Abstract
Since Staphylococcus aureus expresses multiple pathogenic factors, studying their individual roles in single-gene-knockout mutants is difficult. To circumvent this problem, S. aureus clumping factor A (clfA) and fibronectin-binding protein A (fnbA) genes were constitutively expressed in poorly pathogenic Lactococcus lactis using the recently described pOri23 vector. The recombinant organisms were tested in vitro for their adherence to immobilized fibrinogen and fibronectin and in vivo for their ability to infect rats with catheter-induced aortic vegetations. In vitro, both clfA and fnbA increased the adherence of lactococci to their specific ligands to a similar extent as the S. aureus gene donor. In vivo, the minimum inoculum size producing endocarditis in ≥80% of the rats (80% infective dose [ID80]) with the parent lactococcus was ≥107 CFU. In contrast, clfA-expressing and fnbA-expressing lactococci required only 105 CFU to infect the majority of the animals (P < 0.00005). This was comparable to the infectivities of classical endocarditis pathogens such as S. aureus and streptococci (ID80 = 104 to 105 CFU) in this model. The results confirmed the role of clfA in endovascular infection, but with a much higher degree of confidence than with single-gene-inactivated staphylococci. Moreover, they identified fnbA as a critical virulence factor of equivalent importance. This was in contrast to previous studies that produced controversial results regarding this very determinant. Taken together, the present observations suggest that if antiadhesin therapy were to be developed, at least both of the clfA and fnbA products should be blocked for the therapy to be effective.
Staphylococcus aureus is a major pathogen responsible for a wide variety of infections. These range from relatively mild skin and soft-tissue diseases to severe conditions such as osteomyelitis and endocarditis (35, 52). S. aureus also often infects foreign materials (11, 19, 48, 49), thus making it a primary challenge for modern medicine. Finally, S. aureus has successfully collected numerous antibiotic resistance genes and can withstand treatment with the majority of nonexperimental antibacterial drugs, including now glycopeptides (20, 40, 45). This increases the spectrum of untreatable infections and warrants the search for alternative antistaphylococcal strategies.
Understanding the steps of colonization and invasion of the host by the microorganism might greatly help define new targets to interfere with the infective process. However, identifying the key molecular players mediating S. aureus colonization and invasion has been hampered by the multiplicity of pathogenic factors expressed by the organisms. Indeed, S. aureus carries several functionally redundant adhesins binding to both soluble and insoluble components of the host extracellular matrix (39, 53). Moreover, the analysis is complicated by the fact that expression of these determinants is differentially regulated by global regulators agr and sar (3, 38), which promote adhesin expression early in infection and toxin production later on.
In such an intricate context, studying the pathogenic role of individual S. aureus factors using classical gene inactivation has yielded inconclusive results. As an example, four major surface adhesins, i.e., clumping factor A (ClfA), clumping factor B (ClfB), and fibronectin-binding proteins A and B (FnBPA and FnBPB), that demonstrated high-affinity binding to their specific ligands in vitro (14, 31, 36, 50) were of limited relevance in experimental endocarditis in vivo (7, 9, 33). One possible explanation for this in vitro/in vivo paradox was that the inactivated mutants possessed additional, redundant factors that could complement the function of the missing adhesins in the disease process.
This possibility is supported by recent experiments in which the expression of either multiple surface proteins or excreted proteins was hampered at the same time (1, 28, 29). In both cases, the decreased infectivity in animal models of infection was substantially greater than that expected with single-gene-inactivated mutants. However, none of these approaches allowed the pathogenic role of individual factors, the sum of which is responsible for disease, to be identified.
To address this question, we transferred both the clfA and fnbA genes of S. aureus into the less-pathogenic Lactococcus lactis using a previously described method (41). After the adequate expression of these determinants in the recipient bacteria was assessed, the recombinant lactococci were tested in vitro for the gain of function conferred by the transferred genes and in rats with experimental endocarditis. The system should help in building a catalog of staphylococcal determinants important for disease using a variety of different infection models. It is expected to generate information complementary to that from other experimental approaches to enable a better understanding of the complexity of S. aureus pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacteria and plasmids used in this study are listed in Table 1. L. lactis subsp. cremoris was grown at 30°C in M17 medium (Oxoid, Basingstoke, United Kingdom) supplemented with 0.5% glucose (47). S. aureus was grown at 37°C in Mueller-Hinton or tryptic soy medium (Difco Laboratories, Detroit, Mich.); Escherichia coli XL1-blue was grown at 37°C in Luria-Bertani medium (42). Whenever appropriate, antibiotics were added to the media at the following concentrations: erythromycin, 5 μg/ml for L. lactis and 500 μg/ml for E. coli; ampicillin, 50 μg/ml for E. coli. Bacterial stocks were kept at −70°C in liquid medium supplemented with 10% (vol/vol) glycerol.
TABLE 1.
List of the various strains and plasmid used in this study
| Plasmid or strain | Properties | Origin or reference |
|---|---|---|
| L. lactis subsp. cremoris 1363 | Prophage-cured L. lactis subsp. cremoris strain | 12 |
| L. lactis, ClfA positive | L. lactis strain expressing staphylococcal ClfA | 41 |
| L. lactis, FnBPA positive | L. lactis strain expressing staphylococcal FnBPA | This study |
| S. aureus Newman | ClfA-producing strain | 5 |
| S. aureus, ClfA negative | ClfA-negative Newman mutant | 31 |
| S. aureus 8325-4 | Prophage-cured ATCCa strain | 37 |
| S. aureus DU5883 | FnBPA- and FnBPB-negative 8325-4 mutant | 14 |
| S. aureus DU5883, FnBPA positive | FnBPA- and FnBPB-negative 8325-4 mutant, FnBPA complemented | 14 |
| E. coli XL1-blue | 42 | |
| pIL253 | EryR lactococcal plasmid | 44 |
| pOri23-clfA | EryR, L. lactis-E. coli shuttle vector, clfA | 41 |
| pOri23-fnbA | EryR, L. lactis-E. coli shuttle vector, fnbA | This study |
ATCC, American Type Culture Collection.
Cloning experiments.
The staphylococcal fnbA gene was ligated at the BamHI site of the pOri23 vector and expressed in L. lactis as described previously (41). The staphylococcal fnbA gene (44) was amplified by PCR from the chromosome of S. aureus 8325-4, prepared as described previously (27), using forward primer 5′-CC GGATCC GCA TTT AAA GGG AGA TA TTA TA-3′ and reverse primer 5′CC GGATCC CGG GCT TAC TTC ATA TAA TTA TGA A-3′ (BamHI restriction sites are in boldface). The following conditions were applied for 30 cycles: 95°C for 30 s, 55°C for 30 s, and 72°C for 3 min 15 s. The resulting PCR fragment was ligated into pOri23, cloned in E. coli, and transformed by electroporation into L. lactis by described methods (21, 41).
RNA isolation and dot blot analysis.
Liquid cultures (1,000 ml) of L. lactis FnBPA-positive and L. lactis ClfA-positive (Table 1) strains were incubated at 30°C and monitored for growth with a spectrophotometer (Sequoia-Turner, Montainville, Calif.) by measuring optical density at a wavelength of 620 nm. Total RNA was prepared at various time points according to a previously described method (2) using the FastRNA Blue isolation kit (Bio101 Inc., Carlsbad, Calif.) and the FastPrep FP120 machine (Bio101) in accordance with the manufacturer's recommendations. Bacteria were disrupted at a speed rating of 6 for 30 s. Total RNA was then applied to an RNeasy minicolumn (Qiagen GmbH, Hilden, Germany) and isolated as recommended by the manufacturer. Remaining genomic DNA was digested within the column using the RNase-free DNase set (Qiagen). Yields were evaluated both by absorbance at 260 nm and by comparison of the intensities of ethidium bromide-stained 16S and 23S rRNA signals from 1-μg RNA samples electrophoresed through a 1.2% agarose–0.66 M formaldehyde gel in MOPS (morpholinepropanesulfonic acid) running buffer (Sigma, St. Louis, Mo.).
Fifteen micrograms of total RNA was transferred onto Zeta-Probe GT blotting membranes (Bio-Rad Laboratories, Hercules, Calif.) using a Bio-Dot microfiltration apparatus. Membranes were then vacuum baked for 2 h at 80°C. After prehybridization, levels of clfA and fnbA mRNA and 16S rRNA were determined by hybridization with random [α-32P]dCTP-primed DNA probes (Ready-to-Go labeling kit; Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). The clfA, fnbA, and 16S rRNA probes were obtained through PCR. The lactococcal 16S rRNA fragment comprising residues 68 to 633 was amplified at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min using 5′-GCG ATG AAG ATT GGT GCT TGC-3′ as the forward primer and 5′-GGT TGA GCC ACT GCC TTT TAC AC-3′ as the reverse primer. Amplifications of the clfA fragment comprising residues 348 to 581 and the fnbA fragment comprising residues 2311 to 2994 were carried out at 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min using S. aureus Newman genomic DNA as a template. For clfA amplification the forward and reverse primers were 5′-TTG GCG TGG CTT CAG TGC TTG TAG-3′ and 5′-GAT TGT GTC GTT TCC TGT TGT GC-3′, respectively. For fnbA amplification the forward and reverse primers were 5′-AGC CAC GTT GAT ATT AAG AGT-3′ and 5′-AAC AGG TGT TAC TAC TTT ACC-3′, respectively.
Hybridizations were performed overnight at 42°C in the presence of ULTRAhyb solution (Ambion, Austin, Tex.). Blots were washed as recommended. Exposure times of all membranes to radiographic film (medical X-ray film; Fuji Photo Film Europe, Dusseldorf, Germany) were chosen to optimize the signals under conditions preventing saturation. To normalize signal levels, the same filters were rehybridized with probes for constitutively expressed 16S rRNA.
Antibodies.
Anti-ClfA antibodies (30) were kindly provided by T. J. Foster. Peptides corresponding to FnBPA residues 479 to 493 (IQNNKFEYKEDTIKE) were conjugated to keyhole limpet hemocyanin and used to immunize rabbits (Eurogentec, Seraing, Belgium). F(ab′)2 fragments were further prepared from rabbit immune sera using the ImmunoPure F(ab′)2 preparation kit (Pierce, Rockford, Ill.) according to the instructions of the manufacturer.
SDS-PAGE and Western blots.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by standard procedures using 6.5% acrylamide gels (24). Cell wall-associated proteins were extracted, separated by electrophoresis, and transferred onto Immobilon-P polyvinylidene difluoride membranes as previously described (30, 41). The membranes were further incubated for 4 h with a 1:500 dilution of either anti-ClfA or anti-FnBPA F(ab′)2 antibodies prior to a 1-h incubation with a 1:5,000 dilution of goat anti-rabbit antibodies coupled to peroxidase. Bands were revealed with the use of the enhanced chemiluminescence method.
Detection of ClfA and FnBPA proteins by flow cytometry.
The display of ClfA and FnBPA on the surface of L. lactis and S. aureus was monitored by flow cytometry of formaldehyde-fixed log-phase cells prepared and labeled with a 1:100 dilution of either rabbit anti-ClfA F(ab′)2 or anti-FnBPA F(ab′)2 (10, 17). Rabbit antibodies were detected with a 1:500 dilution of goat anti-rabbit F(ab′)2 coupled to fluorescein isothiocyanate. The absence of bacterial autofluorescence was checked using nonlabeled cells. Fluorescence of cells incubated with secondary antibodies alone was used to measure background fluorescence.
Solid-phase adherence assay.
The attachment of bacteria to fibrinogen or fibronectin was measured by a previously described adhesion assay using polymethylmethacrylate (PMMA) coverslips coated with either purified fibrinogen (31) or fibronectin (51). The attachment of bacteria to protein-coated coverslips was averaged and normalized to the number of adherent CFU per square centimeter of polymeric surface (50). The statistical significance of the different attachment properties of the test organisms was evaluated by one-way analysis of variance (ANOVA), and pairwise differences between the means of groups were determined by the t test with the Bonferroni correction. Data were considered significant when P values were ≤0.05 by use of two-tailed significance levels.
Attachment to human platelet-fibrin clots in vitro.
Adherence to human platelet-fibrin clots was measured by a previously described method (33, 34). Platelet-fibrin clots were produced by pouring 0.5 ml of plasma into 30-mm-diameter cell culture plates (Corning Costar, Corning, N.Y.) in the presence of 100 μl of a 500-U/ml National Institutes of Health bovine thrombin solution and 100 μl of a 0.2 mM CaCl2 solution. The clots were then dehydrated overnight at 37°C and kept at 4°C before being used. To determine bacterial adherence, 2 ml of saline containing about 105 CFU/ml was added to the wells, and the plates were agitated for 3 min at 120 rpm on a rotating incubator. The fluid was gently decanted, and the clots were washed twice for 5 min with 2 ml of phosphate-buffered saline to remove nonadherent bacteria. The clots were then overlaid with 3 ml of Columbia agar, which was allowed to solidify before incubation at 30°C. The number of adherent bacteria giving rise to colonies was determined after 24 h of incubation and expressed as a multiple of adherent organisms relative to the original inoculum (adherence ratio = [number of adherent bacteria/inoculum size] × 10,000). Statistical differences between groups were analyzed as described above. In certain experiments, platelet-rich plasma was supplemented with fibronectin (final concentration of either 0.5 or 2 μg/ml, prior to coagulation) to test the effect of an excess of this ligand on bacterial adherence.
Experimental endocarditis in rats.
Sterile aortic vegetations were produced in female Wistar rats as described previously (18). Twenty-four hours after catheterization, groups of 10 rats were inoculated through a tail vein with increasing numbers of bacteria prepared from overnight cultures and sacrificed 24 h later. The rate and severity of valve infection were determined as described previously (33, 46). The minimal inoculum infecting 80% of animals (80% infective dose [ID80]) was assessed 24 h after bacterial challenge. The dilution technique allowed detecting ≥2 log CFU/g of vegetation. Rats with sterile vegetations were considered uninfected. Statistical differences between the frequencies of valve infections were evaluated by the χ2 test with Yates correction. Differences among mean vegetation bacterial densities were evaluated by one-way ANOVA, and pairwise differences between the means of groups were determined by the t test with the Bonferroni correction. Differences were considered significant when P values were ≤0.05 by use of two-tailed significance levels.
RESULTS
Expression of the S. aureus clfA and fnbA genes in lactococci.
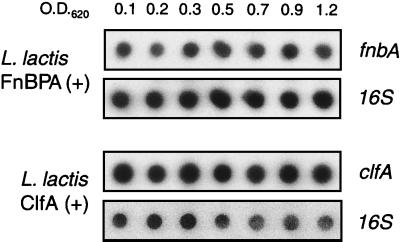
The expression of clfA and fnbA in lactococci was tested both at the transcriptional and translational levels. Figure 1 presents the quantitative analysis of the clfA and fnbA transcripts as a function of bacterial growth. RNA was prepared at various times during growth. For each time point an equal amount of total RNA was blotted and probed for either the 16S rRNA or the clfA or fnbA mRNAs. The proportion of clfA or fnbA mRNA (test genes) to 16S rRNA (constitutively expressed internal control) remained stable during bacterial growth, indicating that clfA or fnbA were constitutively expressed.
FIG. 1.
Transcription of the recombinant cflA and fnbA genes in L. lactis subsp. cremoris. RNA of FnBPA-positive (top) and ClfA-positive (bottom) L. lactis was prepared at various times during bacterial growth. For each time points an equal amount of total RNA was blotted on a membrane, and 16S rRNA and clfA and fnbA mRNAs were detected with specific [α-32P]dCTP-primed DNA probes. The proportion of clfA or fnbA mRNAs (test genes) to 16S rRNA (constitutively expressed internal control) remained stable during bacterial growth, indicating that clfA and fnbA were constitutively expressed. O.D.620, culture optical densities at which bacteria were sampled.
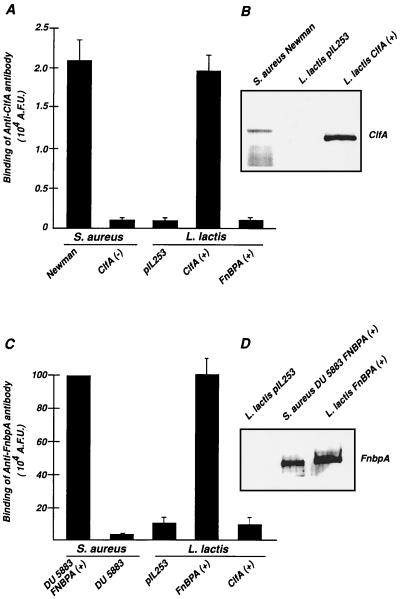
Figure 2 indicates that the ClfA and FnBPA proteins were appropriately expressed and targeted to the lactococcal cell wall. First, the two recombinant proteins migrated in parallel with their native staphylococcal homologues on SDS-PAGE gel and were adequately located in the lactococcal wall fraction as shown by Western blot analysis (Fig. 2B and D). Second, the two proteins were similarly expressed on the surfaces of both the recombinant L. lactis and the S. aureus donor strains, as detected by flow cytometry with specific antiadhesin antibodies (Fig. 2A and C). In contrast, no antibody binding was detected on either lactococci carrying an empty plasmid or S. aureus carrying specific deletions in the clfA and fnbA genes. These experiments provided evidence that the ClfA and FnBPA proteins were functionally anchored to the lactococcal cell wall, making recombinant lactococci suitable for more-sophisticated in vitro and in vivo experiments.
FIG. 2.
Surface expression of ClfA and FnBPA. Surface expression of the adhesins was assessed either by flow cytometry (A and C) or by Western blotting (B and D). ClfA and FnBPA were detected by specific anti-ClfA and anti-FnBPA F(ab′)2 fragments. ClfA (A and B) was detected on both S. aureus Newman and L. lactis ClfA-positive S. aureus but not on ClfA-negative S. aureus or on L. lactis pIL253 carrying an empty vector. Similarly, FnBPA (C and D) was detected on both FnBPA-positive S. aureus DU5883 and FnBPA-positive L. lactis but not on FnBPA negative and FnBPB negative S. aureus DU5883, L. lactis pIL253, or ClfA-positive L. lactis. Data are means ± standard deviations for three separate experiments. A.F.U., arbitrary fluorescence units.
Adhesion properties of ClfA-positive and FnBPA-positive recombinant lactococci on immobilized fibrinogen or fibronectin.
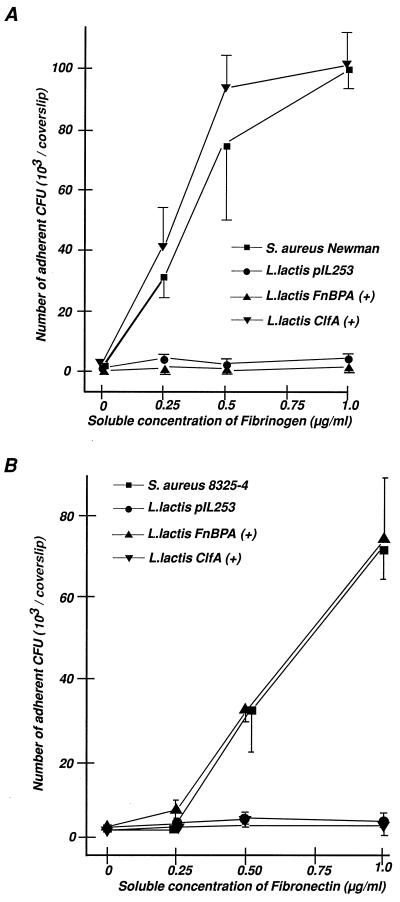
The effect of ClfA and FnBPA on lactococcal adherence to their respective ligands was evaluated (14, 31). Figure 3 indicates that both types of recombinants avidly attached to either fibrinogen-coated (Fig. 3A) or fibronectin-coated (Fig. 3B) surfaces at levels that were similar to those of their staphylococcal gene donor strains (Table 1). In contrast, lactococci carrying an empty plasmid showed virtually no adherence to these surfaces.
FIG. 3.
Adherence of various test organisms to PMMA coverslips coated with increasing concentrations of either fibrinogen (A) or fibronectin (B). The number of adherent bacteria, expressed as CFU (means ± standard errors of the means; n = 3), is shown as a function of the adsorbed ligand. S. aureus Newman was used as a donor for clfA and S. aureus 8325-4 was used as a donor for fnbA because the clfA and fnbA genes were well characterized in these two organisms (14, 31). L. lactis pIL253 carried an empty vector, and ClfA-positive and FnBPA-positive L. lactis expressed the staphylococcal ClfA and FnBPA proteins.
Adherence of lactococci to platelet-fibrin clots.
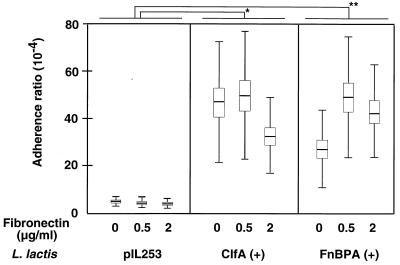
Since cardiac vegetations contain numerous proteins and platelet factors (16), which are not present in the purified ligand assay described above, the adherence experiments were repeated with platelet-fibrin clots, which mimic the cardiac vegetations recovering damaged valves (33, 34). Figure 4 indicates that both ClfA-positive and FnBPA-positive recombinants adhered significantly more than lactococci carrying an empty plasmid in this assay (P < 0.005). Moreover, addition of fibronectin to the system significantly affected the adherence of FnBPA-positive (P = 0.03) lactococci but not that of ClfA-positive or parent lactococci.
FIG. 4.
In vitro adherence of various test organisms to platelet-fibrin clots, supplemented with fibronectin or not supplemented. The adherence ratio was expressed as a function of the original inoculum (see Materials and Methods). Boxes, means ± standard errors of the means and standard deviations for 36 independent determinations in three separate experiments. Asterisk and double asterisks, P < 0.005. Addition of fibronectin to the system significantly affected the adherence of FnBPA-positive lactococci (P = 0.03). The concentration of fibronectin added to the assay mixture and the organisms tested are specified at the bottom.
Experimental endocarditis due to ClfA-positive and FnbA-positive lactococci.
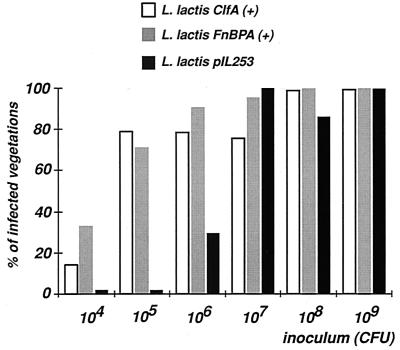
We finally tested the ability of the recombinant lactococci to induce valve infection in rats with catheter-induced aortic vegetations (18). Figure 5 indicates that induction of experimental endocarditis was inoculum dependent. Lactococci carrying an empty plasmid were poorly infective and required up to 107 CFU to infect ca. 80% of the rats (ID80). This was 100 times greater than the ID80s of classical endocarditis pathogens such as Streptococcus spp. (13) and S. aureus (33). In contrast, only 105 CFU of both clfA-positive and fnbA-positive lactococci were required to reach the same infection level (P < 0.00005). This ID80 was comparable to those for typical endocarditis pathogens. Thus, individual expression of both clfA and fnbA determinants was necessary and sufficient to afford high infectivity of lactococci in experimental valve infection.
FIG. 5.
Infectivity titration of recombinant lactococci in rats with experimental endocarditis. The rates of early vegetation infections as a function of inoculum size are shown. Groups of rats were challenged with increasing numbers of lactococci (horizontal axis) carrying either an empty vector (pIL253) or ClfA-encoding [ClfA (+)] or FnBPA-encoding [FnBPA (+)] plasmids. The percentage of infected vegetations (vertical axis) was assessed after 24 h. Asterisk, P < 0.00005 between the parent and the recombinant lactococci at inoculum sizes of 104, 105, and 106 CFU.
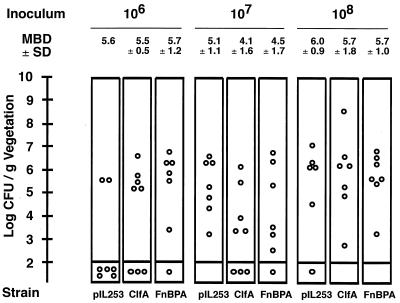
Figure 6 depicts the vegetation bacterial titers at 24 h in rats challenged with increasing inoculum sizes of the various test organisms. All animals with positive valve cultures had comparable bacterial densities in their vegetations (P > 0.05 when compared by ANOVA and t tests). Infection was a yes of no response. Therefore, at this early time point the system discriminated between the intrinsic ability of the bacteria to colonize damaged valves and their ability to grow in situ later on.
FIG. 6.
Vegetation bacterial densities after challenge of rats with large inoculum sizes. Shown is a close-up around the region of ID80 of lactococci carrying an empty plasmid (pIL253), i.e., 107 CFU. Dots, bacterial densities in individual animals. Inoculum sizes are indicated at the top. Mean bacterial densities (MBD) ± standard deviations (SD) of infected vegetations are indicated. Strain abbreviations are as in Fig. 5. Infectivity was a yes or no phenomenon. Once infected, all the animals developed comparable valve disease at 24 h (P > 0.05 when compared by the ANOVA and t tests). This indicates that differences in infectivity were not related to differences in in situ bacterial fitness but rather to the ability of bacteria to colonize the damaged valves early on.
DISCUSSION
The present study identified both ClfA and FnBPA adhesins from S. aureus as critical mediators implicated in the induction of experimental endocarditis in rats. While this was rather confirmatory for ClfA, it is of great importance for FnBPA. Indeed, using genetically engineered S. aureus strains, previously published studies yielded inconclusive results regarding this molecule in a similar model (9, 23).
The colonizing-promoting role of ClfA in endovascular infection was clearly established in specific models, such as in dialysis tubing exposed to circulating blood (10, 50). During short-time blood exposure, the tubing becomes coated with plasma fibrinogen, which promotes avid adherence of ClfA-positive staphylococci. The role of ClfA in promoting cardiac valve infection was also demonstrated in experimental endocarditis but with less clear-cut results. In S. aureus, clfA knockout mutants showed decreased infectivity compared to the clfA-positive parent. However, this difference was only recorded for a limited range of bacterial loads (33). In a reverse experiment, the S. aureus clfA gene was transferred into a surrogate, Streptococcus gordonii, which produced an increase in both bacterial adherence to platelet-fibrin clots and experimental endocarditis (46). As in the S. aureus knockout mutant, the difference between the parent and the transformant organisms was confined to an inoculum window of less than 1 order of magnitude. Thus, in both types of experiments, the involvement of ClfA was limited, and so use of this adhesin as a primary target for new antistaphylococcal treatment strategies is questionable.
However, both the S. aureus and Streptococcus gordonii systems were hampered by the fact that the two organisms were natural endocarditis pathogens (13, 26, 33). This could mask the intrinsic contribution of ClfA to the colonization of endovascular lesions. In the present system, this limitation was overcome by expressing clfA constitutively in a surrogate bacterium that was 100 to 1,000 times less able than staphylococci and streptococci to induce experimental endocarditis in rats. When expressing clfA, the recombinant lactococci acquired the abilities to adhere to fibrinogen and to induce experimental endocarditis at levels comparable to those for the S. aureus gene donor strain. This is an unprecedented phenotypic change in this infection model and identifies ClfA as a critical adhesin capable of mediating endovascular infection on it own.
The implication of the S. aureus fibronectin-binding proteins was established in a few models of foreign-body infection, including colonization of titanium plates removed from the iliac bones of guinea pigs (8) and colonization of PMMA slides explanted from tissue cages in the same animal (14). On the other hand, two studies performed on rats with experimental endocarditis produced conflicting results. Kuypers and Proctor (23) used an S. aureus isolate (879R4S) carrying a single fnb gene and an fnb-inactivated mutant carrying a Tn918 insertion between the unique fnb gene and its promoter (15). They observed that the defective mutant was less able than the parental strain to colonize catheter-induced cardiac vegetation early (1 h) after intravenous inoculation in rats. In a second study, the S. aureus 8325-4 strain expressing both fnbA and fnbB genes was compared to its mutant carrying specific deletions of these two determinants. The two strains showed equivalent abilities to colonize damaged valves both 1 and 24 h after bacterial challenge (9), a result contrasting with the completely defective adherence to fibronectin of the Tn918 mutant strain.
However, in their experiments these authors identified attachment to fibrinogen, which is present along with fibrin in the cardiac vegetation, as a phenotype that might complement the absence of the fibronectin-binding protein for adherence (9). A posteriori, therefore, they could not entirely rule out fibronectin-binding proteins as participating in adherence. A definitive answer would have required a triple fnbA fnbB clfA mutant, but these experiments have not yet been performed. Moreover, additional S. aureus adhesins mediating adherence to fibronectin, such as the major histocompatibility complex analog protein (22), might also be operative in infection but were not considered in these experiments.
As for ClfA, the present system allowed this limitation to be bypassed by expressing only one adhesin at the surface of lactococci. The FnBPA-positive recombinant organism increased both its adherence to immobilized fibronectin and its infectivity in experimental endocarditis to levels similar to those for the S. aureus gene donor. Since no additional adhesins were present on the surrogate lactococci, the recombinant revealed the full effect of the fibronectin-binding protein alone. Like the results of the original experiments by Kuypers and Proctor (23), the present results attribute a critical role to the fibronectin-binding proteins in endovascular infection. Moreover, they support previous observations by Lowrance et al. (26), who described a critical role for fibronectin binding in experimental endocarditis due to Streptococcus sanguis.
In conclusion, expressing individual determinants from S. aureus or other pathogens (4, 6) in less-pathogenic bacteria may help solve some of these complicated issues. It can provide important complementary information to other systems, including systems using inactivation of structural or regulatory genes (1, 28, 29) and systems used for gene discovery such as in vivo expression technology (25) and signature tag mutagenesis (32). Building a catalog of genes important for disease is crucial. Indeed, the present results indicate that, if antiadhesin strategies were to be developed, both ClfA and FnBPA should be targeted in order for the strategies to be effective.
ACKNOWLEDGMENTS
We thank Marlyse Giddey, Jacques Vouillamoz, and Manuela Bento for outstanding technical assistance.
The present work was supported by grants 3200-044099.96/2, 31-52149.97, 3200-045810.95/2, and 31-56689.99 of the Swiss National Foundation.
REFERENCES
- 1.Cheung A L, Eberhardt K J, Chung E, Yeaman M R, Sullam P M, Ramos M, Bayer A S. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Investig. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung A L, Eberhardt K J, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 3.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crago A M, Koronakis V. Binding of extracellular matrix laminin to Escherichia coli expressing the Salmonella outer membrane proteins Rck and PagC. FEMS Microbiol Lett. 1999;176:495–501. doi: 10.1111/j.1574-6968.1999.tb13703.x. [DOI] [PubMed] [Google Scholar]
- 5.Duthie E S, Lorenz L L. Staphylococcal coagulase: mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 6.Elsinghorst E A, Baron L S, Kopecko D J. Penetration of human intestinal epithelial cells by Salmonella: molecular cloning and expression of Salmonella typhi invasion determinants in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:5173–5177. doi: 10.1073/pnas.86.13.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Entenza J M, Foster T J, Ni Eidhin D, Vaudaux P, Francioli P, Moreillon P. Contribution of clumping factor B to pathogenesis of experimental endocarditis due to Staphylococcus aureus. Infect Immun. 2000;68:5443–5446. doi: 10.1128/iai.68.9.5443-5446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer B, Vaudaux P, Magnin M, el Mestikawy Y, Proctor R A, Lew D P, Vasey H. Novel animal model for studying the molecular mechanisms of bacterial adhesion to bone-implanted metallic devices: role of fibronectin in Staphylococcus aureus adhesion. J Orthop Res. 1996;14:914–920. doi: 10.1002/jor.1100140611. [DOI] [PubMed] [Google Scholar]
- 9.Flock J I, Hienz S A, Heimdahl A, Schennings T. Reconsideration of the role of fibronectin binding in endocarditis caused by Staphylococcus aureus. Infect Immun. 1996;64:1876–1878. doi: 10.1128/iai.64.5.1876-1878.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.François P, Schrenzel J, Stoerman-Chopard C, Favre H, Hermann M, Foster T, Lew D, Vaudaux P. Identification of plasma proteins adsorbed on hemodialysis tubing that promote Staphylococcus aureus adhesion. J Lab Clin Med. 2000;135:32–42. doi: 10.1016/s0022-2143(00)70018-7. [DOI] [PubMed] [Google Scholar]
- 11.Francois P, Vaudaux P, Foster T J, Lew D P. Host-bacteria interactions in foreign body infections. Infect Control Hosp Epidemiol. 1996;17:514–520. doi: 10.1086/647358. [DOI] [PubMed] [Google Scholar]
- 12.Gasson M J, Lyndon-Davies F. Prophage-cured derivatives of Streptococcus lactis and Streptococcus cremoris. Appl Environ Microbiol. 1980;40:964–966. doi: 10.1128/aem.40.5.964-966.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glauser M P, Bernard J P, Moreillon P, Francioli P. Successful single-dose amoxicillin prophylaxis against experimental streptococcal endocarditis: evidence for two mechanisms of protection. J Infect Dis. 1983;147:568–575. doi: 10.1093/infdis/147.3.568. [DOI] [PubMed] [Google Scholar]
- 14.Greene C, McDevitt D, Francois P, Vaudaux P E, Lew D P, Foster T J. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol Microbiol. 1995;17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 15.Greene C, Vaudaux P E, Francois P, Proctor R A, McDevitt D, Foster T J. A low-fibronectin-binding mutant of Staphylococcus aureus 879R4S has Tn918 inserted into its single fnb gene. Microbiology. 1996;142:2153–2160. doi: 10.1099/13500872-142-8-2153. [DOI] [PubMed] [Google Scholar]
- 16.Hamill R. Role of fibronectin in infective endocarditis. Rev Infect Dis. 1987;9:S360–S371. doi: 10.1093/clinids/9.supplement_4.s360. [DOI] [PubMed] [Google Scholar]
- 17.Hartford O, Francois P, Vaudaux P, Foster T J. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol Microbiol. 1997;25:1065–1076. doi: 10.1046/j.1365-2958.1997.5291896.x. [DOI] [PubMed] [Google Scholar]
- 18.Heraief E, Glauser M P, Freedman L R. Natural history of aortic valve endocarditis in rats. Infect Immun. 1982;37:127–131. doi: 10.1128/iai.37.1.127-131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrmann M, Vaudaux P E, Pittet D, Auckenthaler R, Lew P D, Schumacher-Perdreau F, Peters G, Waldvogel F A. Fibronectin, fibrinogen, and laminin act as mediators of adherence of clinical staphylococcal isolates to foreign material. J Infect Dis. 1988;158:693–701. doi: 10.1093/infdis/158.4.693. [DOI] [PubMed] [Google Scholar]
- 20.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 21.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jönsson K, McDevitt D, McGavin M H, Patti J M, Höök M. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J Biol Chem. 1995;270:21457–21460. doi: 10.1074/jbc.270.37.21457. [DOI] [PubMed] [Google Scholar]
- 23.Kuypers J M, Proctor R A. Reduced adherence to traumatized rat heart valves by a low-fibronectin-binding mutant of Staphylococcus aureus. Infect Immun. 1989;57:2306–2312. doi: 10.1128/iai.57.8.2306-2312.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 26.Lowrance J M, Baddour L M, Simpson W A. The role of fibronectin binding in the rat model of experimental endocarditis caused by Streptococcus sanguis. J Clin Investig. 1990;86:7–13. doi: 10.1172/JCI114717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marmur J. A procedure for isolation of desoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 28.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick R P, Muir T W. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazmanian S K, Liu G, Jensen E R, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci USA. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDevitt D, Francois P, Vaudaux P, Foster T J. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol Microbiol. 1995;16:895–907. doi: 10.1111/j.1365-2958.1995.tb02316.x. [DOI] [PubMed] [Google Scholar]
- 31.McDevitt D, François P, Vaudaux P, Foster T J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 32.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 33.Moreillon P, Entenza J M, Francioli P, McDevitt D, Foster T J, François P, Vaudaux P. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreillon P, Overholser C D, Malinverni R, Bille J, Glauser M P. Predictors of endocarditis in isolates from cultures of blood following dental extractions in rats with periodontal disease. J Infect Dis. 1988;157:990–995. doi: 10.1093/infdis/157.5.990. [DOI] [PubMed] [Google Scholar]
- 35.Musher D M, Lamm N, Darouiche R O, Young E J, Hamill R J, Landon G C. The current spectrum of Staphylococcus aureus infection in a tertiary care hospital. Medicine. 1994;73:186–208. doi: 10.1097/00005792-199407000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Ni Eidhin D, Perkins S, François P, Vaudaux P, Hook M, Foster T J. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol. 1998;30:245–257. doi: 10.1046/j.1365-2958.1998.01050.x. [DOI] [PubMed] [Google Scholar]
- 37.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 38.Novick R P, Muir T W. Virulence gene regulation by peptides in staphylococci and other Gram-positive bacteria. Curr Opin Microbiol. 1999;2:40–45. doi: 10.1016/s1369-5274(99)80007-1. [DOI] [PubMed] [Google Scholar]
- 39.Patti J M, Hook M. Microbial adhesins recognizing extracellular matrix macromolecules. Curr Opin Cell Biol. 1994;6:752–758. doi: 10.1016/0955-0674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 40.Perl T M. The threat of vancomycin resistance. Am J Med. 1999;106:26S–37S. doi: 10.1016/s0002-9343(98)00354-4. and 48S–52S. [DOI] [PubMed] [Google Scholar]
- 41.Que Y, Haefliger J, Francioli P, Moreillon P. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect Immun. 2000;68:3516–3522. doi: 10.1128/iai.68.6.3516-3522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Signas C, Raucci G, Jonsson K, Lindgren P E, Anantharamaiah G M, Hook M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 45.Smith T L, Pearson M L, Wilcox K R, Cruz C, Lancaster M V, Robinson-Dunn B, Tenover F C, Zervos M J, Band J D, White E, Jarvis W R. Emergence of vancomycin resistance in Staphylococcus aureus. N Engl J Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 46.Stutzmann P, Entenza J M, Vaudaux P, Francioli P, Glauser M P, Moreillon P. Study of Staphylococcus aureus pathogenic genes by transfer and expression in the less virulent organism Streptococcus gordonii. Infect Immun. 2001;69:657–664. doi: 10.1128/IAI.69.2.657-664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Antimicrob Agents Chemother. 1985;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaudaux P, Pittet D, Haeberli A, Huggler E, Nydegger U E, Lew D P, Waldvogel F A. Host factors selectively increase staphylococcal adherence on inserted catheters: a role for fibronectin and fibrinogen or fibrin. J Infect Dis. 1989;160:865–875. doi: 10.1093/infdis/160.5.865. [DOI] [PubMed] [Google Scholar]
- 49.Vaudaux P, Pittet D, Haeberli A, Lerch P G, Morgenthaler J J, Proctor R A, Waldvogel F A, Lew D P. Fibronectin is more active than fibrin or fibrinogen in promoting Staphylococcus aureus adherence to inserted intravascular catheters. J Infect Dis. 1993;167:633–641. doi: 10.1093/infdis/167.3.633. [DOI] [PubMed] [Google Scholar]
- 50.Vaudaux P E, François P, Proctor R A, McDevitt D, Foster T J, Albrecht R M, Lew D P, Wabers H, Cooper S L. Use of adhesion-defective mutants of Staphylococcus aureus to define the role of specific plasma proteins in promoting bacterial adhesion to canine arteriovenous shunts. Infect Immun. 1995;63:585–590. doi: 10.1128/iai.63.2.585-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaudaux P E, Waldvogel F A, Morgenthaler J J, Nydegger U E. Adsorption of fibronectin onto polymethylmethacrylate and promotion of Staphylococcus aureus adherence. Infect Immun. 1984;45:768–774. doi: 10.1128/iai.45.3.768-774.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waldvogel F A. Staphylococcus aureus (including toxic shock syndrome) In: Mandel G L, Bennett J E, Dolin R, editors. Principles and practise of infectious diseases. 4th ed. Vol. 1. New York, N.Y: Churchill Livingstone; 1995. pp. 1754–1777. [Google Scholar]
- 53.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]