Abstract
For decades, bacteria have been exploited as vectors for vaccines and therapeutics. However, the bacterial arsenal used has historically been limited to a few strains. Advancements in immunology, combined with the development of genetic tools, have expanded our strategies and capabilities to engineer bacteria using various delivery strategies. Depending on the application, each delivery strategy requires specific considerations, optimization, and safety concerns. Here, we review various modes of therapeutic delivery used to target or vaccinate against a variety of ailments in preclinical models and in clinical trials. We highlight modes of bacteria-derived delivery best suited for different applications. Finally, we discuss current obstacles in bacteria-derived therapies and explore potential improvements of the various modes of therapeutic delivery.
Bacteria-derived delivery vehicles are promising therapeutic works-in-progress
Long exploited for fermentation processes in food and chemical industries, bacteria are now being developed as vaccines and ‘live biotherapeutic products’ (see Glossary) to deliver antimicrobials and immunomodulating molecules. Through synthetic biology, defined here as the ‘designing and constructing biological modules, biological systems, and biological machines or, re-design of existing biological systems for useful purposes’, bacteria can become mini-factories and distributors of therapeutics and vaccines with a variety of advantages [1,2]. Bacteria administered intranasally or orally produce and deliver therapeutic once inside the body. Probiotics and attenuated or inactivated pathogenic bacteria are used for vaccine delivery by exploiting their immune-stimulating properties [3]. Because live bacterial delivery vehicles deliver the effector molecule in situ, the recombinant therapeutic does not require purification, which is a major advantage.
Across recent decades, researchers have explored multiple strains of bacteria as therapeutic delivery vehicles, including probiotic strains (Table 1). However, few have reached the clinical trial stage. Lactococcus lactis secreting interleukin-10, for example, reached a Phase 2 clinical trial, but only the safety of the vehicle was established. Multiple clinical trials at various phases are currently underway (see Table S1 in the supplemental information online), and we await the first use of a live biotherapeutic delivery vehicle in practice. Bacteria-based vaccine delivery, contrastingly, has enjoyed greater success. Nonliving, bacteria-derived membrane vesicles (MVs) demonstrated success in humans in the form of Bexsero®, a Neisseria meningitidis serogroup B vaccine that recently received FDA approval [4]. Currently, Advaxis, Inc. is testing a Listeria monocytogenes strain producing a listeriolysin-antigen fusion (tLLO fused to HPV-16 E7) in a Phase 3 clinical trial to treat cervical cancer [see Table S1 in the supplemental information online; National Clinical Trial number (NCT): NCT02853604i].
Table 1.
Reports on bacterial therapeutic delivery vehicles (2017–2022)
| Delivery approach | Organism | Therapeutic or vaccine | Target disease | Modela | Refs |
|---|---|---|---|---|---|
| Secretion | E. coli Nissle | Tum-5 and p53 | B16 melanoma tumor | C57BL/6 mice | [82] |
| Cytotoxic compounds (colibactin, glidobactin, or luminmide) | UT-SCC-5, human head and neck squamous cell carcinoma | Specific-pathogen-free female NMRI nude mice | [83] | ||
| Antimicrobials (Enterocin A, Enterocin B, and Hiracin JM79) | Vancomycin-resistant Enterococcus (VRE) | BALB/c mice | [84] | ||
| L. lactis | Proinsulin and interleukin-10 (IL-10) | Type 1 diabetes | NOD mice | [85] | |
| Pancreatitis-associated protein I (PAP) | Intestinal mucositis | BALB/c mice | [86] | ||
| Interleukin-17A (IL-17A) | HPV-induced cancer | C57BL/6 mice | [87] | ||
| Interleukin-35 (IL-35) | Collagen-induced arthritis | C57BL/6 mice | [88] | ||
| Glucagon-like peptide-1 (GLP-1) | Parkinson’s disease | MPTP-treated C57BL/6 mice | [89] | ||
| GLP-1 | Obesity | High-fat diet-fed C57BL/6 mice | [90] | ||
| β-lactamase | Antibiotic-induced gut dysbiosis | C57BL/6 mice | [91] | ||
| L. reuteri | Interleukin-22 (IL-22) | Inflammatory bowel disease (IBD) | Human intestinal enteroids | [8] | |
| IL-22 | Fatty liver disease | Diet-induced obesity in C57BL/6mice | [6] | ||
| Lacticaseibacillus paracasei | ACE2 | Diabetic retinopathy | Diabetic eNOS−/− mice | [92] | |
| Bifidobacterium longum | Manganese superoxide dismutase (MnSOD) or α-Melanocyte-stimulating hormone (α-SH) | Colitis | DSS-treated nonpathogenic SD rats | [93] | |
| Bifidobacterium breve | Interleukin-24 (IL-24) | Head and neck squamous cell carcinoma | BALB/c male nude mice injected with WSU-HN6 cells | [94] | |
| Membrane vesicles (MVs) and BGs | BG derived from E. coli Nissle | Lewis lung carcinoma (LLC) cryo-lysate | Lung cancer tumor | C57BL/6 mice | [29] |
| MVs derived from N. meningitidis NZ98/254 | Vaccine: Bexsero® | N. meningitidis serogroup B | FDA approved for use in humans | [4] | |
| MVs derived from Salmonella enterica serovar Typhimurium and Salmonella Enteritidis | Vaccine | S. Typhimurium and S. Enteritidis | C57BL/6 mice | [33] | |
| MVs derived from Acinetobacter baumannii | Antibiotic | Enterotoxigenic E. coli (ETEC) | C57BL/6 N mice | [26] | |
| BGs derived from E. coli Nissle | Ciprofloxacin | Intracellular Staphylococcus aureus | C57BL/6 mice | [95] | |
| MV derived from Bacteroides thetaiotaomicron | Keratinocyte growth factor-2 (KGF-2) | Acute colitis | DSS-induced colitis in C57BL/6 mice | [96] | |
| MV derived from E. coli | Tumor antigen | Lung melanoma | C57BL/6 mice | [97] | |
| Antigen/toxin display | Various lactobacilli | Vaccine (fusion antigen AgE6) | Mycobacterium tuberculosis | C57BL/6 BomTac mice | [40] |
| Bacillus toyonensis | Vaccine [tetanus toxin (TTFC)] | Tetanus | BALB/c mice | [39] | |
| Lysis | L. reuteri | IL-22 (phage-mediated lysis) | Alcohol-induced liver disease | Ethanol-binge-fed B6 mice | [51] |
| Increase survival of mice exposed to total body irradiation | C57BL/6NTac and C57BL/6 Lgr5+ mice | [52,54,55] | |||
| L. monocytogenes | Ovalbumin protein (lyses following phagocytosis) | Allergy vaccine | C57BL/6 mice | [56] | |
| S. enterica serovar Typhimurium | Caspase-3 | Hepatocellular carcinoma | BALB/c and C57L/J mice | [98] |
Abbreviations: DSS, dextran sulfate sodium; eNOS, endothelial nitric oxide synthase; MPTP, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; NMRI, Naval Medical Research Institute.
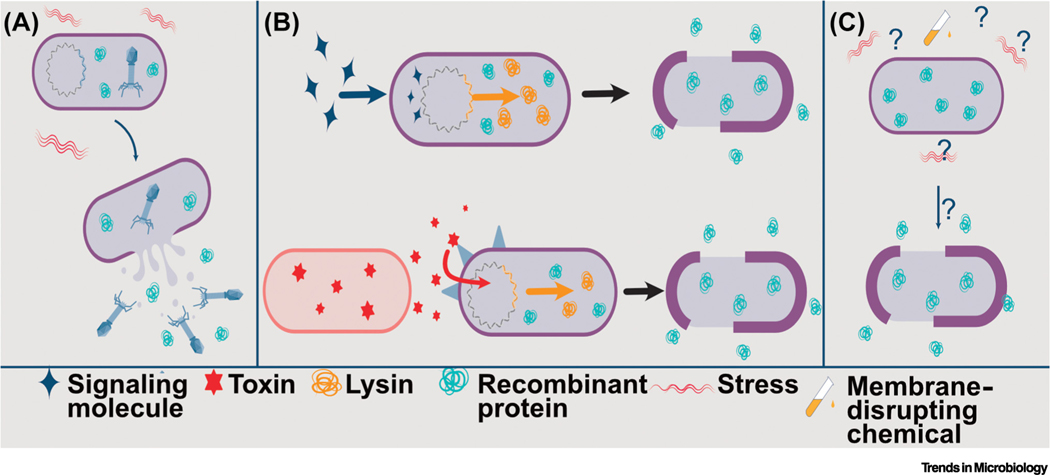
As the bacteria-derived biotherapeutic and vaccine fields have grown, modes of therapeutic delivery have diversified. While various microbial therapeutics have demonstrated success in animal models, optimization studies to increase in vivo performance are mostly lacking. Increasing the efficiency and efficacy of bacteria-based delivery may be critical to narrowing the gap between success in animal trials and the clinic. This review discusses the utility and versatility of four modes of bacteria-based delivery: secretion, membrane vesicles and bacterial ghosts (BGs), surface display, and lysis (Figure 1, Key figure). We examine obstacles to achieving the full potential of bacterial delivery vehicles in humans and provide suggestions on how to approach these challenges.
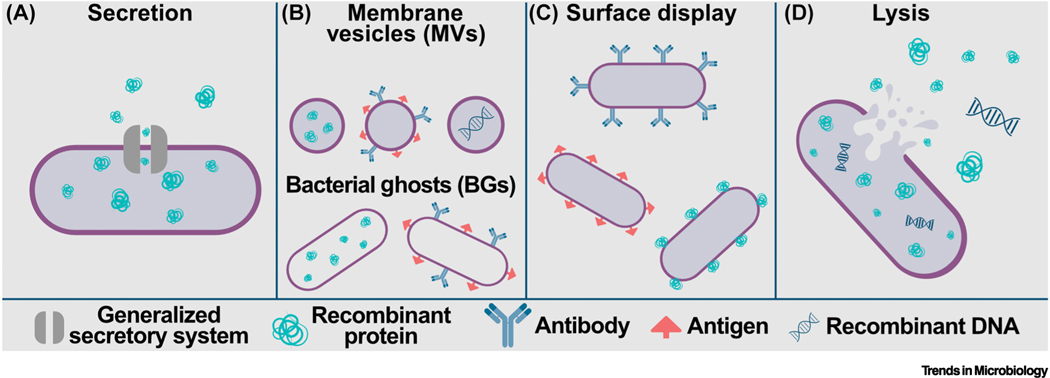
Figure 1.
(A) Secretion systems present in bacteria are exploited as biotherapeutic delivery mechanisms. Secreted products include recombinant proteins with and without fusion to carrier proteins. An overview of secretion systems is given in Figure 2. (B) Membrane vesicles (MVs, top) and bacterial ghosts (BGs, bottom) are nonliving nanobodies derived from both Gram-positive and Gram-negative bacteria that can deliver recombinant proteins when taken up by host cells; or they can display antibody, antigen, or fusion proteins on the surface of the cell envelope. More detailed information about MVs and BGs is given in Figure 3. (C) Surface display by bacteria of enzymes, antigen, or antibody facilitate interactions between recombinant therapeutic and cells or compounds encountered in the host. (D) Bacterial lysis releases intracellularly accumulated recombinant proteins and/or nucleic acids either in the milieu of the gastrointestinal (GI) tract or following bacterial uptake by host cells. Lysis is prophage-mediated, spontaneous, host-driven, or in response to antibiotic exposure. For more detail, see Figure 5.
Key figure
Graphical summary of modes of therapeutic delivery in synthetic biology
Secretion
The native bacterial secretion machinery is the most common method for delivering therapeutics (Table 1 and Figure 2). Therapeutic delivery via secretion maintains the integrity of the bacterial cell to support microbe–host interactions. Close physical associations between the delivery vehicle and host cells at the epithelial barrier have been proposed to contribute to the diffusion of the effector molecules through gaps in tight junctions, thus achieving systemic delivery [5]. While the mechanism of action has not been identified, Oh et al. demonstrated that oral administration of recombinant probiotic-secreting interleukin-22 (IL-22) to mice increased systemic IL-22 levels [6].
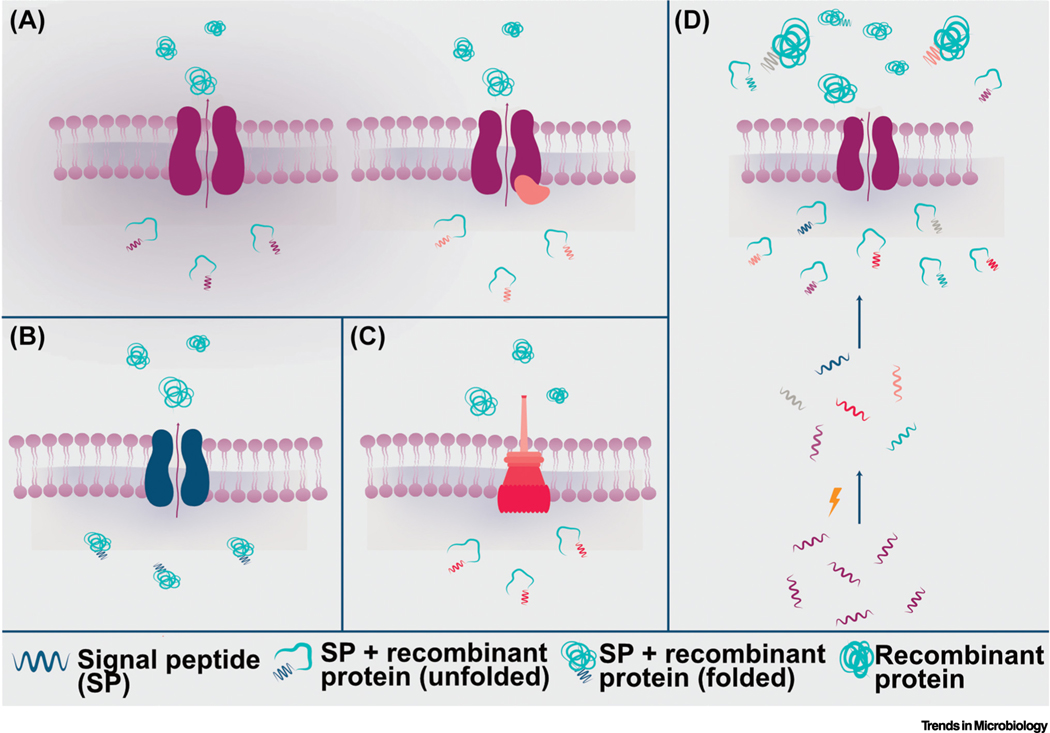
Figure 2. Secretory systems used in biotherapeutic and vaccine delivery.
The most utilized secretion systems for bacterial therapeutic delivery are (A) Sec (+/– SecA2), (B) Tat, and (C) Type III. Briefly, signal peptides target unfolded (Sec, SecA2, and Type III) or folded (Tat) recombinant proteins towards secretory machinery and are cleaved upon translocation of mature protein into the extracellular space. (D) Signal peptide optimization is often required for the secretion of properly cleaved and folded mature recombinant protein, and is a major bottleneck for this mode of delivery. Signal peptide libraries can be created through mutagenesis (lightning bolt), from native signal peptides, and/or from heterologous signal peptides. High-throughput screening of these signal peptides, fused to recombinant proteins, can result in the identification of a signal peptide that is properly cleaved and results in the successful translocation of mature recombinant protein.
Recent clinical trials for bacterial secretion of effector molecules include a Phase 2 study that tested the ability of L. lactis (AG013), secreting human Trefoil Factor 1, to treat oral mucositis, though researchers terminated this study due to lack of efficacy (NCT03234465ii). L. lactis (AG019) secreting hPINS and hIL-10 is currently being tested in a Phase 2a clinical trial to treat type I diabetes (NCT03751007iii, 32). Meanwhile, live vaccine delivery via secretion has made more progress as a mode of delivery, and a L. monocytogenes strain secreting an antigen-adjuvant fusion protein (tLLO-HPV-16 E7) is currently in a Phase 3 clinical trial to treat cervical cancer (NCT02853604i).
Engineering bacteria to secrete recombinant proteins is not trivial. Hijacking the cell’s secretion machinery to secrete the recombinant protein is disadvantageous because it places a burden on an essential system to the cell [7]. The native secretory machinery is responsible for cell envelope biogenesis, energy conversion, and nutrient uptake [7]. Overexpression of recombinant protein that exploits secretion systems can result in fitness defects [8]. Downstream, this can negatively impact the efficiency of delivery and therefore, efficacy. However, some encouraging studies explore methods to circumnavigate the fitness burden imposed by synthetic gene circuits, which are often used to control the expression of therapeutics [9]. For example, orthogonal ribosomes engineered with synthetic 16S rRNA can only translate genes within the synthetic circuit, while host ribosomes can translate both host genes and circuit genes [9,10]. Orthogonal ribosomes dedicated to the gene circuit decrease the cellular burden on the host, and can also be applied to secreted therapeutics [9,10].
The most commonly utilized secretion systems to export recombinant protein (SecA2, Tat and Type III), require a signal peptide to target the protein for secretion (Figure 1) [7]. Signal peptides are not ‘one-size-fits-all’. It is likely that different therapeutic targets require different signal peptides based on the N terminus of the therapeutic [11–13]. For example, some Limosilactobacillus strains encode a signal peptidase that improperly cleaves some therapeutics that begin with a proline [8,14]. Other amino acids and protein structures impede proper signal peptidase cleavage into mature protein, resulting in varying levels of successful production [8,13]. Ortiz et al. illustrated this issue when describing the optimization of secretion of human interleukin-22 (hIL-22) by Limosilactobacillus reuteri. They determined that hIL-22 secreted by L. reuteri was improperly cleaved upon secretion. Mutating the N terminus of hIL-22, and using a signal peptide from Lactobacillus plantarum (Lp_0350), significantly improved hIL-22 processing by L. reuteri. However, based on their observations, and those of Oh et al., appropriate cleavage and production of mature IL-22 require further optimization [6,8]. Therefore, signal peptide design could be a bottleneck for high-throughput development of recombinant therapeutics.
Optimization of the secretion of recombinant proteins
Depending on the therapeutic or vaccine target, various steps can be taken to improve the efficiency and efficacy of recombinant protein secretion. For example, signal peptide optimization can improve the delivery of correctly processed products and increase yield. Screening libraries of mutagenized, native, or heterologous signal peptides is a common strategy for optimizing secretion (Figure 2D) [8,12,13]. A signal peptide from L. plantarum improved the processing and amount of biologically active hIL-22 secreted by L. reuteri even though overall hIL-22 production decreased [8]. Unfortunately, the predictive power of signal peptide analysis for the secretion of recombinant protein is currently limited [11]. Instead, testing a variety of signal peptides for each new recombinant protein is required for the foreseeable future. Recent studies describe tools and suggestions for signal peptide optimization in detail [11,15,16].
Other optimization strategies of recombinant protein secretion involve the use of carrier proteins. For example, random and combinatorial mutagenesis of the E. coli carrier protein OsmY increased secretion of recombinant β-glucosidase threefold [17]. Chaperones, a type of carrier protein, prevent aggregation of recombinant protein and help protein folding by binding to hydrophobic regions. In a detailed review, Mamipour et al. described chaperones with the potential to improve recombinant protein secretion [18]. Carrier proteins may increase the integrity and amount of the secreted therapeutic product, but the effects of the protein fusion on the effector molecule efficacy should be considered and tested.
While the bacterial secretion system appears to be the logical choice to export a protein of interest, findings established in the model organisms Bacillus subtilis or E. coli may not translate to another organism of interest. Through experimentation, a balance will have to be found between efficient secretion and therapeutic production. Combining current secretion optimization strategies, a systematic and logical approach, and preclinical models will be required to ensure biological efficacy of the effector molecule.
Membrane vesicles and bacterial ghosts
MVs and BGs are lipid membranes capable of containing DNA, RNA, and small organic compounds. Various bacterial species (see Table S2 in the supplemental information online) have evolved to naturally emit MVs to traffic signal molecules [19], deliver toxins and anti-growth factors to eukaryotic cells [20,21], and antimicrobials to other bacterial cells [22] (Figure 3). Select pathogenic bacteria employ MVs to transfer virulence factors and antimicrobial resistance proteins between bacterial cells [23]. Whereas MVs are naturally occurring, experimenters must generate BGs. BGs are vacant cell envelopes of Gram-positive and Gram-negative bacteria (see Table S2 in the supplemental information online) generated by disrupting the cell membrane. Expression of a phage-protein lysin (protein E), for example, lyses the bacterial cell, which results in the loss of cytoplasmic contents and generates a BG [24].
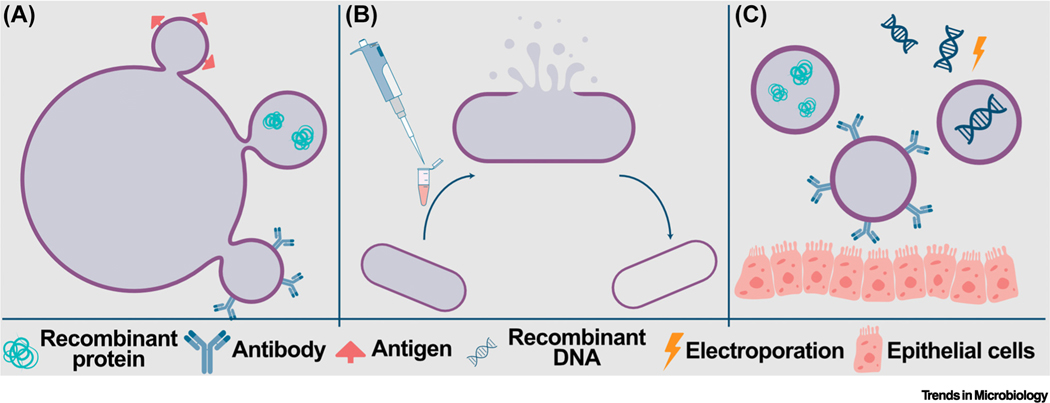
Figure 3. Membrane vesicles (MVs) and bacterial ghosts (BGs).
MVs and BGs can be used to deliver recombinant protein, antigen, antibody, or recombinant DNA (A) MVs are naturally occurring structures that bacteria produce in response to stress, such as antibiotic exposure. (B) BGs form through disruption of the cell membrane that leads to expulsion of the intracellular contents, leaving an empty membrane that can carry proteins and/or nucleic acids. (C) Both MVs and BGs can be engineered to carry recombinant therapeutics and/or DNA (incorporated into the cell envelope via electroporation), or to display antibody, antigen, or fusion proteins that are tethered to the lipid bilayer of the cell envelope. Depending on the design, MVs and BGs can be engineered to facilitate interactions with host cells, such as gut epithelial cells, as depicted here.
An obvious advantage of MVs/BGs as a therapeutic chassis is their stability at – cost-effective – room temperature storage [25]. Depending on the bacteria of origin, MVs/BGs can also contribute to the stability of the payload by protecting it from bile and stomach acids encountered during gastrointestinal transit [26]. In addition to protein, loaded MVs/BGs that enter host cells via phagocytosis (based on the properties of the bacteria of origin) can deliver a DNA cargo intracellularly [27,28]. Jiao et al. engineered a BG (pVAX1-porB) derived from Salmonella Enteritidis to carry DNA encoding the major outer membrane protein of Neisseria gonorrhoeae. Oral immunization of mice with pVAX1-porB conferred bactericidal activity of the serum on N. gonorrhoeae, suggesting that the BG mounted an effective immune response by targeting macrophages [27]. Various groups exploited MVs/BGs to develop treatments for bacterial infections [22,26], cancer therapy [28–30], and vaccines [31,32]. These types of delivery vehicle are versatile in application and have demonstrated success in animal models [26,29].
Another advantage of engineered MVs/BGs is that they can target specific host cells, such as via affibody display [28]. Affibodies are small, robust molecules that, upon fusion to bacterial surface proteins, can target MVs/BGs to specific cells such as tumors [28]. Affibody fusions ensure that MV/BG payloads are only delivered to target cells, and may decrease or eliminate side effects [28]. Additionally, MV/BG vaccines can display immunogenic molecules on their surface, targeting them to immune cells while delivering antigens to mount a more vigorous immune response [33]. Because MVs/BGs are nonliving, there is a relative lack of regulatory hurdles such as antimicrobial sensitivity testing, identification of drug–drug interactions, or virulence factor screening, which, by contrast, apply to live biotherapeutic products [34].
In terms of safety, nonpathogenic-based MV/BG- delivery platforms may be more suitable for patients that are severely immunocompromised compared to live-attenuated vaccines. In contrast to live attenuated vaccines such as Bacillus Calmette–Guérin (BCG), which have diseased patients with primary immunodeficiency diseases (PIDs) [35], it is expected that MV/BG-based vaccines are less likely to cause adverse reactions in patients [36]. Though MVs/BGs carry immunogenic properties, they are nonreplicating and devoid of virulence factors. Notably, an MV vaccine targeting Neisseria meningitidis serogroup B, called Bexsero®, received FDA approval in 2015 [4].
The remaining obstacles to applying MVs/BGs include low membrane vesicle yield and the removal of toxins and lipopolysaccharides (LPS) from Gram-negative bacteria [37]. Some methods to optimize safety include the use of Generally Recognized As Safe (GRAS) strains or genetic modifications to remove LPS and toxins [37]. Unlike live bacterial delivery vehicles, membrane vesicles are strictly carriers and do not produce the therapeutic molecule. Therefore, MVs/BGs require additional engineering steps to enclose the therapeutic payload in the vesicle before administration. We can further optimize MVs/BGs by engineering the bacteria to produce them at high yields, package therapeutic payloads themselves, and release them in specific situations or in response to disease-specific biomarkers. Optimization steps like these would allow for more controlled and efficient delivery of a therapeutic by MVs/BGs.
Surface display
Both pathogenic and nonpathogenic bacteria have immunostimulatory effects, mostly driven by proteins located on the bacterial cell or spore surfaces. Surface display of recombinant antigen or antibody on bacteria can act as potent vaccines by exploiting the immunomodulation ability of bacteria (Figure 4). Generally, vaccines that stimulate both humoral and cell-mediated immunity provoke more robust immune responses than those that stimulate one type of immunity [38]. This is because, when combined humoral immunity targets antigens directly with antibodies, and cell-mediated immunity attacks infected antigen-presenting cells with T cells, this provides a differentiated, two-pronged response. Surface antigen display takes advantage of the natural stimulation of cell-mediated immunity of the bacterial surface while delivering antigen to activate humoral immunity. Therefore, attenuated pathogens and spores are common choices for this type of bacterial therapeutic. In fact, Bacillus spores have historically served as vaccines through surface antigen display due to the spore’s capacity to naturally target immune cells. The hardiness of spores makes Bacillus a valuable chassis for vaccine delivery. For example, oral delivery of B. subtills spores displaying tetanus toxin stimulated both types of immune responses in mice [39].
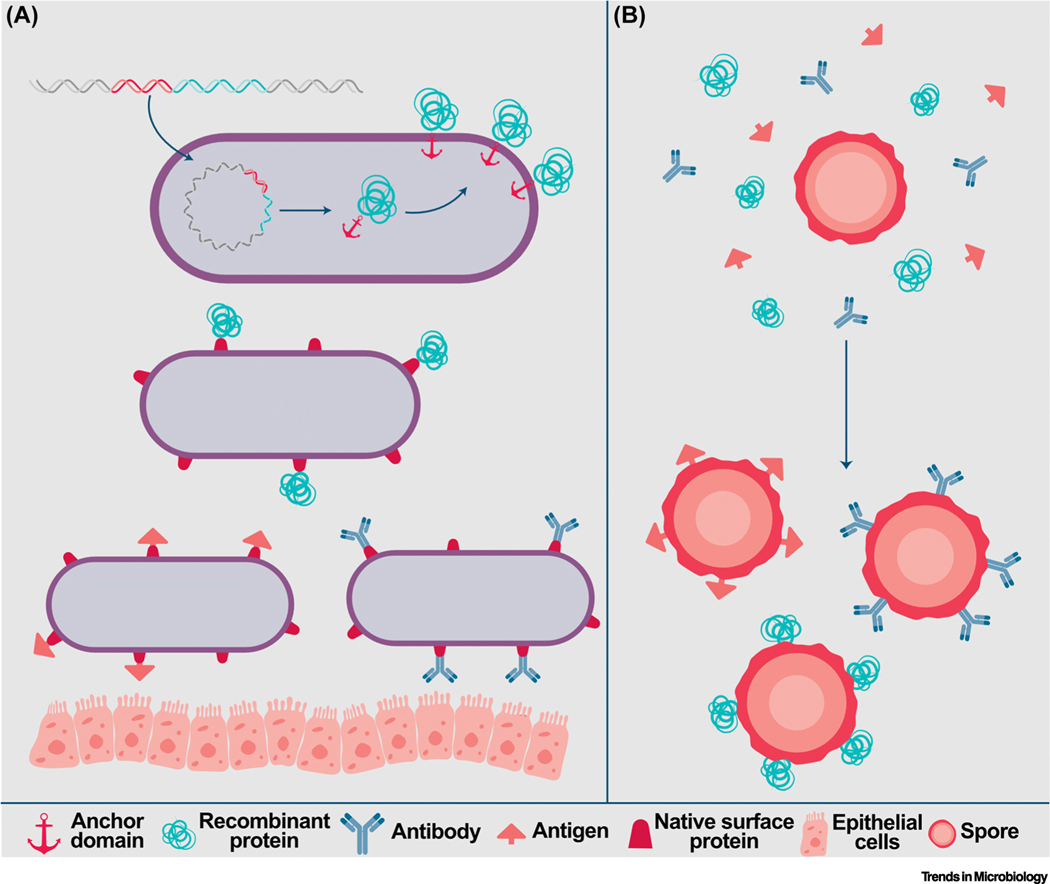
Figure 4. Surface display of recombinant proteins facilitates the delivery of vaccines and therapeutics.
Surface display of fusion of therapeutic proteins to partial or entire surface proteins of (A) non-spore-forming or (B) spore-forming bacteria facilitates their incorporation onto the cell surface and, subsequently, interactions with host cells. Typically, a gene fusion is engineered to encode an anchoring domain or native surface protein attached to a recombinant protein, antibody, or antigen. Through the native bacterial machinery, the resulting fusion protein is incorporated into the cell envelope and exposed to the extracellular space. Alternatively, recombinant protein, antigen, or antibody can be incorporated onto the surface of spore-forming bacteria via adsorption.
Advances in engineering and improved understanding of surface protein anchoring mechanisms allow insertion of exogenous antigens or antibodies into bacterial cell envelopes, enabling surface protein display. Engineered fusion proteins, consisting of full or partial native surface proteins, anchor exogenous antigens to the cell surface, resulting in the production of vaccine candidates in a variety of bacteria [40–42]. Protein fusion design approaches vary from internal modification of existing surface proteins [42], fusion to the C terminus of a recombinant or natural surface protein [41], or fusion to the cell-anchoring domain of a surface protein [40]. Adsorption also allows recombinant antigen attachment to bacterial spores (Figure 4B) [39]. Similarly, nanoliposomes carrying chemotherapeutic agents can be covalently attached to bacteria, such as to Magnetococcus marinus, which was recently used to deliver chemotherapy to colorectal tumors [43]. Cell-wall-anchoring motifs like LPxTG, N-terminal transmembrane helices, or S-layer proteins allow for recombinant antigen and antibody display via protein fusions [40,44].
A potential limitation of surface display, in the case of mucosal vaccines, is that the immunity-stimulating molecule is exposed to harsh conditions caused by bile and stomach acids, which may degrade the recombinant protein [45]. But this may be dependent on the host and type of protein exposed to the surface. For example, in a study comparing L. lactis vaccines targeting human papillomavirus (HPV), researchers demonstrated that surface display of HPV-16 E7 protein evoked a stronger immune response from cytotoxic T lymphocytes than L. lactis either secreting or intracellularly accumulating E7, indicating that degradation of surface-displayed antigen did not impede provocation of an immune response [46]. Continuous production of these cell surface structures possibly mitigates issues related to degradation by constantly replenishing the displayed proteins. Additionally, Bermúdez-Humarán et al. proposed that the L. lactis cell wall components confer immune stimulation, which is the main advantage of bacterial surface display of antigens. Despite this example, antigen or antibody degradation remains a risk in using surface display as a mode of delivery, and should be assessed when testing new chassis candidates.
As genetic tools advance for a broader range of bacteria, robust, nonpathogenic strains that survive gastrointestinal transit should be explored as safer alternatives to pathogenic chassis. Recently, Kuczkowska et al. engineered a variety of Lactobacillus spp. To display antigens of Mycobacterium tuberculosis, and multiple strains demonstrated a protective effect in mice [40]. While relatively narrow in applicability compared to surface display on spores and pathogenic bacteria, probiotic bacteria displaying surface antigens show great promise as vaccine delivery vehicles. Lacticaseibacillus casei displaying HPV E7 protein fused to transmembrane protein PgsA is currently being tested in a Phase 2 clinical trial (see Table S1 in the supplemental information online).
Lysis
Phage-mediated lysis
Prophages are latent bacterial viruses whose genomes incorporate into the bacterial genome. Prophages are abundant in commensal bacteria and probiotics, and they play a dynamic role in bacterial fitness in vivo [47,48]. L. reuteri prophages are activated during gastrointestinal transit, which leads to lysis of a subpopulation of L. reuteri cells to release the bacteriophages (Figure 5A). These bacteriophages provide L. reuteri with a competitive advantage by killing bacteria sensitive to these bacteriophages. [49,50]. To deliver therapeutics, Alexander et al. leveraged the finding that phage lyse part of the L. reuteri population during gastrointestinal (GI) transit. Specifically, L. reuteri engineered to intracellularly accumulate an effector molecule released the recombinant protein upon prophage activation during GI transit [51–53]. This approach has proven successful in different preclinical disease models. Delivery of murine IL-22 by L. reuteri via phage-mediated lysis ameliorated alcohol-induced liver disease in ethanol-binge-fed mice and increased the survival of mice exposed to total body irradiation [51,52,54,55].
Figure 5. Therapeutic delivery via lysis.
(A) Prophage within a bacterial cell can be activated due to stress during gastrointestinal (GI) transit. Bacteria engineered to intracellularly accumulate recombinant therapeutic protein will therefore release therapeutic upon prophage-mediated lysis. (B) (Top) Phage-derived lytic proteins, such as endolysins, can also be exploited to cause cell lysis and subsequent therapeutic release. Lysins cause holes to form in the cell membrane, releasing therapeutic loads. Genes for lytic proteins under the control of an inducible promoter that is activated by the expression of a regulatory or signal molecule can control the timing of lysis that is advantageous for the successful delivery of recombinant protein. (Bottom) Biosensors can also enhance the utility of lysis-mediated delivery of therapeutics by bacteria. For example, the expression of a lytic protein can be activated by toxins produced by pathogenic bacteria via an engineered sensor-response regulatory system. The expression of the lytic protein results in cell lysis and the release of intracellularly accumulated bacteriocin that can then kill the toxin-producing pathogen. (C) A less understood mechanism of bacterial lysis is spontaneous lysis. Stress response(s), the expression of autolysins, or degradation of the cell wall/membrane of the bacterial cell have been proposed as the cause of cell lysis and subsequent release of intracellularly accumulated therapeutic.
In addition to exploiting in vivo activation of native prophage, controlled expression of phage-derived endolysins or holins induce lysis [56–58] (Figure 5B). Induced expression of the lambda lysis gene cluster of E. coli in S. Typhimurium, for example, resulted in lysis and release of intracellularly accumulated cytotoxic Cp53 peptide to kill tumor cells the S. Typhimurium had invaded [57]. Lysis of L. monocytogenes delivering a DNA vaccine was also augmented via expression of a phage-derived lysin (lysA), and a quorum-sensing-driven lysis circuit was used to lyse E. coli Nissle engineered to deliver intracellularly accumulated checkpoint inhibitors to cancer cells [59,60]. Phage and their lytic mechanisms are therefore powerful tools to mediate the release of therapeutics from bacteria.
Biosensors may also enhance the success of therapeutic delivery via phage-mediated-lysis by implementing a sensor-response system, resulting in a ‘smart probiotic’ that can both detect and respond to disease [61]. A ‘smart probiotic’ could induce lysis in response to the presence of a disease marker to release therapeutic based on the disease state. Saeidi et al. demonstrated in vitro efficacy of biosensing E. coli engineered to sense and kill Pseudomonas aeruginosa [61]. Briefly, Saeidi et al. engineered E. coli to respond to quorum sensing molecules (acyl-homoserine lactones; AHLs) derived from P. aeruginosa. In response to the presence of AHLs, the E. coli produces lysin, causing the release of intracellularly accumulated bacteriocin that subsequently kills P. aeruginosa [61]. Inducible quorum sensing systems have also been used to program lysis in response to bacterial population density and to synchronize population-wide lysis and cargo release [60,62].
Prophages are also understood to be relatively specific to their hosts [63]. This is relevant for optimization of a prophage-related delivery mechanism due to the release of virion along with the therapeutic. It is important that the released phages do not disturb the resident microbiota, especially considering that prophages of pathogenic hosts often carry and transfer virulence factors and antimicrobial resistance to other bacteria [64]. With that in mind, natural prophage of pathogenic bacteria should be avoided, and exogenous inducible lysis systems should be used instead when using pathogenic bacteria as delivery vehicles.
Spontaneous lysis
Intracellular accumulation of therapeutics for in situ delivery has demonstrated success in a variety of disease states and as allergen vaccines [65,66]. The mechanism(s) of lysis is in these applications are unknown (Figure 5C). Noncommensal bacteria, such as L. lactis, have not evolved to survive the harsh conditions of the human GI tract [67]. Therefore, it is likely that the release of intracellular therapeutic by L. lactis is due to spontaneous, stress-induced lysis and/or degradation of the cell wall. Prophages do not drive the lysis of L. lactis MG1363, a commonly used strain for biotherapeutic delivery, because L. lactis MG1363 lacks active prophage [68]. Instead, stress-induced expression of autolysin AcmA may cause lysis of L. lactis MG1363 during GI transit [69]. Therefore in addition to prophage, bacterial autolysins are candidates to program lysis [57,58]. While industrial production of molecules has been enhanced by autolysin-mediated lysis, this mechanism has yet to be used for biotherapeutic or vaccine delivery. Further understanding of these underlying stress-response mechanisms will allow similar optimization of lysis as in prophage activation. However, this process currently appears random and fine-tuned control of lysis, regardless of the mechanism, remains a challenge to overcome.
Safety
Regardless of the delivery mechanism employed to deliver the therapeutic, these microbes are considered genetically modified organisms (GMOs). In the USA, GMOs are regulated by the FDA, the US Environmental Protection Agency (EPA), and the US Department of Agriculture (USDA). These agencies ensure that GMOs are safe for human, plant, and animal health, and they monitor the impact of GMOs on the environment [70]. At the same time, most consumers have a limited understanding of GMOs. On the topic of ‘safety of engineered microbes’, consumers typically rely on resources found on the internet, which may lead to misinformation and disinformation, including that GMOs would not be safe or would be less safe than non-GMOs [71]. However, the use of genetic tools to modify the genome of a microbe does not automatically mean that the modified organism is less safe. In fact, modifications can be made to promote safety by, for example, removing or inactivating genes to reduce natural antibiotic resistance [72]. That these examples refer to ‘human-made’ modifications instead of mutations acquired in nature is not relevant to safety. To put this in perspective, here is a hypothetical example. Microbe A has naturally acquired a mutation that increases the production of a valuable protein. Whole-genome sequencing reveals that Microbe A differs by one nucleotide from Microbe B. By genetic modification, scientists alter the single nucleotide in the chromosome of Microbe B. The sequence of the genome of engineered Microbe B is determined and shows that engineered Microbe B is now 100% genetically identical to Microbe A. Further analyses revealed that engineered Microbe B now produces the valuable protein at levels comparable to Microbe A. Based on this example, there is no reason to believe that engineered Microbe B is less safe than the natural isolate Microbe A.
In contrast to genetic engineering, exposing microbes to a chemical that induces mutations in the DNA yields mutated bacteria that are non-GMO [73]. While this approach could be used to screen a library of mutated bacteria to identify a mutation yielding the same phenotype as described for Microbe A, additional mutations are often acquired. Would this non-GMO organism, with multiple mutations throughout the chromosome, be safer than the GMO in which scientists have modified a single base? It is beyond the scope of this work to dive into the details of the different approaches; Pedersen et al. summarize the construction of (non-)genetically modified derivatives of L. lactis along with the analyses of non-GMO and GMO strains [74]. We also refer to the work of Derkx et al., which discusses various strategies to modify food-grade lactic acid bacteria without genetic engineering [75].
Thus, simply because a microbe is engineered, this does not make the engineered microbe less safe than a nonmodified microbe. However, safety could be a concern depending on the therapeutic product delivered combined with the context or ecological framework in which the (microbial) therapeutic is used. Until we have a more comprehensive understanding of the interplay between the host immune system and the microbiome in health and disease, which is complicated by interpersonal variation in the human microbiome [76], one could question approaches to favor or promote long-term colonization of certain GMOs. In the meantime, we must continue to rely on high-quality preclinical research geared towards human applications to reduce the risk to human subjects [77].
Concluding remarks and future perspectives
Modes of bacteria-mediated delivery of vaccines and therapeutics have greatly diversified. The variety of diseases ameliorated in animal models by delivering novel therapeutics has increased as well as the number of clinical trials with microbial therapeutics. Selection of the mode by which recombinant proteins are delivered by the bacteria should be carefully considered and will depend on the disease target and corresponding therapeutic. For transient delivery of therapeutic, intracellular accumulation combined with lysis may be more appropriate, while vaccine delivery may be safely accomplished by bacterial membrane vesicles or ghosts rather than attenuated pathogens.
In addition to the optimization of bacteria-mediated delivery, therapeutic efficacy will also depend on systemic or in situ delivery, or the route of administration (oral, intranasal, intravenous, etc.). In the case of intranasal delivery of therapeutics, secretion may be better suited to achieve an effective dosage than intracellular accumulation. Though there is evidence of systemic delivery of IL-22 produced by L. reuteri, the underlying mechanism for this is unclear [6]. Future studies for mucosal delivery of therapeutics should aim to understand how the systemic delivery of therapeutics is achieved. Examining how bacterial delivery vehicles associate with the epithelial layer of the host gut, perhaps by systematically identifying adhesion proteins, can help to elucidate how the bacteria facilitate therapeutic delivery across the epithelial layer into the bloodstream.
Several challenges remain for the application of bacterial delivery of vaccines and therapeutics in humans (see Outstanding questions). Scalability and batch-to-batch variation are issues in both live biotherapeutics and fermentation processes. Small-scale fermentations often do not progress linearly to large-scale bioreactors and, analogously, dosage effects in mice may not scale linearly in humans [78]. To maximize dosage via the phage-mediated lysis mode of delivery, for example, researchers should consider hijacking the phage regulatory mechanism to increase lysis and subsequent therapeutic release. Optimization steps for fermenter scale-up from the biofuel and fermentation industries can provide guidance for addressing bacterial culture scalability, though applying these principles to a chosen delivery vehicle will likely require further customization [79,80]. In terms of safety, robust biological and environmental containment strategies for recombinant bacteria are still in their infancy [81]. Lastly, while the term ‘personalized medicine’ is often used, we are far removed from its application. Studies aiming to yield a mechanistic understanding of microbial ecology, including taking into consideration differences at the strain level, are expected to advance the field towards therapeutic and vaccine success in humans. Projecting forward, we envision that future application of microbial therapeutics and vaccines will be based on a holistic approach guided by the ecological microbiome footprint, and if needed, combined with traditional medicine.
Outstanding questions.
To what extent does the microbiome composition impact the therapeutic efficacy of (engineered) probiotics? And vice versa, to what extent does a microbial therapeutic alter the composition of the microbiome? Can we increase therapeutic efficacy when microbial therapies are combined with standardized diets?
Can evolutionary and ecological insights into microbe–host interactions lead ultimately to more efficacious microbial therapies? Can this, for example, be accomplished by implementing knowledge from microbial ecological networks? How can this be leveraged towards personalized medicine?
What efforts should be taken to replace fear-based regulation with risk-based regulation with regard to the development and use of engineered microbes in the clinic and environment?
Should we be concerned about the potential for long-term colonization of an engineered probiotic?
How reliable are current bioinformatic pipeline models to predict the secretion and processing of recombinant proteins, and what advances can be made?
What are the factors that contribute to discrepancies in the success of bacteria-derived delivery vehicles in animal models compared to human clinical trials? How can these obstacles be overcome? For example, would the assessment of therapeutic efficacy in multiple animal models help us better understand current discrepancies?
How can the overall scalability and efficacy of bacteria-derived vaccine and therapeutic delivery vehicles be improved?
Supplementary Material
Highlights.
Bacteria-derived therapeutics and vaccines have proven effective in various animal disease models, and several are being used in active clinical trials.
Advances in genetic tools and expression systems have diversified both the bacteria used and the way they are engineered as therapeutic and vaccine delivery vehicles. Secretion, membrane vesicles or bacterial ghosts, surface display, and cell lysis are all modes of delivery by bacteria-derived therapeutics and vaccines; each with its own advantages and disadvantages.
Challenges to overcome are the development and implementation of widely applicable and robust containment strategies, and implementing knowledge on mechanistic microbial ecology – perhaps combined with traditional medicine – towards microbial therapeutic design as a step towards personalized medicine.
Acknowledgments
L.M.A. was awarded the Louis and Elsa Thomsen Wisconsin Distinguished Graduate Fellowship from the College of Agricultural and Life Sciences and the Dissertation Completion Fellowship from the Graduate School, University of Wisconsin-Madison. J-P.V.P. receives funding from the UW-Madison Food Research Institute, the National Institute of Allergy and Infectious Diseases, the National Institute of General Medical Sciences and the National Center for Complementary and Integrative Health of the National Institutes of Health under award numbers R21AI146634, R01GM135483, and R01AT011202, respectively. J-P.V.P also acknowledges support from Animal Health and Production and Animal Products: Animal Health and Disease (grant no. 2021-67015-34316)] from the USDA National Institute of Food and Agriculture.
Glossary
- Adjuvant
a substance which enhances the body’s immune response to an antigen
- Affibody
a small, robust molecule that, upon fusion to a bacterial surface protein, can target MVs/BGs to specific cells such as tumors
- Antibody
a protein produced in response to, and counteracting, a specific antigen. Antibodies combine chemically with substances which the body recognizes as alien, such as pathogens and toxins
- Antigen
a toxin or other foreign substance which induces an immune response in the body, especially the production of antibodies
- Autolysins
endogenous lytic enzymes that break down the cell wall of bacteria
- Bacterial ghost (BG)
vacant cell envelope of Gram-positive and Gram-negative bacteria generated by disrupting the cell membrane; BGs can contain DNA, RNA, and small organic compounds
- Biomarker
a biological molecule, found in the body, that is a sign of a normal or abnormal process, or of a condition or disease
- Biosensor
a living organism or biological molecule used to detect the presence of disease
- Biotherapeutics
therapy products in which the active substance is extracted or produced from a biological source
- Cell-mediated immunity
an immune response that does not involve antibodies and attacks antigen-presenting cells with T cells
- Chaperones
proteins that help other proteins to fold properly and/or to translocate within and outside the cell
- Holin
small protein produced by bacteriophage that leads to the degradation of the cell wall and, eventually, lysis
- Humoral immunity
antibody-mediated immunity; it targets antigens on pathogens directly
- Lipopolysaccharide (LPS)
the major component of the outer membrane of Gram-negative bacteria that often acts as a toxin of pathogenic bacteria
- Live biotherapeutic products
microorganisms that are engineered as delivery systems for antimicrobials, cancer therapy, and immunomodulating molecules
- Lysin
an enzyme that degrades peptidoglycan
- Membrane vesicle (MV)
naturally occurring lipid membrane emitted by bacteria and capable of containing DNA, RNA, and small organic compounds
- Phagocytosis
the ingestion of bacteria or other material by human cells. Probiotics: live microorganisms that, when administered in adequate amounts, confer a health benefit on the host
- Prophage
latent bacterial virus whose genome incorporate into the bacterial genome
- Signal peptide
short peptide located in the N terminus of proteins that carries information for protein secretion
- Smart probiotic
microorganism that can detect and respond to environmental cues in real time within a host
- Surface display
a protein-engineering technique in which a recombinant protein is anchored to the bacterial cell membrane and exposed to the extracellular space
- Tight junctions
barriers between epithelial and endothelial cells that regulate the diffusion of molecules across tissues
- Transmembrane protein
a membrane protein that spans the entirety of the cell membrane
- Virulence factor
bacteria-associated molecules that are required for a bacterium to cause disease
Footnotes
Declaration of interests
J-P.V.P received unrestricted funds from BioGaia AB, a probiotic-producing company. J-P.V.P is the founder of the consulting company Next-Gen Probiotics, LLC.
Supplemental information
Supplemental information associated with this article can be found online at https://doi.org/10.1016/j.tim.2022.09.003.
This study is registered with ClinicalTrials.gov: https://clinicaltrials.gov/ct2/show/NCT02853604
This study is registered with ClinicalTrials.gov: https://clinicaltrials.gov/ct2/show/NCT03234465
This study is registered with ClinicalTrials.gov: https://clinicaltrials.gov/ct2/show/NCT03751007
References
- 1.Nakano T. et al. (2013) Molecular Communication, Cambridge University Press [Google Scholar]
- 2.del Rio B. et al. (2018) Lactic acid bacteria as a live delivery system for the in situ production of nanobodies in the human gastrointestinal tract. Front. Microbiol 9, 3179 [Google Scholar]
- 3.Lebeer S. et al. (2010) Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol 8, 171–184 [DOI] [PubMed] [Google Scholar]
- 4.Gorringe AR and Pajón R. (2012) Bexsero. Hum. Vacc. Immunother. 8, 174–183 [DOI] [PubMed] [Google Scholar]
- 5.Hollander D. and Kaunitz JD (2020) The ‘Leaky Gut’: tight junctions but loose associations? Dig. Dis. Sci 65, 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh J-H et al. (2020) Secretion of recombinant Interleukin-22 by engineered Lactobacillus reuteri reduces fatty liver disease in a mouse model of diet-induced obesity. mSphere 5, e00183–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green ER and Mecsas J. (2016) Bacterial secretion systems: an overview. In Virulence Mechanisms of Bacterial Pathogens (Kudva IT et al., eds), pp. 213–239, ASM; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortiz-Velez L. et al. (2020) Challenges and pitfalls in the engineering of human Interleukin 22 (hIL-22) secreting Lactobacillus reuteri. Front. Bioeng. Biotechnol 8, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grob A. et al. (2021) Experimental tools to reduce the burden of bacterial synthetic biology. Curr. Opin. Syst. Biol 28, 100393 [Google Scholar]
- 10.Darlington APS et al. (2018) Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes. Nat. Commun 9, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freudl R. (2018) Signal peptides for recombinant protein secretion in bacterial expression systems. Microb. Cell Factories 17, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao D. et al. (2019) Enhanced extracellular expression of Bacillus stearothermophilus α-amylase in Bacillus subtilis through signal peptide optimization, chaperone overexpression and α-amylase mutant selection. Microb. Cell Factories 18, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu G. et al. (2018) Systematic screening of optimal signal peptides for secretory production of heterologous proteins in Bacillus subtilis. J. Agric. Food Chem 66, 13141–13151 [DOI] [PubMed] [Google Scholar]
- 14.Duar RM et al. (2015) Identification and characterization of intestinal lactobacilli strains capable of degrading immunotoxic peptides present in gluten. J. Appl. Microbiol 118, 515–527 [DOI] [PubMed] [Google Scholar]
- 15.Peng C. et al. (2019) Factors influencing recombinant protein secretion efficiency in gram-positive bacteria: signal peptide and beyond. Front. Bioeng. Biotechnol 7, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleiner-Grote GRM et al. (2018) Secretion of recombinant proteins from E. coli. Eng. Life Sci 18, 532–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Perez D. et al. (2021) Random and combinatorial mutagenesis for improved total production of secretory target protein in Escherichia coli. Sci. Rep 11, 5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mamipour M. et al. (2017) An overview on molecular chaperones enhancing solubility of expressed recombinant proteins with correct folding. Int. J. Biol. Macromol 102, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toyofuku M. et al. (2017) Membrane vesicle-mediated bacterial communication. ISME J. 11, 1504–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondal A. et al. (2016) Cytotoxic and Inflammatory responses induced by outer membrane vesicle-associated biologically active proteases from Vibrio cholerae. Infect. Immun 84, 1478–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan S. et al. (2021) Oral delivery of a Lactococcus lactis expressing extracellular TGFβR2 alleviates hepatic fibrosis. Appl. Microbiol. Biotechnol 105, 6007–6018 [DOI] [PubMed] [Google Scholar]
- 22.Dean SN et al. (2020) Lactobacillus acidophilus membrane vesicles as a vehicle of bacteriocin delivery. Front. Microbiol 11, 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner T. et al. (2018) Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J. Proteome 187, 28–38 [DOI] [PubMed] [Google Scholar]
- 24.Panthel K. et al. (2003) Generation of Helicobacter pylori ghosts by PhiX Protein E-mediated inactivation and their evaluation as vaccine candidates. Infect. Immun 71, 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeyaram A. and Jay SM (2017) Preservation and storage stability of extracellular vesicles for therapeutic applications. AAPS J. 20, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W. et al. (2020) Development of novel nanoantibiotics using an outer membrane vesicle-based drug efflux mechanism. J. Control. Release 317, 1–22 [DOI] [PubMed] [Google Scholar]
- 27.Jiao H. et al. (2018) Design and immune characterization of a novel Neisseria gonorrhoeae DNA vaccine using bacterial ghosts as vector and adjuvant. Vaccine 36, 4532Outstanding questions –4539 [DOI] [PubMed] [Google Scholar]
- 28.Gujrati V. et al. (2014) Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano 8, 1525–1537 [DOI] [PubMed] [Google Scholar]
- 29.Kraśko JA et al. (2017) Bacterial ghosts as adjuvants in syngeneic tumour cell lysate-based anticancer vaccination in a murine lung carcinoma model. Oncol. Rep 37, 171–178 [DOI] [PubMed] [Google Scholar]
- 30.Youssof AME et al. (2019) Bacterial ghosts carrying 5-fluorouracil: a novel biological carrier for targeting colorectal cancer. AAPS PharmSciTech 20, 48. [DOI] [PubMed] [Google Scholar]
- 31.Pan Q. et al. (2015) Comparative evaluation of the protective efficacy of two formulations of a recombinant Chlamydia abortus subunit candidate vaccine in a mouse model. Vaccine 33, 1865–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rappazzo CG et al. (2016) Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine 34, 1252–1258 [DOI] [PubMed] [Google Scholar]
- 33.Micoli F. et al. (2018) Comparative immunogenicity and efficacy of equivalent outer membrane vesicle and glycoconjugate vaccines against nontyphoidal Salmonella. PNAS 115, 10428–10433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouanet A. et al. (2020) Live biotherapeutic products, a road map for safety assessment. Front. Med 7, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norouzi S. et al. (2012) Bacillus Calmette-Guérin (BCG) complications associated with primary immunodeficiency diseases. J. Infect 64, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farjadian F. et al. (2018) Bacterial components as naturally inspired nano-carriers for drug/gene delivery and immunization: Set the bugs to work? Biotechnol. Adv 36, 968–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bitto NJ and Kaparakis-Liaskos M. (2017) The therapeutic benefit of bacterial membrane vesicles. Int. J. Mol. Sci 18, 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clem AS (2011) Fundamentals of vaccine immunology. J. Global Infect. Dis 3, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos FDS et al. (2020) A probiotic treatment increases the immune response induced by the nasal delivery of spore-adsorbed TTFC. Microb. Cell Factories 19, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuczkowska K. et al. (2019) Comparison of eight Lactobacillus species for delivery of surface-displayed mycobacterial antigen. Vaccine 37, 6371–6379 [DOI] [PubMed] [Google Scholar]
- 41.Li R. et al. (2015) Mucosally administered Lactobacillus surface-displayed influenza antigens (sM2 and HA2) with cholera toxin subunit A1 (CTA1) Induce broadly protective immune responses against divergent influenza subtypes. Vet. Microbiol 179, 250–263 [DOI] [PubMed] [Google Scholar]
- 42.Kajikawa A. et al. (2015) Mucosal immunogenicity of genetically modified Lactobacillus acidophilus expressing an HIV-1 Epitope within the Surface Layer Protein. PLoS One 10, e0141713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felfoul O. et al. (2016) Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nat. Nanotechnol 11, 941–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X. et al. (2016) Heterologous expression of carcinoembryonic antigen in Lactococcus lactis via LcsB-mediated surface displaying system for oral vaccine development. J. Microbiol. Immunol. Infect 49, 851–858 [DOI] [PubMed] [Google Scholar]
- 45.Michon C. et al. (2016) Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb. Cell Factories 15, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bermúdez-Humarán LG et al. (2004) An inducible surface presentation system improves cellular immunity against human papillomavirus type 16 E7 antigen in mice after nasal administration with recombinant lactococci. J. Med. Microbiol 53, 427–433 [DOI] [PubMed] [Google Scholar]
- 47.Kim M-S and Bae J-W (2018) Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J. 12, 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shkoporov AN and Hill C. (2019) Bacteriophages of the human gut: the ‘known unknown’ of the microbiome. Cell Host Microbe 25, 195–209 [DOI] [PubMed] [Google Scholar]
- 49.Oh J-H et al. (2019) Prophages in Lactobacillus reuteri are associated with fitness trade-offs but can increase competitiveness in the gut ecosystem. Appl. Environ. Microbiol 86, e01922–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh J-H et al. (2019) Dietary fructose and microbiota-derived short-chain fatty acids promote bacteriophage production in the gut symbiont Lactobacillus reuteri. Cell Host Microbe 25, 273–284.e6 [DOI] [PubMed] [Google Scholar]
- 51.Hendrikx T. et al. (2018) Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X. et al. (2020) Second-generation probiotics producing IL-22 increase survival of mice after total body irradiation. In Vivo 34, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander LM et al. (2019) Exploiting prophage-mediated lysis for biotherapeutic release by Lactobacillus reuteri. Appl. Environ. Microbiol 85, e02335–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamade DF et al. (2022) Lactobacillus reuteri releasing IL-22 (LR-IL-22) facilitates intestinal radioprotection for whole-abdomen irradiation (wai) of ovarian cancer. Radiat. Res 198, 89–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espinal A. et al. (2022) Intestinal radiation protection and mitigation by second-generation probiotic Lactobacillus-reuteri engineered to deliver Interleukin-22. Int. J. Mol. Sci 23, 5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha S. et al. (2017) A suicidal strain of Listeria monocytogenes is effective as a DNA vaccine delivery system for oral administration. Vaccine 35, 5115–5122 [DOI] [PubMed] [Google Scholar]
- 57.Camacho EM et al. (2016) Engineering Salmonella as intracellular factory for effective killing of tumour cells. Sci. Rep 6, 30591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menacho-Melgar R. et al. (2020) Improved, two-stage protein expression and purification via autoinduction of both autolysis and auto DNA/RNA hydrolysis conferred by phage lysozyme and DNA/RNA endonuclease. Biotechnol. Bioeng 117, 2852–2860 [DOI] [PubMed] [Google Scholar]
- 59.van Pijkeren JP et al. (2010) A novel Listeria monocytogenes-based dna delivery system for cancer gene therapy. Hum. Gene Ther 21, 405–416 [DOI] [PubMed] [Google Scholar]
- 60.Gurbatri CR et al. (2020) Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med 12, eaax0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saeidi N. et al. (2011) Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol. Syst. Biol 7, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miano A. et al. (2020) Inducible cell-to-cell signaling for tunable dynamics in microbial communities. Nat. Commun 11, 1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koskella B. and Meaden S. (2013) Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M. et al. (2020) Role of enterotoxigenic Escherichia coli prophage in spreading antibiotic resistance in a porcine-derived environment. Environ. Microbiol 22, 4974–4984 [DOI] [PubMed] [Google Scholar]
- 65.Sarate PJ et al. (2019) E. coli Nissle 1917 is a safe mucosal delivery vector for a birch-grass pollen chimera to prevent allergic poly-sensitization. Mucosal Immunol. 12, 132–144 [DOI] [PubMed] [Google Scholar]
- 66.Yang G. et al. (2015) Effective treatment of hypertension by recombinant Lactobacillus plantarum expressing angiotensin converting enzyme inhibitory peptide. Microb. Cell Factories 14, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobson A. et al. (2011) Fate and efficacy of lacticin 3147-producing Lactococcus lactis in the mammalian gastrointestinal tract. FEMS Microbiol. Ecol 76, 602–614 [DOI] [PubMed] [Google Scholar]
- 68.Wegmann U. et al. (2007) Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol 189, 3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steen A. et al. (2005) Autolysis of Lactococcus lactis is increased upon d-Alanine depletion of peptidoglycan and lipoteichoic acids. J. Bacteriol 187, 114–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jennings Jerry D. (1986) Coordinated Framework for Regulation of Biotechnology, Office of Science and Technology Policy, Washington, D.C., USA [Google Scholar]
- 71.Wunderlich S. and Gatto KA (2015) Consumer perception of genetically modified organisms and sources of information. Adv. Nutr 6, 842–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Pijkeren J-P and Britton RA (2012) High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 40, e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Breyer D. et al. (2009) Commentary: genetic modification through oligonucleotide-mediated mutagenesis. A GMO regulatory challenge? Environ. Biosaf. Res 8, 57–64 [DOI] [PubMed] [Google Scholar]
- 74.Pedersen MB et al. (2005) The long and winding road from the research laboratory to industrial applications of lactic acid bacteria. FEMS Microbiol. Rev 29, 611–624 [DOI] [PubMed] [Google Scholar]
- 75.Derkx PM et al. (2014) The art of strain improvement of industrial lactic acid bacteria without the use of recombinant DNA technology. Microb. Cell Factories 13, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta VK et al. (2017) Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol 8, 1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kimmelman J. (2007) Ethics at phase 0: clarifying the issues. J. Law Med. Ethics 35, 727–733 [DOI] [PubMed] [Google Scholar]
- 78.Walter J. et al. (2020) Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180, 221–232 [DOI] [PubMed] [Google Scholar]
- 79.Wang G. et al. (2020) Developing a computational framework to advance bioprocess scale-up. Trends Biotechnol. 38, 846–856 [DOI] [PubMed] [Google Scholar]
- 80.Humbird D. and Fei Q. (2016) Chapter 20. Scale-up considerations for biofuels. In Biotechnology for Biofuel Production and Optimization (Eckert CA and Trinh CT, eds), pp. 513–537, Elsevier [Google Scholar]
- 81.Yao M. et al. (2017) Microencapsulation of Lactobacillus salivarious Li01 for enhanced storage viability and targeted delivery to gut microbiota. Food Hydrocoll. 72, 228–236 [Google Scholar]
- 82.He L. et al. (2019) Intestinal probiotics E. coli Nissle 1917 as a targeted vehicle for delivery of p53 and Tum-5 to solid tumors for cancer therapy. J. Biol. Eng 13, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li R. et al. (2019) Expressing cytotoxic compounds in Escherichia coli Nissle 1917 for tumor-targeting therapy. Res. Microbiol 170, 74–79 [DOI] [PubMed] [Google Scholar]
- 84.Geldart KG et al. (2018) Engineered E. coli Nissle 1917 for the reduction of vancomycin-resistant Enterococcus in the intestinal tract. Bioeng. Transl. Med 3, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takiishi T. et al. (2017) Reversal of diabetes in NOD mice by clinical-grade proinsulin and il-10–secreting Lactococcus lactis in combination with low-dose anti-cd3 depends on the induction of foxp3-positive t cells. Diabetes 66, 448–459 [DOI] [PubMed] [Google Scholar]
- 86.Carvalho RD et al. (2017) Secretion of biologically active pancreatitis-associated protein I (PAP) by genetically modified dairy Lactococcus lactis NZ9000 in the prevention of intestinal mucositis. Microb. Cell Factories 16, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jacouton E. et al. (2019) Anti-tumoral effects of recombinant Lactococcus lactis strain secreting IL-17A cytokine. Front. Microbiol 9, 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maddaloni M. et al. (2018) Delivery of IL-35 by Lactococcus lactis ameliorates collagen-induced arthritis in mice. Front. Immunol 9, 2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fang X. et al. (2019) Neuroprotective effects of an engineered commensal bacterium in the 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine Parkinson disease mouse model via producing glucagon-like peptide-1. J. Neurochem 150, 441–452 [DOI] [PubMed] [Google Scholar]
- 90.Wang L. et al. (2021) Engineered bacteria of MG1363-pMG36e-GLP-1 attenuated obesity-Induced by high fat diet in mice. Front. Cell. Infect. Microbiol 11, 595–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cubillos-Ruiz A. et al. (2022) An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis. Nat. Biomed. Eng 6, 910–921 [DOI] [PubMed] [Google Scholar]
- 92.Verma A. et al. (2019) Expression of human ACE2 in Lactobacillus and beneficial effects in diabetic retinopathy in mice. Mol. Ther. Methods Clin. Develop 14, 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu M. et al. (2018) Oral engineered Bifidobacterium longum expressing rhMnSOD to suppress experimental colitis. Int. Immunopharmacol 57, 25–32 [DOI] [PubMed] [Google Scholar]
- 94.Wang L. et al. (2017) Bifidobacterium breve as a delivery vector of IL-24 gene therapy for head and neck squamous cell carcinoma in vivo. Gene Ther. 24, 699–705 [DOI] [PubMed] [Google Scholar]
- 95.Xie S. et al. (2020) Bacterial ghosts for targeting delivery and subsequent responsive release of ciprofloxacin to destruct intracellular bacteria. Chem. Eng. J 399, 125700 [Google Scholar]
- 96.Carvalho AL et al. (2019) Bioengineering commensal bacteria-derived outer membrane vesicles for delivery of biologics to the gastrointestinal and respiratory tract. J. Extracell. Vesicles 8, 1632100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng K. et al. (2021) Bioengineered bacteria-derived outer membrane vesicles as a versatile antigen display platform for tumor vaccination via Plug-and-Display technology. Nat. Commun 12, 2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raman V. et al. (2021) Intracellular delivery of protein drugs with an autonomously lysing bacterial system reduces tumor growth and metastases. Nat. Commun 12, 6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.