Abstract
Coronavirus disease‐2019 (COVID‐19) is pro‐inflammatory disorder characterized by acute respiratory distress syndrome. Interleukin‐6, a cytokine secreted by macrophages, which mediates an inflammatory response, is frequently increased and associated with the severity in COVID‐19 patients. The differential expression of IL6 cytokine in COVID‐19 patients may be associated with the presence of single nucleotide polymorphisms (SNPs) in regulatory region of cytokine genes. The aim of this study is to investigate the role of two promoter polymorphisms of the IL6 gene (–597G > A and –174G > C) with the severity of COVID‐19. The study included 242 patients, out of which 97 patients with severe symptoms and 145 patients with mild symptoms of COVID‐19. Genotyping of two selected SNPs, rs1800795 (–174G > C) and rs1800797 (–597G > A) of promoter region of IL6 gene, was performed by polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP). In our study, individuals with GC genotypes of IL6 (–174G > C) polymorphism showed significantly higher risk of severity [adjusted odds (OR) 3.86, p <.001] but we did not observe any association of COVID‐19 severity with rs1800797 (–597G > A) polymorphism. The COVID‐19 severity was significantly higher in individuals having ‘C’ allele of IL6 (–174G > C) polymorphism (p = .014). Linkage disequilibrium between rs1800795 (–174G > C) and rs1800797 (–597G > A) showed that individuals having AC* haplotype significantly association with COVID‐19 severity (p = .034). Our results suggest that ‘C’ allele of rs1800795 (–174G > C) polymorphism of IL6 may be the risk allele for severity of COVID‐19 in North Indian population.
Keywords: COVID‐19, haplotype analysis, IL6, PCR‐RFLP, severity, SNPs
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) was first time reported in Wuhan, China at the end of 2019 which is considered as a severe threat to global public health and the main cause of the ongoing coronavirus disease 2019 (COVID‐19) pandemic (Wu et al., 2020; Zhou et al., 2020). While many COVID‐19 patients suffer mild‐to‐moderate illness or remain asymptomatic, more than 20% of SARS‐CoV‐2 infections result in acute lung injury (ALI) and severe acute respiratory distress syndrome (ARDS), which can cause severe pneumonia, alveolar injury, and, in the most serious instances, even death (Falahi et al., 2022). The common symptoms of COVID‐19 include fever, headache, shortness of breath, dry cough, sputum production, sore throat, fatigue, myalgia, haemoptysis and diarrhoea (Gong et al., 2022). COVID‐19 infection is associated with overproduction pro‐inflammatory cytokines (also known as cytokine storms) such as interleukins‐6 (IL6), IL1β, IL4, IL10, IL18, interferon (IFN)γ, tumour necrosis factor alpha (TNFα) (Costela et al., 2020; Roshanravan et al., 2020). The cytokine storm is main factor for severe clinical complications in COVID‐19 patients and is the main reason of death (Bhaskar et al., 2020).
Key points
COVID‐19 severity is associated with elevated level of pro‐inflammatory cytokines such as IL6 and differential IL6 cytokine production may be associated with the presence of single nucleotide polymorphisms (SNPs) in regulatory region of cytokine genes.
In this study, we observed that ‘C’ allele of IL6 −174G > C polymorphism of may be the risk allele for severity of COVID‐19.
This study provides the predictive biomarker that will help the clinician to identify early diagnosis of the severity of the patients.
IL6, a key inflammatory cytokine, is secreted by a fraction of immune and non‐immune cells in lung tissue, such as resident alveolar macrophages, T lymphocytes, alveolar type II epithelial cells (ECs) and lung fibroblasts (Zhang et al., 2020). IL6 plays a crucial role to induce lymphocytic apoptosis that leads to the development of lymphopaenia in COVID‐19 patients (Abbasifard & Khorramdelazad, 2020; Tan et al., 2020). The high level of IL6 significantly downregulates the expression of human leukocyte D antigen (HLA‐DR) that substantially impair lymphocyte function, along with the depletion of CD4+ lymphocytes, CD19+ lymphocytes and natural killer (NK) cells. In addition, IL6 is considered to have an impact in the severity of COVID‐19 patients, which is significantly associated with adverse clinical outcomes (Chen et al., 2020; Gong et al., 2020; Liu et al., 2021).
IL6 gene encodes 21 KDa IL6 glycoprotein in human that harbour many single‐nucleotide polymorphisms (SNPs) in the coding and non‐coding regions (Jia et al., 2015). The presence of SNPs in the regulatory regions, such as promoters, introns and the 5′‐ and 3′‐UTR regions, may account for the differences in cytokine production between individuals, whereas genetic polymorphisms in gene‐coding regions may result in the loss or alteration of function in the expressed proteins (Haukim et al., 2002). Several epidemiological studies have been reported that the genetic polymorphisms at rs1800795 (−174G > C), rs1800796 (−572G > C) and rs1800797 (−597G > A) of the IL6 gene promoter are associated with the risk and severity of many diseases, such as pneumonia, asthma, chronic obstructive pulmonary disease (COPD) (Chen et al., 2015; He et al., 2009; Jimenez‐Sousa et al., 2017; Kirtipal et al., 2020). Thus, this study was undertaken to evaluate the possible correlation between genetic polymorphism at rs1800795 (−174G > C), and rs1800797 (−597G > A) of IL6 gene promoter and the severity of susceptibility to COVID‐19 in North Indian population.
2. MATERIALS AND METHODS
2.1. Sample collection and experimental design
This study was approved by the Ethics Committee of Era University, India (ELMC&H/R_Cell/EC/2020/272) and all study's procedures in this investigation involving humans’ patients were carried out according to the ethical guidelines of Era University, India. A total of 242 RT‐PCR confirmed COVID‐19 patients with age more than 20 years, having fever and respiratory symptoms were recruited from Era's Lucknow Medical College and Hospital (ELMC&H) of Era University, Lucknow. Pregnant lady and patients with known malignant disease were excluded.
All clinical and demographic data such as age, sex, hypertension and diabetes mellitus were collected from hospital records under supervision of expert clinician. All patients involved in this study were categorized into mild (145 patients) and severe (97 patients) group (Table 1), as per the guidelines of the Indian Council of Medical Research (ICMR) (Abbas et al., 2021; Verma et al., 2021). Patients with respiratory rate less than 24 per minute and SpO2 > 94% on room air were considered as mild patient while patients with respiratory rate more than 30 per minute OR SpO2 < 90% on room air with pneumonia were categorized into severe patients. After an informed consent form from patients, 5 ml of venous blood were obtained in ethylene diamine tetraacetic acid (EDTA) vials from all subjects and stored in −20°C for further use.
TABLE 1.
Association of COVID‐19 severity with demographic and clinical parameters
| Variables | Mild (n, %) | Severe | p Value |
|---|---|---|---|
| Age | |||
| <45 | 73 (50.3) | 12 (12.4) | <.001 |
| ≥ 45 | 72 (49.7) | 85 (87.6) | |
| Sex | |||
| Male | 94 (64.8) | 66 (68.0) | .605 |
| Female | 51 (35.2) | 31 (32.0) | |
| Diabetes | |||
| Non‐diabetic | 133 (91.7) | 66 (68) | <.001 |
| Diabetic | 12 (8.3) | 31 (32) | |
| Hypertension | |||
| Non‐hypertensive | 138 (95.2) | 79 (81.4) | <.001 |
| Hypertensive | 7 (4.8) | 18 (18.6) | |
2.2. DNA isolation and genotyping
The genomic DNA was isolated from venous blood, using a commercially available kit (NucleoSpin Blood, DNA Mini kit, Macherey‐Nagel, Germany). The quality and quantity of DNA was assessed by Nanodrop (Thermo Fisher, USA) and 1% agarose gel electrophoresis, respectively. The ratio of absorbance at 260 nm and 280 nm (A260/A280) with a range of 1.7−2.0 was used to determine the acceptability criteria for DNA quality/purity.
The genetic polymorphism at rs1800795 (−174G > C), and rs1800797 (−597G > A) of IL6 gene promoter were examined by using polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP). Using gradient master cycler (Bio‐Rad, USA), PCR amplification was carried out in a 25 µl reaction mixture with genomic DNA (150–200 ng), 10 pmol of each primer and 1× master mix (Takara Bio, Japan). A set of the primers, F5’‐GGAGTCACACACTCCACCT‐3′, R5’‐CTGATTGGAAACCTTATTAAG‐3′ for rs1800797 and F‐5′‐CTCTTTGTCA AGACATGCCA‐3′, R‐5′ GGGAAAATCCCACATTTGATAA‐3′ for rs1800795 (Saxena et al., 2020) were used in PCR reaction mixture, with following conditions: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 45 s, annealing at 60°C for 1.15 min, extension at 72°C for 2.30 min and final extension at 72°C for 10 min. The PCR products were examined on a 2% agarose gel stained with ethidium bromide (EtBr), using a gel documentation system (Bio‐Rad, USA).
PCR products were digested with 10 units of restriction enzymes directly for RFLP. To detect rs1800795 (−174G > C) polymorphism, 5 µl of PCR products was digested with 1 µl of NlaIII restriction enzyme (10 U/µl) in 1 µl of 10× buffer [a restriction endonuclease buffer produced by New England Biolabs USA, containing 100 mM of Tris‐HCI (pH 7.5), 100 mM of MgCl2, and 10 mM dithiothreitol] and 3 µl of ddH2O for 16 h at 37°C. To detect rs1800797 (−597G > A) polymorphism, 5 µl of PCR products was digested with 1 µl of FokI (10 U/µl) in 1 µl of 10 × buffer and 3 µl of ddH2O for 16 h at 37°C. Digested products were electrophoresed on 2.5% agarose gels, stained with ethidium bromide, and photographed using a gel documentation system (Bio‐Rad, USA) (Figure 1a and b).
FIGURE 1.
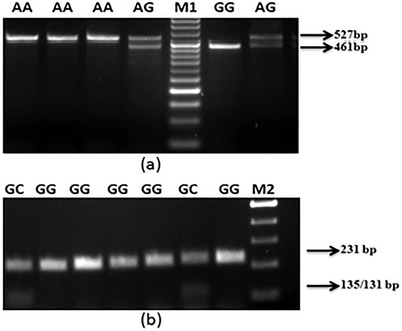
Ethidium bromide stained agarose gels showing different genotypes of IL6 gene. (a) SNP (G > A) showing AA: 527 bp, AG: 527 and 461 bp, GG: 461 bp, M1‐50 bp. (b) SNP (G > C) showing GG: 231 bp, GC: 135/131 bp, M2‐100 bp
2.3. Statistical analysis
Continuous variables were summarized as mean ± SE and compared using the Student's t‐test. Clinical and demographic data were compared by using chi‐square (χ2) analysis and Fisher's exact test. Fisher's exact test was used to compare allele and genotype frequencies in mild and severe COVID‐19 cases using a 2 × 2 contingency table and the respective p values adjusted by using the Bonferroni correction (p < .025). In addition, Hardy Weinberg equilibrium (HWE) for rs1800795 (−174G > C) and rs1800797 (−597G > A) polymorphisms in the severe and mild COVID‐19 groups was accessed by the χ2 test. The strength of the association was determined using the odds ratio (OR) at a 95% confidence interval (CI). All p values were considered statistically significant if p < .05. Most of the analyses were performed by SPSS (Version 21.0). Haplotype association of the two SNPs (single nucleotide polymorphisms) was carried out by using SHEsis software (online version).
3. RESULTS
3.1. Demographic and clinical characteristics of patients
Among 242 COVID‐19 patients, 97 were severe (31 women, 66 men) with a mean age of 58.26 ± 11.54 years and 145 were mild COVID‐19 (51 women, 94 men) with a mean age of 46.01 ± 15.43 years were studied. We found a significant difference between the severe and mild COVID‐19 patients based on age (p < .001). Severe COVID‐19 patients were older than mild COVID‐19 patients. We did not find any association between sex and COVID‐19 severity (p = .605). However, we observed that patients having any other comorbidities such as diabetes and hypertension, showed significantly higher risk of severity (p < .001) (Table 1). [Correction added on 19 November 2022, after first online publication: the probability values have been changed from p < .0001 to p <.001 throughout the text.]
3.2. Genetic analysis
The genotypic pattern at rs1800795 (−174G > C) and rs1800797 (−597G > A) of IL6 gene promoter are showed in Figure 1a and b. Genotypes and allele frequencies among mild and severe patients are shown in Table 2. We did not find homozygous recessive genotypes (CC) of rs1800795 (−174 G > C) polymorphism in our study population (Table 2). The GC genotype frequency was higher in severe patients as compared to mild and showed significant higher risk of COVID‐19 severity (2.26‐folds, p = .009). The risk of severity showed further increased when the data were adjusted for age, gender, hypertension, and diabetes (3.86‐folds, p < .001). The ‘C’ allele frequency of rs1800795 (−174G > C) polymorphism was higher in severe patients as compared to mild patients and statistically significant (p = .014) (Table 2). In case of rs1800797 (−597G > A) polymorphism, the frequency of ‘G’ alleles of rs1800797 (−597G > A) polymorphism is higher in mild cases (21.8%) as compared to severe cases (18.8%) but did not find any significant association (p = .441) (Table 2).
TABLE 2.
Genotypic and allelic frequencies of IL6‐597(G > A) and IL6‐174(G > C) gene polymorphisms in mild (n = 145) and COVID‐19 severe cases (n = 97)
|
Genotypes/alleles 145 (60.0) |
97 (40.0) |
Mild (n, %) Unadjusted OR (95% CI) |
p Value |
Severe (n, %) Adjusted b OR (95% CI) |
p Value | |
|---|---|---|---|---|---|---|
| IL6 −174 G > C | ||||||
| GG | 120 (82.8) | 66 (68) | 1.0 (Ref.) | 1.0 (Ref.) | ||
| GC | 25 (17.2) | 31 (32.0) | 2.26 (1.22–4.13) | .009 | 3.86 (1.81–8.24) | <.001 |
| CC | — | — | — | — | — | |
| G a | 265 (91.3) | 163 (84.0) | 1.0 (Ref.) | |||
| C a | 25 (8.7) | 31 (16.0) | 2.02 (1.149–3.536) | .014 | ||
| HWE | 0.524 | 0.173 | ||||
| IL6 −597 G > A | ||||||
| AA | 84 (57.9) | 61 (62.9) | 1.0 (Ref.) | 1.0 (Ref.) | ||
| AG | 59 (40.7) | 34 (35.1) | 0.79 (0.465–1.356) | .397 | 0.86 (0.47–1.57) | .626 |
| GG | 2 (1.4) | 2 (2.0) | 1.37 (0.189–10.04) | .752 | 1.17 (0.11–11.99) | .889 |
| AG/GG | 61 (42.1) | 36 (37.1) | 0.81 (0.479–1.37) | .441 | 0.87 (0.48–1.57) | .651 |
| A a | 227 (78.2) | 156 (81.2) | 1.0 (Ref.) | |||
| G a | 63 (21.8) | 36 (18.8) | 0.81 (0.47–1.37) | .441 | ||
| HWE | 0.060 | 0.540 | ||||
p < .025 (0.05/2) was significant after Bonferroni's correction.
Allele.
Adjusted for age, gender, diabetes and hypertension.
CI = confidence interval; OR = odds ratio; HWE = Hardy Weinberg equilibrium.
Haplotype analysis of two loci of IL6 gene polymorphisms (rs1800795 and rs1800797) was performed and the linkage disequilibrium between the above two loci are shown in Figure 2 (D: 0.66; R 2: 0.22). The degree of LD among the two SNPs was relatively moderate. The frequency of AC* haplotype was higher in severe patients as compared to mild patients and showed significantly 2.83‐fold higher risk of severity (p = .034) (Figure 2) but the frequency of GG* haplotype was higher in mild patients as compared to severe patients. So, GG* haplotype showed a significant protection against severity of COVID‐19 (p = .0393). In addition, the genotypic frequencies at rs1800795 (−174G > C) of IL6 polymorphism among mild and severe cases were not deviated from the Hardy–Weinberg equilibrium [Pearson's χ2 = 1.290, p = .524 (mild); Pearson's χ2 = 3.508, p = .173 (severe)]. Similarly, the genotypic frequencies at rs1800797 (−597G > A) of IL6 polymorphism was not deviated from the Hardy–Weinberg equilibrium [Pearson's χ2 = 5.594, p = .060 (mild); Pearson's χ2 = 1.231, p = .540 (severe)].
FIGURE 2.
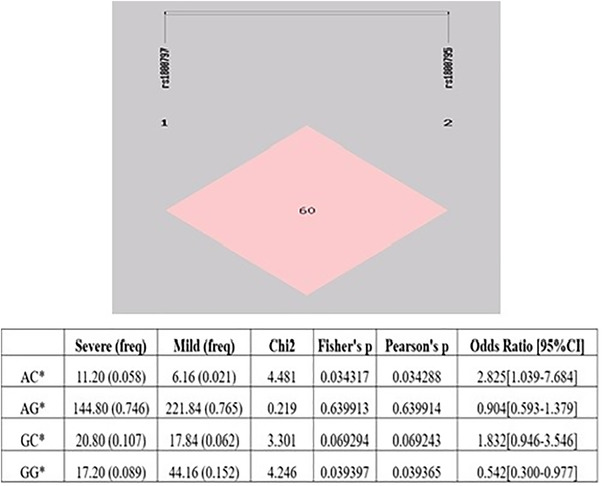
Haplotype analysis of SNPs, namely, of IL6 −597G > A (rs1800797) and IL6 −174G > C (rs1800795) for association with COVID‐19 cases. Linkage disequilibrium (LD) in subjects is represented as pink square for LD (SHEsis Software, ver.21 Online)
4. DISCUSSION
Genetic variations determine the divergent susceptibility to infectious illnesses in different individuals. Several studies have demonstrated that IL6 polymorphisms were associated to the consequence of viral infections through influencing IL6 protein synthesis (Velazquez‐Salinas et al., 2019; Cussigh et al., 2011). SARS‐CoV‐2 viral infection causes COVID‐19 that reflects a variety of severity levels in different individuals such as asymptomatic, moderate, severe and life‐threatening lower respiratory tract infections, including the development of ALI and ARDS (Gralinski & Baric, 2015). According to several studies, increased serum IL6 levels are a key characteristic of severe COVID‐19 and serve as a biomarker for predicting the disease's severity (Liu et al., 2020; Zhu et al., 2020). These clinical data support the hypothesis that that IL6 genotyping determine the degree of susceptibility to SARS‐CoV‐2 infection and severity of COVID‐19 disease in different individuals. Thus, the present study revealed a novel association of between genetic polymorphism at rs1800795 (−174G > C), and rs1800797 (−597G > A) of IL6 gene promoter and the severity of susceptibility to COVID‐19 in North Indian population.
In our study, the haplotype analysis of these two polymorphisms (−597G > A and −174G > C) of IL6 gene suggested that individuals with AC* haplotype showed ∼2‐fold higher risk of COVID‐19 severity (p = .034) but the GG* haplotype protected the patients from the severity (p = .039). Further, our study revealed that C allele and GC genotype of −174G > C (rs1800795) polymorphism of IL6 gene was higher in severe patients as compared to mild patients of COVID‐19 and showed significantly higher risk of severity (p = .009; .014 respectively). Additionally, a Chinese study reported that the CC genotype of −174 G > C (rs1800795) polymorphism showed higher risk pneumonia‐induced sepsis (Mao et al., 2017; Yeh et al., 2010). A meta‐analysis also showed that the C allele of the −174G > C (rs1800795) polymorphism is linked to higher IL6 production and more severe pneumonia (Ulhaq & Soraya, 2020). In contrast, Falahi et al., showed that the frequency of G allele and GG genotype of −174G > C (rs1800795) polymorphism is higher in mild patients as compared to severe (Falahi et al., 2022). Velez et al. (2008) found that the IL6 polymorphism at three regions (rs1800797, rs1800796 and rs1800795) was significantly associated with IL6 amniotic fluid concentration in preterm birth candidates. A recent study in Iranian population has demonstrated −597G > A (rs1800797) polymorphism but they did not find any association with the severity of COVID‐19 (Falahi et al., 2022). Our result also supported to this study, we did not found association of −597G > A (rs1800797) and severity of COVID‐19 (p > .05).
Epidemiological and polymorphism studies observed differential severity among the patients of several lung diseases such as asthma, chronic obstructive pulmonary disease (COPD) etc. (Jin & Wang, 2003; Kirtipal et, al., 2020). Genetic polymorphisms in promoter region of IL6 gene are associated with the risk of pneumonia (Chen et al., 2015; Martinez et al., 2013; Solé‐Violán et al., 2010). Two most common genetic polymorphisms in promoter region of IL6 gene are −597G > A (rs1800797) and −174G > C (rs1800795), that affects the secretion of IL6 cytokine in patients. SARS‐CoV‐2 infection triggers inflammatory response that results in the huge discharge of pro‐inflammatory cytokines and that causes cytokine storm. Several studies analysed the cytokine profiles of COVID‐19 patients and found that the cytokine storm is linked to a poor prognosis in severe COVID‐19 cases (Ragab et al., 2020). Innate and adaptive immunity is activated by viral infections and this infection leads to the production of several cytokines, including IL6. IL6 is a pleotropic cytokine that involved in complex cellular response such as proliferation, differentiation, survival and apoptosis (Scheller et al., 2011). It is produced from many types of cells such as fibroblasts, keratinocytes, macrophages, mast cells, monocytes, DCs and T and B lymphocytes (Mauer et al., 2015; Tanaka et al., 2014). Disturbance in Immune response and cytokine storm formation is due to IL6 and other cytokines, this disturbance causes severe complications and mortality in COVID‐19 patients. Differential disease progression and treatment response of infectious diseases is due to presence of genetic variations. Polymorphisms of IL6 gene were related to the prognosis of viral illness by disturbing IL6 protein synthesis (Chen. et al., 2015) A study on IL6 knockout mouse model showed that IL6 is required for antiviral therapy and T‐ and B‐cell responses against influenza virus (Velazquez‐Salinas et al., 2019). SNPs in the regulatory regions are the cause of variability in cytokine production among individuals (Haukim et, al., 2002). In our study, we also observed that severity of COVID‐19 significantly higher in older cases as compared to younger patients and the presence of other comorbid conditions such as diabetes and hypertension also increased the severity of COVID‐19 (p < .001).
The limited sample size (n = 242) of the current study, which reduced its statistical power, is one of its main limitations. We did also not include the healthy control individuals in our study as Saxena et al. reported the frequency of recessive alleles, ‘G’ at −597A > G and ‘C’ allele at −174G > C of IL6 polymorphisms, were 12.86% and 20.35%, respectively in healthy control population from North India. The frequency of GC* haplotype of IL6 polymorphisms was 4.6% in control group (Saxena et al., 2014). The association between IL6 genetic polymorphisms and disease severity in North Indian COVID‐19 patients has to be further investigated with a bigger sample size in order to provide definitive proof.
5. CONCLUSION
Combining data on the immunogenetic effects of IL6 genetic polymorphism previously reported in lung diseases and viral diseases, we also recommended taking IL6 polymorphism into consideration as a major factor to comprehend the degree of susceptibility to SARS‐CoV‐2 viral infection and severity of COVID‐19 illness in North Indian population. Based on IL6 polymorphism, the treatment response to COVID‐19 in infected human populations could be further investigated to build a population based therapeutic approach for the development of personalized medicine.
AUTHOR CONTRIBUTIONS
The study was conceived and conceptualized by Farzana Mahdi, Mohammad Abbas and Sushma Verma. Shrikant Verma, Mohammad Abbas and Sushma Verma conducted the technical feasibility. The experiments were performed by Shrikant Verma. The data were evaluated by Mohammad Abbas, Zeba Siddiqi and Syed T. Raza, while the study was written by Mohammad Abbas, Faizan H. Khan and Shrikant Verma.
CONFLICT OF INTEREST
The authors declare that they have no competing interest.
ACKNOWLEDGEMENT
We would like to thank Eras Lucknow Medical College and Hospital, Era University for providing the lab space and intramural funding support (ELMC&H/ RCell/EC/2020/272) for this research.
Verma, S. , Verma, S. , Khan, F. H. , Siddiqi, Z. , Raza, S. T. , Abbas, M. , & Mahdi, F. (2023). Genetic polymorphisms of IL6 gene –174G > C and –597G > A are associated with the risk of COVID‐19 severity. International Journal of Immunogenetics, 50, 5–11. 10.1111/iji.12605
[Correction added on 19 November 2022, after first online publication: one of the author affiliations was removed.]
REFERENCES
- Abbasifard, M. , & Khorramdelazad, H. (2020). The bio‐mission of interleukin‐6 in the pathogenesis of COVID‐19: A brief look at potential therapeutic tactics. Life sciences, 257, 118097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas, M. , Verma, S. , Verma, S. , Siddiqui, S. , Khan, F. H. , Raza, S. T. , & Mahdi, F. (2021). Association of GSTM1 and GSTT1 gene polymorphisms with COVID‐19 susceptibility and its outcome. Journal of Medical Virology, 93(9), 5446–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar, S. , Sinha, A. , Banach, M. , Mittoo, S. , Weissert, R. , Kass, J. S. , & Kutty, S. (2020). Cytokine storm in COVID‐19 immunopathological mechanisms, clinical considerations, and therapeutic approaches: The REPROGRAM consortium position paper. Frontiers in Immunology, 11, 1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Li, N. , Wan, H. , Cheng, Q. , Shi, G. , & Feng, Y. (2015). Associations of three well‐characterized polymorphisms in the IL‐6 and IL‐10 genes with pneumonia: a meta‐analysis. Scientific Reports, 5(1), 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Zhao, B. , Qu, Y. , Chen, Y. , Xiong, J. , Feng, Y. , & Li, F. (2020). Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clinical Infectious Diseases, 71(8), 1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela‐Ruiz, V. J. , Illescas‐Montes, R. , Puerta‐Puerta, J. M. , Ruiz, C. , & Melguizo‐Rodríguez, L. (2020). SARS‐CoV‐2 infection: The role of cytokines in COVID‐19 disease. Cytokine & growth Factor Reviews, 54, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cussigh, A. , Falleti, E. , Fabris, C. , Bitetto, D. , Cmet, S. , Fontanini, E. , Bignulin, S. , Fornasiere, E. , Fumolo, E. , Minisini, R. , Pirisi, M. , & Toniutto, P. (2011). Interleukin 6 promoter polymorphisms influence the outcome of chronic hepatitis C. Immunogenetics, 63(1), 33–41. [DOI] [PubMed] [Google Scholar]
- Falahi, S. , Zamanian, M. H. , Feizollahi, P. , Rezaiemanesh, A. , Salari, F. , Mahmoudi, Z. , & Karaji, A. G. (2022). Evaluation of the relationship between IL‐6 gene single nucleotide polymorphisms and the severity of COVID‐19 in an Iranian population. Cytokine, 154, 155889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, B. , Huang, L. , He, Y. , Xie, W. , Yin, Y. , Shi, Y. , Xiao, J. , Zhong, L. , Zhang, Y. , Jiang, Z. , Hao, F. , Zhou, Y. , Li, H. , Jiang, L. , Yang, X. , Song, X. , Kang, Y. , Tuo, L. , Huang, Y. , … Yang, Z. (2022). A genetic variant in IL‐6 lowering its expression is protective for critical patients with COVID‐19. Signal Transduction and Targeted Therapy, 7(1), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, J. , Dong, H. , Xia, Q.‐S. , Huang, Z. , Wang, D. , Zhao, Y. , Liu, W. , Tu, S. , Zhang, M. , Wang, Q. , & Lu, F. (2020). Correlation analysis between disease severity and inflammation‐related parameters in patients with COVID‐19: A retrospective study. BMC Infectious Diseases, 20(1), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski, L. E. , & Baric, R. S. (2015). Molecular pathology of emerging coronavirus infections. The Journal of Pathology, 235(2), 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haukim, N. , Bidwell, J. L. , Smith, A. J. P. , Keen, L. J. , Gallagher, G. , Kimberly, R. , & D'Alfonso, S. (2002). Cytokine gene polymorphism in human disease: On‐line databases, supplement 2. Genes & Immunity, 3(6), 313–330. [DOI] [PubMed] [Google Scholar]
- He, J. Q. , Foreman, M. G. , Shumansky, K. , Zhang, X. , Akhabir, L. , Sin, D. D. , & Sandford, A. J. (2009). Associations of IL6 polymorphisms with lung function decline and COPD. Thorax, 64(8), 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez‐Sousa, M. A. , Medrano, L. M. , Liu, P. , Fernandez‐Rodriguez, A. , Almansa, R. , Gomez‐Sanchez, E. , & Resino, S. (2017). IL‐6 rs1800795 polymorphism is associated with septic shock‐related death in patients who underwent major surgery: A preliminary retrospective study. Annals of Intensive Care, 7(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, W. , Fei, G. H. , Hu, J. G. , & Hu, X. W. (2015). A study on the effect of IL‐6 gene polymorphism on the prognosis of non‐small‐cell lung cancer. OncoTargets and Therapy, 8, 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, P. , & Wang, E. (2003). Polymorphism in clinical immunology–from HLA typing to immunogenetic profiling. Journal of Translational Medicine, 1(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirtipal, N. , Thakur, H. , Sobti, R. C. , & Janmeja, A. K. (2020). Association between IL6 gene polymorphism and the risk of chronic obstructive pulmonary disease in the north Indian population. Molecular Biology Research Communications, 9(2), 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Wang, H. , Shi, S. , & Xiao, J. (2021). Association between IL‐6 and severe disease and mortality in COVID‐19 disease: a systematic review and meta‐analysis. Postgraduate Medical Journal, 98(1165), 871–879. doi: 10.1136/postgradmedj-2021-139939 [DOI] [PubMed] [Google Scholar]
- Liu, T. , Zhang, J. , Yang, Y. , Ma, H. , Li, Z. , Zhang, J. , & Yi, J. (2020). The potential role of IL‐6 in monitoring severe case of coronavirus disease 2019. MedRxiv. [DOI] [PMC free article] [PubMed]
- Mao, Z. R. , Zhang, S. L. , & Feng, B. (2017). Association of IL‐10 (‐819T/C,‐592A/C and‐1082A/G) and IL‐6‐174G/C gene polymorphism and the risk of pneumonia‐induced sepsis. Biomarkers, 22(2), 106–112. [DOI] [PubMed] [Google Scholar]
- Mauer, J. , Denson, J. L. , & Brüning, J. C. (2015). Versatile functions for IL‐6 in metabolism and cancer. Trends in Immunology, 36(2), 92–101. [DOI] [PubMed] [Google Scholar]
- Martinez‐Ocaña, J. , Olivo‐Diaz, A. , Salazar‐Dominguez, T. , Reyes‐Gordillo, J. , Tapia‐Aquino, C. , Martínez‐Hernández, F. , Manjarrez, M. E. , Antonio‐Martinez, M. , Contreras‐Molina, A. , Figueroa‐Moreno, R. , Valdez‐Vazquez, R. , Kawa‐Karasik, S. , Rodríguez‐Zulueta, P. , Flisser, A. , Maravilla, P. , & Romero‐Valdovinos, M. (2013). Plasma cytokine levels and cytokine gene polymorphisms in Mexican patients during the influenza pandemic A (H1N1) pdm09. Journal of Clinical Virology, 58(1), 108–113. [DOI] [PubMed] [Google Scholar]
- Ragab, D. , Salah Eldin, H. , Taeimah, M. , Khattab, R. , & Salem, R. (2020). The COVID‐19 cytokine storm; what we know so far. Frontiers in Immunology, 11, 1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshanravan, N. , Seif, F. , Ostadrahimi, A. , Pouraghaei, M. , & Ghaffari, S. (2020). Targeting cytokine storm to manage patients with COVID‐19: A mini‐review. Archives of Medical Research, 51(7), 608–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, M. , Agrawal, C. G. , Srivastava, N. , & Banerjee, M. (2014). Interleukin‐6 (IL‐6)‐597 A/G (rs1800797) &‐174 G/C (rs1800795) gene polymorphisms in type 2 diabetes. The Indian Journal of Medical Research, 140(1), 60. [PMC free article] [PubMed] [Google Scholar]
- Saxena, M. , Ali, D. , Modi, D. R. , Al Ali, H. , Hussain, S. A. , & Manohrdas, S. (2020). Eleven genetic variants of seven important candidate genes involved in manifestation of type 2 diabetes mellitus. Journal of King Saud University‐Science, 32(5), 2569–2575. [Google Scholar]
- Scheller, J. , Chalaris, A. , Schmidt‐Arras, D. , & Rose‐John, S. (2011). The pro‐and anti‐inflammatory properties of the cytokine interleukin‐6. Biochimica et Biophysica Acta (BBA)‐Molecular Cell Research, 1813(5), 878–888. [DOI] [PubMed] [Google Scholar]
- Solé‐Violán, J. , de Castro, F. R. , García‐Laorden, M. I. , Blanquer, J. , Aspa, J. , Borderías, L. , & Rodriguez‐Gallego, C. (2010). Genetic variability in the severity and outcome of community‐acquired pneumonia. Respiratory Medicine, 104(3), 440–447. [DOI] [PubMed] [Google Scholar]
- Tan, L. , Wang, Q. , Zhang, D. , Ding, J. , Huang, Q. , Tang, Y. Q. , & Miao, H. (2020). Lymphopenia predicts disease severity of COVID‐19: A descriptive and predictive study. Signal Transduction and Targeted Therapy, 5(1), 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T. , Narazaki, M. , & Kishimoto, T. (2014). IL‐6 in inflammation, immunity, and disease. Cold Spring Harbor Perspectives in Biology, 6(10), a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulhaq, Z. S. , & Soraya, G. V. (2020). Anti‐IL‐6 receptor antibody treatment for severe COVID‐19 and the potential implication of IL‐6 gene polymorphisms in novel coronavirus pneumonia. Medicina Clinica, 155(12), 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez‐Salinas, L. , Verdugo‐Rodriguez, A. , Rodriguez, L. L. , & Borca, M. V. (2019). The role of interleukin 6 during viral infections. Frontiers in Microbiology, 10, 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez, D. R. , Fortunato, S. J. , Williams, S. M. , & Menon, R. (2008). Interleukin‐6 (IL‐6) and receptor (IL6‐R) gene haplotypes associate with amniotic fluid protein concentrations in preterm birth. Human Molecular Genetics, 17(11), 1619–1630. [DOI] [PubMed] [Google Scholar]
- Verma, S. , Abbas, M. , Verma, S. , Khan, F. H. , Raza, S. T. , Siddiqi, Z. , & Mahdi, F. (2021). Impact of I/D polymorphism of angiotensin‐converting enzyme 1 (ACE1) gene on the severity of COVID‐19 patients. Infection, Genetics and Evolution, 91, 104801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. , Yu, B. , Chen, Y.‐M. , Wang, W. , Song, Z.‐G. , Hu, Y. , Tao, Z.‐W. , Tian, J.‐H. , Pei, Y.‐Y. , Yuan, M.‐L. , Zhang, Y.‐L. , Dai, F.‐H. , Liu, Y. , Wang, Q.‐M. , Zheng, J.‐J. , Xu, L. , Holmes, E. C. , & Zhang, Y.‐Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, K. Y. , Li, Y. Y. , Hsieh, L. L. , Chen, J. R. , & Tang, R. P. (2010). The −174 G/C polymorphism in interleukin‐6 (IL‐6) promoter region is associated with serum IL‐6 and carcinoembryonic antigen levels in patients with colorectal cancers in Taiwan. Journal of Clinical Immunology, 30(1), 53–59. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Wu, Z. , Li, J. W , Zhao, H. , & Wang, G. Q. (2020). The cytokine release syndrome (CRS) of severe COVID‐19 and Interleukin‐6 receptor (IL‐6R) antagonist Tocilizumab may be the key to reduce the mortality. International Journal of Antimicrobial Agents, 55(5), 105954–105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. , Shen, X. R. , Wang, X. S. , Zheng, K. , Zhao, Q. J. , Chen, F. , Deng, L. L. , Liu, B. , Yan, F. X. , Zhan, Y. Y. , Wang, G. F. , Xiao, & Shi, Z. L. (2020). Regulation of inflammatory cytokines and inhibition of T and B lymphocytes X. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z. , Cai, T. , Fan, L. , Lou, K. , Hua, X. , Huang, Z. , & Ningbo, P. R. (2020). Clinical value of immune‐inflammatory parameters to assess the severity of immune‐inflammatory parameters to assess the severity of coronavirus disease 2019. International Journal of Infectious Diseases, 95, 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]


