Abstract
The family of attaching and effacing (A/E) bacterial pathogens, which includes diarrheagenic enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC), remains a significant threat to human and animal health. These bacteria intimately attach to host intestinal cells, causing the effacement of brush border microvilli. The genes responsible for this phenotype are encoded in a pathogenicity island called the locus of enterocyte effacement (LEE). Citrobacter rodentium is the only known murine A/E pathogen and serves as a small animal model for EPEC and EHEC infections. Here we report the full DNA sequence of C. rodentium LEE and provide a comparative analysis with the published LEEs from EPEC, EHEC, and the rabbit diarrheagenic E. coli strain RDEC-1. Although C. rodentium LEE shows high similarities throughout the entire sequence and shares all 41 open reading frames with the LEE from EPEC, EHEC, and RDEC-1, it is unique in its location of the rorf1 and rorf2/espG genes and the presence of several insertion sequences (IS) and IS remnants. The LEE of EPEC and EHEC is inserted into the selC tRNA gene. In contrast, the Citrobacter LEE is flanked on one side by an operon encoding an ABC transport system, and an IS element and sequences homologous to Shigella plasmid R100 and EHEC pO157 flank the other. The presence of plasmid sequences next to C. rodentium LEE suggests that the prototype LEE resided on a horizontally transferable plasmid. Additional sequence analysis reveals that the 3-kb plasmid in C. rodentium is nearly identical to p9705 in EHEC O157:H7, suggesting that horizontal plasmid transfer among A/E pathogens has occurred. Our results indicate that the LEE has been acquired by C. rodentium and A/E E. coli strains independently during evolution.
Citrobacter rodentium, formerly Citrobacter freundii biotype 4280, is the causative agent of transmissible murine colonic hyperplasia, a naturally occurring disease primarily in suckling mice among laboratory colonies (4, 47, 50). Mice infected by C. rodentium develop mild diarrhea and coat ruffling and exhibit retarded growth. In extreme cases, mice undergo rectal prolapse and show moderate to high rates of mortality depending on the mouse strains studied (32, 47). C. rodentium belongs to a family of bacterial pathogens causing attaching and effacing (A/E) lesions in affected hosts. The histopathological hallmarks of these pathogens are intimate bacterial attachment to the host intestinal epithelial cells and the effacement of brush border microvilli. Enteropathogenic Escherichia coli (EPEC), an important causative agent of infantile diarrhea in developing countries, and enterohemorrhagic E. coli (EHEC), which causes hemorrhagic colitis and hemolytic-uremic syndrome and is the cause of frequent outbreaks of food and water poisoning in the developed world, are the prominent members of the A/E family (41). Various E. coli isolates from rabbits, dogs, cats, and pigs have been associated with enteric infections and diarrhea and have been found to cause A/E lesions (20, 34, 41, 44, 61). Recently, it has been shown that mouse pathogenic E. coli (MPEC), the infectious agent of megaenteron in mice (28), is actually a misclassified C. rodentium isolate (32). Therefore, C. rodentium is presently the only known murine A/E pathogen.
The A/E pathology is determined by a pathogenicity island called the locus of enterocyte effacement (LEE). When cloned on a plasmid, the LEE from EPEC and the rabbit diarrheagenic E. coli strain RDEC-1 can confer the A/E lesion-inducing phenotype upon laboratory E. coli K12 strains (29, 36). Sequences homologous to the LEE from EPEC have been detected in all the A/E pathogens tested so far, including C. rodentium (35). The complete nucleotide sequences for the LEE from EPEC O127:H6 strain E2348/69 and EHEC O157:H7 strain EDL933 as well as the rabbit O15:H− strain RDEC-1 have been published, and they are highly conserved in both linear gene order and nucleotide as well as in predicted protein sequences, suggesting a common origin (17, 45, 61). The LEE contains genes coding for LEE gene expression regulator Ler, a type III secretion system, an outer membrane adhesin intimin and its translocated receptor Tir, and several secreted proteins (EspA, -D, -B, -F, and -G), as well as a number of open reading frames (ORFs) of undetermined functions. Sequence comparison analysis has shown that while most of the genes coding for the type III secretion system show greater than 95% identity among the three sequenced LEEs from EPEC, EHEC, and RDEC-1, the differences among other genes are more than expected for clonal divergence among E. coli strains (45, 61). Many of these highly divergent genes encode proteins that are believed to be involved in interactions with the host. The LEE, similar to pathogenicity islands found in other bacteria, has a GC content less than that of the E. coli K-12 chromosome (17, 45, 61). It has been hypothesized that A/E pathogens acquired the LEE from other sources via horizontal gene transfer, but it is not clear when this happened, where the LEE originated, or what the mechanism of transfer was (22).
Similar to other A/E pathogens, intimin and EspB have been demonstrated to be important virulence factors for C. rodentium pathogenesis (42, 48, 49). Recent work by Higgins et al. (24, 25) has shown that C. rodentium infection in mice elicits a mucosal Th1-type immune response and lesions reminiscent of murine inflammatory bowel disease and that bacterial intimin is responsible for inducing inflammation and crypt hyperplasia. Thus, C. rodentium infection in mice can potentially serve as a murine model for studying not only the molecular pathogenesis of A/E pathogens but also intestinal inflammation and epithelial cell proliferation. Since EPEC and EHEC are human pathogens, it is very difficult to study the molecular interplay between the bacteria and their host. Other than a few limited human volunteer studies on EPEC (5, 12, 54), most of the work on the LEE has been performed in tissue culture cells or in in vitro-cultured biopsies (19). Rabbit, porcine and cattle models exist for EPEC and EHEC (1, 10, 13, 34, 61), but they are expensive and limited by the tools available to study the hosts. In order to probe the role played by host factors during an infection, a murine model, for which the genetics is well studied and numerous spontaneous and induced mutants as well as immunological reagents are available for analysis, is highly desirable.
Present data support a central role for the LEE in the pathogenesis of C. rodentium and other A/E pathogens. However, there are only four LEE genes in C. rodentium (tir/cesT/eae and espB) which have been sequenced and deposited in the GenBank (32, 42, 48, 49). The complete nucleotide sequence of C. rodentium LEE reported in this paper should facilitate studies on the functions of the LEE-encoded proteins in the bacteria and more importantly for those translocated virulence factors and their interactions with and functions inside the host cell. The Citrobacter LEE sequence also advances our ability to use C. rodentium-mouse interactions as an animal model for studying EPEC- and EHEC-mediated disease in humans. In addition, the unique features of Citrobacter LEE have offered an opportunity for a comparative analysis of entire LEEs from different animal A/E pathogens (human, rabbit, and mouse) and shed new light on the acquisition and evolution of the LEE pathogenicity island in enteric bacteria.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
C. rodentium strains, E. coli strains, and plasmids used or constructed in this study are described in Table 1. Bacteria were grown in Luria-Bertani agar or Luria-Bertani broth supplemented with appropriate antibiotics at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description and relevant phenotype | Reference or source |
|---|---|---|
| Strains | ||
| DBS100 | C. rodentium ATCC 51459 | 48, 49 |
| DBS231 | DBS100 cured of 65-kb plasmid | 48, 49 |
| EPEC E2348/69 | Wild-type E. coli O127:H6 isolate | 17 |
| EHEC EDL933 | Wild-type E. coli O157:H7 isolate | 45 |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector, Ampr | Stratagene |
| pCR2.1-TOPO | PCR cloning vector, Ampr Kanr | Invitrogen |
| pCRII-TOPO | PCR cloning vector, Ampr Kanr | Invitrogen |
| pTOPO-eae/espB | DBS100 eae to espB cloned in pCR2.1-TOPO | This study |
| pCRII-ler/cscT | DBS100 ler to escT cloned in pCRII-TOPO | This study |
| pCRII-cscT/C | DBS100 escT to escC cloned in pCRII-TOPO | This study |
| pCRII-cscC/N | DBS100 escC to escN cloned in pCRII-TOPO | This study |
| pCRII-cscN/tir | DBS100 escN to tir cloned in pCRII-TOPO | This study |
| pCRII-cscC | C. rodentium escC clone in pCRII-TOPO | This study |
| pBS-CRP3EV | C. rodentium 3-kb plasmid cloned in pBluescript II SK(+) as EcoRV fragment | This study |
| pBS-CRP3HIII | C. rodentium 3-kb plasmid cloned in pBluescript II SK(+) as HindIII fragment | This study |
Molecular techniques, DNA cloning and sequencing, and sequence analysis.
For PCR and inverse PCR cloning (43), we routinely used the proof-reading ELONGASE Amplification System (GIBCO BRL/Life Technologies) to minimize the error rate by PCR. PCR cloning was done using the TOPO TA Cloning Kit from Invitrogen with either the vector pCR2.1-TOPO or pCRII-TOPO. The DNA sequence was determined at the Nucleic Acid/Protein Service Unit of the University of British Columbia using the Taq Dye terminator method and an automated 373A DNA Sequencer (Applied Biosystems). M13 reverse, −20, and −40 primers complementary to the PCR cloning vectors and subcloning vector pBluescript II SK(+) (Stratagene) as well as primers designed from available DNA sequences were used for sequencing. Sequences were manually edited and analyzed using the sequence analysis programs DNA Strider, Gene Jockey, and ClustalW and tools available at the website of the National Center for Biotechnology Information. Nucleotide and protein sequence homology searches were done at the National Center for Biotechnology Information's BLAST search server with the filter checked off.
Cloning and sequencing of C. rodentium LEE and its flanking regions.
The important primers used for cloning the C. rodentium LEE region in this study are described in Table 2. When we began these studies, the only LEE genes sequenced in C. rodentium were eae (for intimin), cesT (encoding the chaperone for Tir), and the 3′ end of the tir gene (48, 49). By using a forward primer identical to the EPEC tir gene start codon region (Tir-1) and a reverse primer from the C. rodentium tir gene 3′ end (Tir-2), the Citrobacter tir gene was cloned by PCR and sequenced. A 320-bp promoter region of the Citrobacter tir gene was subsequently cloned by inverse PCR and sequenced. The C. rodentium espB gene was cloned by PCR using primers (EspB-F and EspB-R) based on the EPEC espB gene sequence. The C. rodentium eae-to-espB region was cloned by PCR in pCR2.1-TOPO to create pTOPO-eae/espB using a Citrobacter primer annealing to the 3′ flanking region of the intimin gene (Int-5) and a Citrobacter espB primer, Deng18. To clone the downstream flanking region of espB, a series of inverse PCRs were carried out (43). For inverse PCR, total genomic DNA was isolated from C. rodentium using the cetyltrimethylammonium bromide method (60), and the DNA was digested with one of the following restriction enzymes: EcoRI, EcoRV, HhaI, HindIII, HincII, PstI, SalI, XhoI, Sau3A, or BamHI. The digested DNA was self ligated and used as templates for ELONGASE inverse PCR using primers annealing to the sequenced region immediately upstream of the region intended for cloning. This led to the cloning of the espF, rorf1, and rorf2 genes. The same inverse-PCR strategy was used to clone the downstream flanking region of C. rodentium LEE. This completed the cloning of the tir gene to the end of the right-side junction of C. rodentium LEE.
TABLE 2.
Nucleotide primers used in this study
| Primer | Nucleotide sequence (from 5′ to 3′) | Target regiona |
|---|---|---|
| EscC-F | GATATAGGACGAATTGTG | EPEC escC |
| EscC-R | ATTCGCTAGATGCAGATTTTATCGG | EPEC escC |
| EscN-F | GGGAATAATATCGAACTTAAAG | EPEC escN |
| EscN-R | AGGCAACCACTTTGAATAGGC | EPEC escN |
| EscR-F | GATATGTCTCAATTAATGACCATTGGC | EPEC escR |
| EscR-R | CATCAATTCACCACCAACAGAAATTCG | EPEC escR |
| EscT-F | ATGAATGAGATAATGACGGTCATAGTATC | EPEC escT |
| EscT-R | TCACTCATTAATCATGCTCGGTAACG | EPEC escT |
| EscV-F | GAGCGCGTTCGCAGGATG | EPEC escV |
| EscV-R | ATGCTCTGAAATCATTTACCG | EPEC escV |
| EspB-F | ATGAATACTATCGATAATACCAATG | EPEC espB |
| EspB-R | TTACCCAGCTAAGCGAGCCGCTTGC | EPEC espB |
| Ler-F | CATGCGGAGATTATTTATTATGAATATGG | EPEC ler |
| Ler-R | GTTAAATATTTTTCAGCGGTATTATTTC | EPEC ler |
| Tir-1 | ATGCCTATTGGTAACCTTGGT | EPEC tir |
| Binv-1 | CGACTGCTCGTGACCTTAATGACC | CR espB |
| Cler-2 | GCTGGGATATACTAATGTGCCTG | CR ler |
| Cr1-1 | GCTGCCTTTTATCTCTGATGCCG | CR rorf1 |
| CscC-1 | GTTCTCCAGTAAGCGTGATCC | CR escC |
| CscC-2 | CCGGTAAGACTATTCGAGGTG | CR escC |
| CscN-1 | CTGCAAGTTCTCGTGTAAGCAC | CR escN |
| CcsN-2 | CTGGCATGTATTGTGGGCAGTG | CR escN |
| CscT-1 | GCGATTCATCCTCTATTACTGGCG | CR escT |
| CscT-2 | CCCAGCGGTCTGAGTATGCAAAAG | CR escT |
| Deng18 | GAACTGGCGCTGTCAGTCGTGCTAC | CR espB |
| Finv-1 | GGCTGTAGCAGCGGCAAATCATAAGC | CR escF |
| Int-5 | GTCTAATCATATAAACCCGGC | CR eae |
| R2-inv1 | CCAATGAGCAGAAAGCTGCAGTTGTGG | CR rorf2/espG |
| Tir-2 | GCTTATACTACAACTTGGTTC | CR tir |
| Tir-10 | CAAGAAAGAACAAATAACAGGCC | CR tir upstream |
CR, C. rodentium LEE; EPEC, EPEC E2348/69 LEE.
The nucleotide sequences of EPEC and EHEC LEE are highly conserved and nearly identical in certain regions, most of which encode proteins involved in the type III secretion system (45). This conservation also extends to C. rodentium LEE by Southern blot analysis (35). To clone the region upstream of tir to the end of the left-side junction of C. rodentium LEE, primers identical to the EPEC LEE nucleotide sequences were designed for several of the highly conserved LEE genes. These genes included escC, escV, escN, escR, escT, and orf1/ler. These primers were used to clone the related C. rodentium genes by using the ELONGASE PCR Amplification System: primers EscC-F and EscC-R for PCR amplifying C. rodentium escC, EscV-F and EscV-R for escV, EscN-F and EscN-R for escN, EscR-F and EscR-R for escR, EscT-F and EscT-R for escT, and Ler-F and Ler-R for ler. The cloned C. rodentium genes were sequenced, and their internal sequences were used to design new, C. rodentium-specific primers (Table 2) for PCR cloning of the C. rodentium LEE region spanning from ler to tir. This LEE region was cloned by PCR as four overlapping fragments: ler to escT (pCRII-ler/cscT, using primers Cler-2 and CscT-2), escT to escC (pCRII-cscT/C, using primers CscT-1 and CscC-1), escC to escN (pCRII-cscC/N, using primers CscC-2 and CscN-1), and escN to tir (pCRII-cscN/tir, using primers CscN-2 and Tir-10). These PCR products, between 4 and 8 kb, were agarose gel purified using Qiaquick Gel Extraction Kit from Qiagen and were cloned into pCRII-TOPO (TA Cloning Kit; Invitrogen). The PCR clones were restriction mapped with EcoRV, HindIII, PstI, HincII, and HaeIII and were subcloned into pBluescript II SK(+). The subclones were sequenced by using M13 reverse and −20 primers and by primer walking. The left-side LEE junction fragment upstream of C. rodentium ler was cloned by a series of inverse PCRs (43) using ELONGASE, similar to the cloning of the right-side junction of C. rodentium LEE (see above).
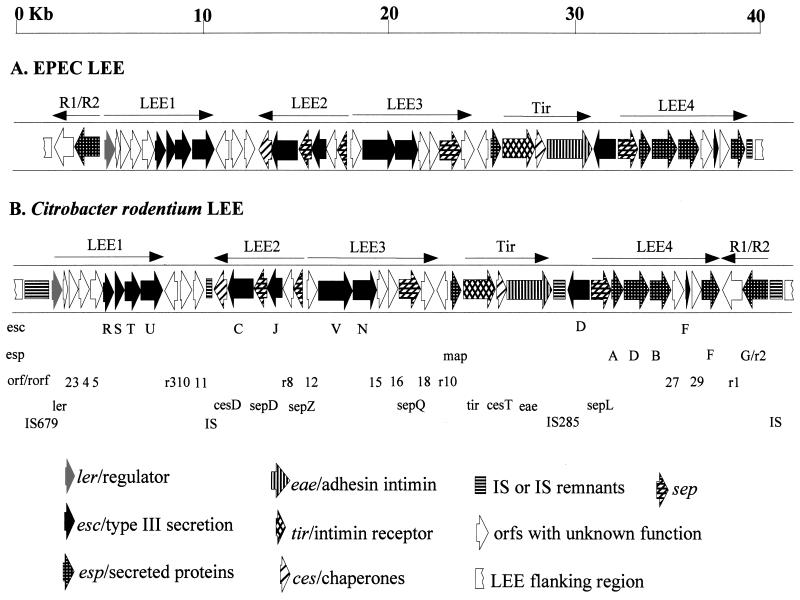
All the clones derived from the PCR and inverse-PCR cloning were subjected to subcloning and DNA sequencing. The linear order and junctions of the subclones were verified by PCR analysis after the sequences were completed and annotated. To facilitate sequence comparison, the presentation and designation of C. rodentium LEE sequences and genes (Fig. 1 and Table 3) followed that of EPEC LEE (17).
FIG. 1.
Gene map for C. rodentium LEE. Gene designation is according to that of EPEC E2348/69 LEE, which is shown for comparison (17). The orientation of each gene is shown by the direction of the arrow. Note the different locations of the rorf1 (r1) and rorf2 (espG/r2) genes in EPEC and C. rodentium LEE, as well as the association of several IS's or IS remnants with the C. rodentium LEE. The major operons encoded by the LEE (LEE1, -2, -3, and -4, Tir, and R1/R2) and their transcriptional directions are shown and adapted from references 16 and 40. Please see Fig. 3 for details of the LEE flanking regions. The map is not drawn completely to scale.
TABLE 3.
ORFs of C. rodentium LEE and their homology with those of EPEC E2348/69, EHEC EDL933, and RDEC-1 LEEs
| ORFs expressed by the LEEs (product length [aa])a
|
Common gene name | Probable protein function | Comparison of protein sequences (% identity/% similarity)
|
|||||
|---|---|---|---|---|---|---|---|---|
| CRb | EPEC | EHEC | EPEC vs EHEC | CR vs EPEC | CR vs EHEC | CR vs RDEC-1 | ||
| rorf1 (272) | L0056 (272) | Unknown | 99/99 | |||||
| rorf2 (398) | L0055 (398) | espG | Secreted protein | 98/98 | ||||
| CR1 (129) | orf1 (129) | L0054 (129) | ler | LEE regulator | 99/99 | 90/95 | 89/94 | 89/94 |
| CR2 (72) | orf2 (72) | L0053 (72) | Type III secretion? | 100/100 | 89/96 | 89/96 | 90/99 | |
| CR3 (107) | orf3 (107) | L0052 (107) | Type III secretion? | 100/100 | 84/91 | 84/91 | RDEC-1: no orf3 | |
| CR4 (199) | orf4 (199) | L0051 (199) | Type III secretion? | 99/100 | 86/95 | 86/94 | 88/95 | |
| CR5 (232) | orf5 (231) | L0050 (231) | Type III secretion? | 98/99 | 76/90 | 76/89 | 76/90 | |
| CR6 (217) | orf6 (217) | L0049 (217) | escR | Type III secretion | 99/100 | 91/95 | 91/96 | 92/96 |
| CR7 (89) | orf7 (89) | L0048 (89) | escS | Type III secretion | 100/100 | 97/98 | 97/98 | 97/99 |
| CR8 (258) | orf8 (258) | L0047 (258) | escT | Type III secretion | 99/100 | 92/97 | 91/97 | 93/97 |
| CR9 (345) | orf9 (345) | L0046 (345) | escU | Type III secretion | 99/100 | 90/96 | 90/96 | 91/97 |
| rCR1 (153) | rorf3 (152) | L0045 (152) | Unknown | 98/99 | 88/93 | 90/95 | 91/96 | |
| CR10 (112) | orf10 (123) | L0044 (123) | Unknown | 98/100 | 88/96 | 88/96 | 92/96 | |
| CR11 (135) | orf11 (137) | L0043 (137) | Unknown | 99/100 | 91/96 | 91/96 | 91/96 | |
| rCR2 (151) | rorf4 (151) | L0042 (151) | cesD | Chaperone for EspD | 100/100 | 93/98 | 93/98 | 95/98 |
| rCR3 (512) | rorf5 (512) | L0041 (512) | escC | Type III secretion | 99/100 | 95/97 | 96/98 | 95/98 |
| rCR4 (151) | rorf6 (151) | L0040 (151) | sepD? | Type III secretion | 98/99 | 88/93 | 87/91 | 85/93 |
| rCR5 (190) | rorf7 (190) | L0039 (190) | escJ | Type III secretion | 100/100 | 95/97 | 95/97 | 95/97 |
| rCR6 (122) | rorf8 (142) | L0038 (142) | Type III secretion? | 93/94 | 79/88 | 77/87 | 79/89 | |
| rCR7 (100) | rorf9 (98) | L0037 (99) | sepZ | Type III secretion | 61/78 | 69/78 | 65/77 | 67/76 |
| CR12 (117) | orf12 (117) | L0036 (117) | Type III secretion? | 100/100 | 92/94 | 92/94 | 92/95 | |
| CR13 (675) | orf13 (675) | L0035 (675) | escV | Type III secretion | 99/99 | 94/98 | 94/98 | 95/98 |
| CR14 (446) | orf14 (446) | L0034 (446) | escN | Type III secretion | 99/100 | 93/96 | 94/97 | 95/97 |
| CR15 (125) | orf15 (125) | L0033 (125) | Type III secretion? | 100/100 | 86/95 | 86/95 | 86/94 | |
| CR16 (103) | orf16 (138) | L0032 (91) | Type III secretion? | 97/99 | 77/91 | 78/91 | 78/85 | |
| CR17 (305) | orf17 (305) | L0031 (305) | sepQ | Type III secretion | 93/97 | 82/90 | 82/90 | 80/87 |
| CR18 (179) | orf18 (176) | L0030 (168) | Type III secretion? | 75/83 | 77/87 | 68/78 | 67/80 | |
| rCR8 (120) | rorf10 (120) | L0029 (127) | Unknown | 79/86 | 86/92 | 78/88 | 77/91 | |
| CR19 (203) | orf19 (203) | L0028 (203) | map | Secreted protein | 89/93 | 79/90 | 72/85 | 77/88 |
| CR20 (547) | orf20 (550) | L0027 (558) | tir | Intimin receptor | 60/70 | 79/86 | 58/71 | 68/79 |
| CR21 (156) | orf21 (156) | L0026 (156) | cesT | Chaperone for Tir | 97/99 | 95/97 | 94/97 | 96/97 |
| CR22 (936) | orf22 (939) | L0025 (934) | eae | Intimin, outer membrane adhesin | 83/89 | 79/88 | 78/86 | 86/92 |
| rCR9 (406) | rorf11 (406) | L0024 (406) | escD | Type III secretion | 98/99 | 91/95 | 93/96 | 93/96 |
| CR23 (351) | orf23 (351) | L0023 (351) | sepL | Type III secretion | 94/99 | 91/97 | 90/96 | 91/96 |
| CR24 (192) | orf24 (192) | L0022 (192) | espA | Secreted protein | 81/91 | 79/90 | 78/91 | 83/92 |
| CR25 (380) | orf25 (380) | L0021 (374) | espD | Secreted protein | 74/84 | 85/91 | 70/81 | 85/92 |
| CR26 (321) | orf26 (321) | L0020 (312) | espB | Secreted protein | 61/77 | 85/92 | 62/77 | 66/80 |
| CR27 (135) | orf27 (135) | L0019 (135) | Unknown | 99/99 | 90/93 | 90/93 | 90/93 | |
| CR28 (73) | orf28 (73) | L0018 (73) | escF | Type III secretion | 100/100 | 60/89 | 60/89 | 60/89 |
| CR29 (98) | orf29 (92) | L0017 (92) | Unknown | 95/97 | 83/93 | 86/93 | 86/93 | |
| CR30 (301) | orf30 (206) | L0016 (248) | espF | Secreted protein | 88/92 | 67/79 | 65/76 | 68/80 |
| rCR10 (272) | rorf1 (272) | L0056 (272) | Unknown | 99/99 | 84/92 | 84/91 | 82/90 | |
| rCR11 (398) | rorf2 (398) | L0055 (398) | espG | Secreted protein | 98/98 | 74/85 | 74/85 | 76/87 |
aa, amino acids.
CR, C. rodentium.
Southern blot analysis of C. rodentium plasmids.
Plasmid purification from C. rodentium was done using the Qiagen Midi Plasmid Purification Kit according to the Qiagen Plasmid Purification Handbook. Plasmid preparations were separated in a 0.7% agarose gel, stained with ethidium bromide before being photographed, depurinated in 250 mM HCl, and transferred to nitrocellulose membrane by standard Southern blotting procedure. The blot was hybridized to an agarose gel-purified escC gene probe from C. rodentium, using the digoxigenin nonradioactive nucleic acid labeling and detection system from Boehringer Mannheim and the Digoxigenin System User's Guide for Filter Hybridization. The escC gene was cloned into pCRII-TOPO after PCR amplification with EscC-F and EscC-R primers to create pCRII-cscC. Plasmid pCRII-cscC was digested with EcoRI, and the escC fragment was gel purified and used as a probe.
Cloning and sequencing of pCRP3, the 3-kb plasmid in C. rodentium.
Plasmid DNA was isolated from C. rodentium using the Qiagen Miniprep Kit. The 3-kb plasmid was gel purified, digested with either EcoRV or HindIII, cloned into pBluescript II SK(+) (Stratagene), and sequenced using M13 −20 and reverse primers and primer walking.
Nucleotide sequence accession number.
The DNA sequences reported in this paper have been submitted to GenBank. The GenBank accession numbers for C. rodentium LEE and the plasmid pCRP3 are AF311901 and AF311902, respectively.
RESULTS AND DISCUSSION
C. rodentium LEE has a different linear gene order for the rorf1 and rorf2/espG genes from that of EPEC, EHEC, and RDEC-1.
Similar to EPEC and EHEC, C. rodentium secretes several proteins (Esps) into minimal culture medium and translocates some of these proteins into host cells. These proteins include EspA, EspB, and EspD (19, 42, 52). We were interested in studying Tir and the Esp proteins and their roles in C. rodentium pathogenesis towards the goal of using C. rodentium mouse infection as an animal model for studying EPEC and EHEC infection of humans. We first cloned and sequenced the LEE region between eae and espB genes in C. rodentium. When we tried to clone the espF gene from C. rodentium by inverse PCR, we found that rorf1 and rorf2/espG were located downstream of espF (Fig. 1). This was unexpected, because in the LEE of EPEC, EHEC, and RDEC-1, there is absolute conservation of gene order and gene number (with RDEC-1 LEE missing only the orf3), and the rorf1 and rorf2/espG genes are located upstream of the ler gene at the opposite end of the LEE (17, 45, 61). To rule out any cloning/sequencing artifacts, PCR analyses were carried out. Primers Binv-1(in espB) and R2-inv1 (in rorf2/espG) amplified a 3.9-kb fragment, and another pair of primers, Finv-1 (in escF) and Cr1–1 (in rorf1), produced a 2.3-kb DNA band in C. rodentium (data not shown), verifying our DNA sequencing result. As expected, the same pairs of primers did not amplify any product from EPEC and EHEC (data not shown). This result suggests that C. rodentium LEE was inserted differently into its bacterial genome and thus represents an evolutionary lineage different from that of EPEC, EHEC, and RDEC-1 LEEs. DNA sequencing of the region between eae and espB genes in C. rodentium revealed two more differences between C. rodentium LEE and the other three LEEs. One is the presence of an insertion sequence (IS) between eae and escD genes in C. rodentium, and the other is that C. rodentium espF has more repeats than its counterparts in other A/E pathogens (see below). These additional differences as well as the unique location of the rorf2/espG and rorf1 operon prompted us to clone and sequence the entire LEE region of C. rodentium.
Sequence overview of C. rodentium LEE.
The nucleotide sequence submitted to GenBank (accession number AF311901) spans 42,001 bp, covering the core region of the C. rodentium LEE and about 3 kb of non-LEE flanking sequences on either side. Based on its homology to the LEE sequences from both EPEC and EHEC (17, 45), C. rodentium LEE is defined as the region from bp 2842 to bp 38967, a total of 36,126 bp. It is slightly larger than that of EPEC and EHEC LEEs, owing to the presence of an IS and some remnants of IS elements (see below). Like most pathogenicity islands and the EPEC and EHEC LEE, C. rodentium LEE has a lower-than-average GC content compared to the E. coli K12 strain average of 50.80% (22). The GC content for the full-length 42,001 bp is 40.07%. The GC content for the core region of C. rodentium LEE from bp 2842 to bp 38967 is even lower at 38.05%, whereas those for the left- and right-side flanking regions are 54.77 and 50.26%, respectively. The GC content of C. rodentium LEE (38.05%) is very similar to those for the LEE from EPEC (38.36%), EHEC (39.59%), and RDEC-1 (41.3%) (17, 45, 61).
Corrections to previous GenBank entries of C. rodentium genes reveal that C. rodentium and MPEC encode identical intimin and Tir.
Only four genes of C. rodentium strain DBS100 LEE have been previously sequenced, eae and orfU/cesT (GenBank accession no. L11691 [48, 49]), espB (GenBank accession no. AF177537 [42]), and tir (GenBank accession nos. L11691, AF301617, and AF301618 [32]). More recently, the eae and tir genes of MPEC have been sequenced (GenBank accession nos. AB040740 and AB026719). MPEC has been shown to be a misclassified C. rodentium strain, and it is clonal to C. rodentium in evolution (32). However, an alignment of the Tir and intimin sequences of C. rodentium DBS100 and MPEC shows some differences, especially for intimin (32; data not shown). We therefore compared our LEE sequence to these preexisting GenBank entries for C. rodentium and MPEC and found some interesting differences. Whenever a discrepancy was found between our sequence and the GenBank entries, an independent PCR was performed to clone the specific region, and at least two independent PCR clones were sequenced to verify our DNA sequences.
Two differences were found between our espB and the espB entry AF177537, resulting in changes in the predicted protein sequence. Our sequence at codon 204 is GAG (Glu) instead of AAG (Lys) and at codon 303 is GCT (Ala) instead of CCT (Pro). Luperchio et al. (32) found that C. rodentium Tir and MPEC Tir show only a single-amino-acid difference at residue 149 (Thr versus Asn). However, our Tir sequence of C. rodentium DBS100, the same strain that they used, has Asn at the 149 position, suggesting that there might be a sequence error in the tir sequence by Luperchio et al. (32) and that MPEC and C. rodentium have identical Tir. For the eae gene, there are eight missense changes between C. rodentium entry L11691 and MPEC entry AB040740, at codons 500 to 505, 526, and 745. In L11691, from nucleotide 2365 to 2394 (eae codons 500 to 505), the sequence is GAA CGC AAA GCG CAC AAC (GluArgLysAlaHisAsn). Our sequence for the same region is GGA AGC CAA AGC GCA CAA (GlySerGlnSerAlaGln). The missing of a G at position 2366 and the addition of a C at position 2394 have created a frameshift in the stretch in L11691 eae. Our eae gene sequence is identical to the MPEC sequence (AB026719) for the codons 500 to 505 and 526 (Ala versus Arg in L11691) but agrees with L11691 at codon 745 (Val versus Gly in AB026719). When all the sequence errors are removed, MPEC and C. rodentium now encode identical Tir and intimin and resemble each other even more closely than previously thought (32). Our results confirmed that C. rodentium and MPEC are indeed clonal and remain the only known A/E pathogen in mice.
Sequence comparison of C. rodentium LEE with that of EPEC, EHEC, and RDEC-1.
A gene map for C. rodentium LEE is presented in Fig. 1. To facilitate sequence comparison, the gene map for EPEC LEE is also included in the figure, and the presentation and designation of C. rodentium LEE genes followed that of EPEC E2348/69 LEE (GenBank accession no. AF022236 [17]). It should be noted that the other two fully sequenced LEE elements of EHEC EDL933 (GenBank accession no. AF071034) and RDEC-1 (GenBank accession no. AF200363) are very similar in size and gene number and order to EPEC LEE (45, 61).
Size of LEE.
The core region of C. rodentium LEE is slightly longer than 36 kb, whereas the LEE of EPEC, EHEC, and RDEC-1 spans about 34 kb (17, 45, 61). The more-than-2-kb difference can be accounted for by a larger intergenic region between orf11 and cesD and the presence of an IS in the intergenic region between eae and escD in C. rodentium. The intergenic region between orf11 and cesD in EPEC and EHEC is 382 bp, while that of C. rodentium is 1,462 bp, of which three ORFs longer than 50 amino acid residues can be deduced. These ORFs show similarities to a hypothetical protein YagA (GenBank accession nos. P37007 and D83536) encoded by the intergenic region of thrW-argF in E. coli (58) and most likely represent remnants of IS or transposable elements. The IS located between eae and escD genes in C. rodentium is 1,136 bp long, has 29-bp terminal inverted repeats, and generates 8-bp duplications at the target site. This IS element shows 83% identity to IS285 on plasmid pMT-1 of Yersinia pestis (GenBank accession no. AF053947). Because of two deletions in the C. rodentium IS element compared to IS285, which is 1,321 bp long, the C. rodentium IS is probably no longer self-mobile. Downstream of the eae gene and immediately before the IS element, there is a succession of eight direct repeats of the sequence CCCGGCA, possibly a result of the insertion event of the IS element. The GC content is 44.70% for the orf11/cesD intergenic region and 47.18% for the IS285-like element. They are higher than that of the LEE (38.05%) but lower than that of the LEE flanking regions (>50%), suggesting that these regions might have been acquired along with the rest of the LEE, although it is also possible that the IS285-like element transposed to the location after acquisition of the LEE. Southern blot analysis indicated that sequences homologous to the IS285-like element located in C. rodentium LEE were present in the genomes of both C. rodentium and EPEC E2348/69 (data not shown).
Another structural difference in the C. rodentium LEE is the lack of the enterobacterial repeat intergenic consensus (ERIC) element found in the upstream region of the ler gene promoter in EPEC and EHEC LEE (17, 45). Interestingly, the RDEC-1 LEE does not have the ERIC element at the location either (61), suggesting that the ERIC element might be a new addition to the EPEC and EHEC LEE. Although the exact function for the widely distributed ERIC family of repeated elements is not known, it has been implicated in gene regulation (27). Since Ler controls LEE gene expression, the absence of the ERIC element in the C. rodentium ler promoter region may mean that C. rodentium LEE gene expression is regulated differently from that of EPEC and EHEC LEE. It is worth noting that the promoter region of EPEC and EHEC ler is more than 98% identical (45), whereas only 194 bp of the nucleotide sequence upstream of C. rodentium ler start codon can be aligned with that of EPEC ler promoter, and that the homology level between the two is less than 79%. Furthermore, the ler (LEE1) operon and the rorf2/rorf1 operon in EPEC and EHEC LEE are transcribed in opposite directions and share the same intergenic region, but in C. rodentium the two operons are located at two different ends of the LEE (see below). The homology level between the promoter region of C. rodentium rorf2/rorf1 operon and that of EPEC and EHEC only extends to about 50 bp upstream of the start codon and drops off sharply after that, implying regulatory differences in rorf2/rorf1 gene expression between C. rodentium and EPEC or EHEC.
Gene organization of LEE.
The number of predicted genes and their linear orientation and order are absolutely conserved for the LEE from EPEC, EHEC, and RDEC-1, with the exception that the orf3 gene of RDEC-1 LEE lacks an obvious start codon (17, 45, 61). As shown in Fig. 1, although C. rodentium shares all the 41 ORFs predicted for the LEE, the location of two of its ORFs, rorf1 and rorf2, is completely different from that for the other LEEs. Instead of being at the beginning (leftmost) of the LEE like in EPEC, EHEC, and RDEC-1, rorf1 and rorf2 in C. rodentium are located downstream of the espF gene to end the LEE at the right side. The other end of C. rodentium LEE is the ler gene, encoding the regulator for LEE expression. Since rorf1 and rorf2 are located so differently in C. rodentium and the E. coli A/E strains, it raises the possibility that these two genes were acquired separately from and later than the rest of the LEE. If that were the case, they may show a different GC content. However, a calculation of the GC content of the C. rodentium rorf1 and rorf2 region showed 38.39%, very similar to that of the rest of the C. rodentium LEE (38.05%). The GC content for EPEC rorf1 and rorf2 region is 37.16%, also similar to the average of 38.36% of the EPEC LEE.
ORFs of LEE.
As shown in Table 3 and Fig. 1, C. rodentium LEE shares all 41 ORFs with EPEC and EHEC LEE. Detailed description of these ORFs is provided in Table 3 and can be found in references 17 and 45. For most of the ORFs, the similarity between EPEC and EHEC is much greater than that between C. rodentium and either EPEC or EHEC. Most of the proteins implicated in the type III secretion system (Esc, Sep, and Ces) are more than 98% homologous, if not identical, between EPEC and EHEC, suggesting the need for strong conservation of function. However, even for these highly conserved proteins, the homology between C. rodentium and EPEC or EHEC rarely surpasses 95% (Table 3). On the other hand, RDEC-1 has more than half of the LEE proteins showing more than 90% identity to those of both EPEC and EHEC, although EPEC and EHEC are somewhat more similar to each other than to RDEC-1 in most cases (61). As the ORFs of C. rodentium LEE show almost identical degrees of divergence from those of EPEC, EHEC, and RDEC-1 LEEs (Table 3), this suggests that the LEEs of E. coli A/E pathogens (EPEC, EHEC, and RDEC-1) are more closely related to each other than to that of C. rodentium and form one lineage, while C. rodentium LEE forms another.
In contrast to the strong conservation of proteins involved in the type III secretion, most of the rest of the ORFs encoded by the LEE of EPEC, EHEC, and RDEC-1, including SepZ, Orf18, rOrf10, Tir, intimin, EspA, EspD, EspB, and EspF, show differences greater than expected for clonal divergence among E. coli strains (17, 61). Many of these highly divergent proteins (Tir and EspB, -D, and -F) are translocated into the host cell by the bacteria and involved in interactions with the host (14, 19). C. rodentium LEE continues this trend, showing considerable divergence in most of these ORFs compared to EPEC, EHEC, and RDEC-1 (Table 3). Often in these cases, the degree of similarity of EPEC versus EHEC, C. rodentium versus EPEC or EHEC, and C. rodentium versus RDEC-1 is comparable and the evolutionary lineage is obscure. In general, with the type III secretion system included, the similarity between C. rodentium and EPEC is about the same as that between C. rodentium and EHEC or RDEC-1. The only obvious exceptions are Tir, EspB, and EspD, which show considerably higher identity between C. rodentium and EPEC (79 to 85%) than between C. rodentium and EHEC (58 to 70%) (32) (Table 3). The significance of this observation is not clear. It is interesting that Tir translocation requires EspB and EspD, both translocated proteins themselves (11, 19, 30). It is possible that there is a structural constraint in the evolution of Tir, EspB, and EspD for each A/E pathogen.
Among the more divergent ORFs between C. rodentium and EPEC, EHEC, or RDEC-1, some of the C. rodentium proteins displayed very interesting features. One of these proteins is EspF, a translocated effector protein recently shown to be involved in altering transepithelial electrical resistance and epithelial barrier function as well as mediating host cell apoptosis (9, 14, 39). EspF carries proline-rich repeats that are similar to SH3-binding domains and when translocated may interact directly with host proteins to exert its effects (38). The size of EspF varies greatly for different A/E pathogens. EHEC EDL933 has 248 amino acid residues, whereas EPEC E2348/69 has 206 and RDEC-1 only has 160 (17, 45, 61). C. rodentium EspF is the largest among the group, at 301 amino acid residues. The size difference in EspF is largely the result of the numbers of a 47-amino-acid repeat. RDEC-1 has two repeats, EPEC three, EHEC four, and C. rodentium five (Fig. 2). While the A/E E. coli strains (EPEC, EHEC, and RDEC-1) show 80% or greater identity and share an absolutely conserved proline-rich APPPPT motif (61), C. rodentium EspF is considerably more divergent, only 65% identical to that of EHEC and 67% to that of EPEC, and its proline-rich motif also varies, being either APSPPT, APQSPT, or APQPPT (Fig. 2). It will be interesting to see whether the number and type of these repeats play a host-specific role in the various diseases incurred by the different bacteria or are simply a result of duplication or deletion by a slippage mechanism during DNA replication.
FIG. 2.
Alignment of EspF proteins from C. rodentium, EHEC O157:H7 strain EDL933, EPEC O127:H6 strain E2348/69, and rabbit diarrheagenic E. coli strain RDEC-1 (O15:H−). Identical amino acids are indicated by dots, while dashes show absent amino acids. Note the different numbers of the 47 amino acid repeats and the different sizes of the proteins.
Other intriguing differences for ORFs of C. rodentium LEE are found in SepZ, EscF, Orf16, rOrf1, and rOrf2/EspG (Table 3). SepZ is considered to be part of the type III secretion system (17). While the rest of the type III proteins show extreme conservation, SepZ shows less than 70% identity between EPEC, EHEC, RDEC-1, and C. rodentium and is one of the most divergent proteins encoded by the LEE, suggesting a possible role in determining the specificity of the type III secretion. EscF, a protein also hypothesized to be involved in the type III secretion, is extremely conserved and is identical in EPEC, EHEC, and RDEC-1. The homolog of EscF in Yersinia enterocolitica has recently been shown to form the needlelike structure of the type III secretion apparatus (26). Surprisingly, C. rodentium EscF is only 60% homologous to that of either EPEC, EHEC, or RDEC-1, although it has the same size (Table 3). Orf16 is encoded in the same operon as some highly conserved proteins constituting the type III secretion system (EscV, EscN, and SepQ). Orf16 varies in size for EPEC (138 amino acids), EHEC (91 amino acids), and C. rodentium (103 amino acids), due to the use of different start codons. Despite their size difference, Orf16 of EPEC and EHEC is 96.7% identical. However, Orf16 of C. rodentium and of RDEC-1 is quite divergent from that of EPEC and EHEC (61; Table 3). The same pattern is also true for rOrf1 and rOrf2/EspG. While EPEC and EHEC encode nearly identical proteins, RDEC-1 and C. rodentium show high divergence, especially for rOrf2/EspG. Sequence comparison indicates that rOrf1 is similar to an outer membrane protein encoded by a gene on the Salmonella enterica serovar Typhimurium virulence plasmid (17, 23). This gene is located immediately upstream of the rck gene, which is required for both serum resistance and cell invasion (8). It has been recently shown that rOrf2/EspG is a translocated protein (15), suggesting that it may be an effector. The divergence seen in rOrf1 and rOrf2/EspG is consistent with the degree of variation existing among the proteins translocated into the host cell (EspB, EspD, and EspF and Tir) or interacting with the host (intimin). Similar degrees of divergence between C. rodentium and EPEC, EHEC, or RDEC-1 were also seen for another LEE-encoded translocated protein, Orf19/Map, which has recently been shown to be targeted to host cell mitochondria (31).
These variations in the LEE-encoded proteins underline the mosaic structure and plasticity of the LEE and also raise questions about the significance of this divergence in bacterial pathogenesis. It has been previously thought that the divergence in intimin, Tir, and the Esp proteins seen between EPEC E2348/69 and EHEC EDL933 LEE, which far exceeds the clonal divergence observed for E . coli strains, is the result of natural selection for adaptation to host and tissue specificity or evasion of the host immune response, since these proteins directly interact with the host (45). However, recent sequencing of the RDEC-1 LEE has revealed that rabbit RDEC-1's intimin, EspA, EspD, and EspB are identical or nearly identical to those in rabbit EPEC O103, human EPEC O111, human EHEC O26, and a pig EPEC O45 strain (61). It has therefore been suggested that the variations seen in the host-interacting proteins are largely a function of evolutionary lineage rather than adaptation to a specific host, since A/E E. coli strains with closely related intimin and Esps can colonize and cause disease in diverse hosts, such as humans, rabbits, dogs, and pigs (61).
Indeed, this seems to agree with the LEE lineages predicted by intimin typing (2, 3, 14, 37, 44). Based on the protein sequences, PCR analysis and serotyping, intimin from different A/E pathogens is classified into at least five groups, intimin α of EPEC E2348/69 (O127) and other EPEC1 (O55 and O142) strains; intimin β, which is a large group having diverse hosts and includes human EPEC2 (O111 and O128), human EHEC2 (O26 and O111), rabbit REPEC (O103) and RDEC-1, and C. rodentium; intimin γ of EHEC1 (O157); intimin δ of EPEC1 O86 and O49 serotypes; and intimin ɛ of EHEC O8, O11, O45, O103, O121, and O165 (2, 44). In this scheme, C. rodentium intimin and RDEC-1 intimin are in the same β group. However, while RDEC-1 has identical or nearly identical intimin to that of human EPEC2 O111, human EHEC2 O26, rabbit REPEC O103, and pig O45 in the same group (61), C. rodentium intimin shows 14 to 16% divergence from these β intimins. This degree of difference is very comparable to that (17%) seen between intimins α (EPEC1) and γ (EHEC1) and that (21 to 22%) between C. rodentium and EPEC1 or EHEC1 (Table 3). This suggests that while it may be true that C. rodentium has a β-type intimin, C. rodentium LEE belongs to a lineage quite different from that of EPEC2, EHEC2, REPEC, and RDEC-1, in agreement with the phylogenetic tree based on intimin typing in which C. rodentium forms a separate branch by itself in the β-intimin group (44). This is further supported by the fact that C. rodentium also possesses quite divergent Esps and has a different LEE gene order (Fig. 1) and a different LEE insertion site (see next section).
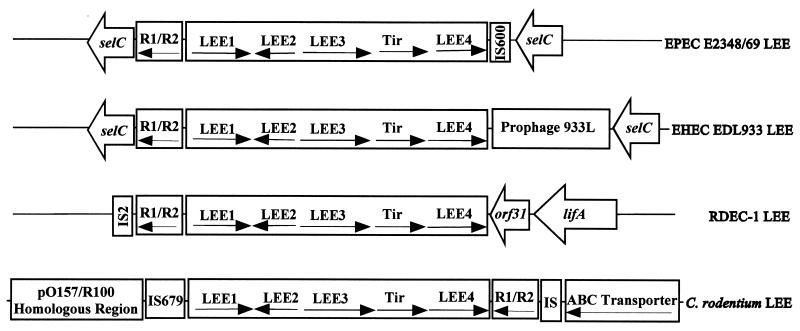
Insertion site of LEE.
Many tRNA genes are frequently used as integration sites for pathogenicity islands, and the LEE has been found to be associated with at least three tRNA loci (22). The LEE from both EPEC E2348/69 and EHEC EDL933 as well as several EPEC1 and EHEC1 serovars is inserted at the selC tRNA gene (35, 45, 59). On the other hand, the LEE from some A/E E. coli isolates of different evolutionary lineages has been shown to be inserted into the pheU tRNA locus, while others have their LEE inserted into a third, yet-to-be-described, site (51, 57, 59). The newly sequenced RDEC-1 LEE is not inserted into either selC or pheU, but its exact insertion site has not been determined (61). The RDEC-1 LEE is flanked by an IS2 element and the lifA toxin gene, but the lifA gene may have inserted next to the RDEC-1 LEE after the acquisition of the LEE, similar to the prophage 933L located next to the LEE in EHEC EDL933 (45, 61). It is interesting that IS and remnants of mobile elements are frequently found at the ends of the LEE (17, 51, 61), suggesting a role for these elements in the LEE dissemination. Although it is believed that the LEE has been acquired at multiple times and inserted into several sites during the evolution of pathogenic A/E E. coli strains (14, 46), the mechanism for its acquisition and dissemination is not clear at all.
As shown in Fig. 3 and by GenBank accession no. AF311901, C. rodentium LEE is most likely inserted into a chromosomal location different from those used by other LEEs identified so far. At the right flank of the C. rodentium LEE, upstream of the rorf2/rorf1 operon, there are three 41- and 40-bp direct repeats and then remnants of several IS elements belonging to the IS600, IS3, and IS30 families. This is then followed by DNA sequences forming an operon encoding a type I/ABC transport system. This operon shows similarity to the leukotoxin secretion system from Pasteurella haemolytica and to both the chromosomal and plasmid hemolysin secretion systems of E. coli, as well as to various ABC transport systems from Pseudomonas spp., Vibrio cholerae, Rickettsia prowazekii, Campylobacter fetus, and S. enterica serovar Typhimurium. However, the substrate for this transport system does not resemble leukotoxins or hemolysins. Rather, it is similar to extracellular matrix binding proteins from gram-positive bacteria from Streptococcus spp. and Staphylococcus spp. (data not shown). We are presently investigating whether this ABC transport system is required for C. rodentium virulence.
FIG. 3.
Insertion sites of the LEE in the chromosomes of C. rodentium, EPEC, EHEC, and RDEC-1. The major operons and their transcriptional directions in the LEE are shown. Note the unique gene organization of rorf2 (espG) and rorf1 genes in C. rodentium LEE. The diagram is not drawn to scale.
At the other end of the C. rodentium LEE, located upstream of the ler gene is the 2.7-kb IS679, an IS element also found in the adherence factor plasmid of EPEC (55). The IS679 is followed immediately by sequences strongly homologous (95% identical at nucleotide level) to plasmid R100 of Shigella flexneri and pO157 of EHEC O157:H7 (7, 33). This homology extends at least for the 6-kb DNA that we have cloned and sequenced so far (data not shown). This homologous region encodes proteins important for plasmid replication and maintenance in plasmids R100 and pO157 (GenBank accession nos. AP000342 for R100 and AF074613 and AB011549 for pO157). This is the first time that plasmid-like sequences have been identified at the flanks of the LEE, although short sequences homologous to plasmids have been found in the pathogenicity islands of uropathogenic E. coli strains J96 and 536 (6, 53). Pathogenicity islands have been found inserted into chromosomes as well as carried on large virulence plasmids (22). It is possible that C. rodentium LEE originally resided on a plasmid before it was acquired and inserted into the C. rodentium genome next to the ABC transport system operon (Fig. 3) and that the plasmid sequences have yet to be completely lost during evolution. The integration and excision of plasmids into and from chromosomes have been described for many bacteria (21).
The LEE is not located in two large plasmids present in C. rodentium.
Due to the lack of genome sequence information for C. rodentium, we cannot be completely sure that the ABC transport system operon located next to the LEE is actually part of the Citrobacter chromosome. The presence of sequences highly homologous to plasmids next to the LEE region as well as the unique location of the rorf2 and rorf1 genes in C. rodentium LEE raises the possibility that the C. rodentium LEE could still be residing on a plasmid and has yet to be integrated into the chromosome.
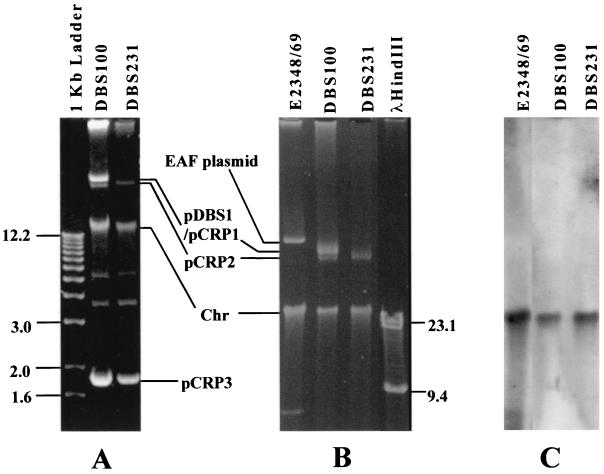
Previously, Schauer and Falkow (49) showed that there are two plasmids in C. rodentium, one 65 kb (pDBS1) long and the other 3 kb. The 65-kb plasmid is smaller than the virulence plasmids of human EPEC adherence factor (EAF) plasmid pB171 (69 kb [55]) and EHEC pO157 (92 kb [7, 33]), and curing the 65-kb plasmid does not affect C. rodentium virulence in mice (49). In light of our finding of sequences homologous to plasmids R100 and pO157 located next to the LEE, we reevaluated the plasmid profile of C. rodentium to see whether there were any other large plasmids. We used the Plasmid Midi Purification Kit, which is capable of purifying plasmids larger than 150 kb (Qiagen). As a control, we used EPEC strain E2348/69, which carries the large EAF plasmid. As shown in Fig. 4A, C. rodentium has at least three plasmids, the 65-kb pDBS1 and the 3-kb plasmid, as reported by Schauer and Falkow (49), as well as a 60-kb plasmid which is present in both the wild-type C. rodentium DBS100 and the 65-kb-plasmid-cured strain DBS231. The reason why Schauer and Falkow (49) did not detect the plasmid could be due to its relatively low copy number and similar size to that of pDBS1. We named the 60- and 3-kb plasmids pCRP2 and pCRP3 (for C. rodentium plasmid), respectively. We propose to rename pDBS1 as pCRP1 for consistency, since pDBS2 and pDBS3 have been used by Schauer and Falkow (49) to name other recombinant plasmids. No other large plasmid was found in C. rodentium by our method.
FIG. 4.
C. rodentium carries at least three plasmids, and the LEE is not located on these plasmids. (A) Ethidium bromide-stained 0.7% agarose gel, with 1-kb DNA ladder from GIBCO BRL as molecular marker (in kilobase pairs); (B) ethidium bromide-stained 0.7% agarose gel, with λHindIII as standard; (C) Southern blot hybridization of gel B using the C. rodentium escC gene as a probe. Locations of the three plasmids, pCRP1/pDBS1, pCRP2, and pCRP3, in C. rodentium and the EAF plasmid are indicated. DBS100, wild-type C. rodentium strain; DBS231, C. rodentium strain cured of the plasmid pDBS1/pCRP1; E2348/69, EPEC O127:H6 strain E2348/69. Chr stands for sheared chromosomal DNA band. Sizes are given in kilobases.
Southern blot analysis using a DNA probe of the C. rodentium escC gene, which is highly conserved between C. rodentium and EPEC, showed that Citrobacter LEE is not located in the 60- and 65-kb plasmids. The probe did not hybridize to the EAF plasmid in EPEC either. Rather, the probe hybridized to the sheared chromosomal DNA band in both C. rodentium and EPEC (Fig. 4B and C), suggesting that C. rodentium LEE is located in the bacterial chromosome, similar to that of EPEC and EHEC. However, it is still possible that C. rodentium LEE is located on a low-copy-number, extremely large plasmid that cannot be detected with our methods.
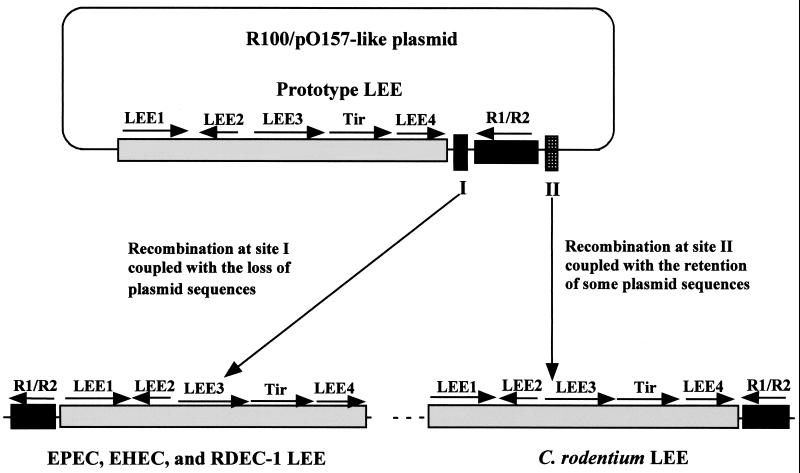
A model for LEE acquisition and dissemination by A/E pathogens.
Based on our characterization of C. rodentium LEE and its comparison to the sequenced LEEs from EPEC, EHEC, and RDEC-1, we propose that before it was introduced into the genome of various A/E pathogens, the prototype LEE was originally on a plasmid similar to plasmid R100 of Shigella spp. and pO157 of EHEC. And the LEE has since been acquired by A/E pathogens several times during evolution (14), possibly via the horizontal transfer of this putative plasmid. Bacteriophages and plasmids are known to transfer pathogenicity islands, and they frequently use tRNA genes as integration sites (22). The origin of the LEE on a plasmid can readily explain why even among E. coli A/E strains, there are at least three different insertion sites (61). The integration of the LEE plasmid into different tRNA loci, followed by the deletion of plasmid sequences unrelated to the A/E phenotype, could theoretically have led to the different lineages of the LEE that are presently seen among the A/E E. coli strains (14).
We propose that in the case of C. rodentium, the putative LEE plasmid was inserted next to the ABC transport system operon (Fig. 3). It is possible that the acquisition of C. rodentium LEE occurred later than that of EPEC and EHEC and that evolution has yet to remove all the plasmid sequences. An interesting complication for this hypothesis is the different location of rorf1 and rorf2 genes in C. rodentium, compared to that in EPEC, EHEC, and RDEC-1 (Fig. 1 and 3). We propose that the prototype LEE had the same gene organization as the C. rodentium LEE, so that the rorf2/espG and rorf1 operon would be located next to the operon LEE4 encoding other secreted proteins. We further hypothesize that the recombination and integration of the putative LEE plasmid into bacterial chromosome can occur at at least two different sites, I and II (Fig. 5). Recombination at site I in the region between espF and rorf1, coupled with subsequent removal of the plasmid sequence during evolution, would generate the gene pattern seen in the LEE of A/E-inducing E. coli strains such as EPEC, EHEC, and RDEC-1, while recombination at site II in the upstream region before the rorf2/1 operon would result in the gene organization in C. rodentium LEE (Fig. 5). The recombination could be mediated by IS elements, repeated sequences, and even short stretches of homologous regions (21). Indeed, bacteriophages and IS elements or their remnants have been found at the flanks of the LEE and other pathogenicity islands from various strains (17, 22, 45, 51, 61), although some of these IS elements and phages may have inserted next to the LEE after its acquisition (45).
FIG. 5.
A schematic model to explain the different gene organization seen for the rorf1 and rorf2/espG genes in the C. rodentium LEE versus that of EPEC, EHEC, and RDEC-1. The model is based on the hypothesis that the prototype LEE was located on an R100- and pO157-like plasmid. When it was disseminated and introduced into different A/E pathogens during evolution, the insertion of the plasmid into the bacterial chromosome can occur at at least two sites, I and II. These different recombination events, coupled with the retention or loss of the plasmid sequences (dashed line for C. rodentium LEE), resulted in the different gene orders for rorf1 and rorf2 (espG) genes and the different LEE lineages observed for EPEC, EHEC, RDEC-1, and C. rodentium, as well as other A/E pathogens. The major operons encoded by the LEE (LEE1, -2, -3, and -4, Tir, and R1/R2) and their transcriptional directions are shown and adapted from references 16 and 40.
Our model implies that plasmid-related sequences used to reside in the intergenic region between ler and rorf2/espG genes in EPEC, EHEC, and RDEC-1, but database searches did not reveal any obvious homology between the intergenic region and any plasmid sequences. However, this intergenic region, at 1,251 bp in EPEC and 1,244 bp in EHEC, is considerably larger than most other intergenic regions in the LEE. Other than the immediate promoter regions for the rorf2 and ler genes, the majority of the EPEC and EHEC ler-rorf2 intergenic region shows no similarity at all to any C. rodentium LEE sequence. The ler-rorf2 intergenic region of RDEC-1 is only 1,139 bp. Although it aligns well with that of EPEC and EHEC in the promoter regions of the ler and rorf2 genes, its middle portion shows considerable divergence. Interestingly, located in this intergenic region in EPEC and EHEC LEE is a copy of the ERIC element, which is absent from RDEC-1 and C. rodentium LEE (17, 45, 61; this study), suggesting that there is considerable sequence plasticity in the region. It is possible that this region has undergone numerous mutations and recombinations during evolution to resemble any known plasmid sequence. It should be noted here that our model is used to explain the different gene organization and LEE lineages seen in C. rodentium and EPEC, EHEC, and RDEC-1. It does not exclude the possibility of LEE undergoing relocation mediated by recombination events or transposable elements once it is introduced into a particular bacterial host.
Sequence of 3-kb plasmid in C. rodentium reveals recent horizontal plasmid transfer between C. rodentium and EHEC.
The wide dissemination of the LEE among A/E pathogens suggests that the original LEE plasmid was capable of being transferred among different bacterial species. The concept that horizontal plasmid transfer has occurred between A/E pathogens is supported by our sequencing of the 3-kb plasmid pCRP3 in C. rodentium. It has been shown previously that pCRP3 is very similar in size to a small plasmid in EHEC O157:H7 strain EDL932 (49). We cloned and sequenced the 3-kb plasmid pCRP3 from C. rodentium (Fig. 4A) (GenBank accession no. AF311902). It is 3,172 bp in length. Database searches revealed that pCRP3 has exactly the same size as a plasmid named p9705 in EHEC O157:H7 strain 9705 (GenBank accession no. AB040037) and that the two plasmids are more than 99.9% identical at nucleotide level, with only three mismatches throughout the entire sequence. This is very surprising, since C. rodentium and E. coli belong to different bacterial genera and should have diverged a long time ago (32, 47, 50). The near-identity between pCRP3 and p9705 indicates that there has been a recent exchange of genetic materials between C. rodentium and EHEC. Sequence analysis shows that there is also considerable, though less striking, similarity between pCRP3 and the 3,306-bp plasmid of an EHEC O157:H7 isolate from the Sakai outbreak as well as the S. enterica serovar Typhimurium plasmid NTP16 (33). Other than coding for a plasmid mobilization function (MobA), pCRP3/p9705 carries no obvious important virulence genes. It will be interesting to see whether this conserved plasmid plays any role in the pathogenesis of C. rodentium and EHEC. It is possible that pCRP3/p9705 represents just a selfish, self-mobilizable plasmid. Nevertheless, its presence in both C. rodentium and EHEC supports the notion of constant sharing of genetic materials among pathogens.
Concluding remarks: C. rodentium-mouse interaction as small-animal model for studying human A/E pathogens EPEC and EHEC.
In the last 10 years or so, tremendous progress has been made in deciphering the pathogenic mechanisms of human EPEC and EHEC. The discovery of the LEE and the subsequent elucidation of the central role played by intimin, Tir, and the Esps in inducing the A/E lesions have made EPEC and EHEC a model system for studying type III secretion and bacterial pathogenesis (14, 19). However, most of these studies have been conducted in tissue and organ culture models. In three human volunteer studies, the role of intimin, bundle-forming pili, and EspB in EPEC pathogenesis has been verified (5, 12, 54), although differences between in vivo and in vitro models were also revealed. No human volunteer studies for EHEC have been carried out due to its much more severe disease and potential complications compared to EPEC. Animal models, such as rabbits, pigs, and cattle, have also been used to study the function of intimin, Tir, EspA, and EspB in EPEC and EHEC disease (1, 10, 13, 34, 56). However, even this type of study has been limited due to the cost and complexity of these model systems. Therefore, for most of the putative virulence factors identified in A/E pathogens, their importance in disease remains to be validated in a relevant and practical animal model. In addition, host responses to infection by an A/E pathogen have been subjected to limited study for the same reasons.
The full C. rodentium LEE sequence reported in this paper should help us study the function of various putative virulence factors encoded by the LEE in C. rodentium-induced A/E pathology and mucosal hyperplasia. We have now established a method for making deletion mutants in C. rodentium using a suicide vector based on the sacB gene (W. Deng, B. A. Vallance, Y. Li, and B. B. Finlay, unpublished data) and are now mutagenizing various putative virulence genes in the Citrobacter LEE and testing their roles in mouse infection. It should be noted that despite the apparent differences in gene sequences and organization, the similarities between C. rodentium LEE and that of the A/E E. coli pathogens are striking, suggesting that the role of these genes in pathogenesis and disease is conserved. For example, it has been previously shown that the intimin gene from EPEC can complement a C. rodentium eae mutant (18, 24). We have evidence that EHEC tir gene can complement a C. rodentium tir deletion mutant (Deng et al., unpublished data). These data have demonstrated the functional conservation of virulence proteins encoded by the LEE from different A/E pathogens and have validated the relevance of using the C. rodentium-mouse interaction as a model for studying human EPEC and EHEC infections. This conservation of function will allow us to study mouse immune responses to EPEC or EHEC LEE-encoded proteins using recombinant C. rodentium strains. It is our hope that the full LEE sequence and new tools available now in C. rodentium, together with the availability of a wide range of genetic and immunological reagents in mice, will allow us to finally probe the in vivo functions of various LEE-encoded virulence proteins as well as host factors in disease and use the mouse model to evaluate different EPEC and EHEC virulence factors for their suitability as targets for disease intervention and vaccine development.
ACKNOWLEDGMENTS
We are grateful to John Brumell for his critical reading of the manuscript and helpful discussions.
Research in our laboratory is supported by a Howard Hughes International Research Scholar Award and operating grants from the CIHR and ID Biomedical Corporation to B.B.F. W.D. and B.A.V. are both recipients of an MRC postdoctoral fellowship. B.B.F. is a Medical Research Council Scientist.
REFERENCES
- 1.Abe A, Heczko U, Hegele R G, Finlay B B. Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J Exp Med. 1998;188:1–10. doi: 10.1084/jem.188.10.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adu-Bobie J, Frankel G, Bain C, Goncalves A, Trabulsi L, Douce G, Knutton S, Dougan G. Detection of intimins α, β, γ and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 1998;36:662–668. doi: 10.1128/jcm.36.3.662-668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agin T S, Wolf M K. Identification of a family of intimins common to Escherichia coli causing attaching-effacing lesions in rabbits, humans, and swine. Infect Immun. 1997;65:320–326. doi: 10.1128/iai.65.1.320-326.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthold S W, Coleman G L, Jacoby R O, Livestone E M, Jonas A M. Transmissible murine colonic hyperplasia. Vet Pathol. 1978;15:223–236. doi: 10.1177/030098587801500209. [DOI] [PubMed] [Google Scholar]
- 5.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 6.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burland V, Shao Y, Perna N T, Plunkett G, Sofia H J, Blattner F R. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo D M, Heffernan E J, Wu L, Harwood J, Fierer J, Guiney D G. Identification of a domain in Rck, a product of the Salmonella typhimurium virulence plasmid, required for both serum resistance and cell invasion. Infect Immun. 1996;64:2019–2023. doi: 10.1128/iai.64.6.2019-2023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crane J K, McNamara B P, Donnenberg M S. Role of EspF in host cell death induced by enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:197–211. doi: 10.1046/j.1462-5822.2001.00103.x. [DOI] [PubMed] [Google Scholar]
- 10.Dean-Nystrom E, Bosworth B, Moon H, O'Brien A. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun. 1998;66:4560–4563. doi: 10.1128/iai.66.9.4560-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deibel C, Kramer S, Chakraborty T, Ebel F. EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol Microbiol. 1998;28:463–474. doi: 10.1046/j.1365-2958.1998.00798.x. [DOI] [PubMed] [Google Scholar]
- 12.Donnenberg M, Tacket C, James S, Losonsky G, Nataro J, Wasserman S, Kaper J, Levine M. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Investig. 1993;92:1412–1417. doi: 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnenberg M, Tzipori S, McKee M, O'Brien A, Alroy J, Kaper J. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Investig. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnenberg M S, Whittam T S. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J Clin Investig. 2001;107:539–548. doi: 10.1172/JCI12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott S J, Krejany E O, Mellies J L, Robins-Browne R M, Sasakawa C, Kaper J B. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with functions similar to VirA of Shigella flexneri. Infect Immun. 2001;69:4027–4033. doi: 10.1128/IAI.69.6.4027-4033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott S J, Sperandio V, Giron J A, Shin S, Mellies J L, Wainwright L, Hutcheson S W, McDaniel T K, Kaper J B. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y K, Lai L C, McNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 18.Frankel G, Phillips A D, Novakova M, Field H, Candy D C A, Schauer D B, Douce G, Dougan G. Intimin from enteropathogenic Escherichia coli restores murine virulence to a Citrobacter rodentium eaeA mutant: induction of an immunoglobulin A response to intimin and EspB. Infect Immun. 1996;64:5315–5325. doi: 10.1128/iai.64.12.5315-5325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 20.Goffaux F, China B, Janssen L, Mainil J. Genotypic characterization of enteropathogenic Escherichia coli (EPEC) isolated in Belgium from dogs and cats. Res Microbiol. 2000;151:865–871. doi: 10.1016/s0923-2508(00)01153-0. [DOI] [PubMed] [Google Scholar]
- 21.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 22.Hacker J, Kaper J B. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 23.Heffernan E J, Harwood J, Fierer J, Guiney D. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol. 1992;174:84–91. doi: 10.1128/jb.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins L M, Frankel G, Connerton I, Goncalves N S, Dougan G, MacDonald T T. Role of bacterial intimin in colonic hyperplasia and inflammation. Science. 1999;285:588–591. doi: 10.1126/science.285.5427.588. [DOI] [PubMed] [Google Scholar]
- 25.Higgins L M, Frankel G, Douce G, Dougan G, MacDonald T T. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect Immun. 1999;67:3031–3039. doi: 10.1128/iai.67.6.3031-3039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoiczyk E, Blobel G. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc Natl Acad Sci USA. 2001;98:4669–4674. doi: 10.1073/pnas.071065798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hulton C S, Higgins C F, Sharp P M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991;5:825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 28.Itoh K, Matsui T, Tsuji K, Mitsuoka T, Ueda K. Genetic control in the susceptibility of germfree inbred mice to infection by Escherichia coli O115a,c:K(B) Infect Immun. 1988;56:930–935. doi: 10.1128/iai.56.4.930-935.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karaolis D K, McDaniel T K, Kaper J B, Boedeker E C. Cloning of the RDEC-1 locus of enterocyte effacement (LEE) and functional analysis of the phenotype on HEp-2 cells. Adv Exp Med Biol. 1997;412:241–245. doi: 10.1007/978-1-4899-1828-4_36. [DOI] [PubMed] [Google Scholar]
- 30.Kenny B, DeVinney R, Stein M, Reinscheid D, Frey E, Finlay B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 31.Kenny B, Jepson M. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell Microbiol. 2000;2:579–590. doi: 10.1046/j.1462-5822.2000.00082.x. [DOI] [PubMed] [Google Scholar]
- 32.Luperchio S A, Newman J V, Dangler C A, Schrenzel M D, Brenner D J, Steigerwalt A G, Schauer D B. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. J Clin Microbiol. 2000;38:4343–4350. doi: 10.1128/jcm.38.12.4343-4350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo C H, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han C G, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 1998;5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Marches O, Nougayrede J P, Boullier S, Mainil J, Charlier G, Raymond I, Pohl P, Boury M, De Rycke J, Milon A, Oswald E. Role of tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect Immun. 2000;68:2171–2182. doi: 10.1128/iai.68.4.2171-2182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDaniel T K, Kaper J B. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 37.McGraw E A, Li J, Selander R K, Whittam T S. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol Biol Evol. 1999;16:12–22. doi: 10.1093/oxfordjournals.molbev.a026032. [DOI] [PubMed] [Google Scholar]
- 38.McNamara B P, Donnenberg M S. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol Lett. 1998;166:71–78. doi: 10.1111/j.1574-6968.1998.tb13185.x. [DOI] [PubMed] [Google Scholar]
- 39.McNamara B P, Koutsouris A, O'Connell C B, Nougayrede J P, Donnenberg M S, Hecht G. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J Clin Investig. 2001;107:621–629. doi: 10.1172/JCI11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellies J L, Elliott S J, Sperandio V, Donnenberg M S, Kaper J B. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 41.Nataro J, Kaper J. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman J V, Zabel B A, Jha S S, Schauer D B. Citrobacter rodentium espB is necessary for signal transduction and for infection of laboratory mice. Infect Immun. 1999;67:6019–6025. doi: 10.1128/iai.67.11.6019-6025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 2.4.1–2.4.2. [Google Scholar]
- 44.Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun. 2000;68:64–71. doi: 10.1128/iai.68.1.64-71.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perna N T, Mayhew G F, Posfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reid S D, Herbelin C J, Bumbaugh A C, Selander R K, Whittam T S. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 47.Schauer D B. Murine colonic hyperplasia. In: Miller V L, Kaper J B, Portnoy D A, Isberg R R, editors. Molecular genetics of bacterial pathogenesis. Washington, D. C.: American Society for Microbiology; 1994. pp. 197–208. [Google Scholar]
- 48.Schauer D B, Falkow S. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect Immun. 1993;61:2486–2492. doi: 10.1128/iai.61.6.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schauer D B, Falkow S. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect Immun. 1993;61:4654–4661. doi: 10.1128/iai.61.11.4654-4661.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schauer D B, Zabel B A, Pedraza I F, O'Hara C M, Steigerwalt A G, Brenner D J. Genetic and biochemical characterization of Citrobacter rodentium sp. nov. J Clin Microbiol. 1995;33:2064–2068. doi: 10.1128/jcm.33.8.2064-2068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sperandio V, Kaper J B, Bortolini M R, Neves B C, Keller R, Trabulsi L R. Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol Lett. 1998;164:133–139. doi: 10.1111/j.1574-6968.1998.tb13078.x. [DOI] [PubMed] [Google Scholar]
- 52.Stein M, Kenny B, Stein M A, Finlay B B. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J Bacteriol. 1996;178:6546–6554. doi: 10.1128/jb.178.22.6546-6554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swenson D L, Bukanov N O, Berg D E, Welch R A. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tacket C O, Sztein M B, Losonsky G, Abe A, Finlay B B, McNamara B P, Fantry G T, James S P, Nataro J P, Levine M M, Donnenberg M S. Role of EspB in experimental human enteropathogenic Escherichia coli infection. Infect Immun. 2000;68:3689–3695. doi: 10.1128/iai.68.6.3689-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tobe T, Hayashi T, Han C G, Schoolnik G K, Ohtsubo E, Sasakawa C. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect Immun. 1999;67:5455–5462. doi: 10.1128/iai.67.10.5455-5462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tzipori S, Gunzer F, Donnenberg M, de Montigny L, Kaper J, Donohue-Rolfe A. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect Immun. 1995;63:3621–3627. doi: 10.1128/iai.63.9.3621-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vieira M A, Andrade J R, Trabulsi L R, Rosa A C, Dias A M, Ramos S R, Frankel G, Gomes T A. Phenotypic and genotypic characteristics of Escherichia coli strains of non-enteropathogenic E. coli (EPEC) serogroups that carry EAE and lack the EPEC adherence factor and Shiga toxin DNA probe sequences. J Infect Dis. 2001;183:762–772. doi: 10.1086/318821. [DOI] [PubMed] [Google Scholar]
- 58.Volkert M R, Loewen P C, Switala J, Crowley D, Conley M. The Δ(argF-lacZ)205(U169) deletion greatly enhances resistance to hydrogen peroxide in stationary-phase Escherichia coli. J Bacteriol. 1994;176:1297–1302. doi: 10.1128/jb.176.5.1297-1302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wieler L H, McDaniel T K, Whittam T S, Kaper J B. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol Lett. 1997;156:49–53. doi: 10.1111/j.1574-6968.1997.tb12704.x. [DOI] [PubMed] [Google Scholar]
- 60.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 219–227. [DOI] [PubMed] [Google Scholar]
- 61.Zhu C, Agin T S, Elliott S J, Johnson L A, Thate T E, Kaper J B, Boedeker E C. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect Immun. 2001;69:2107–2115. doi: 10.1128/IAI.69.4.2107-2115.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]