ABSTRACT
There has been significant increase in the use of molecular tools for the diagnosis of invasive aspergillosis (IA) and mucormycosis. However, their range of detection may be too limited as species diversity and coinfections are increasing. Here, we aimed to evaluate a molecular workflow based on a new multiplex PCR assay detecting the whole Aspergillus genus and the Mucorales order followed by a species-specific PCR or a DNA-sequencing approach for IA and/or mucormycosis diagnosis and species identification on serum. Performances of the MycoGENIE Aspergillus spp./Mucorales spp. duplex PCR kit were analyzed on a broad range of fungal strains and on sera from high-risk patients prospectively over a 12-month period. The kit allowed the detection of nine Aspergillus species and 10 Mucorales (eight genera) strains assessed. No cross-reactions between the two targets were observed. Sera from 744 patients were prospectively analyzed, including 35 IA, 16 mucormycosis, and four coinfections. Sensitivity varies from 85.7% (18/21) in probable/proven IA to 28.6% (4/14) in COVID-19-associated pulmonary aspergillosis. PCR-positive samples corresponded to 21 A. fumigatus, one A. flavus, and one A. nidulans infections. All the disseminated mucormycosis were positive in serum (14/14), including the four Aspergillus coinfections, but sensitivity fell to 33.3% (2/6) in localized forms. DNA sequencing allowed Mucorales identification in serum in 15 patients. Remarkably, the most frequent species identified was Rhizomucor pusillus (eight cases), whereas it is barely found in fungal culture. This molecular workflow is a promising approach to improve IA and mucormycosis diagnosis and epidemiology.
KEYWORDS: invasive aspergillosis, mucormycosis, molecular diagnosis, fungal PCR, DNA sequencing, Aspergillus, Mucorales, Rhizomucor pusillus, Cunninghamella
INTRODUCTION
Invasive mold infections (IMI), including invasive aspergillosis (IA) and mucormycosis, are life-threatening diseases occurring mainly in critically ill patients (1, 2). Although an early targeted antifungal therapy is essential for IMI management and therapeutic success, their diagnoses are still challenging. Indeed, as histopathological analysis of deep-seated biopsies showing hyphae (septate for Aspergillus or nonseptate for Mucorales) is barely performed, the diagnosis relies mainly on a combination of clinical, radiological, and mycological features (3–5). Among the latter, molecular tools, such as real-time PCR, have shown potential on blood-derived samples, respiratory samples, or deep-seated biopsies (6–11). Aspergillus PCR was included in the last version of the criteria of the European Organization of Research and Treatment of Cancer (EORTC)/Mycoses Study Group Education and Research Consortium (MSGERC) for the definition of IA (12). Even though Mucorales PCR was not yet included in these criteria because of a lack of standardization or test availability, it has also greatly improved mucormycosis diagnosis in last years (13). Indeed, because fungal culture often fails and no antigenic biomarkers are prospectively validated yet (14), PCR might provide the sole mycological evidence during mucormycosis. Aspergillus and Mucorales PCR have also shown utility for the disease prognostic (15, 16), or for the detection of Aspergillus fumigatus azole-resistance mutations (17).
Despite undeniable advantages of real-time PCR for IMI diagnosis and management, it has some limitations. Indeed, in contrast to fungal culture, PCR is limited by its individual range of detection. Even if IA are mainly related to the species A. fumigatus, due to the change in the Aspergillus taxonomy and the discover of cryptic species, up to 30 several Aspergillus species have been involved in IA (18). Conversely, mucormycosis, are related to a broad range of genus and species belonging to the Mucorales order, including the most frequent genera Rhizopus, Mucor, or Lichtheimia (2, 19). However, genus repartition differs across geographical sites and up to nine genera have been involved in human disease (19). Consequently, because commercial real-time PCR kits are limited to few species for Aspergillus or certain genera for Mucorales, causative-species may be outside their scope and infections may be underdiagnosed. Moreover, coinfections are increasingly reported, but also may be underdiagnosed if Aspergillus and Mucorales PCR are not combined (20).
Recently, a multiplex real-time PCR assay simultaneously targeting the whole Aspergillus genus and the whole Mucorales order, has been commercialized to overcome these issues (MycoGENIE Aspergillus spp./Mucorales spp. real-time PCR assay, Ademtech). However, with this approach, species or genus identification is not achievable, whereas it could be of interest for antifungal adaptation, especially in mucormycosis given that susceptibility patterns to azole drugs have been shown to be species or genus specific (21).
Here, we first aimed to evaluate analytical performances of the MycoGENIE Aspergillus spp./Mucorales spp. real-time PCR assay. In addition, we also aimed to set-up and evaluate a molecular workflow based on this multiplex PCR assay followed by a species-specific PCR or a DNA-sequencing approach, for the diagnosis of IA and mucormycosis and species identification, on serum.
MATERIALS AND METHODS
Study design and patients.
The study was conducted in a single center at the Bordeaux University hospital (France) and was divided into two steps. The first step consisted in the determination of the analytical performances of the multiplex PCR assay, using a broad range of fungal strains. The second step focused on the clinical evaluation of the workflow using serum samples collected prospectively from patients at high risk for IMI, between September 2020 and August 2021. Cases were classified as proven or probable IMI according to the EORTC/MSGERC criteria (12), putative IA according to the AspICU criteria for IA in intensive care unit patients (22), or COVID-19 Associated Pulmonary Aspergillosis (CAPA) according to the ECMM/ISHAM consensus criteria for severe COVID-19 patients (23). This study complies with the ethical and legal requirements of French law (April 15, 2019) and the Declaration of Helsinki. Written or verbal informed consent from all participants was not required because samples were collected through routine clinical work and patient identifiable information were anonymized prior to analysis.
Sample collection and processing.
A panel of pure fungal strains obtained from clinical practice and accurately identified by MALDI-TOF mass spectrometry using the MSI-2 online platform (24) were used to determine analytical performances. Strains were harvested from Sabouraud agar culture plates, transferred into 800 μL of lysis buffer (Roche), and submitted to ultrasonic lysis. DNA extraction was then performed on the MagNAPure Compact device (Roche), using 400 μL of fungal lysate to an elution volume of 50 μL. For the prospective step, all the sera sent to the parasitology mycology laboratory for the diagnosis of IMI during the study period were included. DNA extraction was performed routinely on the MagNAPure 96 device (Roche), using the Viral NA plasma extraction kit (Roche), from 1 mL of serum to an elution volume of 50 μL. A positive (i.e., pool of negative sera spiked with A. fumigatus DNA [AmpliRun, Vircell]) and negative (nuclease-free water) controls were used during each run to validate the extraction step. DNA extracts were then stored at +4°C less than a week or at −20°C further.
Molecular workflow.
(i) MycoGENIE PCR assays. PCR assays were performed on the LightCycler 480 system (Roche) using the MycoGENIE Aspergillus spp./Mucorales spp. real-time duplex PCR kit (Ademtech, Pessac, France), which simultaneous targets both the 28S rDNA regions of the Aspergillus genus and the Mucorales order as well as an internal PCR control. DNA extracts from positive and negative extraction controls were included in each run and served, respectively, as internal quality control and negative PCR control. External positive controls provided by the manufacturer for each target were also included in each run. Amplifications were performed according to manufacturer’s recommendations and amplification curves were analyzed on the LC480 software. Cycle threshold (Ct) values lower than 40 were considered positive results for both targets, according to manufacturer’s instruction.
Serum samples screened positive for the Aspergillus spp. target were further assessed, testing the same DNA extract, with the MycoGENIE Aspergillus fumigatus/TR34/L98H specific PCR assay (Ademtech, Pessac, France), targeting specifically the species A. fumigatus and the cyp51a mutations TR34 and L98H, associated with azole resistance.
(ii) Mucorales DNA sequence-based identification. DNA extracts from samples screened positive for the pan-Mucorales target and fungal strains were submitted for Sanger DNA sequencing, targeting a part of the 18S rDNA. Briefly, the 18S rDNA fragment was amplified using the primers ZM1mo/ZM3mod as previously described (10 min of initial denaturation at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C. and 30 s at 72°C) (25). Amplicons and fragment size were then checked (from 170 to 190 pb) on the QIAxcel Advanced device (Qiagen), purified (HT ExoSAP-IT High-Throughput PCR Product Cleanup, Affymetrix) and submitted to a sequencing PCR using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Finally, purified products were separated and analyzed on the Genetic Analyzer 3500xL Dx (Applied Biosystems). Negative and positive controls (i.e., nuclease-free water and DNA extract from a Mucorales strain, respectively) were used in each sequencing run.
Resulting DNA sequences were analyzed using the ChromasPro v1.7.1 software (Technelysium Pty Ltd.) and identified using the nucleotide Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (NCBI) (available at https://blast.ncbi.nlm.nih.gov/Blast.cgi), searching the curated 18S rRNA RefSeq database. According to the best identification percentages obtained, the final identification was given at the species level (only one species with the best identification percentage), the species complex level (two or more species belonging to same species complex with the same, or close, identification percentage) or the genus level (two or more species belonging to the same genus, but not to same species complex, with the same, or close, identification percentage). Mucorales species identification was also performed by establishing phylogenetic trees, including a broad range of Mucorales type-strains sequences, using MEGA X and iTOL v6 (https://itol.embl.de).
Statistical analysis.
Statistical analyses were performed using Fischer’s exact, Mann-Whitney, and Wilcoxon matched-pairs signed rank tests as appropriate (Prism 9 software). P < 0.05 (two-tailed) was considered statistically significant.
RESULTS
Analytical performances of the MycoGENIE Aspergillus spp./Mucorales spp. PCR assay.
Limits of detection (LOD), using A. fumigatus and Lichtheimia corymbifera genomic DNA, were determined at 10 copies/mL for both targets. At LOD, mean Ct +/– standard deviation were 36.06 ± 0.31 and 35.56 ± 0.45 for the Aspergillus and Mucorales targets, respectively and 30.66 ± 0.063 and 31.61 ± 0.067 at 100× LOD, respectively.
Regarding fungal strains, 45 species were assessed, including nine Aspergillus species, 10 Mucorales species, 19 fungal species causing bloodstream infection, and seven other molds (Table 1). All Aspergillus and Mucorales species were detected by the corresponding target. Cross reactions were observed for the Aspergillus spp. target with Penicillium, Paecilomyces, and Purpureocillium species, but not for the Mucorales target (Table 1).
TABLE 1.
Results of the MycoGENIE Aspergillus spp./Mucorales spp. multiplex PCR assay obtained on pure fungal strains
| Strains | Aspergillus spp. target | Mucorales spp. target |
|---|---|---|
| Aspergillus genus | ||
| A. fumigatus | + | − |
| A. flavus | + | − |
| A. niger | + | − |
| A. welwitschiae | + | − |
| A. terreus | + | − |
| A. nidulans | + | − |
| A. sublatus | + | − |
| A. nishimurae | + | − |
| A. sydowii | + | − |
| Mucorales order | ||
| Rhizopus arrhizus | − | + |
| Rhizopus microsporus | − | + |
| Rhizomucor pusillus | − | + |
| Mucor indicus | − | + |
| Mucor circinelloides | − | + |
| Lichtheimia corymbifera | − | + |
| Apophysomyces sp. | − | + |
| Cunninghamella sp. | − | + |
| Syncephalastrum sp. | − | + |
| Mycotypha sp. | − | + |
| Other fungi causing bloodstream infection | ||
| Candida albicans | − | − |
| Candida glabrata | − | − |
| Candida auris | − | − |
| Candida parapsilosis | − | − |
| Candida tropicalis | − | − |
| Candida krusei | − | − |
| Cryptococcus neoformans | − | − |
| Saprochaete clavata | − | − |
| Magnusiomyces capitatus | − | − |
| Trichosporon inkin | − | − |
| Exophiala oligosperma | − | − |
| Exophiala spinifera | − | − |
| Fusarium verticilloides | − | − |
| Fusarium oxysporum | − | − |
| Fusarium dimerum | − | − |
| Fusarium falciforme | − | − |
| Fusarium proliferatum | − | − |
| Scedosporium apiospermum | − | − |
| Lomentospora prolificans | − | − |
| Other environmental molds | ||
| Penicillium roqueforti | + | − |
| Paecilomyces variotii | + | − |
| Purpureocillium lilacinum | + | − |
| Acremonium sclerotigenum | − | − |
| Phaeoacremonium parasiticum | − | − |
| Sarocladium kiliense | − | − |
| Trichoderma sp. | − | − |
Clinical evaluation on sera for IMI diagnosis.
During the prospective step of the study, 2,392 serum samples from 744 patients at risk of IMI were sent to the parasitology mycology department for IMI diagnosis and screened by the Mycogenie Aspergillus spp./Mucorales spp. PCR assay. Fifty-five patients were diagnosed as having an IMI, including 35 IA (two proven, 16 probable, three putative, and 14 CAPA), 16 mucormycosis (10 disseminated, three posttraumatic, and three digestive), and four Aspergillus/Mucorales coinfections (three probable and one putative IA coexisting with four mucormycosis). One patient had a mucormycosis relapse 4 months after the first episode. Mycological findings of IMI cases are listed in Tables 2 and 3.
TABLE 2.
Mycological characteristics of the 39 patients with invasive aspergillosisa
| IA patients (n = 39) | IA classification | Serum results |
Other samples results |
||||
|---|---|---|---|---|---|---|---|
| Aspergillus spp./A. fumigatus PCR (Ct) | GM antigenb | Sample type | Aspergillus spp./A. fumigatus PCR (Ct) | GM antigenb | Mycological examination (DE and culture)c | ||
| P8d | Probable IPA | 35.7/34.5 | <0.5 | BAL | >40/>40 | <0.5 | Negative |
| P13 | Proven IA | 39.5/>40 | <0.5 | TA | >40/>40 | <0.5 | Negative |
| Cutaneous biopsy | 39/>40 | NA | Positive DE and culture (A. flavus) | ||||
| P16d | Probable IPA | 32.8/>40 | <0.5 | BAL | NA/NA | NA | Negative DE/Positive culture (A. nidulans) |
| P19d | Probable IPA | 29/28.6 | 3.4 | BAL | >40/>40 | <0.5 | Negative |
| P20 | Proven IPA | 34.7/32 | <0.5 | BAL | 33.4/40 | 3.07 | Negative |
| Pulmonary biopsy | 37.6/36 | NA | Negative DE/Positive culture (A. fumigatus) | ||||
| P21d | Putative IPA | 36.3/36.3 | 1.01 | BAL | 30.7/29.2 | 9.98 | Positive DE/Negative culture |
| P23 | Probable IPA | 33.4/33.6 | <0.5 | TA | 23.6/22.6 | NA | Negative DE/Positive culture (A. fumigatus + A. flavus) |
| P24 | Probable IPA | 33.6/30.8 | 0.87 | BAL | 31.6/30.5 | 5.19 | Negative |
| P25 | Probable IPA | 33.7/32.7 | 4.50 | Induce sputum | NA/NA | NA | Negative DE/Positive culture (A. fumigatus) |
| P26 | Probable IPA | 34/32.9 | 0.62 | Induce sputum | NA/NA | NA | Negative DE/Positive culture (A. fumigatus) |
| P27 | Probable IPA | 34.2/33.2 | 0.69 | BAL | 34.7/34 | 0.73 | Negative DE/Positive culture (A. fumigatus + A. welwitschiae) |
| P28 | Probable IPA | 35.5/31.9 | <0.5 | BAL | > 40/>40 | <0.5 | Negative |
| P29 | Probable IPA | 36/34.2 | 9.57 | NA | NA/NA | NA | NA |
| P30 | Probable IPA | 36/35.4 | 1.85 | BAL | > 40/>40 | <0.5 | Negative |
| P31 | Probable IPA | 38.9/36.4 | 0.67 | BAL | 36.9/35.8 | 9.6 | Negative |
| P32 | Probable IPA | 36.6/34.5 | <0.5 | BAL | 26.5/25.7 | 5.9 | Negative DE/Positive culture (A. fumigatus) |
| P33 | Probable IPA | 36.8/>40 | <0.5 | BAL | > 40/>40 | <0.5 | Negative |
| P34 | Probable IPA | 37.9/36.2 | <0.5 | BAL | 34.3/32.2 | 3.02 | Negative |
| P35 | Probable IPA | 37.1/35.9 | <0.5 | BAL | 37.2/34.9 | <0.5 | Negative DE/Positive culture (A. fumigatus) |
| P36 | Probable IPA | >40/NA | <0.5 | BAL | 34.3/NA | NA | Negative DE/Positive culture (A. fumigatus) |
| P37 | Probable IPA | >40/NA | <0.5 | BAL | 34.7/NA | <0.5 | Negative DE/Positive culture (A. fumigatus) |
| P38 | Probable IPA | >40/>40 | <0.5 | BAL | 35.5/33.6 | 0.58 | Negative |
| Pleural fluid | 33/30.8 | NA | Negative | ||||
| P39 | Putative IPA | 34.2/33.2 | 9.05 | BAL | 29.4/NA | 11.1 | Positive DE and culture (A. fumigatus) |
| P40 | Putative IPA | 34.6/33.2 | <0.5 | BAL | 34.2/32.8 | 1.76 | Positive DE and culture (A. fumigatus) |
| P41 | Putative IPA | >40/>40 | <0.5 | BAL | 28.9/NA | 2.34 | Positive DE and culture (A. fumigatus) |
| P42 | CAPA | 33/30.9 | 1.21 | BAL | 29/28.1 | 5.26 | Positive DE and culture (A. fumigatus + A. citrinoterreus) |
| P43 | CAPA | 33.4/32.2 | 2.54 | TA | NA/NA | NA | Negative DE/Positive culture (A. fumigatus) |
| P44 | CAPA | 33.6/31.9 | 0.52 | NA | NA/NA | NA | NA |
| P45 | CAPA | 33.7/34.1 | 0.51 | TA | NA/NA | NA | Positive DE and culture (A. fumigatus) |
| P46 | CAPA | >40/>40 | <0.5 | BAL | 33.5/33.5 | <0.5 | Negative DE/Positive culture (A. fumigatus) |
| P47 | CAPA | >40/>40 | <0.5 | BAL | 35.2/NA | 2.42 | Negative DE/Positive culture (A. fumigatus) |
| P48 | CAPA | >40/>40 | <0.5 | BAL | 31.6/30.6 | 3.75 | Negative DE/Positive culture (A. fumigatus) |
| P49 | CAPA | >40/>40 | <0.5 | BAL | 32.5/30 | 1.61 | Negative DE/Positive culture (A. fumigatus) |
| P50 | CAPA | >40/>40 | <0.5 | BAL | 33.6/30.5 | 1.36 | Negative DE/Positive culture (A. fumigatus + A. terreus) |
| P51 | CAPA | >40/>40 | <0.5 | BAL | 33.2/30.7 | 0.68 | Negative |
| P52 | CAPA | >40/>40 | <0.5 | BAL | 33.9/31.2 | <0.5 | Negative DE/Positive culture (A. fumigatus) |
| P53 | CAPA | >40/>40 | <0.5 | BAL | 33.2/NA | <0.5 | Positive DE and culture (A. fumigatus + A. felis) |
| P54 | CAPA | >40/>40 | <0.5 | BAL | 27.6/>40 | <0.5 | Negative |
| P55 | CAPA | >40/>40 | <0.5 | TA | 29.9/28.4 | NA | Negative DE/Positive culture (A. fumigatus) |
IA, invasive aspergillosis; IPA, invasive pulmonary aspergillosis; CAPA, COVID-19 associated pulmonary aspergillosis; BAL, bronchioalveolar lavage; TA, tracheal aspirate; GM, galactomannan; DE, direct examination; NA, not available.
GM antigen was detected using the Platelia Aspergillus Ag assay (Bio-Rad, France), according to manufacturer's instruction.
Fungal culture was performed on Sabouraud Chloramphenicol Gentamicine tube (Bio-Rad, France) incubated at 30°C during 21 days.
Indicates patients with Aspergillus/Mucorales coinfection.
TABLE 3.
Mycological characteristics of the 20 patients with mucormycosisa
| Patients (n = 20) | Mucormycosis classification | Serum |
Other samples |
||||
|---|---|---|---|---|---|---|---|
| Mucorales PCR (Ct) | DNA Sequencing | Sample type | Mucorales PCR (Ct) | DNA Sequencing | Mycological examination (DE and culture)b | ||
| P1 | Disseminated | 30 | Rh. pusillus | BAL | 35.3 | Rh. pusillus | Negative |
| P2 | Disseminated | 29.6 | Rh. pusillus | NA | NA | NA | NA |
| P3 | Disseminated | 32.8 | NA | NA | NA | NA | NA |
| P4 | Disseminated | 32.3 | R. microsporus complex | NA | NA | NA | NA |
| P5 | Digestive | >40 | NA | BAL | >40 | NA | Negative |
| Stool | 35 | Failed | Negative DE/Positive culture (Syncephalastrum racemosum) | ||||
| P6 | Digestive | 33.3 | Lichtheimia sp. | BAL | >40 | NA | Negative |
| Stool | 36.1 | Failed | Negative | ||||
| P7 | Disseminated | 34.4 | Rh. pusillus | BAL | 31.7 | Rh. pusillus | Negative |
| P8c | Disseminated | 28.7 | Rh. pusillus | BAL | >40 | NA | Negative |
| P9 | Disseminated | 28.9 | Rh. pusillus | BAL | 30.2 | Rh. pusillus | Positive DE/Negative culture |
| Pericardial fluid | 35.7 | Rh. pusillus | Negative | ||||
| P10 | Digestive | >40 | NA | BAL | >40 | NA | Negative |
| Stool | 38.6 | Failed | Positive DE and culture (M. circinelloides) | ||||
| P11 | Posttraumatic | 32.9 | Mucor circinelloides complex | Cutaneous biopsy | 32 | Mucor circinelloides complex | Negative DE/Positive culture (M. circinelloides) |
| P12 | Disseminated | 33.1 | R. microsporus complex | BAL | 25.7 | R. microsporus complex | Negative DE/Positive culture (R. microsporus) |
| P14 | Posttraumatic | >40 | NA | Cutaneous biopsy | 32.3 | Mucor circinelloides complex | Negative DE/Positive culture (M. circinelloides) |
| P15 | Disseminated | 30.1 | Rh. pusillus | BAL | 32.9 | Rh. pusillus | Negative |
| P16c | Disseminated | 36.9 | R. microsporus complex | BAL | 35.1 | R. microsporus complex | Negative |
| P17 | Posttraumatic | >40 | NA | Cutaneous biopsy | 34.5 | Rh. pusillus | Negative |
| P18d | Disseminated | 33 | Cunninghamella sp. | BAL | 31.7 | Cunninghamella sp. | Negative |
| Pulmonary biopsy | NA | NA | Positive DE/Negative culture | ||||
| P19c | Disseminated | 31.8 | Rh. pusillus | BAL | >40 | NA | Negative |
| P21c | Disseminated | 26.9 | Rh. pusillus | BAL | >40 | NA | Negative |
| P22 | Disseminated | 31.7 | Cunninghamella sp. | BAL | 19.4 | Cunninghamella sp. | Positive DE and culture (Cunninghamella elegans) |
BAL, Bronchioalveolar lavage; DE, direct examination; NA, not available.
Fungal culture was performed on Sabouraud Chloramphenicol Gentamicin tube (Bio-Rad, France) incubated at 30°C during 21 days.
Indicates patients with Aspergillus/Mucorales coinfection.
Indicates the patient with a mucormycosis relapse.
Among the patients suffering from IA (n = 39), Aspergillus spp. PCR was positive in serum in only 25 patients (64.1%) (Table 2). However, the sensitivity varied greatly according to the IA classification. Indeed, the sensitivity in serum was significantly higher in patients with proven/probable/putative IA (21/25, 84%) than in CAPA patients (4/14, 28.5%) (P = 0.0012, Fischer’s exact test). All positive sera were assessed for the MycoGENIE Aspergillus fumigatus/TR34/L98H specific PCR assay. All except three were also positive for the A. fumigatus target, allowing the identification of A. fumigatus as the causative IA agent in 22 patients. Interestingly, Ct values were significantly lower (median of 1.3 Ct) for the A. fumigatus target than the Aspergillus spp. target (P < 0.001, Wilcoxon matched-pairs signed rank test). Among the three A. fumigatus PCR-negative patients, two had a fungal culture positive to another species (one A. flavus and one A. nidulans). Finally, one serum sample was positive for both TR34 and L98H targets, which was in accordance with antifungal susceptibility testing of the A. fumigatus isolate retrieved in BAL and showing pan-azoles resistance.
Regarding mucormycosis, the Mucorales spp. PCR was positive in serum in 16 of the 20 infected patients (80%), including the four Aspergillus coinfections (Table 3). Sensitivity in serum was 100% for the disseminated forms (n = 14), whereas it was only 33.3% in digestive or posttraumatic cutaneous localized forms (n = 6). In contrast, fungal culture was positive in only six patients overall (30%), but on four of the six localized forms (66.7%).
Finally, among patients in whom IMI diagnosis was excluded (n = 684), the PCR assay was positive for the Aspergillus spp. and the Mucorales targets, in 10 and four patients, respectively. Interestingly, these patients exhibited significantly later Ct values than true positive patients for the Aspergillus target (mean of 37.1 versus 35.7 for true positive patients, P = 0.042, Mann-Whitney test) and for the Mucorales target (mean of 35.7 versus 33.2 for true positive patients, P = 0.039, Mann-Whitney test). Positive Ct values in no IMI patients were systematically > 35 and the positive result was not confirmed by complementary analyses. Diagnosis performances of the MycoGENIE Aspergillus/Mucorales assay are summarized in Table 4.
TABLE 4.
Performances of the MycoGENIE Aspergillus spp./Mucorales spp. multiplex PCR assay for the diagnosis of invasive aspergillosis, mucormycosis, or both infectionsa
| PCR targets | Sensitivity % (95% IC) | Specificity % (95% IC) | PPV % (95% IC) | NPV % (95% IC) | LR+ (95% IC) | LR– (95% IC) | DOR (95% IC) |
|---|---|---|---|---|---|---|---|
| Aspergillus target | 64.1 (48.4 to 77.3) | 98.6 (97.4 to 99.2) | 71.4 (55 to 83.7) | 98 (96.7 to 98.8) | 45.19 (18.69 to 100.3) | 0.364 (0.23 to 0.53) | 124.1 (47.72 to 280.4) |
| Mucorales target | 80 (58.4 to 91.9) | 99.5 (98.6 to 99.8) | 80 (58.4 to 91.9) | 99.5 (98.6 to 99.8) | 144.8 (41.42 to 417.9) | 0.201 (0.08 to 0.42) | 720 (153.7 to 2723) |
| Combined target | 67.3 (54.1 to 78.2) | 97.8 (96.6 to 98.8) | 72.6 (59.1 to 82.9) | 97.4 (95.9 to 98.4) | 33.11 (16 to 64.62) | 0.334 (0.22 to 0.48) | 99.11 (45.42 to 214.7) |
IC, interval confidence; PPV, positive predictive value; NPV, negative predictive value; LR+, positive likelihood ratio; LR–, negative likelihood ratio; DOR, diagnostic odds ratio.
Mucorales species identification on serum.
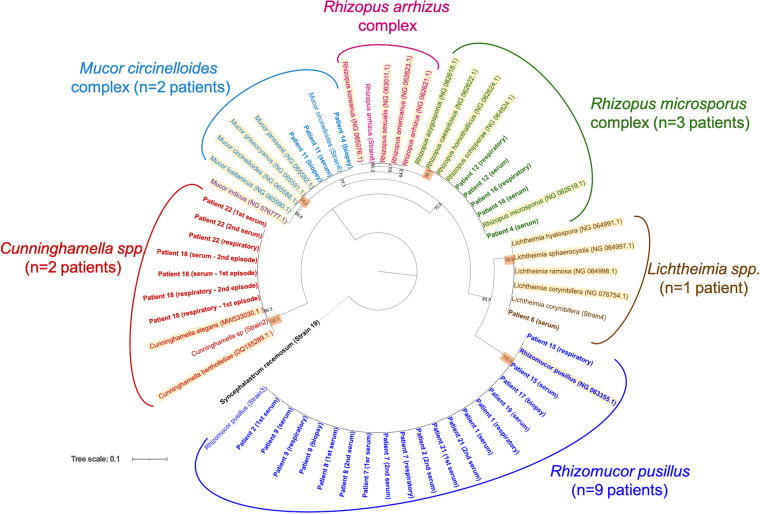
Twenty-four sera positive for the Mucorales PCR target were submitted to DNA sequencing for species identification. They originated from 15 of the 20 patients with mucormycosis and three of the four patients in whom the diagnosis of mucormycosis was excluded. Species or genus identification was achievable for the 15 patients with mucormycosis (21 samples), resulting in eight Rhizomucor pusillus, three Rhizopus microsporus complex, two Cunninghamella sp., one Mucor circinelloides complex, and one Lichtheimia sp. (Fig. 1). For the three patients in whom the diagnosis of mucormycosis was ruled out, the attempted sequencing assay failed suggesting a false positive result of the Mucorales spp. PCR assay.
FIG 1.
Maximum-likelihood phylogenetic tree obtained from the analysis of Mucorales 18S rRNA partial sequences. Data set includes sequences (i) from 17 patients positive for the Mycogenie Mucorales target in serum, in respiratory sample or in biopsy; (ii) from six Mucorales strains obtained from clinical practice and (iii) from 21 Mucorales type strains. Syncephalastrum racemosum was used as an outgroup. Numbers above the nodes correspond to bootstrapping value generated from 1,000 replicates. Only values above 60% are indicated. Patients are represented in bold as follows: patient number (sample type +/episode). “Respiratory” means respiratory sample such as bronchio-alveolar lavage, bronchial aspiration, or induce sputum. “Biopsy” means deep-seated or cutaneous biopsies. Type strains are highlighted as follows: species name (NCBI accession number).
DISCUSSION
In the present study, we evaluated the performances of the MycoGENIE Aspergillus spp./Mucorales spp. multiplex PCR assay. We also evaluated its performances on serum in the diagnostic strategy of IA and mucormycosis, in combination with a species-specific PCR or a DNA-sequencing approach for species identification. This multiplex PCR assay showed a high analytical sensitivity and allowed a broad detection of Aspergillus and Mucorales species (Table 1). Interestingly, these included rare species already involved in IMI (such as A. sublatus, A. welwitschiae, or A. sydowii for IA, and Apophysomyces spp. or Syncephalastrum spp. for mucormycosis), but that were not included in other available commercial or in-house PCR assays (10, 26). Therefore, this multiplex PCR assay may help to improve the diagnosis of these rare infections. Moreover, no cross-reactions nor competition were observed between the two targets, while true Aspergillus/Mucorales clinical coinfections were identified, demonstrating the potential of this PCR assay to detect mixed infections. No cross-reactions with other genera were observed for the Mucorales target, whereas the Aspergillus target cross-reacted with the genera Penicillium, Purpureocillium, and Paecilomyces. This result was not unexpected as these three genera are highly close taxonomically to the genus Aspergillus and it is a consequence of the necessary lowering analytical specificity required to detect the whole Aspergillus genus (27). Cross-reactions with these three genera should not be an issue in serum samples whereas results obtained in respiratory samples must be interpreted with caution. Indeed, these three genera are environmental molds that can be found fortuitously in the respiratory tract, especially in patients suffering from a chronic respiratory disease (28).
The prospective clinical evaluation on sera in high-risk IMI patients showed a high specificity for both targets (Table 4), even if Ct values above 35 should be interpreted with caution. Indeed, false-positive results were systematically > 35 Ct, whereas true positive results varied across this cutoff (especially in beginning infection or after the initiation of targeted antifungal therapy). In contrast, earlier Ct values (<35) in serum are highly suggestive of an active infection.
Regarding the Aspergillus target, sensitivity in serum was relatively low (64.1%) overall. However, the IA population in this study was composed of a high number of CAPA patients (n = 14/39) in whom molecular and antigenic biomarkers have shown a low sensitivity in serum (29). Regarding only proven and probable IA (n = 21), the sensitivity in serum raised up to 85.7%, which is consistent with previous reports (7). Combination with the species-specific A. fumigatus PCR assay for Aspergillus spp. positive sera showed also its importance to confirm specificity. Indeed, the 10 patients with a false-positive Aspergillus spp. PCR in serum were all negative for the A. fumigatus target. In contrast, 22 of the 25 patients with a true IA were positive for both targets. This PCR combination in sera also allowed to identify the species involved in IA (A. fumigatus) in most cases (n = 22), even in the absence of a positive fungal culture (n = 10) (Table 2). Finally, as the A. fumigatus assay exhibited significant lower Ct values than the Aspergillus spp. PCR, using this species-specific PCR may be of interest in case of very high suspected IA with a negative Aspergillus spp. PCR.
Regarding the pan-Mucorales PCR target, sensitivity in serum for the diagnosis of mucormycosis was high (80%), as recently reported in a multicenter study with another Mucorales PCR assay (10). All the disseminated cases (n = 14) were positive in serum, whereas sensitivity was decreased (33.3%) in more localized forms, as previously reported for cutaneous mucormycosis (16). Thanks to this PCR assay, we identified a higher number of mucormycosis cases than with conventional mycological methods (direct examination and/or fungal culture). Indeed, among the 20 mucormycosis cases, only eight were positive by conventional mycological methods (Table 3), whereas PCR in serum was positive in 16 patients. These latter included 11 PCR-positive only patients, in whom the diagnosis of mucormycosis may be discussed, as Mucorales PCR is not yet included in diagnosis criteria. However, among these, eight patients exhibited at least two consecutive positive sera, including four patients also positive by PCR in a respiratory sample or deep biopsy, strengthening so the mucormycosis diagnosis. DNA sequencing performed on Mucorales PCR-positive samples corroborates the specificity of the assay by identifying a broad range of Mucorales species, even for the four Aspergillus coinfections. Interestingly, in patients with multiple PCR-positive sera and/or another PCR-positive site (e.g., respiratory sample or biopsy), identifications were concordant between samples (Fig. 1; Table 3). In patients with positive culture and PCR (n = 3), molecular identification on serum was also concordant with the fungal culture identification (Table 3). Finally, in the patient having suffered from a relapse 4 months later (patient 18, Fig. 1), the same species (Cunninghamella spp.) was identified in both episodes in sera and respiratory samples. Thus, this DNA-sequencing approach greatly improved the identification of the etiologic agent of mucormycosis in comparison to fungal culture and brings new insights in mucormycosis epidemiology. Moreover, it could help for antifungal adaptation as in vitro azole susceptibility has been shown to be genus/species dependent (21). Interestingly, the most prevalent species we identified in sera was Rh. pusillus (eight infections), whereas this species is barely found in culture (none in this study). Reagent contamination was excluded by the use of negative controls during each molecular step. A similar finding was reported in a recent study (10), suggesting an underestimation of this hard-to-grow and highly thermophilic species in mucormycosis. This could be particularly true in very immunocompromised patients in onco-hematology wards, as Rh. pusillus was initially described as less pathogenic than other Mucorales species (30).
In conclusion, this multiplex Aspergillus spp./Mucorales spp. PCR assay followed by sequential specific PCR and/or DNA sequencing is a promising workflow in serum for IMI diagnosis and species identification in high-risk patients. This study also underlines the need to associate mycological examination of the suspected infection site by conventional and/or molecular methods, as serum may lack sensitivity in some situations such as CAPA or cutaneous mucormycosis. Using this combination and workflow, almost all etiologic agents were identified, including in cases of Aspergillus/Mucorales coinfections which may improve therapeutic management.
ACKNOWLEDGMENTS
This study was supported by internal funding.
Authors declare no conflict of interest related to the content of this study.
Part of this study was presented during the 32nd European Congress of Clinical Microbiology and Infectious Diseases (April 23 to 26, 2022, Lisbon, Portugal).
Data (DNA sequences) can be obtained freely by contacting the corresponding author.
Writing original draft: S.I.; writing review & editing: all; conceptualization: S.I., L.D.; investigation: all; formal analysis: S.I., L.P.; supervision: L.D.
Contributor Information
S. Imbert, Email: sebastien.imbert@u-bordeaux.fr.
Kimberly E. Hanson, University of Utah
REFERENCES
- 1.Kosmidis C, Denning DW. 2015. The clinical spectrum of pulmonary aspergillosis. Thorax 70:270–277. 10.1136/thoraxjnl-2014-206291. [DOI] [PubMed] [Google Scholar]
- 2.Petrikkos G, Skiada A, Drogari-Apiranthitou M. 2014. Epidemiology of mucormycosis in Europe. Clin Microbiol Infect 20 Suppl 6:67–73. 10.1111/1469-0691.12563. [DOI] [PubMed] [Google Scholar]
- 3.Cornely OA, Alastruey-Izquierdo A, Arenz D, Chen SCA, Dannaoui E, Hochhegger B, Hoenigl M, Jensen HE, Lagrou K, Lewis RE, Mellinghoff SC, Mer M, Pana ZD, Seidel D, Sheppard DC, Wahba R, Akova M, Alanio A, Al-Hatmi AMS, Arikan-Akdagli S, Badali H, Ben-Ami R, Bonifaz A, Bretagne S, Castagnola E, Chayakulkeeree M, Colombo AL, Corzo-León DE, Drgona L, Groll AH, Guinea J, Heussel C-P, Ibrahim AS, Kanj SS, Klimko N, Lackner M, Lamoth F, Lanternier F, Lass-Floerl C, Lee D-G, Lehrnbecher T, Lmimouni BE, Mares M, Maschmeyer G, Meis JF, Meletiadis J, Morrissey CO, Nucci M, Oladele R, Pagano L. Mucormycosis ECMM MSG Global Guideline Writing Group., et al. 2019. Global guideline for the diagnosis and management of mucormycosis: an initiative of the European Confederation of Medical Mycology in cooperation with the Mycoses Study Group Education and Research Consortium. Lancet Infectious Diseases 19:e405–e421. 10.1016/S1473-3099(19)30312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ullmann AJ, Aguado JM, Arikan-Akdagli S, Denning DW, Groll AH, Lagrou K, Lass-Flörl C, Lewis RE, Munoz P, Verweij PE, Warris A, Ader F, Akova M, Arendrup MC, Barnes RA, Beigelman-Aubry C, Blot S, Bouza E, Brüggemann RJM, Buchheidt D, Cadranel J, Castagnola E, Chakrabarti A, Cuenca-Estrella M, Dimopoulos G, Fortun J, Gangneux J-P, Garbino J, Heinz WJ, Herbrecht R, Heussel CP, Kibbler CC, Klimko N, Kullberg BJ, Lange C, Lehrnbecher T, Löffler J, Lortholary O, Maertens J, Marchetti O, Meis JF, Pagano L, Ribaud P, Richardson M, Roilides E, Ruhnke M, Sanguinetti M, Sheppard DC, Sinkó J, Skiada A, et al. 2018. Diagnosis and management of Aspergillus diseases: executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clin Microbiol Infect 24:e1–e38. 10.1016/j.cmi.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Bassetti M, Bouza E. 2017. Invasive mould infections in the ICU setting: complexities and solutions. J Antimicrob Chemother 72:i39–i47. 10.1093/jac/dkx032. [DOI] [PubMed] [Google Scholar]
- 6.White PL, Alanio A, Brown L, Cruciani M, Hagen F, Gorton R, Lackner M, Millon L, Morton CO, Rautemaa-Richardson R, Barnes RA, Donnelly JP, Loffler J. Fungal PCR Initiative. 2022. An overview of using fungal DNA for the diagnosis of invasive mycoses. Expert Rev Mol Diagn 22:169–184. 10.1080/14737159.2022.2037423. [DOI] [PubMed] [Google Scholar]
- 7.Cruciani M, Mengoli C, Barnes R, Donnelly JP, Loeffler J, Jones BL, Klingspor L, Maertens J, Morton CO, White LP. 2019. Polymerase chain reaction blood tests for the diagnosis of invasive aspergillosis in immunocompromised people. Cochrane Database Syst Rev 9:CD009551. 10.1002/14651858.CD009551.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imbert S, Meyer I, Palous M, Brossas J-Y, Uzunov M, Touafek F, Gay F, Trosini-Desert V, Fekkar A. 2018. Aspergillus PCR in bronchoalveolar lavage fluid for the diagnosis and prognosis of aspergillosis in patients with hematological and non-hematological conditions. Front Microbiol 9:1877. 10.3389/fmicb.2018.01877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imbert S, Brossas J-Y, Palous M, Joly I, Meyer I, Fekkar A. 2017. Performance of Aspergillus PCR in cerebrospinal fluid for the diagnosis of cerebral aspergillosis. Clin Microbiol Infect 23:889. 10.1016/j.cmi.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Millon L, Caillot D, Berceanu A, Bretagne S, Lanternier F, Morio F, Letscher-Bru V, Dalle F, Denis B, Alanio A, Boutoille D, Bougnoux M-E, Botterel F, Chouaki T, Charbonnier A, Ader F, Dupont D, Bellanger A-P, Rocchi S, Scherer E, Gbaguidi-Haore H, Herbrecht R. 2022. Evaluation of serum Mucorales PCR for the diagnosis of Mucormycoses: the MODIMUCOR prospective trial. Clin Infect Dis 75:777–785. 10.1093/cid/ciab1066. [DOI] [PubMed] [Google Scholar]
- 11.Scherer E, Iriart X, Bellanger AP, Dupont D, Guitard J, Gabriel F, Cassaing S, Charpentier E, Guenounou S, Cornet M, Botterel F, Rocchi S, Berceanu A, Millon L. 2018. Quantitative PCR (qPCR) detection of mucorales DNA in bronchoalveolar lavage fluid to diagnose pulmonary mucormycosis. J Clin Microbiol 56:e00289-18. 10.1128/JCM.00289-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnelly JP, Chen SC, Kauffman CA, Steinbach WJ, Baddley JW, Verweij PE, Clancy CJ, Wingard JR, Lockhart SR, Groll AH, Sorrell TC, Bassetti M, Akan H, Alexander BD, Andes D, Azoulay E, Bialek R, Bradsher RW, Bretagne S, Calandra T, Caliendo AM, Castagnola E, Cruciani M, Cuenca-Estrella M, Decker CF, Desai SR, Fisher B, Harrison T, Heussel CP, Jensen HE, Kibbler CC, Kontoyiannis DP, Kullberg B-J, Lagrou K, Lamoth F, Lehrnbecher T, Loeffler J, Lortholary O, Maertens J, Marchetti O, Marr KA, Masur H, Meis JF, Morrisey CO, Nucci M, Ostrosky-Zeichner L, Pagano L, Patterson TF, Perfect JR, Racil Z, et al. 2020. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis 71:1367–1376. 10.1093/cid/ciz1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dannaoui E. 2022. Recent developments in the diagnosis of mucormycosis. JoF 8:457. 10.3390/jof8050457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies GE, Thornton CR. 2022. Development of a monoclonal antibody and a serodiagnostic lateral-flow device specific to Rhizopus arrhizus (Syn. R. oryzae), the principal global agent of mucormycosis in humans. JoF 8:756. 10.3390/jof8070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imbert S, Gauthier L, Joly I, Brossas J-Y, Uzunov M, Touafek F, Brun S, Mazier D, Datry A, Gay F, Fekkar A. 2016. Aspergillus PCR in serum for the diagnosis, follow-up and prognosis of invasive aspergillosis in neutropenic and nonneutropenic patients. Clin Microbiol Infect 22:562. 10.1016/j.cmi.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millon L, Herbrecht R, Grenouillet F, Morio F, Alanio A, Letscher-Bru V, Cassaing S, Chouaki T, Kauffmann-Lacroix C, Poirier P, Toubas D, Augereau O, Rocchi S, Garcia-Hermoso D, Bretagne S. French Mycosis Study Group. 2016. Early diagnosis and monitoring of mucormycosis by detection of circulating DNA in serum: retrospective analysis of 44 cases collected through the French Surveillance Network of Invasive Fungal Infections (RESSIF). Clin Microbiol Infect 22:810.e1-810–e8. 10.1016/j.cmi.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Dannaoui E, Gabriel F, Gaboyard M, Lagardere G, Audebert L, Quesne G, Godichaud S, Verweij PE, Accoceberry I, Bougnoux M-E. 2017. Molecular diagnosis of invasive Aspergillosis and detection of azole resistance by a newly commercialized PCR Kit. J Clin Microbiol 55:3210–3218. 10.1128/JCM.01032-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imbert S, Cassaing S, Bonnal C, Normand A, Gabriel F, Costa D, Blaize M, Lachaud L, Hasseine L, Kristensen L, Guitard J, Schuttler C, Raberin H, Brun S, Hendrickx M, Piarroux R, Fekkar A. 2021. Invasive aspergillosis due to Aspergillus cryptic species: a prospective multicentre study. Mycoses 64:1346–1353. 10.1111/myc.13348. [DOI] [PubMed] [Google Scholar]
- 19.Walther G, Wagner L, Kurzai O. 2019. Updates on the taxonomy of mucorales with an emphasis on clinically important taxa. JoF 5:106. 10.3390/jof5040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramírez-Hinojosa JP, Medrano-Ahumada S, Arenas R, Bravo-Escobar A, Paraguirre-Martínez S, Xicohtencatl-Cortes J, Martínez-Herrera E, Hernández-Castro R. 2021. Fungal invasive co-infection due to Aspergillus fumigatus and Rhizopus arrhizus: a rhino-orbital presentation. JoF 7:1096. 10.3390/jof7121096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borman AM, Fraser M, Patterson Z, Palmer MD, Johnson EM. 2021. In vitro antifungal drug resistance profiles of clinically relevant members of the mucorales (Mucoromycota) especially with the newer triazoles. JoF 7:271. 10.3390/jof7040271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blot SI, Taccone FS, Van den Abeele A-M, Bulpa P, Meersseman W, Brusselaers N, Dimopoulos G, Paiva JA, Misset B, Rello J, Vandewoude K, Vogelaers D. AspICU Study Investigators. 2012. A clinical algorithm to diagnose invasive pulmonary aspergillosis in critically ill patients. Am J Respir Crit Care Med 186:56–64. 10.1164/rccm.201111-1978OC. [DOI] [PubMed] [Google Scholar]
- 23.Koehler P, Bassetti M, Chakrabarti A, Chen SCA, Colombo AL, Hoenigl M, Klimko N, Lass-Flörl C, Oladele RO, Vinh DC, Zhu L-P, Böll B, Brüggemann R, Gangneux J-P, Perfect JR, Patterson TF, Persigehl T, Meis JF, Ostrosky-Zeichner L, White PL, Verweij PE, Cornely OA. Infectious Disease Canada. 2021. Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infectious Diseases 21:e149–e162. 10.1016/S1473-3099(20)30847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Normand A-C, Blaize M, Imbert S, Packeu A, Becker P, Fekkar A, Stubbe D, Piarroux R. 2021. Identification of molds with matrix-assisted laser desorption ionization-time of flight mass spectrometry: performance of the newly developed MSI-2 application in comparison with the Bruker Filamentous Fungi Database and MSI-1. J Clin Microbiol 59:e0129921. 10.1128/JCM.01299-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Springer J, Goldenberger D, Schmidt F, Weisser M, Wehrle-Wieland E, Einsele H, Frei R, Löffler J. 2016. Development and application of two independent real-time PCR assays to detect clinically relevant Mucorales species. J Med Microbiol 65:227–234. 10.1099/jmm.0.000218. [DOI] [PubMed] [Google Scholar]
- 26.Guegan H, Iriart X, Bougnoux M-E, Berry A, Robert-Gangneux F, Gangneux J-P. 2020. Evaluation of MucorGenius mucorales PCR assay for the diagnosis of pulmonary mucormycosis. J Infect 81:311–317. 10.1016/j.jinf.2020.05.051. [DOI] [PubMed] [Google Scholar]
- 27.Alanio A, Bretagne S. 2014. Difficulties with molecular diagnostic tests for mould and yeast infections: where do we stand? Clin Microbiol Infect 20 Suppl 6:36–41. 10.1111/1469-0691.12617. [DOI] [PubMed] [Google Scholar]
- 28.Delhaes L, Touati K, Faure-Cognet O, Cornet M, Botterel F, Dannaoui E, Morio F, Le Pape P, Grenouillet F, Favennec L, Le Gal S, Nevez G, Duhamel A, Borman A, Saegeman V, Lagrou K, Gomez E, Carro M-L, Canton R, Campana S, Buzina W, Chen S, Meyer W, Roilides E, Simitsopoulou M, Manso E, Cariani L, Biffi A, Fiscarelli E, Ricciotti G, Pihet M, Bouchara J-P. 2019. Prevalence, geographic risk factor, and development of a standardized protocol for fungal isolation in cystic fibrosis: results from the international prospective study “MFIP”. J Cyst Fibros 18:212–220. 10.1016/j.jcf.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Dellière S, Dudoignon E, Voicu S, Collet M, Fodil S, Plaud B, Chousterman B, Bretagne S, Azoulay E, Mebazaa A, Dépret F, Mégarbane B, Alanio A. 2022. Combination of mycological criteria: a better surrogate to identify COVID-19-associated pulmonary aspergillosis patients and evaluate prognosis? J Clin Microbiol 60:e0216921. 10.1128/JCM.02169-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribes JA, Vanover-Sams CL, Baker DJ. 2000. Zygomycetes in human disease. Clin Microbiol Rev 13:236–301. 10.1128/CMR.13.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]