Abstract
Therapeutic applications of synthetic mRNA were proposed more than 30 years ago, and are currently the basis of one of the vaccine platforms used at a massive scale as part of the public health strategy to get COVID-19 under control. To date, there are no published studies on the biodistribution, cellular uptake, endosomal escape, translation rates, functional half-life and inactivation kinetics of synthetic mRNA, rates and duration of vaccine-induced antigen expression in different cell types. Furthermore, despite the assumption that there is no possibility of genomic integration of therapeutic synthetic mRNA, only one recent study has examined interactions between vaccine mRNA and the genome of transfected cells, and reported that an endogenous retrotransposon, LINE-1 is unsilenced following mRNA entry to the cell, leading to reverse transcription of full length vaccine mRNA sequences, and nuclear entry. This finding should be a major safety concern, given the possibility of synthetic mRNA-driven epigenetic and genomic modifications arising. We propose that in susceptible individuals, cytosolic clearance of nucleotide modified synthetic (nms-mRNAs) is impeded. Sustained presence of nms-mRNA in the cytoplasm deregulates and activates endogenous transposable elements (TEs), causing some of the mRNA copies to be reverse transcribed. The cytosolic accumulation of the nms-mRNA and the reverse transcribed cDNA molecules activates RNA and DNA sensory pathways. Their concurrent activation initiates a synchronized innate response against non-self nucleic acids, prompting type-I interferon and pro-inflammatory cytokine production which, if unregulated, leads to autoinflammatory and autoimmune conditions, while activated TEs increase the risk of insertional mutagenesis of the reverse transcribed molecules, which can disrupt coding regions, enhance the risk of mutations in tumour suppressor genes, and lead to sustained DNA damage. Susceptible individuals would then expectedly have an increased risk of DNA damage, chronic autoinflammation, autoimmunity and cancer. In light of the current mass administration of nms-mRNA vaccines, it is essential and urgent to fully understand the intracellular cascades initiated by cellular uptake of synthetic mRNA and the consequences of these molecular events.
Keywords: Autoimmunity, Autoinflammation, DNA damage, Endogenous transposable elements, Genomic integration, IFN, LINE-1, mRNA vaccine, nms-mRNA, TREX-1
Introduction
Exogenous mRNA was first proposed in 1990 for use in therapeutic [1] and preventative [2], [3] applications. Since then, there has been marked, and growing, interest in developing novel RNA-based therapeutics. Current clinical applications of two main types of RNA-based technology: messenger RNA (mRNA) and small interfering RNA (siRNA) are focused on cancer immunotherapy, protein replacement therapy, genome editing and vaccination. The central idea surrrounding this technology is simple: mRNA-based applications allow the delivery of genetic instructions to correct a somatic defect by synthesizing the normal version of a missing or altered protein [1], or the delivery of instructions to create an antigenic protein to induce specific immune responses [4]; while siRNA-based applications silence disease-related genes in a sequence-specific manner [5]. Prior to the COVID-19 pandemic, the only RNA-based therapeutics that had received clinical approval from the U.S. Food and Drug Administration (FDA) or the European Medicines Agency (EMA) are four synthetic lipid nanoparticle (LNP)-mRNA or chemically-modified siRNA drugs (patisiran, givosiran, lumasiran, and inclisiran) [5]. These are commercially available to treat uncommon conditions, such as acute hepatic porphyria, type I hyperoxaluria, heterozygous familial hypercholesterolemia, and hereditary transthyretin-mediated amyloidosis.
Despite nearly-three decades since it was first shown that mRNA could be used to generate specific immune responses against a pathogen, prior to the COVID-19 pandemic, vaccines based on mRNA for human use had only been developed and tested in pre-clinical and clinical trials [6]. In 2020, as a result of the COVID-19 pandemic, mRNA-based vaccines were developed at an unprecedented speed. In less than a year, two mRNA-based COVID-19 vaccines were designed, manufactured, tested and authorized for general and widespread use in the human population. An emergency public health situation can oftentimes justify rapid decisions, and some will necessarily be based on less than the minimum desirable information. However, regardless of the emergency, some corners must never be cut, particularly those that, if overlooked, could seriously impact human health. In other words, even emergency public health measures should heed the fundamental premise of primum non nocere, perhaps one of the main precepts of bioethics that all medical students are taught throughout the world [7], [8]. Although it can be argued that every preventative or therapeutic pharmacological intervention is a two-edged sword if we consider every single potential side effect that could be associated with its use [9], before consenting to receiving the COVID-19 mRNA vaccines, recipients must certainly be informed about what is known and what is not known in terms of immediate- and long-term safety. This was explicitly reminded to the medical and scientific community soon after vaccine authorization [10], [11], but has not been done systematically, at least not in most countries.
The safety profile of nucleoside-modified synthetic mRNA (hereafter, nms-mRNA) is far from completely understood. Trials to evaluate the biodistribution, cellular uptake, endosomal escape, translation rates, functional half-life and inactivation kinetics of synthetic mRNA, rates and duration of vaccine-induced antigen expression in different cell types, as well as potential interactions with the host genome were bypassed. One of the major safety concerns of introducing nms-mRNA, such as that contained in the two approved mRNA COVID-19 vaccines, is the possibility that such modifications will ultimately lead to epigenetic and/or genomic modifications in dividing and non-dividing cells. Regrettably, despite the lack of information, most scientific reviews that address risks and benefits of these vaccine platforms underscore their high levels of safety (e.g. [12], [13], [14]) and claim that there is no risk of genomic integration with these vaccines [15], [16], as can occur – albeit with low frequency - with plasmid DNA-based vaccines [17] or some vectorized vaccines [18], [19]. Nevertheless, it must be understood that before the synthetic mRNA COVID-19 vaccine authorization and mass rollout, to the best of our knowledge, not a single published paper had experimentally examined the possibility of occurrence of epigenetic phenomena (such as modifications of the chromatin structure), chromosomal integration of retrotranscribed nms-mRNA, genotoxicity and oncogenesis following mRNA vaccine uptake. In the 14 months following vaccine authorization, only one peer-reviewed study that we know of examined one of these possibilities, showing that vaccine nms-mRNA can activate the expression of endogenous transposable elements (TEs), undergo reverse transcription and enter the cell nucleus [20].
The early 2020 consensus report of the scientific working meeting of the Coalition for Epidemic Preparedness Innovations (CEPI) and the Brighton Collaboration (BC) Safety Platform for Emergency Vaccines (SPEAC) that focused on reducing safety concerns of the COVID-19 vaccines being designed, did not present evidence of any study on the potential genotoxicity of nms-mRNA, nor did they express any concern about the lack of studies on this subject (see [21]). What, then, is the scientific evidence that has sustained the claim that the vaccines based on nms-mRNA used to immunize against COVID-19 cannot insert into the host genome? Which body of scientific evidence has shown that no adverse effects related to genotoxicity or carcinogenicity are to be expected in the cells of vaccine recipients? A thorough review of the peer-reviewed literature on synthetic mRNA vaccine safety shows that all papers mention high levels of safety without providing any citation, or that they provide a citation for a recent review study [22], which states that exogenous mRNA is a non-integrating platform and that there is “no potential risk of [..] or insertional mutagenesis”, without providing any scientific evidence to back this claim. In fact, not one of the 38 studies cited in that review paper to show a list of the mRNA vaccines available for in vivo preclinical use, had investigated genotoxicity or potential oncogenesis. Similarly, for all of the eight mRNA vaccines that were undergoing or had completed human clinical trials cited [22], no such studies had been conducted.
Following their initial deployment in December 2020, COVID-19 mRNA vaccines have been widely distributed to individuals from many countries across the world. At the time of writing this paper, according to data from the World Health Organization,1 more than 782 million doses of mRNA vaccines have been administered to date. If we consider that, according to the WHO, on average, people have received 1.69 doses of these vaccines1, then based on the number of doses inoculated, more than 462 million people have received between at least one dose of an nms-mRNA vaccine. Furthermore, nms-mRNA vaccines are currently under development to protect against other infectious and non-infectious diseases [23]. In light of such a massive administration of this type of vaccines, identifying potential safety signals, understanding the mechanisms that can cause such events, and adjusting recommendations accordingly, is not only expected from the scientific and medical community, it is imperative.
The hypothesis
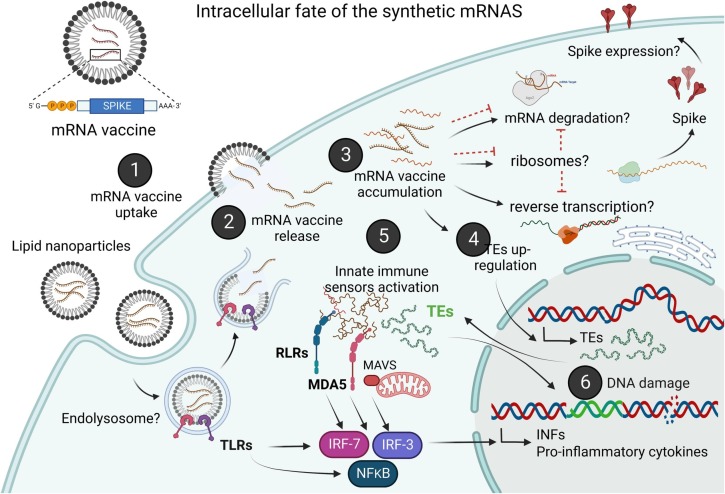
We hypothesize that in genetically- or physiologically-susceptible individuals, clearance of nms-mRNAs is hampered. Sustained presence of nms-mRNA in the cytoplasm deregulates endogenous transposable elements (TEs), leading to reverse transcription of the vaccine-mRNA. Intracellular accumulation of the nms-mRNA and the reverse transcribed cDNA molecules triggers intrinsic cytosolic RNA and DNA sensory pathways. Simultaneous activation of these pathways initiates a coordinated innate response against both types of non-self nucleic acids, prompting type-I interferon and pro-inflammatory cytokine production which if unregulated, leads to autoinflammatory and autoimmune conditions. Activated TEs increase the risk of insertional mutagenesis of the retrotranscribed vaccine mRNA, which can disrupt coding regions, enhance the risk of mutations in tumour suppressor genes, and lead to sustained DNA damage. Our hypothesis is represented graphically in Fig. 1 .
Fig. 1.
Schematic representation of our proposed hypothesis. Following intracellular delivery of the vaccine (1), vaccine nms-mRNA is released from the lipid-nanoparticles into the cytosol (2) and accumulated in the cytosol (3), which may unsilence TE expression (4), leading to the activation of foreign RNA and cytosolic DNA sensors, such as RLRs, RIG-I, MDA-5 and TREX1, and enhancing the expression of proinflammatory cytokines and type-I IFN (5). TE activity can lead to DNA damage via insertional mutagenesis and genomic instability, and enhancing the expression of pro-inflammatory cytokines and type-I IFN (6). Inflammasome activation may also have a regulatory role in preventing cGAS-STING mediated type-I IFN production, thus establishing a chronic regulatory circuit wherein type-I IFNs inhibit the inflammasome and the activated inflammasome also inhibits type I-IFN production (not shown in the figure).
Evaluation of the hypothesis
Innate recognition and intracellular fate of synthetic mRNA
Upon viral infection, the intracellular sensing pathways that mediate innate immune detection of foreign RNA are endosomal toll-like receptors (TLRs) 3, 7 and 8, and cytosolic RIG-I-like receptors (RLRs). The RLR family comprises three members: retinoic acid inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA-5), and the laboratory of genetics and physiology 2 (LGP2) gene [24]. Activated RLRs bind to the mitochondrial antiviral signalling protein (MAVS), in turn up-regulating TBK1 and IKK kinases, which activate transcription factors NF-κB, IRF-3, and IRF-7. When activated, these factors translocate to the nucleus and initiate a potent response that includes the inducible production of type I interferons (IFNs) and pro-inflammatory cytokines. These molecules up-regulate the expression of several other genes, many of which have marked antiviral effects, including degradation of foreign nucleic acids [25], [26]. In addition to inducing type I IFNs, RLRs and MAVS also activate apoptosis, promoting self-destruction of the infected cell [27].
Viral RNA can also trigger cell death and induce inflammatory cytokines such as IL-1β via the activation of the nucleotide-binding domain, leucine-rich repeat–containing proteins (NLRs) NLRP3 inflammasome [28]. In addition, in certain cell lineages there are pre-existing cellular intrinsic antiviral factors, such as RNASE L, the IFN-inducible dsRNA-dependent protein kinase R (PKR), apolipoprotein B mRNA–editing enzyme catalytic polypeptide (APOBEC3G), tripartite motif containing proteins (TRIM) TRIM5α, Tetherin/BST-2, SAMHD1, TREX1, IFITM, and the IFIT family proteins, that can bind to viral components and block viral replication directly, even before the onset of the IFN response, although most of these factors can be further induced by IFNs to amplify their antiviral effects [29], [30], [31].
In contrast to viral RNA, vaccine nms-mRNA has been modified by the incorporation of nucleobase N1-methylpseudouridine (m1Ψ) [32] in order to stabilize and protect from nuclease degradation, prolong cytosolic half-life, promote binding to the small ribosomal subunit, and improve translation efficiency [33]. Whether the accumulation of nms-mRNA within cells can activate cytosolic sensors similarly to what occurs with viral RNA has never been studied in depth. One of the few published studies on this topic showed that RNA molecules with modified nucleotides interrupt early signalling of the RIG-I-like innate immune activation pathway, and RNA containing pseudouridine binds to RIG-I but fails to trigger the canonical RIG-I conformational changes associated with robust innate immune signalling [34]. However, it is reasonable to assume that nms-mRNA can still induce a type-I IFN-dependent immune response via innate immune receptor recognition [35], [36], [37]. MDA-5, for instance, does not only recognize viral RNA; it also recognizes synthetic RNA, and endogenous RNA [38], and can bind to even one single strand of dsRNA [39]. There is evidence that binding of non-modified exogenous mRNA to host RLRs activates innate immune pathways that lead to an “antiviral state” in vaccine-transfected cells, reducing intracellular stability and translation rates of the foreign mRNA [40]. However, this does not appear to be the case for the nms-mRNA of current vaccines, as it is now known that the modified nucleic acid can be detected within the germinal centres of axillary lymph nodes of vaccines for at least 60 days after inoculation [41].
Although much has been learned about patterns that serve as recognition motifs for intracellular sensors of nucleic acids, there is not yet a real understanding of the recognition of nms-mRNA, contained in currently available mRNA vaccines. RIG-I’s recognition signal of cytosolic RNA is a free 5′-triphosphate [42], [43]. Additional studies that used chemically synthesized RNAs found that a base-paired region in the range of 10 to 20 nucleotides proximal to the free 5′-triphosphate end of the RNA ligand is essential for immunostimulatory activity via RIG-I [44], [45], rather than any RNA sequence per se [42]. Similarly, it is RNA length and secondary structures, and not the RNA sequence, that are considered key determinants of MDA5 activation, and of the involvement of the signalling adaptor MAVS [46], [47].
It is possible that instead of activating IFNs directly, cellular restriction factors will detect and inhibit translation of smn-mRNAs even before the onset of the IFN response, making RNA recognition one of the first triggers of the innate immune responses. Among these restriction factors are IFN-induced proteins with tetratricopeptide repeats (IFIT) cytoplasmic proteins (reviewed in [29], [48]). The IFIT proteins can be expressed by IFN-independent pathways, and can recognize viral RNA that contains a 5′-triphosphate (5′-ppp) moiety [49] or lacks 2′-O-methylation [50]. In other words, IFITs recognize specific RNA motifs found in viral RNA but absent in cellular mRNA, and direct binding of IFIT proteins to 5′-ppp viral RNA inhibits viral translation and replication, without the need for IFN activation. Thus, it is reasonable to assume that the intracellular accumulation and persistence of nms-mRNA following vaccine uptake could directly activate MDA-5 and IFIT family proteins, among other RNA sensors, initiating an orchestrated innate immune reaction against the synthetic RNAs and leading to a chronic ‘antiviral cellular state’.
Regardless of what is known about viral genomic and subgenomic RNA motif recognition by RLRs [51], it remains unclear how and for how long the nms-mRNAs interact with intracellular RNA sensors. The fate of highly stabilized nms-mRNA within the cytoplasm is essential to understand; but as yet, remains unexplored and was not considered prior to the emergency use authorization and approval of nms-mRNA vaccines for human use. Similarly, the presence of truncated mRNAs, short dsRNA fragments, and other contaminants within the vaccines [52] that could further alter immune recognition and deregulate immune signalling pathways, has not, to the best of our knowledge, been examined. However, considering the wide biodistribution of mRNA-nanolipid vaccine compounds [53], as well as the increased translational capacity and persistence of synthetic modified mRNA [33], it is not unreasonable to assume that mRNA-based vaccines could induce sustained inflammation and a persistent anti-viral cellular state in various tissues. The organism-level consequences of the herein hypothesized molecular events will be discussed later in this paper.
Reverse-transcription and genomic integration of foreign RNA
It was generally believed that the genome of RNA viruses could not integrate into the host's genome. However, evidence of integration of non-retroviral subgenomic viral RNA into the host cell has been described for some viruses, such as Ebola virus, Marburg virus [54], vesicular stomatitis virus and lymphocytic choriomeningitis virus [55], [56], [57] in humans and other mammalian hosts [58], and it is now thought that human retrotransposons can facilitate reverse transcription of non-retroviral viral RNA genomes and, subsequently, enable their genomic insertion [56]. SARS-CoV-2 RNA has been detected for months in many recovered COVID-19 patients that were not shedding infectious virus, and a pre-print of a study that followed a cohort of 50 individuals that were exhibiting symptoms of long-COVID post vaccination, reported a similar finding [59]. A proposed explanation of this phenomenon was that parts of the SARS-CoV-2 genome could be undergoing reverse-transcription and genomic integration within infected somatic cells, leading to persistent transcription of the integrated sequences. A recent paper confirmed this hypothesis by an in vitro study that detected the presence of reverse-transcribed copies of SARS-CoV-2 sequences in transfected human cells, and by finding active transcription of the integrated subgenomic SARS-CoV-2 segments [60]. Given that the integrated sequences only corresponded to the 3′ end of the SARS-CoV-2 genome, viable infectious virions would not be able to be produced as a result of such genomic insertion, although viral-host chimeric transcripts were observed in various tissues of two COVID-19 patients analysed [60].
When considering mRNA vaccines, the current paradigm is that the synthetic RNA cannot integrate into the genome of the vaccine recipients’ cells. However, a recent study that used hepatic cancer cells [20] showed that the Pfizer/BioNTech vaccine mRNA BNT162b2 can undergo reverse-transcription within the cytoplasm of human cells and enter the nucleus following the activation of LINE-1, a genomic transposable element (TE). Genomic TEs, which comprise endogenous retroviruses (ERVs), long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and DNA transposons, are repetitive sequences that, when activated, copy themselves or other sequences and insert these copies into the genome [61]. Being a source of genomic instability, they are mostly repressed in the majority of mammalian somatic cells, except during early embryogenesis, and aberrant expression of TEs has been associated with various diseases, from cancer to autoimmune disorders [62].
Human TEs are amongst the earliest host elements to be deregulated following the entry of a virus into a cell, and their activity significantly enhances antiviral gene expression, particularly that of IFN-β [63] and IFN-γ [64]. Activity occurs via cis-acting enhancer-promoter or via the erroneous identification of transcribed TE sequences in the cytoplasm as viral RNA by innate immune sensors. Studies conducted in mice and in humans have shown that during the course of a viral infection, expression of ERV and LINE varies, preceding the up-regulation of antiviral genes, immune response genes, and the major histocompatibility complex (MHC) genes, suggesting that up-regulated TEs are a key component of conserved, early intracellular host defence responses [63]. Cells can also aberrantly recognize dsRNA structures adopted by TEs, which resemble viral RNAs, and trigger a type-I IFN response. In humans, LINE-1 components can trigger innate immune signalling via the activation of both RIG-I- and MDA-5-mediated RNA sensing pathways that will up-regulate IFN expression [65] and initiate inflammatory responses via foreign RNA sensing and gene-regulation [66]. TE-mediated IFN activation plays a role in the development of autoimmune diseases characterized by constitutive type-I IFN activation, cancer, and cell senescence (reviewed by [67]).
Given that Zhang et al. [60] detected a consensus recognition site of the human LINE-1 endonuclease component flanking both ends of the integrated SARS-CoV-2 genomic sequences, they proposed that the observed phenomenon could be, at least partly, due to LINE-1 involvement. LINE-1 is highly abundant within the genome of mammals, including humans [68], where it has been amplifying copies for more than 160 million years [69]. Most of the 500,000 copies of LINE-1 contained in the human genome are present as truncated repeats or copies that contain mutations that affect retrotransposition [70]; however, there are roughly 150 full-length copies that are capable of self-copying and transposing into other genomic regions [71]. This cis effect is common in cells where LINE-1 is not silenced, but there is also an, albeit less common, trans effect that can be exerted by LINE-1, that results in the reverse transcription and insertion of other genetic sequences [72].
In the germ line and in most somatic cells of humans, LINE-1 activity is typically suppressed by different molecular mechanisms, including small interfering RNA (siRNA)-mediated gene silencing [73], histone and DNA methylation [74], and enzymatic activity of APOBEC3 [64] and SAMHD1 [75], [76]. However, prior to implantation, the inner cell mass and trophectoderm cells show de novo endogenous LINE-1 insertions [77], and in vitro studies with embryonic stem cells and pluripotent stem cells have described endogenous LINE-1 transcription and translation which lead to retrotransposition [78]. This is because during the initial stages of embryogenesis, the cellular milieu supports active retrotransposition of LINE-1 [79], and embryonic somatic cells down-regulate the molecular restrictions of LINE-1 retrotransposition, most likely because LINE-1 activity plays a key role in generating somatic mosaicism [80]. Interestingly, in post-natal organisms, some cell types, including neurons and glial cells, have fewer restrictions on LINE-1 activity leading to somatic genome variation in the nervous system [70], [81], which contributes to neurogenesis [82] but also to neuropsychiatric diseases when LINE-1 activity is abnormally high [83]. LINE-1 is now also known to be active in mature T-lymphocytes, where it plays a role in controlling T cell quiescence and exhaustion [84].
Based on the results reported by Zhang et al. [60], an independent study addressed whether the same phenomenon of reverse transcription and nuclear entry could be observed for vaccine mRNA. Using a human hepatic cell line (Huh7) and the Pfizer/BioNTech mRNA vaccine, Aldén et al. [20] found that, indeed, the exogenous mRNA activates both ORFs of LINE-1, leading to both reverse transcription and nuclear transposition of the full length vaccine mRNA (BNT162b2) that encodes for the full SARS-CoV-2 Spike glycoprotein [85]. Reverse transcription occurred in as little as six hours post vaccine exposure, and the reverse-transcribed BNT162b2 cDNA was found to enter the cell nucleus [20]. Having chosen a hepatic cell line for the experiment was intentional: a report on the Pfizer/BioNTech mRNA vaccine pharmacokinetics revealed that the highest concentration of vaccine mRNA, second only to the inoculation site, was detected in the liver a few hours after inoculation,2 revealing that the nano-lipid-bound nms-mRNA does not remain in the deltoid muscle or in the axillary lymph nodes, confirming what was reported in an independent study on vaccine nms-mRNA bio-distribution [53]. Furthermore, studies of lipid nanoparticle mRNA delivery systems conducted on rats and mice showed evidence of transient hepatotoxicity [86], [87], [88], suggesting that this organ could be a safety concern for the nms-mRNA vaccines. The results reported by Aldén et al. [20] should not be generalized, given that the study was done using a cancer cell line, and LINE-1 tends to be transcriptionally active in cancer cells [89]. However, LINE-1 transcription and protein expression was higher in the cancer cells exposed to BNT162b2 mRNA than in the cancer cells that received only saline solution [20], suggesting that reverse transcription and nuclear transposition of the vaccine mRNA was not due to LINE-1 already being active in the cancer cell line. Simply put, to date, there are no scientifically valid and biologically relevant reasons to assume that the same phenomenon could not occur in somatic cells of a person that receives the mRNA vaccine.
LINE-1 mediated inflammation
Host genomic silencing of TE expression is critical in most non-embryogenic tissues to prevent not only genome damage, but also untimely or sustained inflammation. Failure to silence LINE-1 retrotransposon expression results in increased expression of locus-specific copies of LINE-1 and is accompanied by an inflammatory signature associated with IFN activation [90]. However, as aforementioned, it is becoming increasingly evident that intracellular exposure to foreign RNA (i.e., viral or synthetic) can reactivate TEs and co-opt them to initiate an “anti-viral state”. Activation of TEs is known to occur in SARS-Cov-2 infections [91], similar to what has been observed during infections with other RNA viruses [92], [93], [94], [95], and DNA viruses [96], in different cell-types and host-species.
Activated TEs may stimulate antiviral gene expression, via cis-acting enhancer functions or via their recognition as viral motifs by pattern recognition receptors [24], such as RIG-I and MDA-5, that can detect ssRNA, dsRNA, synthetic RNA and cellular RNA [97]. Furthermore, transcribed TE sequences are capable of forming dsRNAs that can, in turn, be recognized by pattern recognition receptors as described earlier, and trigger a sustained anti-viral, pro-inflammatory cellular status [98]. This can lead to the development of autoimmune and auto-inflammatory diseases [99]. Thus, it is reasonable to assume that the stabilized and persistent vaccine nms-mRNA could promote and sustain inflammation in tissues exposed to the vaccine following its biodistribution. A chronic state of active innate immune responses due to LINE-1 activity would ensue, and sustain and promote reverse transcription, nuclear importation and genomic integration of retrotranscribed sequences: in other words, a molecular vicious circle with serious clinical consequences could follow reception of vaccine nms-mRNA, most likely worsening with each dose received.
LINE-1 mediated DNA damage and p53 gene mutations
Given that LINE-1 transposition of copied sequences requires cleavage of both strands of the genomic DNA (DSB), unsilencing its activity can cause double-strand DNA breaks in germ-line and somatic cells [100]. In many cancer cells, LINE-1 activity is known to be correlated with p53 mutations and copy number alterations [89] that are key to carcinogenesis, particularly in breast, ovarian, endometrial and colon cancers. Other tissues can be similarly affected. For instance, a study of hepatitis C virus (HCV) cell transformation showed that as a result of sustained inflammation from chronic infection with HCV, LINE-1 expression is activated before oncogenic transformation, and that unsilenced LINE-1 contributes to genomic instability of the hepatocellular carcinoma, even after viral clearance [101]. In vitro induction of LINE-1 expression increased phosphorylation of MRN complex member RAD50 [89], a catalytic protein complex key for coordinating and sensing DSBs and initiating the DNA damage response pathway [102]. Thus, unsilenced LINE-1 in somatic tissues that are expected vaccine targets (i.e. dendritic cells, lymph nodes, muscle cells) and unintended vaccine targets (e.g. liver, adrenal glands, spleen, ovaries, and brain) [53] could conceivably increase the risk of genotoxicity and carcinogenesis in those tissues, and given that newly inserted copies of TE sequences can be transmitted to each successive cellular generation and modify the somatic human genome [103], sustained activity of LINE-1 from persistent vaccine mRNA could be important for carginogenesis. As stated before, the risk would conceivably increase with each dose received. These molecular phenomena would expectedly be more frequent in intended and unintended vaccine target cells with intrinsic high levels of LINE-1 expression, such glial cells [70], [81], T-lymphocytes [84], senescent cells [104], and in cells with reduced DNA damage repair mechanisms. Susceptibility would be particularly high for individuals with suppressed or suboptimal cellular adaptive immune responses, or those with neuropsychiatric diseases, where LINE-1 activity is abnormally high [83]. In addition, it has already been shown that cells transfected with SARS-CoV-2 Spike gene exhibit an increased response to DNA damage, ROS production, and a senescent cellular state, which can, in turn, lead to paracrine senescence in adjacent cells and endothelial dysfunction [105]. Although the authors of that study speculated that the expectedly brief duration of antigenic stimulation following vaccination might be insufficient for vaccinated individuals to exhibit similar effects, it is now known that both nms-mRNA and its encoded protein are viable and expressed for weeks [41], so it is plausible that such effects would, indeed, be prevalent.
Cytosolic accumulation of DNA activates pro-inflammatory responses
In addition to the recognition of foreign RNA, cytosolic DNA can also be detected by a signalling cascade termed the IFN-stimulatory DNA (ISD) response. This sensory pathway activates a potent type-I IFN production via the same transcription factor involved in foreign RNA recognition: interferon regulatory factor 3 (IRF3) [106]. Earlier in our paper, we described the molecular cascades that we hypothesize would arise from the persistent stabilized cytosolic nms-mRNA. We will now describe the hypothesized fate and biological consequences of the cytosolic accumulation of retrotranscribed vaccine DNA.
Cyclic GMP–AMP synthase (cGAS) is the main cytosolic immune sensor that binds cytosolic double-stranded DNAs from viruses, bacteria, mitochondria, micronuclei, as well as DNA from endogenous retroelements. The activation of cGAS generates cyclic dinucleotide cyclic GMP-AMP (cGAMP), which, in turn, activates a type-I Interferon response via Stimulator of Interferon Genes (STING). STING signalling can trigger transcriptional activation of NF-κB, initiating the synthesis of pro-inflammatory cytokines, including type-I IFNs IFN-α and IFN-β [107]. Therefore, the cGAS-STING pathway mediates immune defence against foreign DNA and against tumour-derived DNA [108], [109]. However, aberrant activation of the cGAS-STING pathway by self-DNA leaked into the cytosol or by failure to eliminate accumulated self-DNA can also lead to autoinflammatory and autoimmune disease and promote tumorigenesis [109], [110]. This is why it is essential for the proper removing of accrued cytosolic non-productive reverse-transcribed DNA and fragments derived from endogenous retroelements such as L1 retrotransposons, Long Terminal Repeat (LTR) endogenous retroviruses, and SINE elements, in order to prevent self-DNA-mediated activation of nucleic acid sensors that would otherwise upregulate type-I IFN and pro-inflammatory cytokines.
Han and co-workers [111] recently reported that SARS-CoV-2 ORF9b, encoded by an alternative ORF within the N gene, negatively regulates antiviral immunity by inhibiting the activation of type-I and type-III IFNs that are induced by cytosolic dsRNA-sensing pathways of RIG-I/MDA5-MAVS signalling, and that SARS-CoV-2 infection can also suppress the induction of types I and III IFNs by TRIF and STING, which are proteins of the cytosolic DNA-sensing pathway, and of the cGAS-STING signalling cascade, respectively. Remarkably, the cGAS-STING pathway has been recently reported as a critical driver of aberrant type-I IFN responses in severe cases of COVID-19, a disease caused by an RNA virus, not a DNA virus [112]. Given that strict compartmentalization of cellular DNA in the nucleus and mitochondria is necessary to avoid sensing self-DNA, the source of cytosolic immunostimulatory DNA following SARS-CoV-2 infection remains unknown but could be explained by LINE-1 driven reverse transcription following infection [60]. A not mutually exclusive explanation is that the source of cytosolic DNA in severe COVID-19 patients is fragmented mitochondrial DNA within vascular endothelial cells caused by mitochondrial dysfunction induced by SARS-CoV-2 Spike glycoprotein [113]. When released into the cytosol, fragmented mitochondrial DNA could activate the cGAS–STING pathway within the endothelial cells. Hence, it is reasonable to hypothesize that the reverse transcribed vaccine mRNAs that accumulated in the cytosol following early nms-mRNA activation of TEs, leads to the activation of the cGAS-STING pathway. The cytosolic accumulated DNA could become a self-immunostimulatory molecule leading to cGAS/STING-dependent innate immune activation.
Trex1 polymorphisms and the accumulation of endogenous retroelements may directly be the cause of myocarditis following reception of mRNA vaccines
A widely expressed protein in mammalian cells, the 3′-->5′ DNA exonuclease, 3′ repair exonuclease 1 (TREX1, previously known as DNase III), degrades single-stranded DNA and double-stranded DNA substrates by removing nucleotides from the 3′ ends of DNA molecules [114], [115]. TREX1 helps maintain innate immune tolerance to cytosolic self-DNA by removing DNA substrates to prevent the initiation of autoimmunity. Studies have shown that mutations in TREX1 lead to the accumulation of self-DNA in the cytosol of TREX1-deficient cells, which, as discussed above, triggers systemic inflammation and autoimmunity by chronic activation of a cGAS-STING-mediated type-I interferon response [116]. Examples of autoinflammatory and autoimmune conditions associated with TREX1 mutations are systemic lupus erythematosus, Aicardi-Goutieres syndrome, cryofibrinogenemia, chilblain lupus, Cree encephalitis, and retinal vasculopathy with cerebral leukodystrophy (reviewed by [117]).
Unresolved or excessive accumulation of cytosolic DNA directly activates the ISD pathway inducing robust transcription of the TREX1 gene and initiating IRF3-dependent production of type-I IFN [118]. Increased levels of type-I IFN are typical of autoimmune disorders, likely related to the accumulation of cytosolic self-DNA due to a disrupted TREX1 enzyme which fails to maintain host innate immune tolerance to self-DNA and results in an abnormal innate immune response with clinical consequences. For instance, TREX1 deficient mice develop lethal lymphocytic inflammatory myocarditis with progressive dilated cardiomyopathy and circulatory failure, as well as pathological changes in the lymphoid organs, in both the spleen and the thymus, consistent with an autoimmune cardiomyopathy [119] due to an IFN-dependent response that is characterized by dramatic over-expression of IFN-β mRNA in heart tissue. Consistent with an autoimmune pathology, serum from TREX1-deficient mice contained high concentrations of IgG autoantibodies that strongly stained heart tissue in immunohistochemistry assays. The autoantibodies collected from TREX1-deficient mice were able to bind to knockout and wild-type heart extracts indistinctly, showing that the autoantigens associated with inflammatory myocarditis were not specific to the TREX1 knockout hearts, and broader autoreactivity was observed for sera from older TREX1-deficient mice, as expected with epitope spreading [118].
These findings should not be overlooked in light of the growing number of cases of acute myocarditis and myopericarditis reported in mRNA vaccine recipients, particularly in young males following the second dose [120], [121], [122], which led the FDA to issue a warning on the increased risks of myocarditis and pericarditis following the second dose of a COVID-19 mRNA vaccine.3 Considering that the human TREX1 gene exhibits mutations and that TREX1 gene single nucleotide polymorphisms (SNPs) have been related to severe outcome of infectious disease [123] and autoimmune conditions [124] in humans, it is possible that polymorphisms in TREX1 and in other genes that encode proteins that directly or indirectly regulate cytosolic DNA sensors, may determine susceptibility to nms-mRNA vaccines, influence response to immunization, and influence susceptibility to severe inflammatory disorders, including myocarditis, after mRNA COVID-19 vaccination.
Experimental testing of our hypothesis
Our hypothesis must be tested experimentally using a model animal, such as the rat, with similar immune responses to foreign RNA and TEs, including LINE-1, of humans. Our hypothesis would also need to be tested in a subset of vaccinated and non-vaccinated individuals, using high-resolution genomic comparative analysis of differentially expressed TE and immune gene transcripts. Such an approach would shed light on the connection between the TEs and inflammatory gene regulation networks trigged by cytosolic RNA and DNA, and would contribute to our understanding of the genotoxic, mutagenic, carcinogenic and immunopathogenic risk posed by nms-mRNA vaccines.
The questions that would need to be addressed would be whether: 1) mRNA expression of TEs differs between vaccinated and non-vaccinated individuals, 2) mRNA expression of TREX-1 differs between vaccinated and non-vaccinated individuals, 3) mRNA expression of INFs, IRF3, inflammasome markers differs between vaccinated and non-vaccinated individuals, and 4) whether IgG autoantibodies and detectable reactivity against heart tissue are observed at a higher frequency in vaccinated individuals than in non-vaccinated individuals.
Consequences and discussion
We have argued and presented evidence that vaccine-mRNAs may cause a cascade of cellular innate antiviral responses, eliciting up-regulation of endogenous TEs as well as the activation of RNA and DNA sensors through conserved pathways that involve IFN production, thus promoting the expression of hundreds of IFN target genes that may sustain a chronic antiviral state within the vaccine transfected cells and neighbouring cells. We have also suggested a novel mechanism that could underlie the innate response to nms-mRNAs.
Different host factors may coordinate the responses to nms-mRNA via intrinsic RNA and DNA sensory pathways, and interindividual genetic variations such as SNPs, or splice variants of transcripts of key signalling molecules may hamper the accurate elimination of foreign and self cytosolic nucleic acids, leading to up-regulated sustained pro-inflammatory responses and increasing the risk of autoinflammatory and autoimmune conditions, genomic instability and cancer. Therefore, exposure and subsequent accumulation of nms-mRNA could increase the complexity of intracellular responses to foreign nucleic acids, up-regulating TEs and IFN signalling through an, as yet uncharacterized, TLR-, RIG-I-, MDA-5, IFITs- and cGAS-STING-independent upstream pathway. This would be a new paradigm in our understanding of the cellular responses to synthetic mRNA therapeutic applications. The modifications made to nms-mRNA vaccines confer intracellular mRNA stability [32] and increase translation efficiency [33] but could also be a significant determinant of autoinflammatory and autoimmune responses if, as hypothesized, they activate TEs and other nucleic acid sensors, lead to type-1 interferon and pro-inflammatory cytokine expression, and affect the cell’s innate ability to discriminate non-self vs self cytosolic motifs [125] by disrupting host innate immune tolerance to cytosolic self-DNA.
IFNs are synthesized and secreted by all cell types when their cell surface or cytoplasmic receptors identify viral molecular patterns [126]. Adequate and opportune activation of nucleic acid sensors is essential for the host to eliminate infective viruses. However, the impact of nms-mRNA on nucleic acid sensors and the extent and consequences of intracellular responses to these persistent nucleic acids is still unknown. The induction of a sustained anti-viral cellular status via up-regulation of relevant genes, including those that encode IFN-α and IFN-β, following nms-mRNA vaccination, would likely lead to the chronic upregulation of a pro-inflammatory gene network that could predispose to autoinflammatory and autoimmune conditions. In addition, the pro-inflammatory status and the activation of intracellular RNA and DNA sensors would unsilence endogenous retroelements. These molecular events would increase the risk of genomic, chromosomal, and cellular instability, and carcinogenesis.
Although mouse models of Middle East respiratory syndrome (MERS) [127], SARS-CoV [128], and influenza [129] show vigorous induction of type-I and -III IFNs, the involvement of these cytokines in COVID-19 patients is contentious. Broggi et al. [130] found that mRNA levels of IFN in the naso-oropharyngeal swabs of patients with severe COVID-19 did not differ from those of healthy controls. In contrast, the bronchoalveolar lavage fluid of patients with severe disease presented elevated levels of inflammatory cytokines and type-I (IFN-α and IFN-β) and -III (IFN-λ) IFNs. This is consistent with the involvement of type-III IFNs in the antiviral immune response at epithelial surfaces during early stages of viral infection [130], and suggests coordinated activity by type-I and type-III IFNs during the interplay of innate and adaptive immune responses at the respiratory and gastrointestinal barriers.
In a recent comparative analysis of the immune responses to natural SARS-CoV-2 infection vs. immune responses to COVID mRNA vaccination, phenotypic and transcriptional profiling of immune cells revealed striking up-regulation of type-I and type-II IFNs in COVID-19 patients, but not in vaccinated individuals [131]. These observations were interpreted as consistent with the idea that anti-COVID-19 mRNA vaccines actively suppress type-I IFN signalling while eliciting a robust adaptive immune response. Based on the observations by Ivanova et al. [131] and using reports of adverse events following vaccination from the VAERS database, Seneff et al. [132] argued that SARS-CoV-2 mRNA vaccination impairs type-I IFN signalling, and can affect the regulatory control of protein synthesis and onco-surveillance, paving the way to an increased risk of neurodegeneration, immune thrombocytopenia, myocarditis, Bell's palsy, hepatic disease, suppression of adaptive immune responses, diminished DNA damage repair, and tumorigenesis. We hold a complementary view to that of Seneff et al. [132], as we propose that IFN regulation is different between nms-mRNA vaccinated and naturally-infected individuals owing to differences in the target cells and in the molecular signalling pathways that are activated. Epithelial surfaces of the respiratory and enteric tracts are the main battlefield for natural infections [133], whereas intramuscular administration, systemic distribution, and tissue accumulation of the nms-mRNA vaccine evade natural mucosal barriers, leading to danger sensing signals and non-self DNA and RNA intracellular detection at a systemic level. There is already some experimental evidence for this. Upon intratracheal administration of a synthetic analogue of double-stranded RNA (polyinosine:polycytidylic acid; poly I:C) in mice, which stimulates both TLR3 and the RIG-I–MDA-5 pathway in vivo [134], the lung-resident dendritic cells expressed the highest levels of IFN-λ transcript, during both the early and late phases following the administration of poly I:C. In contrast, epithelial cells, monocytes, and alveolar macrophages expressed type I IFNs and pro-inflammatory cytokines, but not IFN-λ, in response to poly (I:C). Consistent with in vivo data, in vitro TLR7 stimulation only induced up-regulation of pro-inflammatory cytokines while activation of RIG-I and MDA-5 via intracellular delivery of poly (I:C) and of triphosphate hairpin RNA (3p-hpRNA) induced high levels of type-I IFNs, but not type-III IFNs, in a MAVS–dependent manner [130].
We propose that the active suppression of Type-I IFN production that has been observed in vaccinated individuals [131] and discussed by Seneff et al. [132] reflects dysregulation of Type-I IFN-signalling triggered by the NLRP3, AIM2, or MxA inflammasome activation by host-derived molecules recruited upon detection of endogenous indicators of cellular danger or stress [135], as the cytosolic accumulation of either synthetic mRNA, cleavage products of RNAs generated by the antiviral RNAse L pathway or retrotranscribed DNAs. Activation of the inflammasome occurs in response to self and foreign activators (reviewed by [136]), and can be both protective and harmful. Once activated, it can drive immunopathology because IL-1β stimulates systemic inflammation responses by activating NF-κB and c-Jun N-terminal kinase signalling pathways, leading to cytokine storms, which are common in acute inflammatory diseases [28]. Therefore, inflammasome activation and IL-1β production are tightly regulated during viral infection to prevent a damaging hyper-inflammatory response. Type-I IFNs act as potent negative regulators of the NLRP3 inflammasome and have a dual role, both as powerful antiviral agents, and as homeostatic immune regulators. Inflammasome activation can also have a regulatory role in innate antiviral defence, preventing cGAS-STING mediated IFN production during infection with DNA viruses [137], revealing a regulatory circuit wherein type-I IFNs inhibit the inflammasome and the activated inflammasome also inhibits type I-IFN production [138].
SARS-CoV-2 and other human coronaviruses can trigger the inflammasome in infected cells [139], [140], and serum inflammasome markers are related to COVID-19 severity [141]. In peripheral blood mononuclear cells (PBMCs) of moderate to severe COVID-19 patients and tissues of post-mortem patients on autopsy, the NLRP3 inflammasome was activated [141]. However, it is still unknown whether the nms-mRNA vaccines can also, directly or indirectly, activate the inflammasomes in an IFN-dependent or independent way, and following activation, inhibit type-I and type-III IFN production.
So far, the basis of nms-mRNA-induced immunogenicity is still not fully understood. For instance, the cap 2′-O methylation of mRNA prevents recognition by IFN-induced RNA binding proteins [142], and several studies addressing the issue of synthetic RNA 5′ end modifications on RIG-I binding affinity and its impact on the activation of innate immune signalling have suggested that 5́' cap modifications can drive either strong, suboptimal, or abolished IFN signalling (reviewed by [138]), whereas the impact of the synthetic RNA 5́′ modifications on MDA-5 or cGAS activation has yet to be determined. We urgently need controlled experimental studies for a better understanding of mRNA vaccine safety. Specifically, we need to know whether nms-mRNA activates different intracellular nucleic acid sensors to those activated during SARS-Cov-2 infections, whether the nms-mRNA is early and directly detected by intrinsic sensors or cellular targets in an IFN-dependent or an IFN-independent way, and we must understand the molecular mechanisms that underlie early up-regulation and cytosolic accumulation of TEs as a potential source of immune stimulatory self-DNA, genomic instability, and mutagenesis, due to an increase in LINE-1 mediated reverse transcription and integration. Cells with impaired self-DNA removing pathways are known to display genome instability and have a higher risk of malignant transformation. Hence, individuals with particular genetic polymorphisms in genes that encode DNA sensors, such as TREX1, that are exposed to cellular stress and inflammatory stimuli related to the nms-mRNA and to the accumulation of DNA following up-regulation of TEs, may be at more risk of developing severe inflammatory and autoimmune disorders, and carcinogenesis. If this is so, then the ‘one size fits all’ approach of mass vaccination using nms-mRNA technology would not be a safe public health measure for humanity.
If our hypothesis were to be confirmed, the implications for public health would be staggering and appalling in the context of the mass-scale COVID-19 vaccination already taking place, particularly if the nms-mRNA enters brain [82], bone marrow [84], and – if already present in the vaccinee – cancerous or pre-cancerous cells [143], or if the vaccine is administered to females early in their pregnancy and the nms-mRNA transfects embryonic cells [77]. It stands to reason that if our hypothesis is proven to be correct, any other mRNA vaccine candidate should be fully investigated to understand the cytoplasmic and nuclear sensor, intrinsic factors, and signalling pathways activated by every single and combined synthetic 5′ Cap, GC content, polyA tails, and UTRs modifications made to the vaccine mRNA in order to fully elucidate the extent of their downstream signalling mechanisms of action and the potential impacts on health. Knowledge gained from these studies will be crucial for understanding, beyond unproven assumptions, the safety of mRNA vaccines and mRNA-based therapies on human health.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
https://ourworldindata.org/grapher/covid-vaccine-doses-by-manufacturer (accessed on 7 September 2022).
Available online: https://www.ema.europa.eu/en/documents/assessment-report/comirnaty-epar-public-assessment-report_en.pdf (accessed on 24 April 2022).
References
- 1.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., et al. Direct gene transfer into mouse muscle in vivo. Science. 1979;1990(247):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 2.Martinon F., Krishnan S., Lenzen G., Magné R., Gomard E., Guillet J.-G., et al. Induction of virus-specific cytotoxic T lymphocytesin vivo by liposome-entrapped mRNA. Eur J Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 3.Boczkowski D., Nair S.K., Snyder D., Gilboa E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med. 1996;184:465–472. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoerr I., Obst R., Rammensee H., Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., et al. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur S.P., Gupta V. COVID-19 vaccine: a comprehensive status report. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rostenberghe H. Primum Non Nocere. Malaysian J Med Sci. 2021;28:122–124. doi: 10.21315/mjms2021.28.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doulberis M., Papaefthymiou A., Kotronis G., Gialamprinou D., Soteriades E.S., Kyriakopoulos A., et al. Does COVID-19 vaccination warrant the classical principle “ofelein i mi vlaptin”? Medicina (B Aires) 2021;57:253. doi: 10.3390/medicina57030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lecroy K. The lie of primum non nocere. Am Fam Physician. 2001;64:1942. [PubMed] [Google Scholar]
- 10.Cardozo T., Veazey R. Informed consent disclosure to vaccine trial subjects of risk of COVID-19 vaccines worsening clinical disease. Int J Clin Pract. 2021:75. doi: 10.1111/ijcp.13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruno R, McCullough PA, Forcades I Villa T, Henrion-Caude A, García-Gasca T, Zaitzeva GP, et al. SARS-CoV-2 mass vaccination: Urgent questions on vaccine safety that demand answers from international health agencies, regulatory authorities, governments and vaccine developers. Preprint 2021.
- 12.Benteyn D., Heirman C., Bonehill A., Thielemans K., Breckpot K. mRNA-based dendritic cell vaccines. Expert Rev Vaccines. 2015;14:161–176. doi: 10.1586/14760584.2014.957684. [DOI] [PubMed] [Google Scholar]
- 13.Chavda V.P., Hossain M.K., Beladiya J., Apostolopoulos V. Nucleic acid vaccines for COVID-19: a paradigm shift in the vaccine development arena. Biologics. 2021;1:337–356. doi: 10.3390/biologics1030020. [DOI] [Google Scholar]
- 14.Anand P., Stahel V.P. The safety of Covid-19 mRNA vaccines: a review. Patient Saf Surg. 2021;15:20. doi: 10.1186/s13037-021-00291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 16.Kreiter S., Diken M., Pascolo S., Nair S.K., Thielemans K.M., Geall A. RNA vaccination therapy: advances in an emerging field. J Immunol Res. 2016;2016:1–2. doi: 10.1155/2016/9703914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Troilo P.J., Wang X., Griffiths T.G., Pacchione S.J., Barnum A.B., et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11:711–721. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- 18.Harui A., Suzuki S., Kochanek S., Mitani K. Frequency and stability of chromosomal integration of adenovirus vectors. J Virol. 1999;73:6141–6146. doi: 10.1128/JVI.73.7.6141-6146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng C., Baum B.J., Iadarola M.J., O’Connell B.C. Genomic integration and gene expression by a modified adenoviral vector. Nat Biotechnol. 2000;18:176–180. doi: 10.1038/72628. [DOI] [PubMed] [Google Scholar]
- 20.Aldén M., Olofsson Falla F., Yang D., Barghouth M., Luan C., Rasmussen M., et al. Intracellular reverse transcription of Pfizer BioNTech COVID-19 mRNA vaccine BNT162b2 in vitro in human liver cell line. Curr Issues Mol Biol. 2022;44:1115–1126. doi: 10.3390/cimb44030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert P.-H., Ambrosino D.M., Andersen S.R., Baric R.S., Black S.B., Chen R.T., et al. Consensus summary report for CEPI/BC March 12–13, 2020 meeting: Assessment of risk of disease enhancement with COVID-19 vaccines. Vaccine. 2020;38:4783–4791. doi: 10.1016/j.vaccine.2020.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin S., Tang X., Chen Y., Chen K., Fan N., Xiao W., et al. mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduct Target Ther. 2022;7:166. doi: 10.1038/s41392-022-01007-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 25.Goubau D., Deddouche S., Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbalat R., Lau L., Locksley R.M., Barton G.M. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orzalli M.H., Kagan J.C. Apoptosis and necroptosis as host defense strategies to prevent viral infection. Trends Cell Biol. 2017;27:800–809. doi: 10.1016/j.tcb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tate M.D., Mansell A. An update on the NLRP3 inflammasome and influenza: the road to redemption or perdition? Curr Opin Immunol. 2018;54:80–85. doi: 10.1016/j.coi.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Yan N., Chen Z.J. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee S., Basler C.F., Amarasinghe G.K., Leung D.W. Molecular mechanisms of innate immune inhibition by non-segmented negative-sense RNA viruses. J Mol Biol. 2016;428:3467–3482. doi: 10.1016/j.jmb.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nance K.D., Meier J.L. Modifications in an emergency: the role of N1-methylpseudouridine in COVID-19 vaccines. ACS Cent Sci. 2021;7:748–756. doi: 10.1021/acscentsci.1c00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S.C., Sekhon S.S., Shin W.-R., Ahn G., Cho B.-K., Ahn J.-Y., et al. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Mol Cell Toxicol. 2022;18:1–8. doi: 10.1007/s13273-021-00171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durbin A.F., Wang C., Marcotrigiano J., Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. MBio. 2016:7. doi: 10.1128/mBio.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for toll-like receptor 3. J Biol Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 36.Drews K., Tavernier G., Demeester J., Lehrach H., de Smedt S.C., Rejman J., et al. The cytotoxic and immunogenic hurdles associated with non-viral mRNA-mediated reprogramming of human fibroblasts. Biomaterials. 2012;33:4059–4068. doi: 10.1016/j.biomaterials.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 37.Rautsi O., Lehmusvaara S., Salonen T., Häkkinen K., Sillanpää M., Hakkarainen T., et al. Type I interferon response against viral and non-viral gene transfer in human tumor and primary cell lines. J Gene Med. 2007;9:122–135. doi: 10.1002/jgm.997. [DOI] [PubMed] [Google Scholar]
- 38.Dias Junior A.G., Sampaio N.G., Rehwinkel J. A balancing act: MDA5 in antiviral immunity and autoinflammation. Trends Microbiol. 2019;27:75–85. doi: 10.1016/j.tim.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pepini T., Pulichino A.-M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., et al. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185:1025–1040.e14. doi: 10.1016/j.cell.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 1979;2006(314):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 43.Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., et al. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5’-Phosphates. Science. 1979;2006(314):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proceedings of the National Academy of Sciences 2009;106:12067–72. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed]
- 45.Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-I and melanoma differentiation–associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 48.Fensterl V., Sen G.C. Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol. 2015;89:2462–2468. doi: 10.1128/JVI.02744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pichlmair A., Lassnig C., Eberle C.-A., Górna M.W., Baumann C.L., Burkard T.R., et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 50.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt A., Rothenfusser S., Hopfner K.-P. Sensing of viral nucleic acids by RIG-I: From translocation to translation. Eur J Cell Biol. 2012;91:78–85. doi: 10.1016/j.ejcb.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tinari S. The EMA covid-19 data leak, and what it tells us about mRNA instability. BMJ. 2021:n627. doi: 10.1136/bmj.n627. [DOI] [PubMed] [Google Scholar]
- 53.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belyi V.A., Levine A.J., Skalka A.M. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klenerman P., Hengartner H., Zinkernagel R.M. A non-retroviral RNA virus persists in DNA form. Nature. 1997;390:298–301. doi: 10.1038/36876. [DOI] [PubMed] [Google Scholar]
- 56.Geuking M.B., Weber J., Dewannieux M., Gorelik E., Heidmann T., Hengartner H., et al. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science. 1979;2009(323):393–396. doi: 10.1126/science.1167375. [DOI] [PubMed] [Google Scholar]
- 57.Shimizu A., Nakatani Y., Nakamura T., Jinno-Oue A., Ishikawa O., Boeke J.D., et al. Characterisation of cytoplasmic DNA complementary to non-retroviral RNA viruses in human cells. Sci Rep. 2015;4:5074. doi: 10.1038/srep05074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horie M., Honda T., Suzuki Y., Kobayashi Y., Daito T., Oshida T., et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patterson BK, Francisco EB, Yogendra R, Long E, Pise A, Beaty C, et al. SARS-CoV-2 S1 Protein Persistence in SARS-CoV-2 Negative Post-Vaccination Individuals with Long COVID/ PASC-Like Symptoms. 2022.
- 60.Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proceedings of the National Academy of Sciences 2021;118. https://doi.org/10.1073/pnas.2105968118. [DOI] [PMC free article] [PubMed]
- 61.Wells J.N., Feschotte C. A field guide to eukaryotic transposable elements. Annu Rev Genet. 2020;54:539–561. doi: 10.1146/annurev-genet-040620-022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grandi N, Tramontano E. HERV Envelope Proteins: Physiological Role and Pathogenic Potential in Cancer and Autoimmunity. Front Microbiol 2018;9. doi: 10.3389/fmicb.2018.00462. [DOI] [PMC free article] [PubMed]
- 63.Macchietto M.G., Langlois R.A., Shen S.S. Virus-induced transposable element expression up-regulation in human and mouse host cells. Life Sci Alliance. 2020;3:e201900536. doi: 10.26508/lsa.201900536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chuong E.B., Elde N.C., Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 1979;2016(351):1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao K., Du J., Peng Y., Li P., Wang S., Wang Y., et al. LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways. J Autoimmun. 2018;90:105–115. doi: 10.1016/j.jaut.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 66.Tunbak H., Enriquez-Gasca R., Tie C.H.C., Gould P.A., Mlcochova P., Gupta R.K., et al. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat Commun. 2020;11:5387. doi: 10.1038/s41467-020-19170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gázquez-Gutiérrez A., Witteveldt J., Heras S.R., Macias S. Sensing of transposable elements by the antiviral innate immune system. RNA. 2021;27:735–752. doi: 10.1261/rna.078721.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 69.Smit A.F.A., Tóth G., Riggs A.D., Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 70.Sanchez-Luque F.J., Kempen M.-J.-H.-C., Gerdes P., Vargas-Landin D.B., Richardson S.R., Troskie R.-L., et al. LINE-1 evasion of epigenetic repression in humans. Mol Cell. 2019;75:590–604.e12. doi: 10.1016/j.molcel.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 71.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran J v., et al. Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences 2003;100:5280–5. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed]
- 72.Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 73.Soifer H.S. Do Small RNAs Interfere With LINE-1? J Biomed Biotechnol. 2006;2006:1–8. doi: 10.1155/JBB/2006/29049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baba Y., Murata A., Watanabe M., Baba H. Clinical implications of the LINE-1 methylation levels in patients with gastrointestinal cancer. Surg Today. 2014;44:1807–1816. doi: 10.1007/s00595-013-0763-6. [DOI] [PubMed] [Google Scholar]
- 75.Zhao K., Du J., Han X., Goodier J.L., Li P., Zhou X., et al. Modulation of LINE-1 and Alu/SVA retrotransposition by aicardi-goutières syndrome-related SAMHD1. Cell Rep. 2013;4:1108–1115. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu S., Li J., Xu F., Mei S., le Duff Y., Yin L., et al. SAMHD1 inhibits LINE-1 retrotransposition by promoting stress granule formation. PLoS Genet. 2015;11:e1005367. doi: 10.1371/journal.pgen.1005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muñoz-López M, Vilar R, Philippe C, Rahbari R, Richardson SR, Andres-Anton M, et al. LINE-1 Retrotransposition Impacts the Genome of Human Pre-Implantation Embryos and Extraembryonic Tissues. BioRxiv 2019.
- 78.Klawitter S, Fuchs N v., Upton KR, Muñoz-Lopez M, Shukla R, Wang J, et al. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat Commun 2016;7:10286. doi: 10.1038/ncomms10286. [DOI] [PMC free article] [PubMed]
- 79.Protasova M.S., Andreeva T.V., Rogaev E.I. Factors regulating the activity of LINE1 retrotransposons. Genes (Basel) 2021;12:1562. doi: 10.3390/genes12101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Faulkner G.J., Garcia-Perez J.L. L1 mosaicism in mammals: extent, effects, and evolution. Trends Genet. 2017;33:802–816. doi: 10.1016/j.tig.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 81.Bedrosian T.A., Quayle C., Novaresi N., FredH G. Early life experience drives structural variation of neural genomes in mice. Science. 1979;2018(359):1395–1399. doi: 10.1126/science.aah3378. [DOI] [PubMed] [Google Scholar]
- 82.Ormundo L.F., Machado C.F., Sakamoto E.D., Simões V., Armelin-Correa L. LINE-1 specific nuclear organization in mice olfactory sensory neurons. Mol Cell Neurosci. 2020;105 doi: 10.1016/j.mcn.2020.103494. [DOI] [PubMed] [Google Scholar]
- 83.Terry D.M., Devine S.E. Aberrantly high levels of somatic LINE-1 expression and retrotransposition in human neurological disorders. Front Genet. 2020;10 doi: 10.3389/fgene.2019.01244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marasca F., Sinha S., Vadalà R., Polimeni B., Ranzani V., Paraboschi E.M., et al. LINE1 are spliced in non-canonical transcript variants to regulate T cell quiescence and exhaustion. Nat Genet. 2022;54:180–193. doi: 10.1038/s41588-021-00989-7. [DOI] [PubMed] [Google Scholar]
- 85.Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sedic M., Senn J.J., Lynn A., Laska M., Smith M., Platz S.J., et al. Safety evaluation of lipid nanoparticle-formulated modified mRNA in the Sprague-Dawley rat and cynomolgus monkey. Vet Pathol. 2018;55:341–354. doi: 10.1177/0300985817738095. [DOI] [PubMed] [Google Scholar]
- 87.Tanaka H., Takata N., Sakurai Y., Yoshida T., Inoue T., Tamagawa S., et al. Delivery of oligonucleotides using a self-degradable lipid-like material. Pharmaceutics. 2021;13:544. doi: 10.3390/pharmaceutics13040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sato Y., Matsui H., Yamamoto N., Sato R., Munakata T., Kohara M., et al. Highly specific delivery of siRNA to hepatocytes circumvents endothelial cell-mediated lipid nanoparticle-associated toxicity leading to the safe and efficacious decrease in the hepatitis B virus. J Control Release. 2017;266:216–225. doi: 10.1016/j.jconrel.2017.09.044. [DOI] [PubMed] [Google Scholar]
- 89.McKerrow W, Wang X, Mendez-Dorantes C, Mita P, Cao S, Grivainis M, et al. LINE-1 expression in cancer correlates with p53 mutation, copy number alteration, and S phase checkpoint. Proceedings of the National Academy of Sciences 2022;119. doi: 10.1073/pnas.2115999119. [DOI] [PMC free article] [PubMed]
- 90.Tiwari B., Jones A.E., Caillet C.J., Das S., Royer S.K., Abrams J.M. p53 directly represses human LINE1 transposons. Genes Dev. 2020;34:1439–1451. doi: 10.1101/gad.343186.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marston J.L., Greenig M., Singh M., Bendall M.L., Duarte R.R.R., Feschotte C., et al. SARS-CoV-2 infection mediates differential expression of human endogenous retroviruses and long interspersed nuclear elements. JCI Insight. 2021;6 doi: 10.1172/jci.insight.147170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van der Kuyl A.C. HIV infection and HERV expression: a review. Retrovirology. 2012;9:6. doi: 10.1186/1742-4690-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jones R.B., Song H., Xu Y., Garrison K.E., Buzdin A.A., Anwar N., et al. LINE-1 retrotransposable element DNA accumulates in HIV-1-infected cells. J Virol. 2013;87:13307–13320. doi: 10.1128/JVI.02257-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li F., Nellåker C., Sabunciyan S., Yolken R.H., Jones-Brando L., Johansson A.-S., et al. Transcriptional derepression of the ERVWE1 locus following influenza A virus infection. J Virol. 2014;88:4328–4337. doi: 10.1128/JVI.03628-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krischuns T., Günl F., Henschel L., Binder M., Willemsen J., Schloer S., et al. Phosphorylation of TRIM28 enhances the expression of IFN-β and proinflammatory cytokines during HPAIV infection of human lung epithelial cells. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kwun H.J., Han H.J., Lee W.J., Kim H.S., Jang K.L. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res. 2002;86:93–100. doi: 10.1016/S0168-1702(02)00058-8. [DOI] [PubMed] [Google Scholar]
- 97.Brisse M., Ly H. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hale B.G. Antiviral immunity triggered by infection-induced host transposable elements. Curr Opin Virol. 2022;52:211–216. doi: 10.1016/j.coviro.2021.12.006. [DOI] [PubMed] [Google Scholar]
- 99.Funabiki M., Kato H., Miyachi Y., Toki H., Motegi H., Inoue M., et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 100.Gasior S.L., Wakeman T.P., Xu B., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sudhindar P.D., Wainwright D., Saha S., Howarth R., McCain M., Bury Y., et al. HCV activates somatic L1 retrotransposition—a potential hepatocarcinogenesis pathway. Cancers (Basel) 2021;13:5079. doi: 10.3390/cancers13205079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Situ Y., Chung L., Lee C., Ho V. MRN (MRE11-RAD50-NBS1) complex in human cancer and prognostic implications in colorectal cancer. Int J Mol Sci. 2019;20:816. doi: 10.3390/ijms20040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Philippe C., Vargas-Landin D.B., Doucet A.J., van Essen D., Vera-Otarola J., Kuciak M., et al. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife. 2016:5. doi: 10.7554/eLife.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Cecco M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019;566:73–78. doi: 10.1038/s41586-018-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meyer K., Patra T., Vijayamahantesh R.R. SARS-CoV-2 spike protein induces paracrine senescence and leukocyte adhesion in endothelial cells. J Virol. 2021:95. doi: 10.1128/JVI.00794-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stetson D.B., Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 107.Hwang H.S., Lee M.H., Choi M.H., Kim H.A. Induction of pro-inflammatory cytokines by 29-kDa FN-f via cGAS/STING pathway. BMB Rep. 2019;52:336–341. doi: 10.5483/BMBRep.2019.52.5.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen Q., Sun L., Chen Z.J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 109.Barber G.N. STING: infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Motwani M., Pesiridis S., Fitzgerald K.A. DNA sensing by the cGAS–STING pathway in health and disease. Nat Rev Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 111.Han L., Zhuang M., Deng J., Zheng Y., Zhang J., Nan M., et al. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5–MAVS, TLR3–TRIF, and cGAS–STING signaling pathways. J Med Virol. 2021;93:5376–5389. doi: 10.1002/jmv.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ablasser A., Chen Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science. 1979;2019:363. doi: 10.1126/science.aat8657. [DOI] [PubMed] [Google Scholar]
- 113.Lei Y., Zhang J., Schiavon C.R., He M., Chen L., Shen H., et al. SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE 2. Circ Res. 2021;128:1323–1326. doi: 10.1161/CIRCRESAHA.121.318902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mazur D.J., Perrino F.W. Excision of 3′ termini by the Trex1 and TREX2 3′→5′ exonucleases. J Biol Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- 115.Lindahl T., Barnes D.E., Yang Y.-G., Robins P. Biochemical properties of mammalian TREX1 and its association with DNA replication and inherited inflammatory disease. Biochem Soc Trans. 2009;37:535–538. doi: 10.1042/BST0370535. [DOI] [PubMed] [Google Scholar]
- 116.Yan N. Immune diseases associated with TREX1 and STING dysfunction. J Interferon Cytokine Res. 2017;37:198–206. doi: 10.1089/jir.2016.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rice G.I., Rodero M.P., Crow Y.J. Human disease phenotypes associated with mutations in TREX1. J Clin Immunol. 2015;35:235–243. doi: 10.1007/s10875-015-0147-3. [DOI] [PubMed] [Google Scholar]
- 118.Stetson D.B., Ko J.S., Heidmann T., Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morita M., Stamp G., Robins P., Dulic A., Rosewell I., Hrivnak G., et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bautista García J., Peña Ortega P., Bonilla Fernández J.A., Cárdenes León A., Ramírez Burgos L., Caballero D.E. Acute myocarditis after administration of the BNT162b2 vaccine against COVID-19. Revista Española de Cardiología (English Edition) 2021;74:812–814. doi: 10.1016/j.rec.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dionne A., Sperotto F., Chamberlain S., Baker A.L., Powell A.J., Prakash A., et al. Association of Myocarditis With BNT162b2 Messenger RNA COVID-19 Vaccine in a Case Series of Children. JAMA Cardiol. 2021;6:1446. doi: 10.1001/jamacardio.2021.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mevorach D., Anis E., Cedar N., Bromberg M., Haas E.J., Nadir E., et al. Myocarditis after BNT162b2 mRNA Vaccine against Covid-19 in Israel. N Engl J Med. 2021;385:2140–2149. doi: 10.1056/NEJMoa2109730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Silva D de C, Amoras E da SG, Moura TCF, Lopes FT, Gomes STM, Costa CA da, et al. TREX1 531C>T Polymorphism is Associated with High Proviral Load Levels in HTLV-1-Infected Persons. Viruses 2019;12:7. doi: 10.3390/v12010007. [DOI] [PMC free article] [PubMed]
- 124.Nezos A., Makri P., Gandolfo S., de Vita S., Voulgarelis M., Crow M.K., et al. TREX1 variants in Sjogren’s syndrome related lymphomagenesis. Cytokine. 2020;132 doi: 10.1016/j.cyto.2019.154781. [DOI] [PubMed] [Google Scholar]
- 125.Leung D.W., Amarasinghe G.K. When your cap matters: structural insights into self vs non-self recognition of 5′ RNA by immunomodulatory host proteins. Curr Opin Struct Biol. 2016;36:133–141. doi: 10.1016/j.sbi.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Broz P., Monack D.M. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol. 2013;13:551–565. doi: 10.1038/nri3479. [DOI] [PubMed] [Google Scholar]
- 127.Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Investig. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frieman M.B., Chen J., Morrison T.E., Whitmore A., Funkhouser W., Ward J.M., et al. SARS-CoV Pathogenesis Is Regulated by a STAT1 Dependent but a Type I, II and III Interferon Receptor Independent Mechanism. PLoS Pathog. 2010;6:e1000849. doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Major J., Crotta S., Llorian M., McCabe T.M., Gad H.H., Priestnall S.L., et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 1979;2020(369):712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., lo Cascio A., et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 1979;2020(369):706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ivanova E.N., Devlin J.C., Buus T.B., Koide A., Shwetar J., Cornelius A., et al. Discrete immune response signature to SARS-CoV-2 mRNA vaccination versus infection. MedRxiv. 2021:preprint. [Google Scholar]
- 132.Seneff S., Nigh G., Kyriakopoulos A.M., McCullough P.A. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and MicroRNAs. Food Chem Toxicol. 2022;164 doi: 10.1016/j.fct.2022.113008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hermant P., Michiels T. Interferon-λ in the context of viral infections: production, response and therapeutic implications. J Innate Immun. 2014;6:563–574. doi: 10.1159/000360084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 135.Broz P., Dixit V.M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 136.Davis B.K., Wen H., Ting J.-P.-Y. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang Y., Ning X., Gao P., Wu S., Sha M., Lv M., et al. Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity. 2017;46:393–404. doi: 10.1016/j.immuni.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 138.Carty M., Guy C., Bowie A.G. Detection of viral infections by innate immunity. Biochem Pharmacol. 2021;183 doi: 10.1016/j.bcp.2020.114316. [DOI] [PubMed] [Google Scholar]
- 139.Siu K., Yuen K., Castano-Rodriguez C., Ye Z., Yeung M., Fung S., et al. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ahn M., Anderson D.E., Zhang Q., Tan C.W., Lim B.L., Luko K., et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat Microbiol. 2019;4:789–799. doi: 10.1038/s41564-019-0371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rodrigues T.S., de Sá K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2021:218. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Williams G.D., Gokhale N.S., Snider D.L., Horner S.M. The mRNA Cap 2′- O -methyltransferase CMTR1 regulates the expression of certain interferon-stimulated genes. MSphere. 2020:5. doi: 10.1128/mSphere.00202-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kazazian H.H., Moran J.v. Mobile DNA in health and disease. N Engl J Med. 2017;377:361–370. doi: 10.1056/NEJMra1510092. [DOI] [PMC free article] [PubMed] [Google Scholar]