Abstract
Optimizing outcomes in prostate cancer (PCa) requires precision in characterization of disease status. This effort was directed at developing a PCa extracellular vesicle (EV) Digital Scoring Assay (DSA) for detecting metastasis and monitoring progression of PCa. PCa EV DSA is comprised of an EV purification device (i.e., EV Click Chip) and reverse-transcription droplet digital PCR that quantifies 11 PCa-relevant mRNA in purified PCa-derived EVs. A Met score was computed for each plasma sample based on the expression of the 11-gene panel using the weighted Z score method. Under optimized conditions, the EV Click Chips outperformed the ultracentrifugation or precipitation method of purifying PCa-derived EVs from artificial plasma samples. Using PCa EV DSA, the Met score distinguished metastatic (n = 20) from localized PCa (n = 20) with an area under the receiver operating characteristic curve of 0.88 (95% CI:0.78–0.98). Furthermore, longitudinal analysis of three PCa patients showed the dynamics of the Met scores reflected clinical behavior even when disease was undetectable by imaging. Overall, a sensitive PCa EV DSA was developed to identify metastatic PCa and reveal dynamic disease states noninvasively. This assay may complement current imaging tools and blood-based tests for timely detection of metastatic progression that can improve care for PCa patients.
Keywords: Prostate cancer, Liquid biopsy, Extracellular vesicle, Metastatic biomarker, Nanotechnology
1. Introduction
In the United States (US), prostate cancer (PCa) is the most common, non-cutaneous, solid tumor malignancy affecting men. More than 268,490 cases will be diagnosed in 2022. [1] PCa is also the second leading cause of cancer-related death in American men and will claim more than 34,500 lives this year. [1] Advances in PCa care and diagnosis have brought forward several new tests and interventions including molecular imaging, antibody-targeted therapeutics, and immune therapies. [2] Additionally, the role of focal therapies has emerged as a means of controlling limited recurrences in the hope of avoiding protracted toxicity from systemic therapy. Given the cost and specificity of these treatments, [3] there is a growing need for simple, rapid, and affordable tests which can help to improve the triage of diagnostics and therapeutics in the growing pool of patients who need treatment.
This demand has fueled an interest in exploring liquid biopsies that noninvasively provide molecular insights into the underlying disease. [4] While many have turned to circulating tumor DNA, [5,6] these approaches are limited to characterization of fragmented DNA [7] and lack the ability to trace transcriptomic changes which are known to be highly relevant to the evolution of PCa. [8] Other groups have used various technologies to isolate circulating tumor cells (CTCs), [9,10] which provide rich genetic information. However, the time and cost of isolating and preserving the integrity of CTCs are challenging. In addition, clinical applications are limited due to the rarity of CTCs in relatively early-stage disease.
There has been growing interest in exploring the potential of extracellular vesicles (EVs) to address this unmet need. [4] EVs are phospholipid bilayer-enclosed particles released from both normal and cancer cells into body fluids, e.g., urine and blood. [4] These EVs carry membrane-bound markers and genetic materials including miRNA, mRNA, and fragments of DNA inherited from their parental cells. [11] By transferring these biomolecules between cells, EVs can function as a vehicle for intercellular communication. [11] This process has also been shown to prime the pre-metastatic niche at distant organs and facilitate the invasion and dissemination of cancer cells. [4,11] Compared to CTCs, EVs have advantages in their higher frequency in circulation [12] and the simplicity of sample preservation. [13] In addition, considering the relatively low rate of DNA mutations and the frequently altered transcriptomic patterns in PCa [8,14], EV-derived mRNA may provide more biological information relevant to PCa progression, which may not be obtained from ctDNA. As such, profiling mRNA in PCa-derived EVs holds great potential for rapid and accurate characterization of PCa. This approach may offer the opportunity to dynamically monitor the evolving biology of PCa over its natural history. One major challenge of this approach, however, is that PCa-derived EVs typically co-exist with those from non-tumor sources, resulting in complex background signals. [15] Thus, selective purification of PCa-derived EVs, which constitute only a minor portion of total EVs in plasma samples, is a major technical barrier.
To overcome this, many efforts have been dedicated to enriching PCa-derived EVs and analyzing their molecular contents. [16–18] For example, Han et al. introduced an ultrasensitive DNA tetrahedron-based thermophoretic assay to detect mRNA in PCa-derived EVs and demonstrated its excellent performance in distinguishing PCa from benign prostate hyperplasia. [16] Others proposed nanomaterial-based strategies for multiplexed biodetection of potential biomarkers in cancer-derived EVs. [19–21] Recently, our team developed a novel microfluidic device coupled with a unique nanosurface (i.e., EV Click Chip) for purification of tumor-derived EVs. [18] By using reverse transcription-droplet digital polymerase chain reaction (RT-ddPCR) for quantification of the disease relevant mRNAs from purified EVs, we developed an EV Digital Scoring Assay (DSA) (i.e., EV Click Chip plus RT-ddPCR). This technology was initially applied to the identification of early-stage hepatocellular carcinoma within a group of cirrhotic patients. [18] With modifications of this approach, we aimed to apply the EV DSA to PCa and benchmark the performance of this assay in reflecting the clinical states of patients with PCa.
In this study, we optimized the EV DSA for rapid and sensitive analysis of the mRNA contents from PCa-derived EVs. The optimized EV DSA outperformed ultracentrifugation and a commercial EV precipitation assay in purifying PCa-derived EVs. A panel of 11 PCa-relevant mRNA markers was developed through a rigorous screening framework for specific detection of the underlying PCa in the blood background. Finally, a pilot study with 40 PCa patients showed the ability of this EV DSA to distinguish metastatic from localized PCa. In an analysis of serial blood samples from 3 patients, changes in the mRNA signatures detected by the EV DSA associated with clinical behavior observed over the course of treatment. These findings demonstrated the potential utility of this assay in PCa which may provide a novel means of interrogating EVs suitable for deployment into the clinical setting that may be used to improve personalized care.
2. Results
2.1. Optimization of PCa EV Digital Scoring Assay
As illustrated in Fig. 1, the PCa EV DSA was comprised of two major components: EV Click Chip for purification of PCa-derived EVs and RT-ddPCR for quantification of the mRNAs from the purified EVs. Two antibodies were selected for EV Click Chip to facilitate capture of PCa-derived EVs with high sensitivity and specificity: anti-epithelial cell adhesion molecule (EpCAM) and anti-prostate-specific membrane antigen (PSMA). These markers were selected on the basis of their well-characterized and strong expression on the surface of PCa epithelial cells. [22–25] Artificial plasma samples were used (Fig. 2a) to evaluate the performance of EV Click Chip throughout the optimization process. 22Rv1-derived EVs spiked into healthy donor plasma were used for assay optimization focusing on measurements of the AR-V7 transcript, a known androgen receptor variant present in 22Rv1 derived-EVs and absent in healthy donors’ plasma. The counts of AR-V7 transcripts were used to calculate the EV recovery yield (Fig. 2a). The counts of AR-V7 transcripts in the artificial plasma samples and the EV Click Chip-purified EVs were labeled as AR-V7 transcriptspre-purified and AR-V7 transcriptspost-purified, respectively. The EV recovery yield obtained by EV Click Chips was calculated using the following Eq. (1): [18].
Fig. 1.
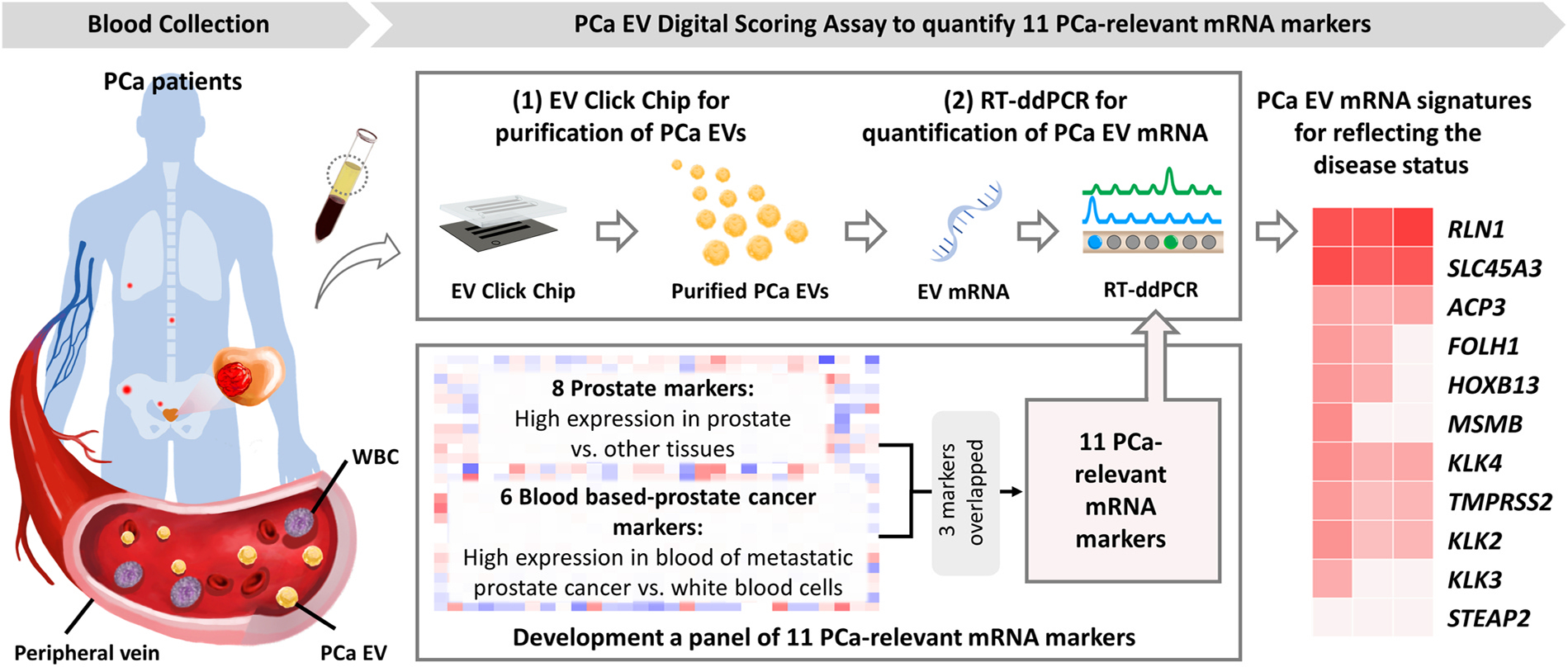
PCa EV Digital Scoring Assay is comprised of two major components: (1) EV Click Chip for purifying PCa-derived EVs from plasma samples, and (2) RT-ddPCR for quantifying mRNA markers from the purified PCa-derived EVs. In parallel, a panel of 11 PCa-relevant genes, comprised of tissue-based prostate markers and blood-based PCa markers, was adopted to reflect disease state. EV, extracellular vesicle; PCa, prostate cancer; RT-ddPCR, reverse transcription-droplet digital polymerase chain reaction.
Fig. 2. Optimization of capture antibody and EV Click Chip using artificial PCa EV samples.
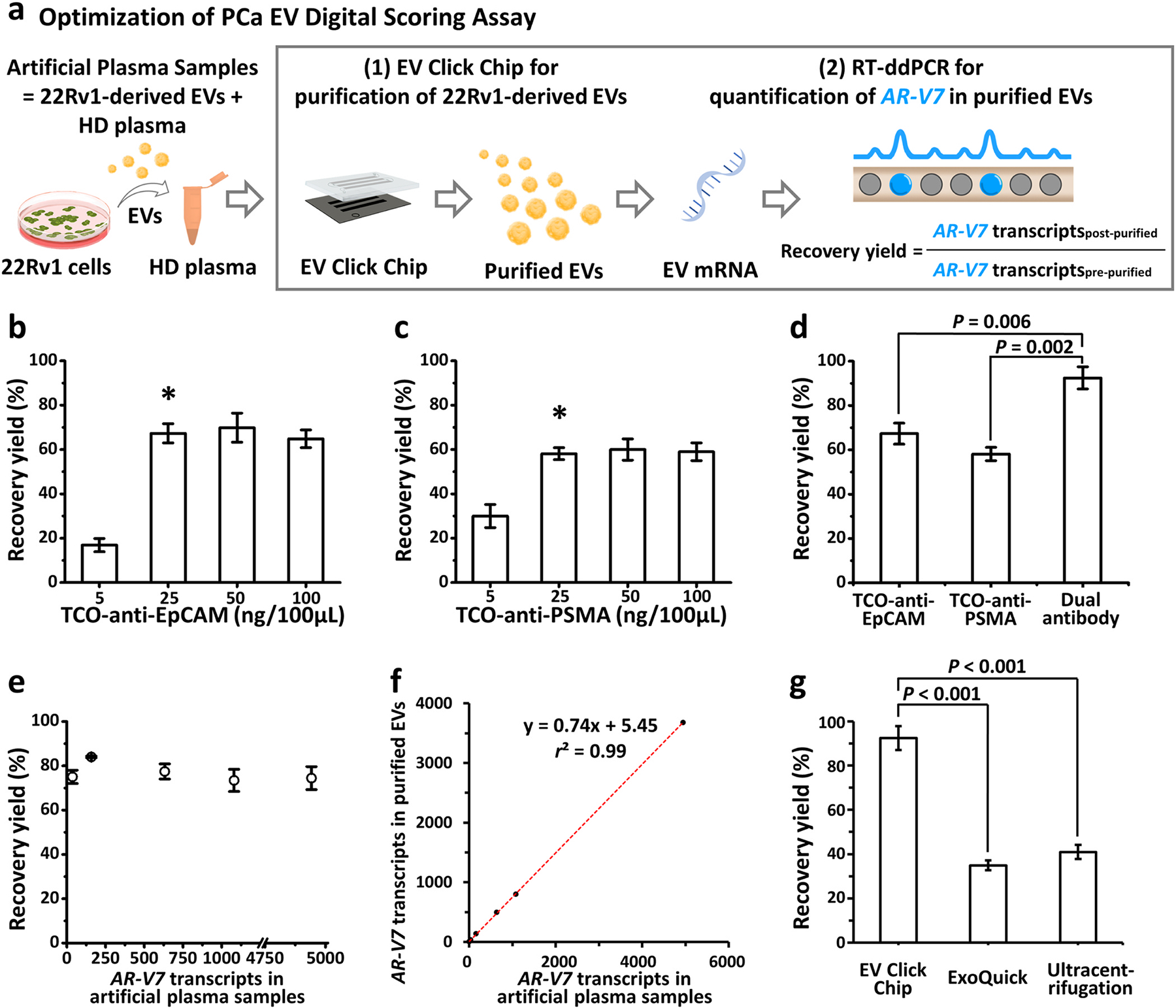
(a) Schematic workflow of a quantitative method to evaluate the performance of EV Click Chip. Artificial plasma samples were prepared by spiking 22Rv1-derived EVs into the plasma from a male healthy donor. EV Click Chips were utilized to purify 22Rv1-derived EVs and RT-ddPCR was applied to quantify the AR-V7 transcripts in the purified EVs to obtain the recovery yield. (b-c) Recovery yields observed for EV Click Chip at different concentrations of TCO-anti-EpCAM and TCO-anti-PSMA. Data are presented as means ± SD of three independent assays. *Indicates the concentration selected for further optimization studies. (d) The recovery yields observed in the presence of individual and dual antibodies, i.e., TCO-anti-EpCAM, TCO-anti-PSMA, and combination of the two antibodies. Data are presented as means ± SD of three independent assays. (e) Capture performance of EV Click Chip using artificial sample containing 0–5000 AR-V7 transcripts per 500-μL volume. (f) Dynamic linearity range of the recovery yields observed for EV Click Chip across artificial sample containing 0–5000 AR-V7 transcripts. (g) The recovery yields observed for optimized EV Click Chip, ExoQuick® ULTRA EV isolation kit and ultracentrifugation. Data are presented as means ± SD of three independent assays. EpCAM, epithelial cell adhesion molecule; EV, extracellular vesicle; PCa, prostate cancer; PSMA, prostate-specific membrane antigen; RT-ddPCR, reverse transcription-droplet digital polymerase chain reaction; SD, standard deviation; TCO, trans-cyclooctene.
| (1) |
The optimal concentrations of trans-cyclooctene (TCO)-anti-EpCAM and TCO-anti-PSMA for PCa DSA were determined by testing the EV recovery yields with a series of different concentrations of these two antibodies individually. As shown in Fig. 2b & 2c, for a 100-μL artificial plasma sample, optimal EV recovery yields were achieved using 25 ng for TCO-anti-EpCAM and TCO-anti-PSMA, respectively. EV recovery yield was substantially higher with the combination of TCO-anti-EpCAM and TCO-anti-PSMA at their optimal concentrations as compared to individual antibody (TCO-anti-EpCAM, P = 0.006; TCO-anti-PSMA, P = 0.002, Fig. 2d).
Using this dual capture antibody combination and the optimized assay parameters from our previous study, [18] the dynamic range of capture performance of EV Click Chip was investigated using artificial plasma samples spiked with different amount of 22Rv1-derived EVs containing 0–5000 AR-V7 transcripts in a 500-μL volume. Our results demonstrated consistent capture (Fig. 2e) across a wide range of concentration of artificial plasma samples. In addition, the results confirmed the linearity of the EV recovery yields by EV Click Chip (y = 0.74x, r2 = 0.99, Fig. 2f). Consistency in the triplicate measurements of the capture performance revealed the high precision of the optimized assay (Supplementary Table 1). We also demonstrated high purity (90.38%) of the captured EVs by EV Click Chip (see Supplementary Methods for measurement and calculation of purity).
Lastly, we compared the EV recovery yields between the optimized EV Click Chip, ultracentrifugation, and ExoQuick® ULTRA EV Isolation Kit for Serum and Plasma (a commercially available assay). All artificial plasma samples were analyzed in triplicate. The EV recovery yield of the optimized EV Click Chip (92.5 ± 5.4%; Fig. 2g) was significantly higher than those from ultracentrifugation (41.0 ± 3.2%; P < 0.001) and the ExoQuick® EV isolation methods (34.9 ± 2.2%; P < 0.001).
Characterization of EVs was carried following guidelines from the International Society for Extracellular Vesicles (ISEV). [26] The morphologies of the 22Rv1-derived EVs before capture, the EVs captured on EV Click Chip, and the EVs released from EV Click Chip were characterized by transmission electron microscopy (TEM; Supplementary Figure 1a, 1b, and 1c). The size of 22Rv1-derived EVs used for optimization ranged from 30 to 450 nm in diameter with a mean of 186.3 nm by nanoparticle tracking analysis (NTA; Supplementary Figure 1d). Size distribution and intactness of EVs were not altered during purification. This was consistent with the findings from our previous study. [18] (Supplementary Figure 1d). Western blot analysis showed the presence of epithelial tissue specific transmembrane protein EpCAM (MISEV2018 [26] Category 1) and cytosolic protein Annexin V (Category 2), as well as the absence of non-EV co-isolated structures apolipoprotein APOA1 (Category 3) and intracellular mitochondria cytochrome C (Category 4) in LNCaP-derived EVs and artificial PCa plasma samples purified by EV Click Chips (Supplementary Figure 1e). Both immunocapture markers i.e., EpCAM and PSMA were identified on the LNCaP-derived EVs before and after purification. None of the above markers were detected in male healthy donor plasma purified by EV Click Chip, confirming the specificity of EV Click Chip for capturing EpCAM and PSMA positive EVs. LNCaP cell lysate and healthy donor plasma were analyzed in parallel as positive controls for these markers.
2.2. Development and validation of a panel of 11 PCa-relevant genes
After the optimization of PCa-derived EV purification, a panel of mRNA markers reflecting the underlying malignancy was developed. The selection framework for these genes is shown in the Fig. 3a. First, to ensure the markers were highly specific to prostate rather than other tissues, 120 candidate genes were nominated on the basis of elevated expression in prostatic epithelium in contrast to 36 other tissues in The Tissue Atlas of The Human Protein Atlas. [27,28] Eight genes specific to prostate tissue were selected by applying a threshold of a greater than 5-fold change when comparing prostate and non-prostate tissues. In parallel, to incorporate markers associated with metastatic PCa, the gene set from Miyamoto et al. [29] was considered given its use in a PCa liquid biopsy setting as blood-based biomarkers for detection of disseminated PCa. The six top expressed genes in their dataset (GEO GSE67980) were incorporated into the final panel. As 3 genes overlapped between the above gene sets, this process yielded 11 PCa-relevant genes, including ACP3, FOLH1, HOXB13, KLK2, KLK3, KLK4, MSMB, RLN1, SLC45A3, STEAP2, and TMPRSS2 (Supplementary Table 2). Since the majority of the background EVs circulating in cancer patient’s blood originated from immune cells, [30] it was crucial to ensure the selected genes are expressed at low levels in immune cells to minimize background noise. The expression level of these 11 genes was further evaluated in PCa tissues (from The Cancer Genome Atlas [TCGA]), [8] PCa cell lines (from Cancer Cell Line Encyclopedia [CCLE]), [31] and immune cells (from Differentiation MAP [DMAP]), [32] As shown in Fig. 3b, these 11 genes were highly expressed in PCa tissues and PCa cell lines, while lowly expressed in immune cells (P < 0.001, respectively), making them potential PCa markers for a blood-based assay.
Fig. 3. Development and validation of a panel of 11 PCa-relevant genes.
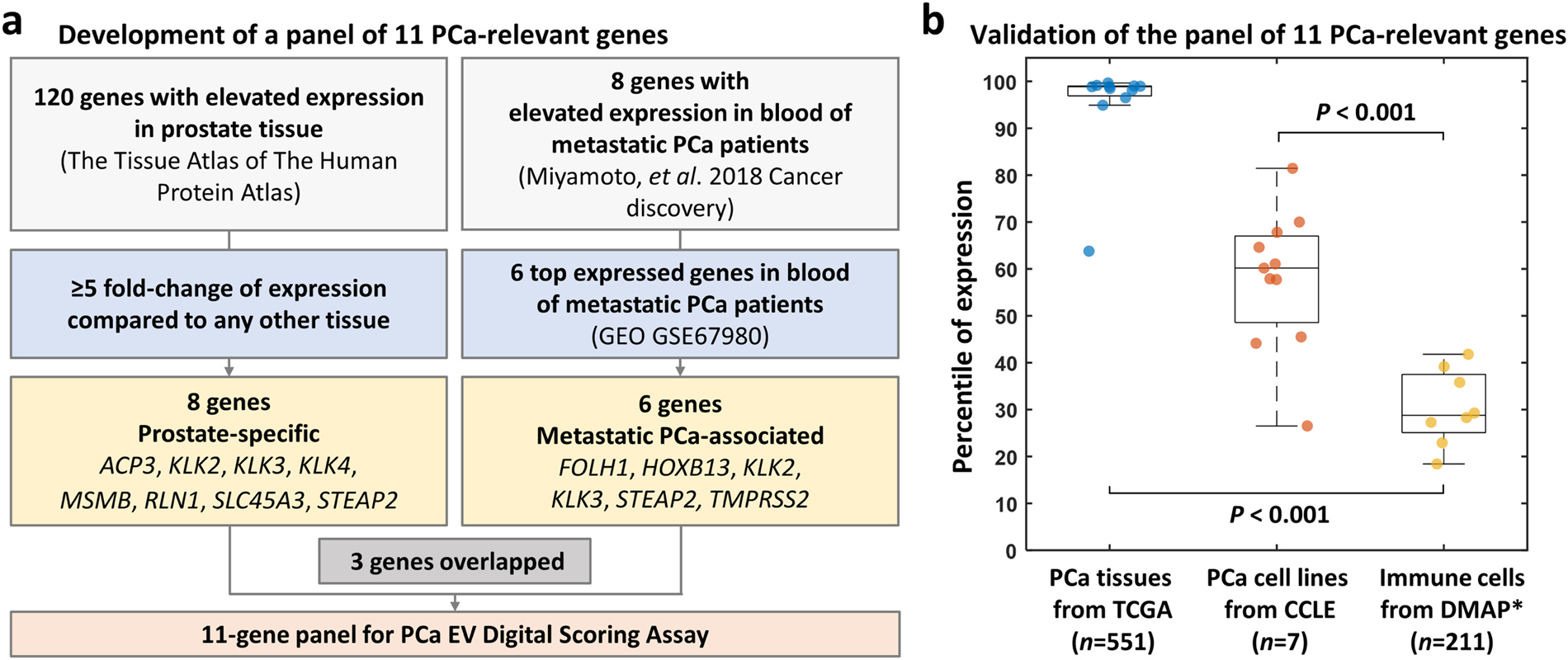
(a) Schematic flow of the selection of 11-gene panel for PCa EV Digital Scoring Assay, including ACP3, FOLH1, HOXB13, KLK2, KLK3, KLK4, MSMB, RLN1, SLC45A3, STEAP2, and TMPRSS2. (b) Box plot of the percentile of expression level of the 11-gene panel in primary PCa patients’ tissues from TCGA, PCa cell lines from CCLE, and immune cells from DMAP. Each dot represents the mean of percentile of each maker. *Expression of STEAP2, SLC45A3, and KLK4 are not available in DMAP. CCLE, Cancer Cell Line Encyclopedia; DMAP, Differentiation MAP dataset; EV, extracellular vesicle; PCa, prostate cancer; TCGA, The Cancer Genome Atlas.
2.3. Performance of the 11-gene panel in PCa cell line-derived EVs
Primers and probes for these 11 genes were validated using RT-ddPCR to confirm specificity (Fig. 4a). Strong signals were detected in PCa cells while no signal was detected in white blood cells (WBCs) from a male healthy donor. To ensure that these genes were also expressed in PCa-derived EVs with limited expression in background EVs on EV Click Chip, the following were tested: (1) PCa cell line-derived EVs without purification of EV Click Chip, (2) PCa cell line-derived EVs purified from artificial plasma samples by EV Click Chip, and (3) EVs isolated from a male healthy donor’s plasma by EV Click Chip (Fig. 4b). Compared to the strong signals in PCa cell line-derived EVs, no signals were detected in healthy donor’s EVs with the exception of low RLN1. In addition, similar expression profiles were observed in PCa cell line-derived EVs before and after purification, which indicated that the mRNA components were not affected by the purification process. Finally, we compared the expression of the 11 genes in PCa cells and their derived EVs. There was a high correlation in gene expression patterns between EVs and their parental cells (LNCaP, r = 0.95, P < 0.001; C4–2B, r = 0.74, P = 0.01; 22Rv1, r = 0.89, P < 0.001). These findings point to the utility of these 11 genes in the context of the EV DSA.
Fig. 4. Performance of the 11-gene panel in PCa cell line-derived EVs.
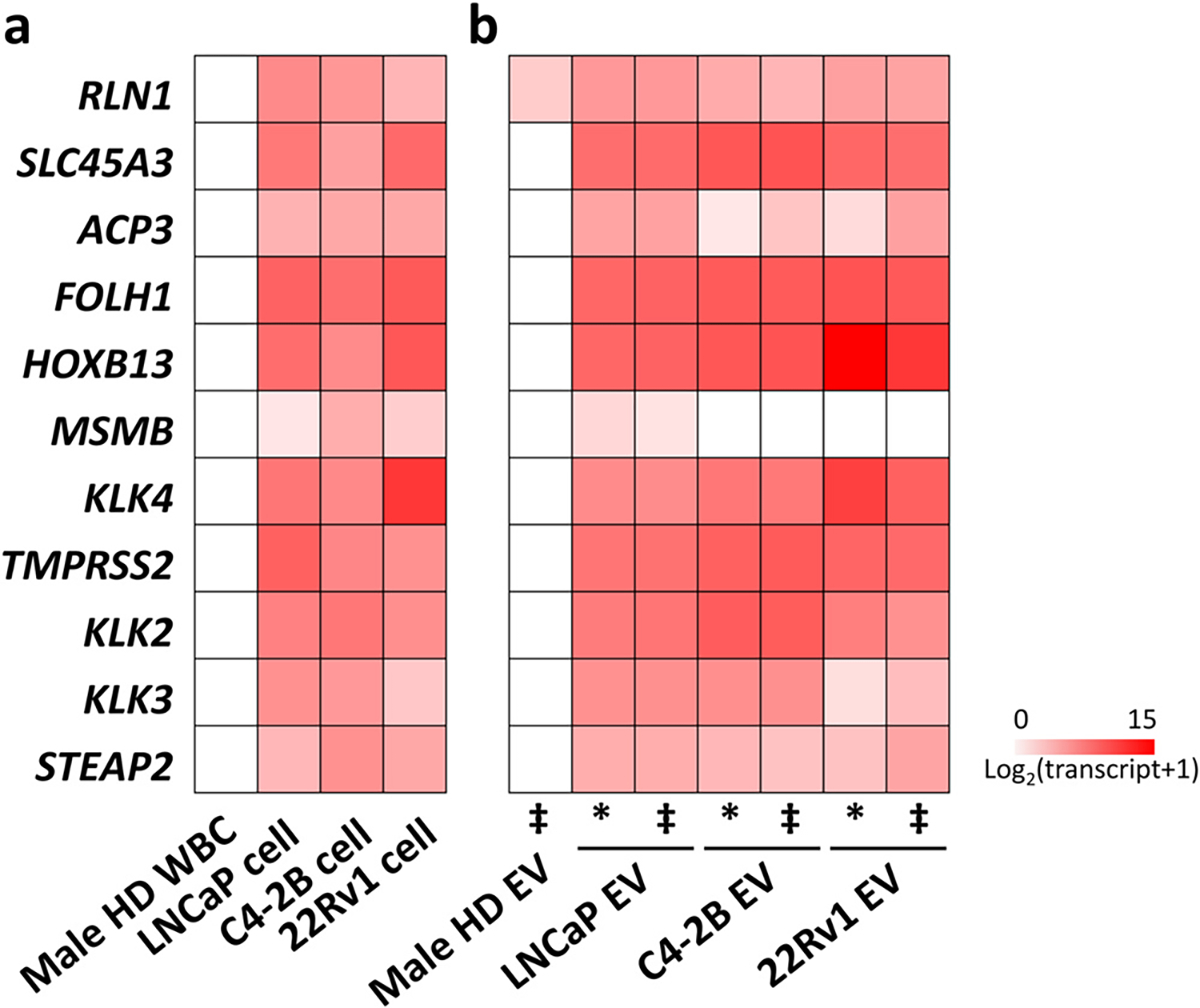
(a) Expression heatmap of the 11 gene-panel with LNCaP, C4–2B, and 22Rv1 cells (positive controls) and male healthy donor (HD) WBCs (negative control). (b) Expression heatmap of the 11 gene-panel in the PCa cell line-derived EVs before purification (labeled as *), purified PCa cell line-derived EVs from artificial plasma samples (i.e., a male HD’s plasma spiked with LNCaP, C4–2B, and 22Rv1-derived EVs, respectively) by EV Click Chip (labeled as ‡), and purified EVs from a male HD’s plasma by EV Click Chip. Transcript counts are log2-transformed as shown in the heatmaps. ‡ after purification by EV Click Chip. * before purification by EV Click Chip. EV, extracellular vesicle; HD, healthy donor; PCa, prostate cancer; RT-ddPCR, reverse transcription-droplet digital polymerase chain reaction; WBC, white blood cell.
2.4. Met scores for differentiating metastatic from localized PCa
Having validated the panel and PCa EV DSA on artificial samples, we designed a pilot study to benchmark the performance of this assay on clinical samples (Fig. 5a). For this pilot study, plasma samples from two groups of patients were utilized: (1) localized PCa (n = 20); (2) metastatic PCa (n = 20). Demographic and clinical information of the patients are presented in Table 1.
Fig. 5. Performance of PCa EV Digital Scoring Assay in distinguishing metastatic PCa patients from localized PCa patients.
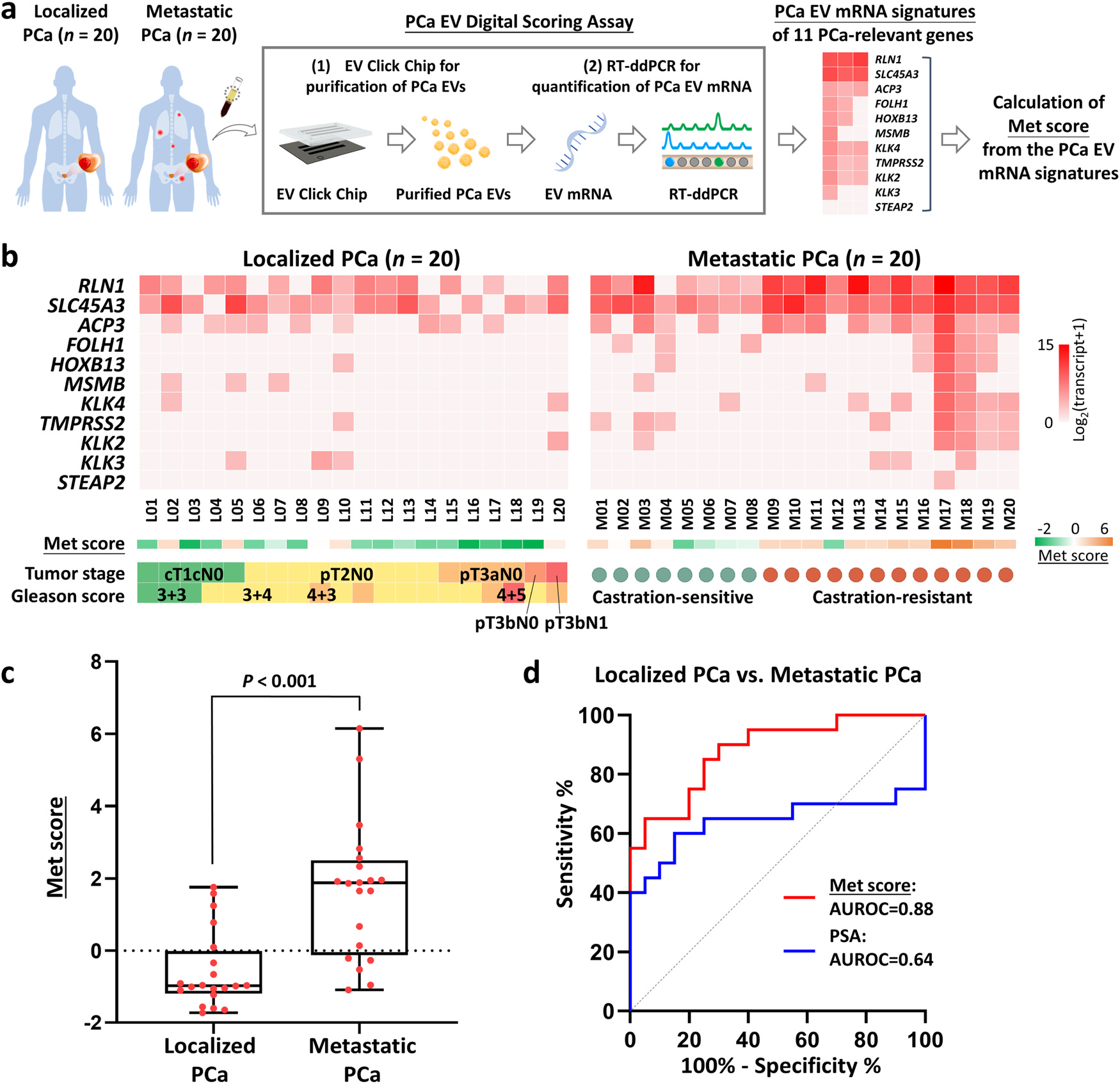
(a) The workflow of PCa EV Digital Scoring Assay for quantifying the 11-gene panel and calculating Met score in purified EVs from patients with localized PCa (n = 20) and metastatic PCa (n = 20). (b) Heatmap of the 11 genes in patients with localized PCa (n = 20) and metastatic PCa (n = 20). Transcript counts were log2-transformed. Corresponding Met scores were calculated from the expression of the 11 genes. (c) Box plot of the distribution of Met scores among localized PCa (n = 20) and metastatic PCa patients (n = 20). (d) ROC curves for Met score and serum PSA concentration to distinguish metastatic from localized PCa (Met score: AUROC=0.88, P < 0.001, 95% CI=0.78–0.98; serum PSA concentration: AUROC=0.64, P = 0.12, 95% CI=0.45–0.84). AUROC, area under the receiver operating characteristic curve; CI, confidence interval; EV, extracellular vesicle; PCa, prostate cancer; PSA, prostate-specific antigen; ROC, receiver operating characteristic.
Table 1.
Clinical characteristics of PCa patients.
| Characteristic | Localized PCa (n = 20) | Characteristic | Metastatic PCa (n = 20) |
|---|---|---|---|
|
| |||
| Median Age, y (IQR) | 64 (60.3–69.8) | Median Age, y (IQR) | 69.5 (60.8–76.5) |
| Age < 60, n (%) | 4 (20.0) | Age < 60, n (%) | 5 (25.0) |
| Age ≥ 60, n (%) | 16 (80.0) | Age ≥ 60, n (%) | 15 (75.0) |
| Race/ethnicity, n (%) | Race/ethnicity, n (%) | ||
| White | 10 (50.0) | White | 13 (65.0) |
| Black | 6 (30.0) | Black | 3 (15.0) |
| Asian | 3 (l5.0) | Asian | 2 (10.0) |
| Hispanic | 1 (5.0) | Hispanic | 2 (10.0) |
| Median PSA, ng/mL (IQR) | 7.4 (5.5–11.4) | Median PSA, ng/mL (IQR) | 22.1 (1.6–222.5) |
| The extent of the primary tumor, n (%) | Castration sensitivity, n (%) | ||
| T1c | 6 (30.0) | Castration-sensitive | 7 (35.0) |
| T2 | 8 (40.0) | Castration-resistant | 13 (65.0) |
| T3 | 6 (30.0) | ||
| Nodal involvement, n (%) | |||
| N0 | 19 (95.0) | ||
| N1 | 1 (5.0) | ||
| 3 + 3 | 3 (15.0) | ||
| 3 + 4 | 12 (60.0) | ||
| 4 + 3 | 4 (20.0) | ||
| 4 + 5 | 1 (5.0) | ||
| Treatment choice, n (%) | |||
| Active surveillance | 6 (30.0) | ||
| Radical prostatectomy | 14 (70.0) | ||
IQR, interquartile range; PCa, prostate cancer; PSA, prostate-specific antigen.
Expression of the 11 genes from the PCa EV DSA is shown as a heatmap in Fig. 5b with log2-transformed transcript counts. A Met score for each patient sample was calculated using the weighted Z score method based on the 11-gene signature. [33] As shown in Fig. 5c, the Met scores of metastatic PCa samples were significantly higher (P < 0.001) than those from patients with localized PCa. Receiver operating characteristic (ROC) analysis further confirmed the ability of the Met score to distinguish metastatic PCa from localized PCa with an area under the ROC curve (AUROC) of 0.88 (95% CI=0.78–0.98; sensitivity=85%, specificity=75%; Fig. 5d) at the optimal cutoff of − 0.30, which significantly surpassed the predictive power of serum prostate-specific antigen (PSA) concentration (AUROC=0.64, 95% CI=0.45–0.84, P = 0.03; sensitivity=65%, specificity=75%) at the cutoff of 10.25 ng/mL. The addition of PSA to Met score did not increase the predictive power (AUROC=0.89, 95% CI=0.79–0.99, P = 0.46; sensitivity=85%, specificity=75%). To investigate the performance of each gene in discriminating metastatic from localized PCa, a Met score was calculated for each individual gene and ROC analyses were performed. Among the 11 genes, the ROC analyses for RLN1, SLC45A3, and ACP3 were statistically significant when being considered as sole predictors for metastatic PCa (Supplementary Figure 2). While FOLH1 (PSMA) and STEAP2 were exclusively seen in EVs from metastatic PCa, their discriminatory power did not meet statistical significance in our limited samples.
To consider the impact of multiple tumor features (macroscopic and microscopic) on the Met scores, a subgroup analysis was conducted. The Met scores calculated from the 11-gene panel did not show a strong correlation with T stage or Gleason score (Supplementary Figure 3a and 3b). No association was identified between Met scores and tumor volume/mass determined at the final pathologic evaluation of radical prostatectomy (Supplementary Figure 3c and 3d). On the other hand, among patients with metastatic PCa, the Met scores were significantly higher in metastatic castration-resistant prostate cancer (mCRPC) than those in metastatic castration-sensitive prostate cancer (mCSPC) (P = 0.04, Supplementary Figure 3e). Lastly, we found the Met scores were not associated with serum PSA concentration in both localized and metastatic PCa (Supplementary Figure 3f and 3g).
2.5. Dynamic performance of the PCa EV Digital Scoring Assay
As noted above, an advantage of blood-based biomarker assays is the capacity to examine the changes in information in the face of clinical events of importance over time. To illustrate the dynamic performance of this assay, we analyzed selected samples from three patients in our cohort over the course of their treatment.
Case A. (Fig. 6a): This was a patient with a pT3bN0 Gleason 3 + 4 PCa who experienced a persistent PSA elevation following radical prostatectomy. Conventional imaging identified an isolated metastasis to the right iliac bone that was treated with metastasis directed radiotherapy without treating the prostatic fossa. After the treatment, his disease became quiescent by biochemical and radiographic surveillance for four years. At this timepoint of low and stable disease burden, his Met score was also the lowest compared to subsequent blood draws. The patient was noted to have a rising serum PSA concentration four years post-prostatectomy and finally had a 68Ga-PSMA positron emission tomography and computed tomography (PET/CT) when his serum PSA concentration rose to 1.4 ng/mL. The PET/CT identified an L4 lesion and periaortic lymph nodes suited for metastasis directed radiotherapy. Two blood draws during this progression phase revealed a consecutive increase of Met score, corresponding to a continuous increase of disease burden and/or activity clinically.
Fig. 6. Dynamic performance of the PCa EV Digital Scoring Assay.
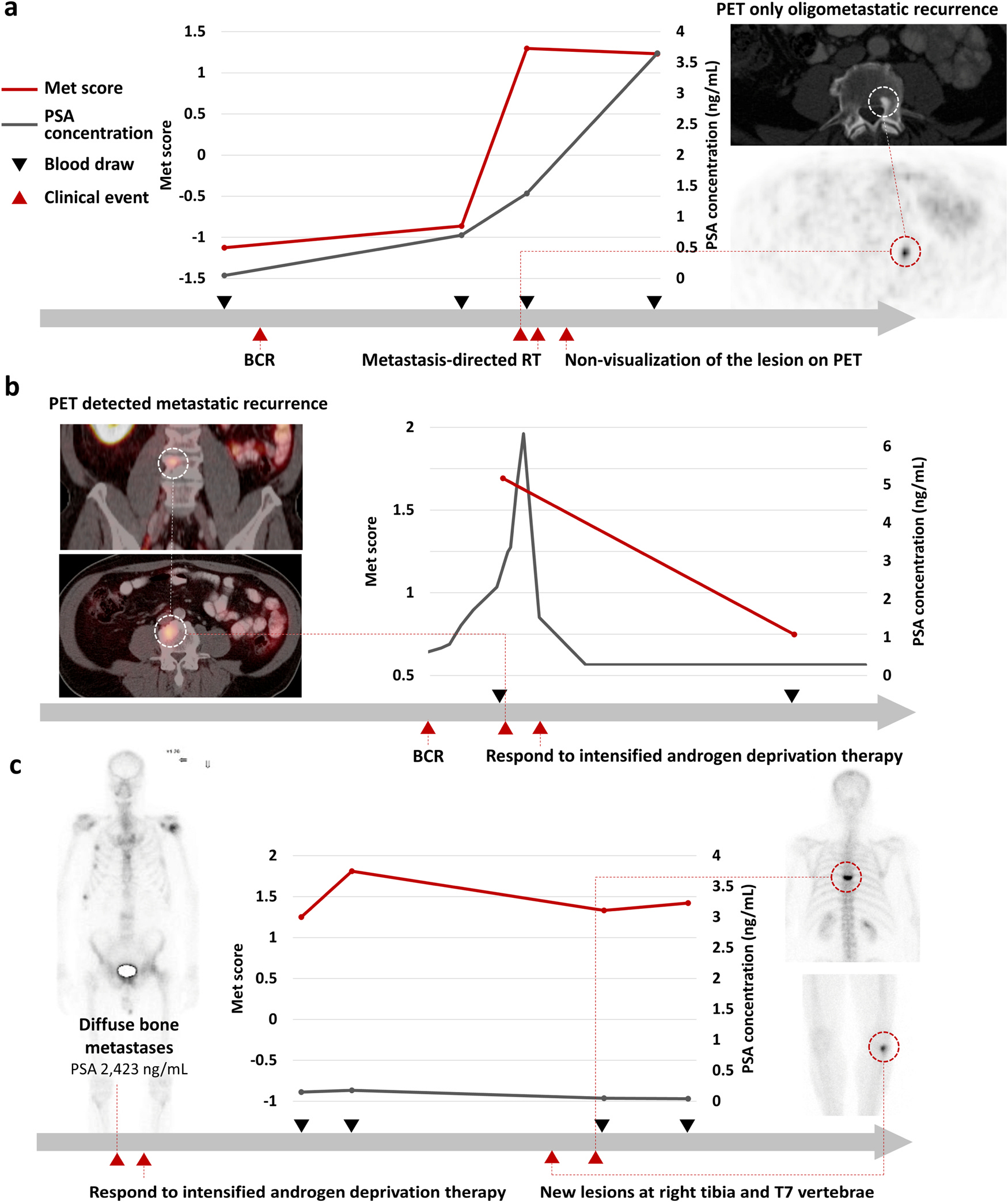
Met scores from the PCa EV DSA were generated for three PCa patients over the course of treatment. (a) Patient undergoing metastasis-directed therapy via stereotactic radiotherapy without subsequent benefit; (b) Patient with clinical response to intensified androgen deprivation therapy; and (c) Patient with suspected progression by radiograph and symptoms later found to be erroneous. BCR, biochemical recurrence; EV, extracellular vesicle; PCa, prostate cancer; PET, positron emission tomography; PSA, prostate specific antigen; RT, radiotherapy.
This patient declined castration therapy and opted for radiation to the metastatic lesion. He did not experience a decline in his serum PSA concentration following radiation but did have a 68Ga-PSMA PET/CT that no longer showed the L4 lesion or the nodes. The Met score after the treatment remained stable, which was commensurate with the clinical picture. The lack of benefit from focal treatment was further confirmed by the emergence of a new rib metastasis emerging on CT and bone scan one year later.
In this clinical case, the Met score reflected the increase of disease burden and their lack of response to focal treatment. Of note, the Met score detected the progression of disease when the disease burden was below the limits of detection even with PSMA PET/CT.
IQR, interquartile range; PCa, prostate cancer; PSA, prostate-specific antigen.
Case B. (Fig. 6b): This patient presented with biochemical recurrence after radical prostatectomy followed by salvage radiotherapy. His serum PSA concentration rose to 6.4 ng/mL. While conventional CT and bone scans failed to identify metastatic deposits, a 68Ga-PSMA-PET/CT identified multiple retroperitoneal lymph nodes and an osseous metastasis at L4. His pattern of spread was deemed as not appropriate for focal therapy. As such, intensified androgen deprivation therapy (ADT) was initiated. His serum PSA declined and has now been undetectable.
In this case, conventional imaging failed to detect the metastatic disease and was not informative in monitoring the patient’s response, whereas the Met score decreased after the initiation of medical castration which mirrored his clinical status.
Case C. (Fig. 6c): This patient presented with advanced, metastatic prostate cancer with a serum PSA concentration in excess of 2000 ng/mL. Imaging showed diffuse bone metastases. As such, the patient was started on intensified ADT with abiraterone acetate and prednisone. He had a favorable biochemical and clinical response. Two years after starting the therapy, he presented with acute back pain and upper extremity weakness. Though his PSA remained stable, imaging showed a new lesion at T7 and in the right tibia noted as concerning for metastatic progression. Due to the neurologic symptoms, a magnetic resonance imaging (MRI) of the spine was obtained and was read as suspicious for a metastatic lesion. The clinical picture raised concerns for neuroendocrine transformation and progression. Upon referral to neurosurgery, the diagnosis was changed to compression fracture resulting from drug-induced osteoporosis. Radiographs of the tibia showed no correlation with the bone scan- i.e., no fracture or sclerosis to indicate metastatic disease. With conservative measures, the patient’s symptoms abated. He has remained clinically well for an additional one year and continues therapy.
Four serial blood samples were evaluated by PCa EV DSA. The first two bloods were drawn while his disease was responding to intensified ADT very well. The last two bloods were drawn after the new suspicious lesions emerged. Met scores of across these four blood samples remained stable regardless of the emergence of new lesions. This case demonstrated the assay was capable of clarifying progression from non-progression events detected by radiographs that could have resulted in dramatically different courses of clinical action – in this case, chemotherapy for progression versus bone modifying agents for osteoporosis.
3. Discussion
In the current study, we have presented the development of a PCa EV DSA, which combines Click Chip technology for purification of PCa-derived EVs with RT-ddPCR for quantification of the disease-relevant mRNAs. In parallel, an 11-gene mRNA panel associated with the underlying PCa was generated through a rigorous selection process (Fig. 1). Using this approach, the Met scores calculated from the expression of the 11 genes had the capacity to distinguish metastatic from localized PCa. Finally, in the presented cases, the changes in the Met scores reflected the changes in disease burden in the face of treatment over its course. This points toward the potential for using this assay to monitor PCa over its disease course.
Blood-based testing has been an important part of PCa clinical care. Since its introduction in the 1980 s, serum PSA measurements have become widely used and have played an important role in advancing the field. [34] It is clear, however, to all practitioners that there are important limitations to the value of this approach. PSA measurements have not been highly accurate as a means of identifying metastatic disease. [35] As a particular example, in the setting of biochemical recurrence following radical prostatectomy, absolute PSA concentration can help risk stratify patients for benefit from salvage radiotherapy, but it cannot be used to identify patients with disease beyond a standard radiation port who are likely to progress through standard salvage radiotherapy. [36] Moreover, while changes in PSA in an individual patient are useful, there is a significant variation in the rate of PSA secretion from prostatic tissues that can obscure the clinical picture. For example, it is recognized that irritation of the prostate gland can lead to elevated serum PSA concentrations not related to cancer. [37] Conversely, anaplastic variants of PCa often produce lower amounts of PSA than more typical acinar adenocarcinomas. [38] As such, there remains a need for a non-PSA based means of assessing disease behavior that can be used to address these clinical scenarios.
The utility of EV transcripts enriched from urine as a noninvasive diagnostic biomarker for detecting high-risk PCa has been investigated over the past decade. [39,40] By measuring PCA3 and ERG transcripts, the ExoDx Prostate (IntelliScore) (EPI) test can noninvasively detect high-grade (Gleason score ≥7) PCa and has been used to help minimize biopsies among patients who likely only have low-grade disease. [39,40] In contrast to numerous urine-based EV-mRNA studies in this field, to our knowledge, few blood-based studies investigated the association between EV-mRNA and the disease behavior of PCa. Reported studies placed emphasis on the prognostic role of AR-V7 and identified castration-resistant PCa patients with AR-V7 in EVs as at risk for disease progression and poor survival. [41–43] While these existing EV assays focus on only one or two transcripts within a single stage of PCa, the clinical relevance and significance of mRNA in EVs have not been widely elucidated across the broad spectrum of PCa despite the growing understanding of aberrant transcription in PCa evolution. [8,44] As such, there has remained a need for an EV assay profiling a panel of multiple PCa-relevant mRNAs with the potential for reflecting the disease state of PCa patients and their evolving biology.
As demonstrated in our previous study, EV Click Chips allow for rapid purification of tumor-derived EVs. [18] Integration of the nanosubstrate-surface, dual-antibody capture, and click chemistry-mediated EV capture/release process not only enhanced purification efficiency of PCa-derived EVs, but also minimized nonspecific binding of particles from the background. [18] It is worthy to note that after optimization, EV Click Chip outperformed ultracentrifugation and commercial ExoQuick Assay in specific purification of PCa-derived EVs. This underscores its advantage in extraction of PCa-specific information from small volumes of patient plasma. In addition, the use of RT-ddPCR further allows for absolute quantification of mRNA transcripts, reducing bias when measuring signals from rare vesicles. [45] Lastly, we successfully applied the established EV DSA [18] to PCa, indicating this assay can be rapidly adapted to different diseases and clinical settings.
In our analysis of clinical samples, the mRNA contents of PCa-derived EV were profiled using PCa EV DSA for a cohort of patients with differing states across the disease spectrum of PCa. With 40 patients being profiled, we noted that Met scores were significantly higher in metastatic patients than in localized patients. This may be attributed to increased EV shedding [46] and alteration in mRNA contents during metastatic progression. Interestingly, we observed that FOLH1 (PSMA) and STEAP2 were exclusively found in EVs from metastatic PCa. Others have reported that FOLH1 (PSMA) and STEAP2 are overexpressed in aggressive PCa cells and tissues. Several studies have also pointed to their roles in promoting angiogenesis, migration, and invasion. [47–49] Our observation highlights the performance of Met scores as means of describing the expression pattern of the 11-gene panel. This approach outperformed individual genes in distinguishing the disease states. In addition, it is important to note that the genes in this panel are involved in multiple pathways associated with PCa progression in addition to androgen receptor signaling. This is also supported by the fact that the Met score was not correlated with serum PSA concentrations in both localized and metastatic PCa patients. This underscores that our approach provides information reflecting PCa disease status that is independent of serum PSA measurements. Overall, our study demonstrated significant changes in gene expression identified from tumor-derived EVs across different stages of PCa. Further investigations are warranted to explore the biological roles of this expression signature in EVs of metastatic PCa.
To further explore the dynamic change of EV-based signatures over the course of disease evolution, we performed analyses of specimens collected over time and across therapies for given patients. The presented cases illustrate the dynamic performance of the PCa EV DSA in the clinical setting.
The first case (Fig. 6a) showed the Met score accords with the persistent increase of micro-metastatic disease below the PET-detection threshold that blossomed into metastatic lesions one year later. This points toward the ability of this assay to detect molecular signals from microscopic deposits of disease which even the PSMA-PET imaging could not identify. Detection of micro-metastatic disease is still a critical unmet need in PCa. Patients with micro-metastatic disease would not benefit from the focal therapy including salvage radiotherapy and metastasis-directed therapy due to the presence of disease outside of treated areas, as shown in this case. The development of a test with sensitivity beyond the current imaging modalities will greatly improve shared-decision making around these focal treatments to optimize outcomes and quality of life.
In the second case (Fig. 6b), the initial high Met score reflected his metastatic disease detected by PSMA-PET/CT, which is in line with our findings that metastatic PCa patients have higher Met scores (Fig. 5c). In addition, the Met score decreased in response to intensified ADT which was also reflected by a decrease in serum PSA concentration and improvement in imaging. This was consistent with our observation that the Met scores of mCSPC patients responding to intensified ADT were lower than those of mCRPC patients progressing under ADT (Supplementary Figure 3e). As many genes in our panel are regulated by androgen receptor activation, [50] we posited that the Met score may reflect the effect of the overall clinical effect of androgen suppression. Further studies that explore the Met score as a biomarker for hormonal responsiveness of PCa are ongoing.
Pathologic fracture due to bone metastasis of PCa can be challenging to distinguish from compression fracture as a result of osteoporosis due to long-term use of ADT. In the third case (Fig. 6c), the PCa EV DSA accurately indicated the 99Tc-bone scan finding was not caused by PCa metastasis. This identification of the cause of the lesion can result in dramatically different clinical action on whether to initiate the next line of PCa treatment. In conclusion, this vignette illustrates the utility of this assay to provide PCa-specific molecular information as an adjunct to imaging.
We do recognize that there are limitations to the study as presented. First, due to the lack of comprehensive PCa EV-derived mRNA data in the current scientific literature or publicly available databases, the genes selected for the initial assay were selected from tissue-based datasets. These may not provide the most suitable candidates for an EV-based study. Thus, we conducted a PCa cell line-derived EV interrogation to confirm the applicability of this gene panel in EVs. As the knowledge of EV cargo continues to evolve, this assay can be easily adapted using alternative genes to identify and reflect specific changes of disease biology- particularly those that may aid in optimizing selection of therapies. Second, the pilot study was a small, single-center experience. This limits the correlation of Met scores with more clinical parameters. However, the results confirm the value and clinical utility of this novel approach–profiling PCa relevant EV-mRNA to depict metastatic status of PCa patients and dynamic change of their underlying PCa. Recent studies [51–53] have demonstrated that machine learning approaches combined with CTC- or EV-based liquid biopsies significantly enhanced sensitivity and specificity for disease detection. Larger cohorts combined with advanced machine learning approaches are warranted for further validating the precision, accuracy, and clinical performance of this assay. Given the stability of EVs in frozen plasma and the rapid processing afforded by the PCa EV DSA, this system is ready for deployment in both current and historic PCa clinical studies to validate the EV-mRNA signatures as biomarkers for disease monitoring.
4. Conclusions
In summary, we developed a novel PCa EV DSA to characterize variations in gene expression within EVs across a spectrum of PCa patients. To our knowledge, this is the first study demonstrating that EV-derived mRNAs can reflect the dynamics of the biological processes correlating to the clinical behavior of PCa. Our results warrant additional validation studies of the PCa EV DSA in a larger, multicenter cohort to facilitate its application in clinical settings. Finally, this assay may offer the opportunity to complement current tools including imaging and PSA to provide greater insights into the active biology of an underlying PCa for better personalized care.
5. Materials and methods
5.1. Cell culture
22Rv1 (ATCC CRL-2505) and LNCaP (ATCC CRL-1740) cells were purchased from American Type Culture Collection. C4–2B cells were generously provided by Dr. Leland W. K. Chung. Cells were cultured in RPMI 1640 media (catalog# 10040CV, Thermo Fisher Scientific, USA) with 10% fetal bovine serum and 100 U mL−1 penicillin-streptomycin in a humidified incubator with 5% CO2. To harvest the PCa cell line-derived EVs, cells were grown to 70% confluency in 18 Nunc™ EasYDish™ Dishes (145 cm2, catalog# 150468, Thermo Fisher Scientific, USA) and incubated in serum-free media for 24–48 h. The serum-free media was then collected for harvesting cell line-derived EVs.
5.2. Harvest of PCa cell line-derived EVs by ultracentrifugation
The collected serum-free media incubated with 22Rv1, C4–2B, and LNCaP cells was first centrifuged at 300 g (4 °C) for 5 min three times and then centrifuged at 2800 g (4 °C) for 10 min to remove cells and cell debris, Afterwards, the supernatant was transferred to Open-Top Thinwall Ultra-Clear Tubes (catalog# 344058, Beckman Coulter, Inc., USA), and ultracentrifuged at 100,000 g (4 °C) for 70 min using SW 32 Ti rotor and Optima L-100 XP Ultracentrifuge (Beckman Coulter, Inc., USA). Finally, the pelleted PCa cell line-derived EVs were resuspended with 200-μL ice-cold PBS and collected.
5.3. Preparation of artificial PCa plasma samples
To prepare the artificial plasma samples mimicking plasma samples collected from PCa patients, a 50-μL aliquot of EVs derived from LNCaP cells was spiked into 200-μL male healthy donors’ plasma. This was used for western blot characterization of PCa EVs throughout the purification process by EV Click Chip. Ten μL aliquots of EV pellets derived from 22Rv1, C4–2B, and LNCaP cells, respectively, were spiked into 90-μL of male healthy donors’ plasma. These were used for optimization of the purification of PCa-derived EVs and validation of the 11-gene panel.
5.4. Purification of EVs from artificial plasma sample
Artificial plasma was centrifuged at 10,000g for 10 min to remove cell debris before EV purification. EV purification using an ExoQuick® ULTRA EV Isolation Kit for Serum and Plasma (category# EQULTRA-20A-1, System Biosciences, USA) was carried out per the manufacturer’s protocol. Ultracentrifugation was performed at 100,000 g (4 °C) for 70 min using an SW 32 Ti rotor and Optima L-100 XP Ultracentrifuge (Beckman Coulter, Inc., USA).
5.5. PCa EV Digital Scoring Assay
PCa EV Digital Scoring Assay is comprised of two major components: (1) EV Click Chip for purifying PCa-derived EVs from plasma samples, and (2) RT-ddPCR for quantifying mRNA markers from the purified PCa-derived EVs.
EV Click chip is a nanostructured microfluidic chip utilizing both immunoaffinity and click chemistry to purify the tumor-derived EVs. [18] The following three major approaches were integrated to purify EVs: (1) nanostructured substrates, (2) microfluidic chaotic mixers, and (3) covalent chemistry-mediated EV capture/release.
First, the densely packed silicon nanowires substrates (SiNWS) of EV Click Chip substantially increase the chip surface area interacting with EVs. [18] In addition, EV Click Chip’s microfluidic chaotic mixer enhances repeated physical contact between the flow-through tumor-derived EVs and SiNWS. [18] These two structures work synergically to increase the capture efficiency of EVs onto the chip. Second, a pair of click chemistry motifs, i.e., tetrazine (Tz) and trans-cyclooctene (TCO), are conjugated onto SiNWS and capture antibodies (anti-EpCAM and anti-PSMA), respectively. Plasma samples are incubated with the TCO-grafted antibodies to label prostate cancer (PCa)-derived EVs. Once plasma samples are introduced to EV Click Chips, the click chemistry reaction between Tz-conjugated SiNWS and TCO-conjugated PCa-derived EVs is extremely fast, selective, irreversible, and bioorthogonal, [54] resulting in immobilization of the PCa-derived EVs onto the chip surface. Subsequent exposure to a disulfide bonds reducing agent, 1,4-dithiothreitol (DTT) [55] leads to the specific release of the PCa-derived EVs from the SiNWS by cleaving the embedded disulfide bond between captured PCa-derived EV and EV Click Chip, which further increases the purity of the harvested EVs. Further detailed mechanisms and complete protocol of EV Digital Scoring Assay (DSA) have been described in our previous study. [18].
5.5.1. Preparation of TCO-antibody conjugates
For capturing PCa-derived EVs, TCO-antibody conjugates (i.e., TCO-anti-EpCAM and TCO-anti-PSMA) were prepared as described below. Goat anti-human EpCAM (catalog#AF960, R&D Systems, Inc., USA) and sheep anti-human PSMA (catalog#AF4234, R&D Systems, Inc., USA) antibodies were incubated with TCO-PEG4-NHS ester (catalog# A137–2, Click Chemistry Tools, USA) in phosphate-buffered saline (PBS) at room temperature for 30 min. All the TCO-antibody conjugates were freshly prepared just prior to use.
5.5.2. EV Click Chip for purification of PCa-derived EVs
First, the artificial plasma samples were incubated with individual capture TCO-antibody (5, 25, 50, 100 ng TCO-anti-EpCAM and TCO-anti-PSMA, respectively, per 100 μL artificial plasma samples, Fig. 2b–c) or dual capture TCO-antibody combination (25 ng TCO-anti-EpCAM and 25 ng TCO-anti-PSMA per 100 μL artificial plasma samples, Fig. 2d) for 30 min at room temperature to determine and validate the optimal concentrations of capture TCO-antibodies for EV Click Chips. The optimal dual capture TCO-antibody combination (25 ng TCO-anti-EpCAM and 25 ng TCO-anti-PSMA per 100 μL plasma samples) was used for clinical plasma samples in the subsequent clinical study. Second, after incubation with individual capture TCO-antibody or dual capture TCO-antibody combination, the artificial plasma samples or clinical plasma samples were then introduced into the tetrazine (Tz)-grafted EV Click Chip microfluidic devices, where the PCa-derived EVs in the plasma samples were rapidly and irreversibly captured through the click reaction between TCO and Tz. Third, to release these PCa-derived EVs from the chips, 100 μL 1,4-dithiothreitol (DTT) (50 mM) was injected into EV Click Chips at the flow rate of 1.0 mL/h to cleave the embedded disulfide bonds linking the PCa-derived EVs and EV Click Chip. Lastly, the released EVs were collected and DTT was removed before the subsequent RNA extraction process.
5.5.3. RT-ddPCR for quantification of mRNA markers from the purified PCa-derived EVs
After the removal of DTT, these purified PCa-derived EVs were then lysed with 700 μL QIAzol Lysis Reagent (catalog#79306, Qiagen, USA). RNA extraction and subsequent cDNA conversion were conducted using miRNeasy Micro Kits (catalog#217084, Qiagen, USA) and Maxima H Minus First Strand cDNA Synthesis Kits (catalog#K1652, Thermo Scientific, USA), respectively, according to the manufacturer’s instructions. The resultant cDNA was aliquoted into 6 portions for the quantification of the 11 genes by duplex ddPCR on a QX200 Droplet Digital PCR System (catalog#1864001, Bio-Rad Laboratories, Inc.) according to the manufacturer’s instructions. Information on primers and probes (catalog#4331182, #4448489, Thermo Scientific, USA) used for each tested gene in the RT-ddPCR assay is provided in Supplementary Table 3. The transcript copies of each gene were calculated based on the counts of positive droplets using the QuantaSoft™ software.
5.6. Characterization of EV before and after purification
Characterization of EVs before and after purification was carried out following the recommendations of the International Society for Extracellular Vesicles (ISEV). [26] In brief, nanoparticle tracking analysis (NTA), western blotting analysis, and transmission electron microscopy (TEM) were conducted. Detailed descriptions of the steps for NTA, western blotting, and TEM are provided in the Supplementary Methods.
5.7. Enrollment of PCa patients
All the participants in this study were enrolled in Cedars-Sinai Medical Center (CSMC) Institutional Review Board (IRB)-approved protocols (IRB #Pro00042197, #Pro00033050, #Pro00051931). Samples were collected only after the participants provided written informed consent. The samples utilized in this study were collected between January 2016 - December 2020. We included two groups of PCa patients (localized PCa [n = 20] and metastatic PCa [n = 20]) with a pathologically proven diagnosis of prostate adenocarcinoma for clinical validation of PCa EV DSA: (1) Patients with clinically localized PCa were defined as who are on active surveillance or prior to radical prostatectomy (prostate intact) without any evidence of cancer metastasis either on next-generation imaging (PSMA PET/CT, 68Ga or 18F-based) or conventional radiographs (CT/MRI, 99Tc-single-photon emission computed tomography [SPECT] bone scan). (2) Patients with metastatic PCa were defined as who had metastatic PCa detected by conventional radiographs. Additionally, three PCa patients with serial blood collections over the course of their natural history were included to evaluate the dynamic performance of PCa EV DSA. For patients classified by PSMA PET/CT, a metastatic lesion was defined by the presence of a PET-detectable lesion with an uptake greater than background and an SUV minimum of 10. This was consistent with previous studies utilizing PSMA PET as a screening modality for detection of metastatic disease.
5.8. Clinical blood sample processing
Peripheral venous blood samples were collected from PCa patients or healthy donors in a BD Vacutainer glass tube (BD, USA) with acid citrate dextrose at CSMC. Blood samples were sequentially centrifuged at 530 g for 10 min and then 4600 g for 10 min to collect plasma within 4 h of collection. Plasma samples were stored at − 80 °C and thawed immediately in a 37 °C water bath before use. After thawed, plasma samples were centrifuged at 10,000g for 10 min before running through the PCa EV DSA described above.
5.9. Statistical analysis
To assess the linearity of the EV recovery yields in artificial plasma samples by the PCa DSA, linear regression analysis was performed to calculate the slope and coefficient of determination (r2). During the optimization process of PCa EV DSA, EV recovery yields were expressed as Mean ± standard deviation. Significant differences between different purification methods were evaluated using two sample t test.
Percentiles of the expression of the 11 selected genes in primary PCa tissues (from The Cancer Genome Atlas [TCGA]), [8] PCa cell lines (from Cancer Cell Line Encyclopedia [CCLE]), [31] and immune cells (from Differentiation MAP dataset [DMAP]) [32] were compared using the Wilcoxon rank-sum test. The correlation of the expression of the selected genes between PCa cell lines and their derived EVs was computed as Pearson’s correlation coefficient.
The Met score of the selected 11 genes, which represents the likelihood estimate of 11-gene upregulation, was computed from the mRNA expression of these genes using a weighted Z score method. [33] After the median centering of expression data across the samples, Met scores were computed by the error-weighted mean of the expression values of the 11 genes in a sample. Briefly, for each sample, if Mean(sig) and Mean(all) are the mean expression of the genes in the signature and all the genes in the genome, respectively, and σ is the standard deviation of all the genes, then the Met score used to measure the relative expression level of the signature in the patient sample is calculated as below:
where n is the number of genes in signature. The value of Met score is represented in units of standard deviation, which measures the differences between the gene expression of the signatures and those from the genome. We also assume that the mean of all the genes in the genome follows standard normal distribution with median-centered data, which is zero. A positive value of Met score from a patient sample indicates collective upregulation of the genes in the signature, while a negative value suggests collective downregulation. The magnitude of the Met score represents the degree of upregulation or downregulation.
ROC curve analysis was used to evaluate the performance of Met score and PSA concentration, respectively, focusing on the capability of each to distinguish localized PCa from metastatic PCa. Youden’s index was used to identify the optimal cutoff of Met score and PSA concentration. The AUROC between Met score and PSA concentration was compared using the paired DeLong’s test. Pearson’s correlation coefficients were computed for correlation between Met score and continuous variables. A Mann–Whitney U test was used to compare the differences of Met score between categorical variables.
To assess statistical power of 40 samples in this pilot study, we performed a power analysis. Briefly, the chance of having a type I error (i.e., false negative) is less than 0.001 in this pilot study consisting of 40 patients. In addition, the sample size of 20 for each group provides 0.998 power for discriminating localized from metastatic patients using a one-sided two-sample t-test, type I error (α) of 0.05. This high power suggests the probability of having a type II error (i.e., false positive) is only 0.002.
All statistical analyses were performed using R statistical software (version 4.1.0; R Foundation, Vienna, Austria), GraphPad Prism (version 9.2.0; GraphPad Software, Inc., CA, USA), and MATLAB (Mathworks, Natick, MA, USA) with two-sided tests and a significance level of 0.05.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Leland Chung for providing C4–2B cells. The results shown in this study are part based upon data from the Human Protein Atlas (http://www.proteinatlas.org), The Cancer Genome Atlas (https://www.cancer.gov/tcga), Cancer Cell Line Encyclopedia (https://sites.broadinstitute.org/ccle/), and Differentiation MAP dataset (https://rdrr.io/github/mchikina/CellCODE/man/DMAP.html). We acknowledge the efforts of all the authors and staffs in the creation of the Human Protein Atlas, The Cancer Genome Atlas, Cancer Cell Line Encyclopedia, and Differentiation MAP dataset.
Financial support and sponsorship
This work is supported by United States Department of Defense (PC180192, PC171066, PC190482); and National Institute of Health (R01 CA218356, U01 CA198900, P01 CA233452, R01 CA255727, R01 CA253651, R01 CA246304, U01 EB026421, R21 CA240887, and R21 CA235340). This work has also received support from developmental funds from Cedars-Sinai Cancer. The funders had no role in the collection of data; the design and conduct of the study; management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Abbreviations:
- DSA
Digital Scoring Assay
- EV
extracellular vesicle
- PCa
prostate cancer
- RT-ddPCR
reverse transcription-droplet digital polymerase chain reaction
Footnotes
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests, Hsian-Rong Tseng reports a relationship with CytoLumina Technologies Corp. that includes: equity or stocks. Hsian-Rong Tseng reports a relationship with Pulsar Therapeutics Corp. that includes: equity or stocks. Hsian-Rong Tseng, Yazhen Zhu, Na Sun has patent Covalent chemistry enables extracellular vesicle purification on nanosubstrates - toward early detection of hepatocellular carcinoma pending to Hsian-Rong Tseng, Yazhen ZHU, Na Sun, Vatche G. AGOPIAN. Corresponding author Dr. Edwin Posadas is an uncompensated advisor to CytoLumina Technologies Corp.
CRediT authorship contribution statement
Jasmine J. Wang: Conceptualization, Data Curation, Formal Analysis, Funding Acquisition, Investigation, Methodology, Visualization, Writing – Original Draft, Writing – Review & Editing. Na Sun: Conceptualization, Formal Analysis, Investigation, Methodology, Visualization, Writing – Review & Editing. Yi-Te Lee: Conceptualization, Formal Analysis, Investigation, Methodology, Visualization, Writing – Original Draft, Writing – Review & Editing. Minhyung Kim: Data Curation, Formal Analysis, Methodology, Writing – Review & Editing. Tatyana Vagner: Methodology, Writing – Review & Editing.Krizia Rohena-Rivera: Methodology, Writing – Review & Editing. Zhili Wang: Investigation, Writing – Review & Editing. Zijing Chen: Data Curation, Writing – Review & Editing. Ryan Y. Zhang: Investigation, Writing – Review & Editing. Tiffany X. Zhang: Visualization, Writing – Review & Editing. Howard M. Sandler: Resources, Writing – Review & Editing. Timothy J. Daskivich: Resources, Writing – Review & Editing. Neil A. Bhowmick: Resources, Writing – Review & Editing. Sungyong You: Conceptualization, Formal Analysis, Funding Acquisition, Methodology, Supervision, Writing – Review & Editing. Dolores Di Vizio: Resources, Supervision, Writing – Review & Editing. Hsian-Rong Tseng: Conceptualization, Funding Acquisition, Methodology, Resources, Supervision, Writing – Original Draft, Writing – Review & Editing. Jie-Fu Chen: Supervision, Writing – Review & Editing. Yazhen Zhu: Conceptualization, Funding Acquisition, Methodology, Resources, Supervision, Writing – Review & Editing. Edwin M. Posadas: Conceptualization, Funding Acquisition, Methodology, Resources, Supervision, Writing – Original Draft, Writing – Review & Editing. All the authors: Writing – Review & Editing.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.nantod.2022.101746.
Data availability
Data will be made available on request.
References
- [1].Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer statistics, 2022, CA Cancer J. Clin. 72 (1) (2022) 7–33, 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- [2].Rebello RJ, Oing C, Knudsen KE, et al. , Prostate cancer, Nat. Rev. Dis. Prim. 7 (1) (2021) 9, 10.1038/s41572-020-00243-0 [DOI] [PubMed] [Google Scholar]
- [3].Trogdon JG, Falchook AD, Basak R, Carpenter WR, Chen RC , Total medicare costs associated with diagnosis and treatment of prostate cancer in elderly men, JAMA Oncol. 5 (1) (2019) 60–66, 10.1001/jamaoncol.2018.3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gao Z, Pang B, Li J, Gao N, Fan T, Li Y, Emerging Role of Exosomes in Liquid Biopsy for Monitoring Prostate Cancer Invasion and Metastasis, Front Cell Dev. Biol. 9 (2021) 679527, 10.3389/fcell.2021.679527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tukachinsky H, Madison RW, Chung JH, et al. , Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms, Clin. Cancer Res 27 (11) (2021) 3094–3105, 10.1158/1078-0432.CCR-20-4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen E, Cario CL, Leong L, et al. , Cell-free DNA concentration and fragment size as a biomarker for prostate cancer, Sci. Rep. 11 (1) (2021) 5040, 10.1038/s41598-021-84507-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Volik S, Alcaide M, Morin RD, Collins C, Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies, Mol. Cancer Res 14 (10) (2016) 898–908, 10.1158/1541-7786.MCR-16-0044 [DOI] [PubMed] [Google Scholar]
- [8].Cancer Genome Atlas Research N, The molecular taxonomy of primary prostate cancer, Cell 163 (4) (2015) 1011–1025, 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dong J, Chen JF, Smalley M, et al. , Nanostructured substrates for detection and characterization of circulating rare cells: from materials research to clinical applications, Adv. Mater. 32 (1) (2020) e1903663, 10.1002/adma.201903663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Miyamoto DT, Zheng Y, Wittner BS, et al. , RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance, Science 349 (6254) (2015) 1351–1356, 10.1126/science.aab0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].van Niel G, D’Angelo G, Raposo G, Shedding light on the cell biology of extracellular vesicles, Nat. Rev. Mol. Cell Biol. 19 (4) (2018) 213–228, 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- [12].Nanou A, Miller MC, Zeune LL, et al. , Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival, Br. J. Cancer 122 (6) (2020) 801–811, 10.1038/s41416-019-0726-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z, miRNA in plasma exosome is stable under different storage conditions, Molecules 19 (2) (2014) 1568–1575, 10.3390/molecules19021568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shtivelman E, Beer TM, Evans CP, Molecular pathways and targets in prostate cancer, Oncotarget 5 (17) (2014) 7217–7259, 10.18632/oncotarget.2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ferguson S, Weissleder R, Modeling EV Kinetics for Use in Early Cancer Detection, Adv. Biosyst. 4 (12) (2020) e1900305, 10.1002/adbi.201900305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Han Z, Wan F, Deng J, et al. , Ultrasensitive detection of mRNA in extracellular vesicles using DNA tetrahedron-based thermophoretic assay, Nano Today (2021) 38, 10.1016/j.nantod.2021.101203 [DOI] [Google Scholar]
- [17].Deng J, Zhao S, Li J, et al. , One-Step Thermophoretic AND Gate Operation on Extracellular Vesicles Improves Diagnosis of Prostate Cancer, Angew. Chem. (2022), 10.1002/ange.202207037 [DOI] [PubMed] [Google Scholar]
- [18].Sun N, Lee YT, Zhang RY, et al. , Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring, Nat. Commun. 11 (1) (2020) 4489, 10.1038/s41467-020-18311-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu X, Wu W, Cui D, Chen X, Li W, Functional Micro-/Nanomaterials for Multiplexed Biodetection, Adv. Mater. 33 (30) (2021) e2004734, 10.1002/adma.202004734 [DOI] [PubMed] [Google Scholar]
- [20].Zou D, Wu W, Zhang J, et al. , Multiplex detection of miRNAs based on aggregation-induced emission luminogen encoded microspheres, RSC Adv. 9 (68) (2019) 39976–39985, 10.1039/c9ra07680h [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yu Z, Lin S, Xia F, et al. , ExoSD chips for high-purity immunomagnetic separation and high-sensitivity detection of gastric cancer cell-derived exosomes, Biosens. Bioelectron. 194 (2021) 113594, 10.1016/j.bios.2021.113594 [DOI] [PubMed] [Google Scholar]
- [22].Went PT, Lugli A, Meier S, et al. , Frequent EpCam protein expression in human carcinomas, Hum. Pathol. 35 (1) (2004) 122–128, 10.1016/j.humpath.2003.08.026 [DOI] [PubMed] [Google Scholar]
- [23].Spizzo G, Fong D, Wurm M, et al. , EpCAM expression in primary tumour tissues and metastases: an immunohistochemical analysis, J. Clin. Pathol. 64 (5) (2011) 415–420, 10.1136/jcp.2011.090274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ross JS, Sheehan CE, Fisher HA, et al. , Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer, Clin. Cancer Res 9 (17) (2003) 6357–6362. [PubMed] [Google Scholar]
- [25].Bostwick DG, Pacelli A, Blute M, Roche P, Murphy GP, Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma, Cancer 82 (11) (1998) 2256–2261, [DOI] [PubMed] [Google Scholar]
- [26].Thery C, Witwer KW, Aikawa E, et al. , Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines, J. Extra Vesicles 7 (1) (2018) 1535ṣ750, 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Uhlen M, Zhang C, Lee S, et al. , A pathology atlas of the human cancer transcriptome, Science 357 (6352) (2017), 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- [28].Human Protein Atlas available from 〈http://www.proteinatlas.org〉. Accessed Aug 11, 2021.
- [29].Miyamoto DT, Lee RJ, Kalinich M, et al. , An RNA-based digital circulating tumor cell signature is predictive of drug response and early dissemination in prostate cancer, Cancer Disco 8 (3) (2018) 288–303, 10.1158/2159-8290.CD-16-1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nanou A, Zeune LL, Terstappen L, Leukocyte-derived extracellular vesicles in blood WITH AND Without EpCAM Enrichment, Cells 8 (8) (2019), 10.3390/cells8080937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Barretina J, Caponigro G, Stransky N, et al. , The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity, Nature 483 (7391) (2012) 603–607, 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Novershtern N, Subramanian A, Lawton LN, et al. , Densely interconnected transcriptional circuits control cell states in human hematopoiesis, Cell 144 (2) (2011) 296–309, 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Levine DM, Haynor DR, Castle JC, et al. , Pathway and gene-set activation measurement from mRNA expression data: the tissue distribution of human pathways, Genome Biol. 7 (10) (2006) R93, 10.1186/gb-2006-7-10-r93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Barry MJ, Clinical practice. Prostate-specific-antigen testing for early diagnosis of prostate cancer, N. Engl. J. Med 344 (18) (2001) 1373–1377, 10.1056/NEJM200105033441806 [DOI] [PubMed] [Google Scholar]
- [35].Thomsen FB, Westerberg M, Garmo H, et al. , Prediction of metastatic prostate cancer by prostate-specific antigen in combination with T stage and Gleason Grade: Nationwide, population-based register study, PLoS One 15 (1) (2020) e0228447, 10.1371/journal.pone.0228447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lee AK, D’Amico AV, Utility of prostate-specific antigen kinetics in addition to clinical factors in the selection of patients for salvage local therapy, J. Clin. Oncol. 23 (32) (2005) 8192–8197, 10.1200/JCO.2005.03.0007 [DOI] [PubMed] [Google Scholar]
- [37].Nadler RB, Humphrey PA, Smith DS, Catalona WJ, Ratliff TL, Effect of inflammation and benign prostatic hyperplasia on elevated serum prostate specific antigen levels, J. Urol. 154 (2 Pt 1) (1995) 407–413, 10.1097/00005392-199508000-00023 [DOI] [PubMed] [Google Scholar]
- [38].Schwartz LH, LaTrenta LR, Bonaccio E, Kelly WK, Scher HI, Panicek DM, Small cell and anaplastic prostate cancer: correlation between CT findings and prostate-specific antigen level, Radiology 208 (3) (1998) 735–738, 10.1148/radiology.208.3.9722854 [DOI] [PubMed] [Google Scholar]
- [39].Donovan MJ, Noerholm M, Bentink S, et al. , A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result, Prostate Cancer Prostatic Dis. 18 (4) (2015) 370–375, 10.1038/pcan.2015.40 [DOI] [PubMed] [Google Scholar]
- [40].McKiernan J, Donovan MJ, O’Neill V, et al. , A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy, JAMA Oncol. 2 (7) (2016) 882–889, 10.1001/jamaoncol.2016.0097 [DOI] [PubMed] [Google Scholar]
- [41].Del Re M, Biasco E, Crucitta S, et al. , The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients, Eur. Urol. 71 (4) (2017) 680–687, 10.1016/j.eururo.2016.08.012 [DOI] [PubMed] [Google Scholar]
- [42].Del Re M, Crucitta S, Sbrana A, et al. , AR-V7 and AR-FL expression is associated with clinical outcome: a translational study in patients with castrate resistant prostate cancer, BJU Int. (2019), 10.1111/bju.14792 [DOI] [PubMed] [Google Scholar]
- [43].Joncas FH, Lucien F, Rouleau M, et al. , Plasma extracellular vesicles as phenotypic biomarkers in prostate cancer patients, Prostate 79 (15) (2019) 1767–1776, 10.1002/pros.23901 [DOI] [PubMed] [Google Scholar]
- [44].Chen S, Huang V, Xu X, et al. , Widespread and functional RNA circularization in localized prostate cancer, e22, Cell 176 (4) (2019) 831–843, 10.1016/j.cell.2019.01.025 [DOI] [PubMed] [Google Scholar]
- [45].Pinheiro LB, Coleman VA, Hindson CM, et al. , Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification, Anal. Chem. 84 (2) (2012) 1003–1011, 10.1021/ac202578x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Biggs CN, Siddiqui KM, Al-Zahrani AA, et al. , Prostate extracellular vesicles in patient plasma as a liquid biopsy platform for prostate cancer using nanoscale flow cytometry, Oncotarget 7 (8) (2016) 8839–8849, 10.18632/oncotarget.6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Whiteland H, Spencer-Harty S, Morgan C, et al. , A role for STEAP2 in prostate cancer progression, Clin. Exp. Metastas-.-. 31 (8) (2014) 909–920, 10.1007/s10585-014-9679-9 [DOI] [PubMed] [Google Scholar]
- [48].Conway RE, Rojas C, Alt J, et al. , Prostate-specific membrane antigen (PSMA)-mediated laminin proteolysis generates a pro-angiogenic peptide, Angiogenesis 19 (4) (2016) 487–500, 10.1007/s10456-016-9521-x [DOI] [PubMed] [Google Scholar]
- [49].Burnell SEA, Spencer-Harty S, Howarth S, et al. , STEAP2 knockdown reduces the invasive potential of prostate cancer cells, Sci. Rep. 8 (1) (2018) 6252, 10.1038/s41598-018-24655-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Stanbrough M, Bubley GJ, Ross K, et al. , Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer, Cancer Res 66 (5) (2006) 2815–2825, 10.1158/0008-5472.CAN-05-4000 [DOI] [PubMed] [Google Scholar]
- [51].Wang B, Zhang S, Meng J, et al. , Evaporation-Induced rGO coatings for highly sensitive and non-invasive diagnosis of prostate cancer in the PSA gray zone, Adv. Mater. 33 (40) (2021) e2103999, 10.1002/adma.202103999 [DOI] [PubMed] [Google Scholar]
- [52].Zarnowski R, Noll A, Chevrette MG, et al. , Coordination of fungal biofilm development by extracellular vesicle cargo, Nat. Commun. 12 (1) (2021) 6235, 10.1038/s41467-021-26525-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Min L, Wang B, Bao H, et al. , Advanced nanotechnologies for extracellular vesicle-based liquid biopsy, Adv. Sci. (Weinh. ) 8 (20) (2021) e2102789, 10.1002/advs.202102789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Blackman ML, Royzen M, Fox JM, Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity, J. Am. Chem. Soc. 130 (41) (2008) 13518–13519, 10.1021/ja8053805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Smith EA, Wanat MJ, Cheng Y, Barreira SVP, Frutos AG, Corn RM, Formation, spectroscopic characterization, and application of sulfhydryl-terminated alkanethiol monolayers for the chemical attachment of DNA onto gold surfaces, Langmuir 17 (8) (2001) 2502–2507, 10.1021/la001064q [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


