Abstract
Background:
Following allogeneic kidney transplantation, a substantial proportion of graft loss is attributed to the formation of donor specific antibodies and antibody-mediated rejection (AMR). B cells infiltrate kidney grafts during AMR; however, the origins, repertoires, and functions of these intrarenal B cells remain elusive.
Methods:
Here we use murine allogeneic kidney transplant models to study the origins, transcriptional programming and B cell receptor (BCR) repertoire of intragraft B cells, and in vitro stimulation assays to evaluate the ability of intragraft B cells to promote CD4+ T cell expansion.
Results:
B cells infiltrate kidney grafts in settings of allogeneic, but not syngeneic, transplantation. Intragraft B cells have characteristics of activation but are transcriptionally distinct from germinal center (GC) B cells and resemble innate-like B cells. BCR sequencing demonstrates that the majority of intragraft B cells do not originate from lymph node GC B cells and are largely germline. Class switched intragraft B cells are rare but can be donor-specific and produce IgG capable of binding to the kidney allograft. Lastly, intrarenal B cells are capable of stimulating naïve T cells but have altered ability to promote T follicular helper (Tfh) expansion.
Conclusions:
Together, these data demonstrate that intrarenal B cells during transplant rejection are transcriptionally distinct from lymph node B cells.
Introduction
Despite progress in understanding kidney transplantation, approximately 3% of kidney transplant patients reject their grafts per year.1 Maintaining long-term survival of kidney grafts after allogeneic transplantation requires a more complete understanding of the processes leading to rejection.2,3 In particular, mechanisms controlling antibody-mediated rejection (AMR) are poorly understood, resulting in a paucity of specific therapeutic treatments.4,5 Most inflammatory and regulatory adaptive immune responses after transplantation have been thought to be generated in secondary lymphoid organs. However, newer paradigms suggest that proinflammatory and regulatory alloimmune responses can also be generated and regulated locally within the graft.6 Early studies have found substantial aggregates of B cells in dysfunctional pediatric kidney grafts,7 and there is growing evidence that B cell infiltrates may have roles in rejection since their presence predicts poor graft survival.8,9 Moreover, B cell depletion mitigates rejection in both mice and humans,10,11 although such studies are confounded by the loss of both pathogenic and regulatory B cells systemically.12
Early studies reported B cell infiltration in the majority of rejected kidney allografts in human patients and that these B cells showed evidence of somatic hypermutation (SHM).13 Beyond the production of allo-specific antibodies, intragraft B cells may contribute to graft rejection by providing/altering cytokines and supporting T cell expansion.11 In support of this, intragraft B cells were observed to form cognate interactions with T follicular helper (Tfh) cells in human biopsies.14 However, the origins and transcriptional programming of graft infiltrating B cells and their roles in controlling AMR remain poorly understood.
To study the origins and transcriptional programming of intrarenal B cells after kidney transplantation, we utilized a preclinical allogeneic kidney transplantation model. Using this model, we found that intragraft B cells are transcriptionally distinct from lymph node B cells and have transcriptional features of innate-like B cells. Using BCR sequencing, we determined that intragraft B cells likely do not originate from lymph node germinal center (GC) B cells and have minimal somatic hypermutation. Using single cell culture systems, we found that although switched intrarenal B cells are infrequent, they can be specific for allo-antigens and secrete antibodies that bind to allogeneic kidneys. Finally, intrarenal B cells had the functional capacity to stimulate T cell expansion but reduced ability to stimulate Tfh cells. Together, these data suggest unique features of intragraft B cells during kidney transplant rejection.
Methods and Materials:
Animals
Balb/c, C57Bl/6 and Ighm−/−(also called uMT) were from Jackson Laboratories. Foxp3IRES-GFP mice on the C57BL/6 background have been published previously.15 All animal studies were performed according to the Brigham and Women’s Hospital Institutional Animal Care and Use Committee and National Institute of Health guidelines.
Kidney Transplantation
Kidney transplantation was performed as previously published.16 In brief, we transplanted donor kidneys into the abdominal cavity of recipient mice. The artery and vein of the donor’s kidney were anastomosed “end-to-side” to the recipient’s abdominal aorta and vena cava. The graft ureter was embedded into the recipient’s bladder. On postoperative days 2–4, the ureter of recipient native kidneys was ligated. The mouse creatinine assay kit (Crystal Chem, USA) was used to assess the graft function.
Histopathology staining
Hematoxylin and eosin (HE) staining was performed based on standard techniques.17 Immunohistochemistry (IHC) and Immunofluorescence (IF) staining were used to detect the expression of B220 (Biolegend, RA3-6B2), CD3(Cell Signaling Technology, D4V8L), C4d (Novus Biologicals, 16D2) and IgG (SouthernBiotech) expression in the graft tissues. The staining process was used as a reference in a previous study.18 Images were taken on a ZEISS Axiolab 5 microscope.
Sorting
Sorting of target populations was performed as described previously.19–21 In brief, the draining lymph nodes and graft tissues digested by collagenase I (Worthington, USA) were smashed and passed through 70-micron filters. Intragraft lymphocytes were separated using ficoll (Thomas Scientific, USA). Cells were stained with anti-CD45 (Biolegend, 30-F11), CD4 (Biolegend, RM4-5), CD19 (Biolegend, 6D5), CD38 (Biolegend, Clone 90), CD138 (Biolegend, 281-2), T-and B-cell activation antigen (GL7, BD Biosciences, GL-7) for 30 minutes at 4 °C and then sorted on an Aria II cell sorter. Total B cells from grafts and lymph nodes were defined as CD45+CD4−CD19+, lymph node GC B cells were defined as CD4−CD19+CD138−CD38lo/−GL-7+, CD4+ T cells were defined as CD45+CD4+CD19−, and Tfh cells were gated as CD4+ICOS+CXCR5+FoxP3−CD19−.
RNA-seq
Bulk RNA-seq was performed as described previously.15 RNA-seq reactions were sequenced on an Illumina NextSeq sequencer (Illumina) utilizing 50-basepair reads. The CLC Genomics Workbench v.8.0.1 RNA-seq analysis software program was used for analysis (Qiagen). RNA-seq data were compared to the innate-B cell gene modules using GSEA analysis software (Broad Institute). The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed on DAIVD website (https://david.ncifcrf.gov/) with the cut-off of p-value were 0.05.
BCR sequencing and clonal assessment
Single cells from lymph node GC B cells (gated as B220+CD45+CD38lo/−GL-7+ CD138− CD4−) and intragraft B cells (gated as CD45+B220+CD4−) were sorted into 96-well PCR plates containing 5 μl/well of TCL lysis buffer (Qiagen) with 1% β-mercaptoethanol for Igh sequencing as previously described.22–24 IMGT HighV-QUEST was used to assign V(D)J rearrangements to Igh sequences. To detect VH mutations, the IMGT and Vbase2 databases were utilized.25 Functional rearrangements were classified as clones if they had the same VH and JH regions, the same CDR-H3 length, and at least 80% similarity in CDR-H3 amino acid sequences.
Flow Cytometry
The following antibodies were used for surface staining at 4°C for 30 minutes: anti-CD45 (Biolegend, 30-F11), anti-CD4 (Biolegend, RM4-5), anti-CD19 (Biolegend, 6D5), T- and B-cell activation antigen (BD Biosciences, GL-7), CD38 (Biolegend, 90), CD138 (Biolegend, 281-2), and anti-CD95 (BD Biosciences, Jo2). Samples were then intracellularly stained with anti-IgG1 (eBiosciences, FJK-16S) after fixation with the Foxp3 Fix/Perm buffer set according to the manufacturer’s instructions (eBioscience). Flow cytometry analysis was performed on a Cytek Aurora spectral analyzer and analyzed with FlowJo v10 (FlowJo LLC).
Donor specific antibody (DSA) measurements
Fresh splenocytes from WT Balb/c or C57Bl/6 mice were plated and incubated with serum (1:50 dilution for IgG, 1:25 dilution for IgM) for 30 minutes on ice, washed, and then stained with anti-CD19 (Biolegend, 6D5) and with FITC-labeled anti-IgG or anti-IgM. Samples were analyzed on a Cytek Aurora flow cytometer. DSA was calculated by measuring the anti-IgG or anti-IgM signal on CD19+ splenocytes which is expressed as a mean fluorescence intensity (MFI).
Single B cell cultures
The single B cell cultures were performed according to previous studies.24 Briefly, individual B cells were sorted from transplanted kidney grafts and cultured with NB21 feeder cells26 for 6 days. Supernatants were screened for IgG positivity by ELISA. IgG positive clones were further screened for DSA reactivity.
In Vitro functional assays
CD4+ T cells, Tfh cells and B cells were isolated from LNs of Foxp3IRES-GFP C57Bl/6 mice challenged with NP-OVA 10 days. After kidney transplantation 20 days, total CD4+ T cells and B cells from grafts were sorted for co-culture. Then, in vitro assays were performed by co-culturing 5×104 B cells with 1.5×104 Tfh or CD4+ T cells and anti-CD3/IgM in 96-well U bottom plates. Cultures were maintained in complete medium for 4 days for CD4+ T-B cells culture and 6 days for Tfh-B cells culture. The cytokines concentration of supernatant was measured by Luminex assay (Cytokine 17-Plex Mouse ProcartaPlex™ Panel, ThermoFisher).
Statistics
To compare the differences across groups, the Student’s 2-tailed unpaired t test was performed using GraphPad Prism, version 9 (GraphPad Software). All tests were 2-sided and P-values less than 0.05 were deemed significant. Values shown in graphs represent the mean standard deviation (SD).
Results
B cells infiltrate grafts during allogeneic kidney transplant rejection
To explore the origins and programming of graft B cells during transplantation, we used an allogeneic kidney transplant model.16 We transplanted a kidney from Balb/c (allogeneic) or C57BL/6 (B6) (syngeneic) mice into B6 recipients followed by ureter occlusion of both native kidneys to ensure a life-sustaining function of the transplanted kidney (Figure 1A). Ureter occlusion was used instead of bilateral nephrectomy to more closely maintain pathological features of rejection and to achieve a more uniform kinetics of rejection (Figure S1). Serum, grafts, and draining lymph nodes (dLNs) of recipients were recovered at day 20 post-transplantation, a time point just before rejection (Figure 1B). Serum creatine levels were substantially increased in the allogeneic transplanted mice, compared to syngeneic transplanted mice (Figure 1C). Moreover, allogeneic kidney recipients had high levels of de novo donor-specific antibodies (dnDSA) for both IgG and IgM isotypes, a feature which was absent in syngeneic transplant recipients (Figure 1D). We also assessed the transplanted kidney grafts histologically to determine the extent of rejection. Compared to untransplanted naïve kidneys, the syngeneic control kidneys showed little signs of inflammatory infiltration and had normal glomerular structure (Figure 1E). In contrast, the allogeneic transplant kidneys displayed infiltration of mononuclear inflammatory cells in the renal interstitium along with variably severe tubular injury and glomerulitis. By immunohistochemistry (IHC), we found substantial CD3 and B220 positive immune cell infiltration in allografts (Figure 1F). The intragraft B cells were mainly located in the renal interstitium, with a small number in the glomeruli and rarely in the renal tubule. We also assessed C4d deposition as an indication of AMR. We found linear C4d sedimentation in peritubular capillaries in the kidneys of allogeneic transplant recipients which was missing from naïve kidneys and kidneys of syngeneic recipients (Figure 1G). These findings suggest that B220+ B cells infiltrate allogeneic kidney allografts undergoing mixed AMR and T-cell mediated rejection (TCMR).
Figure 1. Characterization of rejection after allogeneic kidney transplantation.
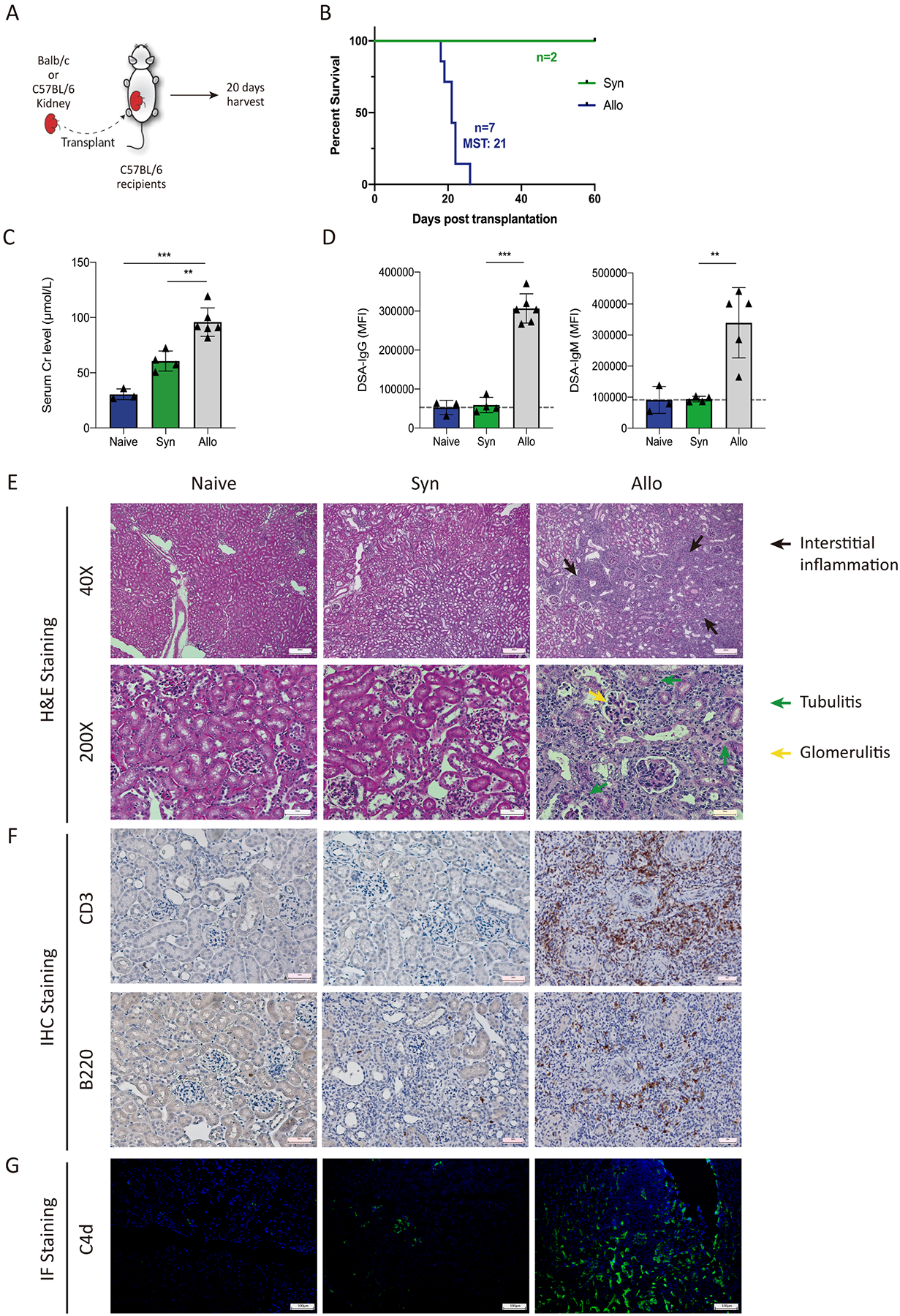
(A) Diagram of the kidney transplantation experiment. C57BL/6 (syngeneic) or Balb/c (allogeneic) kidneys were transplanted into C57BL/6 recipients. The ureters of both native kidneys were ligated to remove native kidney function on day 2–4. 20 days after transplantation recipients were analyzed. (B) Survival of transplanted recipients between the syngeneic and the allogeneic group. MST, median survival time. (C) Serum creatinine levels of recipient mice. (D) DSA IgG and IgM measurements from serum samples. Dotted line indicates background levels using a syngeneic C57BL/6 probe. (E) Representative histological images of transplanted kidneys at postoperative day 20. Hematoxylin and eosin stain. Magnification: upper panels: 40x for renal interstitium observation, scale bars: 200 μm; lower panels: 200X for nephron observation, scale bars: 50 μm. (F) Immunohistochemistry staining for CD3 and B220 in transplanted graft tissues. Magnification: 200×, scale bars: 50 μm. (G) Representative immunofluorescence staining for C4d sedimentation in transplanted grafts. Magnification: 100×, scale bars: 100 μm. Data are from a single experiment and are representative of 3 independent repeats with n=4–5 mice per group. Statistics **: P < 0.01; ***: P < 0.001.
B cells are dispensable for graft CD4 T infiltration after allogeneic kidney transplantation
Since our allogeneic kidney transplant model includes features of both AMR and TCMR, we next tested whether B cells were required for graft inflammation. Ighm−/− (also called “uMT”) mice that have severe deficiencies in peripheral B cells were used.27 These mice were transplanted with allogeneic Balb/c kidneys and kidney grafts were assessed at 20 days after transplantation. The uMT recipients had a lower serum creatine level compared to WT recipients suggesting decreased rejection status (Figure 2A). As expected, uMT recipients had undetectable DSA for IgG and IgM isotypes (Figure 2B). Although uMT mice have reduced B cell populations, small numbers of B cells may persist through use of non-IgM BCRs. Therefore, we next assessed B cell infiltration into grafts by flow cytometry. Absence of B cells was confirmed in dLNs of uMT recipients (Figure 2C). Additionally, we found very few B cells in grafts of uMT recipients. In contrast, there were no substantial differences in CD4+ T cell infiltration in the grafts from uMT and WT recipients (Figure 2D). Substantial rejection, especially intimal arteritis, was seen in both uMT and WT recipients consistent with similar TCMR (Figure 2E). However, there was less substantial peritubular capillaritis and C4d deposition in the uMT group. Moreover, we found the absence of B cells did not result in substantial changes in the infiltration or distribution of CD3+ cells within grafts. These data suggest that B cells may contribute to overall rejection after allogeneic kidney transplantation, but do not substantially control total CD3+ T cell infiltration.
Figure 2. B cells are dispensable for graft CD4 T infiltration after allogeneic kidney transplantation.
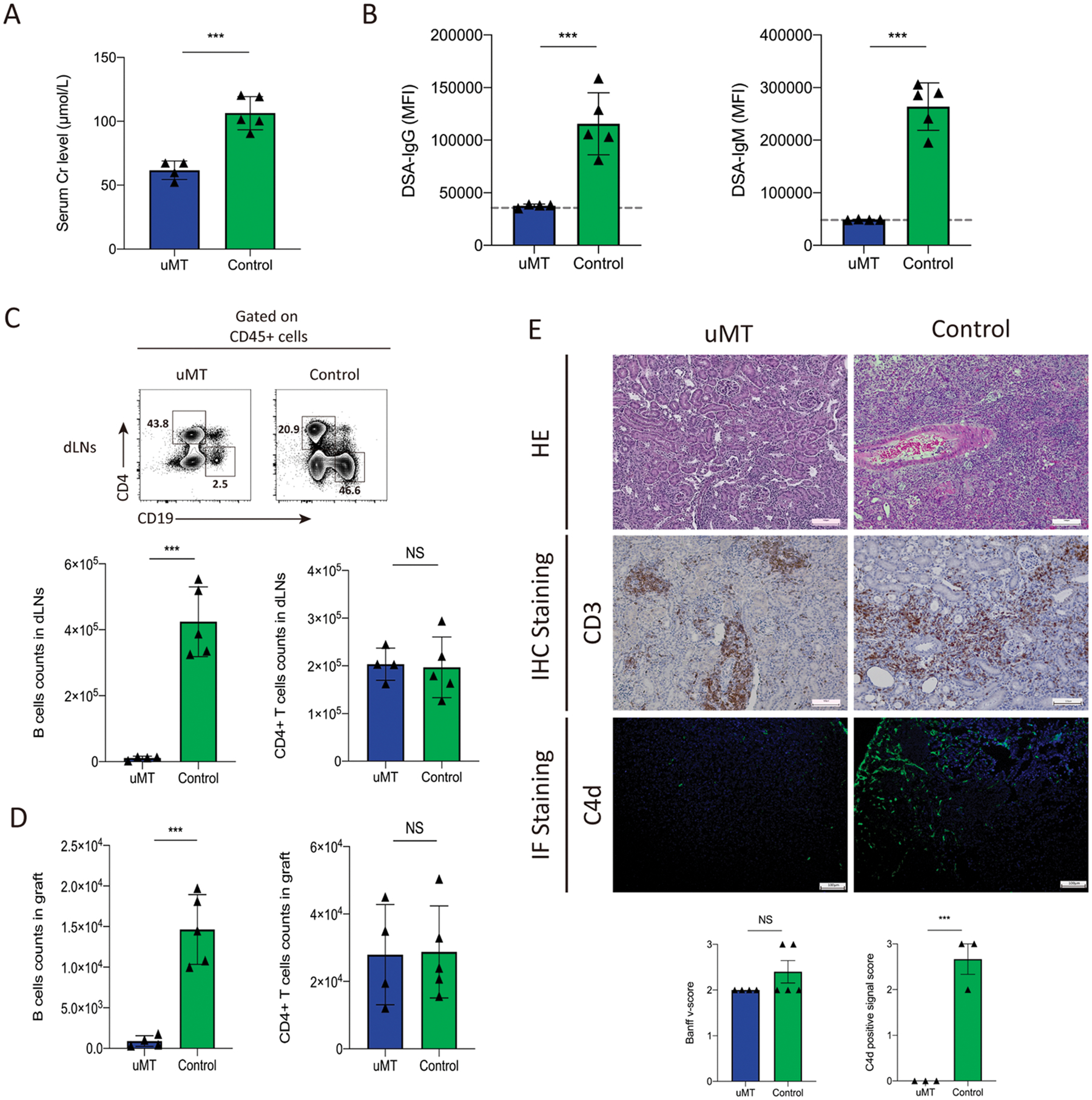
(A) Serum creatinine levels in C57BL/6 WT and uMT recipients of Balb/c kidney transplants at day 20 post-transplantation. (B) DSA measurements for IgG and IgM isotypes expressed as mean fluorescence intensity (MFI). Dotted line indicates the level of detection using a C57BL/6 probe for background. (C, D) Quantification of CD19+ B cells and CD4+ T percentage in draining lymph nodes and count in the grafts by flow cytometry. Representative gating (upper) and quantification (low) are shown. (E) Histological images of transplanted kidneys from HE staining, immunohistochemistry (IHC) staining for CD3 expression and immunofluorescent (IF) staining for C4d expression. Below: is histopathological scoring of intimal arteritis based on the Banff 2017 and the C4d positive signal score (0, −; 1, +; 2, ++; 3, +++). Magnification: 100×, scale bars: 100 μm. Data are from combined experiments with n=4–5 mice per group. Statistics: Student’s 2-tailed unpaired T test, **: P < 0.01; ***: P < 0.001.
Intrarenal B cells have a distinct innate-like transcriptional signature during allogeneic kidney rejection
Recent studies in humans have shown that intrarenal B cells at the time of rejection have features of innate-like B cells.28 Therefore, we next studied the phenotype and transcriptional program of intragraft B cells. Allografts from allogeneic recipients had substantially higher numbers of total lymphocytes, including CD4+ T cells and B cells, compared to grafts from syngeneic recipients (Figure 3A). Although B cells were numerous in the grafts of allogeneic recipients, they comprised only ~10% of all lymphocytes (Figure 3B). Interestingly, when B cells were assessed for a GL7+FAS+ GC-like phenotype we found ~15% of B cells infiltrating the allograft expressed GL7 and FAS, compared to ~5% in the dLN of allogeneic recipients (Figure 3C). Importantly, the frequency of GL7+FAS+ B cells was lower in the dLN of syngeneic recipients compared to allograft recipients, however plasma-like cells (defined as CD138+ cells) were infrequent (<1%) and similar in both groups (Figure 3D). Naive B cells, gated as CD19+CD38+IgG1− B cells were lower in frequency in graft B cells compared to LN B cells (Figure 3E). Class switched B cells (defined as CD19+IgG1+CD38− cells) and memory-like B cells (defined IgG1+CD38+ cells) were present in a higher frequency of B cells in the allograft. These data suggest that a fraction of intragraft B cells have some evidence of either ongoing or previous activation.
Figure 3. Intrarenal B cells have a distinct phenotype and transcriptional profile.
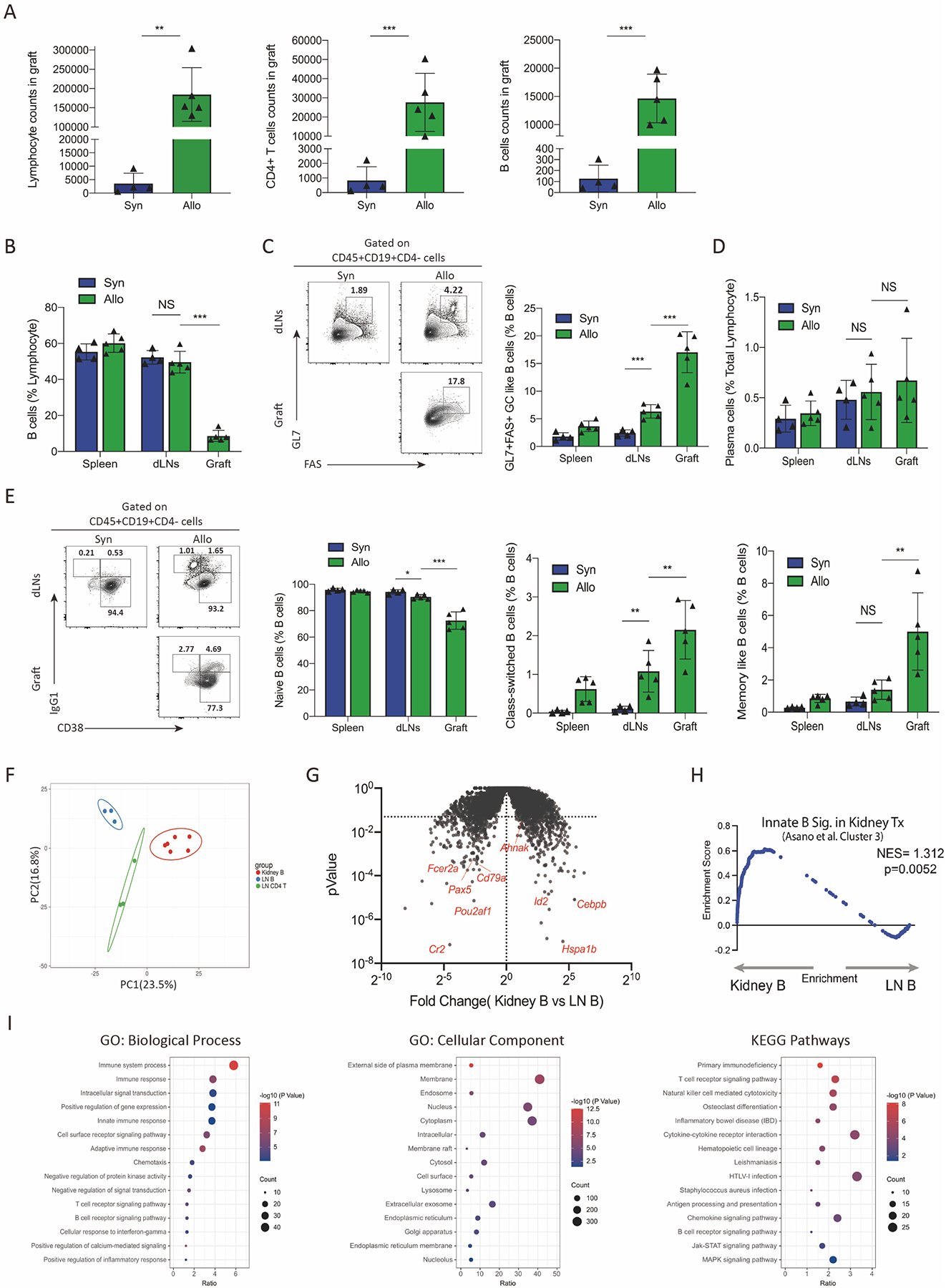
(A) Assessment of total lymphocyte count, CD45+CD4+ T cells count and CD45+CD19+ B cells count in the kidney grafts by flow cytometry. (B) Percentage of CD19+ B cells of total lymphocytes in the spleen, draining lymph nodes and grafts tissues among syngeneic and allogeneic recipients. (C) Gating strategy for GC-like B cells (gated as CD45+CD19+GL7+FAS+CD4−) and the percentage of GC-like B cells out of all CD19+B cells from different tissues. (D) Percentage of plasma cells in total lymphocytes (gated as CD45+CD138+) from spleens, dLNs and grafts. (E) Analysis of Naïve B cells (gated as CD45+CD19+CD38−IgG1−CD4−), class-switched B cells (gated as CD45+CD19+CD38−IgG1+CD4−), and memory-like B cells (CD45+CD19+CD38+IgG1+CD4−) in B cells. Representative gating (left) and quantification (right) are shown. (F) Principal component analysis (PCA) showing the relationship between transcriptional profiles of intrarenal B cells (Kidney B), dLNs B cells (LN B) and dLNs CD4+T cells (LN CD4 T) in allogeneic kidney recipients. (G) Volcano plot showing differentially expressed genes (DEGs) between intrarenal B cells and dLNs B cells. Genes with P value <0.05 were considered significant. (H) Gene set enrichment analysis (GSEA) comparing intrarenal B cells in kidney grafts to an innate-like B cell gene module (Cluster 3 from Asano et al.) (I) Enrichment of GO terms and KEGG pathways of the DEGs between intrarenal B cells and dLNs B cells. Data are from a single experiment and are representative of 3 independent repeats with n=4–5 mice per group (A-E). RNA-seq data are from combined experiments (F-I). Column graphs represent the mean with error bars indicating standard error. P value indicates 2-tailed student’s T test. NS: not significant; **: P < 0.01; ***: P < 0.001.
To understand intragraft B cells in more detail we performed bulk RNAseq transcriptional analyses. We sorted CD45+CD4+CD19− T cells (LN CD4 T) and CD45+CD19+CD4− B cells (LN B) from dLNs as well as CD45+CD19+CD4− B cells (Kidney B) from the kidney graft of allogeneic recipients 20 days after transplantation. By principal component analysis (PCA), intrarenal B cells were phenotypically distinct from both LN B and T cells (Figure 3F). We further analyzed the transcriptional differences and found 850 differentially expressed genes (DEGs)(p<0.05), 486 of which were more highly expressed in intragraft B and 364 of which were more highly expressed in lymph node B cells (Figure 3G). Genes more highly expressed in lymph node B versus intragraft B cells included Cr2, Cd79a, Pou2af1, and Pax5. Genes more highly expressed in intragraft B cells versus lymph node B cells included Ahnak, Id2, and Hspa1b. The elevated expression of Ahnak was of particular interest since a recent report suggested this gene is highly upregulated on intragraft B cells in the setting of human kidney transplantation and may mark innate-like B cells.28 To determine if intragraft B cells have features of innate-like B cells, we performed gene set enrichment analysis (GSEA) using gene signature of “Cluster 3” from Asano et al, which corresponds to the cluster with the highest innate-like B cell signature. We found that intragraft B cells (Kidney B) had a significantly increased enrichment for the innate-like gene module compared to dLNs B cells (LN B) (Figure 3H). We performed additional enrichment analysis based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Figure 3I). We found enrichment for innate immune response and cell receptor signaling pathways. Enrichment analysis at the level of cellular component (CC) showed these DEGs mainly located on membrane, nucleus, and cytoplasm. KEGG enrichment analysis revealed cytokine-cytokine receptor interaction, B cell receptor and Jak-STAT signaling pathways. Together, these data suggest that intragraft B cells during kidney transplantation have some features of activation and antigen experience but are transcriptionally distinct from LN B cells and share features with innate-like B cells.
Graft infiltrating B cells do not originate from lymph node germinal center B cells but can secrete donor specific antibody
Somatic hypermutation (SHM) and affinity maturation typically occur within GCs of secondary lymphoid organs. We next determined whether intragraft B cells originate from a lymph node GC B cell population. We sorted individual B cells and performed BCR sequencing on CD19+CD45+ intragraft B cells (Graft B) and draining lymph node GL7+CD38- GC B cells (LN GC B) on day 20 after transplantation (Figure 4A). In this analysis we included paired LN and graft B cells from 2 separate recipients and also included additional intragraft B cells from other 2 recipients (Figure 4B). We found that LN GC B cells had a wide distribution of immunoglobulin isotypes and only ~25% of B cells were unswitched, with the most prominent switched isotype being IgG1 (~40%) followed by IgG2b (~20%) (Figure 4C). In contrast, 98.3% of intragraft B cells were unswitched. When we assessed the total number of mutations in VH segments, we found LN GC B cells had a high number of mutations with very few germline sequences (Figure 4D). Next, we assessed clonal expansion and evolution by grouping sequences that likely originated from the same founder clone (defined as same VH and JH segment usage, same CDRH-3 length and at least 80% amino acid identity). We found a moderate amount of clonal expansion in LN GC B cells (43 unique clones out of 61 sequences) but very little clonal expansion in intragraft B cells (109 unique clones out of 119 sequences) (Figure 4E). Almost all expanded clones in the LN GC B compartment had evidence of SHM (Figure 4F–G). Interestingly, we found clones (IGHV1-53/IGHJ3 and IGHV1-81/IGHJ4) with mutated unswitched (IgM) and mutated switched sequences from the same recipient suggesting that some class switch recombination may occur in GCs after initial SHM. We also found the same germline unswitched clone (IGHV1-81/IGHJ3) in 2 different recipients suggesting a public clonotype. The expanded clones for intragraft B cells were almost all unswitched and germline with the exception of an expanded IgM clone (IGHV5-17/IGHJ2) with 1 mutation (Figure 4G). Interestingly, both of the nonexpanded but switched sequences we found in intragraft B cells had evidence of somatic hypermutation with 1 IgG1 sequence having 5 mutations and 1 IgG2c sequence having 9 mutations. We did not find any shared clones between the LN GC B cells and intragraft B cells, with 2 exceptions. In 2 cases (IGHV3-6/IGHJ2 and IGHV1-59/IGHJ3) we found evidence of B cells from LN GC and intragraft compartments that met the threshold for clonality. However, since in both cases the LN and intragraft B cells were from different recipients, these did not originate from the same founder B cell. Together these data indicate that although most intragraft B cells are unswitched and germline, the few switched B cells have evidence of SHM.
Figure 4. Switched graft infiltrating B cells are rare but can be alloreactive.
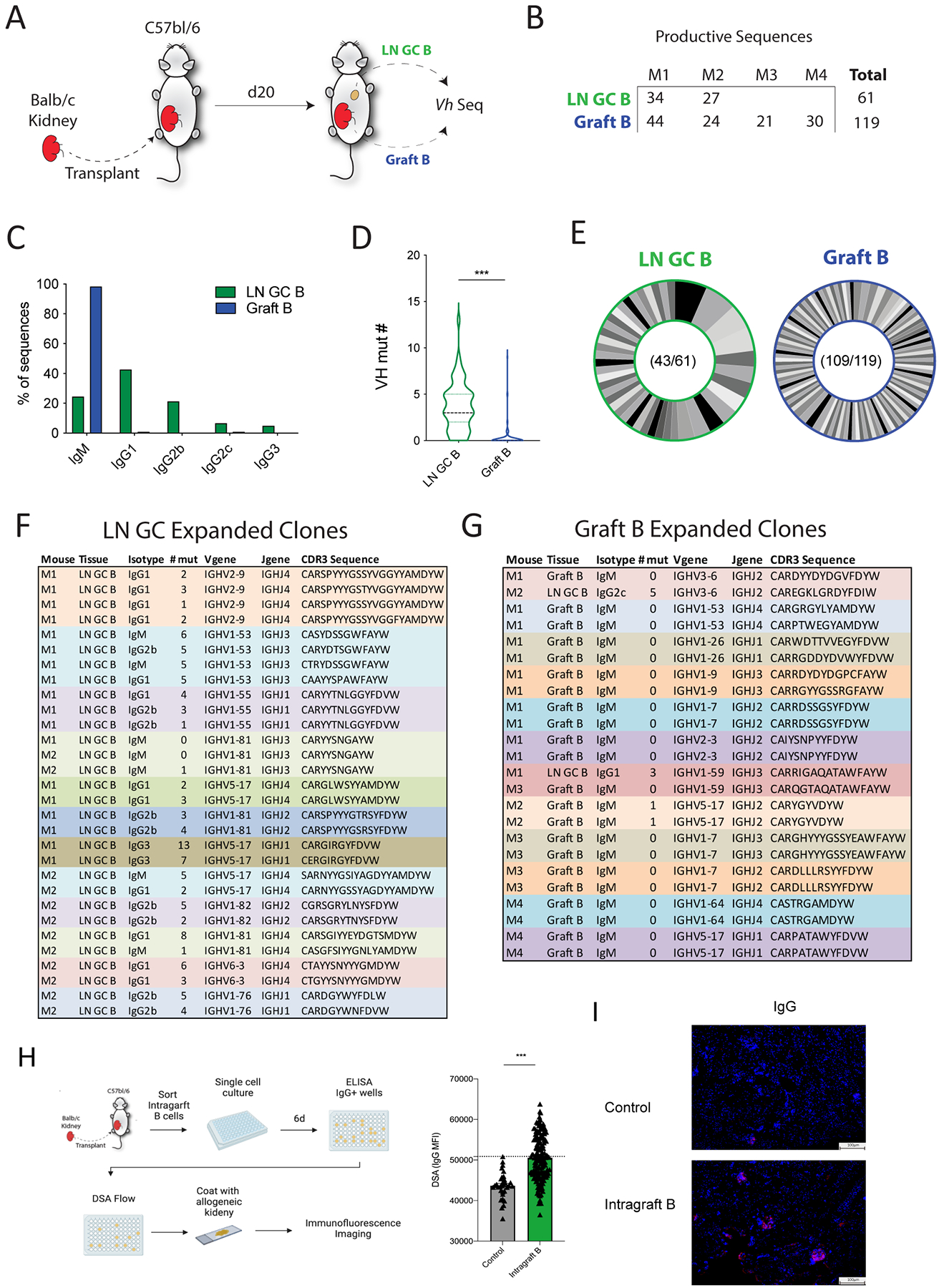
(A) Diagram of the BCR sequencing experiment. LN GC B cells (LN GC B) and intragraft B cells (Graft B) were sorted from allogeneic kidney transplant recipients on day 20 after transplantation and BCR sequencing performed. (B) Number of LN GC B cells and Graft B sequences that passed quality control cutoffs. M1: mouse 1, M2: mouse 2, M3: mouse 3, M4: mouse 4. (C) Distribution of sequences for indicated immunoglobulin isotype. (D) Somatic hypermutation analysis for V-heavy (VH) segment in LN GC B and Graft B cells. (E) Analysis of clonal distribution in B cells. Each slice indicates a unique clone. Numbers indicate the total number of clones and the number of sequences analyzed. (F, G) Details of expanded clones (sequences found 2 or more times) in dLNs GC B cells (F) and intragraft B cells (G). (H) Schematic of single B cell culture assays. Single intragraft B cells from B6 mice transplanted with a Balb/c kidney 20 days prior were cultured for 6 days with NB21 feeder cells. Culture supernatants were screened for IgG positivity, and DSA was measured in IgG switched clones. (I) Immunofluorescence staining of naïve Balb/c kidneys with pooled top 10 alloreactive clones from (H). Positive signal: red; DAPI: blue. Magnification: 100×, scale bars: 100 μm. Data are from a single experiment and are representative of 3 independent repeats Student’s 2-tailed unpaired T test, ***p<0.001.
Next we explored the specificity of intragraft B cells. We used a single B cell culture system in which individual B cells were cultured with NB21 feeder cells allowing for B cell expansion and secretion of antibody into the culture supernatant (without class switching) which can be assessed for specificity.24,26 Since each culture is derived from a single B cell, it represents an individual clone. We sorted GL7+ B cells from kidney grafts and performed single B cells culture assays (Figure 4H). We first screened cultures for IgG+ clones because these would be predicted to be the most pathogenic. The IgG+ clones were assessed for donor reactivity by using a threshold based on both blank wells and IgG-negative clones. We found that 37.8% of intragraft IgG B cell clones had detectable reactivity for alloantigens. To confirm that the DSA produced by these clones were capable of binding to alloantigens, we pooled the supernatant from the highest 10 DSA positive intragraft B cell clones and used them to stain allogeneic kidneys (from naïve Balb/c mice). We found that the pooled antibody from DSA specific intragraft clones was able to bind to kidney alloantigens (Figure 4I). Taken together, these data suggest B cells undergo substantial SHM and clonal evolution in lymph node GCs during AMR. In contrast, graft B cells are mostly unswitched, however when switched B cells occur, a high proportion exhibit alloreactivity.
Intrarenal B cells have diminished ability to expand T follicular helper cells
The altered transcriptional program in intragraft B cells suggests unique functionality. Therefore, we next determined the ability of intrarenal B cells to promote CD4+ T cell expansion compared to lymph node B cells. To this end, we used an in vitro CD4 and B cell stimulation assay. Lymph node CD4+ T cells (LN T) and CD19+ B (LN B) were sorted from NP-OVA immunized mice, and then co-cultured with intrarenal B cells (KD B) or T cells (KD T) for 4 days in the presence of polyclonal activators (Figure 5A). We found that both intrarenal B cells and LN B cells can promote LN CD4+ T cell expansion (Figure 5B). Interestingly, intrarenal B cells promoted kidney CD4+ T cell expansion less well compared to LN B cells. We also assessed GL7 expression on B cells but found no substantial differences in the ability of LN T cells to promote GL7 expression on kidney or LN B cells (Figure 5C), however, kidney CD4+ T cells promoted less GL7 expression on B cells from both LN and kidney.
Figure 5. Intrarenal B cells have diminished ability to expand Tfh cells.
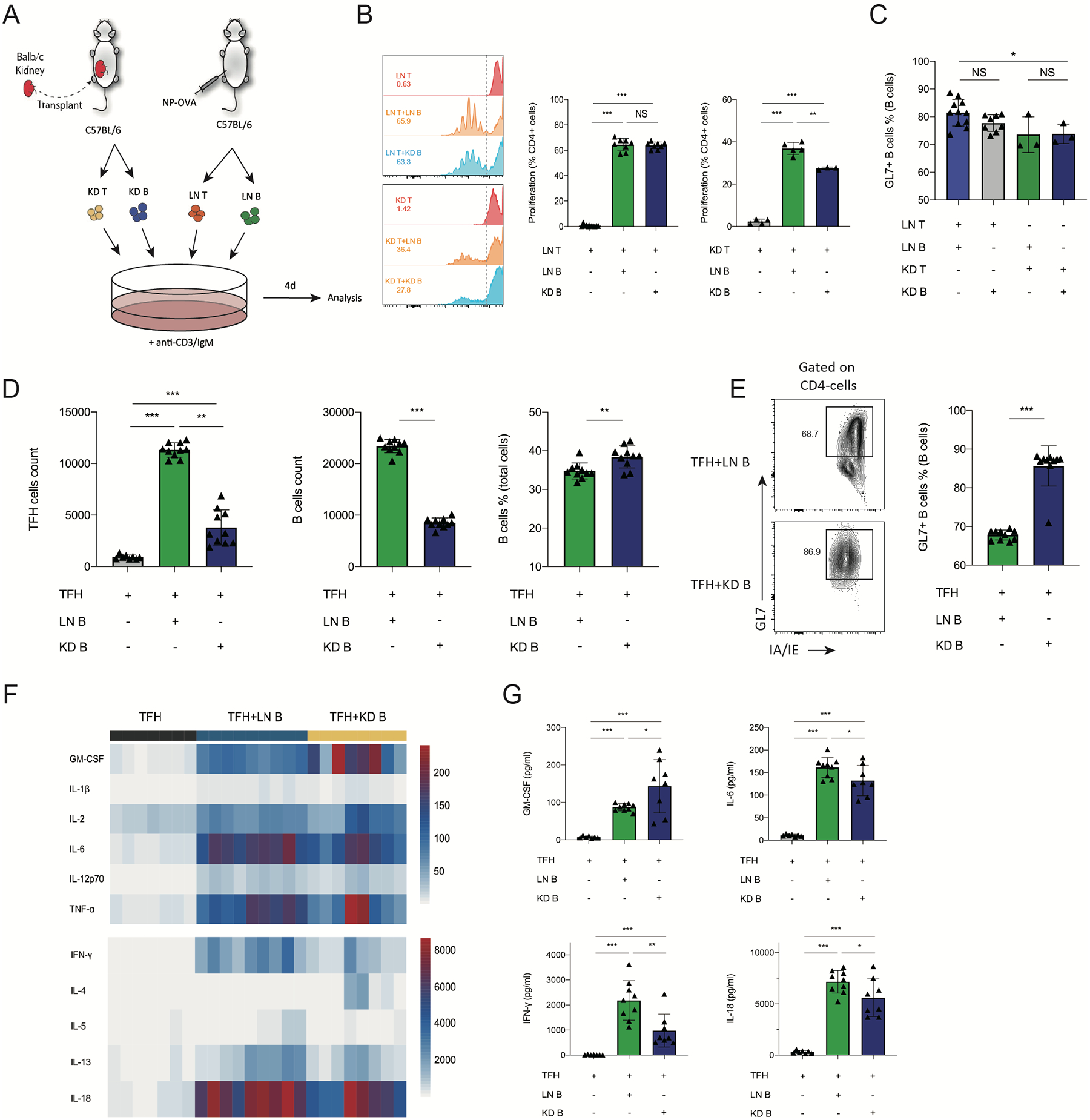
(A) Schematic of in vitro T-B cell stimulation assay. CD4+ T cells (LN T) and CD19+ B (LN B) cells from NP-OVA immunized mice were cultured with intrarenal B (KD B) or CD4+T cells for 4 days. (B) Quantification of T cells proliferation. T cells from LNs and grafts were stained with CellTrace Violet, the peaks represent successive generations of cells. The right side of the dashed line represents nonproliferating cells. Percentage of proliferated cells in total T cells were calculated. (C) Quantification of GL7+IA/IE+ B cells. (D) CD4+ICOS+CXCR5+FoxP3–CD19– cells (TFH) and CD19+CD4− B (LN B) cells from NP-OVA immunized mice were cultured with intrarenal B (CD45+CD19+CD4−) cells (KD B) for 6 days. Quantification of Tfh (CD4+IA/IE−) count, B cells (GL7+IA/IE+) percentage and count in the Tfh-B cells stimulation assays. (E) Quantification of GL7+ B cells. Representative plots (left) and quantification (right) are shown. (F) Heatmap of cytokine concentration of supernatant in Tfh-B cells culture by luminex analysis. (G) The concentration of cytokines GM-CSF, IL-6, IFN-γ, IL-18 in the supernatant in co-culture of Tfh and B cells. Data are from a single experiment and are representative of 3 independent repeats. NS: not significant; **: P < 0.05; **: P < 0.01; ***: P < 0.001.
In our previous research, we found that follicular T cells control B cell effector responses during AMR after kidney transplantation,16 but whether the intragraft B cells can expand follicular T helper cells (Tfh) is poorly understood. Therefore, we next performed a Tfh-mediated B cell stimulation assay in vitro. For this, Foxp3IRES−GFP mice were immunized with NP-OVA and total Tfh (CD4+ICOS+CXCR5+Foxp3−) and B (CD19+CD4−) cells were sorted from LNs after 10 days. Tfh cells were cultured for 6 days along with the intragraft B cells (KD B) sorted from kidney transplant recipients at 20 days postsurgery.
Intrarenal B cells cultured with Tfh cells resulted in a ~60% reduction in Tfh cell expansion compared to LN B cells cultured with Tfh cells (Figure 5D). Moreover, intragraft B cells expanded to a lesser degree compared to lymph node B cells in these assays. However, the expression of the GC activation marker GL7 was higher in intragraft B cells at the end of these assays suggesting B cells do become activated (Figure 5E). In addition, we assessed cytokines typically produced by Tfh cells in these assays. Consistent with LN B cells, intrarenal B cells led to similar stimulation of many cytokines (Figure 5F). However, the concentrations of IL-6, IL-18 and IFN-γ in KD B wells were somewhat lower than LN B cells (Figure 5G). Only 1 cytokine was higher in the intrarenal B cell condition which was GM-CSF. Together, these data suggest intrarenal B cells have the stimulatory capacity to expand CD4+ T cells, but the ability to stimulate Tfh cells is reduced compared to lymph node B cells. Therefore, intrarenal B cells are phenotypically and transcriptionally distinct from lymph node B cell subsets.
Discussion
Although broad immunosuppression has helped to limit graft rejection, AMR is still a significant clinical problem.29,30 Here, we explored the transcriptional program, origins and specificity of graft infiltrating B cells after life-sustaining allogeneic kidney transplantation. To render transplantation life-sustaining we used a ureter obstruction method on native kidneys.16 We assessed the graft infiltrating B cells at a time point just prior to rejection to allow for full clonal expansion and differentiation.16 This transplantation model includes features of AMR, characterized by the presence of circulating DSA and C4d deposition, along with histological signs of acute tissue damage such as glomerulitis and peritubular capillaritis, so TCMR likely plays a role.31 In our model, B cell infiltration into kidney allografts was similar in magnitude to previous kidney transplant studies.8,32–35 In uMT recipients, which lack most B cells, we found decreased rejection (as determined by serum creatinine) without alterations in T cell infiltration. Collectively, these data suggest a mixture of AMR and TCMR in our kidney transplant model and that B cells contribute to the rejection process, although probably not by recruiting and/or expanding donor-specific T cells.
We found that lymph node GC B cells showed substantial evidence of clonal expansion with most expanded clones having substantial amounts of SHM regardless of class switching. While a relatively high proportion of intrarenal B cells had a GC B cell-like phenotype as assessed by expression of GL7, were mostly unswitched and were clonally and transcriptionally distinct. Furthermore, rare switched graft B cells had evidence of SHM and moderate alloreactivity. These data suggest that intragraft B cells may contribute to rejection by secreting alloreactive antibody locally. Interestingly, we found that the gene Ahnak was highly expressed in intragraft B cells, similar to a previous study in humans,28 and also observed an enrichment of an innate-like B cell signature from the human study in our transcriptional dataset.
There is mounting evidence that B cells can perform antibody-independent functions such as antigen presentation in the setting of organ transplantation.33 Zarkhin et al. found B cells colocalized with CD4+ T cells by immunohistochemical (IHC) staining in human graft samples, but little is known about the specific functions of B cells in kidney grafts.35 We previously showed that Tfh cells promote DSA to mediate AMR in kidney transplantation.16 At the level of the kidney, Liarski et al. used novel computer-assisted cell distance mapping to suggest Tfh and B cells have cognate interactions within human renal biopsies.14 By IHC staining we also show similar localization of CD3+ T cell and B cell clusters suggesting interactions. Using in vitro functional experiments, we found that intrarenal B cells had similar stimulatory capacity to promote total CD4 T cell activation and expansion. However, we found that intrarenal B cells had modest defects in the ability to stimulate Tfh responses. Interestingly, the cytokine profile expressed by Tfh cells was distinct during culture with intrarenal B cells compared to lymph node B cells with reduced production of IFN-g, IL-6, and IL18, but increased GM-CSF. The increase in GM-CSF may point to functions in altering M2 macrophages. It is also possible that the intrarenal B cells may polarize Tfh cells to a distinct Tfh subset that has specialized roles in the kidney. Since innate B cells have a capacity for cognate help to T cells this may be a general feature of innate-like B cells or may be specific to intrarenal B cells.36
Taken together, our results suggest that the intragraft B cells are unique with innate-like immune features, and are mostly unswitched and minimally clonally expanded, which is distinct from the B cells in lymph nodes. However, a small population of intragraft B cells that are switched are enriched for alloreactivity. Therefore, intragraft B cells may have roles in controlling rejection by secreting antibody, promoting T cell differentiation/expansion and by having innate-like functions. Future studies to understand these cells in more detail may provide a rationale for treating kidney transplant rejection by targeting B cells.
Supplementary Material
Funding:
This work was supported by the National Institute of Health through grants P01AI056299 and R01AI153124.
Abbreviations:
- AMR
antibody-mediated rejection
- Tfh
T follicular helper
- SHM
somatic hypermutation
- GC
germinal center
- dLN
draining lymph nodes
- DSA
donor-specific antibodies
- IHC
immunohistochemistry
- IF
immunofluorescence
- TCMR
T-cell mediated rejection
- GSEA
gene set enrichment analysis
Footnotes
Disclosure:
The authors declare no conflicts of interest.
References:
- 1.Lamb KE, Lodhi S, Meier-Kriesche HU. Long-term renal allograft survival in the United States: a critical reappraisal. Am J Transplant. 2011;11(3): 450–462. [DOI] [PubMed] [Google Scholar]
- 2.Opelz G, Dohler B, Collaborative Transplant Study R. Influence of time of rejection on long-term graft survival in renal transplantation. Transplantation. 2008;85(5): 661–666. [DOI] [PubMed] [Google Scholar]
- 3.Wojciechowski D, Wiseman A. Long-Term Immunosuppression Management: Opportunities and Uncertainties. Clin J Am Soc Nephrol. 2021;16(8): 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong AS, Rothstein DM, Safa K, et al. Outstanding questions in transplantation: B cells, alloantibodies, and humoral rejection. Am J Transplant. 2019;19(8): 2155–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan P, Li H, Han M, et al. PSMP Is Discriminative for Chronic Active Antibody-Mediated Rejection and Associate With Intimal Arteritis in Kidney Transplantation. Front Immunol. 2021;12: 661911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsiao HM, Li W, Gelman AE, et al. The Role of Lymphoid Neogenesis in Allografts. Am J Transplant. 2016;16(4): 1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349(2): 125–138. [DOI] [PubMed] [Google Scholar]
- 8.Hippen BE, DeMattos A, Cook WJ, et al. Association of CD20+ infiltrates with poorer clinical outcomes in acute cellular rejection of renal allografts. Am J Transplant. 2005;5(9): 2248–2252. [DOI] [PubMed] [Google Scholar]
- 9.Alsughayyir J, Pettigrew GJ, Motallebzadeh R. Spoiling for a Fight: B Lymphocytes As Initiator and Effector Populations within Tertiary Lymphoid Organs in Autoimmunity and Transplantation. Front Immunol. 2017;8: 1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinmetz OM, Lange-Husken F, Turner JE, et al. Rituximab removes intrarenal B cell clusters in patients with renal vascular allograft rejection. Transplantation. 2007;84(7): 842–850. [DOI] [PubMed] [Google Scholar]
- 11.Tse GH, Johnston CJ, Kluth D, et al. Intrarenal B Cell Cytokines Promote Transplant Fibrosis and Tubular Atrophy. Am J Transplant. 2015;15(12): 3067–3080. [DOI] [PubMed] [Google Scholar]
- 12.Clatworthy MR, Watson CJ, Plotnek G, et al. B-cell-depleting induction therapy and acute cellular rejection. N Engl J Med. 2009;360(25): 2683–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferdman J, Porcheray F, Gao B, et al. Expansion and somatic hypermutation of B-cell clones in rejected human kidney grafts. Transplantation. 2014;98(7): 766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liarski VM, Kaverina N, Chang A, et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci Transl Med. 2014;6(230): 230ra246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clement RL, Daccache J, Mohammed MT, et al. Follicular regulatory T cells control humoral and allergic immunity by restraining early B cell responses. Nat Immunol. 2019;20(10): 1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammed MT, Cai S, Hanson BL, et al. Follicular T cells mediate donor-specific antibody and rejection after solid organ transplantation. Am J Transplant. 2021;21(5): 1893–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R, Sage PT, Finn K, et al. B Cells Drive Autoimmunity in Mice with CD28-Deficient Regulatory T Cells. J Immunol. 2017;199(12): 3972–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Wang Z, Zhang J, et al. Combined Immunotherapy With Belatacept and BTLA Overexpression Attenuates Acute Rejection Following Kidney Transplantation. Front Immunol. 2021;12: 618737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sage PT, Sharpe AH. In vitro assay to sensitively measure T(FR) suppressive capacity and T(FH) stimulation of B cell responses. Methods Mol Biol. 2015;1291: 151–160. [DOI] [PubMed] [Google Scholar]
- 20.Sage PT, Francisco LM, Carman CV, et al. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol. 2013;14(2): 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou S, Clement RL, Diallo A, et al. FoxP3 and Ezh2 regulate Tfr cell suppressive function and transcriptional program. J Exp Med. 2019;216(3):605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue): W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mesin L, Schiepers A, Ersching J, et al. Restricted Clonality and Limited Germinal Center Reentry Characterize Memory B Cell Reactivation by Boosting. Cell. 2020;180(1): 92–106.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavazzoni CB, Hanson BL, Podesta MA, et al. Follicular T cells optimize the germinal center response to SARS-CoV-2 protein vaccination in mice. Cell Rep. 2022;38(8): 110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Retter I, Althaus HH, Munch R, et al. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005;33(Database issue): D671–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuraoka M, Schmidt AG, Nojima T, et al. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity. 2016;44(3): 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suah AN, Tran DV, Khiew SH, et al. Pregnancy-induced humoral sensitization overrides T cell tolerance to fetus-matched allografts in mice. J Clin Invest. 2021;131(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asano Y, Daccache J, Jain D, et al. Innate-like self-reactive B cells infiltrate human renal allografts during transplant rejection. Nat Commun. 2021;12(1): 4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chong AS. B cells as antigen-presenting cells in transplantation rejection and tolerance. Cell Immunol. 2020;349: 104061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chong AS. Mechanisms of organ transplant injury mediated by B cells and antibodies: Implications for antibody-mediated rejection. Am J Transplant. 2020;20 Suppl 4: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardinal H, Dieude M, Brassard N, et al. Antiperlecan antibodies are novel accelerators of immune-mediated vascular injury. Am J Transplant. 2013;13(4): 861–874. [DOI] [PubMed] [Google Scholar]
- 32.Tsai EW, Rianthavorn P, Gjertson DW, et al. CD20+ lymphocytes in renal allografts are associated with poor graft survival in pediatric patients. Transplantation. 2006;82(12): 1769–1773. [DOI] [PubMed] [Google Scholar]
- 33.Heidt S, Vergunst M, Anholts JDH, et al. Presence of intragraft B cells during acute renal allograft rejection is accompanied by changes in peripheral blood B cell subsets. Clin Exp Immunol. 2019;196(3): 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiqiu W, Jinsong C, Dongrui C, et al. CD20+ B-cell infiltration is related to the time after transplant and poor prognosis of acute cellular rejection in renal transplant. Exp Clin Transplant. 2013;11(5): 412–417. [DOI] [PubMed] [Google Scholar]
- 35.Zarkhin V, Kambham N, Li L, et al. Characterization of intra-graft B cells during renal allograft rejection. Kidney Int. 2008;74(5): 664–673. [DOI] [PubMed] [Google Scholar]
- 36.Grasseau A, Boudigou M, Le Pottier L, et al. Innate B Cells: the Archetype of Protective Immune Cells. Clin Rev Allergy Immunol. 2020;58(1): 92–106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


