Abstract
Alzheimer's disease (AD) is a chronic neurological disease characterized by memory loss and progressive cognitive impairment. The characteristic AD pathologies include extracellular senile plaques formed by β-amyloid protein deposition, neurofibrillary tangles formed by hyper-phosphorylation of τ protein and neuronal loss caused by glial cell proliferation. However, the pathogenesis of AD is still unclear. Dysregulation of RNA methylation is associated with biological processes, including neurodevelopment and neurodegenerative disease. N6-methyladenosine (m6A) is the main modification in eukaryotic RNA and may be associated with the pathophysiology of AD. Circular RNA (circRNA) is a new type of evolutionarily conserved non-coding RNA without 5′-cap and 3′-polyadenylic acid tail. circRNA undergoes m6A RNA methylation and may be involved in the pathogenesis of AD. In the present study, high-throughput sequencing was performed to assess the degree of circRNA m6A methylation in APP/PS1 AD and C57BL/6 mice. These results suggested that circRNA m6A methylation in AD mice was markedly altered compared to the control group. Furthermore, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis was used to predict associated pathways; genes with different circRNA m6A methylation in AD mice were associated with ‘axon guidance’, ‘long-term potentiation’, ‘glutamatergic synapse’, ‘cholinergic synapse’, ‘GABAergic synapse’ and ‘long-term depression’. Methylated RNA immunoprecipitation reverse transcription-quantitative PCR demonstrated that among the eight selected circRNA m6A genes, there were five genes that demonstrated significantly increased methylation and three demonstrated significantly decreased methylation. In summary, the present study indicated that circRNA m6A methylation may be associated with pathogenesis of AD.
Keywords: Alzheimer's disease, circular RNA, circRNA m6A methylation, Gene Ontology, Kyoto Encyclopedia of Genes and Genomes
Introduction
Alzheimer's disease (AD) is a chronic neurodegenerative disease characterized by memory loss and progressive cognitive decline, which results from neuronal death in numerous brain regions, including the hippocampus, entorhinal areas, temporal and parietal lobes and restricted regions within the frontal cortex and cingulate gyrus (1). According to a previous study, there were 50 million patients with AD worldwide in 2020 and this number is projected to reach 152 million by 2050 (2). AD pathology is characterized by extracellular plaques formed by β-amyloid protein deposition, neurofibrillary tangles formed by τ hyperphosphorylation and neuronal loss caused by proliferation of glial cells (3). Numerous factors may influence the incidence of AD, including aging, genetic and environmental factors (4). However, the pathogenesis of AD is still unclear. Previous studies have reported that aging and environmental factors can affect the incidence of AD via epigenetic modification and evidence suggests that epigenetic mechanisms, including DNA methylation and histone modifications, could serve an important role in the pathogenesis of AD (5–8).
RNA modification is a form of post-transcriptional regulation and methylation is the primary form of this modification (9). m6A is the most common type of RNA modification of mRNA and is involved in the regulation of numerous important biological processes, such as RNA degradation, translation, RNA splicing and nuclear export (10). In the nervous system, m6A methylation affects neuronal function, neurogenesis and neuronal differentiation via regulation of mRNA splicing (11) and participates in the regulation of the transcription of genes associated with brain disease risk (12). Increasing studies have reported that m6A serves an important regulatory role in brain development, synaptic plasticity, learning and memory (13,14). According to a previous report, specific m6A methylation exists in the mouse cerebral cortex and cerebellum tissue (15). A previous study reported that protein expression levels of METTL3, which encodes the m6A methyltransferase, is downregulated in the hippocampus (16). The accumulation of METTL3 has been reported to be positively correlated with levels of insoluble τ protein in postmortem human AD samples (16). Furthermore, overexpression of METTL3 has been reported to have rescued β-amyloid (Aβ)-induced synaptic damage and cognitive impairment in vivo (17). Our previous study reported that total methylation level of m6A RNA in the hippocampus of AD mouse model is significantly higher compared with that of the control group, which indicates that m6A methylation of RNA promotes the development of AD (18).
Circular (circ)RNAs are evolutionarily conserved non-coding RNAs without 5′caps and 3′polyadenylation tails (19). circRNAs have been reported to serve a role in the regulation of neurodegenerative disease through their interaction with disease-related miRNAs in numerous studies (20,21). circRNAs have received growing attention for their involvement in the pathogenesis of AD (22,23). A circRNA microarray study reported that AD mouse models had abnormal circRNA in the hippocampus (24,25). m6A RNA methylation has also been reported in circRNAs (26). A review by Li and Jin (27) reported that circRNAs serve an important role in the production and clearance of Aβ, AD neuroinflammation, oxidative stress and autophagy, and that circRNAs are widely involved in regulation of AD physiological and pathophysiological processes and may have the potential to be new biomarkers and novel therapeutic targets. Furthermore, circRNA ciRS-7 has been reported to act as an endogenous, anticomplementary microRNA (miRNA or miR) ‘sponge’ to adsorb and affect normal miRNA-7 function (28). Downregulation of ciRS-7 might increase endogenous miRNA-7 levels in AD. Due to inhibition by the ‘sponging’ effects of ciRS-7, expression of AD-associated targets, such as ubiquitin protein ligase UBE2A, an autophagic, phagocytic protein essential in the clearance of amyloid peptides in the AD brain, may be downregulated (28).
In the present study, high-throughput sequencing was used to assess dysregulation of the circRNA m6A profile in the brains of APP/PS1 and control mice, and evaluate the differences. Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was used to evaluate the biological roles and signaling pathways of these differentially modified circRNA m6A. The methylation degree of circRNA m6A was detected using methylated RNA immunoprecipitation reverse transcription-quantitative PCR (MeRIP-RT-qPCR).
Materials and methods
Mouse model
APP/PS1 double-transgenic male mice (age, 9 months) were used as AD models (10 per group) and age-matched C57BL/6 wild-type mice (9 months old; n=20) were used as the control group. All 20 mice were purchased from Beijing Huafukang Biotechnology Co., Ltd. All mice were housed at a controlled room temperature (25±2°C), in 50–60% humidity, with 12-h light and dark cycles, and free access to water and food for 2 weeks. The mice were sacrificed using cervical dislocation; lack of breathing and cardiac arrest were considered to confirm death. The cortex, hippocampus and cerebellum of mice were dissected and immediately frozen using liquid nitrogen before being stored at −80°C for use in future experiments. All procedures were performed according to the guidelines of the Ethical Committee for Animal Experiments of Shandong University (Jinan, Shandong, China) and the study protocol was approved by the Ethical Committee for Animal Experiments of Shandong University [approval no. KYLL-2020(KJ)A-0098].
All of the 20 mice were eventually euthanized. The humane endpoints used in the present study were as follows: Labored breathing, nasal discharge, lethargy or persistent recumbency, difficulty with ambulation or inability to obtain food or water.
RNA extraction
Total RNA was isolated from frozen mouse brain samples using TRIzol® reagent (Thermo Fisher Scientific, Inc.). A DeNovix spectrophotometer (DS-11) was used to determine RNA quality. Samples with an A260/A280 ratio 1.9-2.2 were used for further experiments.
MeRIP high-throughput sequencing
circRNA m6A high-throughput sequencing (circRNA m6A-seq) was performed by Cloudseq Biotech, Inc. High-throughput sequencing of circRNA m6A methylation was performed on hippocampal samples from APP/PS1 and C57BL/6 control mice (3 per group). All the following steps were performed by Cloudseq Biotech Inc. Briefly, RNA fragments were treated with anti-m6A polyclonal antibody (cat. no. 202003; Synaptic Systems GmbH) in immunoprecipitation (IP) buffer [150 mM NaCl, 0.1% NP-40 and 10 mM Tris-HCl (pH 7.4)] for 2 h at 4°C. After that, the mixture was immunoprecipitated by incubating it for 2 h at 4°C with 50 µl protein A beads (Thermo Fisher Scientific, Inc.). The bound RNA was eluted from the beads using m6A (Berry & Associates; Biosearch Technologies) in IP buffer and extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.). The NEBNext® Ultra II Directional RNA Library Prep kit (cat. no. NEB#E7760; New England Biolabs, Inc.) was used to obtain purified RNA for the creation of an RNA-seq library. RNA quantification and quality assurance with the NanoDrop ND-1000 (Thermo). The concentration of the control group was 561.6 ng/µl and the concentration of the experimental group was 1,240.23 ng/µl. GenSeq® m6A MeRIP kit (GS-ET-001) via Illumina HiSeq 4000 sequencer was used for 150-bp paired-end sequencing of input samples without IP and the m6A IP samples with 6 ng of total RNA input. The differentially expressed genes of circRNA m6A methylation of the samples from the two groups were then examined. Methylated sites were identified by MACS2 (1.4.2) software. Differentially methylated sites were identified by diffReps (1.55.6). The distinct physiological activities of these genes were analyzed using GO and KEGG pathway analysis, where P<0.05 was considered to indicate a statistically significant difference. The generated data are available under accession number GSE216901 from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/).
MeRIP-RT-qPCR
Total RNA (1 µg) was extracted from mouse hippocampal samples and cDNA was synthesized using the PrimeScript First Strand cDNA Synthesis Kit (cat. no. RR047A; Takara Bio, Inc.) according to the manufacturer's instructions. RT-qPCR was performed using the SYBR-Green assay (cat. no. RR041A; Takara Bio, Inc.) and a StepOnePlus Real-Time PCR system (Mastercycler; Eppendorf) in order to determine the relative RNA levels of target genes. RT-qPCR using gene-specific primers was performed on the input control and the m6A-IP samples. The thermocycling conditions were as follows: 40 cycles of 95°C for 30 sec, 95°C for 15 sec and 55°C for 15 sec. The primers used for RT-qPCR are presented in Table I.
Table I.
Sequences of primers used for reverse transcription-quantitative PCR.
| Target | Sequence (5′-3′) |
|---|---|
| chr16:85056324-85120697- | F: TTAGTGAATGCCGGTGATGC |
| R: ACCACAACCACCACTGAGTC | |
| chr16:85013627-85030365+ | F: AAATGGGCATGCTCGTTCTC |
| R: TGTGTGTGTGTGTGTGTTGG | |
| chr12:16977542-16978102+ | F: CGACAGCTTGCTCCAAACAA |
| R: TTTAATGTGGCACCTACGGC | |
| chr16:8708859-8716619- | F: CCTCTTCTGCGTTGCTGTG |
| R: GCGCCCCATTACATTTTGAAG | |
| chr11:108923152-108931579+ | F: ACTGACCGACGATTCCATGT |
| R: AGGAGAGTCACTAACACGGC | |
| chrX:143709648-143744001+ | F: TCAGGAAGCATTGTGAGTGT |
| R: AGTGAATTCCCCGGTGACTG | |
| chr5:137291102-137291478+ | F: TTTCTTCCTCCACGCCTTGT |
| R: AGGACGGATGAAACCCAGAA | |
| chr1:68305845-68396315- | F: ATGAACCATGACACACAGGC |
| R: AGATGGACTCCTCACATGCC |
F, forward; R, reverse.
Statistical analysis
Data are presented as the mean ± SEM of at least three independent experiments. Unpaired student's t-test was used to assess statistical differences between groups. P<0.05 was considered to indicate a statistically significant difference. SPSS 17.0 software (SPSS Inc.) was used for data analysis.
Results
Differential expression between APP/PS1 and wild-type mice
Compared with the control group, high-throughput sequencing demonstrated that certain circRNA m6A genes were methylated at higher or lower levels in APP/PS1 AD mice. A total of 537 genes demonstrated an increase in methylation in the AD group; however, a decrease in methylation was demonstrated in 1,359 genes. The results demonstrated that there was a total of 4,918 methylation sites; upregulated genes accounted for 10.92% of possible sites and the downregulated genes accounted for 27.63% of possible sites. A total of 50 upregulated and 50 downregulated genes (ranked according to fold-change) are presented in Table SI.
GO and KEGG pathway analysis of differentially methylated circRNA m6A genes
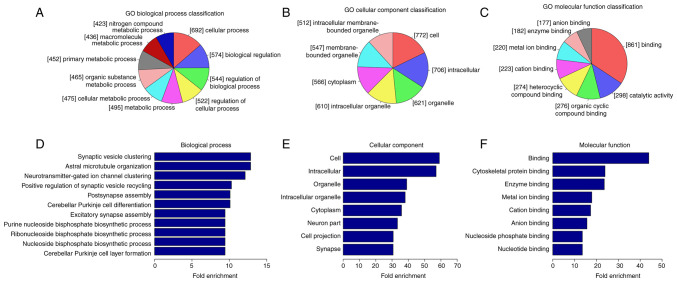
To evaluate the biological role of circRNA m6A methylation in AD mice, the genes containing significantly altered m6A peaks, which indicated differentially methylated genes (DMGs), were analyzed using GO and KEGG pathway analysis. There was a marked difference in degree of methylation between the AD and control groups. The three parts of GO analysis (Fig. 1) are biological process (BP), cell component (CC) and molecular function (MF). BP analysis demonstrated that the genes with lower circRNA m6A methylation in the AD group were significantly linked with ‘synaptic vesicle clustering’, ‘astral microtubule organization’, ‘neurotransmitter-gated ion channel clustering’, ‘positive regulation of synaptic vesicle recycling’, ‘postsynapse assembly’ and ‘cerebellar Purkinje cell differentiation’. Significant GO CC terms for differentially modified RNAs demonstrated that these circRNAs were associated with ‘cell’, ‘intracellular’, ‘organelle’ and ‘cytoplasm’. For MF, these circRNAs were associated with ‘cytoskeletal protein binding’, ‘enzyme binding’, ‘metal ion binding’, ‘cation binding’ and ‘anion binding’.
Figure 1.
Methylation of m6A RNA is associated with specific functions in AD, according to GO analysis. The genes in the AD group with decreased m6A RNA methylation expression are presented for (A) biological process, (B) cell component, (C) molecular function. Fold enrichment of (D) biological process, (E) cell component and (F) molecular function terms. GO, Gene Ontology; AD, Alzheimer's disease; m6A, N6-methyladenosine.
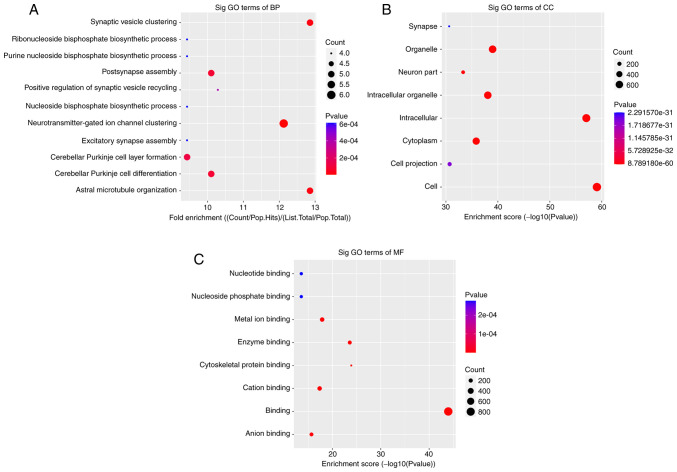
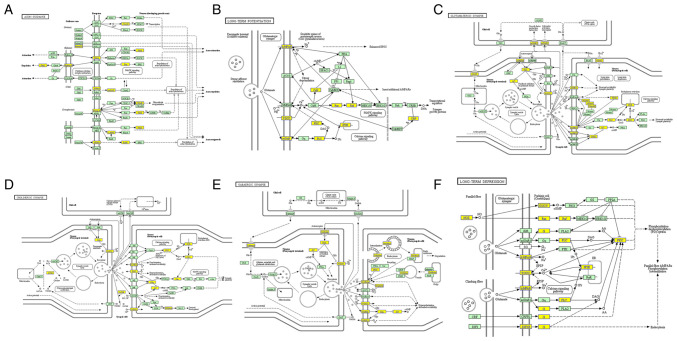
The significant enrichment pathways with the top 10 enrichment scores [log10(P-value)] were assessed using the KEGG pathway dot-plot. The KEGG pathway for BP was associated with ‘synaptic vesicle clustering’, ‘postsynapse assembly’, ‘neurotransmitter-gated ion channel clustering’, ‘cerebellar Purkinje cell layer formation’, ‘cerebellar Purkinje cell differentiation’ and ‘astral microtubule organization’ (Fig. 2A). For CC, the KEGG pathway was associated with ‘organelle’, ‘intracellular part’, ‘intracellular organelle’, ‘cytoplasm’ and ‘cell part’ (Fig. 2B). In MR, the KEGG pathways were enriched for ‘metal ion binding’, ‘enzyme binding’, ‘cation binding’ and ‘anion binding’ (Fig. 2C). Furthermore, the six primary pathways were associated with ‘axon guidance’, ‘long-term potentiation’, ‘glutamatergic synapse, ‘cholinergic synapse’, ‘GABAergic synapse’ and ‘long-term depression’ (Fig. 3).
Figure 2.
Analysis of pathways affected by circRNA m6A in AD brain samples using Kyoto Encyclopedia of Genes and Genomes demonstrated the top 10 genes with differential circRNA m6A methylation. Analysis predicted the pathways affected by circRNA m6A in AD brain samples. (A) Biological process. (B) Cell component. (C) Molecular function. The enrichment score was calculated as-log10(P-value). circRNA, circular RNA; AD, Alzheimer's disease; m6A, N6-methyladenosine; GO, Gene Ontology.
Figure 3.
Pathways associated with synaptic function from which genes to be studied were selected. (A) ‘Axon guidance’. (B) ‘Long-term potentiation’. (C) ‘Glutamatergic synapse’. (D) ‘Cholinergic synapse’. (E) ‘GABAergic synapse’. (F) ‘Long-term depression’. Green, organism-specific pathway; yellow, significantly downregulated circular RNA coding genes. The original source for parts A, B and C in Fig. 3 was the previously published article ‘Abnormality of m6A mRNA Methylation Is Involved in Alzheimer's Disease’ (18) and written permission from the copyright owner of article was obtained and submitted.
circRNA m6A methylation of gene expression is different
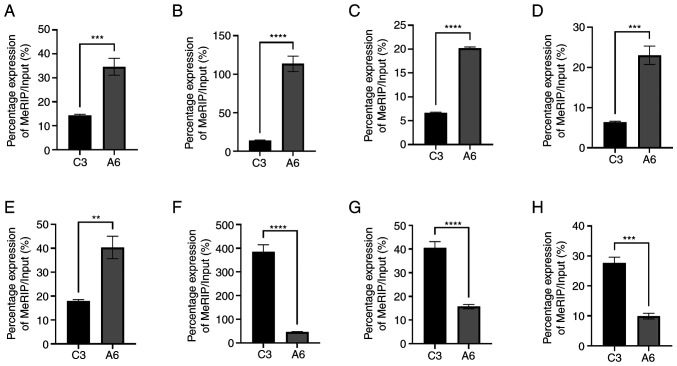
The results of high-throughput sequencing analysis demonstrated that the degree of methylation in the AD group was different from that in the control group. Eight differentially expressed genes were selected and methylation levels were assessed using RT-qPCR. Compared with the control group, among the eight selected genes, 5 demonstrated significantly higher methylation levels, and the remaining three demonstrated significantly lower methylation levels (Fig. 4). Combined with the aforementioned high-throughput sequencing results, these results suggested that circRNA m6A methylation was different in the brain of the AD mice compared with the normal mice.
Figure 4.
Methylated RNA immunoprecipitation reverse transcription-quantitative PCR of eight genes selected from high-throughput sequencing. C3 represents the control group and A6 represents the APP/PS1 mouse group. (A) chr16:85056324-85120697-. (B) chr16:85013627-85030365+. (C) chr12:16977542-16978102+. (D) chr16:8708859-8716619-. (E) chr11:108923152-108931579+. (F) chrX:143709648-143744001+. (G) chr5:137291102-137291478+. (H) chr1:68305845-68396315-. font-size:75%;P<0.005, font-size:75%;P<0.001 and font-size:75%;P<0.0001.
Discussion
A perfect mouse model of AD would develop neuropathology as well as behavioral changes associated with AD (29). Double transgenic mice (APP/PS1) have been widely used to study the pathogenesis of AD and assess the effectiveness of AD treatments (30–32). The APP/PS1 mouse model of AD demonstrates a significant increase in β-amyloid production associated with certain behavioral abnormalities. Behavioral tests have previously demonstrated that compared with wild-type mice, transgenic APP/PS1 mice have significant memory deficits at 6 months of age and that this deficit is exacerbated and more errors occurred at 12 months of age. Cognitive deficits and reference memory impairments of APP/PS1 mouse appear before 6 months of age and lasted for the rest of the life (30,33,34). Deficits in associative learning have also been described in the fear conditioning task at 6–8 months of age and passive avoidance deficits were demonstrated at 12 months of age (35). In the present study, the APP/PS1 mouse was used as an animal model of AD.
In eukaryotes, m6A is the most prevalent internal RNA modification and regulates all aspects of RNA metabolism (16). m6A has been associated with neurogenesis (36), learning and memory (37), brain development (12,38–40) and axon regeneration (41) in previous studies. The presence of specific m6A methylation in mouse cerebral cortex and cerebellum tissue was also reported in a previous study (38). In our previous study, it was reported that the methylation genes in the AD group were associated with presynaptic membranes, postsynaptic membranes and synaptic growth, which suggested that m6A was likely involved in AD (18). All these results indicated that m6A is associated with AD.
circRNAs are a type of RNA made through reverse splicing, in which the 3′ and 5′ ends of a transcript are chemically spliced together to form a continuous loop. In eukaryotes, many circRNAs have been reported and the nervous system and synapses are reported to be particularly rich in these circRNAs (42). A recent study reported aberrant regulation of circRNAs in AD and three circRNAs that may provide clinical insight into AD risk and progression (43). circRNAs have been reported to demonstrate the same m6A modification as mRNAs but are frequently found in distinct places; the m6A readers YTHDF1 and YTHDF2 interact with m6A circRNAs, while the m6A writer METTL3 regulates m6A levels (44).
In the present study, high-throughput sequencing on the degree of circRNA m6A methylation in the hippocampus, cortex and cerebellum in APP/PS1 and normal mice as controls was performed. In comparison to control mice, AD animals demonstrated significantly different levels of m6A methylation of certain circRNAs.
In patients with AD, synapse loss and damage are the most likely causes of cognitive decline (45,46). Furthermore, previous studies have reported that m6A is linked to synaptic plasticity (47,48). According to a recent study, a novel circRNA is linked to psychiatric disorder and regulates synaptic gene expression and cognitive flexibility (49). In the present study, the KEGG pathway results demonstrated that circRNA m6A methylation alteration was associated with glutamatergic, cholinergic and GABAergic synapses. These results indicated that circRNA m6A methylation may have been involved in AD pathogenesis.
To evaluate the aforementioned results, 8 differentially expressed genes were selected from the high-throughput sequencing data and MeRIP-RT-qPCR was performed. These genes were associated with AD. chr16:85056324-85120697- and chr16:85013627-85030365+ are part of the amyloid-β precursor protein (APP) gene and mutations in this gene are linked to the development of early onset (familial) AD (50). chr12: 16977542-16978102+ is a fragment of the Rock2 gene, which has been reported to be correlated with Aβ production (51). ch16:8708859-8716619- is associated with the ubiquitin specific peptidase 7 gene and has an anti-neuroinflammatory effect (52). chr11:108923152-108931579+ is part of the Axin2 gene, which serves an important role in the Wnt/β-catenin signaling pathway, which inhibits Aβ production and τ protein hyperphosphorylation in the brain (53). chrX:143709648-143744001+ is part of the p21 activated kinase gene, which regulates DNA synthesis and neuronal apoptosis caused by familial AD mutants of APP (54). chr5:137291102-137291478+, part of the acetylcholinesterase gene, has been reported to be linked to pathogenesis either by increasing cholinergic deficit or exacerbating Aβ fibril formation and toxicity in AD (55). Furthermore, chr1:68305845-68396315- is reported to be associated with Erbb4, which mediates amyloid β-induced neurotoxicity via JNK/τ signaling pathway activation (56). The results demonstrated that compared with the control group, AD mouse brain samples demonstrated increased methylation levels of five circRNAs and decreased methylation levels of three circRNAs. This suggested that there were differences in levels of methylation in AD mice and that circRNA m6A methylation was related to AD.
In summary, the present study demonstrated that compared with the control group, AD mouse brain samples had different levels of circRNA m6A methylation. MeRIP-RT-qPCR supported these data and indicated that circRNA m6A methylation may have been involved in AD pathogenesis. However, the specific molecular mechanism of the differentially expressed circRNA m6A genes in AD was not thoroughly studied and the potential binding miRNAs were not elucidated. These data suggested that circRNA m6A methylation was associated with AD but more research is needed to confirm this association. In the future, the circRNA of the APP gene associated with Aβ production should be evaluated (chr16:85056324-85120697-) and MeRIP-RT-qPCR verification of methylation level of circAPP m6A in the hippocampus of APP/PS1 mice should be performed. APP/PS1 cells could be used to study the specific regulation mechanism of circAPP m6A on Aβ production in an AD cell model via regulation of APP.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was support by The Special Scientific Research Project of Health Young Medical Science and Technology Talents in Xinjiang Uygur Autonomous Region (grant no. WJWY-202210), The National Natural Science Foundation of China (grant no. 82171410 and 81870848) and The Key Research and Development Program of Shandong Province (grant no. 2017GSF218046).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216901).
Authors' contributions
XZ and YWa designed and performed the experiments, collected and assembled the data, and wrote the manuscript. YS, MH, FL and YWe analyzed data. SH, SY and XLZ performed data analysis and interpretation. YWa and JB were responsible for the conception and design of the study, and the revision and final approval of the manuscript. All authors have read and approved the final manuscript. XZ and YWa confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The present study protocol was reviewed and approved by The Ethical Committee for Animal Experiments of Shandong University [approval no. KYLL-2020(KJ)A-0098].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lane CA, Hardy J, Schott JM. Alzheimer's disease. Eur J Neurol. 2018;25:59–70. doi: 10.1111/ene.13439. [DOI] [PubMed] [Google Scholar]
- 2.Yiannopoulou KG, Papageorgiou SG. Current and future treatments in Alzheimer disease: An update. J Cent Nerv Syst Dis. 2020;12:1179573520907397. doi: 10.1177/1179573520907397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JZ, Wang ZH, Tian Q. Tau hyperphosphorylation induces apoptotic escape and triggers neurodegeneration in Alzheimer's disease. Neurosci Bull. 2014;30:359–366. doi: 10.1007/s12264-013-1415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwok JB. Role of epigenetics in Alzheimer's and Parkinson's disease. Epigenomics. 2010;2:671–682. doi: 10.2217/epi.10.43. [DOI] [PubMed] [Google Scholar]
- 5.Ma X, Liu L, Meng J. MicroRNA-125b promotes neurons cell apoptosis and tau phosphorylation in Alzheimer's disease. Neurosci Lett. 2017;661:57–62. doi: 10.1016/j.neulet.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 6.Patrick E, Rajagopal S, Wong HA, McCabe C, Xu J, Tang A, Imboywa SH, Schneider JA, Pochet N, Krichevsky AM, et al. Dissecting the role of non-coding RNAs in the accumulation of amyloid and tau neuropathologies in Alzheimer's disease. Mol Neurodegener. 2017;12:51. doi: 10.1186/s13024-017-0191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irier HA, Jin P. Dynamics of DNA methylation in aging and Alzheimer's disease. DNA Cell Biol. 2012;31((Suppl 1)):S42–S48. doi: 10.1089/dna.2011.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unnikrishnan A, Freeman WM, Jackson J, Wren JD, Porter H, Richardson A. The role of DNA methylation in epigenetics of aging. Pharmacol Ther. 2019;195:172–185. doi: 10.1016/j.pharmthera.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Chen YS, Sun BF, Yang YG. RNA methylation: Regulations and mechanisms. Yi Chuan. 2018;40:964–976. doi: 10.16288/j.yczz.18-175. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 10.Shafik AM, Zhang F, Guo Z, Dai Q, Pajdzik K, Li Y, Kang Y, Yao B, Wu H, He C, et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer's disease. Genome Biol. 2021;22:17. doi: 10.1186/s13059-020-02249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 12.Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171:877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widagdo J, Anggono V. The m6A-epitranscriptomic signature in neurobiology: From neurodevelopment to brain plasticity. J Neurochem. 2018;147:137–152. doi: 10.1111/jnc.14481. [DOI] [PubMed] [Google Scholar]
- 14.Flamand MN, Meyer KD. The epitranscriptome and synaptic plasticity. Curr Opin Neurobiol. 2019;59:41–48. doi: 10.1016/j.conb.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Du K, Wang J, Nie Y, Lee T, Sun T. Unique and specific m6A RNA methylation in mouse embryonic and postnatal cerebral cortices. Genes (Basel) 2020;11:1139. doi: 10.3390/genes11101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang H, Camats-Perna J, Medeiros R, Anggono V, Widagdo J. Altered expression of the m6A methyltransferase METTL3 in Alzheimer's disease. eNeuro. 2020;7:ENEURO.0125–20.2020. doi: 10.1523/ENEURO.0125-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao F, Xu Y, Gao S, Qin L, Austria Q, Siedlak SL, Pajdzik K, Dai Q, He C, Wang W, et al. METTL3-dependent RNA m6A dysregulation contributes to neurodegeneration in Alzheimer's disease through aberrant cell cycle events. Mol Neurodegener. 2021;16:70. doi: 10.1186/s13024-021-00484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han M, Liu Z, Xu Y, Liu X, Wang D, Li F, Wang Y, Bi J. Abnormality of m6A mRNA methylation is involved in Alzheimer's disease. Front Neurosci. 2020;14:98. doi: 10.3389/fnins.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noureddini M, Bagheri-Mohammadi S. Adult hippocampal neurogenesis and Alzheimer's disease: Novel application of mesenchymal stem cells and their role in hippocampal neurogenesis. Int J Mol Cell Med. 2021;10:1–10. doi: 10.22088/IJMCM.BUMS.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang JL, Su M, Wu DP. Functional roles of circular RNAs in Alzheimer's disease. Ageing Res Rev. 2020;60:101058. doi: 10.1016/j.arr.2020.101058. [DOI] [PubMed] [Google Scholar]
- 24.Huang JL, Qin MC, Zhou Y, Xu ZH, Yang SM, Zhang F, Zhong J, Liang MK, Chen B, Zhang WY, et al. Comprehensive analysis of differentially expressed profiles of Alzheimer's disease associated circular RNAs in an Alzheimer's disease mouse model. Aging (Albany NY) 2018;10:253–265. doi: 10.18632/aging.101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Xu P, Chen B, Zhang Z, Zhang C, Zhan Q, Huang S, Xia ZA, Peng W. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer's disease-like rats using microarray analysis. Aging (Albany NY) 2018;10:775–788. doi: 10.18632/aging.101427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, Liu J, Sun Z. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol Cancer. 2020;19:105. doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Jin G. The circRNA and role in Alzheimer's disease: From regulation to therapeutic and diagnostic targets. Hippocampus. 2022 [Google Scholar]
- 28.Akhter R. Circular RNA and Alzheimer's disease. Adv Exp Med Biol. 2018;1087:239–243. doi: 10.1007/978-981-13-1426-1_19. [DOI] [PubMed] [Google Scholar]
- 29.Lok K, Zhao H, Shen H, Wang Z, Gao X, Zhao W, Yin M. Characterization of the APP/PS1 mouse model of Alzheimer's disease in senescence accelerated background. Neurosci Lett. 2013;557(Pt B):84–89. doi: 10.1016/j.neulet.2013.10.051. [DOI] [PubMed] [Google Scholar]
- 30.Xiong H, Callaghan D, Wodzinska J, Xu J, Premyslova M, Liu QY, Connelly J, Zhang W. Biochemical and behavioral characterization of the double transgenic mouse model (APPswe/PS1dE9) of Alzheimer's disease. Neurosci Bull. 2011;27:221–232. doi: 10.1007/s12264-011-1015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heneka MT, Kummer MP, Stutz A, Delekate A, Schwartz S, Vieira-Saecker A, Griep A, Axt D, Remus A, Tzeng TC, et al. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izco M, Martínez P, Corrales A, Fandos N, García S, Insua D, Montañes M, Pérez-Grijalba V, Rueda N, Vidal V, et al. Changes in the brain and plasma Aβ peptide levels with age and its relationship with cognitive impairment in the APPswe/PS1dE9 mouse model of Alzheimer's disease. Neuroscience. 2014;263:269–279. doi: 10.1016/j.neuroscience.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Denver P, English A, McClean PL. Inflammation, insulin signaling and cognitive function in aged APP/PS1 mice. Brain Behav Immun. 2018;70:423–434. doi: 10.1016/j.bbi.2018.03.032. [DOI] [PubMed] [Google Scholar]
- 34.Götz J, Bodea LG, Goedert M. Rodent models for Alzheimer disease. Nat Rev Neurosci. 2018;19:583–598. doi: 10.1038/s41583-018-0054-8. [DOI] [PubMed] [Google Scholar]
- 35.Webster SJ, Bachstetter AD, Nelson PT, Schmitt FA, Van Eldik LJ. Using mice to model Alzheimer's dementia: An overview of the clinical disease and the preclinical behavioral changes in 10 mouse models. Front Genet. 2014;5:88. doi: 10.3389/fgene.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G, Zhao JC. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206. doi: 10.1038/s41593-018-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, Li J, Hao P, Zhang Y, Zhang F, et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–253. doi: 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang M, Lv H, Zhang W, Ma C, He X, Zhao S, Zhang ZW, Zeng YX, Song S, Niu Y, Tong WM. Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017;7:170166. doi: 10.1098/rsob.170166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 40.Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He X, Wu G, Zhao S, Zhang Y, Wang D, et al. RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19:68. doi: 10.1186/s13059-018-1435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weng YL, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97:313–325.e6. doi: 10.1016/j.neuron.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;22:1903–1912. doi: 10.1038/s41593-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Fan H, Sun J, Ni M, Zhang L, Chen C, Hong X, Fang F, Zhang W, Ma P. Circular RNA expression profile of Alzheimer's disease and its clinical significance as biomarkers for the disease risk and progression. Int J Biochem Cell Biol. 2020;123:105747. doi: 10.1016/j.biocel.2020.105747. [DOI] [PubMed] [Google Scholar]
- 44.Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20:2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kashyap G, Bapat D, Das D, Gowaikar R, Amritkar RE, Rangarajan G, Ravindranath V, Ambika G. Synapse loss and progress of Alzheimer's disease-a network model. Sci Rep. 2019;9:6555. doi: 10.1038/s41598-019-43076-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.John A, Reddy PH. Synaptic basis of Alzheimer's disease: Focus on synaptic amyloid beta, P-tau and mitochondria. Ageing Res Rev. 2021;65:101208. doi: 10.1016/j.arr.2020.101208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chokkalla AK, Mehta SL, Vemuganti R. Epitranscriptomic regulation by m6A RNA methylation in brain development and diseases. J Cereb Blood Flow Metab. 2020;40:2331–2349. doi: 10.1177/0271678X20960033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madugalle SU, Meyer K, Wang DO, Bredy TW. RNA N6-methyladenosine and the regulation of RNA localization and function in the brain. Trends Neurosci. 2020;43:1011–1023. doi: 10.1016/j.tins.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmerman AJ, Hafez AK, Amoah SK, Rodriguez BA, Dell'Orco M, Lozano E, Hartley BJ, Alural B, Lalonde J, Chander P, et al. A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol Psychiatry. 2020;25:2712–2727. doi: 10.1038/s41380-020-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Zhou X, Li G, Zhang Y, Wu Y, Song W. Modifications and trafficking of APP in the pathogenesis of Alzheimer's disease. Front Mol Neurosci. 2017;10:294. doi: 10.3389/fnmol.2017.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herskowitz JH, Feng Y, Mattheyses AL, Hales CM, Higginbotham LA, Duong DM, Montine TJ, Troncoso JC, Thambisetty M, Seyfried NT, et al. Pharmacologic inhibition of ROCK2 suppresses amyloid-β production in an Alzheimer's disease mouse model. J Neurosci. 2013;33:19086–19098. doi: 10.1523/JNEUROSCI.2508-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang XW, Feng N, Liu YC, Guo Q, Wang JK, Bai YZ, Ye XM, Yang Z, Yang H, Liu Y, et al. Neuroinflammation inhibition by small-molecule targeting USP7 noncatalytic domain for neurodegenerative disease therapy. Sci Adv. 2022;8:eabo0789. doi: 10.1126/sciadv.abo0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia L, Piña-Crespo J, Li Y. Restoring Wnt/β-catenin signaling is a promising therapeutic strategy for Alzheimer's disease. Mol Brain. 2019;12:104. doi: 10.1186/s13041-019-0525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McPhie DL, Coopersmith R, Hines-Peralta A, Chen Y, Ivins KJ, Manly SP, Kozlowski MR, Neve KA, Neve RL. DNA synthesis and neuronal apoptosis caused by familial Alzheimer disease mutants of the amyloid precursor protein are mediated by the p21 activated kinase PAK3. J Neurosci. 2003;23:6914–6927. doi: 10.1523/JNEUROSCI.23-17-06914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park D, Choi EK, Cho TH, Joo SS, Kim YB. Human neural stem cells encoding ChAT gene restore cognitive function via acetylcholine synthesis, Aβ elimination, and neuroregeneration in APPswe/PS1dE9 mice. Int J Mol Sci. 2020;21:3958. doi: 10.3390/ijms21113958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Zhang L, Zhou D, Li H, Xu Y. ErbB4 mediates amyloid β-induced neurotoxicity through JNK/tau pathway activation: Implications for Alzheimer's disease. J Comp Neurol. 2021;529:3497–3512. doi: 10.1002/cne.25207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216901).