Abstract
To control the ongoing COVID‐19 pandemic, a variety of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) vaccines have been developed. However, the rapid mutations of SARS‐CoV‐2 spike (S) protein may reduce the protective efficacy of the existing vaccines which is mainly determined by the level of neutralizing antibodies targeting S. In this study, we screened prevalent S mutations and constructed 124 pseudotyped lentiviral particles carrying these mutants. We challenged these pseudoviruses with sera vaccinated by Sinovac CoronaVac and ZF2001 vaccines, two popular vaccines designed for the initial strain of SARS‐CoV‐2, and then systematically assessed the susceptivity of these SARS‐CoV‐2 variants to the immune sera of vaccines. As a result, 14 S mutants (H146Y, V320I + S477N, V382L, K444R, L455F + S477N, L452M + F486L, F486L, Y508H, P521R, A626S, S477N + S698L, A701V, S477N + T778I, E1144Q) were found to be significantly resistant to neutralization, indicating reduced protective efficacy of the vaccines against these SARS‐CoV‐2 variants. In addition, F486L and Y508H significantly enhanced the utilization of human angiotensin‐converting enzyme 2, suggesting a potentially elevated infectivity of these two mutants. In conclusion, our results show that some prevalent S mutations of SARS‐CoV‐2 reduced the protective efficacy of current vaccines and enhance the infectivity of the virus, indicating the necessity of vaccine renewal and providing direction for the development of new vaccines.
Keywords: infectivity, mutation, neutralization, SARS‐CoV‐2, spike, vaccines
1. INTRODUCTION
As of May 2022, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), the causative agent of COVID‐19, has caused over 513 million infections and more than 6.2 million deaths worldwide. 1 SARS‐CoV‐2, as a member of the genus Betacoronavirus, is an enveloped single positive‐stranded RNA virus with four structural proteins that are spike (S), envelope (E), membrane (M), and nucleocapsid (N). 2 The S protein binds to angiotensin‐converting enzyme 2 (ACE2) on the host cell surface and mediates viral internalization. 3 , 4 During the spread of SARS‐CoV‐2, S protein undergoes rapid mutation which may alter the infectivity and transmissibility of SARS‐CoV‐2 variants. 5 As S protein is the primary antigen inducing protective immune response, it is the major target for the development of SARS‐CoV‐2 vaccines, and thus mutations in S may affect the protective efficacy of the vaccines. 6 Therefore, it is critical to closely monitor the antigenic variation of SARS‐CoV‐2 S.
SARS‐CoV‐2 displays particularly high mutation rates as an RNA virus, resulting in an abundance of mutants in its genome. 7 , 8 Over 20,000 mutations have been detected in SARS‐CoV‐2 genome in the past 2 years. 9 During the early stage of the COVID‐19 pandemic, SARS‐CoV‐2 accumulated approximately two prevalent mutations per month. 7 The frequency of SARS‐CoV‐2 mutations increased as the epidemic persisted and the virus spread more widely. Currently, a mass of SARS‐CoV‐2 variants have emerged. The early prevalent variants harbored D614G mutation on their S which alters the conformation of S protein, thereby increasing the infectivity and transmissibility of the virus though not affecting the vaccine efficiency. 10 , 11 Then, the variants Alpha (B.1.1.7), Beta (B.1.351), and Gamma (P.1) carrying N501Y mutation were identified in late 2020 in the United Kingdom, South Africa, and Brazil, respectively. 12 , 13 , 14 The N501Y mutation, located in the receptor‐binding domain (RBD) region, significantly increases the binding affinity of S to ACE2, and has immune escape properties. 15 , 16 In addition, the E484K mutation, which was found in Beta and Gamma variants, reduces the potency of neutralizing antibodies by 10‐fold or more in recovered convalescent individuals. 17 , 18 Later on, variant Delta (B.1.617.2), first detected in India, quickly spread throughout the world. 19 The Delta variant has two new mutations in the RBD region compared to other variants, namely L452R and T478K. The L452R mutation increases spike stability, viral infectivity, and replication, as well as immune evasion. 20 , 21 The T478K mutation enhances viral entry into ACE2‐expressing cells but sensitizes the virus to antibody neutralization. 22 , 23 Now, the Omicron variant, firstly detected in South Africa at the end of 2021, carries more than 30 mutations in its S and has spread around the world at a ferocious rate. Several single mutations in the RBD of the Omicron variant, including G339D, S371L, S375F, N440K, G446S, E484A, Q493K, and G496S, has been reported to impair the antibody neutralization. 24 , 25 Besides, L452Q, F490S, and R346K carried by Lambda (C.37) and Mu (B.1.621) variants have also been proved to enhance the variant's ability to evade neutralization. 26 Aside from RBD, the N‐terminal domain (NTD) is also the viral antigenic epitope subjected to most natural selection pressure. Y145N, K150R, S151P, A222V, and R246I mutations in NTD have been shown to increase neutralization escape. 27
Worldwide population immunity gained through vaccination is an effective way to control the pandemic of COVID‐19. Currently, over 11.6 billion doses of SARS‐CoV‐2 vaccines have been administered worldwide. 1 The four main types of SARS‐CoV‐2 vaccines are mRNA vaccines (e.g., Pfizer/BioNTech and Moderna), adenoviral vector vaccines (e.g., AstraZeneca‐Oxford), inactivated vaccines (e.g., BBIBP‐CorV and Sinovac‐CoV), and recombinant subunit protein vaccines (e.g., Zhifei). 28 Phase III clinical trials of the BBIBP‐CorV and Sinovac‐CoV vaccines showed a population protection efficacy of 72.8% and 50.7%, respectively. 29 , 30 Two doses of inactivated vaccine followed by one dose of mRNA vaccine significantly increased antibody titers and enhanced the neutralization of variants Delta and Omicron. 31 ZF2001, a three‐dose RBD‐based recombinant protein vaccine, demonstrated good population protection in Phase III clinical trials but showed significantly reduced neutralizing effectiveness against four Omicron subtypes which are BA.1, BA.2, BA.3, and BA.4. 32 , 33
Certain mutations in S are closely related to immune evasion and viral infectivity. Despite several most prevalent mutations such as D614G and N501Y, little is known about the potential impact of other high‐frequency mutations in S. Herein, we investigated the influence of natural nonsynonymous mutations of S on the protective efficacy of vaccines. First, we screened a total of 124 high‐frequency S mutants using the bioinformatics tool BioAider. 34 Then, the neutralizing efficiency of vaccinated sera against these mutants and their utilization efficiency of the receptor were analyzed. We found that 8 RBD mutations and 6 non‐RBD mutations on spike protein of SARS‐CoV‐2 were found to be markedly resistant to the neutralization of vaccinated sera, implying that the SARS‐CoV‐2 variants carrying these mutations reduced protective efficacy of the vaccines. In addition, Sinovac CoronaVac vaccine were more effective than ZF2001 vaccine in neutralizing certain mutants with non‐RBD mutation, implying that CoronaVac vaccine induce a broad spectrum of non‐RBD neutralizing antibodies. Furthermore, F486L and Y508H potentially enhance viral infectivity to human cells. In summary, our research provides a broad perspective on how S mutations may affect existing vaccines.
2. MATERIALS AND METHODS
2.1. Cell lines and plasmids
HEK293T (National Collection of Authenticated Cell Cultures; GNHu17) and HeLa‐hACE2 cells (stable expression of human ACE2 gene) were maintained in Dulbecco's modified Eagle's medium (Gibco; Cat#C11995500BT) supplemented with 10% fetal bovine serum (Gibco; Cat#C11995500BT), 100 units of penicillin and 0.1 mg/ml of streptomycin (Gibco; Cat#15140122) in 5% CO2 at 37°C.
Full‐length SARS‐CoV‐2 S (Wuhan‐1 strain, wild type), variants Alpha, Beta, Gamma, Delta, Lambda, Mu, and Omicron spike (GenBank accession number: NC_045512.2, GISAID accession number: EPI_ISL_5253387, EPI_ISL_5195381, EPI_ISL_5254522, EPI_ISL_3558827, EPI_ISL_4348182, EPI_ISL_431119, EPI_ISL_6825398) were synthesized by Sangon Biotech. All synthetic S genes were human codon‐optimized and constructed into pcDNA3.1 vectors with a C‐terminal HA tag.
2.2. Serum sample collection
Serum samples were collected from healthy individuals which vaccinated with the Sinovac CoronaVac and ZF2001 vaccines (Supporting Information: Table S1). The collection was conducted in 14 days, on average, following administration of the second or the third dose. Negative serum samples were collected from unvaccinated healthy individuals before the pandemic. All serum samples were aliquoted and stored at −80°C. Before experiments, aliquots of serum samples were heat‐inactivated at 56°C for 30 min.
2.3. Viral genomes and S mutant analysis
High coverage human SARS‐CoV‐2 strain sequences were downloaded from the GISAID database (https://www.gisaid.org/) with strain submission dates on or before October 20, 2020 (corresponding to sampling dates of December 24, 2019 to October 13, 2020). Following removal of redundant, ambiguous, and incomplete sequence, a total of 88,247 sequences of the S protein were extracted for alignment analyses. Multiple sequence alignment was performed using MAFFT v7.149 program in BioAider v1.314 34 and analyzed the S mutation characteristic, including mutation frequency and proportion of mutants. In total, we have selected 124 mutants, including 20 single mutants and 104 multiple mutants with D614G mutation. (Supporting Information: Table S2).
2.4. Construction of spike mutants
Spike point‐mutated plasmids were constructed by site‐directed mutagenesis. The pcDNA3.1‐SARS‐CoV‐2 S plasmid was used as the prototype to generate the plasmid with mutagenesis in the S gene. Following the procedure of circular PCR, 15−20 nucleotides before and after the target mutation site were selected as forwarding primers, while the reverse complementary sequences were selected as reverse primers. Following PCR, the template chain was digested using DpnI restriction endonuclease (NEB; Cat#R0176S). Afterward, the PCR digested product was directly used to transform E. coli Top 10 competent cells, single clones were selected and then sequenced. The primers designed for the specific mutation sites are listed in Supporting Information: Table S3.
2.5. RBD protein expression and purification
Recombinant SARS‐CoV‐2 RBD was prepared using the Bac‐to‐Bac baculovirus expression system. In brief, the coding sequences for SARS‐CoV‐2 RBD (spike residues 319−541) were cloned into the pFastBac vector. The gp67 signal peptide sequence was added to the protein N terminus for protein secretion, and a Hexa His tag was added to the C terminus to facilitate protein purification.
Transfection, virus amplification, and recombinant protein production were conducted with Sf9 cells. Cell supernatants were collected and centrifuged after infection for 72 h. After removal of most impurities, the recovered proteins were pooled and then purified on a BeyoGoldTM His‐tag Purification Resin column (Beyotime; Cat#P2210). Finally, each collected protein was prepared in a buffer consisting of 20 mM Tris‐HCl (pH 7.2) and 150 mM NaCl and concentrated to approximately 2 mg/ml using an ultrafiltration tube (Millipore; Cat# UFC501096) for further use.
2.6. Enzyme‐linked immunosorbent assay
High‐protein‐binding microtiter plates were coated overnight at 4°C with the purified recombinant SARS‐CoV‐2 RBD at 2 μg/ml in carbonate buffer. The plates were then washed with 1×PBS containing 0.05% Tween 20 and blocked with 1×PBS containing 1% bovine serum albumin (Beyotime; Cat#ST2249) for 1 h at 37°C. Serum samples were initially diluted 1:100 and RBD monoclonal antibodies mAb#1 (ACRO; Cat#S1N‐M122) and mAb#2 (GenScript; Cat#A02109) were initially diluted 1 μg/ml, followed by a threefold serial dilution and incubated for 1 h at 37°C. Horseradish peroxidase‐conjugated goat anti‐human IgG antibody diluted 1:5000 (Abbkine; Cat# A21050) was used to detect the binding of IgG, and plates were subsequently developed with Tetramethylbenzidine Substrate (Beyotime; Cat#P0209). Absorbance was measured at 450 nm on a microplate spectrophotometer (BioRad). To normalize the assays, control antibodies with known binding characteristics were included on each plate and the plates were developed when the absorbance of the negative control was lower than 0.5 OD450 units. All experiments were performed in duplicate two to three times.
2.7. Pseudoviruses production and infection assay
Lentivirus (HIV‐1)‐based pseudoviral particles carrying luciferase reporter and different SARS‐CoV‐2 S variants were prepared in this study. Briefly, HEK293T cells (7 × 106 cells) were cotransfected with 10 μg of a backbone plasmid encoding an Env‐defective (pNL4‐3.luc.R‐E‐) and 10 μg of plasmids expressing parental S or its mutants using Lipofectamine 2000 (Thermo Fisher Scientific; Cat# 11668019) according to the manufacturer's protocol. At 48−72 h posttransfection, the supernatants containing the pseudoviral particles were harvested and centrifuged. The amount of pseudoviruses particle prepared was quantified using the qPCR assay, and the measured value was normalized to the level of HIV‐1 p24 gene. The prepared pseudoviruses were stored at −80°C until use.
To prepare target cells for pseudoviruses infection assay, Hela‐hACE2 cells (2 × 104 cells/100 μl) were seeded in 96‐well plates and infected with 100 μl of the same amount of pseudoviruses. At 48 h postinfection, the infected cells were lysed with a Luciferase Assay System (Promega; Cat# E1501), and the luminescent signal was measured using a GloMax®20/20 Luminometer (Promega). Each infection experiment was carried out three times.
2.8. Pseudoviruses neutralization assay
The 50% tissue culture infective dose (TCID 50 ) of pseudoviruses was evaluated using Hela‐hACE2 cells by threefold serial dilutions. Chemiluminescence signals were detected 48 h after the incubation of cells and pseudoviruses from different S mutants. Then virus neutralization assay was conducted as described previously. 35 Briefly, 50 μl serial dilutions of human sera and RBD monoclonal antibody preparations (positive control) were added into 96‐well plates. After that, 50 μl pseudoviruses with concentration of 1300 TCID50/ml was added into the plates, followed by incubation at 37°C for 2 h. Afterward, Hela‐hACE2 cells were added to the plates (2 × 104 cells/100 μl cells per well), followed by incubation at 37°C in a humidified atmosphere with 5% CO2. Luminescence detection was performed after 48 h of incubation. The 50% effective dilution (EC50) was defined as the dilution of the antiserum that reduced the relative luminescence units by 50%, compared with the relative luminescence units in the virus control wells containing no antiserum.
2.9. Structural models
The hACE2/S complex was used as a template for homology modeling. 36 The mutations in the models were aligned, and the interactions between the SARS‐CoV‐2 S and ACE2 proteins were compared in PyMOL.
2.10. Statistical analysis
GraphPad Prism 9 (GraphPad) was used for plotting and statistical analysis; the values were expressed as mean ± SD. One‐way ANOVA and Holm−Sidak's multiple comparison tests were used for statistical analysis. A p value of less than 0.05 was considered to be significant and the significance was labeled as *p < 0.05, **p < 0.01, and ***p < 0.005.
3. RESULTS
3.1. Screen of high‐frequency mutations of SARS‐CoV‐2 S protein
To identify important mutations of SARS‐CoV‐2 S, we screened three groups of natural mutants by bioinformatics analysis. As shown in Figure 1, compared with the SARS‐CoV‐2 Wuhan‐1 strain (WT), we defined a mutation with a frequency higher than 10 as a high‐frequency mutation. Group A harbored high‐frequency single‐amino‐acid mutants (20 strains). Group B contained all high‐frequency combinations of D614G and mutations in the non‐RBD region (62 strains), including L18F, T29I, H49Y, A222V, Q677H, and A829T mutations which were also included in Group A. Group C included high‐frequency combinations of D614G and mutations in the RBD (42 strains). Based on these variants, we generated 124 pseudoviruses. (Figure 1, Supporting Information: Table S1).
Figure 1.
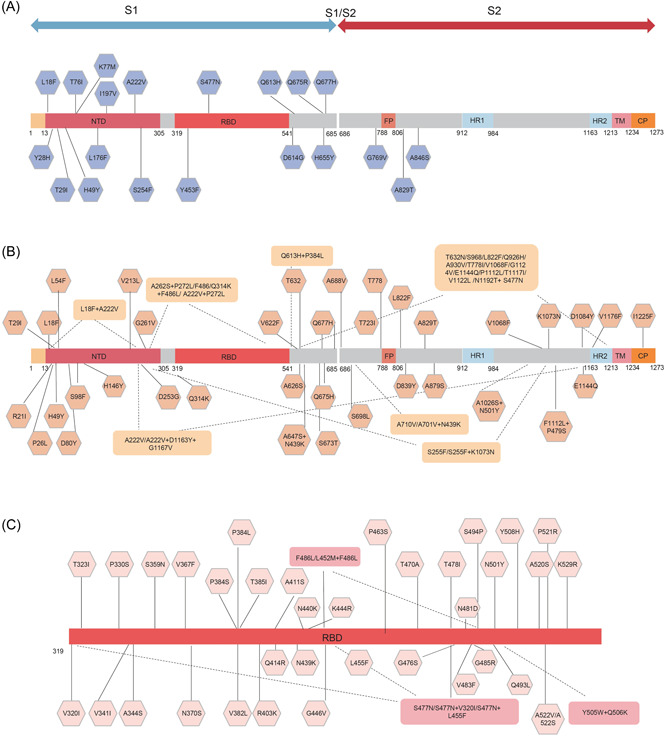
SARS‐CoV‐2 variants selected in this study. (A) Variants with high‐frequency single amino acid mutations across the entire S gene. (B) Variants combined with D614G across the entire S gene excluding RBD region. (C) Variants combinations with D614G in the RBD. RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.2. Vaccination induces RBD‐specific IgG antibodies production
To evaluate whether specific RBD neutralizing antibodies could be elicited after vaccination, we incubated serially diluted sera with RBD protein and then calculated the RBD‐specific antibody titer of each serum (Figure 2). The results showed that RBD‐specific IgG antibodies were present in all SARS‐CoV‐2 vaccinated sera, although antibody titer levels varied.
Figure 2.
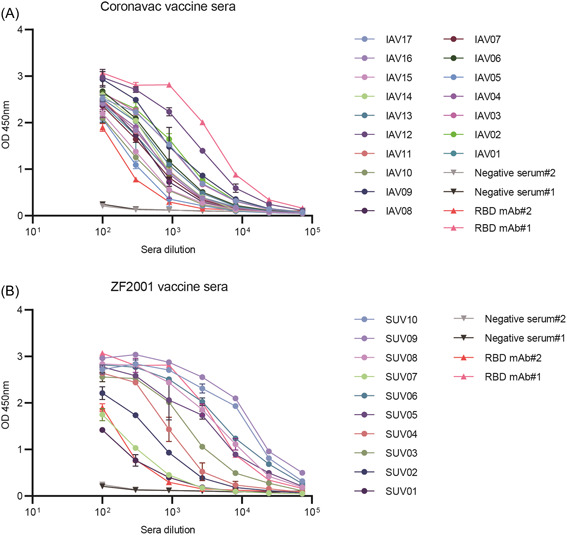
RBD specific antibody titer of CoronaVac and ZF2001 vaccinated sera. (A) RBD‐specific total IgG levels in the CoronaVac vaccinated sera. (B) RBD‐specific total IgG levels in the ZF2001 vaccinated sera. RBD mAb#1 and RBD mAb#2 served as positive controls. Error bars indicate one standard deviation of mean values. RBD, receptor‐binding domain.
3.3. Neutralizing efficiency of sera vaccinated by CoronaVac vaccine against high‐frequency S mutants
To determine whether the vaccinated sera inhibited infection of pseudoviruses carrying different S mutants. First, we verified the protective effect of vaccinated sera against SARS‐CoV‐2 WT using pseudoviruses neutralization assay (Supporting Information: Figure S1). All vaccinated sera inhibited SARS CoV‐2 pseudoviruses entry into HeLa‐hACE2 cells with varying degrees. In addition, vaccinated sera against SARS‐CoV‐2 WT with neutralization titers (EC50) less than 30, such as IAV07, SUV01, SUV02, and SUV07, were eliminated (Supporting Information: Table S4). The remaining 23 highly reactive serum samples, including 16 CoronaVac vaccinated sera and 7 ZF2001 vaccinated sera, were analyzed for their neutralization efficacy against S mutants.
We next explored the sensitivity of the natural S mutant to the 16 CoronaVac vaccinated sera. All sera from CoronaVac recipients suppressed infection with most SARS‐CoV‐2 variants, demonstrating that the CoronaVac vaccine had good neutralizing efficacy against these mutants (Figure 3). Notably, some changes in the RBD region significantly altered sensitivity to sera vaccinated by CoronaVac vaccine. When compared to the reference strain, the neutralization sensitivity of the D614G + F486L mutant to 15 of 16 sera (except for IAV13) was found to be reduced by more than 10‐folds. While D614G + F486L + L452M, D614G + F486L + A262S, and D614G + Q314K + A262S + F486L mutants were observed with 6‐fold, 2.7‐fold, and 3‐fold reduction in susceptibility to neutralization, respectively. These results confirmed that F486 may be a key immunogenic site on S that has a significant impact on antibody binding efficiency. S477N and D614G + S447N mutants showed no change in sensitivity to CoronaVac vaccinated sera compared to the reference strain. However, the other 5 mutants (V320I, L455F, S698L, T778I, and N1192T in combination with D614G + S477N) showed higher resistance to vaccinated sera. Furthermore, the 5 mutants combined with D614G (D614G + V382L, D614G + K444R, D614G + L455F, D614G + Y508H, and D614G + P521R) demonstrated a significantly decreased sensitivity, with a reduction of over 4‐folds, to CoronaVac vaccinated sera.
Figure 3.
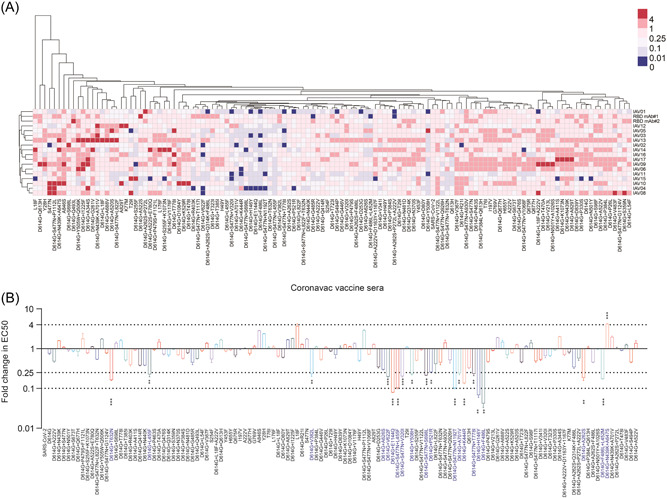
Differential sensitivity of the natural mutants to a panel of CoronaVac vaccinated sera. (A) Serial dilutions of 16 CoronaVac vaccinated sera samples were individually mixed with the pseudoviruses at 37°C for 2 h before adding HeLa‐hACE2 cells for incubation of 48 h to determine the EC50. The experiments were repeated at least two times. Hem I software was used to analyze the data and draw the heatmap. The red and blue boxes indicate the increase or decrease of the neutralization activity compared with SARS‐CoV‐2 WT strain, as shown in the scale bar. RBD mAb#1 and RBD mAb#2 served as positive controls. (B) Summary of the data from 16 CoronaVac vaccinated sera, with the values presented as mean ± SD. The horizontal dashed lines indicate the threshold of 4‐fold or 10‐fold difference. ACE2, angiotensin‐converting enzyme 2; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
In addition, mutations in non‐RBD region of the S protein also significantly affected the neutralization efficiency. It is worth mentioning that some mutants, including A829T, A846S, D614G + A344S, and D614G + A688V (Figure 3A), were even more sensitive to 2 or 3 of the 16 tested sera than the reference strain, whereas more mutants were found to be resistant to all CoronaVac vaccinated sera. These mutants include double amino acid changes such as D614G + H146Y, D614G + A262S, D614G + V622F, D614G + A626S, D614G + T632N, D614G + A701V, and D614G + E1144Q (Table 1).
Table 1.
Characteristics of SARS‐CoV‐2 S mutants
| Group A | Group B | Group C | |
|---|---|---|---|
| Number of variants | 20 | 62 | 42 |
| Increased sensitivity to CoronaVac vaccine sera | D614G + N439K + A647S | ||
| Decreased sensitivity to CoronaVac vaccine sera | D614G + H146Y、D614G + A262S、D614G + V622F、D614G + A626S、D614G + T632N、D614G + A701V、D614G + S477N + S698L、D614G + E1144Q、D614G + S477N + T778I、D614G + S477N + N1192T | D614G + V382L、D614G + K444R、D614G + L455F、D614G + S477N + V320I、D614G + S477N + L455F、D614G + Y508H、D614G + P521R、D614G + L452M + F486L、D614G + F486L | |
| Increased sensitivity to ZF2001 vaccine sera | None | D614G + A829T | D614G + Y505W + Q506K |
| Decreased sensitivity to ZF2001 vaccine sera | T29I、H49Y |
D614G + H146Y、D614G + V213L、D614G + D253G、D614G + S255F、D614G + G261V、D614G + A626S、D614G + A688V、D614G + A701V、D614G + L822F、D614G + E1144QD614G + S477N + T632N、D614G + S477N + S698L、D614G + S477N + T778I、D614G + S477N + Q926H、D614G + S477N + A930V、D614G + S477N + V1122L、D614G + A262S + P272L + A222V |
D614G + V382L、D614G + K444R、D614G + S477N + V320I、D614G + S477N + L455F、D614G + P479S、D614G + Y508H、D614G + P521R、D614G + A520S、D614G + A522S、D614G + L452M + F486L、D614G + F486L、 |
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.4. Neutralizing efficiency of sera vaccinated by ZF2001 vaccine against high‐frequency S mutants
For ZF2001 vaccine, we found SUV03, SUV04, SUV05, SUV06, SUV09, and SUV10 serum were unable to neutralize 6, 6, 1, 2, 3, and 1 SARS‐CoV‐2 mutants, respectively (Figure 4A). These mutants included single‐amino‐acid substitutions such as L176F, Q613H, and A829T, as well as mutants combined with D614G, including D614G + H146Y, D614G + T323I, D614G + K444R, D614G + V622F, D614G + A701V, D614G + L822F, D614G + E1144Q, D614G + S477N + S698L, D614G + S477N + A903V, and D614G + S477N + N1192T.
Figure 4.
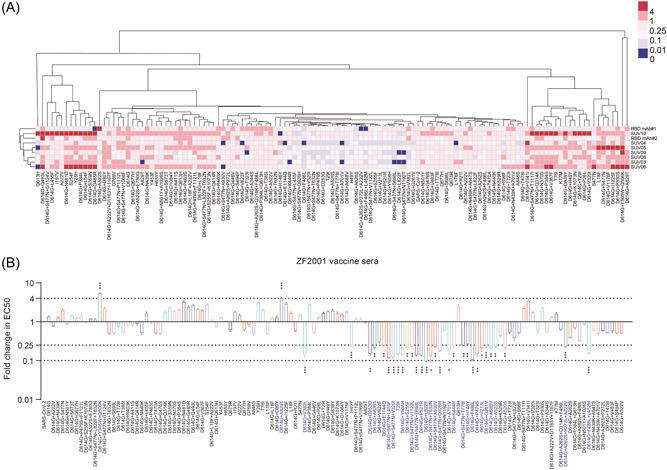
Differential sensitivity of the natural mutants to a panel of ZF2001 vaccinated sera. (A) Serial dilutions of 7 ZF2001 vaccinated sera were individually mixed with the pseudoviruses at 37°C for 2 h before adding HeLa‐hACE2 cells for incubation of 48 h to determine the EC50. The experiments were repeated at least two times. Hem I software was used to analyze the data and draw the heatmap. The red and blue boxes indicate the increase or decrease of the neutralization activity compared with SARS‐CoV‐2 WT strain, as shown in the scale bar. RBD mAb#1 and RBD mAb#2 served as positive controls. (B) Summary of the data from 7 ZF2001 vaccinated sera, with the values presented as mean ± SD. The horizontal dashed lines indicate the threshold of 4‐fold or 10‐fold difference. ACE2, angiotensin‐converting enzyme 2; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Similar to CoronaVac vaccinated sera, the 7 ZF2001 vaccinated sera showed a decreased neutralization efficiency to D614G + V382L, D614G + F486L, D614G + Y508H, and D614G + P521R mutants by more than 4 folds (Figure 4A). These results confirmed that these 4 amino acid sites in RBD were important for receptor binding. However, D614G + A829T and D614G + Y505W + Q506K mutants were more sensitive to 2 or 3 ZF2001 vaccinated sera. As shown in Figure 3B, when the data of individual vaccinated sera were pooled together to analyze the sensitivity of all mutants, a marked difference (>4‐fold) was observed. It is noteworthy that several non‐RBD mutants significantly reduce the neutralizing ability of ZF2001 vaccinated sera compared to CoronaVac vaccine sera. These mutants include D614G + V213L, D614G + D253G, D614G + S255F, D614G + G261V, D614G + P521R, D614G + A688V, and D614G + L822F (Figures 3B and 4B). In addition, more mutants combined with D614G + S477N, including V320I, L455F, T632N, S698L, T778I, Q926H, A930V, and V1122L, do have decreased sensitivity over fourfold to the immune sera of ZF2001 vaccine.
To further investigate the differences in neutralizing resistance of the SARS‐CoV‐2 variants, we analyzed the neutralization capacity of 10 highly reactive sera, including 5 CoronaVac and 5 ZF2001 vaccinated sera against SARS‐CoV‐2 WT and variants VOCs and VOIs (Figure 5). D614G, Alpha, and Lambda variants were more sensitive to the tested sera than SARS‐CoV‐2 WT, suggesting that the vaccine was effective in preventing infection of these variants. Meanwhile, the variant Gamma demonstrated a fourfold decrease in susceptibility to ZF2001 vaccinated sera, however, there was no significant difference with CoronaVac vaccinated sera (Figures 5B,C). In addition, a 4‐fold difference in reactivity to grouped vaccine sera was observed between two variants (Beta and Delta) and SARS‐CoV‐2 WT. In the case of Omicron variants, which are now rapidly evolving around the world, the neutralization activity has been reduced by more than 13‐fold. The variant Mu is also noteworthy, the mean neutralization titer of Mu pseudoviruses was 44, which represented a 4‐fold reduction of neutralization compared to the SARS‐CoV‐2 WT. These findings indicate that the two‐dose CoronaVac vaccine and three‐dose ZF2001 vaccine is insufficient to provide full protection against these newly emerging variants, particularly the Omicron and Mu variants.
Figure 5.
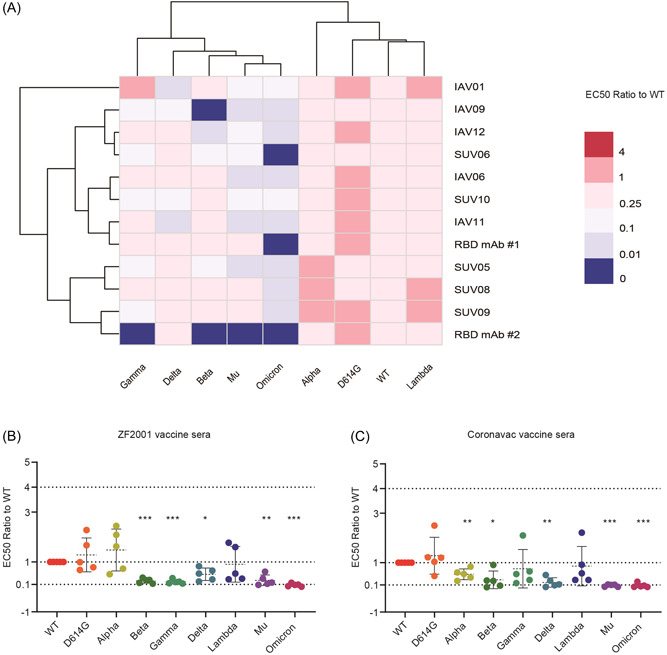
Differential sensitivity of the natural variants to a panel of vaccinated sera. (A) Serial dilutions of 10 serum samples with high RBD antibody titers were individually mixed with pseudoviruses at 37°C for 2 h before adding HeLa‐hACE2 cells for incubation of 48 h to determine the EC50. The experiments were repeated at least two times. Hem I software was used to analyze the data and draw the heatmap. The red and blue boxes indicate the increase or decrease of the neutralization activity compared with SARS‐CoV‐2 WT strain, as shown in the scale bar. RBD mAb#1 and RBD mAb#2 served as positive controls. (B) Summary of the data from 5 CoronaVac vaccinated sera, with the values presented as mean ± SD. (C) Summary of the data from 5 ZF2001 vaccinated sera, with the values presented as mean ± SD. ACE2, angiotensin‐converting enzyme 2; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.5. Alteration of receptor utilization and virus entry by S mutant
Having identified the S mutants with altered neutralization, we next investigated the infectivity of different S mutants using pseudoviruses infection assays. We tested the entry efficiency of 124 pseudoviruses carrying distinct S mutants to Hela‐hACE2 cells, where more than fourfold changes compared to the reference virus were considered as significant. As shown in Figure 6, compared to SARS‐CoV‐2 WT, 12 of the 20 S single mutants significantly reduced viral infectivity, and the other 7 mutants had no significant change in ACE2 utilization ability. Notably, the remaining 104 mutants combined with D614G exhibit different infectivity. Among them, 14 mutants located in the RBD region (D614G + V341I, D614G + N439K, D614G + T385I, D614G + A411S, D614G + Q414R, D614G + N440K, D614G + L455F, D614G + P463S, D614G + T470A, D614G + G476S, D614G + P384L, D614G + Y508H, D614G + F486L, and D614G + P479S), led to more than 4‐fold increase in the utilizing efficiency of ACE2, which means that these mutations are critical for viral receptor binding. Besides, 4 mutants located in the NTD region, including D614G + S98F, D614G + V213L, D614G + D253G, D614G + G261V, and 4 mutants adjacent to the S1/S2 cleavage site, including D614G + A626S, D614G + T632N, D614G + A688V, D614G + S698L, D614G + T778I, significantly enhanced the ability of pseudoviruses to utilize ACE2. It is worth noting that most mutants combined with D614G + S477N did not enhance the utilization of ACE2, among which 2 mutations (D614G + S477N + P1112L and D614G + S477N + V1068F) were determined as low‐infectivity, with relative luciferase units (RLU) decreasing over 10‐fold (Figure 6).
Figure 6.
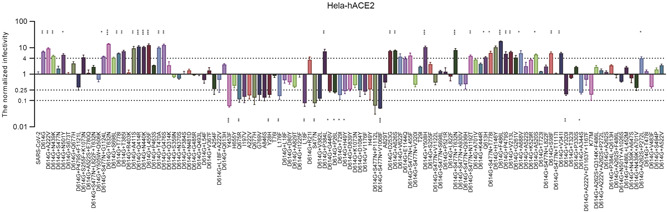
Infectivity analysis of S mutants in Hela‐hACE2 cell. Infectivity of natural variants and experimental mutants conducted in Hela‐ACE2, RLU values generated with the infection of the mutants, as measured by luminescence meter, were compared SARS‐CoV‐2 WT strain. A difference of 4‐fold is considered significant; all experiments were conducted two times (mean ± SD) unless specified. ACE2, angiotensin‐converting enzyme 2; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
4. DISCUSSION
Rapid mutations on SARS‐CoV‐2 S have been altering viral antigenicity and infectivity, thus posing a risk of decreased vaccine efficacy. 37 , 38 Indeed, six epidemic peaks of SARS‐CoV‐2 variants, namely D614G and Alpha, Beta, Gamma, Delta, and Omicron variants defined by WHO as VOC, have occurred in just 2 years. 39 Notably, Omicron, the most prevalent variant currently, has further derived into multiple subtypes (BA.1, BA.2, BA.3, BA.4, and BA.5), which carry more than 30 mutations, including 15 in the RBD region, severely disrupting the immunogenicity of vaccine sera. 33 , 39
In this study, we employed pseudoviruses neutralization assays to examine whether the currently administered CoronaVac vaccine and ZF2001 vaccines efficiently protected HeLa‐ACE2 cells from the entry of wild‐type or mutated S pseudoviruses. Our results show that pseudoviruses carrying the F486L mutation is highly infectious and more resistant to vaccine sera (Supporting Information: Figure S2). F486 is located at the RBD‐ACE2 binding interface and forms van der Waals forces with M82 of human ACE2 (Figure 7). Currently, six amino acid substitutions have appeared in position 486 of the S protein, including leucine (L), valine (V), isoleucine (I), cystine (C), tyrosine (Y), and serine (S), with frequencies of 2,519, 1,733, 1,733, 399, 399, and 399, respectively (Supporting Information: Table S2). The F486V mutation has been identified in Omicron subtypes BA.4 and BA.5, which began their outbreaks in South Africa in January 2022. BA.4 has spread over at least 16 countries with more than 1,800 cumulative cases as of today, while BA.5 has been detected in at least 17 countries with over 1,300 cumulative cases. 1 According to research, BA.4 and BA.5 have the highest degree of immune escape compared to other Omicron subtypes. 40 Second, SARS‐CoV‐2 F486 corresponds to L472 in the SARS‐CoV RBD region, which may explain why SARS‐CoV‐specific RBD antibodies and drugs are virtually inactive against SARS‐CoV‐2. 41 We speculated that both leucine and valine are small side chain non‐polar amino acids with reduced hydrophobic interactions, which results in significantly lower binding activity of vaccine sera or therapeutic antibodies (Figure 7). These results suggested that 486th residue could be a potentially key site of SARS‐CoV‐2 immune escape, raising concerns about the transmission of Omicron BA.4 and BA.5 and variants carrying the F486L mutation. In addition, L452M + F486L mutation markedly weakened the neutralizing effect of vaccinated sera. L452M, L452Q, and L452R are typical mutations of Omicron subtypes BA.2.13, BA.2.12.1, and BA.4/5 and Delta variants, respectively (Supporting Information: Table S2). Despite L452M and L452Q currently occurring less frequently, BA.2.12.1 is potentially more infectious, with a 25% transmission advantage over the previously prevalent Omicron BA.2. 1
Figure 7.
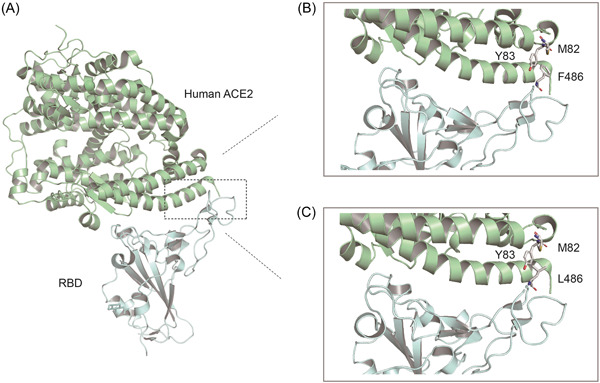
Molecular basis of human ACE2 recognition by the SARS‐CoV‐2 RBD. (A) Cartoon diagram of the crystal structure of RBD in complex with the ACE2 ectodomain (PDB: 6M0J). (B and C) Zoomed‐in views of the RBD‐ACE2 interface with highlighting modulation of interactions due to introduction of the F486L residue substitutions. ACE2, angiotensin‐converting enzyme 2; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Notably, pseudoviruses carrying V382L, K444R, Y508H, P521R, and S477N + L455F reduced the neutralizing activity of both CoronaVac and ZF2001 vaccinated sera. As shown in Supporting Information: Table S2, despite the low frequency of mutations in these S variants (1490, 1580, 849, 262, and 2372, respectively), Y508H mutations significantly increased ACE2 utilization efficiency, indicating that continuous monitoring of mutations in the RBD region of S is required. K440N is the most common feature of the Omicron variant present in all subtypes. G446V/S is identified in Omicron variants BA.1, BA.1.1, and BA.3 which are prevalent in England, Denmark, and other European countries. 24 Our results show that pseudoviruses carrying the N440K or G446V mutations display a decreasing trend, but less than 4‐fold reduction compared to SARS‐CoV‐2 WT, in neutralization to vaccinated sera, indicating that both CoronaVac and ZF2001 vaccines are well protected against the two mutations. The G446V mutation significantly reduced the efficiency of ACE2 utilization, suggesting that the mutation can alter the S‐ACE2 binding interface and thus affect the efficiency of binding to ACE2 protein.
Our findings also suggest that certain mutations in the non‐RBD region of S protein, such as A626S, A701V, and E1144Q, may reduce the binding efficiency of vaccinated sera and alter their ability to use ACE2. Besides, mutations occurring in non‐RBD have also been associated with immune escape. Previous studies have reported that most evidence for immune escape associated with NTD mutations are located in loop N3 (140−156) and loop N5 (246−260), which overlap with the epitope of antibody 4A8. 42 , 43 Our findings show that the H146Y mutant reduces the neutralizing effect of the two vaccines. The D253G mutation significantly reduced the sensitivity of the ZF2001 vaccine compared to the CoronaVac vaccine, with no more than a fourfold difference in sensitivity. These results indicated that the 146th and the 253th amino acids were antigenic epitopes in the NTD region, and that CoronaVac vaccinated could stimulate the body to produce NTD‐terminal neutralizing antibodies. Interestingly, we also found that T29I and H49Y, with occurrences of 11,168 and 8,407, were significantly resistant to the vaccine sera. However, T29I + D614G and H49Y + D614G were more sensitive, probably due to the D614G mutation changing the conformation of S and interacting better with the antibody, improving the vaccine sensitivity (Table 1). In addition, we speculated that due to individual mechanisms, the antigenic epitopes of antibodies produced after vaccination were also different. Therefore, several sera (such as IAV13, SUV06, and SUV10) were more sensitive to some S mutants, ultimately resulting in increased neutralizing activity.
Another important finding is that both CoronaVac and ZF2001 vaccines elicited functional humoral immune responses, though with differences in epitope recognition and antibody‐mediated functional properties. For some S mutations in non‐RBD regions, CoronaVac vaccinated sera were more effective in protecting against infection than ZF2001, implying that CoronaVac vaccine could induce a broader range of neutralizing antibodies and potential differences in the protective immunity provided by the two vaccines. In addition, there are several limitations in our study. First, in vitro pseudoviruses neutralization assays are based on Hela‐ACE2 cell lines as targets that restrict pseudoviruses entry in an ACE2‐dependent manner, lacking other cell surface proteins that may be involved in natural infection, such as NRP1. 44 Thus, further validation based on living SARS‐CoV‐2 variants is needed to confirm these findings. Moreover, the small sample size of the vaccine sera precludes a thorough examination of immunological differences between populations. Despite these limitations, the findings suggest that S mutations may cause subtle differences in the quality of the humoral immune response elicited by CoronaVac and ZF2001 vaccines.
In summary, 124 pseudotyped SARS‐CoV‐2 carrying various S mutants were analyzed for their infectivity and reactivity to a panel of sera from the vaccinated population. 14S mutants markedly resistant to the vaccinated sera among which 8 contained the mutations in RBD, indicating reduced protective efficacy of the vaccines against these SARS‐CoV‐2 variants. In addition, immune sera produced with Sinovac CoronaVac vaccine showed better neutralization efficiency against non‐RBD mutants than those of ZF2001 vaccine, probably due to the lack of non‐RBD epitopes on ZF2001 vaccine. Notably, F486L and Y508H potentially enhance viral infectivity to human cells. Overall, these findings highlight the importance of closely monitoring S protein mutations and developing new vaccines with a broader spectrum and higher neutralizing potency against SARS‐CoV‐2 variants.
AUTHOR CONTRIBUTIONS
Xing‐Yi Ge and Qiong Wang conceived and designed the study. Qiong Wang wrote the manuscript. Zhi‐Jian Zhou and Sheng‐Bao Ye helped with the figures. Sheng‐Bao Ye, A‐Ling Song, Xi Zhu, Jia‐Mei Peng, Rui‐Min Liang, Chen‐Hui Yang, and Xiao‐Wei Yu helped with the experiment. Xing‐Yi Ge, Ye Qiu, Jie Yu, and Xun Huang revised the whole manuscript. All authors contributed to the work.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
This study was performed following the Declaration of Helsinki and Good Clinical Practice principles. The study protocol was reviewed and approved by Medical Ethics Committee of Xiangya Hospital Central South University (IRB number 202207390). All participants signed a written consent before being enrolled. The study was conducted according to the Declaration of Helsinki and the principles of the Good Clinical Practice Guidelines (ICH‐GCP).
Supporting information
Supporting information.
ACKNOWLEDGMENTS
We thank all study participants for their generous participation and contribution to this work. This work was jointly funded by the National Key Research and Development Program of China Research on the Precision Diagnosis, Treatment, and Integrated Prevention, Control for the elderly with common infectious disease (2020YFC2005403), the National Natural Science Foundation of China (81902070 and 32041001), the Natural Science Foundation of Hunan Province (2019JJ50341), and Double‐First Class Construction Funds of Hunan University (521119400156).
Wang Q, Ye S‐B, Zhou Z‐J, et al. Key mutations in the spike protein of SARS‐CoV‐2 affecting neutralization resistance and viral internalization. J Med Virol. 2022;95:e28407. 10.1002/jmv.28407
Contributor Information
Xun Huang, Email: huangxun@mail.csu.edu.cn.
Jie Yu, Email: yujie_hy@hotmail.com.
Ye Qiu, Email: qiuye@hnu.edu.cn.
Xing‐Yi Ge, Email: xyge@hnu.edu.cn.
DATA AVAILABILITY STATEMENT
Data supporting the findings of this study are available from the corresponding author upon request.
REFERENCES
- 1. WHO . Coronavirus (COVID‐19) dashboard. Accessed May 22, 2022. https://covid19.who.int/
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Damas J, Hughes GM, Keough KC, et al. Broad host range of SARS‐CoV‐2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci. 2020;117(36):22311‐22322. 10.1073/pnas.2010146117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Q, Qiu Y, Li JY, Liao CH, Zhou ZJ, Ge XY. Receptor utilization of angiotensin‐converting enzyme 2 (ACE2) indicates a narrower host range of SARS‐CoV‐2 than that of SARS‐CoV. Transbound Emerg Dis. 2021;68(3):1046‐1053. 10.1111/tbed.13792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hacisuleyman E, Hale C, Saito Y, et al. Vaccine breakthrough infections with SARS‐CoV‐2 variants. N Engl J Med. 2021;384(23):2212‐2218. 10.1056/NEJMoa2105000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mengist HM, Kombe Kombe AJ, Mekonnen D, Abebaw A, Getachew M, Jin T. Mutations of SARS‐CoV‐2 spike protein: implications on immune evasion and vaccine‐induced immunity. Sem Immunol. 2021;55:101533. 10.1016/j.smim.2021.101533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu A, Wang L, Zhou HY, et al. One year of SARS‐CoV‐2 evolution. Cell Host Microbe. 2021;29(4):503‐507. 10.1016/j.chom.2021.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bashor L, Gagne RB, Bosco‐Lauth AM, Bowen RA, Stenglein M, VandeWoude S. SARS‐CoV‐2 evolution in animals suggests mechanisms for rapid variant selection. Proc Natl Acad Sci. 2021;118(44):e2105253118. 10.1073/pnas.2105253118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. National Genomics Data Center . Accessed May 5, 2022. 2022.05.22. https://bigd.big.ac.cn/ncov/variation/annotation.
- 10. Zhou B, Thao TTN, Hoffmann D, et al. SARS‐CoV‐2 spike D614G change enhances replication and transmission. Nature. 2021;592(7852):122‐127. 10.1038/s41586-021-03361-1 [DOI] [PubMed] [Google Scholar]
- 11. Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and functional analysis of the D614G SARS‐CoV‐2 spike protein variant. Cell. 2020;183(3):739‐751. 10.1016/j.cell.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gräf T, Bello G, Venas TMM, et al. Identification of a novel SARS‐CoV‐2 P.1 sub‐lineage in Brazil provides new insights about the mechanisms of emergence of variants of concern. Virus Evol. 2021;7(2):veab091. 10.1093/ve/veab091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. La Rosa G, Mancini P, Bonanno Ferraro G, et al. Rapid screening for SARS‐CoV‐2 variants of concern in clinical and environmental samples using nested RT‐PCR assays targeting key mutations of the spike protein. Water Res. 2021;197:117104. 10.1016/j.watres.2021.117104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P.1 SARS‐CoV‐2 lineage in Manaus, Brazil. Science. 2021;372(6544):815‐821. 10.1126/science.abh2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y, Liu J, Plante KS, et al. The N501Y spike substitution enhances SARS‐CoV‐2 infection and transmission. Nature. 2022;602(7896):294‐299. 10.1038/s41586-021-04245-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Q, Nie J, Wu J, et al. SARS‐CoV‐2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184(9):2362‐2371. 10.1016/j.cell.2021.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuzmina A, Khalaila Y, Voloshin O, et al. SARS‐CoV‐2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post‐vaccination sera. Cell Host Microbe. 2021;29(4):522‐528.e2. 10.1016/j.chom.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou D, Dejnirattisai W, Supasa P, et al. Evidence of escape of SARS‐CoV‐2 variant B.1.351 from natural and vaccine‐induced sera. Cell. 2021;184(9):2348‐2361. 10.1016/j.cell.2021.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mishra A, Kumar N, Bhatia S, et al. SARS‐CoV‐2 Delta variant among Asiatic Lions, India. Emerging Infect Dis. 2021;27(10):2723‐2725. 10.3201/eid2710.211500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deng X, Garcia‐Knight MA, Khalid MM, et al. Transmission, infectivity, and neutralization of a spike L452R SARS‐CoV‐2 variant. Cell. 2021;184(13):3426‐3437.e8. 10.1016/j.cell.2021.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Motozono C, Toyoda M, Zahradnik J, et al. SARS‐CoV‐2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29(7):1124‐1136. 10.1016/j.chom.2021.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y, Liu C, Zhang C, et al. Structural basis for SARS‐CoV‐2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Nat Commun. 2022;13(1):871. 10.1038/s41467-022-28528-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ren W, Ju X, Gong M, et al. Characterization of SARS‐CoV‐2 variants B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.618 by cell entry and immune evasion. mBio. 2022;13(2):e0009922. 10.1128/mbio.00099-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan Y, Li X, Zhang L, Wan S, Zhang L, Zhou F. SARS‐CoV‐2 Omicron variant: recent progress and future perspectives. Signal Transduct Target Ther. 2022;7(1):141. 10.1038/s41392-022-00997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS‐CoV‐2 neutralizing antibodies. Nature. 2022;602(7898):657‐663. 10.1038/s41586-021-04385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen J, Gao K, Wang R, Wei GW. Revealing the threat of emerging SARS‐CoV‐2 mutations to antibody therapies. J Mol Biol. 2021;433(18):167155. 10.1016/j.jmb.2021.167155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dai L, Gao GF. Viral targets for vaccines against COVID‐19. Nat Rev Immunol. 2021;21(2):73‐82. 10.1038/s41577-020-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Al Kaabi N, Zhang Y, Xia S, et al. Effect of 2 inactivated SARS‐CoV‐2 vaccines on symptomatic COVID‐19 infection in adults: a randomized clinical trial. JAMA. 2021;326(1):35‐45. 10.1001/jama.2021.8565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGillCOVID19 Vaccine Tracker Team Sinovac: CoronaVac. https://covid19.trackvaccines.org/vaccines/7/.
- 31. Zuo F, Abolhassani H, Du L, et al. Heterologous immunization with inactivated vaccine followed by mRNA‐booster elicits strong immunity against SARS‐CoV‐2 Omicron variant. Nat Commun. 2022;13(1):2670. 10.1038/s41467-022-30340-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dai L, Gao L, Tao L, et al. Efficacy and safety of the RBD‐dimer‐based Covid‐19 vaccine ZF2001 in adults. N Engl J Med. 2022;386:2097‐2111. 10.1056/NEJMoa2202261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao X, Li D, Ruan W, et al. Effects of a prolonged booster interval on neutralization of omicron variant. N Engl J Med. 2022;386(9):894‐896. 10.1056/NEJMc2119426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou ZJ, Qiu Y, Pu Y, Huang X, Ge XY. BioAider: an efficient tool for viral genome analysis and its application in tracing SARS‐CoV‐2 transmission. Sustainable Cities Soc. 2020;63:102466. 10.1016/j.scs.2020.102466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nie J, Li Q, Wu J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS‐CoV‐2. Emerg Microbes Infect. 2020;9(1):680‐686. 10.1080/22221751.2020.1743767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581(7807):221‐224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li JY, Wang Q, Liao CH, Qiu Y, Ge XY. The 442th amino acid residue of the spike protein is critical for the adaptation to bat hosts for SARS‐related coronaviruses. Virus Res. 2021;295:198307. 10.1016/j.virusres.2021.198307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Niu Z, Zhang Z, Gao X, et al. N501Y mutation imparts cross‐species transmission of SARS‐CoV‐2 to mice by enhancing receptor binding. Signal Transduct Target Ther. 2021;6(1):284. 10.1038/s41392-021-00704-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans JP, Zeng C, Qu P, et al. Neutralization of SARS‐CoV‐2 Omicron sub‐lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe. 2022;30:1093‐1102. 10.1016/j.chom.2022.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cao Y, Yisimayi A, Jian F, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608 (7293):593‐602. 10.1101/2022.04.30.489997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382‐385. 10.1080/22221751.2020.1729069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCarthy KR, Rennick LJ, Nambulli S, et al. Recurrent deletions in the SARS‐CoV‐2 spike glycoprotein drive antibody escape. Science. 2021;371(6534):1139‐1142. 10.1126/science.abf6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kombe Kombe AJ, Zahid A, Mohammed A, Shi R, Jin T. Potent molecular feature‐based neutralizing monoclonal antibodies as promising therapeutics against SARS‐CoV‐2 infection. Front Mol Biosci. 2021;8:670815. 10.3389/fmolb.2021.670815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cantuti‐Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin‐1 facilitates SARS‐CoV‐2 cell entry and infectivity. Science. 2020;370(6518):856‐860. 10.1126/science.abd2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author upon request.


