mit.edu |
| FORECasT Allen et al. (2019) | It predicts editing outcomes (possible mutation patterns and predicted frequencies of the mutation patterns and frame shifts) of the CRISPR/Cas9 system with NGG PAM. | Target DNA sequences and the cleavage sites | Most of genomes | Web tool | - | 2018 | https://partslab.sanger.ac.uk/FORECasT |
| CRISPR-GNL Wang et al. (2019b) | It is an algorithm for CRISPR on-target activity prediction | Normalized gene editing activity from 8,101 gRNAs and 2,488 features | human, mouse, zebrafishDrosophila, Cioa intestinalis | Stand alone application | regression models | 2019 | https://github.com/TerminatorJ/GNL_Scorer |
| DeepCRISPR Chuai et al. (2018) | It predicts whole genome on and off-target profiles | sgRNA sequences with an NGG PAM | Human | Web tool | CNN | 2018 | http://www.deepcrispr.net/ |
| TUSCAN Wilson et al. (2018) | It predicts the degree of CRISPR/Cas9 activity and classifies them into active and inactive categories | All genome | Software | Random forest | 2018 | https://github.com/BauerLab/TUSCAN | |
| SgRNAScorer Chari et al. (2017) | It identifies sgRNA sites and their activities for any PAM sequence of interest | Sequence with a defined spacer length and PAM sequence | Human and mouse | Web tool | SVM | 2017 | https://sgrnascorer.cancer.gov/ |
| CRF Wang and Liang. (2017)* | CRF uses a classifier to filter out invalid CRISPR arrays from all putative candidates | DNA/RNA sequence in FASTA format | Bacteria and archaea | Web tool | Random forest | 2017 | http://bioinfolab.miamioh.edu/crf/home.php |
| GE-CRISPR Kaur et al. (2016) | It predicts and analyses sgRNAs efficiency and gives information like secondary structure of sgRNA, PAM, start and end of coordinates, and GC% | Desired gene or genome sequence in FASTA format | In any trained model | SVM | 2016 | http://bioinfo.imtech.res.in/manojk/gecrispr/ | |
| CRISPRscan Moreno-Mateos et al. (2015) | It’s a predictive sgRNA-scoring algorithm that captures the sequence features affecting the activity of CRISPR/Cas9 in vivo | DNA sequence | Fish | Web tool | Linier regression | 2015 | http://www.crisprscan.org/ |
| WU-CRISPR Wong et al. (2015) | It predicts potential sgRNAs and scores of them | Gene IDs | Human and mouse | Web tool and stand-alone software | SVM | 2015 | http://crispr.wustl.edu |
| SSC Xu et al. (2015) | It’s a program for predicting editing activity of SpCas9 and giving all possible targets with the efficiency scores of various editing modes such as knockout, CRISPRi, or CRISPRa | Target sequences with the length of spacers (19 nt or 20 nt) as | Web tool | Elastic Net | 2015 | http://cistrome.org/SSC/ | |
| CRISPRstrand Alkhnbashi et al. (2014) | It determines the crRNA-encoding strand at CRISPR loci by predicting the correct orientation of repeats. It also determines whether repeats lie on the forward or reverse strand | Attribute type, attribute order, size of the terminal regions, number of blocks within the terminal regions | Bacteria and archaea | Integrated in CRISPRmap web server | graph kernels | 2014 | http://rna.informatik.uni-freiburg.de/CRISPRmap |
*Means the tools are free of charge to access.
2 Computational approaches in CRISPR
Computational approaches are an essential part of CRISPR research. The bioinformatics studies have made significant contributions to the initial discovery of CRISPR (Alkhnbashi et al., 2014; Makarova et al., 2015). We summarize some of them in Table 1. Bioinformatics tools play a significant role in these fields: 1) determination of the specific differences between the CRISPR/Cas systems from archaeal and bacterial sources; 2) determination of required repeat spacer sequences for processing the mature CRISPR RNA (crRNA); 3) prediction of the transcribed strand of CRISPR arrays; 4) determination of CRISPR leader sequences; 5) classification of Cas proteins; 6) prediction of proper gRNA; 7) prediction of on-target and off-target effects; and so on (Listgarten et al., 2016; Lin and Wong, 2018; Listgarten et al., 2018; Herai, 2019; Alkhnbashi et al., 2020; Smith et al., 2020).
According to our review, low cleavage efficiency and off-target effects hamper CRISPR development and application. So, prediction of proper gRNA and prediction of on-target and off-target effects is so critical. In the rest of the paper, we will focus on the tools that have been developed for designing optimal gRNA with low off-target effects.
2.1 gRNA design
There are two fundamental questions in CRISPR researches. The first question is: what are the targets of the given gRNA? Some methods, such as CRISPResso (Pinello et al., 2016) and CRISPRTarget (Biswas et al., 2013), try to calculate potential targets by taking a gRNA as input and using computational algorithms (more details are described in Table 3). Tools like CRISPRTarget (Biswas et al., 2013) offer a way to answer this question using a ML-based approach (Table 4 shows more details). The second important question is how to be confident about the accuracy of CRISPR edits. Most of the tools or methods in CRISPR’s field have been developed to answer these two questions. In Tables 2, 3, we tried to collect all of them and describe their details.
Also, we realized that most of researches in CRISPR area mainly focus on increasing cleavage activity (more on-targets) and cleavage efficiency (low off-target sites). As known, low efficiency makes CRISPR editing unreliable and also hampers CRISPR development and application (Wang et al., 2019a). Unfortunately, the high focus on more activity induces more off-target cleavage, which can be toxic. Therefore, we must maintain a balance between these two criteria. These issues can be resolved by designing successful CRISPR gRNA and choosing an appropriate Cas protein (Kuscu et al., 2014; Shen et al., 2018).
As mentioned earlier, cleavage efficiency varies significantly among different target sites and cell lines (Yan et al., 2018). Several features can influence the gRNA binding ability and the Cas enzyme cutting efficacy. Sequence composite features (nucleotide position, GC content), genetic and epigenetic features (chromatin accessibility, gene expression), and energetic properties (RNA secondary structure, melting temperature, free energy) are the most important influential features on cleavage efficiency (Pallarès Masmitjà et al., 2019; Wang et al., 2020). Based on these features, many computational tools have been developed for designing highly efficient gRNAs. In the rest of this section, we will discuss the most popular ones.
Rule set 1 (Liu et al., 2020) is a ML-based model that uses a support vector machine (SVM), a supervised ML method, and contains a linear regression method for classifying gRNAs. Rule set 1 uses sequence-based features, and its predictive data is highly correlated with experimental results (Xu et al., 2015). Rule set 2 (Liu et al., 2020) is an improved version of Rule set 1 and counts the nucleotides independent location of the gRNA target site within the gene to improve results (Doench et al., 2016). It is a powerful model, used for both CRISPR Knock Out (CRISPR KO) and CRISPR activation/interference (CRISPRa/i) experiments. Another powerful model-based package has been developed and implemented at the Broad Institute to predict gRNA efficiency, named sgRNA Designer (Pallarès Masmitjà et al., 2019).
Elastic Net is another ML-based and regularized regression-based method (Li and Lin, 2010). Although there are significant differences in nucleotide preference between CRISPR KO and CRISPRa/I, the Elastic Net algorithm is used to construct models for both CRISPR KO and CRISPRa/i. Also, this practical algorithm has been applied in Spacer Scoring for CRISPR (SSC) software to predict the gRNA efficiency (Qin et al., 2019). Additionally, well-known platforms such as E-CRISP (Heigwer et al., 2014), CHOPCHOP (Labun et al., 2019), and CRISPRFOCUS (Cao et al., 2017) have applied this method.
Moreno and his colleagues designed another logistic regression-based method and integrated it into CRISPRscan to predict the gRNA precision (Moreno-Mateos et al., 2015). Additionally, they have applied extra features such as guanine enrichment and adenine depletion, which increase the gRNA activity (Cui et al., 2018).
Another ML-based method is WU-CRISPR (Wong et al., 2015) which uses sequence composite features like guanine enrichment and adenine depletion, and some other novel features to build a higher precision model. The CRISPR/Cas9 target online predictor (CCTop) (Stemmer et al., 2015), a platform for CRISPR target prediction, takes advantage of this model. The SgRNAScorer is another software that uses SVM to calculate gRNA on-target scores. The new version of this software can predict other Cas systems such as SaCas9 (Qin et al., 2019) and AsCpf1 [94].
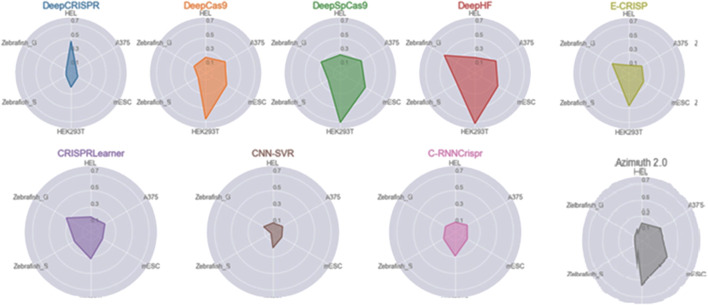
To avoid unwanted effects in other sites except for desired target sites (off-target), researchers try to modify a spacer sequence that does not adopt other sites in the genome. Tools such as CRISPRpred (Hwang and Bae, 2021), DeepSpCas9, and SgRNAScorer are usually limited to the set of preprocessed genomes used when training ML models. To build good gRNAs in genomes other than those used in the training process, researchers can use web-based tools such as CRISPy (Blin et al., 2016). Looking at Tables 1–Tables 4, we have listed the genome in which the editing takes place (named target genome) as a significant feature for all tools. The existence of target genome is even more critical for deep learning-based (DL) methods, because they are usually unpractical in genomes other than the ones from which training data was extracted. Basically, being used in all genomes is a significant strength for ML-based tools. But one tool may not have the same accuracy over all genomes or even all regions of a genome (see Figure 7) (Kim et al., 2021). Furthermore, structural correctness and base-level accuracy of the target genome are important. The accuracy of a genome differs not only between genome sequencing technologies but also across genomic regions, as some stretches of the genome are inherently more difficult to read (Kim et al., 2021). It is commonly known that certain genomic regions are more difficult for sequencing and extracting features. AT-rich or GC-rich regions, which are important for detecting off-target sites, are tough because they respond poorly to the amplification protocols required by some platforms. Palindromic sequences or hairpin structures similar to gRNA structures are difficult to denature, making such regions challenging for sequencing tools (Selvakumar et al., 2022).
FIGURE 7.
Spearman correlation for ML-based tools over the different datasets. Each polygon represents a tool, and the edges illustrate the obtained correlation over the respective dataset.
2.1.1 Selecting the best gRNA
There may be several gRNAs for an experiment, in which case we have to pick the best one. Many computational approaches have been developed for scoring and selecting the best gRNAs. Some of them use experimental data to score a gRNA. According to the different criteria, these methods consider a specific score for each gRNA. The criteria and final score calculation are different in each algorithm. CHOPCHOP (Labun et al., 2019) provides multiple scores for users, such as Rule Set 1 and Rule set 2, SSC (Xu et al., 2015), CRISPRscan [13], and deepCpf1 (Kim et al., 2018). E-CRISP (Heigwer et al., 2014) uses a particular score to determine the quality of each gRNA, named SAE, which combines three scores: specificity, annotation and efficacy. E-CRISP uses Rule Set 1 and SSC too. CCTop (Stemmer et al., 2015) calculates the CRISPRater score to predict the efficiency of gRNAs. CCTop also calculates off-target scores for each sequence. The CRISPOR (Concordet and Haeussler, 2018) ranks gRNAs according to different scores, such as on-target activity and protentional off-targets scores.
To score a gRNA or determine whether it is suitable for the desired genome editing or not, we need to determine potential targets of a gRNA in the selected genome and determine which of these potential targets are desirable. Hence, the number of on-target and off-target sites is critical in gRNA evaluation. In other words, since genomic edits are permanent and very sensitive, it is crucial to determine potential targets before the main editing occurs and then remove or reduce them (Yan et al., 2018). Therefore, many researchers have focused on this issue. Furthermore, many developers have attempted to develop practical tools for this purpose. We will discuss these tools in the next section.
2.2 Prediction of CRISPR specificity (off-target sites)
The prediction of off-target mutations in CRISPR/Cas9 is a hot topic owing to its relevance to gene-editing research. Cas nucleases may cleave unintended genomic sites and cause unexpected mutations called off-target cleavage (Listgarten et al., 2018). Even though the CRISPR/Cas9 system is routinely used in a large variety of tasks, there is also a significant concern that off-target effects may reduce its effectiveness of CRISPR. In response to this concern, researchers have concluded that the best way to mitigate off-target effects is to know when and where they occur and then design guides to avoid them while balancing for on-target efficiency. By predicting CRISPR cutting specificity and designing optimal gRNAs, off-target effects can be effectively relieved. As noted earlier, careful CRISPR target selection and low concentrations of CRISPR components can reduce off-target cleavage (Zetsche et al., 2020).
The off-target predictive modelling problem can be broken down into three main tasks. Given a gRNA to evaluate off-target activity, one needs to (Afzal et al., 2020) search the whole genome for potential targets; in other words, search those regions of the genome matching the guide sequence with up to X number of mismatches (Gaj et al., 2013); score each potential target found in step 1 according to its activity (Jacquin et al., 2019); collect the second stage scores and evaluate the final score of a gRNA. Several solutions have been presented for these tasks, including Cas-OFFinder (Bae et al., 2014), CRISPOR (Concordet and Haeussler, 2018), CHOPCHOP (Labun et al., 2019), and e-CRISPR (Tarasava et al., 2018). These models differ in their search algorithms and the completeness of the search process. Completeness is dictated by options such as the maximum number of mismatches, allowed PAMs, and the search algorithm used.
There are two basic methods to predict the specificity of CRISPR gRNAs: the alignment-based and the scoring-based methods. In the following, we will explain these approaches and give successful examples of each one. Also, the overview of these approaches is depicted in Figure 4.
FIGURE 4.
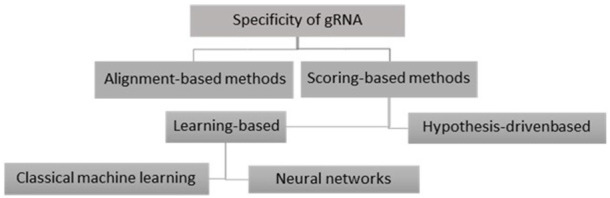
Basic methods to predict the specificity of CRISPR gRNAs.
2.2.1 Alignment-based methods
In the alignment-based method, gRNAs are aligned to a given genome, and off-target sequences and sites are returned. These methods are mainly used to find out all potential off-target sites in silico. Choosing a search engine and setting search parameters plays an important role in evaluating these tools (Liu et al., 2020). For example, if we set the maximum number of mismatches to a large number, like four or more, we will probably find all possible off-targets. The observed rate of off-target activity is about 59% when there is one mismatch between the target DNA and gRNA sequences and decreases toward 0% when four or more mismatches exist (Kim et al., 2021). So, it can be concluded that an increased number of mismatches decreases the likelihood of off-target activity.
Common sequence alignment tools use BLAST, BLAT, Bowtie, Bowtie2, BWA or customized search engines. Table 5 summarizes the search engine of famous alignment-based tools in CRISPR.
TABLE 5.
The most popular alignment-based methods and related search engines.
| Search engine | Methods |
|---|---|
| BLAST | CRISPRTarget (Biswas et al., 2013), CRISPR-P (Liu et al., 2017), and CRISPR-GA (Luyten et al., 2004) |
| BOWTIE | CRISPR-ERA (Liu et al., 2015), CHOPCHOP (Labun et al., 2019), CasFinder (Upadhyay and Sharma, 2014), CCTop (Stemmer et al., 2015), E-CRISP (Heigwer et al., 2014), and CLD (Heigwer et al., 2016) |
| BWA | CRISPR-DO (Ma et al., 2016), CRISPOR (Concordet and Haeussler, 2018), and CRISPETa (Pulido-Quetglas et al., 2017) |
| BRUTE FORCE | GuideScan (Perez et al., 2017), Cas-OFFinder (Bae et al., 2014), FlashFry (McKenna and Shendure, 2018), Crisflash (Jacquin et al., 2019), CRISPRitz (Cancellieri et al., 2020), and CRISPR-SE (Li et al., 2021) |
Compared to methods which use BLAST, Bowtie and BWA as search engine, methods like GuideScan (Perez et al., 2017), Cas-OFFinder (Bae et al., 2014), FlashFry (McKenna and Shendure, 2018), Crisflash (Jacquin et al., 2019), CRISPRitz (Cancellieri et al., 2020), and finally, CRISPR-SE (Li et al., 2021)are faster due to the use of Brute force search engine. In addition, unlike most methods that support only a limited number of mismatches (mostly 3 or 4), Cas-OFFinder, CRISPRitz and CRISPR-SE have more preference due to their support of any number of mismatches.
The Bowtie and BWA are traditional tools for short sequence alignment that can be used for off-target sites detection (de Ruijter and Guldenmund, 2016). However, they cannot identify small PAMs since they were developed for NGS read alignment. Moreover, these tools allow very limited mismatches with default parameters, so they cannot identify all potential off-target sites.
Most tools, like CCTop (Stemmer et al., 2015), modify default algorithms and parameters and utilize Bowtie (de Ruijter and Guldenmund, 2016) to find off-target sites. CCTop follows three main steps. In the first step, CCTop identifies PAM sites; In the second step, it modifies default parameters (up to five mismatches against one in default) of Bowtie, and uses them to search for matches and mismatches in protospacer sequences. In the third step, it evaluates the off-target score for each candidate gRNA.
SeqMAp (Jiang and Wong, 2008) is an ultrafast short sequence mapping tool used in sgRNAcas9 (Xie et al., 2014) to find off-target sites. The sgRNAcas9 classifies all off-target sites into three categories and scores them to choose the best gRNA.
CasOT (Xiao et al., 2014) is another tool that can find Cas9 on-target and off-target sites with up to six mismatches in the seed region (12 nucleotides adjacent to the PAM). This tool can also determine whether off-targets are within a coding exon (Listgarten et al., 2016) or not. FlashFry (McKenna and Shendure, 2018) is another alignment-based method that defines off-targets with high speed. Additionally, it chooses the best gRNA and provides useful information such as annotating off-target sites, on and off-target scores, GC content, etc. FlashFry is a good choice for many applications because of its high speed and comprehensive output. Crisflash (Jacquin et al., 2019) is another one that belongs to the alignment-based approaches group. Crisflash designs gRNAs with a tree-based algorithm and uses user-supplied variant data to optimizes gRNA accuracy. It uses an N-ary tree structure, which searches up to four mismatches. CRISPRitz (Cancellieri et al., 2020) used a four-bit-based encoding to represent each nucleotide to allow for efficient bitwise operations. CRISPRitz supports off-targets with both mismatches and indels.
CALITAS (Fennell et al., 2021) is a new CRISPR-Cas-aware aligner tool which uses a modified and CRISPR-tuned version of the Needleman–Wunsch algorithm, supports an unlimited number of mismatches and gaps, and allows PAM mismatches or PAM-less searches. CALITAS returns a single best alignment for a given off-target site and it enables off-targets to be referenced directly using alignment coordinate.
CHOPCHOP v3.0 (Labun et al., 2019), a well-known model, is another tool that uses Bowtie with parameters–V and–L to detect off-target sites [90]. But, CRISPOR uses BWA to find all potential off-target sites iteratively and can find all validated off-targets as well as Cas-OFFinder (Bae et al., 2014).
Sequence alignment tools like CRISPy (Qin et al., 2019) and CRISPRdirect (Heigwer et al., 2016) rely on a minimum of one K-mer exact match. They are likely to miss some off-targets, spatially with a high number of mismatches and ultra-short gRNAs (20-mer). So, the accuracy of these methods cannot be very high.
In recent years, some tools like GuideScan (Perez et al., 2017), Cas-OfFinder (Bae et al., 2014), and CRISPR-SE (Li et al., 2021) have been developed with Brute force algorithm as their search engine. GuideScan uses a “tree” data structure with a brute-force algorithm that guarantees the search accuracy. Another tool in this category is Cas-OFFinder. Cas-OFFinder is one of the most popular tools for detecting potential off-target sites, with no limit to the number of mismatches, PAM types, or gRNA length. In our opinion, the most significant advantage of Cas-OFFinder is its high running speed due to using GPUs. It can also predict off-target sites with one-bp deletions or insertions.
OffScan (Cui et al., 2020) is the last one we considered in this study that is, belongs to the alignment-based approaches group. OffScan is not limited by the number of mismatches and allows custom PAM. Besides, OffScan adopts the FM-index, which efficiently improves query speed and reduce memory consumption.
Here, we discussed several alignment-based methods for the prediction of the gRNA output and realized that Cas-OFFinder may be the best option for identifying all potential off-targets with any Cas nucleases among these tools. Although users can reduce the number of outputs by restricting the maximum mismatches while exploring off-target cleavage, there are always redundant outputs; many are false positives.
On the whole, all nucleotide positions containing mismatches do not have the same decisive effect on off-target cleavage, but this issue is not considered in alignment-based methods. Because of this problem, and in order to increase the accuracy of the off-target detection methods, adding the features that influence the non-specific binding of CRISPR gRNAs to the methods is essential. As a result, another group of approaches emerged called scoring-based methods, which are discussed in the following sub-section.
2.2.2 Scoring-based methods
In the scoring-based method, the gRNAs identified in the alignment process are scored and ranked, and the sgRNA with the highest score is selected. There are two groups of scoring-based approaches: 1) hypothesis-driven-based approaches, where off-targets are scored based on the contribution of specific genome context factors to gRNA specificity; 2) learning-based approaches, where gRNAs are scored and predicted from a training model that considers the different features affecting specificity.
MIT (Hsu et al., 2013) is the first popular score-based tool for CRISPR off-target evaluation. To score the off-target efficiency of each gRNA, it counts and evaluates the contributions made by different mismatch positions. It also calculates a weight matrix to determine off-target efficiency for each gRNA (Chuai et al., 2017). The MIT score has been integrated into many CRISPR gRNA design tools, such as CHOPCHOP v3.0 CHOP (Labun et al., 2019) and CRISPOR (Concordet and Haeussler, 2018).
Another popular score-based tool for off-target evaluation is CFD (Cutting Frequency Determination). It is noticeable that gRNA can bind genome loci with non-canonical PAMs such as NAG, NCG, and NGA. So, CFD has added PAM features to their scoring metrics (Abadi et al., 2017). Also, for examining correlations between RNAs and off-targets, gRNAs with mismatches and indels in target sequences are added. GUIDE-seq (Tsai et al., 2015) validated the CFD score and proved that it performs better than the MIT score. The CFD score has been integrated into CRISPRscan (Moreno-Mateos et al., 2015), GuideScan (Perez et al., 2017), CRISPOR (Concordet and Haeussler, 2018), and others. CRISPRoff (Carlson-Stevermer et al., 2020) and uCRISPR (Carlson-Stevermer et al., 2020) integrated energetic properties into their scoring metrics. They both yielded better accuracy than MIT and CFD in off-target prediction.
Scoring-based methods consider only a few features, and unfortunately, all practical features cannot be considered. Also, most features are not understood yet, while learning-based methods use combinations of multiple features to build complex models for better prediction of off-target sites. These models are based on ML and, more recently, DL methods.
DL-based methods are attractive for CRISPR gRNA target efficacy prediction. They are mainly based on CNNs. Table 4 introduces some famous models that use MDL models for gRNA on-target prediction. These models used neural networks to extract features from the input genomic sequence. Generally, they are superior to models that use classical ML tools in prediction accuracy.
DeepCRISPR (Chuai et al., 2018) is a DL-based platform that combines gRNA on-target and off-target site predictions. As mentioned, in DL-based models, we do not need to identify all effective features, as they are detected automatically using the deep neural network. DeepCRISPR learns all possible sequence and epigenetic features that may affect gRNA Knock Out (KO) efficacy (Hana et al., 2021) in its learning process with a large dataset that is, gathered for it.
CRISPR-Cpf1 (Kim et al., 2017) is a ML-based model that achieved high efficiency, although it suffers minor off-target effects. DeepCpf1 (Kwon et al., 2019) is another highly used DL-based algorithm, mainly used in predicting Cpf1 activity. It uses chromatin accessibility data. It showed a significant improvement in the accuracy of Cpf1 activity prediction. CRISPR-DT (Zhu and LiangCRISPR-, 2019) is a recently developed platform for predicting the Cpf1 target efficiency. This model has been implemented with the SVM algorithm and displays better performance than the DL-based models such as DeepCpf1.
CRISPOR (Concordet and Haeussler, 2018) may be the best tool for designing gRNAs. CRISPOR combines multiple tools and gathers a large dataset to develop a highly efficient CRISPR gRNA design. CRISPOR contains 417 genomes and 19 PAM types, making it useful in almost all genomes. CRISPOR calculates two specificity scores: MIT and CFD. Additionally, it calculates ten efficiency scores, including Rule Set 2, CRISPRscan, microhomology, Lindel scores (Chen et al., 2019) and others for outcome prediction. CRISPOR designs primers for each gRNA as well as off-target sites. These primers are helpful when conducting on and off-target validation. CRISPOR enables the filtering of gRNAs with genomic variants based on well-known variant databases.
Some computational tools use CNNs for feature extraction or classification of CRISPR Cas. For instance, Seq-deepCpf1 (Kim et al., 2018; Kwon et al., 2019) has used CNN to extract features from the input gRNA sequence. And DeepCRISPR incorporates a CNN for predicting CRISPR/Cas9 gRNA on-target knockout efficiency and whole-genome off-target profiles. Also, DeepCas9 uses CNN to automatically learn the sequence determinants and predict the activities of gRNAs across multiple species genomes (Bhagwat and Khuri, 2021). Deeper-Bind (Hassanzadeh and Wang, 2016) used a LSTM layer to learn the dependencies between sequence features; this helps improve the prediction of protein binding specificity (Zhang et al., 2020). C-RNNCrispr (Zhang et al., 2020) has used a hybrid architecture combining CNN with bidirectional GRU (BGRU) to predict sgRNA cleavage efficacy (Sledzinski et al., 2020).
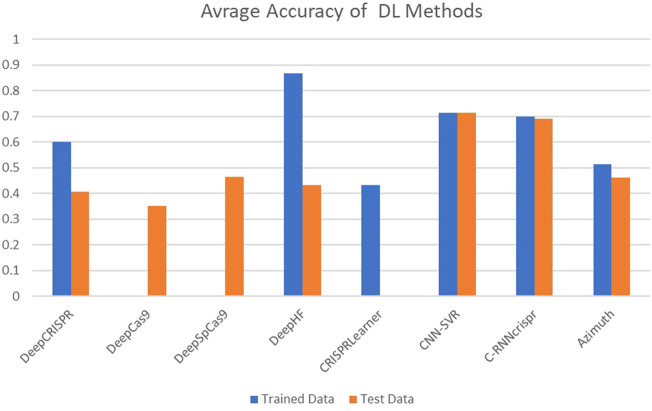
The performance of these tools is quantitatively assessed with two commonly used evaluation metrics, including accuracy and Spearman Correlation Coefficient (SCC) between predicted and real detected off-target activity. However, other evaluation metrics like Precision and Sensitivity (Eqs 2, 3) are used in some research as well. Spearman correlation seems to be a more reliable criterion. Most of these tools achieve promising accuracy in off-target prediction. Figures 5, 6 compare the off-target prediction efficacy of some popular tools. Due to their importance, we compare the accuracy of DL-based tools in separate diagram. The average accuracy of these tools is illustrated in the figures, as their accuracy differs among different genomes. For example, DeepCRISPR was the most accurate tool in the HEL cell line but performed poorly in the others. More details can be found in (Wang et al., 2019a; Zhu and LiangCRISPR-, 2019). Also, as a ML method, the accuracy differs between the train and test datasets. Unfortunately, for DeepCas9 and DeepSpCas9 (Chen et al., 2019), there is no report in their primary reference for the training dataset and the test dataset in CRISPRLearner (Bhagwat and Khuri, 2021). Accuracy, Precision, and Sensitivity are defined as follows, where TP, FP, TN, and FN represent true positive, false positive, true negative, and false negative, respectively.
| (1) |
| (2) |
| (3) |
FIGURE 5.
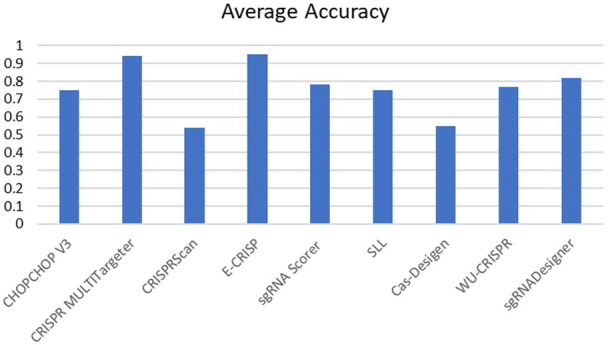
Average accuracy of off-target prediction.
FIGURE 6.
Average accuracy of off-target prediction in DL-based methods.
SCC evaluates the ability of the models to predict the actual efficiency of each gRNA sequence (Konstantakos et al., 2022). While some models are trained to minimize the mean squared error (MSE), the comparison between models on different datasets is necessarily made using Spearman correlation. Figure 7 compares the predictive ability of off-target sites in some ML-based tools over five datasets named Zebrafish_G, Zebrafish_S, HEL, A375, and mESC. In general, the larger the polygon area, the better the overall performance of the tool. Figure 7 clearly illustrates the better and more robust performance of the DeepHF, DeepSpCas9, and DeepCas9 models. As shown, classic ML-based tools such as Azimuth 2.0 achieve comparable performance to DL-based tools. Also, even though E-CRISP is more accurate than some learning-based tools, it does not achieve high enough correlations. However, E-CRISP has stable performance across all datasets. In addition, as it is clear from Figure 7, DeepCRISPR outperforms the other tools on the HEL dataset, and E-CRISP and CRISPRLearner achieve better results based on this metric.
As mentioned, gRNAs are typically designed by computational tools which compare gRNA sequence with a reference genome to predict the activity of on-target and potential off-targets. However, these tools can yield false-positive (FP) or false-negative (FN) results. Furthermore, the DNA in clinical experiments can differ from the reference genome used in the computational modeling, which means they would be more false predictions. Therefore, the accuracy is less than the values shown in Figure 7 in the actual experiment. To resolve this problem, in-vitro based tools have been developed for the experimental detection of off-target sites in a particular DNA sample. Tools like SMRT-OTS and Nano-OTS (Höijer et al., 2020) use long-read single-molecule sequencing.
In this article, we review both traditional and ML-based approaches for gRNA designing and predicting off-target sites. As mentioned before, experimental methods which use third-generation sequencing technology, have a better performance in Cas9 target detection on dark genomic regions (Höijer et al., 2020). This new technology helps us to detect more on-target and off-target sites and to design optimal gRNA. Furthermore, collected data in experimental methods, could improve the accuracy of DL-based tools.
Also, we have presented a comprehensive list of available tools. Each tool has merits and demerits, and the performance of different tools differs in different situations. According to our studies, some tools can be a better choice in some situations; However, others may be more popular in the scientific community. So, choosing the right tool depends on the conditions and limitations of an application.
Among the alignment-based methods, tools like CRISPR-P, Flycrispr, CRISPRseek, Cas-OFFinder, CasOT, sgRNACas9, and Flashfly have high accuracy and efficiency; however, CRISPR-P and Flycrispr are only used in specific genomes. Other tools such as CRISPRseek, Cas-Offinder, and CasOT, are used in almost all genomes. Moreover, they support only particular types of PAMs, while methods such as sgRNACas9 and Flashfly are compatible with all types of PAMs and seem to be a better option for designing gRNAs.
Among the learning-based methods, DL-based methods, including C-RNNCrispr, DeepCpf1, DeepHF, DeepSpCas9, and DeepCRISP, have drawn much interest recently. However, learning-based methods such as CLD, CRISPR-ERA, sgRNA-design, E-CRISP are significant due to their high accuracy and use in all genomes. Finally, based on our study, methods such as CRISPR-SE and E-CRISP are the best options to be used in all genomes with high accuracy.
3 Conclusion
CRISPR systems have been developed for accurate genome editing. Since genomic modifications are permanent (Ding et al., 2018), it is crucial to make precise edits. Most of the tools or methods in CRISPR’s field have been developed to help users design proper gRNA with fewer off-target effects. It is considered that the efficiency of one gRNA may differ among different models and databases. Users must evaluate several gRNAs using multiple models and select the best one for their experiments.
The previous successes of CNN and RNN architectures in bioinformatics motivated other researchers to extend their applications with a DL platform, which we believe is the best solution for predicting off-target effects. DL methods are inexpensive and fast compared to experimental methods. However, their accuracy depends on the amount of available data for a model’s training. Additionally, most of existing methods have three big problems, which means their predictions are not exact. First, they calculate scores based on mismatches to the guide sequence. However, DL-based methods can extract more efficient features hidden in the input data. In other words, DL-based methods can capture features other than gRNA sequence-based features. These features can be utilized and encoded in the input sequence to improve the performance of the existing DL architectures. In addition, most proposed DL-based methods use a one-hot vector representation to encode the input data. (Charlier et al., 2021). The use of newer encoding and embedding methods proposed in the field of DL can enhance the efficiency of existing DL-based methods. Also, the use of gRNA-DNA pair encoding can be helpful. Second, there is a rapid expansion in experimental data in CRISPR research. Most methods cannot scale and improve their performance with this new data. As known, DL-based methods achieve better performance by training on large datasets, but they require a pre-processing step to prepare and aggregate data obtained from diverse sources based on different experimental methods. This step requires enough knowledge about the type of input data, the operation mechanism of CRISPR, and the architecture of the deep neural network. Finally, the most severe issue is that existing DL-based methods still need to be improved in providing sufficient precision for clinical practice usage. NGS-based whole-genome sequencing technologies help to discover almost all off-target sites in the target genome and create a large and more accurate train dataset. As the number of instances in a train dataset increases, the predictions of DL-based methods become closer to experimental observations.
Author contributions
RA: supplied acquisition of data, analysis, interpretation of data and drafting the paper. LS: provided the conception and design of the study, analysis and interpretation of data, revised it critically for important intellectual content, and final approval of the version to be submitted. AK: provided the conception and design of the study, analysis and interpretation of data, revised it critically for important intellectual content, and final approval of the version to be submitted. RA has the first authorship right. LS and AK contributed equally to this work and share senior authorship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abadi S., Yan W. X., Amar D., Mayrose I. (2017). A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput. Biol. 13 (10), e1005807. 10.1371/journal.pcbi.1005807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal S., Sirohi P., Singh N. K. (2020). A review of CRISPR associated genome engineering: Application, advances and future prospects of genome targeting tool for crop improvement. Biotechnol. Lett. 42, 1611–1632. 10.1007/s10529-020-02950-w [DOI] [PubMed] [Google Scholar]
- Ahmed M., He H. H. (2017). SgTiler: A fast method to design tiling sgRNAs for CRISPR/cas9 mediated screening, BioRxiv. 217166. [Google Scholar]
- Alkhnbashi O. S., Costa F., Shah S. A., Garrett R. A., Saunders S. J., Backofen R. (2014). CRISPRstrand: Predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics 30 (17), i489–i496. 10.1093/bioinformatics/btu459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhnbashi O. S., Meier T., Mitrofanov A., Backofen R., Voß B. (2020). CRISPR-Cas bioinformatics. Methods 172, 3–11. 10.1016/j.ymeth.2019.07.013 [DOI] [PubMed] [Google Scholar]
- Alkhnbashi O. S., Mitrofanov A., Bonidia R., Raden M., Tran V. D., Eggenhofer F., et al. (2021). CRISPRloci: Comprehensive and accurate annotation of CRISPR–cas systems. Nucleic Acids Res. 49, W125–W130. 10.1093/nar/gkab456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F., Crepaldi L., Alsinet C., Strong A. J., Kleshchevnikov V., De Angeli P., et al. (2019). Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 37 (1), 64–72. 10.1038/nbt.4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S., Park J., Kim J-S. (2014). Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30 (10), 1473–1475. 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat N., Khuri N. (2021). “Predicting targets for genome editing with long short term memory networks,” in Advances in computer vision and computational biology (Berlin, Germany: Springer; ), 657–670. [Google Scholar]
- Biswas A., Gagnon J. N., Brouns S. J., Fineran P. C., Brown C. M. (2013). CRISPRTarget: Bioinformatic prediction and analysis of crRNA targets. RNA Biol. 10 (5), 817–827. 10.4161/rna.24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A., Staals R. H., Morales S. E., Fineran P. C., Brown C. M. (2016). CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC genomics 17 (1), 356–370. 10.1186/s12864-016-2627-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K., Pedersen L. E., Weber T., Lee S. Y. (2016). CRISPy-web: An online resource to design sgRNAs for CRISPR applications. Synthetic Syst. Biotechnol. 1 (2), 118–121. 10.1016/j.synbio.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel A., Steyaert W., De Rocker N., Menten B., Callewaert B., De Paepe A., et al. (2016). BATCH-GE: Batch analysis of Next-Generation Sequencing data for genome editing assessment. Sci. Rep. 6 (1). 10.1038/srep30330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancellieri S., Canver M. C., Bombieri N., Giugno R., Pinello L. (2020). CRISPRitz: Rapid, high-throughput and variant-aware in silico off-target site identification for CRISPR genome editing. Bioinformatics 36 (7), 2001–2008. 10.1093/bioinformatics/btz867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q., Ma J., Chen C-H., Xu H., Chen Z., Li W., et al. (2017). CRISPR-FOCUS: A web server for designing focused CRISPR screening experiments. PLoS One 12 (9), e0184281. 10.1371/journal.pone.0184281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson-Stevermer J., Kelso R., Kadina A., Joshi S., Rossi N., Walker J., et al. (2020). CRISPRoff enables spatio-temporal control of CRISPR editing. Nat. Commun. 11 (1), 5041–5047. 10.1038/s41467-020-18853-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R., Yeo N. C., Chavez A., Church G. M. (2017). sgRNA Scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth. Biol. 6 (5), 902–904. 10.1021/acssynbio.6b00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier J., Nadon R., Makarenkov V. (2021). Accurate deep learning off-target prediction with novel sgRNA-DNA sequence encoding in CRISPR-Cas9 gene editing. Bioinformatics 37 (16), 2299–2307. 10.1093/bioinformatics/btab112 [DOI] [PubMed] [Google Scholar]
- Chen C-L., Rodiger J., Chung V., Viswanatha R., Mohr S. E., Hu Y., et al. (2020). SNP-CRISPR: A web tool for SNP-specific genome editing. G3 Genes, Genomes, Genet. 10 (2), 489–494. 10.1534/g3.119.400904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., McKenna A., Schreiber J., Haeussler M., Yin Y., Agarwal V., et al. (2019). Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic acids Res. 47 (15), 7989–8003. 10.1093/nar/gkz487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D., et al. (2018). DeepCRISPR: Optimized CRISPR guide RNA design by deep learning. Genome Biol. 19 (1), 80–18. 10.1186/s13059-018-1459-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuai G-h., Wang Q-L., Liu Q. (2017). In silico meets in vivo: Towards computational CRISPR-based sgRNA design. Trends Biotechnol. 35 (1), 12–21. 10.1016/j.tibtech.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Cloney R. (2019). The oracle of inDelphi predicts Cas9 repair outcomes. Nat. Rev. Genet. 20 (1), 4–5. 10.1038/s41576-018-0077-z [DOI] [PubMed] [Google Scholar]
- Concordet J-P., Haeussler M. (2018). Crispor: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic acids Res. 46 (W1), W242–W245. 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. B. T., Platt R. J., Zhang F. (2015). Therapeutic genome editing: Prospects and challenges. Nat. Med. 21 (2), 121–131. 10.1038/nm.3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick T. J., Qiu P., Lee C. M., Fine E. J., Bao G. (2014). COSMID: A web-based tool for identifying and validating CRISPR/cas off-target sites. Mol. Therapy-Nucleic Acids. 3, e214. 10.1038/mtna.2014.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Liao X., Peng S., Tang T., Huang C., Yang C. (2020). OffScan: A universal and fast CRISPR off-target sites detection tool. BMC genomics 21 (1), 872–876. 10.1186/s12864-019-6241-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Xu J., Cheng M., Liao X., Peng S. (2018). Review of CRISPR/Cas9 sgRNA design tools. Interdiscip. Sci. Comput. Life Sci. 10 (2), 455–465. 10.1007/s12539-018-0298-z [DOI] [PubMed] [Google Scholar]
- Dampier W., Chung C-H., Sullivan N. T., Atkins A. J., Nonnemacher M. R., Wigdahl B. (2018). CRSeek: A Python module for facilitating complicated CRISPR design strategies, PeerJ Prepr. Report No, 2167–9843. [Google Scholar]
- de Ruijter A., Guldenmund F. (2016). The bowtie method: A review. Saf. Sci. 88, 211–218. 10.1016/j.ssci.2016.03.001 [DOI] [Google Scholar]
- Ding W., Mao W., Shao D., Zhang W., Gong H. (2018). DeepConPred2: An improved method for the prediction of protein residue contacts. Comput. Struct. Biotechnol. J. 16, 503–510. 10.1016/j.csbj.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34 (2), 184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Ouyang K., Xu X., Xu L., Wen C., Zhou X., et al. (2021). Nanoparticle delivery of CRISPR/Cas9 for genome editing. Front. Genet. 12, 673286. 10.3389/fgene.2021.673286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell T., Zhang D., Isik M., Wang T., Gotta G., Wilson C. J., et al. (2021). CALITAS: A CRISPR-cas-aware ALigner for in silico off-TArget search. CRISPR J. 4 (2), 264–274. 10.1089/crispr.2020.0036 [DOI] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., III (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31 (7), 397–405. 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. (2012). Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. 109 (39), E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R., Mai G., Wang P., Zhou M., Luo Y., Cai Y., et al. (2016). CRISPRdigger: Detecting CRISPRs with better direct repeat annotations. Sci. Rep. 6 (1). 10.1038/srep32942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güell M., Yang L., Church G. M. (2014). Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinformatics 30 (20), 2968–2970. 10.1093/bioinformatics/btu427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hana S., Peterson M., McLaughlin H., Marshall E., Fabian A. J., McKissick O., et al. (2021). Highly efficient neuronal gene knockout in vivo by CRISPR-Cas9 via neonatal intracerebroventricular injection of AAV in mice. Gene Ther. 28, 646–658. 10.1038/s41434-021-00224-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanzadeh H. R., Wang M. D. (2016). “DeeperBind: Enhancing prediction of sequence specificities of DNA binding proteins,” in 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15-18 December 2016 (IEEE; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F., Kerr G., Boutros M. E-C. R. I. S. P. (2014). E-CRISP: Fast CRISPR target site identification. Nat. methods 11 (2), 122–123. 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- Heigwer F., Zhan T., Breinig M., Winter J., Brügemann D., Leible S., et al. (2016). CRISPR library designer (CLD): Software for multispecies design of single guide RNA libraries. Genome Biol. 17 (1), 55–10. 10.1186/s13059-016-0915-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herai R. H. (2019). Avoiding the off-target effects of CRISPR/cas9 system is still a challenging accomplishment for genetic transformation. Gene 700, 176–178. 10.1016/j.gene.2019.03.019 [DOI] [PubMed] [Google Scholar]
- Höijer I., Johansson J., Gudmundsson S., Chin C-S., Bunikis I., Häggqvist S., et al. (2020). Amplification-free long-read sequencing reveals unforeseen CRISPR-Cas9 off-target activity. Genome Biol. 21 (1), 290. 10.1186/s13059-020-02206-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough S. H., Kancleris K., Brody L., Humphryes-Kirilov N., Wolanski J., Dunaway K., et al. (2017). Guide Picker is a comprehensive design tool for visualizing and selecting guides for CRISPR experiments. BMC Bioinforma. 18 (1), 167–210. 10.1186/s12859-017-1581-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 (9), 827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G-H., Bae S. (2021). Computational methods in synthetic biology. Berlin, Germany: Springer, 81–88.Web-based base editing toolkits: BE-Designer and BE-analyzer [DOI] [PubMed] [Google Scholar]
- Hwang G-H., Song B., Bae S. (2021). Current widely-used web-based tools for CRISPR nucleases, base editors, and prime editors. Gene Genome Ed. 1, 100004. 10.1016/j.ggedit.2021.100004 [DOI] [Google Scholar]
- Iyombe J-P. (2019). Correction du gène de la dystrophine avec la méthode CRISPR induced deletion. Québec: CinDel. [Google Scholar]
- Jacquin A. L., Odom D. T., Lukk M. (2019). Crisflash: Open-source software to generate CRISPR guide RNAs against genomes annotated with individual variation. Bioinformatics 35 (17), 3146–3147. 10.1093/bioinformatics/btz019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H-H., Kim S. Y., Rousseaux M. W., Zoghbi H. Y., Liu Z. (2017). CRISPRcloud: A secure cloud-based pipeline for CRISPR pooled screen deconvolution. Bioinformatics 33 (18), 2963–2965. 10.1093/bioinformatics/btx335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Taylor D. W., Chen J. S., Kornfeld J. E., Zhou K., Thompson A. J., et al. (2016). Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351 (6275), 867–871. 10.1126/science.aad8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Wong W. H. (2008). SeqMap: Mapping massive amount of oligonucleotides to the genome. Bioinformatics 24 (20), 2395–2396. 10.1093/bioinformatics/btn429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. science 337 (6096), 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Jiang F., Taylor D. W., Sternberg S. H., Kaya E., Ma E., et al. (2014). Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343 (6176), 1247997. 10.1126/science.1247997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur K., Gupta A. K., Rajput A., Kumar M. (2016). ge-CRISPR-An integrated pipeline for the prediction and analysis of sgRNAs genome editing efficiency for CRISPR/Cas system. Sci. Rep. 6 (1). 10.1038/srep30870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keough K. C., Lyalina S., Olvera M. P., Whalen S., Conklin B. R., Pollard K. S. (2019). AlleleAnalyzer: A tool for personalized and allele-specific sgRNA design. Genome Biol. 20 (1), 167–169. 10.1186/s13059-019-1783-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Kang B. C., Kim J. S. (2021). Identifying genome-wide off-target sites of CRISPR RNA-guided nucleases and deaminases with Digenome-seq. Nat. Protoc. 16 (2), 1170–1192. 10.1038/s41596-020-00453-6 [DOI] [PubMed] [Google Scholar]
- Kim H., Kim S-T., Ryu J., Kang B-C., Kim J-S., Kim S-G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8 (1), 14406–14407. 10.1038/ncomms14406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K., Kim Y., Lee S., Min S., Bae J. Y., Choi J. W., et al. (2019). SpCas9 activity prediction by DeepSpCas9, a deep learning–based model with high generalization performance. Sci. Adv. 5 (11), eaax9249. 10.1126/sciadv.aax9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K., Min S., Song M., Jung S., Choi J. W., Kim Y., et al. (2018). Deep learning improves prediction of CRISPR–Cpf1 guide RNA activity. Nat. Biotechnol. 36 (3), 239–241. 10.1038/nbt.4061 [DOI] [PubMed] [Google Scholar]
- Konstantakos V., Nentidis A., Krithara A., Paliouras G. (2022). CRISPR-Cas9 gRNA efficiency prediction: An overview of predictive tools and the role of deep learning. Nucleic acids Res. 50 (7), 3616–3637. 10.1093/nar/gkac192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32 (7), 677–683. 10.1038/nbt.2916 [DOI] [PubMed] [Google Scholar]
- Kwon K. H., Seonwoo M., Myungjae S., Soobin J., Woo C. J., Younggwang K., et al. (2019). DeepCpf1: Deep learning-based prediction of CRISPR-Cpf1 activity atendogenous sites. Proc. Annu. Meet. Jpn. Pharmacol. Soc. 92, JKL-05. [Google Scholar]
- Labun K., Montague T. G., Krause M., Torres Cleuren Y. N., Tjeldnes H., Valen E. (2019). CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic acids Res. 47 (W1), W171–W174. 10.1093/nar/gkz365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Chen P. B., DiaoCRISPR- Y. S. E. (2021). CRISPR-SE: A brute force search engine for CRISPR design. NAR genomics Bioinforma. 3 (1), lqab013. 10.1093/nargab/lqab013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lin N. (2010). The Bayesian elastic net. Bayesian anal. 5 (1), 151–170. 10.1214/10-ba506 [DOI] [Google Scholar]
- Lin J., Wong K-C. (2018). Off-target predictions in CRISPR-Cas9 gene editing using deep learning. Bioinformatics 34 (17), i656–i663. 10.1093/bioinformatics/bty554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Luo Y. (2019). Tracking CRISPR’s footprints. CRISPR Gene Ed. 1961, 13–28. 10.1007/978-1-4939-9170-9_2 [DOI] [PubMed] [Google Scholar]
- Listgarten J., Weinstein M., Elibol M., Hoang L., Doench J., Fusi N. (2016) Predicting off-target effects for end-to-end CRISPR guide design. bioRxiv.:078253. [Google Scholar]
- Listgarten J., Weinstein M., Kleinstiver B. P., Sousa A. A., Joung J. K., Crawford J., et al. (2018). Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat. Biomed. Eng. 2 (1), 38–47. 10.1038/s41551-017-0178-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zhang Y., Zhang T. (2020). Computational approaches for effective CRISPR guide RNA design and evaluation. Comput. Struct. Biotechnol. J. 18, 35–44. 10.1016/j.csbj.2019.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Ding Y., Zhou Y., Jin W., Xie K., Chen L-L. (2017). CRISPR-P 2.0: An improved CRISPR-cas9 tool for genome editing in plants. Mol. plant 10 (3), 530–532. 10.1016/j.molp.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Liu H., Wei Z., Dominguez A., Li Y., Wang X., Qi L. S. (2015). CRISPR-ERA: A comprehensive design tool for CRISPR-mediated gene editing, repression and activation: Fig. 1. Bioinformatics 31 (22), 3676–3678. 10.1093/bioinformatics/btv423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Chen W., Xue L., Tang B. (2019). Prediction of activity and specificity of CRISPR-Cpf1 using convolutional deep learning neural networks. BMC Bioinforma. 20 (1). 10.1186/s12859-019-2939-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyten H., Plijter J. J., Van Vliet T. (2004). Crispy/crunchy crusts of cellular solid foods: A literature review with discussion. J. texture Stud. 35 (5), 445–492. 10.1111/j.1745-4603.2004.35501.x [DOI] [Google Scholar]
- Ma J., Köster J., Qin Q., Hu S., Li W., Chen C., et al. (2016). CRISPR-DO for genome-wide CRISPR design and optimization. Bioinformatics 32 (21), 3336–3338. 10.1093/bioinformatics/btw476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., Saunders S. J., et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13 (11), 722–736. 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339 (6121), 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manibalan S., Thirukumaran K., Varshni M., Shobana A., Achary A. (2020). Report on biopharmaceutical profile of recent biotherapeutics and insilco docking studies on target bindings of known aptamer biotherapeutics. Biotechnol. Genet. Eng. Rev. 36 (2), 57–80. 10.1080/02648725.2020.1858395 [DOI] [PubMed] [Google Scholar]
- McKenna A., Shendure J. (2018). FlashFry: A fast and flexible tool for large-scale CRISPR target design. BMC Biol. 16 (1), 74–76. 10.1186/s12915-018-0545-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrofanov A., Alkhnbashi O. S., Shmakov S. A., Makarova K. S., Koonin E. V., Backofen R. (2021). CRISPRidentify: Identification of CRISPR arrays using machine learning approach. Nucleic acids Res. 49 (4), e20–e. 10.1093/nar/gkaa1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos M. A., Vejnar C. E., Beaudoin J-D., Fernandez J. P., Mis E. K., Khokha M. K., et al. (2015). CRISPRscan: Designing highly efficient sgRNAs for CRISPR-cas9 targeting in vivo . Nat. methods 12 (10), 982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad Rafid A. H., Toufikuzzaman M., Rahman M. S., Rahman M. S. (2020). CRISPRpred (SEQ): A sequence-based method for sgRNA on target activity prediction using traditional machine learning. BMC Bioinforma. 21. 10.1186/s12859-020-3531-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H., Ui-Tei K. (2015). CRISPRdirect: Software for designing CRISPR/cas guide RNA with reduced off-target sites. Bioinformatics 31 (7), 1120–1123. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien A., BaileyGT-Scan T. L. (2014). GT-scan: Identifying unique genomic targets. Bioinformatics 30 (18), 2673–2675. 10.1093/bioinformatics/btu354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros J. C., Franch M., Tabas-Madrid D., San-León D., Montoliu L., Cubas P., et al. (2016). Breaking-Cas—Interactive design of guide RNAs for CRISPR-cas experiments for ENSEMBL genomes. Nucleic acids Res. 44 (W1), W267–W271. 10.1093/nar/gkw407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallarès Masmitjà M., Knödlseder N., Güell M. (2019). CRISPR gene editing. Berlin, Germany: Springer, 3–11.CRISPR-gRNA design [DOI] [PubMed] [Google Scholar]
- Park J., Bae S., Kim J-S. (2015). Cas-Designer: A web-based tool for choice of CRISPR-cas9 target sites. Bioinformatics 31 (24), 4014–4016. 10.1093/bioinformatics/btv537 [DOI] [PubMed] [Google Scholar]
- Park J., Lim K., Kim J-S., Bae S. (2017). Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 33 (2), 286–288. 10.1093/bioinformatics/btw561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D., Tarleton R. (2015). EuPaGDT: A web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb. genomics 1 (4), e000033. 10.1099/mgen.0.000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A. R., Pritykin Y., Vidigal J. A., Chhangawala S., Zamparo L., Leslie C. S., et al. (2017). GuideScan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 35 (4), 347–349. 10.1038/nbt.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L., Canver M. C., Hoban M. D., Orkin S. H., Kohn D. B., Bauer D. E., et al. CRISPResso: Sequencing analysis toolbox for CRISPR genome editing. bioRxiv. 2016:031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L., Canver M. C., Hoban M. D., Orkin S. H., Kohn D. B., Bauer D. E., et al. (2015). CRISPResso: Sequencing analysis toolbox for CRISPR-cas9 genome editing, bioRxiv. 031203. [Google Scholar]
- Prykhozhij S. V., Rajan V., Gaston D., Berman J. N. (2015). CRISPR multitargeter: A web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PloS onee0119372 10 (3). 10.1371/journal.pone.0119372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido-Quetglas C., Aparicio-Prat E., Arnan C., Polidori T., Hermoso T., Palumbo E., et al. (2017). Scalable design of paired CRISPR guide RNAs for genomic deletion. PLoS Comput. Biol. 13 (3), e1005341. 10.1371/journal.pcbi.1005341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R., Li J., Li H., Zhang Y., Liu X., Miao Y., et al. (2019). Developing a highly efficient and wildly adaptive CRISPR‐SaCas9 toolset for plant genome editing. Plant Biotechnol. J. 17 (4), 706–708. 10.1111/pbi.13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz R., Almog S., Darnell R., Offen D. (2020). CrisPam: SNP-Derived PAM analysis tool for allele-specific targeting of genetic variants using CRISPR-cas systems. Front. Genet. 11, 851. 10.3389/fgene.2020.00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi A., Murik O., Bowler C., Tirichine L. (2016). PhytoCRISP-ex: A web-based and stand-alone application to find specific target sequences for CRISPR/CAS editing. BMC Bioinforma. 17 (1), 261–264. 10.1186/s12859-016-1143-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumar S. C., Preethi K. A., Ross K., Tusubira D., Khan M. W. A., Mani P., et al. (2022). CRISPR/Cas9 and next generation sequencing in the personalized treatment of Cancer. Mol. Cancer 21 (1), 83. 10.1186/s12943-022-01565-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M. W., Arbab M., Hsu J. Y., Worstell D., Culbertson S. J., Krabbe O., et al. (2018). Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563 (7733), 646–651. 10.1038/s41586-018-0686-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skennerton C. T., Imelfort M., Tyson G. W. (2013). Crass: Identification and reconstruction of CRISPR from unassembled metagenomic data. Nucleic acids Res. 41 (10), e105–e. 10.1093/nar/gkt183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledzinski P., Nowaczyk M., Olejniczak M. (2020). Computational tools and resources supporting CRISPR-Cas experiments. Cells 9 (5), 1288. 10.3390/cells9051288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. H., Chen Y-C., Seifuddin F., Hupalo D., Alba C., Reger R., et al. (2020). Genome-wide analysis of off-target CRISPR/Cas9 activity in single-cell-derived human hematopoietic stem and progenitor cell clones. Genes 11 (12), 1501. 10.3390/genes11121501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M., Thumberger T., del Sol Keyer M., Wittbrodt J., Mateo J. L. (2015). CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PloS one 10 (4), e0124633. 10.1371/journal.pone.0124633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Liu H., Liu J., Cheng S., Peng Y., Zhang Q., et al. (2019). CRISPR-local: A local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics 35 (14), 2501–2503. 10.1093/bioinformatics/bty970 [DOI] [PubMed] [Google Scholar]
- Tarasava K., Liu R., Garst A., Gill R. T. (2018). Combinatorial pathway engineering using type I‐E CRISPR interference. Biotechnol. Bioeng. 115 (7), 1878–1883. 10.1002/bit.26589 [DOI] [PubMed] [Google Scholar]
- Tsai S. Q., Zheng Z., Nguyen N. T., Liebers M., Topkar V. V., Thapar V., et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33 (2), 187–197. 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S. K., Sharma S. (2014). SSFinder: High throughput CRISPR-cas target sites prediction tool. BioMed Res. Int. 2014, 1–4. 10.1155/2014/742482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang C., Wang B., Li B., Wang Q., Liu D., et al. (2019). Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat. Commun. 10 (1). 10.1038/s41467-019-12281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xiang X., Cheng L., Zhang X., Luo Y. (2020). CRISPR-GNL: An improved model for predicting CRISPR activity by machine learning and featurization. bioRxiv. 2019:605790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang X., Cheng L., Luo Y. (2020). An overview and metanalysis of machine and deep learning-based CRISPR gRNA design tools. RNA Biol. 17 (1), 13–22. 10.1080/15476286.2019.1669406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liang C. C. R. F. (2017). CRF: Detection of CRISPR arrays using random forest. PeerJ 5, e3219. 10.7717/peerj.3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tilford C., Neuhaus I., Mintier G., Guo Q., Feder J. N., et al. (2017). CRISPR-DAV: CRISPR NGS data analysis and visualization pipeline. Bioinformatics 33 (23), 3811–3812. 10.1093/bioinformatics/btx518 [DOI] [PubMed] [Google Scholar]
- Wilson L. O., Reti D., O'Brien A. R., Dunne R. A., Bauer D. C. (2018). High activity target-site identification using phenotypic independent CRISPR-Cas9 core functionality. CRISPR J. 1 (2), 182–190. 10.1089/crispr.2017.0021 [DOI] [PubMed] [Google Scholar]
- Winter J., Schwering M., Pelz O., Rauscher B., Zhan T., Heigwer F., et al. CRISPRAnalyzeR: Interactive analysis, annotation and documentation of pooled CRISPR screens. BioRxiv. 2017:109967. [Google Scholar]
- Wong N., Liu W., WangWU-Crispr X. (2015). Wu-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16 (1), 218–8. 10.1186/s13059-015-0784-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Cheng Z., Kong L., Zhu Z., Lin S., Gao G., et al. (2014). CasOT: A genome-wide cas9/gRNA off-target searching tool. Bioinformatics 30 (8), 1180–1182. 10.1093/bioinformatics/btt764 [DOI] [PubMed] [Google Scholar]
- Xie S., Shen B., Zhang C., Huang X., Zhang Y. (2014). sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PloS one 9 (6), e100448. 10.1371/journal.pone.0100448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Xiao T., Chen C-H., Li W., Meyer C. A., Wu Q., et al. (2015). Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25 (8), 1147–1157. 10.1101/gr.191452.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Chuai G., Zhou C., Zhu C., Yang J., Zhang C., et al. (2018). Benchmarking CRISPR on-target sgRNA design. Briefings Bioinforma. 19 (4), 721–724. 10.1093/bib/bbx001 [DOI] [PubMed] [Google Scholar]
- Yu S-H., Vogel J., Förstner K. U. (2018). ANNOgesic: A Swiss army knife for the RNA-seq based annotation of bacterial/archaeal genomes. GigaScience 7 (9), giy096. 10.1093/gigascience/giy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Abudayyeh O. O., Gootenberg J. S., Scott D. A., Zhang F. (2020). A survey of genome editing activity for 16 Cas12a orthologs. Keio J. Med. 69 (3), 59–65. 10.2302/kjm.2019-0009-oa [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Dai Z., Dai X. (2020). C-RNNCrispr: Prediction of CRISPR/Cas9 sgRNA activity using convolutional and recurrent neural networks. Comput. Struct. Biotechnol. J. 18, 344–354. 10.1016/j.csbj.2020.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., LiangCRISPR- C. D. T. (2019). CRISPR-DT: Designing gRNAs for the CRISPR-cpf1 system with improved target efficiency and specificity. Bioinformatics 35 (16), 2783–2789. 10.1093/bioinformatics/bty1061 [DOI] [PubMed] [Google Scholar]
- Zhu H., Misel L., Graham M., Robinson M. L., LiangCT-Finder C. (2016). CT-finder: A web service for CRISPR optimal target prediction and visualization. Sci. Rep. 6 (1), 25516–25518. 10.1038/srep25516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Richmond E., LiangCRISPR-Rt C. (2018). CRISPR-RT: A web application for designing CRISPR-C2c2 crRNA with improved target specificity. Bioinformatics 34 (1), 117–119. 10.1093/bioinformatics/btx580 [DOI] [PubMed] [Google Scholar]
- Zhu L. J., Holmes B. R., Aronin N., Brodsky M. H. (2014). CRISPRseek: A bioconductor package to identify target-specific guide RNAs for CRISPR-cas9 genome-editing systems. PloS onee108424 9 (9). 10.1371/journal.pone.0108424 [DOI] [PMC free article] [PubMed] [Google Scholar]