Abstract
Background
Remdesivir is an antiviral drug approved for the treatment of COVID‐19, whose developmental toxicity remains unclear. More information about the safety of remdesivir is urgently needed for people of childbearing potential, who are affected by the ongoing pandemic. Morphogenetic embryoid bodies (MEBs) are three‐dimensional (3D) aggregates of pluripotent stem cells that recapitulate embryonic body patterning in vitro, and have been used as effective embryo models to detect the developmental toxicity of chemical exposures specifically and sensitively.
Methods
MEBs were generated from mouse P19C5 and human H9 pluripotent stem cells, and used to examine the effects of remdesivir. The morphological effects were assessed by analyzing the morphometric parameters of MEBs after exposure to varying concentrations of remdesivir. The molecular impact of remdesivir was evaluated by measuring the transcript levels of developmental regulator genes.
Results
The mouse MEB morphogenesis was impaired by remdesivir at 1–8 μM. Remdesivir affected MEBs in a manner dependent on metabolic conversion, and its potency was higher than GS‐441524 and GS‐621763, presumptive anti‐COVID‐19 drugs that act similarly to remdesivir. The expressions of developmental regulator genes, particularly those involved in axial and somite patterning, were dysregulated by remdesivir. The early stage of MEB development was more vulnerable to remdesivir exposure than the later stage. The morphogenesis and gene expression profiles of human MEBs were also impaired by remdesivir at 1–8 μM.
Conclusions
Remdesivir impaired mouse and human MEBs at concentrations that are comparable to the therapeutic plasma levels in humans, urging further investigation into the potential impact of remdesivir on developing embryos.
Keywords: COVID‐19, developmental toxicity, embryo, gastruloid, pluripotent stem cell, pregnancy, remdesivir
1. INTRODUCTION
Remdesivir (RDV) is the first antiviral drug approved by the U.S. Food and Drug Administration (FDA) for the treatment of coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). RDV is administered intravenously and is converted within cells into the active nucleoside triphosphate form, which inhibits the RNA‐dependent RNA polymerase (RdRp) of RNA viruses, including SARS‐CoV‐2 (Eastman et al., 2020; Kokic et al., 2021). As RdRp is responsible for the replication and transcription of the viral RNA genome, its inhibition is considered the major mechanism of the antiviral action of RDV. The efficacy of RDV to reduce SARS‐CoV‐2 proliferation has been demonstrated in cultured cells, model animals, and humans (Beigel et al., 2020; Goldman et al., 2020; Lin et al., 2021; Pagliano et al., 2022). Regarding the safety of RDV, hepatotoxicity and nephrotoxicity have been implicated in human studies (Gérard et al., 2021; Satsangi, Gupta, & Kodan, 2021). However, the developmental toxicity of RDV, namely the adverse impact on embryos, is still unclear (Budi et al., 2022; Gutierrez et al., 2022; Marzban‐Rad, Ghafarzadeh, Bahmani, & Kazemi, 2022; Saroyo, Rumondang, Febriana, Harzif, & Irwinda, 2021). Due to the ongoing COVID‐19 pandemic, many people, including those who are pregnant or may become pregnant, will continue to be affected. Notably, COVID‐19 appears to affect pregnant people more severely than nonpregnant people (Allotey et al., 2020; Murison, Grima, Simmons, Tuite, & Fisman, 2022; Wang et al., 2022), which underscores the importance of proper treatments, including antivirals, during pregnancy. Thus, it is particularly important to gather more information on the developmental toxicity of RDV through various investigative approaches. Currently, there are registries to monitor pregnancy outcomes in individuals exposed to RDV (FDA, 2022; United Kingdom Teratology Information Service [UKTIS], 2022), which are vital in order to conduct human epidemiologic studies. Animal tests and in vitro assays are also valuable, as they are more amenable for controlled experiments to interrogate the actions of RDV on specific embryological events.
To assess the developmental toxicity of chemicals, in vitro assays using pluripotent stem cells have been explored. Because pluripotent stem cells are capable of self‐renewal and differentiation into various types of tissues, they have been used as investigative tools to model embryo development in vitro (El Azhar & Sonnen, 2021). The basic principle of stem cell‐based assays is that the adverse effects of chemical exposures on in vitro differentiation are interpreted as a sign of developmental toxicity (Riebeling et al., 2012). To date, numerous formats of assays using mouse and human pluripotent stem cells have been reported, each of which utilizes distinct differentiation protocols and endpoints of analyses (Jaklin et al., 2022; Jamalpoor et al., 2022; Lauschke et al., 2021; Luz & Tokar, 2018; Mennen, Oldenburger, & Piersma, 2022; Piersma et al., 2022). Although in vitro assays are not expected to replace animal or human studies, they offer substantial advantages that can facilitate the identification of developmentally toxic chemicals. Stem cell‐based in vitro assays are generally fast, economical, and scalable for high‐throughput screening, and they can serve as ethical alternatives to help reduce animal usage in developmental toxicity testing. Also, stem cell‐based assays allow precise examination of the concentration–effect relationships of specific chemicals, including metabolic products. Such information can be compared with the pharmacokinetic data in humans (e.g., plasma concentrations, metabolic conversions) to provide insights into the potential developmental toxicity in vivo.
The in vitro assay used in the present study consists of morphogenetic embryoid bodies (MEBs), which are three‐dimensional (3D) aggregates of pluripotent stem cells that mimic many features of early development associated with gastrulation (Arias, Marikawa, & Moris, 2022; van den Brink & van Oudenaarden, 2021). During in vitro culture, 3D aggregates of mouse and human stem cells grow in size and transform from a spherical to elongated shape, and exhibit the key characteristics of gastrulation, such as germ layer formation and axial patterning. MEBs, some types of which are referred to as gastruloids (Beccari et al., 2018; Moris et al., 2020), have been used as in vitro assay tools to detect developmental toxicity, and their effectiveness has been tested in reference to a variety of chemical exposures, whose developmental toxicities, or lack thereof, are known from prior animal or human studies (Mantziou et al., 2021; Marikawa, 2022; Marikawa, Chen, Menor, Deng, & Alarcon, 2020; Warkus & Marikawa, 2017). For MEBs made of mouse P19C5 stem cells, the morphological features, namely the size and the extent of elongation, are significantly altered by many developmentally toxic chemical exposures with a concordance of 82.9% (Warkus & Marikawa, 2017). The expression levels of developmental regulator genes, particularly those involved in germ layer formation and axial patterning, are also significantly altered by chemical exposures that impair MEB morphogenesis (Kim & Marikawa, 2018; Kirkwood‐Johnson, Katayama, & Marikawa, 2021; Lau & Marikawa, 2014; Li & Marikawa, 2015, 2016, 2020; Warkus & Marikawa, 2018). For MEBs made of human H9 embryonic stem cells, the expression levels of various regulator genes are altered in response to many developmentally toxic exposures with a concordance of 92.9% (Marikawa et al., 2020). Thus, MEBs are useful in vitro embryo models to assess the developmental toxicity of chemicals in a sensitive and specific manner.
In the present study, MEBs of mouse P19C5 cells were first used to examine the potential adverse effects of RDV, namely by determining the concentrations, timing of exposure, and impact on gene expression profiles that are associated with morphogenesis impairment. The effects of RDV were also compared with those of GS‐441524 (a main metabolite of RDV) and its tri‐ester form GS‐621763, as they are possible anti‐COVID‐19 drugs that may act similarly to RDV (Cox et al., 2021; Rasmussen, Thomsen, & Hansen, 2022; Schäfer et al., 2022; Yan & Muller, 2020). MEBs of human H9 cells were then used to determine the concentrations of RDV that impair the morphology and expression profiles of developmental regulator genes, and were compared with the concentrations that affected the mouse MEBs. The concentration–effect relationships revealed by the MEB‐based assays are discussed in relation to the therapeutic plasma concentrations of RDV in humans to evaluate whether the drug may cause the death or malformation of embryos.
2. MATERIALS AND METHODS
2.1. Test chemicals
RDV (#30354), telaprevir (TVR; #20054), GS‐441524 (#30469), and GS‐621763 (#34125) were commercially obtained from Cayman Chemical (San Diego, CA). These chemicals were dissolved in dimethyl sulfoxide (DMSO; #D2650) from Sigma‐Aldrich (St. Louis, MO) at 5 or 10 mM as stocks, and were stored at −20°C. Chemical structures of RDV, GS‐441524, and GS‐621763, and their conversions into the active nucleoside triphosphate form are depicted in Figure 1a.
FIGURE 1.
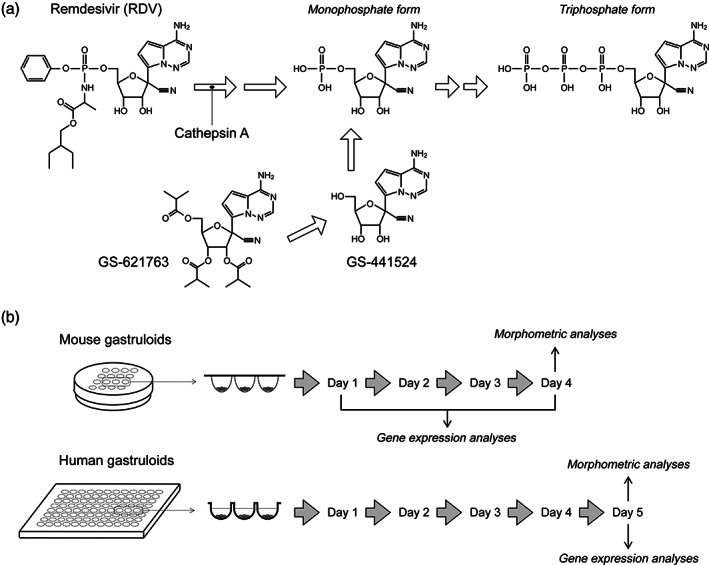
(a) The chemical structures of remdesivir (RDV), GS‐441524, and GS‐621763, and their conversion into the nucleoside triphosphate, which is the therapeutically active form. Cathepsin A catalyzes the first reaction that converts RDV into the alanine metabolite GS‐704277 (not shown), which is then converted to the monophosphate form. (b) The experimental scheme to examine the impact of test chemicals on the morphogenesis and gene expressions in mouse and human morphogenetic embryoid bodies
2.2. Cell culture and generation of MEBs
Mouse P19C5 stem cells, which were derived from the P19 embryonal carcinoma stem cell line (Lau & Marikawa, 2014), were maintained in culture medium consisting of 90% Minimum Essential Medium Alpha with nucleosides and GlutaMAX Supplement (LifeTechnologies, Carlsbad, CA), 7.5% newborn calf serum, 2.5% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. 3D cell aggregates, or embryoid bodies, were generated by hanging drop culture, according to the method described previously (Lau & Marikawa, 2014). Briefly, cells were dissociated with TrypLE Express (LifeTechnologies) and suspended at a density of 10 cells/μl in the culture medium, containing a final concentration of 1% DMSO, with or without a test chemical. Drops (20 μl each) of cell suspension were deposited on the inner surface of Petri dish lids (16 drops per dish), which were then inverted to make hanging drops.
Human H9 embryonic stem cell line (WA09, National Institutes of Health registration number 0062) was obtained from WiCell Research Institute (Madison, WI). H9 cells were maintained in the feeder‐free culture medium mTeSR1 (Stemcell Technologies, Vancouver, BC, Canada) with 40 units/ml penicillin and 40 μg/ml streptomycin in flasks that had been precoated with iMatrix‐511 (Takara Bio, Mountain View, CA). MEBs of H9 cells were generated in round‐bottom 96‐well plates, as described previously (Marikawa et al., 2020). Briefly, cells were dissociated with TrypLE Express and suspended at a density of 20 cells/μl in culture medium consisting of 80% Minimum Essential Medium Alpha with nucleosides and GlutaMAX, 20% 5X Supplement of mTeSR1, 10 μM CHIR99021 (Calbiochem, La Jolla, CA), 2 μM SB431542 (Stemcell Technologies), and 2 nM retinoic acid (Sigma‐Aldrich), with or without a test chemical. Then, 50 μl of cell suspension was placed in each well to generate human embryonic stem cell aggregates with CHIR99021, SB431542, and retinoic acid, which are referred to as HESCA‐CSR (Marikawa et al., 2020). All cells and MEBs were cultured at 37°C in 4.5% CO2 in humidified air.
2.3. Chemical treatment of MEBs
For the experiments to examine the impact on morphogenesis and gene expression, MEBs were exposed to a test chemical at a given concentration from the time of cell aggregation to the end of culture (4 and 5 days for the mouse and human MEBs, respectively) (Figure 1b). The ranges of test chemical concentrations were selected based on a series of pilot experiments to include a low concentration that did not affect the MEB morphogenesis and a high concentration that did. For mouse MEBs, 5 μl of DMSO, containing a test chemical (at 100X concentration), was first prepared, and then mixed with 495 μl of 10 cells/μl cell suspension, creating a desired (1X) concentration of a test chemical with 1% DMSO. This cell suspension was used for hanging drop culture to generate MEBs. MEBs were cultured for 4 days without medium change, and harvested for morphometric analyses (Section 2.4) or gene expression analyses (Section 2.5). Human MEBs were treated with RDV in a manner similar to mouse MEBs with slight differences. Namely, 0.8 μl of DMSO with a test chemical (at 100X concentration) was mixed with 499.2 μl of 20 cells/μl cell suspension, resulting in 0.16% DMSO, which was placed in round‐bottom wells to create MEBs.
In an experiment to examine the impact of timing of exposure, mouse MEBs were treated with RDV (4 μM) at different intervals during the course of a 4‐day culture. In one group, cell aggregates were cultured in RDV‐containing hanging drops from Day 0 to Day 2, followed by transfer (manually using a glass pipet) into control hanging drops for culture until Day 4 (designated as R‐C). In another group, MEBs were exposed in reversed conditions, in which they were cultured in control hanging drops from Day 0 to Day 2, followed by transfer into RDV‐containing hanging drops for culture until Day 4 (designated as C‐R). Simultaneously, two additional groups were also prepared for comparisons, namely cell aggregates that were transferred from control to control hanging drops at Day 2 (designated as C‐C) and those transferred from RDV‐containing to RDV‐containing hanging drops at Day 2 (designated as R‐R).
2.4. Morphometric analyses
MEBs were removed from hanging drops or culture wells, and placed together in a dish filled with phosphate‐buffered saline for photography. Image files in JPG format were opened in the ImageJ program (National Institutes of Health, Bethesda, MD). The circumference of individual MEBs was traced using the polygon selection tool, and three morphometric parameters, namely area, elongation distortion index (EDI), and aspect ratio (AR), were measured. Area was used to evaluate the overall size of the MEBs, and it correlates with the cytotoxic potential (i.e., to reduce cell proliferation or survival) of chemical exposures, as shown in the previous studies for both mouse and human MEBs (Marikawa et al., 2020; Warkus & Marikawa, 2017; Warkus, Yuen, Lau, & Marikawa, 2016). By contrast, EDI and AR were used to gauge the extent of axial elongation. EDI, which is defined as 1/circularity −1, becomes larger when the shape of an MEB is more elongated or distorted (Marikawa, Tamashiro, Fujita, & Alarcón, 2009). AR is the ratio of the major‐to‐minor axis of an ellipse that most tightly fits the circumference of an MEB, and it becomes larger for those that are more elongated along a single axis. For each set of experiments, the morphometric parameters were normalized relative to the average values of control aggregates to calculate relative area, relative EDI, and relative AR, which are expressed as percentages of the control (control = 100%). These relative values were compiled from three or more sets of independent replicates.
2.5. Gene expression analyses
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was performed to determine the relative expression levels of developmental regulator genes. The functional characteristics and PCR primer sequences of the mouse genes examined are listed in Table 1. Previous studies have shown that the expressions of these genes are dysregulated by various chemical exposures that impair the morphogenesis of mouse MEBs (Kim & Marikawa, 2018; Kirkwood‐Johnson et al., 2021; Lau & Marikawa, 2014; Li & Marikawa, 2015, 2016, 2020; Warkus & Marikawa, 2018; Yuan & Marikawa, 2017), and that the presence of morphological impairment in mouse MEBs correlates with the in vivo developmental toxicity of chemical exposures with a concordance of 82.9% (Warkus & Marikawa, 2017). The characteristics and PCR primer sequences of the human genes are described in the previous study (Marikawa et al., 2020). The procedures for RNA extraction, cDNA synthesis, real‐time PCR, and data analysis are described previously (Kirkwood‐Johnson et al., 2021). To assess the impact of RDV on gene expressions in HESCA‐CSR, the Altered Level of Embryogenesis Regulator Transcript (ALERT) score was calculated, which was formulated previously (Marikawa et al., 2020). Briefly, ALERT score represents the number of genes, whose transcript levels were altered by ≥2‐fold compared to the control (either increase or decrease) with statistical significance (p < .05; see Section 2.6) in response to a given chemical exposure. The previous validation study showed that positive ALERT scores correlate with the in vivo developmental toxicity of chemical exposures with a concordance of 92.9% (Marikawa et al., 2020).
TABLE 1.
Mouse developmental regulator genes examined in the present study
| Gene name | Characteristics a | Primer sequences (5′ ➔ 3′) |
|---|---|---|
| Actb |
a. Cytoskeletal actin b. Ubiquitous c. Housekeeping |
F: GAGAGGGAAATCGTGCGTGACATC R: CAGCTCAGTAACAGTCCGCCTAGA |
| Aldh1a2 |
a. Aldehyde dehydrogenase b. Trunk region c. Retinoic acid synthesis |
F: CTTGCCTCACAACAAGTGAGCTTC R: TCACCCAGGTTAGAGACTGGCTTC |
| Brachyury |
a. T‐box transcription factor b. Primitive streak c. Mesendoderm specification |
F: CCTCGGATTCACATCGTGAGAGTT R: AGTAGGTGGGCGGGCGTTATGACT |
| Cdx1 |
a. Homeodomain transcription factor b. Primitive streak c. Axial patterning |
F: TCAGGACTGGACATGAGGTAGAGG R: TGGGAAGGTGGGCATGAGCAGGTA |
| Cyp26a1 |
a. Cytochrome P450 oxidase b. Posterior end c. Retinoic acid catabolism |
F: CGGAGCTGTGTAGGCAAAGAGTTT R: CCTGGAAGTGGGTAAATCTTGCAG |
| Dll1 |
a. NOTCH signaling ligand b. Posterior end c. Somite segmentation |
F: TGCCCACACGTCTATCTTGGATTA R: GTCACATAGACCCGAAGTGCCTTT |
| Fgf8 |
a. FGF signaling ligand b. Primitive streak c. Mesendoderm specification |
F: GTTGCACTTGCTGGTTCTCTGCCT R: AGTCCTTGCCTTTGCCGTTGCTCT |
| Foxc2 |
a. Forkhead transcription factor b. Somitic mesoderm c. Somite segmentation |
F: CCCATAGGGACCCCTAATGACTTC R: GTAACAGTTGGGCAAGACGAAACC |
| Hes7 |
a. HLH transcription factor b. Posterior end c. Somite segmentation |
F: CATACCCTTCTCCCACCTTTAGGC R: AGTGACGAGAAAGCGATTCAAAGG |
| Hoxa1 |
a. HOX transcription factor b. Anterior region c. Axial patterning |
F: CCCTTTCCTTCCACACTGTCTTGT R: AAGACCCGTAAACTCTGCTCTGGA |
| Hoxb9 |
a. HOX transcription factor b. Posterior region c. Axial patterning |
F: AAGCAGGGAGTGGTTTTATGAAGG R: GGGATAGGAATGTATGAATGGGGA |
| Hoxc6 |
a. HOX transcription factor b. Central region c. Axial patterning |
F: TTCGCCACAGGAGAATGTCGTGTT R: CGAGTTAGGTAGCGGTTGAAGTGA |
| Lfng |
a. NOTCH signaling regulator b. Posterior end c. Somite segmentation |
F: AAGCAGGGAGTGGTTTTATGAAGG R: GGGATAGGAATGTATGAATGGGGA |
| Meox1 |
a. Homeodomain transcription factor b. Somitic mesoderm c. Somite differentiation |
F: AAAAATCAGACTTCCCAGCGACAG R: TTCACACGTTTCCACTTCATCCTC |
| Mesp2 |
a. HLH transcription factor b. Somitic mesoderm c. Somite differentiation |
F: CTGCCTTGGAAGTGCCTTTATCTG R: GATACCTAGAAGCGGGGGTGTCTT |
| Mixl1 |
a. Homeodomain transcription factor b. Primitive streak c. Mesendoderm specification |
F: CGACAGACCATGTACCCAGACATC R: TGAGGCTTCAAACACCTAGCTTCA |
| Msgn1 |
a. bHLH domain transcription factor b. Posterior end c. Somite differentiation |
F: CCAGAAAGGCAGCAAAGTCAAGAT R: TCTGTGAGTTCCCCGATGTACTTG |
| Nanog |
a. Homeodomain transcription factor b. Epiblast c. Pluripotency maintenance |
F: GCTTTGGAGACAGTGAGGTGCATA R: GCTACCCTCAAACTCCTGGTCCTT |
| Nodal |
a. NODAL signaling ligand b. Epiblast c. Pluripotency maintenance |
F: GTACATGTTGAGCCTCTACCGAGA R: TCTACAGACAGCTGTCCCTCCTGG |
| Nrarp |
a. NOTCH signaling component b. Posterior end c. Somite segmentation |
F: TGGGAAATAAAAGGGAGGCTGAAT R: GTGCTTGTCTCAGTGTCTGCCATT |
| Notch1 |
a. NOTCH signaling receptor b. Posterior end c. Somite segmentation |
F: GTCTGCAGGCTCCAGTGTTCTGTA R: TCAGTTGGATTTGGATGATGCTGT |
| Sp5 |
a. Zinc finger transcription factor b. Primitive streak c. Wnt signaling transcriptional target |
F: CAGGACAGGAAACTGGGTCGTAGT R: GGCCTAGCAAAAACTTAGGCCTTG |
| Pou5f1 |
a. POU domain transcription factor b. Epiblast c. Pluripotency maintenance |
F: AGGCAGGAGCACGAGTGGAAAGCA R: GGAGGGCTTCGGGCACTTCAGAAA |
| Tbx6 |
a. T‐box transcription factor b. Posterior end c. Axial stem cell differentiation |
F: GGCCTCTCTTCCACCCTTTAGTTC R: CACTAGTAACAAGGCCCCCAGGAG |
| Wnt3 |
a. WNT signaling ligand b. Primitive streak c. Initiation of gastrulation |
F: CAGATGCCCGCTCAGCTATGAACA R: AGCAGCACCAGTGGAAGACGCAAT |
| Wnt3a |
a. WNT signaling ligand b. Posterior end c. Axial stem cell differentiation |
F: GCCACAAGAGCTTCCTGATTGGTA R: CCAGGCAGAAGACAGTCAGTCACC |
Characteristics are: a—molecular function, b—major expression domains around the gastrulation stage (mouse embryonic stages from E5.5 to E8.5), and c—functional significance in early embryo development, which are based on the information available in the Mouse Genome Informatics (www.informatics.jax.org).
2.6. Statistical analyses
All experiments were conducted three times or more, using different batches of cell collections as biological replicates. For morphometric analyses (Section 2.4), compiled data were presented as mean ± 95% CI. Mean values of relative area, relative EDI, and relative AR, were first compared among treatment groups, including controls, by one‐way analysis of variance (ANOVA) to determine whether there were any significant differences, followed by post hoc two‐sample t test to compare between two specific groups. p‐Values of less than .01 were deemed statistically significant. For gene expression analyses (Section 2.5), compiled data were presented as mean ± SD. Mean values of relative expression levels were first compared by one‐way ANOVA, followed by post hoc two‐sample t test to compare between control and chemical‐treated groups. p‐Values of less than .05 were deemed statistically significant.
3. RESULTS
3.1. RDV impairs the morphogenesis of mouse MEBs at therapeutic concentrations
When mouse P19C5 cell aggregates were cultured in the presence of RDV, the morphology of the resulting MEBs at Day 4 was distinctly altered in a concentration‐dependent manner. With increasing concentrations of RDV, MEBs became progressively smaller and less elongated (Figure 2a). All three morphometric parameters (area, EDI, and AR) were significantly reduced by RDV at 2 μM, and they were more markedly reduced at higher concentrations. Strikingly, the EDI and AR (gauging the extent of axial elongation) were significantly reduced even at 1 μM, although the area (measuring the overall growth) was unaffected, suggesting that RDV can impair morphogenesis without cytotoxic impact. These concentrations are close to or lower than the peak plasma concentrations (C max) of RDV in people who received the FDA‐approved therapeutic dose of the drug (4.3–9.0 μM; Humeniuk et al., 2020; Tempestilli et al., 2020).
FIGURE 2.
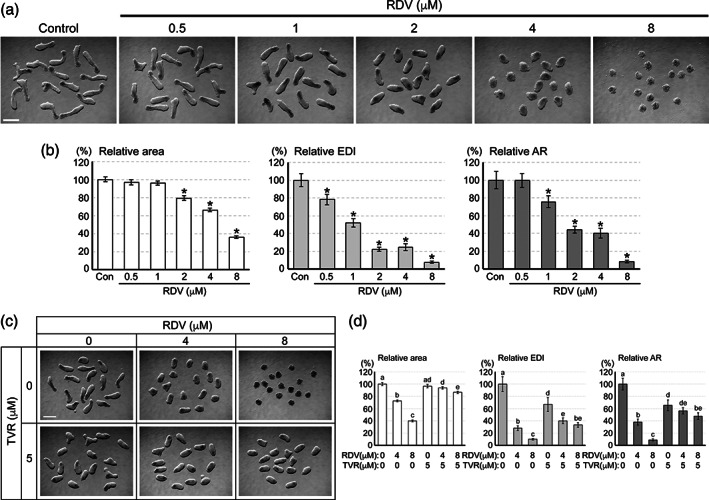
The impact of remdesivir (RDV) on the morphogenesis of mouse morphogenetic embryoid bodies (MEBs). (a) Representative images of mouse MEBs on Day 4 of culture that have been treated with RDV at the indicated concentrations. (b) Morphometric parameters of Day 4 mouse MEBs treated with RDV. Graphs show means ±95% confidence interval (CI) of relative area, relative elongation distortion index (EDI), and relative aspect ratio (AR), based on measurement of 64 aggregates for each condition. Asterisks indicate significant reduction (p < .01) compared to the control. (c) Representative images of Day 4 MEBs that have been co‐treated with RDV and telaprevir (TVR) at the indicated concentrations. (d) Graphs show means ±95% CI of the morphometric parameters of Day 4 MEBs treated with RDV and TVR, based on measurement of 47–48 aggregates for each condition. Different letters indicate statistically significant difference (p < .01) among treatments. Scale bars = 500 μm
To test whether the morphological effects of RDV depend on the intracellular conversion to the triphosphate form, MEBs were co‐treated with RDV and TVR, an inhibitor of Cathepsin A, which mediates the first step of the conversion (Figure 1a). Co‐treatment with TVR significantly alleviated the adverse effects of RDV on all three morphometric parameters (Figure 2c,d). Namely, RDV (8 μM) alone markedly reduced the growth and elongation (area = 40% of control, EDI and AR = 9–10% of control), whereas RDV (8 μM) and TVR (5 μM) together only modestly reduced them (area = 87% of control, EDI and AR = 34–48% of control). Note that even though TVR alone caused a modest reduction in EDI and AR (67% and 65% of control, respectively), it significantly alleviated the adverse effects of RDV on the growth and elongation of MEBs. This suggests that the intracellular conversion to the triphosphate form is required for RDV to impair the morphogenesis of mouse MEBs.
3.2. GS‐441524 and GS‐621763 are less potent than RDV in impairing the morphogenesis of mouse MEBs
GS‐441524 is a main metabolite of RDV in the plasma (C max = 0.5–2.0 μM; Humeniuk et al., 2020; Tempestilli et al., 2020). Because GS‐441524 can also be converted to the triphosphate form through a mechanism distinct from RDV (Figure 1a), its therapeutic application for COVID‐19 has been explored (Rasmussen et al., 2022; Yan & Muller, 2020). Similar to RDV, the growth and elongation of mouse MEBs were diminished by exposure to GS‐441524 (Figure 3a). However, much higher concentrations of GS‐441524 than RDV were needed to elicit similar morphological effects. For example, all three morphometric parameters were reduced by GS‐441524 at 32 μM and above (Figure 3b), whereas they were reduced by RDV at 2 μM and above (Figure 2b).
FIGURE 3.
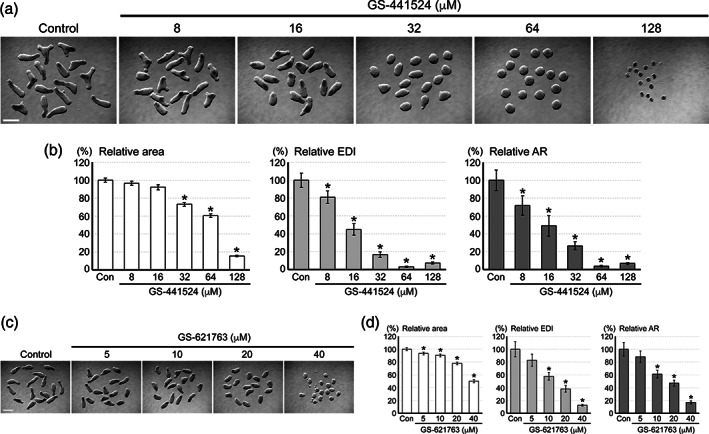
The impact of GS‐441524 and GS‐621763 on the morphogenesis of mouse morphogenetic embryoid bodies (MEBs). (a) Representative images of Day 4 MEBs that have been treated with GS‐441524 at the indicated concentrations. (b) Morphometric parameters of Day 4 MEBs treated with GS‐441524. Graphs show means ±95% confidence interval (CI) of relative area, relative elongation distortion index (EDI), and relative aspect ratio (AR), based on measurement of 47–48 aggregates for each condition. Asterisks indicate significant reduction (p < .01) compared to the control. (c) Representative images of Day 4 MEBs treated with GS‐621763. (d) Graphs show means ±95% CI of the morphometric parameters of Day 4 MEBs treated with GS‐621763. Asterisks indicate significant reduction (p < .01) compared to the control. Scale bars = 500 μm
GS‐621763 is a tri‐ester analog and pro‐drug form of GS‐441524 (Figure 1a), and is also being explored as an oral drug for COVID‐19 (Cox et al., 2021; Schäfer et al., 2022). Exposure to GS‐621763 impaired the morphogenesis of mouse MEBs, and all three morphometric parameters were significantly reduced by it at 10 μM and above (Figure 3c,d). Thus, of the three similar drugs tested, RDV was the most potent in impairing the morphogenesis of mouse MEBs.
3.3. RDV dysregulates the expression of developmental regulators in mouse MEBs
To gain insight into the molecular mechanisms by which RDV impairs the morphogenesis of MEBs, the expression profiles of developmental regulator genes were investigated. The genes listed in Table 1 were selected for analyses, because they are essential regulators of embryonic patterning, and also because their transcriptions are dysregulated by various chemical exposures that impair the morphogenesis of mouse MEBs (Marikawa, 2018, 2022). The transcript levels of the pluripotency maintenance factors (Pou5f1, Nanog, Nodal) were not significantly different between the control and RDV‐exposed (4 μM) MEBs during the course of the 4‐day culture (Figure 4a). However, the expression of many other genes was distinctly altered by RDV exposure. The manner of alterations was variable depending on the genes: some genes were upregulated while others were downregulated by RDV at specific days of culture. Most notably, RDV dysregulated the genes that are involved in somite formation (Tbx6, Dll1, Hes7, Nrarp, Lfng, Mesp2, Meox1, Foxc2) and cranial‐caudal patterning (Hoxa1, Hoxc6, Hoxb9, Aldh1a2, Cyp26a1) (Figure 4a), which may be linked to the morphological impairment of the RDV‐treated MEBs.
FIGURE 4.
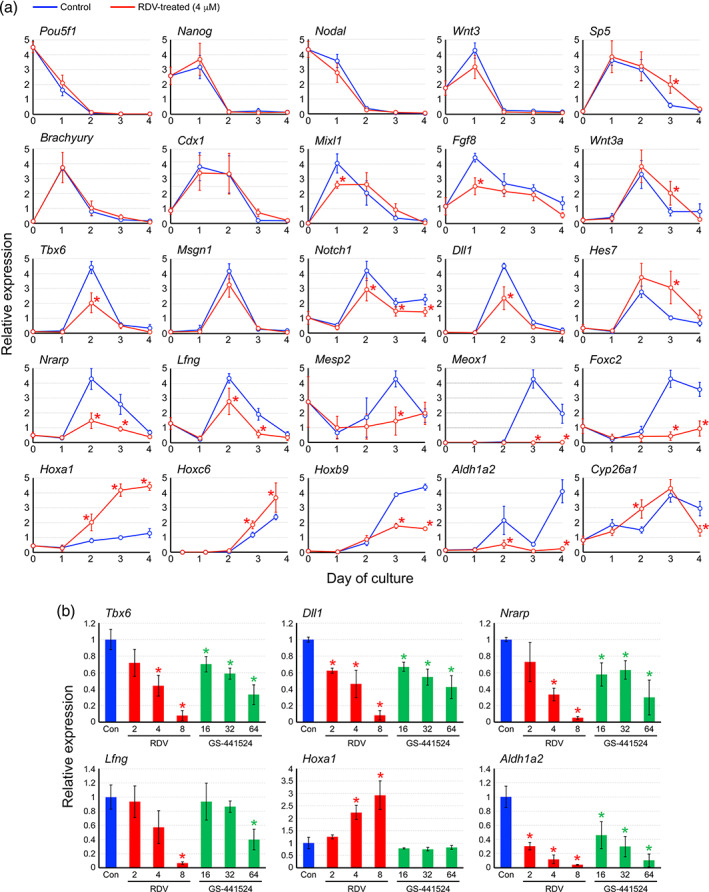
The impact of remdesivir (RDV) on the expressions of developmental regulator genes in mouse morphogenetic embryoid bodies (MEBs). (a) Graphs show means ± SD of relative expression levels in arbitrary unit over the course of 4 days of culture, based on four replicates of experiments. Blue and red lines correspond to control and RDV‐treated (4 μM) MEBs, respectively. Asterisks indicate significant difference (p < .05) compared to the control at the same day of culture. (b) Graphs show means ± SD of relative expression levels (the control value is set as 1) at Day 2 of culture, based on three replicates of experiments. Blue, red, and green bars correspond to control, RDV‐treated, and GS‐441524‐treated MEBs, respectively. Asterisks indicate significant difference (p < .05) compared to the control
The impact of RDV on gene expressions was further investigated in comparison with GS‐441524, the major metabolite of RDV in the plasma. MEBs were exposed to RDV at 2–8 μM or GS‐441524 at 16–64 μM, as these concentrations were comparable in the potency to impair morphogenesis (Figures 2b and 3b). Gene expression was then analyzed at Day 2. Of the six genes examined, five (Tbx6, Dll1, Nrarp, Lfng, Aldh1a2) were downregulated by both RDV and GS‐441524 in a similar manner (Figure 4b). By contrast, one gene (Hoxa1) was upregulated by RDV, but not by GS‐441524 even at the highest concentration tested (64 μM). This suggests that RDV and GS‐441524 exert a similar, but not identical, molecular impact on mouse MEBs.
3.4. Early stages of mouse MEB development are more vulnerable to RDV exposure
The vulnerability of embryos to chemical insults depends on the timing and duration of exposure (Friedman, 2010; Wilson, 1973). To assess the developmental stage‐dependent impact, mouse MEBs were exposed to RDV (4 μM) at different periods during the course of the 4‐day culture (Figure 5a), and their morphology was examined at Day 4. Both the early exposure (Days 0–2; R‐C) and the late exposure (Days 2–4; C‐R) impaired the MEB morphogenesis, as they significantly reduced all three morphometric parameters, compared to the control (C‐C; Figure 5b). However, reductions were more pronounced for the early exposure. Notably, the axial elongation, as measured by EDI and AR, was markedly diminished by R‐C in a manner comparable to the continuous exposure throughout the 4‐day period (R‐R; Figure 5b). Thus, the morphogenesis of MEBs was more sensitively impaired by exposure to RDV during the early (Days 0–2) than the late (Days 2–4) stages.
FIGURE 5.
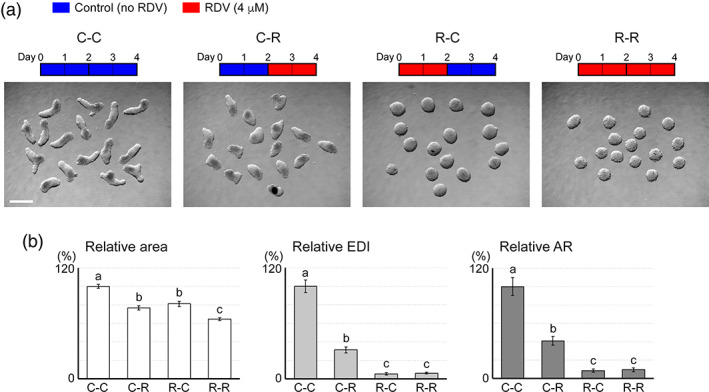
Influence of the timing of remdesivir (RDV) exposure on the morphogenesis of mouse morphogenetic embryoid bodies (MEBs). (a) Representative images of Day 4 MEBs that have been treated with RDV (4 μM) at different intervals, as depicted by the schematic diagrams above. Scale bar = 500 μm. (b) Graphs show means ±95% confidence interval (CI) of relative area, relative elongation distortion index (EDI), and relative aspect ratio (AR) of Day 4 MEBs treated with RDV at different intervals, based on measurement of 48 aggregates for each condition. Different letters indicate statistically significant difference (p < .01) among treatments
3.5. The morphogenesis and gene expression of human MEBs are impaired by RDV
To further assess the potential impact of RDV on human embryos, MEBs made of the human H9 cell line (or HESCA‐CSR as defined previously; Marikawa et al., 2020) were exposed to RDV (0.5–8 μM) and examined at Day 5 of culture. The morphology of HESCA‐CSR was distinctly altered by RDV in a concentration‐dependent manner. Compared to the control, RDV‐exposed HESCA‐CSR appeared smaller and less elongated with higher concentrations (Figure 6a). Specifically, all three morphometric parameters were significantly reduced by RDV at 2 μM, and the extent of reductions was progressively larger with increasing concentrations (Figure 6b). Notably, the concentration–effect relationship of the HESCA‐CSR morphogenesis is similar to that of the mouse MEB morphogenesis (Figure 2b).
FIGURE 6.
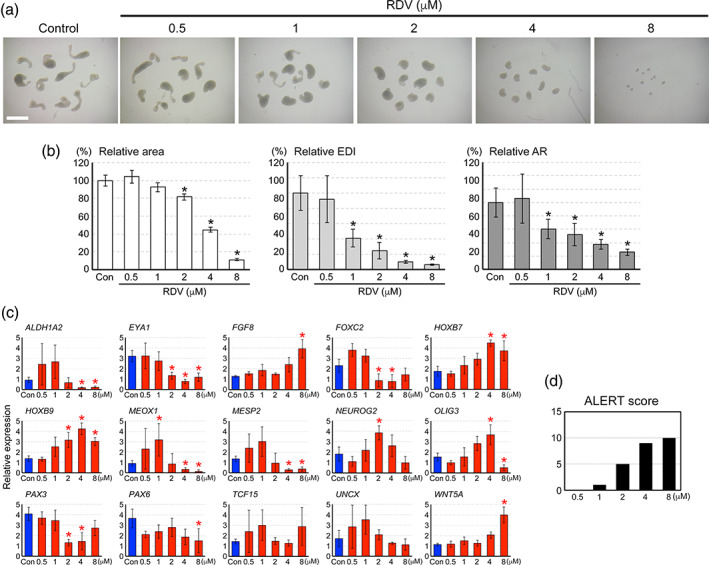
The impact of remdesivir (RDV) on the morphogenesis and gene expression profile in human morphogenetic embryoid bodies (MEBs). (a) Representative images of human MEBs on Day 5 that have been treated with RDV at the indicated concentrations. Scale bar = 1 mm. (b) Morphometric parameters of Day 5 MEBs treated with RDV. Graphs show means ±95% confidence interval of relative area, relative elongation distortion index (EDI), and relative aspect ratio (AR), based on measurement of 39–40 aggregates for each condition. Asterisks indicate a significant reduction (p < .01) compared to the control. (c) Graphs show means ± SD of relative expression levels in arbitrary unit at Day 5 of culture, based on four replicates of experiments. Blue and red bars correspond to control and RDV‐treated MEBs, respectively. Asterisks indicate significant difference (p < .05) with alteration in the expression level by more than twofold (higher or lower) relative to the control. (d) A graph shows the Altered Level of Embryogenesis Regulator Transcript (ALERT) scores of RDV‐treated Day 5 MEBs, based on the gene expression profile in (c)
The transcript levels of the developmental regulator genes were also significantly altered by RDV exposures. The 15 genes examined are essential regulators of embryonic patterning, and are downregulated or upregulated in HESCA‐CSR in response to various developmentally toxic chemical exposures, as described previously (Marikawa et al., 2020). RDV dysregulated the expression of various genes, including those involved in the patterning of the somites (MEOX1, MESP2, PAX3), the cranial‐caudal axis (ALDH1A2, FGF8, HOXB7, HOXB9, WNT5A), and the neuroectoderm (NEUROG2, OLIG3, PAX6) (Figure 6c). ALERT scores, which are the number of altered genes at each concentration, were used to measure the molecular impact (Section 2.5). The ALERT scores of RDV at 0.5, 1, 2, 4, and 8 μM were 0, 1, 5, 9, and 10, respectively (Figure 6d).
4. DISCUSSION
The present study evaluated the potential developmental toxicity of RDV using MEBs made of mouse and human pluripotent stem cells as in vitro models of early embryos. The morphology and gene expression profiles of both mouse and human MEBs were significantly altered by RDV exposures at the concentration range of 1–8 μM, which is comparable to the therapeutic plasma levels (C max = 4.3–9.0 μM; Humeniuk et al., 2020; Tempestilli et al., 2020). Currently, there is no pharmacokinetic information in humans or animals, describing the concentration of RDV in the uterus, where embryo development takes place. However, in the mouse, the concentrations of RDV in various organs (e.g., liver, intestine, kidney, and testis) are found to be 2.2‐ to 20.4‐times higher than in the plasma (Wang & Chen, 2020). If the concentrations of RDV in the uterus after therapeutic administration of the drug are also higher than or similar to the plasma levels, developing embryos may be exposed to adverse effect levels of RDV. Thus, further investigations into the developmental toxicity of RDV are warranted.
The sensitivity of embryos to chemical insults is highly dependent on the developmental stage at the time of exposure (Friedman, 2010; Wilson, 1973). The present study showed that mouse MEBs were impaired by RDV exposure during Days 0–2 more severely than during Days 2–4. This raises the possibility that the impact of RDV is different depending on the time of exposure during pregnancy. Based on the temporal gene expression profile, it is estimated that Days 0–2 of mouse MEB development corresponds to the mouse embryonic stages E5.5–E7.5, when the primitive streak emerges and the body starts to elongate along the cranial‐caudal axis (Li & Marikawa, 2015). These stages are roughly equivalent to the third week of human embryo development, which may be especially sensitive to RDV exposure. However, this does not necessarily mean that other developmental stages are not vulnerable to RDV. Mouse MEBs were still significantly impaired by RDV exposure during Days 2–4 at a therapeutically relevant concentration, even though the severity was less than the Days 0–2 exposure. In addition, a recent study showed that RDV at the therapeutic concentrations (2–8 μM) impairs the in vitro development of mouse embryos at the preimplantation stage, particularly around E2.5–E3.5 (Marikawa & Alarcon, 2022). The same study also showed that the proliferation of undifferentiated human embryonic stem cells, which roughly corresponds to the early second week of development, is diminished by RDV. Thus, it is possible that exposures to RDV may impair embryo development at the stages that are much broader than those represented by MEBs.
TVR, an inhibitor of Cathepsin A, alleviated the morphological effects of RDV on mouse MEBs, suggesting that the action of RDV requires the conversion into the nucleoside triphosphate form (Figure 1a). While the therapeutic target of the triphosphate form is viral RdRp, the endogenous targets that are responsible for the toxic effects of RDV remain unclear (Akinci et al., 2020; Bjork & Wallace, 2021; Xu et al., 2021). Interestingly, GS‐441524 and GS‐621763, which can also be converted into the triphosphate form (Figure 1a), were not as potent as RDV in impairing mouse MEBs. It is possible that these chemicals are not efficiently taken up by MEBs or converted to the triphosphate form, compared to RDV. Also, the present study showed that while many genes were similarly affected by RDV and GS‐441524, the expression of Hoxa1 was upregulated by RDV, but not by GS‐441524. This implies that the mechanisms of the adverse actions are not entirely identical between RDV and GS‐441524. Because GS‐441524 and GS‐621763 are currently being investigated as possible anti‐COVD‐19 drugs (Cox et al., 2021; Rasmussen et al., 2022; Schäfer et al., 2022; Yan & Muller, 2020), it is also important to gather information on their potential developmental toxicity. The concentration–effect relationships obtained from the MEB‐based analyses may provide insight as to whether GS‐441524 and GS‐621763 are developmentally toxic at the concentrations that are therapeutically relevant.
According to the information provided by the manufacturer of Veklury (the brand name of RDV), “remdesivir demonstrated no adverse effect on embryo‐fetal development when administered to pregnant animals at systemic exposures (AUC) of the predominant circulating metabolite of remdesivir (GS‐441524) that were 4 times (rats and rabbits) the exposure in humans at the recommended human dose” (FDA, 2020). Notably, the manufacturer's information also indicates that the plasma C max of RDV itself is 1,580 ng/ml (= 2.62 μM) in rats and 380 ng/ml (= 0.63 μM) in rabbits (FDA, 2020), which are lower than the plasma C max in humans (4.3–9.0 μM; Humeniuk et al., 2020; Tempestilli et al., 2020). This suggests that RDV is more quickly metabolized to GS‐441524 in rats and rabbits than in humans (C max of GS‐441524 in the latter is 0.48–0.52 μM; Humeniuk et al., 2020; Tempestilli et al., 2020). A recent study using pregnant rats has also shown that RDV becomes undetectable in the maternal plasma at 40 min after administration, confirming the rapid metabolism and elimination of RDV (Yang, Lin, Lin, Dalley, & Tsai, 2022). This may also be the case for the mouse, in which the plasma C max of RDV (<1 μM) is considerably lower than that of GS‐441524 (35.8 μM) (Hu et al., 2021). It is possible that in the embryo‐fetal development studies in rats and rabbits, embryos may not have been exposed to RDV at the concentration comparable to the human plasma levels. In the present study, RDV at the human plasma levels (1–8 μM) exerted adverse effects on MEBs, where the conversion of RDV into GS‐441524 is likely to be minimal due to the absence of maternal metabolism. Thus, assays using MEBs, as well as other in vitro systems, may provide insights into the concentration–effect relationships for RDV in a manner corresponding to the human plasma levels.
As the COVID‐19 pandemic persists, many people continue to rely on antiviral treatments, such as RDV. Even though RDV is not generally recommended for pregnant people due to a lack of clinical trial data, it has been administered during pregnancy on a compassionate use basis (Budi et al., 2022; Burwick et al., 2021; Gutierrez et al., 2022; Jorgensen, Davis, & Lapinsky, 2021; Lampejo, 2021; Marzban‐Rad et al., 2022; Saroyo et al., 2021). There are registries to monitor pregnancy outcomes in individuals exposed to RDV (FDA, 2022; UKTIS, 2022), which are likely to provide crucial data for epidemiologic studies. In the near future, there may be sufficient data to allow the definitive assessment of the developmental toxicity of RDV in humans. As shown in the present study, the severity of RDV toxicity on MEB development was dependent on the concentrations and the timing of exposures. Such information may be useful in designing or interpreting epidemiologic studies of RDV treatment during pregnancy, as the drug dosing (which affects the concentration) and the stage of pregnancy (which affects the timing of exposure) are likely to be variable among the patients. If the data of future epidemiologic studies are in line with the results of the present study, it would support the usefulness of MEB‐based assays to study the developmental toxicity of new drugs, not only RDV but also other antivirals that are authorized or being explored for the treatment of COVID‐19.
FUNDING INFORMATION
This work was supported by grants from the Alternatives Research & Development Foundation and the National Institute of Child Health and Human Development at NIH to Y. M. (R03 HD101735 and R03 HD102502) and the NIH Centers of Biomedical Research Excellence Phase 3 to the Institute for Biogenesis Research (P30 GM131944).
CONFLICT OF INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
ACKNOWLEDGMENT
The authors are grateful to Dr Vernadeth B. Alarcon for reading the article and providing valuable comments.
Kirkwood‐Johnson, L. , & Marikawa, Y. (2023). Developmental toxicity of remdesivir, an anti‐COVID‐19 drug, is implicated by in vitro assays using morphogenetic embryoid bodies of mouse and human pluripotent stem cells. Birth Defects Research, 115(2), 224–239. 10.1002/bdr2.2111
Funding information Alternatives Research and Development Foundation; National Institute of Child Health and Human Development, Grant/Award Numbers: R03 HD101735, R03 HD102502; National Institute of General Medical Sciences, Grant/Award Number: P30 GM131944
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Akinci, E. , Cha, M. , Lin, L. , Yeo, G. , Hamilton, M. C. , Donahue, C. J. , … Sherwood, R. I. (2020). Elucidation of remdesivir cytotoxicity pathways through genome‐wide CRISPR‐Cas9 screening and transcriptomics. bioRxiv [Preprint]. Aug 28:2020.08.27.270819. doi: 10.1101/2020.08.27.270819. [DOI]
- Allotey, J. , Stallings, E. , Bonet, M. , Yap, M. , Chatterjee, S. , Kew, T. , … for PregCOV‐19 Living Systematic Review Consortium . (2020). Clinical manifestations, risk factors, and maternal and perinatal outcomes of coronavirus disease 2019 in pregnancy: Living systematic review and meta‐analysis. BMJ, 370, m3320. 10.1136/bmj.m3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias, A. M. , Marikawa, Y. , & Moris, N. (2022). Gastruloids: Pluripotent stem cell models of mammalian gastrulation and embryo engineering. Developmental Biology, 488, 35–46. 10.1016/j.ydbio.2022.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari, L. , Moris, N. , Girgin, M. , Turner, D. A. , Baillie‐Johnson, P. , Cossy, A. C. , … Arias, A. M. (2018). Multi‐axial self‐organization properties of mouse embryonic stem cells into gastruloids. Nature, 562(7726), 272–276. 10.1038/s41586-018-0578-0 [DOI] [PubMed] [Google Scholar]
- Beigel, J. H. , Tomashek, K. M. , Dodd, L. E. , Mehta, A. K. , Zingman, B. S. , Kalil, A. C. , … ACTT‐1 Study Group Members . (2020). Remdesivir for the treatment of Covid‐19—Final report. The New England Journal of Medicine, 383(19), 1813–1826. 10.1056/NEJMoa2007764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork, J. A. , & Wallace, K. B. (2021). Remdesivir; molecular and functional measures of mitochondrial safety. Toxicology and Applied Pharmacology, 433, 115783. 10.1016/j.taap.2021.115783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budi, D. S. , Pratama, N. R. , Wafa, I. A. , Putra, M. , Wardhana, M. P. , & Wungu, C. D. K. (2022). Remdesivir for pregnancy: A systematic review of antiviral therapy for COVID‐19. Heliyon, 8(1), e08835. 10.1016/j.heliyon.2022.e08835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwick, R. M. , Yawetz, S. , Stephenson, K. E. , Collier, A. Y. , Sen, P. , Blackburn, B. G. , … Short, W. R. (2021). Compassionate use of remdesivir in pregnant women with severe coronavirus disease 2019. Clinical Infectious Diseases, 73(11), e3996–e4004. 10.1093/cid/ciaa1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, R. M. , Wolf, J. D. , Lieber, C. M. , Sourimant, J. , Lin, M. J. , Babusis, D. , … Plemper, R. K. (2021). Oral prodrug of remdesivir parent GS‐441524 is efficacious against SARS‐CoV‐2 in ferrets. Nature Communications, 12(1), 6415. 10.1038/s41467-021-26760-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman, R. T. , Roth, J. S. , Brimacombe, K. R. , Simeonov, A. , Shen, M. , Patnaik, S. , & Hall, M. D. (2020). Remdesivir: A review of its discovery and development leading to emergency use authorization for treatment of COVID‐19. ACS Central Science, 6(5), 672–683. 10.1021/acscentsci.0c00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Azhar, Y. , & Sonnen, K. F. (2021). Development in a dish‐in vitro models of mammalian embryonic development. Frontiers in Cell and Development Biology, 9, 655993. 10.3389/fcell.2021.655993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration (FDA) . (2020). VEKLURY, non‐clinical review(s) (09/2020). Retrieved from https://www.accessdata.fda.gov/drugsatfda_docs/nda/2020/214787Orig1s000PharmR.pdf
- Food and Drug Administration (FDA) . (2022). VEKLURY, prescribing information (06/2022). Retrieved from https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/214787Orig1s015lbl.pdf
- Friedman, J. M. (2010). The principles of teratology: Are they still true? Birth Defects Research. Part A: Clinical and Molecular Teratology, 88(10), 766–768. 10.1002/bdra.20697 [DOI] [PubMed] [Google Scholar]
- Gérard, A. O. , Laurain, A. , Fresse, A. , Parassol, N. , Muzzone, M. , Rocher, F. , … Drici, M. D. (2021). Remdesivir and acute renal failure: A potential safety signal from disproportionality analysis of the WHO safety database. Clinical Pharmacology and Therapeutics, 109(4), 1021–1024. 10.1002/cpt.2145 [DOI] [PubMed] [Google Scholar]
- Goldman, J. D. , Lye, D. C. B. , Hui, D. S. , Marks, K. M. , Bruno, R. , Montejano, R. , … GS‐US‐540‐5773 Investigators . (2020). Remdesivir for 5 or 10 days in patients with severe COVID‐19. The New England Journal of Medicine, 383(19), 1827–1837. 10.1056/NEJMoa2015301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, R. , Mendez‐Figueroa, H. , Biebighauser, J. G. , Bhalwal, A. , Pineles, B. L. , & Chauhan, S. P. (2022). Remdesivir use in pregnancy during the SARS‐CoV‐2 pandemic. The Journal of Maternal‐Fetal & Neonatal Medicine, 1–7. 10.1080/14767058.2022.2041595 [DOI] [PubMed] [Google Scholar]
- Hu, W. J. , Chang, L. , Yang, Y. , Wang, X. , Xie, Y. C. , Shen, J. S. , … Liu, J. (2021). Pharmacokinetics and tissue distribution of remdesivir and its metabolites nucleotide monophosphate, nucleotide triphosphate, and nucleoside in mice. Acta Pharmacologica Sinica, 42(7), 1195–1200. 10.1038/s41401-020-00537-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeniuk, R. , Mathias, A. , Cao, H. , Osinusi, A. , Shen, G. , Chng, E. , … German, P. (2020). Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID‐19 in healthy subjects. Clinical and Translational Science, 13(5), 896–906. 10.1111/cts.12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklin, M. , Zhang, J. D. , Schäfer, N. , Clemann, N. , Barrow, P. , Küng, E. , … Kustermann, S. (2022). Optimization of the TeraTox assay for preclinical teratogenicity assessment. Toxicological Sciences, 188(1), 17–33. 10.1093/toxsci/kfac046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamalpoor, A. , Hartvelt, S. , Dimopoulou, M. , Zwetsloot, T. , Brandsma, I. , Racz, P. I. , … Hendriks, G. (2022). A novel human stem cell‐based biomarker assay for in vitro assessment of developmental toxicity. Birth Defects Research, 114, 1210–1228. 10.1002/bdr2.2001 [DOI] [PubMed] [Google Scholar]
- Jorgensen, S. C. J. , Davis, M. R. , & Lapinsky, S. E. (2021). A review of remdesivir for COVID‐19 in pregnancy and lactation. The Journal of Antimicrobial Chemotherapy, 77(1), 24–30. 10.1093/jac/dkab311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I. Q. , & Marikawa, Y. (2018). Embryoid body test with morphological and molecular endpoints implicates potential developmental toxicity of trans‐resveratrol. Toxicology and Applied Pharmacology, 355, 211–225. 10.1016/j.taap.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood‐Johnson, L. , Katayama, N. , & Marikawa, Y. (2021). Dolutegravir impairs stem cell‐based 3D morphogenesis models in a manner dependent on dose and timing of exposure: An implication for its developmental toxicity. Toxicological Sciences, 184(2), 191–203. 10.1093/toxsci/kfab112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokic, G. , Hillen, H. S. , Tegunov, D. , Dienemann, C. , Seitz, F. , Schmitzova, J. , … Cramer, P. (2021). Mechanism of SARS‐CoV‐2 polymerase stalling by remdesivir. Nature Communications, 12(1), 279. 10.1038/s41467-020-20542-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampejo, T. (2021). Remdesivir for the treatment of COVID‐19 in pregnancy. Journal of Medical Virology, 93(7), 4114–4119. 10.1002/jmv.26986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C. G. , & Marikawa, Y. (2014). Morphology‐based mammalian stem cell tests reveal potential developmental toxicity of donepezil. Molecular Reproduction and Development, 81(11), 994–1008. 10.1002/mrd.22423 [DOI] [PubMed] [Google Scholar]
- Lauschke, K. , Treschow, A. F. , Rasmussen, M. A. , Davidsen, N. , Holst, B. , Emnéus, J. , … Vinggaard, A. M. (2021). Creating a human‐induced pluripotent stem cell‐based NKX2.5 reporter gene assay for developmental toxicity testing. Archives of Toxicology, 95(5), 1659–1670. 10.1007/s00204-021-03018-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A. S. , & Marikawa, Y. (2015). An in vitro gastrulation model recapitulates the morphogenetic impact of pharmacological inhibitors of developmental signaling pathways. Molecular Reproduction and Development, 82(12), 1015–1036. 10.1002/mrd.22585 [DOI] [PubMed] [Google Scholar]
- Li, A. S. , & Marikawa, Y. (2016). Adverse effect of valproic acid on an in vitro gastrulation model entails activation of retinoic acid signaling. Reproductive Toxicology, 66, 68–83. 10.1016/j.reprotox.2016.09.015 [DOI] [PubMed] [Google Scholar]
- Li, A. S. W. , & Marikawa, Y. (2020). Methoxyacetic acid inhibits histone deacetylase and impairs axial elongation morphogenesis of mouse gastruloids in a retinoic acid signaling‐dependent manner. Birth Defects Research, 112(14), 1043–1056. 10.1002/bdr2.1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. X. J. , Cho, S. , Meyyur Aravamudan, V. , Sanda, H. Y. , Palraj, R. , Molton, J. S. , & Venkatachalam, I. (2021). Remdesivir in coronavirus disease 2019 (COVID‐19) treatment: A review of evidence. Infection, 49(3), 401–410. 10.1007/s15010-020-01557-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz, A. L. , & Tokar, E. J. (2018). Pluripotent stem cells in developmental toxicity testing: A review of methodological advances. Toxicological Sciences, 165(1), 31–39. 10.1093/toxsci/kfy174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantziou, V. , Baillie‐Benson, P. , Jaklin, M. , Kustermann, S. , Arias, A. M. , & Moris, N. (2021). In vitro teratogenicity testing using a 3D, embryo‐like gastruloid system. Reproductive Toxicology, 105, 72–90. 10.1016/j.reprotox.2021.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa, Y. (2018). Stem‐cell‐based in vitro morphogenesis models to investigate developmental toxicity of chemical exposures. In Rasmussen T. P. (Ed.), Stem cells in birth defects research and developmental toxicology (pp. 71–89). New York, NY: Wiley. [Google Scholar]
- Marikawa, Y. (2022). Toward better assessments of developmental toxicity using stem cell‐based in vitro embryogenesis models. Birth Defects Research, 114, 972–982. 10.1002/bdr2.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa, Y. , & Alarcon, V. B. (2022). Remdesivir impairs mouse preimplantation embryo development at therapeutic concentrations. Reproductive Toxicology, 111, 135–147. 10.1016/j.reprotox.2022.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa, Y. , Chen, H. R. , Menor, M. , Deng, Y. , & Alarcon, V. B. (2020). Exposure‐based assessment of chemical teratogenicity using morphogenetic aggregates of human embryonic stem cells. Reproductive Toxicology, 91, 74–91. 10.1016/j.reprotox.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa, Y. , Tamashiro, D. A. , Fujita, T. C. , & Alarcón, V. B. (2009). Aggregated P19 mouse embryonal carcinoma cells as a simple in vitro model to study the molecular regulations of mesoderm formation and axial elongation morphogenesis. Genesis, 47(2), 93–106. 10.1002/dvg.20473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzban‐Rad, S. , Ghafarzadeh, M. , Bahmani, S. , & Kazemi, A. (2022). The use of remdesivir among pregnant women and associated clinical outcomes in mother and the child. Annals of Medicine and Surgery, 77, 103681. 10.1016/j.amsu.2022.103681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennen, R. H. , Oldenburger, M. M. , & Piersma, A. H. (2022). Endoderm and mesoderm derivatives in embryonic stem cell differentiation and their use in developmental toxicity testing. Reproductive Toxicology, 107, 44–59. 10.1016/j.reprotox.2021.11.009 [DOI] [PubMed] [Google Scholar]
- Moris, N. , Anlas, K. , van den Brink, S. C. , Alemany, A. , Schröder, J. , Ghimire, S. , … Martinez, A. A. (2020). An in vitro model of early anteroposterior organization during human development. Nature, 582(7812), 410–415. 10.1038/s41586-020-2383-9 [DOI] [PubMed] [Google Scholar]
- Murison, K. R. , Grima, A. A. , Simmons, A. E. , Tuite, A. R. , & Fisman, D. N. (2022). Severity of SARS‐CoV‐2 infection in pregnancy in Ontario: A matched cohort analysis. Clinical Infectious Diseases, ciac544. 10.1093/cid/ciac544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliano, P. , Sellitto, C. , Scarpati, G. , Ascione, T. , Conti, V. , Franci, G. , … Filippelli, A. (2022). An overview of the preclinical discovery and development of remdesivir for the treatment of coronavirus disease 2019 (COVID‐19). Expert Opinion on Drug Discovery, 17(1), 9–18. 10.1080/17460441.2021.1970743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma, A. H. , Baker, N. C. , Daston, G. P. , Flick, B. , Fujiwara, M. , Knudsen, T. B. , … Kojima, H. (2022). Pluripotent stem cell assays: Modalities and applications for predictive developmental toxicity. Current Research in Toxicology, 3, 100074. 10.1016/j.crtox.2022.100074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, H. B. , Thomsen, R. , & Hansen, P. R. (2022). Nucleoside analog GS‐441524: Pharmacokinetics in different species, safety, and potential effectiveness against Covid‐19. Pharmacology Research & Perspectives, 10(2), e00945. 10.1002/prp2.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riebeling, C. , Hayess, K. , Peters, A. K. , Steemans, M. , Spielmann, H. , Luch, A. , & Seiler, A. E. (2012). Assaying embryotoxicity in the test tube: Current limitations of the embryonic stem cell test (EST) challenging its applicability domain. Critical Reviews in Toxicology, 42(5), 443–464. 10.3109/10408444.2012.674483 [DOI] [PubMed] [Google Scholar]
- Saroyo, Y. B. , Rumondang, A. , Febriana, I. S. , Harzif, A. K. , & Irwinda, R. (2021). Remdesivir treatment for COVID 19 in pregnant patients with moderate to severe symptoms: Serial case report. Infectious Disease Reports, 13(2), 437–443. 10.3390/idr13020042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satsangi, S. , Gupta, N. , & Kodan, P. (2021). Current and new drugs for COVID‐19 treatment and its effects on the liver. Journal of Clinical and Translational Hepatology, 9(3), 436–446. 10.14218/JCTH.2020.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, A. , Martinez, D. R. , Won, J. J. , Meganck, R. M. , Moreira, F. R. , Brown, A. J. , … Sheahan, T. P. (2022). Therapeutic treatment with an oral prodrug of the remdesivir parental nucleoside is protective against SARS‐CoV‐2 pathogenesis in mice. Science Translational Medicine, 14(643), eabm3410. 10.1126/scitranslmed.abm3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempestilli, M. , Caputi, P. , Avataneo, V. , Notari, S. , Forini, O. , Scorzolini, L. , … COVID 19 INMI Study Group . (2020). Pharmacokinetics of remdesivir and GS‐441524 in two critically ill patients who recovered from COVID‐19. The Journal of Antimicrobial Chemotherapy, 75(10), 2977–2980. 10.1093/jac/dkaa239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Kingdom Teratology Information Service (UKTIS) . (2022). COVID‐19 antivirals pregnancy registry. Retrieved from https://www.medicinesinpregnancy.org/bumps/COVID-19-Antivirals-Pregnancy-Registry/.
- van den Brink, S. C. , & van Oudenaarden, A. (2021). 3D gastruloids: A novel frontier in stem cell‐based in vitro modeling of mammalian gastrulation. Trends in Cell Biology, 31(9), 747–759. 10.1016/j.tcb.2021.06.007 [DOI] [PubMed] [Google Scholar]
- Wang, H. , Li, N. , Sun, C. , Guo, X. , Su, W. , Song, Q. , … Sun, Y. (2022). The association between pregnancy and COVID‐19: A systematic review and meta‐analysis. The American Journal of Emergency Medicine, 56, 188–195. 10.1016/j.ajem.2022.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , & Chen, L. (2020). Tissue distributions of antiviral drugs affect their capabilities of reducing viral loads in COVID‐19 treatment. European Journal of Pharmacology, 889, 173634. 10.1016/j.ejphar.2020.173634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkus, E. L. , Yuen, A. A. , Lau, C. G. , & Marikawa, Y. (2016). Use of In vitro morphogenesis of mouse Embryoid bodies to assess developmental toxicity of therapeutic drugs contraindicated in pregnancy. Toxicological Sciences, 149(1), 15–30. 10.1093/toxsci/kfv209 [DOI] [PubMed] [Google Scholar]
- Warkus, E. L. L. , & Marikawa, Y. (2017). Exposure‐based validation of an In vitro gastrulation model for developmental toxicity assays. Toxicological Sciences, 157(1), 235–245. 10.1093/toxsci/kfx034 [DOI] [PubMed] [Google Scholar]
- Warkus, E. L. L. , & Marikawa, Y. (2018). Fluoxetine inhibits canonical Wnt signaling to impair Embryoid body morphogenesis: Potential teratogenic mechanisms of a commonly used antidepressant. Toxicological Sciences, 165(2), 372–388. 10.1093/toxsci/kfy143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J. G. (1973). Environment and birth defects. New York, NY: Academic Press. [Google Scholar]
- Xu, Y. , Barauskas, O. , Kim, C. , Babusis, D. , Murakami, E. , Kornyeyev, D. , … Feng, J. Y. (2021). Off‐target in vitro profiling demonstrates that remdesivir is a highly selective antiviral agent. Antimicrobial Agents and Chemotherapy, 65(2), e02237–e02220. 10.1128/AAC.02237-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, V. C. , & Muller, F. L. (2020). Advantages of the parent nucleoside GS‐441524 over remdesivir for Covid‐19 treatment. ACS Medicinal Chemistry Letters, 11(7), 1361–1366. 10.1021/acsmedchemlett.0c00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Lin, I. H. , Lin, L. C. , Dalley, J. W. , & Tsai, T. H. (2022). Biotransformation and transplacental transfer of the anti‐viral remdesivir and predominant metabolite, GS‐441524 in pregnant rats. eBioMedicine, 81, 104095. 10.1016/j.ebiom.2022.104095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, C. J. , & Marikawa, Y. (2017). Developmental toxicity assessment of common excipients using a stem cell‐based in vitro morphogenesis model. Food and Chemical Toxicology, 109(Pt 1), 376–385. 10.1016/j.fct.2017.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


