Abstract
Chicken coccidiosis is a disease caused by Eimeria spp. and costs the broiler industry more than 14 billion dollars per year globally. Different chicken Eimeria species vary significantly in pathogenicity and virulence, so the classification of different chicken Eimeria species is of great significance for the epidemiological survey and related prevention and control. The microscopic morphological examination for their classification was widely used in clinical applications, but it is a time-consuming task and needs expertise. To increase the classification efficiency and accuracy, a novel model integrating transformer and convolutional neural network (CNN), named Residual-Transformer-Fine-Grained (ResTFG), was proposed and evaluated for fine-grained classification of microscopic images of seven chicken Eimeria species. The results showed that ResTFG achieved the best performance with high accuracy and low cost compared with traditional models. Specifically, the parameters, inference speed and overall accuracy of ResTFG are 1.95M, 256 FPS and 96.9%, respectively, which are 10.9 times lighter, 1.5 times faster and 2.7% higher in accuracy than the benchmark model. In addition, ResTFG showed better performance on the classification of the more virulent species. The results of ablation experiments showed that CNN or Transformer alone had model accuracies of only 89.8% and 87.0%, which proved that the improved performance of ResTFG was benefit from the complementary effect of CNN's local feature extraction and transformer's global receptive field. This study invented a reliable, low-cost, and promising deep learning model for the automatic fine-grain classification of chicken Eimeria species, which could potentially be embedded in microscopic devices to improve the work efficiency of researchers and extended to other parasite ova, and applied to other agricultural tasks as a backbone.
Key words: chicken Eimeria classification, deep learning, convolutional and transformer structure, complementary effect
INTRODUCTION
The global demand for protein products has been steadily increasing (Cao and Li, 2013). The poultry industry provides a large amount of meat and egg products for human consumption and its production scale is expected to further increase in the next decade (Mottet and Tempio, 2017; Blake et al., 2020). Chicken coccidiosis is a widespread and economically significant disease caused by protozoan parasite of the genus Eimeria (Chapman et al., 2013; Mesa et al., 2021), costing the global broiler industry more than 14 billion dollars per year (Adams et al., 2022). There are 7 recognized Eimeria species, including E. Tenella, E. Acervulina, E. Maxima, E. Brunetti, E. Mitis, E. Necatrix, and E. Praecox. Chickens infected with different Eimeria species may occur clinical or subclinical symptoms because of significant differences in pathogenicity and virulence among different species (Shirley, 1997). Clinical coccidiosis is more harmful, which not only affects the yield and quality of meat and eggs, but also can cause chicken death with a high probability, while subclinical coccidiosis generally does not cause death (Engidaw and Getachew, 2018). In addition, different Eimeria species may have different drug resistance (Fatoba and Adeleke, 2018). Therefore, the successful identification of Eimeria species can provide guidance for treatment measures. And understanding the prevalence of coccidiosis can help the government to formulate macro-control policies, and rapid and accurate identification of Eimeria species can provide convenience for relevant researchers. Overall, it is of practical significance to distinguish Eimeria species for epidemiological survey and related prevention and control.
The molecular biological methods and microscopic morphological examination were widely adopted to identify parasites (Huang et al., 2017; Mattiello et al., 2000; Fotouhi-Ardakani et al., 2021; Hendershot et al., 2021). The former are accurate and sensitive but require sophisticated protocols, and the latter is a very challenging task for naked eyes due to the small morphological differences among chicken Eimeria species. Therefore, there is an urgent need to develop an automatic identification process for chicken Eimeria species. In some studies, The morphological characteristics of Eimeria oocysts were extracted and semi-automatic recognition was carried out by machine learning algirithms (Kucera and Reznicky, 1991; Castañón et al., 2007; Abdalla and Seker, 2017). Castañón et al. (2007) achieved the best overall accuracy of 85.75%. However, the semi-automatic methods requires manually designed features, which is cumbersome and the model accuracy is insufficient. The rapid development of convolutional neural network (CNN) has provided a powerful tool for the image recognition task (Esteva et al., 2017). Due to the superiority of CNN, it has been used for species identification of various parasites with good results and has been embedded in automated devices (Yang et al., 2020; Butploy et al., 2021; Lee et al., 2021; Thevenoux et al., 2021; Abade et al., 2022). However, there are few studies focusing on the classification of chicken Eimeria species using deep learning methods. Monge and Beltrán (2019) proposed a CNN model to classify chicken Eimeria species and the accuracy was improved to 90.42%, which still has room for improvement.
It is observed that the CNN-based models could achieve better results than traditional models that requires manual feature extraction. But these studies did not realize that the species recognition of some parasites, for example, chicken Eimeria, is a fine-grained classification task, which focusing on the classifying objects of similar but different subtypes (Zhao et al., 2020). The Transformer structure, originally proposed for Natural Language Processing (NLP) tasks (Vaswani et al., 2017; Jacob et al., 2019), which has been successfully applied in major computer vision tasks including fine-grained classification (Carion et al., 2020; Chen et al., 2021; Dosovitskiy et al., 2021; Zheng et al., 2021). And Transformer-Fine-Grained model (TransFG) achieved State-of-The-Art (SOTA) performance on five popular fine-grained classification benchmarks (He et al., 2021). The feature of local region connection makes CNN good at capturing local features, but lacks the ability to capture global features. Transformer can capture global features well, but is less capable of capturing local features. Therefore, integrating CNN and Transformer structure could improve the model performance (Dai et al., 2021; Lu et al., 2022).
In this study, a novel model, named Residual-Transformer-Fine-Grained (ResTFG), was proposed for the classification of chicken Eimeria species based on the residual block (He et al., 2016) and TransFG (He et al., 2021). The main objectives of the present study were to 1) investigate the complementary effects of integrating the Transformer and CNN on the model performance; 2) optimize the number of layers and hyperparameters of the new model for better performance. The study eventually designed and validated a high performance and lightweight model suitable for automatic and real-time micrograph classification of seven chicken Eimeria oocysts.
MATERIALS AND METHODS
Dataset
Dataset description
The dataset used in this study was from a publicly available website (http://www.coccidia.icb.usp.br/), created by the Laboratory of Molecular Biology of Coccidia at the Department of Parasitology of the Institute of Biomedical Sciences and the Cybernetic Vision Research Group at the Institute of Physics, the University of Sao Paulo, firstly published in 2007 (Castañón et al., 2007). The generality of data sources was considered. Several samples of each species were used, which were collected from different geographic sources, in order to dilute possible intra-specific variations and maximize inter-specific discrimination (Castañón et al., 2007). The dataset provides RGB digital micrographs of oocysts of seven Eimeria species of domestic fowl. Each micrograph contains multiple oocysts which are of the same species. To construct the classification image dataset, single oocyst was isolated from micrographs manually. In total, there were 7 categories, 4,243 labeled microscopic images of isolated oocysts. Since the morphological differences of seven oocysts, these images were of different sizes, and the maximum size was up to 447 × 642 pixels (width by height) and the minimum size was 177 × 225 pixels (width by height). Figure 1 shows the characteristic morphology of the 7 chicken Eimeria species.
Figure 1.
Typical micrographs of oocysts of the seven chicken Eimeria species. Samples: (a) E. Maxima, (b) E. Brunetti, (c) E. Tenella, (d) E. Necatrix, (e) E. Praecox, (f) E. Acervulina, and (g) E. Mitis.
Dataset augmentation and splitting
There is an originally large difference in the number of different oocyst categories with uneven distribution, which would tend to result in a model with false classification of some uncertain samples into the category with more samples. Appropriate data augmentation was conducted for categories of images with fewer samples. Specifically, all E. Maxima images were flipped horizontally, and 300 E. Brunetti images and 200 E. Necatrix images were randomly selected for horizontal flipping, which is a commonly used method for dataset augmentation (Wang et al., 2019; Ye et al., 2020; Lu et al., 2022). Considering that the imaging environment of micrographs is controllable and consistent, and the original micrographs are sufficiently representative, it is not necessary to augment the original dataset by utilizing some morphological or color adjustment methods. After balancing the dataset, the total number of images increased from 4,243 to 5,103. Seventy percent of the samples of each category were randomly selected as the training set and the rest thirty percent as the test set. To ensure reproducible evaluation results, a constant random seed “2022” was set. The details of the dataset are shown in Table 1.
Table 1.
The number of images of the chicken Eimeria oocyst dataset.
| Class label | Species name | Original | After data augmentation | Partitioning of the dataset (7:3) |
|
|---|---|---|---|---|---|
| Training | Test | ||||
| ACE | E. Acervulina | 742 | 742 | 520 | 222 |
| BRU | E. Brunetti | 442 | 742 | 520 | 222 |
| MAX | E. Maxima | 360 | 720 | 504 | 216 |
| MIT | E. Mitis | 825 | 825 | 578 | 247 |
| NEC | E. Necatrix | 502 | 702 | 492 | 210 |
| PRA | E. Praecox | 676 | 676 | 474 | 202 |
| TEN | E. Tenella | 696 | 696 | 488 | 208 |
| Total number | 4,243 | 5,103 | 3,576 | 1,527 | |
Methods
Proposed model
The framework of the proposed ResTFG model is shown in Figure 2. The left and the right parts are the CNN and Transformer branches, respectively.
Figure 2.
The overview of the proposed Residual-Transformer-Fine-Grained (ResTFG) model. The left and right parts are the convolutional neural network (CNN) and Transformer branch, respectively. The k, s, and p represent the kernel size, stride, and padding of the convolution layer, respectively. When there is only one residual block, it is then connected to the average pooling layer and the flatten layer; when there are two residual blocks, the second one is directly connected to the flatten layer.
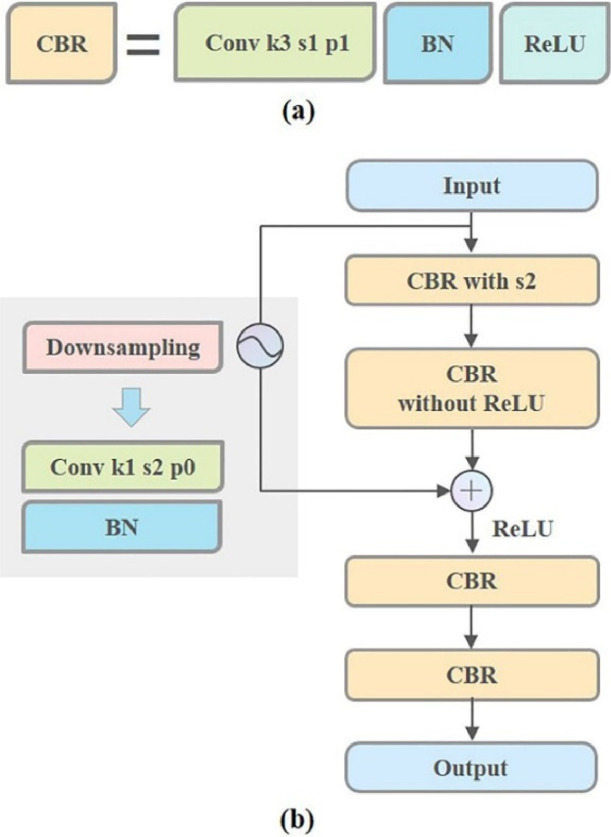
The CNN branch consists of an input layer, a maximum pooling layer, an average pooling layer, a flatten layer, 5 CBR modules (acronym for Convolution Batch-Normalization ReLU (rectified linear unit)), and one or two residual blocks. The k, s, and p in Figure 2 represent the kernel size, stride, and padding of the convolution layer, respectively. As shown in Figure 3(a), the CBR module consists of three layers, a convolution layer with a kernel size of 3 × 3, a stride of 1 × 1 and a padding of 1, followed by a batch normalization (BN) layer and a ReLU layer. The BN layer can speed up the training and convergence of the network, alleviate the gradient dispersion and mitigate the overfitting problem. In order to make the network capable of characterizing nonlinear mappings and further addressing the gradient disappearance problem, the ReLU activation function was added after each BN layer. The residual block is chosen as it can improve the trainability of the deep network with less computation cost. The structure of the residual block is shown in Figure 3(b), composed of an input layer, four CBR modules, a downsampling operation and an output layer. The downsampling operation is implemented by a convolution layer with a kernel size of 1 × 1, a stride of 2 × 2 and no padding, and is connected to a BN layer afterward. The number of the residual block is uncertain (shown in the dashed box in Figure 2) since the structure in the model is subject to optimization. When there is only one residual block, it is then connected to the average pooling layer and the flattening layer. The second residual block is directly connected to the flattened layer when there are two residual blocks. This design aims to match the feature dimensions after the flattened layer with the input dimensions of the Transformer branch.
Figure 3.
The structure of the Convolution Batch-Normalization ReLU (rectified linear unit) module (CBR) (a), and the residual block (b). The k, s, and p represent the kernel size, stride, and padding of the convolution layer, respectively.
The Transformer branch was designed based on a previous study (He et al., 2021). The benefit of this intentionally simplified setting is to reduce the impact of other techniques on model performance (Dai et al., 2021). The final output vector dimension was set to seven to match the number of the chicken Eimeria species in this study. The Transformer branch is described in detail, including token and embedding operations, Transformer encoder block, and the optimization of loss function.
-
(1)Tokens and Embeddings: For the Transformer in NLP, each word in the input sentence is divided into tokens, and a token representing semantic information is a class token. But for the Transformer in computer vision, it is not realistic to regard each pixel as a token due to the limitation of computation, so the image is cut into multiple patches, and each patch is regarded as a token for subsequent processing. In ResTFG models, the patch embedding operation is implemented by the CNN branch. The class token, that is, a learnable vector, is embedded in patch embedding to enable the model for categorization. Different from CNN, the Transformer structure requires position embeddings to encode the location information of patch tokens, so a learnable vector, that is, position token, is added to the patch embedding. Assuming the input image is represented as after being mapped to patch embedding space by the CNN branch, the output after position embedding is:
(1)
where with category and spatial information is the input of the Transformer branch, is the number of image patches, is the class embedding operation, and is the position embedding operation.
-
(2)Transformer Encoder: Transformer Encoder is composed of encoder block layers in total and one Part Select Module (PSM), where is 12 by default.
-
a)Encoder block: As shown in Figure 4, the encoder block is composed of the alternating Layer Normalization (LN) layer, Multi-Head Self-Attention (MSA) layer, and Multi-layer Perceptron (MLP) block. The MLP block contains two linear layers, two dropout layers with a dropout rate of 0.1, and one Gaussian Error Linear Unit (GeLU) activation function layer. For each encoder block, the LN layer is applied before MSA and MLP layer, and the residual connection is applied after each LN layer. Therefore, the output of layer can be expressed as:
(2) (3)
-
a)
where is the output of layer , is the output of MSA layer.
Figure 4.
The structure of the transformer encoder block.
MSA is developed from Self-Attention (SA) mechanism. The image classification task is abstracted into a query task through SA. Every patch token can be linked to the class token through SA. Patches mapped to the embedding space are calculated by matrix (query) and matrix (key) to obtain the attention weight. The weight is then fed into the Softmax function to calculate the dot product of it with matrix (value) for the final output, i.e., the dot-product attention (Vaswani et al., 2017), expressed as follows:
| (4) |
where is the dimension of matrix and . The scaling of dot-product is necessary because the large will result in a large value after , leading to a minimal gradient after the Softmax function, which is not conducive to the network training.
Rich characteristic information can generally be obtained by deepening the number of CNN channels, and each channel can be used to identify a different pattern. Similarly, K attention heads are set in the MSA for better feature capture capability. In this study, the number of attention heads were 12 by default. The output of MSA could be expressed as (Vaswani et al., 2017):
| (5) |
| (6) |
-
b)PSM module: The input of the last encoder block is modified with the PSM layer to take full advantage of the attention information. Previous studies have pointed out that raw attention weights do not necessarily correspond to the relative importance of input tokens, so it is necessary to integrate the attention weights of all previous layers (Abnar and Zuidema, 2020). Specifically, PSM recursively applies matrix multiplication to the raw attention weights in all encoder block layers to get . The tokens corresponding to the index of the maximum K attention heads of each attention weight in are selected and spliced with the class token as the input to the last encoder block (He et al., 2021), which can be expressed as:
(7) (8) (9) (10)
-
b)
-
(3)Loss Function: The contrastive loss is introduced into the model to minimize the similarity of the class tokens of different categories and maximize the similarity of the class tokens of the same category. The total loss is the sum of contrastive loss and cross-entropy loss , which can be expressed as (He et al., 2021):
(11) (12) (13)
where is the batch size, and are pre-processed with L2 normalization, is cosine similarity, is an artificially constant to avoid the loss affected by simple negative samples, is the ground-truth label, and is the predicted label.
Equipment and Environment
To facilitate intensive computation in model training, a professional deep learning platform, SYS-4029GP-TRT was used, equipped with 2 × Intel© Xeon(R) Gold 6147M CPU @ 2.50GHz, a total of 260 GB memory, and 8 graphics cards including 4 × Nvidia TITAN RTX and 4 × Nvidia GeForce RTX 2080 Ti, a total of 140 GB video memory. The testing and inference speed measurement of models were run on a desktop computer with GeForce RTX 3080 GPU and Inter(R) Core (TM) i9-10900KF CPU @3.70GHz. In terms of the software environment, Python-3.8, PyCharm-Professional-2021.2.3, and Pytorch-GPU-1.8.1 framework were used.
Experimental setting
First, 7 existing SOTA models were compared, including VGG11 (Simonyan, 2014), MobilenetNet_V3_Small, MobilenetNet_V3_Large (Howard et al., 2019), ResNet34 (He et al., 2016), DenseNet121 (Huang et al., 2016), Shufflenet_V2_x1_0 (Ma et al., 2018), and TransFG_B16 (He et al., 2021). VGGNet11, ResNet34, and DenseNet121 with their good feature extraction capability and generalization performance have become the preferred backbone of many downstream tasks, and have achieved competitive accuracy in many image recognition applications. MobilenetNet_V3_Small, MobilenetNet_V3_Large, and Shufflenet_V2_x1_0 models are a class of lightweight models with relatively low accuracy but fast inference speed. The TransFG_B16 with a batch size of 16 is one of the TransFG models, which is a Transformer based model specifically designed for fine-grained image classification task. After the comprehensive evaluation of the above 7 SOTA models, a benchmark model was obtained for the subsequent comparison with ResTFG models.
Then a series of experiments were conducted to optimize ResTFG models. First, the effect of 3 hyperparameters and the number of encoder block layers in the Transformer branch on model performance were evaluated. After determining the optimal setting of the Transformer branch, the structure of the CNN branch was further optimized. Furthermore, the effectiveness of the integration of CNN and Transformer structure was proved through ablation experiments.
Image Preprocessing and Hyperparameters setting
The computational cost of the CNN is significantly related to the input image size, which should be sufficiently reduced but without affecting the model performance. In this study, to modify images as little as possible, only two preprocessing steps were adopted before inputting the images into the CNN structure. First, each image was adjusted to a commonly used format for the image classification task with a resolution of 224 × 224 using the resize method in PyTorch deep learning framework. Second, each pixel value was normalized from to [0, 1] to speed up the convergence of the model.
All models used were trained from scratch. The initial learning rate is 0.001, and the decay rate is 0.5 for every 20 epochs. In addition, the number of epochs is 100, the batch size is 64, and the optimizer is Stochastic Gradient Descent (SGD) with a 0.9 momentum and a 1e−4 weight decay. The models proposed in this paper use the sum of cross-entropy loss and contrastive loss as the total loss, while other models only use cross-entropy to calculate the loss.
Evaluation Metrics
In practical application scenarios, the deployment of models will be limited by computational resources. In this study, three metrics, that is, accuracy, the number of parameters, and inference speed (FPS, frames per second), are used for a comprehensive evaluation of model performance to obtain high performance and lightweight model. The receiver operating characteristic (ROC) curve, area under the ROC curve (AUC) and the loss curve of the training stage were also used to compare the performance. In addition, to evaluate the recognition performance of the model for each category of chicken Eimeria species, confusion matrix, precision, recall, and F1 score were adopted. All results are the average value of 3 tests. The calculation formulas of metrics are as follows:
| (14) |
| (15) |
| (16) |
| (17) |
Where is the proportion of the correct sample to the total sample, is the proportion of all samples correctly predicted by the model, is the proportion of all real samples that the model predicts to be correct, is the harmonic mean of and , True Positive () is the number of samples correctly predicted to be positive, True Negative () is the number of samples correctly predicted to be negative, False Positive () is the number of samples falsely predicted to be positive, and False Negative () is the number of samples falsely predicted to be negative.
RESULTS
Performance of Existing Models
The test results of the seven existing SOTA models are shown in Table 2. As expected, the accuracy of MobilenetNet_V3_Small, MobilenetNet_V3_Large and Shufflenet_V2_x1_0 was relatively low. Shufflenet_V2_x1_0 has the lowest number of parameters and accuracy (80.4%), which could not meet the recognition requirement. It was surprising VGG11, with the maximum number of parameters, achieved the fastest inference speed of 409 FPS, but its memory consumption is high. DenseNet121 had a good accuracy, but very slow inference speed. TransFG_B16 also had good accuracy but high memory consumption. ResNet34 was identified as a balanced model with 21.29M parameters, 166FPS and 94.2% accuracy, and therefore, was selected as the benchmark model.
Table 2.
The performance of State-of-The-Art (SOTA) models in the classification of chicken Eimeria species. TransFG_B16 with a batch size of 16 is one of the Transformer-Fine-Grained (TransFG) models.
| Model Name | Parameters (M) | Speed (FPS) | Accuracy (%) |
|---|---|---|---|
| VGG11 | 128.80 | 409 | 86.9 |
| MobilenetNet_V3_Small | 1.53 | 157 | 86.3 |
| MobilenetNet_V3_Large | 4.21 | 107 | 90.7 |
| ResNet34 | 21.29 | 166 | 94.2 |
| DenseNet121 | 6.96 | 56 | 92.7 |
| Shufflenet_V2_x1_0 | 1.26 | 146 | 80.4 |
| TransFG_B16 | 85.80 | 104 | 93.5 |
Optimization of Transformer Branch
Two residual blocks with 13 convolution layers were adopted in ResTFG models by default. The initial value of three hyperparameters in the Transformer branch, that is, hidden size, MLP dimension and the number of multi-attention heads was 768, 3,072, and 12 respectively, which were set according to TransFG_B16. MLP dimension was set to four times of hidden size, so it was not considered a separate hyperparameter. The more multi-attention heads mean the more patterns of correlation between different patches can be learned. The number of the encoder block layer was adjusted at 8 to limit the size of ResTFG models. The initial model was named ResTFG (C13, H12, L8) (a), where [C] represents the number of convolution layers, [H] represents the number of multi attention head, [L] represents the number of the encoder block layer, and the letter suffixes correspond to the different hidden size and MLP dimension.
The hidden size and MLP dimension were first optimized. As shown in Table 3, ResTFG (C13, H12, L8) (a) achieved the highest accuracy of 97.2%, but it had too many parameters. When the hidden size was set to 384, the ResTFG (C13, H12, L8) (b) obtained a tradeoff among parameters, inference speed and accuracy. Then, the hyperparameter H was optimized. It was found that the number of parameters was almost unaffected by H, but the model accuracy was negatively affected by a decreasing H, so H was always set to 12 in subsequent models. Furthermore, the evaluation of hyperparameter L showed the accuracy and inference speed increased with the decrease of L, which might imply a saturation of the model. A similar result occurred in Lu et al.'s study (Lu et al., 2022). Therefore, ResTFG (C13, H12, L2) achieved the best results with 10.86M number of parameters, 216FPS, and 97.1% accuracy. Compared with ResTFG (C13, H12, L8) (a), the number of parameters in ResTFG (C13, H12, L2) was reduced by 86%, and the inference speed was doubled, but the accuracy was only decreased by 0.1%, which is significantly superior to the performance of ResNet34.
Table 3.
The performance comparison of the Residual-Transformer-Fine-Grained (ResTFG) for the Transformer branch with different hyperparameters and the number of the encoder block layer. TransFG_B16 with a batch size of 16 is one of the Transformer-Fine-Grained (TransFG) models.
| Model Name | Hidden size | MLP dimension | Number heads | Number Layers |
Parameters (M) |
Speed (FPS) |
Accuracy (%) |
|---|---|---|---|---|---|---|---|
| TransFG_B16 | 768 | 3,072 | 12 | 12 | 85.80 | 104 | 93.5 |
| ResTFG (C13, H12, L8) (a) | 768 | 3,072 | 12 | 8 | 82.14 | 105 | 97.2 |
| ResTFG (C13, H12, L8) (b) | 384 | 1,536 | 12 | 8 | 21.50 | 112 | 97.0 |
| ResTFG (C13, H12, L8) (c) | 288 | 1,024 | 12 | 8 | 11.90 | 113 | 95.7 |
| ResTFG (C13, H12, L8) (d) | 192 | 768 | 12 | 8 | 6.12 | 116 | 95.7 |
| ResTFG (C13, H8, L8) | 384 | 1,536 | 8 | 8 | 21.50 | 114 | 96.7 |
| ResTFG (C13, H4, L8) | 384 | 1,536 | 4 | 8 | 21.50 | 114 | 96.6 |
| ResTFG (C13, H2, L8) | 384 | 1,536 | 2 | 8 | 21.50 | 114 | 96.0 |
| ResTFG (C13, H12, L6) | 384 | 1,536 | 12 | 6 | 17.96 | 134 | 96.5 |
| ResTFG (C13, H12, L4) | 384 | 1,536 | 12 | 4 | 14.41 | 165 | 96.9 |
| ResTFG (C13, H12, L3) | 384 | 1,536 | 12 | 3 | 12.63 | 183 | 97.0 |
| ResTFG (C13, H12, L2) | 384 | 1,536 | 12 | 2 | 10.86 | 216 | 97.1 |
Optimization of CNN Branch
The convolution layers of the CNN branch were pruned and the parameters of the convolution kernel were adjusted in order to make the model more lightweight. As shown in Table 4, after pruning one residual block, i.e., four convolution layers, the accuracy of ResTFG (C9, H12, L2) (a) only decreased by 0.6%, but there was a 17% improvement in inference speed. It is a good result, but the model is not light enough and there is a possibility to further reduce its memory consumption. Therefore, the kernel size (same value with hidden size) of convolution layers in the residual block was optimized. The results showed that the kernel size strongly influences the number of parameters of the model. When it was reduced from 384 to 156, the number of parameters was reduced from 10.09M to 1.95M which the accuracy was improved by 0.2%. When the number of convolution layers was further reduced, the bonus in the number of model parameters was negligible, but the decrease in accuracy was significant. Therefore, ResTFG (C9, H12, L2) (c) (referred to as optimized ResTFG) obtained the advantages of both high performance and lightweight, with the number of parameters of 1.95M, an inference speed of 256FPS, and an accuracy of 96.9%, which is 10.9 times lighter, 1.5 times faster, and 2.7% higher in accuracy than ResNet34. Table 5 shows the related parameters setting and output shape of each layer of ResTFG (C9, H12, L2) (c).
Table 4.
The performance comparison of the Residual-Transformer-Fine-Grained (ResTFG) for the convolutional neural network (CNN) branch with different kernel parameters and the number of the convolution layer.
| Model Name | Hidden size | MLP dimension | Parameters (M) | Speed (FPS) | Accuracy (%) |
|---|---|---|---|---|---|
| ResTFG (C13, H12, L2) | 384 | 1,536 | 10.86 | 216 | 97.1 |
| ResTFG (C9, H12, L2) (a) | 384 | 1,536 | 10.09 | 254 | 96.5 |
| ResTFG (C9, H12, L2) (b) | 180 | 720 | 2.50 | 256 | 96.7 |
| ResTFG (C9, H12, L2) (c) | 156 | 624 | 1.95 | 256 | 96.9 |
| ResTFG (C5, H12, L2) | 156 | 624 | 1.80 | 299 | 96.2 |
Table 5.
The parameters setting and corresponding output shape of each layer of Residual-Transformer-Fine-Grained (ResTFG) (C9, H12, L2) (c).
| Layer name | Kernel size | Stride | Padding | Output shape |
|---|---|---|---|---|
| Input | 224 × 224 × 3 | |||
| CBR_1 | 7 × 7 | 2 | 3 | 112 × 112 × 64 |
| Max-pooling | 3 × 3 | 2 | 1 | 56 × 56 × 64 |
| CBR_2 | 3 × 3 | 1 | 1 | 56 × 56 × 64 |
| CBR_3 | 3 × 3 | 1 | 1 | 56 × 56 × 64 |
| CBR_4 | 3 × 3 | 1 | 1 | 56 × 56 × 64 |
| CBR_5 | 3 × 3 | 1 | 1 | 56 × 56 × 64 |
| Residual Block | 28 × 28 × 156 | |||
| Avg-pooling | 3 × 3 | 2 | 1 | 14 × 14 × 156 |
| Flatten | 196 × 156 | |||
| Embedding | 197 × 156 | |||
| Encoder blocks | 197 × 156 | |||
| Linear | 7 |
Ablation Studies on ResTFG
To demonstrate that the integration of the CNN and Transformer structure have a positive effect on model performance improvement, the ablation experiments were designed and implemented. Specifically, the Transformer branch and the CNN branch in ResTFG (C9, H12, L2) (c) were removed separately so that only one branch could be used for testing. The test results are shown in Table 6. There is no doubt that the number of parameters would be reduced and the inference speed would increase with only the CNN or Transformer branch. But as Table 6 shows, the accuracy of these two models decreased significantly by 7.1% and 9.9%, respectively. The results showed sufficient evidence that the hybrid model integrating the CNN branch and Transformer branch can fully utilize the advantages of both, which is highly superior to the model with a single branch.
Table 6.
The performance of the Residual-Transformer-Fine-Grained (ResTFG) with only convolutional neural network (CNN) branch or Transformer branch.
| Model Name | Parameters (M) | Speed (FPS) | Accuracy (%) |
|---|---|---|---|
| CNN Only | 0.92 | 549 | 89.8 |
| Transformer Only | 1.03 | 403 | 87.0 |
| ResTFG (C9, H12, L2) (c) | 1.95 | 256 | 96.9 |
DISCUSSION
Balance of Accuracy and Inference Speed
Due to the limited computing resources of embedded and mobile devices, in addition to accuracy, memory consumption and inference speed of the model are also important. If a model has the advantages of high accuracy and low cost at the same time, it has more potential to be applied to the actual scenarios to automatically identify chicken Eimeria species.
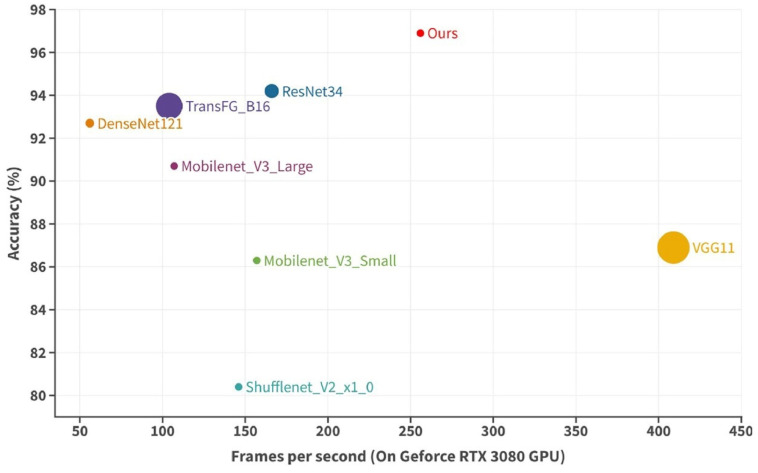
Based on the collected microscopic image dataset of chicken Eimeria oocysts, 7 existing SOTA models were tested. Although VGG11 had the fastest inference speed (409FPS), its parameters reached 128.80M, which could not meet the requirements of lightweight model. Shufflenet_V2_x1_0 has an advantage in the number of parameters (1.26M), but its accuracy is only 80.4%. Compared with other models, ResNet34 achieved good number of parameters (21.29M), inference speed (166FPS) and accuracy (94.2%), but it was still inferior to our proposed model. And there is only two ResTFG models have an accuracy below 96%, with the highest accuracy of 97.2%. Among all the ResTFG models, ResTFG (C5, H12, L2) achieved the fewest number of parameters (1.80M) and the fastest inference speed (299FPS). But ResTFG (C9, H12, L2) (c) (referred to as Ours) is regarded as a balanced model with the advantages of both high performance and lightweight. Figure 5 shows the bubble plots of accuracy, inference speed and the number of parameters of 7 existing SOTA models and Ours, with larger bubbles representing more parameters and vice versa. Ours is in the upper right of the figure, and the bubble size is significantly smaller than ResNet34.
Figure 5.
The bubble plots of accuracy (%), inference speed (frames per second) and the number of parameters of seven State-of-The-Art (SOTA) models and the optimized Residual-Transformer-Fine-Grained (ResTFG) model (Ours).
In addition to the overall accuracy, the ability of models to identify the more virulent Eimeria species is also critical. Table 7 shows the recognition accuracy and recall of each Eimeria species of the benchmark model ResNet34, ResTFG (C13, H12, L8) (b), ResTFG (C13, H12, L2) and ResTFG (C9, H12, L2) (c) (Ours). The comprehensive recognition performance was evaluated by F1 score values, which was calculated by Equation 15. ResTFG (C13, H12, L8) (b) and ResTFG (C13, H12, L2) were selected because of their superior performance in the model optimization process. Among the seven Eimeria species, E. Tenella showed the highest virulence, followed by E. Necatrix (Shirley, 1997). Therefore, the identification performance of models on these two species has been focused on. As can be seen from Table 7, compared with the ResNet34, the optimized model (Ours) has larger F1 score values, that is, the comprehensive recognition performance of E. Tenella and E. Necatrix is better. In addition, compared with ResTFG (C13, H12, L2) and ResTFG (C13, H12, L8) (b), Ours improved the recognition performance of E. Necatrix without affecting the recognition ability of E. Tenella. These results showed that the model proposed in this study is of practical significance, and the optimization strategy is feasible.
Table 7.
Precision, recall, and F1 score of the optimized Residual-Transformer-Fine-Grained (ResTFG) and ResNet34 model for each class.
| Models | ResTFG (C9, H12, L2) (c) (Ours) |
ResNet34 |
ResTFG (C13, H12, L8) (b) |
ResTFG (C13, H12, L2) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class label | Precision | Recall | F1 score | Precision | Recall | F1 score | Precision | Recall | F1 score | Precision | Recall | F1 score |
| ACE | 0.97 | 0.95 | 0.96 | 0.95 | 0.91 | 0.93 | 0.97 | 0.95 | 0.96 | 0.97 | 0.99 | 0.98 |
| BRU | 0.96 | 0.98 | 0.97 | 0.90 | 0.98 | 0.94 | 0.96 | 0.99 | 0.98 | 0.96 | 0.97 | 0.97 |
| MAX | 0.98 | 0.99 | 0.99 | 0.98 | 0.95 | 0.96 | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 | 0.98 |
| MIT | 0.98 | 0.98 | 0.98 | 0.96 | 0.97 | 0.96 | 0.97 | 0.97 | 0.97 | 0.98 | 0.98 | 0.98 |
| NEC | 0.97 | 0.96 | 0.97 | 0.95 | 0.91 | 0.93 | 0.98 | 0.95 | 0.96 | 0.97 | 0.94 | 0.96 |
| PRA | 0.93 | 0.95 | 0.94 | 0.88 | 0.94 | 0.91 | 0.93 | 0.97 | 0.95 | 0.93 | 0.97 | 0.95 |
| TEN | 0.99 | 0.97 | 0.98 | 0.98 | 0.93 | 0.96 | 0.98 | 0.97 | 0.98 | 0.99 | 0.97 | 0.98 |
ROC Curve
The performance of multiple methods on the same task can be easily compared by the ROC curve, the area enclosed by the curve and the coordinate axes is called AUC, which is insensitive to the category distribution. The vertical coordinate of the ROC curve is TPR (True Positive Rate) and the horizontal coordinate is the FPR (False Positive Rate), and the closer the curve is to the upper left as well as the larger the ACU value represents the better model performance.
As shown in Figure 6, the ROC curve of the optimized ResTFG (Ours) is closest to the upper left and the AUC value reaches 0.998, which is better than all other models, where the AUC value of ResNet34 is 0.989. Therefore, our model achieved the best performance.
Figure 6.
The ROC curves of seven State-of-The-Art (SOTA) models and the optimized Residual-Transformer-Fine-Grained (ResTFG) model (Ours).
Convergence Situation
Besides focusing on the performance of the model after being well-trained, the convergence situation of the model during the training stage is also one of the most important concerns for researchers. Figure 7 shows the convergence process of the average loss of seven SOTA models and the optimized ResTFG model with the increase of training iterations. As shown in Figure 7, the average loss value of all the eight models decreased rapidly in the first 20 epochs and converged after about 80 epochs. As can be seen that the convergence of VGG11, MobileNet_V3_Small, MobileNet_V3_Large, and Shufflenet_V2_x1_0 in this task is relatively poor, and of ResNet34 and TransFG_B16 is similar except that the initial loss value of TransFG_B16 is lower. The solid red line represents the optimized ResTFG. Its loss value is always the lowest among all models no matter when the training is stopped. It also has the fastest convergence speed, and it reaches the best performance of ResNet34 after training 50 epochs. In addition, the fluctuation of its loss value is the smallest in the late training period. The results proved that our model has a robust and stable learning capability.
Figure 7.
The loss curves of seven State-of-The-Art (SOTA) models and the optimized Residual-Transformer-Fine-Grained (ResTFG) model (Ours).
Confusion Matrix
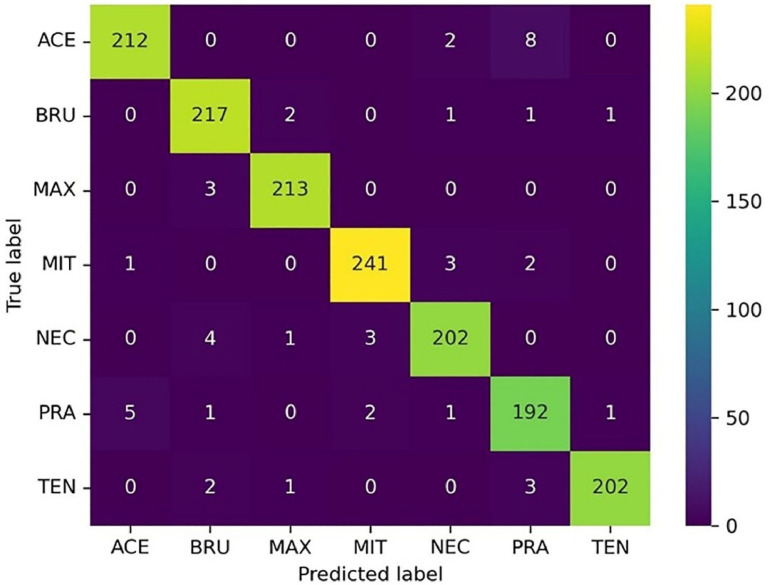
The confusion matrix of the optimized ResTFG is illustrated in Figure 8, which shows the classification performance of the model for each class. According to Equation 14, the overall accuracy is 96.9%.
Figure 8.
Confusion matrix of the optimized Residual-Transformer-Fine-Grained (ResTFG) model (Ours).
The identification accuracy, recall and F1 score of each class are calculated with Equations 15 to 17, and the results are shown in Table 7. As the table shows, the samples of E. Praecox species are more likely to be misclassified, which is the same as in César et al.’s study (2007). In their study, the morphological features and a Bayesian classifier were used to identify seven Eimeria species, and the results showed that E. Praecox species had the worst classification accuracy (74.2%), followed by E. Necatrix species (74.9%). And in this study, E. Praecox species also had the worst classification accuracy of 93%, while E. Necatrix species achieved 97%. It can be seen that the classification accuracy has been greatly improved. The poor performance of different models for the classification of E. Praecox species suggests that E. Praecox is the most difficult to be differentiated correctly among the 7 Eimeria species, which is due to the objective fact of morphological similarities between E. Praecox and the other species (Kucera and Reznicky, 1991)
In this study, the reason preventing the further improvement of the model accuracy is the mutual misclassification between E. Acervulina and E. Praecox species. Eight E. Acervulina samples were misclassified as E. Acervulina and five E. Praecox samples were misclassified as E. Acervulina. The result provides a direction for the further optimization of the mode. On the one hand, the penalty on E. praecox misclassification can be increased during model training to force the model to learn more features of E. praecox species. On the other hand, for data-driven deep learning methods, increasing the number of samples can be tried, especially E. Praecox species which is hard to distinguish, to make the model have better recognition ability and enhance its generalization performance.
CONCLUSIONS
To achieve the automatic fine-grained classification of chicken Eimeria species, a novel deep-learning model, named ResTFG, which integrates the advantages of the CNN and Transformer structure, was proposed in this study. The CNN structure containing residual blocks was used as the backbone, which has a powerful feature extraction ability, and compensated for the defect of CNN lacking a global receptive field through the deployment of the multihead attention mechanism in Transformer. The ablation experiments proved the synergistic effect of integrating the CNN and Transformer structure. Overall, the proposed ResTFG model performs well, achieving an accuracy of 96.9%, an inference speed of 256 FPS, and a memory consumption of 1.95M, which has the advantages of both high accuracy and low cost. This model can improve the work efficiency of researchers. More importantly, for people who do not have the ability to identify Eimeria species with the naked eye, they can obtain species distribution information to infer the severity of the disease with the help of this automatic identification system, which can provide guidance for subsequent medication and the basis for effective control measures. In future work, the ResTFG model would be further optimized and applied to other computer vision and pattern recognition tasks in agricultural engineering.
ACKNOWLEDGMENTS
This work was supported by the Key R&D Program of Zhejiang Province (2021C02026) and the Agriculture Research System of China (CARS-40).
DISCLOSURES
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- Abade A., Porto L.F., Ferreira P.A., Vidal F.D. NemaNet: a convolutional neural network model for identification of soybean nematodes. Biosyst. Eng. 2022;213:39–62. [Google Scholar]
- Abdalla M.A.E., Seker H. Recognition of protozoan parasites from microscopic images: Eimeria species in chickens and rabbits as a case study. 2017 39th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. (EMBC); Jeju, South Korea; 2017. [DOI] [PubMed] [Google Scholar]
- Abnar S., Zuidema W. Proc. 58th Annu. Meeting Assoc. Comput. Linguist. (ACL) 2020. Quantifying attention flow in transformers. Online. [Google Scholar]
- Adams D.S., Kulkarni R.R., Mohammed J.P., Crespo R. A flow cytometric method for enumeration and speciation of coccidia affecting broiler chickens. Vet. Parasitol. 2022;301 doi: 10.1016/j.vetpar.2021.109634. [DOI] [PubMed] [Google Scholar]
- Blake D.P., Knox J., Dehaeck B., Huntington B., Rathinam T., Ravipati V., Ayoade S., Gilbert W., Adebambo A.O., Jatau I.D., Raman M., Parker D., Rushton J., Tomley F.M. Re-calculating the cost of coccidiosis in chickens. Vet. Res. 2020;51:115. doi: 10.1186/s13567-020-00837-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butploy N., Kanarkard W., Intapan P.M. Deep learning approach for Ascaris lumbricoides parasite egg classification. J. Parasitol. Res. 2021;2021:6648038. doi: 10.1155/2021/6648038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li D. Impact of increased demand for animal protein products in Asian countries: implications on global food security. Anim. Front. 2013;3:48–55. [Google Scholar]
- Carion N., Massa F., Synnaeve G., Usunier N., Kirillov A., Zagoruyko S. End-to-end object detection with transformers. 2020 Eur. Conf. Comput. Vision (ECCV); Glasgow; Scottish Event Campus; 2020. [Google Scholar]
- Castañón C.A.B., Fraga J.S., Fernandez S., Gruber A., da F. Costa L. Biological shape characterization for automatic image recognition and diagnosis of protozoan parasites of the genus Eimeria. Pattern Recognit. 2007;40:1899–1910. [Google Scholar]
- Chapman H.D., Barta J.R., Blake D., Gruber A., Jenkins M., Smith N.C., Suo X., Tomley F.M. A selective review of advances in coccidiosis research. Adv. Parasit. 2013;83:93–171. doi: 10.1016/B978-0-12-407705-8.00002-1. [DOI] [PubMed] [Google Scholar]
- Chen H.T., Wang Y.H., Guo T.Y., Xu C., Deng Y.P., Liu Z.H., Ma S.W., Xu C.J., Xu C., Gao W. Pre-trained image processing transformer. 2021 IEEE/CVF Conf. Comput. Vision Pattern Recognit. (CVPR); Nashville, TN, USA; 2021. [Google Scholar]
- Dai Y., Gao Y.F., Liu F.Y. TransMed: transformers advance multi-modal medical image classification. Diagnostics. 2021;11:1384. doi: 10.3390/diagnostics11081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosovitskiy A., Beyer L., Kolesnikov A., Weissenborn D., Zhai X., Unterthiner T., Dehghani M., Minderer M., Heigold G., Gelly S., Uszkoreit J., Houlsby N. An image is worth 16 × 16 words: transformers for image recognition at scale. 2021 Int. Conf. Learn. Represent. (ICLR); Vienna, Austria; 2021. [Google Scholar]
- Engidaw A., Getachew G. A review on poultry coccidiosis. Abyss. J. Sci. Technol. 2018;3:1–12. [Google Scholar]
- Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatoba A.J., Adeleke M.A. Diagnosis and control of chicken coccidiosis: a recent update. J. Parasit. Dis. 2018;42:483–493. doi: 10.1007/s12639-018-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotouhi-Ardakani R., Ghafari S.M., Ready P.D., Parvizi P. Developing, modifying, and validating a TaqMan real-time PCR technique for accurate identification of Leishmania parasites causing most leishmaniasis in Iran. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.731595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Chen J.N., Liu S., Kortylewski A., Yang C., Bai Y.T., Wang C.H. arXiv:2103.07976. 2021. TransFG: a transformer architecture for fine-grained recognition. Accessed August 2022. [DOI] [Google Scholar]
- He K.M., Zhang X.Y., Ren S.Q., Sun J. Deep residual learning for image recognition. 2016 IEEE/CVF Conf. Comput. Vision Pattern Recognit. (CVPR); Las Vegas, NV, USA; 2016. [Google Scholar]
- Hendershot A.L., Esayas E., Sutcliffe A.C., Irish S.R., Gadisa E., Tadesse F.G., Lobo N.F. A comparison of PCR and ELISA methods to detect different stages of Plasmodium vivax in Anopheles arabiensis. Parasite Vector. 2021;14:473. doi: 10.1186/s13071-021-04976-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A., Sandler M., Chu G., Chen L.C., Chen B., Tan M.X., Wang W.J., Zhu Y.K., Pang R.M., Vasudevan V., Le Q.V., Adam H. Searching for MobileNetV3. 2019 IEEE/CVF Conf. Comput. Vision Pattern Recognit. (CVPR); Seoul, South Korea; 2019. [Google Scholar]
- Huang G., Liu Z., Maaten L.V.D., Weinberger K.Q. Densely connected convolutional networks. 2017 IEEE/CVF Conf. Comput. Vision Pattern Recognit. (CVPR); Honolulu, HI, USA; 2016. [Google Scholar]
- Huang Y.Y., Ruan X.C., Li L., Zeng M.H. Prevalence of Eimeria species in domestic chickens in Anhui province, China. J. Parasit. Dis. 2017;41:1014–1019. doi: 10.1007/s12639-017-0927-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob D., Chang M.W., Kenton L., Kristina T. BERT: pre-training of deep bidirectional transformers for language understanding. Proc. 2019 Conf. North Am. Chapt. Assoc. Comput. Linguist.: Hum. Lang. Technol.; Minneapolis, Minnesota; 2019. [Google Scholar]
- Kucera J., Reznicky M. Differentiation of species of Eimeria from the fowl using a computerized image-analysis system. Folia Parasit. 1991;38:107–113. [PubMed] [Google Scholar]
- Lee C.C., Huang P.J., Yeh Y.M., Li P.H., Chiu C.H., Cheng W.H., Tang P. Helminth egg analysis platform (HEAP): an opened platform for microscopic helminth egg identification and quantification based on the integration of deep learning architectures. J. Microbiol. Immunol. 2021;55:395–404. doi: 10.1016/j.jmii.2021.07.014. [DOI] [PubMed] [Google Scholar]
- Lu X.Y., Yang R., Zhou J., Jiao J., Liu F., Liu Y.F., Su B.F., Gu P.W. A hybrid model of ghost-convolution enlightened transformer for effective diagnosis of grape leaf disease and pest. J. King Saud Univ.-Com. 2022;34:1755–1767. [Google Scholar]
- Ma N.N., Zhang X.Y., Zheng H.T., Sun J. ShuffleNet V2: practical guidelines for efficient CNN architecture design. 2018 Eur. Conf. Comput. Vision (ECCV); Munich, Germany; 2018. [Google Scholar]
- Mattiello R., Boviez J.D., McDougald L.R. Eimeria brunetti and Eimeria necatrix in chickens of Argentina and confirmation of seven species of Eimeria. Avian Dis. 2000;44:711–714. [PubMed] [Google Scholar]
- Mesa C., Gomez-Osorio L.M., Lopez-Osorio S., Williams S.M., Chaparro-Gutierrez J.J. Survey of coccidia on commercial broiler farms in Colombia: frequency of Eimeria species, anticoccidial sensitivity, and histopathology. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge D.F., Beltrán C.A. Classification of Eimeria species from digital micrographies using CNNs. 2019 10th Int. Conf. Pattern Recognit. Syst. (ICPRS); Tours, France; 2019. [Google Scholar]
- Mottet A., Tempio G. Global poultry production: current state and future outlook and challenges. Worlds Poult. Sci. J. 2017;73:245–256. [Google Scholar]
- Shirley M.W. Eimeria spp. from the chicken: occurrence, identification and genetics. Acta Vet. Hung. 1997;45:331–347. [PubMed] [Google Scholar]
- Simonyan K., Zisserman A. arXiv:1409.1556. 2024. Very deep convolutional networks for large-scale image recognition. Accessed August 2022. [DOI] [Google Scholar]
- Thevenoux R., Le V.L., Villesseche H., Buisson A., Beurton-Aimar M., Grenier E., Folcher L., Parisey N. Image based species identification of Globodera quarantine nematodes using computer vision and deep learning. Comput. Electron. Agr. 2021;186 [Google Scholar]
- Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser L., Polosukhin I. Attention is all you need. 2017 21st Conf. Neural Inform. Process. Syst. (NIPS); Long Beach, CA, USA; 2017. [Google Scholar]
- Wang J.T., Shen M.X., Liu L.S., Xu Y., Okinda C. Recognition and classification of broiler droppings based on deep convolutional neural network. J. Sensors. 2019;2019 [Google Scholar]
- Yang F., Poostchi M., Yu H., Zhou Z., Silamut K., Yu J., Maude R.J., Jaeger S., Antani S. Deep learning for smartphone-based malaria parasite detection in thick blood smears. IEEE J. Biomed. Health. 2020;24:1427–1438. doi: 10.1109/JBHI.2019.2939121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C.W., Yu Z.W., Kang R., Yousafa K., Qi C., Chen K.J., Huang Y.P. An experimental study of stunned state detection for broiler chickens using an improved convolution neural network algorithm. Comput. Electron. Agric. 2020;170 [Google Scholar]
- Zhao J.J., Peng Y.X., He X.T. Attribute hierarchy based multi-task learning for fine-grained image classification. Neurocomputing. 2020;395:150–159. [Google Scholar]
- Zheng S.X., Lu J.C., Zhao H.S., Zhu X.T., Luo Z.K., Wang Y.B., Fu Y.W., Feng J.F., Xiang T., Torr P.H.S., Zhang L. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. 2021 IEEE/CVF Conf. Comput. Vision Pattern Recognit. (CVPR); Nashville, TN, USA; 2021. [Google Scholar]