Abstract
Pathogens produce virulence factors that interact directly with host molecules, but in many cases the host targets are unknown. The genetic and molecular identification of these orphan targets is often not feasible with mammalian experimental models. However, a substantial number of known targets are molecules and pathways that are conserved among eukaryotes, and therefore the use of nonmammalian model hosts to identify orphan targets may prove useful. To demonstrate the feasibility of this approach, we transformed the nematode Caenorhabditis elegans with a gene encoding the catalytic subunit of pertussis toxin (PTX), which in mammals inactivates Go/iα proteins. Expression of PTX in C. elegans produced phenotypes almost identical to those of a null mutation in the nematode gene encoding Go/iα. Furthermore, PTX suppressed the phenotype of a constitutively active form of nematode Go/iα protein. These results indicate that PTX is functional in nematodes and acts specifically on the C. elegans homologue of the mammalian target.
The biochemical function and host target of many microbial virulence factors remain unknown. Genetic screening using randomly mutagenized organisms is a powerful tool for identifying components of a pathway, but this use of genetics is not feasible for most experimental animals or mammalian cell culture systems used for infectious disease. An alternative strategy for identifying host targets of pathogens is to use a highly tractable genetic model organism, such as the nematode Caenorhabditis elegans.
The advantages of C. elegans as a model organism have been described in detail (6, 22, 30). The microscopic animals are grown at low cost on Petri plates, with a generation time of 3 days and broods numbering about 200 progeny. The cellular anatomy of wild-type C. elegans is essentially invariant and has been described completely. Because the nematodes are transparent, internal tissues of live specimens can be observed by microscopy and the animals can be recovered for further analysis. C. elegans grows as either a self-fertilizing hermaphrodite or in crosses between males and hermaphrodites, facilitating classical genetic analysis and manipulation. The genome of C. elegans has been sequenced (3). Nematodes can be stably transformed by exogenous DNA (18), reverse genetics can be used to create mutations in a specific gene (20), and the functions of many genes can be blocked by RNA interference (7, 9). Transcription throughout the genome can be analyzed using microarrays (21).
These advantages have led a number of researchers to develop experimental systems that use C. elegans to investigate bacterial pathogenesis. A slow-killing infection of C. elegans by Pseudomonas aeruginosa has been described earlier (26, 27); in addition, there are two distinct ways in which P. aeruginosa kills C. elegans rapidly with small-molecule toxins (5, 17). For one mode of toxin-mediated Pseudomonas killing, a mutation in C. elegans that confers strong resistance has been identified (5), suggesting that genetic analysis of the nematode will be useful for understanding the host-pathogen interaction. Two research groups recently reported that Salmonella enterica serovar Typhimurium colonizes the nematode gut and eventually kills the animals (1, 15). Infection of C. elegans by a bacterium that is apparently a natural parasite of nematodes but is not known to be a pathogen of mammals also has been described previously (12).
In the present work, we describe a strategy that allows C. elegans to be used to search for host targets regardless of whether intact bacteria or their secretions have any effect on the nematodes. In this method, C. elegans is transformed with a bacterial virulence gene so that the nematodes themselves manufacture the virulence protein in their cytoplasm. A phenotype resulting from expression of the bacterial gene suggests that the virulence factor has a target that is conserved between mammals and nematodes. In these cases, host factors in the target pathway can be identified by screening C. elegans for mutations that alter the phenotype produced by the virulence gene.
Pertussis toxin (PTX) is well characterized biochemically and has a known target (8). PTX is an ADP-ribosylating enzyme that acts on a subset of heterotrimeric G proteins; the toxin covalently modifies Go/iα subunits and thereby inactivates their signalling. We show that when PTX expression is induced in transgenic C. elegans, phenotypes appear that are almost identical to those of mutations in the nematode gene encoding Go/iα, indicating that the toxin interacts in vivo with its predicted nematode target.
MATERIALS AND METHODS
Nematode strains and culture.
C. elegans strains, genotypes, and sources are listed in Table 1; a standard nomenclature has previously been described (13, 22). Nematodes were grown at room temperature (approximately 22°C) on NGM agar seeded with Escherichia coli strain OP50 (30).
TABLE 1.
C. elegans strainsa
| Strain | Genotype | Source |
|---|---|---|
| N2 | Wild type | CGC |
| CB251 | unc-36(e251) | CGC |
| CB363 | goa-1(n363) | CGC |
| XA1413 | unc-36(e251); qaEx1410[unc-36(+) hsp16-2::ptx] | This study |
| XA1433 | unc-36(e251); qaIs1400[unc-36(+) hsp16-2::ptx] | This study |
| PS1493 | dpy-20(e1362); syIs9[dpy-20(+) goa-1(Q205L)] | P. Sternberg |
CGC, Caenorhabditis Genetics Center, University of Minnesota, St. Paul. Ex, extrachromosomal transgenic array. Is, chromosomally integrated transgenic array.
Construction of PTX expression vector.
The ptx gene, encoding the catalytic subunit S1 of PTX, was amplified from plasmid pPTX42 (16) by PCR using primers 5′-CGGGATCCATGGACGATCCTCCCGCCACCGTATACC and 5′-GGGGTACCCTAGAACGAATACGCGATGCTTTCGTAG. The PCR product lacked all nucleotides encoding the cleavable signal sequence except an ATG to encode an initial methionine. Restriction sites for BamHI and KpnI were included in the primers, and the product was cloned using those enzymes in the C. elegans expression vector pPD49.78 (18). The insert in the resulting plasmid, pCBD14, was verified by DNA sequencing.
Transformation of nematodes.
Standard methods of transformation were used (18). Parent strain CB251 has an uncoordinated (Unc) phenotype because of a mutation in the gene unc-36 (2, 22). Transformation with R1p16, a plasmid containing the wild-type unc-36 gene, restores CB251 to wild-type locomotion (11). A DNA solution containing 50 ng of R1p16 per μl and 50 ng of pCBD14 per μl was injected into the gonad of parental worms, and F1 progeny that were non-Unc were propagated. Strains, including XA1413, were established when the array was transmitted to F2 and subsequent generations. Presence of the ptx gene was confirmed by PCR.
Integration of ptx-carrying array.
In strain XA1413 the transgenic array that includes ptx is transmitted to approximately 16% of progeny. L4 stage XA1413 animals were gamma irradiated at a dose of 4,500 rads. F1 progeny were transferred to individual plates, and their broods were screened for an array transmission frequency of about 75%, indicating heterozygous integration into a chromosome. From these heterozygotes, numerous F2 progeny were picked, allowed to self-fertilize, and screened for 100% transmission, indicating homozygosity of the integrated array. A line thus established was outcrossed six times to unc-36 mutants to remove secondary mutations induced by radiation, and the presence of ptx was confirmed by PCR and Southern hybridization (data not shown). The resulting strain, XA1433, was used for all reported experiments.
Immunoblotting.
Strains N2 and XA1433 were grown to near saturation on duplicate 10-cm-diameter plates of NGM agar and E. coli OP50. One plate of each strain was heat shocked at 33°C for 2 h while the second plate was kept at room temperature. The worms were collected in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) and washed once in TE, and then they were washed twice more in TE to which was added 10 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, and 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) detergent. Samples were brought to 80 μl of densely packed worms, 20 μl of 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added, and the samples were boiled for 10 min. Each lane of a 12.5% gel was loaded with 20 μl. Purified PTX subunit S1 (List Biological Laboratories) was used as a positive control. After electrophoresis and transfer to a membrane, protein was detected using a monoclonal antibody to S1 (gift of R. Rappuoli) diluted 1:500. Donkey anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories) conjugated to horseradish peroxidase was diluted 1:50,000. Signal was detected by chemiluminescence (ECL+; Amersham).
Heat shock assays.
Nematodes were washed from a stock plate into M9 buffer (30) and aliquoted to two microfuge tubes. One tube was incubated in a water bath at 33°C for 20 min, while the control tube was kept at room temperature. Nematodes were pipetted to fresh NGM plates seeded with E. coli OP50. Locomotion was observed and photographed beginning 2 h after heat shock. Measurement of the pharyngeal pumping rate was performed as described previously (10) in the interval 4 to 8 h after heat shock.
Expression of ptx in constitutive goa-1 background.
Hermaphrodites of strain PS1493, carrying the constitutively active goa-1 construct syIs9 (19), were mated either by XA1433 males carrying ptx or by wild-type control males. PS1493 worms have a dominant sluggish phenotype that prevents distinguishing their cross-progeny from self-progeny directly. Therefore, virgin (L4 stage) hermaphrodite progeny were separated from their male siblings; the next day, when the hermaphrodites had matured into adults, they were heat shocked, picked to individual plates, examined, and photographed. Three days later the plates were scored for segregation of the background mutations in the parent strains used to create the transgenic lines, i.e., unc-36 from XA1433 and dpy-20 from PS1493 (19). Only cross-progeny segregated the markers; data from self-progeny were disregarded.
RESULTS
The C. elegans genome contains a single putative target of PTX.
PTX attaches an ADP-ribose moiety to a cysteine residue four amino acids from the carboxyl terminus of target Go/iα proteins (28). The genome of the nematode C. elegans has been sequenced (3), and analysis showed that there are 20 predicted Gα proteins (14). Examination of the predicted amino acid sequences indicated that only one, encoded by the gene goa-1, is in the Go/iα class; it is also the only C. elegans Gα in which cysteine is the fourth amino acid from the carboxyl terminus. GOA-1 protein is expressed in virtually all neurons, many muscles, and some miscellaneous cells of the nematode (19, 23).
Mutations in goa-1 have been characterized previously (19, 23). Animals with the deletion allele goa-1(n363) move hyperactively, with an increase in their body flexion, but their feeding (by contraction of pharyngeal muscles) is slowed (23). Nematodes with a constitutively active goa-1 mutation have an opposite locomotion phenotype, moving sluggishly with little flexion (19). Thus, Go/iα in C. elegans is a modulator of several neuromuscular functions.
Nematodes transformed with the ptx gene express PTX.
We predicted that if PTX functions normally in C. elegans, it would inactivate GOA-1 protein, thereby producing phenotypes similar to those of the deletion mutation goa-1(n363). If PTX affects only GOA-1, expression would not be expected to be lethal, because goa-1 deletion mutants are viable. However, the possibility existed that PTX expression would have pleiotropic effects, possibly leading to lethality. We therefore used an inducible expression system. The ptx gene was cloned downstream of nematode promoter hsp16-2, which is induced by heat shock and, like goa-1, functions in neurons and muscles (18, 25). The construct, pCBD14, encodes a protein identical to the mature form (i.e., after signal sequence cleavage) of the PTX catalytic subunit S1, except for the addition of a methionine residue at the amino terminus.
We chose as a transformation marker the unc-36 gene, which encodes a predicted calcium channel (22). unc-36 mutants are healthy but severely uncoordinated (Unc phenotype), and their locomotion defect is readily distinguished from the hyperactivity of goa-1 mutants. unc-36 mutants transformed with the wild-type unc-36 gene are restored to normal locomotion and are indistinguishable from the wild type. Transgenic nematodes were generated by injecting a DNA mixture of pCBD14 and an unc-36(+) clone into the gonads of parental unc-36 mutants. F1 progeny that inherited the unc-36(+) marker were easily identified because they had wild-type locomotion in a background of Unc animals. Transgenic lines were established in cases in which the marker was inherited by subsequent generations. Eight independent lines were generated, and the presence of the ptx gene was confirmed in each by PCR (data not shown). Preliminary characterization indicated that the phenotype expected after heat shock, hyperactive locomotion, was present (data not shown).
In C. elegans, exogenous transforming DNA assembles into an extrachromosomal tandem array that is unstable mitotically and inherited by less than 100% of progeny; the frequency of transmission varies widely between strains (18). Although strains with unstable arrays can be maintained indefinitely by propagating animals carrying the marker, any given nematode can be a genetic mosaic that has the array in only some cells, and this variability hampers analysis. Arrays can be stabilized, however, by using gamma irradiation to integrate them into the C. elegans genome, after which they are 100% heritable and contained in every cell of the nematode (18). We integrated the ptx array of strain XA1413, outcrossed the nematodes multiple times to remove secondary mutations induced by radiation, and confirmed the presence of ptx by PCR and Southern hybridization (data not shown). All subsequent analyses were performed using the resulting strain, XA1433.
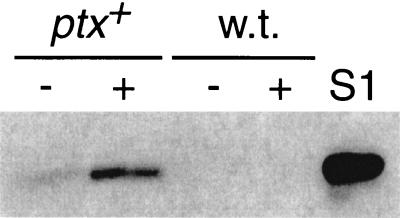
Without heat shock, nematodes carrying the ptx gene expressed the toxin at a level barely detectable by immunoblotting (Fig. 1), but no phenotypes were observed. Heat shocking of transgenic nematodes greatly increased the expression of PTX (Fig. 1), indicating that the heat shock promoter functions as expected.
FIG. 1.
Expression of PTX in transgenic C. elegans. Immunoblot showing the catalytic subunit of PTX in nematodes without (−) or with (+) heat shock. ptx+, nematode strain XA1433 carrying the ptx gene under a heat shock promoter. w.t., wild-type nematode strain N2. S1, 500 pg of purified catalytic subunit S1. Brightness and contrast were adjusted using Adobe Photoshop.
C. elegans expressing PTX mimics Go/iα mutants.
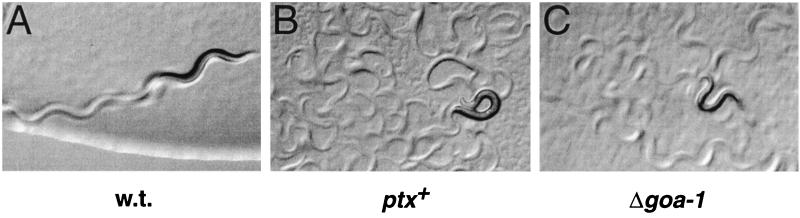
Heat shocked XA1433 animals exhibited dramatic phenotypes. The body flexion of the nematodes was greatly increased compared to that of the wild type, which was observable in both the animals themselves and the tracks they left in a lawn of bacteria (Fig. 2, compare B to A). This phenotype strikingly resembles that of goa-1 loss-of-function mutants (Fig. 2C). Wild-type controls did not display this phenotype after heat shock (Fig. 2A), nor did transgenic animals carrying the unc-36(+) transformation marker but not ptx (data not shown). One difference between PTX-expressing worms and goa-1 mutants was observed. Although both have greatly increased body flexion, goa-1 mutants roam widely across a lawn of bacteria, whereas PTX-expressing worms move back and forth in one spot, thus obliterating their own tracks (Fig. 2B).
FIG. 2.
Phenotypes of PTX expression. Adult nematodes were heat shocked, transferred to NGM agar seeded with E. coli, and photographed. (A) w.t., wild-type strain N2. (B) ptx+, strain XA1433, carrying a heat shock inducible ptx gene. (C) Δgoa-1, strain CB363, a goa-1 deletion mutant. Color transparencies were digitally scanned, converted to grayscale, and adjusted slightly for brightness and contrast with Adobe Photoshop.
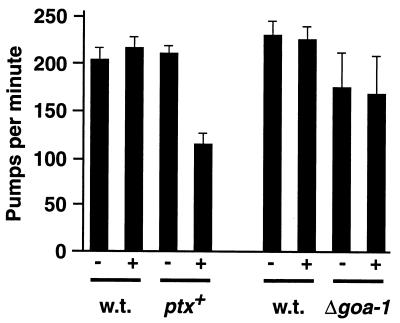
To quantify the effects of PTX expression we measured pumping, the rate of pharyngeal contraction by which C. elegans feeds on bacteria. The pumping rate of a goa-1 deletion mutant is strongly reduced from that of the wild type (23). As shown in Fig. 3, XA1433 transgenic animals also had a markedly reduced pumping rate after heat shock induction of PTX. The transgenic strain pumped identically to the wild type in the absence of heat shock, and heat shock alone did not affect the pumping of either wild-type nematodes or the goa-1 deletion mutants.
FIG. 3.
Rates of pharyngeal pumping without (−) or with (+) heat shock treatment. w.t., wild-type strain N2. ptx+, strain XA1433, carrying a heat shock inducible ptx gene. Δgoa-1, strain CB363 carrying a goa-1 deletion allele, n363. Each cluster of four shows assays on a separate day. Data are mean pumps per minute ± standard deviation for 12 animals.
Expression of PTX suppresses a constitutively active Go/iα mutation.
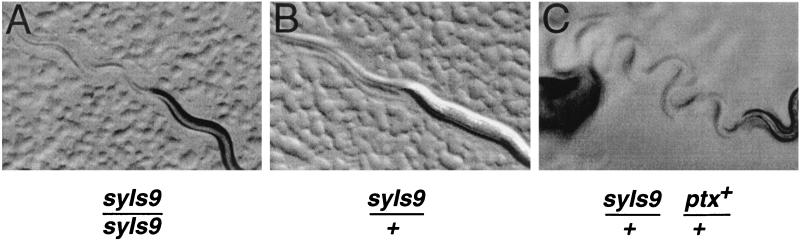
The availability of C. elegans expressing a constitutively active form of GOA-1 provided a further test of the specificity of PTX activity. The transgene syIs9 contains a glutamine-to-leucine mutation that locks GOA-1 protein in the GTP-bound conformation, making it constitutively active (19). Nematodes carrying this construct move sluggishly, with little body flexion (Fig. 4A), a phenotype opposite to that of loss-of-function mutants. We hypothesized that if PTX modifies GOA-1 in C. elegans, it will inactivate the constitutive protein and therefore suppress the sluggish phenotype. We mated hermaphrodites carrying syIs9 to either XA1433 males carrying the ptx transgene or wild-type control males. When progeny were heat shocked, animals heterozygous for both syIs9 and ptx were hyperactive, moved with increased body flexion, and were virtually identical to PTX-expressing worms that lacked syIs9 (compare Fig. 4C to 2B). Control progeny, which were heterozygous for syIs9 alone, retained the sluggish, low-flexion phenotype (Fig. 4B).
FIG. 4.
Rescue of constitutive goa-1 mutation by PTX expression. Nematodes were heat shocked, transferred to seeded NGM agar, and photographed. (A) Parental strain PS1493, carrying the constitutive goa-1 transgene syIs9. (B) Progeny of PS1493 hermaphrodite and wild-type male. (C) Progeny of PS1493 and a ptx male. Genotypes of relevant integrated transgenes are indicated; marker genotypes are omitted for clarity. Images were processed as described in the legend to Fig. 2.
DISCUSSION
The ability to identify bacterial virulence factors outpaces the ability to understand their modes of action. The advent of complete genome sequencing promises to make the full catalog of virulence factors known, but methods are still required to identify their host targets and biochemical functions. Most experimental models for pathogenesis rely on live vertebrates or their cultured cells, and in these systems genetic manipulation of the host is often cumbersome and in some cases impossible.
Some researchers have turned to readily manipulated model organisms, such as C. elegans. Using these nematodes as host, experimental systems have been developed for the human pathogens P. aeruginosa (5, 17, 26, 27) and S. enterica serovar Typhimurium (1, 15) as well as for Microbacterium nematophilum (12), an apparently nematode-specific pathogen. These models require either that whole, live bacteria are pathogenic to C. elegans or that the bacteria produce a diffusible toxic substance.
Despite these successes, it seems unlikely that more than a few mammalian pathogens will affect C. elegans directly. Pathogenesis is a complex collection of sequential phenomena, including host recognition, adherence, virulence gene induction, virulence factor delivery, and evasion of host defenses. Bacteria that have evolved to perform these tasks in a mammal face a different set of challenges with a nematode. Importantly, C. elegans is a natural predator of bacteria, so that before a pathogen can recognize, adhere to, and colonize this host it must avoid lysis and digestion by tissues highly specialized for those purposes.
In another sense, however, the differences between a nematode and a mammal are small. Both possess a collection of fundamental cellular processes that are highly conserved in eukaryotes despite huge evolutionary divergence of other characteristics. Translation of mRNA into protein, signal transduction through G proteins, and regulation of cytoskeletal structure are examples of such processes, and they are also examples of targets of bacterial virulence factors: diphtheria toxin and exotoxin A of P. aeruginosa inactivate elongation factor 2, an essential component of translation (29). PTX and cholera toxin modify heterotrimeric G proteins (8, 24). Yops produced by Yersinia spp. interact with the cytoskeleton (4).
It is therefore expected that the targets of many orphan virulence factors also will be conserved molecules and pathways. In these cases, expression of the virulence factor in nematodes is likely to produce an observable phenotype, thus permitting the use of C. elegans genetics to identify and analyze host targets. To demonstrate the feasibility of this approach, we have made transgenic C. elegans containing a gene for the catalytic subunit S1 of PTX. Toxin expression was made inducible by use of a heat shock promoter. In the absence of the induction signal no phenotype was observed, despite a very low level of basal expression. Induction of PTX produced two dramatic phenotypes: a strong increase in body flexion during locomotion and a strong reduction in pharyngeal contraction rate. These phenotypes are essentially identical to those of mutant nematodes that lack GOA-1, the protein predicted to be the target of PTX. The simplest explanation for these results is that PTX functions in vivo in C. elegans as it does in mammals, i.e., by inactivating Go/iα subunits, thus mimicking the loss-of-function mutation. Because intensive study of C. elegans has provided a wealth of strains and reagents, we were able to conduct an additional test for the activity of PTX in nematodes. Animals expressing a constitutively active form of GOA-1 move sluggishly with little body flexion, a phenotype opposite to that of animals that lack the protein altogether. We found that PTX expression completely suppressed this sluggish phenotype and again mimicked the phenotype of Go/iα deletion.
The most parsimonious interpretation of these results, taken as a whole, is that PTX finds the right target in C. elegans. Although the evolutionary distance between nematodes and mammals is huge, the Go/iα protein is so sufficiently conserved that it functions as a PTX target in both organisms. This is despite the fact that live, virulent Bordetella pertussis cells, the source of PTX, have no effect on C. elegans (C. Darby, unpublished data). These experiments, therefore, establish the feasibility of expressing orphan virulence proteins in the nematode as part of a strategy to identify targets genetically, regardless of whether the bacterium in question is a nematode pathogen. When virulence factor expression produces a phenotype in C. elegans, nematodes can be screened for mutations that either abolish the phenotype or enhance it. Some of these mutations are expected to be in genes that encode either the direct target of the virulence factor or a component of a pathway that includes the target. Using well-developed techniques of C. elegans genetics, these genes can be identified and further characterized.
ACKNOWLEDGMENTS
We thank A. Fire for expression vector pPD49.78, H. R. Horvitz for clone R1p16, S. Kim for the use of a microinjection apparatus, R. Rappuoli for antibody to S1, and P. Sternberg for strain PS1493. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. We are grateful to K. Guilleman and E. Joyce for critical readings of the manuscript.
This work was funded by contracts with Protein Design Labs Inc. and the U.S. Defense Advanced Research Projects Agency.
REFERENCES
- 1.Aballay A, Yorgey P, Ausubel F M. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–1542. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darby C, Cosma C L, Thomas J H, Manoil C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein H F, Shakes D C. Caenorhabditis elegans: modern biological analysis of an organism. San Diego, Calif: Academic Press; 1995. [Google Scholar]
- 7.Fraser A G, Kamath R S, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 8.Gierschik P. ADP-ribosylation of signal-transducing guanine nucleotide-binding proteins by pertussis toxin. Curr Top Microbiol Immunol. 1992;175:69–96. doi: 10.1007/978-3-642-76966-5_4. [DOI] [PubMed] [Google Scholar]
- 9.Gonczy P, Echeverri G, Oegema K, Coulson A, Jones S J, Copley R R, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume A M, Martin C, Ozlu N, Bork P, Hyman A A. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 10.Hajdu-Cronin Y M, Chen W J, Patikoglou G, Koelle M R, Sternberg P W. Antagonism between Goα and Gqα in Caenorhabditis elegans: the RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes Dev. 1999;13:1780–1793. doi: 10.1101/gad.13.14.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman M A, Vassilieva L L, Horvitz H R, Shaw J E, Herman R K. The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell. 1995;83:101–110. doi: 10.1016/0092-8674(95)90238-4. [DOI] [PubMed] [Google Scholar]
- 12.Hodgkin J, Kuwabara P E, Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol. 2000;10:1615–1618. doi: 10.1016/s0960-9822(00)00867-8. [DOI] [PubMed] [Google Scholar]
- 13.Horvitz H R, Brenner S, Hodgkin J, Herman R K. A uniform genetic nomenclature for the nematode Caenorhabditis elegans. Mol Gen Genet. 1979;175:129–133. doi: 10.1007/BF00425528. [DOI] [PubMed] [Google Scholar]
- 14.Jansen G, Thijssen K L, Werner P, van der Horst M, Hazendonk E, Plasterk R H. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- 15.Labrousse A, Chauvet S, Couillault C, Kurz C L, Ewbank J J. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol. 2000;10:1543–1545. doi: 10.1016/s0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- 16.Locht C, Keith J M. Pertussis toxin gene: nucleotide sequence and genetic organization. Science. 1986;232:1258–1264. doi: 10.1126/science.3704651. [DOI] [PubMed] [Google Scholar]
- 17.Mahajan-Miklos S, Tan M W, Rahme L G, Ausubel F M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 18.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 19.Mendel J E, Korswagen H C, Liu K S, Hajdu-Cronin Y M, Simon M I, Plasterk R H, Sternberg P W. Participation of the protein Go in multiple aspects of behavior in C. elegans. Science. 1995;267:1652–1655. doi: 10.1126/science.7886455. [DOI] [PubMed] [Google Scholar]
- 20.Plasterk R H. Reverse genetics: from gene sequence to mutant worm. Methods Cell Biol. 1995;48:59–80. doi: 10.1016/s0091-679x(08)61383-7. [DOI] [PubMed] [Google Scholar]
- 21.Reinke V, Smith H E, Nance J, Wang J, Van Doren C, Begley R, Jones S J, Davis E B, Scherer S, Ward S, Kim S K. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 22.Riddle D L. C. elegans II. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 23.Segalat L, Elkes D A, Kaplan J M. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Science. 1995;267:1648–1651. doi: 10.1126/science.7886454. [DOI] [PubMed] [Google Scholar]
- 24.Serventi I M, Moss J, Vaughan M. Enhancement of cholera toxin-catalyzed ADP-ribosylation by guanine nucleotide-binding proteins. Curr Top Microbiol Immunol. 1992;175:43–67. doi: 10.1007/978-3-642-76966-5_3. [DOI] [PubMed] [Google Scholar]
- 25.Stringham E G, Dixon D K, Jones D, Candido E P. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell. 1992;3:221–233. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan M W, Mahajan-Miklos S, Ausubel F M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan M W, Rahme L G, Sternberg J A, Tompkins R G, Ausubel F M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West R E, Jr, Moss J, Vaughan M, Liu T, Liu T Y. Pertussis toxin-catalyzed ADP-ribosylation of transducin. Cysteine 347 is the ADP-ribose acceptor site. J Biol Chem. 1985;260:14428–14430. [PubMed] [Google Scholar]
- 29.Wilson B A, Collier R J. Diphtheria toxin and Pseudomonas aeruginosa exotoxin A: active-site structure and enzymic mechanism. Curr Top Microbiol Immunol. 1992;175:27–41. doi: 10.1007/978-3-642-76966-5_2. [DOI] [PubMed] [Google Scholar]
- 30.Wood W B. The nematode Caenorhabditis elegans. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]