Abstract
The cytotoxic enterotoxin Act from a diarrheal isolate, SSU, of Aeromonas hydrophila is aerolysin related and crucial to the pathogenesis of Aeromonas infections. To elucidate the role of environmental signals which influence the expression of the cytotoxic enterotoxin gene (act), a portion of the act gene, including the putative promoter region, was fused in frame to a truncated alkaline phosphatase gene (phoA) of Escherichia coli. The act::phoA reporter gene was then introduced into the chromosome of A. hydrophila by using the suicide vector pJQ200SK, allowing the fusion protein to be secreted out into the culture medium. Western blot analysis demonstrated the presence of a correctly size 110-kDa fusion protein in the culture supernatant, which reacted with both anti-Act and anti-alkaline phosphatase antibodies. Based on alkaline phosphatase (PhoA) activity in the culture supernatant, we demonstrated that calcium significantly increased the activity of the act promoter but that glucose and iron repressed its activity in a dose-dependent fashion. The act promoter exhibited optimal activity at pH 7.0 and at 37°C, and maximal PhoA activity was noted when the culture was aerated. Using a Vibrio cholerae iron uptake regulator gene (fur) as a probe, a 2.6-kb SalI/HindIII DNA fragment from an A. hydrophila chromosome was cloned and sequenced. The DNA sequence revealed a 429-bp open reading frame that exhibited 69% homology at the DNA level with the fur gene and 79% homology at the amino acid level with the iron uptake regulator (Fur) protein of V. cholerae. Complementation experiments demonstrated that the A. hydrophila fur gene could restore iron regulation in an E. coli fur-minus mutant. Using the suicide vector pDMS197, we generated a fur isogenic mutant of wild-type A. hydrophila SSU. Northern blot analysis data indicated that the repression in the transcription of the act gene by iron was relieved in the fur isogenic mutant. Further, iron regulation in the fur isogenic mutant of A. hydrophila could be restored by complementation. These results are important in understanding the regulation of the act gene under in vivo conditions.
Aeromonas species cause septicemia and gastroenteritis, and an epidemiological study has implicated Aeromonas spp. in causing food-borne outbreaks and traveler's diarrhea (10). Among various virulence factors produced by Aeromonas spp., the cytotoxic enterotoxin Act may lead to either gastroenteritis or nonintestinal infections, depending upon the route of the infection (10, 11). The cytotoxic enterotoxin gene (act) from a human diarrheal isolate, A. hydrophila SSU, has been cloned, sequenced, and hyperexpressed in our laboratory (11, 12, 20), and an isogenic (act-minus) mutant has been generated. Our data indicated that the act isogenic mutant was significantly attenuated in causing infection in a mouse model (55). Act is a single-chain polypeptide, and the mature form of the toxin exhibits a size of 49 to 52 kDa. Act is aerolysin related, which we have recently shown to activate proinflammatory cytokine and eicosanoid cascades in macrophages, leading to tissue damage and a fluid secretory response (12).
Pathogenesis of bacterial infection requires the interaction of several virulence genes, which are frequently regulated by specific environmental stimuli. While some of these stimuli directly affect the virulence gene, some operate through a regulatory gene (36). At present, little information is available on environmental signals which trigger expression of the act gene during infection of humans and animals with A. hydrophila. To investigate the influence of environmental and nutritional factors on the expression of the act gene, we prepared a reporter gene construct in which the act gene of A. hydrophila SSU was fused in frame to the alkaline phosphatase gene (phoA) of Escherichia coli. The act::phoA reporter gene was then integrated into chromosomal DNA of A. hydrophila SSU by single-crossover homologous recombination, and the resulting mutant was subsequently exposed to different environmental and nutritional stimuli.
Among nutritional factors, iron is essential for cellular metabolism, since it is needed as a cofactor for a great number of enzymes (53). A low iron concentration is the major change when bacteria enter the host, and it has been demonstrated to be a major environmental signal that triggers expression of virulence determinants (31). The mechanism of iron regulation has been shown to be linked to the iron uptake regulator (fur) locus in many bacteria (19). In this study, we examined the environmental and nutritional stimuli that affect act gene expression. Further, we have characterized the fur locus in A. hydrophila and provided evidence that the fur gene is responsible for iron regulation of the act gene.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The sources of A. hydrophila, Vibrio cholerae, and E. coli strains, as well as the plasmids used in this study, are listed in Table 1. Briefly, the suicide vector pJQ200SK contained a P15A origin of replication (ori), a levan sucrase gene (sacB) from Bacillus subtilis, and a gentamicin resistance (Gmr) gene (41). Another suicide vector, pDMS197, has a conditional R6K ori, a sacB gene, and a tetracycline resistance (Tcr) gene (17). The E. coli strains SBC22 and SBC23 contain a chromosomal gene fusion between the iron-regulated promoter of the A subunit of Shiga-like toxin I of E. coli (slt-IA) and the alkaline phosphatase gene from TnphoA. These strains were constructed by integration of the suicide plasmid pSBC48 into the homologous, 3.65-kb, random SmaI fragments of chromosomal DNAs in strains SM796 and SBC796 of E. coli (32). The E. coli strains SBC22 and SBC23 were fur+ and fur-negative mutant, respectively, and both were resistant to ampicillin (Apr), kanamycin (Kmr), and spectinomycin (Smr) (Table 1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Sourcea or reference |
|---|---|---|
| Strains | ||
| A. hydrophila | ||
| SSU | CDC, Atlanta, Ga. | |
| SSU-R | Rifr | Laboratory stock |
| Mutant SSU66 | A. hydrophila SSU-R with chromosomally integrated act::phoA fusion gene; Rifr Gmr, sucrose sensitive | This study |
| Mutant SSU88 | fur isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr, surcrose resistance | This study |
| V. cholerae V86 | Laboratory stock | |
| E. coli | ||
| DH5α | recA gyrA | Laboratory stock |
| SM10 | Kmr λpir | 17 |
| S17-1 | Streptomycin resistance (Strr), trimethoprim resistance (Tmpr), λpir | 25 |
| SBC22 | SM796 with pSBC48 chromosomally integrated via the 3.65-kb SmaI fragment of pSBC40; fur+, Smr Kmr Apr | 32 |
| SBC23 | SBC796 with pSBC48 chromosomally integrated as with SBC22; fur minus, Smr Kmr Apr | 32 |
| C118 | phoA minus | Laboratory stock |
| Plasmids | ||
| pRK2013 | Helper plasmid, Kmr | ATCC, Manassas, Va. |
| pBR322 | Apr Kmr | Amersham |
| pBluescript SK | Apr | Stratagene |
| pUC-4K | Contains a 1.2-kb kanamycin cassette | Amersham |
| pXHC95 | pBluescript recombinant plasmid containing a 2.8-kb BamHI DNA fragment from A. hydrophila SSU chromosomal DNA and harboring the act gene with its promoter region | 55 |
| pUC128 | Contains a phoA gene which lacks its signal sequence and the first 13 codons | Laboratory stock |
| pUCact | Portion of the act gene with its promoter region fused in frame with the phoA gene in plasmid pUC128 | This study |
| pJQ200SK | Suicide vector; P15A sacB Gmr | 41 |
| pDMS197 | Suicide vector; R6K ori sacB Tcr | 17 |
| pJQ200actphoA | Vector pJQ200SK containing a act::phoA fusion gene; Gmr | This study |
| pBfur | pBluescript recombinant plasmid containing a 2.6-kb SalI/HindIII DNA fragment from the A. hydrophila chromosome harboring the fur gene | This study |
| pDMS197fur | Vector pDMS197 containing a truncated fur gene with its flanking sequences for generating a fur isogenic mutant of A. hydrophila | This study |
| pABN203 | E. coli fur gene, cloned in pBR322; Tcr | 49 |
| pBRfur1 | A. hydrophila fur gene, cloned in pBR322 at the SacI site under the control of a promoter which controls the ampicillin resistance gene in the vector; Tcr | This study |
| pBRpfur2 | A. hydrophila fur gene with its putative promoter region cloned in pBR322 at the EcoRI site | This study |
CDC, Centers for Disease Control and Prevention; ATCC, American Type Culture Collection.
Enzymes, chemicals, and recombinant DNA techniques.
The antibiotics ampicillin, gentamicin, tetracycline, kanamycin, spectinomycin, and streptomycin were used at concentrations of 100, 15, 15, 50, 50, and 25 μg/ml, respectively. Rifampin was used at concentrations of 40 μg/ml for bacterial growth and 300 μg/ml during conjugation experiments. All of the antibiotics used were obtained from Sigma (St. Louis, Mo.). Restriction endonucleases and T4 DNA ligase were obtained from Promega (Madison, Wis.). An Advantage cDNA PCR kit was purchased from Clontech (Palo Alto, Calif.). The cyclic AMP (cAMP) analog 8-bromo-cAMP was purchased from Sigma. The stock concentrations of glucose, calcium chloride, ferric sulfate, and the iron chelator 2,2-dipyridyl (Sigma) were 50%, 1 M, 3.6 M, and 0.2 M, respectively. For the alkaline phosphatase (PhoA) assay, the substrate BCIP (5-bromo-4-chloro-3-indolyl phosphate; Sigma) was added at a concentration of 80 μg/ml when Luria-Bertani (LB) medium was used. Alternatively, 0.4% p-nitrophenol (Sigma) was added when T medium (45) was employed for bacterial growth. All of the techniques used in this study were previously described (55).
Construction of an act::phoA reporter gene.
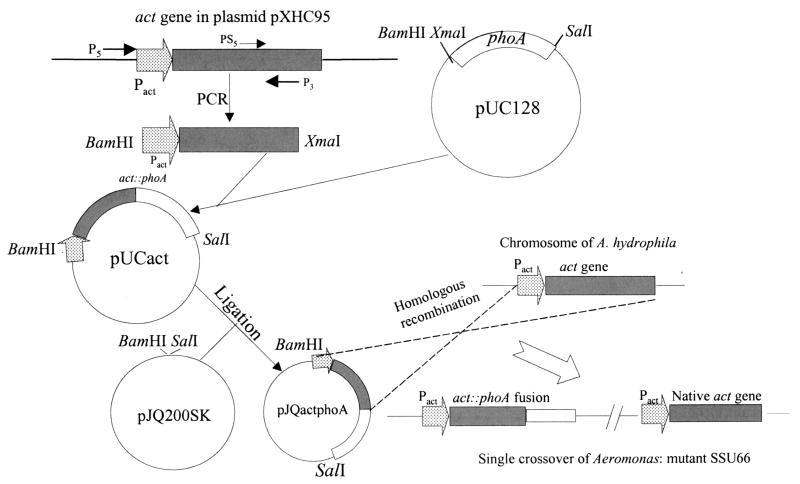
The strategy used to construct an act::phoA reporter gene is shown in Fig. 1. Briefly, we designed two primers; P5 contained a BamHI restriction site 338 bp upstream of the act gene start codon, and P3 contained an XmaI restriction site 1.1 kb downstream from the act gene start codon. The sequences of the P5 and P3 primers were as follows: 5′ CGCGGATCCTAAGAGCCATGTTAT 3′ and 5′ TCACCCGGGTGATGTAACGCTTGTCCCACTG 3′, respectively. The primers were synthesized commercially by Biosynthesis, Inc. (Lewisville, Tex.), and the program used for PCR was as follows: 94°C for 2 min (denaturation) followed by 30 cycles of 94°C for 1 min and 68°C for 3 min. The final extension was performed at 72°C for 7 min. The PCR product was isolated from the agarose gel, purified, and subjected to automated DNA sequence analysis (Protein Chemistry Core Facility, The University of Texas Medical Branch, Galveston). The P3 primer was designed such that the 3′ region of the act gene, which was proteolytically cleaved during processing, was removed during PCR amplification (11).
FIG. 1.
Flow diagram showing act::phoA reporter gene construction. Using primers P5 and P3, a 1.4-kb DNA fragment containing the putative act promoter region and a portion of the act gene was amplified from plasmid pXHC95 and inserted into plasmid pUC128 at BamHI and XmaI restriction sites to generate an in-frame fusion with the phoA gene. The fusion gene was then ligated to the suicide vector pJQ200SK to generate the recombinant plasmid pJQactphoA. By conjugation between E. coli strain S17-1(pJQactphoA) and A. hydrophila, the fusion gene was integrated into the chromosome of A. hydrophila by single-crossover homologous recombination to generate a mutant, SSU66 (Table 1). The solid bar represents the act gene, and the open bar represents the phoA gene. The dotted arrow denotes the putative act promoter. These plasmids are not drawn to scale.
Using primers P5 and P3, a 1.4-kb DNA fragment containing the putative act promoter and a major portion of the act gene was amplified from plasmid pXHC95 (55) and inserted into a plasmid, pUC128, at BamHI and XmaI restriction sites to result in in-frame fusion with the phoA gene (Fig. 1). The resulting recombinant plasmid was designated pUCact and was partially sequenced using primer PS5 to ensure in-frame fusion. The primer PS5 was designed 67 bp upstream of primer P3 (Fig. 1) and had the following sequence: 5′ TCCCCCCGGGATGTAACGCTTGTC 3′. A BamHI/SalI DNA fragment from plasmid pUCact which contained the act::phoA fusion gene was ligated to a suicide vector, pJQ200SK, to generate a recombinant plasmid, pJQactphoA (Fig. 1). The pJQactphoA plasmid in E. coli strain S17-1 was then delivered to A. hydrophila SSU by conjugation. E. coli strains S17-1 and SM10 contain λpir, allowing replication of the suicide vectors only in these strains (55). Briefly, both E. coli strains harboring plasmid pJQactphoA and rifampin-resistant (Rifr) A. hydrophila SSU were grown under static conditions at 37°C overnight. The cultures were mixed (5 ml each) at a concentration of 8 × 106 cells/ml. After 2 h of incubation at 37°C, the mixture was centrifuged (4,000 × g for 10 min), resuspended in 200 μl of LB medium, plated onto LB agar plates without any antibiotic pressure, and incubated for an additional 4 h at 37°C. Subsequently, the culture was removed from the plates and various dilutions (10−4 to 10−9) of the sample were plated onto LB agar plates with rifampin, gentamicin, and the PhoA substrate BCIP. The colonies of A. hydrophila in which act::phoA was integrated into the chromosome exhibited a diffuse blue color around the colonies as a result of the secretion of PhoA. These colonies were identified as Aeromonas by a positive oxidase test to differentiate them from E. coli and by an automated identification system (Clinical Microbiology Laboratory, The University of Texas Medical Branch). The identity of the genuine single-crossover mutant (i.e., A. hydrophila SSU66) (Table 1) was confirmed by using Southern blot analysis with the act gene as a probe (55).
Western blot analysis.
Western blot analysis was performed to detect Act::PhoA fusion protein in culture supernatants of A. hydrophila SSU66. Specific polyclonal antibodies to Act (developed in our laboratory) and mouse monoclonal antibodies to E. coli alkaline phosphatase (Caltag Laboratories, Burlingame, Calif.) were used as primary antibodies, followed by appropriate secondary antibodies, which were labeled with horseradish peroxidase (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). The blots were developed using the SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.). The culture supernatant was prepared as follows. A. hydrophila SSU66 was grown in LB medium containing a proteinase inhibitor tablet (Roche Molecular Biochemical, Indianapolis, Ind.) for 18 h at 37°C with shaking (180 rpm). The culture was harvested and centrifuged at 4,000 × g for 15 min. Subsequently, 20 μl of the supernatant was subjected to sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis and Western blot analysis (12).
Alkaline phosphatase assay to quantitate act::phoA fusion gene expression under different environmental stimuli.
Briefly, a 2-μl aliquot of an A. hydrophila SSU66 culture grown overnight was inoculated into 50-ml sterilized disposable tubes that contained 3 ml of fresh LB medium with suitable antibiotics and different stimuli. For glucose, 0, 0.1, 0.2, 0.5, 1, and 2% concentrations were used, and for calcium, 0, 2, 5, 10 and 25 mM concentrations were selected. The effect of temperature on PhoA activity was studied at 26, 30, and 37°C. The pH values 5.5, 6.0, 7.0, 7.5, 8.5, and 9.0 were chosen to demonstrate the effect of pH on PhoA activity. Likewise, the effect of aeration on PhoA activity was examined using a shaken flask versus static cultures. The experiments were performed in triplicate, and averages of results from three independent experiments were used for data analysis. Unless otherwise indicated, the cultures were grown at 37°C with constant aeration (180 rpm).
After 18 h of growth, the cultures were centrifuged and the supernatants were taken from each tube for PhoA activity measurement. For PhoA activity, the reaction mixture contained the following: 5 to 50 μl of the culture supernatant, 100 μl of 10× reaction buffer (1 M Tris, 1 M NaCl, 50 mM MgCl2, pH 9.5), 2 μl (40 mg/ml) of the PhoA substrate BCIP, and H2O to a final total volume of 1 ml. The mixture was incubated at 37°C with shaking for 1 h. The density of the blue color was measured at 630 nm, and the growth of the culture (diluted 1:20) after 18 h was measured at 600 nm. The PhoA activity was calculated per milliliter of the culture supernatant per unit of growth.
For iron regulation studies, T medium (45) was used instead of LB medium. T medium was supplemented with thiamine (10 μg/ml) and the l-amino acids arginine and leucine (40 μg/ml each). T medium with 36 μM FeSO4 added was considered as having a high iron content, while T medium with a 0.1 mM concentration of an iron chelator represented low-iron medium. For measuring PhoA activity in T medium, the reaction mixture contained the following: 900 μl of the culture supernatant, 100 μl of 10× reaction buffer (as described above), and 100 μl of 0.4% p-nitrophenol. The reaction mixture was incubated at 37°C for 1 h. The PhoA activity was calculated per milliliter of the culture supernatant per unit of growth (6).
Southern blot analysis on the chromosomal DNA of A. hydrophila with the V. cholerae fur gene probe.
Chromosomal DNA was isolated by using a QIAamp DNA Mini Kit (Qiagen Inc., Valencia, Calif.). An aliquot (10 μg) of the chromosomal DNA was digested with suitable enzymes and subjected to 0.8% agarose gel electrophoresis (55). Next, the digested DNA was transferred to a nylon membrane (Gibco BRL, Gaithersburg, Md.) and baked at 80°C for 2 h. The blots were prehybridized and hybridized by using Quikhyb (Stratagene, La Jolla, Calif.) at 68°C as described by the manufacturer. The probe used was a 453-bp V. cholerae fur locus, which was amplified from the chromosomal DNA of V. cholerae V86 by using two specific primers (5′ primer, 5′ ATGTCAGACAATAACCAAGCG 3′, and 3′ primer, 5′ TTATTTCTTCGGCTTGTGAGC 3′). The probe was labeled with [α-32P]dCTP (ICN, Irvine, Calif.) by using a random primer kit (Gibco BRL). The membranes were washed twice at 68°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0) (1) plus 0.1% SDS for 20 min and then twice in 1× SSC plus 0.1% SDS for 20 min at 68°C. The blots were exposed to the X-ray film at −70°C for 2 to 12 h.
Cloning of the fur gene from A. hydrophila SSU.
Based on Southern blot analysis data, the chromosomal DNA of A. hydrophila SSU was digested with SalI and HindIII restriction enzymes. Subsequently, the digested DNA fragments were ligated to a cloning vector, pBluescript SK (Stratagene), at the restriction sites compatible for generation of a plasmid library. Using the V. cholerae fur gene probe, the plasmid library was screened by colony blot hybridization (33). A recombinant plasmid which hybridized with the V. cholerae fur gene probe was designated pBfur (Table 1). The correct identity of the clone was determined by Southern blot and DNA sequence analyses. The conditions used for hybridization and washing of the filters were similar to those described in the previous section on Southern blot analysis. Prior to prehybridization, the colony blots were washed with a buffer (50 mM Tris, 1 M NaCl, 1 mM EDTA, 0.1% SDS, pH 8.0) at 42°C for 2 h to remove cell debris, which resulted in minimal background during exposure to the X-ray film (1, 33).
Complementation of an E. coli fur-minus strain with the cloned fur locus of A. hydrophila SSU and measurement of PhoA activity in E. coli.
Using specific 5′ and 3′ primers, 5′ AAAAGCTTATGGCAGACAACAACCAAGCG 3′ (5′ primer) and 5′ CCAAGCTTCAATCGTCGTGCTTGCAGTC 3′ (3′ primer), the coding region of the fur gene (429 bp) of A. hydrophila was amplified and cloned into the vector plasmid pBR322 at the ScaI site under the control of an ampicillin resistance gene promoter of the vector. The new recombinant plasmid generated was designated pBRfur1 and transformed into an E. coli fur-minus strain, SBC23 (Table 1). T medium supplemented with amino acid mix was used for these experiments. The medium also contained either FeSO4 (36 μM) or an iron chelator (0.1 mM). After overnight growth, cells were centrifuged and 50 μl of 0.1% SDS and 50 μl of chloroform were added to permeabilize the E. coli cells. The PhoA activity then was measured by hydrolysis of p-nitrophenyl phosphate (6).
Construction of fur isogenic mutants of A. hydrophila SSU via double-crossover recombination.
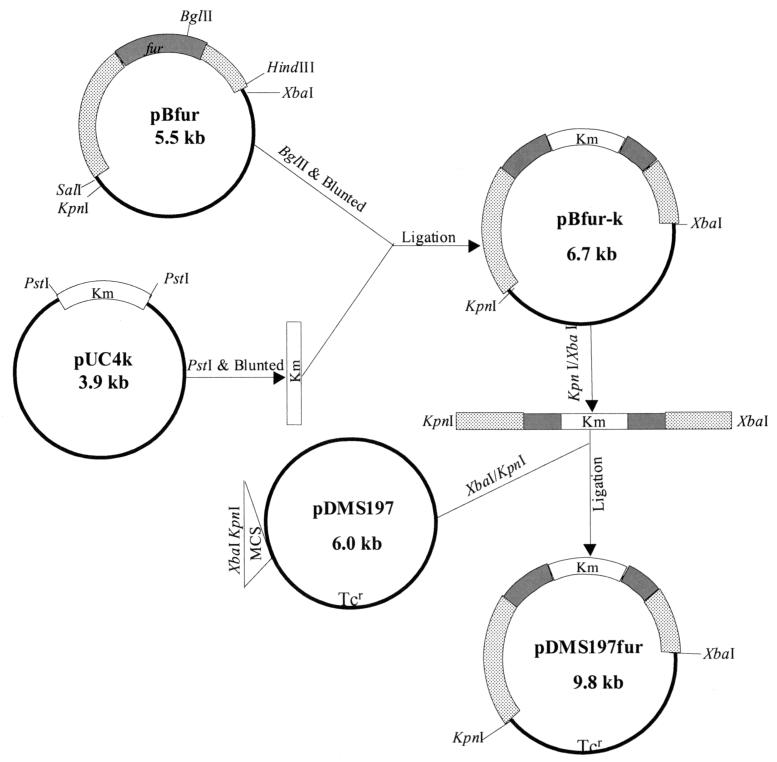
As shown in Fig. 6, plasmid pBfur containing a 2.6-kb SalI/HindIII DNA fragment with the fur gene from the chromosomal DNA of A. hydrophila was used to prepare the fur isogenic mutant. In the fur gene, there was a unique BglII restriction site. By using BglII enzyme, plasmid pBfur was linearized and the ends were made blunt with a PCR polishing kit (Stratagene). A 1.2-kb kanamycin gene cartridge was isolated from plasmid pUC4K (Amersham Pharmacia Biotech, Piscataway, N.J.) by using the restriction enzyme PstI, which bordered the kanamycin gene cassette, and its ends were also made blunt. This kanamycin cassette was ligated to plasmid pBfur at the blunted BglII site to generate a new recombinant plasmid, pBfur-K. By using the restriction enzymes XbaI and KpnI, whose sites existed in the vector, a 3.8-kb DNA fragment, including the 2.6-kb fur locus fragment and the 1.2-kb kanamycin cassette, was removed and ligated to a suicide vector, pDMS197 (tetracycline resistance), at the XbaI and KpnI sites, forming a new recombinant plasmid, pDMS197fur, in E. coli strain SM10 (see Fig. 6). This strategy to prepare isogenic mutants provided, respectively, 2,068 and 568 bp of the 5′ and 3′ DNA sequences flanking the truncated fur gene for double-crossover homologous recombination.
FIG. 6.
Flow diagram showing construction of various recombinant plasmids for the purpose of generating the fur isogenic mutant of A. hydrophila SSU. Plasmid pBfur contained a 2.6-kb DNA fragment from the chromosome of A. hydrophila, which included the fur gene. The fur locus was truncated at the BglII restriction site by introducing a kanamycin resistance cassette from plasmid pUC4K. The truncated fur locus with its flanking sequences was cloned into a suicide vector, pDMS197, forming a recombinant plasmid, pDMS197fur, for the generation of a fur isogenic mutant of A. hydrophila. The shaded bars represent the fur gene, the dotted bars represent sequences flanking the fur gene, and the open bars indicate the kanamycin cassette. These plasmids are not drawn to scale.
The recombinant E. coli SM10(pDMS197fur) (see Fig. 6) strain was conjugated with rifampin-resistant A. hydrophila, as described previously for the development of an act::phoA mutant, and the transconjugants were plated onto LB agar plates with rifampin, kanamycin, and 5% sucrose to select double-crossover transconjugants.
Complementation of the fur isogenic mutant of A. hydrophila SSU88.
By using specific 5′ and 3′ primers (5′ CCAAGCTTATCCACGCTTGCCAGCAC 3′ [5′ primer] and 5′ CCAAGCTTCAATCGTCGTGCTTGCAGT 3′ [3′ primer]), a 1-kb DNA fragment, including the fur gene and its putative promoter region, was amplified from the chromosome of A. hydrophila SSU. It was then ligated to the vector pBR322 at the EcoRI restriction site to generate a recombinant plasmid, pBRpfur2 (Table 1), which was first transformed into E. coli HB101 that carried a helper plasmid, pRK2013 (with kanamycin resistance gene). Subsequently, via conjugation, the recombinant pBRpfur2 plasmid with helper plasmid pRK2013 was transformed into the fur isogenic mutant of A. hydrophila SSU88 (Table 1), which had been generated previously by double crossover. The transconjugants were screened on LB agar plates containing rifampin, kanamycin, and tetracycline. The presence of recombinant plasmid DNA in A. hydrophila SSU88 was confirmed by plasmid isolation and restriction enzyme analysis.
Northern blot analysis.
Wild-type A. hydrophila SSU, the fur isogenic mutant SSU88, and its complemented strain SSU88(pBRpfur2) (Table 1) were grown in LB medium to which 36 μM FeSO4 was added at 37°C overnight. The next morning, 200 μl of the overnight culture was added to 4 ml of the fresh LB medium in 50-ml sterilized disposable tubes with 36 μM FeSO4 and the cultures were allowed to grow for another 3 h. The cells were centrifuged, and the total RNA was isolated by using an RNA isolation kit from Qiagen. The RNA samples (8 μg) were subjected to electrophoresis on a 1.2% formaldehyde agarose gel with 1× MOPS buffer (0.2 M morpholinepropanesulfonic acid [pH 7.0], 0.005 M sodium acetate, 0.01 M EDTA, pH 8.0) (55). A 1.4-kb 32P-labeled act gene from plasmid pXHC95 was used as a probe. The RNA was transferred to the nylon membrane, and after baking, the filters were prehybridized, hybridized, and washed as described for Southern blot analysis. The amount of RNA in each lane was quantitated by scanning 23S or 16S rRNA bands after ethidium bromide staining of the gel, using a Gel Doc 2000 apparatus (Bio-Rad Laboratories, Hercules, Calif.). The abundance of the message for Act was quantitated using a PhosphorImage Storm 860 (Molecular Dynamics, Sunnyvale, Calif.). All of the reagents used for Northern blot analysis were treated with diethylpyrocarbonate (Sigma).
Hemolytic assay.
The wild-type A. hydrophila SSU, its fur isogenic mutant SSU88, the complemented SSU88(pBRpfur2) strain, and other appropriate control cultures (see Table 3) were grown in T medium with or without 36 μM FeSO4 at 37°C overnight. The culture filtrates were first treated with trypsin at a final concentration of 0.05% at 37°C for 1 h and then subjected to hemolytic assay as follows: 100 μl of phosphate-buffered saline (PBS) was added to each of the wells of a 96-well microtiter plate. Next, 100 μl of a culture filtrate was added, followed by twofold dilution, with subsequent addition of 100 μl of 2.5% rabbit erythrocytes (Colorado Serum Company, Denver, Colo.). The plate was incubated at 37°C for 1 h and observed for the lysis of red blood cells. The hemolytic unit was defined as the reciprocal of the highest dilution of Act demonstrating 50% lysis of rabbit erythrocytes. The hemolytic units were presented per unit of growth per milliliter of the culture filtrate. The culture filtrates were treated with trypsin to convert all of the precursor form of Act to a mature form of the toxin (11).
TABLE 3.
The cloned A. hydrophila fur gene complemented Aeromonas fur isogenic mutant SSU88
| Aeromonas strain tested | Hemolytic activity (U/OD600/ml)a
|
|
|---|---|---|
| High iron | Low iron | |
| Wild-type A. hydrophila | 36.5 ± 3.2 | 889 ± 71.5b |
| Wild-type A. hydrophila(pBR322) | 89.5 ± 20.3 | 358 ± 36.9b |
| SSU88 | 582 ± 67.0 | 676 ± 63.8 |
| SSU88(pBR322) | 285 ± 33.1 | 327 ± 11.9 |
| SSU88(pBRpfur1) | 40.5 ± 2.9 | 208.5 ± 30.9b |
| SSU88(pBRfur2) | 20.1 ± 4.0 | 251.3 ± 12.4b |
T medium with or without the addition of 36 μM FeSO4 was used as a high- or low-iron medium. Three independent experiments were performed, and the arithmetic means ± standard deviations are presented. OD600, OD at 600 nm.
Statistically significant value (P ≤ 0.05) under two iron conditions using Student's t test.
Statistical analysis.
Wherever appropriate, the data were analyzed using Student's t test, and P values of ≤ 0.05 were considered significant.
Nucleotide sequence accession number.
The nucleotide sequence of the 2.6-kb SalI/HindIII chromosomal DNA fragment from A. hydrophila SSU, which contained the fur and flavodoxin genes, has been submitted to GenBank with accession number AF349468.
RESULTS
Characterization of single-crossover mutants of A. hydrophila SSU with an act::phoA gene fusion.
The strategy used to develop an act::phoA reporter gene mutant of A. hydrophila is depicted in Fig. 1. The selected A. hydrophila SSU66 mutant (Table 1) was gentamicin resistant and sucrose sensitive, which resulted from the integration of recombinant plasmid pJQactphoA into the chromosomal DNA of wild-type A. hydrophila. The identity of the single crossover was confirmed by Southern blot analysis using the act gene and the pJQ200SK vector as probes (data not shown). The growth rate in LB medium and the hemolytic activity of the single-crossover mutant were similar to those of wild-type A. hydrophila. When the mutant strain (SSU66) and wild-type A. hydrophila were streaked onto the LB plates containing BCIP, the colonies turned blue because of the cytoplasmic phosphatase activity in A. hydrophila. However, the mutant SSU66 showed an additional diffused blue color in the agar around the colonies, indicating secretion of Act::PhoA into the medium. Wild-type A. hydrophila did not exhibit any diffused blue color and served as a negative control. Further, the culture filtrate from the mutant SSU66 was positive for PhoA activity, while the culture filtrate from wild-type A. hydrophila did not exhibit any PhoA activity under similar conditions.
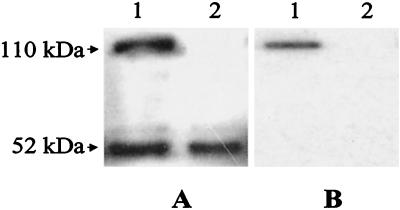
Western blot analysis confirmed that the Act::PhoA fusion protein was secreted out into the culture supernatant of mutant SSU66. As evident from Fig. 2A, when anti-Act antibodies were used, two bands 49 to 52 and 110 kDa in size were observed in the culture supernatant of the mutant strain (lane 1). The 49- to 52-kDa band represented native Act, and the 110-kDa band indicated Act::PhoA fusion protein. In contrast, one band of 49 to 52 kDa was visualized in the supernatant of wild-type A. hydrophila (Fig. 2A, lane 2). When anti-PhoA monoclonal antibodies were used in Western blot analysis, only a 110-kDa band could be detected in the mutant culture supernatant (Fig. 2B, lane 1) and no band was visualized in the culture supernatant of wild-type A. hydrophila (Fig. 2B, lane 2).
FIG. 2.
The Act::PhoA fusion protein was detected in the culture filtrate of A. hydrophila SSU66 based on Western blot analysis. Wild-type A. hydrophila and mutant SSU66 were grown in LB medium containing proteinase inhibitors at 37°C for 18 h. The supernatants were subjected to SDS–12% polyacrylamide gel electrophoresis and Western blot analysis. We used antibodies to Act (A) and monoclonal antibodies to E. coli alkaline phosphatase (B). In each panel, the supernatant from mutant SSU66 is in lane 1 and the supernatant from wild-type A. hydrophila is in lane 2.
To ensure that the expression of the act::phoA fusion gene was controlled only by the act promoter, we also generated an act::phoA reporter gene construct in the pUC128 vector (Table 1) without the act promoter. The correct open reading frame of the fusion gene was confirmed by DNA sequence analysis. Both of the fusion constructs, with and without the act gene promoter, were transformed into a phoA-minus E. coli C118 strain (Table 1) and plated onto LB agar plates which contained the PhoA substrate BCIP (40 μg/ml). Only E. coli colonies with the act promoter exhibited a blue color; the E. coli colonies with the act::phoA fusion gene construct without the act promoter were colorless (data not shown).
Influence of temperature, pH, and aeration on the expression of the act::phoA gene.
The expression of the act::phoA fusion gene in the A. hydrophila mutant SSU66 was examined at 26, 30, and 37°C. The highest level of PhoA activity (60/ml/OD unit) was obtained at 37°C. No apparent difference in the levels of act promoter activity was noted at temperatures of 26 and 30°C (12/ml/OD unit), but the activity was significantly lower than that observed at 37°C.
To determine the effect of pH on act promoter activity, we tested pH values from 5.5 to 9.0. The act promoter had maximum activity at pH 7.0 (60/ml/OD unit), which was reduced to 18/ml/OD unit at pH 5.5. Likewise, at pH values of 8.5 and 9.0, the PhoA activity was reduced to 16 and 6, respectively. At pH values of 6.0 and 7.5, the PhoA activity was 40/ml/OD unit.
The influence of aeration on the act promoter activity was evaluated by growing the mutant A. hydrophila SSU66 under static or shaking (180 rpm) conditions at 37°C. The culture under aeration demonstrated a higher PhoA activity of 57/ml/OD unit, whereas the PhoA activity was only 6/ml/OD unit when the mutant was grown as a nonshaken flask culture. We noted that bacteria grew poorly in nonshaken flask cultures compared to those in shaken flask cultures, indicating that oxygen might be essential for the growth of Aeromonas and for the expression of the act gene.
Influence of glucose, calcium, and iron on the expression of the act::phoA gene.
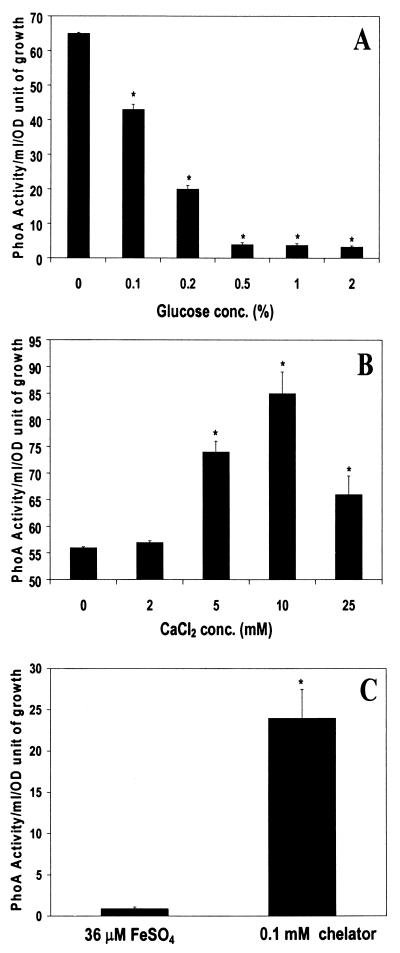
To examine the influence of glucose on act gene expression, the mutant SSU66 was grown in LB medium containing glucose at a concentration ranging from 0 to 2%. As shown in Fig. 3A, glucose repressed act promoter activity in a concentration-dependent fashion. The act promoter activity was abrogated at a glucose concentration of 0.5% and higher. In contrast to glucose (Fig. 3B), calcium increased act promoter activity in A. hydrophila SSU66, with maximum act promoter activity at a concentration of 10 mM. At a higher calcium concentration (25 mM), however, act promoter activity was significantly reduced.
FIG. 3.
Environmental factors affected expression of the act gene of A. hydrophila. A. hydrophila mutant SSU66 was grown in LB medium or T medium (for iron regulation) and exposed to different stimuli. After 18 h of growth, the supernatants were examined for PhoA activity. (A) Various concentrations of glucose were added to the LB medium. (B) LB medium contained different concentrations of calcium. (C) Iron or iron chelator was added to the T medium. ∗ denotes statistically significant values (P ≤ 0.05) by using Student's t test.
For iron regulation experiments, we preferred synthetic T medium to LB medium for bacterial growth, because we could accurately control iron concentration in this medium. It is evident from Fig. 3C that low-iron conditions (with the addition of a 0.1 mM concentration of an iron chelator) increased act promoter activity in A. hydrophila SSU66 by 20-fold compared to that in high-iron medium containing 36 μM FeSO4. Compared to that in LB medium (Fig. 3A and B), the PhoA activity in T medium was less (Fig. 3C). This difference was attributed to the medium and PhoA substrate used in these experiments. For iron regulation experiments, synthetic T medium and a p-nitrophenol substrate was used, whereas for the calcium and glucose experiment, LB medium and the substrate BCIP were employed.
The expression of many bacterial virulence genes is regulated by iron, and an iron uptake regulator gene (fur) has been shown to control this iron regulation (19). Our data with an A. hydrophila SSU66 mutant in which the phoA gene was under the control of the act promoter indeed indicated increased PhoA activity in a medium containing low iron. We therefore performed experiments to demonstrate whether the expression of the act gene was under the control of the fur locus.
Cloning and sequencing of the fur gene of A. hydrophila SSU
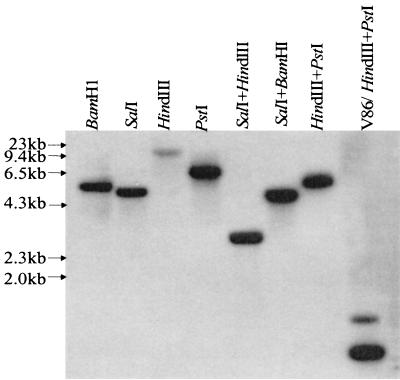
Based on the Southern blot hybridization data using the V. cholerae fur gene as a probe (Fig. 4), we cloned SalI/HindIII DNA fragments in the size range of 2.4 to 2.8 kb in the plasmid vector pBluescript SK. The plasmid library in E. coli DH5α was screened using the V. cholerae fur gene as a probe under high-stringency conditions by colony blot hybridization. Under these hybridization and washing conditions, the fur gene of V. cholerae hybridized with the fur gene of A. hydrophila but not with the E. coli fur gene (32; our unpublished data), allowing easy identification of the positive clones.
FIG. 4.
The fur gene of A. hydrophila SSU hybridized with the V. cholerae fur gene based on Southern blot analysis. The digested chromosomal DNA (10 μg) of A. hydrophila was subjected to 0.8% agarose gel electrophoresis and Southern blot analysis as described in Materials and Methods. Various enzymes or their combinations were used for digestion of the chromosomal DNA. Chromosomal DNA from V. cholerae V86 digested with HindIII and PstI was used as a positive control.
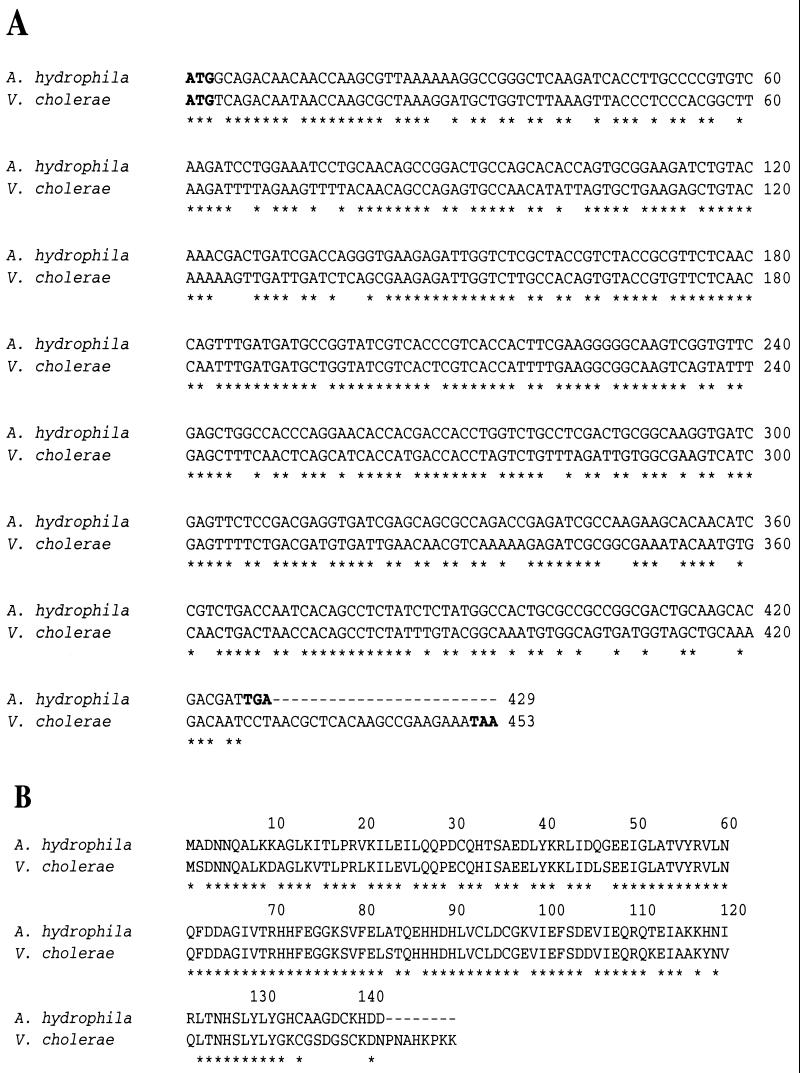
The recombinant E. coli clones, potentially harboring the A. hydrophila fur gene, which reacted with the V. cholerae fur gene were purified, and the identities of the correct recombinant clones were confirmed by Southern blot analysis (data not shown). The DNA sequence analysis of the 2.6-kb SalI/HindIII fragment revealed the presence of a 429-bp open reading frame that had 69% homology at the DNA level and 79% homology at the amino acid level with the V. cholerae fur gene and Fur protein, respectively (Fig. 5). Interestingly, the A. hydrophila Fur protein was missing 8 amino acids at its C terminus compared to that of the V. cholerae Fur protein (Fig. 5B). Based on a BLAST search, the 2.6-kb SalI/HindIII fragment also contained another 525-bp open reading frame, which exhibited 75% homology at the DNA level and 79% homology at the amino acid level with the flavodoxin gene of vibrios (28). The flavodoxin gene was localized upstream of the fur gene at nucleotide positions 650 to 1174. The fur gene was encoded within nucleotide positions 1958 to 2386 of the 2.6-kb SalI/HindIII fragment (data not shown).
FIG. 5.
(A) Comparison of the DNA sequences of the fur genes from A. hydrophila SSU and V. cholerae V86. The start and stop codons of the fur genes are shown in bold. (B) A. hydrophila and V. cholerae Fur protein amino acid sequence comparison. An ∗ indicates either conserved nucleotides or amino acid residues.
Biological function of the A. hydrophila SSU fur gene.
To determine whether the cloned A. hydrophila fur gene was functionally active, an isogenic pair of E. coli strains, SBC22 (fur+) and SBC23 (fur minus) (Table 1), was used. Each of these E. coli strains contained a single copy of the gene fusion between the iron-regulated promoter of the A subunit of Shiga-like toxin I (slt-IA) of E. coli and the phoA gene from TnphoA integrated in the chromosome (32). The original copy of the phoA gene on the chromosomes of these two strains was deleted; the PhoA activity measured in these strains, therefore, was due to expression of the phoA gene from the iron-regulated promoter of the slt-IA gene (32). In E. coli SBC23, the fur gene was also deleted and therefore the strain was fur gene negative. The E. coli strain SBC22, on the other hand, was fur gene positive (32). E. coli strain SBC23 complemented with various plasmids (Tables 1 and 2) were tested in T medium with either 36 μM FeSO4 or a 0.1 mM concentration of an iron chelator. As indicated in Table 2, strain SBC22 had iron regulation ability, resulting in a fourfold increase in the PhoA activity in the medium containing low iron compared to that in the medium containing high iron. Strain SBC23 had lost iron regulation because of the deletion of the fur gene. However, when SBC23 was complemented with either the A. hydrophila fur gene (pBRfur1) or the E. coli fur gene contained in plasmid pABN203, the iron regulation ability of SBC23 was restored, with three- to fourfold-increased PhoA activity in medium containing a low concentration of iron versus that in medium containing a high concentration of iron (Table 2). E. coli strain SBC23 containing the pBR322 vector only did not exhibit any iron regulation and was used as a negative control (Table 2). In both the low- and high-iron media, much higher PhoA activities were observed in E. coli strain SBC23 and E. coli strain SBC23(pBR322) than in other tested E. coli constructs (Table 2). This increased PhoA activity in strain SBC23 was attributed to the deletion of the fur gene. Interestingly, in low-iron medium, the PhoA activity associated with E. coli strain SBC23 was significantly lower than that found in high-iron medium (Table 2).
TABLE 2.
A cloned A. hydrophila fur gene complemented the E. coli fur-minus strain SBC23
| E. coli strain | Alkaline phosphatase activity (U/OD600)a
|
|
|---|---|---|
| High iron | Low iron | |
| SBC22 | 2.2 ± 0.1 | 8.8 ± 1.5b |
| SBC23 | 20.3 ± 0.8 | 10.4 ± 2.2b |
| SBC23(pBR322) | 30.2 ± 3.2 | 26.1 ± 2.3 |
| SBC23(pABN203) | 2.0 ± 0.04 | 6.4 ± 1.4b |
| SBC23(pBRfur1) | 3.3 ± 0.1 | 11.5 ± 2.1b |
T medium with 36 μM FeSO4 was used as a high-iron medium, and T medium with a 0.1 mM concentration of an iron chelator was used as a low-iron medium. OD600, OD at 600 nm. Values are the arithmetic means ± standard deviations of results from three independent experiments.
Statistically significant value (P ≤ 0.05) under both iron conditions using Student's t test.
Generation of a fur isogenic mutant of A. hydrophila SSU.
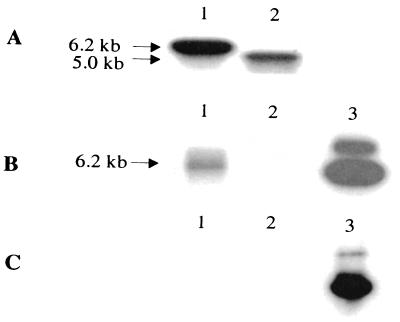
The strategy used to develop a fur isogenic mutant is depicted in Fig. 6. The colonies, which were resistant to rifampin, kanamycin, and sucrose and sensitive to tetracycline, should represent genuine double-crossover mutants, since the suicide vector sequences containing sacB and tetracycline resistance genes should have been lost due to homologous recombination. To confirm the identities of these mutants, the chromosomal DNAs from a selected mutant, SSU88, and wild-type A. hydrophila were isolated and subjected to PCR and Southern blot analysis. Two primers with the sequences 5′ AAAAGCTTATGGCAGACAACAACCAAGCG 3′ and 5′ CCAAGCTTCAATCGTCGTGCTTGCAGTC 3′, which correspond to the 5′ and 3′ ends of the A. hydrophila fur gene, respectively, were used for PCR analysis. Only a 429-bp DNA fragment, which represented the native fur gene, was amplified from wild-type A. hydrophila, and only a 1.7-kb DNA fragment, which represented a truncated fur gene with the kanamycin cassette, was amplified from the double-crossover mutant SSU88 (data not shown). It is also evident from the Southern blot data that, when the fur gene was used as the probe (Fig. 7A), a 6.2-kb band was observed in the mutant SSU88 (lane 1). In the chromosomal DNA of wild-type A. hydrophila, a 5.0-kb band was detected (Fig. 7A, lane 2). Compared to the digested chromosomal DNA of wild-type A. hydrophila, the digested chromosomal DNA fragment of the mutant was larger by 1.2 kb, due to the insertion of a kanamycin cassette. A similarly sized DNA fragment was detected in the digested chromosomal DNA of the mutant strain when the kanamycin cassette was used as a probe (Fig. 7B, lane 1). This probe did not react with the digested DNA from wild-type A. hydrophila (Fig. 7B, lane 2). No band was detected in the digested chromosomal DNAs of both mutant (Fig. 7C, lane 1) and wild-type (Fig. 7C, lane 2) A. hydrophila when the suicide vector pDMS197 was used as a probe. These data indicated that the mutant strain A. hydrophila SSU88 had completely lost the suicide vector sequence as a result of double-crossover homologous recombination. The hemolytic activity of the mutant SSU88 was slightly higher and the growth rate was slightly lower than those of wild-type A. hydrophila.
FIG. 7.
Confirmation of the identity of the fur isogenic mutant of A. hydrophila based on Southern blot analysis. Chromosomal DNAs from wild-type A. hydrophila and mutant SSU88 were isolated and digested with the BamHI and SalI restriction enzymes. Lane 1, digested chromosomal DNA from mutant SSU88; lane 2, digested chromosomal DNA from wild-type A. hydrophila; lane 3, plasmid pDMS197fur digested with XbaI and KpnI as a control. We used an A. hydrophila fur gene (A), a 1.2-kb kanamycin resistance cassette (B), and plasmid pDMS197 (C) as probes. The two bands in panels B and C, lane 3, indicated incomplete digestion of plasmid pDMS197fur.
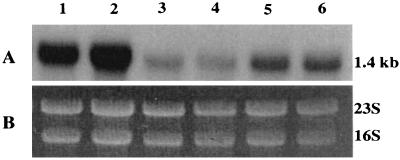
To demonstrate that iron regulation on act gene expression was lost in the fur isogenic mutant SSU88 and that iron regulation could be restored by complementation, Northern blot analysis was performed. The fur isogenic mutant SSU88 and its complemented strain were grown in LB medium with 36 μM FeSO4. Wild-type A. hydrophila SSU was used as a positive control in this experiment. It is evident from Fig. 8 that a 1.4-kb act gene transcript was detected in all of these A. hydrophila strains. However, under high-iron conditions, act mRNA was detected in much smaller amounts in wild-type A. hydrophila (Fig. 8, lanes 3 and 4) and in the complemented strain (lanes 5 and 6) than in the fur isogenic mutant, whose level of act mRNA was fourfold higher (Fig. 8, lanes 1 and 2). These data indicated that the fur isogenic mutant had lost iron regulation due to deletion of the fur gene.
FIG. 8.
Transcription of the act gene was repressed in the A. hydrophila strains, which contained the fur gene under high-iron conditions based on Northern blot analysis. Wild-type A. hydrophila, fur isogenic mutant SSU88, and its complemented strain with the A. hydrophila fur gene in plasmid pBRpfur2 were grown in LB medium with the addition of 36 μM FeSO4. The isolated RNA was subjected to Northern blot analysis as described in Materials and Methods. Lanes 1 and 2, RNA from mutant SSU88; lanes 3 and 4, RNA from wild-type A. hydrophila; lanes 5 and 6, RNA from mutant SSU88 complemented with pBRpfur2. (A) The probe used was a 1.4-kb act gene. (B) The RNA loaded in each lane was quantitated by scanning 16S and 23S rRNA bands after ethidium bromide staining of the gel.
The iron-regulated genes have Fur-binding sites in their promoter regions (19). In the putative promoter region of the act structure gene of A. hydrophila (11), two Fur box-like sequences (TATTA, positions −131 to −135 and positions −178 to −182), starting from the initiation codon of the act structural gene, were detected. These sequences could be the potential sites within the act promoter region to which the Fur protein binds.
Restoration of iron regulation of the hemolytic activity of Act in the complemented strain of A. hydrophila SSU88.
Wild-type A. hydrophila, fur isogenic mutant SSU88, and complemented fur isogenic mutants SSU88(pBRfur1) and SSU88(pBRpfur2) were grown in T medium with or without 36 μM FeSO4. After 18 h of growth at 37°C, the supernatants were taken for measuring hemolytic activity. As shown in Table 3, act gene expression in the wild-type A. hydrophila culture was repressed by 24-fold under high-iron conditions, compared to that under lower-iron conditions. However, the iron regulation of the hemolytic activity of Act was lost in the fur isogenic mutant SSU88. The presence of the vector pBR322 alone in wild-type A. hydrophila SSU reduced the effect of iron regulation of the hemolytic activity of Act from 24-fold in high-iron medium to 4-fold in low-iron medium (Table 3). Like mutant SSU88, no iron regulation of Act hemolytic activity was noted in SSU88 complemented with the pBR322 vector alone. However, the fur gene of A. hydrophila with its putative promoter contained in plasmid pBRpfur2 complemented the fur isogenic mutant SSU88. The hemolytic activity associated with Act in the complemented strain was 13-fold higher in low-iron medium than in high-iron medium. Iron regulation was also noted when the SSU88 mutant was complemented with the fur gene without the putative promoter region (pBRfur1); however, only a fivefold difference in hemolytic activity was noticed in the high- versus that in the low-iron medium.
DISCUSSION
The expression of bacterial virulence genes is frequently influenced by various environmental stimuli. The interaction between the host and pathogen during disease results in a loss of balance between the microbe's clever strategies for survival and multiplication and the formidable defenses of the immune system (36). In this study, the environmental regulation of act gene expression in A. hydrophila was investigated, since Act has been shown to be crucial in Aeromonas-mediated infections (55).
To study regulation of the act gene, it was essential to develop a reporter gene construct in which a portion of the act gene was fused in frame with a reporter gene (e.g., phoA). This gene construct was then integrated into Aeromonas chromosomal DNA via homologous recombination so that the expression of the phoA gene under the control of the act promoter could be measured. This system has three advantages: (i) compared with a multicopy plasmid system, this single-copy act::phoA fusion excludes the undesirable multicopy effects which might counteract the regulatory events; (ii) since the majority of the Act::PhoA is secreted out into the supernatant, it is easier to measure PhoA activity with minimal interference from intracellular PhoA activity; and (iii) since Act is secreted in a precursor form, which requires proteolytic cleavage at its C terminus to be activated, any stimuli that affect the expression of the protease genes would also affect Act-associated hemolytic activity. Therefore, measurement of PhoA activity, instead of hemolytic activity, provided us with an accurate and sensitive method to study act promoter activity under different environmental conditions.
Our experimental data indicated that the act gene from A. hydrophila was optimally expressed at 37°C and at pH 7.0. The temperature-dependent expression of the Pap pilus gene in E. coli (34, 35) and the gene encoding alginate capsule production in Pseudomonas aeruginosa (13) were linked to a nucleoid protein, H-NS, that had histone-like properties (29). The alteration of virulence gene expression in Salmonella enterica serovar Typhimurium by pH is under the control of a two-component phoP-phoQ regulatory system inside the macrophages (22, 37). Likewise, the toxR gene of V. cholerae senses changes in the environment, such as temperature, pH, osmolarity, etc., which alter expression of multiple virulence genes in Vibrio spp. (38). Studies are in progress in our laboratory to identify a regulatory gene(s) which may modulate expression of the act gene and possibly other virulence factors in A. hydrophila under different environmental conditions.
Although addition of glucose to the medium increased the growth rate of A. hydrophila, PhoA activity per unit of growth was significantly repressed. This repression in act promoter activity was specific for glucose only, as galactose and arabinose increased PhoA activity in the culture supernatant. In Vibrio fischeri, the autoinduction of luminescence genes (luxR and luxICDABEG) was found to be repressed by glucose and promoted by iron restriction (15, 16). Although the mechanism(s) of this glucose repression was not clear, it was considered to occur as a result of decreasing cellular levels of cAMP, which retarded synthesis of LuxR protein (15, 16, 44). The transcription of the luxICDABEG gene cluster was proposed to be blocked by iron as a result of binding to an iron-binding repressor protein, resulting in delayed accumulation of the autoinducer (26). Bang et al. (2) similarly reported that glucose repressed V. vulnificus hemolysin production and that glucose altered the interaction of cAMP and cAMP receptor protein. Regassa et al. (43) showed that glucose repressed alpha-hemolysin gene (hla) and staphylococcal enterotoxin C gene (sec+) expression in Staphylococcus aureus through a global regulatory locus, the accessory gene regulator (agr). The addition of cAMP to glucose-grown S. aureus cultures did not relieve repression, and both glucose and galactose down regulated agr expression, which in turn affected expression of the hla and sec+ genes. We also noted that addition of 8-bromo-cAMP to the A. hydrophila culture did not relieve glucose repression of act gene expression. The exact means by which glucose represses act gene expression is under investigation. It is plausible that glucose may alter expression of a regulatory gene which modulates the expression of the act gene.
The promoter activity of the act gene was increased in the presence of calcium, an important environmental signal affecting expression of various bacterial virulence genes. For example, all of the three species of the genus Yersinia possess a virulence characteristic known as the low-Ca2+ response. At temperatures above 34°C, the growth of yersiniae is dependent on a millimolar concentration of calcium. However, the expression of the Yersinia outer membrane protein-encoding genes (yop genes) occurs only in the absence of calcium (4, 46). In Yersinia pestis, the activity of the bacteriocin pesticin was increased by calcium but repressed by iron (7). Further studies revealed that iron and calcium were involved in the synthesis of the pesticin receptor, which was also considered to be the receptor of the siderophore (21, 42). The function of calcium in regulating the synthesis of the pesticin receptor was unclear; however, the role of Fur in regulating the expression of the pesticin receptor was suggested (27, 47, 48). The hemolysin of Actinobacillus pleuropneumoniae is another virulence factor that requires calcium for its expression (46). On the other hand, the expression of a gene encoding a cell surface protein of Arthrobacter photogoniums called LipA (possibly a pilin) was repressed by calcium. Unlike the other known bacterial induction or repression mechanisms that are sensitive to millimolar concentrations of calcium in growth medium, lipA gene expression was shown to be repressed by a calcium concentration of only 1.0 μM. The sensitivity of lipA gene expression to micromolar concentrations of calcium suggests that the regulatory mechanism involves a sensor protein(s) that has very high affinity for calcium (46). That calcium alters the expression of virulence genes through cAMP regulation is an exciting possibility and needs to be explored.
The transcription regulation of several toxin genes has been linked to low iron concentrations (8, 14, 24, 39, 51). The mechanism of iron regulation has been attributed to a fur locus, and the fur genes of different bacteria have been identified (19). In this study, we have shown that the act gene in A. hydrophila was iron regulated. Subsequently, the fur locus of A. hydrophila was cloned and sequenced. The A. hydrophila fur gene exhibited homology with the fur gene of V. cholerae (Fig. 5), and the former also could restore iron regulation in the E. coli fur-minus mutant SBC23 (Table 2).
To further evaluate the role of the fur gene in the expression of the act gene, a fur isogenic mutant of A. hydrophila was generated. Our data indicated that iron regulation of act gene expression was lost in the fur isogenic mutant (Fig. 8 and Table 3) and that iron regulation in this mutant could be restored by complementation (Fig. 8 and Table 3). These experiments indicated that act gene expression was regulated by iron and that the fur locus of A. hydrophila was responsible for this regulation. We also noted that the fur isogenic mutants exhibited a slightly lower growth rate than that of wild-type A. hydrophila, especially in a low-iron medium. Iron is essential for cell growth, as it serves as a cofactor for a large number of enzymes in a cell (9, 53, 54). Bacteria with mutations in the fur gene (e.g., fur isogenic mutants) are also defective in iron uptake regulation, which leads to a relatively low iron level in the cells, resulting in slower cellular metabolism, particularly in a low-iron medium. Interestingly, increased hemolytic activity was noted in the fur isogenic mutant when it was grown in the iron-rich medium. It may have been due to the relief of repression of act expression by iron. Indeed, we demonstrated by Northern blot analysis an increase in act gene transcription in the fur isogenic mutant compared to that in wild-type A. hydrophila in an iron-rich medium (Fig. 8).
Braun et al. (5) reported the sequence 5′ GATAATGATAATCATTATC 3′ as the functional target (Fur box) for the Fur protein, which is a palindromic DNA sequence (40). On the other hand, many iron-regulated promoters appear to have multiple Fur boxes, which could overlap (23, 30, 52) and hence are not compatible with the dimer-palindrome model (40). A recent study (18) suggests that the sequence 5′ NAT(A/T)AT 3′ could be the actual Fur protein-binding site and that three adjacent repeats of this unit would lead to effective binding. While the relative orientations and numbers of these repeats may not be so important, the sequence 5′ NAT(A/T)AT 3′ is considered a consensus sequence for the Fur box (19). Two fur box-like sequences were detected within the putative promoter region of the act gene, but they were not adjacent. These regions may be the potential sites to which the Fur protein might bind and are under investigation. Interestingly, the sequence ATTATTTTT (nucleotides −173 to −181), starting from the start codon of the act structural gene and within the act putative promoter, has also been shown to exist within a Fur-binding sequence (19 bp) in the promoter region of the flbB gene (a transcriptional activator) of E. coli (3, 50). Fur is now being considered a global regulator that coordinates different responses in the cell, rather than a specific transcription factor (18). In this respect, it is reasonable to assume that the sequence of the Fur box should be flexible rather than being a very specific motif.
Intestinal pathogens, such as Aeromonas spp., must overcome numerous host defenses to establish an infection. The results presented in this study revealed that act gene expression was altered by certain environmental stimuli that might contribute to the in vivo virulence of A. hydrophila. In addition, the fur locus of A. hydrophila was identified and its role in iron regulation was established. However, whether this fur gene regulates additional virulence genes in A. hydrophila needs to be investigated.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of Health (AI41611). Jian Sha, a postdoctoral fellow, was supported by a McLaughlin postdoctoral fellowship.
We thank X.-J. Xu and Jana Von Lindren for their work in the initial stages of this study. The editorial assistance of Mardelle Susman is greatly appreciated. We also thank B. Chatuev of our department for providing plasmids pUC128 and pDMS197 and E. coli C118.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1989. [Google Scholar]
- 2.Bang Y B, Lee S E, Rhee J H, Choi S H. Evidence that expression of the Vibrio vulnificus hemolysin gene is dependent on cyclic AMP and cyclic AMP receptor protein. J Bacteriol. 1999;181:7639–7642. doi: 10.1128/jb.181.24.7639-7642.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett D H, Frantz B B, Matsumura P. Flagellar transcriptional activators F1bB and F1aI: gene sequence and 5′ consensus sequence of operons under F1bB and F1aI control. J Bacteriol. 1988;170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barve S S, Straley S C. lcrR, a low-Ca2+-response locus with dual Ca2+-dependent functions in Yersinia pestis. J Bacteriol. 1990;172:4661–4671. doi: 10.1128/jb.172.8.4661-4671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun V, Schaffer S, Hantke K, Troger W. Regulation of gene expression by iron. In: Hauska G, Thauer R, editors. The molecular basis of bacterial metabolism. Berlin, Germany: Springer-Verlag; 1990. pp. 164–179. [Google Scholar]
- 6.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletion and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 7.Brubaker R R, Surgalla M J. Pesticins, I. Pesticin-bacterium interrelationships, and environmental factors influencing activity. J Bacteriol. 1961;82:940–949. doi: 10.1128/jb.82.6.940-949.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calderwood S B, Mekalanos J J. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol. 1987;169:4759–4764. doi: 10.1128/jb.169.10.4759-4764.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chai S, Welch T J, Crosa J H. Characterization of the interaction between Fur and the iron transport promoter of the virulence plasmid in Vibrio anguillarum. J Biol Chem. 1998;273:33841–33847. doi: 10.1074/jbc.273.50.33841. [DOI] [PubMed] [Google Scholar]
- 10.Chopra A K, Houston C W. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1999;1:1129–1137. doi: 10.1016/s1286-4579(99)00202-6. [DOI] [PubMed] [Google Scholar]
- 11.Chopra A K, Houston C W, Peterson J W, Jin G-F. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can J Microbiol. 1993;39:513–523. doi: 10.1139/m93-073. [DOI] [PubMed] [Google Scholar]
- 12.Chopra A K, Xu X-J, Ribardo D, Gonzalez M, Kuhl K, Peterson J W, Houston C W. The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect Immun. 2000;68:2808–2818. doi: 10.1128/iai.68.5.2808-2818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deretic V, Konyecsni W M. A procaryotic regulatory factor with a histone H1-like carboxy-terminal domain: clonal variation of repeats within algP, a gene involved in regulation of mucoidy in Pseudomonas aeruginosa. J Bacteriol. 1990;172:5544–5554. doi: 10.1128/jb.172.10.5544-5554.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dirita V J, Mekalanos J J. Genetic regulation of bacterial virulence. Annu Rev Genet. 1989;23:455–482. doi: 10.1146/annurev.ge.23.120189.002323. [DOI] [PubMed] [Google Scholar]
- 15.Dunlap P V. Iron control of the Vibrio fischeri luminescence system in Escherichia coli. Arch Microbiol. 1992;157:235–241. doi: 10.1007/BF00245156. [DOI] [PubMed] [Google Scholar]
- 16.Dunlap P V, Kuo A. Cell density-dependent modulation of the Vibrio fischeri luminescence system in the absence of autoinducer and LuxR protein. J Bacteriol. 1992;174:2440–2448. doi: 10.1128/jb.174.8.2440-2448.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards R A, Keller L H, Schifferli D M. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 18.Escolar L, Perez-Martin J, de Lorenzo V. Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J Mol Biol. 1998;283:537–547. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]
- 19.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson M R, Xu X-J, Houston C W, Peterson J W, Coppenhaver D H, Popov V L, Chopra A K. Hyperproduction, purification, and mechanism of action of the cytotoxic enterotoxin produced by Aeromonas hydrophila. Infect Immun. 1997;65:4299–4308. doi: 10.1128/iai.65.10.4299-4308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fetherston J D, Lillard J W J R, Perry R D. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields P I, Groisman E A, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1602. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 23.Griggs D W, Konisky J. Mechanism for iron-regulated transcription of Escherichia coli cir gene: metal-dependent binding of Fur protein to the promoter. J Bacteriol. 1989;171:1048–1054. doi: 10.1128/jb.171.2.1048-1054.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hale T L. Genetic basis for virulence in Shigella species. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harayama S, Tsuda M, Lino T. High frequency mobilization of the chromosome of Escherichia coli by a mutant of plasmid RP4 temperature sensitive for maintenance. Mol Gen Genet. 1980;180:47–56. doi: 10.1007/BF00267351. [DOI] [PubMed] [Google Scholar]
- 26.Haygood M G, Nealson K H. Mechanisms of iron regulation of luminescence in Vibrio fischeri. J Bacteriol. 1985;162:209–216. doi: 10.1128/jb.162.1.209-216.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65 000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 28.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L A, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins C F, Hinton J C, Hulton C S, Owen-Hughes T, Pavitt G D, Seirafi A. Protein H1: a role for chromatin structure in regulation of bacterial gene expression and virulence? Mol Microbiol. 1990;4:2007–2012. doi: 10.1111/j.1365-2958.1990.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 30.Hunt M D, Pettis G S, Mcintosh M A. Promoter and operator determinants for Fur-mediated iron regulation in the bidirectional fepA-fes control region of the Escherichia coli enterobactin system. J Bacteriol. 1994;176:3944–3955. doi: 10.1128/jb.176.13.3944-3955.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litwin C M, Boyko S T, Calderwood S B. Cloning, sequencing, and transcriptional regulation of the Vibrio cholerae fur gene. J Bacteriol. 1992;174:1879–1903. doi: 10.1128/jb.174.6.1897-1903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 34.Maurelli A T. Temperature regulation of virulence genes in pathogenic bacteria: a general strategy for human pathogens? Microb Pathog. 1989;7:1–10. doi: 10.1016/0882-4010(89)90106-x. [DOI] [PubMed] [Google Scholar]
- 35.Maurelli A T, Sansonetti P J. Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc Natl Acad Sci USA. 1988;85:2820–2824. doi: 10.1073/pnas.85.8.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller S I, Kukra A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1991;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 39.Pappenheimer A M, Johnson S J. Studies in diphtheria toxin production. I. The effect of iron and copper. Br J Exp Pathol. 1963;17:335–341. [Google Scholar]
- 40.Ptashne M. A genetic switch. Cambridge, Mass: Cell Press and Blackwell Scientific Publications; 1992. [Google Scholar]
- 41.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 42.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 43.Regassa L B, Novick R P, Betley M J. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (arg) in Staphylococcus aureus. Infect Immun. 1992;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruby E G, Nealson K H. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica: a model of symbiosis based on bacterial studies. Biol Bull. 1976;151:574–586. doi: 10.2307/1540507. [DOI] [PubMed] [Google Scholar]
- 45.Simon E H, Tessman R. Thymidine-requiring mutants of phage T4. Genetics. 1963;50:526–532. doi: 10.1073/pnas.50.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith R J. Calcium and bacteria. Adv Microb Physiol. 1995;37:84–131. doi: 10.1016/s0065-2911(08)60144-7. [DOI] [PubMed] [Google Scholar]
- 47.Staggs T M, Fetherston J D, Perry R D. Pleiotropic effects of a Yersinia pestis fur mutation. J Bacteriol. 1994;176:7614–7624. doi: 10.1128/jb.176.24.7614-7624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staggs T M, Perry R D. Fur regulation in Yersinia species. Mol Microbiol. 1992;6:2507–2516. doi: 10.1111/j.1365-2958.1992.tb01427.x. [DOI] [PubMed] [Google Scholar]
- 49.Stoebner J A, Payne S M. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect Immun. 1988;56:2891–2895. doi: 10.1128/iai.56.11.2891-2895.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stojiljkovic I, Baumler A J, Hantke K. Identification and characterization of new iron regulated Escherichia coli gene by Fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 51.Story D G, Frank D W, Farinha M A, Kropinski A M, Iglewski B H. Multiple promoters control the regulation of the Pseudomonas aeruginosa ergA gene. Mol Microbiol. 1990;4:499–503. doi: 10.1111/j.1365-2958.1990.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 52.Tardat B, Touati D. Iron and oxygen regulation in Escherichia coli MnSOD expression: competition between the global regulator Fur and ArcA for binding to DNA. Mol Microbiol. 1993;9:53–63. doi: 10.1111/j.1365-2958.1993.tb01668.x. [DOI] [PubMed] [Google Scholar]
- 53.Wackett L P, Orme-Johnson W H, Walsh C T. Transition metal enzymes in bacterial metabolism. In: Beveridge T J, Doyle R J, editors. Metal ions and bacteria. New York, N.Y: John Wiley & Sons, Inc.; 1989. pp. 165–206. [Google Scholar]
- 54.Watnick P I, Takaahashi E T, Calderwood S B. Purification of Vibrio cholerae Fur and estimation of its intracellular abundance by antibody sandwich enzyme-linked immunosorbent assay. J Bacteriol. 1997;179:243–247. doi: 10.1128/jb.179.1.243-247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X-J, Ferguson M R, Popov V L, Houston C W, Peterson J W, Chopra A K. Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect Immun. 1998;66:3501–3509. doi: 10.1128/iai.66.8.3501-3509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]