ABSTRACT
The standard algorithm for diagnosing hepatitis C virus (HCV) infection has two steps, an HCV antibody test for screening and a nucleic acid amplification test (NAAT) for confirmation. However, the HCV core antigen (HCVcAg) detection assay is an alternative for one-step diagnosis. We aimed to evaluate the diagnostic performance of the Abbott ARCHITECT HCV Ag assay to detect active hepatitis C in serum/plasma in people living with HIV/AIDS (PLWHA), through a systematic review and meta-analysis. PubMed, EMBASE, Scopus, Web of Science, and the Cochrane Library were searched until 20 September 2022 (PROSPERO, CRD42022348351). We included studies evaluating Abbott ARCHITECT HCV Ag assay (index assay) versus NAATs (reference test) in PLWHA coinfected with HCV who did not receive antiviral treatment for HCV. Meta-analysis was performed with the MIDAS module using Stata and random-effects models. The QUADAS-2 tool evaluated the risk of bias. The bivariate analysis was conducted on 11 studies with 2,407 samples. Pooled sensitivity was 0.95 (95% CI = 0.92 to 0.97), specificity 0.97 (95% CI = 0.93 to 0.99), positive likelihood ratio 37.76 (95% CI = 12.84 to 111.02), and negative likelihood ratio 0.06 (95% CI = 0.04 to 0.09). The area under the curve was 0.97 (95% CI = 0.20 to 1.00). For low prevalence (≤5%), the posttest probability that an individual with a positive test was a true positive ranged from 4% to 67%, whereas, at high prevalence (≥10%), the posttest probability was between 81% and 87%, indicating that a confirmatory test should be necessary, particularly with prevalence values of ≤1%. Regardless of prevalence, the probability that an individual with a negative test was a false negative was close to zero, indicating that the individual was not infected with HCV. In conclusion, the accuracy of the Abbott ARCHITECT HCV Ag assay was very good for HCV screening in serum/plasma samples from PLWHA. The clinical utility to confirm HCV infection was acceptable in high-prevalence settings (≥10%) but poor in low-prevalence settings (≤1%). Furthermore, it was excellent in excluding active HCV infection.
KEYWORDS: core antigen, diagnostic accuracy, hepatitis C virus, people living with HIV, screening
INTRODUCTION
Hepatitis C virus (HCV) infection can cause chronic hepatitis C (CHC), which, if not treated in time, can lead to fibrosis, cirrhosis, liver failure, hepatocellular carcinoma, and even death (1). HCV infection is a global health problem since about 58 million people worldwide have CHC, which causes almost 300,000 deaths yearly (2). The majority of cases are concentrated in developing countries, with a higher prevalence in high-risk populations such as men who have sex with men (MSM), female sex workers, people who inject drugs (PWID), and prisoners (3). Direct-acting antivirals (DAAs) against HCV have demonstrated high efficacy rates (>90%). However, HCV infection remains a serious social and public health problem because less than 20% of people infected with HCV are aware that they are infected, as CHC remains asymptomatic for a long time, progressively causing liver damage (2, 4). Therefore, the World Health Organization calls for eliminating viral hepatitis by 2030 by detecting 90% of patients with CHC and treating 80% of them (5).
HCV infection is common in people living with HIV/AIDS (PLWHA) since both viruses share transmission routes (6 to 8). There are around 2.9 million PLWHA coinfected with HCV (4). HCV/HIV coinfection negatively affects the transmission and natural history of HCV infection. HIV infection reduces the chance of HCV spontaneous clearance, increases chronic HCV infection rates, and accelerates the development of HCV-related liver diseases, such as liver cirrhosis, liver failure, and hepatocellular carcinoma (9, 10). Although combined antiretroviral therapy (cART) against HIV can improve these outcomes and decrease HCV-related mortality (11), the primary cause of morbi-mortality among HIV/HCV-coinfected individuals is related to liver diseases (12). As a result, HCV detection and effective treatment should be prioritized for this population.
Currently, the HCV diagnostic algorithm includes two steps: first, detection of antibodies against HCV (anti-HCV), and second, confirmation of active hepatitis C infection by detection of HCV-RNA (13). HCV-RNA detection in clinical practice is performed by a nucleic acid amplification test (NAAT), the gold standard confirmatory assay (14, 15). However, NAATs are time-consuming and involve qualified personnel and advanced equipment, which are expensive (15, 16). Furthermore, RNA is easily degraded, leading to false negative (FN) results (17). The HCV core antigen (HCVcAg) test is an alternative for active HCV infection diagnosis, which is easier, less expensive, and faster than NAATs (16, 18, 19). HCVcAg appears in blood before anti-HCV antibodies, correlates positively with HCV-RNA load, is more stable than HCV-RNA, and is detected within 12 to 15 days after HCV infection, anticipating the window period for diagnosis.
Previous reports have analyzed the diagnostic performance of the Abbott ARCHITECT HCV Ag assay for screening HCV infection in PLWHA (20 to 33), which is today's most widely used HCVcAg test for HCV screening (19). Freiman et al.'s meta-analysis (34) reported high accuracy of the Abbott ARCHITECT HCV Ag assay in the general population, but they did not report data on clinical applications and did not analyze data in PLWHA to diagnose active hepatitis C. To our knowledge, there is no meta-analysis on the diagnostic performance of the HCVcAg test to screen for hepatitis C in PLWHA, a high-risk population with particular characteristics (35).
This study aimed to evaluate the diagnostic performance of the Abbott ARCHITECT HCV Ag assay for detecting active hepatitis C in serum/plasma in PLWHA through a systematic review and meta-analysis of eligible studies published to date (20 September 2022).
MATERIALS AND METHODS
The systematic review was completed according to the Cochrane Handbook for Diagnostic Test Accuracy Reviews (36) and the Preferred Reporting Items for Systematic Reviews and Meta-Analysis of Diagnostic Test Accuracy (PRISMA-DTA) guidelines (37). A completed PRISMA-DTA checklist is available in Supplementary File 1 in the supplemental material.
Search strategy.
Studies were identified using different search commands in Medline/PubMed, EMBASE, SCOPUS, Web of Science, and the Cochrane Library (see Supplementary File 2) between 1 January 1976 and 20 September 2022. This protocol was designed a priori and registered in the online database PROSPERO (CRD42022348351). The literature search was not restricted by language, publication time, study design, or geographic location. Reference lists of included studies were evaluated for further relevant records. Articles from other languages were translated into English with the help of Google Translate.
Study selection.
Study inclusion was determined according to the following criteria: (i) evaluation of diagnostic accuracy in plasma/serum of PLWHA; (ii) comparison between the Abbott ARCHITECT HCV Ag assay and a NAAT (gold standard); and (iii) available data to formulate a 2 × 2 contingency table (true positives [TP], false positives [FP], true negatives [TN], false negatives [FN]). The exclusion criteria were as follows: (i) no available data to estimate sensitivity or specificity (nor from the corresponding author upon reasonable request); (ii) a sample size of less than 10, to avoid bias in the random-effects model; (iii) nonhuman subjects, commercial samples, or off-market assays; (iv) data during and after HCV antiviral therapy; and (v) nonoriginal articles, unpublished data, or data published in case reports, comments, letters to the editor, conference abstracts, book chapters, and review articles. All studies were independently screened by two investigators (A.T.-N. and D.S.-C.), who carefully reviewed the full text of the selected papers to extract all data.
Data extraction.
Diagnostic accuracy of the HCVcAg test and population characteristics were extracted by two investigators (D.S.-C. and A.T.-N.), who carefully reviewed the full text of the selected papers to extract all data and judged each study according to the previous criteria. Disagreement was resolved by consensus and/or consultations with the senior author (S.R.). One investigator (D.S.-C) contacted authors by e-mail for missing data up to three attempts, excluding those without a response.
Assessment of risk of bias.
Study quality was evaluated using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool (38). The risk of bias and concerns over applicability were assessed by two reviewers (A.T.-N. and D.S.-C.). Discrepancies were resolved by a third reviewer (S.R.). The risk of bias was summarized in a table as “low,” “unclear,” or “high” for each domain, whereas the degree of applicability was rated as “low,” “unclear,” or “high” for the first three domains. The “unclear” category was used only when reported data were insufficient (see Supplementary File 3).
Statistical analysis.
All statistical analyses were performed with the MIDAS package in STATA 15.0 (STATA Corp., College Station, TX, USA) (39, 40). Pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and the area under the curve of the summary receiver operating characteristic curve (AUC-SROC) were calculated using a bivariate model, with the studies that had TP+FN > 0 and FP+TN > 0. The bivariate analysis considers the size of the study and uses a random-effects approach to obtain heterogeneity beyond the methodological and clinical differences between studies. We also performed a univariate analysis with all studies, including those with TP+FN = 0 or FP+TN = 0.
The accuracy level was evaluated by plotting the SROC curve and calculating the AUC, considering very good (AUC = 0.90 to 1), good (0.80 to 0.90), fair (0.70 to 0.80), poor (0.60 to 0.70), and failed (0.50 to 0.60) (41). For clinical utility, a four-quadrant likelihood scatter matrix was used as follows (42): (i) left upper quadrant (LUQ; PLR >10, NLR <0.1): confirmation and exclusion; (ii) right upper quadrant (RUQ; PLR >10, NLR >0.1): confirmation only; (iii) left lower quadrant (LLQ; PLR <10, NLR <0.1): exclusion only; and (iv) right lower quadrant (RLQ; PLR <10, NLR >0.1): no confirmation or exclusion. The clinical or patient-relevant utility was also assessed with Fagan’s nomogram, which gave the posttest prediction at the population level, taking into account the prevalence or pretest probability estimation (0.1%, 0.5%, 1%, 5%, 10%, and 15%) and the PLR and NLR values (43).
The Cochran-Q method and inconsistency index (I2) quantified the heterogeneity between the studies, being significant with P of ≤ 0.10 and I2 of ≥50%, respectively (44). Galbraith (radial) (45) and bivariate boxplots (bagplot) (46) were also used to analyze the heterogeneity. Furthermore, meta-regression was performed to define the impact of several factors on diagnostic accuracy measures (P ≤ 0.10): year of publication (Yes: ≤2015; No: >2015), low- or middle-income countries (LMICs; Yes/No), all patients with positive anti-HCV (anti-HCV Ab +) (Yes/No), biological sample type (Yes: only serum; No: plasma or plasma/serum), frozen sample (Yes/No), large sample size (Yes: ≤100; No: >100), and HCV prevalence (≤50; No: >50). We used Deeks’ funnel plot asymmetry test to evaluate publication bias, with a symmetric funnel shape when there is no publication bias (P > 0.10) (47).
Ethics approval and consent to participate.
This study was approved by the Instituto de Salud Carlos III Ethics Committee (ref.: CEI PI 13_2021). This study involves clinical-epidemiological data of patients from published articles, so informed consent signed by patients was unnecessary.
Data availability.
All relevant data are within the article and the supplemental material.
RESULTS
Search results.
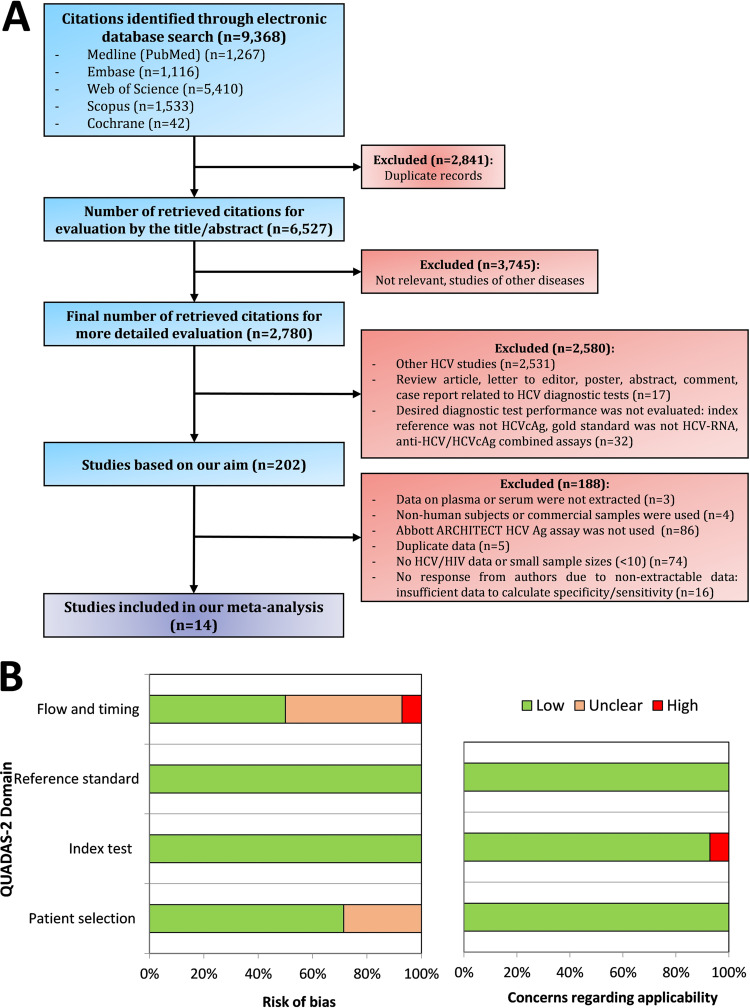
Figure 1A shows the flowchart summarizing the steps to identify the eligible studies. The initial search of electronic databases reported 9,368 articles covering the literature published between 1976 and 2022. Of these, 30% (n = 2,841) were duplicates, and the remaining 6,527 articles were reviewed. Based on the evaluation of titles and abstracts, together with a detailed evaluation, a total of 6,325 studies were filtered out. After evaluating the full texts of 202 studies, 14 were eligible for this meta-analysis because all reported the necessary information and met the inclusion criteria mentioned above (20 to 33).
FIG 1.
(A) Flowchart of the study searching process. (B) Quality assessment of eligible studies according to the QUADAS-2 tool. In panel B, green indicates a low risk, orange stands for unclear risk, and red indicates a high risk. cAg, core antigen; HCV, hepatitis C virus; HIV, human immunodeficiency virus; QUADAS, quality assessment of diagnostic accuracy studies.
Article characteristics.
Table 1 summarizes the main characteristics of the 14 articles included in this meta-analysis. All studies were published between 2012 and 2022, had a cross-sectional design, included adults only, and were published in English, except one published in German (22). Three studies were carried out in LMICs (26, 28, 31). A total of 2,526 individuals were included, with 82.1% male and a mean age of 45. The HCV genotypes ranged from 1 to 6, and only four studies (21, 22, 30, 33) provided information on the HBV status, with a prevalence between 3.8 and 78.3%.
TABLE 1.
Summary of studies included in the meta-analysis detecting HCV core antigen with Abbott ARCHITECT HCVcAg assay in serum and/or plasma samples from PLWHAa
| Author (yr) (reference) | Country | No. | Age (yrs) | Males (%) | HCV genotype | HBV (%) | Sample type | Sample condition | GS cutoff (IU/mL) |
|---|---|---|---|---|---|---|---|---|---|
| Mederacke et al. (2012) (20) | Germany | 71 | N/D | N/D | 1, 3, 4 | N/D | Serum/plasma | Frozen | 15 and 600 |
| Garbuglia et al. (2014) (21) | Italy | 249 | 47 | 69.6 | 1, 3, 4 | 3.8 | Plasma | Frozen | 12 |
| Van Helden et al. (2014) (22) | Germany | 97 | 38.2 | 84.5 | 1, 2, 3 | 9.3 | Serum | Frozen | 15 |
| Cresswell et al. (2015) (23) | UK | 111 | 45.2 | 94.6 | 1, 3, 4 | N/D | Serum | Frozen | N/D |
| Vanhommerig et al. (2015) (24) | Netherlands | 91 | N/D | 100 | 1, 2, 3, 4 | N/D | Serum | Unknown | 615 |
| Arboledas et al. (2017) (25) | Spain | 68 | 49.8 | 80.5 | 1, 3, 4 | N/D | Plasma | Frozen | 15 |
| Duchesne et al. (2017) (26) | Cameroon | 78 | 46.3 | 43.5 | 1, 2, 4 | N/D | Serum | Frozen | 12 |
| Hullegie et al. (2017) (27) | Netherlands | 67 | 41 | 100 | 1 | N/D | Serum/plasma | N/D | 15 and 12 |
| Mohamed et al. (2017) (28) | Tanzania | 65 | 38 | 92.2 | 1, 4 | N/D | Serum | Frozen | 15 |
| Talal et al. (2017) (29) | USA | 19 | 53.8 | 59.6 | 1, 2, 3, 4 | N/D | Serum | Frozen | 15 |
| Alonso et al. (2018) (30) | Spain | 20 | N/D | N/D | 1, 2, 3 | 78.3 | Serum | Frozen | 15 |
| Chayanupatkul et al. (2020) (31) | Thailand | 36 | 41.4 | 83.3 | 1 | N/D | Serum | Frozen | 12 |
| Rossetti et al. (2021) (32) | Italy | 20 | 56 | 80 | 1, 2, 3, 4 | N/D | Serum/plasma | Frozen | 15 and 12 |
| Sun et al. (2022) (33) | Taiwan | 1534 | 40.5 | 97.6 | 1, 2, 6 | 10.3 | Plasma | N/D | 15 |
GS, gold standard; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; IU, international units; No., sample size; N/D, data not available; PLWHA, people living with HIV/AIDS; yrs = years.
HCVcAg quantification was performed using the Abbott ARCHITECT HCV Ag assay, whose lower limit of detection (LLOD) was 3 fmol/L (0.06 pg/mL). The primary gold standard assay to detect HCV-RNA was the COBAS Ampliprep/COBAS TaqMan HCV Real-time PCR (Roche Diagnostics) (n = 9; LLOD: 12, 15, and 600 IU/mL) (20, 22, 25, 27 to 30, 32, 33), followed by the Abbott RealTime HCV Assay (Abbott Diagnostics) (n = 4; LLOD: 12 IU/mL) (21, 23, 26, 31) and the VERSANT HCV RNA Qualitative Assay (Siemens Healthcare Diagnostics) (n = 1; LLOD: 615 IU/mL) (24).
Assessment of risk of bias.
QUADAS-2 (Fig. 1B and Supplementary File 3) showed that the risk of bias in the patient selection domain was unclear in four studies (21, 22, 26, 30) (28.6%) due to the lack of information on whether the study was consecutive, randomized, or neither. The patient sample to evaluate the diagnostic test accuracy was also not documented. Both flow bias and timing bias were unclear in six (42.9%) studies (20, 22, 25, 27, 28, 32) and high in one (7.1%) (33) because the delay between the reference test and the index test was not correctly reported.
All studies had a low risk of bias for the index and reference standard domains. Although the analytical sensitivity of the three NAATs used as a reference standard is different, overall, the tests were highly sensitive, and the variability was minimal. Besides, HCV-RNA loads measured by NAATs correlated well with HCVcAg levels, as is shown in most eligible studies (20 to 23, 26, 28 to 32). Moreover, even if the reference standard resulted in knowledge of the index test result, it is unlikely to introduce bias. The applicability of the included studies was low for all but one study (33), which showed significant concerns about the performance and interpretation of the HCVcAg test.
Diagnostic validity.
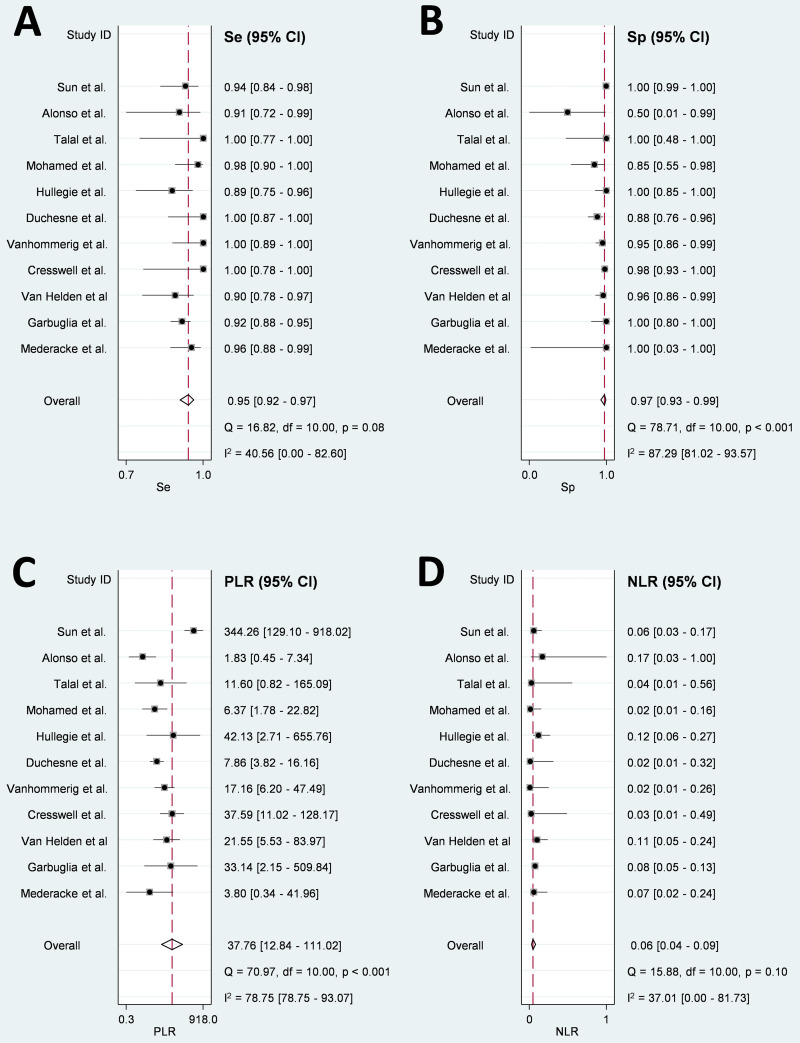
The bivariate analysis was conducted on 11 studies with 2,407 samples. Pooled sensitivity was 0.95 (95% CI = 0.92 to 0.97) (Fig. 2A), specificity was 0.97 (95% CI = 0.93 to 0.99) (Fig. 2B), PLR was 37.76 (95% CI = 12.84 to 111.02) (Fig. 2C), and NLR was 0.06 (95% CI = 0.04 to 0.09) (Fig. 2D). The AUC-SROC was 0.97 (95% CI = 0.20 to 1.00), suggesting that the overall diagnostic performance of HCVcAg in PLWHA was very good (Fig. S1 in the supplemental material). The univariate analysis (n = 14 articles) showed a pooled sensitivity of 0.96 (95% CI = 0.93 to 0.98), slightly higher than the bivariate analysis (Fig. S2).
FIG 2.
Forest plots of pooled Se (A), Sp (B), PLR (C), and NLR (D) for all studies included in the bivariate analysis for detecting active HCV infection with Abbott ARCHITECT HCV Ag assay in PLWHA compared with a confirmatory nucleic acid test. 95% CI, 95% confidence interval; HCV, hepatitis C virus; I2, inconsistency index; NLR, negative likelihood ratio; PLR, positive likelihood ratio; PLWHA, people living with HIV/AIDS; Se, sensitivity; Sp, specificity.
Clinical application.
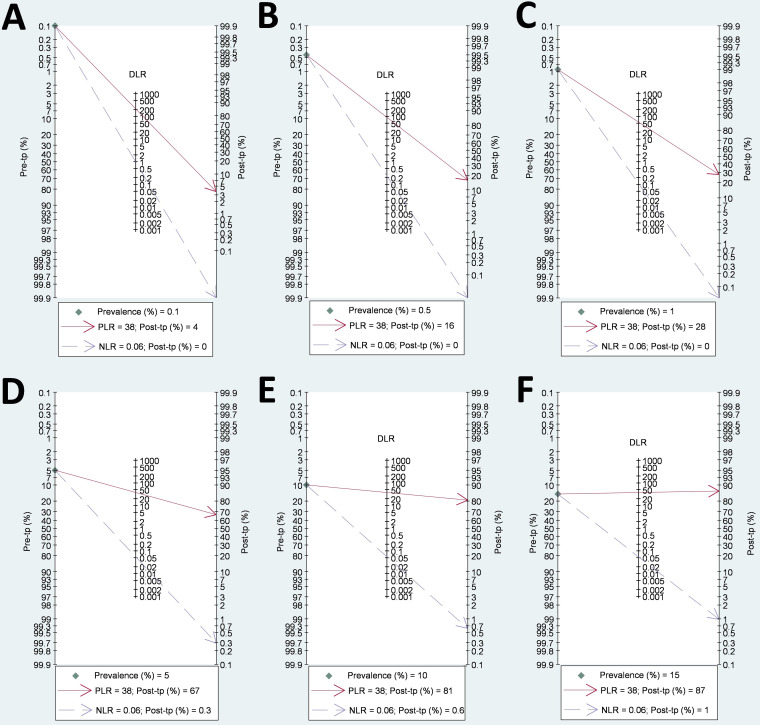
Fagan’s nomograms showed that for low prevalence (≤1%), the posttest probability that an individual with a positive test was a TP ranged from 4% to 28%, while, at high prevalence (≥10%), the posttest probability was between 81% and 87%, indicating that a confirmatory test should be necessary, particularly with prevalence values of ≤1% (Fig. 3). However, the probability that an individual with a negative test was an FN was close to zero, regardless of HCV prevalence (Fig. 3).
FIG 3.
Fagan’s plot of PLR and NLR to evaluate the clinical utility of detecting active HCV infection in PLWHA with Abbott ARCHITECT HCV Ag assay compared with a confirmatory nucleic acid test at different HCV prevalence percentages: 0.1% (A), 0.5% (B), 1% (C), 5% (D), 10% (E), and 15% (F). DLR, diagnostic likelihood ratio; HCV, hepatitis C virus; NLR, negative likelihood ratio; PLR, positive likelihood ratio; PLWHA, people living with HIV/AIDS; Post-tp, posttest probability; Pre-tp, pretest probability.
Exploration of heterogeneity.
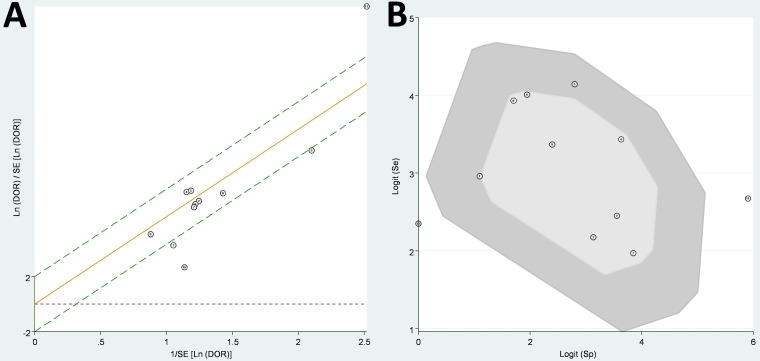
The heterogeneity tests showed I2 = 40.6% (P = 0.08) for sensitivity (moderate heterogeneity), I2 = 87.3% (P < 0.001) for specificity (considerable heterogeneity), I2 = 78.8% (P < 0.001) for PLR (substantial heterogeneity), and no significant heterogeneity (I2 = 37.0% [P = 0.10]) for NLR (Fig. 2). Three studies (22, 29, 30) in the Galbraith plot (Fig. 4A) and two studies (29, 30) in the bivariate box plot fell outside the 95% CI (Fig. 4B), suggesting a source of heterogeneity for these studies, especially for one common to both analyses (29). However, the heterogeneity did not change much after removing this article (29) from the analysis.
FIG 4.
Galbraith plot (A) and bagplot (B) to assess heterogeneity of the bivariate meta-analysis. DOR, diagnostic odds ratio; Se, sensitivity; SE, standard error; Sp, specificity.
Meta-regression analysis showed that four factors (LMICs, biological sample type, sample size, and HCV prevalence) had a significant impact on heterogeneity (P ≤ 0.10) (Table S1). Moreover, year of publication, LMIC, all patients with anti-HCV Ab +, biological sample type, frozen sample, gold standard cutoff, and sample size had a significant effect on sensitivity (P ≤ 0.10), but only LMIC, biological sample type, frozen sample, and sample size had a significant impact on specificity (P ≤ 0.10) (Table S2). However, these significant differences in sensitivity and specificity were minor, with very close values between the groups, and in practice, they were not very relevant.
Based on Deeks’ funnel plot, publication bias was observed (P = 0.01), indicating a potential impact on heterogeneity (Fig. S3).
DISCUSSION
In this bivariate meta-analysis, we found two main findings. (i) The diagnostic validity (accuracy) of the Abbott ARCHITECT HCV Ag assay to diagnose active HCV infection in serum/plasma samples from PLWHA was very high, where the sensitivity, specificity, and AUC-SROC values were very good. (ii) The HCVcAg assay showed limited clinical utility to confirm active HCV infection, mainly in low-prevalence settings (≤1%). Still, it was excellent because the HCVcAg assay excluded active HCV infection regardless of prevalence.
The Abbott ARCHITECT HCV Ag assay is an inexpensive, rapid (~40 min), and easy-to-perform assay with high analytical sensitivity for HCV RNA loads above 10,000 IU/mL, although slightly lower for viral values below this cutoff (17, 48). To our knowledge, this meta-analysis is the first to show the diagnostic performance of the Abbott ARCHITECT HCV Ag assay to diagnose active HCV infection in PLWHA. Freiman et al. (34) published an outstanding meta-analysis that evaluated the Abbott ARCHITECT HCV Ag assay to detect active HCV infection. However, they did not perform a meta-analysis on a subgroup of PLWHA due to the lack of necessary studies. Now, we evaluated the diagnostic performance of the Abbott ARCHITECT HCV Ag assay in PLWHA, collecting data from 14 studies (n = 2,526 individuals), a sample size large enough to extract conclusive results. Our meta-analysis and that of Freiman et al. (34) achieved very similar results in sensitivity (95.0% versus 93.4%), specificity (97.0% versus 98.8%), NLR (both 0.06), and AUC-SROC (both >97%). However, the overall PLR value from our meta-analysis was lower than that of Freiman et al. (34) (37.76 versus 80.6). Despite the latter, both meta-analyses showed an excellent and similar diagnostic validity (accuracy) of the HCVcAg assay for HCV detection. Moreover, Freiman et al. (34) did not show data on clinical application and only highlighted the possible replacement of NAAT by HCVcAg tests in scenarios with a high HCV prevalence. Our meta-analysis found solid evidence to accept or rule out active HCV infection. Fagan’s nomogram showed that the clinical utility of the HCVcAg assay was small in low-prevalence (≤1%) and limited in high-prevalence (≥10%) settings. Thus, NAAT should be necessary to confirm a positive result of the HCVcAg assay. In the best case, the Abbott ARCHITECT HCV Ag test could be an alternative to NAAT in high-prevalence settings (≥10%) where the probability of active HCV infection is greater than 80%. The high prevalence of hepatitis C is usually found in high-risk populations (prisoners, PWID, MSM, or sex workers), where HIV/HCV coinfection is generally high because HCV and HIV share common transmission routes (6). Besides, these risk populations have a higher chance of HCV reinfection after being cured with DAA treatment because they have high-risk behaviors that lead to HCV transmission (49).
The quality of a meta-analysis depends on the quality of the included studies. In our meta-analysis, the overall quality was medium-high because many elements related to the risk of bias were unclear or missing. Some studies did not show information on the use of consecutive or random samples, the design of the studies resembled that of case-control studies, and little information was provided on the flow of patients and time in the study.
Heterogeneity is another factor to consider when assessing a meta-analysis. Our meta-analysis showed moderate-substantial heterogeneity, which is common in meta-analyses of diagnostic tests because all potential confounders are difficult to control. The meta-regression showed that LMIC, biological sample type, sample size, and HCV prevalence significantly impacted heterogeneity. It may also be due to other factors not being analyzed due to insufficient raw data from HCV viral load, HCV subtype, HBV coinfection, or RNA extraction methods. Therefore, a random effects analysis should be used as there is significant heterogeneity between studies, assuming that all studies measure different parameters (50). Furthermore, all analyzed confounders, except HCV prevalence, impacted sensitivity and/or specificity, but their effect was clinically irrelevant.
Limitations.
Finally, some limitations must be considered to interpret our data correctly. (i) We found publication bias in favor of a greater tendency to publish studies with favorable results. Probably, some studies with unfavorable data have not been published. (ii) There was a lack of essential data in some articles. In several works, plasma and serum samples were used in the same study, and the frozen condition of the sample was unknown. Furthermore, as discussed above, we could not analyze by meta-regression the impact of several relevant factors (HCV viral load, HCV subtype, and HBV coinfection, among others) due to the lack of raw data in many articles. (iii) We could not test at what concentration of HCV Ag the RNA is always positive because the ARCHITECT HCV Ag assay uses a cutoff point set by the manufacturer. Besides, we have only aggregated data from each study, not data from the individuals included in these studies. (iv) The same protocols were not applied to extract and determine HCV-RNA, which could affect the results. Additional studies are needed to improve the evaluation of the diagnostic performance of the Abbott ARCHITECT HCV Ag assay in PLWHA.
Conclusions.
In conclusion, the accuracy of the Abbott ARCHITECT HCV Ag assay was very good for HCV screening in serum/plasma samples from PLWHA. The clinical utility to confirm HCV infection was acceptable in high-prevalence settings (≥10%), but poor in low-prevalence settings (≤1%) and excellent in excluding active HCV infection. This HCVcAg assay could help implement HCV screening programs in PLWHA in high-risk populations with difficult access to the health system (PWID, MSM).
ACKNOWLEDGMENTS
We thank the authors of the studies included in this review for providing additional information upon request.
The authors declare no competing interests.
This study was supported by grants from the Instituto de Salud Carlos III (ISCII; grant number PI20CIII/00004 to S.R., and PI19CIII/00009 to I.M.) and Gilead Science (grant number GLD20_0144 to S.R.). This research was also supported by CIBER (Consorcio Centro de Investigación Biomédica en Red) (CB 2021), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación, and Unión Europea – NextGenerationEU (CB21/13/00044). D.S.-C. is a Sara Borrell researcher from ISCIII (grant no. CD20CIII/00001). A.T.-N. is a PhD student in the Program in Biomedical Sciences and Public Health of the UNED International Doctoral School. No funding bodies had any role in study design, data collection, analysis, decision to publish, or manuscript preparation.
Daniel Sepúlveda-Crespo, investigation, resources, writing – original draft; Ana Treviño-Nakoura, investigation, resources, writing – review, and editing; José M. Bellón, investigation, methodology, formal analysis, writing – review, and editing; María A. Jiménez-Sousa, writing – review, and editing; Pablo Ryan, writing – review, and editing. Isidoro Martínez, methodology, writing – reviewing, and editing; Amanda Fernandez-Rodriguez, methodology, writing – reviewing, and editing; Salvador Resino, funding acquisition, conceptualization, formal analysis, writing – original draft, supervision.
Footnotes
Supplemental material is available online only.
Contributor Information
Salvador Resino, Email: sresino@isciii.es.
Alexander J. McAdam, Boston Children's Hospital
REFERENCES
- 1.Spearman CW, Dusheiko GM, Hellard M, Sonderup M. 2019. Hepatitis C. Lancet 394:1451–1466. 10.1016/S0140-6736(19)32320-7. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2022. Hepatitis C. https://www.who.int/news-room/fact-sheets/detail/hepatitis-c.
- 3.Sepulveda-Crespo D, Resino S, Martinez I. 2020. Hepatitis C virus vaccine design: focus on the humoral immune response. J Biomed Sci 27:78. 10.1186/s12929-020-00669-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. 2022. Global health sector strategies: viral hepatitis, 2016–2021. http://apps.who.int/gb/ebwha/pdf_files/WHA69/A69_32-en.pdf?ua=1.
- 5.World Health Organization. 2021. The global health sector strategies on, respectively, HIV, viral hepatitis and sexually transmitted infections 2022–2030. https://www.who.int/publications/i/item/9789240053779.
- 6.Platt L, Easterbrook P, Gower E, McDonald B, Sabin K, McGowan C, Yanny I, Razavi H, Vickerman P. 2016. Prevalence and burden of HCV co-infection in people living with HIV: a global systematic review and meta-analysis. Lancet Infect Dis 16:797–808. 10.1016/S1473-3099(15)00485-5. [DOI] [PubMed] [Google Scholar]
- 7.Nijmeijer BM, Koopsen J, Schinkel J, Prins M, Geijtenbeek TB. 2019. Sexually transmitted hepatitis C virus infections: current trends, and recent advances in understanding the spread in men who have sex with men. J Int AIDS Soc 22(Suppl 6):e25348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin F, Matthews GV, Grulich AE. 2017. Sexual transmission of hepatitis C virus among gay and bisexual men: a systematic review. Sex Health 14:28–41. 10.1071/SH16141. [DOI] [PubMed] [Google Scholar]
- 9.Aisyah DN, Shallcross L, Hully AJ, O'Brien A, Hayward A. 2018. Assessing hepatitis C spontaneous clearance and understanding associated factors: a systematic review and meta-analysis. J Viral Hepat 25:680–698. 10.1111/jvh.12866. [DOI] [PubMed] [Google Scholar]
- 10.Chen JY, Feeney ER, Chung RT. 2014. HCV and HIV co-infection: mechanisms and management. Nat Rev Gastroenterol Hepatol 11:362–371. 10.1038/nrgastro.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gobran ST, Ancuta P, Shoukry NH. 2021. A tale of two viruses: immunological insights into HCV/HIV Coinfection. Front Immunol 12:726419. 10.3389/fimmu.2021.726419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. 2017. Global hepatitis report, 2017. https://www.who.int/publications/i/item/9789241565455.
- 13.Ghany MG, Morgan TR, AASLD-IDSA Hepatitis C Guidance Panel . 2020. Hepatitis C guidance 2019 update: American Association for the Study of Liver Diseases–Infectious Diseases Society of America recommendations for testing. Hepatology 71:686–721. 10.1002/hep.31060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fevery B, Susser S, Lenz O, Cloherty G, Perner D, Picchio G, Sarrazin C. 2014. HCV RNA quantification with different assays: implications for protease-inhibitor-based response-guided therapy. Antivir Ther 19:559–567. 10.3851/IMP2760. [DOI] [PubMed] [Google Scholar]
- 15.Pawlotsky J-M, Negro F, Aghemo A, Berenguer M, Dalgard O, Dusheiko G, Marra F, Puoti M, Wedemeyer H. 2020. EASL recommendations on treatment of hepatitis C: final update of the series. J Hepatology 73:1170–1218. 10.1016/j.jhep.2020.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Galli C, Julicher P, Plebani M. 2018. HCV core antigen comes of age: a new opportunity for the diagnosis of hepatitis C virus infection. Clin Chem Lab Med 56:880–888. 10.1515/cclm-2017-0754. [DOI] [PubMed] [Google Scholar]
- 17.Benito R, Arribas J, Algarate S, Cebollada R, Gude MJ. 2018. Hepatitis C virus core antigen for screening organ donors and recipients. Diagn Microbiol Infect Dis 91:126–129. 10.1016/j.diagmicrobio.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Feld JJ. 2018. Hepatitis C virus diagnostics: the road to simplification. Clin Liver Dis (Hoboken) 12:125–129. 10.1002/cld.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Jie W, Ling J, Yuanshuai H. 2021. HCV core antigen plays an important role in the fight against HCV as an alternative to HCV-RNA detection. J Clin Lab Anal 35:e23755. 10.1002/jcla.23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mederacke I, Potthoff A, Meyer-Olson D, Meier M, Raupach R, Manns MP, Wedemeyer H, Tillmann HL. 2012. HCV core antigen testing in HIV- and HBV-coinfected patients, and in HCV-infected patients on hemodialysis. J Clin Virol 53:110–115. 10.1016/j.jcv.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Garbuglia AR, Monachetti A, Galli C, Sabatini R, Ferreri ML, Capobianchi MR, Bagnarelli P. 2014. HCV core antigen and HCV-RNA in HIV/HCV co-infected patients with different HCV genotypes. BMC Infect Dis 14:222. 10.1186/1471-2334-14-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Helden J, Weiskirchen R. 2014. Hepatitis C diagnostics: clinical evaluation of the HCV-core antigen determination. Z Gastroenterol 52:1164–1170. (In German.) 10.1055/s-0034-1366618. [DOI] [PubMed] [Google Scholar]
- 23.Cresswell FV, Fisher M, Hughes DJ, Shaw SG, Homer G, Hassan-Ibrahim MO. 2015. Hepatitis C core antigen testing: a reliable, quick, and potentially cost-effective alternative to hepatitis C polymerase chain reaction in diagnosing acute hepatitis C virus infection. Clin Infect Dis 60:263–266. 10.1093/cid/ciu782. [DOI] [PubMed] [Google Scholar]
- 24.Vanhommerig JW, van de Laar TJ, Koot M, van Rooijen MS, Schinkel J, Speksnijder AG, Prins M, de Vries HJ, Bruisten SM. 2015. Evaluation of a hepatitis C virus (HCV) antigen assay for routine HCV screening among men who have sex with men infected with HIV. J Virol Methods 213:147–150. 10.1016/j.jviromet.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 25.Arboledas JCA, Guerrero IP, Rodriguez MJB, Martos ET, Perez AB, Leon CC, Sierra Sanchez JF, Prieto MDL, Porcuna NC, Mochon MDO, Macias J, de la Iglesia Salgado A, Granger JR, Fernandez MD, Lozano IG, Ramirez ER, Rivero A, Del Carmen Lozano Dominguez M, Viciana I, Montemayor JCG, Garcia FG. 2017. Hepatitis C virus core antigen in the management of patients treated with new direct-acting antivirals. Diagn Microbiol Infect Dis 89:29–34. 10.1016/j.diagmicrobio.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Duchesne L, Njouom R, Lissock F, Tamko-Mella GF, Rallier S, Poiteau L, Soulier A, Chevaliez S, Vernet G, Rouveau N, Pawlotsky JM, Girard PM, Lacombe K. 2017. HCV Ag quantification as a one-step procedure in diagnosing chronic hepatitis C infection in Cameroon: the ANRS 12336 study. J Int AIDS Soc 20:21446. 10.7448/IAS.20.1.21446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hullegie SJ, GeurtsvanKessel CH, van der Eijk AA, Ramakers C, Rijnders BJA. 2017. HCV antigen instead of RNA testing to diagnose acute HCV in patients treated in the Dutch Acute HCV in HIV Study. J Int AIDS Soc 20:21621. 10.7448/IAS.20.1.21621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohamed Z, Mbwambo J, Shimakawa Y, Poiteau L, Chevaliez S, Pawlotsky JM, Rwegasha J, Bhagani S, Taylor-Robinson SD, Makani J, Thursz MR, Lemoine M. 2017. Clinical utility of HCV core antigen detection and quantification using serum samples and dried blood spots in people who inject drugs in Dar-es-Salaam, Tanzania. J Int AIDS Soc 20:21856. 10.7448/IAS.20.1.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talal AH, Chen Y, Zeremski M, Zavala R, Sylvester C, Kuhns M, Brown LS, Markatou M, Cloherty GA. 2017. Hepatitis C virus core antigen: a potential alternative to HCV RNA testing among persons with substance use disorders. J Subst Abuse Treat 78:37–42. 10.1016/j.jsat.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Alonso R, Perez-Garcia F, Lopez-Roa P, Alcala L, Rodeno P, Bouza E. 2018. HCV core-antigen assay as an alternative to HCV RNA quantification: a correlation study for the assessment of HCV viremia. Enferm Infecc Microbiol Clin (Engl Ed) 36:175–178. 10.1016/j.eimc.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Chayanupatkul M, Chittmittraprap S, Pratedrat P, Chuaypen N, Avihingsanon A, Tangkijvanich P. 2020. Efficacy of elbasvir/grazoprevir therapy in HCV genotype-1 with or without HIV infection: role of HCV core antigen monitoring and improvement of liver stiffness and steatosis. Antivir Ther 25:305–314. 10.3851/IMP3370. [DOI] [PubMed] [Google Scholar]
- 32.Rossetti B, Loggi E, Raffaelli CS, Mercinelli S, Gandolfo C, Savellini GG, Galli S, Vitale G, Di Donato R, Vukotic R, Grandini E, Margotti M, Guarneri V, Furlini G, Re MC, De Luca A, Andreone P, Galli C, Cusi MG. 2021. Hepatitis C Virus Core Antigen (HCVAg): an affordable assay to monitor the efficacy of treatment in DAAs era. New Microbiol 44:89–94. [PubMed] [Google Scholar]
- 33.Sun HY, Liu WD, Wang CW, Wei YJ, Lin KY, Huang YS, Su LH, Chen YT, Liu WC, Su YC, Chen YW, Chuang YC, Lu PL, Hung CC, Yu ML. 2022. Performance of hepatitis C virus (HCV) core antigen assay in the diagnosis of recently acquired HCV infection among high-risk populations. Microbiol Spectr 10:e0034522. 10.1128/spectrum.00345-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freiman JM, Tran TM, Schumacher SG, White LF, Ongarello S, Cohn J, Easterbrook PJ, Linas BP, Denkinger CM. 2016. Hepatitis C core antigen testing for diagnosis of hepatitis C virus infection: a systematic review and meta-analysis. Ann Intern Med 165:345–355. 10.7326/M16-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin NK, Boerekamps A, Hill AM, Rijnders BJA. 2018. Is hepatitis C virus elimination possible among people living with HIV and what will it take to achieve it? J Int AIDS Soc 21(Suppl 2):e25062. 10.1002/jia2.25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cochrane. 2022. Cochrane handbook for systematic reviews of diagnostic test accuracy version 2. Cochrane, London, England. [Google Scholar]
- 37.McInnes MDF, Moher D, Thombs BD, McGrath TA, Bossuyt PM, the PRISMA-DTA Group . 2018. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. JAMA 319:388–396. 10.1001/jama.2017.19163. [DOI] [PubMed] [Google Scholar]
- 38.Easterbrook PJ, Roberts T, Sands A, Peeling R. 2017. Diagnosis of viral hepatitis. Curr Opin HIV AIDS 12:302–314. 10.1097/COH.0000000000000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dwamena B. 2009. Midas: Stata module for meta-analytical integration of diagnostic test accuracy studies (Statistical Software Components S456880). Boston College Department of Economics, Boston, MA. [Google Scholar]
- 40.Macaskill P, Gatsonis C, Deeks J, Harbord R, Takwoingi Y. 2010. Cochrane handbook for systematic reviews of diagnostic test accuracy. Version 1 0 The Cochrane Collaboration, London, England. [Google Scholar]
- 41.Bradley AP. 1997. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Patt Recog 30:1145–1159. 10.1016/S0031-3203(96)00142-2. [DOI] [Google Scholar]
- 42.Rubinstein ML, Kraft CS, Parrott JS. 2018. Determining qualitative effect size ratings using a likelihood ratio scatter matrix in diagnostic test accuracy systematic reviews. Diagnosis (Berl) 5:205–214. 10.1515/dx-2018-0061. [DOI] [PubMed] [Google Scholar]
- 43.Fagan TJ. 1975. Letter: Nomogram for Bayes theorem. N Engl J Med 293:257. 10.1056/NEJM197507312930513. [DOI] [PubMed] [Google Scholar]
- 44.Higgins JP, Thompson SG, Deeks JJ, Altman DG. 2003. Measuring inconsistency in meta-analyses. BMJ 327:557–560. 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galbraith RF. 1988. Graphical display of estimates having differing standard errors. Technometrics 30:271–281. 10.1080/00401706.1988.10488400. [DOI] [Google Scholar]
- 46.Rousseeuw PJ, Ruts I, Tukey JW. 1999. The bagplot: a bivariate boxplot. The American Statistician 53:382–387. 10.2307/2686061. [DOI] [Google Scholar]
- 47.Deeks JJ, Macaskill P, Irwig L. 2005. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 58:882–893. 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Aguilera A, Alados JC, Alonso R, Eiros JM, Garcia F. 2020. Current position of viral load versus hepatitis C core antigen testing. Enferm Infecc Microbiol Clin (Engl Ed) 38(Suppl 1):12–18. 10.1016/j.eimc.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Hosseini-Hooshyar S, Hajarizadeh B, Bajis S, Law M, Janjua NZ, Fierer DS, Chromy D, Rockstroh JK, Martin TCS, Ingiliz P, Hung CC, Dore GJ, Martinello M, Matthews GV. 2022. Risk of hepatitis C reinfection following successful therapy among people living with HIV: a global systematic review, meta-analysis, and meta-regression. Lancet HIV 9:e414–e427. 10.1016/S2352-3018(22)00077-7. [DOI] [PubMed] [Google Scholar]
- 50.The Cochrane Collaboration. 2022. Cochrane handbook for systematic reviews of diagnostic test accuracy version 2.0. https://training.cochrane.org/handbook-diagnostic-test-accuracy. Accessed 6 June 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download jcm.01331-22-s0001.pdf, PDF file, 1.0 MB (1,002.4KB, pdf)
Data Availability Statement
All relevant data are within the article and the supplemental material.