Abstract
The genetic structure of a population of Pseudomonas aeruginosa, isolated from patients with keratitis, endophthalmitis, and contact lens-associated red eye, contact lens storage cases, urine, ear, blood, lungs, wounds, feces, and the environment was determined by multilocus enzyme electrophoresis. The presence and characteristics of virulence factors were determined by restriction fragment length polymorphism analysis with DNA probes for lasA, lasB, aprA, exoS, exoT, exoU, and ctx and by zymography of staphylolysin, elastase, and alkaline protease. These analyses revealed an epidemic population structure of P. aeruginosa, characterized by frequent recombination in which a particular successful clone may increase, predominate for a time, and then disasappear as a result of recombination. Epidemic clones were found among isolates from patients with keratitis. They were characterized by high activity of a hitherto-unrecognized size variant of elastase, high alkaline protease activity, and possession of the exoU gene encoding the cytotoxic exoenzyme U. These virulence determinants are not exclusive traits in strains causing keratitis, as strains with other properties may cause keratitis in the presence of predisposing conditions. There were no uniform patterns of characteristics of isolates from other types of infection; however, all strains from urinary tract infections possessed the exoS gene, all strains from environment and feces and the major part of keratitis and wound isolates exhibited high elastase and alkaline protease activity, and all strains from feces showed high staphylolysin activity, indicating that these virulence factors may be important in the pathogenesis of these infectious diseases.
Pseudomonas aeruginosa has a wide environmental and ecological distribution and a remarkable ability to adapt to hostile environments with sparse nutrients. This versatility is probably due to a comprehensive arsenal of enzymes combined with a fit gene regulation (28, 58). For humans, P. aeruginosa is an opportunistic pathogen able to cause both local and disseminated infections. In the immunocompromised host, bacteremia, pneumonia, burn wounds, and gastrointestinal infections predominate, whereas wounds and infections of the urinary tract, lungs of cystic fibrosis (CF) patients, external ear, and cornea often occur as a result of a moist or special environment or the presence of foreign bodies such as catheters and contact lenses.
P. aeruginosa is a common cause of serious corneal infections and is the most frequently isolated bacterial species in contact-lens wearers with keratitis. It has only low binding affinity to healthy corneal epithelial cells; however, the ability to adhere may be increased by exposure of cryptic receptors or jeopardized local defense as a result of tissue damage. Accordingly, binding to and colonization of the cornea occur only in areas with tissue damage or exposed stroma uncovering receptors for adhesion (21, 57). Important P. aeruginosa adhesins include pilin, alginate, and the ADP-ribosylating toxin exoenzyme S, which also plays a dominant role in invasion of corneal epithelial cells and Madine-Darby canine kidney cells in vitro (13, 15). Conversely, both exoenzyme S and exoenzyme T have an invasive-inhibitory effect on P. aeruginosa strains expressing the cytotoxic exoenzyme U (5).
Pathogenicity is clearly multifactorial, as P. aeruginosa is known to produce a multitude of virulence-associated exoproducts. Among these, elastase (also termed LasB or pseudolysin) and alkaline protease (also termed AprA or aeruginolysin) are known to degrade a large variety of tissue components, such as proteinaceous elements of connective tissue, and to cleave cell surface receptors on neutrophils, resulting in inhibition of chemotaxis, phagocytosis, and oxidative burst. Furthermore, elastase is capable of degrading elastin, transferrin, tumor necrosis factor-α, interleukin 2, components of the complement cascade, immunoglobulin G (IgG), IgA, and secretory IgA (S-IgA); inducing inhibition of binding of natural killer cells to target cells; and producing interferon γ from T-cells (56), thus providing a basis for sustained infection. Another protease, staphylolysin (also known as LasA) appears to play a role in the pathogenesis of corneal and lung infections (9, 47). It renders elastin more susceptible to degradation by elastase and lyses Staphylococcus aureus by cleaving the peptide bonds within the pentaglycine cross-linking peptides of its cell wall peptidoglycan (32).
The cytotoxic exoenzyme U has emerged as an important pathogenicity factor in P. aeruginosa infections. Like exoenzyme S and exoenzyme T, it is secreted by a type III secretion mechanism directly into the cytosol of epithelial cells with ensuing cell death by an unknown mechanism (12). Another virulence-determining factor may be the presence of the ϕ CTX cytotoxin-converting phage that carries the ctx gene, which is thought to encode a pore-forming polypeptide (43).
Epidemiological studies of P. aeruginosa keratitis are sparse. The bacteria have often been traced to contact lens solutions (62), but the source from which P. aeruginosa contaminates is not clear. It is unknown whether this eye infection can be ascribed to a particular clone or subpopulation with a special profile of virulence properties or to random strains from the environment. The population structure of P. aeruginosa has not been extensively studied. Previous analyses of isolates from local epidemics and special habitats (i.e., the lungs of CF patients) and reference strains have been performed; most of these studies have included limited numbers of strains (4, 6, 16, 28, 33, 35, 49, 50). A recent study found a surprisingly low sequence diversity in the citrate synthase gene citS, i.e., 1 order of magnitude lower than in a comparable housekeeping gene in Salmonella, which exhibits a clonal population structure (28). In contrast, the organization of the genome was very diverse, with signs of insertions, deletions, and other genome rearrangements (50).
In the present study, multilocus enzyme electrophoresis (MLEE) was applied to a collection of eye isolates and a variety of clinical and environmental isolates to identify subpopulations or clones of P. aeruginosa associated with disease or properties characteristic of such isolates. Variations in genes encoding staphylolysin (lasA), elastase (lasB), alkaline protease (aprA), exoenzyme S (exoS), exoenzyme U (exoU), and the cytotoxic phage ctx (ctx) were examined, and enzyme activity and size were assessed by zymography for staphylolysin, elastase, and alkaline protease. Finally, the correlation between these properties and the phylogenetic relationships and origin of the strains was determined.
MATERIALS AND METHODS
Bacterial strains.
A collection of 145 isolates of P. aeruginosa was examined (Table 1). Sixty-nine strains were isolated from eyes, including 61 from patients with keratitis, 5 from patients with endophthalmitis, and 2 from a patient with contact lens-associated red eye (CLARE). Forty strains from the eye were isolates from consecutive patients attending Moorfields Eye Hospital in London, England, over a 12-month period. The remaining strains were isolated from contact lens cases belonging to patients with keratitis (CLSCkp) (n = 5), contact lens cases belonging to asymptomatic wearers (CLSCaw) (n = 4), urine (n = 10), ear infections (n = 10), blood (n = 10), lungs (n = 10) (including 1 from a CF patient), wounds (n = 10), feces (n = 10), and the environment (n = 7). Only a single isolate from each patient was included. Finally, strain PAO1, a wound isolate with a recently completed genome sequence first discovered in Australia in 1952, was included (58). For reference purposes in the enzyme assays, elastase- and alkaline protease-deficient mutants of strains PAO1 (PAO1ΔlasB and PAO1ΔaprA) were included. These mutants, constructed by insertion inactivation of the respective genes, were kindly provided by Anastasia Papakonstantinopoulou and Michael A. Curtis, MRC Molecular Pathogenesis Group, St. Bartholomew's and the Royal London School of Medicine and Dentistry, London, United Kingdom.
TABLE 1.
Designation and origin of 145 P. aeruginosa strains
| Designation(s)a | Country | Origin |
|---|---|---|
| AAB1–AAB3, AAB5–7, AAB9, AAB12–AAB15, AAB17, AAB19, AAB20 | Denmark | Keratitis |
| AKH1, Vej1 | Denmark | Keratitis |
| MiK1–MiK4 | Denmark | Keratitis |
| MK2–MK18, MK20–MK22, MK24–MK25, MK27–MK37, MK39–MK40 | United Kingdom | Keratitis |
| Paer9, PAER 10 | Australia | Keratitis |
| 6206, 6294 | Australia | Keratitis |
| Paer31, Paer32 | India | Keratitis |
| ME1, 19, -23, -26, -38 | United Kingdom | Endophthalmitis |
| Paer1, Paer25 | Australia | CLARE |
| Paer2, Paer3, Paer5, Paer7 | Australia | CLSCaw |
| AAB4, AAB8, AAB10, AAB11, AAB16 | Denmark | CLSCkp |
| MiU1–MiU10 | Denmark | Urine |
| MiE1–MiE10 | Denmark | Ear |
| MiB1–MiB10 | Denmark | Blood |
| MiL1–MiL10 | Denmark | Lung |
| MiS1–MiS10 | Denmark | Wound |
| F1–F10 | Denmark | Feces |
| Sv1, 504 | Denmark | Swimming pool |
| ON16 | Denmark | Sea water |
| ON17 | Denmark | Sea sediment |
| 508 | Denmark | Food |
| PJ39 | London, United Kingdom | Sewage |
| PJ329 | Denmark | Dialysis water |
| PAO1 | Australia | Wound |
Each strain was from a different individual.
All the strains were identified by colonial morphology, Gram staining, mobility characteristic of polar flagellation, pigment production, fluorescence, and phenotypic analysis with the API 20 NE identification kit (bioMérieux, Marcy l'Etoile, France).
MLEE.
Harvested cells of broth cultures were resuspended in buffer (50 mM Tris HCl, 5 mM EDTA [pH 7.5]) and sonicated to release intracellular enzymes. The supernatant of the sonicate was stored at −70°C. Each enzyme extract was examined by starch-gel electrophoresis to determine the relative electrophoretic mobilities of 11 housekeeping enzymes by using methods described by Selander et al. (54). The following 11 enzymes were assayed: malate dehydrogenase (MDH), alkaline phosphatase (ALP), glutamate dehydrogenase, glucose-6-phosphate dehydrogenase, adenylate kinase, and carbamate kinase in buffer system D; esterases and phosphoglucose isomerase in buffer system E; leucine aminopeptidase in buffer system H; hexokinase in buffer system I; and alcohol dehydrogenase in buffer system C.
Data Analysis.
Genetic diversity per locus (h) was calculated using the equation
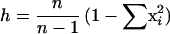 |
where xi is the frequency of the ith allele at the locus and n is the number of electrophoretic types (ETs) in the sample. The genetic distance between ETs was expressed as the proportion of loci at which dissimilar alleles occurred, i.e., the proportion of mismatches.
The multilocus diversity analysis, the dendrogram construction, and the stability of the tree tested by bootstrap analysis were performed with the computer programs ETSTAT, ETDIV, ETCLUS, and ETBOOT (version 2.2) of T. S. Whittam (www.bio.psu.edu/People/Faculty/Whittam/Lab/Programs/). The dendrogram was constructed by computerized cluster analysis performed by the average-linkage method from a matrix of pairwise genetic distances between ETs. The bootstrap procedure was performed with 1,000 average linkage-joining trees with proportional differences as distances.
To test to what extent the P. aeruginosa population was clonal, the index of association (IA) was calculated (38). This is a measure of the variance in the number of pairwise allelic mismatches relative to that expected under the hypothesis of panmixia (i.e., random association of alleles), where the mean number of pairwise allelic mismatches is a measure of genetic distance. IA is defined by the observed variance, V0 of the mean number of loci at which two P. aeruginosa strains differ divided by the expected variance, VE, under the assumption of linkage equilibrium minus 1.
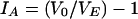 |
If there is random association between loci (linkage equilibrium), V0 approximates VE and hence IA = 0. The index of association was calculated from the MLEE data as outlined by Maynard Smith et al. (38) with the computer program ETLINK of T. S. Whittam (www.foodsafe.msu.edu/whittam /#programs).
Detection of genetic diversity within virulence genes.
Cellular DNA of P. aeruginosa was extracted as previously described, except that lysozyme was omitted (46). Approximately 2 μg of whole-cell DNA was digested with the restriction endonuclease EcoRI, and the fragments were separated by electrophoresis in 1% agarose gels for 16 h at 2 V cm −1 in TAE buffer (0.04 M Tris-acetate, 0.002 M EDTA) and subsequently visualized by ethidium bromide staining. The nucleic acids were transferred and fixed onto Nytran nylon membranes (Schleicher & Schuell, Dassel, Germany), and the hybridizations were carried out as previously described (53) except that the filters were soaked in 1% (wt/vol) Triton X-100 prior to prehybridization and 0.1% sodium pyrophosphate was included in all solutions. The final posthybridization wash was at 60°C in 1× SET (0.15 M NaCl, 0.5 mM EDTA, 20 mM Tris HCl [pH 7.0]) containing 0.1% SDS and 0.1% sodium pyrophosphate. The filters were stripped between hybridizations by being immersed in 1 liter of boiling 0.1% sodium dodecyl sulfate (SDS) and left to cool for 30 min. DNA fragments used as probes in the hybridizations were labeled with [32P]dCTP with the Random Primed DNA Labeling kit (Roche Molecular Biochemicals, Mannheim, Germany), and bands were visualized by autoradiography. As exoS and exoT are 75% identical at the nucleotide sequence level, cross-hybridization between the two chromosomal fragments containing each gene occurs (66). To discriminate between bands representing the two genes, a high-stringency posthybridization wash at 68°C in 0.2× SET containing 0.1% SDS and 0.1% pyrophosphate was performed after hybridization with the exoS probe, whereby hybridization representing the exoT gene disappeared.
Preparation of the DNA probes.
The ctx, lasA, lasB, aprA, exoS, and exoU internal probes (Table 2) were synthesized by PCR. Except for exoU, all PCR amplicons were cloned into Escherichia coli plasmid vector pTA using the Topo TA Cloning kit (Invitrogen, Groningen, The Netherlands). The inserts were verified by partial sequencing. The probes were excised from the vector by digestion with EcoRI, and the DNA fragments were isolated by electrophoresis in 1% agarose gel and extracted with the QIAEX II Agarose Gel Extraction protocol (Qiagen, Hilden, Germany).
TABLE 2.
Source of probes used for Southern hybridization
| Probe | Source | Reference | Primers | Probe size (bp) |
|---|---|---|---|---|
| lasA | PAO1 | http://www.pseudomonas.com | 5′-CGCCATCCAACCTGATGCAAT | 514 |
| 5′-AGGCCGGGGTTGTACAACGGA | ||||
| lasB | PAO1 | http://www.pseudomonas.com | 5′-TGCGATCATGGGTGTTTCGCC | 1,403 |
| 5′-GCCGAGTAGTTGCGGTTC | ||||
| aprA | IFO3455 | 44 | 5′-GTCCTATACCGTCGACCAGGC | 928 |
| 5′-GTCGCTACCCGAGCCGCCGAT | ||||
| exoS | 388 | 31 | 5′-ATCGCTTCAGCAGAGTCCGTC | 1,352 |
| 5′-CAGGCCAGATCAAGGCCGCGC | ||||
| exoU | PA103 | 19 | 5′-GCTACTGCCTCCTCGCTGAAT | 2,015 |
| 5′-AGTCATCTCAACGGTAGTCGA | ||||
| ctx | PA158 | 20 | 5′-ATGAACGATATCGACACGATC | 860 |
| 5′-TCTACTCTGCCGAGCGGACTC |
Zymography of staphylolysin.
To assess staphylolysin activity, a modification of the method of Kessler (27) was used. Samples containing 20 μl of supernatant of log-phase cultures, 1 μl of 10% SDS, 1 μl of glycerol, and 0.3 μl of bromphenol blue were subjected to SDS-polyarcylamide gel electrophoresis (PAGE) at 4°C in a 4 to 20% gradient gel containing 0.1% gelatin. After electrophoresis, staphylolysin was reactivated by immersing the gel in 2.5% Triton X-100, followed by incubation in 20 mM Tris HCl and 0.05% (wt/vol) sodium azide (pH 8.5) for 30 min. The gels were placed over an indicator agar gel (2% [wt/vol] Noble agar [Difco Laboratories, Detroit, Mich.] in 20 mM Tris-HCl, 0.05% [wt/vol] sodium azide [pH 8.5]) containing 3 mg of heat-killed S. aureus/ml and incubated in a humid atmosphere at 37°C. Staphylolysin activity appeared as clearing zones caused by cell lysis of the staphylococci within 6 to 16 h. The zones representing enzymatic activity of staphylolysin were numbered in order of molecular mass in the gel. The results were confirmed by repeating the procedure at least twice for all strains. Staphylolysin activity was determined by the application of 4 μl of supernatant of log-phase cultures into wells in an indicator agar gel as described above. The diameters of clearing zones were measured, and the results were scored as high activity (≥10-mm diameter), low activity (<10-mm diameter), or no activity.
Zymography of elastase and alkaline protease.
Supernatants of log-phase cultures in Mueller-Hinton broth supplemented with 1.3 mM CaCl2 and 0.9 mM MgCl2 were subjected to zymography under reduced and unreduced conditions using 4 to 20% gradient SDS-PAGE gels containing 0.1% gelatin (wt/vol) as a substrate (22, 36). After electrophoresis at 4°C, the proteases were reactivated by immersion of the gel in 2.5% Triton X-100, followed by incubation at 37°C in 0.1 M glycin (pH 8.3) for 4 h, subsequent staining with 1% Coomassie blue, and destaining with 10% acetic acid. Gelatinase activity appeared as clear bands in a blue-stained gel. The zymography assay was confirmed by repeating the procedure at least twice for all the strains. Molecular weight markers (Mark 12 Wide Range Protein Standard; Novex, San Diego, Calif.) were included in each gel. Finally, a collection of strains representing each of the identified protease profiles was analyzed as described, except that either the metalloprotease inhibitor EDTA or one of the serine protease inhibitors, TLCK (N-tolyl-lysine chloromethyl ketone) or BCDS (bathocuproine disulfonate), was added to the incubation buffers at a 1 mM concentration.
Detection of elastase bands in reduced SDS-PAGE gels without gelatin was performed by a Western blot technique with a rabbit antiserum raised against purified recombinant elastase. The antiserum was kindly supplied by A. Lazdunski, Laboratoire d'Ingéniérie et Dynamique des Systèmes Membranaires, Centre National de la Recherche Scientifique, Marseille, France.
RESULTS
MLEE typing.
In the collection of 145 strains of P. aeruginosa, all possessed the 11 enzyme activities. The mobility of the MDH enzyme band was identical in all strains, indicating that the corresponding gene locus was monomorphic. Bands of the remaining 10 enzymes showed variable mobilities indicating that the corresponding gene loci were polymorphic, with 2 to 8 alleles per locus (mean, 4.1) (Table 3). The differences in the mobility of the 11 housekeeping enzymes were unusually small, which made it necessary to perform several reruns to correctly allocate the mobilities to individual alleles (Fig. 1). The exact position of the esterase activity band in four strains and the position of the adenylate kinase activity band in one strain in the gel could not be determined with certainty, in spite of numerous attempts. These alleles were scored as null characters and treated as missing data. The genetic diversity per locus (h) ranged from 0.000 to 0.821 (mean, 0.357) (Table 3). Comparison of the allele profiles identified a total of 88 ETs, among which 24 ETs contained from 2 to 12 isolates and the remaining 64 ETs were represented by a single strain (Fig. 2). ET 29, containing 12 strains including PAO1, and ET 73, containing 6 strains, comprised a disproportionately high number of isolates, the majority of which were from patients with keratitis. Assuming random distribution and calculated on the basis of the relative frequency of the individual alleles, the expected number of strains in ET 29 and ET 73 was 2.10 and 0.04, respectively. Therefore, accumulation of strains in these ETs could not be explained by random association or by recombination of the most common alleles at each locus. Notably, the cluster of 15 isolates assigned to ETs 69 to 73 contained 14 strains from geographically widespread cases of keratitis and 1 strain from sewage in London, England.
TABLE 3.
Genetic diversity (h) at 11 enzyme loci of 145 P. aeruginosa strainsa
| Enzyme locus | No. of alleles | h |
|---|---|---|
| MDH | 1 | 0.000 |
| ALP | 4 | 0.191 |
| EST | 8 | 0.821 |
| GD | 3 | 0.274 |
| G6P | 3 | 0.191 |
| LAP | 2 | 0.221 |
| PGI | 3 | 0.067 |
| HEX | 5 | 0.596 |
| ADH | 6 | 0.591 |
| ADK | 4 | 0.439 |
| CDK | 6 | 0.531 |
| Mean | 4.1 | 0.357 |
Abbreviations: EST, esterases; GD, glutamate dehydrogenase, G6P, glucose-6-phosphate dehydrogenase, LAP, leucine aminopeptidase; PGI, phosphoglucose isomerase; HEX, hexokinase; ADH, alcohol dehydrogenase; ADK, adenylate kinase; CDK, carbamate kinase.
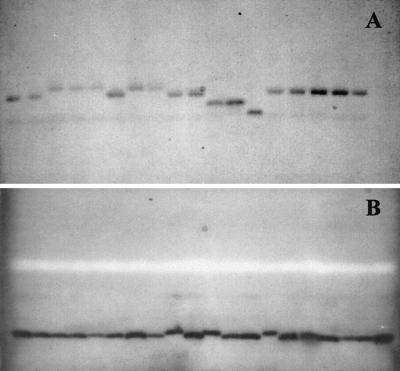
FIG. 1.
Extreme examples of mobility differences observed by MLEE analysis of separate isolates. ALP (A) and glucose dehydrogenase (B) are shown. The strains examined in the two gels are unrelated.
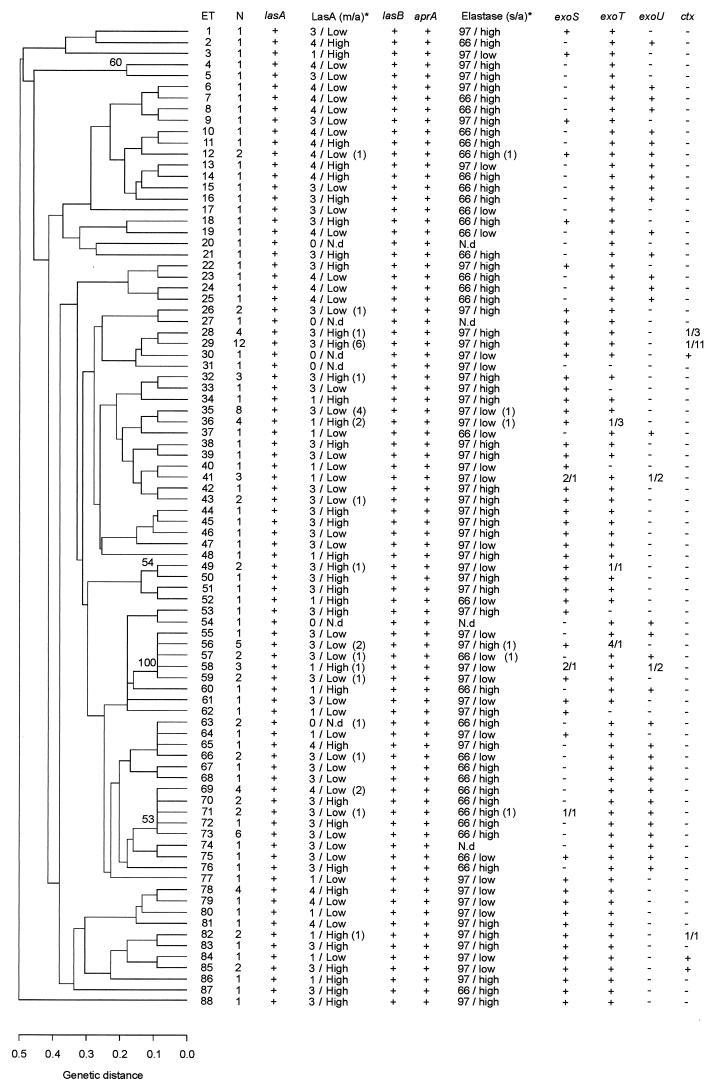
FIG. 2.
Dendrogram based on MLEE analysis of 145 P. aeruginosa strains. Presence of the genes lasA, lasB, aprA, exoS, exoT, exoU, and ctx and presence and activity of the proteases staphylolysin (LasA), elastase, and alkaline protease are indicated for each ET. Numbers in brackets indicate the number of strains differing from the majority characteristics. Numbers divided by an oblique stroke (/) indicate the number of strains positive or negative for the respective genes. Only abberations from majority characteristics of the ET are shown. N, number of isolates in the ET; m/a, electrophoretic mobility of LasA in SDS-PAGE activity of LasA; s/a, size of elastase in SDS-PAGE activity of elastase; N.d, not detectable. Only bootstrap values exceeding 50% are shown in the dendrogram.
The fact that few of the bootstrap values exceeded 50% indicates that limited confidence can be placed on the individual nodes of the tree.
IA.
Analysis of all 145 strains revealed an IA of 0.274 (standard error, 0.114). Thus, IA is significantly different from 0, suggesting linkage disequilibrium in the total population of 145 strains. Analysis of IA with ETs taken as the unit revealed an IA value of 0.148 (standard error, 0.146), implying that the IA value is between 0.294 and 0.002 with 95% confidence limits and thus very close to including 0. The reduction of IA to a value close to 0 when ETs were used as the unit suggests an epidemic population structure (38). The conclusion is supported by the bush-like configuration of the dendrogram combined with the accumulation of strains in ET 29 and cluster ET 69-ET 73 (61).
Polymorphism in putative virulence factors and their relationships to ETs.
To test whether the overall genetic relationships observed in the MLEE analysis or the site of isolation were reflected in putative virulence genes like lasA, lasB, aprA, exoS, exoT, exoU, and ctx, Southern blots of EcoRI-digested DNA from all strains were hybridized with DNA probes specific for each of six genes (Fig. 3). The different restriction fragment length patterns for lasA, lasB, and aprA gene areas were compared to the activity and relative size of the corresponding proteases staphylolysin, elastase, and alkaline protease.
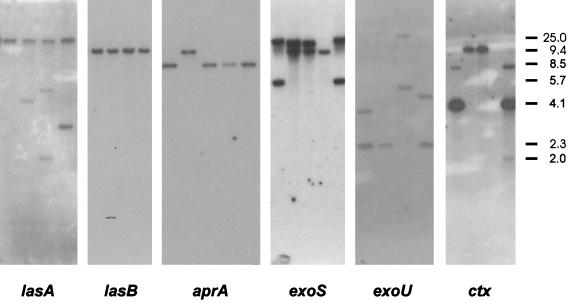
FIG. 3.
Autoradiograms showing Southern blots of EcoRI RFLP typing of whole-cell DNA from representative P. aeruginosa strains hybridized sequentially with each of the six probes lasA, lasB, aprA, exoS, exoU, and ctx. Molecular weight markers in kilobases are indicated to the right of the autoradiograms. The probes and strains examined are as follows: lasA, MK14, MK13, MK9, and PJ39; lasB, AAB1, AAB2, AAB5, and AAB6; aprA, MiE3, MiE2, Mie1, MK40, and MK38; exoS, F6, F7, F8, and F9; exoU, MK11, MK10, MK9, and MK8; and ctx, MiL3, MiL1, MiU2, and MiS3.
lasA and staphylolysin.
Thirteen restriction fragment length polymorphism (RFLP) types of lasA were found among the 145 strains which all hybridized with the probe. Sixty-seven percent of the strains exhibited the same RFLP type of the lasA gene (type 1), 10% were type 2, 5% were type 4, and 3% were type 9. The remaining 15% of the strains belonged to nine different RFLP types. Although there was some congruence of lasA type and ET type, it was of no firm consistency (not shown).
The zymography (Fig. 4) revealed four different staphylolysin activity patterns based on differences in electrophoretic mobility. The frequency of types 1, 2, 3, and 4 were 20, 1, 48, and 19%, respectively. Forty to fifty percent of the isolates from urine, lungs and feces showed type 1, 71 to 73% of the isolates from wounds and the environment exhibited type 3, and 27% of the Danish isolates from patients with keratitis and contact lens storage cases (CLSCkp) were type 4. Despite several attempts with different nutrient media and by harvesting the supernatant in various growth phases, 17 isolates (12%) showed no staphylolysin activity although they showed hybridization with the lasA gene probe. Strikingly, seven of these nonproducing strains were among the 10 isolates from ears. The zymography revealed differences in the activity of staphylolysin. All strains from feces, 40% of isolates from patients with endophthalmitis and keratitis (including CLSCkp), and 30% of blood and lung isolates exhibited high activity of staphylolysin as opposed to the missing or low activity exhibited by the strains from CLARE, CLSCaw, urine, ears, and wounds. No relationship could be found between the RFLP type and the molecular size of staphylolysin measured by SDS-PAGE. Likewise, certain staphylolysin types were accumulated in certain ETs; however, there were no consistent relationships (not shown).
FIG. 4.
Staphylolysin activity of representative strains of P. aeruginosa revealed by clearing zones in an indicator gel containing heat-killed S. aureus. For details of the assay, see the text. The gel demonstrates three of four mobility types recognized among the 88% of strains possessing staphylolysin activity. The three types demonstrated are type 1 (lanes 1, 5, and 6), type 4 (lanes 2 and 4), and type 3 (lane 3). Type 2 (not shown) showed mobility intermediary between types 1 and 3.
lasB and elastase and aprA and alkaline protease.
All isolates hybridized with the lasB probe, including four strains that lacked detectable elastase activity (Fig. 2). The lasB RFLPs were strikingly uniform as all strains, except four, showed an identical pattern. Like lasB, the RFLP for the aprA gene was very similar among all strains. Only nine strains presented a distinct pattern, and of these, six were identical.
SDS-PAGE of unreduced culture supernatants revealed three patterns of gelatinase activity. One type represented by 95 strains was characterized by a combination of activities at 97 and 55 kDa, another type represented by 46 strains was characterized by activities at 66 and 55 kDa, and four strains only exhibited activity corresponding to an apparent molecular weight of 55 kDa (Fig. 2 and Table 4). None of the strains showed gelatinase activity at 97 and 66 kDa simultaneously. The molecular masses of mature elastase and alkaline protease are 33 and 48.4 kDa, respectively, but the enzymes have been reported to migrate under unreduced conditions corresponding to apparent molecular masses of 116 or 163 and 53 kDa, respectively, in SDS-PAGE gels (7.5 to 10% gelatin) (8, 60). Furthermore, the apparent sizes of elastase and alkaline protease in polyacrylamide gels are dependent on the concentration of gelatin and polyacrylamide in the gels (25). By comparison of wild-type strain PAO1 with its elastase- and alkaline protease-deficient mutants, we concluded that the 97-kDa band represented elastase activity and that alkaline protease was represented by the activity band at 55 kDa (Fig. 5). Thus, all 145 strains possessed alkaline protease activity.
TABLE 4.
Genetic and enzyme diversity in strains from different origins
| Isolation sitea | Enzyme
|
Presence of geneb
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staphylolysin
|
Elastase
|
||||||||||||
| Missing | High activity | Molecular mass (kDa)
|
Activity
|
exoU
|
exoS
|
exoT
|
|||||||
| 66 | 97 | Missingc | Low | High | + | − | + | − | + | − | |||
| Keratitis (61) | 3 | 25 | 31 | 29 | 1 | 15 | 46 | 31 | 30 | 31 | 30 | 57 | 4 |
| Endophthalmitis (5) | 0 | 2 | 1 | 4 | 0 | 2 | 3 | 1 | 4 | 4 | 1 | 5 | 0 |
| CLARE (2) | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 2 | 2 | 0 | 2 | 0 |
| CLSCaw (4) | 1 | 0 | 0 | 3 | 1 | 2 | 2 | 0 | 4 | 3 | 1 | 4 | 0 |
| CLSCkp (5) | 0 | 2 | 1 | 4 | 0 | 5 | 0 | 3 | 2 | 2 | 3 | 4 | 1 |
| Urine (10) | 3 | 1 | 2 | 7 | 1 | 7 | 3 | 0 | 10 | 10 | 0 | 10 | 0 |
| Ear (10) | 7 | 0 | 3 | 6 | 1 | 6 | 4 | 5 | 5 | 4 | 6 | 8 | 2 |
| Blood (10) | 0 | 3 | 2 | 8 | 0 | 6 | 4 | 4 | 6 | 6 | 4 | 10 | 0 |
| Lung (10) | 1 | 3 | 2 | 8 | 0 | 7 | 3 | 2 | 8 | 8 | 2 | 10 | 0 |
| Wound (11) | 0 | 0 | 0 | 11 | 0 | 3 | 8 | 0 | 11 | 11 | 0 | 9 | 2 |
| Feces (10) | 0 | 10 | 1 | 9 | 0 | 0 | 10 | 1 | 9 | 9 | 1 | 10 | 0 |
| Environment (7) | 0 | 3 | 2 | 5 | 0 | 0 | 7 | 2 | 5 | 5 | 2 | 5 | 2 |
| Total (145) | 16 | 49 | 46 | 95 | 4 | 57 | 88 | 49 | 96 | 95 | 50 | 134 | 11 |
| % Total | 11 | 34 | 32 | 65 | 3 | 39 | 61 | 34 | 66 | 66 | 34 | 92 | 8 |
Number of isolates are in parentheses.
+, present; −, absent.
Missing activity.
FIG. 5.
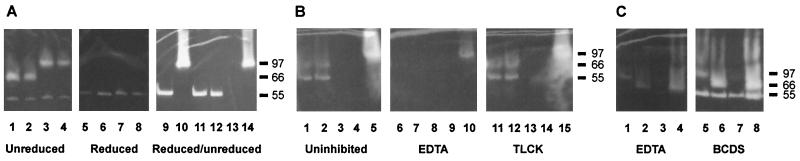
SDS-PAGE gels of culture supernatant from selected strains with clear zones representing gelatinase activity. (A) Lanes 1 to 4 show unreduced samples from four strains producing the two elastase types at 97 and 66 kDa, respectively, and alkaline protease at 55 kDa. Lanes 5 to 8 show the same samples after reduction. Lanes 9 to 14 show gelatinase activity in alternately reduced and unreduced supernatants produced by PAO1 (lanes 9 to 10) and the mutants PAO1ΔlasB (lanes 11 to 12) and PAO1ΔaprA (lanes 13 to 14). (B) Lanes 1 to 5 show uninhibited elastase activity at 66 kDa and alkaline protease activity at 55 kDa; lanes 6 to 10 show inhibition of the same samples by 1 mM EDTA; lanes 11 to 15 show lack of inhibition by TLCK. (C) Lanes 1 to 4 show elastase activity at 97 and 66 kDa and alkaline protease activity at 55 kDa inhibited by 1 mM EDTA. Lanes 5 to 8 represent the same samples and show a lack of inhibition by BCDS. Molecular mass markers in kilodaltons are indicated to the right.
Under reducing conditions, both the 97- and 66-kDa activity bands disappeared, concurrent, in some cases, with a clearly stronger reactivity band at 55 kDa (Fig. 5A). Reduction of culture supernatant from the alkaline protease-deficient mutant of PAO1, likewise, resulted in disappearance of elastase activity at 97 kDa but no emerging activity corresponding to a smaller molecular mass. Thus, it can be concluded that the increased intensity of the 55-kDa activity band was not due to elastase activity showing up corresponding to a smaller molecular mass as a result of reduction.
Gelatinase activity at 66 kDa under nonreducing SDS-PAGE conditions has not previously been described. Addition of protease inhibitors to the incubation buffer showed inhibition of all the gelatinase activity bands by EDTA, but not by TLCK or BCDS (Fig. 5), indicating that the 66-kDa band is a metalloprotease like elastase and alkaline protease (56) and therefore does not represent the recently described serine protease IV (7, 8, 45). The activities at 97 and 66 kDa were mutually exclusive. Western blot analysis of reduced supernatants from two strains representing the two gelatinase forms (strains PAO1 and MK30) revealed a band at 33 kDa reacting with the antiserum specific against P. aeruginosa elastase in both. Accordingly, we refer to the 66-kDa band as a distinct version of elastase.
Sixty-five percent of the strains showed elastase activity at 97 kDa, 32% showed activity at 66 kDa, and 3% lacked detectable activity (Table 4). Apart from showing congruence with the ETs (Fig. 2), the differences in the protease profiles correlated with the site of isolation. All strains from wounds and CLSCaw, 90% of feces isolates, and 80% of the strains from patients with endophthalmitis, CLSCkp, blood, and lungs exhibited activity at 97 kDa, while 50 to 51% of the strains from patients with keratitis and from ears showed elastase activity at 66 kDa (Table 4).
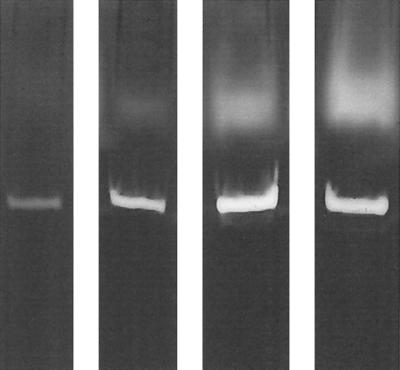
Irrespective of size, the elastase activities could be visually divided into weak or high activities based on the intensity of the bands observed in activity gels. Examples of weak and high enzyme activities are shown in Fig. 6. Repeated blind examination of the activity gels confirmed the reproducibility of scores. Growth curves constructed for representative strains (not shown) revealed that differences in activity were unrelated to differences in growth rate. Interestingly, all environmental and feces isolates and the majority of keratitis (75%) and wound (73%) isolates showed high activity compared to other clinical isolates. The proportion of Danish keratitis isolates with high protease activity was a little lower (60%) than that of English, Australian, and Indian isolates (83%). In comparison, only 30 to 40% of the strains from urine, ear, blood, and lungs showed high protease activity. The majority (76%) of elastase bands at 66 kDa were associated with high activity, compared to 59% of the bands at 97 kDa.
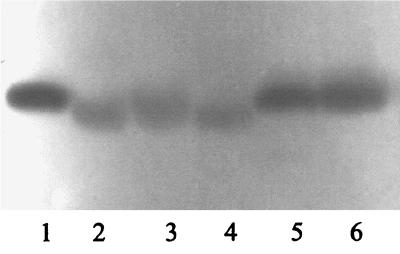
FIG. 6.
Activity differences of elastase (upper bands) and alkaline protease (lower bands) of four strains of P. aeruginosa. The two strains to the left were scored as having low elastase activity and the two strains to the right were scored as having high elastase activity.
There was a positive correlation between the activity levels of alkaline protease and elastase, in accordance with the fact that the two genes are under joint regulation (39). However, there was no association between particular RFLP types of lasB and aprA and molecular size or activity level of the elastase or alkaline protease.
ctx.
The prophage-associated ctx gene was exclusively found in six genetically related Danish strains from urine (n = 2), blood (n = 1), wounds (n = 2), and feces (n = 1) (Fig. 2). Three RFLP types of ctx were found among these strains.
exoS, exoT, and exoU.
The majority of the 145 strains (59%) exhibited a combination of exoS and exoT, 34% possessed only exoT, and 6% possessed only exoS. One strain lacked both exoS and exoT. Strains isolated from patients with keratitis showed a higher percentage of exoT carriers alone (49%), while 80 to 100% of strains from urine, lungs, wounds, and feces exhibited a combination of exoS and exoT (Table 4).
Forty-nine (34%) of the 145 isolates hybridized with the exoU gene probe. The presence or absence of exoU strongly correlated with the ETs. With one exception in ET 41 and ET 58, both containing three strains, there was complete congruence (Fig. 2). Ten RFLP types were identified, with type 7 the most prevalent (46%). However, there was no correlation between the exoU RFLP type and ET affiliation of the strains. The genes exoS and exoU were almost mutually exclusive, as all but three exoU-positive strains lacked exoS.
The distribution of the exoU gene among the clinical isolates was very heterogeneous. Fifty to 51% of the isolates from patients with keratitis and ear infections harbored the gene. In contrast, all strains from CLSCaw, CLARE, urine, and wounds and 10% of strains from feces lacked the gene. There was no relationship between exoU RFLP type and the origin of isolates.
Eye isolate-rich ETs and clusters.
ET 29 and the cluster of ETs 69 to 73 clearly differed from each other in the MLEE analysis and in the presence and types of several virulence factors. All 12 strains in ET 29, which were isolated from 1952 to 1997, possessed type 1 lasA, showed elastase activity at 97 kDa varying from very weak to high, and lacked the exoU gene, whereas the cluster of ETs 69 to 73 exhibited different types of lasA, high elastase activity at 66 kDa, and the presence of exoU. Although the strains from Moorfields Eye Hospital patients dominated ET 29, ET 29 included Danish clinical isolates from urine, ears, lungs, wounds, and blood, whereas the cluster of ETs 69 to 73, except for the isolate from sewage in London, exclusively contained isolates from patients with keratitis (n = 14) with a wide geographical origin including the United Kingdom, India, Australia, and Denmark. No relationships could be found between ETs, diversity in the virulence genes, or protease profiles among the keratitis isolates from contact lens wearers and nonwearers.
Overall relationships between genotype and phenotype.
Apart from a few exceptions, the members of a single ET shared the same zymogram protease profiles and possession or absence of the gene encoding the cytotoxic exoenzyme U, confirming the genetic relationship of strains in the same ET (Fig. 2). A single strain in each of ETs 12, 35, 36, 56, 57, and 71 exhibited a different elastase profile; likewise, a single strain differed in ETs 41 and 58 with regard to the possession of exoU. Apart from ET 29 and the cluster of ETs 69 to 73, the strains of different origins were scattered at random in the dendrogram, and the genetic relationships between the strains seemed to be independent of the site of isolation and spatial and temporal distribution.
DISCUSSION
The genetic diversity of P. aeruginosa has been previously investigated only in local epidemics, in special habitats like the lungs of patients with CF, and among serotype reference strains (2, 4, 16, 28, 33). Combined with the fact that most of these studies included a limited number of strains, it becomes difficult to apply the results to the P. aeruginosa population structure in general. The present study included 145 strains of P. aeruginosa representing clinical and environmental strains from a variety of sources, with isolates from eyes constituting the majority. They were sampled with a wide geographical distribution from four countries including Denmark, the United Kingdom, India, and Australia and were isolated during a 15-year period from 1984 to 1999. This comprehensive strain collection was selected to gain insight into the evolution, population structure, and disease-associated properties of this widespread opportunistic pathogen, with a special emphasis on eye infections.
The low mean genetic diversity (0.357) in the collection of P. aeruginosa strains determined by MLEE analysis is more in accordance with that found in many human pathogens with narrow ecological specificity, such as Haemophilus influenzae and Bordetella pertussis (55), than in related soil bacteria, which range from 0.574 to 0.581 in Burkholderia cepacia (26, 63); 0.718 in a Pseudomonas population consisting of Pseudomonas fluorescens, Pseudomonas putida, Pseudomonas syringae, Pseudomonas viridiflava, and Pseudomonas cichorii (18); and 0.876 in Pseudomonas stutzeri (48). However, the finding is in accordance with observations recently reported by Kiewitz and Tümmler (28) based on comparative sequence analyses of a combination of housekeeping genes (citS and oriC), resistance determinants (ampC), and genes encoding surface exposed proteins (pilA, fliC, and oprI). In several of these genes, the pairwise difference of nucleotide sequences was only 0.25%, which also explains the unusually small electrophoretic mobility differences observed among the alleles of the 11 housekeeping enzymes analyzed in our study. Kiewitz and Tümmler (28) have proposed that this paradoxically high degree of genetic conservation in an ancient and ecologically versatile species may be the result of a strong selection for optimal codon usage.
The genetic structure of a bacterial population is highly influenced by the level of recombination among its members. In recent years, evidence of recombination in housekeeping genes has been found in several bacterial species (11). However, if the rate of recombination is sufficiently low, selection of favorable mutants will occur, giving rise to a clonal population structure characterized by linkage disequilibrium of alleles (37). Statistical analysis of our MLEE data for the 88 ETs revealed an IA value close to 0 (0.148 ± 0.1463), indicating that alleles were in linkage equilibrium and that recombination is frequent enough to break up clonal formation. However, when calculated on the basis of all isolates, IA increased to 0.274 ± 0.1142, which is statistically significantly different from 0 and in support of linkage disequilibrium. Combined, these findings suggest an epidemic population structure (38) with frequent recombination among members of the population and occasional emergence of clones that successfully spread and persist for a while within a limited geographic and temporal span. It is conceivable that this is the scenario that explains the extremely low genetic diversity (0.138) recently reported for a collection of P. aeruginosa isolates from five CF centers in France. Ninety-two percent of 314 CF isolates clustered in two ETs, separated only at the shikimate dehydrogenase locus (35). A recent report of the application of random amplified polymorphic DNA typing to a collection of French P. aeruginosa isolates from patients with pneumonia and bacteremia and from the environment also revealed evidence of recombination combined with repeated isolation of selected random amplified polymorphic DNA types (51).
It is conceivable that epidemic spread of a virulent clone also explains the 14 keratitis isolates in the ET 69 to 73 cluster. Eight of these isolates were recovered from patients attending the Moorfields Eye Hospital in London, and we detected a London sewage isolate in the same cluster. In a survey performed at Moorfields Eye Hospital, Acanthamoeba contamination of cold water taps supplied by roof tanks was identified as the source of infection in 7 of 26 cases of Acanthamoeba keratitis diagnosed. In five cases, the strains showed identical mitochondrial DNA RFLP results for isolates from patients' corneas and those from their home tap water, firmly implicating tap water as the source of infection (29). The special arrangement of water storage tanks in the roof used to supply the bathroom in most homes in the United Kingdom provides an ideal environment for microbes, including P. aeruginosa and Acanthamoeba, to multiply. This may well be the source of the keratitis cases caused by the 14 isolates of P. aeruginosa belonging to the same clone.
As opposed to housekeeping genes, bacterial virulence factors are generally believed to be under strong selection for antigenic diversification. Several previous studies indicate that the P. aeruginosa proteases elastase, alkaline protease, and staphylolysin are implicated in the pathogenesis of human infections (41, 47, 64). The structural genes lasA, lasB, and aprA encoding these proteases were ubiquitous in the studied population. However, the observed RFLP patterns indicate a surprisingly low degree of polymorphism in regions of these virulence-associated genes and suggest that the human habitat and its inherent immunological selection pressure are not sufficiently important to override the apparent need for genetic conservation in P. aeruginosa. The low heterogeneity of lasB confirms the results of other studies (17, 34), but this is the first study to use lasA and aprA as probes in RFLP analyses. The vast majority of strains showed RFLP patterns identical with these probes.
The toxin genes ctx, exoS, exoT, and exoU were notable exceptions to this pattern. Only proportions of the strains possessed these toxin genes (exoT > exoS > exoU > ctx). However, presence of the individual genes correlated, with few exceptions, with particular phylogenetic lineages (Fig. 2). Interestingly, exoU and ctx have a G+C content of 54 and 58%, respectively (GenBank accession numbers X14956 and U97065). This as opposed to the coding regions of lasA, lasB, aprA, exoS, and exoT, which all reflect the overall 66.6% G+C content of the P. aeruginosa genome (58). The genome of strain PAO1 includes 10 regions of 3.0 kb or greater that exhibit significantly lower G+C content (49.2 to 58.5%) and unusual codon usage, possibly indicative of recent horizontal transfer (58). However, neither ctx nor exoU is present in strain PAO1. The aberrant G+C% content of ctx is in accordance with the previous demonstration of the gene as part of a prophage in a Japanese isolate of P. aeruginosa (20, 40). Interestingly, among the 145 isolates examined by us, ctx was present only in 6 Danish isolates from urine, feces, wounds, and blood.
The significantly different G+C content of the exoU gene and its presence in only part of the population, likewise, suggest acquisition by horizontal transfer from an external source. This may also apply to exoS and exoT, although their G+C content does not differ from that of the genome in general. While there are no known homologues of exoU, available genetic data indicate that the exoS and exoT loci are phylogenetically related to the virulence determinant yopE of Yersinia enterocolitica (67). The observed extensive genetic polymorphism of exoU suggests that this gene is exempt from the strict control for conservation that applies to other parts of the P. aeruginosa genome.
Several studies have elucidated the significance of specific proteases and exoenzymes in the pathogenesis of experimental Pseudomonas infections by using deletion mutants. However, there is limited information about the presence and relative activity of these putative virulence factors in clinical isolates from different diseases.
The main focus of this study was on isolates from eye infections. Previous experimental studies with keratitis models have demonstrated that the P. aeruginosa proteases, which are found in all strains, play an important role in pathogenesis. Thus, staphylolysin production increases virulence in animal models of keratitis (9, 47), and both elastase and alkaline protease are known to induce extensive necrosis in corneal tissues in a dose-dependent manner (24, 30, 59, 60). In addition, elastase activates corneal matrix metalloproteinases, resulting in further destruction of the cornea (36, 59), and all three proteases may interfere with the protective functions of both humoral and cellular components of the immune system (56). The 61 keratitis isolates included in this study were distributed across the entire MLEE dendrogram and showed no uniform pattern of putative virulence determinants, apart from the production of these three proteases (Table 4). This may indicate that there is no uniform pathogenesis of P. aeruginosa keratitis. It is conceivable that given the right conditions, such as a corneal scratch combined with wear of a soft contact lens, any strain with protease activity may cause keratitis. Conversely, strains with particular properties may cause keratitis in a previously undamaged eye. We believe that the 14 keratitis strains in the ET 69 to 73 cluster represent such strains with enhanced pathogenic potential. These strains were of a wide geographic origin and exhibited a cytotoxic genotype with both exoU and exoT, in addition to a high elastase activity at 66 kDa. This hypothesis is supported by previous identification of ExoU as an important factor in the pathogenesis of experimental keratitis, possibly due to its ability to induce apoptosis of epithelial cells and macrophages (10, 13, 14). The pathogenic role of ExoU is further supported by our finding that six isolates from the superficial inflammatory eye condition CLARE and CLSCaw lacked the exoU gene (Table 4).
Neither was there any uniform pattern of characteristics of isolates from other types of infection (Table 4). One notable exception was the 10 isolates from urinary tract infections, which all had the exoS gene but lacked exoU. In the whole collection, these two genes were, with the exception of three keratitis isolates, mutually exclusive in accordance with previous observations (10, 13). Increased ExoS activity in urinary tract isolates was recently demonstrated (52) which, combined with our findings, indicates that this exoenzyme may be important in the pathogenesis of urinary tract infections caused by P. aeruginosa. Exoenzyme U production was recently shown to be associated with increased virulence in a murine model of acute pneumonia and systemic spread in accordance with the hypothesis that cytotoxicity plays a role in dissemination of P. aeruginosa (1). However, only 6 of 20 (30%) of our blood and lung isolates possessed the exoU gene (Table 4), suggesting that it is not a crucial factor. This conclusion is in agreement with the low prevalence of exoU found among Japanese P. aeruginosa isolates from blood and lungs (13 and 9%, respectively) (23).
Expression of virulence factors in P. aeruginosa is under comprehensive regulation (58), and expression in vitro does not necessarily reflect expression during infectious processes. Nevertheless, previous studies have demonstrated correlation between relative protease activities in vitro and certain disease associations of clinical isolates. In the present study, the activity of elastase and alkaline protease was determined by a semiquantitative method measuring the specific activity of the two individual proteinases. The activity of the two proteinases was positively correlated in agreement with the fact that both are regulated by the transcriptional activators lasR-lasI and rhlR-rhlI by a quorum-sensing mechanism (39). Dividing the activity into low- and high-activity groups revealed that all strains from the environment and feces and the major part of keratitis and wound isolates exhibited high protease activity in contrast to isolates from urine, ears, blood, lungs, and CLSCkp. A previous study of P. aeruginosa strains indicated that elastase production is highest in isolates from acute lung infections; intermediate in isolates from burns, wounds, and urine; and lowest in isolates from CF sputum and blood (65), thus differing mainly from the present findings for lung isolates. Elastase production by environmental strains has been reported to be similar to that of clinical strains (42). However, the detection of elastase activity in those studies was performed by an elastin-Congo red assay, which, in addition to elastase, detects activity of both staphylolysin and alkaline protease (64).
Variation was also noted in this study in the staphylolysin activity of the isolates. A total of 12% of the 145 isolates lacked detectable activity (Table 4). Most notable, all 10 feces isolates showed high activity, while 7 of 10 ear isolates lacked activity. The significance of this is not clear, but the environment in the gut appears to select for P. aeruginosa strains with generally high protease activity (Table 4).
The molecular mass of mature elastase is 33 kDa. However, for as-yet-unexplained reasons, enzyme activity appears to correspond to a considerably higher molecular size in SDS-PAGE gelatin under nonreducing conditions, the exact position depending on the concentration of gelatin and polyacrylamide in the gels (8, 25, 60). By comparison of wild-type strain PAO1 with its elastase-deficient mutant we concluded that the 97-kDa band observed in strain PAO1 and 94 other strains represented elastase (Fig. 5). However, in the remaining 46 strains the activity corresponded to a molecular mass of 66 kDa. The enzyme activity at both these locations disappeared under reducing condition in contrast to the activity of alkaline protease (Fig. 5). Based on the following evidence, we conclude that the gelatinase activities observed at 97 and 66 kDa represent two different allelic versions of elastase. The two forms were mutually exclusive in the collection of 145 strains and both were recognized by an antiserum specific to elastase. The activity of both forms was lost under reducing conditions, and both were inhibited by EDTA but not by traditional inhibitors of serine proteases (Fig. 5), excluding that the 66-kDa form represents the recently described protease IV (7, 43).
The molecular nature of the two different sizes of elastase, which have not been previously described, is yet unknown. Elastase is known to have two intrachain disulfide bridges linking Cys30 with Cys57 and Cys 270 with Cys297 in the 301-residue mature protein (http://ncbi.nlm.nih.gov/entrez/quiry.fcgi?cmd = Retrieve&db = Protein&list _ uids = 119263&dopt = GenPept), which are necessary for processing and activity of the secreted protease (3). The two mobility variants of elastase observed in nonreducing gels with gelatin is likely to reflect different conformations of the mature protein resulting from differences in the lasB gene sequence. An alternative explanation is the formation of sequence-dependent noncovalently linked oligomers (dimers or trimers).
In conclusion, this study revealed an epidemic population structure of P. aeruginosa in which frequent recombination is combined with occasional epidemic spread of emerging successful clones. Examples of epidemic clones were demonstrated among isolates from cases of keratitis. Such clones were characterized by high activity of a hitherto-unrecognized 66-kDa version of elastase, high alkaline protease activity, and possession of the exoU gene encoding a cytotoxic exoenzyme. The results furthermore suggest that strains with other properties may cause keratitis in the presence of predisposing conditions.
ACKNOWLEDGMENTS
We thank Niels Ehlers, Department of Ophthalmology, Aarhus University Hospital, for advice. The strains used in this study were kindly donated by Melville M. Matheson, Institute of Ophthalmology, London, United Kingdom (strains designated MK and ME); Henrik C. Schønheyder, Department of Clinical Microbiology, Aalborg Sygehus, Aalborg, Denmark (strains designated AAB); Jens K. Møller, Department of Clinical Microbiology, Aarhus University Hospital, Aarhus, Denmark (all strains except ÅKH 1 were isolated by Ole Steen Mikkelsen and designated MiU, MiE, MiB, MiL, MiS, and MiK); Claus Pommerencke, Department of Ophthalmology, Vejle Sygehus, Vejle, Denmark (strains designated Vej); Peter Gerner-Smidt, Statens Serum Institut, Copenhagen, Denmark (strains designated F); Flemming Boisen, Miljø-og levnedsmiddelkontrollen, Odense, Denmark (strains designated 504 and 508); Ole Nybroe, Institut for Økologi, Copenhagen, Denmark (strains designated ON); Mark D. Willcox, Cooperative Research Center for Eye Research and Technology, University of New South Wales, Sydney, Australia (strains designated Paer, 6206, and 6294); and Michael A. Curtis and Anastasia Papakonstantinopoulou, MRC Molecular Pathogenesis Group, St. Bartholomew's and the Royal London School of Medicine and Dentistry, London, United Kingdom (strains PAO1, PAO1Δ lasB, and PAO1Δ aprA). The rabbit antiserum against Pseudomonas elastase was a kind gift from A. Lazdunski, Laboratoire d'Ingéniérie et Dynamique des Systèmes Membranaires, Centre National de la Recherche Scientifique, Marseille, France.
This work was supported by a grant from the Danish Eye Health Society and by the Danish Medical Research Council.
REFERENCES
- 1.Allewelt A, Coleman F T, Grout M, Priebe G P, Pier G B. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect Immun. 2000;68:3998–4004. doi: 10.1128/iai.68.7.3998-4004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingen E, Denamur E, Picard B, Goullet P, Lambert-Zekovsky N, Foucaud P, Navarro J, Elion J. Molecular epidemiologic analysis of Pseudomonas aeruginosa strains causing failure of antibiotic therapy in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1992;11:432–437. doi: 10.1007/BF01961858. [DOI] [PubMed] [Google Scholar]
- 3.Braun P, Ockhuijsen C, Eppens E, Koster M, Bitter W, Thommassen J. Maturation of Pseudomonas aeruginosa elastase: formation of the disulfide bonds. J Biol Chem. 2001;276:26030–26035. doi: 10.1074/jbc.M007122200. [DOI] [PubMed] [Google Scholar]
- 4.Charnock C, Bergan T. Multilocus enzyme electrophoresis of major O-antigen reference strains of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 1992;11:810–816. doi: 10.1007/BF01960880. [DOI] [PubMed] [Google Scholar]
- 5.Cowell B A, Chen D Y, Frank D W, Vallis A J, Fleiszig S M. ExoT of cytotoxic Pseudomonas aeruginosa prevents uptake by corneal epithelial cells. Infect Immun. 2000;68:403–406. doi: 10.1128/iai.68.1.403-406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denamur E, Picard B, Decoux G, Denis J B, Elion J. The absence of correlation between allozyme and rrn RFLP analysis indicates a high gene flow rate within human clinical Pseudomonas aeruginosa isolates. FEMS Lett. 1993;110:275–280. doi: 10.1111/j.1574-6968.1993.tb06334.x. [DOI] [PubMed] [Google Scholar]
- 7.Engel L S, Hill J M, Caballero A R, Green L C, O'Callaghan R J. Protease IV, a unique extracellular protease and virulence factor from Pseudomonas aeruginosa. J Biol Chem. 1998;27:16792–16797. doi: 10.1074/jbc.273.27.16792. [DOI] [PubMed] [Google Scholar]
- 8.Engel L S, Hobden J A, Moreau J M, Callegan M C, Hill J M, O'Callaghan R J. Pseudomonas deficient in protease IV has significantly reduced corneal virulence. Investig Ophthalmol Vis Sci. 1997;38:1535–1542. [PubMed] [Google Scholar]
- 9.Estrellas P S, Alionte L G, Hobden J A. A Pseudomonas aeruginosa strain isolated from a contact lens-induced acute red eye (CLARE) is protease-deficient. Curr Eye Res. 2000;20:157–165. [PubMed] [Google Scholar]
- 10.Evans D J, Frank D W, Finck-Barbançon V, Wu C, Fleizig S M J. Pseudomonas aeruginosa invasion and cytotoxicity are independent events, both of which involve protein tyrosine kinase activity. Infect Immun. 1998;66:1453–1459. doi: 10.1128/iai.66.4.1453-1459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil E J, Holmes E C, Enright M C, Bessen D, Kalia A, Day N P J, Chan M-S, Hood D, Zhou J, Spratt B G. Recombination within natural populations of pathogeneic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci USA. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M J, Wu C, Mende-Mueller L, Frank D W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 13.Fleizig S M J, Wiener-Kronish J P, Miyazaki H, Vallas V, Mostov K E, Kanada D, Sawa T, Yen T S B, Frank D W. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleizig S M J, Zaidi T S, Pier G B. Pseudomonas aeruginosa invades corneal epithelial cells in vitro. Infect Immun. 1995;63:4072–4077. doi: 10.1128/iai.63.10.4072-4077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleizig S M J, Zaidi T S, Preston M J, Grout M, Evans D J, Pier G B. The relationship between cytotoxicity and corneal epithelial invasion by clinical isolates of Pseudomonas aeruginosa. Infect Immun. 1996;64:2288–2294. doi: 10.1128/iai.64.6.2288-2294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith S J, Nathan C, Selander R K, Chamberlin W, Gordon S, Kabins S, Weinstein R A. The epidemiology of Pseudomonas aeruginosa in oncology patients in a general hospital. J Infect Dis. 1989;160:1030–1036. doi: 10.1093/infdis/160.6.1030. [DOI] [PubMed] [Google Scholar]
- 17.Hamood A N, Griswold H. DNA hybridization analysis of the Pseudomonas aeruginosa elastase gene (lasB) from different clinical isolates. Can J Microbiol. 1995;41:910–917. doi: 10.1139/m95-125. [DOI] [PubMed] [Google Scholar]
- 18.Haubold B, Rainey P B. Genetic and ecotypic structure of a fluorescent Pseudomonas population. Mol Ecol. 1996;5:747–761. [Google Scholar]
- 19.Hauser A R, Kang P J, Engel J N. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi T, Kamio Y, Hishinuma F, Usami Y, Titani K, Terawaki Y. Pseudomonas aeruginosa cytotoxin: the nucleotide sequence of the gene and the mechanism for activation of the protoxin. Mol Microbiol. 1989;3:861–868. doi: 10.1111/j.1365-2958.1989.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 21.Hazlet L D, Moon M, Burk R S. In vivo identification of sialic acid as the ocular receptor for Pseudomonas aeruginosa. Infect Immun. 1986;20:25–29. doi: 10.1128/iai.51.2.687-689.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heussen C, Dowdle E B. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal Biochem. 1980;102:196–202. doi: 10.1016/0003-2697(80)90338-3. [DOI] [PubMed] [Google Scholar]
- 23.Hirakata Y, Finlay B B, Simpson D A, Kohno S, Kamihira S, Speert D P. Penetration of clinical isolates of P. aeruginosa through MDCK epithelial cell monolayers. J Infect Dis. 2000;181:765–769. doi: 10.1086/315276. [DOI] [PubMed] [Google Scholar]
- 24.Howe T R, Iglewski B H. Isolation and characterization of alkaline protease-deficient mutants of Pseudomonas aeruginosa in vitro and in a mouse eye model. Infect Immun. 1984;43:1058–1063. doi: 10.1128/iai.43.3.1058-1063.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hummel K M, Penheiter A R, Gathman A C, Lilly W W. Anomalous estimation of protease molecular weights using gelatine-containing SDS-PAGE. Anal Biochem. 1995;233:140–142. doi: 10.1006/abio.1996.0019. [DOI] [PubMed] [Google Scholar]
- 26.Johnson W M, Tyler S D, Rozee K R. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J Clin Microbiol. 1994;32:924–930. doi: 10.1128/jcm.32.4.924-930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler E. β-Lytic endopeptidases. Methods Enzymol. 1995;248:740–756. doi: 10.1016/0076-6879(95)48050-1. [DOI] [PubMed] [Google Scholar]
- 28.Kiewitz C, Tümmler B. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J Bacteriol. 2000;182:3125–3135. doi: 10.1128/jb.182.11.3125-3135.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilvington S. Through a glass darkly—contact lenses and personal hygiene. Microbiol Today. 2000;27:66–69. [Google Scholar]
- 30.Kreger A S, Grey L D. Purification of Pseudomonas aeruginosa proteases and microscopic characterization of pseudomonal protease-induced rabbit corneal damage. Infect Immun. 1978;19:630–648. doi: 10.1128/iai.19.2.630-648.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulich S M, Yahr T L, Mende-Mueller L M, Barbieri J T, Frank D W. Cloning the structural gene for the 49-kDa form of exoenzyme S (exoS) from Pseudomonas aeruginosa strain 388. J Biol Chem. 1994;269:10431–10437. [PubMed] [Google Scholar]
- 32.Larche M, Hearn W R, Syskind J W, Tipper D J, Strominger J L. Specificity of a bacteriolytic enzyme from Pseudomonas aeruginosa. J Bacteriol. 1969;100:254–259. doi: 10.1128/jb.100.1.254-259.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levin M H, Weinstein R A, Selander R K, Ochman H, Kabins S A. Association of infection caused by Pseudomonas aeruginosa serotype O11 with intravenous abuse of pentazocine mixed with tripelennamine. J Clin Microbiol. 1984;20:758–762. doi: 10.1128/jcm.20.4.758-762.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loutit L, Tompkins L S. Restriction enzyme and Southern hybridization analysis of Pseudomonas aeruginosa strains from patients with cystic fibrosis. J Clin Microbiol. 1991;29:2897–2900. doi: 10.1128/jcm.29.12.2897-2900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin C, Boyd E F, Quentin R, Massicot P, Selander R K. Enzyme polymorphism in Pseudomonas aeruginosa strains recovered from cystic fibrosis patients in France. Microbiology. 1999;145:2587–2594. doi: 10.1099/00221287-145-9-2587. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto K, Shams N B K, Hanninen L A, Kenyon K R. Cleavage and activation of corneal matrix metalloproteinases by Pseudomonas aeruginosa proteases. Investig Ophthalmol Vis Sci. 1993;34:1945–1953. [PubMed] [Google Scholar]
- 37.Maynard Smith J. Do bacteria have population genetics? In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Cambridge, United Kingdom: Press Syndicate of the University of Cambridge; 1995. pp. 1–12. [Google Scholar]
- 38.Maynard Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKnight S L, Iglewski B H, Pesci E C. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayami K, Kanaya S, Ohnishi M, Terawaki Y, Hayashi T. The complete nucleotide sequence of phi CTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol Microbiol. 1999;31:399–419. doi: 10.1046/j.1365-2958.1999.01158.x. [DOI] [PubMed] [Google Scholar]
- 41.Nicas T I, Iglewski B H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- 42.Nicas T I, Iglewski B H. Production of elastase and other exoproducts by environmental isolates of Pseudomonas aeruginosa. J Clin Microbiol. 1986;23:967–969. doi: 10.1128/jcm.23.5.967-969.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Callaghan R J, Engel L S, Hobden J A, Callegan M C, Green L C, Hill J M. Pseudomonas keratitis. The role of an uncharacterized exoprotein, protease IV, in corneal virulence. Investig Ophthalmol Vis Sci. 1996;37:534–543. [PubMed] [Google Scholar]
- 44.Ohnishi M, Hayashi T, Terawaki Y. Mechanism of the cytolytic action of Pseudomonas aeruginosa cytotoxin: oligomerization of the cytotoxin on target membranes. FEBS Lett. 1994;356:357–360. doi: 10.1016/0014-5793(94)01311-x. [DOI] [PubMed] [Google Scholar]
- 45.Okuda K, Morihara K, Atsumi Y, Takeuchi H, Kawamoto S, Kawasaki H, Suzuki K, Fukushima J. Complete nucleotide sequence of the structural gene for alkaline protease from Pseudomonas aeruginosa IFO 3455. Infect Immun. 1990;58:4083–4088. doi: 10.1128/iai.58.12.4083-4088.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poulsen K, Hjorth J P, Kilian M. Limited diversity of the immunoglobulin A1 protease gene (iga) among Haemophilus influenzae serotype b strains. Infect Immun. 1988;56:987–992. doi: 10.1128/iai.56.4.987-992.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preston M J, Seed P C, Toder D S, Iglewski B H, Ohman D E, Gustin J K, Goldberg J B, Pier G B. Contribution of proteases and LasR to virulence of Pseudomonas aeruginosa during corneal infections. Infect Immun. 1997;65:3086–3090. doi: 10.1128/iai.65.8.3086-3090.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rius N, Fusté M C, Guasp C, Lalucat J, Lorén J G. Clonal population structure of Pseudomonas stutzeri, a species with exceptional genetic diversity. J Bacteriol. 2001;183:736–744. doi: 10.1128/JB.183.2.736-744.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Römling U, Greipel J, Tümmler B. Gradient of genomic diversity in the P. aeruginosa chromosome. Mol Microbiol. 1995;17:323–332. doi: 10.1111/j.1365-2958.1995.mmi_17020323.x. [DOI] [PubMed] [Google Scholar]
- 50.Römling U, Schmidt K D, Tümmler B. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J Mol Biol. 1997;271:386–404. doi: 10.1006/jmbi.1997.1186. [DOI] [PubMed] [Google Scholar]
- 51.Ruimy R, Genauzeau E, Barnabe C, Beaulieu A, Tibayrenc M, Andremont A. Genetic diversity of Pseudomonas aeruginosa strains isolated from ventilated patients with nosocomial pneumonia, cancer patients with bacteremia, and environmental water. Infect Immun. 2001;69:584–588. doi: 10.1128/IAI.69.1.584-588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rumbaugh K P, Hamood A N, Griswold J. Analysis of P. aeruginosa clinical isolates for possible variations within the virulence genes exotoxin A and exotoxin S. J Surg Res. 1999;82:95–105. doi: 10.1006/jsre.1998.5523. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour J M, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selander R K, Musser J M. Population genetics of bacterial pathogenesis. In: Iglewski B H, Clark V L, editors. Molecular basis of bacterial pathogenesis. New York, N.Y: Academic Press, Inc.; 1990. pp. 11–36. [Google Scholar]
- 56.Steadman R, Heck L W, Abrahamson D R. The role of proteases in the pathogenesis of Pseudomonas aeruginosa infections. In: Campa M, Bendinelli M, Friedman H, editors. Pseudomonas aeruginosa as an opportunistic pathogen. New York, N.Y: Plenum Press; 1993. pp. 129–145. [Google Scholar]
- 57.Stern G A, Weizenkorn D, Valenti J. Adhaerence of Pseudomonas aeruginosa to the mouse cornea. Epithelial v. stromal adhaerence. Arch Ophthalmol. 1982;100:1956–1958. doi: 10.1001/archopht.1982.01030040936014. [DOI] [PubMed] [Google Scholar]
- 58.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbik K, Lim R, Smith K, Spencer D, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosa PAO1: an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 59.Twining S S, Davis S D, Hyndiuk R A. Relationship between proteases and descemetocele formation in experimental Pseudomonas keratitis. Curr Eye Res. 1986;5:503–510. doi: 10.3109/02713688608996372. [DOI] [PubMed] [Google Scholar]
- 60.Twining S S, Kirschner S E, Mahnke L A, Frank D W. Effect of Pseudomonas aeruginosa elastase, alcaline protease, and exotoxin A on corneal proteinases and proteins. Investig Ophthalmol Vis Sci. 1993;34:2699–2712. [PubMed] [Google Scholar]
- 61.Whittam T S. Sex in the soil. Curr Biol. 1992;2:676–678. doi: 10.1016/0960-9822(92)90140-6. [DOI] [PubMed] [Google Scholar]
- 62.Wilson L A, Schlitzer R L, Ahern D G. Pseudomonas corneal ulcers associated with soft contact-lens wear. Am J Ophthalmol. 1981;92:546–554. doi: 10.1016/0002-9394(81)90649-8. [DOI] [PubMed] [Google Scholar]
- 63.Wise M G, Shimkets L J, McArthur J V. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1995;61:1791–1798. doi: 10.1128/aem.61.5.1791-1798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolz C, Helstern E, Haug M, Galloway D R, Vasil M L, Döring G. Pseudomonas aeruginosa lasB mutant constructed by insertional mutagenesis reveals elastolytic activity due to alkaline protease and the lasA fragment. Mol Microbiol. 1991;5:2125–2131. doi: 10.1111/j.1365-2958.1991.tb02142.x. [DOI] [PubMed] [Google Scholar]
- 65.Woods D E, Schaffer M S, Rabin H R, Campbell G D, Sokol P A. Phenotypic comparison of Pseudomonas aeruginosa strains isolated from a variety of clinical sites. J Clin Microbiol. 1986;24:260–264. doi: 10.1128/jcm.24.2.260-264.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yahr T L, Barbieri J T, Frank D W. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yahr T L, Hovey A K, Kulich S M, Frank D W. Transcriptional anlysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol. 1995;177:1169–1178. doi: 10.1128/jb.177.5.1169-1178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]