Abstract
Three genes, ipgD, mxiC, and mxiA, all in the invasion region of the Shigella virulence plasmid, were sequenced from strains representing a range of Shigella serotypes and from two enteroinvasive Escherichia coli (EIEC) isolates. The plasmids can be classified into two relatively homogeneous sequence forms which are quite distinct. pINV A plasmids are found in Shigella flexneri strains F6 and F6A, S. boydii strains B1, B4, B9, B10, B14, and B15, S. dysenteriae strains D3, D4, D6, D8, D9, D10, and D13, and the two EIEC strains (M519 and M520). pINV B plasmids are present in S. flexneri strains F1A, F2A, F3A, F3C, F4A, and FY, two S. boydii strains (B11 and B12), and S. sonnei. The D1 pINV plasmid is a recombinant with ipgD gene more closely related to those of pINV A but with mxiA and mxiC genes more closely related to those of pINV B. The phylogenetic relationships of the plasmid and those of the chromosomal genes of Shigella strains are largely consistent. The cluster 1 and cluster 3 strains tested (G.M. Pupo, R. Lan, and P. R. Reeves, Proc. Natl. Acad. Sci. USA 97:10567–10572, 2000) have pINV A and pINV B plasmids, respectively. However, of the three cluster 2 strains (B9, B11, and B15), B9 and B15 have pINV A while B11 has a pINV B plasmid. Those Shigella (D8 and D10 and S. sonnei) and EIEC strains which do not group with the main body of Shigella strains based on chromosomal genes were found to have plasmids belonging to one or the other of the two types and must have acquired these by lateral transfer.
Molecular evidence derived from studies involving DNA hybridization, multilocus enzyme electrophoresis, and sequencing of housekeeping genes indicates that Escherichia coli and all members of the genus Shigella belong to the same species (7, 22, 25), whereas the current classification scheme recognizes four species within the genus Shigella: S. dysenteriae, S. flexneri, S. boydii, and S. sonnei, representing subgroups A, B, C, and D, respectively. Shigella strains are further divided into 38 serotypes based on O antigen variation: 13 in S. dysenteriae, 18 in S. boydii, 6 in S. flexneri, and 1 in S. sonnei. It appears from molecular data that the Shigella phenotype arose within E. coli. In this paper we look at the relationships of 38 serotypes that are in reality Shigella clones of E. coli.
Shigella is the causative agent of shigellosis or bacillary dysentery, a diarrheal disease of humans. It involves an acute inflammatory colitis which in its worst case is characterized by intestinal cramps, bloody diarrhea, and neurologic symptoms such as lethargy, confusion, severe headache, and convulsions (1). However, since the 1940s, pathogenic forms of E. coli have been recognized, and the enteroinvasive Escherichia coli (EIEC) strains have the same mode of pathogenesis as Shigella spp.
Virulence in all Shigella strains is dependent on the presence of a large 210- to 230-kb plasmid. The virulence plasmids pWR100 in S. flexneri 5, pMYSH6000 in S. flexneri 2a, and pSS120 in S. sonnei and those in other Shigella strains are determinants for invasiveness and the ability to cause disease (13) and are collectively termed pINV plasmids. A common 31-kb core region within pINV, essential for the virulence phenotype (13), includes genes for invasins, molecular chaperones, motility, regulation, and a specialized type III secretion apparatus (23). A similar plasmid is found in EIEC strains.
We recently analyzed the relationships of Shigella strains to each other and to other strains of E. coli by sequencing eight housekeeping genes in four separate regions around the chromosome (26). The study showed that Shigella strains fall into one of three main clusters within E. coli. Cluster 1 contained the majority of the S. dysenteriae and S. boydii serotypes. Cluster 2 consisted of a smaller group of S. boydii serotypes and S. dysenteriae serotype 2. Cluster 3 contains all of the S. flexneri serotypes except serotypes 6 and 6A, which were in cluster 1. S. dysenteriae 1, 8 and 10, and S. sonnei were outside of these three main groups but also clearly within E. coli, while S. boydii 13 is not within E. coli and is apparently the only known representative of a related species. From this it was evident that the Shigella phenotype arose several times in E. coli.
EIEC, like Shigella strains, tend to have a reduction in the number of substrates used relative to commensal E. coli strains and may be Lac−, nonmotile, and lysine decarboxylase negative. EIEC strains have an invasion plasmid similar to that harbored by Shigella strains (33). This raises the question of the relationships and evolution of the invasion plasmids present in these strains, and to better understand pINV evolution, we sequenced three genes within the invasion region from a range of Shigella strains and EIEC strains.
MATERIALS AND METHODS
Strains.
We used 46 Shigella strains to represent the known serotypes and two EIEC strains. Details are given in Pupo et al. (26) for Shigella strains and Pupo et al. (25) for the two EIEC strains. For S. dysenteriae, S. flexneri and S. boydii, for which there are multiple serotypes, we refer to serotypes as D1, F1A, and B1 for S. dysenteriae 1, S. flexneri 1A, and S. boydii 1, respectively, and so forth. As we used only one strain from each serotype, we have used the serotype name alone in the text.
Criteria for gene selection.
Three genes (ipgD, mxiA, and mxiC) were selected for analysis from the pINV genes for which sequence was available. The mxi genes were chosen because mxiA and mxiC genes are involved in export, effectively a housekeeping function (23). The levels of polymorphism in the homologues of the two genes in the inv-spa complex in Salmonella enterica are comparable to those of housekeeping genes (6), and the genes were selected to represent such housekeeping genes. On the other hand ipgD was chosen because the protein is secreted (3) and appears more variable from comparative sequence analysis of a 20-kb segment of the invasion region of the S. sonnei pINV (GenBank accession no. D50601), a 4-kb segment of the S. flexneri 2a invasion region (bases 230 to 4218 of S. sonnei sequence) (GenBank accession no. L04309), and a 10-kb fragment of the S. flexneri invasion region (bases 10702 to 20323 of S. sonnei) (GenBank accession no. D13663). This pattern of levels of variation is found generally in type III secretion systems (15). ipgD has recently been reported to be involved in host cell invasion during infection (21).
PCR and sequencing.
Sequence was obtained directly from PCR product. Primers for PCR and sequencing (Table 1) were based on the published sequences (GenBank accession nos D50601 and L04309) and selected taking into account sequence conservation. Double-stranded PCR product was purified with the Wizard PCR purification system (Promega, Madison, Wis.) to remove excess PCR primer and eluted in 30 μl of sterile distilled water, and the sequence was determined by the dye terminator technique using a thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) and automated 377 DNA sequencer (Applied Biosystems, Burwood, Victoria, Australia), according to the manufacturer's instructions.
TABLE 1.
Oligonucleotides used in this study
| Gene (bp) | Primer name | 5′ start positiona | Primer sequence (5′-3′) | Length (bases) | Annealing temp (°C) |
|---|---|---|---|---|---|
| ipgD (1617) | 1660 | −355 | TATCAGGCTCGGAGTGTTAT | 20 | 51 |
| 1662 | 605 | TCGGCGTCAGAAGAGAAGTC | 20 | ||
| 1661 | 547 | GACCAGAGTTATTATCACAG | 20 | 49 | |
| 1663 | 1474 | TATTAGCACATCATCATCAA | 20 | ||
| mxiC (1067) | 1814 | −11 | GTAGGTGATGTATGCTTG | 18 | 47 |
| 1816 | 1068 | GATCACTTTCATCTCCTG | 18 | ||
| mxiA (2066) | 1817 | −3 | GAGATGAAAGTGATCCAG | 18 | 48 |
| 1818 | 999 | AAATGTACCAGTATAGCC | 18 | ||
| 1819 | 946 | ATTATAAAAAGGTCGTAG | 18 | 46 | |
| 1820 | 2004 | AGCATACGATATAACGAG | 18 |
Refered to the first base (A) in the ATG start codon.
Sequence analysis.
DNA sequences were assembled and edited using programs Phred, Phrap, and Consed (12). Further analysis was undertaken using programs available from the Australian National Genomic Information Service (ANGIS) at the University of Sydney. Sequence comparisons were analyzed using the Multicomp package (29). Multicomp calculates nucleotide diversity (π) by the method described by Nei and Miller (20) and average pairwise percent difference. Calculation of synonymous and nonsynonymous substitution rates was done using the program kindly provided by W-H. Li (16). Molecular evolutionary relationships among each of the genes studied were examined by the neighbor-joining method of tree construction (19, 31), based on distance estimated using the two-parameter method of Kimura (14). Phylogenetic trees and bootstrap analysis to determine the statistical stability of each node were done using Phylip (version 3.5 written by Joseph Felsenstein, Department of Genetics, University of Washington, Seattle, Wash.).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequence determined in this study are AY029074 to AY029154.
RESULTS AND DISCUSSION
Presence of plasmid pINV and amplification of ipgD, mxiC, and mxiA genes.
46 Shigella and two EIEC strains were used in attempts to amplify ipgD, mxiC, and mxiA. PCR-negative strains were screened by DNA hybridization with an ipgD, mxiC, or mxiA probe at low stringency. The F1B, F2B, F3B, F4B, FX, D2, D12, B3, B5, B7, B8, B13, B16, B17, and B18 strains used were negative for all three probes and most likely have lost the plasmid. This is perhaps not surprising, as spontaneous loss of the plasmid, which results in loss of virulence, occurs readily (35). In addition, the ipgD gene and mxi genes could not be amplified from some strains. mxiC only was obtained for FY and D13, and mxiA only for D5 and D7. No ipgD sequence was obtained from D8. No PCR product was obtained from B6 and D11 for any of the genes, but they were positive by probing, which indicates that B6 and D11 may have a divergent form of pINV. Five EIEC strains were tested, but only two (M519 and M520) have plasmids.
Sequence variation in ipgD.
The 1,617-bp ipgD gene is located at the beginning of the mxi operon, and IpgD is secreted and involved in the manipulation of cellular process of entry into host cells (3, 21). Sequences were obtained for 23 of the Shigella strains representing all major Shigella clusters as described by Pupo et al. (26), as well as for the two EIEC strains. The S. sonnei sequence (M1608) has a few differences relative to that reported for S. sonnei isolate HW383 (GenBank accession number D50601), which could be either sequencing errors in the latter or strain differences. We confirmed all bases involved in isolate M1608.
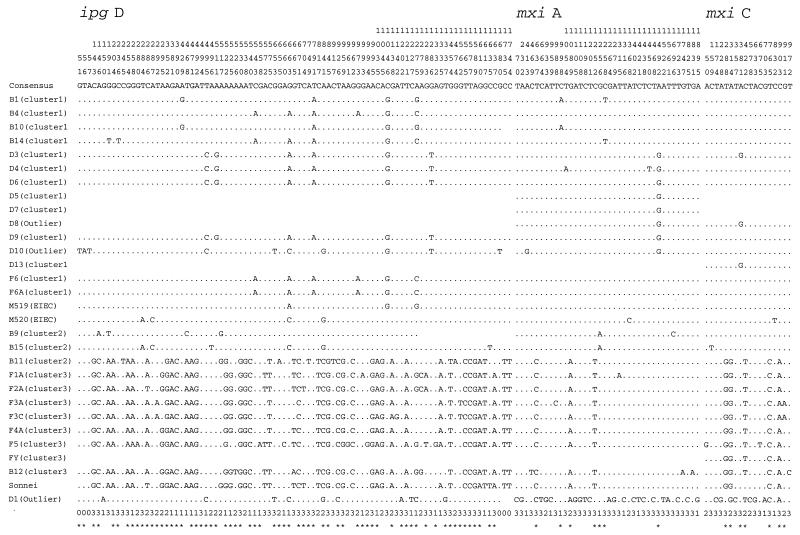
There were 91 (5.19%) polymorphic nucleotide sites in the 1,754 bp sequenced, which included the 1,617 bp of ipgD (Fig. 1), and polymorphic sites were distributed evenly throughout the gene. A comparison of the sequences obtained revealed an average pairwise nucleotide difference of 1.67%, which ranged between 0.0 and 3.39%. This can be compared with pairwise differences in chromosomal genes of the same strains ranging between 0.01 and 1.35% (26).
FIG. 1.
Alignment of the polymorphic sites for the three genes sequenced. Note that for some strains for which no sequence was obtained, the corresponding region appears blank. The numbers at the top of the figure, reading vertically, are base positions. Numbers (1, 2, and 3) at bottom are codon positions of that base (0 indicates a noncoding base), and an asterisk under a base indicates a phylogenetically informative site.
Sequence variation in mxi genes.
The 1,067-bp mxiC gene is located approximately 7.7 kb downstream of ipgD, and MxiC is involved in the transport of bacterial proteins into host cells across the cell membrane (23). A total of 1,000 bp (bases 33 to 1032, 94% of the gene) was obtained from 27 Shigella and two EIEC strains; 18 nucleotide sites (0.20%) were found to be polymorphic (Fig. 1), with an average pairwise difference of 0.36% (range, 0.0 to 1.10%).
The mxiA gene (2006 bp) is located 7 bp downstream of mxiC and is an essential component of the type III secretory machinery (4). The gene product has several potential membrane-spanning regions. We obtained 1,947 bp (bases 36 to 1982, 97% of the gene) sequence for 27 Shigella and two EIEC strains; 38 (1.95%) polymorphic nucleotides were found (Fig. 1). The average pairwise difference was 0.22% and ranged between 0.0 and 1.34%.
The gene products of mxiA and mxiC appear to be membrane bound (5), and variation in mxi genes is lower than in ipgD, which encodes a secreted protein. This pattern of levels of variation fits the general observation that membrane-bound proteins are less variable than secreted proteins in type III secretion systems (6, 15).
Genetic relationship among the plasmids.
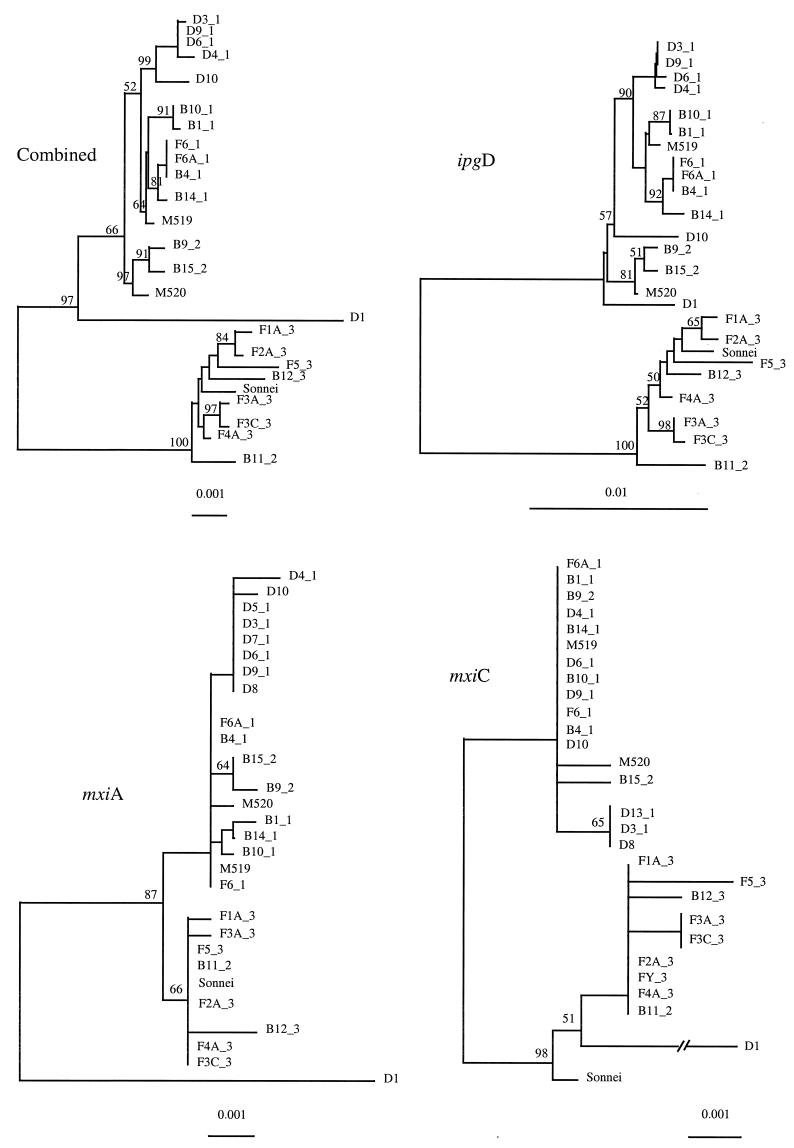
Neighbor-joining trees were constructed for each gene and also for the three genes combined (Fig. 2). In the three individual gene trees, all sequences fell into two clusters, which indicates that there are two sequence forms of the plasmid, which we refer to as pINV A and pINV B. This distinction is supported by data on plasmid incompatibility (17) (see below). The distribution of the two forms correlates well with variation in chromosomal genes (26). All cluster 1 strains have the pINV A form, and cluster 3 strains have pINV B. However, of the three cluster 2 strains for which we obtained sequence (B9, B11, and B15), B9 and B15 have pINV A form, while B11 has the pINV B form. S. sonnei has pINV B, and the two EIEC strains have the pINV A form.
FIG. 2.
Phylogenetic trees generated by the neighbor-joining method for the three genes sequenced. The number after the underscore gives the cluster number identified by chromosomal gene trees (26). Bootstrap values are percentages of 1,000 replications and are indicated at the nodes if the value is greater than 50%. Only strains with sequences for all three genes are included in the combined tree.
We reported previously that in chromosomal gene trees (26) there are five outlier Shigella strains (D1, D8, D10, S. sonnei, and B13) which do not fall into the three clusters. We obtained sequences for all three genes from D1, D10, and S. sonnei and two genes (mxiA and mxiC) from D8, but none for B13. The plasmid gene sequences for all but D1 are typical pINV A or pINV B sequences.
The D1 pINV plasmid must have a complex history. The D1 ipgD gene is more similar to that of pINV A, with a segment (bp 495 to 841) identical to that of D10, while the mxiA and mxiC genes are more similar to the pINV B form, sharing five of the seven informative sites. However, there are many unique polymorphic sites in the two mxi genes.
Distribution of two forms of pINV correlates with previously reported plasmid incompatibility groups.
The two forms of pINV also differ in plasmid incompatibility. The pINV plasmid present in Shigella and EIEC strains has the RepFIC replicon belonging to the RepFIIA family (36). Makino et al. (17) divided pINV plasmids into two groups based on compatibility with an IncF1 plasmid (R386). The incompatible group consisted of S. sonnei and S. flexneri other than S. flexneri serotype 6, while the compatible group included S. flexneri serotype 6, S. boydii (three strains from serotypes 1 and 2), S. dysenteriae (four strains from serotypes 1 and 4), and five EIEC strains of four serotypes. This plasmid incompatibility grouping correlates well with division into two sequence forms. D1, as discussed above, is of uncertain origin. However, the D1 pINV plasmid is most likely a pINV A plasmid with the mxi genes from a distantly related pINV B plasmid, as by plasmid incompatibility D1 is grouped with pINV A plasmids.
Origin of pINV and development of two forms.
The virulence plasmid pWR100 (a pINV B form) from an S. flexneri 5 isolate has recently been sequenced (8). The plasmid consists of regions differing in GC content, with the largest block of genes of the entry region and osp genes of similar GC content and possibly same origin, but the latter were interspersed with insertion sequence elements. It was suggested that the plasmid has been assembled from different sources. Our data on the two forms of pINV relate to three genes of the spa-mxi-spa locus and plasmid compatibility. The correlation in these two properties indicates that they have been associated throughout the period over which the Shigella strains diversified. However, it is not excluded that other parts of the plasmid were added or modified by recombination over this time frame.
Comparative divergence of plasmid and chromosomal genes.
Based on synonymous substitution (Ks) data for chromosomal genes, it was estimated that cluster 1 and cluster 2 are about 50,000 to 270,000 years old, while cluster 3 is about 35,000 to 170,000 years old (26). The pINV genes show similar levels of divergence to housekeeping genes (Ks is 0.0012 and 0.0037 for the three pINV genes of cluster 1 and cluster 3 strains, respectively, compared to 0.0016 and 0.001 for housekeeping genes of cluster 1 and 3 strains). It appears that the common ancestors for the strains in each of Shigella clusters 1 and 3 carried the ancestral A and B forms of the pINV plasmid.
Lateral transfer of pINV.
Of the 25 pINV plasmids in our study, 24 fell into group A or B, the exception being D1. Within the 24 strains were 3 outlier Shigella strains (D8, D10, and S. sonnei) as well as the two EIEC strains. These five strains are outside the major Shigella clusters, as shown by sequence of housekeeping genes (25, 26). But the pINV sequences show that their plasmids are either pINV A or pINV B, and all five were probably acquired by lateral transfer. That S. sonnei has a pINV B plasmid while D8 and D10 have a pINV A plasmid indicates that strains with either plasmid can be donors.
The presence of several outlier Shigella-EIEC strains and their acquisition of plasmids of either plasmid form indicates that transfer of the pINV plasmid into other E. coli strains has occurred several times. There may be many more cases of transfer in EIEC strains. In a previous study, five EIEC strains of four serotypes were found to be distributed among nonpathogenic E. coli, except that the two strains of the same serotype clustered together (25), indicating four independent transfers of the invasion plasmid. The mechanism of such transfer is not known. The pINV plasmid of several S. flexneri strains was found to be unable to initiate conjugation (17, 34), attributed to the presence of only a partial tra operon (27), but this may not be representative. It has also been shown that the pINV plasmid from an F5 strain is mobile in the presence of other functional F plasmids (34). Watanabe and Nakamura (37) demonstrated that with mobilization by an RP4 derivative, pINV for S. sonnei, D1 and F2A could be introduced into previously nonpathogenic E. coli and be stably maintained. It seems clear from our data that with or without mobilization, pINV plasmid transfer has been important in the origin of new Shigella-EIEC clones.
Patterns of evolution in EIEC
The development of EIEC, including the Shigella clones, parallels the evolution of other forms of pathogenic E. coli. Reid et al. (30) recently studied the evolutionary patterns of enteropathogenic (EPEC) and enterohemorrhagic (EHEC) strains by using sequence of seven chromosomal genes and distribution of virulence genes and found that gain of virulence elements has occurred many times in separate lineages. The acquisition of virulence factors in EPEC and EHEC strains is thought to occur in a stepwise fashion, starting with the locus of enterocyte effacement (LEE) pathogenicity island for both and subsequent acquisition of the EHEC plasmid (pO157) and STX phage to give an EHEC strain or the EAF plasmid only for EPEC strains (30). In contrast to the stepwise model of EPEC and EHEC strains, EIEC and Shigella strains seem to have only the pINV plasmid as a required virulence element, although there are loci on the chromosome encoding genes which may enhance virulence, e.g., the SHE pathogenicity island (2, 28). The other identified characteristics are loss of chromosomal properties, e.g., lysine decarboxylation and curli production, although we do not know which if any are prerequisites for it to become an invasive organism. However, development of EIEC, as for EPEC and EHEC, has occurred many times.
Shigella strains lack several catabolic pathways widely present in E. coli. EIEC and Shigella strains spend much of their time within eukaryotic cells and clearly have a very different nutrient supply from most E. coli. The loss of catabolic functions is presumably related to this, but in most cases it is not clear if it is simply that functions are lost only because they are redundant or if the functions are deleterious. In the case of lysine decarboxylase (LDC), it seems that the function is deleterious. Maurelli et al. (18) reported that the introduction of cadA, the gene encoding LDC, into S. flexneri 2a resulted in attenuation of virulence by inhibition of iron-regulated enterotoxins. Deletions in the region of cadA in Shigella and EIEC strains were found, and it was concluded that LDC is detrimental to the particular pathogenic lifestyle. LDC activity, present in the majority of E. coli strains, is absent from Shigella and EIEC strains and was used as a characteristic to differentiate E. coli and Shigella. There is also circumstantial evidence that there is selection against production of curli fimbriae (32).
There are losses of other functions in Shigella strains, including motility, lactose fermentation, gas production, and utilization of acetate, mucate, and xylose. It is not clear how these losses are related to virulence. They are more likely be a tradeoff of “use it or lose it” in adaptation to the specific niche that Shigella occupies. Cooper and Lenski (9) reported that during the evolution of E. coli populations in a minimal-glucose environment, unused catabolic functions decayed while the evolving populations adapted to the glucose. It was argued that this is the effect of antagonistic pleiotropy (tradeoff) in ecological specialization rather than mutation accumulation for the decay of the unused catabolic functions by accumulation of mutations by random genetic drift. It remains to be shown if this applies to most cases of loss of function in Shigella and EIEC clones.
Virulence variation of EIEC and Shigella.
Little is known of variation in the level of virulence of Shigella clones except for D1. D1, the type form of Shigella, is the primary agent of epidemic shigellosis and is isolated from around 30% of dysentery cases in areas where Shigella spp. are endemic (13). The Shiga toxin, carried on a phage in D1 but no other Shigella or EIEC strains, contributes significantly to the clinical manifestation of D1 infections (24).
EIEC strains are generally perceived to be less virulent than Shigella clones, with a higher infectious dose of EIEC in volunteers (10, 11). We have now shown that the virulence plasmids of the two EIEC strains are one of the two forms of Shigella plasmid. Therefore, virulence differences between EIEC and Shigella may not lie in the virulence plasmid, unless the difference is in regulation of virulence genes or interaction of chromosomal genes with plasmid-encoded genes. It seems necessary to reassess the variation in virulence within and between EIEC and Shigella strains using a widely representative set of strains based on the emerging phylogenetic classification of Shigella and EIEC that will help elucidate their mechanisms of virulence and pathogenesis.
Taxonomic status of Shigella.
The data presented in this paper add more sequence-based evidence to the already overwhelming evidence that the four Shigella species and E. coli are in reality one species. We are in the position of having phylogenetic trees based on either housekeeping genes (26) or pINV plasmid genes (this paper) in which sequences of five named species are intermingled. This is very confusing unless one understands that the five species are in fact a single species. To avoid further confusion, it is important that this issue of nomenclature be resolved.
Conclusions.
The evolutionary relationships of the pINV (virulence) plasmids of Shigella and EIEC were analyzed using sequence of three genes in the invasion region of the plasmid. The plasmids can be classified into two distinct but closely related forms on this basis. The clustering of Shigella strains by plasmid forms is largely consistent with that based on chromosomal gene sequences. Moreover, a number of Shigella strains which were found to be unrelated to the main body of Shigella strains appear to have acquired their virulence plasmid by lateral transfer from other pINV-carrying strains. This situation also applies to the two EIEC strains studied: both were found to have plasmids identical to those found in Shigella strains. Shigellae are forms of EIEC, with the three main clusters representing three successful clones. The evolution of EIEC and Shigella forms of E. coli involves gain of a pINV plasmid and loss of catabolic functions, and we postulate that the loss of most catabolic functions is not related to virulence but due to adaptation to the niche that they occupy.
S. dysenteriae 1 (D1), the type form for Shigella and prominent in outbreaks, is unusual in all aspects. D1 is distinct from other Shigella strains in both chromosomal genes and pINV genes. D1 is also unique among Shigella and other enteroinvasive strains of E. coli in the production of Shiga toxin. The combination of the two virulence factors results in its distinctive clinical and pathogenetic features.
ACKNOWLEDGMENTS
The research was supported by a grant from the National Health and Medical Research Council of Australia.
We are grateful to Johanne Lefebvre and Pierre Harbec (Laboratoire de Santé Publique du Quebec, Canada) for providing Shigella strains. We thank the anonymous referees for suggestions on improvement of the manuscript.
REFERENCES
- 1.Acheson D W K, Keusch G T. Shigella and enteroinvasive Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 763–784. [Google Scholar]
- 2.Al-Hassan K, Rajakumar K, Bulach D, Robbins-Browne R, Adler B, Sakellaris H. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb Pathog. 2001;30:1–8. doi: 10.1006/mpat.2000.0404. [DOI] [PubMed] [Google Scholar]
- 3.Allaoui A, Menard R, Sansonetti P J, Parsot C. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect Immun. 1993;61:1707–1714. doi: 10.1128/iai.61.5.1707-1714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews G P, Hromockyj A E, Coker C, Maurelli A T. Two novel virulence loci, mxiA and mxiB, in Shigella flexneri 2a facilitate excretion of invasion plasmid antigens. Infect Immun. 1991;59:1997–2005. doi: 10.1128/iai.59.6.1997-2005.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews G P, Maurelli A T. mxiA of Shigella flexneri 2a, which facilitates export of invasion plasmid antigens, encodes a homolog of the low-calcium-response protein, LcrD, of Yersinia pestis. Infect Immun. 1992;60:3287–3295. doi: 10.1128/iai.60.8.3287-3295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd E F, Li J, Ochman H, Selander R K. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J Bacteriol. 1997;179:1985–1991. doi: 10.1128/jb.179.6.1985-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner D J, Fanning G R, Miklos G V, Steigerwalt A G. Polynucleotide sequence relatedness among Shigella species. Int J Syst Bacteriol. 1973;23:1–7. [Google Scholar]
- 8.Buchrieser C, Glaser P, Rusniok C, Nedjari H, D'Hauteville H, Kunst F, Sansonetti P, Parsot C. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 9.Cooper V S, Lenski R E. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- 10.DuPont H L, Formal S B, Hornick R B, Snyder M J, Libonati J P, Sheahan D J, LaBrec E H, Kala J P. Pathogenesis of Escherichia coli diarrhea. N Engl J Med. 1971;285:1–9. doi: 10.1056/NEJM197107012850101. [DOI] [PubMed] [Google Scholar]
- 11.Dupont H L, Levine M M, Hornick R B, Formal S B. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 12.Gordon D, Abajian C, Green P. CONSED - A graphical tool for sequence finishing. PCR Methods Appl. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 13.Hale T L. Genetic basis of virulence in Shigella species. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;116:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 15.Kresse A U, Beltrametti F, Muller A, Ebel F, Guzman C A. Characterization of SepL of enterohemorrhagic Escherichia coli. J Bacteriol. 2000;182:6490–6498. doi: 10.1128/jb.182.22.6490-6498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W-H. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 17.Makino S, Sasakawa C, Yoshikawa M. Genetic relatedness of the basic replicon of the virulence plasmid in shigellae and enteroinvasive Escherichia coli. Microb Pathog. 1988;5:267–274. doi: 10.1016/0882-4010(88)90099-x. [DOI] [PubMed] [Google Scholar]
- 18.Maurelli A T, Fernandez R E, Bloch C A, Rode C K, Fasano A. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nei M. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. [Google Scholar]
- 20.Nei M, Miller J C. A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics. 1990;125:873–879. doi: 10.1093/genetics/125.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niebuhr K, Jouihri N, Allaoui A, Gounon P, Sansonetti P J, Parsot C. IpgD, a protein secreted by the type III secretion machinery of Shigella flexneri, is chaperoned by IpgE and implicated in entry focus formation. Mol Microbiol. 2000;38:8–19. doi: 10.1046/j.1365-2958.2000.02041.x. [DOI] [PubMed] [Google Scholar]
- 22.Ochman H, Whittam T S, Caugant D A, Selander R K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 23.Parsot C, Sansonetti P J. Invasion and the pathogenesis of Shigella infections. Curr Top Microbiol Immunol. 1996;209:25–42. doi: 10.1007/978-3-642-85216-9_2. [DOI] [PubMed] [Google Scholar]
- 24.Prado D, Cleary T G, Pickering L K, Ericsson C D, Bartlett A V, III, DuPont H L, Johnson P C. The relation between production of cytotoxin and clinical features in shigellosis. J Infect Dis. 1986;154:149–155. doi: 10.1093/infdis/154.1.149. [DOI] [PubMed] [Google Scholar]
- 25.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pupo G M, Lan R, Reeves P R. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radnedge L, Davis M A, Youngren B, Austrin S J. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J Bacteriol. 1997;179:3670–3675. doi: 10.1128/jb.179.11.3670-3675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri SHE pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeves P R, Farnell L, Lan R. MULTICOMP: a program for preparing sequence data for phylogenetic analysis. Comput Appl Biol Sci. 1994;10:281–284. doi: 10.1093/bioinformatics/10.3.281. [DOI] [PubMed] [Google Scholar]
- 30.Reid S D, Herbelin C J, Bumnaugh A C, Selander R K, Whittam T S. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 31.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Sakellaris H, Hannink N K, Rajakumar K, Bulach D, Hunt M, Sasakawa C, Adler B. Curli loci of Shigella spp. Infect Immun. 2000;68:3780–3783. doi: 10.1128/iai.68.6.3780-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sansonetti P J, d'Hautevitte H, Ecobichon C, Pourcel C. Molecular comparison of virulence plasmids Shigella and enteroinvasive Escherichia coli. Ann Microbiol. 1983;134A:295–318. [PubMed] [Google Scholar]
- 34.Sansonetti P J, Kopecko D J, Formal S B. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun. 1982;35:852–860. doi: 10.1128/iai.35.3.852-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasakawa C, Kamata K, Sakai T, Murayama S Y, Makino S, Yoshikawa M. Molecular alteration of the 140-megadalton plasmid associated with loss of virulence and Congo red binding activity in Shigella flexneri. Infect Immun. 1986;51:470–475. doi: 10.1128/iai.51.2.470-475.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva R M, Saadi S, Maas W K. A basic replicon of virulence-associated plasmids of Shigella spp. and enteroinvasive Escherichia coli is homologous with a basic replicon in plasmids of IncF groups. Infect Immun. 1988;56:836–842. doi: 10.1128/iai.56.4.836-842.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe H, Nakamura A. Large plasmids associated with virulence in Shigella species have a common function necessary for epithelial cell penetration. Infect Immun. 1985;48:260–262. doi: 10.1128/iai.48.1.260-262.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]