Abstract
Public Health Genomics (PHG) is a relatively new field. The wide application of genomic technologies played a pivotal role in elucidating the full genomic sequence of the SARS‐CoV‐2 virus. This breakthrough proved to be the starting point in the manufacture of diagnostic kits and the subsequent making of vaccines. Beyond the COVID‐19 pandemic, many African countries can take advantage of the various investments in genomic technologies to introduce and intensify the use of genomics for public health gain. Public Health Genomics effectively monitors, prevents, and manages non‐communicable and infectious diseases. However, there are several challenges to implementing PHG in Africa. In this perspective article, we discuss the utilization of PHG during the COVID‐19 pandemic, the lessons learned from using PHG to manage and contain the COVID‐19 pandemic, as well as potential challenges Africa may face when putting PHG into practice compared to challenges of other regions. We also discuss our recommendations for overcoming these challenges.
Keywords: Africa, benefits, challenges, COVID‐19, Public Health Genomics, recommendations
1. INTRODUCTION
Public Health Genomics (PHG) is a relatively new multidisciplinary field concerned with the effective and responsible translation of genome‐based knowledge and technologies to improve population health. It assesses the impact of genes and their interaction with behavior, diet, and the environment on population health. 1 , 2 Diseases are multifactorial and are influenced by heredity, lifestyle choices, and environmental factors. 3 Genetics and genomics are different from one another in that one gene at a time is usually examined in genetics whereas genomics studies all the genes present in an organism's cells, as well as how they interact with one another and the environment to influence the organism's growth and development. Genetic research into disorders usually concentrates on those that are caused by variations in just one gene, such as Cystic Fibrosis, Muscular Dystrophy, and Fragile X Syndrome. 4 Genomics has made understanding the causes, risks, prevention, diagnosis, and treatment of various diseases easier. 5 Table 1 shows some of the genomic studies that have improved the understanding of diseases.
Table 1.
Genomic studies that have improved the understanding of diseases
| Study | Authors | Reference | The aspect of the disease improved |
|---|---|---|---|
| New insights into the genetic etiology of Alzheimer's disease and related dementias | Bellenguez et al. (2022) | 6 | Etiology |
| The quest to find genes that drive severe COVID | Callaway (2021) | 7 | Risk |
| Integrating lipidomics and genomics: emerging tools to understand cardiovascular diseases | Tabassum and Ripatti (2021) | 8 | Prevention |
| The role of radiogenomics in the diagnosis of breast cancer: a systematic review | Darvish et al. (2022) | 9 | Diagnosis |
| Diagnosis and treatment of type 1 diabetes at the dawn of the personalized medicine era | Akil et al. (2021) | 10 | Treatment |
| Improving prediction performance of colon cancer prognosis based on the integration of clinical and multi‐omics data | Tong et al. (2020) | 11 | Prognosis |
Clinical professionals can use genomics to examine a patient's genes and make a molecular diagnosis. 5 Numerous genetic tests are available for targeted tumor treatment, noninvasive prenatal screening, and the diagnosis of children and uncommon diseases. In Africa, these tests are still not widely accessible. 12 Additionally, genomics enables better knowledge of the roles played by gene expression, the interactions between genes, and how various genetic variants affect the phenotype of disease. 13 New and better treatments have been created as a result of the study of pharmacogenomics, a subfield of genomics. Pharmacogenomics is anticipated to greatly enhance clinicians' capacity to evaluate treatment responses, including how various persons metabolize medications and which individuals are more prone to develop adverse reactions. Clinicians should be able to use genetic profiles to determine the proper medications and dosages for various groups. 14 Although the implementation of PHG has several advantages, it also has some disadvantages. One of the disadvantages is that new‐generation sequencing may generate incidental findings which may be of uncertain value to both patients and clinicians. 15 It is also difficult to convey the results to patients in accurate and easily understandable terms. In addition, individuals receiving positive results may suffer from psychological and social harm. Furthermore, genetic susceptibility test results may potentially be used by insurers and employers to discriminate against those whose results are positive. 15
The National Institutes of Health and the Wellcome Trust established the Human Hereditary and Health in Africa (H3Africa) initiative in 2010 to facilitate the study of genomics to improve the health of Africans. This initiative has been instrumental in providing genomic resources, capacitating laboratories, and training genomics experts in the continent. 16 PHG played a pivotal role in the fight against the Corona Virus disease of 2019 (COVID‐19) pandemic. The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), is the etiological agent of COVID‐19. Apart from airborne and droplet transmission, the virus can be transmitted through contact with contaminated surfaces. 17 We believe that PHG adoption will benefit Africa given the potential benefits of genomics to public health. In this article, we discuss what PHG was used for during the COVID‐19 pandemic and lessons learned from the application of PHG during the COVID‐19 pandemic, the challenges Africa may face when implementing PHG compared to other regions, and our recommendations for overcoming those challenges.
2. METHODOLOGY
For this study, we conducted a literature review of the uses of PHG in general, its history, its use during the COVID‐19 pandemic, the lessons Africa learned from the COVID‐19 pandemic concerning PHG, the challenges to implementing PHG in Africa, and the challenges that were faced by other regions that implemented PHG earlier than Africa. We searched for articles published in English on the WHO website, and peer‐reviewed articles on Google Scholar and PubMed. The keywords we used for the literature search were PHG, Africa, the COVID‐19 pandemic, lessons, and challenges. We used the Boolean operators “AND” and “OR” to retrieve articles that have both terms or either term, thereby delimiting the search. Wildcard symbols and truncation symbols were also used to expand a search term to include all forms of a root word. Truncation symbols were put at the end of the root word while wildcard symbols were put into the middle of the search term to allow for different spellings. Since this study was not a systematic review, we did not appraise the quality of the studies we used.
3. USES OF PHG DURING THE COVID‐19 PANDEMIC
PHG played a variety of roles in the fight against the COVID‐19 pandemic. It was employed in the determination of the causal agent and its source, comprehension of SAR‐CoV‐2 characteristics, surveillance, and the development of molecular diagnostics, treatments, and vaccines. 18 Interindividual variability regarding the incidence, severity and mortality rate of COVID‐19 was recently recorded. Different genetic polymorphisms of specific genes might account for higher susceptibility and unexpected outcomes of COVID‐19 infections in different populations. It was found that both Angiotensin Converting Enzyme 2 (ACE2) and Trans‐membrane protease serine type 2 (TMPRSS2) are playing a crucial role in virus entry into host cells. 19 The role and effect of variants alleles of Human Leukocyte Antigen (HLA) on the severity and incidence of infection with COVID‐19 and its management as well were investigated. HLA‐DQA1 may induce the production of anti‐drug antibodies against anti‐TNF drugs like infliximab and adalimumab, which are used as options in the management of COVID‐19, and therefore might result in management failure. Also, HLA‐B*46:01 carrier individuals were found to be more vulnerable to COVID‐19. 20
Understanding viral entrance processes, host‐virus interactions at the molecular level, and the viral‐specific host components required for infection have all benefited from genomic research. Additionally, PHG was able to give insight into the variations in immune responses between individuals with severe disease and those with mild disease, and the disease's natural history. Being able to define more precisely which populations were most at risk of infection through genomic risk prediction allowed preventative measures to be targeted to the most at‐risk subgroups, thereby significantly impacting population health. 21
Genomic surveillance was used to track SARS‐CoV‐2 variants and their spread. Variants that were of public concern due to their increased transmissibility and increased severity were monitored, and this increased the precision of public health responses since scarce resources could be targeted toward the most virulent variant. 21 The practice of PHG in Africa helped in the discovery of new the Omicron variant in South Africa and Botswana. 22 Genetic epidemiology research located the second wave's origin in Victoria, Australia, which led to the swift implementation of stringent and comprehensive public health measures. 23 In China, SARS‐CoV‐2 monitoring of wastewater allowed for the early detection of the COVID‐19 waves and the successful implementation of public health measures. 24 Phylogenomic analyses were used to track transmission of the virus within hospitals and the contribution of travel‐associated strain introductions to the establishment of the epidemic in certain countries. 25 Genomic sequencing was utilized to identify the virus causing the outbreaks more quickly and accurately, which enabled quick responses to the outbreaks and more effective surveillance. 18
The development of novel COVID‐19 diagnostics, treatments, and vaccines was greatly aided by the integration of genomics with other medical technologies. This came about as a result of improved high‐throughput pathogen sequencing techniques, like the CoronaHiT method for SARS‐CoV‐2. 26 The identification of therapeutic targets for drug development also employed genomic data, and this enabled scientists to develop vaccines to prevent SARS‐CoV‐2 infection. 18
4. LESSONS LEARNED BY AFRICA FROM THE COVID‐19 PANDEMIC
Africa gained important lessons from the use of PHG during the COVID‐19 pandemic. These included the need to collaborate, capacitate its institutions, develop a workforce skilled in genomics, and raise enough funds from local sources to cover the cost of its healthcare. 27 , 28
There were few genetic sequencing resources available on the continent. By the end of November 2020, only 2% of the SARS‐CoV‐2 sequences that were submitted to the Global Initiative on Sharing Avian Influenza Data (GISAID) were from the WHO Africa region. Additionally, the sequences came from 23 different nations in the region, with 56% of them coming from South Africa. 29 African nations also saw a larger collection to submission time gap than other regions. African nations took 50 days to sequence 7000 genomes as of May 2021, compared to 25 days for 590 000 genomes in Europe and 26 days for more than 498 000 genomes in the USA. Africa only sequenced 0.36% of its positive samples, compared to the US sequencing of 1.5% and the UK sequencing of 9.3%. This demonstrated that there was a lack of genome sequencing capabilities on the continent. This highlighted the importance of having a strong national and continental capability for genome sequencing. 28 The antimicrobial resistance (AMR) crisis was exacerbated by COVID‐19, and whole genome sequencing was used to gather data on the early onset and transmission of AMR. This allowed scientists to track the sources of drug‐resistant genes, that is, whether they were from animals, humans, or the environment, and to formulate timely policies to combat AMR. 21
Due to the delays in genome sequencing, new variants spread without detection. The lack of laboratories that could handle COVID‐19 samples, a lack of funding or restrictions on importing reagents and equipment, a lack of genomic experts to analyze the genomes and produce useful results, and the use of older sequencing technologies with low throughput were all blamed for the delays in sequence submission to GISAID. 28 African countries realized the value of producing genomic tools and reagents on the continent since doing so would assist to mitigate supply issues brought on by delays in international trade due to lockdowns. Furthermore, COVID‐19 highlighted the continent's excessive reliance on external donors, who are usually unavailable during pandemics due to obligations in their nations. This could have served as a warning to African nations to invest more in healthcare to have enough laboratory capacity to address disease outbreaks that require genetic sequencing, as well as having the capacity to develop vaccines to mitigate the overreliance on external manufacturers who prioritized HICs in the distribution of the vaccines. 27 Since the sequencing program required the participation of various experts, the continent realized that cooperation between experts with various skills and the mobilization of resources were necessary to produce prompt, precise, and efficient results that could have an impact on public health. Additionally, the continent realized that exchanging sequencing data would maximize the impact of genetic sequencing on its public health response. 30
5. STEPS REQUIRED FOR THE IMPLEMENTATION OF PHG IN DEVELOPING COUNTRIES
Several steps are required for the implementation of PHG in developing countries. Some of the steps include building genomic infrastructure capacity and developing PHG policies; building strategic partnerships with developed countries and engaging communities; building technical capacity; and developing knowledge bases. 31 The steps are summarized in Figure 1.
Figure 1.
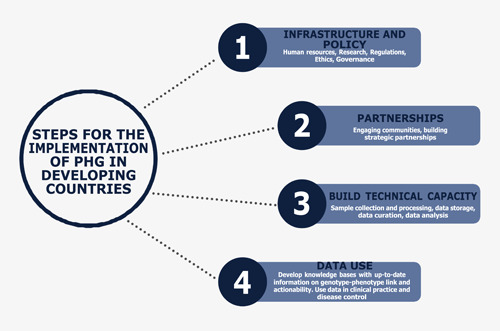
Steps for implementation of PHG in developing countries
6. CHALLENGES TO THE IMPLEMENTATION OF PHG IN AFRICA COMPARED TO OTHER REGIONS
The implementation of PHG in Africa faces many challenges including a lack of genomic technology and computing infrastructure, an absence of genetic banks, high degrees of genetic diversity among the populations, a lack of skilled HCWs, a lack of genomic literacy in the population, and a lack of policies on ethical issues that can be brought about by PHG practice. The challenges will be compared to challenges faced by other regions in the world.
6.1. Lack of genomic technology, infrastructure, and skilled HCWs
Africa has a disproportionately higher disease burden and other health problems than other regions of the world, yet it also has limited access to the majority of emerging genomic technologies that are available in high‐income countries (HICs). Global disparities in the accessibility, quality, and application of genetic and genomic technology are mainly due to a lack of funding and genomic infrastructure, and the presence of more pressing health issues like HIV and TB. 31 Public Health Genomics requires complex data housing infrastructure, a lot of processing power and computing infrastructure, and data analytics which are not available in most African nations. The majority of African nations lack access to the necessary computational tools and bioinformatics infrastructures, such as the Cloud Infrastructure for Microbial Bioinformatics (CLIMB), an online tool for medical microbiologists. 32 As no company manufactures genomic tests in Africa, the reagents and equipment are still expensive. The reagents must be transported under particular circumstances from other continents at a significant expense. 31 Additionally, Africa has a shortage of HCWs with formal expertise in data science and computational genomics. 33 Although the United States has a well‐developed genomic infrastructure, it is still facing several challenges to the implementation of PHG. Some of the challenges include unclear organizational policies or criteria for use, inconsistent modes of integrating genomic information into the electronic health record system, and concerns about the costs of the tests by patients and healthcare institutions. In addition, many HCWs in the USA still lack genomic knowledge. 34 In Hong Kong, the costs of genetic tests are still high, and as a result, only a few selected genetic tests are offered in the public health system. Moreover, there is also a shortage of HCWs trained in genomics. 35
6.2. Lack of genetic banks and the genetic diversity of the population
Population data on genome‐disease associations and genome‐environment interactions form the basis of the benefits of PHG. 36 By 2020, about 2 decades after the office of PHG was established in the USA, only five countries in Africa had started or were planning to start national genomic programs. These countries were South Africa, Zimbabwe, Tunisia, Egypt, and Rwanda. Although many genomic tools and technologies have been developed, a few of them have been translated into healthcare and public health practice. 33 The possible reason for this is that genetic variants that contribute to a particular disease may vary among different people and populations, meaning that their relative significance may also vary. Therefore, the results of research conducted on genomics in HICs cannot be put into practice in African countries because of the differences in the populations. Worsening the situation is also the high degree of genetic diversity and population stratification across Africa. 33 There is also a lack of investment in the infrastructure required to evaluate genomic tools and techniques for validity and utility in the African setting. 31 Although genetic banks are available in Europe, some banks do not share their data and samples with other genetic banks and researchers. This has been reported to result in the unnecessary collection of samples and inefficient genetic research, which negatively impacts the practice of PHG. 37
6.3. Lack of policies on ethical issues related to PHG
The practice of PHG also raises several ethical issues. Since genetic information can be obtained in the absence of clinical symptoms, its use in isolation may have a weaker predictive association with health outcomes compared to other health information. There is a potential to under‐, over‐, or misdiagnose people. In addition, PHG may also lead to psychosocial harm. 38 Uncovering genomic determinants of health have implications not only for the individual tested but for genetic relatives as well. It is also challenging to determine conditions that will need to be screened in a particular population and who should be tested. Ethical issues such as patient autonomy and privacy, consent to participate in sequencing and storage of personal data, and access to genomic information considering some of the information's limitations in predictive value and clinical utility will also need to be addressed. 15 Although most countries in Asia have PHG regulations, the regulations are inconsistent. While some countries like Japan and Singapore have regulations to protect personal information, China and Indonesia have little control over privacy issues. As a result, this may affect collaboration among them, affecting the successful implementation of PHG in the region. 39
6.4. Lack of genomic literacy in the population
Considering that most of the people in the continent lack genomic literacy, it will be difficult to convince them that these genomic tests are useful, especially if they are not sick, or in situations where they will not be able to get treatment for the diseases due to high costs or unavailability of treatment options. 40 There is still a lack of understanding of genetic testing and the implication of the results among patients in the USA, more than 2 decades after the country started implementing PHG. 35
7. RECOMMENDATIONS TO ADDRESS THE CHALLENGES
To address challenges to PHG implementation in Africa, we recommend prioritization of a few diseases in the initial stages of implementation, collaboration with HICs and academic institutions, increased health funding, the introduction of PHG courses for HCWs, community engagement, and dissemination of PHG information, and recognizing PHG as a medical specialty.
7.1. Prioritization of a few diseases
Given that most African countries have limited financial resources to implement health programs, we recommend that they prioritize a few diseases when they start PHG programs. Prioritization of diseases should be based on the disease burden in the population, the availability of behavioral and environmental interventions to reduce the risk of the disease, the cost‐effectiveness of a range of clinical and environmental interventions for the diseases, and the availability and accessibility of treatment options. 31 Family health history may also be an important but cheap source of information to identify people at risk of a particular disease who may require genetic testing. Diseases that should be prioritized are infectious diseases such as influenza, tropical diseases, and AIDS; chronic diseases like diabetes, obesity, cancer, and cardiovascular diseases; and birth defects like hemoglobinopathies, Down's syndrome, neural tube defects, congenital heart defects, and cleft lip and palate. 41
7.2. Collaboration, increased funding, and research
PHG can potentially reduce global health inequalities. This is because it can provide African countries with efficient, cost‐effective, and robust means of preventing, diagnosing, and treating major diseases that affect their populations. 42 However, to achieve this, genomic health disparities must be reduced through equitable economic investment, clinical research, and the provision of genomic services and technologies globally. This can be achieved by the exchange of genomic information, expertise, and technologies between HICs and African countries. Exchange programs for healthcare workers should be organized so that African healthcare workers acquire skills in PHG from HICs who have more experience in PHG. Collaborations between HICs and African countries should be encouraged as this may allow African countries to access bioinformatics infrastructures such as CLIMB.
Since the success of PHG requires population‐based data, African countries should work together with academic institutions and other stakeholders so that research is carried out on the genomics of their populations. African countries should also increase funding for health to support the development of infrastructure for conducting genomic‐related population research. Some of the research costs can also be shared among countries since some populations in neighboring countries share the same genetic traits.
7.3. Training of a genomic workforce, improving genomic literacy, and recognizing PHG as a medical specialty
Considering that most HCWs and the general population in Africa have limited genomic literacy, we recommend that genomics is taught in schools and colleges. Courses in Genomics and Bioinformatics should be introduced from diploma to doctorate levels. This will ensure that literacy on genomics improves in the continent. There should be a restructuring of existing healthcare workers' training curricula to help develop competencies that are specific to Africa. Furthermore, genomics education can also be incorporated into continuous professional development activities for HCWs. Policymakers in the continent should also be lobbied to recognize PHG as a specialty field and to come up with ethical policies that govern the practice of PHG.
7.4. Community engagement and information dissemination
For PHG to be accepted by the population, countries should make sure that there is community engagement and participation during PHG policy formulation. Objective evaluation of the potential benefits against the potential harms of PHG should be carried out transparently so that communities can support the programs. Furthermore, guidelines on PHG should take into consideration the religions and cultures of the different populations. Information on genomics can also be distributed through mass media and other platforms.
8. CONCLUSION
Public Health Genomics is a relatively new discipline in medicine compared to other medical specialties. Its benefits in the surveillance, prevention, and control of communicable diseases like COVID‐19 are well documented. PHG is also important for the prevention of birth defects. However, its implementation in Africa has been slow due to a lack of financial resources, infrastructure, genetic literacy, shortage of a professionally trained genomic workforce, and little genetic research. We, therefore, recommend the exchange of genetic knowledge and techniques between HICs and African countries, training of a genomic workforce, recognition of PHG as a specialty field, and prioritization of a few diseases during the initial phases of PHG programs' implementation.
AUTHOR CONTRIBUTIONS
Enos Moyo: Conceptualization; writing original draft. Perseverance Moyo: Conceptualization, writing original draft. Tapfumanei Mashe: Writing review and editing. Mathias Dzobo: Writing review and editing. Itai Chitungo: Writing review and editing. Tafadzwa Dzinamarira: Conceptualization; supervision; writing review and editing.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Moyo E, Moyo P, Mashe T, Dzobo M, Chitungo I, Dzinamarira T. Implementation of Public Health Genomics in Africa: lessons from the COVID‐19 pandemic, challenges, and recommendations. J Med Virol. 2022;95:e28295. 10.1002/jmv.28295
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no data sets were generated or analysed during the current study.
REFERENCES
- 1. Bartoshesky LE. Public health genetics/genomics. Dela J Public Health. 2021;7(5):4‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zimmern R, Stewart A. 2006. public health genomics: origins and basic concepts. Ital J Public Health. 2006;3(3–4):9‐15. [Google Scholar]
- 3. Stolk RP, Rosmalen JG, Postma DS, et al. Universal risk factors for multifactorial diseases. Eur J Epidemiol. 2008;23:67‐74. [DOI] [PubMed] [Google Scholar]
- 4. Marchant G, Barnes M, Evans JP, LeRoy B, Wolf SM. From genetics to genomics: facing the liability implications in clinical care. J Law Med Ethics. 2020;48(1):11‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCormick K, Calzone K. The impact of genomics on health outcomes, quality, and safety. Nurs Manage. 2016;47(4):23‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellenguez C, Küçükali F, Jansen IE, et al. New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat Genet. 2022;54:412‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Callaway E. The quest to find genes that drive severe COVID. Nature. 2021;595:346‐348. [DOI] [PubMed] [Google Scholar]
- 8. Tabassum R, Ripatti S. Integrating lipidomics and genomics: emerging tools to understand cardiovascular diseases. Cell Mol Life Sci. 2021;78:2565‐2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Darvish L, Bahreyni‐Toossi MT, Roozbeh N, Azimian H. The role of radiogenomics in the diagnosis of breast cancer: a systematic review. Egypt J Med Hum Genet. 2022;23:99. 10.1186/s43042-022-00310-z [DOI] [Google Scholar]
- 10. Akil AAS, Yassin E, Al‐Maraghi A, et al. Diagnosis and treatment of type 1 diabetes at the dawn of the personalized medicine era. J Transl Med. 2021;19:137. 10.1186/s12967-021-02778-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tong D, Tian Y, Zhou T, et al. Improving prediction performance of colon cancer prognosis based on the integration of clinical and multi‐omics data. BMC Med Inform Decis Mak. 2020;20(1):22. 10.1186/s12911-020-1043-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lumaka A, Carstens N, Devriendt K, et al. Increasing African genomic data generation and sharing to resolve rare and undiagnosed diseases in Africa: a call‐to‐action by the H3Africa rare diseases working group. Orphanet J Rare Dis. 2022;17(230):1‐6. 10.1186/s13023-022-02391-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mattick JS, Dziadek MA, Terrill BN, Kaplan W, Spigelman AD, Bowling FG. The impact of genomics on the future of medicine and health. Med J Aust. 2014;201(1):17‐20. [DOI] [PubMed] [Google Scholar]
- 14. Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet. 2019;394(10197):521‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roberts J, Dolinoy D, Tarini B. Emerging issues in public health genomics. Annu Rev Genomics Hum Genet. 2014;15:461‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adebamowo SN, Francis V, Tambo E, et al. Implementation of genomics research in Africa: challenges and recommendations. Glob Health Action. 2018;11(1):1419033. 10.1080/2F16549716.2017.1419033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. PriyankaChoudhary OP, Singh I, Patra G. Aerosol transmission of SARS‐CoV‐2: the unresolved paradox. Travel Med Infect Dis. 2020;37:101869. 10.1016/2Fj.tmaid.2020.101869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saravanan KA, Panigrahi M, Kumar H, et al. Role of genomics in combating COVID‐19 pandemic. Gene. 2022;823:146387. 10.1016/2Fj.gene.2022.146387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alshahawey M, Raslan M, Sabri N. Sex‐mediated effects of ACE2 and TMPRSS2 on the incidence and severity of COVID‐19; The need for genetic implementation. Curr Res Transl Med. 2020;68(4):149‐150. 10.1016/2Fj.retram.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raslan MA, Alshahawey M, Shehata EM, Sabri NA. Does human leukocyte antigen gene polymorphism affect management of COVID‐19 patients? A review article. Scientific J Genet Gene Ther. 2020;6(1):001‐003. 10.17352/sjggt.000018 [DOI] [Google Scholar]
- 21. Khoury M, Holt K. The impact of genomics on precision public health: beyond the pandemic. Genome Med. 2021;13(67):1‐4. 10.1186/s13073-021-00886-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baberjee I, Robinson J, Banerjee I, Sathian B. Omicron: the pandemic propagator and lockdown instigator—what can be learnt from South Africa and such discoveries in future. Nepal J Epidemiol. 2021;11(4):1126‐1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lane CR, Sherry NL, Porter AF, et al. Genomics‐informed responses in the elimination of COVID‐19 in Victoria, Australia: an observational, genomic epidemiological study. Lancet Public Health. 2021; 6(8):547‐556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deng Y, Xu X, Zheng X, et al. Use of sewage surveillance for COVID‐19 to guide public health response: a case study in Hong Kong. Sci Total Environ. 2022;821:153250. 10.1016/j.scitotenv.2022.153250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lucey M, Macori G, Mullane N, et al. Whole‐genome sequencing to track severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) transmission in nosocomial outbreaks. Clin Infect Dis. 2021;72(1):727‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baker DJ, Aydin A, Le‐Viet T, et al. CoronaHiT: high‐throughput sequencing of SARS‐CoV‐2 genomes. Genome Med. 2021;13(1):21. 10.1186/s13073-021-00839-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adepoju P. Challenges of SARS‐CoV‐2 genomic surveillance in Africa. Lancet Microbe. 2021;2(4):E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalia K, Saberwal G, Sharma G. The lag in SARS‐CoV‐2 genome submissions to GISAID. Nat Biotechnol. 2021;39:1058‐1060. [DOI] [PubMed] [Google Scholar]
- 29. Lu L, Lycett S, Ashworth J, Mutapi F, Woolhouse M. What are SARS‐CoV‐2 genomes from the WHO Africa region member states telling us? BMJ Glob Health. 2021;6(1):e004408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. World Health Organization . SARS‐CoV‐2 genomic sequencing for public health goals: interim guidance. 2021. Accessed August 29, 2022. https://apps.who.int/iris/handle/10665/338483
- 31. The African Academy of Sciences . Policy paper: A framework for the implementation of Genomic Medicine for Public Health in Africa. 2021. Accessed August 31, 2022. https://www.aasciences.africa/publications/policy-paper-framework-implementation-genomic-medicine-public-health-africa
- 32. Connor TR, Loman NJ, Thompson S, et al. CLIMB (the Cloud Infrastructure for Microbial Bioinformatics): an online resource for the medical microbiology community. Microb Genom. 2016;2(9):e000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jongeneel VC, Kotze MJ, Bhaw‐Luximon A, et al. A view on genomic medicine activities in Africa: implications for policy. Front Genet. 2022;13:769919. 10.3389/2Ffgene.2022.769919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sperber NR, Carpenter JS, Cavallari LH, et al. Challenges and strategies for implementing genomic services in diverse settings: experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med Genomics. 2017;10:35. 10.1186/s12920-017-0273-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chu AT, Fung JL, Tong AH, et al. Potentials and challenges of launching the pilot phase of Hong Kong genome project. J Transl Genet Genom. 2022;6:290‐303. [Google Scholar]
- 36. Pang T, Oestergaard M. Creating policy frameworks for public health genomics to benefit developing countries. Per Med. 2014;11(5):487‐496. 10.2217/pme.14.37 [DOI] [PubMed] [Google Scholar]
- 37. Tupasela A. Data hugging in European Biobank Networks. Sci Cult. 2021;30(4):513‐534. [Google Scholar]
- 38. Prince A, Berkman B. Reconceptualizing harms and benefits in the genomic age. Per Med. 2018;15(5):419‐428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen H, Pang T. Human Genomics in Asia. International Encyclopedia of the Social & Behavioral Sciences. Vol. 11. 2nd ed. 2015:318. 10.1016/2FB978-0-08-097086-8.82041-3 [DOI] [Google Scholar]
- 40. Mboowa G, Sserwadda I. Role of genomics literacy in reducing the burden of common genetic diseases in Africa. Mol Genet Genomic Med. 2019;7(7):e00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zimmern R, Khoury M. The impact of genomics on public health practice: the case for change. Public Health Genom. 2012;15(3‐4):118‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pang T. The impact of genomics on global health. Am J Public Health. 2002;92(7):1077‐1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no data sets were generated or analysed during the current study.


