Abstract
BACKGROUND:
When aortic cells are under stress, such as increased hemodynamic pressure, they adapt to the environment by modifying their functions, allowing the aorta to maintain its strength. To understand the regulation of this adaptive response, we examined transcriptomic and epigenomic programs in aortic smooth muscle cells (SMCs) during the adaptive response to angiotensin II (AngII) infusion and determined its importance in protecting against aortic aneurysm and dissection (AAD).
METHODS:
We performed single-cell RNA sequencing (scRNA-seq) and single-cell sequencing assay for transposase-accessible chromatin (scATAC-seq) analyses in a mouse model of sporadic AAD induced by AngII infusion. We also examined the direct effects of YAP on the SMC adaptive response in vitro. The role of YAP in AAD development was further evaluated in AngII-infused mice with SMC-specific Yap deletion.
RESULTS:
In wild-type mice, AngII infusion increased medial thickness in the thoracic aorta. ScRNA-seq analysis revealed an adaptive response in thoracic SMCs characterized by upregulated genes with roles in wound healing, elastin and collagen production, proliferation, migration, cytoskeleton organization, cell-matrix focal adhesion, and AKT and TGF-β signaling. ScATAC-seq analysis showed increased chromatin accessibility at regulatory regions of adaptive genes and revealed the mechanical sensor YAP/TEADs as a top candidate transcription complex driving the expression of these genes (e.g., Lox, Col5a2, Tgfb2). In cultured human aortic SMCs, cyclic stretch activated YAP, which directly bound to adaptive gene regulatory regions (e.g., Lox) and increased their transcript abundance. SMC-specific Yap deletion in mice compromised this adaptive response in SMCs, leading to an increased AAD incidence.
CONCLUSIONS:
Aortic stress triggers the systemic epigenetic induction of an adaptive response (e.g., wound healing, proliferation, matrix organization) in thoracic aortic SMCs that depends on functional biomechanical signal transduction (e.g., YAP signaling). Our study highlights the importance of the adaptive response in maintaining aortic homeostasis and preventing AAD in mice.
Keywords: aortic aneurysm and dissection, biomechanical environment, extracellular matrix, smooth muscle cell
Graphic Abstract
INTRODUCTION
The aortic wall is exposed to a constantly changing environment. Under stress such as increased hemodynamic pressure, the aortic wall senses mechanical signals, undergoes remodeling, and increases its thickness to maintain aortic strength and sustain pressure.1,2 This adaptive response is orchestrated by multiple cell types, particularly aortic smooth muscle cells (SMCs), that receive mechanical signals and activate signaling and gene expression to increase aortic wall strength.3–5 Various genetic defects and environmental risk factors may compromise this adaptive response,6–10 resulting in aortic failure, aortic aneurysm and dissection (AAD) development, and, ultimately, rupture.11 However, the molecular and cellular processes of this adaptive response and its regulation are poorly understood.
To characterize the responses of SMCs to aortic stress, we performed single-cell RNA sequencing (scRNA-seq) and single-cell sequencing assay for transposase-accessible chromatin (scATAC-seq) analyses of aortas from a mouse model of sporadic AAD induced by challenging wild-type mice with a high-fat diet (HFD) and angiotensin II (AngII) infusion. We observed that AngII infusion induced an increase in medial thickness that was associated with a comprehensive and coordinated adaptive gene expression program in thoracic aortic SMC populations to promote adaptive features such as cell proliferation, extracellular matrix (ECM) production, and wound healing. Yes-associated protein (YAP) was identified as a key transcription factor (TF) driving this adaptive response that is critical for maintaining aortic homeostasis and protecting the aorta from medial degeneration and the development of aneurysm and dissection.
MATERIALS AND METHODS
Animal Studies
Mice were maintained in the Center for Comparative Medicine at Baylor College of Medicine, and all animal procedures were performed according to a protocol (AN-4195) approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine. All animal experiments complied with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (NIH Publication No. 8023, revised 1978). The mice were maintained on a 12-hour light/12-hour dark cycle at a controlled temperature of 21 ± 1 °C and had free access to water and either a standard laboratory diet (Mouse Diet, no. 5015, LabDiet, St. Louis, MO) or a Western HFD (D12108C, Research Diets Inc., New Brunswick, NJ).
Mice
Male wild-type (WT; C57BL/6J), Myh11-CreERT2 (B6.FVB-Tg [Myh11-cre/ERT2]1Soff/J), and Yap1fl/fl (STOCK Yap1tm1.1Dupa/J) mice (Table S1 in the Supplemental Material) were obtained from The Jackson Laboratory (Bar Harbor, ME) and genotyped by using polymerase chain reaction (PCR). In our modified12 mouse model of sporadic AAD, sporadic AAD was induced by challenging WT mice with a combination of a HFD and AngII infusion, as described previously.13–15 Briefly, four-week-old male WT mice were fed with a HFD (4.5 kcal/g, 20% protein, 40% carbohydrate, 40% fat, and 1.25% cholesterol; Research Diets, Inc., D12108C, New Brunswick, NJ) for 8 weeks and AngII infusion (2,000 ng/kg/min AngII; cat# A2900-50MG, Sigma-Aldrich Corp., St. Louis, MO, USA) during the last 4 weeks through an osmotic minipump (Model 2004; ALZA Scientific Products, Mountain View, CA, USA). Mice fed a standard rodent laboratory diet for 8 weeks and infused with saline during the last 4 weeks were used as controls.
To study mice with the SMC-specific deletion of Yap, we used four-week-old male Yap1f/f; Myh11-CreERT2+ mice in which the Cre allele was inserted into the Y chromosome (X/YCre+). When the mice were 6 weeks old, they were given 2 mg/day intraperitoneal injections of tamoxifen (Sigma T-5648) for 5 consecutive days to induce the SMC-specific deletion of Yap (SMC-Yap−/−; n=23). Littermates of Yap1f/f; Myh11-CreERT2+ mice injected with corn oil (vehicle) were used as a control (SMC-Yap+/+; n=20). After the last injection, the mice were rested for 7 days for Cre recombination. Eight-week-old male SMC-Yap−/− or SMC-Yap+/+ mice were infused with either saline or a low-rate of AngII infusion (1000 ng/kg/min) for 4 weeks and fed with either a standard rodent laboratory diet or HFD. Mice were euthanized by CO2 asphyxiation and were then perfused with 10 ml ice-cold phosphate-buffered saline (PBS) via the left ventricle. Mice were allocated to experimental groups on the basis of genotypes and were then randomized for various experimental procedures.
For the scRNA-seq and scATAC-seq studies, we collected aortic tissues after 7 days of AngII infusion to capture the early response to aortic stress. Only male mice were studied because male mice have a higher incidence of aortic dissection,16 and the Cre allele of Myh11-CreERT2 mice was inserted into the Y chromosome (X/YCre+).
Mice Inclusion and Exclusion Criteria
Animals demonstrating sickness or severe stress before osmotic pump implantation were euthanized and excluded. Buprenorphine-SR 1mg/kgBW and Meloxicam 4mg/kgBW were given to each mice 1 hour before the surgery. After pump implantation, animals’ health condition was evaluated by authors and veterinarian every day. Reduce pain medication Buprenorphine-SR was given to mouse as 1mg/kgBW every 48 hours if necessary. The mice died before the 7 days/ 28 days AngII infusion postoperative time point, aorta samples will be collected for gross image and survival curve analysis, but excluded from diameter measurement, staining and western blotting study.
Aortic Cell Suspension Preparation
Samples of ascending aorta were harvested from each group (n=3) and were pooled in Hanks’ Balanced Salt Solution (HBSS, #14175095, Thermo Fisher Scientific, Waltham, MA) with 10% fetal bovine serum. For AngII infusion groups, 3 non-dissected mice aortas were randomly selected for single cell suspension preparation. Extra-aortic tissues, such as periaortic fat, were removed, and the aortic tissues were cut into small pieces and incubated with enzyme cocktail in HBSS containing Ca2+/Mg2+ (#14025092, Thermo Fisher) for 60 minutes at 37 °C. The cell suspension was filtered through a 40-μm cell strainer (CLS431750-50EA, Sigma-Aldrich), centrifuged at 350 g for 5 minutes, and resuspended by using cold HBSS (#14175095) with fetal bovine serum (5% vol/vol). Cells were stained with 4′, 6-diamidino-2-phenylindole (DAPI) and were sorted to select viable cells (≥95% viability) by using flow cytometry (FACS Aria III, BD Biosciences, Franklin Lakes, NJ).
scRNA-seq and scATAC-seq
Single-cell suspensions were dispensed separately onto the Chromium Controller (10x Genomics, Pleasanton, CA) with a target of 10,000 cells/sample. The scRNA-seq library was constructed by using the Chromium Single-Cell 3′ v3 Reagent Kit (10x Genomics). Cells were mixed with barcoded primer-linked gel beads. Each cell was barcoded with a unique index, and every transcript within a single cell was barcoded with a unique molecular identifier (UMI). cDNAs were pooled, truncated, and amplified to generate cDNA libraries that were sequenced using a Next Generation sequencer NovaSeq 6000 (Illumina, Inc., San Diego, CA) in a pair-end fashion to obtain more than 80,000 reads per cell.
Single-nuclei suspensions were prepared according to the manufacturer’s instructions (10x Genomics). Nuclei were loaded with a capture target of 10,000 nuclei per sample. ScATAC-seq libraries were prepared for sequencing according to the 10x Genomics scATAC-seq solution protocol. ScATAC-seq libraries were sequenced by using PE150 sequencing on an Illumina NovaSeq with a target depth of 25,000 reads per nucleus.
Processing and analyses of scRNA-seq and scATAC-seq data are presented in the Materials and Methods of the Supplemental Material.
Aortic Diameter Measurement
In euthanized mice, the aorta was exposed and rinsed with cold phosphate-buffered saline (PBS), and the periaortic tissue was removed. Then, the aorta was excised and further cleaned and rinsed with cold PBS to remove any residual blood in the lumen. In a mouse model of sporadic AAD, we evaluated the ascending, arch, descending thoracic, suprarenal abdominal, and infrarenal segments of the excised aorta. Images of the aorta were obtained by using an Olympus SZX7 microscope at a magnification of 0.4X (scale bar = 2 mm), and the diameter of each aortic segment was measured with the use of DP2-BSW software (Olympus Life Science Solutions, Center Valley, PA) by two independent observers who were blinded to the identity of the groups. The median diameter of the different regions was calculated and compared among the groups.
Definition of Aortic Dilatation, Aortic Aneurysm, and Aortic Dissection
For each aortic segment of WT or SMC-Yap−/− or SMC-Yap+/+ mice, dilatation was defined as an aortic diameter ≥1.25 but <1.5 times the mean aortic diameter of the segment in saline-infused mice with the same genetic background. Aneurysm was defined as an aortic diameter ≥1.5 times the mean aortic diameter of the segment in saline-infused mice with the same genetic background. Aortic dissection was defined as the presence of hematoma within the aortic wall detected on gross examination or as the presence of layer separation within the aortic media or medial-adventitial boundary (with a false lumen hematoma) detected upon histologic examination of the aorta. Aortic rupture and premature death were documented.
Classification of AAD Severity
The severity of AAD was classified as we have reported previously 14 modification of the classification system described by Daugherty and colleagues12: normal aorta, dilated aorta, aortic aneurysm without dissection, aortic dissection (as indicated by intramural thrombus) without aneurysm, aortic aneurysm with dissection, or ruptured aorta. We defined AAD as the presence of aneurysm, dissection, or rupture. Severe AAD was defined as the presence of dissection or rupture. Aneurysmal tissue was evaluated by 3 independent observers who were blinded to the experimental groups. In the case of a discrepancy, the observers discussed the case and agreed on the classification.
Medial Thickness Measurement
Ascending aortas from WT_Saline/CD mice (n=7) and WT_AngII/HFD mice (n=7) were harvested and freshly embedded in optimal cutting temperature (OCT) compound and snap-frozen in liquid nitrogen. They were sectioned at 5-um thickness at the maximal ascending aortic diameter area. Ascending aorta sections were stained with anti-SM22a primary antibody and DAPI. Tissue sections were examined by using a Leica microscope (Leica Microsystems Inc., Buffalo Grove, IL) or a Leica SP5 confocal microscope (Leica), and digital images were acquired (Leica Application Suite X, version 2.6.3). All quantitative image analyses were performed using Leica Application Suite X, version 2.6.3. Aortic medial thickness was measured from the subendothelial border (internal elastic lamina) to the external elastic lamina. To account for local variations in thickness, we determined the mean of 5 measurements from randomly chosen high-magnification fields for each scanned slide.
In-vivo Assay for Cell Proliferation
WT Saline/CD mice (n=5) and WT AngII/HFD mice (n=5) mice were injected daily with EdU (50 mg/kgBW) intraperitoneally for 5 days, starting on day 2 after pump implantation. Ascending aortas were harvested at day 7, freshly embedded in OCT, and snap-frozen in liquid nitrogen. EdU incorporation was detected using the Click-iT® Plus EdU Alexa Fluor® 647 Imaging kit (Life Technologies) according to the manufacturer’s instructions. In addition, we performed EdU staining with immunofluorescence co-staining. Frozen sections of aorta or cells were fixed with Cytofix (BD Biosciences), permeabilized with Perm/Wash (BD Biosciences), and labelled with Click-iT™ EdU. After EdU labeling, sections were blocked with 10% donkey serum at room temperature for 1 hour and stained with anti-SM22α antibody at 4 °C overnight. Samples were then stained with secondary antibody at room temperature for 1 hour. Tissue sections/cells were observed by using a Leica SP5 confocal microscope (Leica). For each mouse, 3 different sections were analyzed. For each section, images were captured from 3 randomly selected views. For each image, the number of EdU+/DAPI+ double-positive cells and the number of total SM22a+/DAPI+ double-positive cell nuclei were quantified, and the percentage of EdU+ smooth muscle cells was calculated.
Aortic Contractility
We examined aortic contractility in aortas from saline- or AngII-infused SMC-Yap+/+ mice and SMC-Yap−/− mice. In euthanized mice, the aorta was exposed, perfused with ice-cold PBS, and transferred to a physiologic salt solution (PSS) containing 130mM NaCl, 4.7mM KCl, 1.17mM MgSO4·7H2O, 14.9mM NaHCO3, 1.18mM KH2PO4, 0.026mM EDTA, 5.5mM glucose, and 1.6mM CaCl2·7H2O. Aortas were cleaned without damaging the endothelium. The ascending aorta was excised and cut into 2-mm segments by using a ruler under the microscope as a guide. The harvested ascending aortic segment was mounted onto metal stirrups of a wire myograph machine (DMT, Hinnerup, Denmark) and incubated at 37 °C for 60 minutes in equilibrated carbogen buffer (95% O2, 5% CO2) that was refreshed every 20 minutes. The aortic segment was then incubated with phenylephrine in high potassium PSS solution containing 74.7mM NaCl, 60mM KCl, 1.17mM MgSO4·7H2O, 14.9mM NaHCO3, 1.18mM KH2PO4, 0.026mM EDTA, 5.5mM glucose, and 1.6mM CaCl2·7H2O. The segment was then washed with PSS. The process was repeated 4 times with increasing concentrations of phenylephrine (10−9 M to 10−5 M). A cumulative concentration-response curve to phenylephrine was constructed with LabChart to evaluate the contractile function of the ascending aorta.
Hematoxylin and Eosin Staining
Aortic sections were stained with hematoxylin and eosin (Sigma-Aldrich) and Verhoeff–van Gieson (Sigma-Aldrich) according to the manufacturer’s instructions. The aortic sections were examined by 2 independent observers who were blinded to the experimental groups.
Immunofluorescence Staining and Imaging
Optimal cutting temperature (OCT)-embedded aortic sections or treated cells on slides were fixed with Cytofix (BD Biosciences, San Jose, CA) and permeabilized with Perm/Wash (BD Biosciences). Nonspecific staining was reduced by blocking with 10% donkey serum. The sections or cells were then incubated with primary antibody at room temperature for 2 hours or at 4 °C overnight, washed with PBS, and incubated with secondary antibody. Antibodies used for immunostaining are listed in Table I in the Data Supplement. Nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI). The slides were mounted with Dako Fluorescence Mounting Medium (Dako North America, Inc., Carpinteria, CA). Slides of sections or treated cells incubated with secondary antibody alone were used as negative controls. Tissue sections were examined by using a Leica microscope (Leica Microsystems Inc., Buffalo Grove, IL) or a Leica SP5 confocal microscope (Leica).
Terminal Deoxynucleotidyl Transferase dUTP Nick End Labelling (TUNEL) Assay and Immunofluorescence Staining
To study apoptosis by using an in situ cell death detection kit (Roche Applied Science, Indianapolis, IN, USA), we performed TUNEL staining according to the manufacturer’s instructions. In addition, we performed TUNEL staining with immunofluorescence costaining. First, frozen sections of aorta or cells were fixed with Cytofix (BD Biosciences), permeabilized with Perm/Wash (BD Biosciences), and subjected to TUNEL staining. After TUNEL staining, sections or cells were blocked with 10% donkey serum at room temperature for 1 hour and stained with anti-SM22-α antibody at 4 °C overnight. Sections or cells were then stained with secondary antibody at room temperature for 1 hour. Tissue sections or cells were observed by using a Leica SP5 confocal microscope (Leica). For each aorta or cell treatment condition, images were captured from 3 randomly selected views. For each image, the number of positive cells and the number of total cell nuclei were quantified, and the percentage of positive cells was calculated.
Western Blot Analyses
Protein lysates from treated cells or aortic tissues were prepared as described previously.13,14 Protein samples (15 μg per lane) were subjected to sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis and were transferred to polyvinyl difluoride membranes. The membranes were blocked for 1 hour in blocking solution of Tris-buffered saline containing 5% nonfat dried milk and 0.5% Tween 20 and then were incubated with a primary antibody. Next, the membranes were washed and incubated with horseradish peroxidase–conjugated anti-rabbit or anti-mouse secondary antibody. Antibodies used for western blotting are listed in Table I in the Data Supplement. Protein bands were visualized by using Clarity Enhanced Chemiluminescence (Bio-Rad Laboratories, Inc., Hercules, CA) and were exposed with HyBlot ES autoradiography film (Denville Scientific Inc., Holliston, MA). The blots were quantified with densitometry by using the Quantity One imaging program (Bio-Rad). Protein levels were normalized to those of β-actin and were expressed as a percentage of the untreated control.
Cell Culture and Transfection
Human thoracic aortic SMCs (ATCC, Manassas, VA) were cultured in SMC medium (Cell Applications, Inc. San Diego, CA) with 5% FBS (Thermo Fisher). SMCs were transfected with siRNA by using Lipofectamine RNAMAX (Thermo Fisher) or plasmid DNA by using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s instructions at the given concentrations for 24 hours. YAP siRNAs were purchased from Thermo Fisher, and YAP-WT and YAP-S127A plasmids were purchased from Addgene.
Application of Mechanical Stretch
For the application of mechanical cyclic stretch, vascular SMCs were plated on type I collagen–coated flexible silicone-bottomed BioFlex plates (Flexercell International) at an initial density of 5 × 105 cells per well. After seeding the cells for 24 h, they were incubated with 1% FBS/SMC medium for 12 hours before the cyclic stretch experiments to arrest the growth of cells. The cells were then subjected to cyclic stretch produced by a computer-controlled vacuum (FX-5000T Strain Unit; Flexcell International) at a 10% strain magnitude and constant frequency of 1 Hz for 24 h. Cells cultured under the same conditions but not subjected to mechanical strain were used as the static control.
Real-time Quantitative RT-PCR
Total RNA from the aortic SMC was extracted with a commercial kit (Invitrogen) according to the manufacturer’s instructions. The mRNA was reverse-transcribed into cDNA by using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed by using the Real-Time PCR System (Bio-Rad). The primers for human LOX were 5-TGT ACCGCTATGGTTACACTC G-3 (forward) and 5-GGCAGGGACAGTTGCTTCT-3 (reverse); the primers for 18S rRNA were 5- GTAACCCGTTGAACCCCATT -3 (forward) and 5- CCATCCAATCGGTAGCG -3 (reverse). The mRNA levels were acquired from the value of the threshold cycle (Ct) of LOX and were normalized against the Ct of GAPDH.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation assay was performed as described previously 17,18 by using the EZ-ChIP Kit according to the manufacturer’s protocol (EMD Millipore Corp., Billerica, MA, USA). Briefly, aortic SMCs were incubated with 1% formaldehyde at room temperature for 10 minutes to cross-link the DNA-protein complexes. Glycine was added to each sample to quench the unreacted formaldehyde. The cells were washed, harvested, and lysed. Cell lysates were sonicated and centrifuged to produce chromatin fragments that were 200 to 1000 base pairs long. Immunoprecipitation was performed with an anti-YAP antibody–protein A-agarose slurry (IgG was used as the negative control). The immunocomplex beads were washed sequentially with low-salt wash buffer, high-salt wash buffer, LiCl wash buffer, and Tris-EDTA buffer. The immunocomplex was eluted with elution buffer (100 mmol/L NaHCO3, 1% SDS). The eluted immunocomplex and the input were incubated with 200 mM NaCl at 65 °C overnight to reverse the cross-linking and then were incubated with proteinase K to digest the remaining proteins. The DNA was recovered by performing phenol/chloroform/isoamyl alcohol extraction and was used as a template for PCR.19 To quantify the DNA, we performed real-time quantitative PCR and analyzed the results by using the comparative delta Ct method. Gel electrophoresis in 1.5% agarose gel was used to examine the size and purity of the PCR products. The primers used for the YAP/TEAD binding site in the 5’-flanking region of the human LOX gene were 5’- GATTCCCAGCTTCCACTGAA −3’ (forward) and 5’- TGCTGATTTTGCTGACAAGC-3’ (reverse).
Availability of Data
The raw data for experiments presented, including quantification for Western blotting, flow cytometry, immunofluorescence staining, aortic diameter, and wire myography, are available from the corresponding author upon reasonable request. All sequencing data have been made publicly available at the Gene Expression Omnibus (GEO) and can be accessed at GSE213735 (single cell RNA-seq), and GSE214082 (single cell ATAC-seq). The computer code used in this study is available upon request.
Statistical Analysis
All quantitative data are presented as the mean ± standard deviation or as the median and interquartile range, as appropriate. Data were analyzed with SPSS software, version 11.0 (SPSS Inc., Chicago, IL, USA). Normality of the data was examined with the Kolmogorov-Smirnov test. For two-group comparisons, Student’s t tests were used to compare normally distributed values, and the Mann-Whitney test was used to compare data that failed the normality test. Multiple groups were compared using one-way analysis of variance with the Holm-Sidak method for normally distributed datasets, or the Kruskal-Wallis test with Dunn’s post hoc test for data that did not pass the normality test. For all statistical analyses, 2-tailed probability values were used.
RESULTS
Aortic Stress Triggered a Systemic Induction of Genes in Aortic Remodeling and Adaptation in SMCs
We first examined the adaptive response in WT mice fed with a high-fat diet and infused with AngII (2000 ng/kg/min) (AngII/HFD). Mice fed with a standard rodent laboratory diet and infused with saline (saline/control diet [CD]) were used as controls. Administering AngII/HFD has been shown to induce aortic degeneration in thoracic and abdominal aortic segments in C57BL/6J WT mice,15 along with the development of aortic dilatation (98%), AAD (including aortic aneurysm, dissection, and rupture; 82%), severe AAD (including dissection and rupture; 58%), and rupture (24%). AngII infusion increased systolic blood pressure from 100 to 110 mmHg to 160 to 180 mmHg. No atherosclerosis was observed in this model.15
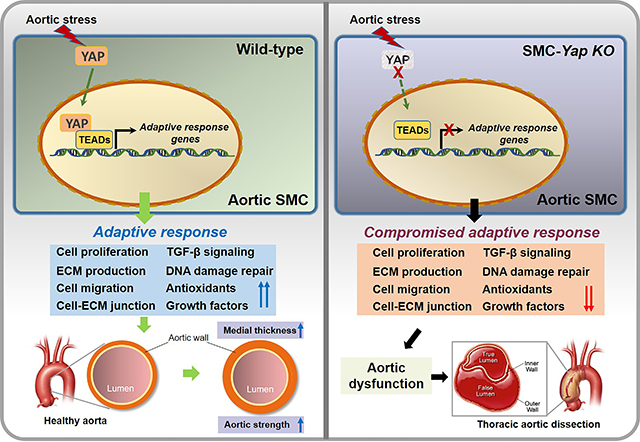
Compared with ascending aortas of saline/CD-treated WT mice, those of AngII/HFD-treated WT mice showed a significant increase in aortic medial thickness (Figure 1A) and SMC proliferation (Figure 1B). To understand the gene regulation underlying this response, we performed scRNA-seq analysis of ascending aortas from saline/CD-treated WT mice and AngII/HFD-treated WT mice (n=3 per group) (Figure S1A in the Supplemental Material). Aortas were collected after day 7 of AngII infusion to capture the early response to aortic stress. To avoid the effect of aortic dissection, we used nondissected tissues from the AngII/HFD-treated group. From a total of 16,811 qualified cells (8698 cells from the WT saline/CD-treated group and 8113 cells from the WT AngII/HFD-treated group), unsupervised clustering20 identified 11 distinct clusters (Figure S1B in the Supplemental Material) that were classified into 6 major cell types including vascular SMCs, fibroblasts, endothelial cells, pericytes, immune cells, and cycling cells21 (Figure 1C, and Figure S1C in the Supplemental Material). SMC clusters were further clustered into 3 subsets (Figure S1D, S1E, and S1F in the Supplemental Material).
Figure 1. Single-cell RNA sequencing (scRNA-seq) analysis showing a dynamic smooth muscle cell (SMC) adaptive response to angiotensin II infusion and a high-fat diet (AngII/HFD).
A, Representative immunofluorescence staining of SMCs (SM22-α) showing that AngII/HFD increased medial thickness in the aortic wall (M, media) of wild-type (WT) mice. Box and whisker plots showing that the extent of medial thickness was increased in the ascending aorta of AngII/HFD-treated WT mice compared with saline/control diet (CD) (standard rodent diet)-treated mice (n=7 per group). B, Immunostaining showing an increase in EdU-labeled proliferating SMCs (SM22-α+) in the ascending aortic tissue of AngII/HFD-treated WT mice and the depletion of SMCs in aneurysm and dissection (AAD) tissues of AngII/HFD-treated mice. Box and whisker plots showing that the quantity of EdU+ SMCs was increased in the ascending aorta of AngII/HFD-treated WT mice (n=5) compared with saline/CD-treated WT mice (n=5). C, A 2-dimensional uniform manifold approximation and projection (UMAP) plot showing all cells colored according to the 9 major cell clusters. D-E, Dot plots showing gene ontology (GO) enrichment terms (top 20) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of differentially expressed genes (DEGs) induced by AngII infusion in all SMCs. F, Violin plots showing the mRNA abundance of select adaptive response genes in all SMCs. *FDR (false discovery rate) <0.001, SMCs in AngII/HFD-treated WT mice vs. SMCs in saline/CD-treated WT mice in scRNA-seq. An unpaired Student’s t-test was used in (A) and (B). P<0.001. Data are shown as box and whisker plots with the first quartile, minimum, median, third quartile, and maximum.
We next examined the changes in the gene expression profile of SMCs in response to AngII infusion. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (Figure 1D, 1E, and Figure S1G in the Supplemental Material) of the combined SMC clusters showed that, compared with aortic SMCs of saline/CD-treated WT mice, SMCs of AngII/HFD-treated WT mice showed the increased expression of genes involved in wound healing, elastic fiber assembly, ECM organization, actin cytoskeleton organization, and cell-substrate-matrix/focal adhesion. The enriched features also included response to oxidative stress, response to unfolded protein, and angiogenesis. Several signaling pathways such as P13K-Akt signaling and transforming growth factor (TGF)-β signaling were also upregulated in SMCs of AngII/HFD-treated mice (Figure 1E). Consistent with the GO and KEGG analyses, the analysis of differentially expressed genes (DEGs) (Figure 1F) showed that AngII/HFD increased the mRNA abundance of adaptive response genes associated with proliferation,22,23 collagen production and collagen fibril organization, elastic fiber organization, cell-matrix junctions, cell migration,23–26 cell survival, antioxidants, DNA damage repair, growth factor secretion, and TGF-β signaling. The induction of adaptive genes was also observed in WT mice that were infused with AngII (1000 ng/kg/min) alone (Figure S1H in the Supplemental Material), suggesting that AngII infusion was the main trigger for this adaptive response. Together, these data suggest that exposure of the aorta to AngII/HFD triggers the systemic induction of a gene set involved in SMC function and ECM remodeling that may be important for maintaining aortic wall strength and sustaining increased aortic pressure.
Aortic Stress Increased the Chromatin Accessibility of Adaptive Genes in SMCs
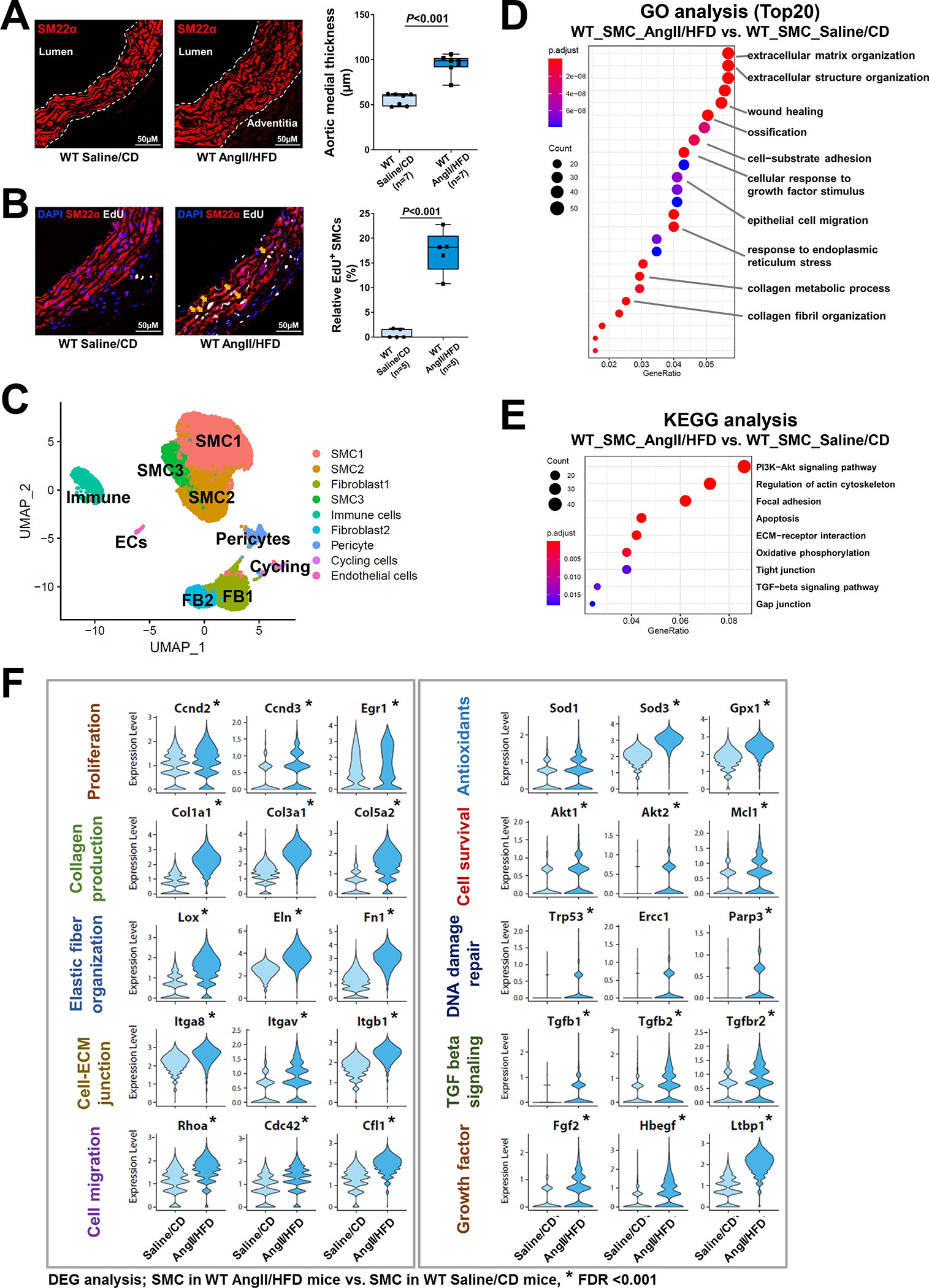
To understand the control of gene expression in response to aortic stress, we performed scATAC-seq analysis to examine the chromatin accessibility of the adaptive response genes in aortic SMCs of AngII/HFD-treated mice (i.e., infused with AngII for 7 days) and of saline/CD-treated mice (n=3 per group). Gene activity scores (corresponding to the chromatin accessibility peaks in the regulatory regions of a given gene) were calculated for all of the genes, and gene activity profiles were analyzed for a total of 19,810 qualified single-nuclei (11,054 single-nuclei from the WT saline/CD-treated group, 8756 single-nuclei from the WT AngII/HFD-treated group). A total of 18 clusters (Figure 2A) were identified on the basis of similarities in gene activity profiles. The identities of these clusters were determined on the basis of the chromatin accessibility/gene activity of cell marker genes (Figure 2B, Figure S2 in the Supplemental Material). There were 3 SMC clusters that showed SMC-specific gene activity.
Figure 2. Single-cell sequencing assay for transposase-accessible chromatin (scATAC-seq) analysis showing that aortic stress triggers chromatin accessibility dynamics of smooth muscle cells (SMCs).
A, Ascending aortas of AngII/HFD-treated mice (i.e., infused with AngII for 7 days) and of saline/CD-treated mice (n=3 per group) were used for single live-cell isolation, single nuclei preparation, and scATAC-seq analysis. ScATAC-seq data projected on a 2-dimensional uniform manifold approximation and projection (UMAP). Cells are colored by cluster. Maph: macrophage, FB: fibroblast, EC: endothelial cell. B, Genome browser views (IGV) of cell-cluster combined scATAC-seq cell type marker peaks. Each row is a cluster (left), and each column is a locus within 50 kb of an ATAC-seq cell type marker gene. The colored peaks represent the normalized read count coverage. C, Dot plot showing the gene ontology (GO) enrichment terms of genes activated by AngII infusion. D, Violin plots showing the Cicero gene activity of select adaptive response genes in all SMCs. *Adjusted P-value <0.05, SMCs in AngII/HFD-treated mice vs. SMCs in saline/CD-treated mice in scATAC-seq.
Next, we compared gene activity profiles in SMCs of saline/CD-treated and AngII/HFD-treated mice by performing analyses of GO (Figure 2C) and DAGs (Figure 2D) in SMCs (the combined SMC clusters). As expected, the gene activities in cytokine production and the inflammatory response were increased in SMCs of AngII/HFD-treated mice. Consistent with their gene expression profiles, SMCs of AngII/HFD-treated mice also exhibited gene activity enrichment in wound healing, angiogenesis, proliferation, ECM organization, actin cytoskeleton organization, cell migration, oxidative stress response, and TGF-β signaling. Together, these data indicate that aortic exposure to AngII/HFD induces adaptive gene expression, partially by increasing the chromatin accessibility of the gene regulatory regions.
YAP/TEADs Were Identified as Top Candidate Transcription Factors in the Increased Chromatin Accessibility of Adaptive Genes
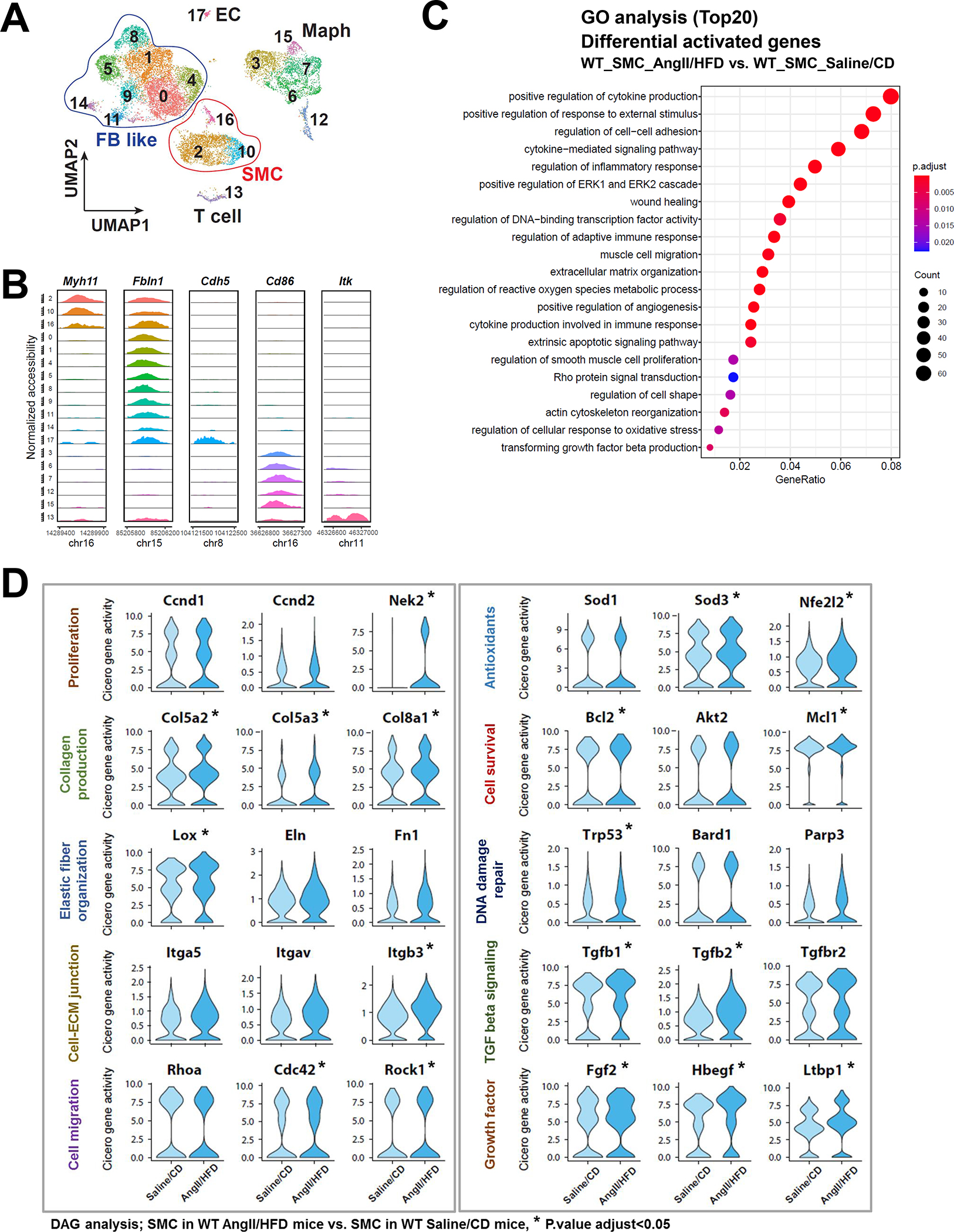
To further understand the regulation of the adaptive response, we sought to identify the transcription factors (TFs) involved in the induction of adaptive genes. We started by analyzing the correlation between the gene activities of 80 adaptive genes (Table S2 in the Supplemental Material) and the motif activities of more than 600 TFs. Motif activities of MEF2s and SRF, known master regulators of SMCs,27–29 showed the strongest positive correlation with the gene activities of the adaptive response genes (Figure 3A). This finding confirmed the reliability of this approach. Interestingly, transcriptional enhanced associate domains (TEAD) protein family members were another group of TFs with motif activities that strongly and positively correlated with the activities of adaptive genes (Figure 3A).
Figure 3. TEADs involved in the adaptive response of smooth muscle cells (SMCs).
A, Heat map showing correlation coefficients between selected adaptive response genes and SMC-related transcription factor (TF) motif activity. B-C, Selected enriched gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations for potential TEAD targets involved in the SMC adaptive response.
TEADs are transcriptional partners of YAP, a key signaling molecule in sensing mechanical signals and promoting cell proliferation and organ development. To understand the broader role of YAP/TEADs in SMC gene expression during AngII infusion, we examined the chromatin accessibility of YAP/TEAD targets that contain TEAD motifs. A total of 7094 TEAD motif–containing genes were found in aortic wall that had counts higher than expected (fold enrichment >1). Of these genes, 2383 showed enriched chromatin accessibility/gene activities in SMCs from AngII/HFD-treated mice compared with those from saline/CD-treated mice. GO analysis of these 2383 genes showed features in muscle cell proliferation, actin filament organization, and ECM organization in SMCs (Figure 3B), and the KEGG analysis of these genes showed enrichment in Wnt, TGF-β and PI3K-Akt signaling pathways in SMCs (Figure 3C). Together, these data support a potential role of YAP/TEADs in enhancing the chromatin accessibility/gene activity of the adaptive genes in SMCs in response to AngII/HFD.
A paralog of YAP is the transcriptional regulator TAZ (WW domain containing transcription regulator 1, or WWTR1), which can also bind to TEADs. Our sc-RNAseq data showed that, although the abundance of both Yap1 and Taz (Wwtr1) mRNA was increased after infusion with AngII, the level of Yap1 mRNA was much higher than that of Taz mRNA (Figure S3A and S3B in the Supplemental Material), suggesting that YAP may play a more important role than TAZ in this setting.30–32 For this reason, we focused our study on the role of YAP in the adaptive response.
YAP/TEADs Were Involved in Open Chromatin Remodeling of Adaptive Genes and Lox
For the 80 adaptive genes, we examined local and co-accessible distal peaks. TEAD motifs were detected in the significantly enriched peaks of 37 adaptive genes including Lox, Eln, Fn1, Col3a1, Col5a2, Col5a3, Tgfb2, Tgfbr2, and Smad3 (Table S2 in the Supplemental Material). Furthermore, TEAD motifs were detected in constant opening or inducible peaks in Lox, Fn1, Col5a2, Sod3, Fgf2, Tgfb2, Tgfbr2, and Smad3 (Figure 4A, and Figure S4A in the Supplemental Material).
Figure 4. Chromatin accessibility landscape and TEAD motif distribution at the locus of Lox and other adaptive genes in smooth muscle cell (SMC) clusters.
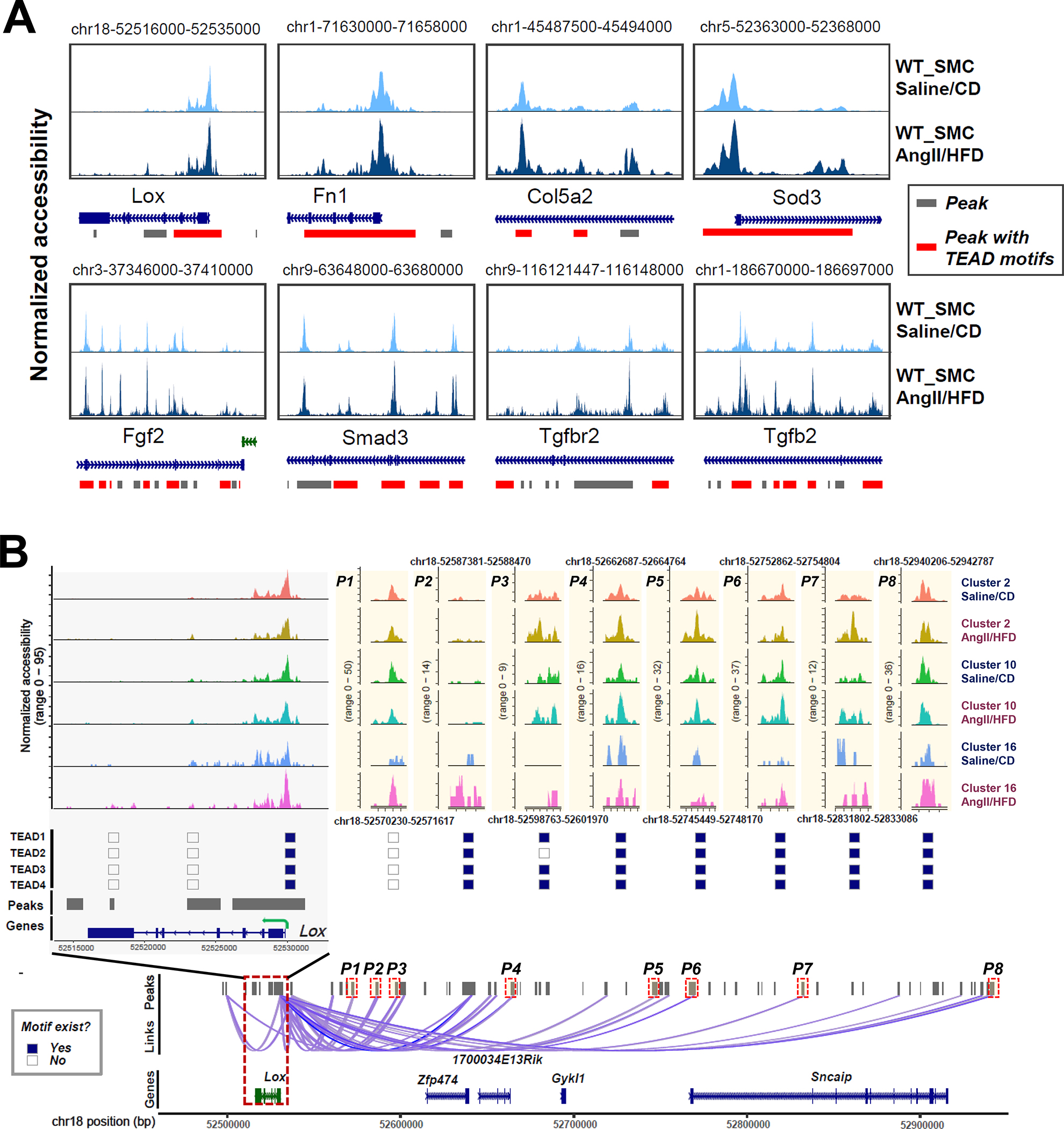
A, Accessibility and peaks detected at the chromatin regions of 8 selected adaptive genes. Accessibility was shown for SMCs under different conditions. Peaks harboring TEAD motifs were highlighted in red. B, Aortic stress–inducible peaks at the Lox gene locus and the distal peaks that were co-accessible with peaks at the Lox gene locus in SMC clusters are shown. The distribution of TEAD motifs on aortic stress–inducible Lox peaks in SMC clusters is shown.
We further studied lysyl oxidase (LOX), a critical component in elastic fiber that maintains aortic wall integrity.33,34 Lox expression and gene activity in SMCs was significantly increased (Figure 1F, and Figure 2D) in AngII/HFD-treated mice compared with saline/CD-treated mice. We identified 23 peaks as cis-regulatory elements of Lox (Figure 4B, and Figure S4B in the Supplemental Material). Interestingly, these peaks showed different patterns of change. Some peaks showed constant opening with minimal differences between saline/CD-treated mice and AngII/HFD-treated mice, suggesting that these elements may be involved in the constitutive expression of Lox. In contrast, several peaks were closed in most cells in saline/CD-treated mice and were opened in AngII/HFD-treated mice, suggesting that these regions may be important for AngII/HFD-induced Lox expression. Importantly, we found that TEAD1/2/3/4 motifs were detected not only in the constantly opening peaks, but also in the inducible peaks.
By using ENCODE and JASPAR databases as references, we identified several enhancers of Lox (Figure S4C in the Supplemental Material). Two proximal enhancers were identified in the peak in the Lox gene body (Figure S4C in the Supplemental Material, highlighted peak 1), and one of those contained TEAD binding sites. Two enhancers were identified in the peak 3.7 kb upstream of Lox (Figure S4C in the Supplemental Material, highlighted peak 2), and one of those contained TEAD binding motifs. This peak showed increased accessibility in the SMCs of AngII/HFD-treated mice and was co-accessible with the first highlighted peak. A distal enhancer that contained TEAD binding motifs was identified in a peak at about 220 kb upstream of Lox (Figure S4C in the Supplemental Material, highlighted peak 3). That peak showed increased accessibility in the SMCs of AngII/HFD-treated mice and was co-accessible with the second highlighted peak. Together, these data implied that YAP/TEAD may bind to the regulatory regions of adaptive genes and participate in the constitutive and inducible expression of these genes.
YAP Was Bound to the Regulatory Regions of the Adaptive Genes (e.g., LOX) and Induced Their Expression in Response to Cyclic Stretch
To further determine the direct role of YAP in response to mechanical stress in SMCs, we examined the activation of YAP and its effects on SMC gene expression in response to cyclic stretch, which mimics the mechanical force created by blood pressure on aortic cells. Human thoracic aortic SMCs (in low serum) underwent cyclic stretch (10%, 1 Hz) for 24 hours. Cyclic stretch significantly increased the percentage of EdU (5-ethynyl-2’-deoxyuridine)-labelled proliferating SMCs (Figure 5A) and the production of collagen I, collagen III, elastin, and LOX (pre-LOX and active-LOX) (Figure 5B, and Figure S5A in the Supplemental Material).
Figure 5. Critical role of YAP in promoting smooth muscle cell (SMC) proliferation and the expression of extracellular matrix (ECM) genes in response to cyclic stretch.
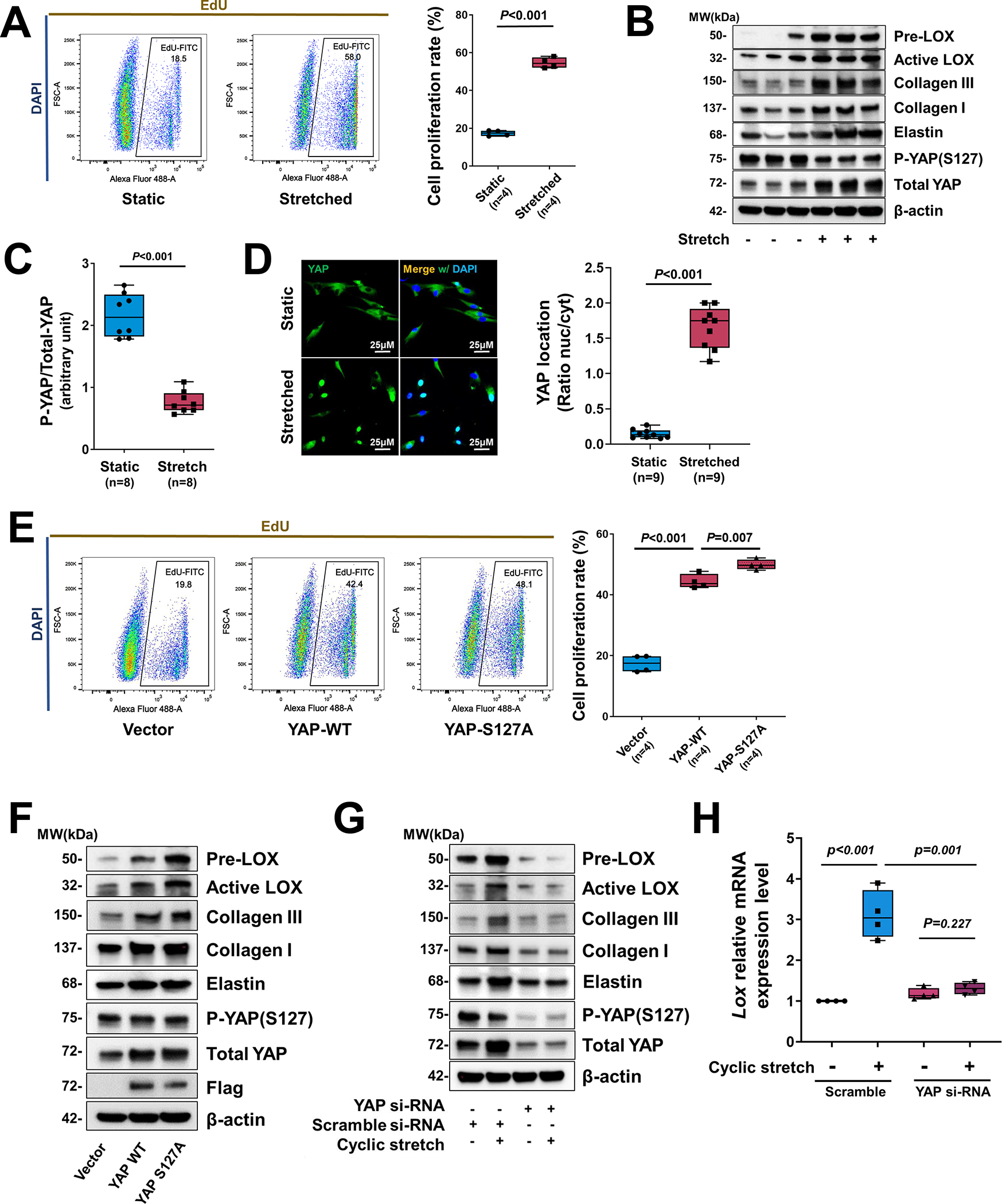
A, Human aortic SMCs (in low serum) underwent cyclic stretch for 24 h. Flow cytometry analysis showing that cyclic stretch induced cell proliferation (indicated by EdU labeling) in cultured SMCs (n=4 biologic repeats). B, Western blotting results showing that cyclic stretch induced ECM protein production in cultured SMCs and YAP expression and dephosphorylation in cultured SMCs that underwent cyclic stretch (n=8 biologic repeats). C, Western blotting quantification showing that cyclic stretch induced a marked dephosphorylating of YAP in cultured SMCs. D, Representative immunofluorescence staining (images and quantification data) showing the cyclic stretch–induced nuclear translocation of YAP (n=3 biologic repeats and 3 replicates). E-F, SMCs were transfected with wild-type (WT) YAP or constitutively-active YAP (S127A) (n=5 biologic repeats). Overexpression of YAP induced cell proliferation (E) and ECM protein production (F). G, Silencing YAP with siRNA prevented ECM protein production. H, LOX mRNA levels (n=5 biologic repeats) were increased by cyclic stretch, and silencing Yap with siRNA partially prevented LOX expression induced by cyclic stretch. Two-way analysis of variance (ANOVA) with Bonferroni’s post hoc test for pairwise comparisons was used in (H). A paired Student’s t-test was used in (C). Data are shown as box and whisker plots with the first quartile, minimum, median, third quartile, and maximum in (A), (C), (D), (E), and (H).
Cyclic stretch also increased YAP expression, YAP dephosphorylation/activation (Figure 5B, C), and YAP translocation into the nucleus (Figure 5D). In addition, YAP nuclear translocation was observed in ascending aortic cells of AngII/HFD-treated mice (Figure S5B in the Supplemental Material). Importantly, overexpression of WT YAP or constitutively active YAP (S127A, mutated at the large tumor suppressor kinase LATS1/2 phosphorylation site) induced cell proliferation (Figure 5E) and the production of LOX, elastin, collagen I, and collagen III (Figure 5F, and Figure S5C in the Supplemental Material). Conversely, deleting YAP prevented the cyclic stretch–induced increase in the abundance of LOX protein and cleavage (Figure 5G, and Figure S5D in the Supplemental Material) and Lox mRNA (Figure 5H).
To identify YAP’s direct targets in SMCs and examine the effects of stretch on YAP binding to targets, we performed YAP-based ChIP-seq analysis (chromatin immunoprecipitation followed by high-throughput sequencing). A total of 581 binding sites/motifs were enriched (over matching inputs) in stretched SMCs. These included known YAP targets (Table S3 in the Supplemental Material), such as Ctgf,35 Ccnd1,36 Itgb2,37 and Ankrd1.38 By overlapping our YAP-based ChIP-seq data with publicly available YAP/TEAD ChIP-seq data,39 we identified a novel set of YAP targets, such as ECM-associated genes (Col12a1, Fn1, Lox, Mfap5, and Fstl1), angiogenesis-associated genes (Tnfrsf12a, Jag1,40 Notch2, and Srpx2), cell migration genes (Enah41 and Anln), growth factor gene Igfbp2, and TGF-β signaling genes (Smad2 and Rasl11b).
Collectively, our findings suggest a critical role for YAP in sensing mechanical signals and inducing ECM production and cell proliferation in SMCs.
SMC-specific YAP Played a Critical Role in Adaptive Gene Expression in Response to AngII Infusion
To further determine the role of YAP in SMC adaptive gene expression during aortic stress, we performed scRNA-seq analysis of ascending aortas from mice with the SMC-specific deletion of Yap (SMC-Yap−/−, n=3) and from littermate controls (SMC-Yap+/+, n=3) that were infused with either saline or AngII (1000 ng/kg/min infusion for 7 days) alone (Figure 6A). The efficiency of Yap knockout in ascending aortas was confirmed with Western blotting (Figure S6A in the Supplemental Material). The high AAD incidence (>75%) in the sporadic AAD model (i.e., mice fed a HFD and infused with AngII at a rate of 2000 ng/kg/min) resulted in too small of a window in which to detect further increased AAD induction in SMC-Yap−/− mice. Thus, for the SMC-specific Yap deficient mice study, we used a lower infusion rate of AngII (1000 ng/kg/day), and mice were fed a standard laboratory diet.
Figure 6. The YAP-mediated adaptive response identified by performing single-cell transcriptomic analysis of smooth muscle cell (SMC)-specific Yap knockout mouse aortas.
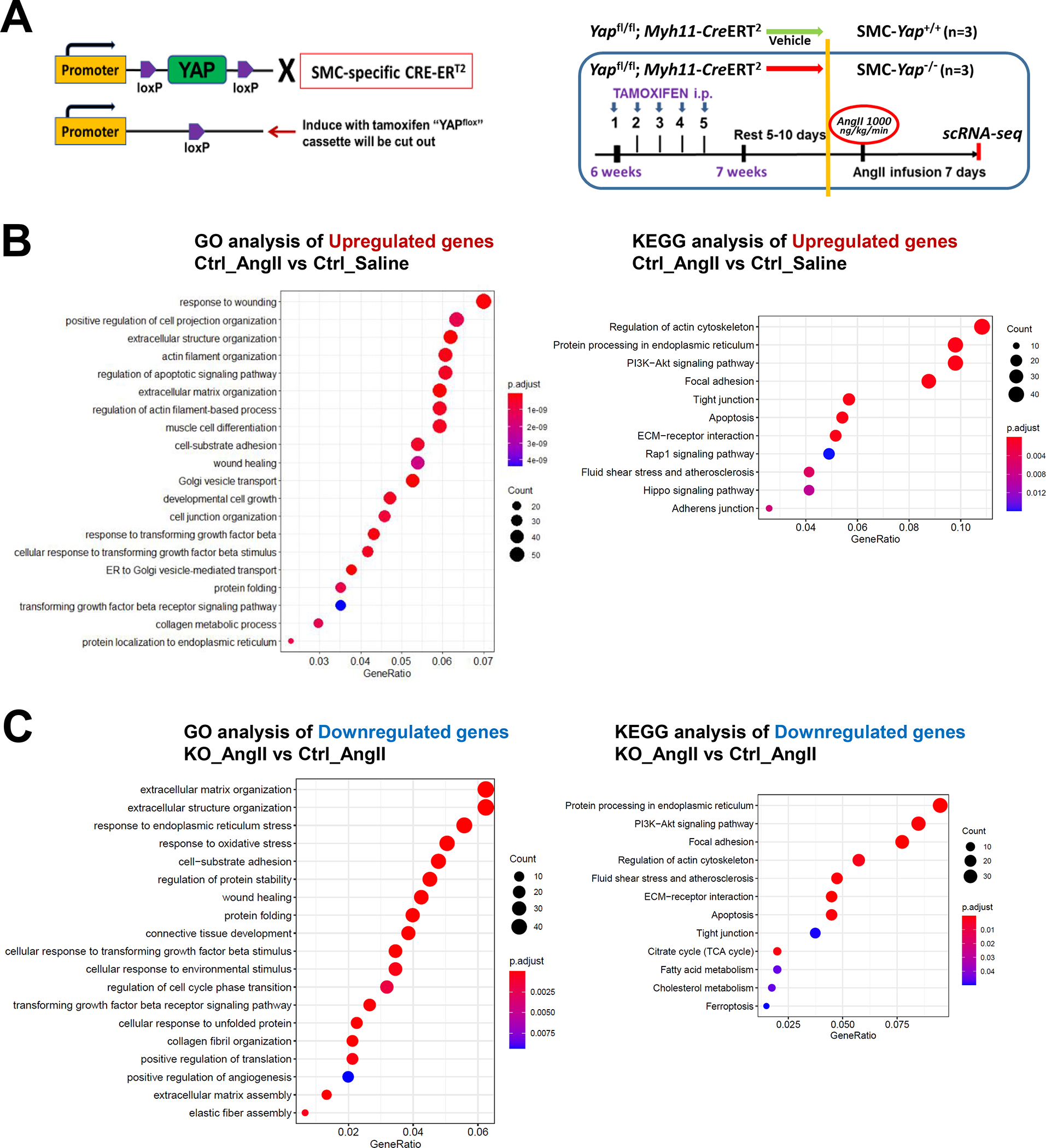
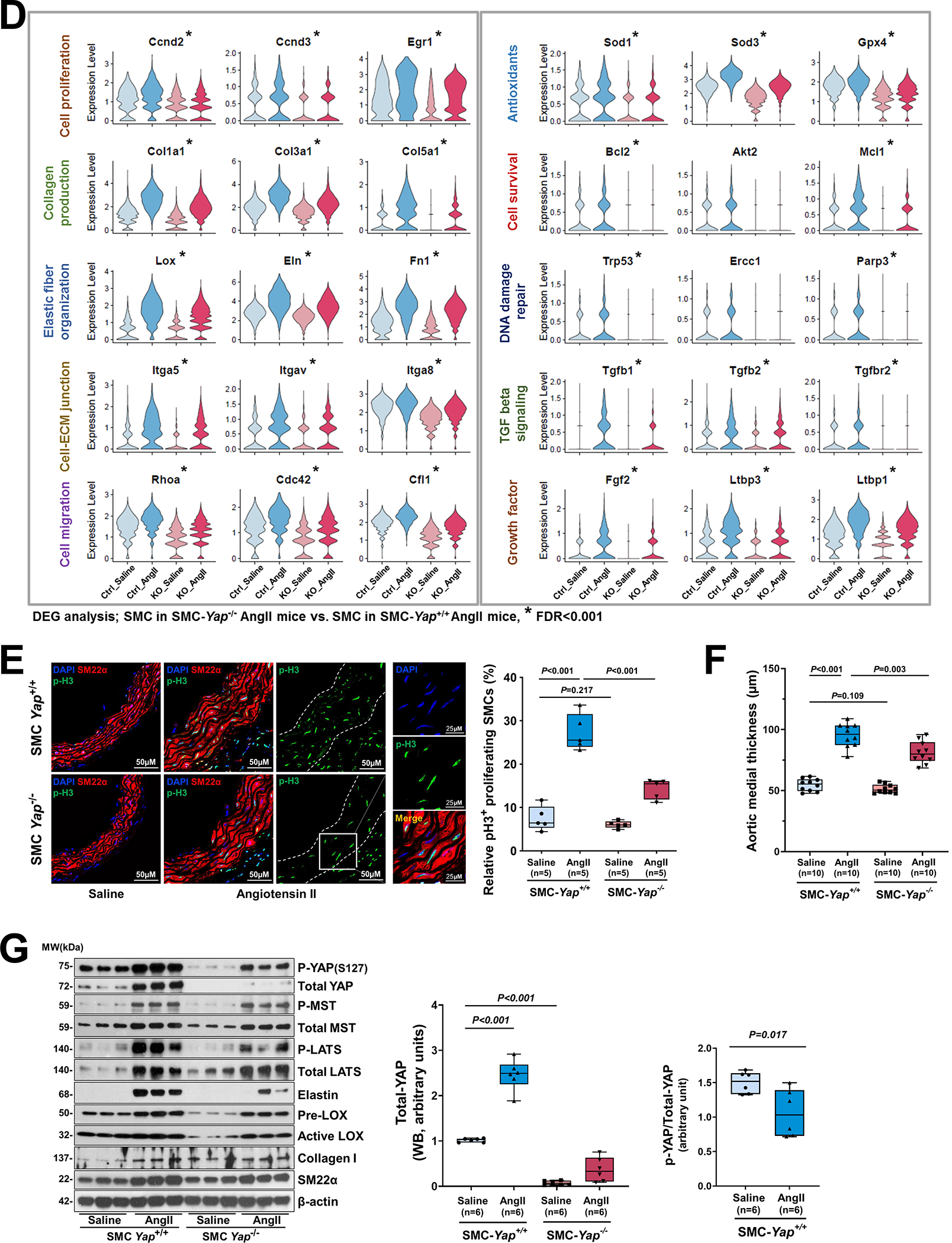
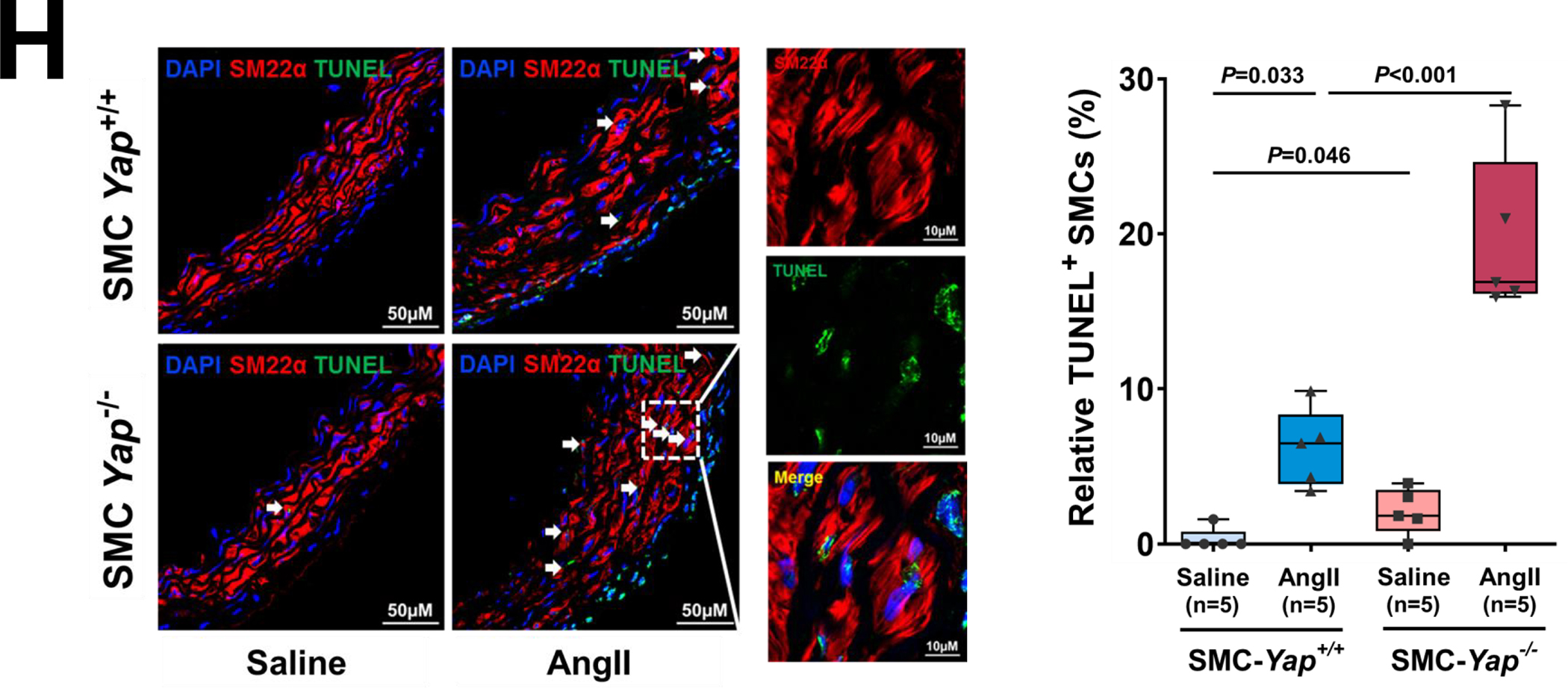
A, Six-week-old male Yap1fl/fl; Myh11-CreERT2 mice were given tamoxifen or vehicle (corn oil) via daily intraperitoneal injection for 5 days. Seven days later, mice were infused with angiotensin II (AngII 1,000 ng/kg/min; n=3) or saline (n=3; control) for 7 days, and ascending aortas were collected for single-cell RNA-seq. B, Dot plot of the top 20 enriched gene ontology (GO) biologic process terms and top-enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms identified in SMCs of AngII-infused SMC-Yap+/+ mice (Ctrl_AngII) compared with SMCs of saline-infused SMC-Yap+/+ mice (Ctrl_Saline). Associated genes were significantly upregulated in SMC-Yap+/+ AngII mice (logFC< −0.3 & FDR<0.05). The size of the dots represents the number of genes that are on the list of significant differentially expressed genes associated with GO and KEGG terms. The color of the dots represents the P-adjusted values. C, Dot plot of the top GO biologic process terms and KEGG pathway terms identified in SMCs of AngII-infused SMC-Yap−/− mice (KO_AngII) compared with SMCs of AngII-infused SMC-Yap+/+ mice (Ctrl_AngII) showing the significant downregulation of the associated genes in SMC-Yap−/− AngII-infused mice (logFC< −0.3 & FDR<0.05). D, Violin plots showing the mRNA abundance changes of adaptive response genes in all Myh11+ SMCs of 4 groups of mice (Ctrl_Saline, Ctrl_AngII, KO_Saline, and KO_ AngII). *FDR <0.001, SMCs in SMC-Yap−/− AngII-treated mice vs. SMCs in SMC-Yap+/+ AngII-treated mice in scRNAseq. E, Immunofluorescence staining and quantification of the cell proliferation marker phospho-histone H3 indicated that less SMC proliferation occurred in the ascending aortic medial wall of SMC-Yap−/− AngII-treated mice than in that of SMC-Yap+/+ AngII-treated mice. F, Box and whisker plots showing that medial thickness was reduced in the ascending aorta of AngII-treated SMC-Yap−/− mice compared with AngII-treated SMC-Yap+/+ mice (n=10 per group). G, Western blot analysis of ascending aorta lysates showing that levels of ECM proteins elastin, collagen I, and LOX were markedly lower in AngII-treated SMC-Yap−/− mice than in AngII-treated SMC-Yap+/+ mice. Box and whisker plots showing the quantification of the fold change in total-Yap and the quantification of the p-YAP/total-Yap ratio in Ctrl_Saline and Ctrl_AngII. H, Representative images and corresponding quantification of TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) staining showing a significant increase in the number of TUNEL-positive cells in the ascending aortas of AngII-treated SMC-Yap−/− mice compared with AngII-treated SMC-Yap+/+ mice. Nuclei were counterstained with DAPI (blue). Two-way analysis of variance with the Bonferroni post hoc test was used for pairwise comparisons in (E), (F), (G), and (H). Data are shown as box and whisker plots with the first quartile, minimum, median, third quartile, and maximum.
We obtained a total of 27,582 qualified cells from 4 groups of mice and identified 16 distinct clusters, including 12,878 Myh11-positive SMCs and SMC-like cells (Figure S6B in the Supplemental Material). These SMCs were further clustered into 3 SMC subclusters including contractile SMCs, proliferative SMCs, and ECM-producing SMCs (Figure S6C in the Supplemental Material), which matched the SMC clusters identified in WT mice (Figure 1C).
We then compared gene expression profiles between SMC-Yap+/+ and SMC-Yap−/− mice. In the absence of AngII infusion (Figure S6D in the Supplemental Material), SMC-Yap−/− mice (KO_Saline) showed a lower mRNA abundance of genes with roles in ATP metabolic processes, muscle cell proliferation, tight and adherens junctions, cell-substrate adhesion, ECM organization, elastic fiber assembly, and regulation of the actin cytoskeleton than did SMC-Yap+/+ mice (Ctrl_Saline). Results of GO and KEGG analyses showed that AngII infusion in SMC-Yap+/+ mice (Ctrl_AngII) increased the mRNA abundance of genes with roles in wound healing, SMC proliferation, migration, ECM production/organization, cell-substrate adhesion/focal adhesion, and the cellular response to TGF-β stimulus. However, this upregulation was compromised in AngII-infused SMC-Yap−/− mice (KO_AngII) (Figure 6B and 6C). Further DEG analysis also showed that AngII infusion induced the expression of adaptive response genes in SMC-Yap+/+ mice and that this induction was compromised in SMC-Yap−/− mice (Figure 6D, and Figure S6E in the Supplemental Material), supporting a role for YAP in promoting the expression of genes in the adaptive response. Furthermore, scRNA-seq analysis showed that AngII infusion–induced TGF-β signaling (e.g., Tgfb2, Tgfb3, Gdf6, Inhba, Tgfbr1, and Igf1) was compromised in SMC-Yap−/− mice (Figure S6F in the Supplemental Material).
These gene expression profiles were confirmed by further analyses (Figure 6E through 6G). AngII infusion induced YAP activation, as indicated by increased YAP expression (Figure 6G), a decreased p-YAP/total-YAP ratio, and increased nuclear translocation of YAP (Figure S5B in the Supplemental Material) in the SMCs of AngII-infused SMC-Yap+/+ mice compared with saline-infused SMC-Yap+/+ mice. Compared with AngII-infused SMC-Yap+/+ mice (n=10), AngII-infused SMC-Yap−/− mice (n=10) showed significantly reduced SMC proliferation (Figure 6E), medial thickness (Figure 6F), and production of ECM proteins (elastin, LOX, collagen I, and collagen III) (Figure 6G) but increased SMC apoptosis (Figure 6H). Together, these findings support a critical role for YAP in promoting the adaptive response under aortic stress.
SMC-specific YAP Played a Critical Role in Aortic Protection
Finally, we evaluated the role of SMC YAP in aortic protection by comparing disease development in SMC-Yap−/− and SMC-Yap+/+ littermate mice that were infused with AngII (1000 ng/kg/min) (Figure S7A in the Supplemental Material). Compared with the AngII-infused SMC-Yap+/+ littermate mice, AngII-infused SMC-Yap−/− mice showed markedly increased aortic degeneration/destruction (Figure 7A), aortic enlargement in most aortic segments (Figure 7B), and increased incidences of dilatation (100% vs 85%, P=0.05), AAD (including all aneurysm, dissection, and rupture; 83% vs 30%; P<0.001), and severe AAD (including dissection and rupture; 83% vs 30%; P<0.001) (Figure 7C). Furthermore, AngII-infused SMC-Yap−/− mice showed a higher incidence of premature death, but this was not statistically significant (17% vs 5%, P=0.21) (Figure S7B in the Supplemental Material). The increased incidences of AAD and severe AAD observed in AngII-infused SMC-Yap−/− mice were observed particularly in the ascending aorta, aortic arch, and suprarenal aorta (Figure 7D–G, and Figure S7C in the Supplemental Material).
Figure 7. Protective role of smooth muscle cell (SMC)-specific YAP in the aortic wall.
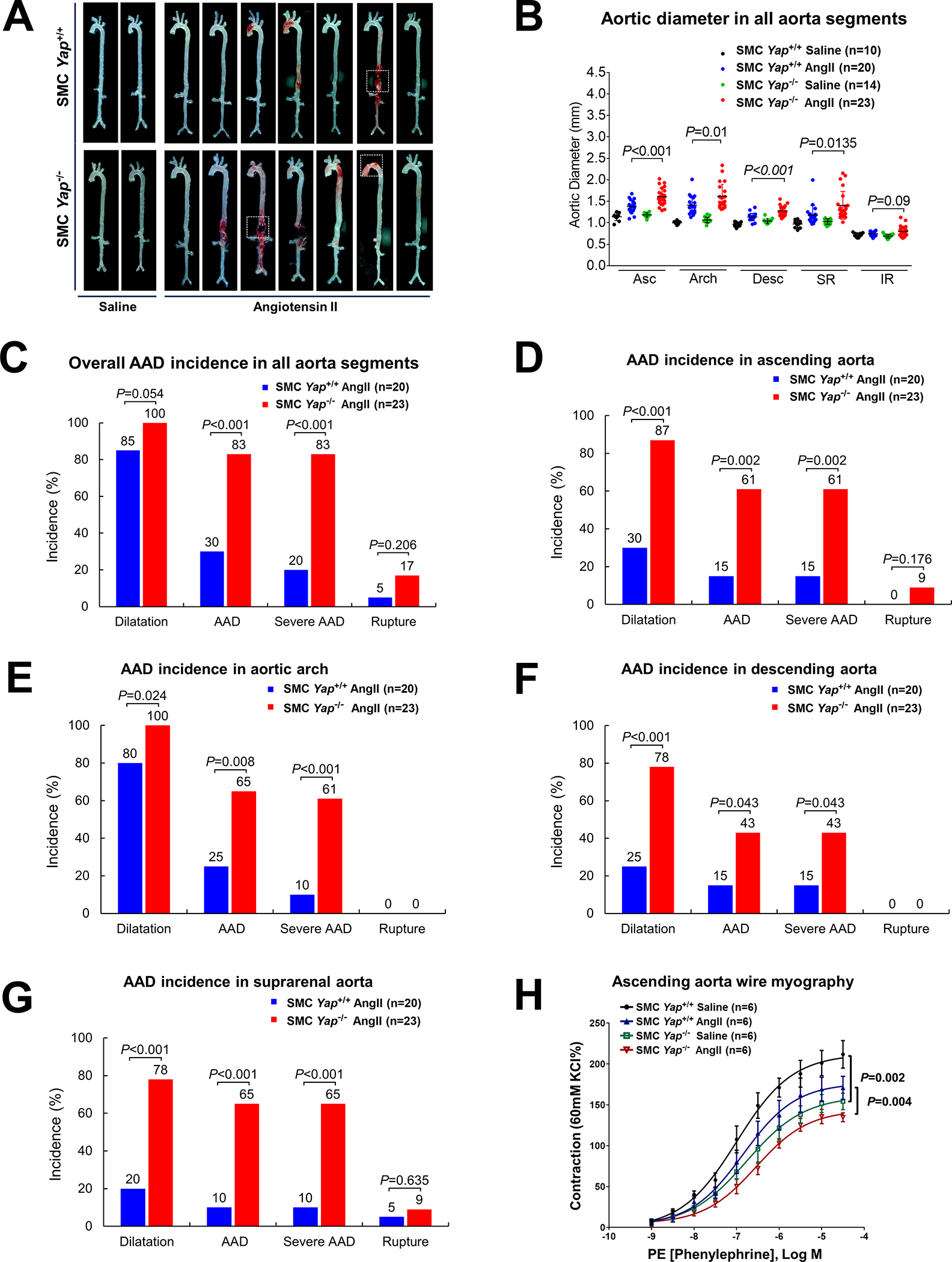
A, Representative images of excised aortas showing more aortic damage in AngII-infused SMC-specific Yap-deficient mice (SMC-Yap−/− AngII; n=23) than in AngII-infused littermate control mice (SMC-Yap+/+ AngII; n=20). Boxes indicate rupture area. B, Median aortic diameters of SMC-Yap−/− AngII mice were enlarged in various aortic segments compared with those of SMC-Yap+/+ AngII mice. Asc, ascending; Desc, descending; SR, suprarenal; IR, infrarenal. C, The overall incidence of AAD was significantly higher in SMC-Yap−/− AngII mice than in SMC-Yap+/+ AngII mice. D-G, The incidence of AAD in different aortic segments was significantly lower in SMC-Yap−/− AngII mice than in SMC-Yap+/+ AngII mice. H, Wire myography analysis of ascending thoracic aortic rings showing that the contractile response to phenylephrine was significantly reduced in saline-infused SMC-Yap−/− mice (SMC-Yap−/− Saline, n=6) compared with saline-infused littermate control mice (SMC-Yap+/+ Saline, n=6). AngII infusion decreased contractile ability in both SMC-Yap−/− AngII mice (n=6) and SMC-Yap+/+ AngII mice (n=6). Multi-way analysis of variance with the Holm-Sidak test was used for pairwise comparisons in (H). Data are presented as mean ± standard deviation of the mean.
We also examined contractile function in aortas from SMC-Yap−/− mice and SMC-Yap+/+ mice by performing wire myography analysis of phenylephrine-induced contraction. To exclude the effects of aortic fibrotic remodeling and stiffness from advanced aortic disease, we tested the biomechanical function of ascending thoracic aortas without significant gross destruction. In conditions without AngII infusion, contractile function was significantly compromised in SMC-Yap−/− mice compared with SMC-Yap+/+ mice. AngII infusion (1000 ng/kg/min for 7 days) reduced contractility in SMC-Yap+/+ mice that was even further reduced in SMC-Yap−/− mice (Figure 7H, and Figure S7D in the Supplemental Material), indicating that the loss of Yap caused contractile dysfunction in both conditions with or without AngII infusion. Together, these findings support that SMC YAP plays a critical role in maintaining aortic structural and functional integrity under aortic stress (Figure S7E in the Supplemental Material).
DISCUSSION
In this study, we investigated adaptive gene expression in aortic SMCs in response to aortic stress induced by AngII infusion in mice. We show that AngII infusion activated a comprehensive epigenetic induction of chromatin remodeling and gene expression associated with proliferation, migration, ECM production/organization, antioxidants, DNA repair, and survival (Figure VS7E in the Supplemental Material). Furthermore, we show that YAP in SMCs plays a critical role in this adaptive response, given that the adaptive response was compromised by Yap deficiency in SMCs and resulted in aortic destruction, biomechanical dysfunction/failure, and development of aortic aneurysm and dissection. Our findings support the importance of the adaptive response for aortic homeostasis/protection and reveal YAP as a key factor driving this response.
Aortic cells are exposed to a constantly changing environment with respect to hemodynamic forces42,43 (e.g., shear stress, cyclic stretch, and hydrostatic pressure) and aortic architecture44–46 (e.g., cell shape and density, ECM rigidity, and cell-ECM interactions). These changes alter gene expression in aortic cells, particularly SMCs, and modify aortic structure and function to maintain aortic strength.47 However, the molecular mechanisms of this adaptive response are poorly understood. Recent advances in single-cell techniques48,49 have enabled the investigation of this adaptive response in diverse aortic cell populations at single-cell resolution. By using sc-RNAseq analysis, we discovered that AngII infusion induced a comprehensive gene expression program in SMCs of the thoracic aortic wall that promotes cell proliferation, migration, angiogenesis, ECM production and organization, cell-matrix interactions, antioxidation, DNA repair, and survival. These changes collectively function as an adaptive response that increases aortic wall thickness and strength and its ability to sustain pressure and prevent dilatation, dissection, and rupture. Interestingly, aside from some commonly shared gene expression patterns, each SMC subcluster exhibited its own unique pattern of adaptive gene expression, suggesting that these SMC subclusters contribute to the aortic adaptive response in a specific, yet coordinated manner.
To understand the control of gene expression and identify TFs that drive the induction of gene expression in the SMC adaptive response,50–52 we performed scATAC-seq analysis to investigate the genome-wide chromatin accessibility landscape of genes in SMC populations and the dynamic changes that occurred after AngII infusion. GO analysis of enriched peaks in AngII/HFD-treated mice showed that AngII infusion induced systemic chromatin opening in genes involved in proliferation, migration, and ECM organization, suggesting that the induction of gene expression was partially controlled at the epigenetic level. Analysis of the regulatory regions of these adaptive genes provided further mechanistic insights into their regulation. For example, in Lox, some chromatin regions/peaks showed constant opening with minimal changes after AngII infusion, whereas other chromatin regions/peaks showed significant enrichment in AngII/HFD-treated WT mice, suggesting differential roles of these peaks under the constitutive or inducible expression of target genes. Similarly, different patterns of peaks were also observed in other adaptive genes.
In our search for TFs that drive this adaptive gene expression, we identified YAP/TEADs as central TFs that orchestrate adaptive gene expression. YAP/TEADs are critical for the induction of genes encoding ECM proteins including Lox, Eln, Fn1, and Col3a1. YAP/TEADs motifs were detected in the constant opening peaks and enriched peaks of Lox, suggesting a role for YAP/TEADs in constitutive and inducible Lox expression. In SMCs, YAP/TEADs were involved in stretch-induced Lox expression by directly binding to the promoter of Lox and inducing its expression. SMC-Yap deficiency compromised the induction of Lox in AngII-infused mice. Similarly, YAP/TEADs are involved in promoting other adaptive responses such as SMC proliferation. In SMC clusters, YAP/TEADs motifs were present in genes involved in cell proliferation (e.g., Birc5, Ccnl1, and Klf5), and Yap deficiency compromised the AngII infusion–induced expression of genes involved in cell cycle progression (e.g., Cdkn3, Foxm1, and Nek2),53 thus preventing cell proliferation. Therefore, YAP may promote the aortic adaptive response by directly inducing the expression of adaptive genes in ECM production and SMC proliferation. Furthermore, our study suggests a critical role of YAP/TEADs in the induction of TGF-β signaling54–56 during AngII infusion. ScATAC-seq analysis revealed TEAD motifs in the regulatory regions of the TGF-β signaling molecules Smad2, Smad3, and Tgfbr2, and SMC-specific Yap deficiency compromised AngII infusion–induced TGF-β signaling in mice. TGF-β signaling is known to play a critical role in ECM production, SMC differentiation, and aortic homeostasis.57,58 Thus, by activating TGF-β signaling, YAP/TEADs may effectively amplify the adaptive response in the aortic wall.
Of note, YAP, which lacks a DNA-binding domain, regulates the expression of diverse target genes by binding to DNA-binding TF partners such as TEAD1-4,59 MRTF–SRF signaling,60 SMAD2/3, RUNXs, TCF4, TBX5, EGR1, GATA4, p73, and KLF4.37,61,62 These co-TFs bind to and control the expression of different target genes. Myocardin-related TFs are key regulators in SMC gene expression and have been shown to interact with YAP.63,64 Interestingly, we observed the induced expression of myocardin-related TFs in AngII-infused mice (Figure S8A in the Supplemental Material). The possibility remains that, by partnering with different cofactors, YAP coordinately and simultaneously induces the expression of different target adaptive genes, thereby collectively contributing to the adaptive response.
AngII infusion may affect the aortic wall through multiple mechanisms. For example, AngII infusion has been shown to increase blood pressure and hemodynamic stress that can lead to YAP activation and wall remodeling. AngII signaling can also activate YAP through AngII’s receptor AT1R and induce cell injury in target cells.65,66 Furthermore, AngII increases inflammation and oxidative stress.67,68 Although the combination of these factors creates aortic stress in the AAD model, mechanical force is likely to be a major factor in triggering the observed adaptive response. Results of our in vitro experiments showed that mechanical stretch can trigger YAP activation and the expression of adaptive genes in human aortic SMCs, which is similar to what we observed in mice with AngII-induced AAD.
Increasing evidence suggests a central role for YAP in mechanotransduction.69 YAP senses an array of mechanical and geometric signals70,71 in the microenvironment. Once activated, YAP is involved in tissue repair, regeneration, and remodeling, which is part of the adaptive response to the environment. Our findings indicate that aortic stress such as increased hemodynamic pressure in the aortic wall activates YAP in SMCs. YAP, together with its partner TFs, induces a set of transcriptional programs that promote the adaptive response including the expression of genes involved in SMC proliferation and ECM production. We found that SMC-specific Yap deficiency in mice compromised this adaptive response and rendered the aortic wall vulnerable to aortic stress, resulting in aortic destruction, biomechanical failure, aneurysm, and dissection. We also observed that, even under the basal condition, contractile gene expression was lower in SMC-Yap−/− mice than in SMC-Yap+/+ mice (Figure S8B in the Supplemental Material), indicating potential defects in the aortic function of SMC-Yap−/− mice. Although spontaneous AAD did not develop in SMC-Yap−/− mice without AngII infusion (monitored for up to 5 months), defects in the aortic function of SMC-Yap−/− mice were likely to compromise the aortic response to stress. The aortic dysfunction observed under the basal condition may make the aorta vulnerable and more susceptible to stress-induced degeneration and disease formation. Our study not only underscores the critical role of mechanotransduction in the aortic adaptive response, but it also highlights the importance of the adaptive response in maintaining aortic homeostasis. Risk factors such as genetic factors, aging, metabolic stress, and inflammation can compromise this response and increase susceptibility to aortic degeneration and destruction.
Our study has limitations. First, we performed our study in male mice only. Myh11-CreERT2 is inserted in the Y chromosome; thus, Cre is expressed in male mice only. Sex dimorphism in AAD development has been well established.72 It is not clear whether there is sex-based difference in adaptive response that leads to different susceptibility to disease formation in males and females. Future study with SMC-specific Cre in both male and female mice73 is needed to address this question. Additionally, we demonstrated the adaptive response in AngII infusion-induced disease model (with or without HFD). It is not clear whether this adaptive also exists in other disease models, such as the BAPN-induced AAD model.74 If so, further studies are needed to understand the upstream signals that activate this adaptive response, and the extent this adaptive response protects aorta against disease formation.
In conclusion, aortic stress induces an epigenetic program in SMCs that promotes the adaptive response to maintain aortic homeostasis. YAP plays a central role in this adaptive response by sensing mechanical signals and inducing adaptive gene expression (Figure S9 in the Supplemental Material). A compromised adaptive response may represent an important mechanism underlying aortic disease development.
Supplementary Material
Highlights.
Aortic stress including increased hemodynamic pressure triggers an adaptive response in aortic smooth muscle cells (SMCs) characterized by the induction of genes associated with wound healing, proliferation, migration, extracellular matrix production/organization, actin cytoskeleton organization, and cell-matrix focal adhesion.
These changes in gene expression are controlled partially by chromatin remodeling and increased accessibility at the enhancers and promoters of the adaptive response genes.
The mechanical sensor YAP/TEADs is a top candidate transcription factor driving the induction of adaptive gene expression in aortic SMCs.
By activating TGF-β signaling, YAP may effectively amplify the adaptive response in the aortic wall.
Acknowledgments
We gratefully acknowledge Nicole Stancel, PhD, ELS (D), of the Department of Scientific Publications at the Texas Heart Institute, for editorial support.
Sources of Funding
This work was supported by NIH grants R01HL131980, R01HL143359, R01HL158157, R01 HL159988, R35HL155649 and American Heart Association grant 18SFRN33960114. Transcriptome analyses were performed at the Department of Molecular and Human Genetics Functional Genomics Core and Single Cell Genomics Core at Baylor College of Medicine, which are partially supported by grants from the NIH (grants S10OD023469, S10OD018033, S10OD025240, CA125123, and 1P30ES030285) and the Cancer Prevention Research Institute of Texas (core grant RP170005). Dr. Yanming Li is supported by the Victor A. McKusick Fellowship Grant from The Marfan Foundation. Dr. LeMaire’s work is supported in part by the Jimmy and Roberta Howell Professorship in Cardiovascular Surgery at Baylor College of Medicine.
Non-Standard Abbreviations and Acronyms
- AAD
Aneurysm and dissection
- ANOVA
Analysis of variance
- DAG
Differentially activated genes
- DEG
Differentially expressed genes
- ECM
Extracellular matrix
- GO
Gene ontology
- HBSS
Hanks’ Balanced Salt Solution
- HFD
High-fat diet
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NIH
National Institutes of Health
- PBS
Phosphate-buffered saline
- PCR
Polymerase chain reaction
- SMC
Smooth muscle cell
- TEAD
Transcriptional enhanced associate domains
- TF
Transcription factor
- TUNEL
Transferase dUTP nick end labeling
- UMAP
Uniform manifold approximation and projection
- UMI
Unique molecular identifier
- WT
Wild-type
- YAP
Yes-associated protein
Footnotes
Disclosures
Dr. LeMaire serves as a consultant for Terumo Aortic and Cerus; and serves as a principal investigator for clinical studies sponsored by Terumo Aortic and CytoSorbents.
Supplemental Materials
Online Figures S1–S9
Expanded Materials and Methods
Online Table S1 (Major Resources Table)
Online Table S2 (TEAD Motif-enriched Adaptive Genes)
Online Table S3 (YAP-based ChIP-seq All Peaks Annotation)
Contributor Information
Chen Zhang, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX; Texas Heart Institute, Houston, TX.
Yanming Li, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX; Texas Heart Institute, Houston, TX.
Abhijit Chakraborty, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX; Texas Heart Institute, Houston, TX.
Yang Li, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX; Texas Heart Institute, Houston, TX.
Kimberly R. Rebello, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX Texas Heart Institute, Houston, TX.
Pingping Ren, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX; Texas Heart Institute, Houston, TX.
Wei Luo, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX; Texas Heart Institute, Houston, TX.
Lin Zhang, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX; Texas Heart Institute, Houston, TX.
Hong S. Lu, Saha Cardiovascular Research Center, University of Kentucky, Lexington, KY Department of Physiology, University of Kentucky, Lexington, KY.
Lisa A. Cassis, Department of Pharmacology and Nutritional Sciences, University of Kentucky, Lexington, KY
Joseph S. Coselli, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX Texas Heart Institute, Houston, TX; Cardiovascular Research Institute, Baylor College of Medicine, Houston, TX.
Alan Daugherty, Saha Cardiovascular Research Center, University of Kentucky, Lexington, KY; Department of Physiology, University of Kentucky, Lexington, KY.
Scott A. LeMaire, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX Texas Heart Institute, Houston, TX; Cardiovascular Research Institute, Baylor College of Medicine, Houston, TX.
Ying H. Shen, Division of Cardiothoracic Surgery, Michael E. DeBakey Department of Surgery, Baylor College of Medicine, Houston, TX Cardiovascular Research Institute, Baylor College of Medicine, Houston, TX.
REFERENCES
- 1.Humphrey JD, Schwartz MA, Tellides G, Milewicz DM. Role of mechanotransduction in vascular biology: focus on thoracic aortic aneurysms and dissections. Circ Res. 2015;116:1448–1461. doi: 10.1161/CIRCRESAHA.114.304936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherifova S, Holzapfel GA. Biomechanics of aortic wall failure with a focus on dissection and aneurysm: A review. Acta Biomater. 2019;99:1–17. doi: 10.1016/j.actbio.2019.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humphrey JD, Milewicz DM, Tellides G, Schwartz MA. Cell biology. Dysfunctional mechanosensing in aneurysms. Science. 2014;344:477–479. doi: 10.1126/science.1253026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen YH, LeMaire SA, Webb NR, Cassis LA, Daugherty A, Lu HS. Aortic Aneurysms and Dissections Series. Arterioscler Thromb Vasc Biol. 2020;40:e37–e46. doi: 10.1161/ATVBAHA.120.313991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anwar MA, Shalhoub J, Lim CS, Gohel MS, Davies AH. The effect of pressure-induced mechanical stretch on vascular wall differential gene expression. J Vasc Res. 2012;49:463–478. doi: 10.1159/000339151 [DOI] [PubMed] [Google Scholar]
- 6.Pinard A, Jones GT, Milewicz DM. Genetics of Thoracic and Abdominal Aortic Diseases. Circ Res. 2019;124:588–606. doi: 10.1161/CIRCRESAHA.118.312436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michel JB, Jondeau G, Milewicz DM. From genetics to response to injury: vascular smooth muscle cells in aneurysms and dissections of the ascending aorta. Cardiovasc Res. 2018;114:578–589. doi: 10.1093/cvr/cvy006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogunovic N, Meekel JP, Micha D, Blankensteijn JD, Hordijk PL, Yeung KK. Impaired smooth muscle cell contractility as a novel concept of abdominal aortic aneurysm pathophysiology. Sci Rep. 2019;9:6837. doi: 10.1038/s41598-019-43322-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milewicz DM, Trybus KM, Guo DC, Sweeney HL, Regalado E, Kamm K, Stull JT. Altered Smooth Muscle Cell Force Generation as a Driver of Thoracic Aortic Aneurysms and Dissections. Arterioscler Thromb Vasc Biol. 2017;37:26–34. doi: 10.1161/ATVBAHA.116.303229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karimi A, Milewicz DM. Structure of the Elastin-Contractile Units in the Thoracic Aorta and How Genes That Cause Thoracic Aortic Aneurysms and Dissections Disrupt This Structure. Can J Cardiol. 2016;32:26–34. doi: 10.1016/j.cjca.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo DC, Papke CL, He R, Milewicz DM. Pathogenesis of thoracic and abdominal aortic aneurysms. Ann N Y Acad Sci. 2006;1085:339–352. doi: 10.1196/annals.1383.013 [DOI] [PubMed] [Google Scholar]
- 12.Daugherty A, Manning MW, Cassis LA. Angiotensin II promotes atherosclerotic lesions and aneurysms in apolipoprotein E-deficient mice. J Clin Invest. 2000;105:1605–1612. doi: 10.1172/JCI7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren P, Hughes M, Krishnamoorthy S, Zou S, Zhang L, Wu D, Zhang C, Curci JA, Coselli JS, Milewicz DM, et al. Critical Role of ADAMTS-4 in the Development of Sporadic Aortic Aneurysm and Dissection in Mice. Sci Rep. 2017;7:12351. doi: 10.1038/s41598-017-12248-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu D, Ren P, Zheng Y, Zhang L, Xu G, Xie W, Lloyd EE, Zhang S, Zhang Q, Curci JA, et al. NLRP3 (nucleotide oligomerization domain-like receptor family, pyrin domain containing 3)-caspase-1 inflammasome degrades contractile proteins: Implications for aortic biomechanical dysfunction and aneurysm and dissection formation. Arterioscler Thromb Vasc Biol. 2017;37:694–706. doi: 10.1161/ATVBAHA.116.307648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo W, Wang Y, Zhang L, Ren P, Zhang C, Li Y, Azares AR, Zhang M, Guo J, Ghaghada KB, et al. Critical Role of Cytosolic DNA and Its Sensing Adaptor STING in Aortic Degeneration, Dissection, and Rupture. Circulation. 2020;141:42–66. doi: 10.1161/CIRCULATIONAHA.119.041460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinet P, Milewicz DM, Cassis LA, Leeper NJ, Lu HS, Smith JD. Consideration of Sex Differences in Design and Reporting of Experimental Arterial Pathology Studies-Statement From ATVB Council. Arterioscler Thromb Vasc Biol. 2018;38:292–303. doi: 10.1161/ATVBAHA.117.309524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao Y, Luo W, Zhang L, Wu W, Yuan L, Xu H, Song J, Fujiwara K, Abe JI, LeMaire SA, et al. STING-IRF3 Triggers Endothelial Inflammation in Response to Free Fatty Acid-Induced Mitochondrial Damage in Diet-Induced Obesity. Arterioscler Thromb Vasc Biol. 2017;37:920–929. doi: 10.1161/ATVBAHA.117.309017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen YH, Zhang L, Ren P, Nguyen MT, Zou S, Wu D, Wang XL, Coselli JS, LeMaire SA. AKT2 confers protection against aortic aneurysms and dissections. Circ Res. 2013;112:618–632. doi: 10.1161/CIRCRESAHA.112.300735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan L, Mao Y, Luo W, Wu W, Xu H, Wang XL, Shen YH. Palmitic acid dysregulates the Hippo-YAP pathway and inhibits angiogenesis by inducing mitochondrial damage and activating the cytosolic DNA sensor cGAS-STING-IRF3 signaling mechanism. J Biol Chem. 2017;292:15002–15015. doi: 10.1074/jbc.M117.804005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao CJ, Tung P, Blischak JD, Burnett JE, Barr KA, Dey KK, Stephens M, Gilad Y. Characterizing and inferring quantitative cell cycle phase in single-cell RNA-seq data analysis. Genome Res. 2020;30:611–621. doi: 10.1101/gr.247759.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun SG, Zheng B, Han M, Fang XM, Li HX, Miao SB, Su M, Han Y, Shi HJ, Wen JK. miR-146a and Kruppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 2011;12:56–62. doi: 10.1038/embor.2010.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghavan A, Zhou G, Zhou Q, Ibe JC, Ramchandran R, Yang Q, Racherla H, Raychaudhuri P, Raj JU. Hypoxia-induced pulmonary arterial smooth muscle cell proliferation is controlled by forkhead box M1. Am J Respir Cell Mol Biol. 2012;46:431–436. doi: 10.1165/rcmb.2011-0128OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh T, Gordon RE, Clemmons DR, Busby WH Jr., Duan C Regulation of vascular smooth muscle cell responses to insulin-like growth factor (IGF)-I by local IGF-binding proteins. J Biol Chem. 2003;278:42886–42892. doi: 10.1074/jbc.M303835200 [DOI] [PubMed] [Google Scholar]
- 25.Gerthoffer WT. Mechanisms of vascular smooth muscle cell migration. Circ Res. 2007;100:607–621. doi: 10.1161/01.RES.0000258492.96097.47 [DOI] [PubMed] [Google Scholar]
- 26.Faix J, Weber I. A dual role model for active Rac1 in cell migration. Small GTPases. 2013;4:110–115. doi: 10.4161/sgtp.23476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Firulli AB, Miano JM, Bi W, Johnson AD, Casscells W, Olson EN, Schwarz JJ. Myocyte enhancer binding factor-2 expression and activity in vascular smooth muscle cells. Association with the activated phenotype. Circ Res. 1996;78:196–204. doi: 10.1161/01.res.78.2.196 [DOI] [PubMed] [Google Scholar]
- 28.Long X, Creemers EE, Wang DZ, Olson EN, Miano JM. Myocardin is a bifunctional switch for smooth versus skeletal muscle differentiation. Proc Natl Acad Sci U S A. 2007;104:16570–16575. doi: 10.1073/pnas.0708253104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–81. doi: 10.1152/ajpcell.00386.2006 [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J. Defining the protein-protein interaction network of the human hippo pathway. Mol Cell Proteomics. 2014;13:119–131. doi: 10.1074/mcp.M113.030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G, Pandolfi PP, Givol D, Strano S, Lu X, et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell. 2008;32:803–814. doi: 10.1016/j.molcel.2008.11.019 [DOI] [PubMed] [Google Scholar]
- 32.Haskins JW, Nguyen DX, Stern DF. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci Signal. 2014;7:ra116. doi: 10.1126/scisignal.2005770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu D, Shen YH, Russell L, Coselli JS, LeMaire SA. Molecular mechanisms of thoracic aortic dissection. J Surg Res. 2013;184:907–924. doi: 10.1016/j.jss.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jana S, Hu M, Shen M, Kassiri Z. Extracellular matrix, regional heterogeneity of the aorta, and aortic aneurysm. Exp Mol Med. 2019;51:1–15. doi: 10.1038/s12276-019-0286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizuno T, Murakami H, Fujii M, Ishiguro F, Tanaka I, Kondo Y, Akatsuka S, Toyokuni S, Yokoi K, Osada H, et al. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31:5117–5122. doi: 10.1038/onc.2012.5 [DOI] [PubMed] [Google Scholar]
- 37.Kim MK, Jang JW, Bae SC. DNA binding partners of YAP/TAZ. BMB Rep. 2018;51:126–133. doi: 10.5483/bmbrep.2018.51.3.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, Zhu LJ, Goel HL, Mercurio AM, Park JS, et al. Tead and AP1 Coordinate Transcription and Motility. Cell Rep. 2016;14:1169–1180. doi: 10.1016/j.celrep.2015.12.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albig AR, Becenti DJ, Roy TG, Schiemann WP. Microfibril-associate glycoprotein-2 (MAGP-2) promotes angiogenic cell sprouting by blocking notch signaling in endothelial cells. Microvasc Res. 2008;76:7–14. doi: 10.1016/j.mvr.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee MY, Garvey SM, Baras AS, Lemmon JA, Gomez MF, Schoppee Bortz PD, Daum G, LeBoeuf RC, Wamhoff BR. Integrative genomics identifies DSCR1 (RCAN1) as a novel NFAT-dependent mediator of phenotypic modulation in vascular smooth muscle cells. Hum Mol Genet. 2010;19:468–479. doi: 10.1093/hmg/ddp511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riha GM, Lin PH, Lumsden AB, Yao Q, Chen C. Roles of hemodynamic forces in vascular cell differentiation. Ann Biomed Eng. 2005;33:772–779. doi: 10.1007/s10439-005-3310-9 [DOI] [PubMed] [Google Scholar]
- 43.Albinsson S, Hellstrand P. Integration of signal pathways for stretch-dependent growth and differentiation in vascular smooth muscle. Am J Physiol Cell Physiol. 2007;293:C772–782. doi: 10.1152/ajpcell.00622.2006 [DOI] [PubMed] [Google Scholar]
- 44.Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urbanczyk M, Layland SL, Schenke-Layland K. The role of extracellular matrix in biomechanics and its impact on bioengineering of cells and 3D tissues. Matrix Biol. 2020;85–86:1–14. doi: 10.1016/j.matbio.2019.11.005 [DOI] [PubMed] [Google Scholar]
- 46.Gilbert PM, Weaver VM. Cellular adaptation to biomechanical stress across length scales in tissue homeostasis and disease. Semin Cell Dev Biol. 2017;67:141–152. doi: 10.1016/j.semcdb.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lacolley P, Regnault V, Segers P, Laurent S. Vascular smooth muscle cells and arterial stiffening: Relevance in development, aging, and disease. Physiol Rev. 2017;97:1555–1617. doi: 10.1152/physrev.00003.2017 [DOI] [PubMed] [Google Scholar]
- 48.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, Pokholok D, Aryee MJ, Steemers FJ, Lebofsky R, et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat Biotechnol. 2019;37:916–924. doi: 10.1038/s41587-019-0147-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander MR, Owens GK. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu Rev Physiol. 2012;74:13–40. doi: 10.1146/annurev-physiol-012110-142315 [DOI] [PubMed] [Google Scholar]
- 51.Owens GK. Molecular control of vascular smooth muscle cell differentiation and phenotypic plasticity. Novartis Found Symp. 2007;283:174–191; discussion 191–173, 238–141. doi: 10.1002/9780470319413.ch14 [DOI] [PubMed] [Google Scholar]
- 52.Das S, Senapati P, Chen Z, Reddy MA, Ganguly R, Lanting L, Mandi V, Bansal A, Leung A, Zhang S, et al. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nat Commun. 2017;8:1467. doi: 10.1038/s41467-017-01629-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engeland K Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018;25:114–132. doi: 10.1038/cdd.2017.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goumans MJ, Liu Z, ten Dijke P. TGF-beta signaling in vascular biology and dysfunction. Cell Res. 2009;19:116–127. doi: 10.1038/cr.2008.326 [DOI] [PubMed] [Google Scholar]
- 55.Takeda N, Hara H, Fujiwara T, Kanaya T, Maemura S, Komuro I. TGF-beta signaling-related genes and thoracic aortic aneurysms and dissections. Int J Mol Sci. 2018;19. doi: 10.3390/ijms19072125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu JH, Wei H, Jaffe M, Airhart N, Du L, Angelov SN, Yan J, Allen JK, Kang I, Wight TN, et al. Postnatal deletion of the type II transforming growth factor-beta receptor in smooth muscle cells causes severe aortopathy in mice. Arterioscler Thromb Vasc Biol. 2015;35:2647–2656. doi: 10.1161/ATVBAHA.115.306573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003 [DOI] [PubMed] [Google Scholar]
- 58.Goumans MJ, Ten Dijke P. TGF-beta signaling in control of cardiovascular function. Cold Spring Harb Perspect Biol. 2018;10. doi: 10.1101/cshperspect.a022210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim T, Hwang D, Lee D, Kim JH, Kim SY, Lim DS. MRTF potentiates TEAD-YAP transcriptional activity causing metastasis. EMBO J. 2017;36:520–535. doi: 10.15252/embj.201695137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pocaterra A, Romani P, Dupont S. YAP/TAZ functions and their regulation at a glance. J Cell Sci. 2020;133. doi: 10.1242/jcs.230425 [DOI] [PubMed] [Google Scholar]
- 62.Zhu C, Li L, Zhao B. The regulation and function of YAP transcription co-activator. Acta Biochim Biophys Sin (Shanghai). 2015;47:16–28. doi: 10.1093/abbs/gmu110 [DOI] [PubMed] [Google Scholar]
- 63.Foster CT, Gualdrini F, Treisman R. Mutual dependence of the MRTF-SRF and YAP-TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics. Genes Dev. 2017;31:2361–2375. doi: 10.1101/gad.304501.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim T, Yang SJ, Hwang D, Song J, Kim M, Kyum Kim S, Kang K, Ahn J, Lee D, Kim MY, et al. A basal-like breast cancer-specific role for SRF-IL6 in YAP-induced cancer stemness. Nat Commun. 2015;6:10186. doi: 10.1038/ncomms10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wennmann DO, Vollenbroker B, Eckart AK, Bonse J, Erdmann F, Wolters DA, Schenk LK, Schulze U, Kremerskothen J, Weide T, et al. The Hippo pathway is controlled by Angiotensin II signaling and its reactivation induces apoptosis in podocytes. Cell Death Dis. 2014;5:e1519. doi: 10.1038/cddis.2014.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forrester SJ, Booz GW, Sigmund CD, Coffman TM, Kawai T, Rizzo V, Scalia R, Eguchi S. Angiotensin II Signal Transduction: An Update on Mechanisms of Physiology and Pathophysiology. Physiol Rev. 2018;98:1627–1738. doi: 10.1152/physrev.00038.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das S, Zhang E, Senapati P, Amaram V, Reddy MA, Stapleton K, Leung A, Lanting L, Wang M, Chen Z, et al. A Novel Angiotensin II-Induced Long Noncoding RNA Giver Regulates Oxidative Stress, Inflammation, and Proliferation in Vascular Smooth Muscle Cells. Circ Res. 2018;123:1298–1312. doi: 10.1161/CIRCRESAHA.118.313207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.St Paul A, Corbett CB, Okune R, Autieri MV. Angiotensin II, Hypercholesterolemia, and Vascular Smooth Muscle Cells: A Perfect Trio for Vascular Pathology. Int J Mol Sci. 2020;21. doi: 10.3390/ijms21124525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 70.Dupont S Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp Cell Res. 2016;343:42–53. doi: 10.1016/j.yexcr.2015.10.034 [DOI] [PubMed] [Google Scholar]
- 71.Wang L, Luo JY, Li B, Tian XY, Chen LJ, Huang Y, Liu J, Deng D, Lau CW, Wan S, et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature. 2016;540:579–582. doi: 10.1038/nature20602 [DOI] [PubMed] [Google Scholar]
- 72.Mosca L, Barrett-Connor E, Wenger NK. Sex/gender differences in cardiovascular disease prevention: what a difference a decade makes. Circulation. 2011;124:2145–2154. doi: 10.1161/CIRCULATIONAHA.110.968792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao M, Zhou J, Wang F, Ali YH, Chan KL, Zou F, Offermanns S, Jiang Z, Jiang Z. An X-linked Myh11-CreER(T2) mouse line resulting from Y to X chromosome-translocation of the Cre allele. Genesis. 2017;55. doi: 10.1002/dvg.23054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sawada H, Beckner ZA, Ito S, Daugherty A, Lu HS. beta-Aminopropionitrile-induced aortic aneurysm and dissection in mice. JVS Vasc Sci. 2022;3:64–72. doi: 10.1016/j.jvssci.2021.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Risso D, Perraudeau F, Gribkova S, Dudoit S, Vert JP. A general and flexible method for signal extraction from single-cell RNA-seq data. Nat Commun. 2018;9:284. doi: 10.1038/s41467-017-02554-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data for experiments presented, including quantification for Western blotting, flow cytometry, immunofluorescence staining, aortic diameter, and wire myography, are available from the corresponding author upon reasonable request. All sequencing data have been made publicly available at the Gene Expression Omnibus (GEO) and can be accessed at GSE213735 (single cell RNA-seq), and GSE214082 (single cell ATAC-seq). The computer code used in this study is available upon request.


