To the Editor,
SARS‐CoV‐2 RNA in the bloodstream, despite a very low viral load level, has been associated with COVID‐19 severity. 1 The duration of viral shedding, sometimes longer than in immunocompetent patients, could be associated to relapse of viral replication with symptoms re‐exacerbations, and increase in the risk of severe infection and death. 2
This study reports a case of severe persistent SARS‐CoV‐2 infection in a 56‐years old immunocompromised male patient presenting an increasing viral load in the bloodstream over a period of 45 days. It was also possible to perform whole genome sequencing of the virus collected from both bronchoalveolar lavage (BAL) and plasma specimens.
The patient had a history of low‐grade non‐Hodgkin lymphoma treated firstly with bendamustine plus rituximab and then with maintenance therapy with rituximab every 45 days. The patient, affected by ischemic heart disease and COPD, received the third dose of COVID‐19 vaccine on 9th January 2022.
On 25th January 2022, he was diagnosed with asymptomatic SARS‐CoV‐2 infection in another hospital. After a few days, the patient developed a persistent fever with mild respiratory distress and was treated with oral molnupiravir and intravenous sotrovimab, without any benefit. No evidence of anti‐SARS‐CoV‐2 IgG antibodies was detected.
On 14th March 2022, the nasopharyngeal swab (NPS) confirmed the persistence of SARS‐CoV‐2.
On 22nd March, upon admittance to the Emergency Room of our hospital, he presented high fever and dyspnoea. The NPS resulted negative for SARS‐CoV‐2 RNA.
Three days later, despite the persistence of symptoms and the detection of anti‐SARS‐CoV‐2 IgG antibodies, the patient still tested negative for SARS‐CoV‐2 RNA on NPS, while RT‐PCR was positive in plasma sample (Cycle threshold—Ct—values: 33 for RdRP, 35 for ORF8 targets).
The chest X‐ray showed evidence of COPD and mild interstitial pneumonia. The chest CT scan revealed signs of centrilobular emphysema and mild parenchymal consolidations (Figure 1A). As the clinical conditions deteriorated, he received a broad‐spectrum antibiotic therapy, with ineffective results.
Figure 1.
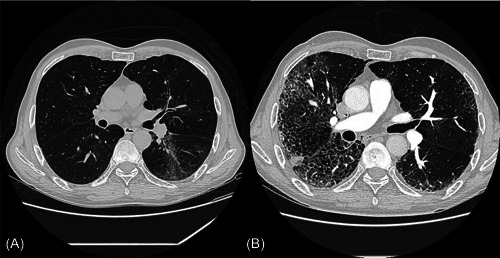
(A) Chest computer tomography (CT) performed upon entrance at the Emergency Department, showing signs of centrilobular emphysema and mild parenchymal consolidations. (B) Chest CT performed 27 days after, showing severe interstitial bilateral pneumonia and dystrophic‐bullous areas.
On 28th March, BAL resulted positive for SARS‐CoV‐2 RNA (Ct values: 21 ORF1ab and 23N). Viral RNA was detected in the bloodstream confirming the Ct values previously detected. Whole genome sequencing (WGS) performed on BAL showed that the patient was infected by Omicron variant sub‐lineage BA.1.1 (accession ID: EPI_ISL_14479735).
His respiratory conditions worsened requiring high‐flow oxygen support. Therefore, the patient was treated with intravenous remdesivir for 5 days in addiction to intravenous dexamethasone 6 mg/day with a rapid clinical improvement: remission of fever and progressive reduction of oxygen therapy.
On 9th April, despite steroid treatment, the fever increased, and he reported dyspnoea. From 11th to 27th April, an increasing SARS‐CoV‐2 viral load was observed in the bloodstream (Ct values: 29 and 30 vs. 24 and 26 for RdRP and ORF8, respectively).
Considering the comorbidities and the persistence of SARS‐CoV‐2, AIFA (Italian Drugs Agency) approved the off‐label treatment with oral nirmatrelvir/ritonavir plus a second course of intravenous sotrovimab. After a few days, as the patient's conditions furtherly worsened despite antiviral and monoclonal therapy, hyperimmune serum was administrated without any benefit (Figure 1B shows severe interstitial pneumonia progressed despite all treatments).
On 5th May 2022, SARS‐CoV‐2 RNA was detected in NPS (Ct values: 27 RdRP and 29 ORF8) and plasma samples (Ct values: 22 RdRP and 24 ORF8). Given the low Ct value detected in the plasma specimen, both culture and sequencing of the viral genome were attempted. Cell culture did not show virus‐induced cytopathic effect, while WGS confirmed the presence in the bloodstream of the same Omicron sublineage BA.1.1 (EPI_ISL_14596883) as identified in the BAL.
The full‐genome sequence was assessed by iSeq 100 instrument (Illumina), using the CleanPlex SARS‐CoV‐2 Flex kit (Paragon Genomics, Arrow); data analysis was performed by means of SOPHiA‐DDM‐v4 platform and viral lineage was assigned using Pangolin tool (v4.1.1, data‐v1.13). Regarding sequencing data, for BAL and plasma specimens 275 749 and 605 704 read pairs were sequenced, of these 272 045 and 603 313 were mapped corresponding to 96.7% and 99.6% of all reads, respectively, providing an observed coverage of 491x and 1,261x, respectively.
GISAID showed two quite similar viral strains harboring a comparable mutations pattern in both samples (Supporting Information: Table S1). The patient, unfortunately, died on 6th May 2022. Figure 2 shows the viral dynamics.
Figure 2.
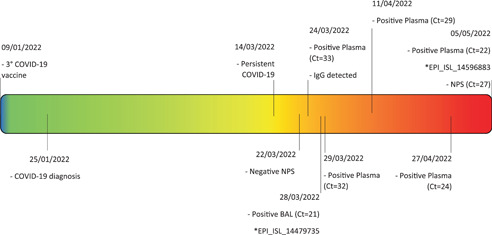
Patient's clinical course and laboratory findings. The figure summarizes the evolution of patient's clinical picture, underling the timespan between COVID‐19 vaccination on 9th January 2022, COVID‐19 diagnosis and the persistence of SARS‐CoV‐2 RNA until decease on 6th May 2022. * indicates genome sequence Accession ID (GISAID). BAL, bronchoalveolar lavage; Ct, real‐time PCR Cycle threshold values; NPS, nasopharyngeal swab.
This study reports the case of a patient with prolonged shedding of SARS‐CoV‐2 RNA in the bloodstream associated with an unusual high viral load that could be associated with viable and replication‐competent SARS‐CoV‐2 strain. Despite the absence of a cytopathic effect in cell cultures, the whole genome sequencing on plasma sample highlighted the presence of a complete viral genome even after 90 days from diagnosis.
Today therapeutic options against COVID‐19 are represented by antivirals and monoclonal antibodies (mAbs). SARS‐CoV‐2‐specific mAbs and antiviral treatments are approved for early postexposure treatment of patients with risk of severe COVID‐19 disease, especially immunocompromised patients with a poor host response. Unfortunately, all these drugs need to be administered within a very short time from symptoms onset to achieve the best efficacy against the virus. 3 , 4 People with lymphoid malignancies, especially those on B‐cell‐depleting therapies such as anti‐CD20 drugs, are particularly susceptible to having persistent infections and impaired response to vaccination due to defective humoral immunity and a poor ability to produce neutralizing antibodies.
Our patient was treated with the correct antiviral and monoclonal timing during the first phase of the disease but, probably because of his immune system status, he didn't manage to completely clear SARS‐CoV‐2. During hospitalization in our ward, the patient was not within the recommended timeframe for treatment, but without other available treatment options, two specific antiviral drugs (remdesivir and nirmatrelvir/ritonavir), and one monoclonal treatment (sotrovimab) were administered. Specifically, after administration of remdesivir plus intravenous corticosteroids, the fever subsided for a few days. However, it relapsed in association with worsening respiratory failure and didn't respond to a second course of sotrovimab in combination with nirmatrelvir/ritonavir. In particular, the administration of remdesivir was ineffective probably due to the immunocompromised status and the consequent prolonged viral shedding and nirmatrelvir/ritonavir failed because it was administered too late, when “cytokine storm” was already activated. 5 The inefficacy of mAb administration might be explained by the presence of S371L/F mutation that affected most of the RBD‐directed antibodies. 6 Tada et al. observed that the presence of the single mutation S371L affects sotrovimab activity destabilizing the antibody nearby structure. 7
In our case, the abundance of SARS‐CoV‐2 RNA in the bloodstream allowed the genomic analysis, which revealed two similar viral genomes in plasma and airways. Confirming the evidence by Andersson et al. about the lack of association between virions detected in the bloodstream and viral infectiousness, 8 the prolonged persistence of SARS‐CoV‐2 RNA at high levels in the bloodstream of our patient might be the consequence of massive viral dissemination from the pulmonary district; this hypothesis is confirmed by the absence of intra‐host viral evolution as revelated in other immunosuppressed long COVID‐19 cases (phylogenetic tree in Supporting Information: Figure S1), as well as the absence of viral growth despite the low Ct values. 9 , 10 Further studies, especially in immunocompromised patients with persistence of SARS‐CoV‐2, should be conducted to investigate the viral viability in different anatomical districts and the possible emergence of specific mutations related to viral compartmentalization.
AUTHOR CONTRIBUTIONS
Alberto Rizzo, Fiorenza Bracchitta, Federica Salari, and Davide Mileto contributed to writing the article and performed virologic activities. Antonella Foschi, Ivano Faggion, Fabio Borgonovo, and Chiara Fusetti contributed to writing the article and visited the patient. Giuliano Rizzardini coordinated clinical activities and Maria R. Gismondo coordinated laboratory activities. Alessandra Lombardi and Valeria Micheli coordinated virologic activities and reviewed the article. All authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Table S1. Frequencies of aminoacidic changes detected in S gene: bronchoalveolar lavage (BAL) versus plasma sample.
Figure S1. Phylogenetic tree showing relationship between patient's sequences and other viral isolates. The phylogenetic analysis revealed a close relationship between viral sequences from bronchoalveolar lavage (BAL) and plasma samples (red circle). They form a small cluster with no evidence of viral evolution. The black squares refer to Delta variant while the black triangles to the Omicron one. The white circle is related to the Italian patient 1 while the white square to SARS‐CoV‐2 reference genome (NC_045512.2). The light blue diamond shape is related to the published case of intra‐host evolution [7].
DATA AVAILABILITY STATEMENT
The data are not publicly available due to privacy or ethical restrictions. The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Cardeñoso Domingo L, Roy Vallejo E, Zurita Cruz ND, et al. Relevant SARS‐CoV‐2 viremia is associated with COVID‐19 severity: prospective cohort study and validation cohort. J Med Virol. 2022;94(11):5260‐5270. 10.1002/jmv.27989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martínez‐Chinchilla C, Vazquez‐Montero L, Palazón‐Carrión N, et al. Persistence of SARS‐CoV‐2 infection in severely immunocompromised patients with complete remission B‐cell lymphoma and anti‐CD20 monoclonal antibody therapy: a case report of two cases. Front Immunol. 2022;13:860891. 10.3389/fimmu.2022.860891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Molnupiravir COVID‐19 Treatment Guidelines . National Institutes of Health NIH; 2022. https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/molnupiravir
- 4. Ertesvåg NU, Sakkestad ST, Zhou F, et al. Persistent fever and positive PCR 90 days post‐SARS‐CoV‐2 infection in a Rituximab‐Treated patient: a case of late antiviral treatment. Viruses. 2022;14(8):1757. 10.3390/v14081757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carlin AF, Clark AE, Chaillon A, et al. Virologic and immunologic characterization of coronavirus disease 2019 recrudescence after nirmatrelvir/ritonavir treatment. Clin Infect Dis. Published online June 20, 2022:ciac496. 10.1093/cid/ciac496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iketani S, Liu L, Guo Y, et al. Antibody evasion properties of SARS‐CoV‐2 omicron sublineages. Nature. 2022;604(7906):553‐556. 10.1038/s41586-022-04594-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tada T, Zhou H, Dcosta BM, et al. Increased resistance of SARS‐CoV‐2 omicron variant to neutralization by vaccine‐elicited and therapeutic antibodies. EBioMedicine. 2022;78:103944. 10.1016/j.ebiom.2022.103944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andersson MI, Arancibia‐Carcamo CV, Auckland K, et al. SARS‐CoV‐2 RNA detected in blood products from patients with COVID‐19 is not associated with infectious virus. Wellcome Open Res. 2020;5:181. 10.12688/wellcomeopenres.16002.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mancon A, Rizzo A, Mileto D, et al. Viro‐immunological evaluation in an immunocompromised patient with long‐lasting SARS‐CoV‐2 infection. Emerg Microbes Infect. 2022;11(1):786‐789. 10.1080/22221751.2022.2045877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mileto D, Foschi A, Mancon A, et al. A case of extremely prolonged viral shedding: could cell cultures be a diagnostic tool to drive COVID‐19 patient discharge? Int J Infect Dis. 104:631. 10.1016/j.ijid.2020.11.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Frequencies of aminoacidic changes detected in S gene: bronchoalveolar lavage (BAL) versus plasma sample.
Figure S1. Phylogenetic tree showing relationship between patient's sequences and other viral isolates. The phylogenetic analysis revealed a close relationship between viral sequences from bronchoalveolar lavage (BAL) and plasma samples (red circle). They form a small cluster with no evidence of viral evolution. The black squares refer to Delta variant while the black triangles to the Omicron one. The white circle is related to the Italian patient 1 while the white square to SARS‐CoV‐2 reference genome (NC_045512.2). The light blue diamond shape is related to the published case of intra‐host evolution [7].
Data Availability Statement
The data are not publicly available due to privacy or ethical restrictions. The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


