Abstract
A hallmark of herpes simplex virus (HSV) infection is the establishment of latent virus in peripheral sensory ganglia of the latently infected host. We and others originally reported that the latency-associated transcript (LAT) is the only abundantly expressed viral gene in neurons within trigeminal ganglia (TG) of a latently infected host. Here, we investigated the possible contribution of various cells [i.e., B cells, dendritic cells (DCs), fibroblasts, glial cells, innate lymphoid cells (ILCs), macrophages, microglia, monocytes, natural killer cells, neurons, neutrophils, and T cells] isolated from TG of latently infected mice. Our results demonstrated that all of these cell types contain LAT, with DCs, neurons, and ILCs having the most LAT+ cells. These results suggest that HSV-1 can establish a quiescent/latent infection in a subset of nonneuronal cells, which enhances the chances that the virus will survive in its host.
Immune cells in trigeminal ganglia are the culprits in hiding and protecting HSV during latency.
INTRODUCTION
Originally using immunofluorescence, electron microscopic, autoradiographic, and in situ nucleic acid hybridization, neurons of latently infected mice were shown to contain herpes simplex virus type 1 (HSV-1) (1, 2). Subsequently, expression of more than 80 HSV-1 genes that occurs during lytic infection is drastically modified, and the latency-associated transcript (LAT) is the only gene product consistently detected in abundance during latency in neurons of mice, rabbit, and human (3–7). In both animals and humans, HSV-1 travels via anterograde and retrograde transport between the site of infection and the ganglia of infected hosts (8, 9). Thus, a cardinal characteristic of infection with HSV-1 and other members of the α-herpesviruses is their ability to establish latency in sensory neurons of an infected host (5, 6, 10, 11). Once acquired, a latently infected host has a lifelong pattern of episodic recurrence, and the virus may reactivate at various times throughout the life of a latently infected host, travel back to the original site of infection, and cause recurrent disease. Thus, infected individuals serve as permanent carriers who are intermittently infectious (12–16).
The LAT gene is located in the viral long repeats and is, therefore, present in two copies per genome (17). The primary LAT transcript is 8.3 kb and unstable, while a very stable 2-kb LAT transcript, also referred to as the “stable” or “major” LAT, is derived from the primary transcript by splicing (6, 7, 18–20). The 2-kb LAT accumulates in the nucleus of latently infected neurons but can also be detected in the cytoplasm (6, 7, 20–22). A second 1.3- to 1.5-kb LAT RNA is apparently derived from the 2-kb LAT by splicing and is also abundant during latency (4, 23). Since the original reports of LAT detection in neurons of latently infected mice, rabbits, and humans due to its abundance (4, 6, 7, 20, 24), there has been a general belief that neurons are the only site that harbor latent virus. However, in addition to neurons, sensory tissues contain many immune and nonimmune cell types, and development of more sensitive techniques, such as single-cell analysis, allowed us to determine whether nonneuronal cells also carry the virus during latency. In this study, we looked at the contribution of both neuronal and nonneuronal cells to HSV-1 latency using a combination of reverse transcription polymerase chain reaction (RT-PCR), PCR, and single-cell analysis of total cell populations of trigeminal ganglia (TG). We found that in addition to neurons, LAT is also expressed in immune cells in the TG of latently infected mice. These results suggest that the current lack of success developing a vaccine strategy against HSV-1 infection may be due to host immune cells that protect against virus clearance, or these cells have different functions compared to the same cell type that is not latently infected.
RESULTS
Detection of LAT in TG nonneuronal cells of latently infected mice
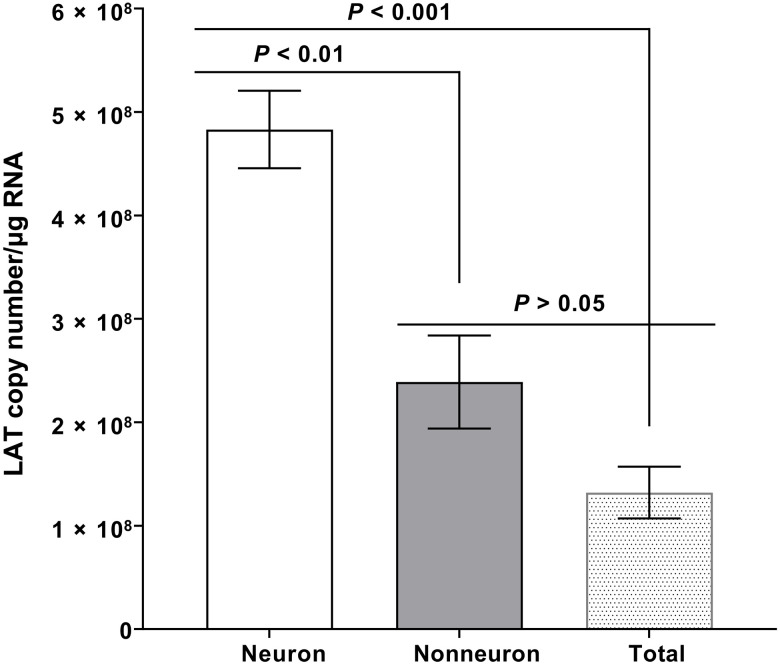
In neurons of latently infected mice, rabbits, and humans, the 2-kb LAT is the only HSV-1 gene product consistently detected in abundance during latency (3–7). No study has shown the presence of LAT during latency in nonneuronal cells. Thus, in this study, we asked whether nonneuronal cells also carry LAT. Wild-type (WT) C57BL/6 mice (n = 24) were infected ocularly with 2 × 105 plaque-forming units (PFUs) per eye of HSV-1 strain McKrae and TG from infected mice were isolated on day 35 post infection (PI) when the infection becomes latent, and virus can no longer be recovered from the cell-free homogenates of the affected ganglia. Neurons and nonneuronal cells were isolated by cell extraction, and total RNA was isolated from TG of some infected mice. Isolated neuronal RNA, nonneuronal RNA, and total TG RNA were used to quantify LAT RNA copy number, as we described previously (25, 26). Cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA was used as an internal control. The amount of LAT RNA during latency in isolated neurons was significantly higher than in nonneuronal RNA (Fig. 1; P < 0.01) and total TG RNA (Fig. 1; P < 0.001), while levels of LAT RNA in nonneuronal and total TG RNA were similar (Fig. 1; P > 0.05). These results suggest that nonneuronal cells contain LAT RNA but at lower levels. Previously, in both mouse and human TG, approximately 1% of the cells were shown to be neurons (27–29). The lower levels of LAT RNA in total nonneuronal cell RNA or in total TG RNA is probably due to higher expression and/or higher stability of LAT in isolated neurons, and LAT promoter is neuronal specific compared with its presence in isolated nonneuronal cells or total isolated cells in the TG.
Fig. 1. Detection of LAT in the nonneuronal fraction of latently infected TG.
Wild-type (WT) mice were ocularly infected with 2 × 105 PFU per eye of HSV-1 strain McKrae without corneal scarification. TG were harvested on day 35 PI. Four latently infected TG from two mice were pooled together and dissociated into a single-cell suspension by digesting with collagenase D. Neuronal and nonneuronal fractions were isolated using the Miltenyi Biotec neuron isolation kit, as described in the company protocol. Briefly, dissociated cells were stained with the antibody cocktail, mixed with magnetic beads, and neuronal and nonneuronal cells were collected. Total RNA was extracted from each fraction, and unfractionated RNA from individual TG was used as a control. LAT copy number was measured by qRT-PCR using a standard curve generated from pGEM5317, as we described previously (73). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used to normalize relative levels of LAT RNA expression. Each bar represents the mean LAT copy number/μg RNA ± SEM from 12 mice for nonneuronal RNA and neuronal RNA and from 12 mice for total TG RNA.
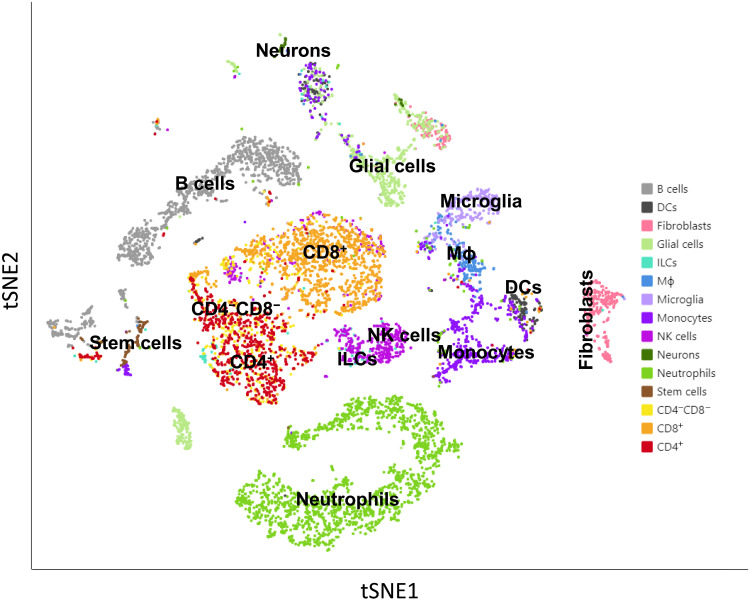
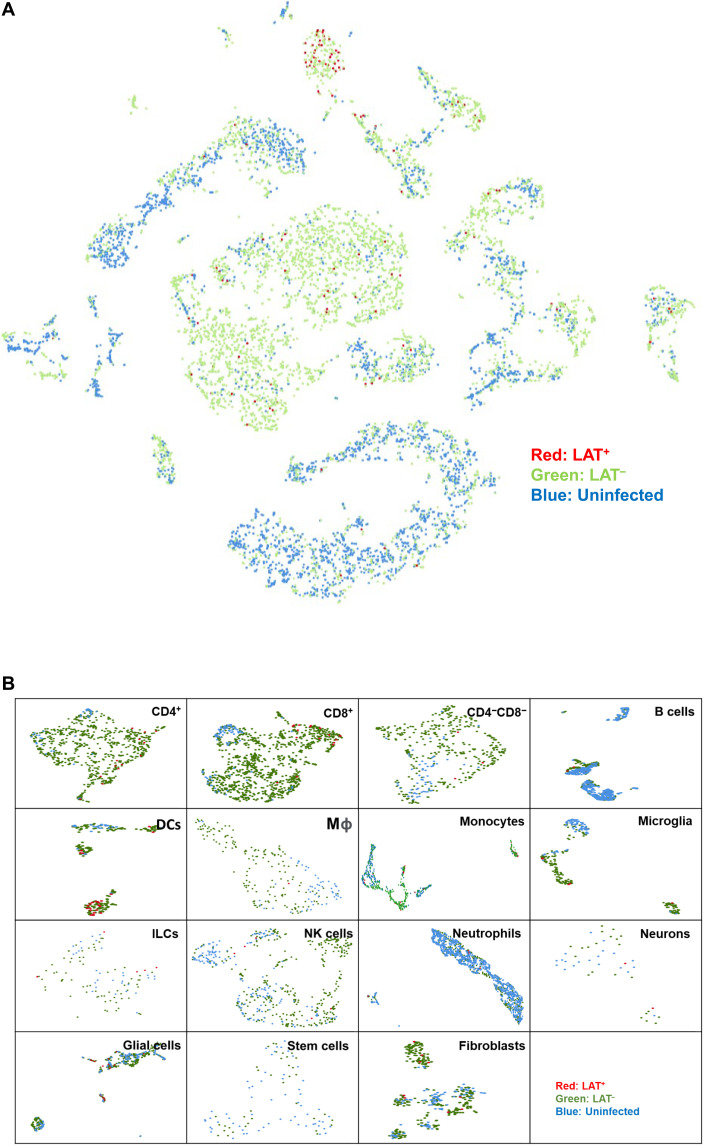
Nonneuronal cells in the TG consist of different cell types, including several types of immune cells (30, 31). Our results described in Fig. 1 suggested that nonneuronal cells also contain LAT. To determine what cell types are carrying LAT, mice were infected ocularly, as above, and TG from latently infected mice were isolated on day 35 PI. TG from mock-infected mice were used as a control. TG from 15 infected mice were pooled together and dissociated into single cells. Dead cells were removed using a dead cell removal kit, and CD45+ immune cells were pulled down using anti-CD45 beads. Purified cells from 15 latently infected mice were mixed back with the original single-cell suspension at a 1:1 ratio and then used for single-cell sequencing. Multiple MouseRNAseq Data and ImmGenData from package celldex were used as described previously to determine the cell types isolated from TG (32–34). Single-cell analysis identified B cells, dendritic cells (DCs), fibroblasts, glial cells, innate lymphoid cells (ILCs), macrophages, microglia, monocytes, natural killer (NK) cells, neurons, neutrophils, stem cells, CD4−CD8− T cells, CD4+ T cells, and CD8+ T cells in cells isolated from TG of latently infected mice (Fig. 2). Next, using an aggregated cell separation map of three combined TG cell samples (one from uninfected mice and two from infected mice), each dot on the map was traced back to its origins in Loupe software to identify it as being from the uninfected or infected group (Fig. 3A). Thus, in Fig. 3A, we looked at LAT+, LAT−, and uninfected isolated cells. Infected LAT+ cells are shown in red, infected LAT− cells are shown in green, and uninfected cells are shown in blue (Fig. 3A). There were fewer infected LAT+ cells than infected LAT− cells (Fig. 3A). As expected, more LAT+ cells were detected in neurons of infected mice than in other cell types, although more LAT− cells were detected than LAT+ cells in infected samples, and the red LAT+ cells spread across the aggregated t-distributed stochastic neighbor embedding (tSNE) map (Fig. 3A; compare red cells versus green cells). These results suggest that many of the cells in TG of latently infected mice are LAT-; thus, not all cells from latently infected TG are infected or express LAT during latency. We next analyzed each individual cell type using UMAP (Uniform Manifold Approximation and Projection) (Fig. 3B). This analysis showed that LAT+ cell populations in DCs, CD4+ T cells, and CD8+ T cells are well separated from uninfected controls by UMAP, but LAT+ cell populations in neurons and glial cells are not well separated (Fig. 3B). However, in nearly all cell types, LAT+ cells are not well separated from LAT−-infected cells when using either UMAP (Fig. 3B) or tSNE (Fig. 3A) methods.
Fig. 2. tSNE map of individual cell types from mouse TG.
Mice were infected with HSV-1 McKrae. TG were harvested 35 days PI, pooled together (30 TG), and dissociated to single cells. Dead cells were removed using a dead cell removal kit, and CD45+ immune cells were isolated as described in Materials and Methods. Purified cells were mixed back with the original single-cell suspension for single-cell sequencing. ImmGen reference was used to determine immune cell types, while multiple mouse single-cell sequence references were used to determine neurons.
Fig. 3. Detection of LAT+ cells in TG of latently infected mice.
(A) Aggregated tSNE map of single-cell sequencing from uninfected (blue), infected (green), or LAT+ cells (red). As described in Fig. 2 legend, the aggregated cell separation map was created by Loupe to visualize LAT+ cells. Cells expressing LAT (expression level greater than 1) were marked as red. (B) Individual UMAP of each cell type containing LAT. Each cell type was divided into three groups according to infection status or LAT expression status (greater than 1). Red, infected and LAT+; green, infected and LAT−; and blue, uninfected and LAT−.
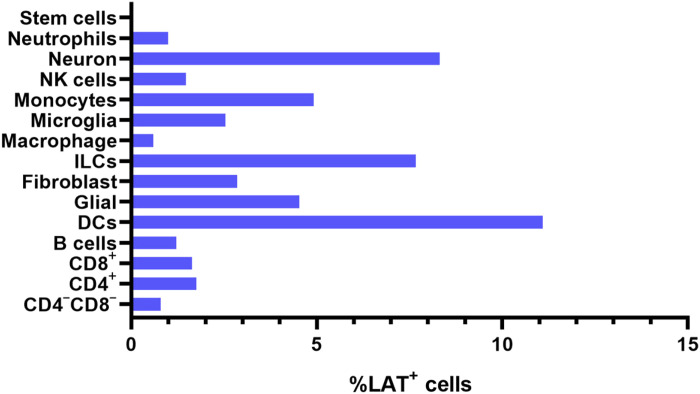
Because the results above indicate the presence of LAT in most of the isolated cell types (Fig. 3, A and B), we next calculated the percentage of LAT+ cells in each cell type (Fig. 4). LAT was detected in neutrophils, neurons, NK cells, monocytes, microglia, macrophages, ILCs, fibroblasts, glial cells, DCs, B cells, CD8+ T cells, CD4+ T cells, and CD4−CD8− T cells but not in stem cells (Fig. 4). DCs, neurons, and ILCs had the highest percentage of LAT+ cells followed by monocytes and glial cells, which had a similar percentage of LAT+ cells, while microglia and fibroblasts had a similar percentage of LAT+ cells (Fig. 4). Furthermore, neutrophils, NK cells, B cells, CD8+ T cells, CD4+ T cells, and CD4−CD8− T cells had similar levels of LAT+ cells, with macrophages and stem cells having the lowest levels of LAT+ cells (Fig. 4). This result suggests that in addition to neurons, cells including neutrophils, NK cells, monocytes, microglia, macrophages, ILCs, fibroblasts, glial cells, DCs, B cells, CD8+ T cells, CD4+ T cells, and CD4−CD8− cells also contain LAT. Similar to this study, T cells and B cells were detected in TG of latently infected mice and humans, but the presence of LAT in immune cells of latently infected mice and humans was not evaluated (35, 36). The percentage of LAT+ cells in infected neurons was nearly identical to the percentage of LAT+ DCs and ILCs (Fig. 4). This result is consistent with our previous study in which we showed that the absence of DCs is associated with reduced levels of latency in infected mice (37, 38). We previously showed that CD8α−/− latently infected mice had significantly less LAT expression than did WT mice or CD8β−/− mice, that adoptive transfer of CD8α+ DCs, but not CD8α+ cells, to CD8α−/− mice rescued LAT levels compared to that of WT mice (37, 38), and that DCs do not have protective roles against HSV-1 infection (39, 40).
Fig. 4. Percentage of LAT+ cells in different cell populations.
The percentage of LAT+ cells in different infected cell populations are shown. In addition, number of LAT+/total cells for CD4−CD8− T cells, 3 of 372; CD4+ T cells, 13 of 737; CD8+ T cells, 17 of 1029; B cells, 5 of 408; DCs, 24 of 216; glial cells, 26 of 572; fibroblasts, 9 of 214; ILCs, 6 of 78; macrophages, 1 of 164; microglia, 5 of 196; monocytes, 17 of 345; NK cells, 5 of 337; neurons, 2 of 24; and neutrophils, 5 of 498.
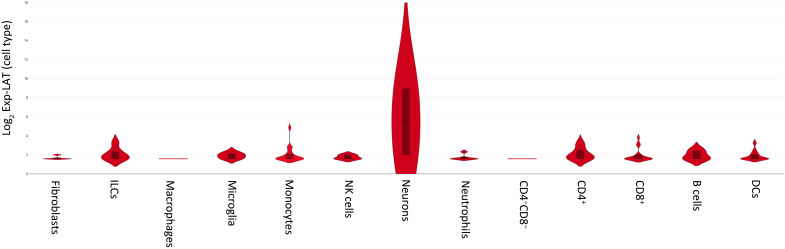
Results in Figs. 3 and 4 showed that in addition to neurons, LAT was detected in nearly all cell types, except in stem cells. Thus, we created violin plots to determine the relative levels of LAT expression in each cell type (Fig. 5). The violin plot showed that neurons had the highest level of LAT expression, while LAT expression was similar in ILCs, CD4+ T cells, and B cells, followed by microglia, monocytes, NK cells, CD8+ T cells, and DCs (Fig. 5). Last, fibroblasts and neutrophils had similar LAT levels with macrophages, with CD4−CD8− cells having the lowest levels of LAT expression (Fig. 5). These results confirm previous studies by showing that higher levels of LAT expression in neurons is the reason for its relatively easy detection in latently infected ganglia of mice, rabbits, and humans by in situ hybridization and northern (RNA) blotting compared with other cell types (4, 6, 7, 41). The LAT promoter is active in most cell types, especially in neuronal cells (42–44). HSV-1 is a neurotropic virus, and the unique properties of the LAT promoter produces efficient, long-term LAT expression in vivo (44). Thus, higher LAT expression in neurons of latently infected mice as compared to other cell types is probably due to the neuronal specificity of the LAT promoter. Our RT-PCR and single-cell analyses showed that LAT expression in isolated neurons was higher than in nonneuronal cells, which could be due to the neuronal specificity of the LAT promoter or to higher viral DNA copy numbers in neurons than in other cell types.
Fig. 5. LAT expression levels in different cell types.
Violin plots using Loupe software for single-cell sequencing analysis were used to measure LAT expression levels in LAT+ cells from each cell type.
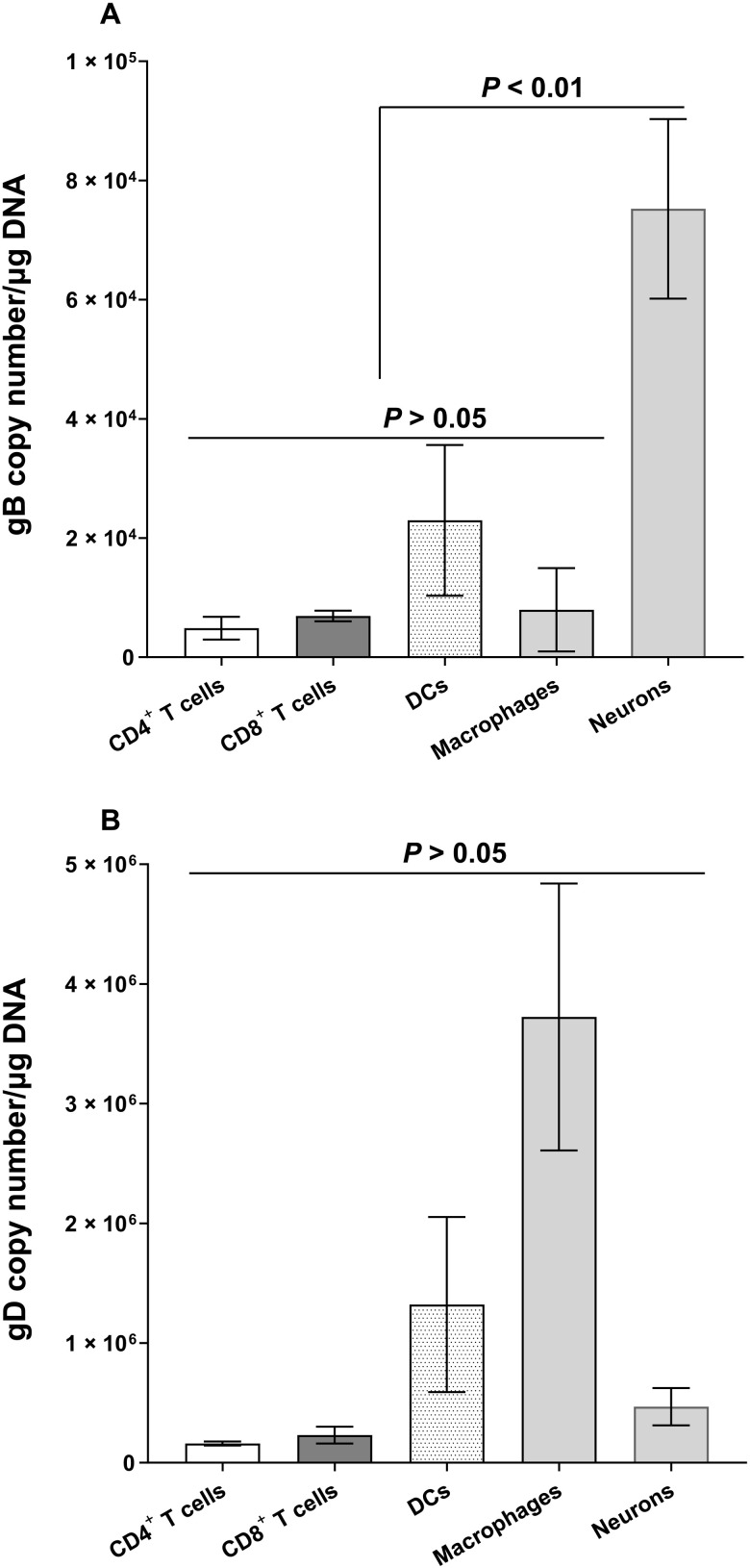
Detection of LAT in nonneuronal cell types may suggest that we are detecting virus fragments rather than intact virus, although, in contrast to RNA viruses, such as influenza (45), defective-interfering particles are not common in HSV-1. To rule out this possibility, we infected additional mice as above, and CD4+ T cells, CD8+ T cells, DCs, macrophages, and neurons were isolated from TG of these latently infected mice on day 35 PI. DNA from each cell type was isolated, and we performed PCR for glycoprotein B (gB) and glycoprotein D (gD) DNA as these two genes are essential HSV-1 glycoproteins and are separated by more than 80,000 nucleotides (17). gB DNA was detected in isolated CD4+ T cells, CD8+ T cells, DCs, and macrophages. Differences in gB expression between these cells was not statistically significant (Fig. 6A; P > 0.05), while levels of gB DNA in neurons was significantly higher than in CD4+ T cells, CD8+ T cells, DCs, or macrophages (Fig. 6A; P < 0.01). Similar to gB DNA (Fig. 6A), gD DNA was also detected in CD4+ T cells, CD8+ T cells, DCs, and macrophages, and no significant differences were detected between them (Fig. 6B; P > 0.05). In contrast to the levels of gB DNA in neurons (Fig. 6A), no significant differences were detected between levels of gD DNA in neurons, CD4+ T cells, CD8+ T cells, DCs, and macrophages (Fig. 6B; P > 0.05). Differences between gB and gD DNA levels in infected mouse neurons may be caused by lower stability of gD DNA than gB DNA. These results confirm and extend the above results, showing that in addition to neurons, HSV-1 DNA is detected in different cell types of the TG with higher LAT RNA levels that are independent of viral DNA levels in neurons. Higher LAT expression in neurons than in nonneurons could be due to neuronal specificity of the LAT promoter or to the nondividing nature of neurons, which may cause constant levels of input and output virus in latently infected neurons, while cell division of nonneuronal cells may cause LAT levels to decrease.
Fig. 6. Detection of gB and gD DNA in neuronal and nonneuronal fractions of latently infected TG.
WT mice were infected ocularly with HSV-1 McKrae as described in Fig. 1 legend. TG were isolated on day 35 PI, and eight TG from four mice were pooled and dissociated to single cells. Immune cells enriched by Percoll density gradient were stained with CD45, CD3, CD4, CD8, F4/80, and CD11c antibodies. CD4, CD8, F4/80, and CD11c populations were sorted, and genomic DNA from each population was purified. gB (A) and gD (B) copy numbers were measured by qPCR, and GAPDH was used as internal control. Experiments were repeated three times.
DISCUSSION
It is estimated that 70 to 90% of American adults have antibodies to HSV-1 and/or HSV-2, and about 25% of these individuals have clinical symptoms upon routine doctor visit, with HSV-1 being responsible for >90% of ocular HSV infections (15, 16, 46–49). A significant proportion (15 to 50%) of primary genital herpes is caused by HSV-1, and recent studies indicate that the proportion of first clinical episode of genital herpes due to HSV-1 is increasing (50–52). Annually, in the United States, approximately 500,000 people suffer from recurrent ocular HSV episodes requiring doctor visits, medication, and, in severe cases, corneal transplants (15, 16, 46–49, 53, 54). Ocular HSV-1 infection can cause eye disease ranging in severity from blepharitis, conjunctivitis, and dendritic keratitis to disciform stromal edema and necrotizing stromal keratitis (15, 16, 46, 55). Despite the seriousness of herpes infection, no vaccine has been developed to prevent HSV infection or recurrences. Thus, development of a safe vaccine to prevent and control serious HSV infection is essential. Eliciting neutralizing antibody alone can completely protect immunized mice from eye disease and death but will not protect immunized mice from virus replication in the eye or the establishment of latency and reactivation (56, 57). Thus, protection against eye disease and death is much easier to achieve than protection against virus replication and establishment of latency during HSV-1 infection. Because of latent infection, eye disease is much more likely to occur following recurrent, rather than primary, HSV infection. Because of the problems associated with recurrent ocular infection, a major goal of any anti-HSV vaccine must be to prevent and/or reduce latency establishment and thus reactivation.
Multiple approaches have been used to develop a vaccine against HSV infection ranging from using attenuated virus to using recombinantgB+ gD or gD alone (58–60). However, none of these vaccines were effective in humans. Our current study suggests that the absence of any viable vaccine against HSV infection is probably due to the fact that the virus is not cleared by the immune system because it is hiding in the immune cells and is thus protected from clearance by neutralizing antibodies and by both innate and adaptive immune responses since these nonneuronal sites for latency have biological relevance with respect to virus transmission. This study may help us look outside the box to design a different type of vaccine that can penetrate immune cells to clear the virus and to further reduce and preferentially eliminate the latent virus reservoir. Our findings in this study may also be relevant to the other 14 members of the simplexvirus genus (61), especially HSV-2, which is the causative agent of genital herpes, and all human vaccine trials involving development of a vaccine against genital herpes have failed (58, 59, 62–64). Similar to this study, it was previously shown that equid herpesvirus 1, bovine herpesvirus 1, and pseudorabies virus can establish a latent/quiescent infection in immune cells in pharyngeal tonsil and to the lymphoid tissue (65–67). In summary, this observation was made in a murine model of HSV-1 infection, which, unlike humans, the virus does not spontaneously reactivate to cause recurrent eye disease, and the findings are needed to be verified in human TG.
MATERIALS AND METHODS
Ethics statement
All animal procedures were performed in strict accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and the NIH Guide for the Care and Use of Laboratory Animals (ISBN 0-309-05377-3). The animal research protocol was approved by the Institutional Animal Care and Use Committee of Cedars-Sinai Medical Center (Protocol nos. 8837 and 9129).
Mice, virus, and ocular infection
Plaque-purified virulent HSV-1 strain McKrae was used in this study. Both male and female 6- to 8-week old C57BL/6 mice (the Jackson Laboratory, Bar Harbor, ME) were used. Mice were ocularly infected with 2 × 105 pfu per eye of McKrae virus without corneal scarification, as we described previously (68). Mock-infected control mice were ocularly treated with media only.
Isolation of neurons and nonneuronal cells from latently infected TG
To isolate neurons and nonneuronal cells from TG, TG from latently infected or mock-infected mice were harvested and pooled. Pooled TG were then incubated in 1× Hanks’ balanced salt solution (HBSS) with collagenase D (20 mg/ml) and deoxyribonuclease 1 at 37°C for 1 hour to create a single-cell suspension. Neuronal cells were then isolated by negative selection, while nonneuronal cells were isolated by positive selection using a neuron isolation kit (Miltenyi Biotec, no. 130-115-389) following the company’s instructions. Briefly, cells were stained with the nonneuronal cell biotin antibody cocktail (containing antibodies that bind to nonneuronal cells, such as astrocytes, oligodendrocytes, microglia, endothelial cells, and immune cells, except erythrocytes) at 4°C for 5 min and then washed with 1× phosphate-buffered saline (PBS) and 0.5% bovine serum albumin (BSA). Cells were spun down, resuspended in 1× Dulbecco’s PBS plus 0.5% BSA, and incubated with anti-biotin microbeads for 10 min at 4°C. Neuronal cells were separated by passage through an LS column under a magnetic field, while nonneuronal cells were attached, washed, and collected. Isolated cells were used for RNA extraction and single-cell analysis.
Cell sorting
Mice were infected with HSV-1 McKrae, and TG were harvested on day 35 PI. Four TG were pooled and incubated with collagenase D to dissociate them into single cells. Immune cells were enriched by Percoll gradient. Enriched immune cells were stained with BV510-CD45, BV421-CD3, fluorescein isothiocyanate–CD4, phycoerythrin-CD11c, allophycocyanin (APC)–F4/80, APC-Fire750-CD8a, and live-dead staining dye 7-AAD. CD4 T cells (CD45+CD3+CD4+), CD8 T cells (CD45+CD3+CD8+), CD11c+ DCs (CD45+CD3−CD11c+), and F4/80+ macrophages (CD45+CD3−F4/80+) were sorted using a BD Aria 3 sorter.
RNA extraction and quantitative RT-PCR
After ocular infection with 2 × 105 PFU per eye of McKrae virus, intact TG or specific populations of cells isolated from TG of latently infected mice were collected on day 35 PI. Isolated tissues were immersed in TRIzol reagent and processed for RNA extraction, as we described previously (69, 70). LAT RNA levels were determined in isolated neurons, isolated nonneuronal cells, or total TG using custom-made LAT primers and probes as follows: forward primer, 5′-GGGTGGGCTCGTGTTACAG-3′; reverse primer, 5′-GGACGGGTAAGTAACAGAGTCTCTA-3′; and probe, 5′-FAM-ACACCAGCCCGTTCTTT-3′ [amplicon length, 81 base pairs (bp)]. GAPDH was used to normalize transcripts, ABI Mm999999.15_G1—amplicon length, 107 bp. Relative LAT copy number was calculated using standard curves generated from pGem-LAT5317.
DNA extraction and TaqMan quantitative PCR analysis of gB and gD DNA in isolated neurons and nonneuronal cells from TG of latently infected mice
WT mice were infected as described above. TG were isolated on day 35 PI, and eight TG from four mice were pooled and dissociated into single cells using collagenase D. Immune cells were enriched by Percoll density gradient and then stained with CD45, CD3, CD4, CD8, F4/80, or CD11c antibodies. CD4, CD8, F4/80, and CD11c populations were sorted, and neuronal cells were isolated as described above. DNA was isolated from individual cell populations using the DNeasy Blood & Tissue Kit (Qiagen, Stanford, CA, catalog no. 69506) according to the manufacturer’s instructions. Sequences of the gB custom TaqMan primer sets used in this study are as follows: 5′-AACGCGACGCACATCAAG-3′ (forward), 5′-CTGGTACGCGATCAGAAAGC-3′ (reverse), and 5′-FAM-CAGCCGCAGTACTACC-3′ (probe). The gB amplicon length is 72 bp. Relative gB DNA copy numbers were calculated using standard curves generated from plasmid pAc-gB1 (71). Sequences of the gD custom TaqMan primer sets used in this study are as follows: 5′-GCGGCTCGTGAAGATAAACG-3′ (forward), 5′-CTCGGTGCTCCAGGATAAACT G-3′ (reverse), and 5′-FAM-CTGGACGGAGATTACA-3′ (probe). The gD amplicon length is 59 bp. Relative gD DNA copy numbers were calculated using standard curves generated from plasmid pAc-gD1 (72). GAPDH was used to normalize transcripts in all experiments.
Single-cell analysis of cells isolated from TG of latently infected mice
Mice were infected ocularly with HSV-1 McKrae and TG were harvested on day 35 PI. Thirty TG from 15 latently infected mice were pooled and washed with cold 1× HBSS six times and then incubated with collagenase D (2 mg/ml) at 37°C for 1 hour to generate single cells. Dead cells were removed using a dead cell removal kit from STEMCELL (Seattle, WA, USA) according to the company protocol. CD45-positive cells were isolated using a mouse CD45-positive cell selection kit from STEMCELL according to company protocols. Isolated CD45-positive cells were mixed back with the original single-cell suspension at a 1:1 ratio and used for single-cell sequencing library preparation. Single-cell RNA-seq libraries were prepared according to the Single Cell 3′ v3.1 Reagent Kits User Guide (10x Genomics, Pleasanton, CA). Cell suspensions were loaded on a Chromium Controller instrument (10x Genomics) to generate single-cell Gel Bead-In-EMulsions (GEMs). Gel Bead-In-Emulsion-reverse transcription (GEM-RT) was performed in a Veriti 96-well thermal cycler (Thermo Fisher Scientific, Waltham, MA). GEMs were then harvested, and the cDNA was amplified and cleaned up using a SPRIselect Reagent Kit (Beckman Coulter, Brea, CA). Indexed sequencing libraries were constructed using a Chromium Single-Cell 3′ Library Kit for enzymatic fragmentation, end repair, A tailing, adapter ligation, ligation cleanup, sample index PCR, and PCR cleanup. The barcoded sequencing libraries were quantified by quantitative PCR using a Collibri Library Quantification Kit (Thermo Fisher Scientific). Libraries were sequenced on a NovaSeq 6000 (Illumina, San Diego, CA) according to the Single Cell 3′ v3.1 Reagent Kits User Guide, with a sequencing depth of ~40,000 reads per cell.
Data analysis
Raw sequencing data were demultiplexed and converted to FASTQ format using bcl2fastq v2.20. Cell Ranger v6.0.0 (10x Genomics) was used for barcode identification, read alignment, and Unique Molecular Identifier (UMI) quantification with default parameters and aligning to the mouse reference genome GRCm38. In addition to the empty droplet filtering performed by Cell Ranger, poor-quality cells were removed by setting a maximum mitochondrial read of 15%. For LAT− cells, additional filtering was done, and LAT− cells with an acceptable range of 300 to 9000 detected genes and 500 to 9000 UMI counts were kept for downstream analysis. The samples were integrated together using Seurat v4.0.5 (32). To obtain two-dimensional projections of the population dynamics, principal components analysis (PCA) was run on the integrated gene-barcode matrix to reduce the number of feature dimensions. After running PCA, both UMAP and tSNE algorithms were applied to the top 20 principal components to further reduce these components and visualize cells in a two-dimensional space. Louvain clustering of the cells was done with resolution 0.1. Clusters were annotated using the R package SingleR, which annotates each cell by comparing its transcriptome to reference datasets (34). We used two reference datasets, MouseRNAseqData and ImmGenData from package celldex (34). For glial cells, annotations were additionally checked and confirmed with common marker gene expression. Differential expression analyses were conducted with Seurat using the FindMarkers function on the normalized RNA expression matrix. Subsequent Gene Ontology (GO) enrichment analyses were performed using the R package ClusterProfileR with term annotations drawn from the R package org.Mm.eg.db (33). Single-cell tSNE and UMAP embeddings GO enrichment plots were created with custom R scripts.
Statistical analysis
Fisher’s exact test and Mann-Whitney test were performed using the computer program Instat (GraphPad, San Diego, CA). Results were considered statistically significant at a P value of <0.05.
Acknowledgments
Funding: This work was supported by NIH grants RO1EY13615, RO1EY024649, RO1EY026944, RO1EY029160, and RO1EY029677 to H.G.
Author contributions: Conceptualization: S.W. and H.G. Methodology: S.W., X.S., A.R., and C.S. Investigation: S.W. and H.G. Visualization: S.W., A.R., and H.G. Funding acquisition: H.G. Project administration: H.G. Supervision: H.G. Writing—original draft: S.W. and H.G. Writing—review and editing: S.W., X.S., A.R., C.S., and H.G.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper.
REFERENCES AND NOTES
- 1.M. L. Cook, V. B. Bastone, J. G. Stevens, Evidence that neurons harbor latent herpes simplex virus. Infect. Immun. 9, 946–951 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. L. Cook, J. G. Stevens, Latent herpetic infections following experimental viraemia. J. Gen. Virol. 31, 75–80 (1976). [DOI] [PubMed] [Google Scholar]
- 3.J. M. Hill, F. Sedarati, R. T. Javier, E. K. Wagner, J. G. Stevens, Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174, 117–125 (1990). [DOI] [PubMed] [Google Scholar]
- 4.D. L. Rock, A. B. Nesburn, H. Ghiasi, J. Ong, T. L. Lewis, J. R. Lokensgard, S. L. Wechsler, Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J. Virol. 61, 3820–3826 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.J. G. Stevens, Human herpesviruses: A consideration of the latent state. Microbiol. Rev. 53, 318–332 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S. L. Wechsler, A. B. Nesburn, R. Watson, S. Slanina, H. Ghiasi, Fine mapping of the major latency-related RNA of herpes simplex virus type 1 in humans. J. Gen. Virol. 69, 3101–3106 (1988). [DOI] [PubMed] [Google Scholar]
- 7.S. L. Wechsler, A. B. Nesburn, R. Watson, S. M. Slanina, H. Ghiasi, Fine mapping of the latency-related gene of herpes simplex virus type 1: Alternative splicing produces distinct latency-related RNAs containing open reading frames. J. Virol. 62, 4051–4058 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.R. J. Diefenbach, M. Miranda-Saksena, M. W. Douglas, A. L. Cunningham, Transport and egress of herpes simplex virus in neurons. Rev. Med. Virol. 18, 35–51 (2008). [DOI] [PubMed] [Google Scholar]
- 9.B. Feierbach, M. Bisher, J. Goodhouse, L. W. Enquist, In vitro analysis of transneuronal spread of an alphaherpesvirus infection in peripheral nervous system neurons. J. Virol. 81, 6846–6857 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.N. W. Fraser, T. Valyi-Nagy, Viral, neuronal and immune factors which may influence herpes simplex virus (HSV) latency and reactivation. Microb. Pathog. 15, 83–91 (1993). [DOI] [PubMed] [Google Scholar]
- 11.D. Phelan, E. R. Barrozo, D. C. Bloom, HSV1 latent transcription and non-coding RNA: A critical retrospective. J. Neuroimmunol. 308, 65–101 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Y. J. Gordon, Pathogenesis and latency of herpes simplex virus type 1 (HSV-1): An ophthalmologist’s view of the eye as a model for the study of the virus-host relationship. Adv. Exp. Med. Biol. 278, 205–209 (1990). [DOI] [PubMed] [Google Scholar]
- 13.H. E. Kaufman, A. M. Azcuy, E. D. Varnell, G. D. Sloop, H. W. Thompson, J. M. Hill, HSV-1 DNA in tears and saliva of normal adults. Invest. Ophthalmol. Vis. Sci. 46, 241–247 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.I. Steiner, Human herpes viruses latent infection in the nervous system. Immunol. Rev. 152, 157–173 (1996). [DOI] [PubMed] [Google Scholar]
- 15.B. A. Barron, L. Gee, W. W. Hauck, N. Kurinij, C. R. Dawson, D. B. Jones, K. R. Wilhelmus, H. E. Kaufman, J. Sugar, R. A. Hyndiuk, P. R. Laibson, R. D. Stulting, P. A. Asbell, C. R. Dawson, T. P. Margolis, R. A. Nozik, H. B. Ostler, B. A. Barron, M. S. Insler, H. E. Kaufman, D. B. Jones, A. Y. Matoba, K. R. Wilhelmus, R. D. Stulting, G. O. Waring III, L. A. Wilson, R. A. Hyndiuk, S. B. Koenig, B. M. Massaro, P. A. Asbell, A. P. Davis, M. J. Newton, J. Sugar, S. Lam, J. B. Robin, H. H. Tessler, P. R. Laibson, E. J. Cohen, K. G. Leavitt, C. J. Rapuano, W. W. Hauck, L. Gee, J. E. Hidayat, C. R. Dawson, W. W. Hauck, D. B. Jones, H. E. Kaufman, N. Kurinij, S. Bangdiwala, W. E. Barlow, J. W. Chandler, L. J. Nelson, A. B. Nesburn, C. R. Dawson, L. Gee, W. W. Hauck, J. E. Hidayat, N. Kurinij, K. R. Wilhelmus, L. M. Merin, L. A. Todaro, Herpetic eye disease study. Ophthalmology 101, 1871–1882 (1994). [DOI] [PubMed] [Google Scholar]
- 16.K. R. Wilhelmus, C. R. Dawson, B. A. Barron, P. Bacchetti, L. Gee, D. B. Jones, H. E. Kaufman, J. Sugar, R. A. Hyndiuk, P. R. Laibson, R. D. Stulting, P. A. Asbell, Risk factors for herpes simplex virus epithelial keratitis recurring during treatment of stromal keratitis or iridocyclitis. Herpetic Eye Disease Study Group. Br. J. Ophthalmol. 80, 969–972 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D. J. McGeoch, M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, P. Taylor, The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69, 1531–1574 (1988). [DOI] [PubMed] [Google Scholar]
- 18.E. K. Wagner, W. M. Flanagan, G. Devi-Rao, Y. F. Zhang, J. M. Hill, K. P. Anderson, J. G. Stevens, The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J. Virol. 62, 4577–4585 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. J. Farrell, A. T. Dobson, L. T. Feldman, Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. U.S.A. 88, 790–794 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.J. G. Spivack, N. W. Fraser, Expression of herpes simplex virus type 1 latency-associated transcripts in the trigeminal ganglia of mice during acute infection and reactivation of latent infection. J. Virol. 62, 1479–1485 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M. Nicosia, J. M. Zabolotny, R. P. Lirette, N. W. Fraser, The HSV-1 2-kb latency-associated transcript is found in the cytoplasm comigrating with ribosomal subunits during productive infection. Virology 204, 717–728 (1994). [DOI] [PubMed] [Google Scholar]
- 22.M. Ahmed, N. W. Fraser, Herpes simplex virus type 1 2-kilobase latency-associated transcript intron associates with ribosomal proteins and splicing factors. J. Virol. 75, 12070–12080 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N. W. Fraser, J. G. Spivack, Z. Wroblewska, T. Block, S. L. Deshmane, T. Valyi-Nagy, R. Natarajan, R. M. Gesser, A review of the molecular mechanism of HSV-1 latency. Curr. Eye Res. 10, 1–13 (1991). [DOI] [PubMed] [Google Scholar]
- 24.J. G. Stevens, E. K. Wagner, G. B. Devi-Rao, M. L. Cook, L. T. Feldman, RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235, 1056–1059 (1987). [DOI] [PubMed] [Google Scholar]
- 25.S. Wang, U. Jaggi, J. Yu, H. Ghiasi, Blocking HSV-1 glycoprotein K binding to signal peptide peptidase reduces virus infectivity in vitro and in vivo. PLOS Pathog. 17, e1009848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.K. R. Mott, C. J. Bresee, S. J. Allen, L. BenMohamed, S. L. Wechsler, H. Ghiasi, Level of herpes simplex virus type 1 latency correlates with severity of corneal scarring and exhaustion of CD8+ T cells in trigeminal ganglia of latently infected mice. J. Virol. 83, 2246–2254 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.N. M. Sawtell, Comprehensive quantification of herpes simplex virus latency at the single-cell level. J. Virol. 71, 5423–5431 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R. L. Thompson, N. M. Sawtell, Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75, 6660–6675 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. J. LaGuardia, R. J. Cohrs, D. H. Gilden, Numbers of neurons and non-neuronal cells in human trigeminal ganglia. Neurol. Res. 22, 565–566 (2000). [DOI] [PubMed] [Google Scholar]
- 30.K. Messlinger, A. F. Russo, Current understanding of trigeminal ganglion structure and function in headache. Cephalalgia 39, 1661–1674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S. Thalakoti, V. V. Patil, S. Damodaram, C. V. Vause, L. E. Langford, S. E. Freeman, P. L. Durham, Neuron-glia signaling in trigeminal ganglion: Implications for migraine pathology. Headache 47, 1008–1023 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck III, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. Wu, E. Hu, S. Xu, M. Chen, P. Guo, Z. Dai, T. Feng, L. Zhou, W. Tang, L. Zhan, X. Fu, S. Liu, X. Bo, G. Yu, clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D. Aran, A. P. Looney, L. Liu, E. Wu, V. Fong, A. Hsu, S. Chak, R. P. Naikawadi, P. J. Wolters, A. R. Abate, A. J. Butte, M. Bhattacharya, Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. Shimeld, J. L. Whiteland, S. M. Nicholls, E. Grinfeld, D. L. Easty, H. Gao, T. J. Hill, Immune cell infiltration and persistence in the mouse trigeminal ganglion after infection of the cornea with herpes simplex virus type 1. J. Neuroimmunol. 61, 7–16 (1995). [DOI] [PubMed] [Google Scholar]
- 36.M. van Velzen, L. Jing, A. D. M. E. Osterhaus, A. Sette, D. M. Koelle, G. M. G. M. Verjans, Local CD4 and CD8 T-cell reactivity to HSV-1 antigens documents broad viral protein expression and immune competence in latently infected human trigeminal ganglia. PLOS Pathog. 9, e1003547 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.K. R. Mott, S. J. Allen, M. Zandian, B. Konda, B. G. Sharifi, C. Jones, S. L. Wechsler, T. Town, H. Ghiasi, CD8α dendritic cells drive establishment of HSV-1 latency. PLOS ONE 9, e93444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.K. R. Mott, D. Gate, H. H. Matundan, Y. N. Ghiasi, T. Town, H. Ghiasi, CD8+ T cells play a bystander role in mice latently infected with Herpes Simplex Virus 1. J. Virol. 90, 5059–5067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.K. R. Mott, H. Ghiasi, Role of dendritic cells in enhancement of herpes simplex virus type 1 latency and reactivation in vaccinated mice. Clin. Vaccine Immunol. 15, 1859–1867 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.K. R. Mott, D. UnderHill, S. L. Wechsler, H. Ghiasi, Lymphoid-related CD11c+CD8α+dendritic cells are involved in enhancing herpes simplex virus type 1 latency. J. Virol. 82, 9870–9879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.W. J. Mitchell, I. Steiner, S. M. Brown, A. R. MacLean, J. H. Subak-Sharpe, N. W. Fraser, A herpes simplex virus type 1 variant, deleted in the promoter region of the latency-associated transcripts, does not produce any detectable minor RNA species during latency in the mouse trigeminal ganglion. J. Gen. Virol. 71, 953–957 (1990). [DOI] [PubMed] [Google Scholar]
- 42.G. C. Perng, J. C. Zwaagstra, H. Ghiasi, R. Kaiwar, D. J. Brown, A. B. Nesburn, S. L. Wechsler, Similarities in regulation of the HSV-1 LAT promoter in corneal and neuronal cells. Invest. Ophthalmol. Vis. Sci. 35, 2981–2989 (1994). [PubMed] [Google Scholar]
- 43.J. Zwaagstra, H. Ghiasi, A. B. Nesburn, S. L. Wechsler, In vitro promoter activity associated with the latency-associated transcript gene of herpes simplex virus type 1. J. Gen. Virol. 70, 2163–2169 (1989). [DOI] [PubMed] [Google Scholar]
- 44.J. C. Zwaagstra, H. Ghiasi, S. M. Slanina, A. B. Nesburn, S. C. Wheatley, K. Lillycrop, J. Wood, D. S. Latchman, K. Patel, S. L. Wechsler, Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: Evidence for neuron specificity and for a large LAT transcript. J. Virol. 64, 5019–5028 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S. G. Rabinowitz, J. Huprikar, The influence of defective-interfering particles of the PR-8 strain of influenza A virus on the pathogenesis of pulmonary infection in mice. J Infect Dis 140, 305–315 (1979). [DOI] [PubMed] [Google Scholar]
- 46.C. R. Dawson, Ocular herpes simplex virus infections. Clin. Dermatol. 2, 56–66 (1984). [DOI] [PubMed] [Google Scholar]
- 47.T. J. Liesegang, Classification of herpes simplex virus keratitis and anterior uveitis. Cornea 18, 127–143 (1999). [DOI] [PubMed] [Google Scholar]
- 48.T. J. Liesegang, Herpes simplex virus epidemiology and ocular importance. Cornea 20, 1–13 (2001). [DOI] [PubMed] [Google Scholar]
- 49.T. J. Hill, Ocular pathogenicity of herpes simplex virus. Curr. Eye Res. 6, 1–7 (1987). [DOI] [PubMed] [Google Scholar]
- 50.C. M. Roberts, J. R. Pfister, S. J. Spear, Increasing proportion of herpes simplex virus type 1 as a cause of genital herpes infection in college students. Sex. Transm. Dis. 30, 797–800 (2003). [DOI] [PubMed] [Google Scholar]
- 51.B. A. Auslander, F. M. Biro, S. L. Rosenthal, Genital herpes in adolescents. Semin. Pediatr. Infect. Dis. 16, 24–30 (2005). [DOI] [PubMed] [Google Scholar]
- 52.A. E. Singh, B. Romanowski, T. Wong, S. Gourishankar, L. Myziuk, J. Fenton, J. K. Preiksaitis, Herpes simplex virus seroprevalence and risk factors in 2 Canadian sexually transmitted disease clinics. Sex. Transm. Dis. 32, 95–100 (2005). [DOI] [PubMed] [Google Scholar]
- 53.D. Pavan-Langston, Ocular viral infections. Med. Clin. North Am. 67, 973–990 (1983). [DOI] [PubMed] [Google Scholar]
- 54.L. Corey, The current trend in genital herpes. Progress in prevention. Sex Transm Dis 21, S38–S44 (1994). [PubMed] [Google Scholar]
- 55.P. S. Binder, A review of the treatment of ocular herpes simplex infections in the neonate and immunocompromised host. Cornea 3, 178–182 (1984). [PubMed] [Google Scholar]
- 56.K. Mott, D. J. Brick, N. van Rooijen, H. Ghiasi, Macrophages are important determinants of acute ocular HSV-1 infection in immunized mice. Invest. Ophthalmol. Vis. Sci. 48, 5605–5615 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Y. Osorio, H. Ghiasi, Recombinant herpes simplex virus type 1 (HSV-1) codelivering interleukin-12p35 as a molecular adjuvant enhances the protective immune response against ocular HSV-1 challenge. J. Virol. 79, 3297–3308 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.L. Corey, A. G. Langenberg, R. Ashley, R. E. Sekulovich, A. E. Izu, J. M. Douglas Jr., H. H. Handsfield, T. Warren, L. Marr, S. Tyring, R. DiCarlo, A. A. Adimora, P. Leone, C. L. Dekker, R. L. Burke, W. P. Leong, S. E. Straus, Recombinant glycoprotein vaccine for the prevention of genital HSV-2 infection: Two randomized controlled trials. Chiron HSV Vaccine Study Group. Jama 282, 331–340 (1999). [DOI] [PubMed] [Google Scholar]
- 59.R. B. Belshe, P. A. Leone, D. I. Bernstein, A. Wald, M. J. Levin, J. T. Stapleton, I. Gorfinkel, R. L. Morrow, M. G. Ewell, A. Stokes-Riner, G. Dubin, T. C. Heineman, J. M. Schulte, C. D. Deal; Herpevac Trial for Women , Efficacy results of a trial of a herpes simplex vaccine. N. Engl. J. Med. 366, 34–43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D. M. Koelle, H. Ghiasi, Prospects for developing an effective vaccine against ocular herpes simplex virus infection. Curr. Eye Res. 30, 929–942 (2005). [DOI] [PubMed] [Google Scholar]
- 61.P. J. Walker, S. G. Siddell, E. J. Lefkowitz, A. R. Mushegian, E. M. Adriaenssens, D. M. Dempsey, B. E. Dutilh, B. Harrach, R. L. Harrison, R. C. Hendrickson, S. Junglen, N. J. Knowles, A. M. Kropinski, M. Krupovic, J. H. Kuhn, M. Nibert, R. J. Orton, L. Rubino, S. Sabanadzovic, P. Simmonds, D. B. Smith, A. Varsani, F. M. Zerbini, A. J. Davison, Changes to virus taxonomy and the statutes ratified by the International Committee on Taxonomy of Viruses (2020). Arch. Virol. 165, 2737–2748 (2020). [DOI] [PubMed] [Google Scholar]
- 62.L. R. Stanberry, S. L. Spruance, A. L. Cunningham, D. I. Bernstein, A. Mindel, S. Sacks, S. Tyring, F. Y. Aoki, M. Slaoui, M. Denis, P. Vandepapeliere, G. Dubin; GlaxoSmithKline Herpes Vaccine Efficacy Study Group , Glycoprotein-D-adjuvant vaccine to prevent genital herpes. N. Engl. J. Med. 347, 1652–1661 (2002). [DOI] [PubMed] [Google Scholar]
- 63.A. B. Kern, B. L. Schiff, Vaccine therapy in recurrent herpes simplex. Arch. Dermatol. 89, 844–845 (1964). [DOI] [PubMed] [Google Scholar]
- 64.D. I. Bernstein, F. Y. Aoki, S. K. Tyring, L. R. Stanberry, C. S. Pierre, S. D. Shafran, G. L. Roels, K. van Herck, A. Bollaerts, G. Dubin; The GlaxoSmithKline Herpes Vaccine Study Group , Safety and immunogenicity of glycoprotein D-adjuvant genital herpes vaccine. Clin. Infect. Dis. 40, 1271–1281 (2005). [DOI] [PubMed] [Google Scholar]
- 65.P. M. Chesters, R. Allsop, A. Purewal, N. Edington, Detection of latency-associated transcripts of equid herpesvirus 1 in equine leukocytes but not in trigeminal ganglia. J. Virol. 71, 3437–3443 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. T. Winkler, A. Doster, C. Jones, Persistence and reactivation of bovine herpesvirus 1 in the tonsils of latently infected calves. J. Virol. 74, 5337–5346 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.A. Sabo, J. Rajcani, Latent pseudorabies virus infection in pigs. Acta Virol. 20, 208–214 (1976). [PubMed] [Google Scholar]
- 68.H. Ghiasi, R. Kaiwar, A. B. Nesburn, S. Slanina, S. L. Wechsler, Expression of seven herpes simplex virus type 1 glycoproteins (gB, gC, gD, gE, gG, gH, and gI): Comparative protection against lethal challenge in mice. J. Virol. 68, 2118–2126 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.U. Jaggi, H. H. Matundan, K. Tormanen, S. Wang, J. Yu, K. R. Mott, H. Ghiasi, Expression of murine CD80 by herpes simplex virus 1 in place of latency-associated transcript (LAT) can compensate for latency reactivation and anti-apoptotic functions of LAT. J. Virol. 94, e01798–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.S. J. Allen, A. Rhode-Kurnow, K. R. Mott, X. Jiang, D. Carpenter, J. I. Rodriguez-Barbosa, C. Jones, S. L. Wechsler, C. F. Ware, H. Ghiasi, Interactions between herpesvirus entry mediator (TNFRSF14) and latency-associated transcript during herpes simplex virus 1 Latency. J. Virol. 88, 1961–1971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.H. Ghiasi, R. Kaiwar, A. B. Nesburn, S. L. Wechsler, Expression of herpes simplex virus type 1 glycoprotein B in insect cells. Initial analysis of its biochemical and immunological properties. Virus Res. 22, 25–39 (1992). [DOI] [PubMed] [Google Scholar]
- 72.H. Ghiasi, A. B. Nesburn, S. L. Wechsler, Cell surface expression of herpes simplex virus type 1 glycoprotein H in recombinant baculovirus-infected cells. Virology 185, 187–194 (1991). [DOI] [PubMed] [Google Scholar]
- 73.K. R. Mott, G. C. Perng, Y. Osorio, K. G. Kousoulas, H. Ghiasi, A recombinant herpes simplex virus type 1 expressing two additional copies of gK is more pathogenic than wild-type virus in two different strains of mice. J Virol. 81, 12962–12972 [DOI] [PMC free article] [PubMed] [Google Scholar]