Abstract
Background: Histone acetylations acting as active hallmarks for gene transcription is involved in regulating numerous developmental and stress-responsive gene expression.
Methods: The data from chromatin immunoprecipitation sequencing (ChIP-seq) was performed by using histone H3 lysine 9 acetylation (H3K9ac) antibody, and RNA sequencing (RNA-seq) utilizing rice seedlings inoculated by Magnaporthe oryzae (M. oryzae) were integrated.
Results: RNA-seq data revealed that 422, 460 and 466 genes were up-regulated at 12h, 24h and 48h after inoculation. ChIP-seq data showed that 60%-80% of blast up-regulated genes at different time points were marked with H3K9ac, which was prone to be enriched in both TSS and gene body region. However, the H3K9ac level at a rather small proportion of the up-regulated genes was elevated after M. oryzae inoculation. We found that seven WRKY genes induced by rice blast fungus harbor H3K9ac. For different WRKY genes, blast fungus induction led to the increase of H3K9ac in distinct regions, including promoter, TSS or gene body, indicating that histone acetylation may play diverse roles in the activation of defense-related genes. By searching DNA-binding motifs of transcription factors in the promoter of genes with increased H3K9ac after M. oryzae infection, we found that ERF family protein-binding motifs were enriched with high -log P-value (>20), including ERF1, DEAR3, DREB2C, RAP2.6, RRTF1_3ARY, all of which contain GCC-box (GCCGCC).
Conclusion: In this study, we revealed that the vast majority of genes induced by fungus M. oryzae were marked with H3K9ac preferring both TSS and gene body regions. However, H3K9ac enrichment was increased, responding to M. oryzae inoculation only at a low proportion of these genes, including several WRKY genes. Besides, for different genes, the increment of H3K9ac occurred in different regions. Finally, ERF proteins that have been proved to bind GCC-box might be one of the potential transcription factors for recruiting histone acetyltransferases to deposit histone acetylation at defense-related genes in rice.
Keywords: Rice, Magnaporthe oryzae, histone acetylation, ERF, WRKY, H3K9ac
1. INTRODUCTION
Both animals and plants evolve immune systems to cope with microbial infection. Animal immune systems are more advanced and could be enhanced specifically by multiple strategies [1-4]. Plants are immobile organisms that evolve a flexible system to adapt to environmental changes. Plant-endophytic fungi interactions also contribute to abiotic and biotic stress tolerance [5]. Rice (Oryza sativa) is one of the most important food crops. Rice blast, caused by the fungus M. oryzae, is known as the cancer of rice, and can lead to reduced rice yields, or even no grains in severe cases. During the long-term co-evolution with M. oryzae, rice has formed a precise signal sensing and defense mechanism [6-8]. After recognizing pathogen attacks by cell surface receptors, the defense response is triggered immediately to activate plant immunity. Pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) are the most acceptable models used to describe the mechanism of plant-microbe interaction. Although PTI and ETI employ distinct receptors, they seem to share the signaling network, which is constituted by mitogen-activated protein kinase (MAPK) cascades, calcium flux, reactive oxygen species (ROS) burst, transcriptional reprograming and phytohormone signalling [9]. At the terminal of defense signaling, six major transcription factor (TF) families (AP2/ERF (APETALA2/ethylene responsive factor), bHLH (basic helix-loop-helix), MYB (myeloblastosis related), NAC (no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF1/2), and cup-shaped cotyledon (CUC2)), WRKY, and bZIP (basic leucine zipper)) are dedicated to establish a regulatory network for activating downstream defense genes [10].
In eukaryotes, transcriptional regulation involves both TFs and epigenetic regulators. TFs regulate transcription efficiency by binding to specific DNA motifs and recruiting or blocking RNA polymerase II (Pol II) to or from the core promoter [11]. Epigenetic regulators directly change chromatin structure, such as histone modifications, DNA methylation, and chromatin remodeling, to affect Pol II or TFs binding to DNA [12]. TFs and epigenetic regulators are interconnected to cooperatively regulate gene transcription [11]. Histone acetylation refers to the addition of acetyl group to the lysine residues mainly located at the N-terminal tail of histone. Histone acetylation is balanced by opposite actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs). It is generally accepted that histone acetylations serve as active marks for gene activation, participating not only in developmental processes but also in response to biotic and abiotic stresses in plant [13]. Genome-wide analysis of histone acetylation enrichment unravels that they are mainly enriched surrounding transcription start site (TSS) [14], suggesting that they are possibly related to transcription initiation.
HATs are more likely to be recruited by TFs to target loci to deposit histone acetylation. The homologs of GCN5 or its partner ADA2 in several plant species have been revealed to interact with multiple kinds of TFs. In Arabidopsis, ADA2 interacts with an ERF TF CBF1 to target cold-induced genes [15]. In wheat, TaGCN5 and TaADA2 form a ternary protein complex with a MYB TF TaEPBM1 to activate TaECR transcription [16]. In Populus, a bZIP TF PtrAREB1-2 recruits PtrGCAN5 and Ptr ADA2b to three NAC TFs for gene activation [17]. In rice, the recruitment of GCN5-ADA2 is required for a WOX TF WOX11 to regulate root-specific gene expression [18].
Several HATs and HDACs have been characterized to play a crucial role in plant biotic stress responses [13]. In soybean, the HAT General Control Non-depressive 5 (GCN5) positively regulates defense-related genes by depositing H3 lysine9 acetylation (H3K9ac) at these genes in collaboration with its partner Alteration/Deficiency in Activation 2 (ADA2) [16]. However, GCN5 in Arabidopsis negatively regulates plant immune response by repressing salicylic acid (SA) accumulation [19]. In addition, GCN5 homologs in both Arabidopsis and wheat contribute to cuticular wax biosynthesis, and stimulation of the cuticular wax biosynthesis in wheat can result in the accumulation of VLC aldehydes, which triggers the germination of Bgt conidia [16, 19, 20]. HAC1 and HAC5 form a complex with NPR1 and TGA to activate PR gene transcription by catalyzing histone H3 acetylation [21]. Overexpression of the HDAC HDA19 in Arabidopsis enhances plant resistance to the pathogen, but the mechanism of how HDA19 is involved has not been revealed in the study despite those genes related to jasmonic acid and ethylene signaling are up-regulated in HDA19-overexpression plants [22]. In another study, it was demonstrated that HDA19 inhibited transcription activation activity of two WRKY TFs WRKY38 and WRKY62, which function as negative regulators of plant defense [23]. By contrast, HDA6 negatively regulates plant immunity by repressing pathogen-responsive genes by removing H3 acetylation at corresponding loci [24]. Similarly, a wheat homolog of HDA6 and TaHDA6 also targets defense-related genes to deacetylate H3 and H4 [25]. In addition, TaHDA6 represses target gene expression in concert with TaHOS15 and TaHDT701 [26]. The latter protein is the homolog of rice HDT701, a histone deacetylase 2 (HD2) type HDAC that is responsible for H4 deacetylation [27]. The function of rice HDT701 in innate immunity has been characterized. Overexpression of HDT701 in transgenic rice enhances susceptibility to the pathogens, whereas silencing of HDT701 in transgenic rice enhances resistance to the pathogens [27]. HDT701 directly binds to defense-related genes to repress their expression [27]. The above discoveries uncover the common role of histone acetylation in the regulation of plant immune response.
The development of next-generation sequencing (NGS) technologies increases not only the efficiency of genome sequencing but also transcriptomic and epigenomic analysis [28-30]. To date, the investigation into genome-wide H3K9ac enrichment responding to disease is only performed in Paulownia fortunei [31]. A small proportion of genes with differential H3K9ac levels showed changes in expression in response to phytoplasma infection [31]. In this paper, to explore the potential role of H3K9ac in the activation of blast fungus-induced genes in rice, we performed chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) experiment to correlate genome-wide H3K9ac profile with gene expression before and after M. oryzae infection. We found that H3K9ac was enriched around TSS, in line with the previous studies in rice [32]. Interestingly, for M. oryzae up-regulated genes, H3K9ac tends to be deposited both at TSS and in gene body region. However, H3K9ac at most of these genes did not change after M. oryzae inoculation, similar to the result studied in Paulownia fortunei. Furthermore, GCC-box was identified by analyzing motifs of promoter of genes with differential H3K9ac, suggesting ERF TFs may serve as potential proteins for recruiting HATs to defense-related genes for histone acetylation. Finally, from ChIP-seq and RNA-seq data, we discovered that seven WRKY genes was transcriptionally induced and carry altered H3K9ac after M. oryzae infection, which was confirmed by ChIP-qPCR and RT-qPCR. We used epigenomic and transcriptomic approaches to reveal the possible role of H3K9ac in the induction of defense-related genes and the candidate TFs mediating the deposition of H3K9ac, which have not been reported before in rice. These results lay the foundation for improvement of epigenetic mechanism during plant immune response.
2. MATERIALS AND METHODS
2.1. Plant Materials and Growth Conditions
Rice variety Oryza sativa ssp japonica cv Hwayoung was used for all experiments. Rice seeds are sterilized with 75% alcohol for 40 seconds, then 20% sodium hypochlorite for 40 minutes, rinsed with ultrapure water and soaked in water for 24 hours. After the seeds are fully absorbed, they are placed in a 32°C light incubator to accelerate germination. The germinated seeds were grown in a growth room kept at 26°C and 70% relative humidity with a 12-h light/12-h dark cycle [6].
2.2. Rice Blast Inoculation
M. oryzae JC2 isolates were used for inoculation. Four-leaf-stage rice seedlings were sprayed with a spore suspension (2.0 x 105 spores/mL) containing 0.4% gelatin. After inoculation the plants were placed inside a dark room (100% humidity at 26°C for 24 h) and then transferred to a high humidity room kept at 26°C and 12-h light/12-h dark cycle.
2.3. RNA-Seq and Data Analysis
Rice seedlings without roots were harvested at 12 hours, 24 hours, and 48 hours after inoculation and RNAs were extracted using TRNzol Universal Reagent (TIANGEN). The RNA quality for RNA-seq and RT-qPCR was examined using a Bioanalyzer 2100 (Agilent).To generate the RNA-seq libraries, the UltraTM RNA Library Preparation Kit (NEB, USA)for Illumina was used. A total of 18 libraries were sequenced on an Illumina platform and 150 bp paired-end read were generated [33]. RNA-seq data were obtained by removing contaminations and low-quality reads with Trimmomatic (version 0.33) [33]. Then, clean reads were mapped to the reference genome of rice (MSU7.0) with Hisat2 v2.2.0 [33]. The R package of DESeq2 (v4.0.3) were used to normalize counts and analyze the difference of RNA expression level with adjusted P-value<0.05 and |log2 (Fold change)| ≥1. Heat maps were generated by Heml tools. KEGG analyses were conducted using the online site (http://structuralbiology.cau.edu.cn/PlantGSEA/analysis.php).
2.4. Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) experiment was conducted as described in Hu et al. [32]. Briefly, 2g of rice seedling without roots was cross-linked by 1% (v/v) formaldehyde and used to isolate nuclei and chromatin. The chromatin was broken into 200- to 250-bp fragments by sonication and immunoprecipitated with the H3K9ac antibody (Abcam, ab10812). After extensive washing, immunoprecipitated chromatin was de-cross-linked to release the DNA for ChIP-qPCR or ChIP-seq library construction, and unimmunoprecipitated chromatin was used as input.
2.5. ChIP-seq and Data Analysis
After the ChIP-seq library was constructed, sequencing was performed on the Illumina platform. Fastp (version 0.20.1) was used to filter out low-quality reads. Clean reads were mapped to the rice genome (MSU7.0) by BWA (version 0.7.17), allowing up to two mismatches. SAMtools (version 1.11) was used to remove potential PCR duplicates. MACS2 software (version 2.2.7.1) was used to call peaks by default parameters, and the input sample was used as a control. The R package of DiffBind (v 3.2.0) was used to analyze the difference in peak level. The differential peaks were identified according to the criteria p-value<0.05 and |log2 (Fold change)| ≥1. AME (version 5.4.1) was used for motif analysis of the promoter of genes with increased H3K9ac. Differential gene expression data and H3K9ac peaks data were integrated to reveal correlations between H3K9ac and gene expression using R scripts [17].
2.6. RT-qPCR and ChIP-qPCR
cDNAs were synthesized by reverse transcription with HiScript II QRT SuperMix for qPCR (+gDNA wiper) (Vazyme) according to the manufacturer’s protocol. RT-qPCR and ChIP-qPCR were performed using SuperReal PreMix Plus (SYBR Green) (TIANGEN) on an ABI System (Stepone). The PCR reactions were performed under the following conditions: preincubation at 95 °C for 15 min, then 40 cycles of 95 °C for 10 s, and 60 °C for 1min. In addition, the expression levels were calculated using the 2−∆∆Ct method for each sample. The Actin gene was used as an internal control for RT-qPCR. The primers used in this study are listed in Table S1.
3. RESULTS
3.1. Transcriptomic Changes at Different Time Points in Response to M. oryzae Infection
More and more attention has been paid to the research and application of rice blast resistance. Up to now, about 100 rice blast resistance genes, about 500 quantitative trait loci (QTL) and 77 defense-regulator (DR) genes have been identified [34]. In order to determine transcriptomic profiles responding to M. oryzae infection, rice seedlings with four leaves were inoculated with M. oryzae JC2. Seedlings without roots were harvested at 12h, 24h and 48h after M. oryzae inoculation. RNAs were extracted for library construction and illumina sequencing. Three biological repeats were performed for RNA-seq. 28-50 million reads per sample were obtained, and the mapping rate of all the samples was more than 98% (Table S2). The results indicate that 422, 460 and 466 genes were up-regulated (termed as up-DEGs, fold change >2, FDR<0.05) at 12h, 24h and 48h, while 46, 178 and 159 genes were down-regulated (termed as down-DEGs fold change <0.5, FDR<0.05) in response to blast fungus infection (Fig. 1A, Table S3 and Table S4). This suggests that more genes are required to be activated than repressed for blast resistance. We found that over 50% of the genes up-regulated at each time point were specifically induced at that time point, including 251, 216 and 264 genes at 12h, 24h and 48h, respectively (Fig. 1B and 1D), demonstrating that those genes were transiently and successively activated during blast fungus infection. Similarly, temporary repression was also observed for the down-regulated genes (Fig. 1C and 1E). However, 59 genes were induced at three-time points (Fig. 1B, Table 1). Many of these genes have been reported to be defense-related genes, such as OsPR1, OsPR4b, chitinase (Table 1). And we also found two genes encoding hormone metabolism enzymes: Gn1a/OsCKX2 and ACS1. OsCKX2 is a cytokinin oxidase, the key enzyme for maintaining cytokinin homeostasis. Overexpression of this gene in young panicle dramatically reduces grain number and vice versa, suggesting OsCKX2 to be a key regulator of rice grain yield [35]. Up-regulation of OsCKX2 by blast infection indicates that it may be involved in balancing defense response and plant growth. ACS1 encodes an enzyme for catalyzing biosynthesis of the ethylene, which has been reported to be induced by rice blast fungus [36].
Fig. (1).

Differential gene expression after M. oryzae inoculation. (A) Volcano plot of differential gene expression at 12h, 24h and 48h after M. oryzae inoculation. Red dots indicate genes with unchanged expression, and blue dots indicate differentially expressed genes (DEGs). Overlapping of up-DEGs (B) and down-DEGs (C) at 12h, 24h and 48h after M. oryzae inoculation is presented by venn diagram. Heat maps show log2 (Fold change) of up-DEGs (D) and down-DEGs (E) at 12h, 24h and 48h respectively after M. oryzae inoculation.
Table 1.
The list of 59 genes induced at all three time points after M. oryzae infection.
|
Locus Number
(MSU) |
Gene Name |
log2FC
(12h) |
log2FC
(24h) |
log2FC
(48h) |
|---|---|---|---|---|
| LOC_Os07g03730 | OsPR1 | 1.17 | 1.66 | 1.55 |
| LOC_Os11g37960 | OsPR4b | 1.11 | 1.12 | 1.54 |
| LOC_Os08g40690 | C10122 | 1.25 | 1.59 | 3.01 |
| LOC_Os06g51050 | Cht3 | 2.72 | 4.25 | 3.70 |
| LOC_Os10g39680 | Cht8 | 2.09 | 2.46 | 2.88 |
| LOC_Os04g10060 | OsKS4 | 2.08 | 3.15 | 3.34 |
| LOC_Os11g17440 | OsRLCK322 | 2.55 | 1.40 | 2.74 |
| LOC_Os04g29580 | OsWAK37 | 1.97 | 1.38 | 2.52 |
| LOC_Os04g28780 | SDRLK-25 | 1.83 | 1.66 | 1.96 |
| LOC_Os08g09080 | GLP8-11 | 1.33 | 1.43 | 1.50 |
| LOC_Os08g13440 | GLP8-12 | 2.30 | 3.17 | 5.17 |
| LOC_Os04g09900 | OsCPS4 | 1.70 | 1.52 | 2.28 |
| LOC_Os03g13210 | poxN | 1.56 | 1.73 | 2.76 |
| LOC_Os12g30824 | OsKSL10 | 1.67 | 2.75 | 4.64 |
| LOC_Os03g51740 | ACS1 | 3.41 | 3.20 | 2.61 |
| LOC_Os09g25070 | WRKY62 | 1.41 | 2.36 | 1.06 |
| LOC_Os05g47650 | AP2/EREBP#096 | 1.12 | 1.01 | 1.10 |
| LOC_Os01g64310 | ENAC1 | 1.33 | 1.47 | 1.79 |
| LOC_Os03g10210 | HOX12 | 1.46 | 1.74 | 1.07 |
| LOC_Os09g27500 | CYP76L1 | 1.45 | 1.20 | 1.04 |
| LOC_Os10g37160 | - | 2.88 | 6.02 | 7.91 |
| LOC_Os01g10110 | Gn1a/OsCKX2 | 3.38 | 3.73 | 3.41 |
| LOC_Os01g51570 | Gns10 | 1.80 | 2.20 | 2.49 |
| LOC_Os07g35560 | - | 1.06 | 1.00 | 1.07 |
| LOC_Os07g35480 | Gns11 | 2.07 | 1.54 | 1.98 |
| LOC_Os07g48020 | POX22.3 | 2.72 | 3.02 | 2.46 |
| LOC_Os07g48050 | POX3006 | 8.45 | 6.97 | 5.87 |
| LOC_Os02g14430 | prx30 | 1.31 | 1.96 | 2.57 |
| LOC_Os05g04490 | prx70 | 1.50 | 1.42 | 2.49 |
| LOC_Os06g32990 | prx83 | 1.46 | 1.40 | 1.77 |
| LOC_Os04g36670 | OsARGOS | 1.12 | 1.70 | 1.53 |
| LOC_Os01g11860 | OsDJ-1A | 2.30 | 2.22 | 1.60 |
| LOC_Os07g23430 | OsFAD2-2 | 1.46 | 2.00 | 2.51 |
| LOC_Os07g23410 | OsFAD2-3 | 1.37 | 1.85 | 1.62 |
| LOC_Os11g24070 | OsLTP1.14 | 3.53 | 5.16 | 2.84 |
| LOC_Os06g49190 | OsLTP2.6 | 3.04 | 6.34 | 4.48 |
| LOC_Os10g38350 | OsGSTU20 | 2.92 | 3.44 | 4.64 |
| LOC_Os06g44300 | OsGL1-3 | 3.42 | 2.82 | 2.68 |
| LOC_Os07g34260 | OsPKS15 | 1.99 | 2.16 | 2.35 |
| LOC_Os01g43890 | OsSCP4 | 1.54 | 2.29 | 1.60 |
| LOC_Os04g10000 | OsSDR110C-MS2 | 1.84 | 1.76 | 2.48 |
| LOC_Os01g58280 | OsSub8 | 1.34 | 1.04 | 2.08 |
| LOC_Os03g50160 | OsUCL9 | 1.06 | 1.11 | 1.04 |
| LOC_Os08g10310 | SHR5 | 1.45 | 1.26 | 4.28 |
| LOC_Os09g37200 | - | 2.86 | 1.78 | 3.98 |
| LOC_Os12g24320 | - | 1.22 | 1.30 | 1.46 |
| LOC_Os02g48200 | - | 1.47 | 1.77 | 2.53 |
| LOC_Os02g56860 | - | 3.69 | 3.11 | 3.41 |
| LOC_Os03g48540 | - | 1.01 | 1.09 | 1.34 |
| LOC_Os04g39300 | - | 1.29 | 1.22 | 1.31 |
| LOC_Os04g39380 | - | 2.61 | 1.56 | 2.32 |
| LOC_Os06g11450 | - | 1.15 | 1.10 | 1.05 |
| LOC_Os06g21210 | - | 1.75 | 1.20 | 1.06 |
| LOC_Os10g42040 | - | 2.70 | 2.75 | 2.67 |
| LOC_Os11g05800 | - | 1.13 | 1.17 | 1.08 |
| LOC_Os02g50470 | - | 1.29 | 1.38 | 1.21 |
| LOC_Os04g49950 | - | 4.13 | 1.77 | 1.30 |
| LOC_Os11g28530 | - | 1.28 | 1.21 | 1.79 |
| LOC_Os06g39120 | - | 1.61 | 1.72 | 2.63 |
To understand the induced genes activity at each time point, we categorized the induced genes into four classes according to the fold change of induction (I: 1≤log2FC≤2; II: 2<log2FC≤3; III: 3<log2FC≤4; IV: log2FC>4). The proportion of class I genes at 12h is higher than that at 24h and 48h, whereas the proportion of class IV genes at 12h is much lower than at 24h and 48h (Fig. S1A). This suggests that genes with strong induction is induced later after M. oryzae infection. We also categorized them into five classes according to gene transcription levels before induction (I: 0≤normalized count≤10; II:10<normalized count≤100; III:100< normalized count≤300; IV: 300< normalized count≤500; V: normalized count>500). The percentage of genes in each class in relation to all the induced genes at each time point was calculated. The results showed that the proportions of class I and V genes at 24h (18% and 15%) and 48h (21% and 12%) were higher than those at 12h (8% and 10%) (Fig. S1B), which demonstrates that the genes with extremely lower or higher expression levels are prone to be induced in the later stage after infection while more moderately expressed genes were induced in the early stage.
3.2. Biosynthesis of Secondary Metabolites are Activated by Blast Infection
Synthesis of some secondary metabolites, such as “Phenylpropanoid biosynthesis”, “Phenylalanine metabolism” and “Phenylalanine, tyrosine and tryptophan biosynthesis”, are mainly involved in the synthesis of secondary antibacterial metabolites (serotonin, lignin and antitoxin, etc.), which play an important role in the process of rice resisting pathogenic infection [37, 38]. Considering that many genes were induced specifically at a certain time point, we would like to learn whether the induced genes could be involved in specific biological pathways. The blast fungus-induced genes at each time point were used for KEGG analysis. We found that genes involved in metabolic pathways and biosynthesis of secondary metabolites were enriched at all the three-time points. Secondary metabolites such as terpenoids and Phenylalanine-derived phytoalexins, coined as Phytoalexins, have indeed been reported to contribute to plant innate immunity and their de novo synthesis is triggered by pathogen infection [39-41]. Our analysis revealed that Phenylalanine metabolism and phenylpropanoid biosynthesis genes were induced at 12h while diterpenoid metabolism genes were induced at 24h and 48h (Fig. 2A). These results demonstrate that blast induces synthesis of different secondary metabolites successively in rice. To further confirm the observation, we use cytoscape to construct the network of pathways. MAPK signaling pathway was identified in addition to diterpenoid biosynthesis and phenylpropanoid biosynthesis (Fig. 2B). No connection was established among the three pathways. However, genes related to synthesizing Phytoalexins such as OsCPS2, OsCPS4, OsKS4 and OsKSL10 were found in diterpenoid biosynthesis pathway [42-44]. And Lignin biosynthesis genes exemplified by CYP84A5, 4CL5, poxN and poxA were included in phenylpropanoid biosynthesis pathway [45-47].
Fig. (2).

Pathway enrichment of up-DEGs. (A) The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of up-DEGs at 12h, 24h and 48h after M. oryzae inoculation. (B) Pathway network diagram of up-regulated DEGs with P-value < 0.05 were visualized by Cytoscape.
3.3. Genome-wide Changes of H3K9ac in Response to Blast Fungus Inoculation
In order to explore the role of H3K9ac in the response of blast infection, we used the same tissues as RNA-seq to conduct ChIP-seq experiment at 48h after blast infection. Two biological repeats were performed. Over 30 million reads per sample were obtained and the mapping rate of all the samples was more than 83% (Table S5). Sharp peak of H3K9ac enrichment at TSS was observed by metaplots analysis (Fig. 3A). The Overall H3K9ac level was slight increased by rice blast fungus infection compared to the mock (Fig. 3A). Integration of ChIP-seq data and RNA-seq data revealed that no correlation was found between H3K9ac change and gene induction, although for some induced genes H3K9ac level was increased (Fig. 3B). However, metaplots analysis indicated that H3K9ac levels at up-DEGs at three-time points were dramatically increased compared to the change of overall H3K9ac level (Fig. 3C). Moreover, we found that H3K9ac enrichment peaks at TSS of up-DEGs at three-time points (Fig. 3C), especially at 12h, were apparently less sharp than genome-wide, while H3K9ac enrichment peaks of down-DEGs were not (Fig. S2). This suggests that H3K9ac at up-DEGs is more likely to be enriched at TSS as well as in the gene body region. And H3K9ac level in both regions was increased in the process of gene induction after blast infection.
Fig. (3).

H3K9ac alteration responding to M. oryzae infection. (A) Metaplots of genome-wide H3K9ac profile in genic regions before (H2O) and after (JC2) M. oryzae infection. (B) Correlation analysis of gene expression and H3K9ac enrichment responding to M. oryzae infection at 12h, 24h and 48h. (C) Metaplots of H3K9ac profiles of up-DEGs at 12h, 24 and 48h after inoculation.
Histone acetylation marks may represent the permissive state of chromatin for defense priming. To understand how many genes are primed before blast induction, we investigated the proportions of the induced genes carrying H3K9ac in different classes categorized according to gene transcription level before induction at a three-time point. 79%, 72%, and 64% of genes transcriptionally induced at 12h, 24h and 48h possess H3K9ac (Table S6). With the increase of gene transcription level, the proportion of genes marked with H3K9ac rises, which coincide with the active role of H3K9ac in gene transcription. Specifically, we found that nearly half of class I genes which were expressed in an extremely low level (47% at 12h, 55% at 24h and 54% at 48h) possess H3K9ac (Table S6). To further learn whether the permissive state conferred by H3K9ac is related to gene induction strength, the proportions of the induced genes carrying H3K9ac in different classes categorized according to induction fold change were calculated. Interestingly, the result showed that the percentage of the genes marked with H3K9ac was decreased with the growth of fold change (Table S7), suggesting that the permissive state of M. oryzae induced genes is not an essential prerequisite for strong induction.
3.4. GCC Box was Identified in Promoters of the Genes with Increased H3K9ac
It has been evidenced that HATs can be recruited to target genes by TFs that bind specific DNA sequences or motifs. The ABRE motif bound by AREBs (bZIP TFs) has been identified from Populus trichocarpa by analyzing promoters of genes with differential H3K9ac levels responding to drought stress [17]. Molecular and genetic evidences indicate that GCN5 is responsible for H3K9ac at three NAC genes [17], which carry the ABRE motif in the promoter, by interacting with AREB1. Similarly, in order to identify TFs that may guide H3K9ac at blast-responding genes, we scanned the promoter of genes with increased H3K9ac after blast infection to find the most enriched DNA motifs. The result showed that many ERF family protein-binding motifs were enriched with high -log P-value (>20), including ERF1, DEAR3, DREB2C, RAP2.6, RRTF1_3ARY, all of which contain GCC-box (GCCGCC) (Fig. 4A and 4B). Rice genome contains 139 ERF family genes divided into 15 groups [48]. ERF1, DEAR3, DREB2C, RAP2.6 and RRTF1 belong to groups IX, III, IV and X, respectively. Forty-nine ERF genes covering these groups were found to be induced by responding to blast fungus infection disclosed by RNA-seq data (Fig. S3). This demonstrates that ERF TFs might mediate histone acetylation at blast fungus induced genes, which is consistent with activation of the biosynthesis of ethylene by a blast fungus infection.
Fig. (4).
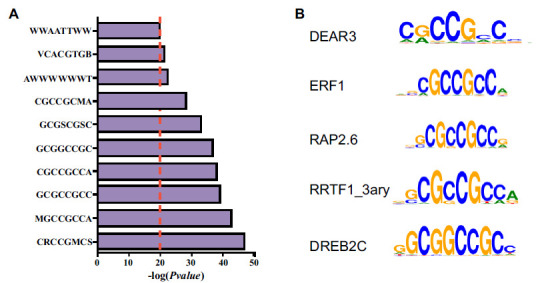
Identification of transcription factor binding motifs related to H3K9ac. (A) Analysis of motif enrichment of the promoters with increased H3K9ac [log2 (Fold change)≥1]. B represents C or G or T, V represents A or C or G, W represents A or T, S represents C or G, R represents A or G, and M represents A or C. (B) The top five motifs in the promoters of genes with increased H3K9ac [log2 (Fold change) ≥1].
3.5. Many WRKY Genes are Transcriptionally Induced by M. oryzae Infection
Rice experience four stages after infected by M. oryzae to activating its autoimmune response: Perception, Signal transduction, Transcription activation and Defense response [49]. WRKY genes encode a large family of TFs which play a crucial role in immune signal transduction to induce defense genes. Many of WRKY genes have been characterized as key regulators of disease resistance in rice [50]. The RNA-seq data revealed that 42 WRKY genes were up-regulated and 5 WRKY genes were down-regulated responding to blast infection (Fig. 5A). When we chose seven WRKY genes for confirmation of the induction by qRT-PCR, we found that they were indeed induced by blast fungus infection (Fig. S5A). However, the induced time point for some of these genes does not conform well to the RNA-seq data. For example, OsWRKY70 was induced at 48h in the RNA-seq data while up-regulation was detected at 12h by qRT-PCR (Fig. S5A). OsWRKY50 was up-regulated at 12h in the RNA-seq data while dramatic induction was observed until 24h by qRT-PCR (Fig. S5A).
Fig. (5).

Changes of expression and H3K9ac levels of WRKY genes. (A) Heat map of log2(fold change) of WRKY genes at 12h, 24h and 48h after M. oryzae inoculation. The data were extracted from our RNA-seq data. (B) Genome browser screen shots of H3K9ac at seven WRKY genes before (H2O) and after (JC2) M. oryzae infection.
3.6. H3K9ac is Involved in the Induction of Seven WRKY Genes
Involvement of histone acetylation in the regulation of WRKY genes has been reported in chickpea and pear [51, 52]. Our ChIP-seq data revealed that H3K9ac at seven of 42 induced WRKY genes (OsWRKY28, OsWRKY40, OsWRKY45, OsWRKY50, OsWRKY62, OsWRKY70, OsWRKY76) were increased after rice blast fungus infection (Fig. 5B). Four of the seven WRKY genes (OsWRKY28, OsWRKY45, OsWRKY62, OsWRKY76) have been reported to be essential regulators of rice immune response [34, 53-55]. Interestingly, we found that H3K9ac was enriched both at TSS and in gene body region of the seven WRKY genes (Fig. 5B), consistent with the above genome-wide observation (Fig. 3C). However, H3K9ac was increased in different regions of seven WRKY genes. For OsWRKY40 and OsWRKY50, H3K9ac was elevated only in the gene body region, whereas for OsWRKY70, H3K9ac was increased only around TSS. Both TSS and gene body-enriched H3K9ac were increased for OsWRKY28 and OsWRKY76. The increment of H3K9ac enrichment in the promoter of OsWRKY45 and OsWRKY62 was also observed in addition to TSS and gene body region (Fig. 5B). These results were testified by ChIP-qPCR (Fig. S5B). Furthermore, H3K9ac enrichment in the gene body region at the seven WRKY genes appears to have a bias towards exons (Fig. 5B). To investigate if these WRKY genes could be potential targets of ERF TFs, we searched the GCC-box motif in the promoter of these genes. Three and five GCC-box motifs were found in the promoter of OsWRKY45 and OsWRKY76, respectively (Fig. S4), suggesting the possible involvement of ERF TFs in mediating epigenetic regulation of these two genes.
4. DISCUSSION
It has long been accepted that histone acetylation positively correlates with gene activation, possibly by neutralizing positive charges of histone to impair the interaction between histones and negative-charged DNA. Disruption of the interaction between histones and DNA gives rise to a permissive state of chromatin prepared for transcription activation. In this study, we could not find de novo histone acetylation at any rice blast fungus induced gene, which was defined as the gene with no H3K9ac peak before induction and marked by H3K9ac after induction. This suggests that activation of defense response genes does not require the transition of chromatin state coupled by H3K9ac. However, the possibility could not be ruled out that in certain cell types de novo, H3K9ac deposition does occur due to mixed tissue sample used for our ChIP experiment, which may be validated by the future application of the single-cell ChIP-seq in plant sample. Furthermore, 20-30% of induced genes did not possess the H3K9ac mark both before and after the induction indicating that H3K9ac is not necessary for activating these genes (Table S6). Maybe other active histone marks such as H3K4me3 are involved in the induction of these genes. Next, we would like to know if histone acetylation is related to the strength of induction. Unexpectedly, we found that the more strongly genes are induced, the lower proportion of genes harbor H3K9ac (Table S7), excluding the correlation of histone acetylation with the strong gene induction. Less strongly induced genes enriching H3K9ac are hypothesized to result from the extremely low expression of these genes before induction which is observed from the heat map in Fig. S1B.
H3K9ac is mainly enriched downstream TSS at most genes, which is conserved in animals and plants [14, 32]. The location of H3K9ac determines that it may play a role in transcription initiation. The SAGA complex containing GCN5 and the TAFIID complex containing TAF1 are both required for the recruitment of Pol II to core promoter, further supporting the view [56, 57]. It has also been proved that H3K9ac acts as an anchor for the binding of super elongation complex (SEC), through which RNA polymerase II (pol II) pause release that is important for the switch from transcription initiation to elongation takes place [58]. Our ChIP-seq data also showed enrichment of H3K9ac surrounding TSS in genome-wide (Fig. 3A). Nevertheless, our data revealed that H3K9ac at blast up-DEGs is inclined to occupy both TSS and gene body region (Fig. 3C). Genome browser screenshots of H3K9ac at seven WRKY genes exhibit a similar inclination (Fig. 5B). Intriguingly, in the gene body region of these WRKY genes, H3K9ac is more concentrated in exons than introns. It has been postulated that the distribution bias of histone marks towards exons might be involved in co-transcriptional splicing by slowing down pol II [9]. It is of great interest to verify if H3K9ac might play roles in modulating both transcription initiation and elongation as well as splicing. However, we found that H3K9ac in distinct regions (promoters, TSSs or gene body) was increased for the different WRKY genes responding to rice blast fungus infection, suggesting that this mark maybe play divergent roles for the induction of specific genes.
Interaction between HATs and various TFs that bind specific cis-element is an important way for the recruitment of HATs to target loci. Thus, the identification of transcription factor binding motifs in the promoter of genes with dynamic change of histone acetylation is probably a useful method to predict HAT-interacting TFs. A bZIP TF AREB1 has been successfully identified in Populus to associate with GCN5 by obtaining ABRE motif enriched in drought-responsive genes with altered H3K9ac level [17]. By using a similar method, we identified the GCC-box as the most enriched motif in the promoter of genes with increased H3K9ac after blast fungus inoculation. Therefore, GCC-box-binding ERF family TFs could be potential candidates for recruiting HATs to defense-related genes. The association of ERF TF (CBF1) with GCN5 and ADA2 has been proved in Arabidopsis [15]. Functional studies of ERF TFs carried out in several plant species reveal the importance of ERF TFs in the regulation of defense-related gene expression [59]. Our RNA-seq data also showed that more than one-third of ERF TFs are up-regulated by M. oryzae in rice (Fig. S3). Future work for identifying ERF TFs and HATs responsible for coupling histone acetylation of defense-related genes is required. We narrow the ERF gene list down from 139 to 49 genes, since they are induced by defense response. However, as some ERF genes could also be post-transcriptionally regulated by defense signaling [59], these genes can not be removed from the list. In addition, only two of seven WRKY genes characterized in this study have GCC-box in the promoter, suggesting additional TFs are needed for deposition of histone acetylation. Since the complex containing TGA and HAC1/5 is also essential for plant immunity and PR induction in Arabidopsis [21], their homologs in rice should be taken into account in addition to ERF proteins and GCN5.
CONCLUSION
In this study, we performed RNA-seq to reveal blast-induced genes, and the result showed that 422, 460 and 466 genes were up-regulated at 12h, 24h and 48h after M. oryzae inoculation, many of which have been functionally studied for the role in rice immune response. ChIP-seq data demonstrated that most of these up-regulated genes carried H3K9ac before induction, but H3K9ac at a minority of them was increased after induction, including seven WRKY genes, four of which are the essential regulators of rice immune response. This suggests that H3K9ac may serve as a priming chromatin state for the induction of most defense-related genes. However, whether H3K9ac is necessary for the induction requires further confirmation by studying the function of histone acetyltransferases. In addition, H3K9ac at blast-induced genes preferred to be enriched in both TSS and gene body regions, echoing that the priming chromatin state is possibly important for transcriptional initiation and elongation in the process of induced expression.
By searching DNA-binding motifs of transcription factors in the promoter of genes with increased H3K9ac after M. oryzae infection, we found that ERF family protein-binding motifs were enriched with high -log P-value (>20), including ERF1, DEAR3, DREB2C, RAP2.6, RRTF1_ 3ARY, all of which contain GCC-box (GCCGCC). This demonstrates that ERF proteins might be responsible for recruiting HATs to acetylate histone at many defense-related genes. Both functional studies of ERF genes on the induction of defense-related genes and protein-protein interaction between ERFs and HATs are necessitated in the future to confirm the candidate ERF proteins.
ACKNOWLEDGEMENTS
Declared none.
AUTHORS' CONTRIBUTIONS
Xu Y: Investigation; Methodology; Data curation; Miao YX: Data curation; Formal analysis; Tian XJ: Data curation; Wang QH: Formal analysis; Hu YF: Conceptualization; Funding acquisition; Writing - original draft; Writing - review &editing. Luo Q: Writing - review & editing; Supervision.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The authors confirm that the data supporting the findings of this study are available within the article.
FUNDING
This work was supported by The Scientific Research Program key Project of Hubei Provincial Department of Education (D20204301).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
REFERENCES
- 1.Soleymani S., Hadi A., Asgari F., Haghighipour N., Bolhassani A. Combination of mechanical and chemical methods improves gene delivery in cell-based HIV vaccines. Curr. Drug Deliv. 2019;16(9):818–828. doi: 10.2174/1567201816666190923152914. [DOI] [PubMed] [Google Scholar]
- 2.Bolhassani A., Shahbazi S., Agi E., Haghighipour N., Hadi A., Asgari F. Modified DCs and MSCs with HPV E7 antigen and small Hsps: Which one is the most potent strategy for eradication of tumors? Mol. Immunol. 2019;108:102–110. doi: 10.1016/j.molimm.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Hadi A., Rastgoo A., Haghighipour N., Bolhassani A., Asgari F., Soleymani S. Enhanced gene delivery in tumor cells using chemical carriers and mechanical loadings. PLoS One. 2018;13(12):e0209199. doi: 10.1371/journal.pone.0209199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadi A., Rastgoo A., Eskandarian M., Haghighipour N., Bolhassani A. Development of delivery systems enhances the potency of cell-based HIV-1 therapeutic vaccine candidates. J. Immunol. Res. 2021;2021:5538348. doi: 10.1155/2021/5538348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu H., Wei T., Lou H., Shu X., Chen Q. A critical review on communication mechanism within plant-endophytic fungi interactions to cope with biotic and abiotic stresses. J. Fungi (Basel) 2021;7(9):719. doi: 10.3390/jof7090719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., Zhang H., Yang R., Zeng Q., Han G., Du Y., Yang J., Yang G., Luo Q. Identification of miRNAs contributing to the broad-spectrum and durable blast resistance in the yunnan local rice germplasm. Front. Plant Sci. 2021;12:749919. doi: 10.3389/fpls.2021.749919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abd. Kharim M.N. Wayayok, A.; Abdullah, A.F.; Shariff, A.R.M. Effect of variable rate application on rice leaves burn and chlorosis in system of rice intensification. Malaysian J. Sustain. Agric. 2020;4(2):66–70. doi: 10.26480/mjsa.02.2020.66.70. [DOI] [Google Scholar]
- 8.Nicholas H. 2017. An economic analysis of anthropogenic climate change on rice production in malaysia. Masters Thesis, Universiti Utara Malaysia: Kedah July. [Google Scholar]
- 9.Dhami P., Saffrey P., Bruce A.W., Dillon S.C., Chiang K., Bonhoure N., Koch C.M., Bye J., James K., Foad N.S., Ellis P., Watkins N.A., Ouwehand W.H., Langford C., Andrews R.M., Dunham I., Vetrie D. Complex exon-intron marking by histone modifications is not determined solely by nucleosome distribution. PLoS One. 2010;5(8):e12339. doi: 10.1371/journal.pone.0012339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuda K., Somssich I.E. Transcriptional networks in plant immunity. New Phytol. 2015;206(3):932–947. doi: 10.1111/nph.13286. [DOI] [PubMed] [Google Scholar]
- 11.Spitz F., Furlong E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012;13(9):613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 12.Dawson M.A., Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Kumar V., Thakur J.K., Prasad M. Histone acetylation dynamics regulating plant development and stress responses. Cell. Mol. Life Sci. 2021;78(10):4467–4486. doi: 10.1007/s00018-021-03794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.Y., Peng W., Zhang M.Q., Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stockinger E.J., Mao Y., Regier M.K., Triezenberg S.J., Thomashow M.F. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 2001;29(7):1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong L., Qiu X., Kang J., Wang Y., Chen H., Huang J., Qiu M., Zhao Y., Kong G., Ma Z., Wang Y., Ye W., Dong S., Ma W., Wang Y. A phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr. Biol. 2017;27(7):981–991. doi: 10.1016/j.cub.2017.02.044. [DOI] [PubMed] [Google Scholar]
- 17.Li S., Lin Y.J., Wang P., Zhang B., Li M., Chen S., Shi R., Tunlaya-Anukit S., Liu X., Wang Z., Dai X., Yu J., Zhou C., Liu B., Wang J.P., Chiang V.L., Li W. The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell. 2019;31(3):663–686. doi: 10.1105/tpc.18.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou S., Jiang W., Long F., Cheng S., Yang W., Zhao Y., Zhou D-X. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell. 2017;29(5):1088–1104. doi: 10.1105/tpc.16.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S., Piquerez S.J.M., Ramirez-Prado J.S., Mastorakis E., Veluchamy A., Latrasse D., Manza-Mianza D., Brik-Chaouche R., Huang Y., Rodriguez-Granados N.Y., Concia L., Blein T., Citerne S., Bendahmane A., Bergounioux C., Crespi M., Mahfouz M.M., Raynaud C., Hirt H., Ntoukakis V., Benhamed M. GCN5 modulates salicylic acid homeostasis by regulating H3K14ac levels at the 5′ and 3′ ends of its target genes. Nucleic Acids Res. 2020;48(11):5953–5966. doi: 10.1093/nar/gkaa369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T., Xing J., Liu X., Yao Y., Hu Z., Peng H., Xin M., Zhou D.X., Zhang Y., Ni Z. GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exp. Bot. 2018;69(12):2911–2922. doi: 10.1093/jxb/ery077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin H., Choi S.M., Kang M.J., Yun S.H., Kwon D.J., Noh Y.S., Noh B. Salicylic acid-induced transcriptional reprogramming by the HAC-NPR1-TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res. 2018;46(22):11712–11725. doi: 10.1093/nar/gky847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi S.M., Song H.R., Han S.K., Han M., Kim C.Y., Park J., Lee Y.H., Jeon J.S., Noh Y.S., Noh B. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012;71(1):135–146. doi: 10.1111/j.1365-313X.2012.04977.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim K-C., Lai Z., Fan B., Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20(9):2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Hu Q., Wu Z., Wang H., Han S., Jin Y., Zhou J., Zhang Z., Jiang J., Shen Y., Shi H., Yang W. Histone Deacetylase 6 represses pathogen defence responses in Arabidopsis thaliana. Plant Cell Environ. 2017;40(12):2972–2986. doi: 10.1111/pce.13047. [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Zhi P., Wang X., Fan Q., Chang C. Wheat WD40-repeat protein TaHOS15 functions in a histone deacetylase complex to fine-tune defense responses to Blumeria graminis f.sp. tritici. J. Exp. Bot. 2019;70(1):255–268. doi: 10.1093/jxb/ery330. [DOI] [PubMed] [Google Scholar]
- 26.Zhi P., Kong L., Liu J., Zhang X., Wang X., Li H., Sun M., Li Y., Chang C. Histone deacetylase TaHDT701 functions in TaHDA6-TaHOS15 complex to regulate wheat defense responses to Blumeria graminis f.sp. tritici. Int. J. Mol. Sci. 2020;21(7):E2640. doi: 10.3390/ijms21072640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding B., Bellizzi M.R., Ning Y., Meyers B.C., Wang G.L. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell. 2012;24(9):3783–3794. doi: 10.1105/tpc.112.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yumni R.M.I., Karim M.F., Midin M.R. Genome size determination of cucumber (Cucumis sativus), honeydew (Cucumis melo inodorus) and rock melon (Cucumis melo cantalupensis) via flow cytometry. Sci. Heritage J. 2021;5(1):14–16. doi: 10.26480/gws.01.2021.14.16. [DOI] [Google Scholar]
- 29.Zhang C., Liu X., Liu C., Luo X. Characterization of the Complete Mitochondrial Genome of Acanthacorydalis fruhstorferi van der Weele (Megaloptera: Corydalidae). J. Kans. Entomol. Soc. 2021;93(4):267. doi: 10.2317/0022-8567-93.4.267. [DOI] [Google Scholar]
- 30.Nwankwoala H.O., Harry M.T., Warmate T. Assessing aquifer vulnerability and contaminant plume at artisanal refining sites in parts of okrika and ogu-bolo local government areas, rivers state, Nigeria. Water Conservation and Management. 2020;4(2):68–72. [Google Scholar]
- 31.Yan L., Zhai X., Zhao Z., Fan G. Whole-genome landscape of H3K4me3, H3K36me3 and H3K9ac and their association with gene expression during Paulownia witches' broom disease infection and recovery processes. 3 Biotech. 2020;10(8):336. doi: 10.1007/s13205-020-02331-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y., Lai Y., Chen X., Zhou D.X., Zhao Y. Distribution pattern of histone marks potentially determines their roles in transcription and RNA processing in rice. J. Plant Physiol. 2020;249:153167. doi: 10.1016/j.jplph.2020.153167. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y., Tan F., Zhao Y., Zhou S., Chen X., Hu Y., Zhou D.X. A chromodomain-helicase-DNA-binding factor functions in chromatin modification and gene regulation. Plant Physiol. 2020;183(3):1035–1046. doi: 10.1104/pp.20.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W., Chern M., Yin J., Wang J., Chen X. Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 2019;50:114–120. doi: 10.1016/j.pbi.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 35.Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. Cytokinin oxidase regulates rice grain production. Science. 2005;309(5735):741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 36.Iwai T., Miyasaka A., Seo S., Ohashi Y. Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 2006;142(3):1202–1215. doi: 10.1104/pp.106.085258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W., Gao S., Zhou X., Chellappan P., Chen Z., Zhou X., Zhang X., Fromuth N., Coutino G., Coffey M., Jin H. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 2011;75(1-2):93–105. doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M., Zhang S., Hu J., Sun W., Padilla J., He Y., Li Y., Yin Z., Liu X., Wang W., Shen D., Li D., Zhang H., Zheng X., Cui Z., Wang G.L., Wang P., Zhou B., Zhang Z. Phosphorylation-guarded light-harvesting complex II contributes to broad-spectrum blast resistance in rice. Proc. Natl. Acad. Sci. USA. 2019;116(35):17572–17577. doi: 10.1073/pnas.1905123116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piasecka A., Jedrzejczak-Rey N., Bednarek P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 2015;206(3):948–964. doi: 10.1111/nph.13325. [DOI] [PubMed] [Google Scholar]
- 40.Ji X., Hou C., Shi M., Yan Y., Liu Y. An Insight into the research concerning panax ginseng c. a. meyer polysaccharides: A review. Food Rev. Int. 2020;2020:1771363. doi: 10.1080/87559129.2020.1771363. [DOI] [Google Scholar]
- 41.Ji X., Peng B., Ding H., Cui B., Nie H., Yan Y. Purification, structure and biological activity of pumpkin polysaccharides: A review. Food Rev. Int. 2021;2021:1904973. doi: 10.1080/87559129.2021.1904973. [DOI] [Google Scholar]
- 42.Peters R.J. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry. 2006;67(21):2307–2317. doi: 10.1186/s12870-019-2156-5. [DOI] [PubMed] [Google Scholar]
- 43.Toyomasu T. Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci. Biotechnol. Biochem. 2008;72(5):1168–1175. doi: 10.1007/s00425-021-03625-0. [DOI] [PubMed] [Google Scholar]
- 44.Toyomasu T., Usui M., Sugawara C., Otomo K., Hirose Y., Miyao A., Hirochika H., Okada K., Shimizu T., Koga J., Hasegawa M., Chuba M., Kawana Y., Kuroda M., Minami E., Mitsuhashi W., Yamane H. Reverse-genetic approach to verify physiological roles of rice phytoalexins: Characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiol. Plant. 2014;150(1):55–62. doi: 10.1111/ppl.12066. [DOI] [PubMed] [Google Scholar]
- 45.Takeda Y., Koshiba T., Tobimatsu Y., Suzuki S., Murakami S., Yamamura M., Rahman M.M., Takano T., Hattori T., Sakamoto M., Umezawa T. Regulation of Coniferaldehyde 5-Hydroxylase expression to modulate cell wall lignin structure in rice. Planta. 2017;246(2):337–349. doi: 10.1007/s00425-017-2692-x. [DOI] [PubMed] [Google Scholar]
- 46.Chen H.C., Song J., Wang J.P., Lin Y.C., Ducoste J., Shuford C.M., Liu J., Li Q., Shi R., Nepomuceno A., Isik F., Muddiman D.C., Williams C., Sederoff R.R., Chiang V.L. Systems biology of lignin biosynthesis in Populus trichocarpa: Heteromeric 4-coumaric acid:coenzyme A ligase protein complex formation, regulation, and numerical modeling. Plant Cell. 2014;26(3):876–893. doi: 10.1105/tpc.113.119685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hiroyuki Ito; Hiraga, S.; Tsugawa, H.; Matsui, H.; Honma, M.; Otsuki, Y.; Murakami, T.; Ohashi, Y., Xylem-specific expression of wound-inducible rice peroxidase genes in transgenic plants. Plant Sci. 2000;155:85–100. doi: 10.1111/pbi.12951. [DOI] [PubMed] [Google Scholar]
- 48.Sharoni A.M., Nuruzzaman M., Satoh K., Shimizu T., Kondoh H., Sasaya T., Choi I.R., Omura T., Kikuchi S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011;52(2):344–360. doi: 10.1093/pcp/pcq196. [DOI] [PubMed] [Google Scholar]
- 49.Delteil A., Zhang J., Lessard P., Morel J-B. Potential candidate genes for improving rice disease resistance. Rice (N. Y.) 2010;3(1):56–71. doi: 10.1007/s12284-009-9035-x. [DOI] [Google Scholar]
- 50.Viana V.E., Busanello C., da Maia L.C., Pegoraro C., Costa de, Oliveira A. Activation of rice WRKY transcription factors: An army of stress fighting soldiers? Curr. Opin. Plant Biol. 2018;45(Pt B):268–275. doi: 10.1016/j.pbi.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Chakraborty J., Ghosh P., Sen S., Das S. Epigenetic and transcriptional control of chickpea WRKY40 promoter activity under Fusarium stress and its heterologous expression in Arabidopsis leads to enhanced resistance against bacterial pathogen. Plant Sci. 2018;276:250–267. doi: 10.1016/j.plantsci.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 52.Li X., Guo W., Li J., Yue P., Bu H., Jiang J., Liu W., Xu Y., Yuan H., Li T., Wang A. Histone acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in pear fruit. Plant Physiol. 2020;182(4):2035–2046. doi: 10.1104/pp.20.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chujo T., Miyamoto K., Shimogawa T., Shimizu T., Otake Y., Yokotani N., Nishizawa Y., Shibuya N., Nojiri H., Yamane H., Minami E., Okada K. OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 2013;82(1-2):23–37. doi: 10.1007/s11103-013-0032-5. [DOI] [PubMed] [Google Scholar]
- 54.Inoue H., Hayashi N., Matsushita A., Xinqiong L., Nakayama A., Sugano S., Jiang C-J., Takatsuji H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. USA. 2013;110(23):9577–9582. doi: 10.1073/pnas.1222155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokotani N., Sato Y., Tanabe S., Chujo T., Shimizu T., Okada K., Yamane H., Shimono M., Sugano S., Takatsuji H., Kaku H., Minami E., Nishizawa Y. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013;64(16):5085–5097. doi: 10.1093/jxb/ert298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonnet J., Wang C.Y., Baptista T., Vincent S.D., Hsiao W.C., Stierle M., Kao C.F., Tora L., Devys D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014;28(18):1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhuiyan T., Timmers H.T.M. Promoter Recognition: Putting TFIID on the Spot. Trends Cell Biol. 2019;29(9):752–763. doi: 10.1016/j.tcb.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 58.Gates L.A., Shi J., Rohira A.D., Feng Q., Zhu B., Bedford M.T., Sagum C.A., Jung S.Y., Qin J., Tsai M.J., Tsai S.Y., Li W., Foulds C.E., O’Malley B.W. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J. Biol. Chem. 2017;292(35):14456–14472. doi: 10.1074/jbc.M117.802074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang P.Y., Catinot J., Zimmerli L. Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 2016;67(5):1231–1241. doi: 10.1093/jxb/erv518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.


