Abstract
The majority of deep learning (DL) based deformable image registration methods use convolutional neural networks (CNNs) to estimate displacement fields from pairs of moving and fixed images. This, however, requires the convolutional kernels in the CNN to not only extract intensity features from the inputs but also understand image coordinate systems. We argue that the latter task is challenging for traditional CNNs, limiting their performance in registration tasks. To tackle this problem, we first introduce Coordinate Translator, a differentiable module that identifies matched features between the fixed and moving image and outputs their coordinate correspondences without the need for training. It unloads the burden of understanding image coordinate systems for CNNs, allowing them to focus on feature extraction. We then propose a novel deformable registration network, im2grid, that uses multiple Coordinate Translator’s with the hierarchical features extracted from a CNN encoder and outputs a deformation field in a coarse-to-fine fashion. We compared im2grid with the state-of-the-art DL and non-DL methods for unsupervised 3D magnetic resonance image registration. Our experiments show that im2grid outperforms these methods both qualitatively and quantitatively.
Keywords: Deformable image registration, Deep learning, Magnetic resonance imaging, Template matching
1. Introduction
Deformable registration is of fundamental importance in medical image analysis. Given a pair of images, one fixed and one moving, deformable registration warps the moving image by optimizing the parameters of a nonlinear transformation so that the underlying anatomies of the two images are aligned according to an image dissimilarity function [11,16,32,34,37]. Recent deep learning (DL) methods use convolutional neural networks (CNNs) whose parameters are optimized during training; at test time, a dense displacement field that represents the deformable transformation is generated in a single forward pass.
Although CNN-based methods for segmentation and classification are better than traditional methods in both speed and accuracy, DL-based deformable registration methods are faster but usually not more accurate [4,8,13,15,39]. Using a CNN for registration requires learning coordinate correspondences between image pairs, which has been thought to be fundamentally different from other CNN applications because it involves both extracting and matching features [14,25]. However, the majority of existing works simply rely on CNNs to implicitly learn the displacement between the fixed and moving images [4,13,15].
Registration involves both feature extraction and feature matching, but to produce a displacement field, matched features need to be translated to coordinate correspondences. We argue that using convolutional kernels for the latter two tasks is not optimal. To tackle this problem, we introduce Coordinate Translator, a differentiable module that matches features between the fixed and moving images and identifies feature matches as precise coordinate correspondences without the need for training. The proposed registration network, named im2grid, uses multiple Coordinate Translator’s with multi-scale feature maps. These produce multi-scale sampling grids representing coordinate correspondences, which are then composed in a coarse-to-fine manner to warp the moving image. im2grid explicitly handles the task of matching features and establishing coordinate correspondence using Coordinate Translator’s, leaving only feature extraction to our CNN encoder.
Throughout this paper, we use unsupervised 3D magnetic resonance (MR) image registration as our example task and demonstrate that the proposed method outperforms the state-of-the-art methods in terms of registration accuracy. We think it is important to note that because producing a coordinate location is such a common task in both medical image analysis and computer vision, the proposed method can be impactful on a board range of applications.
2. Related Works
Traditional registration methods solve an optimization problem for every pair of fixed, If, and moving, Im, images. Let ϕ denote a transformation and let the best transformation be found from
| (1) |
where Im ○ ϕ yields the warped image Iw. The first term focuses on the similarity between If and Im ○ ϕ whereas the second term—weighted by the hyper-parameter λ—regularizes ϕ. The choice of Lsim is application-specific. Popular methods using this framework include spline-based free-form deformable models [32], elastic warping methods [11,27], biomechanical models [16], and Demons [34,37]. Alternatively, learning-based methods have also been used to estimate the transformation parameters [9,19].
Recently, deep learning (DL) methods, especially CNNs, have been used for solving deformable registration problems. In these methods, ϕ is typically represented as a map of displacement vectors that specify the voxel-level spatial offsets between If and Im; the CNN is trained to output ϕ with or without supervision [4,6,13,15,20]. In the unsupervised setting, the displacement field is converted to a sampling grid and the warped image is produced by using a grid sampler [26] with the moving image and the sampling grid as input. The grid sampler performs differentiable sampling of an image (or a multi-channel feature map) using a sampling grid; it allows the dissimilarity loss computed between the warped and fixed images to be back-propagated so the CNN can be trained end-to-end. In past work, [4] used a U-shaped network to output the dense displacement; [12,13] used an encoder network to produce a sparse map of control points and generated the dense displacement field by interpolation; and [8] replaced the bottleneck of a U-Net [31] with a transformer structure [36]. Several deep learning methods also demonstrate the possibility of using a velocity-based transformation representation to enforce a diffeomorphism [10,39].
Our method represents the transformation using a sampling grid G, which can be directly used by the grid sampler. For N-dimensional images (N = 3 in this paper), G is represented by an N-channel map. Specifically, for a voxel coordinate (where contains all the voxel coordinates in If), G(x) should ideally hold a coordinate such that the two values If(x) and Im(G(x)) represent the same anatomy. Note that the displacement field representation commonly used by other methods can be found as G − GI, where GI is the identity grid GI(x) = x.
3. Method
For the image pair If and Im, the proposed method produces a sampling grid G0 that can be used by the grid sampler to warp Im to match If. Similar to previous DL methods, we use a CNN encoder to extract multi-level feature maps from If and Im. Instead of directly producing a single displacement field from the CNN, G0 is the composition of multi-level sampling grids, generated from the multi-level feature maps with the proposed Coordinate Translator’s.
3.1. Coordinate Translator
Let F and M denote the multi-channel feature maps that are individually extracted from If and Im, respectively. The goal of a Coordinate Translator is to take as input both F and M, and produce a sampling grid G that aligns M interpolated at coordinate G(x) with F(x) for all .
As the first step, for every x, cross-correlation is calculated between F(x) and M(ci) along the feature dimension, where for i ∈ [1,K] are a set of candidate coordinates. The results are a K-element vector of matching scores between F(x) and every M(ci): >
| (2) |
The choice of ci’s determines the search region for the match. For example, defining ci to be every coordinates in will compare F(x) against every location in M; these matches can also be restricted within the 3 × 3 × 3 neighborhood of x. We outline our choices of ci’s in Sect. 4. The matching scores are normalized using a softmax function to produce a matching probability pi,
| (3) |
We interpret the matching probabilities as the strength of attraction between F(x) and the M(ci)’s. Importantly, we can calculate a weighted sum of ci’s to produce a coordinate , i.e., , which represents the correspondence of x in the moving image Im. This is conceptually similar to the combined force in the Demons algorithm [34]. For every the corresponding x′ forms the Coordinate Translator output, G.
Coordinate Translator can be efficiently implemented as the Scaled Dot-Product Attention introduced in the Transformer [36] using matrix operations. For 3D images with spatial dimension H × W × S and C feature channels, we reshape F and M to and the identity grid GI to . Thus Coordinate Translator with can be readily computed from,
| (4) |
with the softmax operating on the rows of FMT.
Positional Encoding Layer.
In learning transformations, it is a common practice to initialize from (or close to) an identity transformation [4,8,26]. As shown in Fig. 1, we propose a positional encoding layer that combines position information with F and M such that the initial output of Coordinate Translator is an identity grid. Inside a positional encoding layer, for every x = (x1,… ,xN) with xi’s on an integer grid (xi ∈ {0,…,di−1}), we add a positional embedding (PE),
to the input feature map, where di is the pixel dimension along the ith axis. Trigonometric identities give the cross-correlation of PEs at x1 and x2 as
where Δxi is the difference in the i th components of x1 and x2. This has maximum value when x1 = x2 and decreases with the L1 distance between the two coordinates. We initialize the convolutional layer to have zero weights and bias and the learnable parameter α = 1 (see Fig. 1) such that only the PEs are considered by Coordinate Translator at the beginning of training. As a result, among all , M(x) will have the highest matching score with F(x), thus producing GI as the initial output. Coordinate Translator also benefits from incorporating the position information as it allows the relative distance between ci and x to contribute to the matching scores, similar as the positional embedding in the Transformer [36].
Fig. 1.
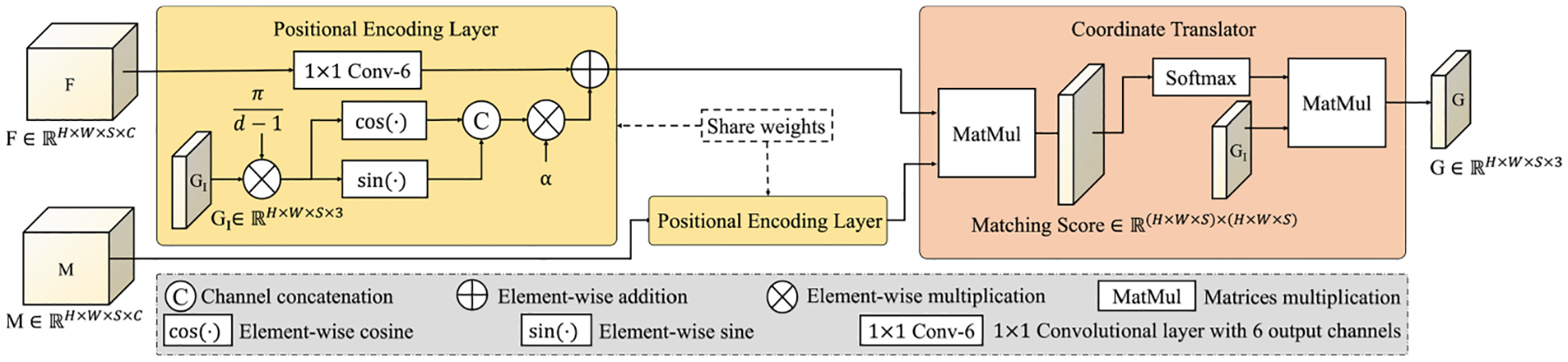
Structure of the proposed positional encoding layer and Coordinate Translator.
3.2. im2grid Network Architecture
The proposed im2grid network is shown in Fig. 2. Similar to previous methods, im2grid produces a sampling grid to warp Im to Iw. Our CNN encoder uses multiple pooling layers to extract hierarchical features from the intensity images. In the context of intra-modal registration, it is used as a Siamese network that processes If and Im separately. For clarity, Fig. 2 only shows a three level im2grid model with three level feature maps F1/F2/F3 and M1/M2/M3 for If and Im, respectively. In our experiment, we used a five level structure. Our grid decoder uses the common coarse-to-fine strategy in registration. Firstly, coarse features F3 and M3 are matched and translated to a coarse sampling grid G3 using a Coordinate Translator. Because of the pooling layers, this can be interpreted as matching downsampled versions of If and Im, producing a coarse displacement field. G3 is then used to warp M2, resolving the coarse deformation between M2 and F2 so that the Coordinate Translator at the second level can capture more detailed displacements with a smaller search region. Similarly, M1 is warped by the composed transformation of G3 and G2 and finally the moving image is warped by the composition of the transformations from all levels. A visualization of a five-level version of our multi-scale sampling grids is provided in Fig. 3. In contrast to previous methods that use CNNs to directly output displacements, our CNN encoder only needs to extract similar features for corresponding anatomies in If and Im and the exact coordinate correspondences are obtained by Coordinate Translator’s. Because our CNN encoder processes If and Im separately, it is guaranteed that our CNN encoder only performs feature extraction.
Fig. 2.
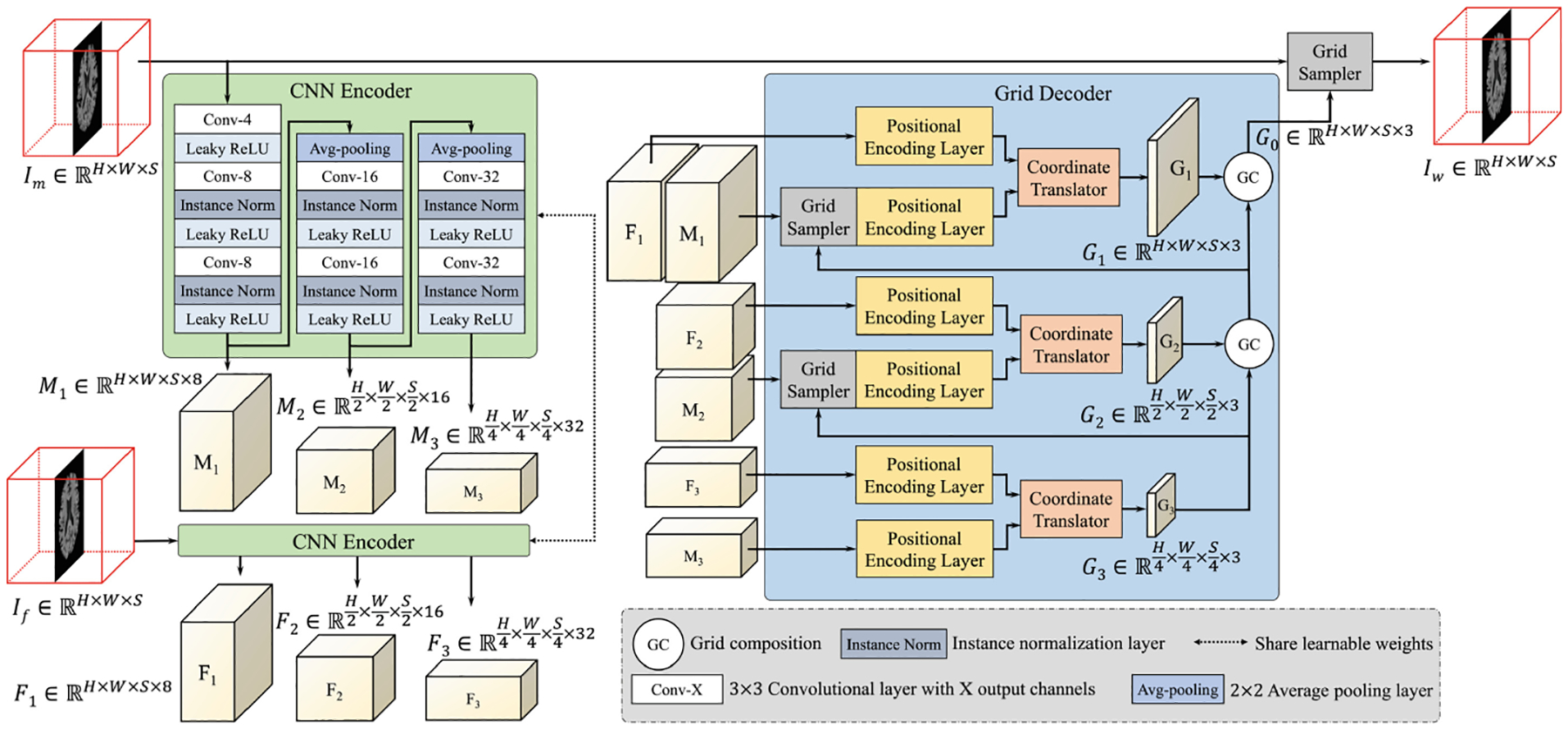
Example of the proposed im2grid network structure with a 3-level CNN encoder. The grid composition operation can be implemented using the grid sampler with two grids as input.
Fig. 3.
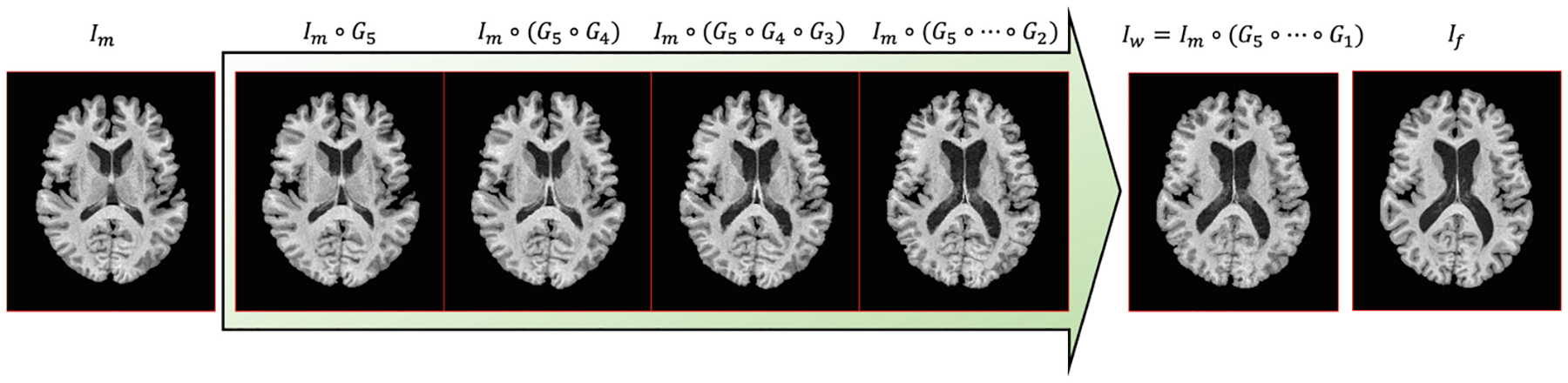
Visualization of the multi-scale sampling grids by sequentially applying finer grids to the moving image. Here we used a five-level CNN encoder and G5,…,G1 are coarse to fine sampling grids produced from the five-level feature maps.
The proposed network is trained using the mean squared difference between If and Iw(= Im ○ϕ) and a smoothness loss that regularizes the spatial variations of the G’s at every level,
| (5) |
where is the cardinality of and all Gi’s and GI are normalized to [−1, 1].
4. Experiments
Datasets.
We used the publicly available OASIS3 [28] and IXI [1] datasets in our experiments. 200, 40, and 100 T1-weighted (T1w) MR images of the human brain from the OASIS3 dataset were used for training, validation, and testing, respectively. During training, two scans were randomly selected as If and Im, while validation and testing used 20 and 50 pre-assigned image pairs, respectively. For the IXI dataset, we used 200 scans for training, 20 and 40 pairs for validation and testing, respectively. All scans underwent N4 inhomogeneity correction [35], and were rigidly registered to MNI space [18] with 1 mm3 (for IXI) or 0.8 mm3 (for OASIS3) isotropic resolution. A white matter peak normalization [30] was applied to standardize the MR intensity scale.
Evaluation Metrics.
First, we calculated the Dice similarity coefficient (DSC) between segmentation labels of If and the warped labels of Im. An accurate transformation should align the structures of the fixed and moving images and produces a high DSC. We obtained a whole brain segmentation for the fixed and moving images using SLANT [24] and combined the SLANT labels (133 labels) to TOADS labels (9 labels) [5]. The warped labels were produced by applying each methods deformation field to the moving image labels. Second, we measured the regularity of the transformations by computing the determinant of the Jacobian matrix, which should be globally positive for a diffeomorphic transformation.
Implementation Details.
Our method was implemented using PyTorch and trained using the Adam optimizer with a learning rate of 3 ×10−4, a weight decay of 1 × 10−9, and a batch size of 1. Random flipping of the input volumes along the three axes were used as data augmentation. We used a five-level structure and tested different choices of ci’s for each Coordinate Translator. We found that given the hierarchical structure, a small search region at each level is sufficient to capture displacements presented in our data. Therefore, we implemented two versions of our method: 1) im2grid which used a 3 × 3 search window in the axial plane for producing G1 and a 3 × 3 ×3 search window at other levels; and 2) im2grid-Lite which is identical to im2grid except that the finest grid G1 is not used.
Baseline Methods:
We compared our method with several state-of-the-art DL and non-DL registration methods: 1) SyN: Symmetric image normalization method [2], implemented in the Advanced Normalization Tools (ANTs) [3]; 2) voxelmorph: A deep learning based unsupervised method trained with the mean squared error loss [4]; 3) ViT-V-Net: A transformer [36] based network structure proposed in [8].
For SyN, a wide range of hyper-parameters were tested on the OASIS3 validation set and the best performing parameters were used for generating the final results. For voxelmorph and ViT-V-Net, we adopted the same training strategies as the proposed method, including the loss function and data augmentation. We optimize the parameters of each method for performance on the OASIS3 validation set and then used those parameters in testing on both datasets.
Results.
For both OASIS3 and IXI test datasets, we registered the moving to the fixed image and report the averaged DSC for all labels in Table 1. In both datasets, the proposed methods outperform the comparison methods for DSC. For each individual anatomic label, we also conducted a paired, two-sided Wilcoxon signed rank test (null hypothesis: the difference between paired values comes from a distribution with zero median, α = 10−3) between our methods and the comparison methods. Both proposed methods show significant DSC improvements for seven of nine labels and comparable DSC performance to the best comparison method for the remaining two labels (thalamus and putamen). Visual examples on OASIS3 data are shown in Fig. 4. It can be seen, especially from the highlighted regions, that the warped image produced by the proposed methods have a better agreement with the fixed image.
Table 1.
The Dice coefficient (DSC), the average number of voxels with negative determinant of Jacobian (# of |Jϕ| < 0) and the percentage of voxels with negative determinant of Jacobian (%) for affine transformation, SyN, Voxelmorph, ViT-V-Net, and the proposed methods. The results of the initial alignment by the preprocessing steps are also included. Bold numbers indicate the best DSC for each dataset.
| OASIS3 | IXI | |||||
|---|---|---|---|---|---|---|
| DSC | # of |Jϕ| < 0 | % | DSC | # of |Jϕ| < 0 | % | |
| Initial | 0.651 ± 0.094 | – | 0% | 0.668 ± 0.107 | – | 0% |
| Affine | 0.725 ± 0.068 | – | 0% | 0.748 ± 0.052 | – | 0% |
| SyN [2] | 0.866 ± 0.029 | 223 | <0.002% | 0.845 ± 0.035 | 613 | 0.008% |
| Voxelmorph [4] | 0.883 ± 0.040 | 85892 | <0.7% | 0.842 ± 0.068 | 21574 | <0.3% |
| ViT-V-Net [8] | 0.872 ± 0.042 | 110128 | <0.9% | 0.845 ± 0.068 | 21298 | <0.2% |
| im2grid-Lite | 0.909 ± 0.021 | 38915 | <0.4% | 0.870 ± 0.043 | 14917 | <0.2% |
| im2grid | 0.908 ± 0.023 | 11880 | <0.1% | 0.865 ± 0.050 | 3235 | <0.04% |
Fig. 4.
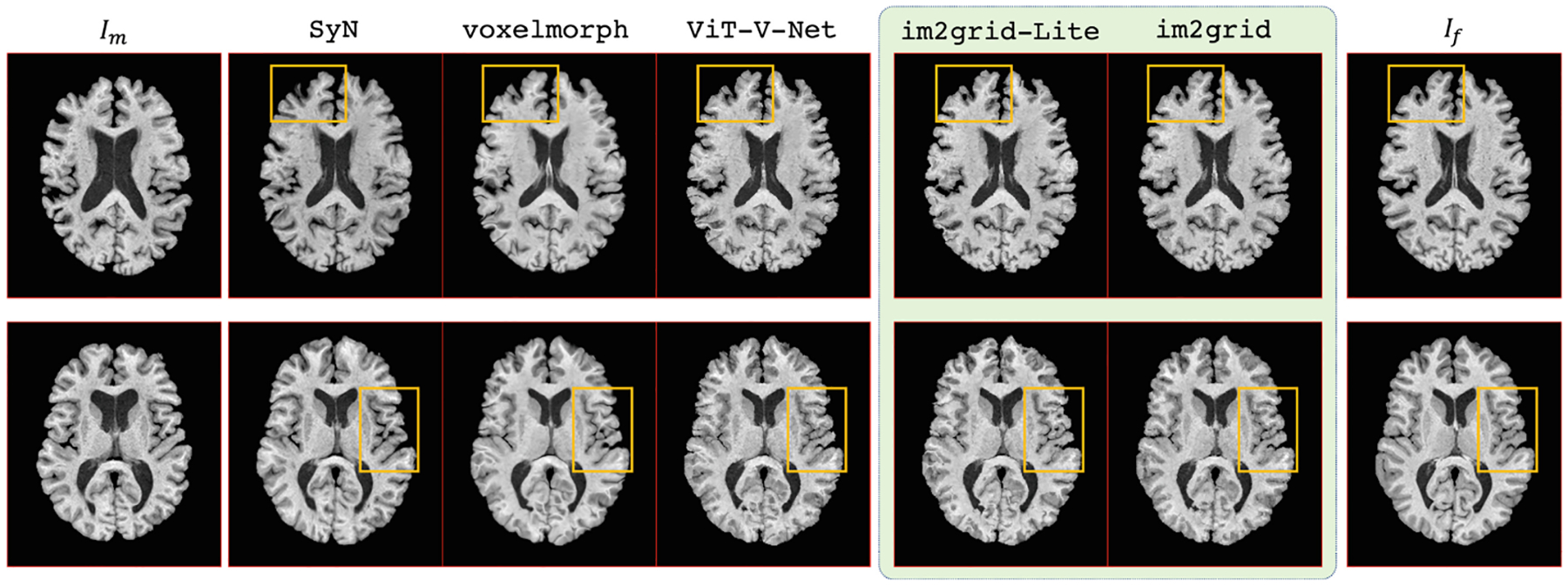
Examples of registering the moving image (the first column) to the fixed image (the last column) using SyN, voxelmorph, ViT-V-Net, and our proposed methods.
Evaluation on Learn2Reg Validation Dataset.
We also test the proposed method on the inter-subject brain MRI registration task from the Learn2Reg challenge [22] (L2R 2021 Task 3). All scans from the challenge have been pre-processed following [23], and for evaluation purpose segmentation maps of 35 labels were generated using FreeSurfer [17]. We choose the im2grid-Lite version for this task because the challenge evaluation is done on the ×2 downsampled images. During training, two scans were randomly selected from the training set and used as input to the proposed method. The performance is evaluated by comparing the warped segmentation of the moving image and the segmentation of the fixed image. The results are summarized in Table 2, where the DSC represents the average Dice coefficient of all segmented labels; DSC30 is the lowest 30% DSC among all cases, which measures the robustness of the methods; SDlogJ is the standard deviation of the log of the Jacobian determinant of the deformation field; and HD95 represents the 95% percentile of Hausdorff distance of segmentations. The results of several state-of-the-art methods from the challenge leaderboard are also included. The proposed method shows better accuracy as well as robustness among the comparison methods. Although adopting the instance-specific optimization as described in [4] can potential boost the performance on the validation set, our method only used the training set because we assume that such fine tuning process is not available during deployment.
Table 2.
Results of the proposed method and several state-of-the-art methods on the Learn2Reg 2021 Task 3 validation dataset.
5. Discussion
In this paper, we proposed Coordinate Translator for producing coordinate correspondences from two feature maps. Additionally, we proposed the im2grid network that uses Coordinate Translator’s for deformable image registration. For unsupervised 3D magnetic resonance registration, im2grid outperforms the state-of-the-art methods in accuracy with a similar training and testing speed as other deep learning based registration methods. Although im2grid has no explicit guarantee of being diffeomorphic, the deformation fields it generated contains fewer voxels with negative determinant of Jacobian compared with other deep learning methods that output deformation fields directly from feature maps. We believe this comes from our design decision to restrict the candidate voxels to the immediate neighborhood of a voxel, which yields a locally smooth deformation field at each scale. We note that even a diffeomorphic algorithm with theoretical guarantees (e.g., SyN) can produce non-diffeomorphic transformations because of errors introduced during interpolation [38].
For registration, we demonstrated that using Coordinate Translator for matching features and establishing coordinate correspondences together with the convolutional networks for feature extraction can significantly boost the performance. Coordinate Translator is a general module that can be incorporated in many existing network structures and therefore is not limited to the registration task. We believe that many tasks that involve image input and coordinate output can benefit from the use of the Coordinate Translator module.
Acknowledgement.
This work was supported in part by the NIH/NEI grant R01-EY032284 and the Intramural Research Program of the NIH, National Institute on Aging.
References
- 1.IXI Brain Development Dataset. https://brain-development.org/ixi-dataset/
- 2.Avants BB, Epstein CL, Grossman M, Gee JC: Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal 12(1), 26–41 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avants BB, Tustison N, Song G, et al. : Advanced normalization tools (ANTS). Insight J. 2(365), 1–35 (2009) [Google Scholar]
- 4.Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV: VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38(8), 1788–1800 (2019) [DOI] [PubMed] [Google Scholar]
- 5.Bazin PL, Pham DL: Topology-preserving tissue classification of magnetic resonance brain images. IEEE Trans. Med. Imaging 26(4), 487–496 (2007) [DOI] [PubMed] [Google Scholar]
- 6.Cao X, et al. : Deformable image registration based on similarity-steered CNN regression. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 300–308. Springer, Cham: (2017). 10.1007/978-3-319-66182-7_35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Frey EC, He Y, Segars WP, Li Y, Du Y: TransMorph: transformerfor unsupervised medical image registration. arXiv preprint arXiv:2111.10480 (2021) [Google Scholar]
- 8.Chen J, He Y, Frey EC, Li Y, Du Y: ViT-V-Net: vision transformer for unsupervised volumetric medical image registration. arXiv preprint arXiv:2104.06468 (2021) [Google Scholar]
- 9.Chou CR, Frederick B, Mageras G, Chang S, Pizer S: 2D/3D image registration using regression learning. Comput. Vis. Image Underst 117(9), 1095–1106 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalca AV, Balakrishnan G, Guttag J, Sabuncu MR: Unsupervised learning for fast probabilistic diffeomorphic registration. In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G (eds.) MICCAI 2018. LNCS, vol. 11070, pp. 729–738. Springer, Cham: (2018). 10.1007/978-3-030-00928-1_82 [DOI] [Google Scholar]
- 11.Davatzikos C: Spatial transformation and registration of brain images using elastically deformable models. Comput. Vis. Image Underst 66(2), 207–222 (1997) [DOI] [PubMed] [Google Scholar]
- 12.De Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I: A deep learning framework for unsupervised affine and deformable image registration. Med. Image Anal 52, 128–143 (2019) [DOI] [PubMed] [Google Scholar]
- 13.de Vos BD, Berendsen FF, Viergever MA, Staring M, Išgum I: End-to-end unsupervised deformable image registration with a convolutional neural network. In: Cardoso MJ, et al. (eds.) DLMIA/ML-CDS −2017. LNCS, vol. 10553, pp. 204–212. Springer, Cham: (2017). 10.1007/978-3-319-67558-9_24 [DOI] [Google Scholar]
- 14.Dosovitskiy A, et al. : FlowNet: learning optical flow with convolutional networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2758–2766 (2015) [Google Scholar]
- 15.Fan J, Cao X, Yap PT, Shen D: BIRNet: brain image registration using dual-supervised fully convolutional networks. Med. Image Anal 54, 193–206 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrant M, Warfield SK, Nabavi A, Jolesz FA, Kikinis R: Registration of 3D intraoperative MR images of the brain using a finite element biomechanical model. In: Delp SL, DiGoia AM, Jaramaz B (eds.) MICCAI 2000. LNCS, vol. 1935, pp. 19–28. Springer, Heidelberg: (2000). 10.1007/978-3-540-40899-4_3 [DOI] [Google Scholar]
- 17.Fischl B: FreeSurfer. NeuroImage 62(2), 774–781 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonov V, Evans A, McKinstry R, Almli C, Collins D: Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage 47, S102 (2009) [Google Scholar]
- 19.Gutiérrez-Becker B, Mateus D, Peter L, Navab N: Learning optimization updates for multimodal registration. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds.) MICCAI 2016. LNCS, vol. 9902, pp. 19–27. Springer, Cham: (2016). 10.1007/978-3-319-46726-9_3 [DOI] [Google Scholar]
- 20.Han R, et al. : Deformable MR-CT image registration using an unsupervised end-to-end synthesis and registration network for endoscopic neurosurgery. In: Medical Imaging 2021, vol. 11598, p. 1159819. International Society for Optics and Photonics (2021) [Google Scholar]
- 21.Han R, et al. : Deformable MR-CT image registration using an unsupervised end-to-end synthesis and registration network for endoscopic neurosurgery. In: Medical Imaging 2021: Image-Guided Procedures, Robotic Interventions, and Modeling, vol. 11598, p. 1159819. International Society for Optics and Photonics (2021) [Google Scholar]
- 22.Hering A, et al. : Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning. arXiv preprint arXiv:2112.04489 (2021) [DOI] [PubMed] [Google Scholar]
- 23.Hoopes A, Hoffmann M, Fischl B, Guttag J, Dalca AV: HyperMorph: amortized hyperparameter learning for image registration. In: Feragen A, Sommer S, Schnabel J, Nielsen M (eds.) IPMI 2021. LNCS, vol. 12729, pp. 3–17. Springer, Cham: (2021). 10.1007/978-3-030-78191-0_1 [DOI] [Google Scholar]
- 24.Huo Y, et al. : 3D whole brain segmentation using spatially localized atlas network tiles. Neuroimage 194, 105–119 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilg E, et al. : FlowNet 2.0: evolution of optical flow estimation with deep networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2462–2470 (2017) [Google Scholar]
- 26.Jaderberg M, Simonyan K, Zisserman A, et al. : Spatial transformer networks. In: Advances in Neural Information Processing Systems, vol. 28 (2015) [Google Scholar]
- 27.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP: Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29(1), 196–205 (2009) [DOI] [PubMed] [Google Scholar]
- 28.LaMontagne PJ, et al. : OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. MedRxiv (2019) [Google Scholar]
- 29.Lv J, et al. : Joint progressive and coarse-to-fine registration of brain MRI via deformation field integration and non-rigid feature fusion. IEEE Trans. Med. Imaging (2022) [DOI] [PubMed] [Google Scholar]
- 30.Reinhold JC, et al. : Evaluating the impact of intensity normalization on MR image synthesis. In: Medical Imaging 2019: Image Processing, vol. 10949, p. 109493H. International Society for Optics and Photonics (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronneberger O, Fischer P, Brox T: U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham: (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 32.Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ: Non-rigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18(8), 712–721 (1999) [DOI] [PubMed] [Google Scholar]
- 33.Siebert H, Hansen L, Heinrich MP: Fast 3D registration with accurate optimisation and little learning for Learn2Reg 2021. In: Aubreville M, Zimmerer D, Heinrich M (eds.) MICCAI 2021. LNCS, vol. 13166, pp. 174–179. Springer, Cham: (2022). 10.1007/978-3-030-97281-3_25 [DOI] [Google Scholar]
- 34.Thirion JP: Image matching as a diffusion process: an analogy with Maxwell’s demons. Med. Image Anal 2(3), 243–260 (1998) [DOI] [PubMed] [Google Scholar]
- 35.Tustison NJ, et al. : N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29(6), 1310–1320 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaswani A, et al. : Attention is all you need. In: Advances in Neural Information Processing Systems, vol. 30 (2017) [Google Scholar]
- 37.Vercauteren T, Pennec X, Perchant A, Ayache N: Diffeomorphic demons: efficient non-parametric image registration. Neuroimage 45(1), S61–S72 (2009) [DOI] [PubMed] [Google Scholar]
- 38.Wyburd MK, Dinsdale NK, Namburete AIL, Jenkinson M: TEDS-Net: enforcing diffeomorphisms in spatial transformers to guarantee topology preservation in segmentations. In: de Bruijne M, et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 250–260. Springer, Cham: (2021). 10.1007/978-3-030-87193-2_24 [DOI] [Google Scholar]
- 39.Yang X, Kwitt R, Styner M, Niethammer M: Quicksilver: fast predictive image registration–a deep learning approach. Neuroimage 158, 378–396 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]


