Abstract
Background: Next-generation sequencing (NGS) technologies are being continuously used for high-throughput sequencing data generation that requires easy-to-use GUI-based data analysis software. These kinds of software could be used in-parallel with sequencing for the automatic data analysis. At present, very few software are available for use and most of them are commercial, thus creating a gap between data generation and data analysis.
Methods: GAAP is developed on the NodeJS platform that uses HTML, JavaScript as the front-end for communication with users. We have implemented FastQC and trimmomatic tool for quality checking and control. Velvet and Prodigal are integrated for genome assembly and gene prediction. The annotation will be done with the help of remote NCBI Blast and IPR-Scan. In the back- end, we have used PERL and JavaScript for the processing of data. To evaluate the performance of GAAP, we have assembled a viral (SRR11621811), bacterial (SRR17153353) and human genome (SRR16845439).
Results: We have used GAAP software to assemble, and annotate a COVID-19 genome on a desktop computer that resulted in a single contig of 27994bp with 99.57% reference genome coverage. This assembly predicted 11 genes, of which 10 were annotated using annotation module of GAAP. We have also assembled a bacterial and human genome 138 and 194281 contigs with N50 value 100399 and 610, respectively.
Conclusion: In this study, we have developed freely available, platform-independent genome assembly and annotation (GAAP) software (www.deepaklab.com/gaap). The software itself acts as a complete data analysis package with quality check, quality control, de-novo genome assembly, gene prediction and annotation (Blast, PFAM, GO-Term, pathway and enzyme mapping) modules.
Keywords: NGS, software, GUI, genome assembly, gene prediction, annotation
1. INTRODUCTION
High-throughput next-generation sequencing technologies result in the exponential growth of the whole genome and transcriptome level projects [1-3]. Continuous decline in the cost of next-generation sequencing and emergence of highly efficient sequencing technologies transformed data analysis from single gene/protein to genome/pan-genome level [4-6]. In the past, numerous tools have been developed varying from genome assembly (de-novo and reference), annotation, variant calling, etc. to analyse the high-throughput sequencing data [7, 8]. Most of these tools are developed for Linux-based command-line systems that need bioinformatics expertise to execute them.
To overcome this problem, few platform independent tools such as CLC-GWB (https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench/), Blast2GO (https://www.blast2go.com/), VAGUE (https://bioinformaticshome.com/tools/wga/descriptions/VAG- UE.html), Strand NGS (https://www.strand-ngs.com/), etc. have been developed, but most of them required a licence. For example, Blast2GO, a commercial software has gained popularity due to its graphical-user interface, but it can only be used for genome or transcriptome annotation purposes [9, 10]. Similarly, CLC-GWB, OmicsBox, and Strand NGS have been developed for genome/transcriptome analysis, but the major limitation is that they are also not free for academic use. In 2013, a graphical user interface of velvet i.e. VAGUE has been developed for the de-novo genome assembly purpose [11]. But, the major pitfall is that it can only be run on a Linux-based system and can only be used for assembly, not for the annotation purpose.
Therefore, the present situation demands a cross-platform, GUI-based, comprehensive genome assembly and annotation package that could be used without any restrictions. Here, we described “GAAP”, a fast, automated, memory efficient, open source software package that could be freely used for genome assembly and annotation purposes without the requirement of installation of any kind of dependencies.
2. METHODS
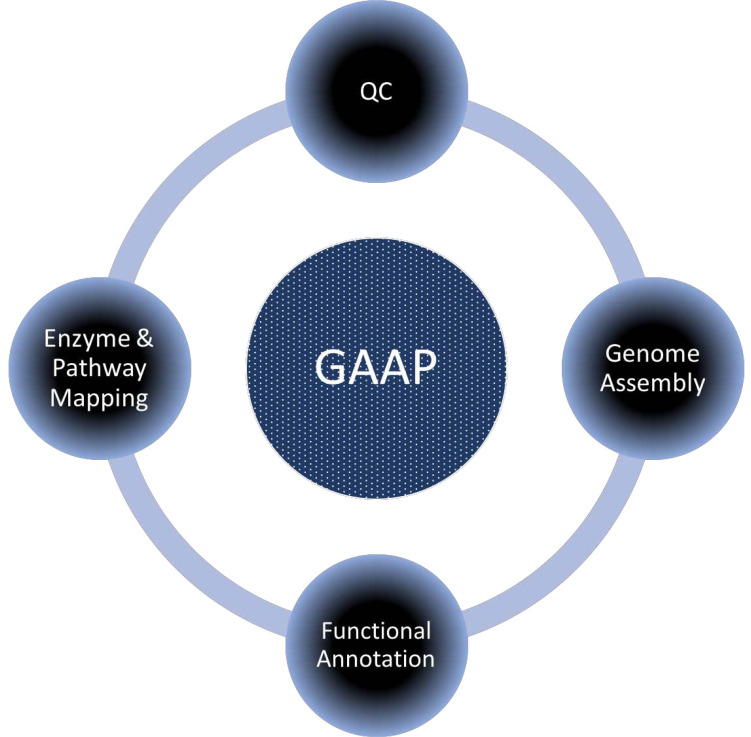
GAAP basically has four different modules that are required for complete genome assembly and annotation process (Fig. 1). Here, we briefly describe each module.
Fig. (1).
Depicting different module of the GAAP software.
2.1. Data Pre-processing
In this section, we have provided the facility to download fastq files from NCBI-SRA by providing the SRA-ID in the input [3, 12]. Quality check and quality control are the two preliminary steps before any kind of NGS data analysis. In this case, we have implemented the FastQC (http:// www.bioinformatics.babraham.ac.uk/projects/fastqc) for quality check and Trimmomatic software for filtering the low quality contaminated and adapter containing reads [13]. Further, we have provided the facility to generate an inter-leaved fastq file from two separate (forward and reverse) paired-end fastq files using in-house PERL script.
2.2. Genome Assembly
For de-novo assembly, the command-line Velvet tool is implemented in the GAAP software [14]. The window binaries of Velvet are taken from Applied Math (https://www.applied-maths.com/velvet-1104-windows-executables-zip). We have provided the different options to users such as hash value (K-mer), minimum contig length, expected coverage and unused reads file. Finally, the software will create a contig fasta file in the result directory.
2.3. Gene Prediction
Once the assembly is completed, the next step is to find the genes in the assembled genome. For this, we have implemented prodigal software in which users have to select the assembly fasta file [15]. The software will generate a fasta file for predicted genes, proteins as well as pseudogenes.
2.4. Annotation
Genome annotation is one of the most crucial steps to find the function of a gene/protein based on different kinds of analysis. In this case, we have used the remote Blast facility of NCBI in which the query sequence is directly submitted to NCBI Blast server without the need to maintain any local copy of databases [16]. We have provided the options to select the e-value, percent identity and query coverage cuff-off to get meaningful results. Furthermore, we have also provided the facility to run remote IPR-Scan to assign the domains and families as well as gene ontology (GO) terms [17, 18]. In addition to that, facility to map the KEGG enzymes and pathways [19, 20] has also been provided based on the GO-term [21, 22].
2.5. Results
Finally, the entire results file will be compiled and visualized in the form of HTML table. Users can also sort the results in ascending or descending order by clicking on any of the selected columns.
3. RESULTS & DISCUSSION
In this study, we have downloaded a viral genome dataset from NCBI SRA (SRR11621811) using GAAP software. The SRA data represented single-end sequencing of COVID-19 genome using Illumina NextSeq500 platform that contained 383155 reads of size 31-75bp length. The raw dataset was processed for quality checking and control using fastQC and trimmomatics, respectively (http://ncbi.github. io/sra-tools/). We obtained 369407, 366054 and 380289 high-quality reads from GAAP, CLC-GWB and OmicsBox, which were subsequently used for de-novo genome assembly. As shown in Table 1, the performance of GAAP software in-term of N50, total assembly length is nearly similar to OmicsBox and better than CLC-GWB. Furthermore, GAAP is able to identify a single long contig of 29774bp, which covers 99.57% of the reference genome (Table 1). Furthermore, we were able to predict 11, 18, and 11 genes using GAAP, CLC-GWB and OmicsBox, respectively. Further, remote-blast is able to annotate 8 genes using CLC-GWB, 10 genes from OmicsBox as well as GAAP software. We have also extracted the associated protein domains, GO-term, enzyme and pathways associated with the predicted genes. We have analysed this dataset and compared it with other existing software such as CLC-GWB, OmicsBox using QUAST webserver [23]. Similarly, we have performed the de-novo genome assembly on a bacterial and human genome with about 1.4 GB paired-ends and 102GB single-end reads, respectively. As mentioned in Table 1, for bacterial genome (SRR17153353) assembly GAAP resulted in 138 contigs with N50 value 100399. However, CLC-GWB resulted in 304 contigs with N50 value 215940. Likewise, in case of human genome (SRR16845439), CLCGWB and GAAP resulted in 3494959 contigs with 842 N50 value and 194281 contigs with 610 N50 value. As shown in Fig. (2) and Table 2, GAAP has several advantages over other existing software that could help the researchers to analyse their data on a simple desktop computer. In comparison to VAUGE, GAAP offers QC and genome annotation to analyse the genome. Similarly, except remote blast, CLC-GWB does not offer remote protein domains, GO-term, enzyme and pathway analysis. However, OmicsBox (recently upgraded from Blast2GO) offers all these services, but it is a licenced software. Thus, as compared to these, we offer a platform independent genome assembly and analysis tool that can perform all the analysis steps without any dependency on the bioinformatics experts.
Table 1.
Comparative statistics of genome assembly using CLC-GWB, OmicsBox and GAAP.
| CLC-GWB | OmicBox | GAAP | |
|---|---|---|---|
| COVID Genome (SRR11621811) | |||
| Total Reads | 383155 | ||
| Quality Reads | 366054 | 380289 | 369407 |
| No. of Assembled Contigs | 2 | 2 | 1 |
| N50 | 25659 | 29928 | 29774 |
| Length of Largest Contig (bp) | 25659 | 29928 | 29774 |
| Length of Smallest Contig (bp) | 4000 | 56 | - |
| Total Assembly Length (bp) | 29659 | 29984 | 29774 |
| Genome Fraction (%) | 99.04 | 99.83 | 99.57 |
| GC Content (%) | 37.96 | 37.99 | 37.98 |
| Bacterial Genome (SRR17153353) | |||
| Total Reads | 9796952 | ||
| Quality Reads | 9452768 | - | 8049822 |
| No. of Assembled Contigs | 304 | - | 138 |
| N50 | 215940 | - | 100399 |
| Length of Largest Contig (bp) | 474481 | - | 320965 |
| Length of Smallest Contig (bp) | 151 | - | 200 |
| Total Assembly Length (bp) | 6461008 | - | 6398876 |
| GC Content (%) | 66.46 | - | 66.44 |
| Human Genome (SRR16845439) | |||
| Total Reads | 540730909 | ||
| Quality Reads | 486776483 | - | 472635344 |
| No. of Assembled Contigs | 3494959 | - | 194281 |
| N50 | 842 | - | 610 |
| Length of Largest Contig (bp) | 8220 | - | 1812 |
| Length of Smallest Contig (bp) | 154 | - | 200 |
| Total Assembly Length (bp) | 1673103862 | - | 46572102 |
| GC Content (%) | 41.16 | - | 38.06 |
Fig. (2).
Homepage of the GAAP software.
Table 2.
Comparison of GAAP with other genome assembly and annotation software.
| Software | GAAP | VAUGE | CLC GWB | Galaxy | OmicsBox |
|---|---|---|---|---|---|
| Licence | Free | Free | Licenced | Free | Licenced |
| Standalone | Yes | Yes | Yes | No | Yes |
| Quality Control | Yes | No | Yes | Yes | Yes |
| Assembly | Yes | Yes | Yes | Yes | Yes |
| Gene Prediction | Yes | No | Yes | Yes | Yes |
| Blast Annotation | Yes | No | Yes | Yes* | Yes |
| Domain & Family Annotation | Yes | No | Yes* | Yes* | Yes |
| GO Annotation | Yes | No | No | No | Yes |
| Pathway Mapping | Yes | No | No | Yes | Yes |
| Enzyme Mapping | Yes | No | No | No | Yes |
Note: *Need locally installed database.
4. IMPLEMENTATION, REQUIREMENTS AND AVAILABILITY
GAAP is developed on the NodeJS platform that uses the HTML, JavaScript as the front-end for communication with users. In the back-end, PERL and JavaScript were used for processing the user data. GAAP is available for all three major platforms i.e. Windows, Linux and MacOS. As shown in Fig. (1), GAAP basically has four independent (Quality control, Genome assembly, Gene Prediction, and Annotation) modules that could be used for the complete analysis of a dataset independently. GAAP can be run on any OS with PERL, JAVA already installed. We recommend a minimum of 8GB RAM for small genomes and un-interrupted internet connection, particularly during SRA fastq data download as well as in the annotation process. Furthermore, depending upon the complexity of genome and data size, the software might require higher computational power. GAAP software is freely available at www.deepaklab.com/gaap.
CONCLUSION
Exponential growth in the sequencing data required GUI-based tools that could simplify the process of data handling and analysis. These kinds of tools will benefit the scientific community working on genome/transcriptome sequencing projects with little knowledge of programming and executing command-line tools. Presently, GAAP software only supports de-novo based genome assembly, which is helpful in the absence of a reference genome. However, if the reference genome is available, researchers more often focus on reference-based assembly, which is quite fast and more helpful in analysing the re-sequencing data. Therefore, in the future we will overcome this limitation by adding a separate module for reference-based assembly. Also, current blast based search does not support local databases, which is most of the times required for some specific analysis. In the future, we will also provide support for a custom database using local blast. These facilities will help the users in fast data analysis in the absence of any powerful computer resources. In a nutshell, GAAP acts as a unique kind of open source, platform independent software package that will serve as a backbone in the field of genomics.
ACKNOWLEDGEMENTS
The authors are thankful to the Department of Biotechnology, Government of India for funding the project “Application of Bioinformatics and Computational Biology in Agriculture-BIC” (BT/PR40193/BTIS/137/23/2021).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
The study is supported by the project “Application of Bioinformatics and Computational Biology in Agriculture-BIC” (BT/PR40193/BTIS/137/23/2021).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Tripathi R., Sharma P., Chakraborty P., Pritish, Varadwaj K. Next-generation sequencing revolution through big data analytics. Front. Life Sci. 2016;9(2):119–149. doi: 10.1080/21553769.2016.1178180. [DOI] [Google Scholar]
- 2.Giani A.M., Gallo G.R., Gianfranceschi L., Formenti G. Long walk to genomics: History and current approaches to genome sequencing and assembly. Comput. Struct. Biotechnol. J. 2019;18:9–19. doi: 10.1016/j.csbj.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kodama Y., Shumway M., Leinonen R. International Nucleotide Sequence Database Collaboration. The sequence read archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao Y., Zhao X., Mace E., Henry R., Jordan D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant. 2019;12(2):156–169. doi: 10.1016/j.molp.2018.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Rouli L., Merhej V., Fournier P.E., Raoult D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 2015;7:72–85. doi: 10.1016/j.nmni.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer P.E., Golicz A.A., Scheben A., Batley J., Edwards D. Plant pan-genomes are the new reference. Nat. Plants. 2020;6(8):914–920. doi: 10.1038/s41477-020-0733-0. [DOI] [PubMed] [Google Scholar]
- 7.Esposito A., Colantuono C., Ruggieri V., Chiusano M.L. Bioinformatics for agriculture in the next-generation sequencing era. Chem. Biol. Technol. Agric. 2016;3:1–12. doi: 10.1186/s40538-016-0054-8. [DOI] [Google Scholar]
- 8.Roumpeka D.D., Wallace R.J., Escalettes F., Fotheringham I., Watson M. A review of bioinformatics tools for bio-prospecting from metagenomic sequence data. Front. Genet. 2017;8:23. doi: 10.3389/fgene.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 10.Conesa A., Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powell D.R., Seemann T. VAGUE: A graphical user interface for the Velvet assembler. Bioinformatics. 2013;29(2):264–265. doi: 10.1093/bioinformatics/bts664. [DOI] [PubMed] [Google Scholar]
- 12.Leinonen R., Sugawara H., Shumway M. International nucleotide sequence database collaboration. The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11(1):119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H-Y., Dosztányi Z., El-Gebali S., Fraser M., Gough J., Haft D., Holliday G.L., Huang H., Huang X., Letunic I., Lopez R., Lu S., Marchler-Bauer A., Mi H., Mistry J., Natale D.A., Necci M., Nuka G., Orengo C.A., Park Y., Pesseat S., Piovesan D., Potter S.C., Rawlings N.D., Redaschi N., Richardson L., Rivoire C., Sangrador-Vegas A., Sigrist C., Sillitoe I., Smithers B., Squizzato S., Sutton G., Thanki N., Thomas P.D., Tosatto S.C.E., Wu C.H., Xenarios I., Yeh L-S., Young S-Y., Mitchell A.L. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones P., Binns D., Chang H-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A.F., Sangrador-Vegas A., Scheremetjew M., Yong S-Y., Lopez R., Hunter S., Valencia A. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Gene Ontology Consortium. The gene ontology resource: 20 Years and still going strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. The Gene Ontology Consortium. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.