Abstract
Bat‐origin RshSTT182 and RshSTT200 coronaviruses (CoV) from Rhinolophus shameli in Southeast Asia (Cambodia) share 92.6% whole‐genome identity with SARS‐CoV‐2 and show identical receptor‐binding domains (RBDs). In this study, we determined the structure of the RshSTT182/200 receptor binding domain (RBD) in complex with human angiotensin‐converting enzyme 2 (hACE2) and identified the key residues that influence receptor binding. The binding of the RshSTT182/200 RBD to ACE2 orthologs from 39 animal species, including 18 bat species, was used to evaluate its host range. The RshSTT182/200 RBD broadly recognized 21 of 39 ACE2 orthologs, although its binding affinities for the orthologs were weaker than those of the RBD of SARS‐CoV‐2. Furthermore, RshSTT182 pseudovirus could utilize human, fox, and Rhinolophus affinis ACE2 receptors for cell entry. Moreover, we found that SARS‐CoV‐2 induces cross‐neutralizing antibodies against RshSTT182 pseudovirus. Taken together, these findings indicate that RshSTT182/200 can potentially infect susceptible animals, but requires further evolution to obtain strong interspecies transmission abilities like SARS‐CoV‐2.
Keywords: ACE2, interspecies transmission, RBD, RshSTT182/200, SARS‐CoV‐2
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction; Structural Biology
Coronaviruses found in Rhinolophus shameli bats can broadly recognize ACE2 orthologs but require further evolution to support interspecies transmission.
Introduction
There are seven CoVs reported to infect humans including severe acute respiratory syndrome coronavirus (SARS‐CoV), Middle East respiratory syndrome coronavirus (MERS‐CoV), human coronavirus NL63 (HCoV‐NL63), human coronavirus OC43 (HCoV‐OC43), human coronavirus 229E (HCoV‐229E), human coronavirus HKU1 (HCoV‐HKU1), and severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) (Huynh et al, 2012; Zaki et al, 2012; Su et al, 2016; Zhu et al, 2020). In the last 20 years, three public health emergencies were caused by CoVs: the SARS‐CoV epidemic (2002), MERS‐CoV epidemic (2012), and SARS‐CoV‐2 pandemic (2019).
For many viral infectious diseases, the discovery of viruses often lags by several decades or more (Tong et al, 2021). For instance, HCoV‐HKU1 was first discovered in a patient with pneumonia in Hong Kong, China, in 2004. However, frozen nasopharyngeal swab samples from children in Brazil from 1995 tested positive for HCoV‐HKU1, indicating that HCoV‐HKU1 was circulating in the population for a long time (Sloots et al, 2006; Goes et al, 2011; Tong et al, 2021). MERS‐CoV was first discovered in Saudi Arabia in 2012, whereas 81% of dromedary camel serum samples collected in Egypt in 1997 and in Sudan and Somalia in 1983–1984 were positive for MERS‐CoV‐neutralizing antibodies (Muller et al, 2014). Evaluating the potential interspecies transmission of viruses is an effective method to raise the alarm about infectious diseases as early as possible.
Bats are considered as the reservoir hosts of HCoV‐NL63, HCoV‐229E, SARS‐CoV, and MERS‐CoV (Guan et al, 2003; Pfefferle et al, 2009; Huynh et al, 2012; Reusken et al, 2014; Corman et al, 2015; Zhou et al, 2020a). At the start of SARS‐CoV‐2 pandemic, several SARS‐CoV‐2‐related CoVs were detected in bats. The overall genome of the bat‐origin CoV RaTG13 shares 96.2% nucleic acid sequence identity with SARS‐CoV‐2 (Zhou et al, 2020b). RmYN02, another bat‐origin CoV, contains a 3‐amino‐acid residue (PAA) insertion at the S1/S2 cleavage site of its spike protein, similar to that of SARS‐CoV‐2, which has not been observed in other SARS‐CoV‐2‐related CoVs except for RacCS203 (Zhou et al, 2020a; Wacharapluesadee et al, 2021). The furin cleavage site (FCS) plays a critical role in SARS‐CoV‐2 infection, and the loss of this site attenuates entry efficiency (Lau et al, 2020; Johnson et al, 2021). Recently, several bat‐origin CoVs, BANAL‐52, BANAL‐103, and BANAL‐236, were detected in bats in North Laos (Temmam et al, 2022). The receptor‐binding domains (RBDs) of the spike protein of BANAL‐52, BANAL‐103, and BANAL‐236 are > 95% identical to SARS‐CoV‐2 and show similar binding affinity as the SARS‐CoV‐2 RBD to hACE2 (Temmam et al, 2022), indicating that these viruses could potentially infect humans. RshSTT182 and RshSTT200 were identified from the Rhinolophus shameli (R. shameli) bat samples in 2010, which are the first SARS‐CoV‐2‐related CoVs identified in Southeast Asia (Cambodia), with 92.6% whole‐genome identity to SARS‐CoV‐2 (Delaune et al, 2021). The RshSTT182 and RshSTT200 RBDs have identical amino acid sequences and share 84.3% identity with the SARS‐CoV‐2 RBD.
The interspecies receptor recognition of SARS‐CoV, SARS‐CoV‐2, RaTG13, GX/P2V/2017, and GD/1/2019 has been evaluated (Wu et al, 2020a; Niu et al, 2021; Liu et al, 2021a). All of these CoVs broadly recognize different ACE2 orthologs, indicating that they are potentially transmitted among different animals. In addition, several natural SARS‐CoV‐2 infections have been reported (Gao & Wang, 2021). Cats, dogs, lions, and tigers in zoos, as well as minks, ferrets, snow leopards, pumas, and gorillas in nature, have been found to be infected with SARS‐CoV‐2 through contacting with COVID‐19 patients (Gao & Wang, 2021). Moreover, mink‐related SARS‐CoV‐2 strains have been transmitted back to humans and caused further community transmission. Notably, SARS‐CoV‐2 variants were detected in infected minks, suggesting that interspecies transmission may give rise to novel variants (Su et al, 2022; Yen et al, 2022). The RshSTT182/200 RBD is highly conserved with the SARS‐CoV‐2 RBD and may also have a broad host range. Host cell receptor binding is a prerequisite for virus infection. Therefore, characterizing the interaction between the RBD of a CoV and the ACE2 orthologs from a broad range of species is an efficient method to screen for potential hosts.
Receptor binding is the first step in CoV infection. A series of protein and carbohydrate receptors/co‐factors for CoVs have been identified. ACE2 is the main receptor for SARS‐CoV, SARS‐CoV‐2, and their related CoVs, such as RaTG13, GX/P2V/2017, and GD/1/2019 (Wang et al, 2020; Zhou et al, 2020b; Niu et al, 2021; Liu et al, 2021a; Han et al, 2022; Li et al, 2022b). The structures of the SARS‐CoV‐2 RBD in complex with human, cat, bat, dog, mink, and pangolin ACE2s have been determined (Wang et al, 2020; Wu et al, 2020b; Ren et al, 2021; Zhang et al, 2021; Liu et al, 2021b). In addition, the structures of hACE2 bound by the RaTG13, GX/P2V/2017, and GD/1/2019 RBDs were solved (Niu et al, 2021; Liu et al, 2021a). The GX/P2V/2017 RBD and GD/1/2019 RBD utilize a similar binding mechanism as SARS‐CoV‐2 and display similar binding affinities as the SARS‐CoV‐2 RBD for hACE2 (Niu et al, 2021). Similar to other SARS‐CoV‐2‐related CoVs, RshSTT182/200 also uses ACE2 as its receptor (Delaune et al, 2021). However, the molecular details of the RshSTT182/200 RBD binding to ACE2 are yet unknown.
In this study, we found that the RshSTT182/200 RBD binds to hACE2 with a substantially lower binding affinity than that of the SARS‐CoV‐2 RBD. The structure of the RshSTT182/200 RBD in complex with hACE2 was determined, and the key residues were identified. As a bat‐origin CoV, the receptor‐binding spectrum of RshSTT182/200 in 18 bat species was narrower than that of SARS‐CoV‐2. We also evaluated the receptor‐binding spectrum of the RshSTT182/200 RBD with 21 animal ACE2 orthologs (including hACE2). The RshSTT182/200 RBD broadly recognized 16 of 21 ACE2 orthologs with significantly lower binding affinities than that of SARS‐CoV‐2. Furthermore, the human, fox, and Rhinolophus affinis (R. affinis) ACE2 receptors supported VSV‐based RshSTT182 pseudovirus transduction, but the entry efficiency mediated by hACE2 was low. Moreover, the sera from SARS‐CoV‐2 convalescent patients or vaccinees could cross‐neutralize VSV‐based RshSTT182 pseudovirus, and the SARS‐CoV‐2 monoclonal antibodies S309, BD‐368‐2, and BD‐604 could cross‐recognize RshSTT182/200 RBD. These results indicate that the surveillance of RshSTT182/200 and its related CoVs carried by potential animal reservoirs should be enhanced to prevent potential pandemics.
Results
Binding of the RshSTT182/200 RBD to hACE2 or R. shameli ACE2
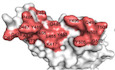
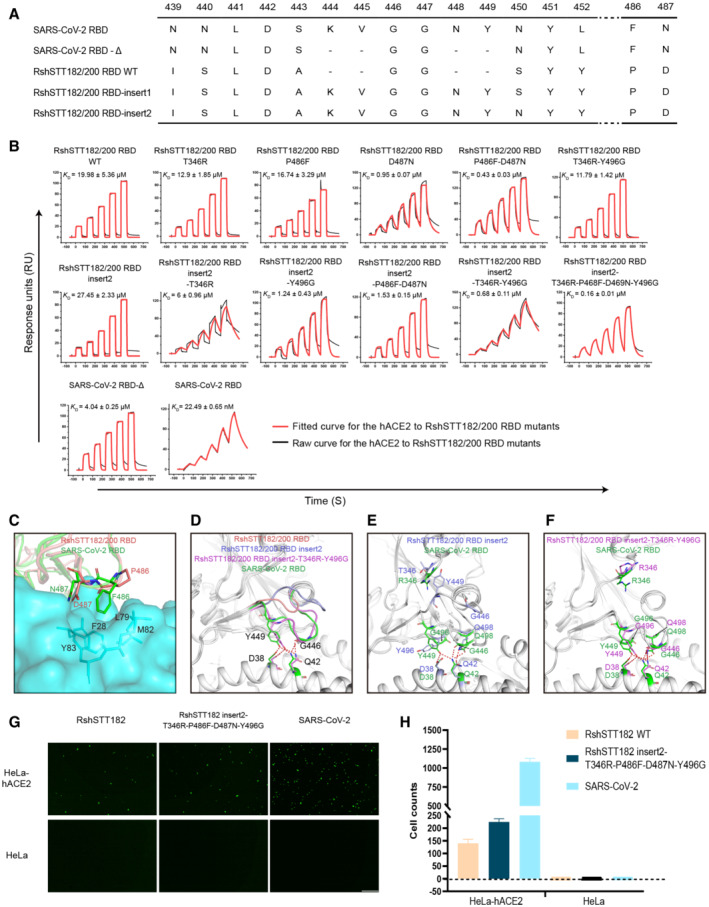
Amino acid sequence alignments indicate that the RshSTT182/200 RBD shares 84.3% identity with the SARS‐CoV‐2 RBD. Among the 21 residues involved in receptor binding in the SARS‐CoV‐2 RBD, five vary in the RshSTT182/200 RBD (Fig EV1A). Compared to SARS‐CoV‐2, four amino acid deletions (K444, V445, N448, and Y449) in the loop between β4 and β5 (named the β4β5 loop) of the RshSTT182/200 RBD domain were observed, which is similar to other bat‐origin SARS‐CoV‐2‐related viruses (Figs EV1B and EV2). The G446 and Y449 residues on the identical loop in the SARS‐CoV‐2 RBD form hydrogen bonds with D38 and Q42 of hACE2 (Wang et al, 2020). To better understand these differences, we adjusted the sequence position of the RshSTT182/200 RBD according to the amino acid sequence of SARS‐CoV‐2 RBD.
Figure EV1. The overall structure of the RshSTT182/200 RBD bound to hACE2.
-
AStructure‐based sequence alignment of the SARS‐CoV‐2 and RshSTT182/200 RBDs. The secondary structure elements were defined based on an ESPript (Robert & Gouet, 2014) algorithm and are labeled based on the SARS‐CoV‐2 RBD/hACE2 complex structure (PDB: 6LZG). Coils indicate α helices, and black arrows indicate β strands. Conserved residues are highlighted in red and residues in blue boxes are highly (80%) conserved. Residues of the SARS‐CoV‐2 RBD and RshSTT182/200 RBD that contact hACE2 through van der Waals interactions are marked with red regular triangles and blue inverted triangles, respectively. The sequence alignment was generated with SnapGene and ESPript.
-
BThe secondary structural elements of the RshSTT182/200 RBD are defined.
-
CFlow cytometry characterization of the RshSTT182/200 or SARS‐CoV‐2 RBDs binding to hACE2. The SARS‐CoV‐2 NTD was used as a negative control. The frequency of RBD‐positive cells in the hACE2‐eGFP‐positive cells is labeled in the upper right corner.
-
DGel filtration profiles of hACE2 (blue), the RshSTT182/200 RBD (saffron yellow), and the RshSTT182/200 RBD/hACE2 complex (red) were analyzed and are displayed. The separation profiles of each of the pooled samples on SDS–PAGE are shown.
-
EThe comparison of the overall structure of the RshSTT182/200 RBD/hACE2 complex compared to the SARS‐CoV‐2 RBD/hACE2 complex structure.
-
FStructural alignment of hACE2 in the RBD/ACE2 complexes. hACE2 in complex with the RshSTT182/200 RBD and SARS‐CoV‐2 RBD (PDB: 6LZG) is shown in salmon and green, respectively. The black lines show the difference between the open and closed states of the α4‐helix.
-
GThe α2‐helix and α4‐helix of hACE2 in the RshSTT182/200 RBD/hACE2 complex (closed state) and SARS‐CoV‐2 RBD/hACE2 complex (open state) are shown.
-
HStructural alignment of free hACE2 and hACE2 in the RBD/ACE2 complex. Free hACE2 (PDB: 1R42) is shown in teal.
-
I–LComparison of the overall structure of the RshSTT182/200 RBD/hACE2 complex with the SARS‐CoV RBD/hACE2 (PDB: 2AJF), GX/P2V/2017 RBD/hACE2 (PDB: 7DDP), GD/1/2019 RBD/hACE2 (PDB: 7DDO), or RaTG13 RBD/hACE2 (PDB: 7DRV) complex.
Source data are available online for this figure.
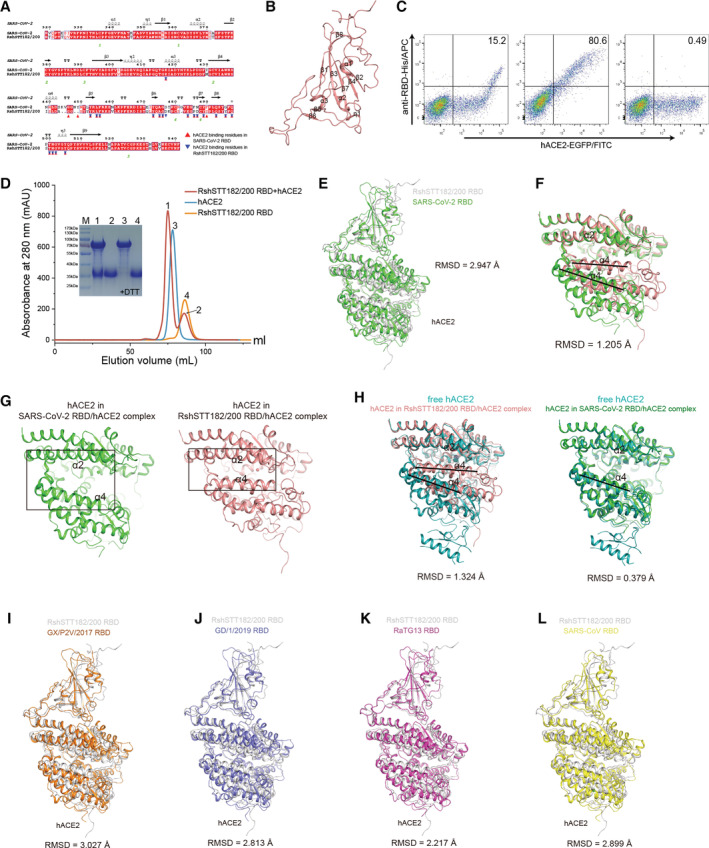
Figure EV2. Structure‐based sequence alignment of the RBD of CoVs.
The figure label is as described in Fig EV1A.
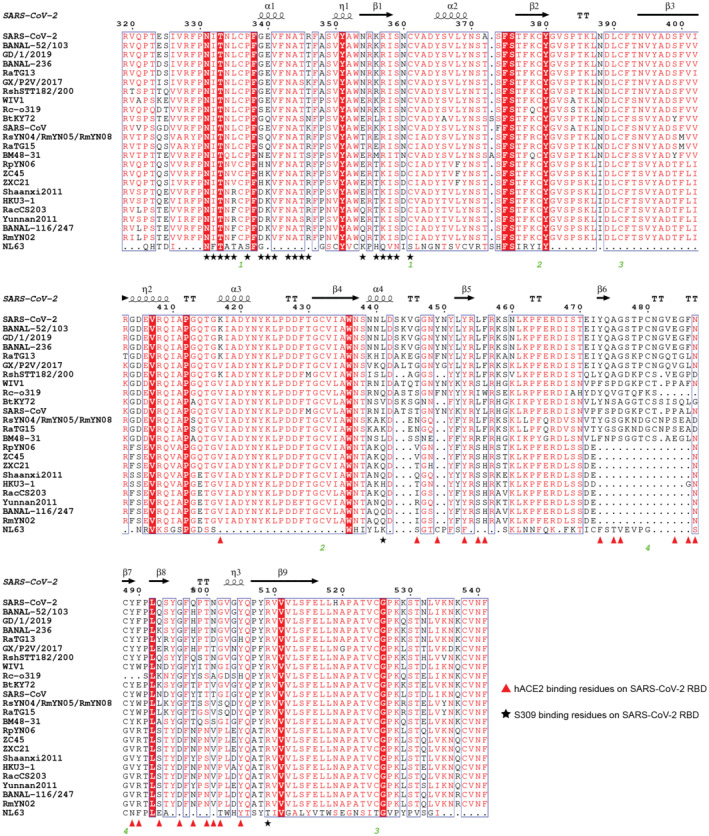
The binding of the RshSTT182/200 RBD to hACE2 was evaluated by flow cytometry (Fig EV1C). The RshSTT182/200 RBD bound to hACE2‐expressing BHK cells at a 15.2% positive rate, which was substantially lower than that of the SARS‐CoV‐2 RBD binding to hACE2‐expressing BHK cells (80.6% positive). Next, the binding affinity of the RshSTT182/200 RBD to hACE2 was determined by surface plasmon resonance (SPR) assays. Consistent with the flow cytometry, the equilibrium dissociation constant (K D) of the RshSTT182/200 RBD binding to hACE2 was 18 μM, which was ~3 orders of magnitude lower than that of the SARS‐CoV‐2 RBD to hACE2 (18.9 nM) (Fig 1A). Furthermore, we determined the binding affinity of R. shameli (the natural reservoir of RshSTT182 and RshSTT200) ACE2 to the RshSTT182/200 and SARS‐CoV‐2 RBDs via SPR. The binding affinity of the RshSTT182/200 RBD for R. shameli ACE2 (K D = 0.93 μM) was higher than that of the SARS‐CoV‐2 RBD for R. shameli ACE2 (K D = 6.64 μM) and was ~1 order of magnitude higher than that of the RshSTT182/200 RBD for hACE2 (Fig 1B).
Figure 1. Binding or structural characterization of the RshSTT182/200 RBD bound to hACE2 or R. shameli ACE2.
- SPR characterization of the RshSTT182/200 and SARS‐CoV‐2 RBDs interacting with hACE2‐hFC. Actual and fitted curves are represented by black lines and red lines, respectively.
- SPR characterization of the RshSTT182/200, SARS‐CoV‐2, and RshSTT182/200 RBDs interacting with R. shameli ACE2‐mFC. Actual and fitted curves are represented by black lines and red lines, respectively.
- The complex structure of the RshSTT182/200 RBD bound to hACE2. The red and orange boxes represent the patches of interacting residues in the RshSTT182/200 RBD/hACE2 complex. The complex structure is shown as a cartoon, and the residues involved in the hydrogen bond interactions are shown as sticks. The β4β5 loop is labeled with a black box.
- The hACE2‐binding interface of the RshSTT182/200 RBD (left) or SARS‐CoV‐2 RBD (right).
- The different interacting residues of the RshSTT182/200 RBD (left) and SARS‐CoV‐2 RBD (right) when bound to hACE2.
Molecular basis of the RshSTT182/200 RBD binding to hACE2
To better understand the molecular basis of the RshSTT182/200 RBD binding to hACE2, the RshSTT182/200 RBD/hACE2 complex was prepared with RshSTT182/200 RBD expressed in Expi293F™ cells and hACE2 expressed in Hi5 insect cells for crystal screening (Fig EV1D). Ultimately, the complex structure of the RshSTT182/200 RBD bound to hACE2 was determined at a resolution of 3.0 Å (Table 1).
Table 1.
Crystallographic data collection and refinement statistics.
| RshSTT182/200 RBD WT/hACE2 | RshSTT182/200 RBD‐insert2 hACE2 | RshSTT182/200 RBD‐ insert2‐T346R‐Y496G and hACE2 | |
|---|---|---|---|
| Data collection | |||
| Space group | C 1 2 1 | P 21 | P 21 |
| Cell dimensions | |||
| a, b, c (Å) | 155.41, 155.23, 61.00 | 81.39, 123.89, 112.79 | 81.13, 121.44, 110.49 |
| α, β, γ (°) | 90.00, 103.66, 90.00 | 90.00, 93.26, 90.00 | 90.00, 92.01, 90.00 |
| Resolution (Å) | 75.51–3.02 (3.18–3.02) | 50.00–3.52 (3.65–3.52) | 50.00–3.38 (3.50–3.38) |
| Unique reflections | 27,642 (4,040) | 28,025 (2,812) | 30,335 (3,024) |
| R merge | 0.099 (0.614) | 0.133 (1.060) | 0.151 (1.444) |
| I/σI | 12.8 (2.7) | 9.0 (1.1) | 10.480 (1.0) |
| Completeness (%) | 99.8 (100.0) | 99.9 (99.9) | 100.0 (100.0) |
| CC1/2 | 0.996 (0.799) | 0.991 (0.370) | 0.990 (0.536) |
| Redundancy | 4.8 (3.8) | 4.8 (5.0) | 5.5 (5.7) |
| Refinement | |||
| Resolution (Å) | 29.64–3.02 | 40.63–3.51 | 46.40–3.37 |
| No. of reflections | 27,508 | 27,972 | 30,163 |
| R work/R free | 0.2137/0.2403 | 0.1993/0.2327 | 0.2692/0.2736 |
| No. of atoms | |||
| Protein | 6,601 | 13,212 | 13,022 |
| Ligand/ion | 1 | 2 | 2 |
| Water | 5 | 0 | 0 |
| B‐factors | |||
| Protein | 83.0 | 129.0 | 156.0 |
| Ligand/ion | 83.0 | 96.5 | 134.0 |
| Water | 49.2 | ||
| RMSDs | |||
| Bond length (Å) | 0.003 | 0.002 | 0.003 |
| Bond angles (°) | 0.590 | 0.500 | 0.606 |
| Ramachandran Statistics (%) | |||
| Favored (%) | 95.89 | 96.63 | 96.70 |
| Allowed (%) | 4.11 | 3.37 | 3.30 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 |
Values in parentheses are for the highest‐resolution shell. WT, wild‐type; RBD, receptor‐binding domain; CC1/2, Pearson correlation coefficient; RMSD, root‐mean‐square deviation.
Overall, the structure of the RshSTT182/200 RBD with hACE2 showed significant conformational differences from the SARS‐CoV‐2 RBD/hACE2 complex (PDB: 6LZG), with a root‐mean‐square deviation (RMSD) of 2.947 Å over 786 equivalent Cα atoms (Fig EV1E). In detail, we found that this difference was due to hACE2 adopting a different conformation. The α2 and α4 helices of hACE2 in the RshSTT182/200 RBD/hACE2 complex are in the closed state, but in other RBD/hACE2 complex structures and free hACE2, α2 and α4 of hACE2 are in the open state (Fig EV1F–L).
As with SARS‐CoV‐2‐ and SARS‐CoV‐2‐related viruses, the residues participating in hACE2 binding in the RshSTT182/200 RBD are distributed into two patches. In patch 1, residues Q24 and Y83 on the N‐terminus of α1 and α2 of hACE2 form a hydrogen bond (H‐bond) network with residues D487 and Y489 in the RshSTT182/200 RBD. The D30 residue of hACE2 also contacts K417 of the RshSTT182/200 RBD by a salt bridge. In patch 2, Y41, K353, and R393 in hACE2 form an H‐bond network with Y496, Q498, T500, N501, G502, and Y505 in the RshSTT182/200 RBD (Fig 1C and Table 2).
Table 2.
Comparison of the binding between hACE2 and the RshSTT182/200 or SARS‐CoV‐2 RBDs.
| hACE2 | RshSTT182/200 RBD | SARS‐CoV‐2 RBD |
|---|---|---|
| S19 | A475 (2), D477 (4) | A475 (3, 1 ), G476 (4) |
| Q24 | A475 (3), G476 (2), D487 (11, 1 ), Y489 (1) | A475 (4), G476 (5), N487 (15, 1 ) |
| T27 | F456 (6), Y473 (1), A475 (2), Y489 (10) | F456 (5), Y473 (1), A475 (2), Y489 (7) |
| F28 | Y489 (7) | Y489 (7) |
| D30 | K417 (7, 1 ), L455 (2), F456 (3) | K417 (4, 1 ), L455 (2), F456 (4) |
| K31 | L455 (2), F456 (6), E484 (1), Y489 (5), Q493 (7) | L455 (2), F456 (5), E484 (1), Y489 (6), F490 (2), Q493 (3) |
| H34 | Y453 (7), L455 (9) | Y453 (5, 1 ), L455 (9), Q493 (6) |
| E35 | Q493 (1), Y496 (1) | Q493 (8) |
| E37 | Y505 (9) | Y505 (7) |
| D38 | Y496 (18), Q498 (2) | Y449 (9, 1 ), G496 (5), Q498 (1) |
| Y41 | Q498 (5), T500 (8, 1 ), N501 (10, 1 ) | Q498 (8), T500 (7, 1 ), N501 (8, 1 ) |
| Q42 | Q498 (4) | G446 (4, 1 ), Y449 (4, 1 ), Q498 (8, 1 ) |
| L45 | T500 (2) | Q498 (3), T500 (1) |
| L79 | P486 (3) | F486 (2) |
| M82 | – | F486 (9) |
| Y83 | P486 (1), D487 (7, 1 ), Y489 (1, 1 ) | F486 (11), N487 (8, 1 ), Y489 (1) |
| N330 | T500 (6) | T500 (8) |
| K353 |
Y496 (6, 1 ), Q498 (4, 1 ), N501 (17), G502 (3, 1 ), Y505 (27) |
G496 (7, 1 ), N501 (11), G502 (4, 1 ), Y505 (28) |
| G354 | G502 (7), Y505 (4) | G502 (7), Y505 (4) |
| D355 | T500 (10), G502 (1) | T500 (8), G502 (1) |
| R357 | T500 (3) | T500 (3) |
| R393 | Y505 (3, 1 ) | Y505 (1) |
| Total | 261, 10 | 288, 13 |
The numbers in parentheses of the RBD residues represent the number of van der Waals contacts between the indicated residue and hACE2. The numbers underlined in bold indicate the number of potential H‐bonds and salt bridge between pairs of residues. The van der Waals contacts were analyzed at a cutoff of 4.5 Å and polar interactions at a cutoff of 3.5 Å.
Different binding characteristics of the RshSTT182/200 RBD and SARS‐CoV‐2 RBD to hACE2
The buried surface area of the RshSTT182/200 RBD (1,711.5 Å2) bound to hACE2 was smaller than that of the SARS‐CoV‐2 RBD in complex with hACE2 (1,767.1 Å2) (Fig 1D). The total contacts of the RshSTT182/200 RBD with hACE2 (261) were lower than that of the SARS‐CoV‐2 RBD with hACE2 (288). In patch 1, there were 111 contacts, including 107 van der Waal (vdw) contacts, 3 H‐bonds, and a salt bridge between the RshSTT182/200 RBD and hACE2, which is fewer than those in the SARS‐CoV‐2 RBD bound to hACE2 (158 contacts, including 150 vdw contacts, 7 H‐bonds, and a salt bridge) (Table 2). On the buried surface area of the RshSTT182/200 RBD, residues D477, P486, D487, Y490, and Y496 differ from the S477, F486, N487, F490, and G496 residues (respectively) of the SARS‐CoV‐2 RBD (Figs 1E and EV1A). G446 and Y449 on the β4β5 loop of the SARS‐CoV‐2 RBD form H‐bonds with hACE2, but in the RshSTT182/200 RBD, the deletion of residues K444, V445, N448, and Y449 renders the β4β5 loop too short to contact hACE2 (Fig 1C).
Key RshSTT182/200 RBD residues for binding to hACE2
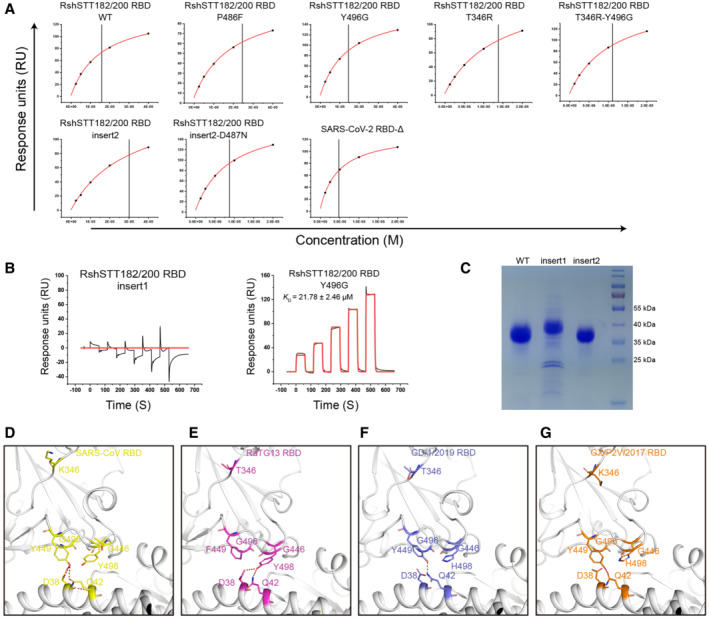
Further analysis was performed to identify the key residues used by the RshSTT182/200 RBD to bind to hACE2. In the SARS‐CoV‐2 RBD/hACE2 complex, F486 forms a small patch of hydrophobic interactions with F28, L79, M82, and Y83 in hACE2, and forms a π‐π interaction with Y83 in hACE2 (Wang et al, 2020). In the RshSTT182/200 RBD/hACE2 complex, F486 is substituted by P486, which does not form a hydrophobic patch with hACE2 in the structure (Fig 2A and C and Table 2). Unexpectedly, when we introduced the P486F substitution into the RshSTT182/200 RBD, the binding affinity of the RshSTT182/200 RBD‐P486F mutant with hACE2 was similar to that of the wild‐type RshSTT182/200 RBD (Figs 2B and EV3A). When D487 in the RshSTT182/200 RBD was substituted with N487, the binding affinity of the RshSTT182/200 RBD‐D487N mutant with hACE2 increased approximately ~21.0‐fold compared to the wild‐type RshSTT182/200 RBD (Fig 2B). The P486F and D487N double mutation further enhanced the binding of the RshSTT182/200 RBD‐D487N with hACE2 (~ 2.2‐fold) (Fig 2B).
Figure 2. Mutation analysis of key residues of the RshSTT182/200 RBD interaction with hACE2.
-
AInsertion or deletion mutation sites of the RshSTT182/200 RBD or SARS‐CoV‐2 RBD mutants.
-
BThe binding affinities of the wild‐type RshSTT182/200 RBD, insert2, and their mutants for hACE2‐hFc.
-
CThe hydrophobic interaction regions of the RshSTT182/200 and SARS‐CoV‐2 RBDs are shown.
-
DThe β4β5 loop comparison of the RshSTT182/200 RBD/hACE2 complex structure with the RshSTT182/200 RBD‐insert2/hACE2 and SARS‐CoV‐2 RBD/hACE2 complexes (PDB: 6LZG). The backbone of the RBD is colored in gray. The β4β5 loop of the RshSTT182/200 RBD, RshSTT182/200 RBD‐insert2, RshSTT182/200 RBD‐insert2‐T346R‐Y486G, and SARS‐CoV‐2 RBD are shown as salmon, light blue, violet, and green, respectively. The RBD residues interacting with hACE2 and their corresponding ACE2 residues are shown as sticks in the corresponding colors.
-
E, FComparison of hydrogen‐bonding interactions in the β4β5 loop region between RshSTT182/200 RBD‐insert2 or RshSTT182/200 RBD‐insert2‐T346R‐Y496G and the SARS‐CoV‐2 RBD.
-
GThe entry efficiency of SARS‐CoV‐2 and RshSTT182 pseudoviruses in HeLa‐hACE2 cells. Green fluorescence indicates the entry of the pseudoviruses into the Hela‐hACE2 cells. Unaltered HeLa cells were used as a negative control. The scale bar indicates 400 μm.
-
HStatistics for transduction of pseudoviruses. Data represent the results of five replicates in one representative assay. All data are presented as mean ± SD. Pseudovirus infection assays were performed at least twice.
Source data are available online for this figure.
Figure EV3. SPR characterization of hACE2 binding to the RshSTT182/200 RBD and its mutants.
-
AThe steady‐state fitting curve is shown when the K D was calculated by the steady‐state model.
-
BSPR analysis of hACE2 binding to the RshSTT182/200 RBD insert1 and Y496G. The raw and fitted curves are displayed as black and red lines, respectively.
-
CSDS–PAGE showing the difference in molecular weight among RshSTT182/200 RBD‐WT, RBD‐insert1, and RBD‐insert2.
-
D–GThe H‐bond interactions of the β4β5 loop region of the SARS‐CoV RBD, RaTG13 RBD, GD/1/2019 RBD, and GX/P2V/2017 RBD are shown.
Source data are available online for this figure.
To evaluate the influence of the deleted amino acids (AAs) in the β4β5 loop, we inserted four AAs (K444, V445, N448, and Y449) into the RshSTT182/200 RBD (RshSTT182/200 RBD‐insert1 mutant construction) (Fig 2A). Unexpectedly, the RshSTT182/200 RBD‐insert1 protein failed to interact with hACE2 (Fig EV3B). Because an N‐linked glycosylation motif was introduced (N448‐Y449‐S450), SDS–PAGE indicated that the molecular weight of RshSTT182/200 RBD‐insert1 was greater than the wild‐type RshSTT182/200 RBD (Fig EV3C). We hypothesized that N448 glycosylation in the RshSTT182/200 RBD‐insert1 may inhibit the interaction of the RBD with ACE2. Therefore, we mutated S450 to N450 (RshSTT182/200 RBD‐insert2 mutant construction) to eliminate the effect of this N‐linked glycosylation and found that the interaction between the RshSTT182/200 RBD‐insert2 protein and hACE2 was re‐established, with a similar binding affinity to that of the wild‐type RshSTT182/200 RBD (Figs 2B and EV3A).
To further analyze why the insertion mutation in RshSTT182/200 RBD‐insert2 did not increase hACE2 binding, we solved the complex structure of RshSTT182/200 RBD‐insert2 with hACE2 and found that the β4β5 loop of RshSTT182/200 RBD‐insert2 was too flexible to interact with hACE2 (Fig 2D). Furthermore, R346 in the SARS‐CoV‐2 RBD has a longer side chain than T346 in the RshSTT182/200 RBD, which may introduce repulsion to assist with the β4β5 loop interacting with hACE2 (Fig 2E). In addition, Y496 in the RshSTT182/200 RBD may clash with Y449 and disrupt the interaction of Y449 with residues on hACE2, whereas G496 of the SARS‐CoV‐2 RBD would exert no such effect (Fig 2E). To validate our hypothesis, we constructed several RshSTT182/200 RBD‐insert2 mutants and evaluated their binding to hACE2. When T346 was substituted with R346, the binding affinity of the RshSTT182/200 RBD‐insert2 T346R increased ~4.6‐fold, and Y496G substitution in RshSTT182/200 RBD‐insert2 increased affinity ~22.1‐fold compared to RshSTT182/200 RBD‐insert2 (Fig 2B). Relative to the RshSTT182/200 RBD, the binding affinity of RshSTT182/200 RBD‐T346R‐Y496G was similar to that of hACE2 (Figs 2B and EV3A). When K444, V445, N448, and Y449 were inserted, the binding affinity of RshSTT182/200 RBD‐insert2‐T346R‐Y496G was ~17.3‐fold higher than that of RshSTT182/200 RBD‐T346R‐Y496G (Figs 2B and EV3A), which is consistent with our hypothesis.
Subsequently, we solved the structure of the RshSTT182/200 RBD‐insert2‐T346R‐Y496G in complex with hACE2. Like SARS‐CoV‐2, both G446 and Y449 in the β4β5 loop of the RshSTT182/200 RBD‐insert2‐T346R‐Y496G protein form contacts with hACE2 (Fig 2F). Other than the SARS‐CoV‐2 RBD, K346 is also observed in the SARS‐CoV and GX/P2V/2017 RBDs, and G496 is found in the SARS‐CoV, RaTG13, GD/1/2019, and GX/P2V/2017 RBDs. Y449 in the β4β5 loop of the SARS‐CoV, GD/1/2019, and GX/P2V/2017 RBDs forms H‐bonds with the corresponding residues in hACE2 (Fig EV3D–G). In the RaTG13 RBD, F449 in the β4β5 loop forms a vdw with hACE2 (Fig EV3E).
The binding affinity of RshSTT182/200 RBD‐insert2‐T346R‐P486F‐D487N‐Y496G with hACE2 (K D = 0.16 ± 0.01 μM) was ~124.9‐fold stronger than that of the wild‐type RshSTT182/200 RBD (K D = 19.98 ± 5.36 μM) (Fig 2B). To evaluate the role of the β4β5 loop in the interaction between the SARS‐CoV‐2 RBD and hACE2, we deleted K444, V445, N448, and Y449 in the SARS‐CoV‐2 RBD (SARS‐CoV‐2 RBD‐Δ) and tested its binding affinity for hACE2. SARS‐CoV‐2 RBD‐Δ (K D = 4.04 ± 0.25 μM) displayed ~179.6‐fold weaker binding affinity for hACE2 than the SARS‐CoV‐2 RBD (K D = 22.49 ± 0.65 nM) (Figs 2B and EV3A).
Next, we tested the entry efficiency of RshSTT182 and RshSTT182 insert2‐T346R‐P486F‐D487N‐Y496G using a VSV‐based pseudovirus assay, with VSV‐based SARS‐CoV‐2 pseudoviruses used as a positive control. Equivalent doses of pseudoviruses (determined by qPCR) were used to infect HeLa cells overexpressing hACE2 (HeLa‐hACE2 cells), with HeLa cells (no hACE2 overexpression) used as a negative control. We found that all three pseudoviruses could enter HeLa‐hACE2 cells (Fig 2G and H). However, the RshSTT182 and RshSTT182 insert2‐T346R‐P486F‐D487N‐Y496G pseudoviruses displayed significantly lower transduction efficiency than SARS‐CoV‐2 pseudovirus. Consistent with their binding affinities, RshSTT182 insert2‐T346R‐P486F‐D487N‐Y496G pseudovirus showed relatively higher transduction efficiency than RshSTT182 pseudovirus (Fig 2G and H).
Narrower receptor‐binding spectrum of bat‐origin RshSTT182/200 virus in bat species than SARS‐CoV‐2
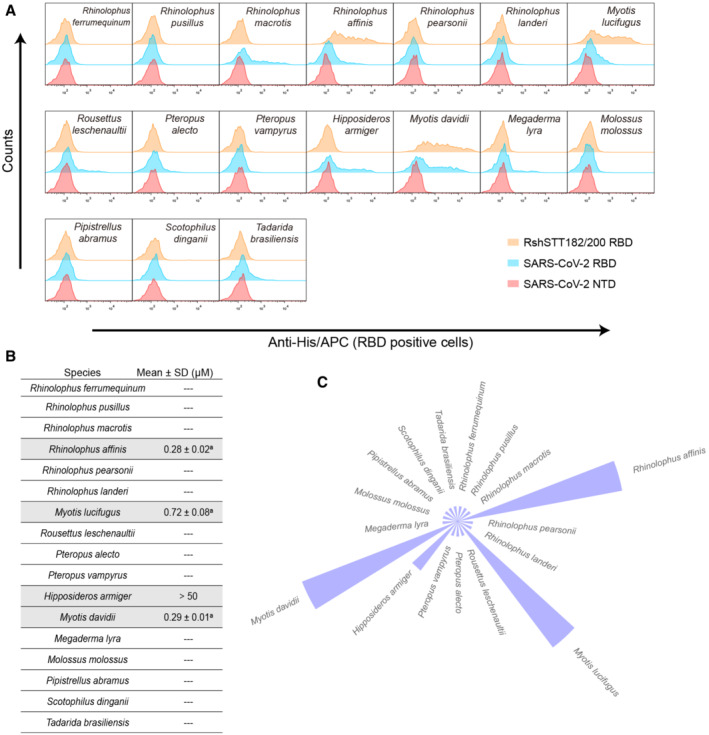
Because RshSTT182/200 is a bat‐origin virus, we evaluated the binding of the RshSTT182/200 RBD to ACE2s from another 17 bat species (including 6 Rhinolophus species and 11 non‐Rhinolophus species). Flow cytometry analysis revealed that R. affinis, Myotis lucifugus, and Myotis davidii were positive for both the RshSTT182/200 RBD and SARS‐CoV‐2 RBD (Fig 3A). In addition, Rhinolophus macrotis, Rhinolophus landeri, Rousettus leschenaultii, Pteropus alecto, Hipposideros amiger, and Megaderma lyra were bound by the SARS‐CoV‐2 RBD but not the RshSTT182/200 RBD. We then quantified the binding affinity of the RshSTT182/200 RBD to 17 bat ACE2 orthologs using SPR (Fig EV4A and B). Consistent with the flow cytometry results, ACE2s from R. affinis (K D = 0.28 ± 0.02 μM), M. lucifugus (K D = 0.72 ± 0.08 μM), and M. davidii (K D = 0.29 ± 0.01 μM) were bound more tightly by the RshSTT182/200 RBD (Figs 3B and C, and EV4A) than that of R. shameli. In addition, the H. armiger ACE2 was also bound by the RshSTT182/200 RBD but with a binding affinity < 50 μM (Figs 3B and C, and EV4A and B). Among the other ACE2 orthologs from bats, the SARS‐CoV‐2 RBD could bind to 9 of 17 (Ma et al, 2021; Niu et al, 2021), whereas the RshSTT182/200 RBD could only interact with the 4 ACE2 orthologs. Thus, RshSTT182/200, a bat‐derived SARS‐CoV‐2‐related CoV, had a narrower bat ACE2‐binding spectrum than SARS‐CoV‐2.
Figure 3. Comparison of binding spectrum of bat‐ACE2 orthologs with the RshSTT182/200 or SARS‐CoV‐2 RBDs.
- Flow cytometry characterization of binding between bat‐ACE2 orthologs and the SARS‐CoV‐2 or RshSTT182/200 RBDs. BHK cells expressing eGFP‐fused ACE2s were stained with the indicated His‐tagged proteins. The SARS‐CoV‐2 NTD was used as a negative control.
- The binding affinities between bat ACE2s and the RshSTT182/200 RBD are shown with the mean ± SD. Mean ± SD represents the average values and standard deviation of the K D across three independent experiments. The a or b superscript of affinity data indicates that the K D value was calculated by kinetics model fitting or steady‐state model fitting, respectively. “‐‐‐” indicates not determined.
- The ring histogram shows the comparison of the binding affinity among bat ACE2 orthologs to the RshSTT182/200 RBD. The size of the column indicates the strength of the interaction.
Figure EV4. SPR characterization of ACE2s binding to the RshSTT182/200 RBD, as well as the key residue contacts between hACE2 and the RshSTT182/200 or SARS‐CoV‐2 RBDs.
- Representative example of sensorgrams and binding kinetics model fitting of ACE2 (mFc‐tagged ACE2) binding to the RshSTT182/200 RBD by SPR analysis. ACE2s in supernatants were captured by anti‐mIgG Fc antibodies immobilized on the CM5 chip, and binding to serially diluted RshSTT182/200 RBD was sequentially tested. The raw and fitted curves are displayed as black and red lines, respectively.
- The saturation curve fitted to the response (RU) at equilibrium was plotted against the protein concentration when K D was estimated by steady‐state fitting. The binding affinity with rapid dissociation and association rates (K d and K a) makes the kinetic fitting inaccurate.
- Residues in hACE2 that form contacts with the RBD are listed. The numbers with asterisks indicate that the key residue only interacts with the SARS‐CoV‐2 RBD. The conserved residues among the 39 ACE2 orthologs are shown as black letters, and the residue substitutions in the ACE2 of 38 species relative to hACE2 are shown as red letters. The gray background represents the binding of the RshSTT182/200 RBD to the ACE2 of this species.
The cross‐species recognition of the RshSTT182/200 RBD to ACE2 orthologs from 21 different animal species
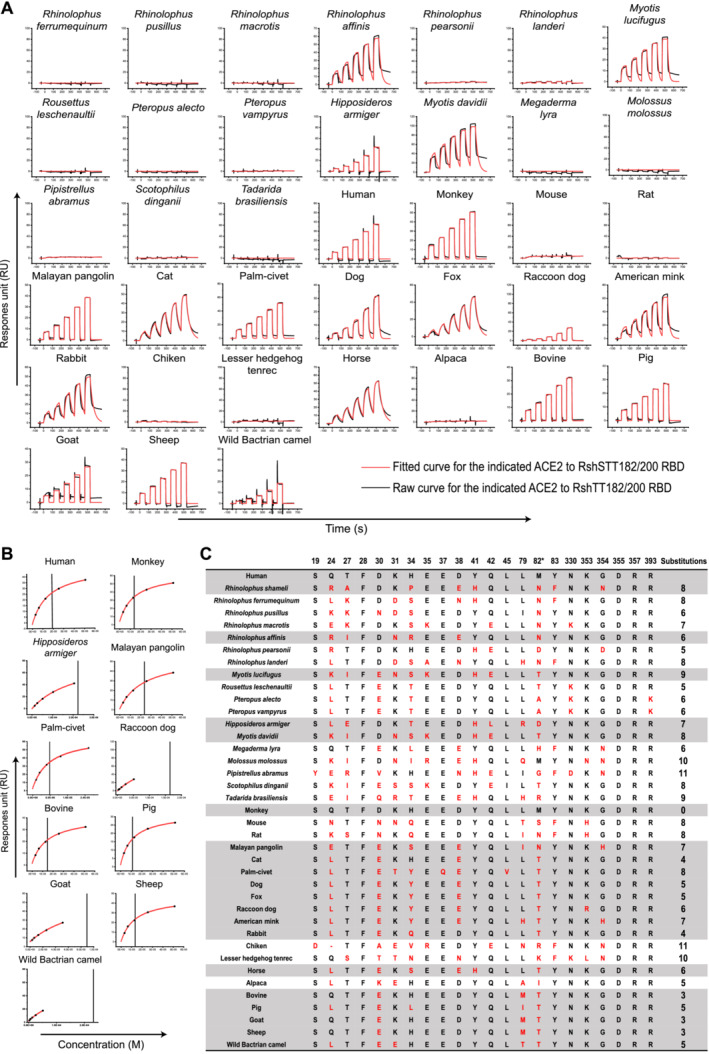
To better understand the potential host range of RshSTT182/200, the binding of the RBD to the ACE2 of 21 species, including humans, domestic animals, and wild animals that belong to nine orders (Primates, Lagomorpha, Rodentia, Pholidota, Perissodactyla, Artiodactyla, Chiroptera, Afrotheria, and Galliformes), was evaluated. We tested the binding of the RshSTT182/200 and SARS‐CoV‐2 RBD proteins to eGFP‐fused ACE2s expressed on the BHK cell surface via flow cytometry. The SARS‐CoV‐2 NTD protein was used as a negative control (Fig 4A). In parallel, we quantified their binding affinity to further verify the binding of the RshSTT182/200 RBD and ACE2 orthologs via SPR (Fig EV4A and B).
Figure 4. SPR characterization of the binding affinities of ACE2 (except bat) for the RshSTT182/200 or SARS‐CoV‐2 RBDs.
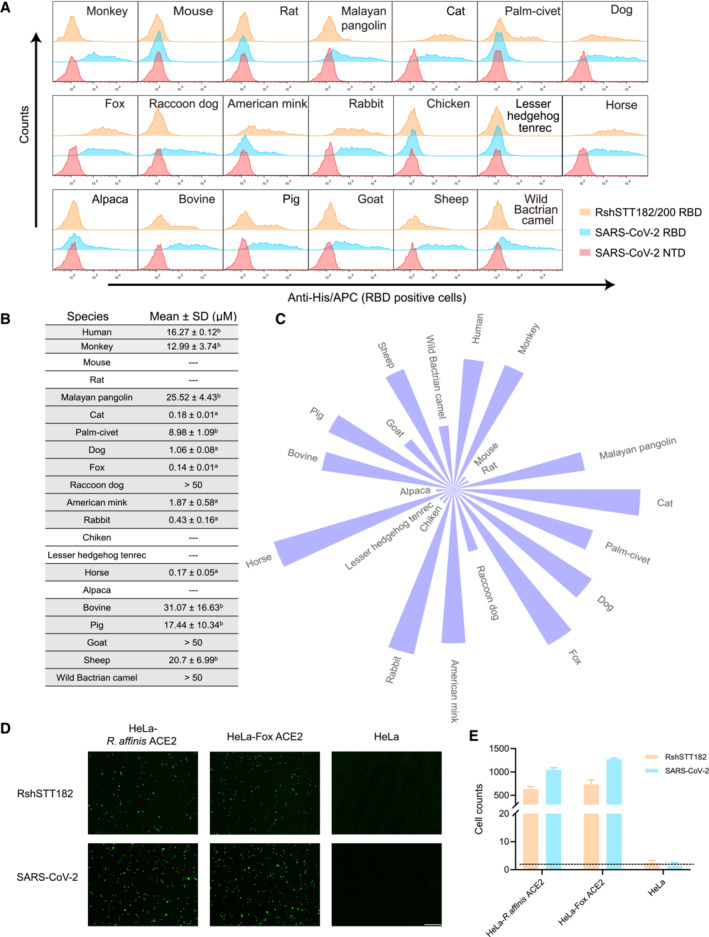
- Flow cytometry characterization of binding between ACE2s and the RshSTT182/200 or SARS‐CoV‐2 RBDs. Cells stained with the RshSTT182/200 RBD, the SARS‐CoV‐2 RBD, and the SARS‐CoV‐2 NTD proteins are shown in orange, marine, and red, respectively. The SARS‐CoV‐2 NTD was used as a negative control.
- The binding affinities between ACE2s and the RshSTT182/200 RBD are shown with the mean ± SD of three independent experiments. The “a” superscript of affinity data represents that the K D value was calculated by kinetics model fitting; “b” represents that the K D value was calculated by steady‐state model fitting. “‐‐‐” indicates not determined.
- The ring histogram shows the comparison of the binding affinities of ACE2 orthologs with the RshSTT182/200 RBD. The size of the column indicates the strength of the interaction.
- The entry efficiency of SARS‐CoV‐2 and RshSTT182 pseudoviruses into HeLa‐hACE2 cells. Green fluorescence indicates the entry of the pseudoviruses into the Hela‐hACE2 cells. Unaltered HeLa cells were used as a negative control. The scale bar indicates 400 μm.
- Statistics for transduction of pseudoviruses. Data represent the results of five replicates in one representative assay. All data are presented as mean ± SD. Pseudovirus infection assays were performed at least twice.
Source data are available online for this figure.
The RshSTT182/200 RBD interacted with cells expressing ACE2 orthologs from Primates (human and monkey), Pholidota (Malayan pangolin), all Carnivora (cat, palm civet, fox, dog, raccoon dog, and American mink), Lagomorpha (rabbit), Perissodactyla (horse), and most Artiodactyla (pig, wild Bactrian camel, bovine, goat, and sheep) (Fig 4A). Compared to the SARS‐CoV‐2 RBD, the binding affinity of the RshSTT182/200 RBD to ACE2 orthologs was substantially lower. The binding affinities of the RshSTT182/200 RBD to cat, fox, rabbit, and horse ACE2s (0.14–0.43 μM) were higher than that of the RshSTT182/200 RBD for R. shameli ACE2 (Figs 1B, 4B and C, and EV4A). Fox ACE2 was bound with the highest affinity by the RshSTT182/200 RBD (Figs 4B and C, and EV4A). The monkey, palm civet, dog, Malayan pangolin, bovine, pig, and sheep ACE2s positively interacted with the RshSTT182/200 RBD, with binding affinities of 1.06–31 μM (Figs 4B and C, and EV4A and B). Raccoon dog, goat, and wild Bactrian camel ACE2s could bind to the RshSTT182/200 RBD but with binding affinities > 50 μM (Figs 4B and C, and EV4A and B). Similar to the SARS‐CoV‐2 RBD, the RshSTT182/200 RBD did not interact with ACE2 orthologs from Rodentia (mouse and rat), Afrotheria (Lesser hedgehog tenrec), or Galliformes (chicken) (Figs 4B and C, and EV4A). From the residue comparison of ACE2s, we found that the F28, D355, and R357 residues are completely conserved among these 39 species (Fig EV4C).
To evaluate whether RshSTT182 efficiently enters into cells expressing ACE2 orthologs with high‐affinity, RshSTT182 and SARS‐CoV‐2 pseudoviruses were used to infect HeLa cells overexpressing fox ACE2 and R. affinis ACE2 (HeLa‐fox ACE2 cells and HeLa‐R. affinis ACE2 cells). The RshSTT182 pseudovirus could effectively enter HeLa‐fox ACE2 cells and HeLa‐R. affinis ACE2 cells (Fig 4D and E).
The cross‐reactive immune response of SARS‐CoV‐2 to RshSTT182/200
To determine if there is a cross‐reactive immune response of SARS‐CoV‐2 to RshSTT182/200, we tested the cross‐recognition of antibodies in COVID‐19 convalescent and vaccinee sera to the RshSTT182/200 RBD with enzyme‐linked immunosorbent assays (ELISAs). The SARS‐CoV‐2 antibodies in donors' sera cross‐recognized the RshSTT182/200 RBD but were lower than for the SARS‐CoV‐2 RBD (Fig 5A). Sera from five healthy donors showed no binding to the RshSTT182/200 RBD or SARS‐CoV‐2 RBD (Fig 5A). We then evaluated the neutralization activity of serum samples from COVID‐19 convalescent and vaccinee against SARS‐CoV‐2 and RshSTT182 pseudoviruses in HeLa‐R. affinis ACE2 cells. The six sera displayed effective neutralization against the pseudoviruses, but the titer of neutralizing antibodies against RshSTT182 was lower than that against SARS‐CoV‐2 (Fig 5B).
Figure 5. The cross‐reactive immune response of SARS‐CoV‐2 antibodies to the RshSTT182/200 RBD.
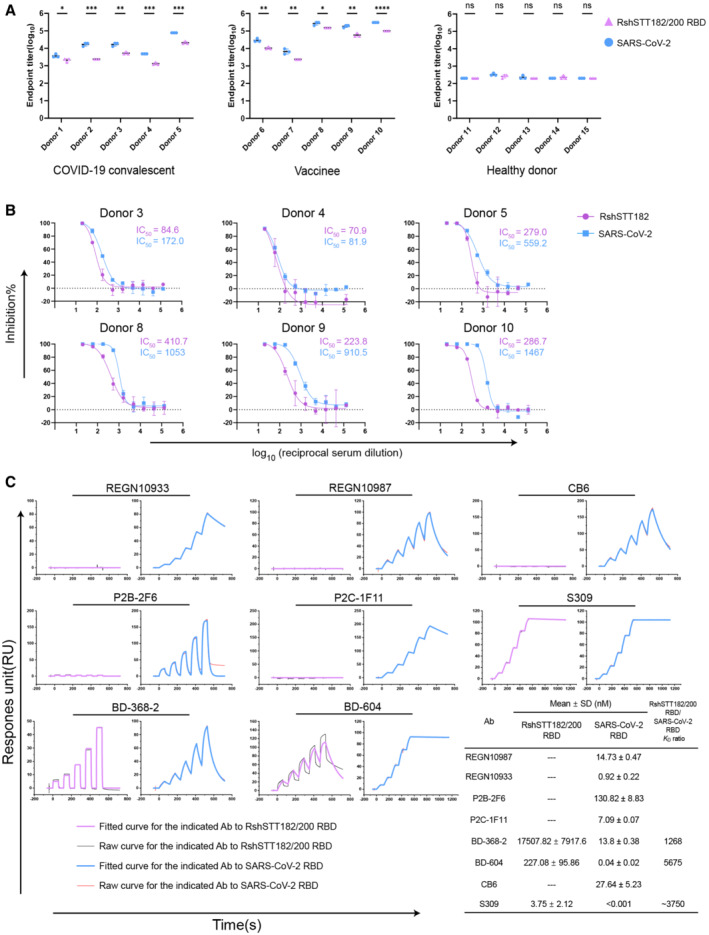
- ELISA measurement of the titers of SARS‐CoV‐2‐ and RshSTT182/200 RBD‐specific immunoglobulin G (IgG) in serum samples of COVID‐19 convalescents and vaccinees or healthy donors. Data showed the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (two‐tailed unpaired t‐test).
- The neutralization curve of serum samples in COVID‐19 convalescents and vaccinees against RshSTT182 and SARS‐CoV‐2 are shown. Bar represents the SD of two replicates.
- SPR characterization of the binding affinities of the SARS‐CoV‐2 RBD or RshSTT182/200 RBD with clinical antibodies. The K D values shown are the mean ± SD of three independent experiments. “‐‐‐” Indicates not determined.
Source data are available online for this figure.
To evaluate the cross‐recognition of SARS‐CoV‐2 RBD isolated monoclonal antibodies (MAbs) for the RshSTT182/200 RBD, we tested the binding affinity of several clinical SARS‐CoV‐2 antibodies (REGN10933, REGN10987, CB6, P2C‐1F11, P2B‐2F6, BD‐368‐2, BD‐604, and S309) to RshSTT182/200 RBD by SPR. Among the eight antibodies, BD‐604, S309, and BD‐368‐2 successfully bound to the RshSTT182/200 RBD with a substantially decreased binding affinity than that of SARS‐CoV‐2 RBD (Huang et al, 2022) (Fig 5C). Both BD‐604 and BD‐368‐2 bind to the receptor‐binding motif (RBM) on the SARS‐CoV‐2 RBD, which is diverse among different CoVs (Fig EV5A and B) (Du et al, 2020). S309 (VIR‐7831) showed the highest binding affinity to RshSTT182/200 RBD among these MAbs (K D = 3.75 ± 2.12 nM). S309 binds to the core region of the RBD, which is conserved among different CoVs, so that it has a broad cross‐neutralizing effect (Figs 5C, EV2, and EV5A and B) (Pinto et al, 2020; Huang et al, 2022). This suggests that the core region of the RBD is a hotspot for cross‐neutralizing MAb screening.
Figure EV5. Structural comparison of antibody‐binding epitope and amino acid conservation in the RBD.
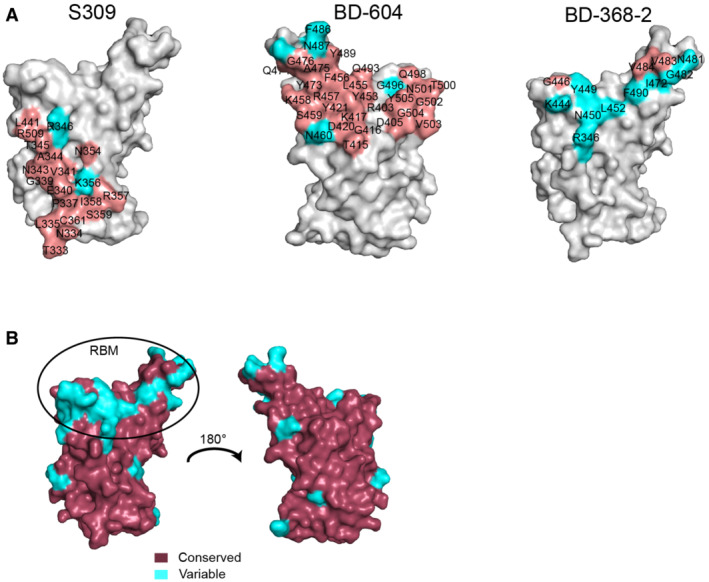
- Comparison of the binding interface between the SARS‐CoV‐2 antibody and the RshSTT182/200 RBD. In S309 (PDB: 7R6X), BD‐604, and BD‐368‐2 (PDB: 7CHF) binding to the SARS‐CoV‐2 RBD site, the conserved residues between the SARS‐CoV‐2 and RshSTT182/200 RBDs are shown in salmon, while variable residues are shown in cyan.
- Conserved residues and variable residues between the SARS‐CoV‐2 and RshSTT182/200 RBDs are shown in raspberry and cyan, respectively.
Discussion
Similar to other CoVs such as SARS‐CoV‐2, SARS‐CoV, RaTG13, GX/P2V/2017, and GD/1/2019, RshSTT182/200 has a broad receptor‐binding spectrum (Wu et al, 2020a; Niu et al, 2021; Liu et al, 2021a). In general, the binding spectrum of the RshSTT182/200 RBD to ACE2 orthologs was different from that of the SARS‐CoV‐2 RBD. Although the RshSTT182/200 RBD broadly recognized most of the ACE2 orthologs, its binding affinities for the orthologs were weaker than those of the SARS‐CoV‐2 RBD. Palm civet is the host of SARS‐CoV, but it is difficult to detect the binding of palm civet ACE2 to the prototype SARS‐CoV‐2 RBD (Guan et al, 2003; Wu et al, 2020a). Herein, we found that it can bind to the RshSTT182/200 RBD. In contrast, the SARS‐CoV‐2 RBD‐sensitive alpaca ACE2 failed to interact with the RshSTT182/200 RBD. The binding affinity of the RshSTT182/200 RBD for fox ACE2 was the highest among the tested ACE2 orthologs. Furthermore, fox ACE2 is broadly bound by the RBDs of many sarbecovirus, such as the SARS‐CoV‐2 prototype strain and omicron variant, SARS‐CoV, RaTG13, GX/P2V/2017, and GD/1/2019 (Wu et al, 2020a; Niu et al, 2021; Liu et al, 2021a; Li et al, 2022a). Therefore, surveillance for sarbecovirus in foxes should be stepped up to prevent their spillover. RshSTT182/200 is a bat‐origin virus, but the binding spectrum of the RshSTT182/200 RBD to bat ACE2 orthologs was much narrower than the SARS‐CoV‐2 RBD. Furthermore, human, fox, and R. affinis ACE2 could mediate RshSTT182 pseudovirus entry into HeLa cells expressing these proteins.
In the RshSTT182/200 RBD/hACE2 complex, we captured the closed state of hACE2, which is also observed in the inhibitor‐bound hACE2 complex and the structure of catalytically inactive hACE2 in complex with the SARS‐CoV‐2 RBD (Towler et al, 2004; Chen et al, 2022). An intermediate state is observed in the crystal structure of the SARS‐CoV RBD/hACE2 complex (Li et al, 2005). The functions of the different conformations of hACE2 in RBD binding and virus infection need to be further studied.
In previous reports, several key substitutions in the RBD, such as Q493Y, Q498Y, Q498H, and N501Y, are shown to increase the receptor binding and host range of CoVs (Niu et al, 2021; Sun et al, 2021; Zhang et al, 2021; Liu et al, 2021a; Tang et al, 2022; Xu et al, 2022a). In the RshSTT182/200 RBD, the D487N substitution significantly enhanced the binding affinity of the RshSTT182/200 RBD to hACE2. CoVs are RNA viruses that can be easily mutated (Xu et al, 2022b). SARS‐CoV‐2 has evolved into several variants since its outbreak in 2019. RshSTT182/200 is also evolving in its natural hosts and a substitution mutation may occur in its RBD. When the D487N substitution occurs, the risk of RshSTT182/200 interspecies transmission to humans will increase. However, the molecular mechanism by which the RshSTT182/200 RBD D487N substitution enhances hACE2 receptor binding is still not fully elucidated. The structure of RshSTT182/200 RBD D487N in complex with hACE2 needs to be determined in the future.
The deletion of amino acids in the β4β5 loop of the RshSTT182/200 RBD is also observed in RsYN04/RmYN05/RmYN08, RaTG15, and BM48‐31 RBDs (Drexler et al, 2010; Guo et al, 2021; Zhou et al, 2021). Similar to RshSTT182/200, the RsYN04/RmYN05/RmYN08 RBD can bind to hACE2 with a significantly lower binding affinity than the SARS‐CoV‐2 RBD (Zhou et al, 2021). Structural analysis revealed that this deletion shortens the β4β5 loop and then breaks the H‐bonds of G446 and Y449 in the β4β5 loop of the RBD with D38 and Q42 of hACE2. In addition, the conformation of this loop was influenced by T346 and Y496. After the deletion was repaired, T346 and Y496 were substituted with R346 and G496, and the binding affinity of the RshSTT182/200 RBD for hACE2 increased significantly. These data indicate that this deletion may not completely abolish the binding of the RBD to hACE2 but substantially decreases binding. Whether this deletion narrows the binding spectrum of the RBD requires further study.
The SARS‐CoV and MERS‐CoV epidemics and the SARS‐CoV‐2 pandemic indicate that CoVs are potential threats to humans. Screening for broadly neutralizing antibodies is an effective way to treat infectious diseases caused by new CoVs. Our previous work indicates that serum samples from COVID‐19 convalescents can efficiently cross‐neutralize RaTG13 (Liu et al, 2021a). Herein, we found that SARS‐CoV‐2 antibodies in COVID‐19 convalescents and vaccinees sera also cross‐neutralize RshSTT182. It indicates that there are some broadly neutralizing antibodies in sera of COVID‐19 convalescents and vaccinees that prevent other CoV infections. A MAb that recognizes the core region of the SARS‐CoV‐2 RBD (S309) is better at cross‐binding the RshSTT182/200 RBD than MAbs that bind to the RBM (BD‐604, CB6, P2C‐1F11, REGN10933, BD‐368‐2, P2B‐2F6, and REGN10987). Structural analysis indicates that the core region of the SARS‐CoV‐2 RBD is a conserved region that is a good target for cross‐neutralizing MAb screening.
In summary, our data indicate that RshSTT182/200 has a broad receptor‐binding spectrum and revealed the molecular basis for RshSTT182/200 RBD binding to hACE2. Moreover, we found that the binding affinities between the RshSTT182/200 RBD and ACE2 orthologs are generally lower than SARS‐CoV‐2, indicating that RshSTT182/200 requires further evolution to obtain strong interspecies transmission abilities like SARS‐CoV‐2.
Materials and Methods
Gene cloning
The coding sequence of the RBD of the RshSTT182/200 spike protein (residues 306–523, GISAID: EPI_ISL_852604 and EPI_ISL_852605) with a 6 × HisTag was synthesized with codon optimization and cloned into the EcoRI and BglII restriction sites of the pCAGGS vector. The RshSTT182/200 RBD mutants were also cloned into the pCAGGS vector. Recombinant proteins were used for crystallization screening, SPR assays, flow cytometry, or ELISAs.
The pCAGGS plasmids containing the sequence of the SARS‐CoV‐2 RBD (residues 319–541, GISAID: EPI_ISL_402119), SARS‐CoV‐2 NTD (residues 20–305, GISAID: EPI_ISL_402119), hACE2 tagged with the Fc domain of human IgG1 (hFc) (residues 19–615, accession number: NP_001358344.1), and the pFastBac plasmids containing the ectodomains of hACE2 (residues 19–615, accession number: NP_001358344.1) were constructed as previously described (Wang et al, 2020; Niu et al, 2021; Zheng et al, 2022).
The coding sequences of full‐length ACE2 orthologs from 39 animals and their extracellular domain fused with the Fc domain of mouse IgG (mFc) were synthesized and cloned into the pEGFP‐N1 and pCAGGS expression vectors, respectively, as previously reported (Wu et al, 2020a; Han et al, 2021; Ma et al, 2021; Niu et al, 2021).
The variable regions of eight MAb heavy chains were cloned into the pCAGGS‐ heavy vector fused to the constant region of IgG1. The variable regions of eight MAb light chains were cloned into the pCAGGS‐kappa/lambda vector fused to the constant region of light chains. The above sequences were cloned into the SacI and KpnI restriction sites.
Protein expression and purification
The SARS‐CoV‐2 RBD, SARS‐CoV‐2 NTD, hACE2‐hFC, RshSTT182/200 RBD, and their mutants were expressed in Expi293F™ Cells (Thermo, A14527) using Sinofection Transfection Reagent (Sino Biological Inc., Cat: STF02). These pCAGGS plasmids were transiently transfected into cells at a density of 2 × 106 cells/ml, and the cells were cultured in an incubator with appropriate culture conditions (37°C; 5% CO2). Cell culture supplement (Sino Biological Inc., Cat: M293‐SUPI) was added to the medium on the 1st and 3rd day after transfection. On the 5th day, the culture supernatant was centrifuged, filtered through a 0.22 μm membrane, and purified using a His‐Trap Excel column (GE Healthcare). Finally, the protein eluted from the His column was further purified by size exclusion using a SuperdexTM200 Increase 10/300 GL column (GE Healthcare) in protein (20 mM Tris–HCl and 150 mM NaCl, pH 8.0) or PBS buffer (1.8 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4).
The hACE2 protein was expressed using the Bac‐to‐Bac baculovirus expression system (Invitrogen), as described in our previous work (Wang et al, 2020). Briefly, the recombinant baculovirus containing the corresponding hACE2‐coding sequence was amplified in Sf9 cells, and the protein was expressed in Hi5 cells. The expression supernatant was collected, centrifuged, and filtered 48 h post‐infection. The protein was purified by affinity chromatography using a HisTrap HP 5 ml column (GE Healthcare) and further purified by size exclusion chromatography using a Superdex 200 column (GE Healthcare) to obtain a uniform preparation. The protein was stored in a buffer (20 mM Tris–HCl and 150 mM NaCl buffer, pH 8.0).
The mFc‐fused ACE2 plasmid was transiently transformed into HEK293T cells (ATCC, CRL‐3216). After 48 h, the culture supernatant was filtered through 0.22 μm membranes and concentrated approximately 25 times. The plasmids of the light and heavy chains of an antibody were co‐transfected into HEK293T cells at a ratio of 3:2, and the culture supernatant was collected after 48 h and centrifuged. These protein supernatants were used for SPR experiments.
Flow cytometry
The ACE2‐pEGFP‐N1 plasmid was transfected into BHK cells (ATCC, CCL‐10) using polyethylenimine (Alfa). When cell fluorescence could be observed with a fluorescence microscope within 24–48 h, the cells were collected, washed twice with PBS, and distributed into 96‐well plates. The cells were incubated with test proteins (SARS‐CoV‐2 RBD, SARS‐CoV‐2 NTD, and RshSTT182/200 RBD with histidine tag) at a concentration of 30 μg/ml at 37°C for 30 min. Then, the cells were washed three times with PBS and stained with anti‐His/APC antibody (1:500, Miltenyi Biotec) at 37°C for 30 min. Finally, the cells were analyzed using a BD FACSCanto (BD Biosciences) after washing them three times with PBS. All data were analyzed using FlowJo V10.
SPR analysis
The binding affinity of the RshSTT182/200 RBD for ACE2‐mFC was measured by SPR in single‐cycle mode using a BIAcore® 8K (GE Healthcare). First, anti‐mIgG antibody (GE Healthcare, Cat: BR100836) was immobilized in flow cells 1 and 2 of the CM5 biosensor chip (GE Healthcare) through an amine coupling reaction. Subsequently, the indicated mFc‐fused protein concentrated supernatant was captured on flow cell 2 with approximately 150–800 response units. Flow cell 1 was used as a negative control. The serially diluted RshSTT182/200 RBD or its mutants then flowed through the chip, and the real‐time response was recorded. Finally, the chip channels were regenerated to remove the ACE2‐mFC ligand using glycine (pH 1.7) after each reaction. The interactions between the RshSTT182/200/200 RBD and antibodies (or RshSTT182/200 RBD mutants and hACE2‐hFC) were measured under the same conditions via direct capture using a Protein A chip.
The running buffer was PBST (1.8 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, and 0.05% (v/v) Tween 20, pH 7.4) in the SPR assay. The equilibrium dissociation constants (binding affinity, K D) of the interactions were calculated using BIAcore® 8K evaluation software (GE Healthcare). The K D values were calculated using the 1:1 (Langmuir) binding model, while the K D values of some weak interactions were calculated using the steady‐state affinity model (shown in Figs EV3A and EV4B) to make the values more accurate. Graphics were prepared using Origin2018.
Serum samples
Serum samples of convalescents were provided by Ditan Hospital, Beijing, China. Serum samples of vaccinees were from volunteers who received the third immunization with the SARS‐CoV‐2 protein subunit vaccines, ZF2001®. Studies were approved by the Ethics Committee of the Institute of Microbiology, Chinese Academy of Sciences (Project Number: SQIMCAS2021149). All candidates signed the written informed consent.
ELISA
After dilution with 0.05 M carbonate–bicarbonate buffer (pH 9.6), the RshSTT182/200 and SARS‐CoV‐2 RBDs were coated in 96‐well ELISA plates (Corning, USA) at 200 ng per well overnight at 4°C. The protein solution was then removed, and the wells were blocked with 5% skim milk in PBST at 37°C for 1 h. After washing with PBST using a plate washer, serum samples at a twofold serial dilution (starting from 1:200) were added to the wells at 37°C for 1 h. PBST was used as a negative control. After washing with PBST, goat anti‐human IgG/HRP (ZSGB‐BIO) diluted to 1:3,000 with PBST was added to the wells and incubated at 37°C for 1 h. After washing again with PBST, 3,3′,5,5′‐tetramethylbenzidine substrate (Beyotime Biotechnology) was added to form a colored product. After 10 min, the reaction was stopped with 2 M HCl, and the absorbance of the colored product was measured by spectrophotometry at 450 nm. Bar charts were generated using GraphPad Prism 8.
Pseudovirus assay
The construction of VSV‐ΔG‐GFP‐based SARS‐CoV‐2 and RshSTT182 pseudoviruses is described in our previous work (Zhao et al, 2021). The pCAGGS vector was used to construct recombinant plasmids for codon‐optimized spike proteins of SARS‐CoV‐2 prototype (Wuhan‐1 reference strain) or RshSTT182 and its mutant, with an 18 amino acid truncation at the C‐terminus of the spike protein. Then, 40 μg of the construct was transfected into HEK 293T cells to prepare corresponding packaging cell lines of each strain. VSV‐ΔG‐G‐GFP pseudovirus was added 24 h after the transfection. After 2 h of infection, VSV‐ΔG‐GFP was removed by changing the medium into fresh complete DMEM medium with anti‐VSV‐G antibody (I1HybridomaATCC® CRL2700™). Supernatants were collected after another 30 h of incubation, passed through a 0.45 μm filter (Millipore, Cat#SLHP033RB), aliquoted, and stored at −80°C. The RshSTT182 supernatants were concentrated 20 times.
For the infection assays, the pseudovirus particles were first normalized to the same amounts by qRT–PCR quantitation. The pseudoviruses were treated with 0.5 U/ml BaseMuncher endonuclease (Abcam, ab270049) for 1 h at 37°C to remove nucleic acid before quantitation. Viral RNA was extracted (Bioer Technology, Cat# BYQ6.6.101711‐213) and quantified by quantitative RT–PCR (qPCR) using a 7500 fast Real‐Time PCR System (Applied Biosystems) with the primers and probe for detecting the L gene of VSV. Then, equivalent amounts of pseudovirus were added to each well of 96‐well plates containing the HeLa‐ACE2 cell line. HeLa cells were used as a control. Plates were incubated 15–18 h post‐infection. The numbers of fluorescent cells and imaging were determined on a CQ1 confocal image cytometer (Yokogawa). Each group contains five parallel experiments.
For the neutralization assays, heat‐inactivated (56°C for 30 min) sera samples were threefold serially diluted starting from 1:10. Pseudovirus was incubated with the sera in a 1:1 ratio at 37°C for 1 h, and the mixture was then added to pre‐plated HeLa‐R. affinis ACE2 cells in 96‐well plates. The numbers of fluorescent cells were calculated on a CQ1 confocal image cytometer (Yokogawa) after 15–18 h of incubation. Software provided with the CQ1 confocal image cytometer was used to calculate the IC50 of the neutralization assay, and statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software Inc.).
Crystallization
The RshSTT182/200 RBD was mixed with hACE2 at a molar ratio of 2:1 and incubated overnight on ice. The mixture was then subjected to gel filtration (Superdex 200 column, GE Healthcare) in protein buffer (20 mM Tris–HCl and 50 mM NaCl buffer, pH 8.0). The RBD/hACE2 complex was fractionated and concentrated to 5 and 10 mg/ml. Crystallization screening was performed by the sitting‐drop method with 1 μl protein mixing with 1 μl reservoir solution at 4 or 18°C. Commercially available kits were used for the initial crystallization screening. High‐resolution crystals were finally obtained in a solution comprised of 0.01 M sodium borate (pH 8.5) and 1.0 M sodium citrate (Kit MD1‐20, Molecular Dimensions) for the RshSTT182/200 RBD/hACE2 complex, 0.1 M MES (pH 6.0), 0.2 M magnesium chloride, and 20% (w/v) PEG 6000 (Kit MD1‐29, Molecular Dimensions) for the RshSTT182/200 RBD‐insert2/hACE2 complex and 0.2 M imidazole malate (pH 7.0) and 20% (w/v) PEG 4000 (Kit MD1‐20, Molecular Dimensions) for the RshSTT182/200 RBD‐insert2‐T346R‐Y496G/hACE2 complex.
Data collection and structure determination
A reservoir solution supplemented with 20% (v/v) glycerol was prepared as a cryoprotectant anti‐freezing buffer for frozen crystals. The crystals were picked from the plate groove using a mini loop and then soaked in the anti‐freezing buffer for a few seconds. Subsequently, the crystals were collected and exposed to soaking in liquid nitrogen for freezing. All diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) BL02U1. The dataset was processed using HKL2000 software (Otwinowski & Minor, 1997). The complex structures were determined by the molecular replacement method using Phaser (Read, 2001) with the previously reported SARS‐CoV‐2 RBD/hACE2 complex structures (PDB: 6LZG). The atomic models were completed with Coot (Emsley & Cowtan, 2004), refined with phenix.refine in Phenix (Adams et al, 2010), and the stereochemical qualities of the ultimate models were assessed using MolProbity45 (Williams et al, 2018). Data collection and structure refinement statistics are summarized in Table 1. All of the structural figures were generated using PyMol (https://pymol.org/2/).
Author contributions
Yu Hu: Data curation; formal analysis; validation; investigation; visualization; methodology; writing – original draft; writing – review and editing. Kefang Liu: Conceptualization; funding acquisition; visualization; writing – original draft; writing – review and editing. Pu Han: Formal analysis; visualization. Zepeng Xu: Writing – review and editing. Anqi Zheng: Investigation. Xiaoqian Pan: Investigation. Yunfei Jia: Investigation. Chao Su: Investigation. Lingfeng Tang: Investigation; writing – review and editing. Lili Wu: Formal analysis; investigation. Bin Bai: Investigation. Xin Zhao: Resources; methodology. Di Tian: Resources. Zhihai Chen: Resources. Jianxun Qi: Formal analysis; supervision; validation. Qihui Wang: Conceptualization; supervision; funding acquisition; methodology; writing – review and editing. George F Gao: Conceptualization; supervision; funding acquisition; project administration; writing – review and editing.
Disclosure and competing interests statement
GFG is an editorial advisory board member.
Supporting information
Expanded View Figures PDF
Source Data for Expanded View
PDF+
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
We thank Yuanyuan Chen, Zhenwei Yang (Institute of Biophysics, Chinese Academy of Sciences), and Zheng Fan (Institute of Microbiology, Chinese Academy of Sciences) for technical help with the BIAcore experiments. We are grateful to Tong Zhao (Institute of Microbiology, Chinese Academy of Sciences) and the Pathogenic Microbiology and Immunology Public Technology Service Center for their support in flow cytometry assays. We thank the staff of the BL02U1 and BL19U1 beamlines at the Shanghai Synchrotron Radiation Facility (SSRF) for their assistance during data collection. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010202 and XDB29040203), the National Natural Science Foundation of China (82225021 and 81922044), and the Ministry of Science and Technology of the People's Republic of China. Kefang Liu was supported by the Postdoctoral Science Foundation of China (2021M700161), Young Elite Scientists Sponsorship Program by CAST (2021QNRC001), and funded by the Open Project Program of CAS Key Laboratory of Pathogen Microbiology and Immunology (CASPMI202204).
The EMBO Journal (2023) 42: e111737
Contributor Information
Jianxun Qi, Email: jxqi@im.ac.cn.
Qihui Wang, Email: wangqihui@im.ac.cn.
George F Gao, Email: gaof@im.ac.cn.
Data availability
The atomic coordinates of the crystal structures of the RshSTT182/200 RBD/hACE2 complex (PDB: 7XBH), RshSTT182/200 RBD‐insert2/hACE2 complex (PDB: 7XBF), and RshSTT182/200 RBD‐insert2‐T346R‐Y496G/hACE2 complex (PDB: 7XBG) were deposited in the Protein Data Bank (www.rcsb.org).
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sun L, Ullah I, Beaudoin‐Bussières G, Anand SP, Hederman AP, Tolbert WD, Sherburn R, Nguyen DN, Marchitto L et al (2022) Engineered ACE2‐Fc counters murine lethal SARS‐CoV‐2 infection through direct neutralization and Fc‐effector activities. Sci Adv 8: eabn4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu‐Sarkodie Y et al (2015) Evidence for an ancestral association of human coronavirus 229E with bats. J Virol 89: 11858–11870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune D, Hul V, Karlsson EA, Hassanin A, Ou TP, Baidaliuk A, Gambaro F, Prot M, Tu VT, Chea S et al (2021) A novel SARS‐CoV‐2 related coronavirus in bats from Cambodia. Nat Commun 12: 6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Gloza‐Rausch F, Glende J, Corman VM, Muth D, Goettsche M, Seebens A, Niedrig M, Pfefferle S, Yordanov S et al (2010) Genomic characterization of severe acute respiratory syndrome‐related coronavirus in European bats and classification of coronaviruses based on partial RNA‐dependent RNA polymerase gene sequences. J Virol 84: 11336–11349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Cao Y, Zhu Q, Yu P, Qi F, Wang G, Du X, Bao L, Deng W, Zhu H et al (2020) Structurally resolved SARS‐CoV‐2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell 183: 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gao GF, Wang L (2021) COVID‐19 expands its territories from humans to animals. China CDC Wkly 3: 855–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes LG, Durigon EL, Campos AA, Hein N, Passos SD, Jerez JA (2011) Coronavirus HKU1 in children, Brazil, 1995. Emerg Infect Dis 17: 1147–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ et al (2003) Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302: 276–278 [DOI] [PubMed] [Google Scholar]
- Guo H, Hu B, Si H‐R, Zhu Y, Zhang W, Li B, Li A, Geng R, Lin H‐F, Yang X‐L (2021) Identification of a novel lineage bat SARS‐related coronaviruses that use bat ACE2 receptor. Emerg Microbes Infect 10: 1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Su C, Zhang Y, Bai C, Zheng A, Qiao C, Wang Q, Niu S, Chen Q, Zhang Y et al (2021) Molecular insights into receptor binding of recent emerging SARS‐CoV‐2 variants. Nat Commun 12: 6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Li L, Liu S, Wang Q, Zhang D, Xu Z, Han P, Li X, Peng Q, Su C et al (2022) Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS‐CoV‐2. Cell 185: 630–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Wu L, Zheng A, Xie Y, He Q, Rong X, Han P, Du P, Han P, Zhang Z et al (2022) Atlas of currently available human neutralizing antibodies against SARS‐CoV‐2 and escape by Omicron sub‐variants BA.1/BA.1.1/BA.2/BA.3. Immunity 55: 1501–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE et al (2012) Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86: 12816–12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Xie X, Bailey AL, Kalveram B, Lokugamage KG, Muruato A, Zou J, Zhang X, Juelich T, Smith JK et al (2021) Loss of furin cleavage site attenuates SARS‐CoV‐2 pathogenesis. Nature 591: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SY, Wang P, Mok BW, Zhang AJ, Chu H, Lee AC, Deng S, Chen P, Chan KH, Song W et al (2020) Attenuated SARS‐CoV‐2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect 9: 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li W, Farzan M, Harrison SC (2005) Structure of SARS coronavirus spike receptor‐binding domain complexed with receptor. Science 309: 1864–1868 [DOI] [PubMed] [Google Scholar]
- Li L, Han P, Huang B, Xie Y, Li W, Zhang D, Han P, Xu Z, Bai B, Zhou J et al (2022a) Broader‐species receptor binding and structural bases of Omicron SARS‐CoV‐2 to both mouse and palm‐civet ACE2s. Cell Discov 8: 65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liao H, Meng Y, Li W, Han P, Liu K, Wang Q, Li D, Zhang Y, Wang L et al (2022b) Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS‐CoV‐2 sub‐variants BA.2 and BA.1.1. Cell 185: 2952–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Pan X, Li L, Yu F, Zheng A, Du P, Han P, Meng Y, Zhang Y, Wu L et al (2021a) Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell 184: 3438–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Tan S, Niu S, Wang J, Wu L, Sun H, Zhang Y, Pan X, Qu X, Du P et al (2021b) Cross‐species recognition of SARS‐CoV‐2 to bat ACE2. Proc Natl Acad Sci USA 118: e2020216118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hu Y, Xia B, Du P, Wu L, Liang M, Chen Q, Yan H, Gao GF, Wang Q et al (2021) Machine learning approach effectively predicts binding between SARS‐CoV‐2 spike and ACE2 across mammalian species—worldwide, 2021. China CDC Wkly 3: 967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MA, Corman VM, Jores J, Meyer B, Younan M, Liljander A, Bosch BJ, Lattwein E, Hilali M, Musa BE et al (2014) MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983‐1997. Emerg Infect Dis 20: 2093–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S, Wang J, Bai B, Wu L, Zheng A, Chen Q, Du P, Han P, Zhang Y, Jia Y et al (2021) Molecular basis of cross‐species ACE2 interactions with SARS‐CoV‐2‐like viruses of pangolin origin. EMBO J 40: e107786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pfefferle S, Oppong S, Drexler JF, Gloza‐Rausch F, Ipsen A, Seebens A, Muller MA, Annan A, Vallo P, Adu‐Sarkodie Y et al (2009) Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis 15: 1377–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A et al (2020) Cross‐neutralization of SARS‐CoV‐2 by a human monoclonal SARS‐CoV antibody. Nature 583: 290–295 [DOI] [PubMed] [Google Scholar]
- Read RJ (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr 57: 1373–1382 [DOI] [PubMed] [Google Scholar]
- Ren W, Lan J, Ju X, Gong M, Long Q, Zhu Z, Yu Y, Wu J, Zhong J, Zhang R et al (2021) Mutation Y453F in the spike protein of SARS‐CoV‐2 enhances interaction with the mink ACE2 receptor for host adaption. PLoS Pathog 17: e1010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken CB, Farag EA, Jonges M, Godeke GJ, El‐Sayed AM, Pas SD, Raj VS, Mohran KA, Moussa HA, Ghobashy H et al (2014) Middle East respiratory syndrome coronavirus (MERS‐CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels, Qatar, April 2014. Euro Surveill 19: 20829 [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research 42: W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM (2006) Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol 35: 99–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, He J, Han P, Bai B, Li D, Cao J, Tian M, Hu Y, Zheng A, Niu S et al (2022) Molecular basis of mink ACE2 binding to SARS‐CoV‐2 and its mink‐derived variants. J Virol 96: e0081422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Gu H, Cao L, Chen Q, Ye Q, Yang G, Li RT, Fan H, Deng YQ, Song X et al (2021) Characterization and structural basis of a lethal mouse‐adapted SARS‐CoV‐2. Nat Commun 12: 5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Zhang D, Han P, Kang X, Zheng A, Xu Z, Zhao X, Wang VY, Qi J, Wang Q et al (2022) Structural basis of SARS‐CoV‐2 and its variants binding to intermediate horseshoe bat ACE2. Int J Biol Sci 18: 4658–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chretien D, Sanamxay D et al (2022) Bat coronaviruses related to SARS‐CoV‐2 and infectious for human cells. Nature 604: 330–336 [DOI] [PubMed] [Google Scholar]
- Tong Y, Liu W, Liu P, Liu WJ, Wang Q, Gao GF (2021) The origins of viruses: discovery takes time, international resources, and cooperation. Lancet 398: 1401–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T, Ryan D, Fisher M, Williams D, Dales NA et al (2004) ACE2 X‐ray structures reveal a large hinge‐bending motion important for inhibitor binding and catalysis. J Biol Chem 279: 17996–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, Lim BL et al (2021) Evidence for SARS‐CoV‐2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun 12: 972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY et al (2020) Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell 181: 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB et al (2018) MolProbity: more and better reference data for improved all‐atom structure validation. Protein Sci 27: 293–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chen Q, Liu K, Wang J, Han P, Zhang Y, Hu Y, Meng Y, Pan X, Qiao C et al (2020a) Broad host range of SARS‐CoV‐2 and the molecular basis for SARS‐CoV‐2 binding to cat ACE2. Cell Discov 6: 68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Su J, Niu S, Chen Q, Zhang Y, Yan J, Shi Y, Qi J, Gao GF, Wang Q (2020b) Molecular basis of pangolin ACE2 engaged by COVID‐19 virus. Chin Sci Bull 66: 73–84 [Google Scholar]
- Xu Z, Kang X, Han P, Du P, Li L, Zheng A, Deng C, Qi J, Zhao X, Wang Q et al (2022a) Binding and structural basis of equine ACE2 to RBDs from SARS‐CoV, SARS‐CoV‐2 and related coronaviruses. Nat Commun 13: 3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Liu K, Gao GF (2022b) Omicron variant of SARS‐CoV‐2 imposes a new challenge for the global public health. Biosaf Health 4: 147–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H‐L, Sit THC, Brackman CJ, Chuk SSY, Gu H, Tam KWS, Law PYT, Leung GM, Peiris M, Poon LLM et al (2022) Transmission of SARS‐CoV‐2 delta variant (AY.127) from pet hamsters to humans, leading to onward human‐to‐human transmission: a case study. Lancet 399: 1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367: 1814–1820 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang Y, Liu K, Li Y, Lu Q, Wang Q, Zhang Y, Wang L, Liao H, Zheng A et al (2021) The molecular basis for SARS‐CoV‐2 binding to dog ACE2. Nat Commun 12: 4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Zheng A, Li D, Zhang R, Sun H, Wang Q, Gao GF, Han P, Daia L (2021) Neutralisation of ZF2001‐elicited antisera to SARS‐CoV‐2 variants. Lancet Microbe 2: e494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A, Wu L, Ma R, Han P, Huang B, Qiao C, Wang Q, Tan W, Gao GF, Han P (2022) A binding‐enhanced but enzymatic activity‐eliminated human ACE2 efficiently neutralizes SARS‐CoV‐2 variants. Signal Transduct Target Ther 7: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC et al (2020a) A novel bat coronavirus closely related to SARS‐CoV‐2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 30: 2196–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang X‐L, Wang X‐G, Hu B, Zhang L, Zhang W, Si H‐R, Zhu Y, Li B, Huang C‐L et al (2020b) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Ji J, Chen X, Bi Y, Li J, Wang Q, Hu T, Song H, Zhao R, Chen Y et al (2021) Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS‐CoV‐2 and related viruses. Cell 184: 4380–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R et al (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]


