ABSTRACT
The genus Mycobacterium contains several slow-growing human pathogens, including Mycobacterium tuberculosis, Mycobacterium leprae, and Mycobacterium avium. Mycobacterium smegmatis is a nonpathogenic and fast growing species within this genus. In 1990, a mutant of M. smegmatis, designated mc2155, that could be transformed with episomal plasmids was isolated, elevating M. smegmatis to model status as the ideal surrogate for mycobacterial research. Classical bacterial models, such as Escherichia coli, were inadequate for mycobacteria research because they have low genetic conservation, different physiology, and lack the novel envelope structure that distinguishes the Mycobacterium genus. By contrast, M. smegmatis encodes thousands of conserved mycobacterial gene orthologs and has the same cell architecture and physiology. Dissection and characterization of conserved genes, structures, and processes in genetically tractable M. smegmatis mc2155 have since provided previously unattainable insights on these same features in its slow-growing relatives. Notably, tuberculosis (TB) drugs, including the first-line drugs isoniazid and ethambutol, are active against M. smegmatis, but not against E. coli, allowing the identification of their physiological targets. Furthermore, Bedaquiline, the first new TB drug in 40 years, was discovered through an M. smegmatis screen. M. smegmatis has become a model bacterium, not only for M. tuberculosis, but for all other Mycobacterium species and related genera. With a repertoire of bioinformatic and physical resources, including the recently established Mycobacterial Systems Resource, M. smegmatis will continue to accelerate mycobacterial research and advance the field of microbiology.
KEYWORDS: distributive conjugal transfer, mycobacterial systems resource, cell envelope, drug discovery, efficient plasmid transformation, leaderless mRNA, mycobacteriophage, mycolic acid, phasmid, virulence factors
INTRODUCTION
Despite the discovery of Mycobacterium tuberculosis in 1882 by Robert Koch (1) and its global health burden (2, 3), it is only in the last 30 years that the secrets of its genetic make-up have begun to be revealed and characterized. For example, in 1990, we did not know: (i) the targets of tuberculosis (TB)-specific drugs, (ii) the molecular genetic basis of attenuation of the vaccine strain, Mycobacterium bovis BCG, (iii) which genes were essential for in vitro and in vivo growth, and if these genes were similar to those of other bacteria. The pathogenicity and slow growth − 3 to 4 weeks to form colonies – make M. tuberculosis extremely difficult to work with in the laboratory. The seminal breakthrough toward developing genetic approaches to study pathogenic mycobacteria was the isolation of a transformable derivative of Mycobacterium smegmatis, designated mc2155. M. smegmatis is a nonpathogenic and fast-growing species (colonies in 3 days) and was historically used as a mycobacteriophage host. The development of mc2155 made M. smegmatis the model for studying properties of all mycobacteria, including pathogens, such as M. tuberculosis and the nontuberculous mycobacteria (NTM) pathogens, Mycobacterium abscessus and Mycobacterium avium (4). This review describes the history of mc2155, its use in developing genetic tools for pathogenic mycobacteria, and its application to characterizing biological mechanisms and targets of TB drugs (Fig. 1). Moreover, this review describes current “omic” approaches pioneered in M. smegmatis and now used for unbiased genome-wide discovery of complex multi-faceted processes in medically important mycobacteria.
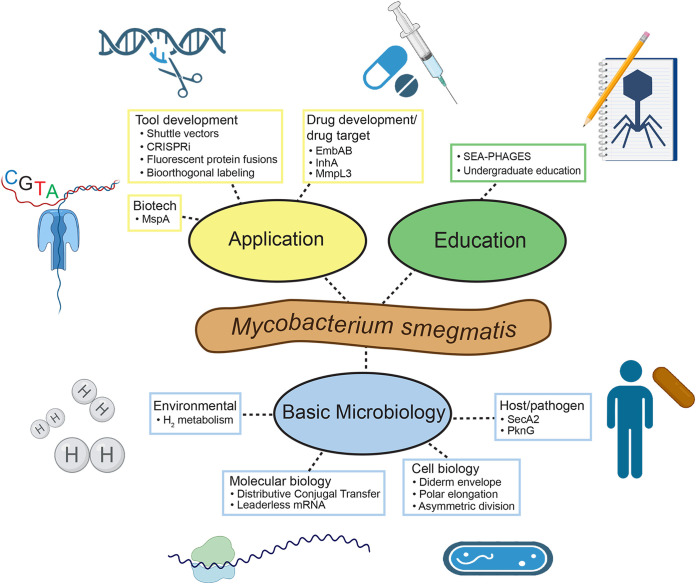
FIG 1.
Web diagram of M. smegmatis’ role as a model organism. M. smegmatis serves applied, educational, and basic science in myriad ways. Colored ovals represent key fields that M. smegmatis contributes to, while terminal rectangular nodes represent examples of scientific contributions in each field, most of which were discussed in this review. Illustrations were created with BioRender.com.
Note on the proposal to rename Mycobacterium smegmatis. We disagree with the recent proposal to rename Mycobacterium smegmatis as Mycolicibacterium smegmatis (5). We support the editorial by Tortoli et al. (6): (i) the split of the Mycobacterium genus has caused unnecessary confusion in health care settings, compromising patient safety, and costing time and money on reeducating health care professionals; (ii) the proposed split was incomplete, not accounting for ~40 species; and (iii) all previously verified taxonomic names are considered valid for use in publication. In addition, the new taxonomic classification was based on conserved signature indels (CSIs) and conserved signature proteins (CSPs). Many CSIs and CSPs may be the result of horizontal gene transfer, which are poor traits for taxonomic classification. Most researchers have chosen to continue using the conventional genus name after the new classification proposal, indicating that Mycobacterium smegmatis is still the preferred taxonomic name.
THE ORIGIN OF M. SMEGMATIS MC2155
While Crawford and Bates had isolated plasmids from mycobacteria in 1979 (7), subsequent attempts to transform mycobacteria had been unsuccessful (8). Foreign DNA was first introduced into mycobacteria in 1987 with a shuttle phasmid (9). This chimeric vector had an E. coli bacteriophage lambda cosmid inserted in a non-essential region of the lytic mycobacteriophage TM4 (9, 10). The shuttle phasmid replicates in E. coli as a plasmid and in mycobacteria as a phage (9, 11). The generation of viable phage that infected both M. smegmatis and M. bovis BCG demonstrated the feasibility of the approach and established that DNA from E. coli was not degraded by mycobacteria. A second shuttle phasmid based on the temperate mycobacteriophage, L1, was generated and shown to lysogenize M. smegmatis mc26, the progenitor strain of M. smegmatis mc2155 (12). Subsequently, a gene encoding kanamycin-resistance (KmR) was cloned into the L1-based shuttle phasmid and successfully transfected into mc26, establishing kanamycin as an effective selection in mycobacteria (12). With this knowledge, a library of plasmids was made by cloning random fragments of the mycobacterial plasmid pAL5000 (13, 14), which replicates in Mycobacterium fortuitum, in an E. coli plasmid encoding KmR (12). This library of chimeric episomal plasmids was electroporated into M. smegmatis mc26 and a few rare transformants were isolated and thus, establishing a plasmid-transformation system (12). These rare transformants turned out to be a mutant of M. smegmatis that was defective in preventing plasmid establishment (15). The plasmid in the original transformant was cured and the resulting plasmid-free strain was designated mc2155. Whereas mc26 remained virtually un-transformable, mc2155 routinely yielded up to a million transformants per microgram of plasmid DNA; it had acquired an efficient plasmid transformation (ept-1) phenotype. In 2014, whole-genome sequence comparisons between mc2155 and its parental strain revealed that the efficient plasmid transformation (ept) mutation mapped to eptC, encoding a structural-maintenance-of-the-chromosome (SMC) protein (16). A similar SMC protein-mediated plasmid restriction has been found in other bacteria as well (17). This ept mutant of M. smegmatis revolutionized approaches to mycobacterial biology and established mc2155 as the workhorse of mycobacterial genetics; fast-growing, nonpathogenic, and transformable.
FEATURES OF M. SMEGMATIS THAT MAKE IT A GREAT MODEL
Shuttle phasmids and plasmids not only enabled the isolation of mc2155, but were used to generate key molecular genetic tools, including integration-proficient vectors (18), expression vectors (19), luciferase-reporter phages (20), and suicide- (21), and recombineering-vectors (22). Furthermore, the ability of M. smegmatis to grow in temperature ranges up to 55°C allowed the isolation of temperature-sensitive TM4 phage mutants (23, 24), providing for both efficient delivery and counter-selection for specialized transduction and Tn- and targeted-mutagenesis (20, 25–27). These early technologies allowed the generation of insertion mutant libraries and the identification of essential genes using Tn-site hybridization and Tn-seq technologies (28–31). As these genetic tools flourished, mc2155 became broadly accepted as the genetically tractable model Mycobacterium.
Its non-pathogenicity was a great entry point for researchers not equipped with a biosafety level (BSL)-3 facility and for novice microbiologists, such as undergraduate students, who cannot work with pathogens. Fast growth is not only ideal for genetics and biochemistry, but also for studying single cells, their growth, and cell architecture. In particular, high-throughput screenings involving single-cell analyses, such as microscopy, are nearly impossible with the pathogens because of the slow growth and need for specialized equipment in BSL-3 facilities. In this section, we highlight the notable features of M. smegmatis, and how newer, shared resources have further enhanced mycobacterial research.
Comparative genomics validate the choice of M. smegmatis as the model Mycobacterium.
Of the ~4,000 protein-coding genes in the genome of M. tuberculosis, > 2,800 have orthologs in M. smegmatis with >50% amino acid identity (32). The average protein identities between protein orthologs in M. smegmatis and their pathogenic cousins are >70% in most cases (33). Most importantly, the similarities extend beyond the M. tuberculosis complex (MTBC). As a conservative estimate, there is a core set of ~1,150 M. smegmatis proteins that share >50% amino acid identity, which are encoded by many species including the MTBC, M. abscessus, Mycobacterium marinum, M. avium, and M. leprae (32). The core proteins almost certainly perform the same functional role in each species. This level of conserved function is also highlighted by transposon mutagenesis studies: 96% of the genes identified as essential in M. smegmatis have orthologs in M. tuberculosis, and 90% of which are essential in M. tuberculosis (34). The genetic organization of each species is also well conserved, with similar patterns of gene co-localization around the chromosome, suggesting conserved mechanisms of gene regulation.
Centralized resources for the study of mycobacteria.
A good model organism is accompanied by community-shared resources of strains, defined knockouts, and plasmids expressing individual genes (e.g., https://biocyc.org/, https://bgsc.org/). These physical resources, integrated with data sets (protein-protein interactions, phenotypic profiling, etc.), permit a systems biology approach to gene function and provide insights on cellular processes. While still in their infancy, similar resources are becoming available for mycobacteria, most importantly in M. smegmatis. For example, bioinformatic resources such as Mycobrowser (https://mycobrowser.epfl.ch/) and BioCyc (https://mycobacterium.biocyc.org/) provide an interactive space to analyze genes of interest, which are cross-referenced to orthologs in other sequenced genomes, with links to key sites describing BLAST searches, putative gene functions, and structures. The Wadsworth Center’s Interactive Genomics browser displays RNA-seq, Ribo-seq, and transcription start site data for M. smegmatis and M. tuberculosis, which allow the user to accurately determine gene boundaries, identify novel genes, leadered and leaderless transcripts, and non-coding RNAs (https://www.wadsworth.org/research/scientific-resources/interactive-genomics). MSRdb (https://msrdb.org/) describes a collection of M. smegmatis gene knockouts, knockdowns, and strains expressing genes fused to a fluorescent reporter for protein localization. It was created as part of the Mycobacterial Systems Resource (MSR) (32), which is based on the 1,153 core proteins described above, most having no assigned function. The physical resource contains: a total of 569 precise gene knockouts of non-essential genes; a collection of plasmids that encode small guide RNAs for CRISPRi-mediated targeted suppression of 843 genes; and 1,116 genes cloned in a vector to express a fluorescent protein (Dendra2)-tagged fusion protein. Like other method breakthroughs, the CRISPRi technology was developed and optimized in M. smegmatis (35) and its application to fine-tune gene repression elegantly demonstrated in M. tuberculosis (36). The pooled collection of CRISPRi plasmid libraries targeting genes and ncRNAs in both M. smegmatis and M. tuberculosis are available through Addgene (https://www.addgene.org/) (36).
We highlight 2 recent studies that exemplify the benefit of using M. smegmatis for microscopy-based approaches. The first example is high-throughput imaging on the library of conserved, fluorescently tagged, core proteins generated in the MSR (37). The spatiotemporal analysis of over 700 proteins demonstrated that sub-cellular protein localization often correlates with function. For example, ribosomal proteins were clustered, but excluded from the nucleoid and the polar and subpolar regions of the cell. The co-localization of proteins with both known and unknown functions provides immediate insight (guilty by association) on the possible function of many hypothetical core proteins, which can be directly translated to other species.
The second study applied CRISPRi gene suppression in M. smegmatis to specifically inhibit 263 essential genes with direct orthologs in M. tuberculosis (38). Single cells were visualized following induction of the CRISPRi system and the impact of essential gene suppression on cell morphotypes captured by high-throughput quantitative imaging, which generated a morphological atlas of phenotypes (cell length, curvature, bulging). Strikingly, suppression of genes with related biological function (e.g., cell division, cell wall synthesis) clustered together according to their morphology, allowing an educated prediction of gene function of hypothetical proteins. The conservation of these essential genes throughout the genus allows their characterization in M. smegmatis to provide a more directed experimental approach in the less experimentally amenable mycobacteria.
NOTABLE DISCOVERIES DRIVEN BY M. SMEGMATIS
Studies with M. smegmatis have revealed many unusual features of the genus. In this section, we highlight a few examples of notable discoveries, while acknowledging the countless other remarkable contributions, which cannot be included due to space limitations.
Distributive conjugal transfer and genome evolution.
Distributive conjugal transfer (DCT) is a novel form of horizontal gene transfer (HGT) first described in M. smegmatis (39, 40). DCT is conceptually equivalent to conjugation in that it requires direct contact between a donor and recipient cell, but the products of DCT are distinct from that of any other form of HGT; the progeny genomes are mosaic blends of the parental genomes (41). Mosaicism results from a single DCT event, dramatically shortening the time to combine parental traits that could confer a competitive evolutionary advantage (42). DCT is not limited to laboratory strains and conditions, as the genomes of independent environmental isolates of M. smegmatis also exhibit the hallmark genome mosaicism (43).
In the past, the concept of HGT within the MTBC was considered almost heretical. While there are abundant mycobacteriophages, no plasmids have been described within the MTBC, and given the pathogen’s intracellular “solitary” lifestyle, it was not surprising there was little evidence for HGT (44). Members of the MTBC are very closely related (>99.9% genome identity), with speciation reflecting host adaptations (e.g., M. bovis causes TB in cattle) (45–47). That concept was challenged when the comparative analysis of multiple genome sequences of Mycobacterium canettii (a smooth colony outlier of the MTBC) revealed thousands of SNPs: the striking similarity to the experimentally produced mosaic genomes of M. smegmatis provided the first indication that HGT had occurred in the MTBC (48–51). Subsequently, direct evidence for DCT between 2 isolates of M. canettii was demonstrated by isolating transconjugant progeny that contained the hallmark, blended parental genomes (52). These studies provided experimental support for the proposal that DCT was a major evolutionary driving force among M. canettii, which ultimately led to the evolution of a progenitor M. tuberculosis species and subsequent speciation into the animal-adapted MTBC (41). Thus, DCT in M. smegmatis has forced a rethinking on HGT and genome evolution among mycobacteria and provided a novel mechanism of bacterial HGT (42).
Type VII (ESX) secretion systems are found in all mycobacteria and are encoded, primarily, in a large, multi-gene locus, esx (53–57). Mycobacteria can contain multiple esx loci (up to 5), which are functionally non-redundant. Functions for most ESX systems and their secreted substrates remain poorly defined, but ESX-1- and ESX-5-mediated secretion is essential for pathogenesis and survival during infection. Remarkably, DCT has provided alternative roles for ESX secretion systems. RNA-seq analysis demonstrated that donor and recipient cells have specific transcriptional responses to direct contact with the opposite mating type. Notably, the ESX-4 secretion system is transcriptionally activated in the recipient on donor contact and is essential for DCT (58). This contact-dependent activation of ESX-4 in the recipient is regulated by ESX-1 activity in the donor providing the first direct evidence for mycobacterial cell-cell communication (58). ESX-4 has since been shown to be important for macrophage infection in M. abscessus (59) and contributes to phagosome permeabilization and protein trafficking in M. tuberculosis (60). Thus, DCT in M. smegmatis has provided the first model system for mycobacterial communication and indicates that ESX systems are likely to mediate unexpected functions beyond virulence.
Leaderless gene expression and small proteins in mycobacteria.
The application of RNA-profiling techniques (RNA-seq and Ribo-seq) in M. smegmatis, M. tuberculosis, and M. abscessus (61–64) has emphasized a startling difference in the gene architecture of mycobacteria compared with the textbook model organisms; the presence of leaderless mRNAs (LLmRNA). LLmRNAs lack a 5′-UTR; the 5′ end of the mRNA begins at the start codon of the gene and, thus, the mRNA lacks a ribosome-binding site (RBS) (65). In E. coli LLmRNAs are extremely rare (3 have been characterized) and poorly expressed (66), but they are abundant in Actinobacteria (25 to 30% of mRNAs in mycobacteria) and Archaea (65, 67–69). M. smegmatis was used in its “model” role, using lacZ and luciferase-gene reporters to demonstrate that: (i) these LLmRNAs were robustly expressed in M. smegmatis but not E. coli; (ii) the level of gene expression was comparable to that of leadered mRNA (LDmRNA); and (iii) any mRNA beginning with a 5′ AUG or GUG (RUG) is translated (64, 70). The consequences of this fundamental difference in gene architecture are many. First, the rationale behind genome annotations in mycobacteria had to be reevaluated. The assumed requirement for a gene to have a promoter and RBS resulted in many mis-annotations. For example, of the 1,285 genes transcribed as LLmRNAs in M. tuberculosis, 338 were incorrectly annotated and required (in-frame) modifications of the start sites, and a further 370 were not annotated (64, 70). In addition, most of the novel genes identified by transcriptional profiling (from both LDmRNAs and LLmRNAs) encoded small proteins (sproteins, <50 amino acids). In the past, small ORFs (sORFs) encoding sproteins had been overlooked by genome annotation pipelines, which use “50 codons” as a cut off to ensure confidence in gene prediction. However, growing experimental evidence in mycobacteria and other prokaryotes has shown that many sproteins are functional, requiring a rethinking of gene definitions (71–75). The addition of hundreds of novel sproteins to the mycobacterial proteome will demand new genetic, biochemical, and proteomic studies to determine their function and M. smegmatis is the ideal organism for those studies, especially for conserved sproteins. As an example, a subclass of conserved sproteins encode 2 to 8 consecutive cysteine residues. These poly-cysteine sORFs act as cis-acting attenuators of downstream genes required for cysteine uptake and biosynthesis (76). Finally, ribosomes must recognize LLmRNAs by a mechanism fundamentally different from that of LDmRNAs. The abundance of LLmRNA indicates that efficient translation mechanisms for both LDmRNAs and LLmRNAs have evolved in-parallel in mycobacteria. Thus, the best bacterial model for the mechanistic study of leaderless translation is M. smegmatis.
Virulence factor secretion.
One prominent example of an M. smegmatis discovery enlightening host-pathogen interactions involves virulence factor secretion. Secretion analyses in M. smegmatis revealed a non-essential, but non-redundant SecA2 pathway, in addition to the housekeeping SecA1 (77). SecA2 is found in all species of Mycobacterium, other Actinobacteria, and also in several Gram-positive pathogens (78, 79). SecA1 interacts with the SecYEG complex, the membrane-spanning channel that translocates nascent unfolded polypeptides into the periplasm. Mycobacteria do not encode an additional secY paralog, in contrast to other bacteria with a second SecA. Instead, structural studies in M. tuberculosis have revealed that SecA2 likely interacts with the canonical SecYEG channel to mediate secretion of SecA2-specific proteins (80, 81).
M. tuberculosis ΔsecA2 is attenuated in SCID and C57BL/6 mice (82, 83). Attenuation of the ΔsecA2 strain stemmed from its inability to export two proteins involved in blocking phagosome maturation: PknG and SapM (84–86). PknG is one of 11 serine-threonine protein kinases (STPK) encoded by M. tuberculosis (87); most of the SPTKs are membrane proteins, are not SecA2 substrates, and function as mediators of trans-membrane signaling. PknG is an exception; it is secreted in a SecA2-dependent manner (88, 89), in addition to having cytoplasmic roles regulating metabolism, redox balance, and biofilm formation (90–94). The first evidence suggesting PknG was a secreted virulence factor was obtained by heterologously expressing M. tuberculosis PknG in M. smegmatis. This M. smegmatis strain secreted PknG and was more resistant to macrophage killing than the wild-type parent (95). This was subsequently validated in M. tuberculosis where PknG, in conjunction with the SapM phosphatase (86), blocks host phagosome maturation and modulates the host immune response (88, 96–98), establishing PknG and SapM as bona fide virulence factors.
Cell envelope, polar elongation, and asymmetric division.
Mycobacterial envelopes are unlike those of any other bacteria. The unusually lipid-rich diderm structure of the mycobacterial cell envelope, initially proposed by Minnikin in 1982 (99), was visualized in 2008 by cryo-electron microscopy of M. smegmatis and M. bovis BCG, indicating a conserved outer membrane (termed the mycomembrane) (100, 101). While some early studies used Mycobacterium phlei, mc2155 inevitably became the preferred model to determine the structure, biosynthesis, and function of the mycobacterial cell envelope (see reviews [102–105]). Here, we discuss the roles M. smegmatis has played in uncovering the spatiotemporal coordination of mycobacterial cell envelope elongation and division, which are distinct from other rod-shaped model bacteria.
Pathogenic mycobacteria persist in the host for decades in chronic infections. Thus, it is important to understand how growth and division are regulated and how cell envelope integrity is maintained in hostile host environments. Early genome analyses revealed that mycobacteria lack conserved cell division proteins, such as MreB and the Min-family proteins, implying fundamental differences in the mechanisms of cell envelope elongation and division compared with other bacteria (106). Using fluorescein-labeled vancomycin, it was shown that de novo growth of the mycobacterial cell occurred at the cell poles and not along its length, unlike other rod-shaped bacteria (107). While mycobacteria lack MreB, they do encode a bacterial homolog of the eukaryotic cytoskeletal protein tropomyosin, named DivIVA (Wag31). DivIVA is localized to the poles where it has been shown to promote peptidoglycan biosynthesis and cell elongation in M. smegmatis (108–111). Intriguingly, the M. smegmatis plasma membrane partitions into two domains. One, termed the intracellular membrane domain (IMD), contains many envelope synthases that localize to sub-polar regions (i.e., regions directly adjacent to the growing poles) (112, 113). There is a positive-feedback loop between IMD formation and peptidoglycan biosynthesis: IMD formation promotes polar-peptidoglycan synthesis, and the synthesized peptidoglycan maintains the IMD (114, 115). These cytoskeletal proteins, envelope synthases, and specialized membrane domains are proposed to form a mycobacterial “elongasome” that coordinates polar growth. As expected for a process fundamental to cell growth, the elongasome is conserved in M. tuberculosis, as evidenced by the polar localization of DivIVA (109), peptidoglycan biosynthesis (109, 116), and the subpolar enrichment of the IMD (117). Proteomic analysis of the M. tuberculosis IMD indicates that it serves as the biosynthetic site of pathogen-specific lipid virulence factors, including phthiocerol dimycolate (117).
Mycobacteria divide asymmetrically by a fast, mechanical-snapping mechanism (107, 118–121). While the mechanism of asymmetric septal placement is unknown, studies largely performed in M. smegmatis have revealed that FtsZ and other cell division proteins assemble a protein complex at the new division site similar to other bacteria (for review [122, 123]). Strikingly, asymmetric division is an actively regulated process, not the result of random septal placement; a mutant of M. smegmatis lacking lamA elongates and divides symmetrically (124). LamA is a mycobacteria-specific, membrane protein of unknown function. Though the physiological significance of asymmetric cell division is not fully understood, loss of asymmetry in lamA-deficient M. tuberculosis was accompanied by increased sensitivity to rifampicin (RIF) (102). A second study that tracked single cells through multiple rounds of asymmetric cell divisions in a microfluidic system also demonstrated that long-birth length and mature-growth poles are associated with RIF tolerance (125), independently supporting the role of asymmetric cell division in producing heterogenous populations of cells with different drug susceptibilities. Stress and antibiotics were later shown to influence cell length heterogeneity in M. tuberculosis clinical isolates, reinforcing the evolutionarily conserved link between morphological heterogeneity and mycobacterial fitness first characterized in M. smegmatis (126).
APPLICATIONS OF M. SMEGMATIS FOR DRUG DISCOVERY AND DEVELOPMENT
Elucidating mechanisms of action for Isoniazid, Ethionamide, and Ethambutol.
Chemotherapy of mycobacterial infections is a lengthy process requiring a cocktail of drugs, usually including Isoniazid (INH), RIF, Ethambutol (EMB), and Pyrazinamide. Of these, M. smegmatis has played a central role in determining the mechanisms of action for INH and EMB. An INH-resistant strain of mc2155 was shown to become sensitive upon expression of the M. tuberculosis catalase-peroxidase gene, katG, suggesting that susceptibility to INH requires the expression of katG (127). KatG was subsequently discovered to modify INH into its active form, demonstrating that INH is a prodrug and explaining why katG mutations are a prevalent mechanism of INH resistance in M. tuberculosis (128). inhA was identified as the target of activated INH by an M. smegmatis missense mutation that conferred co-resistance to both INH and Ethionamide (ETH) (129). inhA encodes an NADH-specific enoyl-acyl carrier protein (ACP) reductase (130, 131). The ability of M. smegmatis to grow at 30°C enabled the discovery of mutants that elucidated the mechanisms of action of INH and ETH (reviewed in [132]). Briefly, when M. smegmatis mutants were isolated for co-resistance to INH and ETH on rich media, half of the mutants were temperature-sensitive. These mutations mapped to ndh, which encodes an NADH oxidase (133). Altered NADH/NAD+ ratios of these mutants were consistent with a model that INH- or ETH-NAD+ adducts inhibit InhA. This model was later verified by X-ray crystallography of an INH-NAD+ or ETH-NAD+ adduct bound to the active site of InhA (134–136). To our knowledge, this is the first example of a prodrug that can be activated to form adducts with an enzyme co-factor (NADH).
The first attempts to determine the mechanism of EMB-mediated anti-mycobacterial activity were carried out using M. smegmatis as far back as the 1960s (e.g., [137, 138]), but more than 2 decades were needed to elucidate the correct mechanism of action. EMB-treated M. smegmatis cells were defective in incorporating radioactive d-glucose into arabinogalactan (139), and accumulated decaprenyl-P-arabinose (140), demonstrating that EMB blocks the transfer of arabinose from its lipid carrier to arabinogalactan in the cell wall. The targets of EMB were discovered through an overexpression screen using M. smegmatis. A plasmid library of M. avium chromosomal fragments were screened for clones exhibiting resistance to EMB. This screen identified 2 arabinosyltransferases, EmbA and B, as mediating resistance to EMB (141). Genetic studies in M. tuberculosis corroborated these results (142, 143). Cryo-EM and X-ray crystallography structures of M. tuberculosis and M. smegmatis EmbA and EmbB complexed with either substrate or EMB confirmed that EMB inhibits arabinosyltransferase activity by binding a region adjacent to the catalytic site (144). Mutations in genes such as katG, inhA, embA, and embB, that confer drug resistance are now rapidly detected by whole-genome sequencing, reducing time to diagnose multidrug-resistant and extensively drug-resistant TB (145, 146).
Drug discovery and development.
Early efforts to screen TB drugs were inefficient due to the use of slow-growing M. tuberculosis. In more recent years, the generalizability of most anti-TB drugs across all mycobacteria has led to the appreciation of M. smegmatis as a pragmatic model organism for initial drug screening and further drug optimization. M. smegmatis was used for the initial compound screening that identified diarylquinoline (later named Bedaquiline) as a specific inhibitor of mycobacterial ATP synthase in 2005, which ended a 40-year dearth in anti-TB drug discovery (147). Furthermore, a cryo-EM study determined the binding site of Bedaquiline in the M. smegmatis ATP synthase (148). WHO recommended the use of Bedaquiline for the treatment of RIF-resistant and multidrug-resistant adult TB in 2013 and, more recently, recommended replacement of injectable second-line drugs with Bedaquline for oral, short-course regimens (149). Bedaquiline has also been included in newer combination regimens in several ongoing clinical trials.
M. smegmatis has been instrumental in the development of inhibitors of the essential mycolic acid transporter protein, MmpL3. Knockdown of the mmpL3 gene in M. smegmatis is lethal, and results in reduced levels of outer membrane mycolic acids and the accumulation of the mycolate carrier, trehalose monomycolate (TMM), suggesting a role in mycolic acid transport (150, 151). Early inhibitors of MmpL3 were discovered by screening chemical libraries for activity against M. tuberculosis growth and mapping resistance mutations back to the mmpL3 gene (151). Using a novel M. smegmatis spheroplast assay, one inhibitor was used to show MmpL3 is a “floppase,” that flips TMM from the cytoplasmic to the periplasmic leaflet of the inner membrane (152). While this study established the first, specific platform for validating other potential MmpL3 inhibitors, the technical difficulty involved with creating spheroplasts prompted development of a more high-throughput approach (153). M. smegmatis was used as a surrogate to express different drug-resistant M. tuberculosis mmpL3 alleles. Unexpectedly, the mmpL3 mutations conferred cross-resistance to different drugs, suggesting that most inhibitors bind a common active site, and predicting that undiscovered inhibitors would most likely bind this same site. Inspired by this insight, the authors developed fluorescent MmpL3 probes based on known inhibitors, which were successfully used in a competitive-binding assay in M. smegmatis to identify new MmpL3 inhibitors (153). This live M. smegmatis-based platform has extraordinary potential for both new drug discovery and drug optimization.
To gain a molecular view of the MmpL3 floppase and the mechanism of drug inhibition, X-ray crystallography structures of M. smegmatis MmpL3 alone and complexed with its ligand TMM and known inhibitors were solved (154, 155). These structures revealed the mechanism by which the protein utilizes a proton gradient to drive TMM translocation and how inhibitors block this proton translocation channel to arrest TMM translocation. As expected, the cryo-EM structure of M. tuberculosis MmpL3 (61% amino acid identity) closely matched that of M. smegmatis MmpL3 (156). The recent discovery of 2 conserved accessory proteins in M. smegmatis, which stabilize MmpL3, add further insight on the mechanisms of MmpL3 function (157). Thus, our increased understanding of the MmpL3-drug mechanism of action promises to accelerate development of second-generation MmpL3 inhibitors, improving upon first-generation inhibitors such as SQ109, which is in phase 2 clinical trials (158). The application of M. smegmatis to isolate inhibitors of ATP synthase and MmpL3 demonstrates many of the key attributes M. smegmatis offers as a model for M. tuberculosis drug-target research.
M. SMEGMATIS WILL CONTINUE TO BE A MODEL FOR ALL MYCOBACTERIA
The development of genetic tools and resources over the past 3 decades has made M. smegmatis the model organism for mycobacterial pathogens, fast- and slow-growing. Here, we highlight the potential of some new tools and technologies, the limitations of M. smegmatis as a model and its creative use in education.
M. smegmatis will inevitably continue serving as a primary incubator space for mycobacterial tool development. Determining the functions of the many unannotated small (and large) proteins will be a priority. Identifying protein-protein interactions can provide enormous insight on protein function, as demonstrated by new in vivo proximity-labeling technologies. These use photo-cross-linking of unnatural amino acids incorporated into mycobacterial proteins (159, 160), or fuze the test protein to peroxidase to mediate biotinylation of nearby proteins (161). The interacting proteins are then identified by quantitative proteomics (160). The continued growth of these technologies requires improved specificity of cross-linking combined with robust proteome-wide MS approaches that encompass small proteins, protein modifications, and protein functions. The optimization of proteomics and metabolomics in M. smegmatis will establish systems-level approaches for high-throughput assignment of protein functions that can be subsequently extrapolated to less biochemically amenable mycobacteria (162).
Reporting metabolic reactions at the subcellular scale is crucial for correlating subcellular localizations of proteins with their activities. Genetic tools are less effective in manipulating and visualizing non-proteinaceous cellular structures such as lipids and glycans, but non-genetic approaches are challenging due to the low permeability and unusual architecture of the mycobacterial envelope. Nevertheless, there are emerging efforts to apply modern (click chemistry) approaches of bioorthogonal metabolic labeling in both M. smegmatis and M. tuberculosis, allowing the biosynthesis of trehalose-containing molecules, peptidoglycans, and arabinan to be visualized in situ (116, 163–165). These chemical reporters can document activities of different metabolic pathways in M. smegmatis (e.g., de novo peptidoglycan synthesis versus remodeling reactions [111]), which can be correlated with subcellular enzyme localizations. These chemical-biology reporters can also be used to determine the metabolic state of cells and have potential as diagnostic and therapeutic tools for mycobacterial infections.
Mycobacterium species are ubiquitous in the soil and include the disease-causing NTMs (e.g., M. abscessus and M. avium) (166). As many of these environmental mycobacteria are poorly characterized and difficult to culture, M. smegmatis provides the model system to characterize their gene products. For example, Tn-seq, recombineering and CRISPR tools applied to M. abscessus have begun to define those genes that make M. abscessus intrinsically resistant to many antibiotics (167–170). Not surprisingly as a soil saprophyte, M. smegmatis encodes novel enzymes, which allow it to survive environmental stress. Mycobacteria are obligate aerobes and, thus, need strategies to combat hypoxia, antimicrobials and toxins, such as carbon monoxide, that inhibit terminal oxidases. Studies using the M. smegmatis toolbox have shown that mycobacteria can metabolize hydrogen gas, detoxify carbon monoxide, and encode multiple flavin/deazaflavin oxidoreductases and hydride transferases to survive these stresses (171–174). These genes are conserved among many mycobacteria and other actinobacteria, indicating the usefulness of M. smegmatis as a model for environmental microbiology.
M. smegmatis has been a favorite host for isolating mycobacteriophages. Although it is beyond the scope of this review, isolated phages have provided many important genetic tools such as site-specific integration systems, gene delivery by transduction, and recombineering (175, 176). Phages have also been explored as diagnostic tools, and successfully used to treat chronic, drug-resistant infections of M. abscessus (177–179). The genomes of bacteriophages are filled with genes of unknown functions, foretelling more surprises, and new tools. Notably, M. smegmatis has also become a prominent educational tool for the future of science; as a host for isolating mycobacteriophages. Through the Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program, high school and college students engage in isolating mycobacteriophages from their local environment, characterizing the phage, and sequencing and annotating its genome (180, 181). It is an inclusive, community-oriented, course-based research experience that has served the education of thousands of students worldwide, while also providing the research community with a cornucopia of novel genes and functions. The collective efforts of students around the world have demonstrated that mycobacteriophages and, thus, mycobacteria, are ubiquitous.
M. smegmatis, like other model organisms, has limitations; it is more distantly related to the slow-growing species and is a poor model for host-pathogen interactions and pathogenesis. Two alternative, slow-growing mycobacteria that are more closely related to M. tuberculosis and used in a BSL-2 laboratory are M. bovis BCG, the attenuated vaccine strain, and M. marinum, a fish pathogen, each with advantages for host infection studies over M. smegmatis (182, 183). Unfortunately, M. bovis BCG has a complex genealogy with lineage-specific mutations (184), making it a problematic model for systematic genetic studies. To address this drawback, genetically defined, attenuated mutants of virulent M. tuberculosis have been created by deleting 2 or more genes (185–187). These attenuated strains can be used safely in a BSL-2 lab, providing a more feasible alternative for M. tuberculosis genetics and biochemistry. Obviously, in vivo studies require virulent strains and here we rely on the M. tuberculosis-mouse and M. marinum-zebrafish models among others as the appropriate platforms to study the process of pathogenesis caused by mycobacterial pathogens (188). The M. marinum-zebrafish is particularly amenable to in vivo studies because of the translucent nature of the fish and the ability to genetically manipulate both the host and the bacterium (thanks to M. smegmatis!) (189–191).
M. smegmatis has been at the vanguard of mycobacterial research, not only revealing new strategies to tackle mycobacterial diseases but also contributing unexpected processes and methods that other model bacteria and textbooks fail to offer. Why and how do mycobacteria conjugate through DCT; translate proteins from leaderless transcripts; elongate from the polar ends; and divide asymmetrically? Mycobacteria rely on machineries and molecules that are more commonly found in eukaryotes, such as STPKs for signaling, proteasomes for protein degradation, and phosphatidylinositols as a major component of the plasma membrane. Why? These unique and unexpected characteristics will surely continue to spark the curiosities of microbiologists and inspire the mycobacterial research community and we foresee that mc2155 will continue its place at the front of this research.
ACKNOWLEDGMENTS
Our laboratories are funded by NIH R21 AI144748 to Y.S.M., R21 AI14760802 and R01 GM13927703 to K.M.D., and R01 AI026170 to W.R.J.
Biographies
Ian L. Sparks earned his B.Sc. in Microbiology from the University of Wisconsin, Madison (2017). As an undergraduate researcher with Douglas Weibel, he studied the impact of iron chelation on divisome assembly arrest in Caulobacter crescentus. As a current graduate student working with Yasu Morita at the University of Massachusetts Amherst, he is studying the function of M. smegmatis lipoglycans in maintaining cell envelope integrity during cell division and characterizing a functional membrane domain of M. tuberculosis previously discovered in M. smegmatis.
Keith M. Derbyshire earned his B.Sc. (Hons) in Microbiology at the University of Liverpool, England (1980), and a Ph.D. in Molecular Biology from the University of Edinburgh, Scotland (1983). As a graduate student with Neil Willetts, he studied plasmids and bacterial conjugation in Escherichia coli. He was a postdoctoral fellow with Nigel Grindley at Yale University, where he worked on the mechanisms of bacterial transposition. He joined the Wadsworth Center, NYS Department of Health in 1992 and is currently the Director of the Division of Genetics and a Professor in Biomedical Sciences at the University at Albany. He switched from working on transposition in E. coli to conjugation in Mycobacterium smegmatis following many discussions with Bill Jacobs, who persuaded him that M. smegmatis was a better model organism! His laboratory uses this great model bacterium to study the molecular biology of mycobacteria with a focus on gene expression, mycobacterial conjugation, and small mycobacterial proteins.
William R. Jacobs, Jr. earned his MA in Mathematics from Edinboro State College in Pennsylvania (1977). As a graduate student with Josie Clark-Curtiss and Roy Curtiss III, he constructed the first genomic libraries of Mycobacterium leprae bacilli, which they had isolated from nine-banded armadillos with Charles Shepard. He embraced recombinant DNA developments by making in vivo cosmid packaging strains and graduated from the University of Alabama in Birmingham in 1985. As a postdoctoral fellow with Barry Bloom at the Albert Einstein College of Medicine (AECOM), he set out to develop genetic systems for M. tuberculosis and M. bovis BCG. In 1987, he successfully introduced foreign DNA into mycobacteria for the first time. Shuttle phasmids were used as tools that allowed his lab to isolate M. smegmatis mc2155, the first plasmid-transformable mutant of M. smegmatis. He was an Investigator in the Howard Hughes Medical Institute for 30 years and was elected to the National Academy of Sciences in 2013. His lab, in the Department of Microbiology and Immunology at the AECOM, continues to use genetics to investigate Leprosy, Tuberculosis, and Herpes viruses.
Yasu S. Morita earned his B.A. from International Christian University in Tokyo, Japan in 1992 and Ph.D. from Johns Hopkins School of Medicine in Baltimore, Maryland in 2000. For his Ph.D. thesis, he studied the biosynthesis of fatty acids and glycosylphosphatidylinositols (GPIs) in Trypanosoma brucei, the etiologic agent of African sleeping sickness, under the mentorship of Paul Englund. He started working on mycobacteria as a postdoctoral fellow at the University of Melbourne in Australia in the lab of Malcolm McConville. He investigated the biosynthesis and physiological significance of GPI-like glycolipids, namely phosphatidylinositol mannosides, lipomannan, and lipoarabinomannan, and continued his study in Taroh Kinoshita’s lab at Osaka University in Japan. He joined the Department of Microbiology at University of Massachusetts Amherst in 2012. He is currently an associate professor and continues exploring the pathways of cell envelope biosynthesis and membrane dynamics in mycobacteria.
Contributor Information
Yasu S. Morita, Email: ymorita@umass.edu.
Patricia A. Champion, University of Notre Dame
REFERENCES
- 1.Koch R. 1882. Die Aetiologie der Tuberculose. (Nach einem in der physiologischen Gesellschaft zu Berlin am 24.März cr. gehaltenem Vortrage). Berliner Klin Wochenschr 19:221–230. [Google Scholar]
- 2.Glaziou P, Floyd K, Raviglione MC. 2018. Global epidemiology of tuberculosis. Semin Respir Crit Care Med 39:271–285. 10.1055/s-0038-1651492. [DOI] [PubMed] [Google Scholar]
- 3.Furin J, Cox H, Pai M. 2019. Tuberculosis. Lancet 393:1642–1656. 10.1016/S0140-6736(19)30308-3. [DOI] [PubMed] [Google Scholar]
- 4.Johansen MD, Herrmann J-L, Kremer L. 2020. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol 18:392–407. 10.1038/s41579-020-0331-1. [DOI] [PubMed] [Google Scholar]
- 5.Gupta RS, Lo B, Son J. 2018. Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol 9:67. 10.3389/fmicb.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tortoli E, Brown-Elliott BA, Chalmers JD, Cirillo DM, Daley CL, Emler S, Floto RA, Garcia MJ, Hoefsloot W, Koh W-J, Lange C, Loebinger M, Maurer FP, Morimoto K, Niemann S, Richter E, Turenne CY, Vasireddy R, Vasireddy S, Wagner D, Wallace RJ, Wengenack N, van Ingen J. 2019. Same meat, different gravy: ignore the new names of mycobacteria. Eur Respir J 54:1900795. 10.1183/13993003.00795-2019. [DOI] [PubMed] [Google Scholar]
- 7.Crawford JT, Bates JH. 1979. Isolation of plasmids from mycobacteria. Infect Immun 24:979–981. 10.1128/iai.24.3.979-981.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford JT, Bates JH. 1986. Analysis of plasmids in Mycobacterium avium-intracellulare isolates from persons with acquired immunodeficiency syndrome. Am Rev Respir Dis 134:659–661. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs WR, Tuckman M, Bloom BR. 1987. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 327:532–535. 10.1038/327532a0. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs WR. 2014. Gene transfer in Mycobacterium tuberculosis: Shuttle phasmids to enlightenment. Microbiol Spectr 2. 10.1128/microbiolspec.MGM2-0037-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs WR, Snapper SB, Tuckman M, Bloom BR. 1989. Mycobacteriophage vector systems. Rev Infect Dis 11 Suppl 2:S404–S410. 10.1093/clinids/11.Supplement_2.S404. [DOI] [PubMed] [Google Scholar]
- 12.Snapper SB, Lugosi L, Jekkel A, Melton RE, Kieser T, Bloom BR, Jacobs WR. 1988. Lysogeny and transformation in mycobacteria: stable expression of foreign genes. Proc Natl Acad Sci USA 85:6987–6991. 10.1073/pnas.85.18.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labidi A, Dauguet C, Goh KS, David HL. 1984. Plasmid profiles of Mycobacterium fortuitum complex isolates. Curr Microbiol 11:235–240. 10.1007/BF01567167. [DOI] [Google Scholar]
- 14.Rauzier J, Moniz-Pereira J, Gicquel-Sanzey B. 1988. Complete nucleotide sequence of pAL5000, a plasmid from Mycobacterium fortuitum. Gene 71:315–321. 10.1016/0378-1119(88)90048-0. [DOI] [PubMed] [Google Scholar]
- 15.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 16.Panas MW, Jain P, Yang H, Mitra S, Biswas D, Wattam AR, Letvin NL, Jacobs WR. 2014. Noncanonical SMC protein in Mycobacterium smegmatis restricts maintenance of Mycobacterium fortuitum plasmids. Proc Natl Acad Sci USA 111:13264–13271. 10.1073/pnas.1414207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deep A, Gu Y, Gao Y-Q, Ego KM, Herzik MA, Zhou H, Corbett KD. 2022. The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA. Mol Cell 82:4145–4159. 10.1016/j.molcel.2022.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MH, Pascopella L, Jacobs WR, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc Natl Acad Sci USA 88:3111–3115. 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs WR, Barletta RG, Udani R, Chan J, Kalkut G, Sosne G, Kieser T, Sarkis GJ, Hatfull GF, Bloom BR. 1993. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 260:819–822. 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 21.Pelicic V, Reyrat JM, Gicquel B. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol Microbiol 20:919–925. 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 22.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 23.Carrière C, Riska PF, Zimhony O, Kriakov J, Bardarov S, Burns J, Chan J, Jacobs WR. 1997. Conditionally replicating luciferase reporter phages: improved sensitivity for rapid detection and assessment of drug susceptibility of Mycobacterium tuberculosis. J Clin Microbiol 35:3232–3239. 10.1128/jcm.35.12.3232-3239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pope WH, Ferreira CM, Jacobs-Sera D, Benjamin RC, Davis AJ, DeJong RJ, Elgin SCR, Guilfoile FR, Forsyth MH, Harris AD, Harvey SE, Hughes LE, Hynes PM, Jackson AS, Jalal MD, MacMurray EA, Manley CM, McDonough MJ, Mosier JL, Osterbann LJ, Rabinowitz HS, Rhyan CN, Russell DA, Saha MS, Shaffer CD, Simon SE, Sims EF, Tovar IG, Weisser EG, Wertz JT, Weston-Hafer KA, Williamson KE, Zhang B, Cresawn SG, Jain P, Piuri M, Jacobs WR, Hendrix RW, Hatfull GF. 2011. Cluster K mycobacteriophages: insights into the evolutionary origins of mycobacteriophage TM4. PLoS One 6:e26750. 10.1371/journal.pone.0026750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam RA, Bloom BR, Hatfull GF, Jacobs WR. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci USA 94:10961–10966. 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardarov S, Bardarov S, Pavelka MS, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology (Reading) 148:3007–3017. 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 27.Jain P, Hsu T, Arai M, Biermann K, Thaler DS, Nguyen A, González PA, Tufariello JM, Kriakov J, Chen B, Larsen MH, Jacobs WR. 2014. Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. mBio 5:e01245-14. 10.1128/mBio.01245-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA 100:12989–12994. 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 30.Sassetti CM, Boyd DH, Rubin EJ. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA 98:12712–12717. 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Judd JA, Canestrari J, Clark R, Joseph A, Lapierre P, Lasek-Nesselquist E, Mir M, Palumbo M, Smith C, Stone M, Upadhyay A, Wirth SE, Dedrick RM, Meier CG, Russell DA, Dills A, Dove E, Kester J, Wolf ID, Zhu J, Rubin ER, Fortune S, Hatfull GF, Gray TA, Wade JT, Derbyshire KM. 2021. A mycobacterial systems resource for the research community. mBio 12:e02401-20. 10.1128/mBio.02401-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malhotra S, Vedithi SC, Blundell TL. 2017. Decoding the similarities and differences among mycobacterial species. PLoS Negl Trop Dis 11:e0005883. 10.1371/journal.pntd.0005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dragset MS, Ioerger TR, Zhang YJ, Mærk M, Ginbot Z, Sacchettini JC, Flo TH, Rubin EJ, Steigedal M. 2019. Genome-wide phenotypic profiling identifies and categorizes genes required for mycobacterial low iron fitness. Sci Rep 9:11394. 10.1038/s41598-019-47905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, Pritchard JR, Church GM, Rubin EJ, Sassetti CM, Schnappinger D, Fortune SM. 2017. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol 2:16274. 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosch B, DeJesus MA, Poulton NC, Zhang W, Engelhart CA, Zaveri A, Lavalette S, Ruecker N, Trujillo C, Wallach JB, Li S, Ehrt S, Chait BT, Schnappinger D, Rock JM. 2021. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell 184:4579–4592. 10.1016/j.cell.2021.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Wolf ID, Dulberger CL, Won HI, Kester JC, Judd JA, Wirth SE, Clark RR, Li Y, Luo Y, Gray TA, Wade JT, Derbyshire KM, Fortune SM, Rubin EJ. 2021. Spatiotemporal localization of proteins in mycobacteria. Cell Rep 37:110154. 10.1016/j.celrep.2021.110154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Wet TJ, Winkler KR, Mhlanga M, Mizrahi V, Warner DF. 2020. Arrayed CRISPRi and quantitative imaging describe the morphotypic landscape of essential mycobacterial genes. Elife 9:e60083. 10.7554/eLife.60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizuguchi Y, Suga K, Tokunaga T. 1976. Multiple mating types of Mycobacterium smegmatis. Jpn J Microbiol 20:435–443. 10.1111/j.1348-0421.1976.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 40.Parsons LM, Jankowski CS, Derbyshire KM. 1998. Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol Microbiol 28:571–582. 10.1046/j.1365-2958.1998.00818.x. [DOI] [PubMed] [Google Scholar]
- 41.Gray TA, Krywy JA, Harold J, Palumbo MJ, Derbyshire KM. 2013. Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol 11:e1001602. 10.1371/journal.pbio.1001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray TA, Derbyshire KM. 2018. Blending genomes: distributive conjugal transfer in mycobacteria, a sexier form of HGT. Mol Microbiol 108:601–613. 10.1111/mmi.13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark RR, Lapierre P, Lasek-Nesselquist E, Gray TA, Derbyshire KM. 2022. A polymorphic gene within the Mycobacterium smegmatis esx1 locus determines mycobacterial self-identity and conjugal compatibility. mBio 13:e0021322. 10.1128/mbio.00213-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA 94:9869–9874. 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brosch R, Philipp WJ, Stavropoulos E, Colston MJ, Cole ST, Gordon SV. 1999. Genomic analysis reveals variation between Mycobacterium tuberculosis H37Rv and the attenuated M. tuberculosis H37Ra strain. Infect Immun 67:5768–5774. 10.1128/IAI.67.11.5768-5774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith NH, Hewinson RG, Kremer K, Brosch R, Gordon SV. 2009. Myths and misconceptions: the origin and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 7:537–544. 10.1038/nrmicro2165. [DOI] [PubMed] [Google Scholar]
- 47.Smith NH, Gordon SV, de la Rua-Domenech R, Clifton-Hadley RS, Hewinson RG. 2006. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat Rev Microbiol 4:670–681. 10.1038/nrmicro1472. [DOI] [PubMed] [Google Scholar]
- 48.Gutierrez MC, Brisse S, Brosch R, Fabre M, Omaïs B, Marmiesse M, Supply P, Vincent V. 2005. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog 1:e5. 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie A-S, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez M-C, Leclerc C, Bentley SD, Stinear TP, Brisse S, Médigue C, Parkhill J, Cruveiller S, Brosch R. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45:172–179. 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mortimer TD, Pepperell CS. 2014. Genomic signatures of distributive conjugal transfer among mycobacteria. Genome Biol Evol 6:2489–2500. 10.1093/gbe/evu175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Derbyshire KM, Gray TA. 2014. Distributive conjugal transfer: new insights into horizontal gene transfer and genetic exchange in mycobacteria. Microbiol Spectr 2. 10.1128/microbiolspec.MGM2-0022-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boritsch EC, Khanna V, Pawlik A, Honoré N, Navas VH, Ma L, Bouchier C, Seemann T, Supply P, Stinear TP, Brosch R. 2016. Key experimental evidence of chromosomal DNA transfer among selected tuberculosis-causing mycobacteria. Proc Natl Acad Sci USA 113:9876–9881. 10.1073/pnas.1604921113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bitter W, Houben ENG, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao L-Y, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog 5:e1000507. 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaziri F, Brosch R. 2019. ESX/type VII secretion systems-an important way out for mycobacterial proteins. Microbiol Spectr 7. 10.1128/microbiolspec.PSIB-0029-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gröschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. 2016. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol 14:677–691. 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
- 56.Bunduc CM, Bitter W, Houben ENG. 2020. Structure and function of the mycobacterial type VII secretion systems. Annu Rev Microbiol 74:315–335. 10.1146/annurev-micro-012420-081657. [DOI] [PubMed] [Google Scholar]
- 57.Lagune M, Petit C, Sotomayor FV, Johansen MD, Beckham KSH, Ritter C, Girard-Misguich F, Wilmanns M, Kremer L, Maurer FP, Herrmann J-L. 2021. Conserved and specialized functions of Type VII secretion systems in non-tuberculous mycobacteria. Microbiology (Reading) 167. 10.1099/mic.0.001054. [DOI] [PubMed] [Google Scholar]
- 58.Gray TA, Clark RR, Boucher N, Lapierre P, Smith C, Derbyshire KM. 2016. Intercellular communication and conjugation are mediated by ESX secretion systems in mycobacteria. Science 354:347–350. 10.1126/science.aag0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laencina L, Dubois V, Le Moigne V, Viljoen A, Majlessi L, Pritchard J, Bernut A, Piel L, Roux A-L, Gaillard J-L, Lombard B, Loew D, Rubin EJ, Brosch R, Kremer L, Herrmann J-L, Girard-Misguich F. 2018. Identification of genes required for Mycobacterium abscessus growth in vivo with a prominent role of the ESX-4 locus. Proc Natl Acad Sci USA 115:E1002–E1011. 10.1073/pnas.1713195115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pajuelo D, Tak U, Zhang L, Danilchanka O, Tischler AD, Niederweis M. 2021. Toxin secretion and trafficking by Mycobacterium tuberculosis. Nat Commun 12:6592. 10.1038/s41467-021-26925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. 2013. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep 5:1121–1131. 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miranda-CasoLuengo AA, Staunton PM, Dinan AM, Lohan AJ, Loftus BJ. 2016. Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence. BMC Genomics 17:553. 10.1186/s12864-016-2868-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawyer EB, Phelan JE, Clark TG, Cortes T. 2021. A snapshot of translation in Mycobacterium tuberculosis during exponential growth and nutrient starvation revealed by ribosome profiling. Cell Rep 34:108695. 10.1016/j.celrep.2021.108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA. 2015. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet 11:e1005641. 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beck HJ, Moll I. 2018. Leaderless mRNAs in the spotlight: ancient but not outdated!. Microbiol Spectr 6. 10.1128/microbiolspec.RWR-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moll I, Grill S, Gualerzi CO, Bläsi U. 2002. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol Microbiol 43:239–246. 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- 67.de Groot A, Roche D, Fernandez B, Ludanyi M, Cruveiller S, Pignol D, Vallenet D, Armengaud J, Blanchard L. 2014. RNA sequencing and proteogenomics reveal the importance of leaderless mRNAs in the radiation-tolerant bacterium Deinococcus deserti. Genome Biol Evol 6:932–948. 10.1093/gbe/evu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hering O, Brenneis M, Beer J, Suess B, Soppa J. 2009. A novel mechanism for translation initiation operates in haloarchaea. Mol Microbiol 71:1451–1463. 10.1111/j.1365-2958.2009.06615.x. [DOI] [PubMed] [Google Scholar]
- 69.Vockenhuber M-P, Sharma CM, Statt MG, Schmidt D, Xu Z, Dietrich S, Liesegang H, Mathews DH, Suess B. 2011. Deep sequencing-based identification of small non-coding RNAs in Streptomyces coelicolor. RNA Biol 8:468–477. 10.4161/rna.8.3.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith C, Canestrari JG, Wang AJ, Champion MM, Derbyshire KM, Gray TA, Wade JT. 2022. Pervasive translation in Mycobacterium tuberculosis. Elife 11:e73980. 10.7554/eLife.73980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Storz G, Wolf YI, Ramamurthi KS. 2014. Small proteins can no longer be ignored. Annu Rev Biochem 83:753–777. 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meydan S, Marks J, Klepacki D, Sharma V, Baranov PV, Firth AE, Margus T, Kefi A, Vázquez-Laslop N, Mankin AS. 2019. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol Cell 74:481–493.e6. 10.1016/j.molcel.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sberro H, Fremin BJ, Zlitni S, Edfors F, Greenfield N, Snyder MP, Pavlopoulos GA, Kyrpides NC, Bhatt AS. 2019. Large-scale analyses of human microbiomes reveal thousands of small, novel genes. Cell 178:1245–1259. 10.1016/j.cell.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gray T, Storz G, Papenfort K. 2022. Small proteins; big questions. J Bacteriol 204:e0034121. 10.1128/JB.00341-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hemm MR, Weaver J, Storz G. 2020. Escherichia coli small proteome. EcoSal Plus 9. 10.1128/ecosalplus.ESP-0031-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Canestrari JG, Lasek-Nesselquist E, Upadhyay A, Rofaeil M, Champion MM, Wade JT, Derbyshire KM, Gray TA. 2020. Polycysteine-encoding leaderless short ORFs function as cysteine-responsive attenuators of operonic gene expression in mycobacteria. Mol Microbiol 114:93–108. 10.1111/mmi.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Braunstein M, Brown AM, Kurtz S, Jacobs WR. 2001. Two nonredundant SecA homologues function in mycobacteria. J Bacteriol 183:6979–6990. 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Braunstein M, Bensing BA, Sullam PM. 2019. The two distinct types of SecA2-dependent export systems. Microbiol Spectr 7. 10.1128/microbiolspec.PSIB-0025-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bensing BA, Seepersaud R, Yen YT, Sullam PM. 2014. Selective transport by SecA2: an expanding family of customized motor proteins. Biochim Biophys Acta 1843:1674–1686. 10.1016/j.bbamcr.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Swanson S, Ioerger TR, Rigel NW, Miller BK, Braunstein M, Sacchettini JC. 2015. Structural similarities and differences between two functionally distinct SecA proteins, Mycobacterium tuberculosis SecA1 and SecA2. J Bacteriol 198:720–730. 10.1128/JB.00696-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ligon LS, Rigel NW, Romanchuk A, Jones CD, Braunstein M. 2013. Suppressor analysis reveals a role for SecY in the SecA2-dependent protein export pathway of mycobacteria. J Bacteriol 195:4456–4465. 10.1128/JB.00630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kurtz S, McKinnon KP, Runge MS, Ting JP-Y, Braunstein M. 2006. The SecA2 secretion factor of Mycobacterium tuberculosis promotes growth in macrophages and inhibits the host immune response. Infect Immun 74:6855–6864. 10.1128/IAI.01022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol Microbiol 48:453–464. 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- 84.Miller BK, Zulauf KE, Braunstein M. 2017. The Sec pathways and exportomes of Mycobacterium tuberculosis. Microbiol Spectr 5. 10.1128/microbiolspec.TBTB2-0013-2016. [DOI] [PubMed] [Google Scholar]
- 85.Sullivan JT, Young EF, McCann JR, Braunstein M. 2012. The Mycobacterium tuberculosis SecA2 system subverts phagosome maturation to promote growth in macrophages. Infect Immun 80:996–1006. 10.1128/IAI.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zulauf KE, Sullivan JT, Braunstein M. 2018. The SecA2 pathway of Mycobacterium tuberculosis exports effectors that work in concert to arrest phagosome and autophagosome maturation. PLoS Pathog 14:e1007011. 10.1371/journal.ppat.1007011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 88.van der Woude AD, Stoop EJM, Stiess M, Wang S, Ummels R, van Stempvoort G, Piersma SR, Cascioferro A, Jiménez CR, Houben ENG, Luirink J, Pieters J, van der Sar AM, Bitter W. 2014. Analysis of SecA2-dependent substrates in Mycobacterium marinum identifies protein kinase G (PknG) as a virulence effector. Cell Microbiol 16:280–295. 10.1111/cmi.12221. [DOI] [PubMed] [Google Scholar]
- 89.Feltcher ME, Gunawardena HP, Zulauf KE, Malik S, Griffin JE, Sassetti CM, Chen X, Braunstein M. 2015. Label-free quantitative proteomics reveals a role for the Mycobacterium tuberculosis SecA2 pathway in exporting solute binding proteins and mce transporters to the cell wall. Mol Cell Proteomics 14:1501–1516. 10.1074/mcp.M114.044685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wolff KA, de la Peña AH, Nguyen HT, Pham TH, Amzel LM, Gabelli SB, Nguyen L. 2015. A redox regulatory system critical for mycobacterial survival in macrophages and biofilm development. PLoS Pathog 11:e1004839. 10.1371/journal.ppat.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khan MZ, Bhaskar A, Upadhyay S, Kumari P, Rajmani RS, Jain P, Singh A, Kumar D, Bhavesh NS, Nandicoori VK. 2017. Protein kinase G confers survival advantage to Mycobacterium tuberculosis during latency-like conditions. J Biol Chem 292:16093–16108. 10.1074/jbc.M117.797563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ventura M, Rieck B, Boldrin F, Degiacomi G, Bellinzoni M, Barilone N, Alzaidi F, Alzari PM, Manganelli R, O'Hare HM. 2013. GarA is an essential regulator of metabolism in Mycobacterium tuberculosis. Mol Microbiol 90:356–366. 10.1111/mmi.12368. [DOI] [PubMed] [Google Scholar]
- 93.Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, Betts JC, Mizrahi V, Smith DA, Stokes RW, Av-Gay Y. 2004. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol 52:1691–1702. 10.1111/j.1365-2958.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- 94.Rieck B, Degiacomi G, Zimmermann M, Cascioferro A, Boldrin F, Lazar-Adler NR, Bottrill AR, Le Chevalier F, Frigui W, Bellinzoni M, Lisa M-N, Alzari PM, Nguyen L, Brosch R, Sauer U, Manganelli R, O'Hare HM. 2017. PknG senses amino acid availability to control metabolism and virulence of Mycobacterium tuberculosis. PLoS Pathog 13:e1006399. 10.1371/journal.ppat.1006399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. 2004. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304:1800–1804. 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 96.Wang J, Ge P, Lei Z, Lu Z, Qiang L, Chai Q, Zhang Y, Zhao D, Li B, Su J, Peng R, Pang Y, Shi Y, Zhang Y, Gao GF, Qiu X-B, Liu CH. 2021. Mycobacterium tuberculosis protein kinase G acts as an unusual ubiquitinating enzyme to impair host immunity. EMBO Rep 22:e52175. 10.15252/embr.202052175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ge P, Lei Z, Yu Y, Lu Z, Qiang L, Chai Q, Zhang Y, Zhao D, Li B, Pang Y, Liu CH, Wang J. 2022. M. tuberculosis PknG manipulates host autophagy flux to promote pathogen intracellular survival. Autophagy 18:576–594. 10.1080/15548627.2021.1938912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pradhan G, Shrivastva R, Mukhopadhyay S. 2018. Mycobacterial PknG targets the Rab7l1 signaling pathway to inhibit phagosome-lysosome fusion. J Immunol 201:1421–1433. 10.4049/jimmunol.1800530. [DOI] [PubMed] [Google Scholar]
- 99.Minnikin DE. 1982. Lipids: complex lipids, their chemistry, biosynthesis and role, p 95–184. In The biology of mycobacteria. Academic Press, London, UK. [Google Scholar]
- 100.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA 105:3963–3967. 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffé M. 2008. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol 190:5672–5680. 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rahlwes KC, Sparks IL, Morita YS. 2019. Cell walls and membranes of Actinobacteria. Subcell Biochem 92:417–469. 10.1007/978-3-030-18768-2_13. [DOI] [PubMed] [Google Scholar]
- 103.Jackson M, Stevens CM, Zhang L, Zgurskaya HI, Niederweis M. 2021. Transporters involved in the biogenesis and functionalization of the mycobacterial cell envelope. Chem Rev 121:5124–5157. 10.1021/acs.chemrev.0c00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Batt SM, Minnikin DE, Besra GS. 2020. The thick waxy coat of mycobacteria, a protective layer against antibiotics and the host’s immune system. Biochem J 477:1983–2006. 10.1042/BCJ20200194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dulberger CL, Rubin EJ, Boutte CC. 2020. The mycobacterial cell envelope - a moving target. Nat Rev Microbiol 18:47–59. 10.1038/s41579-019-0273-7. [DOI] [PubMed] [Google Scholar]
- 106.Hett EC, Rubin EJ. 2008. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev 72:126–156. 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thanky NR, Young DB, Robertson BD. 2007. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb) 87:231–236. 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 108.Melzer ES, Sein CE, Chambers JJ, Siegrist MS. 2018. DivIVA concentrates mycobacterial cell envelope assembly for initiation and stabilization of polar growth. Cytoskeleton (Hoboken) 75:498–507. 10.1002/cm.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. 2014. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA 111:E3243–E3251. 10.1073/pnas.1402158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. 2008. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology (Reading) 154:725–735. 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 111.García-Heredia A, Pohane AA, Melzer ES, Carr CR, Fiolek TJ, Rundell SR, Chuin LH, Wagner JC, Morita YS, Swarts BM, Siegrist MS. 2018. Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. Elife 7:e37243. 10.7554/eLife.37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hayashi JM, Luo C-Y, Mayfield JA, Hsu T, Fukuda T, Walfield AL, Giffen SR, Leszyk JD, Baer CE, Bennion OT, Madduri A, Shaffer SA, Aldridge BB, Sassetti CM, Sandler SJ, Kinoshita T, Moody DB, Morita YS. 2016. Spatially distinct and metabolically active membrane domain in mycobacteria. Proc Natl Acad Sci USA 113:5400–5405. 10.1073/pnas.1525165113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morita YS, Velasquez R, Taig E, Waller RF, Patterson JH, Tull D, Williams SJ, Billman-Jacobe H, McConville MJ. 2005. Compartmentalization of lipid biosynthesis in mycobacteria. J Biol Chem 280:21645–21652. 10.1074/jbc.M414181200. [DOI] [PubMed] [Google Scholar]
- 114.García-Heredia A, Kado T, Sein CE, Puffal J, Osman SH, Judd J, Gray TA, Morita YS, Siegrist MS. 2021. Membrane-partitioned cell wall synthesis in mycobacteria. Elife 10:e60263. 10.7554/eLife.60263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kado T, Akbary Z, Motooka D, Sparks IL, Melzer ES, Nakamura S, Rojas ER, Morita YS, Siegrist MS. 2022. The cell wall polymer initiates plasma membrane partitioning in mycobacteria. bioRxiv. 10.1101/2022.06.12.495848. [DOI] [PMC free article] [PubMed]
- 116.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. 2013. (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol 8:500–505. 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Puffal J, Sparks IL, Brenner JR, Li X, Leszyk JD, Hayashi JM, Shaffer SA, Morita YS. 2022. Compartmentalized cell envelope biosynthesis in Mycobacterium tuberculosis. bioRxiv. 10.1101/2022.01.07.475471. [DOI]
- 118.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104. 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. 2013. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun 4:2470. 10.1038/ncomms3470. [DOI] [PubMed] [Google Scholar]
- 120.Logsdon MM, Ho P-Y, Papavinasasundaram K, Richardson K, Cokol M, Sassetti CM, Amir A, Aldridge BB. 2017. A parallel adder coordinates mycobacterial cell-cycle progression and cell-size homeostasis in the context of asymmetric growth and organization. Curr Biol 27:3367–3374. 10.1016/j.cub.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou X, Halladin DK, Theriot JA. 2016. Fast mechanically driven daughter cell separation is widespread in Actinobacteria. mBio 7:e00952-16. 10.1128/mBio.00952-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Puffal J, García-Heredia A, Rahlwes KC, Siegrist MS, Morita YS. 2018. Spatial control of cell envelope biosynthesis in mycobacteria. Pathog Dis 76: 10.1093/femspd/fty027. [DOI] [PubMed] [Google Scholar]
- 123.Baranowski C, Rego EH, Rubin EJ. 2019. The dream of a Mycobacterium. Microbiol Spectr 7. 10.1128/microbiolspec.GPP3-0008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rego EH, Audette RE, Rubin EJ. 2017. Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 546:153–157. 10.1038/nature22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Richardson K, Bennion OT, Tan S, Hoang AN, Cokol M, Aldridge BB. 2016. Temporal and intrinsic factors of rifampicin tolerance in mycobacteria. Proc Natl Acad Sci USA 113:8302–8307. 10.1073/pnas.1600372113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vijay S, Vinh DN, Hai HT, Ha VTN, Dung VTM, Dinh TD, Nhung HN, Tram TTB, Aldridge BB, Hanh NT, Thu DDA, Phu NH, Thwaites GE, Thuong NTT. 2017. Influence of stress and antibiotic resistance on cell-length distribution in Mycobacterium tuberculosis clinical isolates. Front Microbiol 8:2296. 10.3389/fmicb.2017.02296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Y, Heym B, Allen B, Young D, Cole S. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591–593. 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 128.Telenti A, Honoré N, Bernasconi C, March J, Ortega A, Heym B, Takiff HE, Cole ST. 1997. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol 35:719–723. 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230. 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 130.Dessen A, Quémard A, Blanchard JS, Jacobs WR, Sacchettini JC. 1995. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 267:1638–1641. 10.1126/science.7886450. [DOI] [PubMed] [Google Scholar]
- 131.Quémard A, Sacchettini JC, Dessen A, Vilcheze C, Bittman R, Jacobs WR, Blanchard JS. 1995. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry 34:8235–8241. 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 132.Vilchèze C, Jacobs WR. 2007. The mechanism of isoniazid killing: clarity through the scope of genetics. Annu Rev Microbiol 61:35–50. 10.1146/annurev.micro.61.111606.122346. [DOI] [PubMed] [Google Scholar]
- 133.Miesel L, Rozwarski DA, Sacchettini JC, Jacobs WR. 1998. Mechanisms for isoniazid action and resistance. Novartis Found Symp 217:209–220. 10.1002/0470846526.ch15. [DOI] [PubMed] [Google Scholar]
- 134.Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Sacchettini JC. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279:98–102. 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 135.Vilchèze C, Wang F, Arai M, Hazbón MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC, Jacobs WR. 2006. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med 12:1027–1029. 10.1038/nm1466. [DOI] [PubMed] [Google Scholar]
- 136.Vilchèze C, Weisbrod TR, Chen B, Kremer L, Hazbón MH, Wang F, Alland D, Sacchettini JC, Jacobs WR. 2005. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob Agents Chemother 49:708–720. 10.1128/AAC.49.2.708-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gale GR, Mclain HH. 1963. Effect of ethambutol on cytology of Mycobacterium smegmatis. J Bacteriol 86:749–756. 10.1128/jb.86.4.749-756.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Forbes M, Kuck NA, Peets EA. 1965. Effect of ethambutol on nucleic acid metabolism in Mycobacterium smegmatis and its reversal by polyamines and divalent cations. J Bacteriol 89:1299–1305. 10.1128/jb.89.5.1299-1305.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takayama K, Kilburn JO. 1989. Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob Agents Chemother 33:1493–1499. 10.1128/AAC.33.9.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wolucka BA, McNeil MR, de Hoffmann E, Chojnacki T, Brennan PJ. 1994. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J Biol Chem 269:23328–23335. 10.1016/S0021-9258(17)31657-5. [DOI] [PubMed] [Google Scholar]
- 141.Belanger AE, Besra GS, Ford ME, Mikusová K, Belisle JT, Brennan PJ, Inamine JM. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci USA 93:11919–11924. 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sreevatsan S, Stockbauer KE, Pan X, Kreiswirth BN, Moghazeh SL, Jacobs WR, Telenti A, Musser JM. 1997. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother 41:1677–1681. 10.1128/AAC.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Telenti A, Philipp WJ, Sreevatsan S, Bernasconi C, Stockbauer KE, Wieles B, Musser JM, Jacobs WR. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med 3:567–570. 10.1038/nm0597-567. [DOI] [PubMed] [Google Scholar]
- 144.Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS, Veerapen N, Batt SM, Zhao W, Qin L, Yang X, Wang M, Zhu Y, Zhang B, Bi L, Zhang X, Yang H, Guddat LW, Xu W, Wang Q, Li J, Besra GS, Rao Z. 2020. Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368:1211–1219. 10.1126/science.aba9102. [DOI] [PubMed] [Google Scholar]
- 145.Cohen KA, Manson AL, Desjardins CA, Abeel T, Earl AM. 2019. Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges. Genome Med 11:45. 10.1186/s13073-019-0660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dousa KM, Kurz SG, Bark CM, Bonomo RA, Furin JJ. 2020. Drug-resistant tuberculosis: a glance at progress and global challenges. Infect Dis Clin North Am 34:863–886. 10.1016/j.idc.2020.06.001. [DOI] [PubMed] [Google Scholar]
- 147.Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 148.Guo H, Courbon GM, Bueler SA, Mai J, Liu J, Rubinstein JL. 2021. Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. Nature 589:143–147. 10.1038/s41586-020-3004-3. [DOI] [PubMed] [Google Scholar]
- 149.Ndjeka N, Hughes J, Reuter A, Conradie F, Enwerem M, Ferreira H, Ismail N, Kock Y, Master I, Meintjes G, Padanilam X, Romero R, Schaaf HS, Riele JT, Maartens G. 2020. Implementing novel regimens for drug-resistant TB in South Africa: what can the world learn? Int J Tuber Lung Dis 24:1073–1080. 10.5588/ijtld.20.0174. [DOI] [PubMed] [Google Scholar]
- 150.Varela C, Rittmann D, Singh A, Krumbach K, Bhatt K, Eggeling L, Besra GS, Bhatt A. 2012. MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem Biol 19:498–506. 10.1016/j.chembiol.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Grzegorzewicz AE, Pham H, Gundi VAKB, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SEM, Korduláková J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8:334–341. 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu Z, Meshcheryakov VA, Poce G, Chng S-S. 2017. MmpL3 is the flippase for mycolic acids in mycobacteria. Proc Natl Acad Sci USA 114:7993–7998. 10.1073/pnas.1700062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li W, Stevens CM, Pandya AN, Darzynkiewicz Z, Bhattarai P, Tong W, Gonzalez-Juarrero M, North EJ, Zgurskaya HI, Jackson M. 2019. Direct inhibition of MmpL3 by novel antitubercular compounds. ACS Infect Dis 5:1001–1012. 10.1021/acsinfecdis.9b00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang B, Li J, Yang X, Wu L, Zhang J, Yang Y, Zhao Y, Zhang L, Yang X, Yang X, Cheng X, Liu Z, Jiang B, Jiang H, Guddat LW, Yang H, Rao Z. 2019. Crystal structures of membrane transporter MmpL3, an anti-TB drug target. Cell 176:636–648. 10.1016/j.cell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 155.Su C-C, Klenotic PA, Bolla JR, Purdy GE, Robinson CV, Yu EW. 2019. MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine. Proc Natl Acad Sci USA 116:11241–11246. 10.1073/pnas.1901346116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Adams O, Deme JC, Parker JL, Fowler PW, Lea SM, Newstead S, CRyPTIC Consortium . 2021. Cryo-EM structure and resistance landscape of M. tuberculosis MmpL3: an emergent therapeutic target. Structure 29:1182–1191. 10.1016/j.str.2021.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fay A, Czudnochowski N, Rock JM, Johnson JR, Krogan NJ, Rosenberg O, Glickman MS. 2019. Two accessory proteins govern MmpL3 mycolic acid transport in mycobacteria. mBio 10:e00850-19. 10.1128/mBio.00850-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Boeree MJ, Heinrich N, Aarnoutse R, Diacon AH, Dawson R, Rehal S, Kibiki GS, Churchyard G, Sanne I, Ntinginya NE, Minja LT, Hunt RD, Charalambous S, Hanekom M, Semvua HH, Mpagama SG, Manyama C, Mtafya B, Reither K, Wallis RS, Venter A, Narunsky K, Mekota A, Henne S, Colbers A, van Balen GP, Gillespie SH, Phillips PPJ, Hoelscher M, PanACEA consortium . 2017. High-dose rifampicin, moxifloxacin, and SQ109 for treating tuberculosis: a multi-arm, multi-stage randomised controlled trial. Lancet Infect Dis 17:39–49. 10.1016/S1473-3099(16)30274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang F, Robbins S, Guo J, Shen W, Schultz PG. 2010. Genetic incorporation of unnatural amino acids into proteins in Mycobacterium tuberculosis. PLoS One 5:e9354. 10.1371/journal.pone.0009354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Touchette MH, Van Vlack ER, Bai L, Kim J, Cognetta AB, Previti ML, Backus KM, Martin DW, Cravatt BF, Seeliger JC. 2017. A screen for protein-protein interactions in live mycobacteria reveals a functional link between the virulence-associated lipid transporter LprG and the mycolyltransferase antigen 85A. ACS Infect Dis 3:336–348. 10.1021/acsinfecdis.6b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ganapathy US, Bai L, Wei L, Eckartt KA, Lett CM, Previti ML, Carrico IS, Seeliger JC. 2018. Compartment-specific labeling of bacterial periplasmic proteins by peroxidase-mediated biotinylation. ACS Infect Dis 4:918–925. 10.1021/acsinfecdis.8b00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rhee KY, Jansen RS, Grundner C. 2022. Activity-based annotation: the emergence of systems biochemistry. Trends Biochem Sci 47:785–794. 10.1016/j.tibs.2022.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Swarts BM, Holsclaw CM, Jewett JC, Alber M, Fox DM, Siegrist MS, Leary JA, Kalscheuer R, Bertozzi CR. 2012. Probing the mycobacterial trehalome with bioorthogonal chemistry. J Am Chem Soc 134:16123–16126. 10.1021/ja3062419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Backus KM, Boshoff HI, Barry CS, Boutureira O, Patel MK, D'Hooge F, Lee SS, Via LE, Tahlan K, Barry CE, Davis BG. 2011. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat Chem Biol 7:228–235. 10.1038/nchembio.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Marando VM, Kim DE, Calabretta PJ, Kraft MB, Bryson BD, Kiessling LL. 2021. Biosynthetic glycan labeling. J Am Chem Soc 143:16337–16342. 10.1021/jacs.1c07430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Walsh CM, Gebert MJ, Delgado-Baquerizo M, Maestre FT, Fierer N. 2019. A global survey of mycobacterial diversity in soil. Appl Environ Microbiol 85:e01180-19. 10.1128/AEM.01180-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rifat D, Chen L, Kreiswirth BN, Nuermberger EL. 2021. Genome-wide essentiality analysis of Mycobacterium abscessus by saturated transposon mutagenesis and deep sequencing. mBio 12:e0104921. 10.1128/mBio.01049-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Akusobi C, Benghomari BS, Zhu J, Wolf ID, Singhvi S, Dulberger CL, Ioerger TR, Rubin EJ. 2022. Transposon mutagenesis in Mycobacterium abscessus identifies an essential penicillin-binding protein involved in septal peptidoglycan synthesis and antibiotic sensitivity. Elife 11:e71947. 10.7554/eLife.71947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hurst-Hess K, Rudra P, Ghosh P. 2017. Mycobacterium abscessus WhiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob Agents Chemother 61:e01347-17. 10.1128/AAC.01347-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rudra P, Hurst-Hess KR, Cotten KL, Partida-Miranda A, Ghosh P. 2020. Mycobacterial HflX is a ribosome splitting factor that mediates antibiotic resistance. Proc Natl Acad Sci USA 117:629–634. 10.1073/pnas.1906748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Berney M, Greening C, Conrad R, Jacobs WR, Cook GM. 2014. An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc Natl Acad Sci USA 111:11479–11484. 10.1073/pnas.1407034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Greening C, Berney M, Hards K, Cook GM, Conrad R. 2014. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci USA 111:4257–4261. 10.1073/pnas.1320586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jirapanjawat T, Ney B, Taylor MC, Warden AC, Afroze S, Russell RJ, Lee BM, Jackson CJ, Oakeshott JG, Pandey G, Greening C. 2016. The redox cofactor F420 protects mycobacteria from diverse antimicrobial compounds and mediates a reductive detoxification system. Appl Environ Microbiol 82:6810–6818. 10.1128/AEM.02500-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bayly K, Cordero PRF, Kropp A, Huang C, Schittenhelm RB, Grinter R, Greening C. 2021. Mycobacteria tolerate carbon monoxide by remodeling their respiratory chain. mSystems 6:e01292-20. 10.1128/mSystems.01292-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hatfull GF. 2022. Mycobacteriophages: from petri dish to patient. PLoS Pathog 18:e1010602. 10.1371/journal.ppat.1010602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hatfull GF. 2018. Mycobacteriophages. Microbiol Spectr 6. 10.1128/microbiolspec.GPP3-0026-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Little JS, Dedrick RM, Freeman KG, Cristinziano M, Smith BE, Benson CA, Jhaveri TA, Baden LR, Solomon DA, Hatfull GF. 2022. Bacteriophage treatment of disseminated cutaneous Mycobacterium chelonae infection. Nat Commun 13:2313. 10.1038/s41467-022-29689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dedrick RM, Smith BE, Cristinziano M, Freeman KG, Jacobs-Sera D, Belessis Y, Whitney Brown A, Cohen KA, Davidson RM, van Duin D, Gainey A, Garcia CB, Robert George CR, Haidar G, Ip W, Iredell J, Khatami A, Little JS, Malmivaara K, McMullan BJ, Michalik DE, Moscatelli A, Nick JA, Tupayachi Ortiz MG, Polenakovik HM, Robinson PD, Skurnik M, Solomon DA, Soothill J, Spencer H, Wark P, Worth A, Schooley RT, Benson CA, Hatfull GF. 2022. Phage therapy of Mycobacterium infections: compassionate-use of phages in twenty patients with drug-resistant mycobacterial disease. Clin Infect Dis 10.1093/cid/ciac453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nick JA, Dedrick RM, Gray AL, Vladar EK, Smith BE, Freeman KG, Malcolm KC, Epperson LE, Hasan NA, Hendrix J, Callahan K, Walton K, Vestal B, Wheeler E, Rysavy NM, Poch K, Caceres S, Lovell VK, Hisert KB, de Moura VC, Chatterjee D, De P, Weakly N, Martiniano SL, Lynch DA, Daley CL, Strong M, Jia F, Hatfull GF, Davidson RM. 2022. Host and pathogen response to bacteriophage engineered against Mycobacterium abscessus lung infection. Cell 185:1860–1874. 10.1016/j.cell.2022.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hanauer DI, Graham MJ, Phages SEA, Betancur L, Bobrownicki A, Cresawn SG, Garlena RA, Jacobs-Sera D, Kaufmann N, Pope WH, Russell DA, Jacobs WR, Sivanathan V, Asai DJ, Hatfull GF, SEA-PHAGES . 2017. An inclusive Research Education Community (iREC): impact of the SEA-PHAGES program on research outcomes and student learning. Proc Natl Acad Sci USA 114:13531–13536. 10.1073/pnas.1718188115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SCR, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatfull GF. 2014. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio 5:e01051-13. 10.1128/mBio.01051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tobin DM, Ramakrishnan L. 2008. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol 10:1027–1039. 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- 183.Shiloh MU, Champion PAD. 2010. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol 13:86–92. 10.1016/j.mib.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Brosch R, Gordon SV, Garnier T, Eiglmeier K, Frigui W, Valenti P, Dos Santos S, Duthoy S, Lacroix C, Garcia-Pelayo C, Inwald JK, Golby P, Garcia JN, Hewinson RG, Behr MA, Quail MA, Churcher C, Barrell BG, Parkhill J, Cole ST. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci USA 104:5596–5601. 10.1073/pnas.0700869104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sampson SL, Dascher CC, Sambandamurthy VK, Russell RG, Jacobs WR, Bloom BR, Hondalus MK. 2004. Protection elicited by a double leucine and pantothenate auxotroph of Mycobacterium tuberculosis in guinea pigs. Infect Immun 72:3031–3037. 10.1128/IAI.72.5.3031-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sambandamurthy VK, Derrick SC, Hsu T, Chen B, Larsen MH, Jalapathy KV, Chen M, Kim J, Porcelli SA, Chan J, Morris SL, Jacobs WR. 2006. Mycobacterium tuberculosis ΔRD1 ΔpanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 24:6309–6320. 10.1016/j.vaccine.2006.05.097. [DOI] [PubMed] [Google Scholar]
- 187.Sambandamurthy VK, Derrick SC, Jalapathy KV, Chen B, Russell RG, Morris SL, Jacobs WR. 2005. Long-term protection against tuberculosis following vaccination with a severely attenuated double lysine and pantothenate auxotroph of Mycobacterium tuberculosis. Infect Immun 73:1196–1203. 10.1128/IAI.73.2.1196-1203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Myllymäki H, Niskanen M, Oksanen KE, Rämet M. 2015. Animal models in tuberculosis research - where is the beef? Expert Opin Drug Discov 10:871–883. 10.1517/17460441.2015.1049529. [DOI] [PubMed] [Google Scholar]
- 189.Prouty MG, Correa NE, Barker LP, Jagadeeswaran P, Klose KE. 2003. Zebrafish-Mycobacterium marinum model for mycobacterial pathogenesis. FEMS Microbiol Lett 225:177–182. 10.1016/S0378-1097(03)00446-4. [DOI] [PubMed] [Google Scholar]
- 190.Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. 2002. Real-time visualization of Mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17:693–702. 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- 191.van Leeuwen LM, van der Sar AM, Bitter W. 2014. Animal models of tuberculosis: zebrafish. Cold Spring Harb Perspect Med 5:a018580. 10.1101/cshperspect.a018580. [DOI] [PMC free article] [PubMed] [Google Scholar]