Abstract
Emergence of SARS‐CoV‐2 variants warrants sustainable efforts to upgrade both the diagnostic and therapeutic protocols. Understanding the details of cellular and molecular basis of the virus–host cell interaction is essential for developing variant‐independent therapeutic options. The internalization of SARS‐CoV‐2, into lung epithelial cells, is mediated by endocytosis, especially clathrin‐mediated endocytosis (CME). Although vaccination is the gold standard strategy against viral infection, selective inhibition of endocytic proteins, complexes, and associated adaptor proteins may present a variant‐independent therapeutic strategy. Although clathrin and/or dynamins are the most important proteins involved in CME, other endocytic mechanisms are clathrin and/or dynamin independent and rely on other proteins. Moreover, endocytosis implicates some subcellular structures, like plasma membrane, actin and lysosomes. Also, physiological conditions, such as pH and ion concentrations, represent an additional factor that mediates these events. Accordingly, endocytosis related proteins are potential targets for small molecules that inhibit endocytosis‐mediated viral entry. This review summarizes the potential of using small molecules, targeting key proteins, participating in clathrin‐dependent and ‐independent endocytosis, as variant‐independent antiviral drugs against SARS‐CoV‐2 infection. The review takes two approaches. The first outlines the potential role of endocytic inhibitors in preventing endocytosis‐mediated viral entry and its mechanism of action, whereas in the second computational analysis was implemented to investigate the selectivity of common inhibitors against endocytic proteins in SARS‐CoV‐2 endocytosis. The analysis revealed that remdesivir, methyl‐β‐cyclodextrin, rottlerin, and Bis‐T can effectively inhibit clathrin, HMG‐CoA reductase, actin, and dynamin I GTPase and are more potent in inhibiting SARS‐CoV‐2 than chloroquine. CME inhibitors for SARS‐CoV‐2 infection remain understudied.
Keywords: clathrin‐mediated endocytosis, endocytic inhibitors, mutations, SARS‐CoV‐2 variants
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- AP2
adaptor protein complex
- ARF1
ADP‐ribosylation factor 1
- ARP2/3
actin nucleation factor
- BAR
bin–amphiphysin–Rvs
- CALM
clathrin assembly lymphoid myeloid leukemia
- CCP
clathrin‐coated pits
- CCVs
clathrin‐coated vesicles
- Cdc42
cell division control protein 42 homolog
- CIE
clathrin‐independent endocytosis
- CLICs
clathrin‐independent carriers
- CME
clathrin‐mediated endocytosis
- CvME
caveolae‐mediated endocytosis
- Drp‐1
dynamin‐related protein‐1
- DynII
dynamin II
- Eps15
EGF‐receptor phosphorylation substrate
- ENDOA2
endophilin A2
- FCHo2
fer/Cip4 homology domain‐only proteins 1/2
- FEME
fast endophilin‐mediated endocytosis
- GEEC CLIC/GEEC
glycophosphatidyl‐inositol anchored protein‐enriched endosomal compartments
- GPCRs
G protein‐coupled receptors
- GPI‐APs
glycosylphosphatidylinositol‐anchored proteins
- GRAF1
GTPase activating factor
- GSK3β
serine/threonine protein kinase
- Hsc70
heat shock protein 70
- OCRL
oculocerebrorenal lowe syndrome protein
- PI3KC2α
phosphatidylinositol 3‐kinase C2α
- SARS‐CoV‐2
coronavirus 2
- TfR
transferrin receptor
- TMPRSS2
transmembrane serine protease 2
- N‐WASP
wiskott–aldrich syndrome protein
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) entry into the host cell is crucial for viral infectivity. It is well‐established that the SARS‐CoV‐2 spike (S) glycoprotein interacts with the host cell surface receptors. Many previous studies have used purified spike (S) glycoprotein and lentivirus pseudo‐typed with spike glycoprotein and found that SARS‐CoV‐2 undergoes rapid clathrin‐mediated endocytosis (CME) to gain entry into cells. 1 , 2 , 3 , 4 This scenario is quite similar to the paradigms of receptor‐mediated endocytosis (RME), in which cells internalize their own liganded surface receptors for signal management or to uptake nutrients, such as iron, and cholesterol, from their microenvironment. Although CME is regarded as the most common pathway for virus entry, some reports suggested that different viruses may adopt other routes to infect host cells, such as the transmembrane serine protease 2 (TMPRSS2) pathways and caveolae‐mediated endocytosis (CvME). 5 Organs, especially the lung and intestines, for example, are highly susceptible to viral infection owing to the large amounts of TMPRSS2 found in their cell membranes. 6 , 7 Moreover, the virus may utilize an endocytic pathway when TMPRSS2 is absent. 7 These mechanisms provide efficient routes that enable the virus to evade host immune surveillance, gain sustainable intracellular replication, and develop the associated pathogenesis. The recent SARS‐CoV‐2 pandemic, with its associated high mortality rate, elicited tremendous research efforts to explore the biology, viral–host cell interactions, and screening methods for the virus and to suggest therapeutic approaches via either vaccination or the use of antiviral drugs. The rapid appearance and transmission of new SARS‐CoV‐2 variants as well as the associated high mortality rate have imposed more challenges on research communities to modify existing detection methods and therapeutic protocols. 8 , 9 , 10 , 11 Although vaccination strategies have become the gold standard for preventing SARS‐CoV‐2 infections, challenges have emerged because of mutational variants gaining increased infectivity potential and becoming less responsive to vaccines. 12 , 13 Recent studies compared the titer of neutralizing antibodies of natural SARS‐CoV‐2 infection and Sinovac vaccination to evaluate the efficacy of vaccination. People vaccinated with Sinovac were found to have a lower titer of neutralizing antibodies than the naturally infected patients, 14 indicating that live SARS‐CoV‐2 virus may induce a stronger immune response than the inactivated virus. Heterogeneous vaccination may be required to induce a strong immune response against the virus. Several vaccines that are effective against SARS‐CoV‐2 variants have been developed, such as molnupiravir. Molnupiravir can inhibit the replication of SARS‐CoV‐2 by inducing lethal mutagenesis. 15 Its bioavailability and tolerability have been confirmed in clinical trials. Although molnupiravir has been shown to be effective against SARS‐ CoV‐2, its genotoxicity is concerning; thus, longer‐term clinical trials are needed to complete the safety evaluation. 15 This increases the importance of developing variant‐independent therapeutic strategies. Because of the close resemblance between ligand–receptor CME and the proposed SARS‐CoV‐2 entry route, endocytic proteins serve as potential anti‐SARS‐CoV‐2 targets. Even before the emergence of SARS‐CoV‐2, this infection route was determined to be the preferred route for other viruses such as human hepatitis C virus (HCV), 16 enterovirus 71, 17 Ebola virus, Dengue‐3 virus, 18 and many others. The clathrin‐independent endocytosis (CIE) has also been proposed as an additional route of SARS‐CoV‐2 cell entry. 19 The SARS‐CoV‐2 outbreak accelerated the use of endocytic proteins involved in CME, making endocytic proteins and other cellular entry pathways antiviral targets for potentially blocking SARS‐CoV‐2 entry into host cells. 20 CME inhibitors have been shown to inhibit spike (S) protein endocytosis and pseudo‐virus infectability in recent studies. This is accomplished by preventing the formation and disassembly of clathrin lattices on cytoplasmic surfaces and endosomes. 5 In the same context, destruction of clathrin heavy chains decreases the internalization of viral spike protein. 20 Additionally, inhibitors targeting dynamins I and II and dynamin‐related protein‐1 (Drp‐1) disrupted the endocytic pathway and prevented viral entry through CME pathways. 21 Other studies revealed that inhibition of β‐arrestin blocked viral host cell entry. 20 , 22 Accordingly, this review summarized endocytic protein inhibitors that potentially block SARS‐CoV‐2 entry through endocytic pathways, with a focus on the most common CME‐related proteins. Additionally, molecular docking analysis of these inhibitors was conducted to provide a predictive model of their relative potential as anti‐SARS‐CoV‐2 therapeutic drugs.
2. ENDOCYTIC PATHWAYS AND TRAFFICKING
2.1. Phagocytosis and pinocytosis
Phagocytosis is a mechanical processes that allows the ingestion of particles larger than 0.5 μm, such as apoptotic cells, foreign substances, and microorganisms. 23 , 24 Phagocytosis occurs via a series of well‐studied steps, which include phagocyte detection, recognition of foreign molecules, and movement toward the target cargo for elimination. After physically binding to the targets, pseudopodia formation is initiated, and the pseudopodial membrane fuses to form a vesicle with the targeted material inside, thus forming a phagosome. 24 Finally, the phagosome is merged with a lysosome, where its components are enzymatically hydrolyzed.
Conversely, pinocytosis is used to uptake small particles (less than 0.1 μm) and to ingest fluid by modulating the cellular plasma membrane without pseudopodia formation. 25 Pinocytosis occurs through the formation of vesicles, followed by endocytosis into the cell and fusion with lysosomes to achieve complete digestion Figure 1. Based on particle size, pinocytosis is classified into micropinocytosis and macropinocytosis. The former occurs via the formation of a vesicle bud from the cell to engulf a small particle with a diameter of ≤0.1 μm, whereas the latter occurs indirectly without the formation of vesicles and is commonly observed in white blood cells that engulf larger particles (0.5–5 μm).
FIGURE 1.
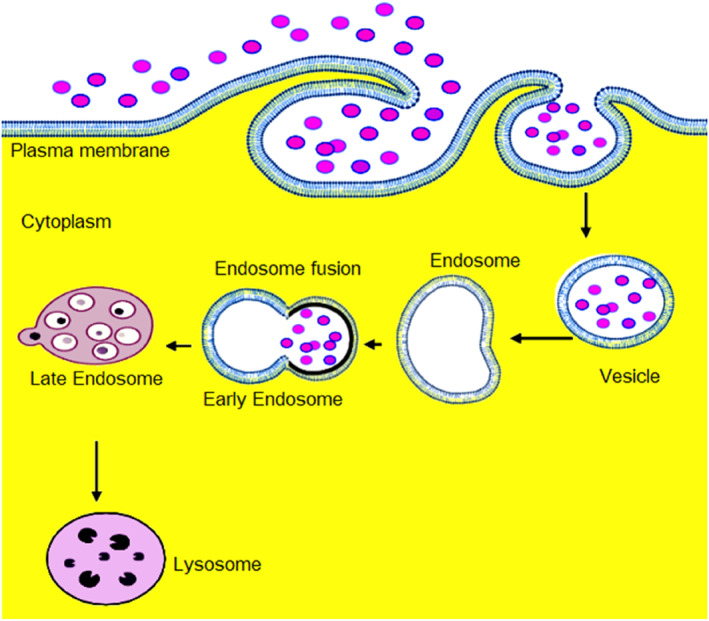
Pinocytosis pathway in which the cell absorbs fluid by invagination and formation of a separate tiny vacuole around each droplet called a vesicle
2.2. CME involves a machinery of proteins, adaptors, and modifiers
In response to extracellular stimuli, such as hormones and growth factors, and to changes in the cellular microenvironment containing nutrients and oxygen, the relative abundance of cell surface receptors is accommodated to perform a range of functions, such as nutrient uptake and signaling. The relative abundance of these receptors is mainly controlled by their endogenous expression, trafficking, and internalization via CME. 26 , 27 The latter mechanisms which include endogenous expression, trafficking, and internalization regulates receptor internalization, recycling, and degradation. These processes require several endocytic cellular proteins that sequentially function to engulf liganded receptors into a fully formed endosome containing ligand–receptor complexes.
CME occurs through two approaches, clathrin‐coated pits (CCPs) and clathrin‐coated vesicles (CCVs). 20 , 28 CME is primarily a clathrin‐ and dynamin‐dependent mechanism and includes several other proteins, such as β‐arrestin, and adaptor proteins, such as adaptor protein‐2 (AP2). The process involves a series of steps, including initiation of CCPs, followed by their stabilization and maturation, and finally, membrane fission. The AP2 heterotetramer, comprised of α, β2, μ2, and σ2 subunits, plays a fundamental role in the initiation of CCPs. 29 Thereafter, scaffolding molecules, including FCHo and eps15, are recruited to participate in either nucleating or stabilizing nascent CCP. This process aids the newly formed CCP to develop into a CCV. 28 The second stage of CCP is maturation and fission and is dependent on regulatory proteins such as amphiphysin, endophilin, Wiskott–Aldrich Syndrome Protein (N‐WASP), cortactin, myosin 1E, sorting nexin 9 (SNX9), synaptojanin, phosphatidylinositol 3‐kinase C2α (PI3KC2α), and phosphatidylinositol phosphate (PIP5K). 30 , 31 Dynamin, a large GTPase, is activated at low levels in nascent CCPs and regulates CCP initiation and maturation. 32 It then forms short helical rings around the necks of deeply invaginated CCPs to catalyze their membrane fission. In CCVs, dynamin is used by proteins with Bin–Amphiphysin–Rvs (BAR) lipid specificity domains, including amphiphysin, endophilin, and SNX9. 33 Subsequently, GTP hydrolysis occurs to drive membrane fusion. After the detachment of the vesicle from the plasma membrane, both heat shock cognate 70 (HSC70) and its cofactor auxilin disassemble the clathrin coat. 28 This allows the uncoated vesicle to progress to lysosomal fusion or be recycled to the cell membrane. For lysosomal fusion, multiple homotypic fusion events occur. This step involves cyclin G‐associated kinase (GAK), HSC70, and oculocerebrorenal Lowe syndrome protein (OCRL). 34 Lysosomal fusion transports the CCV cargo to early endosomes. The internalization of low‐density lipoprotein (LDL) receptors and iron‐binding protein are the best examples of CME, which enable the cellular uptake of LDL cholesterol and iron from the microenvironment Figure 2.
FIGURE 2.
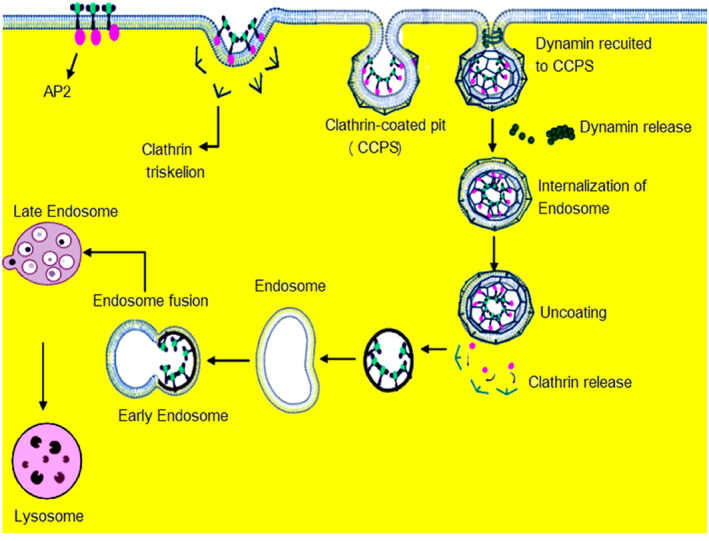
Clathrin‐dependent endocytosis pathways in which the metabolites, proteins and viruses transported into the cell through binding to cell surface receptor. The molecule‐receptor complex enters the cell by in‐folding of the plasma membrane, which eventually gets pinched off into a vesicle. During receptor‐mediated endocytosis (RME), endocytic proteins including, AP2, and clathrin triskelion coats the budding vesicle give spherical form called clathrin‐coated pits (CCPs) and the dynamin recruited to the CCPS. Then the dynamin releases from CCPS forming endosome and un‐coating the clathrin release and bind to the lysosome.
2.3. Caveolae‐mediated endocytosis (CvME)
Caveolae/glycolipid raft‐mediated endocytosis is the best‐characterized clathrin‐independent but dynamin‐dependent endocytic pathway. 35 Caveolae are flask‐shaped plasma membrane invaginations with a diameter of 50–80 nm. Caveolins, considered the main structural proteins, are members of the caveolin gene family and comprise caveolins 1, 2, and 3. Caveolar membranes are enriched with sphingolipid, cholesterol, signaling proteins, and clustered glycosylphosphatidylinositol‐anchored proteins (GPI‐APs). 36 Caveolar‐mediated endocytosis occurs when foreign particles, such as viruses, adhere to the membrane and are trapped in the caveolae. This event initiates a signal transduction cascade in which certain proteins are phosphorylated, resulting in the depolymerization of the cortical actin cytoskeleton of the caveolae cargo, which is followed by the recruitment of actin monomers, forming an actin patch. Concurrently, dynamin is recruited to the caveolae cargo, causing it to be detached from the cell membrane and translocated to the cytosol. 37 According to some reports, endocytic cargo can be internalized by various mechanisms in different cell types or it can switch pathways in a single cell type under certain conditions. In human skin fibroblasts and endothelial cells, for example, albumin is internalized by caveolae, whereas in Chinese hamster ovary cells, it is internalized via a RhoA‐dependent mechanism. 35 , 38
2.4. CLIC/GEEC endocytic pathway
The involvement of crescent‐shaped tubular clathrin‐independent carriers (CLICs) that evolve into glycosylphosphatidylinositol (GPI)‐anchored protein‐enriched early endocytic compartments (GEECs) is referred to as the CLIC/GEEC pathway, which is CIE. This pathway is regulated by some proteins including small GTPases, such as Arf1 and CDC42, 39 , 40 GTPase activating factor GRAF1, 41 actin nucleation factor ARP2/3, 42 and the BAR domain protein IRSp53. These regulators are involved in the uptake of cargo such as hyaluronic acid receptor/CD44, GPI‐anchored proteins (CD59 and Thy‐1), and cholera toxin. 43 Notably, CLIC/GEEC endocytosis is dynamin‐independent for endogenous cargo and not strictly dynamin‐dependent for exogenous cargo such as toxins. 44 , 45 The CLIC/GEEC pathway, unlike micropinocytosis, is a high‐capacity pathway in which the internalized fluid phase is delivered to the cell. Furthermore, in several cell types, the CLIC/GEEC pathway is responsible for the rapid recycling of cell membranes. For instance, the entire cell membrane of mouse embryonic fibroblasts is recycled in less than 15 min via the CLIC/GEEC pathway. In addition to nutrient and toxin uptake, the CLIC/GEEC pathway is recognized as a portal for viral infection. 46
2.5. Flotillin‐dependent endocytosis
Flotillin proteins are ubiquitous in cells and play several roles, including cell adhesion, signal transduction through tyrosine kinases receptors, and cellular trafficking via pathways involving flotillin‐1/reggie‐2 and flotillin‐2/reggie‐1. 47 Flotillins are acylated at the N‐terminal stomatin/prohibitin/flotillin/HflK/C (SPFH) domain and oligomerized at the C‐terminal (flotillin domain). Therefore, they constitutively associate with cholesterol‐enriched lipid microdomains, which participate in endocytosis and other cellular trafficking processes. Flotillin microdomains are dynamic, and upon stimulation, specifically cluster cargo molecules, such as Alzheimer's amyloid precursor protein, dopamine transporter, and epidermal growth factor receptor, at the plasma membrane into flotillin rafts for pre‐endocytic cluster formation before endocytosis. Notably, following their clustering, cargo molecules are endocytosed without any involvement of flotillin. Flotillins have been shown to be functionally dependent on each other, where deletion of one perturbs the stability of the other. Flotillin‐1 seems to be more dependent on flotillin‐2. 48 For instance, in neuroblastoma cells and primary hippocampal neurons, depletion of flotillin‐2 impairs amyloid precursor protein endocytosis, 49 whereas flotollin‐1 remains intact.
2.6. Post‐endocytic events and factors controlling endocytosis
Typically, after macromolecules are internalized by one of the above‐described pathways, they are sorted by endosomes, 50 where they may either be recycled to the plasma membrane, degraded by lysosomes, or sent across the epithelium, endothelium, and blood–brain barrier through endocytosis. 50 , 51 Moreover, the endocytic process is involved in cell signaling from surface receptors such as G protein‐coupled receptors (GPCRs), neurotransmission, and viral infections. Endocytic pathways are multistep mechanisms and are controlled at several stages including scission mechanisms, coat‐associated protein assembly, and in the integration of these steps. 52 In such ordered events, the cell membrane and the soluble endocytic machinery are involved in endocytosis and endosome maturation and trafficking. Endocytic trafficking relies on membrane structural heterogeneity, including the presence of specific lipid and derived lipid components as well as protein domains. This structural pattern plays a pivotal role in defining a region of the membrane to initiate the endocytic event. 52 Endocytic pathways participate in the propagation of membrane heterogeneity, which switches phosphoinositide species throughout the endocytic trafficking process. These metabolic changes are important for the regulation of each step of the intracellular sorting process following endocytosis. 53
3. TYPES OF ENDOCYTIC PROTEIN INHIBITORS
Endocytic inhibitors include various low‐molecular‐weight compounds and peptides. A standardized classification system is yet to be established; however, these inhibitors may be subgrouped according to their chemical nature, therapeutic use, sources, or mode of action as follows: chemical, pharmacological, natural, and genetic inhibitors. 54 , 55 This primitive categorization system is based on the available literature in which various endocytic pathways were targeted. Other classification approaches may consider their targeted endocytic pathway or their selective cellular proteins. This may include inhibitors known to interfere with CME, lipid rafts, CIE, macropinocytosis, phagocytosis, and β‐arrestin. 54 Additional suggested classification approaches may differentiate between clathrin‐ and/or dynamin‐dependent pathways, in which case other intervening factors should be considered, such as evidence‐based selectivity (specific vs. non‐specific), sustainable effects (reversibility), and the affected subcellular organelles.
3.1. Chemical inhibitors
Chemical inhibitors include various molecules and peptides known to inhibit CME, CvME, CIE, CLIC/GEEC, phagocytosis, and micropinocytosis Table 1. Chemical inhibitors may alter the cellular physiology (physiological modifiers) or directly interact with integral endocytic proteins. Physiological modifiers non‐specifically inhibit the endocytic pathway by altering the normal physiological conditions of cells and/or subcellular organelles. For example, clathrin lattices on the plasma membrane may be inhibited by chemical inhibitors such as hypertonic sucrose and 2‐deoxy‐d‐glucose/sodium azide, which trap clathrin in microcages. 56 Endocytosis may also be inhibited by chemical inhibitors that reduce ATP and NADH levels by inhibiting glycolysis. 57 Another chemical inhibitor, monensin, inhibits the endocytic pathway through disruption of the proton gradient. 58 Other chemical inhibitors directly and specifically interact with specific endocytic proteins, including arrestin and dynamins. Barbadin, for example, inhibits the molecular interaction between β‐arrestin and AP2. 59 , 60 Additionally, the GTPase activity of dynamins can be targeted by several compounds including Rhodadyn‐D10, Naphthaladyn 29, Dynole, Dyngo, and Dynasor. These compounds act as competitive inhibitors in the GTP domain and inhibit the GTPase activity of Dyn 1. 61 Similarly, bisphosphonates can inhibit the effect of Dyn II by inhibiting GTPase activity. 62 Disrupting cell membrane structural integrity may be another mechanism for inhibiting endocytosis via a different route, considering the critical role of bisphosphonates in the plasma membrane in the endocytic pathway. Other chemical inhibitors, such as methyl‐β‐cyclodextrin and nystatin, remove cholesterol from the plasma membrane and interfere with the fluid phase, thereby inhibiting CvME. 63 Additionally, the chemical inhibitors filipin, LG186, and 7‐keto‐cholesterol interact with membrane cholesterol and prevent the close packing of acyl chains, resulting in inhibition of the CLIC/GEEC pathway.
TABLE 1.
Summary of chemical inhibitors with their mode of actions
| No | Endocytosis of inhibitors | Mode of action | Target for inhibition | Summary | References |
|---|---|---|---|---|---|
| Chemical inhibitor | |||||
| 1 | Hypertonic sucrose | Inhibition takes place by the dispersion of clathrin lattices on the plasma membrane. It also traps clathrin in microcages. | CME | Nonspecific inhibitor. affects non‐clathrin‐mediated internalization pathway and interferes with fluid phase macropinocytosis. | 57 , 160 , 161 , 162 |
| 2 | Cytosol acidification | Inhibits the scission of the clathrin pits from the membrane. | CME | Nonspecific inhibitor interferes with macropinocytosis and the actin cytoskeleton. | 99 , 163 , 164 |
| 3 | Potassium depletion | Activates the aggregation of clathrin. | CME | Nonspecific inhibitor and affects the actin cytoskeleton. | 164 , 165 |
| 3 | 2‐Deoxy‐d‐glucose/Sodium azide | Decreases ATP and NADH by inhibiting glycolysis. | CME | ‐ | 57 , 166 |
| 4 | Barbadin | Inhibits β‐arrestin/AP2. | CME | Blocks agonist‐promoted endocytosis of the prototypical β2‐adrenergic (β2AR), V2‐vasopressin (V2R) and angiotensin‐II type‐1 (AT1R) receptors. | 60 , 61 , 168 |
| 5 | Monodansylcadaverin | Stabilizes clathrin‐coated vesicles. | CME | Causes changes in actin dynamics. | 168 , 169 , 170 |
| 6 | Phenylarsine oxide | Inhibits O2 consumption and decreases cellular ATP content overlap with those used to inhibit protein internalization. | CME | Inhibits macropinocytosis and phagocytosis. | 172 , 173 , 174 |
| 7 | Bolinaquinone | Affects the function of clathrin‐coated vesicles. | CME | − | 175 , 176 |
| 8 | Peptide inhibitors Wbox 2 | Wbox2 is non‐specific and inhibits clathrin‐independent endocytosis; proposed to be due to either upregulation of clathrin‐independent endocytosis (CIE) during clathrin terminal domain overexpression or that SNX9 is required for CIE, and inhibition by Wbox2 sequesters it from CIE. | CME | − | 176 |
| 9 | Bis‐Ts | Most potent of the dimeric tyrphostins (tyrosine phosphorylation inhibitors and inhibits dynamin). | CME | − | 178 , 179 , 180 |
| 10 | Iminodyns | Interacts with and inhibits dynamin. | CME | − | 181 , 182 |
| 11 | Rhodadyn D10 | Inhibits dynamin 1 by blocking GTPase activity. | CME | − | 183 , 184 |
| 12 | Pthaladyn 23 | Inhibits dynamin 1 by blocking GTPase activity. | CME | − | 185 , 186 |
| 13 | Naphthaladyn 29 | Competitive inhibitor in GTP domain and blocks GTPase activity of dynamin 1. | CME | − | 61 |
| 14 | Pyrimidyn 6–7 | Most potent dynamin 1 inhibitors. | CME | − | 186 |
| 15 | Bisphosphonates | Blocks dynamin 2. | CME | Inhibits the uptake of transferrin as well as adenovirus and simian virus 40. | 63 |
| 16 | Monensin | Affects proton gradient. | CME | Inhibits SARS‐COV‐2 infection. | 59 , 188 |
| 17 | ES9‐17 | It is a potent and selective CME that binds to N‐terminal domain of clathrin heavy chain. | CME | Acts as an inhibitor of the CHC function; cytosol acidification | 33 |
| 18 | Methyl‐β‐cyclodextrin | Removes cholesterol from plasma membrane. | Caveolae‐mediated endocytosis/lipid rafts | Interferes with fluid‐phase endocytosis and clathrin‐mediated endocytosis. | 63 , 188 , 189 |
| 19 | Filipin | Binds to cholesterol in the membrane. | CIE | Toxic at higher concentrations. Inhibits clathrin‐mediated endocytosis. | 190 , 191 , 192 |
| 20 | Nystatin | Binds to cholesterol. | Lipid rafts/cholesterol‐enriched microdomains/caveolae | Interferes with other uptake mechanisms because of changes in membrane fluidity. | 194 , 195 |
| 21 | 7‐keto‐cholesterol | Prevents the close packing of acyl chains. | CLIC/GEEC | − | 141 |
| 22 | LG186 | Reversible inhibitor of Arf‐GEF function. | CLIC/GEEC | − | 140 , 196 |
| 23 | Cytochalasin D, latrunculin | Depolymerizes F‐actin. | Phagocytosis macropinocytosis | Affects most endocytic pathways. | 197 , 198 , 199 |
| 24 | Amiloride derivatives (EIPA and HOE‐694) | Inhibits Na+/H+ exchange | Macropinocytosis | May affect actin. | 65 , 66 , 200 |
| 25 | Dynasore | Inhibits dynamin by blocking GTPase activity. | CME | Interferes with actin. | 27 , 62 , 138 , 201 |
| 26 | Dynole, Dyngo | Inhibits dynamin by blocking the GTPase activity of dynamin I. | CME | Interferes with actin. | 202 , 203 |
| 27 | Pitstop 2 | Interferes with binding of proteins to the N‐terminal domain of clathrin. | CME | Affects most forms of clathrin‐independent endocytosis and causes decreasing in PM mobility. | 204 , 205 |
In another context, although the principles governing the activation and organization of the actin cytoskeleton during CME are not fully understood, there is a spatiotemporal relationship between the generation of actin platform and the formation of CCVs. Accordingly, a few studies investigated the inhibition of CME by disrupting actin formation, where amiloride derivatives (EIPA and HOE‐694), cytochalasin D, and latrunculin were found to inhibit Na+/H+ exchange and disrupt actin formation. 65 , 66
3.2. Pharmacological inhibitors
Pharmacological inhibitors include U.S. Food and Drug Administration (FDA)‐approved medications used for therapeutic purposes that simultaneously affect endocytic pathways as shown in Table 2. Chlorpromazine (CPZ), for example, is used to treat psychotic disorders, such as schizophrenia or manic‐depression, in adults. 66 Some studies reported that phenothiazine derivatives, including CPZ, may affect the formation of CCVs and then inhibit CME. 67 , 68 Similarly, during viral infection, chloroquine inhibits the formation and functioning of CCVs and its analogs inhibit the acidification of endosomes. 69 , 70 Other pharmacological inhibitors, including itraconazole, vinblastine, imipramine, auranofin, terfenadine, and flubendazole, inhibit plasma membrane ruffle formation, which is an early step in macropinocytosis. 27 , 65 Furthermore, the PI3K inhibitors wortmannin and LY294002 inhibit the activity of protein kinase C delta, thereby inhibiting phagocytosis and macropinocytosis. 71 Lipid rafts are rich in glycosphingolipids, cholesterol, and signaling proteins, all of which play a vital role in the internalization and disruption of parasitic, bacterial, and viral infectious particles. 72 Statins, which are commonly used to treat hyperlipidemia, block cholesterol synthesis by inhibiting the activity of 3‐hydroxy‐3‐methylglutaryl CoA (HMG‐CoA) reductase, which is used for the therapeutic reduction of cholesterol‐containing plasma lipoproteins, and then affect the CvME pathway. 73 Another pharmacological inhibitor, genistein, interferes with dynamin and may affect endocytic pathways such as CME. 74
TABLE 2.
Summary of pharmacological inhibitors with mode of action
| No | Endocytosis inhibitors | Mode of action | Target of inhibition | Summary | References |
|---|---|---|---|---|---|
| Pharmacological inhibitors | |||||
| 1 | Chlorpromazine | Translocates clathrin and AP2 adaptor protein from the cell surface to intracellular endosomes. | CME | Inhibits clathrin‐independent endocytosis in some cells. | 64 , 189 , 206 |
| 2 | Phenothiazines | Affects formation of clathrin‐coated vesicles. | Phagocytosis CME | Inhibits dynamin II (dynII) at similar concentrations. | 68 , 69 , 207 |
| 3 | Chloroquine | Affects the function of clathrin‐coated vesicles. | CME | Chloroquine and its analogs inhibit SARS‐COV‐2 acidification of endosomes during the events of viral replication and infection. | 70 , 71 , 208 |
| 3 | Itraconazole | Inhibits plasma membrane ruffle formation, a critical early step in macropinocytosis. | Macropinocytosis | − | 66 , 209 |
| 4 | Vinblastine | Macropinocytosis | − | 66 , 153 , 209 | |
| 5 | Imipramine | Macropinocytosis | Inhibits phagocytosis. | 66 , 209 , 210 | |
| 6 | Auranofin | Macropinocytosis | − | 65 , 210 | |
| 7 | Terfenadine | Macropinocytosis | − | 66 , 212 | |
| 8 | Flubendazole | Macropinocytosis | − | 66 , 212 | |
| 9 | Genistein | Interferes with dynamin and may affect other endocytic processes. | CME | Inhibits several tyrosine kinases. Genistein interferes with caveolae/raft‐mediated endocytosis by blocking the Src kinase‐dependent phosphorylation of caveolin‐1 and preventing vesicle fusion. | 64 , 75 , 193 , 213 |
| 10 | Statins | Affects most endocytic mechanisms. | Caveolae‐mediated endocytosis/lipid rafts | Blocks cholesterol synthesis. Inhibitors of 3‐hydroxy‐3‐methylglutaryl CoA (HMG‐CoA) reductase used for the therapeutic reduction of cholesterol‐containing plasma lipoproteins. | 213 , 214 , 215 |
| 11 | Wortmannin and LY294002 | Affects most endocytic mechanisms | Phagocytosis, macropinocytosis | Inhibits the activity of phosphatidylinositol 3‐kinase. | 72 , 217 , 218 |
| 12 | Rottlerin | Inhibits the activity of protein kinase C delta. | 218 , 219 | ||
3.3. Natural products as anti‐endocytic factors
Some natural inhibitors can interact with lipid raft components, reducing cholesterol and glycosphingolipids in the cell membrane and thus inhibiting viral endocytosis. 73 , 76 , 77 Phytosterols, for example, interact with the components of lipid rafts, thereby reducing cholesterol content in the cell membrane 76 , 78 Table 3. Flavonoids also play a role in endocytic inhibition as they can fluidize the cell membrane (raft‐breaking effect), which leads to inhibition of both membrane fusion and CvME Table 3. Other reports demonstrated that oleuropein and hydroxytyrosol, which are phenolic compounds with antioxidant properties found in the leaves and fruits of olive plants, can inhibit the fusion of viruses with host cell membranes 76 , 79 Table 3. Furthermore, ikarugamycin, isolated from Streptomyces phaeochromogenes, affects CME, 80 whereas brefeldin A, TENin1, and auxin inhibit the formation of clathrin coats pits and ion transport. 81 , 82 , 83
TABLE 3.
Summary of natural products with mode of action
| No | Endocytosis inhibitors | Mode of action | Target of inhibition | Summary | References |
|---|---|---|---|---|---|
| Natural products | |||||
| 1 | Phytosterols | Interact with lipid raft components of the cell membrane resulting in lowering of cell membrane cholesterol or the destabilization of lipid raft structure. | Caveolae‐mediated endocytosis/lipid rafts | The phytosterol‐lipid raft interaction can affect biochemical signaling taking place downstream of the lipid rafts. | 76 , 78 |
| 2 | Flavonoids | It can initiate raft‐like domain formation, called the raft‐making effect, whereas flavonoids located at the polar interface of the lipid bilayer tend to fluidize the membrane, called the raft‐breaking effect, or initiate the formation of micellar or interdigitated structures. | Caveolae‐mediated endocytosis/lipid rafts | Induce prevention of membrane fusion is one way in which flavonoids protect an organism against viral infection. | 76 , 221 , 222 |
| 3 | Hydroxytyrosol | Phenolic compound with antioxidant properties found in the leaves and fruits of the olive (Olea europaea). | Caveolae‐mediated endocytosis/lipid rafts | Oleuropein and hydroxytyrosol inhibit the fusion of viruses with cell membranes. | 76 , 79 |
| 4 | Ikarugamycin | Affects clathrin‐mediated endocytosis. | CME | Isolated from Streptomyces phaeochromogenes and has antiprotozoal activity. | 80 , 146 |
| 5 | Brefeldin A | Inhibits the formation of clathrin coats and ion transport. | CME | Inhibitor of vesicle trafficking, on internodal cells of Chara australis. | 82 , 83 |
| 6 | TENin1 | Inhibits endocytosis, causes endomembrane protein accumulation at the pre‐vacuolar compartment, and impairs gravitropic response in Arabidopsis thaliana. | CME | − | 223 , 224 |
| 7 | Auxin | Promotes efflux from cells by a vesicle‐trafficking‐dependent mechanism. | CME | − | 81 , 225 |
3.4. Genetic inhibitors
Although chemical inhibitors are the most commonly identified inhibitors, the absolute selectivity of their actions remain a major concern. Selective CME inhibitors that target specific molecular targets may either directly or indirectly interfere with other molecules Table 4. Barbadin, for example, is a selective inhibitor of β‐arrestin/AP2 interaction, 83 ERK1/2, and cAMP accumulation in human embryonic kidney cells. 59 Barbadin also affects cellular viability and induces both apoptosis and autophagy in breast cancer cells. 84 , 85 , 86 , 87 , 88 This may give genetic inhibitors more leverage to avoid the anticipated adverse effects of chemical inhibitors. Genetic inhibitors are used to inhibit CME by suppressing the expression of specific endocytic proteins. 75 The expression of mutant forms of critical proteins involved in endocytosis and siRNA‐mediated depletion of these proteins are the most commonly used methods. 89 The mutant dynamin, K44A, is an example of a genetic inhibitor of CME. 90 Dyn2K44A was also used to inhibit CME under conditions of increased clathrin‐independent fluid endocytosis. 91 Clathrin adaptor proteins including AP180 and Epidermal growth factor receptor substrate 15 (Eps15) target regulatory proteins of the CME pathway and thus inhibit endocytosis. The C‐terminal clathrin‐binding domain of AP180 or the truncated form of Eps15 inhibits the initial step (formation of CCPs) of CME. 92 , 93 Furthermore, the C‐terminal (Hub) region of clathrin expression effectively inhibits CME. 94 Endophilin A2 (ENDOA2) depletion prevents the reshaping of the membrane before scission of the vesicle and inhibits fast endophilin‐mediated endocytosis (FEME). 95 The same approach was used to inhibit other endocytic pathways. Caveolin‐1 (CAV1) and caveolae‐associated protein 1 (CAVIN1), for example, prevented caveola formation. 96 , 97 Similarly, CtBP1 and Rabankyrin‐5 (ANKFY1) inhibited macropinosome fission from the plasma membrane and blocked macropinocytosis. 98 , 99
TABLE 4.
Summary of genetic approaches with mode of action
| No | Endocytosis inhibitors | Mode of action | Target of inhibition | Summary | References |
|---|---|---|---|---|---|
| Genetic approachs | |||||
| 1 | Dynamin mutant, Dyn K44A | Causes defects in GTP hydrolysis. | CME | Enhances fluid‐phase uptake. | 90 , 91 , 226 |
| 2 | AP180C | Causes clathrin sequestration. | CME | May cause changes in gene expression. | 94 , 226 |
| 3 | Eps15 mutant | Inhibits clathrin pit assembly. | CME | − | 228 , 229 , 230 |
| 4 | Clathrin Hub mutant | Acts as a dominant‐negative mutant of clathrin. | CME | − | 231 , 232 |
| 5 | siRNA of clathrin | Blocks the formation of clathrin pits. | CME | − | 233 , 234 |
| 6 | siRNA of AP2 | Blocks formation of AP2‐dependent clathrin pits. | CME | − | 232 , 234 |
| 7 | Clathrin (CLTC) | Clathrin depletion prevents the formation of clathrin‐coated pits. | CME | Changes in gene expression are observed due to overexpression and knocking down. | 236 , 237 |
| 8 | Endophilin A2 (ENDOA2) | Endophilin A2 depletion prevents the reshaping of the membrane before scission of the vesicle. | (FEME) | Changes in gene expression are observed due to overexpression and knocking down. | 45 , 95 |
| 9 | Caveolin‐1 (CAV1) | Caveolin‐1 depletion prevents caveola formation. | CAV | Changes in gene expression are observed due to overexpression and knocking down. | 97 , 98 |
| 10 | Caveolae associated protein 1 (CAVIN1) | Cavin‐1 depletion prevents caveola formation. | CAV | Changes in gene expression are observed due to overexpression and knocking down. | 238 , 239 |
| 11 | IRSp53 | IRSp53 depletion interferes with actin dynamics. | CLIC/GEEC | Changes in gene expression are observed due to overexpression and knocking down. | 240 , 241 |
| 12 | PICK1 | PICK1 depletion interferes with actin dynamics, specifically the Arp2/3 complex. | CLIC/GEEC | Changes in gene expression are observed due to overexpression and knocking down. | 242 , 243 |
| 13 | CtBP1 (CTBP1) | CtBP1 depletion inhibits macropinosome fission from the cell surface. | Macropinocytosis | Changes in gene expression are observed due to overexpression and knocking down. | 100 , 244 |
| 14 | Rabankyrin‐5 (ANKFY1) | Depletion of rabankyrin‐5 inhibits the formation of macropinosomes. | Macropinocytosis | Changes in gene expression are observed due to overexpression and knocking down. | 245 , 246 |
4. ENDOCYTOSIS AND TRANSCYTOSIS OF SARS‐CoV‐2
Viral entry into host cells occurs through two distinct pathways: viruses deliver their nucleic acid material into the cytosol after their envelopes fuse with the plasma membrane (pH‐independent entry), 101 or they use the host cell's endocytic machinery (pH‐dependent entry), such as CME and CvME, in addition to poorly characterized routes such as clathrin‐ and caveolae‐independent endocytic pathways Figure 3. A previous study reported that SARS‐CoV‐2 is similar to SARS‐CoV, which can fuse with host cell membranes and internalize into the host cell. Following endocytosis‐mediated entry at an acidic pH, viral and endosomal membranes fuse, and subsequently, the viral genome is released into the cytosol. 102 Initially, the viral spike (S) protein binds to the host cell receptor angiotensin‐converting enzyme 2 (ACE2). 19 This binding triggers endocytic infection similar to the GPCR internalization process. During the entry process, CCPs are formed by AP2 complexes, 2 following which virus‐containing CCVs are released. During recycling, structural proteins, including the viral protein capsid, are moved to late endosomes containing lysosomes for degradation; however, viral nucleic acid RNA initiates the formation of a new virus that egresses from lysosomes. 2
FIGURE 3.
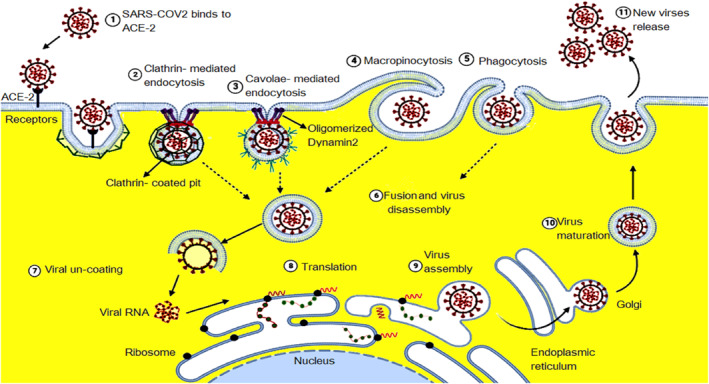
Mechanisms of SARS‐CoV‐2 entrance through binding to the surface receptor (1) and enter to the cell by clathrin‐mediated endocytosis (CME) (2), or caveolae‐mediated endocytosis (3), macropinocytosis (4), and phagocytosis (5) depending on the types of endocytic proteins forming endosome (6) and then viral uncoating (7) releasing viral RNA and then viral translation (8) and eventually virus assembly (9) and maturation (10) then viral release (11).
In contrast to CME‐mediated entry, macropinocytosis‐mediated entry follows a non‐specific internalization pathway that does not require a receptor. 102 , 103 Although macropinocytosis is not the key entry pathway of SARS‐CoV‐2, 103 , 104 the virus may activate the signaling pathways that trigger macropinocytosis and enter the cell by promoting actin‐mediated membrane formation and lamellipodia formation at membrane perturbation sites to close them and form large, irregular vesicles. 105 , 106 Other pathways, such as CIE, could be used by SARS‐CoV‐2 to infect host cells. This pathway is regarded as a cholesterol‐sensitive mechanism. CIE is divided into dynamin‐dependent CvME and dynamin‐dependent or ‐independent non‐clathrin non‐CvME. 105 , 107 , 108 Wang et al. clarified that the coronavirus can infect host cells through various mechanisms, including RME, CIE, caveolae‐independent endocytosis, and lipid raft formation. 19 , 105 The plasma membrane contains sphingolipids and cholesterol (lipid rafts), which are domains enriched with signaling and transport proteins. 105 Entry of SARS‐CoV‐2 by endocytic pathways involving lipid rafts provides new insight into the pathogenesis of acute respiratory distress syndrome and the elevated levels of proinflammatory cytokines associated with it. 105 , 106 Wang et al. demonstrated that coronaviruses, including SARS‐CoV‐2, can infect host cells deficient in CME. 19
On the other hand, Cholesterol‐mediated, dynamin‐dependent, and receptor‐mediated pathways include CvME. CvME pathway forms a specific type of flask‐shaped structure called caveola existing on the cell membrane enriched in cholesterol and sphingolipid as shown in Figure 3. Bastiani, Parton, Cheng, and Nichols clarified that caveolae are microdomains of membrane rafts that can bud into cells as caveosomes, which fuse to form early endosomes for delivery. 109 , 110 The caveolar coat consists of an inner layer containing caveolins and an outer membrane containing cavins, which help to support the caveolin scaffold, promote membrane curvature, and caveolae budding. 111 , 112 SARS‐CoV‐2 is internalized via the ACE‐2 receptor into the host cell by CvME, forming caveosomes and undergoing degradation after accumulation of overexpressed Cav1. 113 The endocytic vesicles, which are formed in lipid rafts, can be stabilized by flotillin (flotillin‐dependent endocytosis). 105 Many studies do not support viral entry by the flotillin‐dependent endocytic pathway because the mechanisms remain unclear. 114 , 115 Glebov showed that SARS‐CoV‐2 can infect host cells via the flotillin‐dependent endocytic pathway, which should be supported with an in vivo clinical study. 106 Furthermore, viruses such as adenoviruses can enter via non‐clathrin non‐CvME in lipid rafts. These viruses can infect host cells through GRAF1‐dependent endocytosis, which is also named as the CLIC/GEEC pathway. 116 The CLIC/GEEC pathway is sensitive to cholesterol depletion and may decrease viral entry when it is cholesterol‐dependent but raft‐independent. 105 , 117 Glebov showed that SARS‐CoV‐2 can infect host cells via the CLIC/GEEC pathway by forming GEECs, which are free of clathrin, caveolae, dynamin, and Rac1. 106 SARS‐CoV‐2 is internalized with ACE‐2 into host cells by forming GEECs from the fusion of smaller CLICs. 105 CLICs and GEECs are formed after actin polymerization, which is regulated by Cdc42, Arf1, and GRAF1.
Transcytosis is the transport of macromolecules, supramolecular complexes, and microorganisms through the cell via membrane‐bound carriers. It is mediated by two major transportation mechanisms. The first mechanism employs CCVs to internalize transported molecules, which enter cell compartments and are then released into the cell via exocytosis. The second mechanism employs caveolae, without their entry into cell compartments. 118 , 119 Notably, the former mechanism is used in intestinal cells and the latter in endothelial cells. SARS‐CoV‐2 enters the intestinal epithelial and M cells by binding to ACE‐2. Thereafter, ACE‐2 is split by cellular proteases such as TMPRSS2. 120 Following entry into the cell with common recycling endosomes, SARS‐CoV‐2 is translocated into the multivesicular bodies/late endosomes. In these endosomes, the viral envelope fuses with endolysosomal membranes, which is triggered by cathepsins, and ultimately releases the genetic material in the cell. 121
5. ENDOCYTIC PROTEINS AS ANTIVIRAL TARGETS AGAINST SARS‐CoV‐2 ENTRY
Since the emergence of SARS‐CoV‐2, various efforts have been made to present new medications for inhibiting its infection with minimal adverse effects. 122 , 123 To achieve this goal, two strategies were adopted. The first was to target the biology of the virus, such as inhibiting its replication in host cells, whereas the second involved targeting the biology of host cells. 106 In both strategies, complete elimination or minimizing the associated undesirable effects remains the primary challenge. Although both approaches have some limitations, drugs targeting the host cell might be advantageous owing to the relative structural and physiological stability of the host cells compared with drugs targeting the virus, which is constantly mutating, resulting in new variants and escalating drug resistance. 124 , 125 A study showed that disruption of the Na+/K+‐ATPase of the host cell's plasma membrane by cationic steroid inhibitors, including ouabain and bufalin, led to the inhibition of the CME pathway and inhibited coronavirus entry. 126 Similarly, ouabain and bufalin inhibited SARS‐CoV‐2's entry by the same mechanism. 127 , 128 Other studies targeted endosomes to inhibit coronavirus‐2 infection. 129 It has also been discovered that endosomal acidification factors, such as bafilomycin A1, inhibit endosomal SARS‐CoV‐2 via the CLIC/GEEC endocytic pathway, which is pH dependent 3 Figure 5. Additionally, Prabhakara et al. showed that niclosamide is considered as a probable SARS‐CoV‐2 entry inhibitor. 93
FIGURE 5.
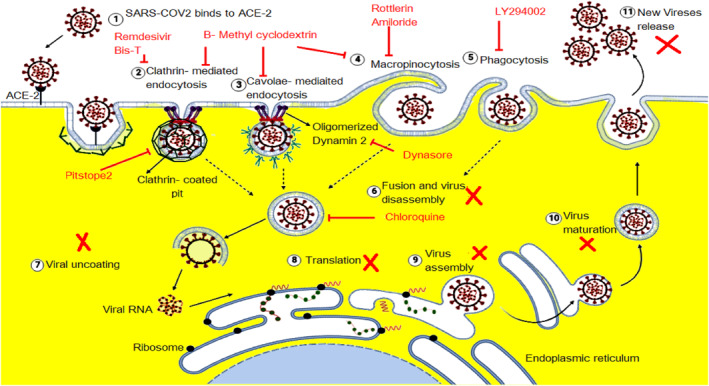
Inhibition of SARS‐CoV‐2 entrance by the large scale of endocytic proteins inhibitors including Remdesivir, Bis‐T inhibits clathrin‐mediated endocytosis (CME), β‐methyl cyclodextrin inhibits caveolae‐mediated endocytosis, CME, and macropinocytosis. Rottlerin and Amilorides inhibit the macropinocytosis pathway. LY294002 inhibits the phagocytosis process, and dynasore inhibits oligomerized dynamin2. And chloroquine inhibits the entrance of viral endosomes.
6. SARS‐CoV‐2 ENTRY INHIBITION
SARS‐CoV‐2 uses endocytic pathways to deliver its virion components into the cytoplasm. The virus infects lung cells through CME in addition to clathrin‐independent processes. 19 , 104 To achieve this, the viral spike protein binds to human ACE2 through its receptor‐binding domain. This binding is activated by human proteases. Previous studies have shown that other receptors and cellular proteases, such as CD147, neuropilin‐1, dipeptidyl peptidase 4, alanyl amino peptidase, 129 and transferrin receptor, 130 also facilitate viral entry and transmission into host cells. During tRNA‐derived RNA fragment (tRF)‐mediated infection, the viral membrane fuses with the lumen of the endosomal membrane, allowing viral RNA to enter the cytosol to infect the host cell. 2 , 100 The viral components undergo the tRF recycling pathway, which exposes SARS‐CoV‐2 capsid proteins to degradation by lysosomes in late endosomes. During this process, SARS‐CoV‐2 RNA drives the formation of a new virus that egresses from lysosomes. 2 These scenarios predict the direct involvement of endocytic machinery in SARS‐CoV‐2 infection. Accordingly, because of their potential as promising broad‐spectrum antiviral targets, endocytic pathway inhibitors have been proposed as a potential treatment Figure 4 and 5. Although the endocytic machinery includes more than 50 proteins and adaptor proteins, some of these proteins serve as key factors; clathrin, for example, is an integral member of the CME and other clathrin‐dependent endocytic pathways. This makes clathrin useful as a primary target. Additionally, clathrin is involved in cell signaling and other activities that may affect viral replication before the stage of nucleocapsid assembly. Chlorpromazine, 2‐deoxy‐d‐glucose/sodium azide, bolinaquinone, and Pitstop 2 inhibit the scission of CCVs from the plasma membrane and affect their subsequent functioning 21 , 67 , 131 , 132 Figure 5 as well as block entry of SARS‐CoV‐2 via the CME pathway. 133 Unfortunately, clinical trials have shown that these drugs have harmful adverse effects, including retinal damage when used in the long term, photosensitivity, liver damage, seizures, headaches, stomach pain, and muscle or nerve damage. 134 , 135 Inhibitor‐mediated complete inhibition of CME is not fully effective, where many studies demonstrated a partial decrease in the number of endosomes Figure 5. Nevertheless, according to clinical trials, even partial virus blockade is of significance, as viral load is related to severity of disease progression. 105 This increases the importance of using multiple endocytic protein inhibitors, such as combinations of dynasore and Pitstop 2, which can result in a substantial reduction in virus titers without affecting early to late viral gene expression 136 Figure 5.
FIGURE 4.
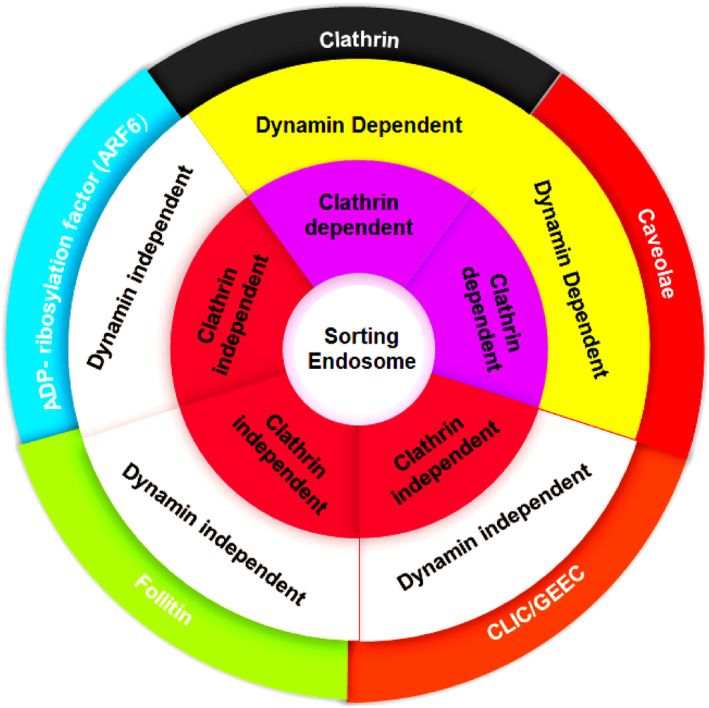
Classification of endocytic pathways according to dynamin and clathrin involvement
Membrane rafts, which are enriched with cholesterol and sphingolipids, are considered portals for viral entry. 104 Cholesterol chelating agents, including methyl‐β‐cyclodextrin, phytosterols, and flavonoids, can block the entry of SARS‐CoV‐2 through CvME/lipid rafts. 63 , 74 They reduce the cholesterol content of the host cell membrane and destabilize the lipid raft structure by interacting with lipid raft components. Additionally, many agents, such as dynasore, Dynoles, and dyngoes, can inhibit the CME pathway and CvME/lipid rafts by inhibiting dynamin GTPase activity. 137 Furthermore, there are two types of macropinocytosis inhibitors. The first type, which includes flubendazole, terfenadine, itraconazole, vinblastine, and imipraminecan, inhibits plasma membrane ruffle formation, a crucial early step in macropinocytosis, thereby inhibiting viral entry into host cells. 65 The other type, which includes amiloride and its derivatives (EIPA and HOE‐694), inhibits Na+/H+ exchange, thus affecting macropinocytotic pathways. 65 SARS‐CoV‐2 can use FEME for entry and infection. 138 ENDOA2 has been identified as a FEME inhibitor whose depletion prevents the remodeling and reshaping of the plasma membrane before vesicle scission, thereby preventing viral entry. 138 Furthermore, viral entry can occur via the CLIC/GEEC pathway, which can be inhibited by LG186 and 7‐keto‐cholesterol, thereby preventing the close packing of acyl chains and affecting the structure of the cell membrane. 139 , 140 Some pharmacological inhibitors including chlorpromazine, nystatin, amiloride, methyl‐β‐cyclodextrin, vinblastine, and itraconazole approved by FDA for human use, although others are still being evaluated. 105 In addition to improving the efficacy of endosomal inhibitors, scientists are investigating natural products for use as potential endosomal inhibitors in an effort to reduce the adverse effects caused by chemical and pharmaceutical endosomal inhibitors.
7. COMPUTATIONAL AND PRECLINICAL STUDIES OF ENDOCYTIC INHIBITORS
We conducted a molecular docking analysis of some of the endocytic protein inhibitors discussed in this review to provide more insight into their potential inhibitory effect against SARS‐CoV‐2. These computational analysis‐based investigations were performed to compare the inhibitory activity of the tested substances to that of the standard drug chloroquine by determining the plausible modes of binding between these substances and their target binding sites. The interactions of the substances with the protein hot spots (key amino acids) were also recorded. Docking studies were performed using the Molecular Operating Environment (MOE 2010) software 141 according to a previously reported procedure 142 and are described in the Supporting Information. X‐ray crystallographic structures of target proteins were obtained from the Protein Data Bank (PDB). 143
For clathrin inhibitors (Group 1), the clathrin terminal domain co‐crystallized with Pitstop 1 (IC50 = 1.80 × 104 nM) 145 as an inhibitor (PDB ID: 2XZG) 146 was used for the docking studies. Remdesivir (docking score; S = −14.46 kcal/mol) was found to have the highest inhibitory activity within the group (Figure 6, Supporting Information Figure S1, and Table 5), which was also higher than those of the co‐crystalized ligand Pitstop 1 (S = −11.23 kcal/mol) and the reference drug chloroquine (S = −11.09 kcal/mol). Remdesivir interacted with the clathrin domain active site via three hydrogen bonding interactions: two with the key amino acid Arg64 backbone using its phosphoryl group's oxygen atom and one with the key amino acid Ile93 through its amino group. It also formed three arene–H bonds with these two key amino acids. Remdesivir interacted with the active site through hydrophobic interactions with the hydrophobic side chains of Arg64, Phe91, and Ile66.
FIGURE 6.
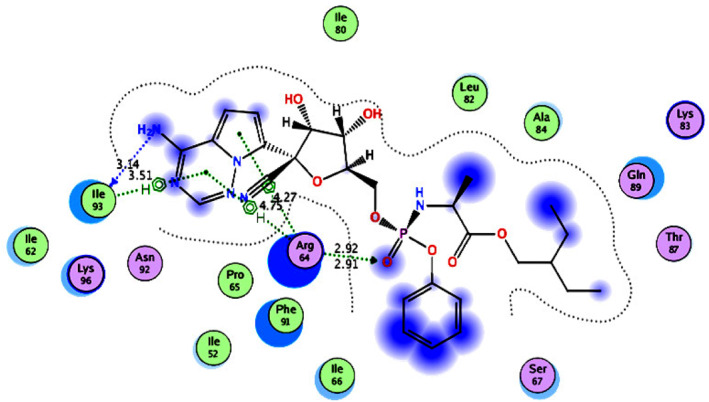
2D diagram (Distances in Å) of remdesivir (Group 1 inhibitor) showing its interaction with the Clathrin domain active site
TABLE 5.
Docking energy scores and amino acids involved in binding for pitstop 1, chloroquine, and Group 1 inhibitors docked with clathrin terminal domain
| Compound | Docking score (kcal/mol) | Amino acids involved in binding |
|---|---|---|
| Pitstop 1 | −11.23 | Phe91, Arg64, Gln89, and Lys96 |
| Chloroquine | −11.09 | Phe91 |
| Sucrose | −11.75 | Phe91, Arg64, and Ile93 |
| Chlorpromazine | −10.73 | Arg64 and Ile93 |
| Lopinavir | −14.42 | Arg64 and Lys96 |
| Remdesivir | −14.46 | Arg64 and Ile93 |
| Monodansylcadaverine | −12.11 | Arg64, Ile93, and Lys96 |
| Phenylarsine oxide | −6.45 | Phe91 |
| Bolinaquinone | −11.61 | Phe91 and Arg64 |
| ES9‐17 | −9.47 | Phe91, Arg64, Ile93, and Ile62 |
| Pitstop 2 | −12.48 | Phe91, Ile93, and Ile62 |
| Ikarugamycin | −11.29 | Lys98 |
| Brefeldin A | −9.62 | Ile93 and Lys96 |
| TENin1 | −11.17 | Arg64 and Ile93 |
| Clathrin A | −11.03 | Ser67 |
| Clathrin B | −10.42 | Ser67 |
| Clathrin C | −10.06 | Phe91 |
For 3‐hydroxy‐3‐methylglutaryl CoA inhibitors (Group 2), the catalytic portion of human HMG‐CoA reductase co‐crystallized with simvastatin (IC50 = 4.3 nM) 147 as an inhibitor (PDB ID: 1HW9) 148 was used for the docking studies. The results (Figure 7, Supporting Information Figure S2, and Table 6) revealed that methyl‐β‐cyclodextrin (S = −13.07 kcal/mol) had the highest inhibitory activity within the group, which was also higher than those of simvastatin (S = −10.55 kcal/mol) and chloroquine (S = −8.38 kcal/mol). Methyl‐β‐cyclodextrin interacted with the catalytic portion of the human HMG‐CoA reductase active site via two hydrogen bonding interactions with the key amino acid Glu559 backbone using two hydroxyl groups. It also interacted with the active site through hydrophobic interactions with the hydrophobic side chains of Gly860, Ala856, Leu857, and Leu853.
FIGURE 7.
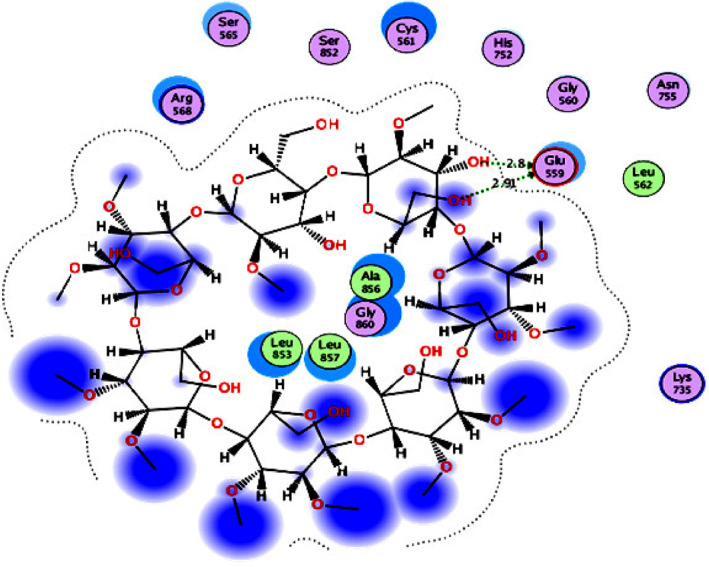
2D diagram (Distances in Å) of methyl‐β‐cyclodextrin (Group 2 inhibitor) showing its interaction with the catalytic portion of the human HMG‐CoA reductase active site
TABLE 6.
Docking energy scores and amino acids involved in binding for simvastatin, chloroquine, and Group 2 inhibitors docked with the catalytic portion of human HMG‐CoA reductase
| Compound | Docking score (kcal/mol) | Amino acids involved in binding |
|---|---|---|
| Simvastatin | −10.55 | Glu559, Lys735, and Asn755 |
| Chloroquine | −8.38 | Cys561 |
| Methyl‐β‐cyclodextrin | −13.07 | Glu559 |
| Filipin | −11.23 | Glu559 and Asn755 |
| Nystatin | −12.72 | Glu559 |
| 2‐Deoxy‐d‐glucose | −7.91 | Glu559 and Ser565 |
| Chlorpromazine | −7.40 | His752 |
| Betulinic acid | −7.63 | Lys735, Asn750, and Asn755 |
| Kaempferol | −9.42 | Cys561 |
| Hydroxytyrosol | −7.90 | Gly560 |
For actin protein inhibitors (Group 3), a monomer of actin cytoskeleton from the dimer complexed with swinholide A (IC50 = 0.04 μg/ml) 149 (PDB ID: 1YXQ chain A) 150 was used for the docking studies. The results (Figure 8, Supporting Information Figure S3, and Table 7) revealed that rottlerin (S = −13.89 kcal/mol) was the most potent inhibitor within the group and had a higher inhibitory activity than those of swinholide A (S = −12.35 kcal/mol) and chloroquine (S = −10.79 kcal/mol). Rottlerin interacted with the actin cytoskeleton active site via one arene–H bond with the key amino acid Tyr133 backbone using the phenyl ring. It also interacted with the active site through hydrophobic interactions with the hydrophobic side chains of Met355, Thr148, Tyr169, and Tyr143.
FIGURE 8.
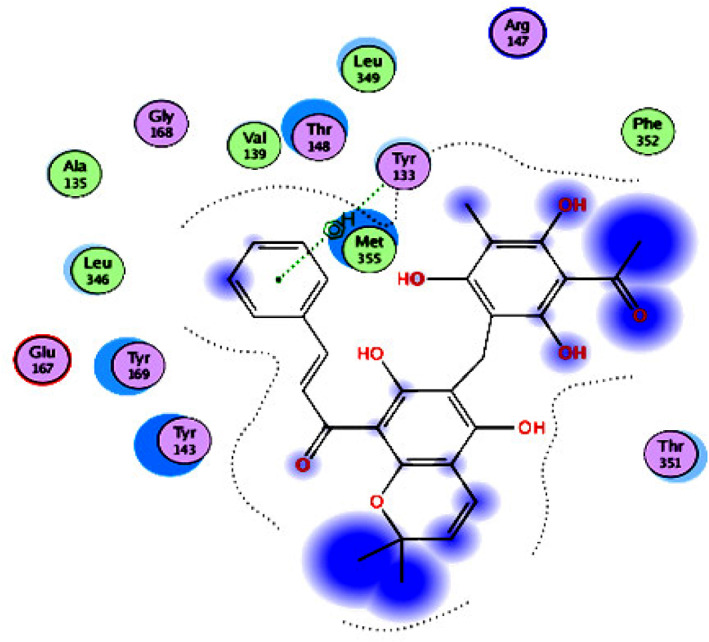
2D diagram of rottlerin (Group 3 inhibitor) showing its interaction with the actin cytoskeleton active site
TABLE 7.
Docking energy scores and amino acids involved in binding for swinholide A, chloroquine, and Group 3 inhibitors docked with actin cytoskeleton
| Compound | Docking score (kcal/mol) | Amino acids involved in binding |
|---|---|---|
| Swinholide A | −12.35 | Ser145, Arg147, and Ala331 |
| Chloroquine | −10.79 | Tyr133 |
| 2‐Deoxy‐d‐glucose | −9.76 | Tyr133, Ile136, and Gly168 |
| Cytochalasin D | −12.38 | Gly168 |
| Latrunculin A | −9.83 | Met355 |
| Latrunculin B | −12.14 | Tyr169 |
| Flubendazole | −10.00 | Ile136 |
| Terfenadine | −11.13 | Met355 and Ile136 |
| Itraconazole | −12.25 | Ile136 |
| Vinblastine | −8.73 | Met355 |
| Wortmannin | −10.31 | Thr351 |
| LY294002 | −10.59 | Tyr133 and Tyr143 |
| Rottlerin | −13.89 | Tyr133 |
| Imipramine | −9.81 | Tyr133 |
For dynamin I GTPase inhibitors (Group 4), molecular docking was performed using the crystal structure of dynamin I GTPase domain complexed with phosphomethylphosphonic acid guanylate ester (GMPPCP) ligand (PDB ID: 3ZYC). 151 The results (Figure 9, Supporting Information Figure S4, and Table 8) showed that Bis‐T (S = −25.73 kcal/mol) had the highest inhibitory activity within the group, which was also higher than those of GMPPCP (S = −23.73 kcal/mol) and chloroquine (S = −14.12 kcal/mol). Bis‐T interacted with the dynamin I GTPase active site through hydrogen bonding with Asp208, Ser46, Arg59, Leu137, and Gly62 amino acids in addition to that with the Mg2+ ion. It also formed hydrophobic interactions with the allosteric hydrophobic back pocket of the hydrophobic side chains of Gln239, Ser41, Lys206, Leu209, Ser238, and Asn236, thereby maximizing the binding interaction with the active site.
FIGURE 9.
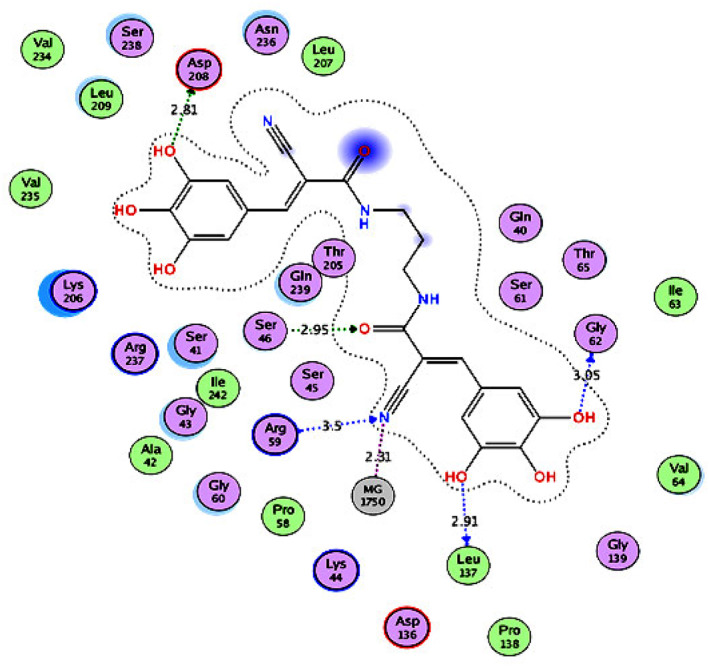
2D diagram (Distances in Å) of Bis‐T (Group 4 inhibitor) showing its interaction with the dynI GTPase domain active site
TABLE 8.
Docking energy scores and amino acids involved in binding for GMPPCP, chloroquine, and Group 4 inhibitors docked with dynI GTPase domain
| Compound | Docking score (kcal/mol) | Amino acids involved in binding |
|---|---|---|
| GMPPCP | −23.73 | Gly60, Ser41, Arg237, Lys206, Asn236, and Asp208 |
| Chloroquine | −14.12 | Lys206 |
| Dynole 25 | −14.67 | Lys206 |
| Dyngo 4a | −18.16 | Gln239 |
| Dyngo 6a | −18.70 | Gly60, Ser46, Lys206, and Lys44 |
| Dynasore | −18.52 | Gly60 |
| Iminodyn 21 | −22.23 | Lys206 and Asp208 |
| Iminodyn 22 | −24.16 | Gly60, Arg237, Lys206, Leu137, Asn236, and Asp208 |
| Iminodyn 23 | −21.93 | Thr65, Gly139, Asn236, and Lys206 |
| Rhodadyn D10 | −17.42 | Gly139, Asn236, and Lys206 |
| Bis‐T | −25.73 | Gly62, Leu137, Arg59, Gln239, Ser46, and Asp208 |
| Pyrimidyn 6 | −16.88 | Gly60, Gly139, and Thr65 |
| Pyrimidyn 7 | −14.18 | Ser41, Val64, and Thr65 |
| Chlorpromazine | −12.3 | Ser46 and Lys206 |
| Genistein | −16.95 | Gly60 and Lys206 |
| 7‐Keto‐cholesterol | −17.2 | Ser45 and Ser46 |
| LG186 | −18.68 | Gly60, Gln239, and Lys206 |
Overall, it was concluded from the docking simulations that remdesivir, methyl‐β‐cyclodextrin, rottlerin, and Bis‐T can effectively inhibit clathrin, HMG‐CoA reductase, actin, and dynamin I GTPase, respectively, and therefore, considered potent inhibitors of SARS‐CoV‐2.
8. CYTOTOXIC EFFECT OF ENDOCYTIC PROTEINS INHIBITORS
Not all drugs have been proven to have an antiviral effect against SARS‐CoV‐2, but some can block various endocytic pathways in cell culture and potentially inhibit SARS‐CoV‐2 endocytosis. 151 However, there are many issues associated with the use of these drugs/inhibitors, such as non‐specificity, affecting normal cellular processes, and cytotoxicity. 152 Although various FDA‐approved drugs, such as chlorpromazine, amiloride, nystatin vinblastine, and itraconazole, are currently available, there use is restricted. Chlorpromazine may have adverse effects in patients, such as allergic reactions, agranulocytosis, severe hypotension, and blockade action of the receptor of chlorpromazine (D2 receptor), which may reduce the efficacy of other medications such as levodopa or cabergoline. Patients with cerebrovascular and cardiovascular diseases should take chlorpromazine with caution. 153 Nystatin also has adverse effects, such as those affecting the gastrointestinal tract including nausea, vomiting, diarrhea, stomach upset and stomach pain, as well as rare adverse effects such as mouth irritation, skin redness, and breathing difficulties. Amiloride has adverse effects such as nausea, vomiting, diarrhea, headache, increased reabsorption of uric acid in the proximal tubule, and potassium retention, which can be critical in patients with compromised renal function. Moreover, amiloride affects the central nervous system, gastrointestinal tract, endocrine system, musculoskeletal system, dermatological system, and hematological systems. 154 , 155 , 156 Vinblastine, a chemotherapeutic agent, can cause hematological adverse effects, such as leukopenia, anemia, and thrombocytopenia, which can lead to intestinal bleeding; gastrointestinal symptoms, such as nausea, vomiting, and abdominal pain; and neurological adverse effects, such as peripheral neuritis, mental depression, headache, and convulsions. Itraconazole is used to treat dermatomycosis and onychomycosis using continuous therapy and may have serious adverse effects, including hepatotoxicity, liver failure and death, granulocytopenia, leukopenia, cardiac failure, congestive heart failure, neutropenia, thrombocytopenia, and immune system disorders. 157 Itraconazole may also cause vertigo, gastrointestinal symptoms, vomiting, pyrosis, constipation, fatigue, fever, allergic reaction, and skin rash.
Vercauteren et al. found that chlorpromazine and methyl‐β‐cyclodextrin, which are supposed to be SARS‐CoV‐2 endocytic inhibitors, exhibited cytotoxic effects on different cell types. 64 Therefore, continuous efforts are being made to develop endocytic protein inhibitors with less cytotoxicity that are safer for human use. FDA approved chlorpromazine, nystatin, amiloride, vinblastine, and itraconazole as safe drugs for human use worldwide, whereas others are under evaluation. 106 Table 1 summarizes different types of inhibitors and their cellular targets, with the underlying mechanisms. Understanding the membrane‐trafficking mechanisms of the cell can aid in the development of new promising drugs against SARS‐CoV‐2 and other viruses. 159
9. SUMMARY AND FUTURE PERSPECTIVES
The coronavirus disease 2019 pandemic is still a global health threat. Despite the rapid spread of the virus and the viral‐related morbidity decreased, research community still struggling in the detection and treatment of SARS‐CoVs variants. The endocytic pathways are possible mechanisms involved in the entry of many viruses like SARS‐CoVs, MERS‐CoVs, and possibly SARS‐CoV‐2 into the host cell. In this review, we summarized the efficacy of different endocytic inhibitors, such as Dyngo 4a, Dyngo 6a, dynasore, Iminodyn 21, Iminodyn 22, Rhodadyn D10, remdesivir, and rottlerin, in combating SARS‐CoV‐2 by inhibiting the CME pathway and CME/lipid rafts. This could be achieved by inhibiting the GTPase activity of dynamin such as dynasore, dynoles, and dyngoes. The virus can also invade the cells via the CLIC/GEEC pathway, which can be blocked by LG186 and 7‐keto‐cholesterol that prevent the close packing of acyl chains and affect the structure of the cell membrane. The molecular docking simulations we carried out, revealed that remdesivir (S = −14.46 kcal/mol), methyl‐β‐cyclodextrin (S = −13.07 kcal/mol), rottlerin (S = −13.89 kcal/mol), and Bis‐T (S = −25.73 kcal/mol) are more potent inhibitors of SARS‐CoV‐2 than chloroquine and act by inhibiting clathrin, HMG‐CoA reductase, actin, and dynamin I GTPase, respectively. Although endocytic inhibitors can potentially inhibit endocytosis‐mediated viral entry, they still have some limitations, such as the lack of information about their specificity and cytotoxicity on the non‐endocytic targets in host cells. Therefore, additional computational analyses along with basic and clinical studies are required to achieve maximum drug‐based variant independent antiviral effects associated with less side complications. Finally, there are a huge number of investigations are still running to investigate the potential antiviral activity of small molecules including endocytic inhibitors against COVID‐19.
AUTHOR CONTRIBUTIONS
Samar Sami Alkafaas: Conceptualization; supervision; software; data curation; writing – original draft; editing and validation. Abanoub Mosaad Abdallah: Molecular docking; data curation; and writing – original draft. Soumya Ghosh: Data curation; writing – original draft and editing. All authors read and approved the final manuscript. Samah A Loutfy: Data curation; writing – original draft and editing. Sara Sami ElKaffas: Data curation; writing – original draft and editing. Nasra F. Abdel Fattah: Data curation; writing – original draft and editing. Mohamed Hessien: Conceptualization; data curation; and writing – original draft.
CONFLICT OF INTEREST
The authors declare that they have no known competing financial interests or personal relationships.
CONSENT FOR PUBLICATION
All the authors read and agreed to publish this article.
Supporting information
Supporting Information S1
ACKNOWLEDGEMENTS
The authors would like to thank the EKB facility for proofreading the manuscript.
Alkafaas SS, Abdallah AM, Ghosh S, et al. Insight into the role of clathrin‐mediated endocytosis inhibitors in SARS‐CoV‐2 infection. Rev Med Virol. 2023;33(1):e2403. 10.1002/rmv.2403
Contributor Information
Samar Sami Alkafaas, Email: samar.alkafas@science.tanta.edu.eg, Email: samarsamy2017@yahoo.com.
Soumya Ghosh, Email: soumyaghosh@yahoo.com, Email: GhoshS@ufs.ac.za.
Mohamed Hessien, Email: mohamed.hussien1@science.tanta.edu.eg, Email: mohamed.hessien@fulbrightmail.org.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Ramadan AA, Mayilsamy K, McGill AR, et al. Identification of SARS‐CoV‐2 spike palmitoylation inhibitors that results in release of attenuated virus with reduced infectivity. Viruses. 2022;14(3):531. 10.3390/v14030531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bayati A, Kumar R, Francis V, McPherson PS. SARS‐CoV‐2 infects cells after viral entry via clathrin‐mediated endocytosis. J Biol Chem. 2021;296. 10.1016/j.jbc.2021.100306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prabhakara C, Godbole R, Sil P, et al. Strategies to target SARS‐CoV‐2 entry and infection using dual mechanisms of inhibition by acidification inhibitors. PLoS Pathog. 2021;17(7):e1009706. 10.1371/journal.ppat.1009706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meskini M, Rami MR, Maroofi P, Ghosh S, Siadat SD, Sheikhpour M. An overview on the epidemiology and immunology of COVID‐19. J InF Public Health. 2021;14(10):1284‐1298. 10.1016/j.jiph.2021.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Makvandi P, Chen M, Sartorius R, et al. Endocytosis of abiotic nanomaterials and nanobiovectors: inhibition of membrane trafficking. Nano Today. 2021;40:101279. 10.1016/j.nantod.2021.101279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koch J, Uckeley ZM, Doldan P, Stanifer M, Boulant S, Lozach PY. TMPRSS2 expression dictates the entry route used by SARS‐CoV‐2 to infect host cells. EMBO J. 2021;40(16):e107821. 10.15252/embj.2021107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zeng C, Evans JP, King T, et al. SARS‐CoV‐2 spreads through cell‐to‐cell transmission. Proc Natl Acad Sci USA. 2022;119(1):119. 10.1073/pnas.2111400119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alhouri A, Salloum A, Harfouch RM, Soumya G. Possible side effects of using detergents during the Covid19 pandemic in Syria. Ann Clin Cases. 2020;1:1023. [Google Scholar]
- 9. Abosheasha MA, El‐Gowily AH. Superiority of cilostazol among antiplatelet FDA‐approved drugs against COVID 19 Mpro and spike protein: drug repurposing approach. Drug Dev Res. 2021;82(2):217‐229. 10.1002/ddr.21743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abosheasha MA, El‐Gowily AH, Elfiky AA. Potential antiviral properties of antiplatelet agents against SARS‐CoV‐2 infection: an in silico perspective. J Thromb Thrombolysis. 2022;53(2):273‐281. 10.1007/s11239-021-02558-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghosh S, Al‐Sharify ZT, Maleka MF, et al. Propolis efficacy on SARS‐COV viruses: a review on antimicrobial activities and molecular simulations. Environ Sci Pollut Control Ser. 2022;29(39):1‐20. 10.1007/s11356-022-21652-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hodgson SH, Mansatta K, Mallett G, Harris V, Emary KR, Pollard AJ. What defines an efficacious COVID‐19 vaccine? a review of the challenges assessing the clinical efficacy of vaccines against SARS‐CoV‐2. Lancet Infect Dis. 2021;21(2):e26‐e35. 10.1016/s1473-3099(20)30773-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rella SA, Kulikova YA, Dermitzakis ET, Kondrashov FA. Rates of SARS‐CoV‐2 transmission and vaccination impact the fate of vaccine‐resistant strains. Sci Rep. 2021;11:1‐10. 10.1038/s41598-021-95025-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Surawan DP, Sumohadi D, Budhitresna AA, et al. Titer disparity of anti‐Spike receptor binding domain SARS‐CoV‐2 antibody between vaccinated and naturally infected individuals. Narrative J. 2022;2(1):2. 10.52225/narra.v2i1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Masyeni S, Iqhrammullah M, Frediansyah A, et al. Molnupiravir: a lethal mutagenic drug against rapidly mutating SARS‐CoV‐2‐A narrative review. J Med Virol. 2022;94(7):3006‐3016. 10.1002/jmv.27730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blanchard E, Belouzard S, Goueslain L, et al. Hepatitis C virus entry depends on clathrin‐mediated endocytosis. J Virol. 2006;80(14):6964‐6972. 10.1128/jvi.00024-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leong KLJ, Ng MM‐L, Chu JJH. The essential role of clathrin‐mediated endocytosis in the infectious entry of human enterovirus 71. J Biol Chem. 2011;286(1):309‐321. 10.1074/jbc.m110.168468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Piccini LE, Castilla V, Damonte EB. Dengue‐3 virus entry into vero cells: role of clathrin‐mediated endocytosis in the outcome of infection. PloS One. 2015;10:e0140824. 10.1371/journal.pone.0140824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H, Yang P, Liu K, et al. SARS coronavirus entry into host cells through a novel clathrin‐and caveolae‐independent endocytic pathway. Cell Res 2008;18(2):290‐301. 10.1038/cr.2008.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tobys D, Kowalski LM, Cziudaj E, et al. Inhibition of clathrin‐mediated endocytosis by knockdown of AP‐2 leads to alterations in the plasma membrane proteome. Traffic. 2021;22(1‐2):6‐22. 10.1111/tra.12770 [DOI] [PubMed] [Google Scholar]
- 21. Prichard KL, O'Brien NS, Murcia SR, Baker JR, McCluskey A. Role of clathrin and dynamin in clathrin mediated endocytosis/synaptic vesicle recycling and implications in neurological diseases. Front Cell Neurosci. 2021:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu B, Guo H, Zhou P, Shi Z‐L. Characteristics of SARS‐CoV‐2 and COVID‐19. Nat Rev Microbiol. 2021;19(3):141‐154. 10.1038/s41579-020-00459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng M, Jiang W, Kim BY, Zhang CC, Fu Y‐X, Weissman IL. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer. 2019;19(10):568‐586. 10.1038/s41568-019-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uribe‐Querol E, Rosales C. Phagocytosis: our current understanding of a universal biological process. Front Immunol. 2020;11:1066. 10.3389/fimmu.2020.01066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foroozandeh P, Jusoh SAM, Shamsuddin S. Passive drug delivery, mechanisms of uptake, and intracellular trafficking. In: Organelle and Molecular Targeting. CRC Press; 2021:129‐152. [Google Scholar]
- 26. O’Sullivan MJ, Lindsay AJ. The endosomal recycling pathway—at the crossroads of the cell. Int J Mol Sci. 2020;21(17):6074. 10.3390/ijms21176074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Al Kafaas SS, Loutfy SA, Diab T, Hessien M. Prestimulation of breast cancer cells with arginine vasopressin accentuates the anticancer effect induced by Dynamin 2 inhibitor; 2022.
- 28. Mettlen M, Chen P‐H, Srinivasan S, Danuser G, Schmid SL. Regulation of clathrin‐mediated endocytosis. Annu Rev Biochem. 2018;87(1):871‐896. 10.1146/annurev-biochem-062917-012644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rappoport JZ, Taha BW, Lemeer S, Benmerah A, Simon SM. The AP‐2 complex is excluded from the dynamic population of plasma membrane‐associated clathrin. J Biol Chem. 2003;278(48):47357‐47360. 10.1074/jbc.c300390200 [DOI] [PubMed] [Google Scholar]
- 30. Boulant S, Kural C, Zeeh J‐C, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin‐mediated endocytosis. Nat Cell Biol. 2011;13(9):1124‐1131. 10.1038/ncb2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mettlen M, Stoeber M, Loerke D, Antonescu CN, Danuser G, Schmid SL. Endocytic accessory proteins are functionally distinguished by their differential effects on the maturation of clathrin‐coated pits. Mol Biol Cell. 2009;20(14):3251‐3260. 10.1091/mbc.e09-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ehrlich M, Boll W, Van Oijen A, et al. Endocytosis by random initiation and stabilization of clathrin‐coated pits. Cell. 2004;118(5):591‐605. 10.1016/j.cell.2004.08.017 [DOI] [PubMed] [Google Scholar]
- 33. Dejonghe W, Sharma I, Denoo B, et al. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat Chem Biol. 2019;15(6):641‐649. 10.1038/s41589-019-0262-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saheki Y, De Camilli P. Synaptic vesicle endocytosis. Cold Spring Harb Perspect Biol. 2012;4(9):a005645. 10.1101/cshperspect.a005645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mayor S, Pagano RE. Pathways of clathrin‐independent endocytosis. Nat Rev Mol Cell Biol. 2007;8:603‐612. 10.1038/nrm2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lemaitre G, Gonnet F, Vaigot P, et al. CD98, a novel marker of transient amplifying human keratinocytes. Proteomics. 2005;5(14):3637‐3645. 10.1002/pmic.200401224 [DOI] [PubMed] [Google Scholar]
- 37. Pelkmans L, Helenius A. Endocytosis via caveolae. Traffic. 2002;3(5):311‐320. 10.1034/j.1600-0854.2002.30501.x [DOI] [PubMed] [Google Scholar]
- 38. Schnitzer JE, Oh P. Albondin‐mediated capillary permeability to albumin. Differential role of receptors in endothelial transcytosis and endocytosis of native and modified albumins. J Biol Chem. 1994;269(8):6072‐6082. 10.1016/s0021-9258(17)37571-3 [DOI] [PubMed] [Google Scholar]
- 39. Kumari S, Mayor S. ARF1 is directly involved in dynamin‐independent endocytosis. Nat Cell Biol. 2008;10(1):30‐41. 10.1038/ncb1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sabharanjak S, Sharma P, Parton RG, Mayor S. GPI‐anchored proteins are delivered to recycling endosomes via a distinct cdc42‐regulated, clathrin‐independent pinocytic pathway. Dev Cell. 2002;2(4):411‐423. 10.1016/S1534-5807(02)00145-4 [DOI] [PubMed] [Google Scholar]
- 41. Lundmark R, Doherty GJ, Howes MT, et al. The GTPase‐activating protein GRAF1 regulates the CLIC/GEEC endocytic pathway. Curr Biol. 2008;18(22):1802‐1808. 10.1016/j.cub.2008.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sathe M, Muthukrishnan G, Rae J, et al. Small GTPases and BAR domain proteins regulate branched actin polymerisation for clathrin and dynamin‐independent endocytosis. Nat Commun. 2021;12(1):5158. 10.1038/s41467-021-25458-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shafaq‐Zadah M, Dransart E, Johannes L. Clathrin‐independent endocytosis, retrograde trafficking, and cell polarity. Curr Opin Cell Biol. 2020;65:112‐121. 10.1016/j.ceb.2020.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kirkham M, Fujita A, Chadda R, et al. Ultrastructural identification of uncoated caveolin‐independent early endocytic vehicles. JCB (J Cell Biol). 2005;168(3):465‐476. 10.1083/jcb.200407078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Renard H‐F, Simunovic M, Lemière J, et al. Endophilin‐A2 functions in membrane scission in clathrin‐independent endocytosis. Nature. 2015;517(7535):493‐496. 10.1038/nature14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumari S, Swetha M, Mayor S. Endocytosis unplugged: multiple ways to enter the cell. Cell Res 2010;20(3):256‐275. 10.1038/cr.2010.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meister M, Tikkanen R. Endocytic trafficking of membrane‐bound cargo: a flotillin point of view. Membranes. 2014;4(3):356‐371. 10.3390/membranes4030356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Solis GP, Hoegg M, Munderloh C, et al. Reggie/flotillin proteins are organized into stable tetramers in membrane microdomains. Biochem J. 2007;403(2):313‐322. 10.1042/bj20061686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schneider A, Rajendran L, Honsho M, et al. Flotillin‐dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J Neurosci. 2008;28(11):2874‐2882. 10.1523/Jneurosci.5345-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alipoor SD, Mortaz E, Jamaati H, et al. COVID‐19: molecular and cellular response. Front Cell Infect Microbiol. 2021;11. 10.3389/fcimb.2021.563085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Traub LM, Bonifacino JS. Cargo recognition in clathrin‐mediated endocytosis. Cold Spring Harb Perspect Biol. 2013;5(11):a016790. 10.1101/cshperspect.a016790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Redpath GM, Betzler VM, Rossatti P, Rossy J. Membrane heterogeneity controls cellular endocytic trafficking. Front Cell Dev Biol. 2020;8:757. 10.3389/fcell.2020.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Picas L, Gaits‐Iacovoni F, Goud B. The emerging role of phosphoinositide clustering in intracellular trafficking and signal transduction. F1000Research. 2016;5. 10.12688/f1000research.7537.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nicola AV, Aguilar HC, Mercer J, Ryckman B, Wiethoff CM. Virus entry by endocytosis. Advances in virology. 2013;2013. 10.1155/2013/469538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78(1):857‐902. 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- 56. Inal J, Miot S, Schifferli JA. The complement inhibitor, CRIT, undergoes clathrin‐dependent endocytosis. Exp Cell Res. 2005;310(1):54‐65. 10.1016/j.yexcr.2005.07.003 [DOI] [PubMed] [Google Scholar]
- 57. Subramanya S, Hardin CF, Steverding D, Mensa‐Wilmot K. Glycosylphosphatidylinositol‐specific phospholipase C regulates transferrin endocytosis in the African trypanosome. Biochem J. 2009;417(3):685‐694. 10.1042/bj20080167 [DOI] [PubMed] [Google Scholar]
- 58. Mollenhauer HH, Morré DJ, Rowe LD. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim Biophys Acta (BBA)‐Reviews Biomembr. 1990;1031(2):225‐246. 10.1016/0304-4157(90)90008-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beautrait A, Paradis JS, Zimmerman B, et al. A new inhibitor of the β‐arrestin/AP2 endocytic complex reveals interplay between GPCR internalization and signalling. Nat Commun. 2017;8:1‐16. 10.1038/ncomms15054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sundqvist M, Holdfeldt A, Wright SC, et al. Barbadin selectively modulates FPR2‐mediated neutrophil functions independent of receptor endocytosis. Biochim Biophys Acta (BBA)‐Molecular Cell Res. 2020;1867(12):118849. 10.1016/j.bbamcr.2020.118849 [DOI] [PubMed] [Google Scholar]
- 61. Abdel‐Hamid MK, Macgregor KA, Odell LR, et al. 1, 8‐Naphthalimide derivatives: new leads against dynamin I GTPase activity. Org Biomol Chem. 2015;13(29):8016‐8028. 10.1039/c5ob00751h [DOI] [PubMed] [Google Scholar]
- 62. Masaike Y, Takagi T, Hirota M, et al. Identification of dynamin‐2‐mediated endocytosis as a new target of osteoporosis drugs, bisphosphonates. Mol Pharmacol. 2010;77(2):262‐269. 10.1124/mol.109.059006 [DOI] [PubMed] [Google Scholar]
- 63. Vercauteren D, Vandenbroucke RE, Jones AT, et al. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol Ther. 2010;18(3):561‐569. 10.1038/mt.2009.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Koivusalo M, Welch C, Hayashi H, et al. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. JCB (J Cell Biol). 2010;188(4):547‐563. 10.1083/jcb.200908086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lin HP, Singla B, Ghoshal P, et al. Identification of novel macropinocytosis inhibitors using a rational screen of Food and Drug Administration‐approved drugs. Br J Pharmacol. 2018;175(18):3640‐3655. 10.1111/bph.14429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mann SK, Marwaha R. Chlorpromazine. StatPearls. 2021. [PubMed] [Google Scholar]
- 67. Daniel JA, Chau N, Abdel‐Hamid MK, et al. Phenothiazine‐derived antipsychotic drugs inhibit dynamin and clathrin‐mediated endocytosis. Traffic. 2015;16(6):635‐654. 10.1111/tra.12272 [DOI] [PubMed] [Google Scholar]
- 68. Wu C‐H, Bai L‐Y, Tsai M‐H, et al. Pharmacological exploitation of the phenothiazine antipsychotics to develop novel antitumor agents–a drug repurposing strategy. Sci Rep. 2016;6:1‐11. 10.1038/srep27540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shang C, Zhuang X, Zhang H, et al. Inhibitors of endosomal acidification suppress SARS‐CoV‐2 replication and relieve viral pneumonia in hACE2 transgenic mice. Virology J. 2021;18:1‐9. 10.1186/s12985-021-01515-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gies V, Bekaddour N, Dieudonné Y, et al. Beyond anti‐viral effects of chloroquine/hydroxychloroquine. Front Immunol. 2020;11:1409. 10.3389/fimmu.2020.01409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen X, Wang Z. Regulation of epidermal growth factor receptor endocytosis by wortmannin through activation of Rab5 rather than inhibition of phosphatidylinositol 3‐kinase. EMBO Rep. 2001;2(9):842‐849. 10.1093/embo-reports/kve179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tarahovsky YS, Muzafarov EN, Kim YA. Rafts making and rafts braking: how plant flavonoids may control membrane heterogeneity. Mol Cell Biochem. 2008;314(1‐2):65‐71. 10.1007/s11010-008-9766-9 [DOI] [PubMed] [Google Scholar]
- 73. Danesh FR, Sadeghi MM, Amro N, et al. 3‐Hydroxy‐3‐methylglutaryl CoA reductase inhibitors prevent high glucose‐induced proliferation of mesangial cells via modulation of Rho GTPase/p21 signaling pathway: implications for diabetic nephropathy. Proc Natl Acad Sci USA. 2002;99(12):8301‐8305. 10.1073/pnas.122228799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang F, Guo H, Zhang J, Chen Q, Fang Q. Identification of the caveolae/raft‐mediated endocytosis as the primary entry pathway for aquareovirus. Virology. 2018;513:195‐207. 10.1016/j.virol.2017.09.019 [DOI] [PubMed] [Google Scholar]
- 75. Kiani AK, Dhuli K, Anpilogov K, et al. Natural compounds as inhibitors of SARS‐CoV‐2 endocytosis: a promising approach against COVID‐19. Acta Biomed Atenei Parm. 2020:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Le Roy C, Wrana JL. Clathrin‐and non‐clathrin‐mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6(2):112‐126. 10.1038/nrm1571 [DOI] [PubMed] [Google Scholar]
- 77. Rezanka T, Siristova L, Sigler K. Sterols and triterpenoids with antiviral activity. Anti‐Infective Agents Med Chem Former Curr Med Chemistry‐Anti‐Infective Agents. 2009;8(3):193‐210. 10.2174/187152109788680207 [DOI] [Google Scholar]
- 78. Yamada K, Ogawa H, Hara A, et al. Mechanism of the antiviral effect of hydroxytyrosol on influenza virus appears to involve morphological change of the virus. Antivir Res. 2009;83(1):35‐44. 10.1016/j.antiviral.2009.03.002 [DOI] [PubMed] [Google Scholar]
- 79. Elkin SR, Oswald NW, Reed DK, Mettlen M, MacMillan JB, Schmid SL. Ikarugamycin: a natural product inhibitor of clathrin‐mediated endocytosis. Traffic. 2016;17(10):1139‐1149. 10.1111/tra.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kleine‐Vehn J, Morris D, Emans N, Jurgens G, Geldner N, Friml J. Auxin inhibits endocytosis and promotes its own efux from cells. Nature 435: 12511256Perera IY, Hung CY, Brady S, Muday GK, Boss WF (2006) A universal role for inositol 1, 4, 5‐trisphosphate‐mediated signaling in plant gravitropism. Plant Physiol. 2005;140:746760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Foissner I, Hoeftberger M, Hoepflinger MC, Sommer A, Bulychev AA. Brefeldin A inhibits clathrin‐dependent endocytosis and ion transport in Chara internodal cells. Biol Cell. 2020;112(11):317‐334. 10.1111/boc.202000031 [DOI] [PubMed] [Google Scholar]
- 82. Bulychev AA, Foissner I. Inhibition of endosomal trafficking by brefeldin A interferes with long‐distance interaction between chloroplasts and plasma membrane transporters. Physiol Plantarum. 2020;169(1):122‐134. 10.1111/ppl.13058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Donia T, Abouda M, Kelany M, Hessien M. β‐Arrestin inhibition induces autophagy, apoptosis, G0/G1 cell cycle arrest in agonist‐activated V2R receptor in breast cancer cells. Med Oncol. 2021;38(4):1‐11. 10.1007/s12032-021-01484-z [DOI] [PubMed] [Google Scholar]
- 84. Diab T, AlKafaas SS, Shalaby TI, Hessien M. Dexamethasone simulates the anticancer effect of nano‐formulated paclitaxel in breast cancer cells. Bioorg Chem. 2020;99:103792. 10.1016/j.bioorg.2020.103792 [DOI] [PubMed] [Google Scholar]
- 85. Alkafaas SS, Diab T, Shalaby T, Hessien M. Dexamethasone improves the responsiveness of hepatoma cells for both free and solvent containing paclitaxel in vitro. Egyptian J Biochem Mol Biol. 2019:37. [Google Scholar]
- 86. Diab T, Alkafaas SS, Shalaby TI, Hessien M. Paclitaxel nanoparticles induce apoptosis and regulate txr1, cyp3a4 and cyp2c8 in breast cancer and hepatoma cells. Anti‐Cancer Agents Med Chem Former Curr Med Chemistry‐Anti‐Cancer Agents. 2020;20(13):1582‐1591. 10.2174/1871520620666200504071530 [DOI] [PubMed] [Google Scholar]
- 87. Donia T, Khedr S, Salim EI, Hessien M. Trichostatin A sensitizes hepatoma cells to Taxol more than 5‐Aza‐dC and dexamethasone. Drug Metabol Person Therapy. 2021. 10.1515/dmdi-2020-0186 [DOI] [PubMed] [Google Scholar]
- 88. El‐Gowily AH, Abosheasha MA. Differential mechanisms of autophagy in cancer stem cells: emphasizing gastrointestinal cancers. Cell Biochem Funct. 2021;39(2):162‐173. 10.1002/cbf.3552 [DOI] [PubMed] [Google Scholar]
- 89. Sundborger AC, Fang S, Heymann JA, Ray P, Chappie JS, Hinshaw JE. A dynamin mutant defines a superconstricted prefission state. Cell Rep 2014;8(3):734‐742. 10.1016/j.celrep.2014.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Damke H, Binns DD, Ueda H, Schmid SL, Baba T. Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages. Mol Biol Cell. 2001;12(9):2578‐2589. 10.1091/mbc.12.9.2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dutta D, Donaldson JG. Search for inhibitors of endocytosis: intended specificity and unintended consequences. Cell Logist. 2012;2(4):203‐208. 10.4161/cl.23967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Prabhakara C, Godbole R, Sil P, et al. Niclosamide inhibits SARS‐CoV2 entry by blocking internalization through pH‐dependent CLIC/GEEC endocytic pathway. bioRxiv. 2020. [Google Scholar]
- 93. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS‐CoV‐2 entry into cells. Nat Rev Mol Cell Biol. 2022;23(1):3‐20. 10.1038/s41580-021-00418-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Granseth B, Fukushima Y, Sugo N, Lagnado L, Yamamoto N. Regulation of thalamocortical axon branching by BDNF and synaptic vesicle cycling. Front Neural Circ. 2013;7:202. 10.3389/fncir.2013.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Genet G, Boyé K, Mathivet T, et al. Endophilin‐A2 dependent VEGFR2 endocytosis promotes sprouting angiogenesis. Nat Commun. 2019;10:1‐15. 10.1038/s41467-019-10359-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nwosu ZC, Ebert MP, Dooley S, Meyer C. Caveolin‐1 in the regulation of cell metabolism: a cancer perspective. Mol Cancer. 2016;15:1‐12. 10.1186/s12943-016-0558-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. de Almeida CJG. Caveolin‐1 and caveolin‐2 can be antagonistic partners in inflammation and beyond. Front Immunol. 2017;8:1530. 10.3389/fimmu.2017.01530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ivanov AI, Nusrat A, Parkos CA. Endocytosis of epithelial apical junctional proteins by a clathrin‐mediated pathway into a unique storage compartment. Mol Biol Cell. 2004;15(1):176‐188. 10.1091/mbc.e03-05-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liberali P, Kakkonen E, Turacchio G, et al. The closure of Pak1‐dependent macropinosomes requires the phosphorylation of CtBP1/BARS. EMBO J. 2008;27(7):970‐981. 10.1038/emboj.2008.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pelkmans L, Helenius A. Insider information: what viruses tell us about endocytosis. Curr Opin Cell Biol. 2003;15(4):414‐422. 10.1016/s0955-0674(03)00081-4 [DOI] [PubMed] [Google Scholar]
- 101. Sieczkarski SB, Whittaker GR. Dissecting virus entry via endocytosis. J Gen Virol. 2002;83(7):1535‐1545. 10.1099/0022-1317-83-7-1535 [DOI] [PubMed] [Google Scholar]
- 102. Donaldson JG. Macropinosome formation, maturation and membrane recycling: lessons from clathrin‐independent endosomal membrane systems. Philosophical Trans R Soc B. 2019;374(1765):20180148. 10.1098/rstb.2018.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Desplanques AS, Nauwynck HJ, Vercauteren D, Geens T, Favoreel HW. Plasma membrane cholesterol is required for efficient pseudorabies virus entry. Virology. 2008;376(2):339‐345. 10.1016/j.virol.2008.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ripa I, Andreu S, López‐Guerrero JA, Bello‐Morales R. Membrane rafts: portals for viral entry. Front Microbiol. 2021;12:120. 10.3389/fmicb.2021.631274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Glebov OO. Understanding SARS‐CoV‐2 endocytosis for COVID‐19 drug repurposing. FEBS J. 2020;287(17):3664‐3671. 10.1111/febs.15369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Van Hamme E, Dewerchin HL, Cornelissen E, Verhasselt B, Nauwynck HJ. Clathrin‐and caveolae‐independent entry of feline infectious peritonitis virus in monocytes depends on dynamin. J Gen Virol. 2008;89(9):2147‐2156. 10.1099/vir.0.2008/001602-0 [DOI] [PubMed] [Google Scholar]
- 107. Spear ED, Ng DT. Single, context‐specific glycans can target misfolded glycoproteins for ER‐associated degradation. J Cell Biol. 2005;169(1):73‐82. 10.1083/jcb.200411136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bastiani M, Parton RG. Caveolae at a glance. J Cell Sci. 2010;123(22):3831‐3836. 10.1242/jcs.070102 [DOI] [PubMed] [Google Scholar]
- 109. Cheng JP, Nichols BJ. Caveolae: one function or many? Trends Cell Biol. 2016;26(3):177‐189. 10.1016/j.tcb.2015.10.010 [DOI] [PubMed] [Google Scholar]
- 110. Kovtun O, Tillu VA, Ariotti N, Parton RG, Collins BM. Cavin family proteins and the assembly of caveolae. J Cell Sci. 2015;128(7):1269‐1278. 10.1242/jcs.167866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Briand N, Dugail I, Le Lay S. Cavin proteins: new players in the caveolae field. Biochimie. 2011;93(1):71‐77. 10.1016/j.biochi.2010.03.022 [DOI] [PubMed] [Google Scholar]
- 112. Parton RG, Howes MT. Revisiting caveolin trafficking: the end of the caveosome. J Cell Biol. 2010;191(3):439‐441. 10.1083/jcb.201009093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mayor S, Parton RG, Donaldson JG. Clathrin‐independent pathways of endocytosis. Cold Spring Harb Perspect Biol. 2014;6:a016758. 10.1101/cshperspect.a016758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Cossart P, Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol. 2014;6(8):6:a016972. 10.1101/cshperspect.a016972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Nonnenmacher M, Weber T. Adeno‐associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell host microbe. 2011;10(6):563‐576. 10.1016/j.chom.2011.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Rojek JM, Perez M, Kunz S. Cellular entry of lymphocytic choriomeningitis virus. J Virol. 2008;82(3):1505‐1517. 10.1128/jvi.01331-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tuma PL, Hubbard AL. Transcytosis: crossing cellular barriers. Physiol Rev. 2003;83(3):871‐932. 10.1152/physrev.00001.2003 [DOI] [PubMed] [Google Scholar]
- 118. Knyazev E, Nersisyan S, Tonevitsky A. Endocytosis and transcytosis of SARS‐CoV‐2 across the intestinal epithelium and other tissue barriers. Front Immunol. 2021;12. 10.3389/fimmu.2021.636966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11:1‐12. 10.1038/s41467-021-22614-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shang J, Wan YS, Luo CM, et al. Cell entry mechanisms of SARS‐CoV‐2. In: Proceedings of the National Academy of Sciences of the United States of America. 2020;117(21):11727‐11734. 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Pachetti M, Marini B, Benedetti F, et al. Emerging SARS‐CoV‐2 mutation hot spots include a novel RNA‐dependent‐RNA polymerase variant. J Transl Med. 2020;18:1‐9. 10.1186/s12967-020-02344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chand GB, Banerjee A, Azad GK. Identification of novel mutations in RNA‐dependent RNA polymerases of SARS‐CoV‐2 and their implications on its protein structure. Peer J. 2020;8:e9492. 10.7717/peerj.9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Annu Rev Biochem. 2010;79(1):803‐833. 10.1146/annurev-biochem-060208-104626 [DOI] [PubMed] [Google Scholar]
- 124. Mazzon M, Marsh M. Targeting viral entry as a strategy for broad‐spectrum antivirals. F1000Research. 2019;8. 10.12688/f1000research.19694.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Burkard C, Verheije MH, Haagmans BL, et al. ATP1A1‐mediated Src signaling inhibits coronavirus entry into host cells. J Virol. 2015;89(8):4434‐4448. 10.1128/jvi.03274-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Owens B. Excitement around hydroxychloroquine for treating COVID‐19 causes challenges for rheumatology. Lancet Rheumatol. 2020;2(5):e257. 10.1016/s2665-9913(20)30089-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Borba M, de Val FA, Sampaio VS, et al. Chloroquine diphosphate in two different dosages as adjunctive therapy of hospitalized patients with severe respiratory syndrome in the context of coronavirus (SARS‐CoV‐2) infection: preliminary safety results of a randomized, double‐blinded, phase IIb clinical trial (CloroCovid‐19 Study). MedRxiv. 2020. [Google Scholar]
- 128. Khan N, Chen X, Geiger JD. Role of endolysosomes in severe acute respiratory syndrome coronavirus‐2 infection and coronavirus disease 2019 pathogenesis: implications for potential treatments. Front Pharmacol. 2020;11:1739. 10.3389/fphar.2020.595888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Fan C, Wu Y, Rui X, et al. Animal models for COVID‐19: advances, gaps and perspectives. Signal Transduct Targeted Ther. 2022;7:1‐24. 10.1038/s41392-022-01087-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tang X, Wu C, Li X, et al. On the origin and continuing evolution of SARS‐CoV‐2. Natl Sci Rev. 2020;7(6):1012‐1023. 10.1093/nsr/nwaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Onelli E, Prescianotto‐Baschong C, Caccianiga M, Moscatelli A. Clathrin‐dependent and independent endocytic pathways in tobacco protoplasts revealed by labelling with charged nanogold. J Exp Bot. 2008;59(11):3051‐3068. 10.1093/jxb/ern154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Stahlschmidt W. Dissection of Clathrin Function Using the Small Molecule Inhibitor Pitstop®‐2; 2014.
- 133. Hu TY, Frieman M, Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID‐19. Nat Nanotechnol. 2020;15(4):247‐249. 10.1038/s41565-020-0674-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID‐19 associated pneumonia in clinical studies. Bioscience Trends. 2020;14(1):72‐73. 10.5582/bst.2020.01047 [DOI] [PubMed] [Google Scholar]
- 135. Stokkermans TJ, Goyal A, Bansal P, Trichonas G. Chloroquine and hydroxychloroquine toxicity. StatPearls. 2021. [PubMed] [Google Scholar]
- 136. Archer MA, Brechtel TM, Davis LE, Parmar RC, Hasan MH, Tandon R. Inhibition of endocytic pathways impacts cytomegalovirus maturation. Sci Rep. 2017;7:1‐12. 10.1038/srep46069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Tsai C‐C, Lin C‐L, Wang T‐L, et al. Dynasore inhibits rapid endocytosis in bovine chromaffin cells. Am J Physiol Cell Physiol. 2009;297(2):C397‐C406. 10.1152/ajpcell.00562.2008 [DOI] [PubMed] [Google Scholar]
- 138. Casamento A, Boucrot E. Molecular mechanism of fast endophilin‐mediated endocytosis. Biochem J. 2020;477(12):2327‐2345. 10.1042/bcj20190342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Thottacherry JJ, Kosmalska AJ, Kumar A, et al. Mechanochemical feedback control of dynamin independent endocytosis modulates membrane tension in adherent cells. Nat Commun. 2018;9:1‐14. 10.1038/s41467-018-06738-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chaudhary N, Gomez GA, Howes MT, et al. Endocytic crosstalk: cavins, caveolins, and caveolae regulate clathrin‐independent endocytosis. PLoS Biol. 2014;12(4):e1001832. 10.1371/journal.pbio.1001832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Moe V. Chemical Computing Group Inc. Canada; 2010. [Google Scholar]
- 142. Al‐Qaisi ZH, Al‐Garawi ZS, Al‐Karawi AJM, et al. Antiureolytic activity of new water‐soluble thiadiazole derivatives: spectroscopic, DFT, and molecular docking studies. Spectrochimica Acta Part A Mol Biomol Spectrosc. 2022;272:120971. 10.1016/j.saa.2022.120971 [DOI] [PubMed] [Google Scholar]
- 143. Menu TMSMO . info@ rcsb. org• www. rcsb. org.
- 144. MacGregor KA, Robertson MJ, Young KA, et al. Development of 1, 8‐naphthalimides as clathrin inhibitors. J Med Chem. 2014;57(1):131‐143. 10.1021/jm4015263 [DOI] [PubMed] [Google Scholar]
- 145. Von Kleist L, Stahlschmidt W, Bulut H, et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell. 2011;146(3):471‐484. 10.1016/j.cell.2011.06.025 [DOI] [PubMed] [Google Scholar]
- 146. Ahmad S, Madsen CS, Stein PD, et al. (3 R, 5 S, E)‐7‐(4‐(4‐fluorophenyl)‐6‐isopropyl‐2‐(methyl (1‐methyl‐1 H‐1, 2, 4‐triazol‐5‐yl) amino) pyrimidin‐5‐yl)‐3, 5‐dihydroxyhept‐6‐enoic acid (BMS‐644950): a rationally designed orally efficacious 3‐hydroxy‐3‐methylglutaryl coenzyme‐A reductase inhibitor with reduced myotoxicity potential. J Med Chem. 2008;51(9):2722‐2733. 10.1021/jm800001n [DOI] [PubMed] [Google Scholar]
- 147. Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG‐CoA reductase. Science. 2001;292(5519):1160‐1164. 10.1126/science.1059344 [DOI] [PubMed] [Google Scholar]
- 148. Kobayashi M, Kawazoe K, Okamoto T, Sasaki T, Kitagawa I. Marine natural products. XXXI. Structure‐activity correlation of a potent cytotoxic dimeric macrolide swinholide A, from the Okinawan marine sponge Theonella swinhoei, and its isomers. Chem Pharmaceut Bull. 1994;42(1):19‐26. 10.1248/cpb.42.19 [DOI] [PubMed] [Google Scholar]
- 149. Klenchin VA, King R, Tanaka J, Marriott G, Rayment I. Structural basis of swinholide A binding to actin. Chem Biol. 2005;12(3):287‐291. 10.1016/j.chembiol.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 150. Chappie JS, Mears JA, Fang S, et al. A pseudoatomic model of the dynamin polymer identifies a hydrolysis‐dependent powerstroke. Cell. 2011;147(1):209‐222. 10.1016/j.cell.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID‐19). Drug Discove Therape. 2020;14(1):58‐60. 10.5582/ddt.2020.01012 [DOI] [PubMed] [Google Scholar]
- 152. Alduhaidhawi AHM, AlHuchaimi SN, Al‐Mayah TA, et al. Prevalence of CRISPR‐cas systems and their possible association with antibiotic resistance in Enterococcus faecalis and Enterococcus faecium collected from hospital wastewater. Infect Drug Resist. 2022;15:1143‐1154. 10.2147/idr.s358248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Stip E, Rizvi TA, Mustafa F, et al. The large action of chlorpromazine: translational and transdisciplinary considerations in the face of COVID‐19. Front Pharmacol. 2020;11:577678. 10.3389/fphar.2020.577678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sun Q, Sever P. Amiloride: a review. J Renin‐Angiotensin‐Aldosterone Syst JRAAS. 2020;21(4):1470320320975893. 10.1177/1470320320975893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Akinyemi KO, Al‐Khafaji NS, Al‐Alaq FT, et al. Extended‐spectrum beta‐lactamases encoding genes among Salmonella enterica serovar typhi isolates in patients with typhoid fever from four academic medical centers in lagos, Nigeria. Rev Invest Clin. 2022;74:165‐171. [DOI] [PubMed] [Google Scholar]
- 156. Raheem HQ, Hussein EF, Ghosh S, AlKafaas SS, Bloemfontein SA. Resistance of Klebsiella pneumoniae from different clinical samples to penicillin, cephalosporin, carbapenem and fluoroquinolone 2021; 44: 06:1–8. [Google Scholar]
- 157. Lestner J, Hope WW. Itraconazole: an update on pharmacology and clinical use for treatment of invasive and allergic fungal infections. Expet Opin Drug Metabol Toxicol. 2013;9(7):911‐926. 10.1517/17425255.2013.794785 [DOI] [PubMed] [Google Scholar]
- 158. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS‐CoV‐2 and other lineage B betacoronaviruses. Nature Microbiol. 2020;5(4):562‐569. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Hansen SH, Sandvig K, Van Deurs B. Molecules internalized by clathrin‐independent endocytosis are delivered to endosomes containing transferrin receptors. J Cell Biol. 1993;123(1):89‐97. 10.1083/jcb.123.1.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Tulapurkar M, Schäfer R, Hanck T, et al. Endocytosis mechanism of P2Y 2 nucleotide receptor tagged with green fluorescent protein: clathrin and actin cytoskeleton dependence. Cell Mol Life Sci. 2005;62(12):1388‐1399. 10.1007/s00018-005-5052-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yao D, Ehrlich M, Henis YI, Leof EB. Transforming growth factor‐β receptors interact with AP2 by direct binding to β2 subunit. Mol Biol Cell. 2002;13(11):4001‐4012. 10.1091/mbc.02-07-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Cooper S, Shedden K, Vu‐Phan D. Invariant mRNA and mitotic protein breakdown solves the Russian Doll problem of the cell cycle. Cell Biol Int. 2009;33(1):10‐18. 10.1016/j.cellbi.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 163. Cosson P, De Curtis I, Pouysségur J, Griffiths G, Davoust J. Low cytoplasmic pH inhibits endocytosis and transport from the trans‐Golgi network to the cell surface. J Cell Biol. 1989;108(2):377‐387. 10.1083/jcb.108.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Carpentier JL, Sawano F, Geiger D, Gorden P, Perrelet A, Orci L. Potassium depletion and hypertonic medium reduce ‘non‐coated’ and clathrin‐coated pit formation, as well as endocytosis through these two gates. J Cell Physiol. 1989;138(3):519‐526. 10.1002/jcp.1041380311 [DOI] [PubMed] [Google Scholar]
- 165. Larkin JM, Brown MS, Goldstein JL, Anderson RG. Depletion of intracellular potassium arrests coated pit formation and receptor‐mediated endocytosis in fibroblasts. Cell. 1983;33(1):273‐285. 10.1016/0092-8674(83)90356-2 [DOI] [PubMed] [Google Scholar]
- 166. Sato K, Nagai J, Mitsui N, Yumoto R, Takano M. Effects of endocytosis inhibitors on internalization of human IgG by Caco‐2 human intestinal epithelial cells. Life Sci. 2009;85(23‐26):800‐807. 10.1016/j.lfs.2009.10.012 [DOI] [PubMed] [Google Scholar]
- 167. Wang G, Jiang L, Wang J, et al. The G protein‐coupled receptor FFAR2 promotes internalization during influenza A virus entry. J Virol. 2020;94(2):e01707‐e01719. 10.1128/jvi.01707-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Guo S, Zhang X, Zheng M, et al. Selectivity of commonly used inhibitors of clathrin‐mediated and caveolae‐dependent endocytosis of G protein–coupled receptors. Biochim Biophys Acta (BBA)‐Biomembranes. 2015;1848(10):2101‐2110. 10.1016/j.bbamem.2015.05.024 [DOI] [PubMed] [Google Scholar]
- 169. Kuhn DA, Vanhecke D, Michen B, et al. Different endocytotic uptake mechanisms for nanoparticles in epithelial cells and macrophages. Beilstein J Nanotechnol. 2014;5:1625‐1636. 10.3762/bjnano.5.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Naim R, Iida T, Takahashi A, Honda T. Monodansylcadaverine inhibits cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin on cultured rat embryonic fibroblast cells. FEMS Microbiol Lett. 2001;196(2):99‐105. 10.1111/j.1574-6968.2001.tb10548.x [DOI] [PubMed] [Google Scholar]
- 171. Gibson AE, Noel RJ, Herlihy JT, Ward WF. Phenylarsine oxide inhibition of endocytosis: effects on asialofetuin internalization. Am J Physiol Cell Physiol. 1989;257(2):C182‐C184. 10.1152/ajpcell.1989.257.2.c182 [DOI] [PubMed] [Google Scholar]
- 172. Kim B, Park YS, Sung JS, Lee JW, Lee SB, Kim YH: Clathrin mediated endocytosis as a potential therapeutic strategy for overcoming refractoriness of EGFR TKI. AACR2019. [DOI] [PMC free article] [PubMed]
- 173. Zeng X, Zhang Y, Nystrom AM. Nyström AM. Endocytic uptake and intracellular trafficking of bis‐MPA‐based hyperbranched copolymer micelles in breast cancer cells. Biomacromolecules. 2012;13(11):3814‐3822. 10.1021/bm301281k [DOI] [PubMed] [Google Scholar]
- 174. Margarucci L, Monti MC, Fontanella B, Riccio R, Casapullo A. Chemical proteomics reveals bolinaquinone as a clathrin‐mediated endocytosis inhibitor. Mol Biosyst. 2011;7(2):480‐485. 10.1039/c0mb00126k [DOI] [PubMed] [Google Scholar]
- 175. Abdel‐Hamid MK, McCluskey A. In Silico docking, molecular dynamics and binding energy insights into the bolinaquinone‐clathrin terminal domain binding site. Molecules. 2014;19(5):6609‐6622. 10.3390/molecules19056609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Chen Z, Mino RE, Mettlen M, et al. Wbox2: a clathrin terminal domain–derived peptide inhibitor of clathrin‐mediated endocytosis. JCB (J Cell Biol). 2020;219(9). 10.1083/jcb.201908189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Hill T, Odell LR, Edwards JK, et al. Small molecule inhibitors of dynamin I GTPase activity: development of dimeric tyrphostins. J Med Chem. 2005;48(24):7781‐7788. 10.1021/jm040208l [DOI] [PubMed] [Google Scholar]
- 178. Hill TA, Gordon CP, McGeachie AB, et al. Inhibition of dynamin mediated endocytosis by the Dynoles– synthesis and functional activity of a family of indoles. J Med Chem. 2009;52(12):3762‐3773. 10.1021/jm900036m [DOI] [PubMed] [Google Scholar]
- 179. Hill TA, Mariana A, Gordon CP, et al. Iminochromene inhibitors of dynamins I and II GTPase activity and endocytosis. J Med Chem. 2010;53(10):4094‐4102. 10.1021/jm100119c [DOI] [PubMed] [Google Scholar]
- 180. Muranen T, Iwanicki MP, Curry NL, et al. Starved epithelial cells uptake extracellular matrix for survival. Nat Commun. 2017;8:1‐12. 10.1038/ncomms13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Marie‐Anaïs F, Mazzolini J, Herit F, Niedergang F. Dynamin‐actin cross talk contributes to phagosome formation and closure. Traffic. 2016;17(5):487‐499. 10.1111/tra.12386 [DOI] [PubMed] [Google Scholar]
- 182. Prichard KL, O'Brien NS, Murcia SR, Baker JR, McCluskey A. Role of clathrin and dynamin in clathrin mediated endocytosis/synaptic vesicle recycling and implications in neurological diseases. Front Cell Neurosci. 2022;15. 10.3389/fncel.2021.754110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Robertson MJ, Hadzic G, Ambrus J, et al. The rhodadyns, a new class of small molecule inhibitors of dynamin GTPase activity. ACS Med Chem Lett. 2012;3(5):352‐356. 10.1021/ml200284s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Daniel JA, Malladi CS, Kettle E, McCluskey A, Robinson PJ. Analysis of synaptic vesicle endocytosis in synaptosomes by high‐content screening. Nat Protoc. 2012;7(8):1439‐1455. 10.1038/nprot.2012.070 [DOI] [PubMed] [Google Scholar]
- 185. Odell LR, Howan D, Gordon CP, et al. The pthaladyns: GTP competitive inhibitors of dynamin I and II GTPase derived from virtual screening. J Med Chem. 2010;53(14):5267‐5280. 10.1021/jm100442u [DOI] [PubMed] [Google Scholar]
- 186. McGeachie AB, Odell LR, Quan A, et al. Pyrimidyn compounds: dual‐action small molecule pyrimidine‐based dynamin inhibitors. ACS Chem Biol. 2013;8(7):1507‐1518. 10.1021/cb400137p [DOI] [PubMed] [Google Scholar]
- 187. Yang N, Shen H‐M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID‐19. Int J Biol Sci. 2020;16(10):1724‐1731. 10.7150/ijbs.45498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Sasso L, Purdie L, Grabowska A, Jones AT, Alexander C. Time and cell‐dependent effects of endocytosis inhibitors on the internalization of biomolecule markers and nanomaterials. J Interdiscipl Nanomedic. 2018;3(2):67‐81. 10.1002/jin2.39 [DOI] [Google Scholar]
- 189. Wayteck L. Effects of endocytosis inhibitors on transfection mediated by polyplexes and lipoplexes carrying mRNA. Master thesis, Ghent University; 2010. [Google Scholar]
- 190. Wang H, Liu W, Yu F, Lu L. Disruption of clathrin‐dependent trafficking results in the failure of grass carp reovirus cellular entry. Virology J. 2016;13:1‐10. 10.1186/s12985-016-0485-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Sandvig K, Kavaliauskiene S, Skotland T. Clathrin‐independent endocytosis: an increasing degree of complexity. Histochem Cell Biol. 2018;150(2):107‐118. 10.1007/s00418-018-1678-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Rejman J, Bragonzi A, Conese M. Role of clathrin‐and caveolae‐mediated endocytosis in gene transfer mediated by lipo‐and polyplexes. Mol Ther. 2005;12(3):468‐474. 10.1016/j.ymthe.2005.03.038 [DOI] [PubMed] [Google Scholar]
- 193. Bolard J. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta (BBA)‐Reviews Biomembr. 1986;864(3‐4):257‐304. 10.1016/0304-4157(86)90002-x [DOI] [PubMed] [Google Scholar]
- 194. Hao M, Mukherjee S, Sun Y, Maxfield FR. Effects of cholesterol depletion and increased lipid unsaturation on the properties of endocytic membranes*[boxs]. J Biol Chem. 2004;279(14):14171‐14178. 10.1074/jbc.m309793200 [DOI] [PubMed] [Google Scholar]
- 195. Thottacherry JJ, Kosmalska AJ, Pradhan S, et al. Mechanochemical feedback control of dynamin independent endocytosis modulates membrane tension in adherent cells. Biophysical J. 2019;116(3):92a‐93a. 10.1016/j.bpj.2018.11.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Fujimoto LM, Roth R, Heuser JE, Schmid SL. Actin assembly plays a variable, but not obligatory role in receptor‐mediated endocytosis. Traffic. 2000;1(2):161‐171. 10.1034/j.1600-0854.2000.010208.x [DOI] [PubMed] [Google Scholar]
- 197. Lamaze C, Fujimoto LM, Yin HL, Schmid SL. The actin cytoskeleton is required for receptor‐mediated endocytosis in mammalian cells. J Biol Chem. 1997;272(33):20332‐20335. 10.1074/jbc.272.33.20332 [DOI] [PubMed] [Google Scholar]
- 198. Puthenveedu MA, Lauffer B, Temkin P, et al. Sequence‐dependent sorting of recycling proteins by actin‐stabilized endosomal microdomains. Cell. 2010;143(5):761‐773. 10.1016/j.cell.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Kälin S, Amstutz B, Gastaldelli M, et al. Macropinocytotic uptake and infection of human epithelial cells with species B2 adenovirus type 35. J Virol. 2010;84(10):5336‐5350. 10.1128/jvi.02494-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Kirchhausen T, Macia E, Pelish HE. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008;438:77‐93. 10.1016/s0076-6879(07)38006-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Robertson MJ, Deane FM, Robinson PJ, McCluskey A. Synthesis of dynole 34‐2, dynole 2‐24 and Dyngo 4a for investigating dynamin GTPase. Nat Protoc. 2014;9(4):851‐870. 10.1038/nprot.2014.046 [DOI] [PubMed] [Google Scholar]
- 202. Basagiannis D, Zografou S, Goula E, Gkeka D, Kolettas E, Christoforidis S. Chemical inhibitors of dynamin exert differential effects in VEGF signaling. Cells. 2021;10(5):997. 10.3390/cells10050997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Dutta D, Williamson CD, Cole NB, Donaldson JG. Pitstop 2 is a potent inhibitor of clathrin‐independent endocytosis; 2012. [DOI] [PMC free article] [PubMed]
- 204. Willox AK, Sahraoui YM, Royle SJ. Non‐specificity of Pitstop 2 in clathrin‐mediated endocytosis. Biol Open. 2014;3(5):326‐331. 10.1242/bio.20147955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Francia V, Reker‐Smit C, Boel G, Salvati A. Limits and challenges in using transport inhibitors to characterize how nano‐sized drug carriers enter cells. Nanomedicine. 2019;14(12):1533‐1549. 10.2217/nnm-2018-0446 [DOI] [PubMed] [Google Scholar]
- 206. Hawtrey A, Joubert D, van Jaarsveld P, Pieterse A, van Zyl J, Ariatti M. Low concentrations of chlorpromazine and related phenothiazines stimulate gene transfer in HeLa cells via receptor‐mediated endocytosis. Drug Deliv. 2002;9(1):47‐53. 10.1080/107175402753413172 [DOI] [PubMed] [Google Scholar]
- 207. Al‐Bari MAA. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol Res Perspecti. 2017;5(1):e00293. 10.1002/prp2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Kanlaya R, Sintiprungrat K, Chaiyarit S, Thongboonkerd V. Macropinocytosis is the major mechanism for endocytosis of calcium oxalate crystals into renal tubular cells. Cell Biochem biophysics. 2013;67(3):1171‐1179. 10.1007/s12013-013-9630-8 [DOI] [PubMed] [Google Scholar]
- 209. Preservation AIMEB . Cell Drinking: A Closer Look at How Macropinocytosis Drives Cholesterol Uptake in Atherosclerotic Vessels.
- 210. Chen X, Zhou HJ, Huang Q, Lu L, Min W. Novel action and mechanism of auranofin in inhibition of vascular endothelial growth factor receptor‐3‐dependent lymphangiogenesis. Anti‐Cancer Agents Med Chem Former Curr Med Chemistry‐Anti‐Cancer Agents. 2014;14(7):946‐954. 10.2174/1871520614666140610102651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Knyazev E, Nersisyan S, Tonevitsky A. Endocytosis and transcytosis of SARS‐CoV‐2 across the intestinal epithelium and other tissue barriers. Front Immunol. 2021;12:3639. 10.3389/fimmu.2021.636966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Chang C‐C, Wu M, Yuan F. Role of specific endocytic pathways in electrotransfection of cells. Mol Ther Methods Clin Dev. 2014;1:14058. 10.1038/mtm.2014.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Sidaway JE, Davidson RG, McTaggart F, et al. Inhibitors of 3‐hydroxy‐3‐methylglutaryl–CoA reductase reduce receptor‐mediated endocytosis in opossum kidney cells. J Am Soc Nephrol. 2004;15(9):2258‐2265. 10.1097/01.asn.0000138236.82706.ee [DOI] [PubMed] [Google Scholar]
- 214. Ward NC, Watts GF, Eckel RH. Statin toxicity: mechanistic insights and clinical implications. Circulation Res. 2019;124(2):328‐350. 10.1161/circresaha.118.312782 [DOI] [PubMed] [Google Scholar]
- 215. Verhulst A, D’Haese PC, De Broe ME. Inhibitors of HMG‐CoA reductase reduce receptor‐mediated endocytosis in human kidney proximal tubular cells. J Am Soc Nephrol. 2004;15(9):2249‐2257. 10.1097/01.asn.0000136778.32499.05 [DOI] [PubMed] [Google Scholar]
- 216. Araki N, Johnson MT, Swanson JA. A role for phosphoinositide 3‐kinase in the completion of macropinocytosis and phagocytosis by macrophages. JCB (J Cell Biol). 1996;135(5):1249‐1260. 10.1083/jcb.135.5.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Fernando LP, Kandel PK, Yu J, McNeill J, Ackroyd PC, Christensen KA. Mechanism of cellular uptake of highly fluorescent conjugated polymer nanoparticles. Biomacromolecules. 2010;11(10):2675‐2682. 10.1021/bm1007103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Sarkar K, Kruhlak MJ, Erlandsen SL, Shaw S. Selective inhibition by rottlerin of macropinocytosis in monocyte‐derived dendritic cells. Immunology. 2005;116:513‐524. 10.1111/j.1365-2567.2005.02253.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Kang Y‐L, Oh C, Ahn S‐H, et al. Inhibition of endocytosis of porcine reproductive and respiratory syndrome virus by rottlerin and its potential prophylactic administration in piglets. Antivir Res. 2021;195:105191. 10.1016/j.antiviral.2021.105191 [DOI] [PubMed] [Google Scholar]
- 220. Zakaryan H, Arabyan E, Oo A, Zandi K. Flavonoids: promising natural compounds against viral infections. Archives Virol. 2017;162(9):2539‐2551. 10.1007/s00705-017-3417-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Ferrer J‐L, Austin M, Stewart C, Jr , Noel J. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Biochem. 2008;46(3):356‐370. 10.1016/j.plaphy.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Doyle SM, Vain T, Robert S. Small molecules unravel complex interplay between auxin biology and endomembrane trafficking. J Exp Bot. 2015;66(16):4971‐4982. 10.1093/jxb/erv179 [DOI] [PubMed] [Google Scholar]
- 223. Paudyal R, Jamaluddin A, Warren JP, et al. Trafficking modulator TENin1 inhibits endocytosis, causes endomembrane protein accumulation at the pre‐vacuolar compartment and impairs gravitropic response in Arabidopsis thaliana. Biochem J. 2014;460(2):177‐185. 10.1042/bj20131136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Paciorek T, Zažímalová E, Ruthardt N, et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435(7046):1251‐1256. 10.1038/nature03633 [DOI] [PubMed] [Google Scholar]
- 225. Song BD, Leonard M, Schmid SL. Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin‐mediated endocytosis. J Biol Chem. 2004;279(39):40431‐40436. 10.1074/jbc.m407007200 [DOI] [PubMed] [Google Scholar]
- 226. Dutta D, Donaldson JG. Sorting of clathrin‐independent cargo proteins depends on Rab35 delivered by clathrin‐mediated endocytosis. Traffic. 2015;16(9):994‐1009. 10.1111/tra.12302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Peng M, Yin N, Zhang W. Endocytosis of FcαR is clathrin and dynamin dependent, but its cytoplasmic domain is not required. Cell Res 2010;20(2):223‐237. 10.1038/cr.2009.120 [DOI] [PubMed] [Google Scholar]
- 228. Aït‐Slimane T, Galmes R, Trugnan G, Maurice M. Basolateral internalization of GPI‐anchored proteins occurs via a clathrin‐independent flotillin‐dependent pathway in polarized hepatic cells. Mol Biol Cell. 2009;20(17):3792‐3800. 10.1091/mbc.e09-04-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Confalonieri S, Salcini AE, Puri C, Tacchetti C, Di Fiore PP. Tyrosine phosphorylation of Eps15 is required for ligand‐regulated, but not constitutive, endocytosis. J Cell Biol. 2000;150(4):905‐912. 10.1083/jcb.150.4.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Larson ER, Van Zelm E, Roux C, Marion‐Poll A, Blatt MR. Clathrin heavy chain subunits coordinate endo‐and exocytic traffic and affect stomatal movement. Plant Physiol. 2017;175(2):708‐720. 10.1104/pp.17.00970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Kitakura S, Vanneste S, Robert S, et al. Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell. 2011;23(5):1920‐1931. 10.1105/tpc.111.083030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Vocelle D, Chan C, Walton SP. Endocytosis controls siRNA efficiency: implications for siRNA delivery vehicle design and cell‐specific targeting. Nucleic Acid Therapeut. 2020;30(1):22‐32. 10.1089/nat.2019.0804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Motley A, Bright NA, Seaman MN, Robinson MS. Clathrin‐mediated endocytosis in AP‐2–depleted cells. J Cell Biol. 2003;162(5):909‐918. 10.1083/jcb.200305145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Kadlecova Z, Spielman SJ, Loerke D, Mohanakrishnan A, Reed DK, Schmid SL. Regulation of clathrin‐mediated endocytosis by hierarchical allosteric activation of AP2. JCB (J Cell Biol). 2017;216(1):167‐179. 10.1083/jcb.201608071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Gilleron J, Querbes W, Zeigerer A, et al. Image‐based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31(7):638‐646. 10.1038/nbt.2612 [DOI] [PubMed] [Google Scholar]
- 236. Moulay G, Lainé J, Lemaître M, et al. Alternative splicing of clathrin heavy chain contributes to the switch from coated pits to plaques. JCB (J Cell Biol). 2020;219(9):e201912061. 10.1083/jcb.201912061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Le PU, Guay G, Altschuler Y, Nabi IR. Caveolin‐1 is a negative regulator of caveolae‐mediated endocytosis to the endoplasmic reticulum. J Biol Chem. 2002;277(5):3371‐3379. 10.1074/jbc.m111240200 [DOI] [PubMed] [Google Scholar]
- 238. Hernández‐Deviez DJ, Howes MT, Laval SH, Bushby K, Hancock JF, Parton RG. Caveolin regulates endocytosis of the muscle repair protein, dysferlin. J Biol Chem. 2008;283(10):6476‐6488. 10.1074/jbc.m708776200 [DOI] [PubMed] [Google Scholar]
- 239. Bisi S, Marchesi S, Rizvi A, et al. IRSp53 controls plasma membrane shape and polarized transport at the nascent lumen in epithelial tubules. Nat Commun. 2020;11:1‐23. 10.1038/s41467-020-17091-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Jenkins LM, Singh P, Varadaraj A, et al. Altering the proteoglycan state of transforming growth factor β type III receptor (TβRIII)/betaglycan modulates canonical Wnt/β‐catenin signaling. J Biol Chem. 2016;291(49):25716‐25728. 10.1074/jbc.m116.748624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Fiuza M, Rostosky CM, Parkinson GT, et al. PICK1 regulates AMPA receptor endocytosis via direct interactions with AP2 α‐appendage and dynamin. JCB (J Cell Biol). 2017;216(10):3323‐3338. 10.1083/jcb.201701034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Bell J, Park E, Ai J, Baker A. PICK1‐mediated GluR2 endocytosis contributes to cellular injury after neuronal trauma. Cell Death Differ. 2009;16(12):1665‐1680. 10.1038/cdd.2009.106 [DOI] [PubMed] [Google Scholar]
- 243. Ivanova D, Imig C, Camacho M, et al. CtBP1‐mediated membrane fission contributes to effective recycling of synaptic vesicles. Cell Rep 2020;30(7):2444‐2459.e2447. 10.1016/j.celrep.2020.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Schnatwinkel C, Christoforidis S, Lindsay MR, et al. The Rab5 effector Rabankyrin‐5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2004;2(9):e261. 10.1371/journal.pbio.0020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Hermle T, Schneider R, Schapiro D, et al. GAPVD1 and ANKFY1 mutations implicate RAB5 regulation in nephrotic syndrome. J Am Soc Nephrol. 2018;29(8):2123‐2138. 10.1681/asn.2017121312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ghosh S, Al-Sharify ZT, Maleka MF, et al. Propolis efficacy on SARS‐COV viruses: a review on antimicrobial activities and molecular simulations. Environmental Science and Pollution Research, 2022;29(39):58628‐58647. 10.1007/s11356-022-21652-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information S1
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


