ABSTRACT
Whether long non-coding RNA Mir-99a-Let-7c Cluster Host Gene (LncRNA MIR99AHG) is involved in osteoporosis (OP) remains vague, so we hereby center on its implication. Old C57BL/6J mice were injected with the silencing lentivirus of MIR99AHG and subjected to microCT analysis and immunohistochemistry on osteogenic cells. The osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) with or without transfection was determined by alkaline phosphatase (ALP) and Alizarin Red S staining. Total N(6)-methyladenosine (m6A) on the bone marrow mesenchymal stem cells (BMSCs) was quantified. The potential methylation site and the complementary binding sites with candidate microRNA (miR) were predicted via bioinformatic analyses, with the latter being confirmed via dual-luciferase reporter, RNA immunoprecipitation and RNA pull-down assays. Quantitative real-time PCR and Western blot were used for quantification assays. MIR99AHG was decreased during the osteogenic differentiation of BMSCs, where increased Osterix (OSX), Collagen, Type I, Alpha 1 (Col1A1), Osteocalcin (OCN) and RUNX Family Transcription Factor 2 (RUNX2) as well as more color-stained areas were found. Also, silencing MIR99AHG relieved the OP in mice and reduced the loss of osteogenic cells. M6A methylation in undifferentiated BMSCs was low and MIR99AHG overexpression abolished the effects of overexpressed METTL3 on promoting osteogenic differentiation. MiR-4660, which was downregulated in BMSCs without differentiation but increased during osteogenic differentiation, could bind with MIR99AHG. Furthermore, miR-4660 promoted osteogenic differentiation and reversed the effects of overexpressed MIR99AHG. The present study demonstrated that METTL3-mediated LncRNA MIR99AHG methylation enhanced the osteogenic differentiation of BMSCs via targeting miR-4660.
KEYWORDS: Osteoporosis, Mir-99a-Let-7c cluster host gene, osteogenic differentiation, bone marrow mesenchymal stem cells, microRNA-4660
Introduction
Osteoporosis (OP) is a systemic and highly prevalent bone disease, characterized by low bone marrow density and increased risk of fracture. It is defined as osteoporotic fracture, which, currently, affects massive populations worldwide, especially the elderly and the postmenstrual women. Besides, it sometimes even causes death owing to lack of proper therapy, which, thus, remains as one of the health issues [1,2]. At present, for the patients who have already suffered from a fragility fracture, pharmacotherapy for OP is recommended as the primary option due to the evidence of pharmacological efficacy targeting bone metabolism [3]. Although the medication for current OP has shown a significant reduction in fractures, some serious adverse reactions may restrain their safety and efficacy for the long-term use [4]. Therefore, it is of great significance and urgency to better understand OP pathogenesis at a molecular level, so as to work out a viable therapy for OP in clinical practice.
Over the last decades, evidence has been emerging to demonstrate the efficacy of bone marrow mesenchymal stem cells (BMSCs) in bone tissue engineering process and some regenerative medicine-based applications, as well as the promotive effects of osteogenic differentiation of BMSCs in the therapy for OP [5,6]. In addition, increasing evidences have not only proved the existence and emergence of long non-coding RNAs (lncRNAs) as modulators on the functions of BMSCs, especially for osteogenic differentiation, but also demonstrated the implication of lncRNAs in the development and progression of OP, where dysregulated lncRNAs in BMSCs may disrupt osteo-adipogenesis differentiation and impair bone homeostasis [7]. Mir-99a-Let-7c Cluster Host Gene (MIR99AHG) is another lncRNA being involved in different cancers [8–10], gastric cancer for instance [11]. However, its implication is currently under-discussed in both the osteogenic differentiation of human BMSCs and OP development and progression, so additional researches are required to fill the gap.
Apart from the discoveries above, the regulatory role of RNA N(6)-methyladenosine (m6A) modification within the osteobiology and OP development has been discovered and discussed as well [12]. M6A has been defined as methylation modification on RNA catalyzed by the methyltransferase at the 6th position of nitrogen atom of adenine (A), with accumulating evidence indicating the participation of M6A RNA methylation in cancer [13–15]. Meanwhile, as one of the most common RNA modifications in eukaryotes, m6A RNA methylation exerts pivotal roles in several processes, including the regulation on cell differentiation, angiogenesis, immune and inflammatory responses and carcinogenesis, with an emphasis on methyltransferase Like 3 (METTL3) as a mediator for mammalian m6A methylation and an executer of m6A-dependent modification on the ncRNAs involved in carcinogenesis [16]. The interaction between METTL3 and lncRNAs has been additionally revealed as well [17,18]. However, from our perspective, whether MIR99AHG can also interact with METTL3 and its role in both BMSCs’ osteogenic differentiation and OP development are inadequately discussed. On these grounds, our current study would mainly center on the two aspects above, along with a proposal of a possible therapeutic method for diagnosis and prognosis on OP in vivo and in vitro.
Materials and methods
Ethics statement
The current study has also obtained the ethic approval of Nanfang Hospital (Serial Number: NF2021110902) and all animal experiments were performed in Nanfang Hospital strictly following the guidance of the China Council on Animal Care and Use. Every effort has been devoted to minimize the pain and discomfort to the animal.
Animal and lentiviruses administration
Female C57BL/6J mice (15-month-old, n = 30) were ordered from Shanghai Laboratory Animal Center (Shanghai, China) and randomly allocated to the following groups: Control, shNC, and shMIR99AHG (n = 10 for each group). All animals were kept in specific pathogen-free (SPF) animal cages at the specified conditions: 12-hour light/dark cycle, temperature (23 ± 2℃), and humidity (55%).
To evaluate the effects of MIR99AHG on OP in vivo, the lentivirus carriers of both short hairpin RNA against MIR99AHG (shMIR99AHG) and its negative control (shNC) ordered from GenePharma (Shanghai, China) were injected into the tail vein of mice twice a week (100 μl for each mouse, 3 months in total) at 1 × 108 PFU/ml [19]. Specifically, the mice were first fixed on a fixator and their tail was wiped using ethanol (A507050, Sangon Biotech, Shanghai, China) for the exposure of the tail vein. A 1 ml syringe was connected to the needle and inserted to the tail vein, and the needle was pumped back prior to the injection so as to confirm the insertion. The investigator was blinded when assessing the outcome.
Micro-CT analyses
Following the sacrifice of mice via inhalation of 5% isoflurane (I40690, Acmec Biochemical, Shanghai, China) and cervical dislocation, the isolated bone tissues, including femur, tibia, and vertebra, were fixed in 10% paraformaldehyde (A500684, Sangon Biotech, Shanghai, China) and stored in 70% ethanol at 4℃ for subsequent analyses. A µCT 50 microCT imaging system (SCANCO Medical, Brüttisellen, Switzerland) was employed for the evaluation, where the samples were placed in the cylindrical holder (diameter: 14 mm) with the long axis of the bone perpendicular to the X-ray beam and the spatial resolution of 8 µm (55 kV, 114 mA and an integration time: 500 ms). Then the built-in software was used for volumetric analyses. The anatomical structure of bone tissues was employed to select the region of interest (ROI). Approximately, 300–400 slices of the vertebrae (L4), that is, 160 µm below the cranial and above the region of caudal growth plate were additionally chosen for the ROIs. The trabecular bone volume fraction (Tb.BV/TV), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), trabecular number (Tb.N), and cortical thickness (Ct.Th) within the ROIs were determined [20].
Immunohistochemistry
Bone samples of mice were fixed in 4% paraformaldehyde (E672002, Sangon Biotech), decalcified, dehydrated and embedded in paraffin (A601801, Sangon Biotech) in sequence. The paraffin-embedded bone section of mice (5-μm-thick) was first treated for antigen retrieval following the digestion in 0.05% trypsin (A003702, Sangon Biotech) at 37℃ for 15 minutes and then incubated with the primary antibody against osteocalcin (ab93876, Abcam, Cambridge, UK) at 4℃ overnight. Then, the counterstain was performed with hematoxylin (A600701, Sangon Biotech) after the detection of immunoactivity using an HRP-streptavidin detection system (N100, ThermoFisher Scientific, Waltham, MA, USA). The results were finally seen in a phase-contrast optical microscope (×100, Eclipse E200, Nikon, Tokyo, Japan) [21].
Bioinformatic analyses
Firstly, based on GSE153829 [22] from NCBI Gene Expression Omnibus (GOE) database (http://www.ncbi.nlm.nih.gov/geo/), the study analyzed the aberrantly expressed lncRNA participated in the formation of ossification in the posterior longitudinal ligament.
Secondly, data concerning the potential m6A methylation sites of MIR99AHG were gathered from SRAMP (http://www.cuilab.cn/sramp/#predSRAMP), and the possible candidate microRNA (miRNA, miR) that could competitively bind with MIR99AHG were downloaded from both LncBase (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php) and LncRNASNP2 (http://bioinfo.life.hust.edu.cn/lncRNASNP/) and summarized. The common miRNAs were sorted via a Venn diagram drawn by Venny online software (v. 2.1.0, http://bioinfogp.cnb.csic.es/tools/venny/), as shown in Figure 6a.
Figure 6.
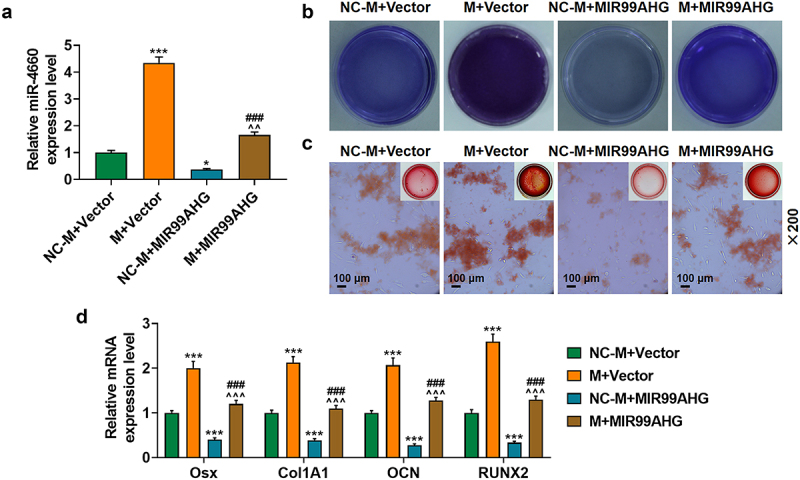
Upregulated miR-4660 reversed the effects of overexpressed MIR99AHG on miR-4660 expression and the osteogenic differentiation of BMSCs.(a) Relative miR-4660 expression following miR-4660 upregulation and MIR99AHG overexpression was measured via qRT-PCR. U6 was the internal reference. (b-c) Effects of overexpressed MIR99AHG and miR-4660 overexpression on osteogenic differentiation of BMSCs were evaluated by both ALP staining (b) and Alizarin Red S staining (c). Magnification: × 200. Scale bar = 100 μm. (d) Relative mRNA expression of Osx, Col1A1, OCN and RUNX2 after overexpressed MIR99AHG and miR-4660 upregulation was quantified with qRT-PCR. β-actin was the internal reference. All experiments have been performed independently in triplicate and data were expressed as mean ± standard deviation (SD). *p<0.05, ***p<0.001, vs. NC-M+Vector; ###p<0.001, vs. M+Vector; ^^^p<0.001, vs. NC-M+MIR99AHG. M: miR-4660 mimic; NC-M: negative control for mimic.
Osteogenic differentiation induction
All procedures in regard to the induction on osteogenic differentiation of BMSCs were confirmed in accordance with a prior study [23]. BMSCs (HUXMA -01,001, Cyagen Biosciences, Nanjing, China) were firstly maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, E600010, Sangon Biotech, Shanghai, China) with 10% fetal bovine serum (FBS, E600001, Sangon Biotech, China) and 100 Unit/mL-0.1 mg/mL Penicillin-Streptomycin (B540732, Sangon Biotech, China) in a Model NU-5700 CO2 Incubator (NU-5700, NuAire, Plymouth, MN, USA) at the designated incubation condition (37℃, 5% CO2).
Then, for osteogenic differentiation, BMSCs were cultured in α-Minimum Essential Medium (α-MEM, SH30205.04, Cytiva lifesciences, Marlborough, MA, USA) which has blended with 10% FBS and 100 Unit/L-100 mg/L Penicillin-Streptomycin at 37℃, 5% CO2. The medium was replaced with a fresh one every 3–4 days. For osteogenic differentiation induction, the medium above was additionally supplemented with 10 mmol/L β-glycerophosphate sodium (J62121, Alfa Aesar, Haverhill, MA, USA), 100 nmol/L Dexamethasone (A17590, Alfa Aesar, USA), and 200 μmol/L ascorbic acid (HY-B0166, MedChemExpress, Princeton, NJ, USA) when cells grew 80–90% confluent, and the induction was conducted for 14 days, with the osteogenic differentiation induction medium being changed every 3 days.
Cell transfection
Before transfection, short hairpin RNA against MIR99AHG (sh-MIR99AHG), small interfering RNA targeting METTL3 (siMETTL3) and their negative control (shNC and siNC) were ordered from Gene Pharma (Shanghai, China) to silence MIR99AHG and METTL3 expression and serve as the controls. For MIR99AHG and METTL3 overexpression, pcDNA 3.1 plasmid (V790–20, Invitrogen, Carlsbad, CA, USA) was employed and the empty plasmid was used for the negative control (NC) (Vector represented the NC of MIR99AHG). Additionally, based on the results from bioinformatic analysis, miR-4660 was selected as the candidate miRNA, and its mimic (M, miR10019728-1-5) and its negative control (NC-M, miR1N0000001-1-5), its inhibitor (miR2171129030231-1-5) and its inhibitor negative control (NC-inhibitor, miR2N0000001-1-5) were purchased from RiboBio (Guangzhou, China). All sequences used are available in Table 1.
Table 1.
Sequences for transfection.
| Gene | Sequence (5’->-3’) |
|---|---|
| sh-MIR99AHG sense obligo | CCGGCTGGGATAATTATCCAAATAACTCGAGTTATTTGGATAATTATCCCAGTTTTTG |
| sh-MIR99AHG antisense obligo | CCGGCTGGGATAATTATCCAAATAACTCGAGTTATTTGGATAATTATCCCAGTTTTTG |
| shNC sense obligo | CCGGATTTGGATAATATAATTATCTAAATAGGGCATCCTTCGAGTTAGACTCCTTTTG |
| shNC antisense obligo | CCGGCTTTAAATTGAAATAACTCGAGTTATTCGGAAGTCCTGCCAGTTTTATTAATTG |
| siMETTL3 | UCUAACUCAGGAUCUGUAGCU |
| siNC | UUUUGAUCCUGAGCAACUCAG |
| miR-4660 M | UGCAGCUCUGGUGGAAAAUGGAG |
| miR-4660 NC-M | UGCGAGAGAAAAUGGCUCUGGUG |
2.5 × 105 cells/well BMSCs were seeded in 6-well plates at 37℃ and transfection was performed using Lipofectamine 3000 reagent (L3000–001, Invitrogen, Carlsbad, CA, USA) as instructed by the manufacturer when cell grew 70–90% confluent. 48 hours later, all cells were collected for subsequent studies.
Alkaline phosphatase (ALP) staining
ALP staining was conducted as illustrated previously [24]. Following the transfection and osteogenic differentiation induction, on day 14, all media were removed and BMSCs were fixed within 4% paraformaldehyde (PFA, A11313, Alfa Aesar, USA). Then, BMSCs were subjected to the staining process with the ALP staining kit (K2035, BioVision, Milpitas, CA, USA) at 37℃ for 30 minutes. All stained BMSCs were finally photographed with an E-M5 Mark IV digital camera (Olympus, Tokyo, Japan) after being washed with both phosphate buffered saline (PBS, J75889, Alfa Aesar, USA) and the washing buffer provided within the kit.
Alizarin red S staining
All processes within Alizarin Red S staining performed in the light of the previous study [25]. After transfection and osteogenic differentiation induction, BMSCs were maintained within a 24-well plate. Then, on day 14, BMSCs were firstly washed with PBS, fixed with 4% PFA at room temperature (RT) for 15 minutes, and finally stained with 1 mL of 2% Alizarin Red S staining solution (#0223, ScienCell, Carlsbad, CA, USA) at 37℃ for 30 minutes. All stained BMSCs were finally observed under an inverted light microscope (GX53, Olympus, Japan) under × 200 magnification.
Total m6A content quantitation
All processes with regards to the quantification of total m6A content were conducted as illustrated previously [26]. In detail, the differentiated or undifferentiated BMSCs were cultured within DMEM in 96-well culture dishes overnight. Total RNA was extracted from these cells using Trizol (T9424, Sigma-Aldrich, USA), and total m6A content was quantified using the m6A RNA Methylation Quantification Kit (P-9005-96, EpigenTek, Farmingdale, NY, USA) according to the manuals provided by the manufacturer. To begin with, 80 μL binding solutions provided by the kits were added within each well. Then, negative control, diluted m6A (which served as the positive control) and sample RNA (2 μL for each) were transferred into the wells, followed by the mixture of the solution to make sure that the solution coats the bottom of the well. The strip plate was covered with plate seal and the mixture was allowed to incubate at 37℃ for 90 minutes. The binding solution was removed and 150 μL of diluted washing buffer was used to wash each well, the processes of which were repeated for 3 times in total. Then, 50 μL of diluted capture antibody offered by the kits was added to each well respectively, and the wells were covered and incubated at RT for an hour. Subsequently, the diluted capture antibody solution was removed via a sterile pipette and the well was washed with 150 μL diluted washing buffer for 3 times. 50 μL of pre-diluted detection antibody in the kits was added into the wells which were incubated at RT for 30 minutes, followed by the washing process of 150 μL diluted washing buffer for 4 times. A total volume of 50 μL diluted enhancer solution affiliated with the kits was added into each well, maintaining at RT for another 30 minutes. The solution was removed then and the wells were washed with 150 μL diluted washing buffer for 5 times in total. The developer solution in the kits was added into the wells, which were incubated at RT for 10 minutes without light. 100 μL of stop solution was finally added to terminate the enzyme reaction when the color in the wells that were served as positive control turned blue. Within 15 minutes, the detected signal was enhanced and quantified via the reading of optical density (OD) value at 450 nm utilizing an iMark microplate reader (Bio-Rad, Hercules, CA, USA) and the amount of total m6A was proportional to the OD value.
Dual-luciferase reporter assay
In accordance with the results from bioinformatic analysis in our study, miR-4660 had the highest prediction score in LncBase. Therefore, we speculated that miR-4660 might be the candidate miRNA for MIR99AHG in our study.
Before the dual-luciferase reporter assay, putative sequences of MIR99AHG containing miR-4660 binding sites were cloned into luciferase reporter vector pMiR-GLO (PAE1330, VWR International, Radnor, PA, USA) to construct the wild-type MIR99AHG reporter plasmid (MIR99AHG WT, sequence: 5’-AUCAGAGAGCAUUUUACUACUGAGCUGCA-3’). As for the mutated MIR99AHG reporter plasmid (MIR99AHG MUT, sequence: 5’-AUCAGAGAGCGUAUUACUACUGCGCAGCA-3’), a site-directed mutagenesis kit (E0554, New England BioLabs, Ipswich, MA, USA) was employed.
3 × 105 cells/well BMSCs were cultured in 48-well plates and co-transfected with miR-4660 M or NC-M and reporter plasmids by Lipofectamine 3000 reagent at 37℃. After 48 hours, the luciferase activities of firefly and Renilla were detected within dual-luciferase reporter system kit (E1910, Promega, USA) by a luminometer (E5311, Promega, USA), and the luciferase activity of Renilla was used as normalization on that of firefly.
RNA immunoprecipitation and RNA pull down assays
All processes concerning the RNA immunoprecipitation and RNA pull down assays were confirmed as described previously [27]. For RNA immunoprecipitation assay, the binding of MIR99AHG and miR-4660 to the Argonaute 2 (Ago2) protein was detected and determined via an RNA-Binding Protein immunoprecipitation kit (17–700, Millipore Corporation, Billerica, MA, USA) based on the manufacturer’s protocols. In the beginning, BMSCs were washed with pre-chilled PBS, lysed using a lysis buffer containing 25 mmol/L Tris-hydrochloride (Tris-HCl, pH7.5, B548124, Sangon Biotech, China), 150 mmol/L Potassium Chloride (KCl, A501159, Sangon Biotech, China), 2 mmol/L ethylene diamine tetraacetic acid (EDTA, B540625, Sangon Biotech, China), 0.5% nonidet P-40 (NP-40, MP1RIST1315, Fisher Scientific, Hampton, NH, USA), 1 mmol/L sodium fluoride (NaF, A500850, Sangon Biotech, China), 1 mmol/L dithiothreitol (DTT, A100281, Sangon Biotech, China), and 100 U/mL ribonuclease inhibitor (R1158, Sigma-Aldrich, USA) within an ice bath for 5 minutes, followed by being centrifugated at 14,000 rpm at 4℃ for 10 minutes. M-280 streptavidin magnetic beads (11205D, Invitrogen, USA) in a total volume of 50 μL were resuspended within 100 μL RIP washing buffer provided by the kit and swirled. Then Eppendorf tubes were placed within a magnetic stand and rotated to allow the magnetic beads align straightly. After the supernatant was removed, the magnetic beads were resuspended in 100 μL RIP washing buffer and added with 5 μg antibodies including rabbit anti-human Ago2 antibody (MA5–23515, Invitrogen, USA) and rabbit anti-human IgG antibody (SA5–10197, Invitrogen, USA), with the ratio for dilutions setting at 1:500. Then, the complex of magnetic beads-antibody was washed and resuspended in 900 μL RIP washing buffer prior to being incubated with cell lysate, one portion of which was taken and designated as the Input group, whilst another portion was co-precipitated by being incubated with the complex at 4℃ overnight. Both the co-precipitated complex and Input were separately detached via proteinase K (A004220, Sangon Biotech, China) for the extraction of RNA used in sequential expression analysis on both MIR99AHG and miR-4660 expressions.
With regard to RNA pull down assay, as per the guidelines of the Magnetic RNA-Protein Pull-Down Kit (20164, ThermoFisher Scientific, Waltham, MA, USA), Eppendorf tubes were firstly added with 1 μg biotin-labeled miR-4660 or its NC (designed and purchased from Pierce Biotech (Rockford, IL, USA) and named as Bio-miR-4660 and Bio-miR-NC respectively) and 500 μL structure buffer, followed by being incubated within a water bath at 95℃ for 2 minutes and ice bath for 3 minutes. The magnetic beads were then resuspended in 50 μL bead suspension and incubated at 4℃ overnight, followed by being centrifuged at 3,000 rpm for 3 minutes. The precipitate was rinsed three times using 500 μL washing buffer after discarding the supernatant and incubated with 10 μL cell lysate at RT for an hour. The incubated magnetic beads-RNA-protein mix was centrifuged again, after which the supernatant was available and washed for 3 times using a washing buffer. Besides, an additional 10 μL cell lysate was used as Control, and MIR99AHG expression was quantified.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA in isolated BMSCs with or without differentiation was extracted with Trizol reagent and preserved in −80℃, while miRNA was extracted via a microRNA isolation kit (KS341025, BioChain, Newark, CA, USA). The concentration of all harvested RNA was measured within a NanoDrop 2000 spectrophotometer (ND-2000, ThermoFisher Scientific, USA), and all procedures with qRT-PCR were performed by a One-Step RT-PCR kit (B639277, Sangon Biotech, China) in CFX384 Touch real-time PCR system (Bio-Rad, USA) under the conditions: the reverse transcription (50℃ for 3 minutes) and the pre-denaturation (95℃ for 3 minutes), followed by 40 cycles of the denaturation (95℃, 10 seconds) and the final extension (60℃ for 30 seconds). Relative expressions were calculated via the 2−ΔΔCT method, while β-actin and U6 were used as internal references [28]. Primer sequences were listed in Table 2.
Table 2.
Primers for qRT-PCR.
| Gene | Primers (5’->-3’) |
|
|---|---|---|
| Forward | Reverse | |
| MIR99AHG | GTACCAAGAGCTGGTATTTC | ATAGTAGTGGCACCCTAAGA |
| Osx | GTCTACACCTCTCTGGACAT | ACCATGGAGTAGGAGTGTT |
| Col1A1 | AGCTGTCTTATGGCTATGAT | CCAGTAGCACCATCATTT |
| OCN | ATGAGAGCCCTCACACTC | CAGCCATTGATACAGGTAG |
| RUNX2 | ATCAAACAGCCTCTTCAG | GTTATGGTCAAGGTGAAACT |
| METTL3 | AATTCTGTGACTATGGAACC | GCTACGATCACATCACAAT |
| METTL14 | CATGTACTTACAAGCCGATA | CATTAGCATGAATGAAGTCC |
| FTO | ATAATGAGGTCGAGTTTGAG | GGAGACATAAGTCCTAGCTC |
| ALKBH | CTATGGACTCAAAGGCTATC | GTGTGTAATGATCTGCTGAG |
| miR-4660 | GCAGCTCTGGTGGAAAATGG | TGTCGTGGAGTCGGCAATTG |
| GAPDH | TTTTTGGTTTTAGGGTTAGTTAGTA | AAAACCTCCTATAATATCCCTCCTC |
| U6 | TACAGAGAAGATTAGCATGGCCC | ACGAATTTGCGTGTCATCCT |
Western blot
Relative protein expression of METTL3 was measured via Western blot as described in a previous study [29]. Total protein in transfected BMSCs was sequentially lysed and extracted with RIPA lysis buffer (C500005, Sangon Biotech, China) after the collection of used cells. The concentration was determined using bicinchoninic acid (BCA) protein kit (C503021, Sangon Biotech, China). 20 μL of total protein lysates was firstly electrophoresed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, A100227, Sangon Biotech, USA), and then transferred into polyvinylidene fluoride (PVDF) membrane (F019533, Sangon Biotech, China). The membrane was blocked using 5% fat-free milk for 2 hours at RT and incubated in those specific primary antibodies for both METTL3 (ab195352, 64kDa) and internal control β-actin (ab8226, 42kDa) bought from Abcam (Cambridge, UK) at 4℃ overnight, following by the incubation process with the horseradish peroxidase (HRP)-conjugated secondary antibodies: goat anti-rabbit IgG H&L (ab205718, Abcam, Cambridge, UK) and goat anti-mouse IgG H&L (ab205719, Abcam, UK) at RT for 1 hour. The dilution ratio of primary antibodies was set at 1:1000 and that of secondary antibodies was 1:2000. The membrane was washed three times with tris-buffer saline tween (TBST, C520009, Sangon Biotech, China), and visualized with an Enhanced chemiluminescence (ECL) western blotting substrate (D601039, Sangon Biotech, China). Besides, the data were analyzed within Odyssey® CLX Imaging System (LI-COR Biosciences, Lincoln, NE, USA) and gray values were calculated by ImageJ (v. 5.0, National Institute of Health, Bethesda, MD, USA).
Statistical analysis
Every experiment in our study was repeated at least three times in an independent manner. All data were expressed as mean ± standard deviation (SD), and calculated and analyzed by SPSS (v. 20, IBM Corporation, Endicott, NY, USA). Kolmogorov–Smirnov test was used to verify normality. Statistical significance was determined with one-way ANOVA followed by Bonferroni post hoc test and paired t test, which was defined as P-value that was lower than 0.05.
Results
MIR99AHG expression quantification and osteogenic differentiation evaluation
In the beginning, we used the lentivirus to knockdown MIR99AHG expression in aged mice, and a reduction in the expression of MIR99AHG was first evidenced (p < 0.001; Figure 1a). It was observable in the results of micro-CT analyses that the knockdown of MIR99AHG could evidently relieve OP in aged mice, with an increase on the osteogenic cells, based on the results of immunohistochemistry analysis (Figure 1b-c).
Figure 1.
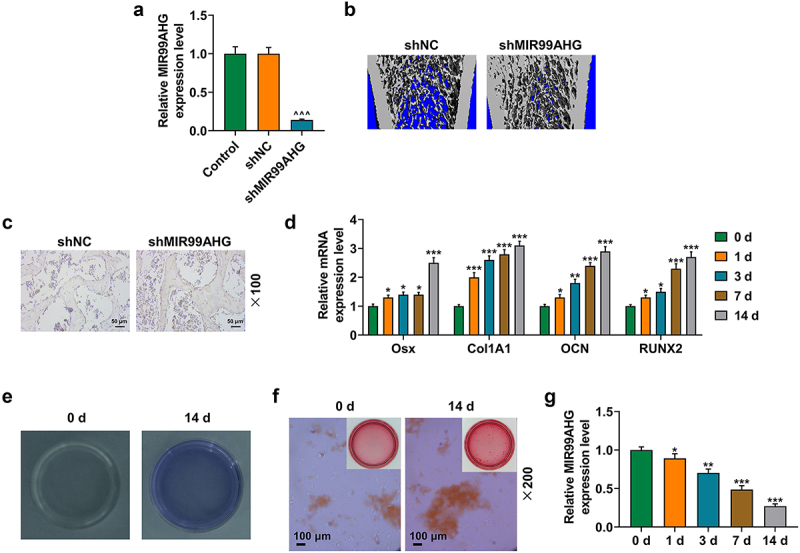
Osteogenic differentiation evaluation and MIR99AHG expression quantitation.(a) Relative MIR99AHG expression in senile mice injected with the lentiviruses of shNC or shMIR99AHG and those without injection was quantified via qRT-PCR. β-actin was used as internal reference. (b) Micro-CT analysis on senile mice injected with the lentiviruses of shNC or shMIR99AHG was conducted. (c) Immunohistochemistry on the bone tissue of mice injected with the lentiviruses of shNC or shMIR99AHG was performed. Magnification: × 100. Scale bar = 50 μm. (d) Relative mRNA expression of Osx, Col1A1, OCN, and RUNX2 in BMSCs on day 1, 3, 7 and 14 post osteogenic differentiation was also quantified with qRT-PCR. β-actin was the internal reference. (e-f) Osteogenic differentiation of BMSCs was evaluated by both ALP and Alizarin Red S staining. Magnification: × 200. Scale bar = 100 μm. (g) Relative MIR99AHG expression in BMSCs on day 1, 3, 7 and 14 post osteogenic differentiation induction was further measured with qRT-PCR. β-actin was the internal reference.
All experiments have been performed independently in triplicate and data were expressed as mean ± standard deviation (SD).
^^^p<0.001, vs. shNC; *p<0.05, **p<0.01, ***p<0.001, vs. 0 d.
MIR99AHG: Mir-99a-Let-7c Cluster Host Gene; BMSCs: bone marrow mesenchymal stem cells; OP: osteoporosis; qRT-PCR: quantitative real-time PCR; shRNA: short hairpin RNA; NC: negative control; ALP: Alkaline phosphatase; Osx: Osterix; Col1A1: Collagen, Type I, Alpha 1; OCN: Osteocalcin; RUNX2: RUNX Family Transcription Factor 2.
By comparing lncRNAs in BMSCs between healthy and posterior longitudinal ligament ossification patients using GEO set (GSE153829), the results showed that MIR99AHG was significantly poorly expressed in BMSCs in patients with ossification of the posterior longitudinal ligament. It suggested that MIR99AHG downregulation may be associated with osteogenic differentiation of BMSCs. As osteogenic differentiation was implicated in the progression of OP, where Osterix (OSX), Collagen, Type I, Alpha 1 (Col1A1), Osteocalcin (OCN) and RUNX Family Transcription Factor 2 (RUNX2) were those osteogenic differentiation-related factors [30,31], we subsequently measured their expressions on day 1, 3, 7 and 14 post osteogenic differentiation induction, and correspondingly, their expressions were all increased following the induction (p < 0.05; Figure 1d).
Since both ALP staining and Alizarin Red S staining could be employed to evaluate the osteogenic differentiation [32], then these two staining were conducted in our study as well, with more colored areas discovered on day 14 post the osteogenic differentiation induction (Figure 1e,f, magnification: × 200). In addition, to confirm the role of MIR99AHG during osteogenic differentiation induction, we measured its expression on day 1, 3, 7 and 14 as well, from which a reduced MIR99AHG expression were found (p < 0.05; Figure 1g).
Overexpressed MIR99AHG suppressed the osteogenic differentiation of BMSCs, while MIR99AHG silence exerted contrary results
To determine the effects of MIR99AHG exerted on BMSCs, we used shRNA against MIR99AHG and constructed MIR99AHG overexpression plasmid to silence or overexpress MIR99AHG, and the successful transfection was evidenced by the increased MIR99AHG expression following the transfection of MIR99AHG overexpression plasmid, whereas shRNA against MIR99AHG repressed the expression of MIR99AHG (p < 0.001; Figure 2a). Then, we used ALP and Alizarin Red S staining again to confirm the effects of MIR99AHG on the osteogenic differentiation of BMSCs, where we found that overexpressed MIR99AHG was associated with an evident reduction on colored area, whereas MIR99AHG silencing did conversely (Figures 2b–2c, magnification: × 200). Furthermore, we discovered that all four osteogenic differentiation-related factors were decreased following MIR99AHG overexpression; however, MIR99AHG silencing exerted promotive effects on their expressions (p < 0.001; Figure 2d).
Figure 2.
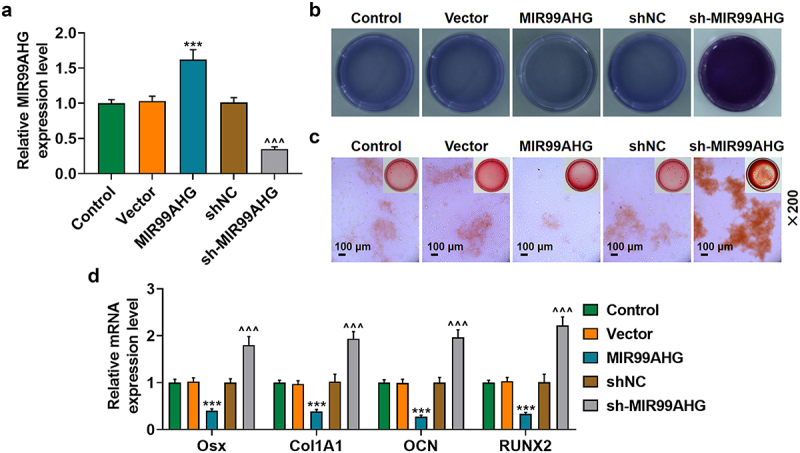
Overexpressed MIR99AHG promoted MIR99AHG expression yet suppressed osteogenic differentiation, while MIR99AHG silence exerted contrary results. (a) Transfection efficiency of MIR99AHG overexpression plasmid and shRNA against MIR99AHG into BMSCs was measured. β-actin was used as internal reference. (b-c) Effects of overexpressed or silenced MIR99AHG on osteogenic differentiation of BMSCs were evaluated by both ALP staining (b) and Alizarin Red S staining (c). Magnification: × 200. Scale bar = 100 μm. (d) Relative mRNA expressions of Osx, Col1A1, OCN and RUNX2 after overexpressed or silenced MIR99AHG were also quantified with qRT-PCR. β-actin was the internal reference. All experiments have been performed independently in triplicate and data were expressed as mean ± standard deviation (SD). ***p<0.001, vs. Vector; ^^^p<0.001, vs. shNC. shRNA: short hairpin RNA; NC: negative control.
M6A methylation and methylation-associated factor METTL3 were upregulated in BMSCs during osteogenic differentiation
To confirm the degree of methylation in BMSCs, we measured m6A content in BMSCs with or without the induction for osteogenic differentiation and found a increased m6A content in the BMSCs with differentiation (p < 0.001; Figure 3a). Additionally, as METTL3, METTL14, FTO Alpha-Ketoglutarate Dependent Dioxygenase (FTO), and AlkB Homolog 1 (ALKBH1) were all associated with methylation or demethylation [33–35], we then measured their expressions in BMSCs with or without the induction for osteogenic differentiation, and confirmed a significant METTL3 and METTL14 downregulation in BMSCs without osteogenic differentiation but no significant change on FTO and ALKBH1 expressions (p < 0.001; Figure 3b). As METTL3 was suggested to be implicated in the osteogenic differentiation and to affect the m6A modification in ncRNAs more dominantly [36,37], METTL3 was chosen for subsequent studies. To determine its role in osteogenic differentiation, we measured its expression on day 1, 3, 7 and 14 post osteogenic differentiation with an increased expression being discovered (p < 0.05; undefined Figures 3c–3e).
Figure 3.
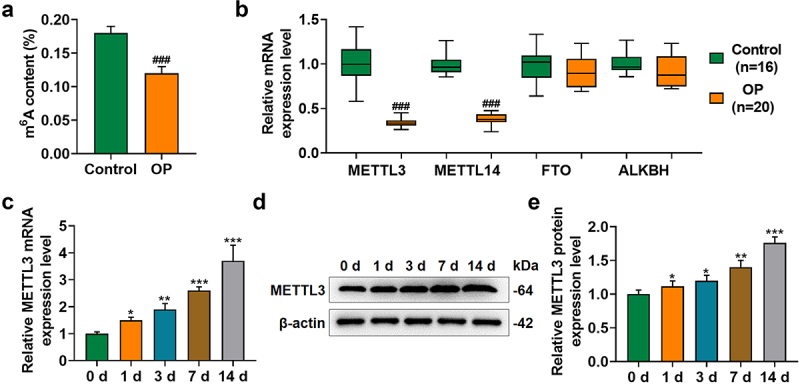
M [6]a methylation and methylation-associated factors upregulated in BMSCs during osteogenic differentiation.(a) M6A content in BMSCs with or without the induction for osteogenic differentiation was determined. (b) Relative mRNA expression of METTL3, METTL14, FTO and ALKBH in BMSCs with or without the induction for osteogenic differentiation was measured via qRT-PCR. β-actin was the internal reference. (c-e) Relative METTL3 mRNA (c) and protein (d-e) expression in BMSCs on day 1, 3, 7 and 14 post osteogenic differentiation was further measured with qRT-PCR and Western blot. β-actin was the internal reference. All experiments have been performed independently in triplicate and data were expressed as mean ± standard deviation (SD). *p<0.05, **p<0.01, ***p<0.001, vs. 0 d. M6a: N6-Methyladenosine; METTL3: methyltransferase like 3, METTL14: methyltransferase like 14, FTO: FTO Alpha-Ketoglutarate Dependent Dioxygenase, ALKBH1: AlkB Homolog 1.
Overexpressed MIR99AHG abolished the effects of METTL3 on BMSCs’ osteogenic differentiation
With the help of the SRAMP database, we successfully confirmed the m6A methylation sites within MIR99AHG sequences, where one site was of extremely high confidence and two sites were of high confidence (Supplementary Fig. S1). And these sequence contents were shown in Supplementary Fig. 2. Based on the results above, we subsequently set out to determine the interaction between METTL3 and MIR99AHG in BMSCs, which was firstly transfected with METTL3 overexpression and siMETTL3. The expression analysis then showed an increased METTL3 expression by METTL3 overexpression plasmid and a decrease via METTL3 silencing (p < 0.001; undefined Figures 4a–4c), indicating successful transfection.
Figure 4.
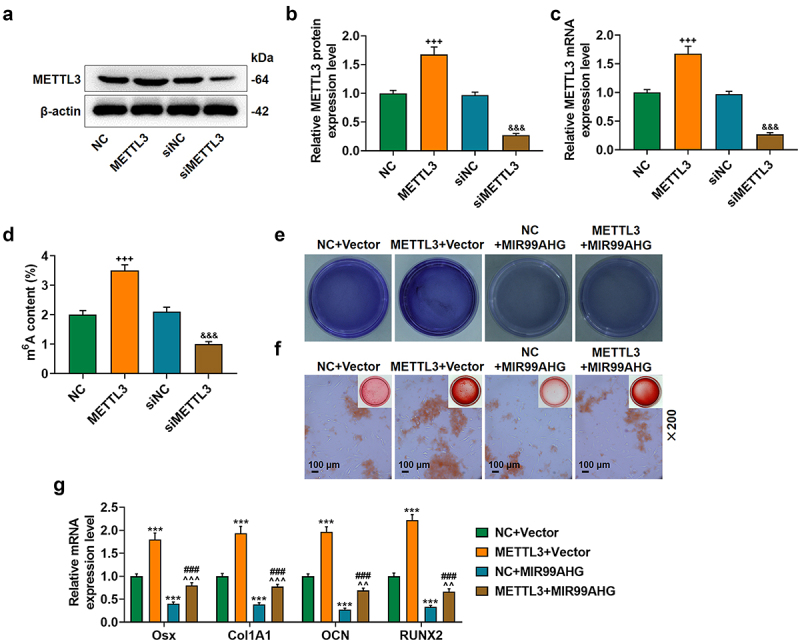
Overexpressed MIR99AHG abolished the effects of METTL3 on BMSCs osteogenic differentiation.(a-c) Relative METTL3 protein (a-b) and mRNA (c) expression after transfection was measured via Western blot and qRT-PCR. β-actin was used as internal reference. (d) M6A content in BMSCs after knocking down and overexpressing METTL3. (e-f) Effects of overexpressed METTL3 and MIR99AHG on osteogenic differentiation of BMSCs were evaluated by both ALP staining (e) and Alizarin Red S staining (f). Magnification: × 200. Scale bar = 100 μm. (g) Relative mRNA expression of Osx, Col1A1, OCN and RUNX2 after overexpressed METTL3 and MIR99AHG transfection was quantified with qRT-PCR. β-actin was the internal reference. All experiments have been performed independently in triplicate and data were expressed as mean ± standard deviation (SD). +++p<0.001, vs. NC; &&&p<0.001, vs. siNC; ***p<0.001, vs. NC+Vector; ###p<0.001, vs. METTL3+Vector; ^^p<0.01, ^^^p<0.001, vs. NC+MIR99AHG.
To confirm methylation level of MIR99AHG in BMSCs after knocking down and overexpressing METTL3, m6A content was measured in BMSCs with or without transfection of METTL3 overexpression plasmid and METTL3 silencing. The results showed a increased m6A content in the BMSCs with transfection of METTL3 overexpression plasmid (p < 0.001; Figure 4d), and a decreased m6A content in the BMSCs with transfection of METTL3 silencing (p < 0.001; Figure 4d).
The interaction between METTL3 and MIR99AHG on the osteogenic differentiation of BMSCs was confirmed. In light of the results from both ALP staining and Alizarin Red S staining, overexpressed METTL3 resulted in increased colored area in BMSCs, whereas MIR99AHG overexpression caused a reduction. Moreover, we further demonstrated that the promotive effects of METTL3 overexpression were abolished by overexpressed MIR99AHG (Figures 4e–4f, magnification: × 200). In consistent with the results above, we noted that METTL3 overexpression upregulated all four osteogenic differentiation-associated factors (OSX, Col1A1, OCN, and RUNX2), whilst the reduction on these four factors expression was found after overexpressed MIR99AHG (p < 0.001; Figure 4g). Besides, we confirmed that overexpressed MIR99AHG reversed the effects of METTL3 overexpression on osteogenic differentiation-related factors in BMSCs (p < 0.01; Figure 4g). Silence MIR99AHG abolished the effects of METTL3 downregulation on BMSCs’ osteogenic differentiation (Supplementary Fig. S3).
MiR-4660, the candidate miRNA, could bind with MIR99AHG and was downregulated in OP yet upregulated during osteogenic differentiation induction
To further confirm the mechanism of MIR99AHG acting on BMSCs, we used two datasets, LncBase and LncRNASNP2, to sort and identify the candidate miRNA for our study, where we found 50 miRNAs in LncBase database and 25 in LncRNASNP2 database. Besides, there were 7 miRNAs included in both LncBase and LncRNA SNP2 databases, that is, hsa-miR-202-5p, hsa-miR-493-3p, hsa-miR-4660, hsa-miR-1227-5p, hsa-miR-544a, hsa-miR-1236-3p and hsa-miR-4517 (Figure 5a), among which miR-4660 was selected for our study, with its highest prediction score in LncBase. Furthermore, in accordance with the results from dual-luciferase reporter assay, we successfully confirmed that miR-4660 could competitively bind with MIR99AHG (p < 0.01; undefined Figures 5b–5c). Moreover, we further concluded that miR-4660 could bind with MIR99AHG, as depicted in results from both RNA immunoprecipitation assay and RNA pull down assay (p < 0.001; undefined Figures 5d–5e). In addition, to confirm the involvement of miR-4660 in the osteogenic differentiation, we measured its expression in BMSCs during the induction for the osteogenic differentiation, where we found that miR-4660 was increased during the osteogenic differentiation induction in BMSCs (p < 0.05; Figure 5f).
Figure 5.
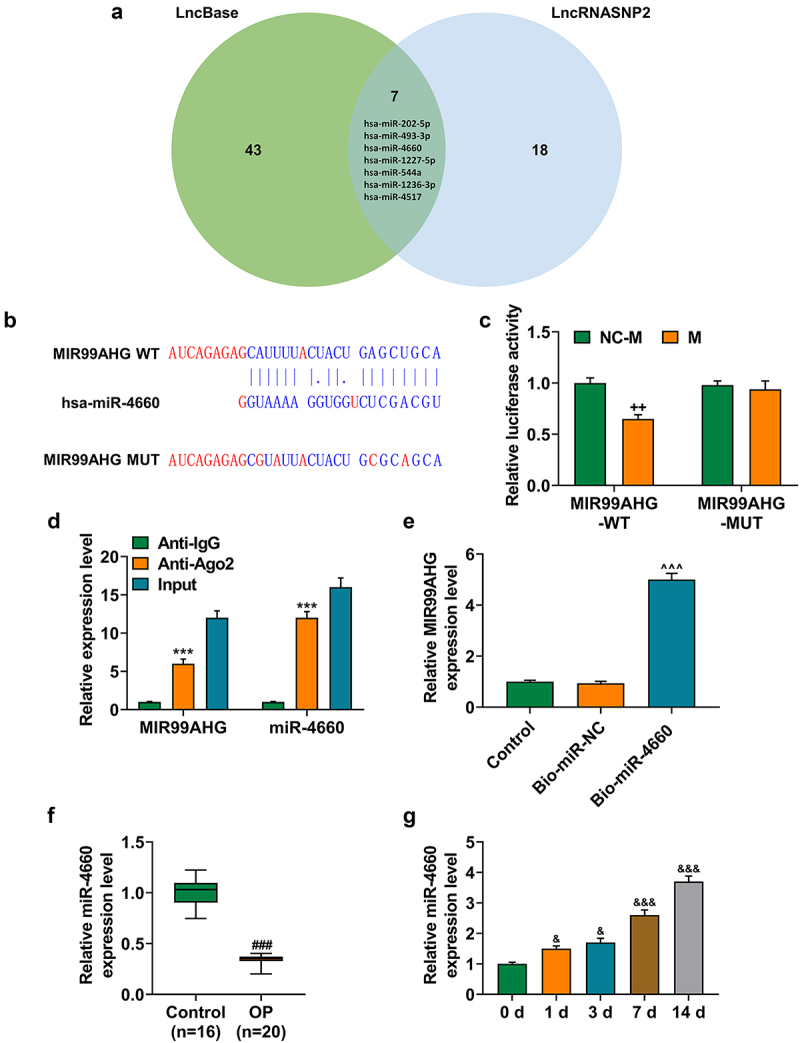
MiR-4660, the candidate miRNA, could bind with MIR99AHG and was downregulated in BMSCs yet upregulated during osteogenic differentiation induction. (a) LncBase (http://carolina.Imis.athena-innovation.Gr/diana_tools/web/index.php) and LncRNASNP2 (http://bioinfo.Life.hust.edu.cn/lncrnasnp#!/) were used to sort and identify the candidate miRNA which could bind with MIR99AHG, and the results were summarized via a Venn diagram. (b-c) Putative binding sites between miR-4660, the candidate miRNA and MIR99AHG were downloaded from LncBase (b), and dual-luciferase reporter assay was used to confirm the results (c). (d-e) Both RNA immunoprecipitation assay (d) and RNA pull down assay (e) confirmed that miR-4660 could bind with MIR99AHG. (f) Relative miR-4660 expression in BMSCs on days 1, 3, 7, and 14 post osteogenic differentiation was further measured with qRT-PCR. U6 was the internal reference. All experiments have been performed independently in triplicate and data were expressed as mean ± standard deviation (SD). ++p<0.001, vs. NC-M; ***p<0.001, vs. anti-IgG; ^^^p<0.001, vs. Bio-miR-NC; &p<0.05, &&&p<0.001, vs. 0 d. miR, miRNA: microRNA; WT: wild-type; MUT: mutated; M: miR-4660 mimic; NC-M: negative control for mimic; IgG: Immunoglobulin G; Ago2: Argonaute 2.
Upregulated miR-4660 reversed the effects of overexpressed MIR99AHG on miR-4660 expression and the osteogenic differentiation of BMSCs
To determine the interaction between miR-4660 and MIR99AHG on BMSCs, we transfected both miR-4660 M and MIR99AHG overexpression plasmid into BMSCs, revealing that miR-4660 M evidently increased miR-4660 expression, yet MIR99AHG overexpression caused a reduction (p < 0.05; Figure 6a). It was also found that the suppressive effects of MIR99AHG on miR-4660 expression was reversed by upregulated miR-4660 (p < 0.01; Figure 6a). Meanwhile, based on the results from the evaluation of osteogenic differentiation of BMSCs with ALP staining and Alizarin Red S staining, less color-stained area was found in BMSCs following MIR99AHG overexpression; however, upregulated miR-4660 not only increased these colored areas but also reversed the effects of MIR99AHG overexpression (Figures 6b–6c, magnification: × 200). As for those osteogenic differentiation factors (OSX, Col1A1, OCN and RUNX2), MIR99AHG overexpression was associated with their reduction yet upregulated miR-4660 both increased their expression and reversed the effects of overexpressed MIR99AHG (p < 0.001; Figure 6d). Silence miR-4660 reversed the effects of MIR99AHG downregulation on miR-4660 expression and the osteogenic differentiation of BMSCs (Supplementary Fig. S4).
Discussion
Accumulating evidence has pointed out and discussed the role of lncRNAs, i.e. those RNA molecules that have no protein-encoding capabilities, in regulating the expression of protein-coding genes via recruiting or sequestering those protein-coding genes, and the participation of lncRNAs as either tumor suppressor or initiator in cancers, where MIR99AHG, as illustrated, has been found to have a prognostic value and a promotive effect in cancer development and progression [11,38–40]. Additionally, the regulatory role of lncRNAs in the biological functions of BMSCs, namely the osteogenic differentiation, has been well discovered and discussed [8–10]. Based on the dataset GSE153829, MIR99AHG was proved to be lower-expressed in the BMSCs of patients with OPLL. Nevertheless, its participation in the osteogenic differentiation of BMSCs, so far as we were concerned, was poorly understood. Here, in our study, we, for the first time, confirmed the suppressive role of MIR99AHG in the osteogenic differentiation of BMSCs. To be specific, under the mediation with a methylation-associated factor METTL3, MIR99AHG, an lncRNA increased in BMSCs of patients with OP, was associated with the suppression on the osteogenic differentiation of BMSCs in vitro and the attenuation on the progression of OP in vivo, providing novel insights into the potential role of MIR99AHG in OP.
Increasing discoveries have underlined the role of BMSCs in osteogenic differentiation and OP, with suggestions referring to the implication of lncRNAs in BMSCs’ osteogenic differentiation [5–7]. As a group of multipotent cells residing within the stroma of BM, BMSCs are those cells which, with the increase on the age, may differentiate into adipocytes, instead of osteoblasts, and undergo the senescence process, inducing osteogenesis decrease and OP. Thus, it is urgent to further unveil the molecular mechanism concerning the functional changes of BMSCs [41,42]. OSX, Col1A1, OCN, and RUNX2 are osteogenic differentiation-related factors upregulated during the osteogenic differentiation [31]. OSX, acting as a factor of both key osteogenic transcription and the Sp family of C2H2-type zinc finger transcription factors, can strengthen the expression on those transcripts which encodes osteoblastic markers, while the increase of Col1A1, an early marker for osteoblast is evidenced during transformation from osteo-progenitor to pre-osteoblast cells [43,44]. OCN, also named as Bone Gamma-Carboxyglutamate Protein (BGLAP), is a calcium-binding protein that is also perceived as the most specific marker of mature or advanced osteoblast. It accumulates within mineralized bone and actively binds to those hydroxyapatite crystals to promote bone crystal growth [45]. In addition, RUNX2 is a pivotal transcription factor with multiple functions. During the stem cells’ osteogenic differentiation, it also regulates the transcription of other osteoblast-associated factors, like Col1A1 and OCN [46]. Besides, due to the fact that ALP is one of the most reliable osteogenic differentiation-associated markers and Alizarin Red S is used to analyze the pluripotent differentiations of BMSCs as confirmed by the appearance of calcium nodules after the staining [32,47,48], ALP staining and Alizarin Red S staining are two staining methods for evaluating osteogenic differentiation, and further confirming the results in our study, from which we conclude that these four factors, namely OSX, Col1A1, OCN, and RUNX2, are all upregulated with more color-stained areas. However, MIR99AHG, the candidate lncRNA employed in our study, is downregulated during the osteogenic differentiation. Besides, we affirm that overexpressed MIR99AHG represses the osteogenic differentiation by decreasing the expression on those four osteogenic differentiation markers and the colored areas in BMSCs, whereas its suppression did conversely. These results, therefore, drive us to conclude that overexpressed MIR99AHG may inhibit the osteogenic differentiation in BMSCs.
Chemical nucleobases’ modifications, including gene reactivation by cytosine methylation and demethylation within promoter regions, are crucial for controlling gene expression, while RNA methylation, as one kind of nucleobases’ modifications, has been interpreted as pivotal in various biological processes [49,50]. Recent studies have demonstrated that m6A, as one of the most abundant modifications, can be detected in mRNA internally and therefore affect the metabolism and functions of mRNAs [51]. Ever since its discovery in the 1970s, m6A RNA methylation, as a predominant internal RNA modification in higher eukaryotes, has gained prodigious attention and participates in all stages of the life cycle of RNA. METTL3 is a critical methyltransferase firstly identified, which can catalyze m6A methylation on ncRNA within mammals to affect the metabolism on RNA [52,53]. Accumulating evidence has also put forward the regulatory roles of METTL3 on the fate of BMSCs and the osteogenic differentiation, in addition to the prior discussions which have provided evidence concerning the interaction between METTL3 and lncRNAs in other biological processes [17,18,36,54]. Nevertheless, the interaction between METTL3 and lncRNA MIR99AHG in BMSCs is rarely discussed, so we, hereby, firstly detect their interactions in BMSCs. Specifically, we firstly find out the downregulation of METTL3-mediated m6A methylation in undifferentiated BMSCs and the upregulation of METTL3 during osteogenic differentiation. Bioinformatic analysis is then performed to ascertain the m6A methylation site of MIR99AHG. In addition, we found that the promotive effects of overexpressed METTL3 on the osteogenic differentiation of BMSCs and its potential interaction with MIR99AHG. To be more specific, increased METTL3 promoted the m6A methylation, the sites of which were evidenced in MIR99AHG. We further speculated that METTL3 overexpression promoted m6A methylation in MIR99AHG to cause a reduction in MIR99AHG, thereby promoting the osteogenic differentiation. These results suggest that MIR99AHG methylation mediated by METTL3 may have promotive effects on the osteogenic differentiation of BMSCs. Nevertheless, the detailed mechanism needs to be additionally addressed.
As a highly conserved endogenous ncRNA with a total length of approximately 19–25 nucleotides, miRNAs have been uncovered to exert their regulatory functions on cell proliferation, differentiation, and apoptosis via binding to the 3’-untranslated regions on target mRNAs. Recent studies have also unveiled the important role of miRNAs during the osteogenic differentiation of BMSCs [55]. As illustrated previously, miR-204 suppresses the osteogenic differentiation of BMSCs, while miR-483-3p and miR-27a-3p exert their promotive effects [56–58]. However, from our point of view, there is poor discussion regarding the effects on miR-4660, a miRNA that has regulatory effects on the pathogenesis of mutation-negative idiopathic oxalosis and the biological functions of ovary [59,60], as well as on the osteogenic differentiation of BMSCs. Here, with bioinformatic tools, we predict miR-4660, which is upregulated during the induction of osteogenic differentiation, as the candidate miRNA that can bind with MIR99AHG. Then, we discover the promotive effects of miR-4660 on BMSCs’ osteogenic differentiation and its capability on reversing the effects of overexpressed MIR99AHG on BMSCs, based on which the conclusion can be drawn that the promotive effects of METTL3-mediated MIR99AHG methylation on the osteogenic differentiation of BMSCs are achieved via targeting miR-4660. Additionally, it is well known that lncRNA, miRNA, and mRNA interactions are related to the formation of competing endogenous RNA (ceRNA). A ceRNA network (lncRNA-miRNA-mRNA) may be constructed through bioinformatics analysis to further find the downstream of miR-4660 in the future research.
Collectively, in our study, to our best knowledge, we, for the first time, systematically introduce and interpret the implication of MIR99AHG, a LncRNA participating in other human cancers [11,39,40], in the osteogenic differentiation of BMSCs. Concretely speaking, we firstly confirmed the overexpression of MIR99AHG in undifferentiated BMSCs, which was also associated with the suppression on the osteogenic differentiation. Meanwhile, we additionally found that MIR99AHG underwent methylation in response to the meditation of overexpressed METTL3, thereby causing a promotion in osteogenic differentiation of BMSCs via sponging miR-4660. We hope that results from our work can bring further insights into the pathogenesis of OP and the roles and functions of ncRNAs involved in both the osteogenic differentiation of BMSCs and in OP development and progression, so as to provide the possible viable therapeutic methods for the diagnosis and prognosis of OP in clinical practice in the future.
Supplementary Material
Funding Statement
Nanjing Key Specialty Supporting Funds [SZDZK201600].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2125751
Data availability statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
References
- [1].Oryan A, Sahvieh S.. Effects of bisphosphonates on osteoporosis: focus on zoledronate. Life Sci. 2021;264:118681. [DOI] [PubMed] [Google Scholar]
- [2].Yang TL, Shen H, Liu A, et al. A road map for understanding molecular and genetic determinants of osteoporosis. Nat Rev Endocrinol. 2020;16(2):91–103. DOI: 10.1038/s41574-019-0282-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Iolascon G, Moretti A, Toro G, et al. Pharmacological therapy of osteoporosis: what’s new? Clin Interv Aging. 2020;15:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cheng C, Wentworth K, Shoback DM. New frontiers in osteoporosis therapy. Annu Rev Med. 2020;71(1):277–288. [DOI] [PubMed] [Google Scholar]
- [5].Arthur A, Gronthos S. Clinical application of bone marrow mesenchymal stem/stromal cells to repair skeletal tissue. Int J Mol Sci. 2020;21(24):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang A, Yu C, You F, et al. Mechanisms of zuogui pill in treating osteoporosis: perspective from bone marrow mesenchymal stem cells. Evid Based Complement Alternat Med. 2018;2018:3717391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Guo Q, Guo Q, Xiao Y, et al. Regulation of bone marrow mesenchymal stem cell fate by long non-coding RNA. Bone. 2020;141:115617. [DOI] [PubMed] [Google Scholar]
- [8].Yang X, Yang J, Lei P, et al. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/satb2 axis. Aging (Albany NY). 2019;11:8777–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang X, Zhao D, Zhu Y, et al. Long non-coding RNA GAS5 promotes osteogenic differentiation of bone marrow mesenchymal stem cells by regulating the miR-135a-5p/foxo1 pathway. Mol Cell Endocrinol. 2019;496:110534. [DOI] [PubMed] [Google Scholar]
- [10].Jin C, Jia L, Tang Z, et al. Long non-coding RNA MIR22HG promotes osteogenic differentiation of bone marrow mesenchymal stem cells via PTEN/AKT pathway. Cell Death Dis. 2020;11(7):601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Meng Q, Wang X, Xue T, et al. Long noncoding RNA MIR99AHG promotes gastric cancer progression by inducing EMT and inhibiting apoptosis via miR577/FOXP1 axis. Cancer Cell Int. 2020;20(1):414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen X, Hua W, Huang X, et al. Regulatory role of RNA N(6)-methyladenosine modification in bone biology and osteoporosis. Front Endocrinol (Lausanne). 2019;10:911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhong H, Tang HF, Kai Y. N6-Methyladenine RNA modification (m 6 A): an emerging regulator of metabolic diseases. Curr Drug Targets. 2020;21(11):1056–1067. [DOI] [PubMed] [Google Scholar]
- [14].Sun T, Wu R, Ming L. The role of m6a RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613. [DOI] [PubMed] [Google Scholar]
- [15].Liu J, Yue Y, Han D, et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. DOI: 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yi YC, Chen XY, Zhang J, et al. Novel insights into the interplay between m(6)a modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xue L, Li J, Lin Y, et al. M6 a transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer. J Cell Physiol. 2020. m 6;236(4):2649–2658. doi: 10.1002/jcp.30023 [DOI] [PubMed] [Google Scholar]
- [18].Su Y, Xu R, Zhang R, et al. N6-Methyladenosine methyltransferase plays a role in hypoxic preconditioning partially through the interaction with lncRNA H19. Acta Biochim Biophys Sin (Shanghai). 2020;52(12):1306–1315. DOI: 10.1093/abbs/gmaa130 [DOI] [PubMed] [Google Scholar]
- [19].Liu ZZ, Hong CG, Hu WB, et al. Autophagy receptor OPTN (optineurin) regulates mesenchymal stem cell fate and bone-fat balance during aging by clearing FABP3. Autophagy. 2021;17(10):2766–2782. DOI: 10.1080/15548627.2020.1839286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang Y, Deng P, Liu Y, et al. Alpha-Ketoglutarate ameliorates age-related osteoporosis via regulating histone methylations. Nat Commun. 2020;11:5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lin Z, He H, Wang M, et al. MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif. 2019;52:e12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cai Z, Liu W, Chen K, et al. Aberrantly expressed lncRnas and mRnas of osteogenically differentiated mesenchymal stem cells in ossification Of The posterior longitudinal ligament. Front Genet. 2020;11:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zheng J, Guo H, Qin Y, et al. Snhg5/mir-582-5p/runx3 feedback loop regulates osteogenic differentiation and apoptosis of bone marrow mesenchymal stem cells. J Cell Physiol. 2020. DOI: 10.1002/jcp.29527. [DOI] [PubMed] [Google Scholar]
- [24].Li J, Xin Z, Cai M. The role of resveratrol in bone marrow-derived mesenchymal stem cells from patients with osteoporosis. J Cell Biochem. 2019;120(10):16634–16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen XJ, Shen YS, He MC, et al. Polydatin promotes the osteogenic differentiation of human bone mesenchymal stem cells by activating the BMP2-Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2019;112:108746. [DOI] [PubMed] [Google Scholar]
- [26].Xia T, Wu X, Cao M, et al. The RNA m6a methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol Res Pract. 2019;215(11):152666. DOI: 10.1016/j.prp.2019.152666 [DOI] [PubMed] [Google Scholar]
- [27].Xie P, Liu M, Chen F, et al. Long non-coding RNA AGAP2-AS1 silencing inhibits PDLIM5 expression impeding prostate cancer progression via up-regulation of MicroRNA-195-5p. Front Genet. 2020;11:1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. methods (San Diego. Calif). 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- [29].Xu K, Yang Y, Feng GH, et al. Mettl3-Mediated m(6)a regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27(9):1100–1114. DOI: 10.1038/cr.2017.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhao J, Liu S, Zhang W, et al. MiR-128 inhibits the osteogenic differentiation in osteoporosis by down-regulating SIRT6 expression. Biosci Rep. 2019;39(9). DOI: 10.1042/BSR20191405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fu L, Jin P, Hu Y, et al. Kr‑12‑a6 promotes the osteogenic differentiation of human bone marrow mesenchymal stem cells via BMP/SMAD signaling. Mol Med Rep. 2020;21(1):61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bou Assaf R, Fayyad-Kazan M, Al-Nemer F, et al. Evaluation of the osteogenic potential of different scaffolds embedded with human stem cells originated from schneiderian membrane: an in vitro study. Biomed Res Int. 2019;2019:2868673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang X, Feng J, Xue Y, et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature. 2016;534(7608):575–578. DOI: 10.1038/nature18298 [DOI] [PubMed] [Google Scholar]
- [34].Yang S, Wei J, Cui YH, et al. M(6)a mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10(1):2782. DOI: 10.1038/s41467-019-10669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li H, Zhang Y, Guo Y, et al. ALKBH1 promotes lung cancer by regulating m6a RNA demethylation. Biochem Pharmacol. 2020;189:114284. [DOI] [PubMed] [Google Scholar]
- [36].Tian C, Huang Y, Li Q, et al. Mettl3 regulates osteogenic differentiation and alternative splicing of vegfa in bone marrow mesenchymal stem cells. Int J Mol Sci. 2019:20(3):551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Han J, Wang JZ, Yang X, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6a-dependent manner. Mol Cancer. 2019;18(1):110. DOI: 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].McCabe EM, Tp R. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol. 2020;75:38–48. [DOI] [PubMed] [Google Scholar]
- [39].Yang B, Shen J, Xu L, et al. Genome-Wide identification of a novel eight-lncRNA signature to improve prognostic prediction in head and neck squamous cell carcinoma. Front Oncol. 2019;9:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Emmrich S, Streltsov A, Schmidt F, et al. LincRnas MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer. 2014;13(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Abdallah BM, Jafari A, Zaher W, et al. Skeletal (stromal) stem cells: an update on intracellular signaling pathways controlling osteoblast differentiation. Bone. 2015;70:28–36. [DOI] [PubMed] [Google Scholar]
- [42].Qadir A, Liang S, Wu Z, et al. Senile osteoporosis: the involvement of differentiation and senescence of bone marrow stromal cells. Int J Mol Sci. 2020;21(1):349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hoshikawa S, Shimizu K, Watahiki A, et al. Phosphorylation-Dependent osterix degradation negatively regulates osteoblast differentiation. Faseb J. 2020;34(11):14930–14945. DOI: 10.1096/fj.202001340R [DOI] [PubMed] [Google Scholar]
- [44].Kannan S, Ghosh J, Dhara SK. Osteogenic differentiation potential of porcine bone marrow mesenchymal stem cell subpopulations selected in different basal media. Biol Open. 2020;9(10). DOI: 10.1242/bio.053280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kaur G, Valarmathi MT, Potts JD, et al. Regulation of osteogenic differentiation of rat bone marrow stromal cells on 2D nanorod substrates. Biomaterials. 2010;31:1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhou P, Shi JM, Song JE, et al. Establishing a deeper understanding of the osteogenic differentiation of monolayer cultured human pluripotent stem cells using novel and detailed analyses. Stem Cell Res Ther. 2021;12(1):41. DOI: 10.1186/s13287-020-02085-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Westhauser F, Karadjian M, Essers C, et al. Osteogenic differentiation of mesenchymal stem cells is enhanced in a 45S5-supplemented β-TCP composite scaffold: an in-vitro comparison of vitoss and vitoss BA. PLoS One. 2019;14(2):e0212799. DOI: 10.1371/journal.pone.0212799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jiao J, Feng G, Wu M, et al. MiR -140-5p promotes osteogenic differentiation of mouse embryonic bone marrow mesenchymal stem cells and post-fracture healing of mice. Cell Biochem Funct. 2020;38(8):1152–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Traube FR, Carell T. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 2017;14(9):1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ovcharenko A, Rentmeister A. Emerging approaches for detection of methylation sites in RNA. Open Biol. 2018;8(9):180121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Brocard M, Ruggieri A, Locker N. M6a RNA methylation, a new hallmark in virus-host interactions. J Gen Virol. 2017;98(9):2207–2214. [DOI] [PubMed] [Google Scholar]
- [52].Pan Y, Ma P, Liu Y, et al. Multiple functions of m(6)a RNA methylation in cancer. J Hematol Oncol. 2018;11(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wang Q, Geng W, Guo H, et al. Emerging role of RNA methyltransferase METTL3 in gastrointestinal cancer. J Hematol Oncol. 2020;13(1):57. DOI: 10.1186/s13045-020-00895-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wu Y, Xie L, Wang M, et al. Mettl3-Mediated m(6)a RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9(1):4772. DOI: 10.1038/s41467-018-06898-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang J, Liu S, Li J, et al. Roles for miRnas in osteogenic differentiation of bone marrow mesenchymal stem cells. Stem Cell Res Ther. 2019;10(1):197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jiang X, Zhang Z, Peng T, et al. miR‑204 inhibits the osteogenic differentiation of mesenchymal stem cells by targeting bone morphogenetic protein 2. Mol Med Rep. 2020;21(1):43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zhou B, Peng K, Wang G, et al. miR‑483‑3p promotes the osteogenesis of human osteoblasts by targeting Dikkopf 2 (DKK2) and the Wnt signaling pathway. Int J Mol Med. 2020;46(4):1571–1581. DOI: 10.3892/ijmm.2020.4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Fu YC, Zhao SR, Zhu BH, et al. MiRNA-27a-3p promotes osteogenic differentiation of human mesenchymal stem cells through targeting ATF3. Eur Rev Med Pharmacol Sci. 2019;23(3 Suppl):73–80. [DOI] [PubMed] [Google Scholar]
- [59].Tu X, Zhao Y, Li Q, et al. Human MiR-4660 regulates the expression of alanine–glyoxylate aminotransferase and may be a biomarker for idiopathic oxalosis. Clin Exp Nephrol. 2019;23(7):890–897. DOI: 10.1007/s10157-019-01723-8 [DOI] [PubMed] [Google Scholar]
- [60].Cai H, Li Y, Li H, et al. Identification and characterization of human ovary-derived circular RNAs and their potential roles in ovarian aging. Aging (Albany NY). 2018;10:2511–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.


