Abstract
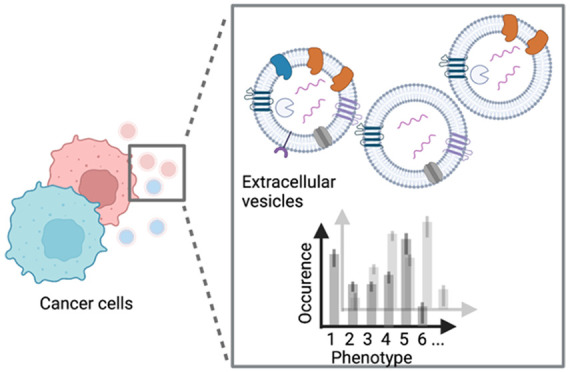
Extracellular vesicles (EVs) are considered as valuable biomarkers to discriminate healthy from diseased cells such as cancer. Passing cytosolic and plasma membranes before their release, EVs inherit the biochemical properties of the cell. Here, we determine protein profiles of single EVs to understand how much they represent their cell of origin. We use a microfluidic platform which allows to immobilize EVs from completely isolated single cells, reducing heterogeneity of EVs as strongly seen in cell populations. After immunostaining, we employ four-color total internal reflection fluorescence microscopy to enumerate EVs and determine their biochemical fingerprint encoded in membranous or cytosolic proteins. Analyzing single cells derived from pleural effusions of two different human adenocarcinoma as well as from human embryonic kidney (SkBr3, MCF-7 and HEK293, respectively), we observed that a single cell secretes enough EVs to extract the respective tissue fingerprint. We show that overexpressed integral plasma membrane proteins are also found in EV membranes, which together with populations of colocalized proteins, provide a cell-specific, characteristic pattern. Our method highlights the potential of EVs as a diagnostic marker and can be directly employed for fundamental studies of EV biogenesis.
Introduction
In 2020, breast cancer was the most often diagnosed cancer and caused most cancer-related cases of death among women worldwide.1 An early diagnosis increases the patients’ chances for a successful and customized treatment.2−5 As breast cancer types vary, malignant cells are identified and differentiated after a biopsy using specific proteins, like Human Epidermal Growth Factor Receptor (HER) 2 and Estrogen Receptor (ER) α. Both proteins are also present in cell membranes and are among the primary target analytes in biochemical and histological breast cancer diagnosis.2,6 However, for early screening without the need of biopsies or imaging, the search for potent biomarkers is of great importance.
All cells, both healthy and diseased, constantly secrete membrane-limited nanovesicles, referred to as extracellular vesicles (EVs), which vary in size (approximately ≤50 nm to 1 μm), lipid composition, and enclosed content such as nucleic acids, carbohydrates, lipids, and proteins, depending on the cell of origin and their biogenesis.7,8 Originating from the plasma membrane or intracellular compartments, EVs carry information about the cell they derive from, in contrast to tissue-unrelated, dissolved proteins.9 EVs have received much attention as possible biomarkers as they circulate in most body fluids like urine,10 saliva,11 or blood12,13 and are therefore easily accessible in non- or minimally invasive diagnostic samples using small liquid biopsies. Indeed, several studies confirmed their use in the diagnosis of pancreatic cancer,13 breast cancer,14 and glioblastoma.15 EVs provide an enormous potential for sensitive diagnosis at an early point of time or for monitoring the progress of a therapy.16 Therefore, detailed knowledge of the relation between EVs and their cells of origin is necessary and requires advanced methods for EV profiling.
Most available methods for EV analysis rely on a large amount of EVs17 and therefore bulk enrichment,18,19 e.g., achieved by differential or density gradient ultracentrifugation,20 filtration,15 or precipitation kits.21 These approaches, however, are often unspecific as large numbers of vesicles are averaged, and they are prone to artifacts, e.g., by introducing damage to the sample.22 In recent years, microfluidic platforms allowed for the efficient enrichment and analysis of EVs from cell culture supernatant or blood and integrated analysis of membrane proteins or nucleic acids.23,24 In particular, combined methods could be realized for capturing EVs, e.g., by means of functionalized magnetic beads25,26 and subsequent analysis by immunolabeling27 or other label-free methods such as SPR,28 SERS,29 and μNMR.30 To account for the large heterogeneity of EVs, methods for profiling single EVs are of particular interest since they enable the identification of subpopulations of EVs with common protein profiles.15 A step further into a more detailed understanding of EV biogenesis was recently achieved by single-cell-derived single EV analysis.20,31−34 For example, optical discrimination of EVs from single cells was possible by means of super-resolution microscopy.33 Other approaches use microfabricated platforms with microwells, which enclose cells and the secreted EVs in the same small compartment.31,34 While very useful to accumulate and capture secreted EVs, these platforms are usually not allowing for washing steps without interfering with the patterned surface coatings, the captured analyte, and the cells themselves unless the cells are confined separately from the EV capturing area. Moreover, previous platforms face challenges in preventing cross-contamination of EVs from different cells. We could overcome these shortcomings by introducing a microfluidic platform that forms two compartments by use of ring-shaped valves.32 We thereby separate the compartments for single-cell capturing and EV capturing and analysis. Surface coating in the two compartments is specific for cell adhesion and EV capturing, respectively. For analysis, washing buffers and solutions for immunolabeling can be exchanged without affecting the isolated cells.
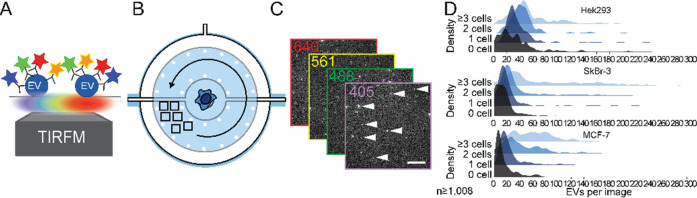
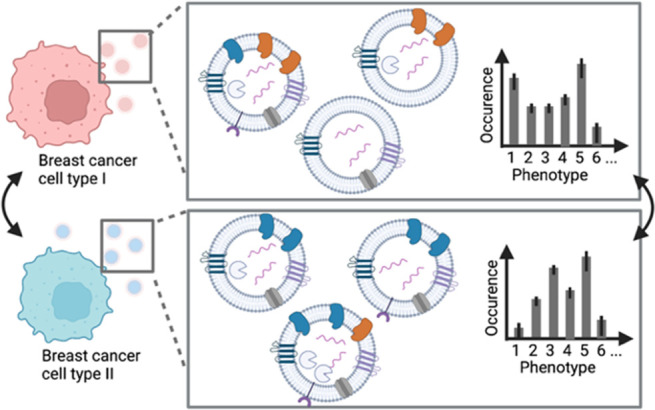
Inspired by the potential of EVs in cancer diagnostics, we further implemented technical improvements to our previously developed microfluidic platform for EV analysis to obtain more fundamental insights into cancer-cell derived EVs.30 Here, we employ the new platform to compare EVs from two breast cancer cell lines (MCF-7 and SkBr-3) and a control cell line (HEK293). We harvest EVs from individual, isolated cells, which allows us to characterize and compare the EV phenotypes in relation to their original cell in an unbiased way (Figure 1). EVs are captured on the surface near the cell of origin. Subsequent immunostaining, four-color total internal reflection fluorescence microscopy (TIRFM) and a custom-made image analysis workflow facilitated the determination of up to 15 EV populations.
Figure 1.
Cells secrete a heterogeneous population of EVs, e.g., with different proteins in the membrane and lumen. In this study, we determine the phenotypic fingerprint of single-cell derived EVs from different breast cancer cells. Image created with BioRender.com.
Our results show that EVs from single cells are sufficient to reveal the phenotypic fingerprint and to differentiate healthy from diseased cells. Furthermore, we could discriminate the two cancer cell lines by using the membrane markers HER2 and ERα. Finally, we obtained a cell type-specific pattern of the EV subpopulations with variations of colocalized HER2, HSP70, CD81, and ERα signals.
Experimental Section
Microfluidic Device Fabrication
The PDMS devices were produced using a 10:1 mass ratio of silicone elastomer base and curing agent (Sylgard 184, Dowsil, formerly Dow Corning). A total of 40 g PDMS was mixed, degassed, and poured onto the silicon master mold with the upper pressure layer, and cured at 80 °C for >2 h. The flexible membranes of the lower pressure and central fluid layer were produced to heights of approximately 25 μm thickness by spin-coating 5 g of PDMS (20 s at 500 rpm and 40 s at 2800 rpm) onto the master mold, which were cured for 1 h at 80 °C. Next, the pressure layer was peeled off and 1 mm inlet holes were punched using 1 mm biopsy puncher. To bond pressure and fluid layers, we spin-coated 2–3 mL of curing agent onto a blank 4-in. silicon wafer at 6000 rpm for 1 min. The cut devices were dropped onto the spin-coated curing-agent layer on the blank silicon wafer, peeled off, and positioned on the PMDS-coated fluid layer. We then sealed the combined layers with a degassed PDMS mixture and cured the assembled devices for 2 h at 80 °C. The assembled devices were peeled off, and inlet holes were punched using a 1.5 mm biopsy puncher. We further spin-coated approximately 1 g of PDMS mixture onto No. 0 microscopy glass slides (6000 rpm for 60 s), resulting in a 10 μm thick PDMS layer. We allowed the uncured PDMS to reflow for over 30 min at room temperature (RT), before curing at 80 °C overnight. To bond the triple-layer PDMS devices and PDMS-coated glass slides, we activated the surfaces of both items using plasma (PDC-32G, Harrick Plasma, U.S.A.) at ∼0.77 mbar for 45 s (18 W). Bonded devices were kept on a heating plate at 100 °C for 10 min and stored at RT for later use.
Cell Culture
Michigan Cancer Foundation 7 (MCF-7), Sloan Kettering Breast Cancer 3 (SkBr-3) cells, and Huma Embryonic Kidney (HEK293) were obtained from ATCC. The cells were cultured in standard cell culture flasks in Dulbecco’s Modified Eagle’s Medium (DMEM; MCF-7, HEK293) or F12/DMEM (SkBr-3) supplemented with 1 g L–1 glucose, pyruvate, 10% (v/v) fetal bovine serum (both Thermo Fisher Scientific) at 37 °C, 5% (v/v) CO2 atmosphere and a relative humidity of 95%. For passaging, cells were trypsinized with Trypsin-EDTA (0.05%; Thermo Fisher Scientific) and passaged in 1:5 ratios twice a week. On-chip medium contained 1× penicillin–streptomycin (Thermo Fisher Scientific) and 2% (v/v) exosome-free fetal bovine serum (System Biosciences).
Experimental Preparation and Surface Functionalization
At the beginning of all experiments, the devices were filled with Milli-Q water by centrifugation at 800g for 10 min, and incubated at 37 °C, 95% relative humidity, and 5% (v/v) CO2 for >30 min on a microscope. We then connected the devices to polytetrafluoroethylene (PTFE) tubing (PKM SA, Switzerland), silicon tubing (Gobatec, Switzerland), and polyether ether ketone (PEEK) microfittings to 10 mL syringes (Becton Dickinson, Switzerland). The syringes were loaded onto syringe pumps (either NE-1002X-ES, World Precision Instruments, Friedberg, Germany, or NanoJet, Chemyx Inc., U.S.A.). Custom bent metal pins were used to connect the pressure channels to a silicon tubing and to pressurized air via a customized masterflex polycarbonate manifold (Cole-Parmer, U.S.A.). The devices were then flushed with sterile phosphate-buffered saline without Ca2+ and Mg2+ (PBS −/–, Sigma-Aldrich). To promote cell adhesion in the cell capturing area, we flushed the entire devices with 100 ng mL–1 fibronectin (Sigma-Aldrich) and incubated them for 30 min (inner valves closed), which then was washed out with PBS −/–. After incubation, we flushed the chips with exosome-depleted fetal calf serum-supplemented medium (2% (v/v) FCS, 1× penicillin–streptomycin, 1 g L–1 glucose DMEM or F12/DMEM). The cells were filtered (pore size 35 μm), flushed into the device, trapped in the chambers, isolated by closing the inner ring valve, and incubated at 37 °C, 5% (v/v) CO2. Cell ratios of analyzed empty wells, single occupied wells, double occupied wells, and wells containing three or more cells were 19.28%, 22.89%, 27.71%, and 30.12%, respectively. The EV-capturing area was incubated for 30 min with 2 mg mL–1 biotinylated bovine serum albumin (biotin-BSA; Thermo Fisher Scientific, Waltham U.S.A.), 100 g mL–1 NeutrAvidin, and 5 ng mL–1 biotinylated antibody against CD63 (Biolegend, London, U.K.). We then flushed the area for EV capture with exosome-free FCS and penicillin–streptomycin-supplemented DMEM. Flow rates were adjusted empirically and kept at 5 uL/min for all flushing and washing steps. At this relatively low flow rates the chip was flushed within less than 2 min while neither trapped cells were washed (“pushed”) out of the traps, nor did pneumatically valves (actuated at 1.6–1.8 bar) open.
Immunocytochemistry
Before staining, cells and EVs were washed in PBS with Ca2+ and Mg2+, fixed in 4% (v/v) paraformaldehyde in PBS with Ca2+ and Mg2+ (pH 7.2), blocked in 4 mass-% heat-shock denatured BSA. Staining was done in 0.1 mass-% heat-shock denatured BSA in PBS with Ca2+ and Mg2+. Respectively used antibodies were biotinylated anti-CD63 (Biolegend), ERα-CF405M, HER2CF640R (both Biotium), HSP70-FITC (Santa Cruz Biotechnology), and CD81-PE (Biolegend).
Data Analysis and Statistics
We collected TIRFM images from empty wells (0-cell, n ≥ 840), single occupied wells (1-cell, n ≥ 1008), double occupied wells (2-cells, n ≥ 1176), and wells with three or more cells (≥3 cells, n ≥ 1344). Following the image analysis described in detail in Nikoloff et al. 2021,32 we determined the frequency distributions of the detected signals per image within an area of 4356 μm2. In total, 42 images were analyzed per chamber. Differences in distributions were tested with two-sided Kolmogorov–Smirnov tests and are given in the Supporting Information as Table SI 1. To test for Gaussian distributions, we used the Shapiro-Wilk test.
Results and Discussion
Design and Operation of the Microfluidic Device
The device is modified from a previously published design.32 We increased then number of wells (from 72 to 12 × 8 = 96 wells) for single cell capture and EV analysis (Figures 2A and S1). Each of these sites has a central hydrodynamic trap of 10 μm to capture an individual cell, which is surrounded by two nested concentric pneumatic valves (Figure 2B,D). While the earlier described device consisted of two PDMS-layers,32 we introduced a third PDMS layer which increased the PDMS–PDMS contact site by 50%, thereby improving the mechanic stability of the device and reduced the risk of damages during manufacturing as we could avoid the previously used fine structures of a few micrometer for separating the inner and outer pneumatic valves. As the third PDMS layer, however, also increased collapsing, we introduced PDMS columns in the central fluid layer to separate the upper and lower pneumatic layer. The inner volume with the cell trap allows us to maintain the cells in a shear stress-free environment. The space between the two valves is used for EV capturing (see below). In contrast to previous similar devices, the chip is manufactured by using three instead of two layers of PDMS, assembled on top of each other. The top layer defines the inner valves (top control layer), the middle layer defines the microfluidic channels including the cell trap (fluid layer), and the bottom layer carries the control lines for the outer valves (bottom control layer; Figure 2C). The use of two control layers maximizes the PDMS–PDMS contact surface to the fluid layer, which results in a robust and reliable chip fabrication. Additionally, we implemented supporting pillars to improve the stability of the valves. The use of the valves is visualized in the SI, Figure S2.
Figure 2.
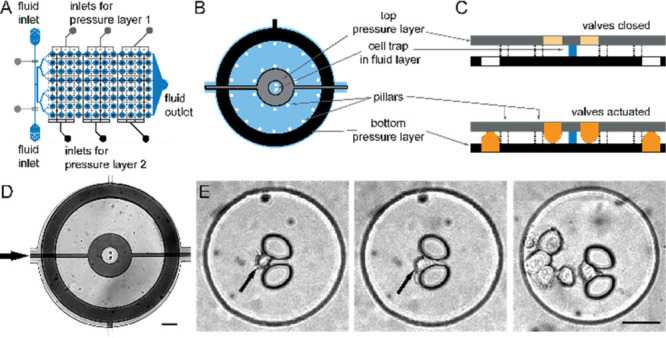
Microfluidic platform for single-cell isolation and EV detection. (A) The three-layer microfluidic device comprises two pressure layers (gray, black) and a fluid layer (blue). (B) Circular cell chambers contain a central hydrodynamic cell trap, which is surrounded by two concentric pneumatic valves in the top (gray) and bottom pressure layer (black), which are separated by pillars. (C) Actuation of the pneumatic results in volumetric separation and allows for specific and spatially restricted surface coatings. (D) Micrograph of one chamber (scale bar: 100 μm). (E) Bright-field images of single trapped cells in the central chamber (black arrows). Occasionally, we have cell clusters of several cells in the chamber (right image, scale bar: 50 μm).
The experimental procedure for single-cell-derived single EV analysis starts with a fibronectin coating in the inner chamber to enhance cell adhesion. Subsequently, we flushed the cell suspension into the chip, to immobilize single cells at the hydrodynamic traps. After isolating the cells by lowering the inner concentric valve, we functionalized the area between the inner and the outer valves with biotinylated mAb against CD63 to immobilize EVs. Next, cells were incubated for 24 h. During this time, the inner valve is open, while the outer valve is closed to prevent diffusion of segregated EVs out of the chamber. Afterward, we continued with immunostaining of the EVs with four differently tagged antibodies and analyzed the population by four-color TIRFM (Figure 3). On captured images we applied a custom image processing pipeline32 to quantify the frequency distribution of detected EVs and to identify and assign each EV to 1 of 15 phenotype subpopulations.
Figure 3.
Workflow for detecting and identifying EV populations form single cells. (A) Using fluorescently conjugated mAbs and four-color TIRFM, we image immobilized EVs (B) in the direct neighborhood of the cells, which are prevented from cross-contamination with EVs from other cells. (C) Multicolor TIRFM images of secreted and immobilized EVs. (D) Customized Matlab imaging processing allows for quantifying and identifying EVs per image. Note that the histograms display raw data (detected signals per image).
Characterization of the Selected Cell Lines
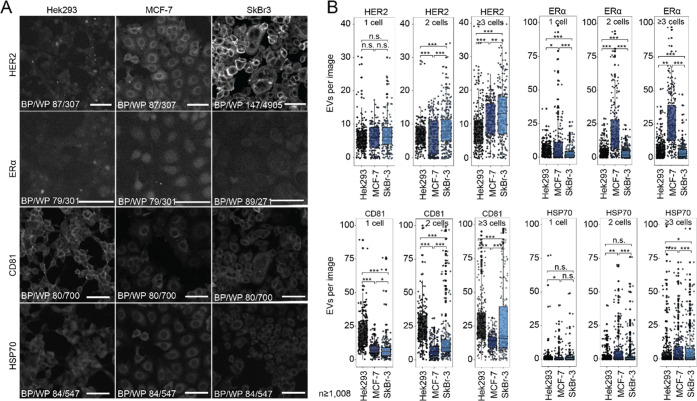
We selected three human cell lines (Michigan Cancer Foundation (MCF)-7 cells, Sloan Kettering Breast Cancer (SkBr)-3 cells, and Human Embryonic Kidney (HEK) 293 cells and confirmed their specific phenotype compositions by immunostaining (Figure 4A). While HEK293 cells are known for their fast proliferation, robustness, and susceptibility for genetic editing, they presumably express lower levels of cancer biomarkers,35 but elevated levels of HSPA1A (HSP70 family) and CD81. The epithelial-like MCF-7 cell line, which overexpresses ERα (ERα+), is sensitive to ER-stimuli, and expresses low HER2 levels (HER2–). SkBr-3 cells are also epithelium-derived and demonstrate higher levels of HER2, but lower ERα expression (ERα–HER2+). Both cell lines are commonly used in fundamental or pharmaceutical cancer research.3,36−41 We chose these cell lines as they feature unique protein expression patterns: MCF-7 cells are ERα+HER2–, SkBr-3 cells are ERα–HER2+, and HEK 293 are ERα–HER2–. Both HEK293 and SkBr-3 express comparatively higher levels of the membranous CD81 than MCF-7, visible in the fluorescence images (Figure 4A) and previous transcriptomics results (HEK293:517.8 normalized transcripts per million (nTPM), SkBr-3:514.4 nTPM, MCF-7:257.2 nTPM).42 In addition, the cytoplasmic chaperone Heat Shock Protein 70 (HSP70), which is a marker for EVs,17 is expressed in all three cell lines.
Figure 4.
EVs represent the phenotype of the cell of origin. (A) Immunocytochemical staining showing distinct expression profiles in SkBr-3, MCF-7, and HEK293 for HER2, of ERα, CD81, and HSP70. Scale bar 100 μm. (B) Dot plots depicting the absolute number of detected EVs per image when immobilized on anti-CD63 antibody. Separated by cell type, we detect EVs from wells with single, two, or three and more cells for all four investigated markers.
On-Chip Analysis of EVs
Next, we captured the cells on-chip and collected the EVs. After cell incubation for 24 h, we fixed, and stained the immobilized EVs with the mAb-conjugates against ERα, HER2, HSP70, and CD81. We occasionally trapped more than one cell (Figure 2E) and used these data with two as well as three and more cells per chamber for comparison. Figure 3D displays the histogram for the number of EVs that we found per image, here a sum of all EVs, regardless of their phenotypes. More entrapped cells led to larger EV numbers per image as well as a broader distribution, as expected. For HEK293 cells, we detected more vesicles (single cell trapping: median of 34 EVs in an image area of 4356 μm2), than for the cancer cells lines (16 EVs for SkBr-3 and 18 EVs for MCF-7). The results show that EV secretion rates are not necessarily higher in cancer cells than in noncancer tissue43,44 and might also depend on higher proliferation rates, along with increased overall cell metabolism.
We further quantified the number of EVs that carry the marker HER2, ERα, CD81, or HSP70 on the CD63-immobilized EVs (Figures 4B and 5, and SI, Table S1). Again, we enumerated more EVs when more than one cell is captured in the chamber. Notably, all markers can be found in the EVs of all cell lines. However, the number of EVs with a selected marker shows large variations. In line with the determined tissue phenotypes, we observed that two and more SkBr-3 cells release more HER2+-EVs than HEK293 and MCF-7 cells. The latter cancer cell line sheds also HER2+-EVs, as most obvious for chambers, where 3 or more cells were trapped, where the number of EVs is significantly higher than in the HEK293 cell line. In contrast, MCF-7 released most ERα+-EVs compared to the other cell lines. For CD81 displaying EVs, the expected larger EV number for HEK293 and SkBr-3 is visible for three and more trapped cells. In line with the CD81 expression levels in HEK293 and SkBr-3 cells, the medians of detected EVs from larger cell populations amounted for 25 and 20 EVs per image from HEK293 and SkBr-3 cells, respectively, in contrast to 14 EVs per image in MCF-7 wells. For HSP70, we found similarly low numbers of EVs in all three cell lines. In summary, we showed that EVs reflect largely the markers of the cell lines they are derived from. However, the large EV heterogeneity makes any single marker insufficient for determining the cell of origin.
Figure 5.
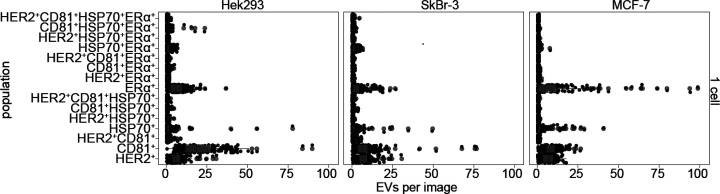
Phenotype analysis of EVs secreted from single cells. Data points (black) and resulting dot plots (in gray) depicting the absolute number of detected EVs, grouped by the cell type and the 15 possible phenotypes, when immobilized on anti-CD63 antibody.
Determination of EV Populations
Next, we analyzed the subpopulations by clustering EVs with colocalized markers (Figure 5). In this analysis of single-cell data, we use the median number of EVs per image for all chambers for a specified combination of markers. The pattern of all 15 possible populations is unique for a given cell line, although again for each subpopulation the variations are large. The additional analysis of EV subpopulations for 2 and ≥3 cells is given in the SI, Figure S3. Most striking is the obvious difference for MCF-7 cells. Most EVs from this cell line carry only one marker, while subpopulations with colocalized markers are less abundant than for the other cell lines. Interestingly, we often detected the subpopulation HSP70+ERα+ in all cell lines. HSP70, in contrast to HER2, ERα, and CD81, is a cytosolic chaperone, which stabilizes ligand-unbound membranous ERα as previously reported.45 Our data suggests that this copresence is maintained in the segregated EVs.
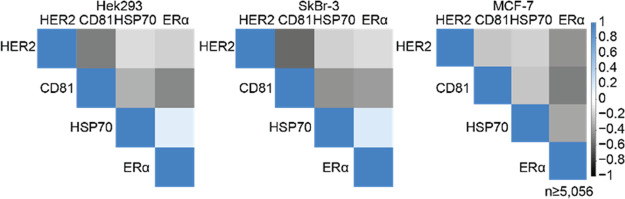
We further performed a correlation analysis (Figure 6). Similar correlation patterns between HEK293 and SkBr-3 cells are visible. HER2 correlated negatively with CD81, and CD81 correlated less with HSP70 and ERα. Again, we observed a weak positive correlation between HSP70 and ERα. The observations contrasted with the correlation patterns observed in MCF-7. Here, HER2, CD81 and HSP70 negatively associate with ERα, which MCF-7 cells overexpress. The observed negative correlations for CD81 and HER2 in Sk-Br-3 and HEK293, similar to ERα and the other target epitopes in MCF-7 cells, are in line with the hypothesis and demonstrate that the overexpression of proteins prevents the analysis of protein-interactions which can be observed at endogenous levels but are not detectable due to the abundance of overexpressed protein.
Figure 6.
Correlation analysis of analyzed markers on detected EV populations. Correlation analysis, showing a weak positive association of HSP70 to integral ERα. Overexpressed membrane proteins, like CD81, HER2, and ERα show negative correlation with most other proteins due to its abundant integration into membranes. Protein overexpression disguises protein colocalization in EV membranes.
Conclusion
Here, we show a robust microfluidic method for the analysis of single EVs derived from single cells. We immobilized and enumerated EVs, as well as characterized their phenotypes. Our results confirm that EVs reflect the unique molecular fingerprint of each tissue of origin. In particular, we conclude that membrane proteins serve as reliable markers, while cytosolic proteins like HSP70 are ubiquitously detected in all cells, therefore, cell-type unspecific and unreliable as standalone biomarkers. Together, we provide strong quantitative evidence that phenotypic analysis of EVs from a single cell are informative enough for discovering differences in their origin and embody a strong potential as diagnostic biomarkers from liquid biopsies. Besides, we believe that our device is very powerful to study fundamental questions in the biogenesis of EVs.
Acknowledgments
We thank André Kling, Maximilian Breitfeld, and Claudius Dietsche (D-BSSE, ETH Zürich) for the wafer fabrication, and Barbara Treutlein (D-BSSE, ETH Zürich) for providing the HEK293 cells. We acknowledge the European Research Council (ERC) for funding (ERC Consolidator Grant No. 681587).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.2c04106.
Photograph of the device, micrographs of the trap and pneumatic valves, phenotype analysis for one, two and multiple cells, table with statistical data (Figures S1–S3 and Table SI 1) (PDF)
Author Contributions
J.M.N. and P.S.D. conceptualized the work and wrote the manuscript. J.M.N. performed the experiments and analyzed the data. M.A.S.-E. did the correlation analysis. P.S.D. provided the funding and supervised the study.
The authors declare no competing financial interest.
Supplementary Material
References
- Ferlay J.; Colombet M.; Soerjomataram I.; Parkin D. M.; Piñeros M.; Znaor A.; Bray F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149 (4), 778–789. 10.1002/ijc.33588. [DOI] [PubMed] [Google Scholar]
- Zubair M.; Wang S.; Ali N. Advanced Approaches to Breast Cancer Classification and Diagnosis. Front. Pharmacol. 2021, 11, 2487. 10.3389/fphar.2020.632079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford C. H. J.; Maie A. L. B.; Bushra A. L. A.; Issam F. Reassessment of Estrogen Receptor Expression in Human Breast Cancer Cell Lines. Anticancer Res. 2011, 31 (2), 521–527. [PubMed] [Google Scholar]
- Diaz L. K.; Sneige N. Estrogen Receptor Analysis for Breast Cancer: Current Issues and Keys to Increasing Testing Accuracy. Adv. Anat. Pathol. 2005, 12 (1), 10–19. 10.1097/00125480-200501000-00003. [DOI] [PubMed] [Google Scholar]
- Cooke T.; Reeves J.; Lanigan A.; Stanton P. HER2 as a Prognostic and Predictive Marker for Breast Cancer. Ann. Oncol. 2001, 12, S23–S28. 10.1093/annonc/12.suppl_1.S23. [DOI] [PubMed] [Google Scholar]
- Gunduz M., Gunduz E., Eds. Heterogeneity of Phenotype in Breast Cancer Cell Lines. Breast Cancer - Carcinogenesis, Cell Growth and Signalling Pathways; InTech, 2011; Vol. 2. 10.5772/21984. [DOI] [Google Scholar]
- Mathieu M.; Martin-Jaular L.; Lavieu G.; Théry C.. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol.. Nature Publishing Group, 2019; pp 9–17. 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- Jeppesen D. K.; Fenix A. M.; Franklin J. L.; Higginbotham J. N.; Zhang Q.; Zimmerman L. J.; Liebler D. C.; Ping J.; Liu Q.; Evans R.; Fissell W. H.; Patton J. G.; Rome L. H.; Burnette D. T.; Coffey R. J. Reassessment of Exosome Composition. Cell 2019, 177 (2), 428–445. 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larios J.; Mercier V.; Roux A.; Gruenberg J. ALIX-and ESCRT-III-Dependent Sorting of Tetraspanins to Exosomes. J. Cell Biol. 2020, 219 (3), na. 10.1083/jcb.201904113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Hill S.; Luther J. M.; Hachey D. L.; Schey K. L. Proteomic Analysis of Urine Exosomes by Multidimensional Protein Identification Technology (MudPIT). Proteomics 2012, 12 (2), 329–338. 10.1002/pmic.201100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanisamy V.; Sharma S.; Deshpande A.; Zhou H.; Gimzewski J.; Wong D. T. Nanostructural and Transcriptomic Analyses of Human Saliva Derived Exosomes. PLoS One 2010, 5 (1), e8577. 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.; Ouyang Y.; Wang Z.; Zhang R.; Huang P.-H.; Chen C.; Li H.; Li P.; Quinn D.; Dao M.; Suresh S.; Sadovsky Y.; Huang T. J. Isolation of Exosomes from Whole Blood by Integrating Acoustics and Microfluidics. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 10584–10589. 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.; Wang J. J.; Gao M.; Wang J. J.; Ma Y.; Gu Y. Membrane Feature-Inspired Profiling of Extracellular Vesicles for Pancreatic Cancer Diagnosis. Anal. Chem. 2021, 93 (28), 9860–9868. 10.1021/acs.analchem.1c01712. [DOI] [PubMed] [Google Scholar]
- Kreger B. T.; Johansen E. R.; Cerione R. A.; Antonyak M. A. The Enrichment of Survivin in Exosomes from Breast Cancer Cells Treated with Paclitaxel Promotes Cell Survival and Chemoresistance. Cancers (Basel) 2016, 8 (12), 111. 10.3390/cancers8120111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.; Fraser K.; Ghaddar B.; Yang K.; Kim E.; Balaj L.; Chiocca E. A.; Breakefield X. O.; Lee H.; Weissleder R. Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano 2018, 12 (1), 494–503. 10.1021/acsnano.7b07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi S.; Mäger I.; Breakefield X. O.; Wood M. J. A. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nature Reviews Drug Discovery. 2013, 12, 347–357. 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Kowal J.; Arras G.; Colombo M.; Jouve M.; Morath J. P.; Primdal-Bengtson B.; Dingli F.; Loew D.; Tkach M.; Théry C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (8), E968–E977. 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli R.; Lässer C.; Lötvall J. Isolation and Characterization of Extracellular Vesicle Subpopulations from Tissues. Nat. Protoc. 2021, 16 (3), 1548–1580. 10.1038/s41596-020-00466-1. [DOI] [PubMed] [Google Scholar]
- Thierry; Théry C.; Amigorena S.; Raposo G.; Clayton A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 3, 1–29. 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- Ji Y.; Qi D.; Li L.; Su H.; Li X.; Luo Y.; Sun B.; Zhang F.; Lin B.; Liu T.; Lu Y. Multiplexed Profiling of Single-Cell Extracellular Vesicles Secretion. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (13), 5979–5984. 10.1073/pnas.1814348116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D.; Ho S.-L.; Chan H.-N.; Wang H.; Hai L.; He X.; Wang K.; Li H.-W. Molecular-Recognition-Based DNA Nanodevices for Enhancing the Direct Visualization and Quantification of Single Vesicles of Tumor Exosomes in Plasma Microsamples. Anal. Chem. 2019, 91, 2768–2775. 10.1021/acs.analchem.8b04509. [DOI] [PubMed] [Google Scholar]
- Gyorgy B.; Szabo T. G.; Pasztoi M.; Pal Z.; Misjak P.; Aradi B.; Laszlo V.; Pallinger E.; Pap E.; Kittel A.; Nagy G.; Falus A.; Buzas E. I. Membrane Vesicles, Current State-of-the-Art: Emerging Role of Extracellular Vesicles. Cell. Mol. Life Sci. 2011, 68 (16), 2667–2688. 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H.; Im H.; Castro C. M.; Breakefield X.; Weissleder R.; Lee H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118 (4), 1917–1950. 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F.; Wang C. C.; Wang C. C.; Gao J.; Liu H.; Zhang Y.; Han L. Enrichment-Detection Integrated Exosome Profiling Biosensors Promising for Early Diagnosis of Cancer. Anal. Chem. 2021, 93 (11), 4697–4706. 10.1021/acs.analchem.0c05245. [DOI] [PubMed] [Google Scholar]
- He M.; Crow J.; Roth M.; Zeng Y.; Godwin A. K. Integrated Immunoisolation and Protein Analysis of Circulating Exosomes Using Microfluidic Technology. Lab Chip 2014, 14, 3773–3780. 10.1039/C4LC00662C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho J.; Chung J.; Im H.; Liong M.; Shao H.; Castro C. M.; Weissleder R.; Lee H. Magnetic Nanosensor for Detection and Profiling of Erythrocyte-Derived Microvesicles. ACS Nano 2013, 7 (12), 11227–11233. 10.1021/nn405016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.; Ji Y.; Deng J.; Li L.; Bai X.; Liu X.; Lin B.; Lu Y. Microfluidics-Based Technologies for the Analysis of Extracellular Vesicles at the Single-Cell Level and Single-Vesicle Level. Chin. Chem. Lett. 2022, 33, 2893. 10.1016/j.cclet.2021.09.058. [DOI] [Google Scholar]
- Im H.; Shao H.; Park Y. Il; Peterson V. M.; Castro C. M.; Weissleder R.; Lee H. Label-Free Detection and Molecular Profiling of Exosomes with a Nano-Plasmonic Sensor. Nat. Biotechnol. 2014, 32 (5), 490–495. 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S.; Wang Y.; Liu Z.; Zhang W.; Yi K.; Zhang X.; Zhang X.; Jiang C.; Yang S.; Wang F.; Xiao X. Beehive-Inspired Macroporous SERS Probe for Cancer Detection through Capturing and Analyzing Exosomes in Plasma. ACS Appl. Mater. Interfaces 2020, 12 (4), 5136–5146. 10.1021/acsami.9b21333. [DOI] [PubMed] [Google Scholar]
- Shao H.; Chung J.; Balaj L.; Charest A.; Bigner D. D.; Carter B. S.; Hochberg F. H.; Breakefield X. O.; Weissleder R.; Lee H. Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med. 2012, 18 (12), 1835–1840. 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son K. J.; Rahimian A.; Shin D.-S. S.; Siltanen C.; Patel T.; Revzin A. Microfluidic Compartments with Sensing Microbeads for Dynamic Monitoring of Cytokine and Exosome Release from Single Cells. Analyst 2016, 141 (2), 679–688. 10.1039/C5AN01648G. [DOI] [PubMed] [Google Scholar]
- Nikoloff J. M.; Saucedo-Espinosa M. A.; Kling A.; Dittrich P. S. Identifying Extracellular Vesicle Populations from Single Cells. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (38), e2106630118. 10.1073/pnas.2106630118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechantsreiter S.; Ambrose A. R.; Worboys J. D.; Lim J. M. E.; Liu S.; Shah R.; Montero M. A.; Quinn A. M.; Hussell T.; Tannahill G. M.; Davis D. M. Heterogeneity in Extracellular Vesicle Secretion by Single Human Macrophages Revealed by Super-resolution Microscopy. J. Extracell. Vesicles 2022, 11 (4), na. 10.1002/jev2.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi M.; Joseph R.; Adolacion J. R. T.; Martinez-Paniagua M.; An X.; Gabrusiewicz K.; Mani S. A.; Varadarajan N. Single-Cell Cloning of Breast Cancer Cells Secreting Specific Subsets of Extracellular Vesicles. Cancers (Basel). 2021, 13 (17), 4397. 10.3390/cancers13174397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaandou L.; Quan D.; Shiloach J. Affecting Hek293 Cell Growth and Production Performance by Modifying the Expression of Specific Genes. Cells 2021, 10 (7), 1667. 10.3390/cells10071667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleensang A.; Vantangoli M. M.; Odwin-DaCosta S.; Andersen M. E.; Boekelheide K.; Bouhifd M.; Fornace A. J.; Li H.-H.; Livi C. B.; Madnick S.; Maertens A.; Rosenberg M.; Yager J. D.; Zhao L.; Hartung T. Genetic Variability in a Frozen Batch of MCF-7 Cells Invisible in Routine Authentication Affecting Cell Function. Sci. Rep. 2016, 6, 1. 10.1038/srep28994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.; Leitner J. W.; Solomon S.; Golovchenko I.; Goalstone M. L.; Draznin B. Effect of Insulin on Cell Cycle Progression in MCF-7 Breast Cancer Cells. Direct and Potentiating Influence. J. Biol. Chem. 2001, 276 (41), 38023–38028. 10.1074/jbc.M104416200. [DOI] [PubMed] [Google Scholar]
- Miglietta A.; Panno M. L.; Bozzo F.; Gabriel L.; Bocca C. Insulin Can Modulate MCF-7 Cell Response to Paclitaxel. Cancer Lett. 2004, 209 (2), 139–145. 10.1016/j.canlet.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Comşa Ş.; Cîmpean A. M.; Raica M. The Story of MCF-7 Breast Cancer Cell Line: 40 Years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [PubMed] [Google Scholar]
- Olsauskas-Kuprys R.; Zlobin A.; Osipo C. Gamma Secretase Inhibitors of Notch Signaling. Onco. Targets. Ther. 2013, 6, 943–955. 10.2147/OTT.S33766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper L. N.; Glathe S.; Vaisman N.; Hynes N. E.; Andrews G. C.; Sela M.; Yarden Y. The ErbB-2/HER2 Oncoprotein of Human Carcinomas May Function Solely as a Shared Coreceptor for Multiple Stroma-Derived Growth Factors. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (9), 4995–5000. 10.1073/pnas.96.9.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul P. J.; Akesson L.; Wiking M.; Mahdessian D.; Geladaki A.; Ait Blal H.; Alm T.; Asplund A.; Björk L.; Breckels L. M.; Bäckström A.; Danielsson F.; Fagerberg L.; Fall J.; Gatto L.; Gnann C.; Hober S.; Hjelmare M.; Johansson F.; Lee S.; Lindskog C.; Mulder J.; Mulvey C. M.; Nilsson P.; Oksvold P.; Rockberg J.; Schutten R.; Schwenk J. M.; Sivertsson A.; Sjöstedt E.; Skogs M.; Stadler C.; Sullivan D. P.; Tegel H.; Winsnes C.; Zhang C.; Zwahlen M.; Mardinoglu A.; Pontén F.; Von Feilitzen K.; Lilley K. S.; Uhlén M.; Lundberg E. A Subcellular Map of the Human Proteome. Science 2017, 356 (6340), na. 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Bebelman M. P.; Janssen E.; Pegtel D. M.; Crudden C. The Forces Driving Cancer Extracellular Vesicle Secretion. Neoplasia 2021, 23 (1), 149. 10.1016/j.neo.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier C. P. R.; Caires H. R.; Barbosa M. A. G.; Bergantim R.; Guimarães J. E.; Vasconcelos M. H. The Role of Extracellular Vesicles in the Hallmarks of Cancer and Drug Resistance. Cells 2020, 9, 1141. 10.3390/cells9051141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamad A. E.; Zhou Z.; Zhou J.; Du Y. Systematic Proteomic Identification of the Heat Shock Proteins (Hsp) That Interact with Estrogen Receptor Alpha (ERα) and Biochemical Characterization of the ERα-Hsp70 Interaction. PLoS One 2016, 11 (8), e0160312. 10.1371/journal.pone.0160312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.