Abstract
We introduce and evaluate the oblique random survival forest (ORSF). The ORSF is an ensemble method for right-censored survival data that uses linear combinations of input variables to recursively partition a set of training data. Regularized Cox proportional hazard models are used to identify linear combinations of input variables in each recursive partitioning step. Benchmark results using simulated and real data indicate that the ORSF’s predicted risk function has high prognostic value in comparison to random survival forests, conditional inference forests, regression, and boosting. In an application to data from the Jackson Heart Study, we demonstrate variable and partial dependence using the ORSF and highlight characteristics of its 10-year predicted risk function for atherosclerotic cardiovascular disease events (ASCVD; stroke, coronary heart disease). We present visualizations comparing variable and partial effect estimation according to the ORSF, the conditional inference forest, and the Pooled Cohort Risk equations. The obliqueRSF R package, which provides functions to fit the ORSF and create variable and partial dependence plots, is available on the comprehensive R archive network (CRAN).
MSC 2010 subject classifications: Primary 60K35, 60K35, secondary 60K35
Keywords: Random Forest, Survival, Machine Learning, Penalized Regression, Cardiovascular Disease
1. Introduction.
Since Breiman (2001) introduced the random forest (RF), it has become recognized as a flexible and accurate tool for classification, regression, and right-censored time-to-event (i.e., survival) analysis (Strobl, Malley and Tutz, 2009). RFs are characterized by an ensemble of decision trees, each of which are grown through recursive partioning (Breiman, 1984). Recursive partitioning is performed in steps, where each step involves splitting a set of data into two descendant subsets, beginning with the full set. The data are split based on an input variable X and a splitting value c such that one descendant set contains observations in the current data with X < c and the other set contains observations in the current data with X ≥ c. The splitting variable and value are chosen to maximize the difference between the descendant sets in an outcome variable, and the splitting variable can be either a single input variable or a linear combination of input variables (LCIVs). Descendant data sets that are split into smaller sets are referred to as ‘non-terminal nodes’ in the tree. Descendant data sets that are too small to split further (i.e., data sets with less than a pre-specified minimum number of unique observations or events) are not split any further and are referred to as ‘terminal nodes’ (i.e., leaves) of the tree.1 Predictions for a single test observation are computed by determining which terminal node the observation falls to and then aggregating the outcomes of participants in the training data who were mapped to the same node.
Breiman (2001) showed that RFs often achieve lower generalization error when LCIVs are used instead of a single variable (i.e., oblique splitting versus axis-based splitting) to split the training data in each recursive partitioning step. Hothorn and Lausen (2003) incorporated LCIVs by applying linear discriminant analysis to out-of-bag data and using predicted values from the discriminant model for in-bag data as a candidate splitting variable. Menze et al. (2011) introduced oblique RFs, and showed that Breiman’s method of constructing LCIVs could be improved by fitting regularized regression models (Friedman, Hastie and Tibshirani, 2001, Sec. 3.4) to the data in non-terminal nodes and synthesizing a splitting variable using the model’s predictions. Zhu, Zeng and Kosorok (2015) proposed to construct LCIVs using the most important K variables, which they identified by fitting ensembles of extremely randomized trees (Geurts, Ernst and Wehenkel, 2006) to the data in each non-terminal node. For classification and regression problems, RFs with LCIVs have achieved excellent generalization error (Breiman, 2001, Sec. 5) for several data sets in public repositories (Dheeru and Karra Taniskidou, 2017).
Random survival forests (RSFs) (Ishwaran et al., 2008) and conditional inference forests (CIF) (Hothorn, Hornik and Zeileis, 2006) extend Breiman’s RF to right-censored survival data, but neither of these recursive partitioning algorithms incorporate LCIVs. Additionally, to our knowledge, there are no studies examining the use of LCIVs for recursive partitioning in the context of right-censored survival outcomes (Bou-Hamad et al., 2011). Therefore, we developed and evaluated a method to incorporate LCIVs in binary decision trees for right-censored survival data. Zhu (2013) found that extremely randomized survival trees did not produce optimal multiplicative coefficients for LCIVs. Thus, we extended the approach of Menze et al. (2011) by using regularized (Zou and Hastie, 2005) Cox proportional hazards (PH) (Cox, 1992; Simon et al., 2011) models to identify multiplicative coefficients for LCIVs in each recursive partitioning step. Following the notation of Menze et al. (2011), we refer to our method as the oblique RSF (ORSF).
The purpose of this article is to describe the ORSF and assess the prognostic value of its predicted risk fucntion. After a brief summary of RSFs and CIFs in Section 2, ORSFs are described in Section 3. Simulated and real data are used in Sections 4 and 5, respectively, to assess performance of the ORSF in comparison to the RSF, CIF, gradient boosted decision trees, and Cox PH models. Performance and mean time required for computation over the entire collection of data from Sections 4 and 5 is assessed in Section 6. In Section 7, we apply the obliqueRSF R package to conduct exploratory analyses using data from participants in the Jackson Heart Study (JHS). Our analysis of the JHS data focuses on atherosclerotic cardiovascular disease (ASCVD) and its dependence on four key variables: age, systolic blood pressure, estimated glomerular filtration rate, and left ventricular mass. We conclude with a summary and discussion of our results in Section 8.
2. Ensemble tree methods for right-censored survival data.
Here we summarize two separate implementations of ensemble tree methods for right-censored survival data:
Random survival forest (RSF) (Ishwaran et al., 2008).
Conditional inference forest (CIF) (Hothorn, Hornik and Zeileis, 2006).
We present an ad-hoc summary of the steps taken to grow ensembles of binary decision trees for right-censored survival data (i.e., survival trees) in Section 2.1. Section 2.2 contains notation to describe the framework of survival ensembles. Last, we present summaries of the RSF and CIF in Sections 2.3 and 2.4, respectively.
2.1. How to grow ensembles of survival trees.
Step 1 Draw B subsamples2 from the data: {D1, …, DB}
- Step 2 For b = 1, …, B, grow a survival tree using Db:
- Initiate 𝒩G = {Db}, the set of ‘nodes to grow.’
- If 𝒩G = ∅, go to (c). Otherwise, for each node in 𝒩G, do the following:
- Define the set of candidate splitting variables for the current node by selecting a random subset of the available predictor variables.3
- 4If there is no evidence of statistical association between the survival outcome and any of the candidate splitting variables, then (1) remove the current node from 𝒩G, (2) label the current node as ‘terminal’, and (3) skip steps (iii) and (iv) below.
- Split the current node into descendant nodes,5 A1 and A2, using the candidate splitting variable that maximizes a log-rank statistic comparing survival outcomes between A1 and A2.
- Remove the current node from 𝒩G. If A1 has at least a minimum number of unique observations (i.e., minsplit or nodesize), add A1 to 𝒩G. Otherwise, label A1 as ‘terminal’. Do the same for A2.
- Define a predicted survival or cumulative hazard function for each terminal node based on the observed survival times in the node.
Step 3 Aggregate predicted survival6 or cumulative hazard functions from the B survival trees to compute ensemble predictions.
2.2. Notation.
Consider right-censored survival data from N participants: (y1, x1, δ1), …, (yN, xN, δN). For participant i, 1 ≤ i ≤ N, yi represents survival time if δi = 1 and time to censoring if δi = 0, xi is a length p vector of predictors: (xi,1, …, xi,p). Denote the jth column of the matrix x = [x1, …, xN]′ as x(j). Let t1 < … < tm denote the m unique event times (assume there are no ties). Denote the bth survival tree as 𝒯b, let 𝒯b(xi) identify the terminal node in 𝒯b participant i is mapped to, and let cib denote the number of times the data from participant i occurs in the bth bootstrap sample. Following the counting process notation of Andersen et al. (2012), define
| (2.1) |
where 0 ≤ s ≤ tm and I(·) is the indicator function. Define
| (2.2) |
and
| (2.3) |
as the number of uncensored events prior to and number of participants at risk at time s, respectively, in 𝒯b(x).
2.3. Random survival forests (RSF).
The RSF extends the prescription described by Breiman and Cutler (2003) to the context of survival analyses. The log-rank statistic (Segal, 1988, Sec. 3.2) is applied to determine which candidate variable and cut-point should be used. A Nelson-Aalen estimator of the cumulative hazard function is formed in each terminal node based on the survival times of observations in the node:
| (2.4) |
In turn, the ensemble survival function is
| (2.5) |
2.4. Conditional inference forests (CIF).
The CIF avoids variable selection bias (Breiman, 1984, pg. 42) by applying permutation tests (Strasser and Weber, 1999) to compare candidate splitting variables. During each recursive partitioning step, if there is no evidence of association between any of the candidate variables and the response, the current node is not split and is labeled terminal. For ensemble prediction, the CIF applies a weighted Kaplan-Meier estimate (Hothorn et al., 2004) based on all training observations in the B leaves containing the new observation:
| (2.6) |
3. The oblique random survival forest (ORSF).
Here we describe the ORSF, beginning with the use of regularized Cox PH models (Section 3.1) and synthesis of candidate splitting variables using regularized Cox PH models (Section 3.2). Additional details related to sampling and prediction are provided in Section 3.3.
3.1. Regularized Cox PH models.
The ORSF embeds regularized Cox PH models into the non-terminal nodes of its survival trees. Suppose there are K observations in the current non-terminal node. The embedded Cox PH model assumes a semi-parametric form for the hazard:
| (3.1) |
where k = 1, …, K, hk(t) is the hazard for observation k at time t, h0(t) is the baseline hazard function, and β is a vector of mtry ≤ p coefficients, where mtry is the number of randomly selected predictor variables for the current node and p is the total number of predictor variable. Estimation of β is carried out using the partial likelihood,
| (3.2) |
where Ri is the set of indices, j, with yj ≥ ti (i.e., those still at risk at time ti), and j(i) is the index of the observation for which an event occurred at time ti. Simon et al. (2011) proposed to maximize (3.2) subject to the elastic net penalty (Zou and Hastie, 2005):
| (3.3) |
where s ≥ 0 is a constant that controls shrinkage and has one-to-one correspondence with the complexity parameter, λ ≥ 0. Setting α = 1 and α = 0 in (3.3) results in the classic ‘Ridge’ and ‘Lasso’ penalties, respectively (Friedman, Hastie and Tibshirani, 2001, Sec. 3.4). Each pair of values for α and λ corresponds to a different solution for β. Notably, with a ridge penalty, every node will develop a solution for β with non-zero regression coefficients for all mtry randomly selected predictor variables. On the other hand, a lasso penalty will usually identify sparse solutions that set most of the mtry coefficients in β to zero.
3.2. Linear combinations of input variables (LCIVs).
LCIVs are synthesized using embedded Cox PH models and used as splitting variables for internal nodes in the ORSF. The LCIVs are synthesized using , where is the stacked matrix of input vectors and is selected from a number of candidate solutions given by the embedded Cox PH model. The maximum number of predictor variables with non-zero regression coefficients in these candidate solutions is mtry. The process that identifies candidate solutions for is governed by (1) whether cross-validation is applied, and (2) the value of α.
If cross-validation is applied (use.cv=TRUE in the ORSF function), then 5-fold cross-validation will be used in each non-terminal node to identify the following λ values:
λCV: The value of λ maximizing cross-validated partial likelihood (van Houwelingen et al., 2006).
λSE: The highest value of λ within one standard error of λCV.
The choice of α affects the candidate solutions identified by λCV and λSE. For example, when α = 1, both λCV and λSE identify solutions that use all of the randomly selected candidate splitting variables; however, the solution identified by λSE will impose a stronger penalty on regression coefficients. If α is set at a value close but not equal to 1, e.g. 0.90, then λCV and λSE will both identify ‘ridge-like’ solutions that estimate non-zero regression coefficients for nearly all of the candidate splitting variables.
If the analyst chooses not to use cross-validation, a regularization path is fitted to the current node’s data to identify λ1, …, λmtry, where λi is the maximum value of lambda such that the model has i effective degrees of freedom (Efron et al., 2004). To ensure these values of λ can be found, we require α < 1 when cross-validation is not used. In this context, the choice of α affects how quickly the regularization path transitions from a minimally to maximally complex solution. For example, when α is close to 0, the regularization path will be ‘lasso-like’ and identify more solutions with a small number of non-zero regression coefficients compared to a ‘ridge-like’ regularization path, where all or nearly all solutions have non-zero regression coefficients for all candidate variables.
Regardless of whether or not cross-validation is used, each candidate solution for is evaluated as follows:
is computed for each observation in the current node.
nsplit candidate cut-points, c1 …, cnsplit are selected at random from the unique values of .
For i = 1, …, nsplit, a log-rank statistic comparing survival curves between observations with and observations with is computed.
When cross-validation is not applied, log-rank statistics are penalized by a scaling factor of (1+γ · df), where γ > 0 is a tuning parameter and df is the effective degrees of freedom of the model that generated the candidate LCIV. Larger values of γ will result in selection of less complex LCIVs with fewer variables. Setting γ = 0 imposes no penalty on the complexity of LCIVs and will result in selection of LCIVs with a larger number of variables. If the maximal log-rank statistic does not exceed a pre-specified threshold, early stopping is applied (i.e., the node is not split and is labeled terminal). Otherwise, the cut-point and candidate solution maximizing the log-rank statistic are used to split the node.
3.3. Additional details.
The ORSF applies subsampling rather than bootstrap sampling with replacement. Ensemble predictions from an ORSF are formed using the same weighted aggregation scheme as in the CIF.
4. Simulation Study.
Here we describe and summarize results from a simulation study following the protocol described by Morris, White and Crowther (2017). The primary aim of the simulation is to compare the prognostic value of the ORSF’s predicted risk function with a variety of competing learning algorithms in three general settings. Data generating mechanisms are summarized in Section 4.1. The primary estimands of the simulation study are described in Section 4.3. Competing learning algorithms and their corresponding hyper-parameter settings are introduced in Section 4.2. Tabular summaries of results are presented in Section 4.5. Computational strategies are reported in Section 4.4. Source codes for the current simulation study are available from the first author’s GitHub site and supplemental material Jaeger et al. (2019).
4.1. Data generating mechanisms.
We generated right-censored survival data from three scenarios using a training sample of N = 500 and a testing sample of M = 1000. The number of predictor variables used to generate survival outcomes, p, was set at 25 and 50 for each scenario. For i = 1, …, N + M, xi = (xi,1, …, xi,p) was generated from a zero mean, unit variance multivariate normal distribution with cov(xi,u, xi,v) = (1/3)|u−v|. Multiplicative coefficient vectors (i.e., β) for variables 1 through p were generated as a uniform sequence of length p, beginning from −1 and ending at +1. The three scenarios were as follows:
The length p vector β contained one effect for each predictor. Simulated outcomes followed a Weibull distribution.
The length 2p vector β contained one main effect for each predictor and, additionally, each predictor was used in two bi-variate interaction terms. The multiplicative coefficients for interaction variables were generated as a uniform sequence of length p, beginning from −1 and ending at +1.7 Simulated outcomes followed the same distribution as in scenario A.
The same β vector was used as described scenario B, and each (yi, δi) followed one of three distinct Weibull distributions, depending on the value of xiβ.
Scenario A is designed to favor the PH model, whereas scenario B will favor survival tree ensembles. Scenario C is of particular interest as the proportional hazards assumption does not hold.
4.2. Competing methods and tuning parameters.
4.2.1. Decision tree ensembles.
ORSF, RSF, and CIF ensembles were composed of 1000 survival trees. The minimum number of unique observations needed to split a node (i.e., minsplit or nodesize) was 10. For the ORSF and RSF, the minimum number of events per terminal node was 1, splitting values were selected by comparing 25 randomly selected cut-points, and the number of variables tried at each node (i.e., mtry) was set to the smallest integer ≥ . The CIF used tuning parameters consistent with an unbiased recursive partitioning framework (Strobl et al., 2007; Hothorn et al., 2010, 2019). Two ORSF models were fitted as follows:
ORSF: The ORSF algorithm is applied with α = 0.50 and without using cross-validation to synthesize candidate splitting variables (i.e., setting use.cv=FALSE as described in Section 3.2).
ORSFCV: The ORSF algorithm is applied with α = 0.05 (a ridge-like penalty) and using cross-validation to synthesize candidate splitting variables (i.e., setting use.cv=TRUE as described in Section 3.2).
Gradient boosted decision trees were fitted using cross-validation to determine (1) the optimal number of boosting steps and (2) an optimal set of hyper-parameter values (Chen and Guestrin, 2016; Friedman, 2001). For (2), we generated 10 sets of hyper-parameter values randomly (i.e., values for tree depth, column subsampling, row subsampling, minimum child weight, and minimum loss reduction required to make a further partition on a leaf node). The hyper-parameter set that maximized cross-validated partial log-likelihood was used to fit an ensemble of gradient boosted decision trees to the training data.
4.2.2. Cox proportional hazards (PH) models.
Regularized Cox PH models were fitted using Lasso-like (α = 0.90) and Ridge-like (α = 0.10) penalties, and using the value of λ that maximized cross-validated partial likelihood (Friedman, Hastie and Tibshirani, 2010a). Unregularized Cox PH models were fitted using stepwise variable selection based on Akaike’s information criteria (Burnham and Anderson, 2004). Boosted Cox PH models were fitted using cross-validation to determine the optimal number of boosting steps and penalized score statistics to determine parameter updates in each boosting step (Tutz and Binder, 2007).
4.3. Prediction Error.
Predicted risk for right-censored survival outcomes can be assessed using the time dependent concordance index and Brier score (Gerds et al., 2013; Graf et al., 1999). For t > 0, the inverse probability of censoring weighted concordance index and Brier score are estimated using
and
respectively, where for participant i in the testing data, is the estimated probability of survival at time t, xi is the input information, and is the estimated probability of censoring. Additionally, defines the probability of censoring weights for concordance.
The concordance index measures the probability at time t that a randomly selected participant who has experienced an event has a higher model-based prediction than a randomly selected subject who has not experienced an event. Concordance indices of 1.00 and 0.50 correspond to perfect and worthless discrimination, respectively. Gerds et al. (2013) demonstrated asymptotic bias of the concordance index introduced by Harrell et al. (1982) and proposed a method to estimate a time-dependent concordance index for models with covariate dependent censoring (i.e., ). Additionally, Blanche, Kattan and Gerds (2018) showed that the time dependent area under the receiver operating characteristic curve rather than the concordance index introduced by Harrell et al. (1982) should be used to assess t-year predicted risk.
For a single observation at time t, the Brier score is the squared difference between observed survival status (e.g., 1 = alive at time t and 0 = dead at time t) and a model-based prediction of survival at time t. The expected Brier score of a prediction model which ignores all predictor variables (i.e. the Kaplan-Meier estimate of survival calculated with all training samples) is a reference value that can be used as a benchmark Brier score for prediction models. For clarity and ease of interpretation, we tabulate both the unscaled and scaled Brier score estimates, where the scaled Brier score is computed as 1 minus the ratio of the unscaled Brier score to the reference Brier score. Similar to the R2 statistic, a scaled Brier score of 1 and 0 indicate a perfect and worthless model, respectively.
4.3.1. Summary measures of prediction error.
The concordance index proposed by Gerds et al. (2013) uses survival time as response and differs conceptually from the the time-dependent receiver-operator characteristic curve proposed by Heagerty, Lumley and Pepe (2000). The latter incident statistic measures a prediction model’s ability to classify survival status at a given time point, while the former cumulative statistic measures a prediction model’s ability to order the survival times. Heagerty and Zheng (2005) have established a formal relationship between these two measures. Following the precedent of Ishwaran et al. (2008), we define concordance error as .
As is time dependent, integration from baseline to a specified follow-up time provides a summary measure of performance that can be tabulated for direct comparisons between competing learning methods. The integrated BS is defined as
| (4.1) |
where T is the specified follow-up time. In our results, we set T equal to the median event time in the testing data. We also present values of and that are scaled by a factor of 100 to avoid an unnecessary amount of leading zero’s (i.e., we present 0.5 instead of 0.005).
4.4. Computational notes and software.
All analyses were performed in R version 3.5.0. Right-censored survival outcomes were generated using the simsurv R package (Brilleman, 2018). We used the RandomForestSRC (Ishwaran and Kogalur, 2019), party (Hothorn et al., 2010), and obliqueRSF (Jaeger, 2018) R packages to fit RSFs, CIFs, and ORSFs, respectively, the xgboost (Chen et al., 2019) package to fit gradient boosted decision trees, the glmnet (Friedman, Hastie and Tibshirani, 2010b) package to fit regularized Cox PH models, the survival (Therneau, 2015) package to fit classical Cox PH models, the MASS (Venables and Ripley, 2002) package to perform forward stepwise selection using Akaike’s information criteria, and the CoxBoost package to fit boosted Cox PH models (Binder, 2013). To compute and , we used the pec package (Mogensen, Ishwaran and Gerds, 2012). Unadjusted Kaplan-Meier estimates were used to estimate inverse probability of censoring weights throughout (Mogensen, Ishwaran and Gerds, 2012, Sec. 6.2).
4.5. Results.
The mean rankings according to for ORSFCV and ORSF were 2.17 and 2.67, respectively, out of the 9 learning algorithms we applied (Table 1). As expected, the ORSF and ORSFCV provided the lowest values of in scenario B. Notably, the ORSF and ORSFCV also provided the lowest values of in scenario C, despite the invalidity of the PH assumption. In comparison to the RSF and CIF, both the ORSF and ORSFCV provided lower values of in each of the six simulated analyses. The absolute (percent) reduction in the mean value of from using the ORSFCV instead of the RSF and using the ORSFCV instead of the CIF was 3.13 (9.62%) and 0.52 (1.58%), respectively.
Table 1.
Mean concordance errors for competing learning methods, aggregated over 100 simulations in three scenarios. The minimum concordance error for each simulated analysis is written in bold text.
| Ensemble Survival Trees§ | Proportional Hazards | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Scenario† | p ‡ | ORSF | ORSFCV | CIF | RSF | Xgboost | CoxBoost | Lasso | Ridge | Step |
| Concordance error: 100 · | ||||||||||
| A | 25 | 30.79 | 30.69 | 31.45 | 33.90 | 31.32 | 30.35 | 30.31 | 30.13 | 30.68 |
| B | 25 | 31.32 | 31.14 | 31.92 | 35.69 | 32.41 | 31.65 | 31.32 | 30.90 | 32.10 |
| C | 25 | 29.88 | 29.86 | 30.49 | 32.52 | 30.64 | 31.87 | 31.87 | 31.63 | 32.35 |
| A | 50 | 31.00 | 30.97 | 31.33 | 34.06 | 31.60 | 33.06 | 32.90 | 32.43 | 33.64 |
| B | 50 | 35.13 | 35.20 | 35.71 | 37.75 | 36.02 | 38.35 | 38.39 | 38.01 | 38.98 |
| C | 50 | 37.36 | 37.39 | 37.44 | 40.11 | 38.15 | 39.71 | 39.80 | 39.14 | 40.49 |
| Monte-Carlo Standard Error of 100 · | ||||||||||
| A | 25 | 0.103 | 0.106 | 0.107 | 0.103 | 0.106 | 0.116 | 0.118 | 0.119 | 0.111 |
| B | 25 | 0.098 | 0.102 | 0.104 | 0.093 | 0.104 | 0.115 | 0.118 | 0.114 | 0.108 |
| C | 25 | 0.057 | 0.063 | 0.068 | 0.072 | 0.097 | 0.081 | 0.077 | 0.081 | 0.080 |
| A | 50 | 0.099 | 0.087 | 0.082 | 0.090 | 0.100 | 0.091 | 0.092 | 0.096 | 0.088 |
| B | 50 | 0.085 | 0.088 | 0.081 | 0.061 | 0.098 | 0.093 | 0.093 | 0.096 | 0.088 |
| C | 50 | 0.069 | 0.070 | 0.063 | 0.095 | 0.076 | 0.101 | 0.096 | 0.086 | 0.104 |
| Percent increase in , relative to minimum (0.00) | ||||||||||
| A | 25 | 2.2 | 1.9 | 4.4 | 12.5 | 4.0 | 0.8 | 0.6 | 0.0 | 1.8 |
| B | 25 | 1.4 | 0.8 | 3.3 | 15.5 | 4.9 | 2.4 | 1.4 | 0.0 | 3.9 |
| C | 25 | 0.0 | 0.0 | 2.1 | 8.9 | 2.6 | 6.7 | 6.7 | 5.9 | 8.3 |
| A | 50 | 0.1 | 0.0 | 1.2 | 10.0 | 2.0 | 6.7 | 6.2 | 4.7 | 8.6 |
| B | 50 | 0.0 | 0.2 | 1.7 | 7.5 | 2.5 | 9.2 | 9.3 | 8.2 | 11.0 |
| C | 50 | 0.0 | 0.1 | 0.2 | 7.4 | 2.1 | 6.3 | 6.5 | 4.8 | 8.4 |
| Rankings based on | ||||||||||
| A | 25 | 6 | 5 | 8 | 9 | 7 | 3 | 2 | 1 | 4 |
| B | 25 | 4 | 2 | 6 | 9 | 8 | 5 | 3 | 1 | 7 |
| C | 25 | 2 | 1 | 3 | 9 | 4 | 6 | 7 | 5 | 8 |
| A | 50 | 2 | 1 | 3 | 9 | 4 | 7 | 6 | 5 | 8 |
| B | 50 | 1 | 2 | 3 | 5 | 4 | 7 | 8 | 6 | 9 |
| C | 50 | 1 | 2 | 3 | 8 | 4 | 6 | 7 | 5 | 9 |
| 2.67 | 2.17 | 4.33 | 8.17 | 5.17 | 5.67 | 5.50 | 3.83 | 7.50 | ||
Descriptions of Scenarios A, B, and C are provided in Section 4.1.
p represents the number of predictor variables in each simulated data set.
RSF = random survival forest; CIF = conditional inference forest; ORSF = oblique random survival forest; ORSFCV = oblique random survival forest with internal cross-validation (see Section 3.2).
Apparent ties in concordance errors are a result of rounding errors.
The mean rankings according to for ORSFCV and ORSF were 4.67 and 4.17, respectively, out of the 9 learning algorithms we applied (Table 2). In comparison to the RSF and CIF, both the ORSF and ORSFCV recorded lower mean values of in each of the 6 simulated analyses. The absolute (percent) increase in the mean scaled value (see Section 4.3) of using the ORSFCV instead of the RSF and using the ORSFCV instead of the CIF were 1.57 (42.39%) and 0.45 (9.37%), respectively.
Table 2.
Mean integrated Brier scores for competing learning methods, aggregated over 100 simulations in three scenarios. The minimum Brier score for each simulated analysis is written in bold text.
| Scenario† | p ‡ | Ensemble Survival Trees§ | Proportional Hazards | Reference* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORSF | ORSFCV | CIF | RSF Xgboost | CoxBoost | Lasso | Ridge | Step | ||||
| Integrated Brier score: 100 · | |||||||||||
| A | 25 | 8.89 | 8.89 | 8.93 | 9.07 | 8.82 | 8.73 | 8.73 | 8.71 | 8.78 | 9.48 |
| B | 25 | 8.93 | 8.93 | 9.02 | 9.17 | 8.87 | 8.81 | 8.78 | 8.74 | 8.93 | 9.43 |
| C | 25 | 8.91 | 8.92 | 8.96 | 9.00 | 8.78 | 8.93 | 8.93 | 8.94 | 8.95 | 9.67 |
| A | 50 | 9.15 | 9.16 | 9.24 | 9.27 | 8.99 | 9.13 | 9.11 | 9.09 | 9.22 | 9.73 |
| B | 50 | 11.15 | 11.16 | 11.16 | 11.31 | 11.14 | 11.32 | 11.33 | 11.31 | 11.37 | 11.61 |
| C | 50 | 11.38 | 11.39 | 11.40 | 11.58 | 11.38 | 11.47 | 11.48 | 11.45 | 11.61 | 11.68 |
| Monte Carlo Standard Error of 100 · | |||||||||||
| A | 25 | 0.014 | 0.014 | 0.015 | 0.012 | 0.012 | 0.011 | 0.010 | 0.010 | 0.009 | 0.016 |
| B | 25 | 0.008 | 0.008 | 0.008 | 0.007 | 0.008 | 0.008 | 0.008 | 0.008 | 0.009 | 0.010 |
| C | 25 | 0.012 | 0.012 | 0.012 | 0.011 | 0.013 | 0.013 | 0.012 | 0.012 | 0.013 | 0.014 |
| A | 50 | 0.019 | 0.019 | 0.020 | 0.018 | 0.018 | 0.019 | 0.019 | 0.019 | 0.018 | 0.021 |
| B | 50 | 0.017 | 0.017 | 0.017 | 0.016 | 0.016 | 0.017 | 0.018 | 0.018 | 0.018 | 0.018 |
| C | 50 | 0.016 | 0.016 | 0.016 | 0.014 | 0.014 | 0.017 | 0.017 | 0.016 | 0.016 | 0.017 |
| Scaled values: 100 · [1 − /Reference] | |||||||||||
| A | 25 | 6.26 | 6.28 | 5.80 | 4.37 | 6.93 | 7.88 | 7.91 | 8.07 | 7.35 | 0.00 |
| B | 25 | 5.31 | 5.35 | 4.42 | 2.84 | 5.97 | 6.61 | 6.94 | 7.33 | 5.38 | 0.00 |
| C | 25 | 7.91 | 7.81 | 7.35 | 6.92 | 9.24 | 7.64 | 7.65 | 7.61 | 7.45 | 0.00 |
| A | 50 | 5.96 | 5.85 | 5.05 | 4.71 | 7.62 | 6.17 | 6.40 | 6.62 | 5.30 | 0.00 |
| B | 50 | 3.95 | 3.85 | 3.85 | 2.54 | 4.03 | 2.45 | 2.42 | 2.53 | 2.04 | 0.00 |
| C | 50 | 2.60 | 2.50 | 2.46 | 0.84 | 2.61 | 1.78 | 1.74 | 1.96 | 0.65 | 0.00 |
| Rankings based on | |||||||||||
| A | 25 | 7 | 6 | 8 | 9 | 5 | 3 | 2 | 1 | 4 | 10 |
| B | 25 | 7 | 6 | 8 | 9 | 4 | 3 | 2 | 1 | 5 | 10 |
| C | 25 | 2 | 3 | 8 | 9 | 1 | 5 | 4 | 6 | 7 | 10 |
| A | 50 | 5 | 6 | 8 | 9 | 1 | 4 | 3 | 2 | 7 | 10 |
| B | 50 | 2 | 4 | 3 | 5 | 1 | 7 | 8 | 6 | 9 | 10 |
| C | 50 | 2 | 3 | 4 | 8 | 1 | 6 | 7 | 5 | 9 | 10 |
| 4.17 | 4.67 | 6.50 | 8.17 | 2.17 | 4.67 | 4.33 | 3.50 | 6.83 | 10.00 | ||
Descriptions of Scenarios A, B, and C are provided in Section 4.1.
p represents the number of predictor variables in each simulated data set.
RSF = random survival forest; CIF = conditional inference forest; ORSF = oblique random survival forest; ORSFCV = oblique random survival forest with internal cross-validation (see Section 3.2).
The reference values are expected Brier scores of a prediction model that ignores all predictor variables, i.e., the Kaplan-Meier estimate calculated using the training data.
Apparent ties in Brier scores are a result of rounding errors.
5. Application to Real Data.
Here we describe and summarize results from a resampling experiment using data from six independent studies. We summarize each study, separately, in Section 5.1. We describe tuning parameters and the resampling procedure we applied in Sections 5.2 and 5.3, respectively. We present and summarize results in Section 5.5.
5.1. Data Sets.
For each of the data sets described in the following sections, study participants who were lost to follow-up or died from causes unrelated to the primary event(s) were censored at time of last contact or time of death, respectively. Characteristics (e.g., number of participants, number of predictors, percent censored) of each data set are tabulated and presented alongside the characteristics of simulated datasets in Table 3.
Table 3.
Data set summary
| Follow-up times§ | ||||||||
|---|---|---|---|---|---|---|---|---|
| Data | Event Type | 25% | 50% | 75% | % Censored | N | M | p |
| A25 | Simulated | 3.9 | 10.0 | 10.0 | 55.4 | 500 | 1000 | 25 |
| B25 | Simulated | 3.9 | 10.0 | 10.0 | 57.5 | 500 | 1000 | 25 |
| C25 | Simulated | 2.8 | 10.0 | 10.0 | 50.6 | 500 | 1000 | 25 |
| A50 | Simulated | 3.8 | 10.0 | 10.0 | 55.2 | 500 | 1000 | 50 |
| B50 | Simulated | 3.5 | 10.0 | 10.0 | 56.2 | 500 | 1000 | 50 |
| C50 | Simulated | 2.6 | 10.0 | 10.0 | 49.5 | 500 | 1000 | 50 |
| PBC | Death | 3.0 | 4.7 | 7.2 | 61.5 | 209 | 209 | 17 |
| GBSG2 | Death or relapse | 1.6 | 3.0 | 4.6 | 56.4 | 342 | 342 | 8 |
| GEBC | Death or relapse | 1.8 | 2.8 | 4.1 | 78.2 | 307 | 307 | 1690 |
| MBC | Death or relapse | 2.3 | 5.3 | 8.8 | 56.4 | 39 | 39 | 4707 |
| JHS1 | CHD or stroke | 10.8 | 11.7 | 12.6 | 90.8 | 915 | 914 | 58 |
| JHS2 | Heart Failure | 9.2 | 10.0 | 10.0 | 93.8 | 904 | 904 | 58 |
| JHS3 | Death | 13.0 | 13.9 | 14.7 | 81.3 | 944 | 943 | 58 |
| REGARDS1 | CHD or stroke | 5.2 | 8.1 | 9.9 | 92.2 | 4485 | 4485 | 67 |
| REGARDS2 | Heart Failure | 5.0 | 8.1 | 9.9 | 97.3 | 4485 | 4484 | 67 |
| REGARDS3 | Death | 6.0 | 8.4 | 10.0 | 87.2 | 4485 | 4485 | 67 |
All follow-up times are in years.
5.1.1. Primary biliary cirrhosis (PBC) data.
The PBC data and their description are taken from Appendix D of Fleming and Harrington (2011). PBC of the liver is a rare and fatal disease of unknown cause. These data were collected for the Mayo Clinic trial in PBC of the liver conducted between January 1974 and May 1984 comparing the drug D-penicillamine with a placebo.
5.1.2. German breast cancer study group (GBSG2) data.
The GBSG2 data and their description are taken from Schumacher et al. (1994). In 1984, the GBSG2 started a multicenter randomized clinical trial to compare the effectiveness of three versus six cycles of 500 mg/m2 cyclophosphamide, 40 mg/m2 methotrexate, and 600 mg/m2 fluorouracil on day 1 and 8 starting perioperatively with or without tamoxifen (3 × 10 mg/d for 2 years).
5.1.3. Gene expression breast cancer (GEBC) data.
The GEBC data and their description are taken from Gene Expression Omnibus database and the analysis by Ternès et al. (2017), respectively. to identify treatment-effect modifiers in 614 breast cancer patients (Hatzis et al., 2011; Desmedt et al., 2011) receiving anthracycline-based adjuvant chemotherapy with (n = 507) or without (n = 107) taxane. For the current analysis, we use pre-processed data from Ternès et al. (2017), who applied frozen robust multiarray (McCall, Bolstad and Irizarry, 2010) and cross-platform normalization (Shabalin et al., 2008), and standardized the remaining 1689 genes.
5.1.4. Microarray breast cancer (MBC) data.
The MBC data and their description are taken from (Van’t Veer et al., 2002). Gene expression profiling was conducted on 78 sporadic lymph-node-negative patients to search for a prognostic signature in their gene expression profiles. Forty-four patients remained free of breast cancer after their initial diagnosis for an interval of at least 5 years (good prognosis group, mean follow-up of 8.7 years), and 34 patients developed distant metastases within 5 years (poor prognosis group, mean time to metastases 2.5 years).
5.1.5. Jackson Heart Study (JHS) data.
The JHS is a population-based prospective cohort study designed to examine the etiology of cardiovascular disease (CVD) (e.g., stroke, coronary heart disease, heart failure) and related risk factors among African Americans.(Taylor Jr et al., 2005) In brief, 5,306 non-institutionalized African-American participants aged ≥ 20 years were recruited from the Jackson, Mississippi, metropolitan area between 2000 and 2004. For the current analyses, we incorporated data from JHS participants who provided complete records of age, sex, anthropometric measures, alcohol/smoking/dietary/exercise habits, medication use, zip code, blood pressure, diabetes, cholesterol, high and low density lipoproteins, triglycerides, electrocardiograms, estimated glomerular filtration rate (eGFR) (Levey et al., 2009), insurance, and history of CVD at baseline. We conducted three separate analyses using these data; JHS1 considered composite events of stroke or coronary heart disease, JHS2 considered heart failure events, and JHS3 considered all-cause mortality events.
5.1.6. The REasons for Geographic And Racial Differences in Stroke (REGARDS) Study.
The REGARDS study was designed to investigate reasons underlying the higher rate of stroke mortality among blacks compared with whites, and among residents of the Southeastern US compared with other US regions (Howard et al., 2005). A total of 30,239 adults from the 48 contiguous US states and the District of Columbia were enrolled between January 2003 and October 2007. For the current analysis, we incorporated data from 8,970 participants who provided complete records of age, sex, race, anthropomorphic measures, alcohol/smoking/dietary/exercise habits, quality of life, self-reported history of CVD, echocardiogram, medication use, blood measures, blood pressure, urine albumin and creatinine, and eGFR. We conducted three separate analyses using these data; REGARDS1 considered composite events of stroke or coronary heart disease (Safford et al., 2012), REGARDS2 considered heart failure events , and REGARDS3 considered all-cause mortality events.
5.2. Tuning Parameters.
We applied the same learning algorithms and corresponding tuning parameter specifications as described in Section 4.2.
5.3. Resampling.
Results from the current resampling experiment are based on 250 replicates of bootstrap cross-validation (Mogensen, Ishwaran and Gerds, 2012, Sec. 4.2). In each replicate, for each analysis (e.g., PBC, GBSG2, JHS1, etc.), we
randomly allocated roughly one half of the data to a training set, and used the rest of the data for testing.
trained each competing method using the training data set.
computed and using each method’s predicted survival curves for observations in the testing data.
5.4. Monte-Carlo Error Relative to Forest Size.
Using the PBC data, we assessed the Monte-Carlo error (seed effect) of the predicted survival curves relative to the number of trees in the ORSF. We first identified three classes of patients using the protoclust package Bien and Tibshirani (2019). We identified one prototype patient in each class, and used these three patients as a hold-out set. We developed an ORSF with 10, 100, and 1000 trees, and then predicted survival curves for patients in the hold-out set. We replicated this step 250 times and plotted each survival curve to visualize Monte-Carlo error for the three separate forest sizes.
5.5. Results.
The three methods that achieved the lowest values of were the ORSFCV, the ORSF, and the CIF (mean ranks of 1.70, 1.80 and 3.20, respectively) (Table 4). Distributions of were consistent among competing learning methods with the exception of the penalized Cox PH models, which had higher variability of in analyses of JHS and REGARDS data (Figure 1). The difference in between the ORSFCV and the RSF was minimal in the GBSG2 analysis (27.86 and 28.71, respectively) and maximal in the GEBC analysis (25.08 and 31.19, respectively). The absolute (percent) increase in the mean value of from using the RSF instead of the ORSFCV or the CIF instead of the ORSFCV was 1.79 (9.08%) and 0.16 (0.82%), respectively. The variability and overall shape of time-dependent trajectories of were similar among the RSF, CIF, and ORSF (Figure 3).
Table 4.
Mean concordance index error for competing learning methods, aggregated over 100 replicates of bootstrap cross-validation in five independent data sets. The minimum concordance error value for each analysis is written in bold text.
| Ensemble Survival Trees§ | Proportional Hazards | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Scenario† | ORSF | ORSFCV | CIF | RSF | Xgboost | CoxBoost | Lasso | Ridge | Step |
| Concordance error: 100 · | |||||||||
| GEBC | 24.98 | 25.08 | 25.79 | 31.19 | 27.08 | 28.62 | 28.59 | 26.47 | – |
| GBSG2 | 28.10 | 27.86 | 28.40 | 28.71 | 28.09 | 30.24 | 30.30 | 29.99 | 29.88 |
| JHS1 | 17.67 | 17.58 | 17.74 | 19.31 | 17.84 | 18.69 | 18.68 | 18.37 | 18.74 |
| JHS3 | 17.61 | 17.57 | 17.91 | 18.40 | 17.63 | 18.54 | 18.90 | 19.66 | 18.07 |
| JHS2 | 13.13 | 13.16 | 13.42 | 13.45 | 13.61 | 17.51 | 18.86 | 14.43 | 14.83 |
| PBC | 12.25 | 12.29 | 12.80 | 13.90 | 12.87 | 13.53 | 13.72 | 12.82 | 15.15 |
| REGARDS1 | 21.81 | 21.83 | 21.84 | 22.69 | 22.15 | 22.68 | 22.57 | 22.35 | 24.58 |
| REGARDS3 | 18.69 | 18.80 | 18.82 | 19.01 | 18.55 | 19.27 | 20.33 | 30.62 | 19.32 |
| REGARDS2 | 13.91 | 13.86 | 14.21 | 16.08 | 14.01 | 15.64 | 17.87 | 29.38 | 16.93 |
| MBC | 29.41 | 29.27 | 27.99 | 32.47 | 31.28 | 40.62 | 39.79 | 35.14 | – |
| Percent increase in , relative to minimum (0.00) | |||||||||
| GEBC | 0.0 | 0.4 | 3.3 | 24.9 | 8.4 | 14.6 | 14.5 | 6.0 | – |
| GBSG2 | 0.9 | 0.0 | 1.9 | 3.0 | 0.8 | 8.5 | 8.7 | 7.6 | 7.2 |
| JHS1 | 0.5 | 0.0 | 0.9 | 9.8 | 1.5 | 6.3 | 6.3 | 4.5 | 6.6 |
| JHS3 | 0.2 | 0.0 | 1.9 | 4.7 | 0.3 | 5.5 | 7.5 | 11.9 | 2.8 |
| JHS2 | 0.0 | 0.2 | 2.2 | 2.5 | 3.7 | 33.4 | 43.7 | 10.0 | 13.0 |
| PBC | 0.0 | 0.3 | 4.5 | 13.5 | 5.1 | 10.5 | 12.0 | 4.7 | 23.7 |
| REGARDS1 | 0.0 | 0.1 | 0.1 | 4.1 | 1.6 | 4.0 | 3.5 | 2.5 | 12.7 |
| REGARDS3 | 0.7 | 1.4 | 1.5 | 2.5 | 0.0 | 3.9 | 9.6 | 65.0 | 4.2 |
| REGARDS2 | 0.4 | 0.0 | 2.6 | 16.1 | 1.1 | 12.8 | 29.0 | 112.0 | 22.2 |
| MBC | 5.1 | 4.6 | 0.0 | 16.0 | 11.7 | 45.1 | 42.2 | 25.5 | – |
| Rankings based on | |||||||||
| GEBC | 1 | 2 | 3 | 8 | 5 | 7 | 6 | 4 | – |
| GBSG2 | 3 | 1 | 4 | 5 | 2 | 8 | 9 | 7 | 6 |
| JHS1 | 2 | 1 | 3 | 9 | 4 | 7 | 6 | 5 | 8 |
| JHS3 | 2 | 1 | 4 | 6 | 3 | 7 | 8 | 9 | 5 |
| JHS2 | 1 | 2 | 3 | 4 | 5 | 8 | 9 | 6 | 7 |
| PBC | 1 | 2 | 3 | 8 | 5 | 6 | 7 | 4 | 9 |
| REGARDS1 | 1 | 2 | 3 | 8 | 4 | 7 | 6 | 5 | 9 |
| REGARDS3 | 2 | 3 | 4 | 5 | 1 | 6 | 8 | 9 | 7 |
| REGARDS2 | 2 | 1 | 4 | 6 | 3 | 5 | 8 | 9 | 7 |
| MBC | 3 | 2 | 1 | 5 | 4 | 8 | 7 | 6 | – |
| Mean | 1.80 | 1.70 | 3.20 | 6.40 | 3.60 | 6.90 | 7.40 | 6.40 | 7.25 |
RSF = random survival forest; CIF = conditional inference forest; ORSF = oblique random survival forest; ORSFCV = oblique random survival forest with internal cross-validation (see Section 3.2).
GEBC = gene expression breast cancer; MBC = microarray breast cancer; GBSG2 = German breast cancer study group; PBC = primary biliary cirrhosis; JHS = Jackson heart study; REGARDS = REasons for Geographic And Racial Differences in Stroke.
Apparent ties in concordance errors are a result of rounding errors.
Fig 1.
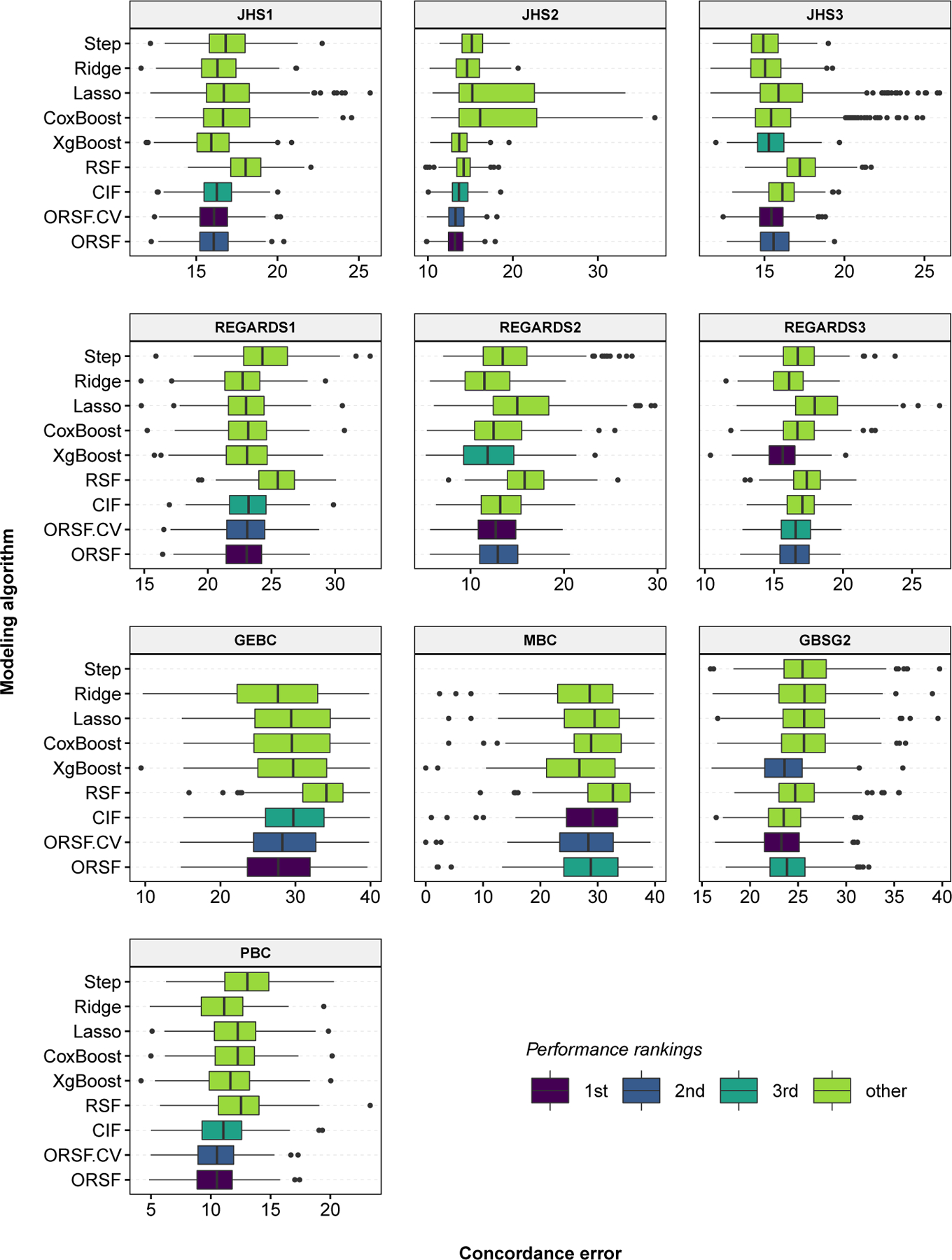
Concordance error values, aggregated over 250 replications of bootstrap cross-validation, for competing methods in 10 analyses of data with right-censored time-to-event outcomes.
Fig 3.
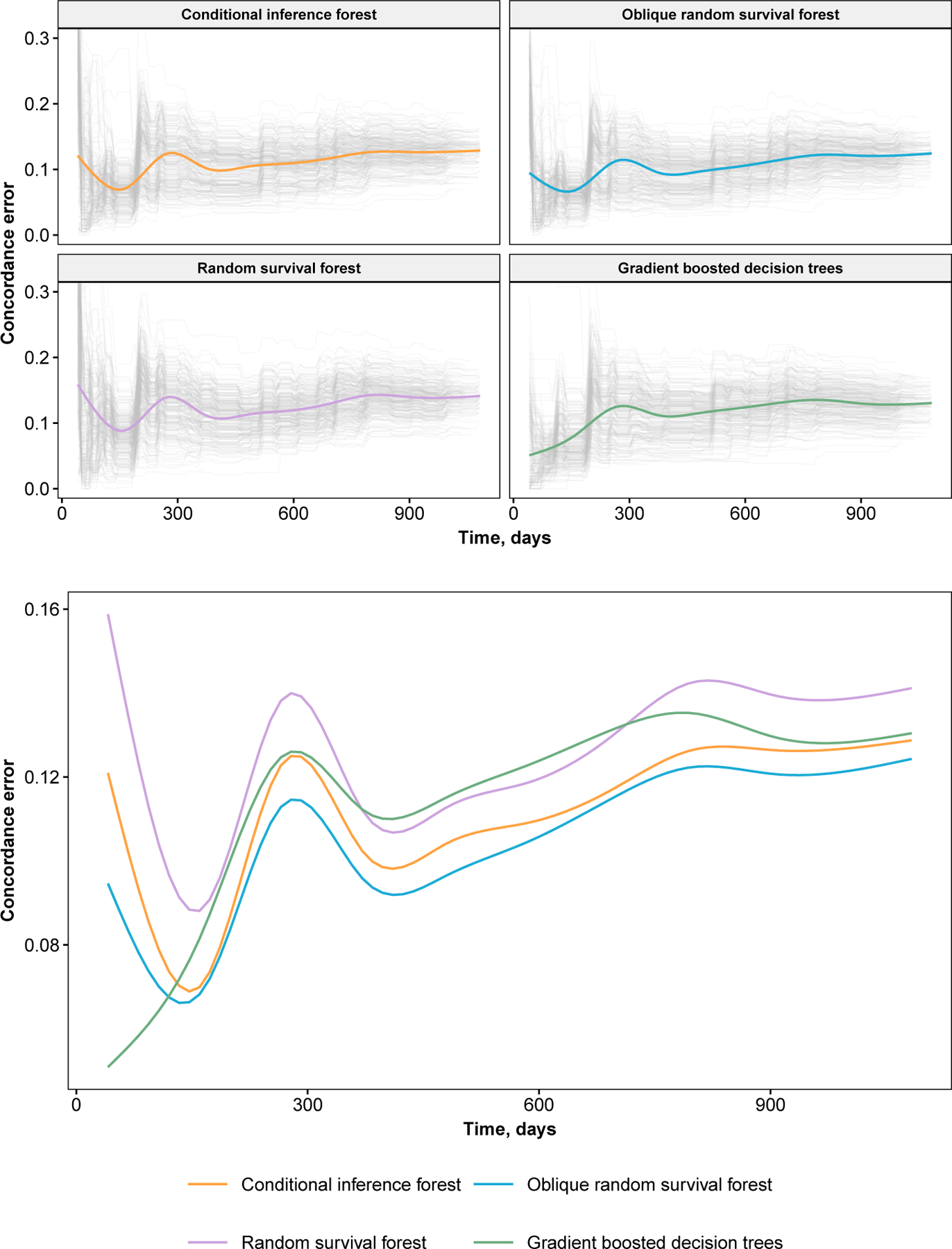
Concordance error values for the primary biliary cirrhosis analysis. Results from individual replications of bootstrap cross-validation are shown as grey trajectories. Smoothed estimates of average values are colored. Error values are plotted from baseline to median event time.
The three methods that achieved the lowest values of were the ORSF, gradient boosted decision trees, and the ORSFCV (mean ranks of 2.30, 2.50 and 3.90, respectively) (Table 5). Distributions of were consistent among the competing learning methods with the exception of the penalized Cox PH models, which had higher variability of in analyses of JHS and REGARDS data (Figure 2). The difference in between the ORSF and the RSF was minimal in the JHS3 analysis (3.228 and 3.232, respectively) and maximal in the GEBC analysis (4.20 and 4.44, respectively). The absolute (percent) increase in the mean scaled value (see Section 4.3) of using the ORSF instead of the RSF or instead of the CIF was 1.54 (21.67%) and 0.77 (9.79%), respectively.
Table 5.
Mean integrated Brier scores for competing learning methods, aggregated over 100 replicates of bootstrap cross-validation in five independent data sets. The minimum Brier score for each analysis is written in bold text.
| Scenario† | Ensemble Survival Trees§ | Proportional Hazards | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ORSF | ORSFCV | CIF | RSF | Xgboost | CoxBoost | Lasso | Ridge | Step | ||
| Integrated Brier score: 100 · | ||||||||||
| GEBC | 4.20 | 4.20 | 4.23 | 4.44 | 4.25 | 4.27 | 4.27 | 4.22 | – | 4.36 |
| GBSG2 | 7.10 | 7.12 | 7.10 | 7.21 | 7.20 | 7.51 | 7.51 | 7.49 | 7.47 | 7.77 |
| JHS1 | 2.31 | 2.31 | 2.33 | 2.32 | 2.31 | 2.35 | 2.36 | 2.34 | 2.35 | 2.49 |
| JHS3 | 3.23 | 3.23 | 3.28 | 3.23 | 3.15 | 3.20 | 3.23 | 3.20 | 3.17 | 3.67 |
| JHS2 | 2.30 | 2.30 | 2.33 | 2.29 | 2.31 | 2.41 | 2.41 | 2.37 | 2.38 | 2.55 |
| PBC | 5.71 | 5.82 | 5.79 | 5.77 | 5.73 | 6.08 | 6.12 | 5.92 | 6.45 | 8.09 |
| REGARDS1 | 1.69 | 1.69 | 1.69 | 1.70 | 1.68 | 1.70 | 1.70 | 1.70 | 1.73 | 1.76 |
| REGARDS3 | 2.21 | 2.22 | 2.23 | 2.20 | 2.16 | 2.18 | 2.21 | 2.26 | 2.21 | 2.43 |
| REGARDS2 | 0.63 | 0.63 | 0.64 | 0.64 | 0.63 | 0.64 | 0.64 | 0.64 | 0.66 | 0.66 |
| MBC | 9.07 | 9.08 | 9.11 | 9.62 | 9.16 | 9.65 | 9.67 | 9.33 | – | 8.91 |
| Scaled values: 100 · [1 − /Reference] | ||||||||||
| GEBC | 3.58 | 3.53 | 3.03 | −1.94 | 2.51 | 1.96 | 1.95 | 3.20 | – | 0.00 |
| GBSG2 | 8.60 | 8.36 | 8.68 | 7.18 | 7.33 | 3.40 | 3.29 | 3.55 | 3.82 | 0.00 |
| JHS1 | 7.46 | 7.37 | 6.75 | 6.84 | 7.46 | 5.67 | 5.53 | 6.21 | 5.59 | 0.00 |
| JHS3 | 12.12 | 12.05 | 10.79 | 12.01 | 14.27 | 12.84 | 12.10 | 12.95 | 13.80 | 0.00 |
| JHS2 | 9.95 | 9.74 | 8.54 | 10.39 | 9.40 | 5.45 | 5.33 | 7.01 | 6.63 | 0.00 |
| PBC | 29.40 | 28.03 | 28.42 | 28.59 | 29.10 | 24.81 | 24.29 | 26.83 | 20.28 | 0.00 |
| REGARDS1 | 3.99 | 3.79 | 3.58 | 3.11 | 4.27 | 3.20 | 3.23 | 3.43 | 1.41 | 0.00 |
| REGARDS3 | 8.93 | 8.50 | 7.85 | 9.16 | 11.04 | 10.24 | 8.86 | 6.63 | 8.88 | 0.00 |
| REGARDS2 | 4.38 | 4.14 | 3.55 | 3.78 | 5.31 | 3.74 | 2.97 | 2.47 | 0.76 | 0.00 |
| MBC | −1.76 | −1.85 | −2.27 | −7.90 | −2.73 | −8.22 | −8.49 | −4.66 | – | 0.00 |
| Ranks in each data set of competing methods based on | ||||||||||
| GEBC | 1 | 2 | 4 | 9 | 5 | 6 | 7 | 3 | – | 8 |
| GBSG2 | 2 | 3 | 1 | 5 | 4 | 8 | 9 | 7 | 6 | 10 |
| JHS1 | 2 | 3 | 5 | 4 | 1 | 7 | 9 | 6 | 8 | 10 |
| JHS3 | 5 | 7 | 9 | 8 | 1 | 4 | 6 | 3 | 2 | 10 |
| JHS2 | 2 | 3 | 5 | 1 | 4 | 8 | 9 | 6 | 7 | 10 |
| PBC | 1 | 5 | 4 | 3 | 2 | 7 | 8 | 6 | 9 | 10 |
| REGARDS1 | 2 | 3 | 4 | 8 | 1 | 7 | 6 | 5 | 9 | 10 |
| REGARDS3 | 4 | 7 | 8 | 3 | 1 | 2 | 6 | 9 | 5 | 10 |
| REGARDS2 | 2 | 3 | 6 | 4 | 1 | 5 | 7 | 8 | 9 | 10 |
| MBC | 2 | 3 | 4 | 7 | 5 | 8 | 9 | 6 | – | 1 |
| Mean | 2.30 | 3.90 | 5.00 | 5.20 | 2.50 | 6.20 | 7.60 | 5.90 | 6.88 | 8.90 |
RSF = random survival forest; CIF = conditional inference forest; ORSF = oblique random survival forest; ORSFCV = oblique random survival forest with internal cross-validation (see Section 3.2).
GEBC = gene expression breast cancer; MBC = microarray breast cancer; GBSG2 = German breast cancer study group; PBC = primary biliary cirrhosis; JHS = Jackson heart study; REGARDS = REasons for Geographic And Racial Differences in Stroke.
The reference values are expected Brier scores of a prediction model that ignores all predictor variables, i.e., the Kaplan-Meier estimate calculated using the training data.
Apparent ties in Brier scores are a result of rounding errors.
Fig 2.
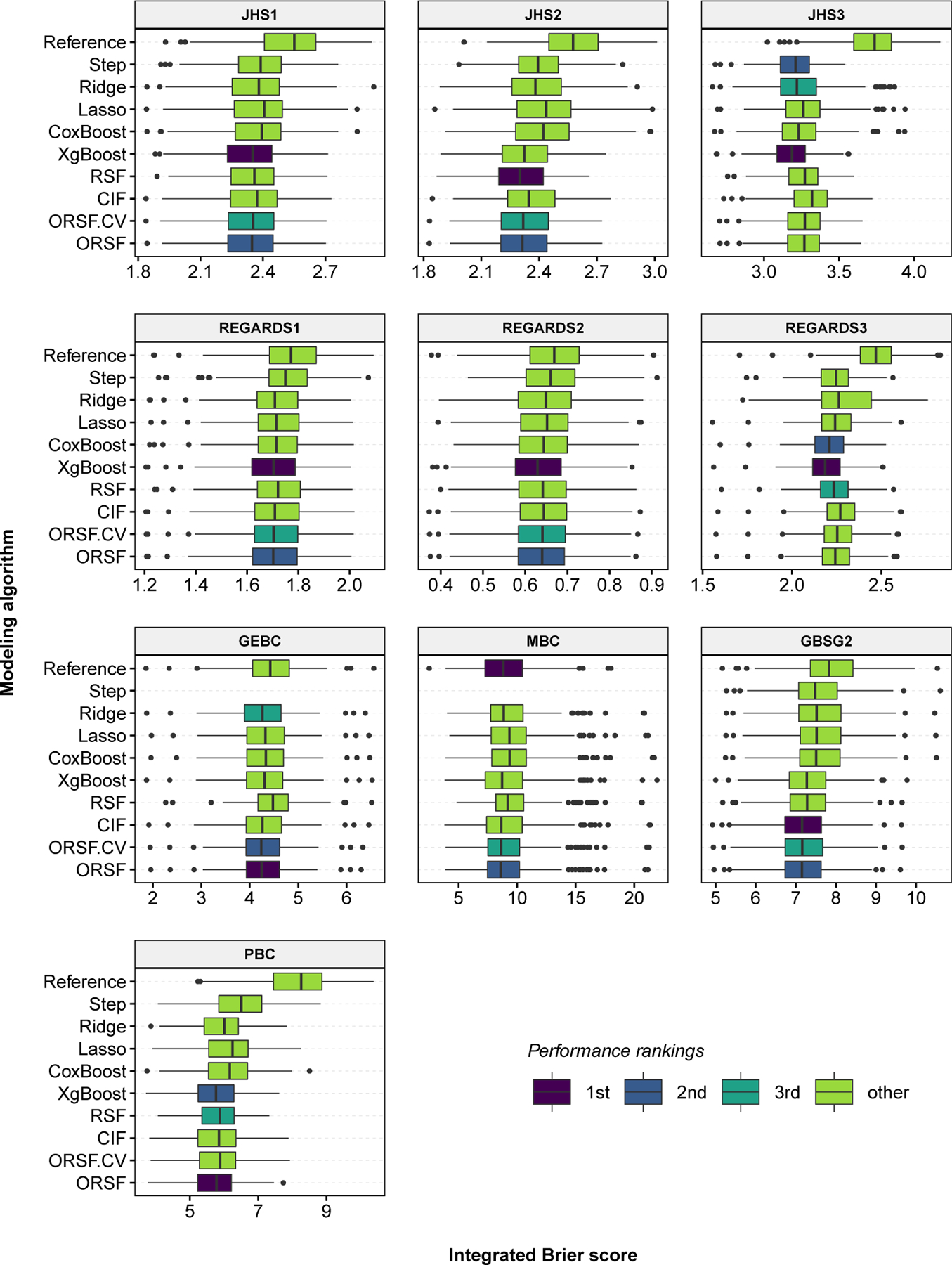
Integrated Brier score values, aggregated over 250 replications of bootstrap cross-validation, for competing methods in 10 analyses of data with right-censored time-to-event outcomes.
Monte-Carlo error was adequate when 1000 trees were used to fit the ORSF (Figure 4). The variability in predicted survival curves was noticeably higher when 10 or 100 trees were used to fit an ORSF.
Fig 4.
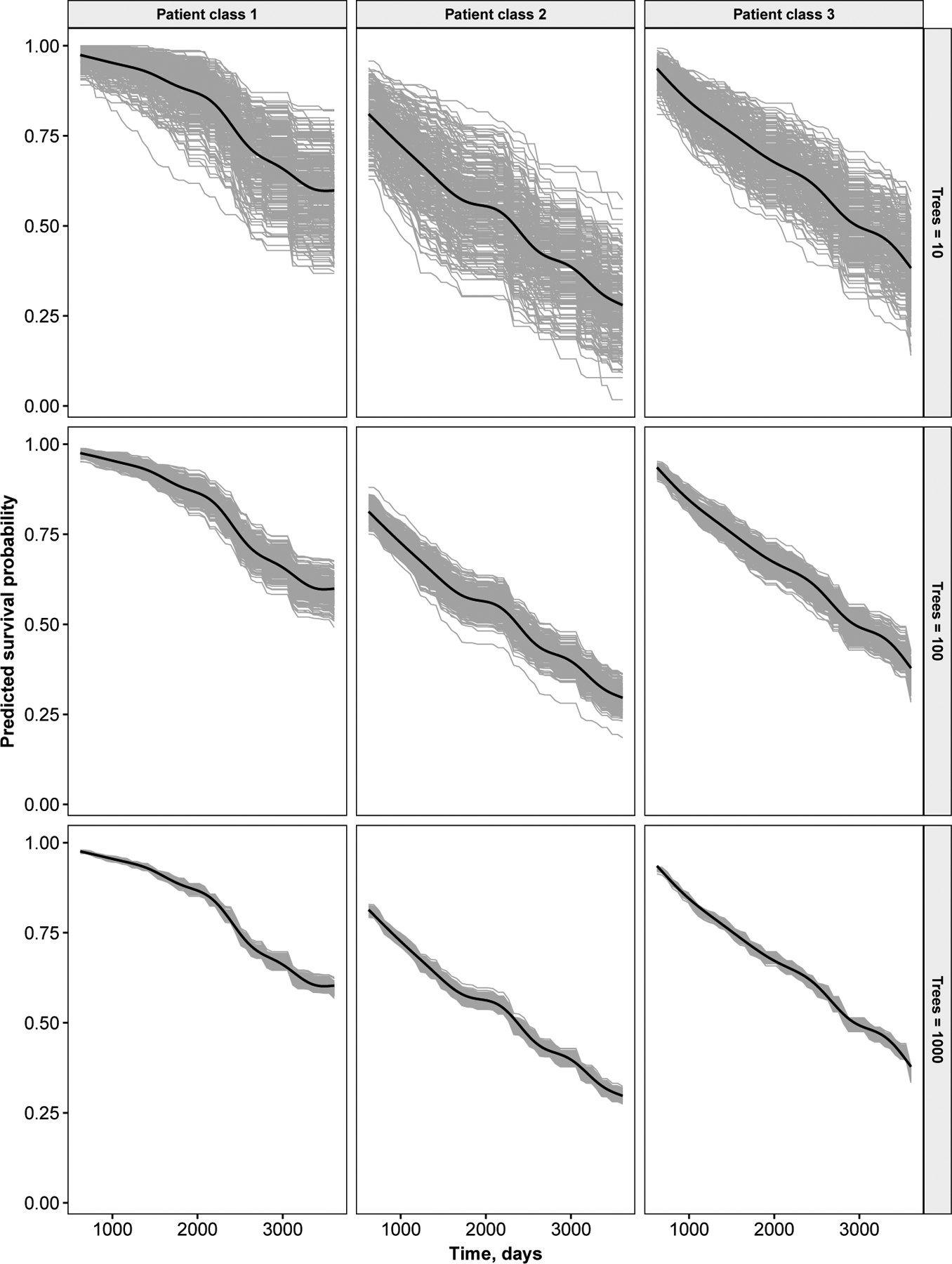
Monte-Carlo error (seed effect) for the primary biliary cirrhosis analysis. Results are shown for three prototypical patients. Grey curves are individual predictions from an individual forest. Smoothed estimates of average values are drawn as black curves.
6. Overall Performance Comparisons.
Here we describe a formal comparison of the performance and computational requirements of each method that was applied in Section 5, incorporating results from each of the real data sets described. To assess the relative performance of each method, we ranked the methods in each data set, separately, giving a rank of 1 to the method with the lowest error, a rank of 2 to the method with the second lowest error, and so on. We recorded the rankings in this manner using both and as the metric for error. In Section 6.1, we describe the procedure used to draw inferences from these rankings. We summarize performance of the learning algorithms in Section 6.2 and the computational resources required to run each algorithm in Section 6.3.
6.1. Statistical comparisons of ranks.
We applied a modification of Friedman’s non-parametric rank test (Friedman, 1937), which compares the average ranks among a set of classifiers over a collection of data sets. Let be the rank of the jth of k classifiers on the ith of D datasets. Friedman’s test compares the average ranks of the competing classifiers, . Under the null hypothesis of equivalent performance, each classifier will have a mean rank of (k + 1)/2, with variance (k2 − 1)/(12D) and
| (6.1) |
where Fp,q denotes an F distribution with p and q numerator and denominator degrees of freedom, respectively, and
| (6.2) |
Iman and Davenport (1980) derived FF to correct the overly conservative statistic (Demšar, 2006). If the null hypothesis of overall equivalent performance is rejected, post-hoc pairwise comparisons can be conducted using
| (6.3) |
to compare the average ranking of the ith and jth classifiers.
6.2. Results.
Among the nine learning algorithms we compared, the hypothesis of equivalence in performance was rejected (FF = 74.1 [p < 0.001] and 17.2 [p < 0.001] for concordance error and integrated Brier scores, respectively) using the overall FF test statistic proposed by Iman and Davenport (1980). Figure 5 shows a color-coded comparison matrix. Diagonal cells are split in two, with the mean rank according to printed in the upper left and the mean rank according to printed in the lower right. The upper left and lower right sections of the matrix show two sided p-values corresponding to pairwise comparisons of the mean ranks between two methods using the rankings according to and , respectively. For example, the mean rankings according to of the ORSFCV and the CIF were 1.70 and 3.20, respectively, and the p-value corresponding to a test of rank equivalence between these two methods was 0.121.
Fig 5.
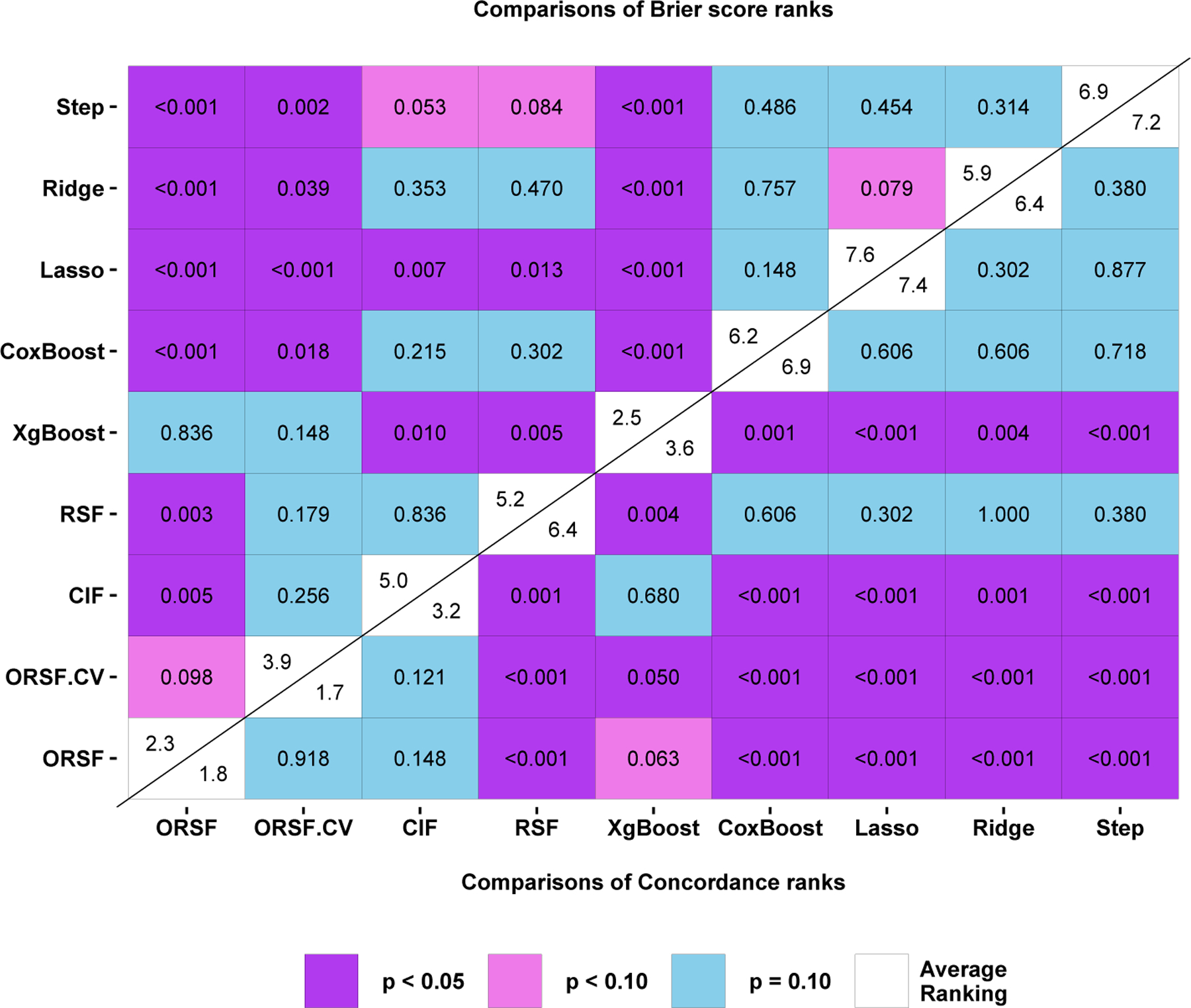
Overall performance comparison between oblique random survival forests and competing learning methods for prediction of the data described in Sections 4 and 5.
After ranking each method in each analysis according to , the ORSFCV had the lowest mean ranking: 1.70. The ORSF and CIF had the second and third lowest mean rankings, 1.80 and 3.20, respectively. For all pairwise comparisons involving for the ORSFCV, excluding comparison with the ORSF (p = 0.918) and CIF, a statistically significant (p < 0.05) difference in mean rankings was observed.
After ranking each method in each analysis according to , the ORSF had the lowest mean ranking (2.30) followed by gradient boosted decision trees (mean rank of 2.50) and the ORSFCV (mean rank of 3.90). For all pairwise comparisons involving for the ORSF, excluding comparisons with the ORSFCV (p = 0.098) and gradient boosted decision trees (p = 0.836), a statistically significant (p < 0.05) difference in mean rankings was observed.
6.3. Computing time.
We recorded the mean amount of time required to fit each learning algorithm in each analysis using real and simulated data (Table 6). As fitting an ORSF requires fitting regularized Cox PH models in each non-terminal node, the mean time of computation for the ORSF was roughly 192 and 32 times that of the RSF and CIF, respectively. When nested cross-validation was used, the mean time of computation for the ORSF was roughly 605 and 102 times that of the RSF and CIF, respectively.
Table 6.
Mean computation time, in seconds, required to fit each learning algorithm in each analysis
| Analysis | ORSF | ORSF.CV | CIF | RSF | Xgboost | CoxBoost | Lasso | Ridge | Step |
|---|---|---|---|---|---|---|---|---|---|
| GEBC | 1265.7 | 5848.1 | 9.8 | 6.5 | 275.1 | 126.3 | 20.0 | 14.4 | 0.0 |
| GBSG2 | 401.6 | 1033.3 | 6.0 | 1.9 | 234.4 | 24.9 | 0.3 | 0.3 | 0.8 |
| PBC | 204.0 | 693.1 | 2.1 | 0.8 | 276.5 | 8.2 | 0.2 | 0.3 | 4.2 |
| MBC | 101.8 | 291.9 | 19.2 | 0.8 | 316.7 | 27.7 | 3.7 | 4.0 | 0.0 |
| JHS1 | 1621.8 | 5005.0 | 56.3 | 6.4 | 293.6 | 203.0 | 3.3 | 3.5 | 41.5 |
| JHS3 | 2556.5 | 7592.1 | 62.2 | 10.6 | 431.6 | 413.3 | 2.9 | 3.1 | 49.5 |
| JHS2 | 1291.9 | 4385.3 | 50.5 | 5.4 | 265.3 | 159.3 | 3.3 | 3.3 | 36.4 |
| REGARDS1 | 1852.3 | 5142.9 | 132.4 | 11.8 | 533.6 | 267.7 | 8.4 | 8.0 | 52.6 |
| REGARDS3 | 2655.5 | 6873.5 | 140.6 | 19.1 | 525.7 | 476.2 | 4.7 | 5.1 | 67.5 |
| REGARDS2 | 909.3 | 3127.5 | 120.1 | 6.3 | 342.7 | 109.5 | 16.2 | 12.8 | 48.9 |
| SIM3.25 | 1448.9 | 4316.8 | 8.5 | 7.1 | 239.7 | 79.8 | 0.3 | 0.3 | 5.6 |
| SIM3.50 | 1593.7 | 5123.4 | 7.9 | 7.7 | 191.1 | 92.9 | 0.4 | 0.4 | 15.0 |
| SIM2.25 | 1084.2 | 3605.3 | 6.7 | 4.4 | 158.1 | 56.8 | 0.3 | 0.3 | 6.1 |
| SIM2.50 | 1311.2 | 4360.8 | 7.8 | 6.6 | 199.7 | 82.5 | 0.4 | 0.5 | 28.3 |
| SIM1.25 | 1176.6 | 3735.7 | 7.8 | 5.8 | 242.5 | 68.7 | 0.3 | 0.3 | 9.6 |
| SIM1.50 | 1342.1 | 4583.2 | 8.3 | 7.5 | 313.8 | 87.0 | 0.5 | 0.5 | 30.7 |
| Overall | 1301.1 | 4107.4 | 40.4 | 6.8 | 302.5 | 142.7 | 4.1 | 3.6 | 24.8 |
7. Application to Jackson Heart Study.
Here we apply the ORSF and the CIF in a comparative example using data from the Jackson Heart Study (JHS). A description of the JHS is given in Section 5.1.5. In the current example, we included 5126 JHS participants who consented to provide follow-up information on ASCVD events. We randomly split the baseline data from these JHS participants into a training set (N = 3000) and a testing set (M = 2126). We imputed missing values in both sets, separately, using a k-nearest neighbors algorithm (Kowarik and Templ, 2016). Using the training data, we developed prediction rules based on the ORSF and CIF, separately. These prediction rules were applied to compute 10-year predicted risk of ASCVD events for JHS participants in the testing data. Using these values of predicted risk, we created variable dependence and partial dependence plots (Section 7.1) that directly compare expected predictions from the ORSF and CIF without and with adjustment for confounding effects, respectively (Friedman, 2001). For variable dependence plots, we included 10-year predicted risk of ASCVD events according to the Pooled Cohort risk equations, a well-known risk prediction equation that is recommended by Whelton et al. (2018) for clinical assessment of 10-year predicted risk for ASCVD events. Last, we compared predicted risk curves according to the ORSF and CIF for four individual JHS participants from the testing data.
7.1. Variable dependence and partial dependence.
Variable dependence plots render the expected value of an outcome (y-axis) relative to the observed values of an input variable (x-axis), using the unaltered training data. Although variable dependence plots illustrate observed relationships, they do not account for variation in covariates of interest apart from the input variable. Partial dependence plots show predicted risk as a function of the variables in a designated subset of input variables by averaging effects of the designated variables over the observed distribution of variables not in the designated set. Let
denote the designated subset of r < p predictor variables. Define the complement set of predictor variables z{s} such that
The predicted risk function, , depends on variables in both subsets. Friedman (2001) shows that conditioning on the variables in z{s} allows to be considered as a function of only the variables chosen in z{r}. Moreover, the partial dependence of on variables in z{r} can be computed by averaging over the observed values of z{s}, i.e.
7.2. Results.
We identified four explanatory variables to analyze for the current application of the ORSF and CIF to data from the JHS: (1) left ventricular mass (LVM) in g/m2.7, (2) age in years, (3) estimated glomerular filtration rate (eGFR) (Levey et al., 2009) in ml/min/1.73m2, and (4) systolic blood pressure (SBP) in mm Hg, measured in a clinical setting. We performed routine descriptive analyses of these variables in the training and testing datasets, separately (Table 7 and Figure 6).
Table 7.
Descriptive statistics for four variables in the Jackson Heart Study.
| Summary measures§ | Percentile | ||||||
|---|---|---|---|---|---|---|---|
| Min. | 25% | 50% | 75% | Max. | Mean | SD | |
| Testing data | |||||||
| Age, years | 20.60 | 45.10 | 55.20 | 64.40 | 93.10 | 55.04 | 12.77 |
| eGFR, ml/min/1.73 m2 | 4.29 | 81.48 | 96.90 | 110.25 | 153.46 | 94.94 | 22.24 |
| Left ventricular mass, g/m2 | 20.84 | 59.55 | 69.17 | 81.33 | 215.50 | 72.88 | 20.27 |
| Systolic blood pressure, mm Hg | 77.98 | 115.58 | 125.66 | 136.67 | 228.36 | 127.35 | 16.91 |
| Training data | |||||||
| Age, years | 21.00 | 45.70 | 56.10 | 65.20 | 95.50 | 55.58 | 12.93 |
| eGFR, ml/min/1.73 m2 | 4.14 | 80.47 | 95.24 | 109.09 | 151.88 | 93.59 | 21.81 |
| Left ventricular mass, g/m2 | 5.28 | 60.16 | 69.26 | 82.56 | 225.45 | 73.30 | 20.10 |
| Systolic blood pressure, mm Hg | 86.24 | 116.26 | 125.66 | 136.67 | 221.02 | 127.60 | 16.90 |
Min = minimum; Max = maximum; SD = standard deviation.
Fig 6.
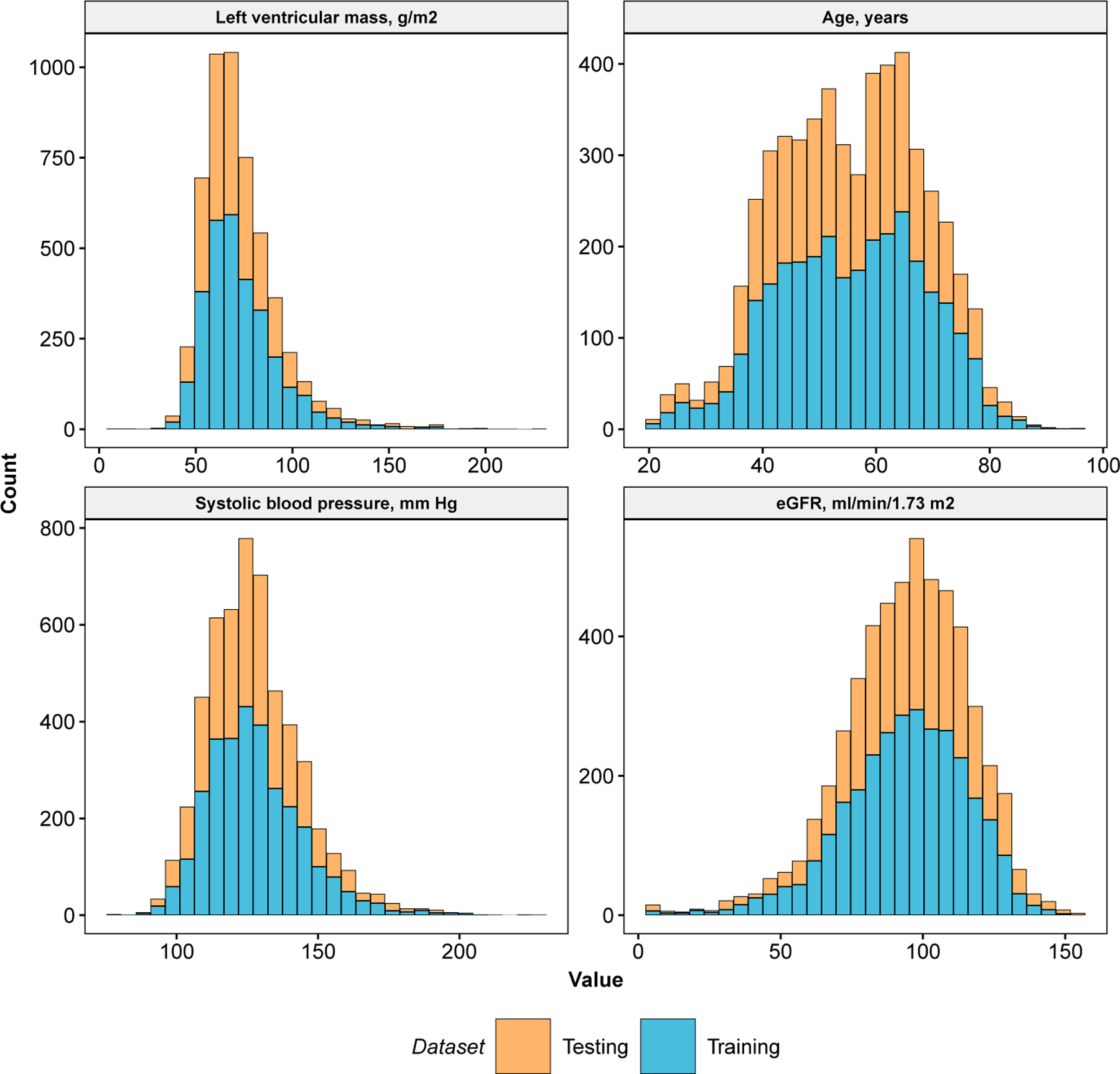
Descriptive summary of left ventricular mass, age, systolic blood pressure, and estimated glomerular filtration rate. Data are from the Jackson Heart Study.
We created variable and partial dependence plots for each explanatory variable and ASCVD risk, separately, using the ORSF and the CIF. Variable dependence plots indicated that predictions from the ORSF demonstrate stronger alignment than the CIF with the Pooled Cohort Risk equations (Figure 7). Partial dependence plots show that the ORSF’s predicted risk function generally differs from the CIF in the upper or lower range of values for each explanatory variable (Figure 8). This pattern of difference in predicted risk may explain the observed differences (Figure 9; top row) and lack of differences (Figure 9; bottom row) in survival curves for four JHS participants in the testing data.
Fig 7.
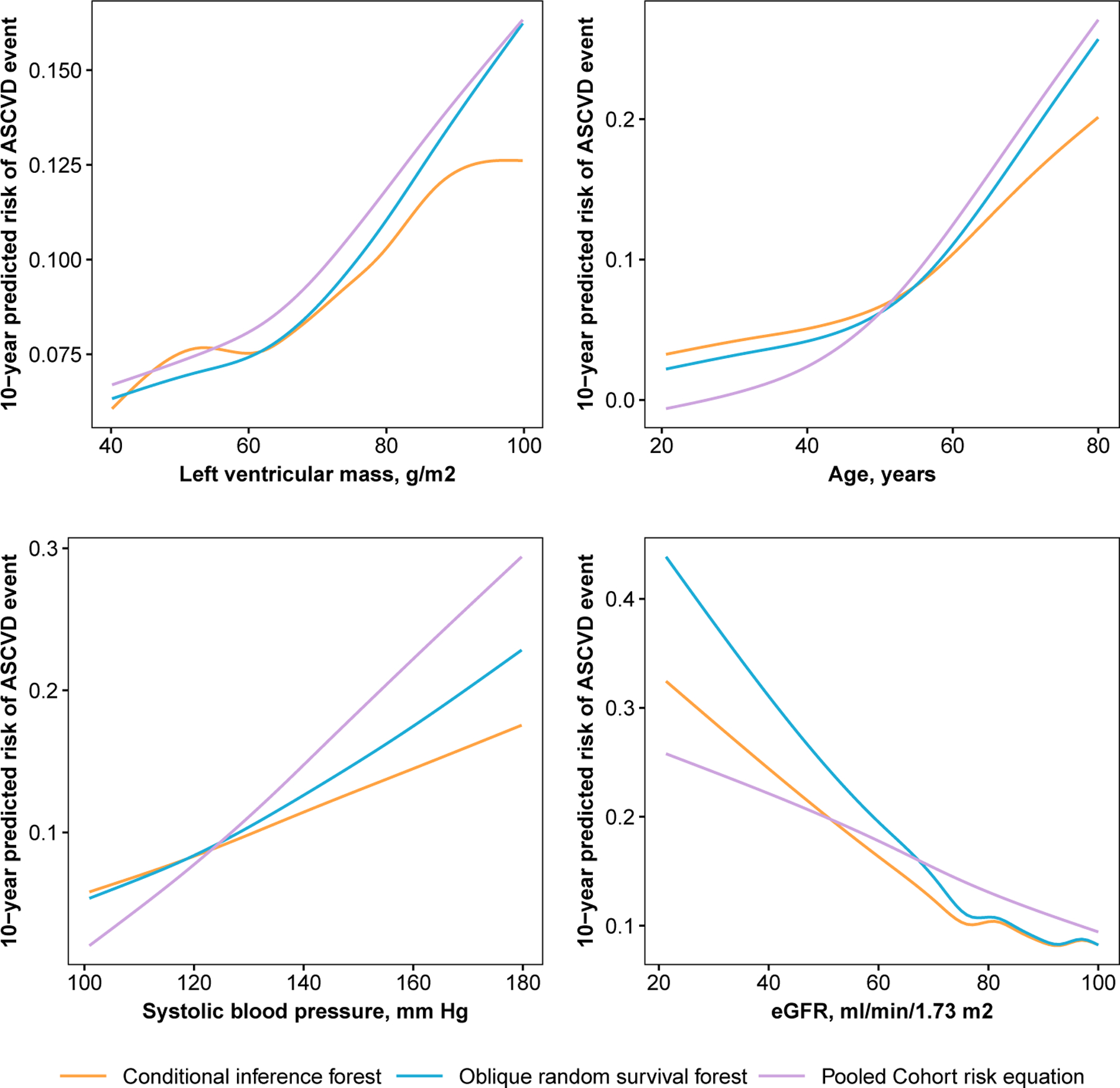
Variable dependence plots for 10-year risk of stroke or coronary heart disease as a function of left ventricular mass, age, systolic blood pressure, and estimated glomerular filtration rate, separately, according to data from participants in the Jackson Heart Study.
Fig 8.
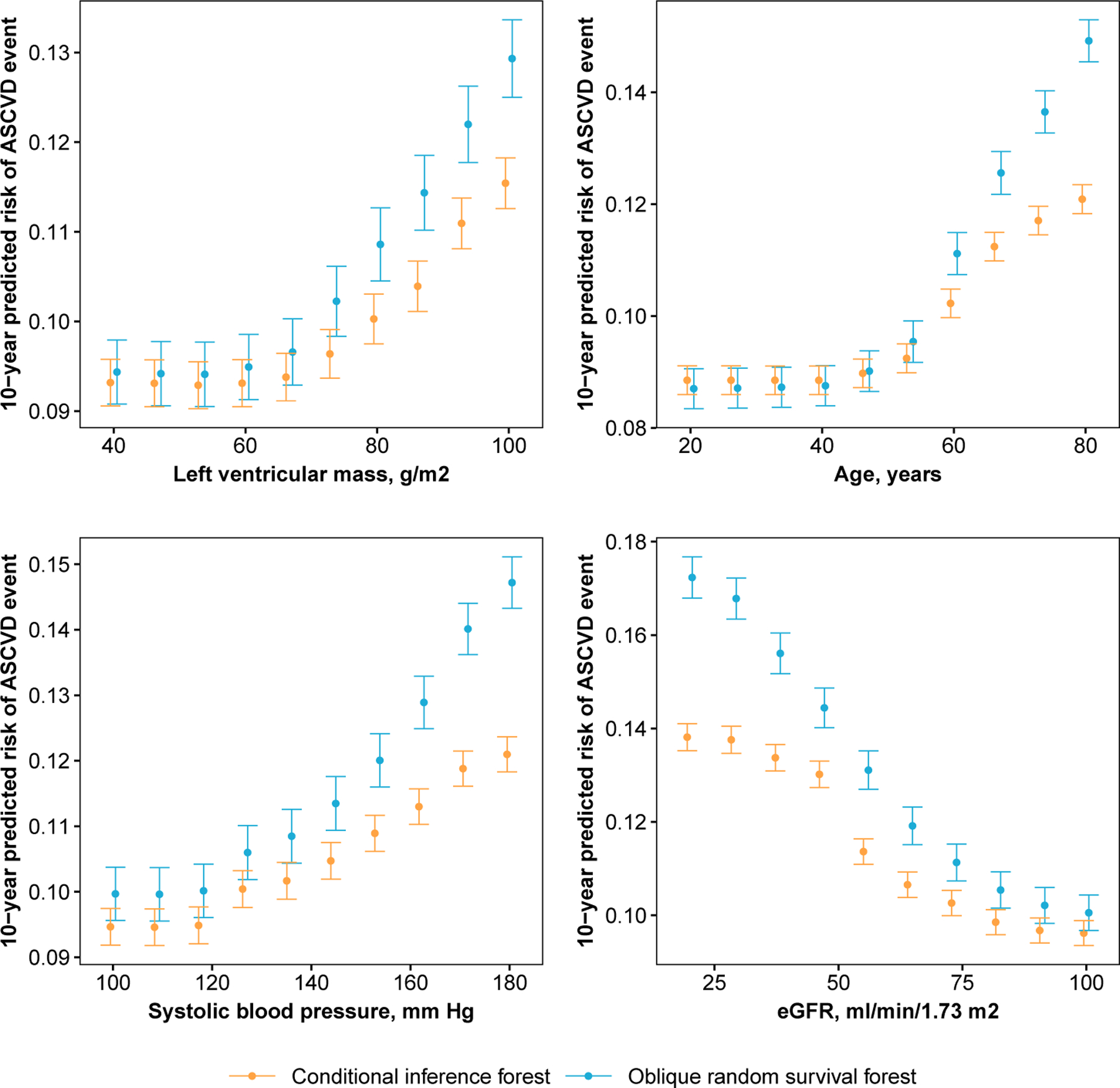
Partial dependence plots for 10-year risk of stroke or coronary heart disease as a function of left ventricular mass, age, systolic blood pressure, and estimated glomerular filtration rate, separately, according to data from participants in the Jackson Heart Study.
Fig 9.
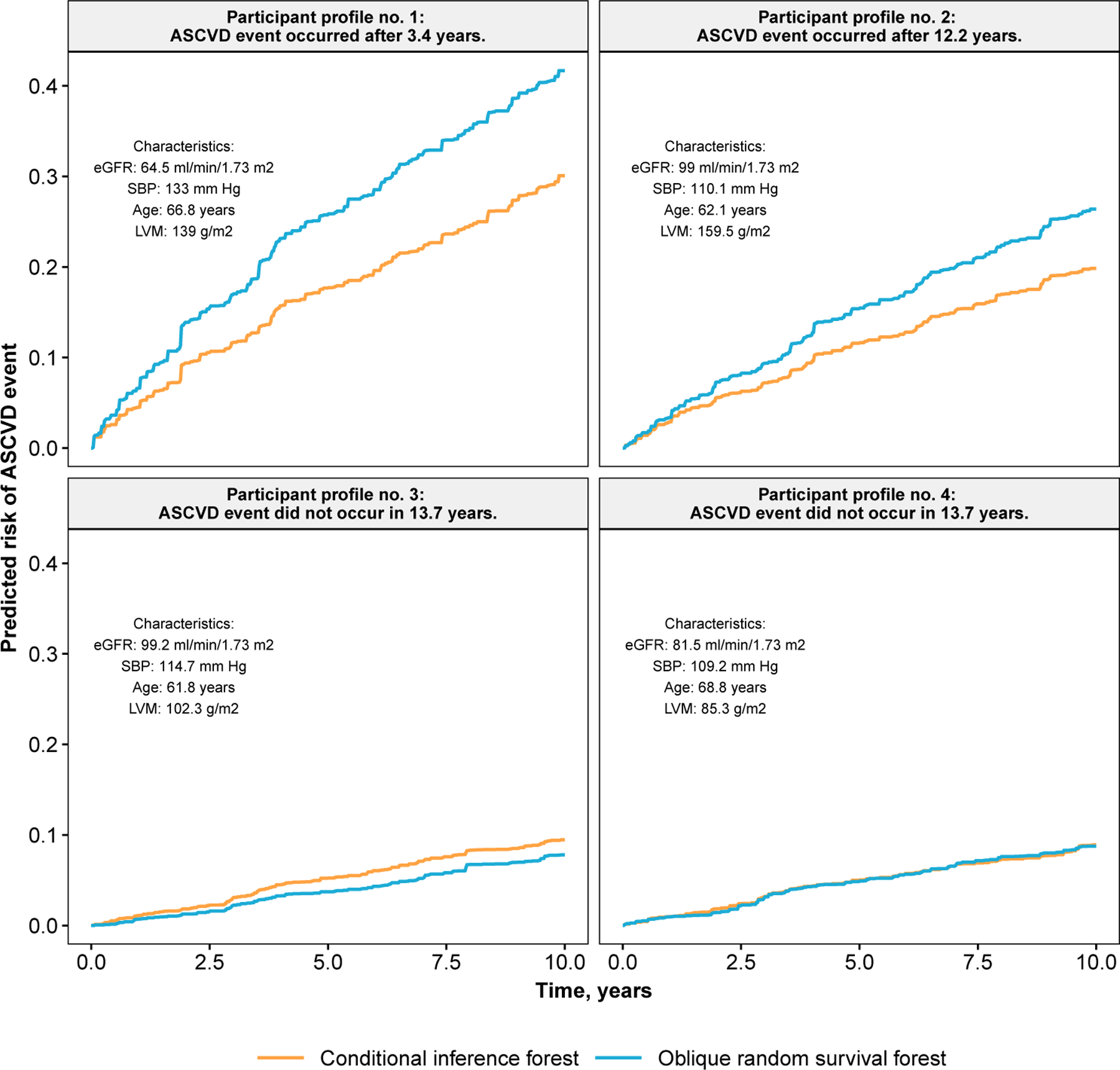
Predicted survival curves for four JHS participants in the testing data.
8. Discussion.
8.1. Summary.
In this article, we have introduced the ORSF and assessed its predictive accuracy. The ORSF extends current implementations of ensemble methods for right-censored time-to-event analyses by applying a recursive partitioning algorithm that can incorporate LCIVs. Our results indicated that the ORSF may provide substantial improvement in discrimination (i.e., lower concordance error) and minor improvement in Brier scores compared to state-of-the-art learning algorithms. Using data from participants in the JHS, we compared dependence plots from the ORSF and CIF using four explanatory variables, separately, for 10-year predicted risk of ASCVD events. Our results demonstrated differences between predicted risk for ASCVD according to the ORSF and CIF. Results also showed that, according to the four variables we analyzed, the ORSF’s predicted risk function had stronger alignment with the Pooled Cohort Risk equations compared to the CIF.
8.2. Why the ORSF works (and when it doesn’t).
In our application to real data, we found that the ORSF’s predicted risk function may substantially lower concordance error compared to the RSF and Brier score compared to the CIF. Given the similarity in these three algorithms, the most likely explanation for the differences we observed is the use of LCIVs, which has been shown to result in additional accuracy and diversity of individual trees in the ensemble (Rainforth and Wood, 2015; Menze et al., 2011; Breiman, 2001). As an informal demonstration (not a comprehensive comparison), we computed predicted survival curves for one observation generated from Scenario C in our simulation study using the ORSF and RSF. Figure 10 illustrates predicted survival curves for this observation from each survival tree in the fitted ORSF and RSF ensembles, separately. In the RSF, many trees give nearly identical survival curves for this participant. On the other hand, there are many more types of survival curves in the ORSF ensemble (diversity). Additionally, survival curves in the ORSF tend to be more aligned with the true survival curve (accuracy). This figure shows one instance where predictions from trees in the ORSF exhibit a clear increase in diversity and accuracy compared to survival trees in the RSF ensemble. However, this is clearly not a claim that this pattern holds in general.
Fig 10.
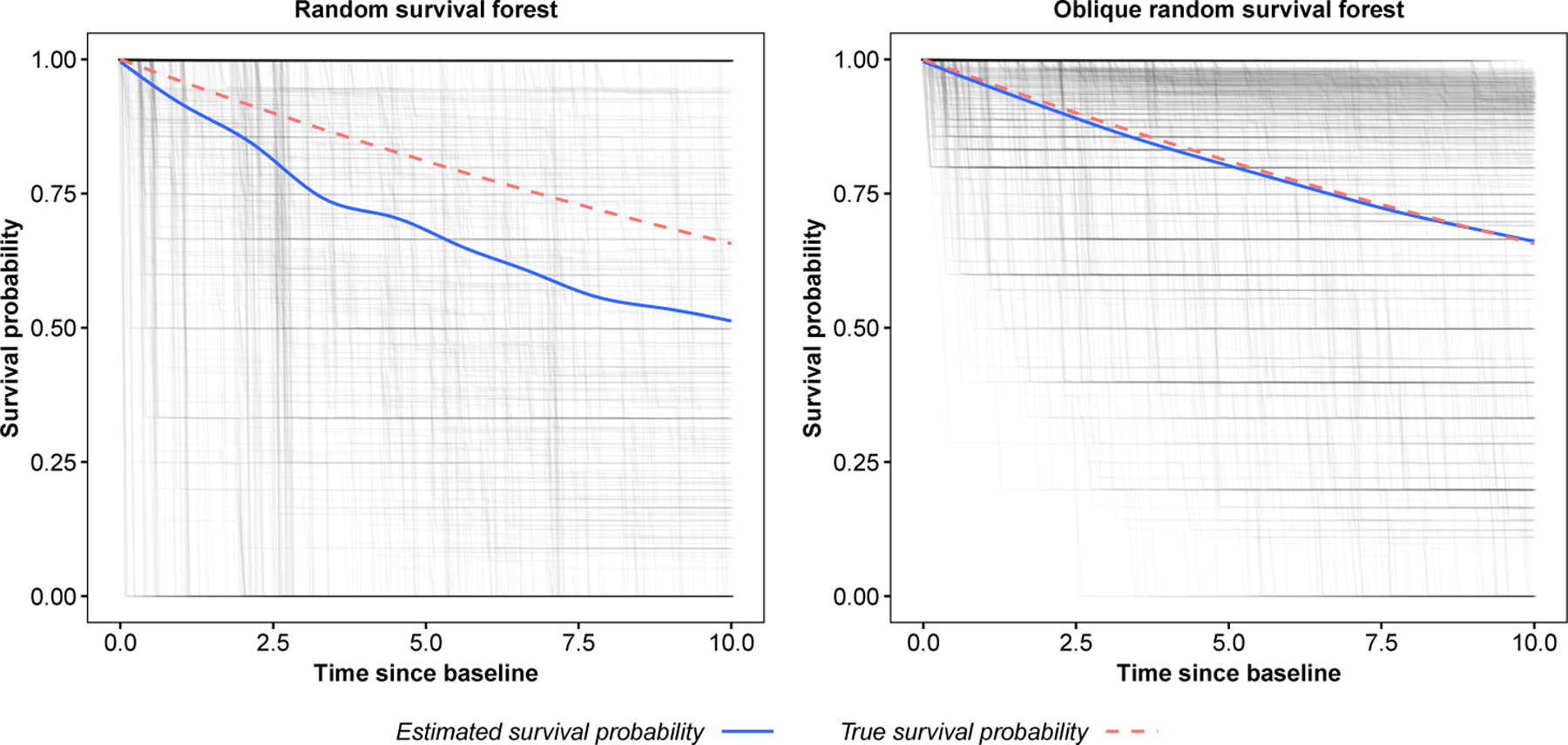
Tree and ensemble predicted survival functions for a single observation from simulated data (Scenario C with p = 50; see Section 4.1). Estimates are from the random survival forest (left) and oblique random survival forest (right).
In our simulated trials, the ORSF recorded higher Brier scores than gradient boosted decision trees. However, this pattern was reversed in the analysis of real data. This is likely due to the invalidity of the proportional hazards assumption in many of the real data problems we considered. These results suggest that in analyses where the proportional hazards assumption is entirely valid, the ORSF and other implementations of RFs for survival analyses may not be able to compete with gradient boosted decision trees or penalized Cox PH models.
In addition to LCIVs, early stopping and class-specific shrinkage of categorical predictors are important components of the ORSF that are based on the CIF (Nasejje et al., 2017). The protocol described by Breiman (1984) is known to result in overfitting and a selection bias towards covariates with many possible splits (i.e., unordered categorical variables) (Hothorn, Hornik and Zeileis, 2006). The ORSF and CIF each apply a mechanism for early stopping if a certain level of statistical association is not measured, and this may explain why both the ORSF and the CIF provided relatively low concordance error. We speculate that early stopping prevents these ensembles from growing decision trees that overfit the training data. However, the ORSF and CIF also provided relatively unimpressive Brier scores in our simulation study and real data analysis compared to gradient boosted decision trees. Therefore, we speculate that early stopping may also prevent decision trees from partitioning a training with a granularity that estimates survival probability as accurately as gradient boosting, especially when the proportional hazard assumption is entirely valid (i.e., scenarios A and B in our simulation study).
8.3. Limitations and Future research.
The additional computational resources required by the ORSF are a clear limitation. While future development and optimization of the C++ routines in the obliqueRSF package may alleviate this limitation, there are several additional topics of interest. For example, the ORSF could be extended to assess the importance of individual variables or clusters of variables. Multiple imputation could be incorporated into the ORSF by imputing missing values separately for each tree in the ensemble using a shared imputation model or a shared set of imputation models. To generate additional diversity in the ensemble, a stochastic error term could be added to the imputation model’s predictions. Predictions from the the ORSF could also be linked to programs that operate on the predicted values of a tree-based ensemble learning algorithm, such as the method of conducting statistical inference introduced by Mentch and Hooker (2016) or the efficient method to compute Shapley additive explanatory values discussed in Lundberg, Erion and Lee (2018). Last, a hybrid algorithm for ORSF that applies cross-validation conditional on sufficient sample size in the current node could be developed. In summary, the obliqueRSF R package offers a powerful learning algorithm, the ORSF (and optionally, the ORSFCV), which may exceed the performance of state-of-the-art learning methods for right-censored time-to-event analyses.
Supplementary Material
Acknowledgements.
The REasons for Geographic and Racial Differences in Stroke (REGARDS) study is supported by a cooperative agreement (U01 NS041588) from the National Institute of Neurological Disorders and Stroke, National Institutes of Health, Department of Health and Human Service. Additional support was provided by grant R01 HL080477 from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke or the National Institutes of Health. Representatives of the funding agency have been involved in the review of the manuscript but not directly involved in the collection, management, analysis or interpretation of the data. The authors thank the other investigators, the staff, and the participants of the REGARDS study for their valuable contributions. A full list of participating REGARDS investigators and institutions can be found at http://www.regardsstudy.org.
The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I/HHSN26800001) and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS. More information about the JHS can be found at https://www.jacksonheartstudy.org/.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke, the National Institutes of Health, The National Heart, Lung, and Blood Institute, The National Institute on Minority Health and Health Disparities, or the Department of Health and Human Services.
Footnotes
The term ‘node’ refers to a descendant data set in the decision tree. The term ‘splitting’ a node refers to partitioning the descendant data into two non-overlapping subset
The RSF applies bootstrap sampling with replacement, whereas the CIF uses sampling without replacement to achieve unbiasedness
Software packages generally use the term mtry to denote the size of the random subset of predictors
This step only occurs in the CI
For survival trees, descendant nodes must have at least nmin unique observations that are not censored.
The CIF applies a nearest neighbor aggregation scheme to aggregate predictions from each tree, whereas the RSF weights all trees equally.
For example, consider p = 4. Variable 1 interacts with variable 2 (effect size: −1), variable 2 interacts with variable 3 (effect size: −1/3), variable 3 interacts with variable 4 (effect size: 1/3), and variable 4 interacts with variable 1 (effect size: 1) giving four main effects and four interaction effects, i.e. 2p terms in β.
References.
- Andersen PK, Borgan O, Gill RD and Keiding N (2012). Statistical Models Based on Counting Processes. Springer Science & Business Media. [Google Scholar]
- Bien J and Tibshirani R (2019). protoclust: Hierarchical Clustering with Prototypes R package version 1.6.3.
- Binder H (2013). CoxBoost: Cox Models by Likelihood Based Boosting for a Single Survival Endpoint or Competing Risks R package version 1.4, available at https://CRAN.R-project.org/package=CoxBoost.
- Blanche P, Kattan MW and Gerds TA (2018). The C-index is not Proper for the Evaluation of t-year Predicted Risks. Biostatistics. [DOI] [PubMed] [Google Scholar]
- Bou-Hamad I, Larocque D, Ben-Ameur H et al. (2011). A Review of Survival Trees. Statistics Surveys 5 44–71. [Google Scholar]
- Breiman L (1984). Classification and Regression Trees. Routledge. [Google Scholar]
- Breiman L (2001). Random Forests. Machine Learning 45 5–32. [Google Scholar]
- Breiman L and Cutler A (2003). Setting up, Using, and Understanding Random Forests V4. 0. University of California, Department of Statistics. [Google Scholar]
- Brilleman S (2018). simsurv: Simulate Survival Data R package version 0.2.2, available at https://CRAN.R-project.org/package=simsurv.
- Burnham KP and Anderson DR (2004). Multimodel Inference: Understanding AIC and BIC in Model Selection. Sociological Methods & Research 33 261–304. [Google Scholar]
- Chen T and Guestrin C (2016). Xgboost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM International Conference on Knowledge Discovery and Data Mining 785–794. ACM. [Google Scholar]
- Chen T, He T, Benesty M, Khotilovich V, Tang Y, Cho H, Chen K, Mitchell R, Cano I, Zhou T, Li M, Xie J, Lin M, Geng Y and Li Y (2019). xgboost: Extreme Gradient Boosting R package version 0.81.0.1, available at https://CRAN.R-project.org/package=xgboost. [Google Scholar]
- Cox DR (1992). Regression Models and Life-tables. In Breakthroughs in Statistics 527–541. Springer. [Google Scholar]
- Demšar J (2006). Statistical Comparisons of Classifiers over Multiple Data Sets. Journal of Machine Learning Research 7 1–30. [Google Scholar]
- Desmedt C, Di Leo A, de Azambuja E, Larsimont D, Haibe-Kains B, Selleslags J, Delaloge S, Duhem C, Kains J-P, Carly B et al. (2011). Multifactorial Approach to Predicting Resistance to Anthracyclines. Journal of Clinical Oncology 29 1578–1586. [DOI] [PubMed] [Google Scholar]
- Dheeru D and Karra Taniskidou E (2017). UCI Machine Learning Repository.
- Efron B, Hastie T, Johnstone I, Tibshirani R et al. (2004). Least Angle Regression. The Annals of Statistics 32 407–499. [Google Scholar]
- Fleming TR and Harrington DP (2011). Counting Processes and Survival Analysis 169. John Wiley & Sons. [Google Scholar]
- Friedman M (1937). The Use of Ranks to Avoid the Assumption of Normality Implicit in the Analysis of Variance. Journal of the American Statistical Association 32 675–701. [Google Scholar]
- Friedman JH (2001). Greedy Function Approximation: a Gradient Boosting Machine. Annals of Statistics 1189–1232. [Google Scholar]
- Friedman J, Hastie T and Tibshirani R (2001). The Elements of Statistical Learning 1. Springer series in statistics New York. [Google Scholar]
- Friedman J, Hastie T and Tibshirani R (2010a). Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software 33 1. [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T and Tibshirani R (2010b). Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software 33 1–22. Available at http://www.jstatsoft.org/v33/i01/. [PMC free article] [PubMed] [Google Scholar]
- Gerds TA, Kattan MW, Schumacher M and Yu C (2013). Estimating a Time-dependent Concordance Index for Survival Prediction Models with Covariate Dependent Censoring. Statistics in Medicine 32 2173–2184. [DOI] [PubMed] [Google Scholar]
- Geurts P, Ernst D and Wehenkel L (2006). Extremely Randomized Trees. Machine Learning 63 3–42. [Google Scholar]
- Graf E, Schmoor C, Sauerbrei W and Schumacher M (1999). Assessment and Comparison of Prognostic Classification Schemes for Survival Data. Statistics in Medicine 18 2529–2545. [DOI] [PubMed] [Google Scholar]
- Harrell FE, Califf RM, Pryor DB, Lee KL and Rosati RA (1982). Evaluating the Yield of Medical Tests. JAMA 247 2543–2546. [PubMed] [Google Scholar]
- Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H et al. (2011). A Genomic Predictor of Response and Survival Following Taxane-anthracycline Chemotherapy for Invasive Breast Cancer. Jama 305 1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heagerty PJ, Lumley T and Pepe MS (2000). Time-dependent ROC Curves for Censored Survival Data and a Diagnostic Marker. Biometrics 56 337–344. [DOI] [PubMed] [Google Scholar]
- Heagerty PJ and Zheng Y (2005). Survival Model Predictive Accuracy and ROC Curves. Biometrics 61 92–105. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Hornik K and Zeileis A (2006). Unbiased Recursive Partitioning: A Conditional Inference Framework. Journal of Computational and Graphical statistics 15 651–674. [Google Scholar]
- Hothorn T and Lausen B (2003). Double-bagging: Combining Classifiers by Bootstrap Aggregation. Pattern Recognition 36 1303–1309. [Google Scholar]
- Hothorn T, Lausen B, Benner A and Radespiel-Tröger M (2004). Bagging Survival Trees. Statistics in Medicine 23 77–91. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Hornik K, Strobl C and Zeileis A (2010). Party: a Laboratory for Recursive Partytioning.
- Hothorn T, Hornik K, Strobl C and Zeileis A (2019). party: A Laboratory for Recursive Partytioning R package version 1.3.3, available at https://CRAN.R-project.org/package=party.
- Howard VJ, Cushman M, Pulley L, Gomez CR, Go RC, Prineas RJ, Graham A, Moy CS and Howard G (2005). The Reasons for Geographic and Racial Differences in Stroke Study: Objectives and Design. Neuroepidemiology 25 135–143. [DOI] [PubMed] [Google Scholar]
- Iman RL and Davenport JM (1980). Approximations of the Critical Region of the Fbietkan Statistic. Communications in Statistics-Theory and Methods 9 571–595. [Google Scholar]
- Ishwaran H and Kogalur UB (2019). Random Forests for Survival, Regression, and Classification (RF-SRC) R package version 2.8.0, available at https://cran.r-project.org/package=randomForestSRC.
- Ishwaran H, Kogalur UB, Blackstone EH and Lauer MS (2008). Random Survival Forests. The Annals of Applied Statistics 841–860. [Google Scholar]
- Jaeger B (2018). obliqueRSF: Oblique Random Forests for Right-Censored Time-to-Event Data R package version 0.1.0, available at https://CRAN.R-project.org/package=obliqueRSF.
- Jaeger BC, Long LD, Long DM, Sims M, Szychowski JM, Min Y-I, Mcclure LA, Howard G and Simon N (2019). Supplement to Oblique Random Survival Forests. The Annals of Applied Statistics 00–00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarik A and Templ M (2016). Imputation with the R Package VIM. Journal of Statistical Software 74 1–16. [Google Scholar]
- Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T et al. (2009). A New Equation to Estimate Glomerular Filtration Rate. Annals of Internal Medicine 150 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg SM, Erion GG and Lee S-I (2018). Consistent Individualized Feature Attribution for Tree Ensembles. arXiv preprint arXiv:1802.03888. [Google Scholar]
- McCall MN, Bolstad BM and Irizarry RA (2010). Frozen Robust Multiarray Analysis (fRMA). Biostatistics 11 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentch L and Hooker G (2016). Quantifying Uncertainty in Random Forests via Confidence Intervals and Hypothesis Tests. The Journal of Machine Learning Research 17 841–881. [Google Scholar]
- Menze BH, Kelm BM, Splitthoff DN, Koethe U and Hamprecht FA (2011). On oblique random forests. In Joint European Conference on Machine Learning and Knowledge Discovery in Databases 453–469. Springer. [Google Scholar]
- Mogensen UB, Ishwaran H and Gerds TA (2012). Evaluating Random Forests for Survival Analysis Using Prediction Error Curves. Journal of Statistical Software 50 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TP, White IR and Crowther MJ (2017). Using Simulation Studies to Evaluate Statistical Methods. arXiv preprint arXiv:1712.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasejje JB, Mwambi H, Dheda K and Lesosky M (2017). A Comparison of the Conditional Inference Survival Forest Model to Random Survival Forests Based on a Simulation Study as well as on two Applications with Time-to-Event Data. BMC Medical Research Methodology 17 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainforth T and Wood F (2015). Canonical Correlation Forests. arXiv preprint arXiv:1507.05444. [Google Scholar]
- Safford MM, Brown TM, Muntner PM, Durant RW, Glasser S, Halanych JH, Shikany JM, Prineas RJ, Samdarshi T, Bittner VA et al. (2012). Association of Race and Sex with Risk of Incident Acute Coronary Heart Disease Events. JAMA 308 1768–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M, Bastert G, Bojar H, Huebner K, Olschewski M, Sauerbrei W, Schmoor C, Beyerle C, Neumann R and Rauschecker for the German Breast Cancer Study Group, H. (1994). Randomized 2 × 2 Trial Evaluating Hormonal Treatment and the Duration of Chemotherapy in Node-positive Breast Cancer Patients. Journal of Clinical Oncology 12 2086–2093. [DOI] [PubMed] [Google Scholar]
- Segal MR (1988). Regression Trees for Censored Data. Biometrics 35–47. [Google Scholar]
- Shabalin AA, Tjelmeland H, Fan C, Perou CM and Nobel AB (2008). Merging Two Gene-expression Studies via Cross-platform Normalization. Bioinformatics 24 1154–1160. [DOI] [PubMed] [Google Scholar]
- Simon N, Friedman J, Hastie T and Tibshirani R (2011). Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. Journal of Statistical Software 39 1–13. Available at http://www.jstatsoft.org/v39/i05/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser H and Weber C (1999). On the Asymptotic Theory of Permutation Statistics.
- Strobl C, Malley J and Tutz G (2009). An Introduction to Recursive Partitioning: Rationale, Application, and Characteristics of Classification and Regression Trees, Bagging, and Random Forests. Psychological Methods 14 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl C, Boulesteix A-L, Zeileis A and Hothorn T (2007). Bias in Random Forest Variable Importance Measures: Illustrations, Sources and a Solution. BMC bioinformatics 8 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HA Jr, Wilson JG, Jones DW, Sarpong DF, Srinivasan A, Garrison RJ, Nelson C and Wyatt SB (2005). Toward Resolution of Cardiovascular Health Disparities in African Americans: Design and Methods of the Jackson Heart Study. Ethn Dis 15 S6–4. [PubMed] [Google Scholar]
- Ternès N, Rotolo F, Heinze G and Michiels S (2017). Identification of Biomarker-by-Treatment Interactions in Randomized Clinical Trials with Survival Outcomes and High-dimensional Spaces. Biometrical Journal 59 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM (2015). A Package for Survival Analysis in S. R package version 2.38, available at https://CRAN.R-project.org/package=survival.
- Tutz G and Binder H (2007). Boosting Ridge Regression. Computational Statistics & Data Analysis 51 6044–6059. [Google Scholar]
- van Houwelingen HC, Bruinsma T, Hart AA, van’t Veer LJ and Wessels LF (2006). Cross-validated Cox Regression on Microarray Gene Expression Data. Statistics in Medicine 25 3201–3216. [DOI] [PubMed] [Google Scholar]
- Van’t Veer LJ, Dai H, Van De Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, Van Der Kooy K, Marton MJ, Witteveen AT et al. (2002). Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature 415 530. [DOI] [PubMed] [Google Scholar]
- Venables WN and Ripley BD (2002). Modern Applied Statistics with S, Fourth ed. Springer, New York. ISBN 0-387-95457-0. [Google Scholar]
- Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, Himmelfarb CD, DePalma SM, Gidding S, Jamerson KA, Jones DW et al. (2018). 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: a Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Journal of the American College of Cardiology 71 e127–e248. [DOI] [PubMed] [Google Scholar]
- Zhu R (2013). Tree-based Methods for Survival Analysis and High-dimensional Data, PhD thesis, The University of North Carolina at Chapel Hill. [Google Scholar]
- Zhu R, Zeng D and Kosorok MR (2015). Reinforcement Learning Trees. Journal of the American Statistical Association 110 1770–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H and Hastie T (2005). Regularization and Variable Selection via the Elastic Net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 67 301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


