Abstract
Background
Multiple studies have reported on classification of raw electrocardiograms (ECGs) using convolutional neural networks (CNNs).
Objective
We investigated an application-specific CNN using a custom ensemble of features designed based on characteristics of the ECG during atrial fibrillation (AF) to reduce inappropriate AF detections in implantable cardiac monitors (ICMs).
Methods
An ensemble of features was developed and combined to form an input signal for the CNN. The features were based on the morphological characteristics of AF, incoherence of RR intervals, and the fact that AF begets more AF. A custom CNN model and the RESNET18 model were trained using ICM-detected AF episodes that were adjudicated to be true AF or false detections. The trained models were evaluated using a test dataset from independent patients.
Results
The training and validation datasets consisted of 31,757 AF episodes (2516 patients) and 28,506 false episodes (2126 patients). The validation set (20% randomly chosen episodes of each type) had an area under the curve of 0.996 for custom CNN (0.993 for RESNET18). Thresholds were chosen to obtain a relative sensitivity and specificity of 99.2% and 92.8%, respectively (99.2% and 87.9% for RESNET18, respectively). The performance in the independent test set (4546 AF episodes from 418 patients; 5384 false episodes from 605 patients) showed an area under the curve of 0.993 (0.991 for RESNET18) and relative sensitivity and specificity of 98.7% and 91.4%, respectively, at chosen thresholds (98.9% and 88.2% for RESNET18, respectively).
Conclusion
An ensemble of features-based CNNs was developed that reduced inappropriate AF detection in ICMs by over 90% while preserving sensitivity.
Keywords: Atrial fibrillation, Implantable cardiac monitors, Convolutional neural networks, Electrocardiogram, Residual neural network, Deep learning, Artificial intelligence
Key Findings.
-
▪
An ensemble of features-based custom convolutional neural network was developed that reduced inappropriate atrial fibrillation detection in implantable cardiac monitors by over 90% while preserving true atrial fibrillation detection sensitivity.
-
▪
The novel approach incorporates features that were constructed based on known electrocardiogram characteristics during true atrial fibrillation and concatenated into a 2-dimensional array used as input to the custom convolutional neural network.
-
▪
The small custom convolutional neural network performed similarly to the widely used larger RESNET18 network when using the same ensemble of features-based 2-dimensional images as input.
Introduction
Subcutaneous implantable cardiac monitors (ICMs) have been used for automatic detection of cardiac arrhythmias and patient symptom–triggered storage of loop-recorded recent electrocardiograms (ECGs).1,2 ICMs have been used for diagnosing the cause of unexplained syncope,3,4 for monitoring of recurrent atrial fibrillation (AF) after ablation of AF,5 and in patients with history of cryptogenic stroke.6 In most of these cases, the main objective is to deliver therapeutic interventions to patients in a timely manner to reduce clinical morbidity associated with these clinical conditions in a safe and cost efficient manner.4 While ICMs are very sensitive to detecting AF, the high sensitivity comes at the cost of reduced specificity. Multiple iterations of enhancement of detection algorithms inside the ICM improved specificity while preserving sensitivity. However, inappropriate AF detections in these ICMs and the associated clinic burden for review of these episodes are still some of the main concerns related to broader use of ICMs for AF management.
Deep learning 2-dimensional (2D) convolutional neural networks (CNNs) have been used extensively for image classification. Multiple studies have used 1D or 2D CNNs in ECG classification problems.7, 8, 9, 10, 11, 12, 13, 14 Further, application-specific CNN has been used to reduce inappropriate AF detection in ICMs while preserving sensitivity for AF detection.14 These techniques directly feed the raw ECG signal into a CNN, which automatically derives features as it trains the deep learning network over multiple iterations and epochs. ECG during AF is characterized by atrial fibrillatory waves or multiple P waves between 2 R waves, or the absence of P waves. Further, ventricular response during AF is primarily controlled by the atrioventricular node, which leads to incoherence of the RR interval time series during AF. ICMs measure a single lead ECG with a 4-cm dipole most often implanted in the fourth intercostal space in a 45° orientation but may be implanted at different locations and orientations. P-wave visibility during sinus rhythm depends on the location and orientation of the ICM. The objective of this study was to transform the ECG measured by ICM and derive features that were specifically related to the ICM-measured ECG characteristics during AF and to develop an application-specific deep learning model to reduce inappropriate AF detection in ICMs while preserving sensitivity for AF detection.
Methods
AF detection in ICM
AF detection in ICMs is primarily based on looking for incoherence of RR intervals over a 2-minute period.15,16 Once AF is detected, there are several additional layers of algorithms that reduce inappropriate detection while preserving sensitivity of AF detection. These include rejection of noise using short interval counts (RR interval <220 ms), bigeminy and trigemini rejection using specific RR interval sequence logic, ectopy with irregular coupling interval and sinus arrhythmia rejection using detection of single P waves between R waves,17,18 and a self-learning algorithm that personalizes detection thresholds in each patient based on device-detected RR irregularity and single P-wave incidences.19 Once an episode is detected by the ICM, the ECG from the first 2 minutes of the detection period is stored in the device, and a proportion of episode ECGs are transmitted to remote monitoring systems for provider’s review. Device-based algorithms are limited in computational complexity due to constraints of battery drain. To further reduce inappropriate detection, advanced algorithms using cloud computing capabilities are used in remote monitoring systems to filter out inappropriately detected episodes prior to provider review.
Deep learning–based episode classification
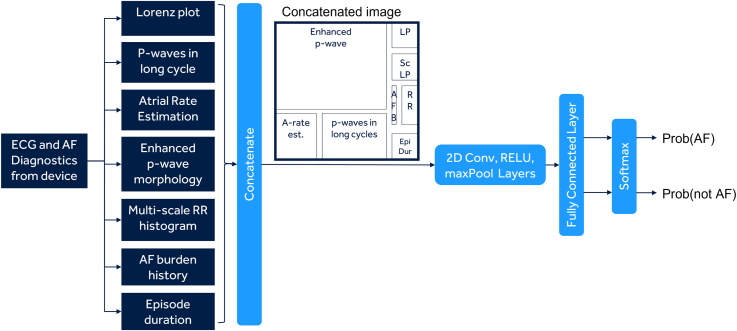
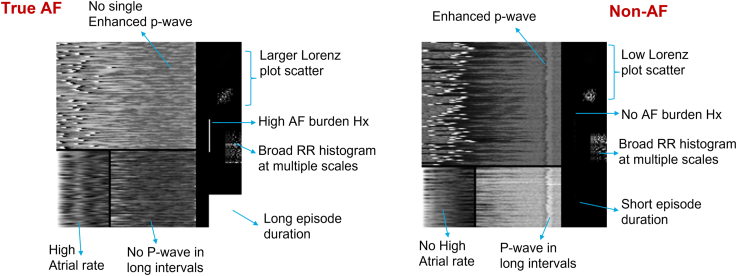
The ECG recorded in a detected AF episode is transformed into different features, each of which can be represented as a 2D array (Figure 1). This includes the Lorenz plot encoding RR interval incoherence (both scaled and nonscaled), enhanced P-wave, P waves in long cycle and atrial rate information encoding atrial components of ECG, and AF burden and episode duration encoding the information that longer episodes and episodes from high AF burden patients are both more likely to be true. The features are concatenated into a 2D array (or image) to form an ensemble of features. The 2D image with an ensemble of features derived from the ECG is used as an input to the deep learning network. Examples of images for AF episodes and non-AF episodes are shown in Figure 2.
Figure 1.
The basic schematic of the formation of the ensemble of features-based 2-dimensional (2D) input array and the deep learning neural network. AF = atrial fibrillation; ECG = electrocardiogram.
Figure 2.
Examples of the 2-dimensional image input in the case of an atrial fibrillation (AF) episode and a non-AF episode.
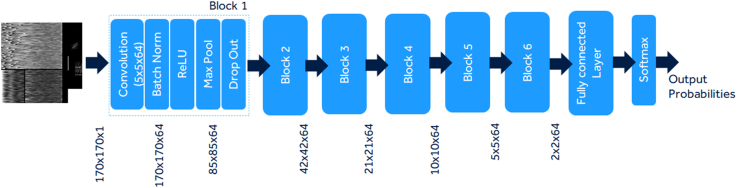
A basic sequential deep learning custom network was trained with 6 blocks with each block including layers of 2D convolution, batch normalization, rectified linear unit, max pooling, and dropout layers along with 1 fully connected and softmax layer (Figure 3). Additionally, publicly available residual neural network (RESNET18) architecture20,21 was also trained using the dataset to serve as a comparison to a widely used deep learning model. The MATLAB (The MathWorks, Inc) deep learning toolbox (version 9.11 - R2021b) was used to train the 2 networks.
Figure 3.
The basic schematic of the custom network with 6 blocks of 2-dimensional convolution, rectified liner unit, batch normalization, and dropout layers.
Data cohort and data analysis
An annotated dataset was created from a real-world dataset of AF episodes detected by the Reveal LINQ™ ICM. The data used in this retrospective analysis was derived from a de-identified real-world dataset stored in the Medtronic Discovery Link data warehouse. All patients provided consent to use their de-identified device data for research purposes when they signed up for Medtronic CareLink™ Network. The centers that allowed use of their patient data for research purposes then consented to storage of patient data in a de-identified Medtronic Discovery Link data warehouse. The Reveal LINQ ICM stored AF episodes were transmitted to the CareLink network and then de-identified and stored in this Discovery Link data warehouse for patients from centers who had consented for deidentified data use. Institutional Review Board evaluations for prior publications22 judged using the de-identified Medtronic Discovery Link data warehouse to fall into the category of nonhuman research; therefore, no Institutional Review Board approval was indicated for use of these deidentified data. This was a retrospective real-world data analysis and not a clinical study and hence is not registered in ClinicalTrials.gov.
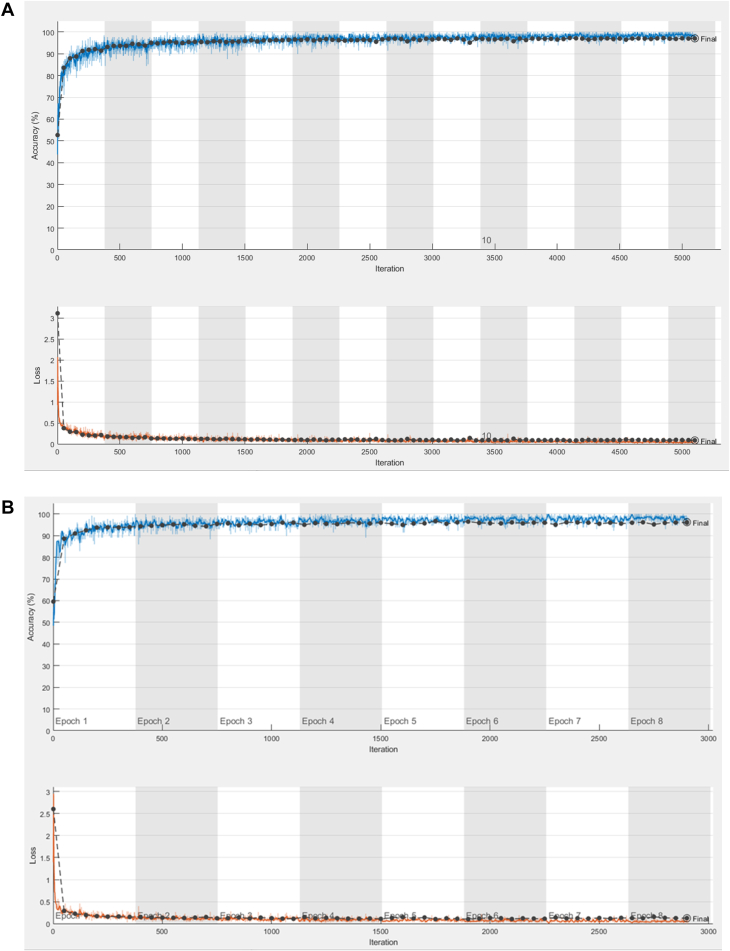
Patients with AF ablation, AF management, cryptogenic stroke, and unexplained syncope as reason for monitoring were included. Over 60,000 ICM-detected AF episodes from over 4000 patients were used to train the deep learning networks. The episodes were adjudicated by a single reviewer (S.S.) following a review process that was validated in an earlier study.23 Multiple randomized 80%–20% splits of training and validation datasets were performed to estimate the extent of potential generalization error. The model trained with the most balanced result with respect to sensitivity and specificity in the validation set was chosen as the final model. The validation set comprised of 20% of independent episodes but not necessarily from independent patients. Training process for the custom network and RESNET18 network are shown in Figure 4. A probability threshold was chosen to classify AF vs non-AF in the validation set to obtain a sensitivity above 99%. Additionally, an independent test dataset was created from independent patients not included in the training and validation datasets. This independent test dataset included ICM-detected AF episodes from consecutive patients who were implanted with an ICM for AF ablation, AF management, and cryptogenic stroke as reason for monitoring and were not included in the training and validation datasets. Classification accuracy as measured by sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and misclassification of true AF and false AF is reported along with receiver-operating characteristic curves. Generalized estimating equation (GEE) estimates for sensitivity, specificity, PPV, and NPV are also reported to adjust for multiple episodes per patient.
Figure 4.
The accuracy and loss function during the training process in the training (blue/red lines) and validation (black lines) datasets using the custom network (A) and the RESNET18 network (B).
Results
A total of 60,263 detected AF episodes from 4007 patients (31,757 true AF episodes from 2516 patients and 28,506 false AF episodes from 2126 patients) were used to train the networks from initial random weights with a split of 80% of episodes used for training and 20% for validation. Transfer learning was not used in the training process. The validation set receiver-operating characteristic curve was used to choose a probability threshold for classification into AF vs non-AF episodes. For the custom network, an area under the curve (AUC) of 0.996 was obtained, and a threshold of 0.95 was chosen for a sensitivity and specificity of 99.2% and 92.8%, respectively. The threshold of 0.95 indicated the probability of the episode being false being higher than 95%. For the RESNET18 network, AUC was 0.993, and a threshold of 0.90 was chosen to obtain a sensitivity and specificity of 99.2% and 87.9%, respectively.
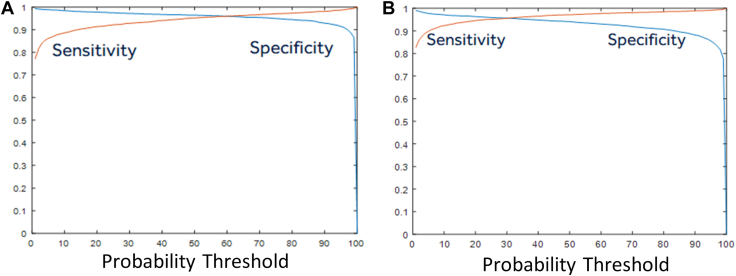
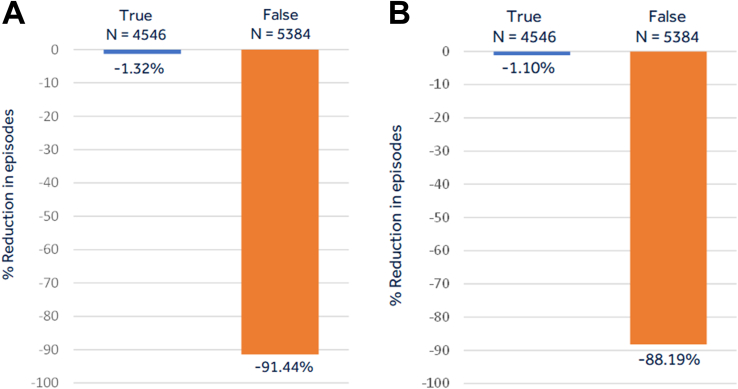
The independent patient test dataset from 898 patients included 4546 true AF episodes from 418 patients and 5384 false AF episodes from 605 patients. Sensitivity, specificity, PPV, and NPV derived using raw proportion of episodes in the independent patient test dataset as well as GEE estimates for the custom CNN and the RESNET18 networks are shown in Table 1. Figure 5 shows the sensitivity and specificity curve as a function of the probability threshold in this independent patient test dataset. The AUC was 0.993 for the custom network performance in the independent patient test dataset (Figure 5A) and 0.991 for the RESNET18 network (Figure 5B). As shown in Figure 6, at the chosen threshold from the validation set, the custom network was able to accurately classify 91.4% of inappropriate detections (ie, achieve a specificity of 91.4%) in the independent patient test set while also inaccurately misclassifying true AF in 1.3% of the episodes (ie, achieve a sensitivity of 98.7%). For the RESNET18 network, 88.2% of inappropriate detections were reduced with loss of sensitivity of 1.1%. The GEE estimates, adjusting for multiple episodes in patients, and the 95% confidence intervals (Table 1) further confirmed that a significantly improved specificity and marginal reduction in sensitivity was obtained, as shown by metrics derived using raw proportion of episodes.
Table 1.
Performance metrics for the custom CNN and the RESNET18 models reported for the independent patient test dataset as raw proportion of episodes and the GEE estimates adjusting for multiple episodes per patient
| Custom CNN |
RESNET18 |
|||
|---|---|---|---|---|
| Raw | GEE (95% CI) | Raw | GEE (95% CI) | |
| Sensitivity | 4486/4546 (98.7) | 98.6 (97.8–99.1) | 4496/4546 (98.9) | 99.0 (98.2–99.4) |
| Specificity | 4923/5384 (91.4) | 90.0 (88.1–91.7) | 4748/5384 (88.2) | 87.8 (85.8–89.7) |
| PPV | 4486/4947 (90.7) | 81.3 (78.1–84.1) | 4496/5132 (87.6) | 76.5 (73.0–79.6) |
| NPV | 4923/4983 (98.8) | 98.1 (97.0–98.8) | 4748/4798 (99.0) | 98.4 (97.2–99.0) |
Values are n/n (%), unless otherwise indicated.
CI = confidence interval; CNN = convolutional neural network; GEE = generalized estimating equation; NPV = negative predictive value; PPV = positive predictive value.
Figure 5.
The sensitivity and specificity as a function of the probability output from the deep learning neural network in the independent patient test dataset for the custom network (A) and the RESNET18 network (B).
Figure 6.
The classification of episodes as non–atrial fibrillation (AF) by the deep learning network showing the proportion of false episodes that are correctly classified as false as well as the proportion of true AF episodes that are misclassified as non-AF episodes for the custom network (A) and the RESNET18 network (B).
Discussion
Deep learning CNNs were utilized to classify ICM-detected AF episodes with an objective to reduce inappropriate AF detections while preserving sensitivity for AF detection. Rather than using the conventional method of feeding in the raw ECG signal into a CNN, the ECG data were transformed into an ensemble of features that emphasized ECG characteristics during AF. An application-specific custom network was designed that used the ensemble of features as a 2D input and had only 6 convolution layers. The performance of the custom network was compared with the publicly available RESNET18 network. Both networks were able to reduce inappropriate ICM-based AF detections by around 90% while also reducing sensitivity for AF detection by around 1%. These performance results were similar or better than results seen using raw ECG as a 1D input into a larger (or deeper) 1D CNN based on the RESNET50 architecture.24 Each method, the raw ECG method and the ensemble of features method, has advantages and disadvantages, and combination using ensemble neural networks may lead to better performance.
The feature selection for the creation of the 2D input was primarily focused on the ECG characteristics during AF. Also, these features were used to detect AF in the ICM in a computationally simplified form. For example, the Lorenz plot technique uses a simple indexing and counting operation inside the ICM device, whereas in this CNN approach it is used as a 2D numerical histogram, and the CNN can potentially extract various advanced features within this 2D array. Similarly, the ICM looks for the presence of single P-wave between 2 R waves using simple difference operations inside the device. In this CNN approach, the raw P-wave–enhanced ECG segment prior to the R-wave was used as input, and thus the CNN could identify various morphologic features in the P waves to identify AF or atrial flutter. Additionally, AF is an atrial arrhythmia, and the CNN was made to focus on the atrial part of the ECG, rather than focusing on the R-wave morphology, which has large changes but very little information about AF. The incoherence of the RR interval sequence was incorporated in multiple features to make the CNN focus on those aspects of the ECG. Finally, the CNN focused on filters over a few samples, so the relationship between information across larger samples was not inherently incorporated into the network architecture. The P-wave enhanced ICM ECG segments were arranged in a 2D format synchronized to the R-wave location in such a way such that CNN filters could compare morphology between multiple beats in the second dimension.
The novel aspect of the methodology described in this study is the method to generate the ensemble of features as a 2D input to a single sequential 2D CNN network. A single sequential CNN network will have the advantage of a small number of trainable parameters, thus reducing the risk of overfitting, but provides less flexibility in terms of having unique filters for specific features in the ensemble of input features. The custom CNN used in this study only had 517,954 trainable parameters, compared with over 8.8 million for the RESNET18 architecture. Alternative approaches have been to use multiple inputs into separate CNN networks and then concatenate the output of each individual network into an ensemble network. The latter approach does provide more flexibility, as the individual networks can be designed differently to tailor them to their different input features. However, that likely would lead to a larger network architecture and a larger number of trainable parameters. Further, most published literature on ECG classification using deep learning networks has focused on classifying all kinds of possible arrhythmia using a single trained network. An application-specific approach, like classifying only episodes detected by the AF detection algorithm resident in the device,15, 16, 17, 18, 19, 20 significantly reduces the degrees of freedom that the network will have to fit to, thereby reducing the required size and depth of the network needed to fit to the problem. This is verified by the fact that the small custom network performed as well as a much larger RESNET18 network. The RESNET50 network was also evaluated, but it ran into overfitting issues, and hence the RESNET18 was evaluated in this study.
The performance of a deep learning network is dependent on the consistency of the ICM ECG adjudication process. In this study, a single reviewer was used to maintain the consistency of the adjudication process. The process followed by the adjudicator was validated against adjudications done by electrophysiologists is a small subset of the data as described previously.20 The adjudicator was found to have <1% error compared with the electrophysiologist adjudications. It should be noted that about 10% of ICM-labeled AF episodes are difficult to adjudicate, as has been found in other smaller studies. Additionally, in the training and validation datasets, discordant analysis was performed multiple times to evaluate reasons for mismatches between adjudication and CNN-predicted class labels. In some proportion of mismatches in the training and validation datasets, erroneous adjudications due to manual error were corrected. The independent test set was independently adjudicated twice by the same adjudicator and mismatches were reviewed a third time to ascertain the final adjudication. Further, the larger the size of the training dataset is, the more generalizable the performance of any trained network is. Data augmentation was not used, and instead additional episodes that were more likely to generate discordant results were included until no further improvement in performance was observed in the training/validation set.
The primary limitation of the study was that the deep learning network was trained on ECG obtained by a specific device with a single-lead ECG vector with electrodes separated by 4 cm and implanted at various locations and orientations. Thus, the trained network is not generalizable to other forms of ECG with different electrode configurations, such as 12-lead ECG systems. However, the same methodology can be used to train a similar network using data collected using the monitoring mode of choice. As mentioned previously, the network performance is only as good as the adjudications and the generalizability of data used to train the network. Because a single reviewer was used for this study, the artificial intelligence was broadly trained to reflect that reviewer’s accuracy in interpretation of presence or absence of AF in the ICM ECG.
Conclusion
A custom ensemble of features arranged as a 2D input to a CNN network using a small number of 2D convolution layers was able to reduce over 90% of inappropriate AF detections while also reducing appropriate detections by a little over 1%. The small custom network performed similarly compared with the widely used larger RESNET18 network when using the same ensemble of features-based 2D image as input.
Acknowledgments
Funding Sources
Funding for this project was received from Medtronic Inc.
Disclosures
Shantanu Sarkar, Shubha Majumder, Jodi L. Koehler, and Sean R. Landman are employees and shareholders of Medtronic Inc.
Authorship
All authors attest they meet the current ICMJE criteria for authorship.
Patient Consent
All patients provided consent to use their de-identified device data for research purposes.
Ethics Statement
No institutional review board approval was indicated for use of this de-identified data.
References
- 1.Krahn A.D., Klein G.J., Yee R., Takle-Newhouse T., Norris C. Use of an extended monitoring strategy in patients with problematic syncope. Circulation. 1999;99:406–410. doi: 10.1161/01.cir.99.3.406. [DOI] [PubMed] [Google Scholar]
- 2.Pürerfellner H., Sanders P., Pokushalov E., Di Bacco M., Bergemann T., Dekker L.R. Miniaturized Reveal LINQ insertable cardiac monitoring system: first-in-human experience. Heart Rhythm. 2015;12:1113–1119. doi: 10.1016/j.hrthm.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 3.Krahn A.D., Klein G.J., Yee R., Skanes A.C. Detection of asymptomatic arrhythmias in unexplained syncope. Am Heart J. 2004;148:326–332. doi: 10.1016/j.ahj.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Farwell D.J., Freemantle N., Sulke A.N. Use of implantable loop recorders in the diagnosis and management of syncope. Eur Heart J. 2004;25:1257–1263. doi: 10.1016/j.ehj.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Verma A., Champagne J., Sapp J., et al. Discerning the incidence of symptomatic and asymptomatic episodes of atrial fibrillation before and after catheter ablation (DISCERN AF): a prospective, multicenter study. JAMA Intern Med. 2013;173:149–156. doi: 10.1001/jamainternmed.2013.1561. [DOI] [PubMed] [Google Scholar]
- 6.Sanna T., Diener H.C., Passman R.S., et al. Cryptogenic stroke and underlying atrial fibrillation. N Engl J Med. 2014;370:2478–2486. doi: 10.1056/NEJMoa1313600. [DOI] [PubMed] [Google Scholar]
- 7.Hannun A.Y., Rajpurkar P., Haghpanahi M., et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siontis K.C., Noseworthy P.A., Attia Z.I., Friedman P.A. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat Rev Cardiol. 2021;18:465–478. doi: 10.1038/s41569-020-00503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siontis K.C., Friedman P.A. The role of artificial intelligence in arrhythmia monitoring. Card Electrophysiol Clin. 2021;13:543–554. doi: 10.1016/j.ccep.2021.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Attia Z.I., Harmon D.M., Behr E.R., Friedman P.A. Application of artificial intelligence to the electrocardiogram. Eur Heart J. 2021;42:4717–4730. doi: 10.1093/eurheartj/ehab649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes J.W., Olgin J.E., Avram R., et al. Performance of a convolutional neural network and explainability technique for 12-lead electrocardiogram interpretation. JAMA Cardiol. 2021;6:1285–1295. doi: 10.1001/jamacardio.2021.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attia Z.I., Noseworthy P.A., Lopez-Jimenez F., et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;394:861–867. doi: 10.1016/S0140-6736(19)31721-0. [DOI] [PubMed] [Google Scholar]
- 13.Marinucci D., Sbrollini A., Marcantoni I., Morettini M., Swenne C.A., Burattini L. Artificial neural network for atrial fibrillation identification in portable devices. Sensors (Basel) 2020;20:3570. doi: 10.3390/s20123570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittal S., Oliveros S., Li J., Barroyer T., Henry C., Gardella C. AI filter improves positive predictive value of atrial fibrillation detection by an implantable loop recorder. J Am Coll Cardiol EP. 2021;7:965–975. doi: 10.1016/j.jacep.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Sarkar S., Ritscher D., Mehra R. A detector for a chronic implantable atrial tachyarrhythmia monitor. IEEE Trans Biomed Eng. 2008;55:1219–1224. doi: 10.1109/TBME.2007.903707. [DOI] [PubMed] [Google Scholar]
- 16.Hindricks G., Pokushalov E., Urban L., et al. Performance of a new leadless implantable cardiac monitor in detecting and quantifying atrial fibrillation: results of the XPECT trial. Circ Arrhythm Electrophysiol. 2010;3:141–147. doi: 10.1161/CIRCEP.109.877852. [DOI] [PubMed] [Google Scholar]
- 17.Pürerfellner H., Pokushalov E., Sarkar S., et al. P-wave evidence as a method for improving algorithm to detect atrial fibrillation in insertable cardiac monitors. Heart Rhythm. 2014;11:1575–1583. doi: 10.1016/j.hrthm.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Sanders P., Pürerfellner H., Pokushalov E., et al. Performance of a new atrial fibrillation detection algorithm in a miniaturized insertable cardiac monitor: results from the Reveal LINQ Usability Study. Heart Rhythm. 2016;13:1425–1430. doi: 10.1016/j.hrthm.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Pürerfellner H., Sanders P., Sarkar S., et al. Adapting detection sensitivity based on evidence of irregular sinus arrhythmia to improve atrial fibrillation detection in insertable cardiac monitors. Europace. 2018;20:f321–f328. doi: 10.1093/europace/eux272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He K., Zhang X., Ren S., Sun J. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; New York, NY: 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 21.Jing E., Zhang H., Li Z., Liu Y., Ji Z., Ganchev I. ECG heartbeat classification based on an improved ResNet-18 model. Comput Math Methods Med. 2021;2021 doi: 10.1155/2021/6649970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan R.M., Koehler J., Ziegler P.D., Sarkar S., Zweibel S., Passman R.S. Stroke risk as a function of atrial fibrillation duration and CHA2DS2-VASc score. Circulation. 2019;140:1639–1646. doi: 10.1161/CIRCULATIONAHA.119.041303. [DOI] [PubMed] [Google Scholar]
- 23.Mittal S., Rogers J., Sarkar S., et al. Real-world performance of an enhanced atrial fibrillation detection algorithm in an insertable cardiac monitor. Heart Rhythm. 2016;13:1624–1630. doi: 10.1016/j.hrthm.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Radtke A.P., Ousdigian K.T., Haddad T.D., Koehler J.L., Colombowala I.K. Artificial intelligence enables dramatic reduction of false atrial fibrillation alerts from insertable cardiac monitors. Heart Rhythm. 2021;18:S47. [Google Scholar]