Abstract
The coronavirus disease 2019 (COVID-19) is a highly contagious and fatal disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In general, the diagnostic tests for COVID-19 are based on the detection of nucleic acid, antibodies, and protein. Among different analytes, the gold standard of the COVID-19 test is the viral nucleic acid detection performed by the quantitative reverse transcription polymerase chain reaction (qRT-PCR) method. However, the gold standard test is time-consuming and requires expensive instrumentation, as well as trained personnel. Herein, we report an ultrasensitive electrochemical biosensor based on zinc sulfide/graphene (ZnS/graphene) nanocomposite for rapid and direct nucleic acid detection of SARS-CoV-2. We demonstrated a simple one-step route for manufacturing ZnS/graphene by employing an ultrafast (90 s) microwave-based non-equilibrium heating approach. The biosensor assay involves the hybridization of target DNA or RNA samples with probes that are immersed into a redox active electrolyte, which are detectable by electrochemical measurements. In this study, we have performed the tests for synthetic DNA samples and, SARS-CoV-2 standard samples. Experimental results revealed that the proposed biosensor could detect low concentrations of all different SARS-CoV-2 samples, using such as S, ORF 1a, and ORF 1b gene sequences as targets. This microwave-synthesized ZnS/graphene-based biosensor could be reliably used as an on-site, real-time, and rapid diagnostic test for COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42114-023-00630-7.
Keywords: DNA biosensor, COVID-19, Microwave, Graphene, Zinc sulfide, Nanocomposite
Introduction
The coronavirus disease 2019 (COVID-19) has been striking the world in the past 2 years [1–5]. People who got infected will have various symptoms including fever, cough, headache, loss of smell, loss of taste, fatigue, and breathing difficulties [6–8]. The major diagnostic methods to detect the disease include computed tomography (CT) scan, nucleic acid amplification test (NAAT), and serological techniques [9]. Today, NAAT using the quantitative reverse transcription polymerase chain reaction (qRT-PCR) is the most widely used method for detecting the COVID-19. However, the qRT-PCR is a time-consuming and expensive test which also requires trained personnel. Therefore, the development of highly sensitive and quick detection methods is urgently needed. And the electrochemical DNA biosensors have received a lot of attraction for being a good detecting device [10–13].
Biosensor is an analytical device detecting biological or chemical reactions and then generating reliable data for several fields such as gene analysis [12, 14], food safety [15, 16], disease diagnosis [17], environmental monitoring [18], drug discovery [19], and water quality screening [20]. A typical biosensor consists of biological receptor, transducer, amplification, and signal processing. Based on different transducers, biosensors can be divided into electrochemical, optical, thermal, electronic, acoustic, and mass-based biosensor [21]. Among different transducers, the electrochemical biosensor is the most widely studied and applied biosensor due to the advantages of low-cost, simplicity, portability, sensitivity, and rapid detection [14, 22]. It is critical to develop an appropriate composite for the signal amplification platform.
In practice, materials such as metals, metal oxides, graphene, conducting polymer, and their composite have been studied as signal amplifier for the electrochemical biosensors [23, 24]. For instance, Saber et al. fabricated a gold electrode decorated by gold nanorods (GNRs) for detecting hepatitis B virus [25]. Due to the large surface area of the GNRs, the modified gold electrode can assemble more probe DNA and exhibit higher sensitivity with a detection limit of 2 × 10−12 M. Similarly, a hybrid nanocomposite containing copper oxide nanowires and carboxyl-functionalized single-walled carbon nanotubes was developed by Chen et al. for detecting the target DNA [26]. The performance of the modified electrode was benefited by the nanocomposite which provides a large surface area, fast electron transfer, and good electrical conductivity with the limit detection of 3.5 × 10−15 M. Graphene also shows a great potential in DNA detection due to its astonishing properties in electrical, optical, mechanical, and chemical aspects [27]. Pumera et al. reported a graphene-based electrode immobilized with hairpin-shaped probe DNA for detecting Alzheimer’s disease [28]. The triple and quadruple layer graphene electrodes presented the best sensitivity with the detection limit of 6.6 × 10−12 M. All cases above show that the modified electrode provides a larger surface area and low charge transfer resistance for superior sensitivity compared to the raw electrode.
In the detection of COVID-19, the antibody testing, also known as serology testing, can check the antibodies to COVID-19 after a person has been infected or vaccinated. For instance, a paper-based biosensor modified by ZnO nanowires showed enhanced sensitivity for detecting antibody of COVID-19 with human serum samples [29]. In addition, Seo et al. reported a graphene-based biosensor functionalized with SARS-CoV-2 spike antibody as a receptor which demonstrated highly sensing detection for the target SARS-CoV-2 antigen protein with a detection limit of 1 × 10−15 g/mL [30]. On the other hand, the molecular test detects the virus’s genetic material to determine if a person has an active infection of COVID-19. Song et al. utilized polyaniline nanowires to modify the glassy carbon electrode for the detection of the COVID-19 N gene, resulting in a detection limit of 3.5 × 10−15 M [31]. This demonstrated that the electrochemical biosensor has the potential as a detection method for COVID-19. In many cases, the process of making the composite takes multiple steps with long preparation time and high cost. Therefore, developing a quick and one-step process is in need.
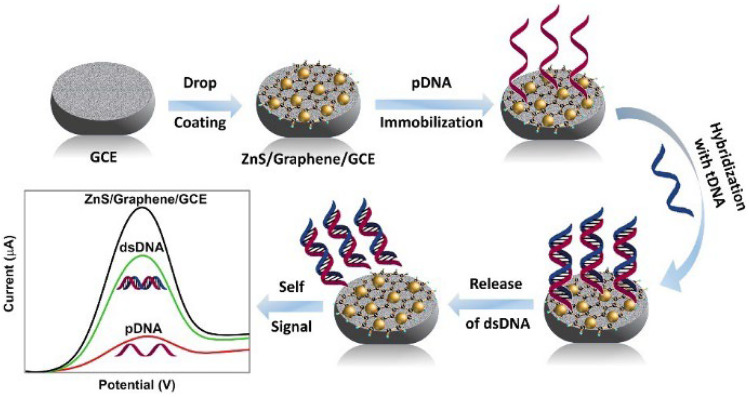
In this paper, we present a one-step and ultrafast microwave-initiated manufacturing process to prepare the electrical interface containing zinc sulfate and graphene (ZnS/graphene). ZnS has attracted a lot of interest due to their superior chemical, physical, and electrochemical properties [32, 33]. For example, a system with urease and CdSe/ZnS quantum dots (QDs) had been developed for the detection of urea by Huang et al. [34]. By the enhancement of QD photoluminescence, the novel system can analyze the urea concentration ranging from 0.01 to 100 mM with good sensitivity, and no enzyme immobilization is required. In addition, Rajesh et al. studied the electrical and DNA sensing properties of ZnS decorated single-walled carbon nanotube (SWNT) to investigate the sensing mechanism by measuring the transfer characteristics of I-V and field-effect transistors (FET) [35]. The result demonstrated that the improvement of the sensitivity is attributed to the large surface-to-volume ratio and the superior charge transfer ability of the ZnS. The graphene which accelerates the reaction rate by absorbing the electromagnetic energy acts as a heat resource during the microwave preparation. Besides, graphene has been applied for different biosensors due to its superior sensing performance including good conductivity, large surface area, excellent thermal conductivity, and mechanical strength [36–39]. Compared with the conventional heating method under thermodynamic equilibrium conditions, the microwave reaction method under non-equilibrium conditions can provide low-cost and time-saving process [40–42]. Most importantly, the ZnS/graphene DNA biosensor provides highly sensitivity. Different ratios of zinc precursors were investigated to obtain the optimized conditions and compared with commercial ZnS mixed with graphene without microwave reaction. The obtained ZnS/graphene sample was drop cast onto a glassy carbon electrode (GCE) surface for further electrochemical experiment. The probe DNA (pDNA) was successfully immobilized with GCE and then hybridized with different concentrations of target DNA (tDNA) to perform self-signal regeneration during the releasing process (Scheme 1). Three-electrode system was utilized while the ZnS/graphene modified electrode acted as the working electrode. The performance of the modified electrode was characterized by cyclic voltammetry (CV) and differential pulse voltammetry (DPV).
Scheme 1.
Schematic illustration of detecting the SARS-CoV-2 by microwave-synthesized ZnS/graphene nanocomposite
Methodology
Materials
Zinc acetate (GR, 99.5% purity) was obtained from BeanTown Chemical Inc. Graphene substrate was obtained from Magnolia Ridge Inc. Sulfur powder (GR, 99.5% purity) was provided by Alfa Aesar, America. The detection samples included artificial DNA probes, artificial DNA targets, and SARS-CoV-2 standard were obtained from Eurofins Genomics LLC (Louisville, Kentucky).
Preparation of ZnS/graphene composites
For the preparation of ZnS/graphene nanocomposites, ZnAc2.2H2O, graphene, and sulfur powder were transferred into a 20-mL scintillation vial and mixed thoroughly to get a homogeneous mixture. The precursor ratios and synthesis parameters are described in Table S1. Subsequently, the vial was placed inside microwave oven and irradiated with a high-power level (∼ 1250 W) for 90 s. After the vial was cooled down to room temperature, the obtained powder was taken out. For comparison, the mixture of graphene and commercial ZnS nanoparticles were also prepared under the same microwave experimental conditions.
The surface of glassy carbon electrode (GCE) was sequentially polished with 0.3- and 0.05-mm alumina slurries, then immersed in Piranha solution (conc. H2SO4:30% H2O2; 3:1) for 30 min, followed by rinsing with DI water, and vacuum dried at 60 ºC. To prepare the coating paste, 100 mg of hybrid powder was suspended in 5 mL of isopropyl alcohol. Then, 0.1 mL of Nafion was added into the suspension following 20 min of sonication. Finally, 20 μL of the suspension was added to the prepared GCE surface and vacuum dried at 60 ºC.
Nanocomposite-based DNA biosensor probe and target sequences
The entire genetic sequence of SARS-CoV-2 was uploaded to the Global Initiative on Sharing All Influenza Data (GISAID) platform on January 10, 2020. The sequence data was used as reference for the design of oligos/probes needed for the development of SARS-CoV-2-specific testing. As shown in Table 1, the probe and target DNA oligonucleotides were complementary to the specific target gene sequences and purchased from Eurofin Genomics.
Table 1.
List of probe and target DNA sequences
| Gene (s) | Synthetic base-pairs of DNA templates |
|---|---|
| S |
Probe GACATGTATAGCATGGAACC Target GGTTCCATGCTATACATGTCTC |
| ORF 1b |
Probe CTTTAATGTTTTATTCTCTA Target TAGAGAATAAAACATTAAAG |
| Mis-matched negative control |
Probe TTCACAAGTGCCGTGCCTAC Target GGGTACTTAACAATGATTATT |
Immobilization and hybridization of DNA
A 20 μL of probe-DNA solution (1.0 × 10−6 M pDNA) was added dropwise to the surface of modified GCE and dried at temperature below the melting Tm (temperature of melting) of corresponding probe sequences. After pDNA immobilization, following by rinsing with DI water, the GCE was dried again. Then, 20 μL of target DNA (tDNA) was added onto the pDNA-modified GCE surface, and the corresponding electrode was dried at room temperature to form the hybrid double-stranded DNA (dsDNA). Subsequently, the electrode was kept at − 0.5 V for 300 s in 1.0 M KCl solution containing 0.2 M K3[Fe(CN)6] to release the dsDNA into the electrolyte. After rinsing the surface of GCE with DI water, the electrode was ready for electrochemical tests.
Material characterization
The surface morphology of the ZnS/graphene nanocomposite was characterized using scanning electron microscope (SEM) on JEOL JSM-7000F coupled with an energy dispersive X-ray spectrometer (EDS, EDAX Instruments). The crystalline phase characteristics of the microwave samples were examined using the X-ray diffraction (XRD) (Philips X’pert MPD diffractometer) under Cu Kα radiation (λ = 1.5405 Å) at 2θ range of 20 − 70°.
Electrochemical measurements
All the electrochemical tests were conducted in a mixture of 1.0 M KCl and 0.2 M K3[Fe(CN)6] at room temperature. Cyclic voltammetry (CV) experiments were carried out at a scan rate of 100 mV s−1 from − 0.4 to 0.8 V for different ZnS/graphene composites. The differential pulse voltammetry (DPV) tests were taken by using parameters as follows: pulse amplitude, 0.05 V; pulse width, 0.05 s; and pulse period, 0.5 s. The peak current difference (ΔIp) was calculated according to the relation of ΔIp = I − I0, where I0 represents the peak current for the modified electrodes immobilized with pDNA and I represents the peak current for the resulting dsDNA after being released into the electrolyte. The calibration plot, ΔIp vs. − log C was drawn by calculating the negative logarithmic values of target samples concentrations, where C is the concentration of target sample. Furthermore, the LOD (limit of detection) is measured using the following equation:
| 1 |
In Eq. (1), the standard deviation of blank solution was measured by running the DPV tests on pDNA-modified GCE for 10 times.
Results and discussion
Optimizing the precursor ratio
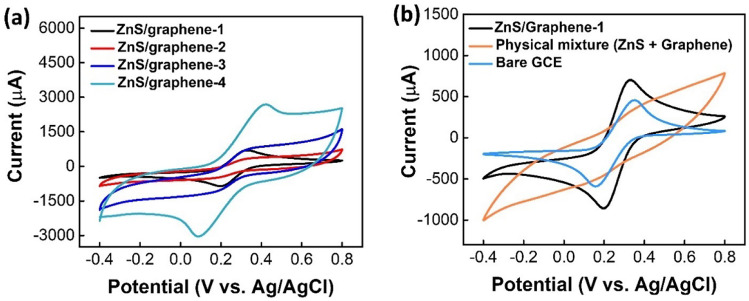
The electrochemical properties of different modified electrodes with different mass of precursors were investigated by CV technique. Among the four different mass ratios (see Table S1), as displayed in Fig. 1a, ZnS/graphene-1 has the prominent redox peaks with the anodic peak to cathodic peak separation of potential, ΔEp (ΔEp = Eanodic peak − Ecathodic peak) of 130 mV. For the samples of ZnS/graphene-2 and ZnS/graphene-3, the redox peaks are not prominent and may be due to the incomplete reaction taking place during the microwave-assisted synthesis with the less amount of Zn-precursor. In addition, although ZnS/graphene-4 provides much higher current due to the presence of high amount of conductive graphene, the ΔEp is as much as 330 mV, which indicates a sluggish kinetic transfer process of electrons.
Fig. 1.
a CVs of different electrodes in a solution containing 1.0 M KCl and 0.2 M K3[Fe(CN)6] as a redox probe at scan rate of 100 mVs−1. Zn(Ac)2⋅2H2O, graphene, and S precursors have a mass ratio of 6:1:1, 4:1:1, 2:1:1, and 6:2:1, respectively. b CV curves of different electrodes in a solution containing 1.0 M KCl and 0.2 M K3[Fe(CN)6]
Moreover, the electrochemical property of ZnS/graphene-1 was compared with a physical mixture of ZnS and graphene with a mass ratio of 6:1, and with the bare GCE. From Fig. 1b, it can be clearly observed that the physical mixture does not show prominent redox peaks. For ZnS/graphene-1 and bare GCE, ΔEp values are 130 mV and 190 mV, respectively. Moreover, the ZnS/graphene-1 yields the maximum peak current, possibly because of the accelerated [Fe(CN)6]3− diffusion towards the surface by a layer of graphene. Apparently, the ZnS/graphene-1 sample exhibited much higher electrochemical activity and faster electron-transfer kinetics (reflected in the lower peak to peak separation) compared to all other modified electrodes including bare GCE. Therefore, for further studies, the detection of DNA samples was carried out on the surface of ZnS/graphene-1 modified GCE which was denoted as ZnS/graphene/GCE thereafter.
Characterization of the modified electrode
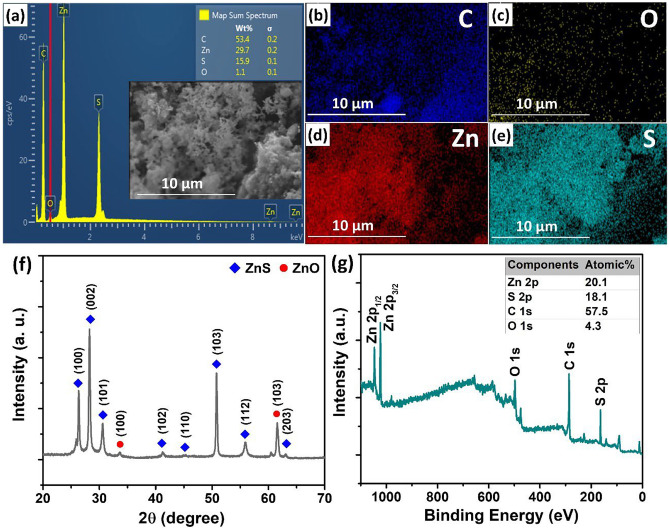
The morphology and distribution of the ZnS/graphene composite are shown in Fig. 2a − e. The SEM image was utilized to characterize the prepared ZnS/graphene composite (Fig. 2a). It can clearly be seen that the graphene substrate was fully decorated by ZnS. The EDS spectrum of ZnS/graphene exhibited the existence of the Zn and S elements. Figure 2b − e illustrated the EDS elemental mapping of ZnS/graphene composite which clearly showed the evenly dispersed elements of Zn and S on the graphene surface. In addition, the C, Zn, and S components were closely stacked, indicating the formation of the hybrid composite. The XRD diffractogram of ZnS/graphene is shown in Fig. 2f. The strong peaks appeared at 2θ around 26.68°, 28.56°, 30.86°, and 51.1° are assigned to (100), (002), (101), and (103) reflections of hexagonal wurtzite ZnS (JCPDS 36–1450), respectively. The overall XPS spectrum for the ZnS/graphene composite is shown in Fig. 2g. The spectrum showed the presence of C 1 s, O 1 s, Zn 2p, and S 2p peaks. The characterized results confirmed the successful synthesis of ZnS/graphene composite. Since microwave-initiated synthesis does not have any inert gas protection during the heating process, the EDS spectrum showed the presence of ZnO. However, the ZnO does not contribute much to DNA detection. The CV and DPV comparison between ZnO and ZnS have been discussed and shown in Fig. S1.
Fig. 2.
a SEM image and EDS spectrum; b − e the EDS elemental mapping; f XRD pattern and g XPS spectrum of ZnS/graphene composite
Detection of synthetic target DNA samples
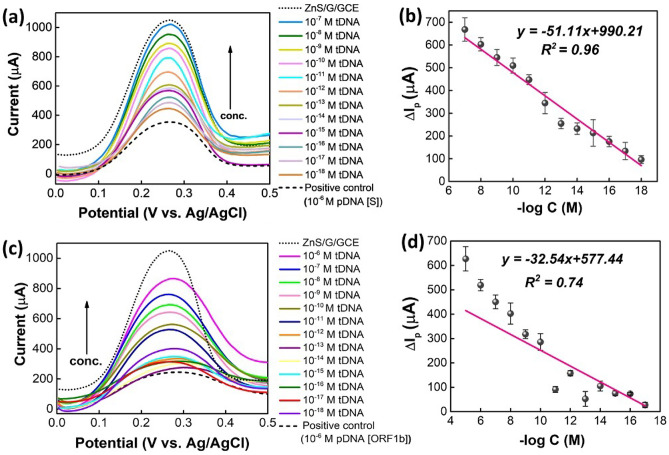
To explore the analytical characteristics of the optimized ZnS/graphene/GCE for the detection of synthetic gene DNA sequences, DPVs were performed in an electrolyte solution of 1.0 M KCl containing 0.2 M K3[Fe(CN)6]. Initially, the sensor electrode of ZnS/graphene/GCE was attached with the 1.0 × 10−6 M probe DNA (pDNA) and treated for different concentrations of target DNA (tDNA) solutions as depicted in Fig. 3a. After immobilizing the pDNA on the surface of ZnS/graphene/GCE, the oxidation or anodic peak current (Ipa) decreases significantly because the nonconductive pDNA blocks the effective electron transfer through the coating surface. However, when the pDNA was hybridized with the tDNA, the peak currents were enhanced linearly with the enhancing concentrations of tDNA from 1.0 × 10−18 M to 1.0 × 10−7 M. The release of the resulted dsDNA from the ZnS/graphene/GCE surface occurs because of hybridization with complementary targets, accompanied with the self-signal regeneration of the ZnS/graphene/GCE. With an increase in tDNA concentration, more of the pDNA can get hybridized and then release into the electrolyte. Therefore, the electron transfers as well as the peak currents increase. Furthermore, the regression calibration plot was drawn from ∆Ip vs. − log C with the correlation coefficient estimated as R2 = 0.96 as shown in Fig. 3b. Thus, the limit of detection (LOD = 3σ/slope) was determined, which was found to be 4.453 × 10−20 M.
Fig. 3.
a DPV plots and b the regression calibration plots (note: probe and target DNA samples are for S-gene). c DPV plots and d the regression calibration plots (Note: probe and complementary DNA samples are for ORF 1b-gene)
In addition, the DPV tests were performed for the ORF 1b-gene. As displayed in Fig. 3c, the results follow a similar trend like S-gene. However, at the lower concentrations of cDNA samples from 10−12 M to 10−18 M, results showed arbitrary currents, and therefore the regression coefficient is only 0.74 (Fig. 3d). Based on these findings, the LOD was measured to be 2.013 × 10−18 M.
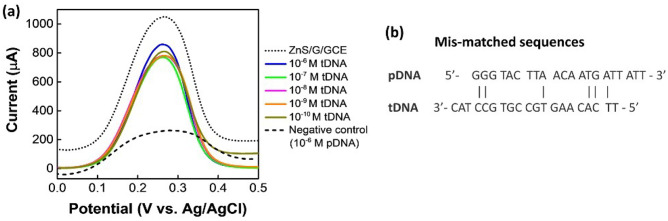
Following the same steps and procedures, DPV tests were also performed for mismatched negative control sample, as displayed in Fig. 4a. Since there is only a few of matched sequences present in these samples (Fig. 4b), the tDNA cannot hybridize successfully with the pDNA. Therefore, although after hybridization the currents get increased from the pDNA-modified electrode, there is no particular trend found for these samples. The matched sequence of S and ORF-1b genes are shown in Fig. S2.
Fig. 4.
a DPV plots of synthetic samples. b Mismatched sequences of pDNA and tDNA (Note: probe and target DNA samples are negative control)
Detection of SARS-CoV-2 standard samples
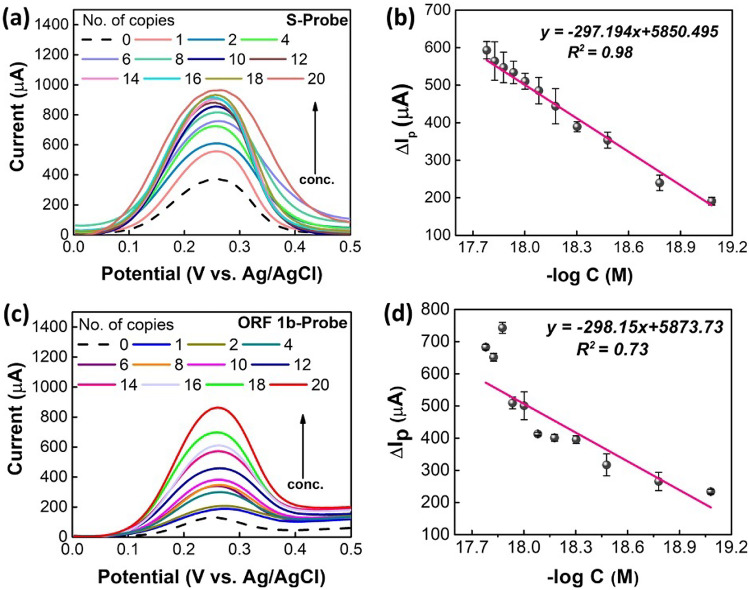
Following the similar DPV characterization technique, the detection of SARS-CoV-2 standard samples was performed. The positive control from TaqPath™ COVID-19 Fast PCR Combo Kit 2.0 was diluted to a working stock of 20 copies/µL on ice (ThermoFisher). This Fast PCR kit Positive Control consists of several different gene templates (200,000 copies/1 mL), such as S, N, and ORF1ab, which is widely used by CDC CLIA certified laboratories. Any of the probe templates targeting these genes can be used to perform the validation of the detection tests; therefore, we tested the S and ORF1b probes used in Fig. 2. As shown in Fig. 5a, S-probe can detect from 20 copies up to 1 copy. The corresponding concentrations of different number of copies are displayed in supporting information, Table S2. From these results, one thing is noticeable that the low concentrations at the range of 10−18 M to 10−20 M can be detected. Despite of the presence of all different genes in a standard solution, only a single probe template was used to detect the hybridized dsDNA. Hence, there is a huge chance of some target templates remain immobilized on the surface of ZnS/graphene/GCE even after rinsing with DI water, which eventually may hinder the electron transfer process resulting arbitrary change in peak currents. Similar to the results for synthetic base-pair DNA samples, the detection of SARS-CoV-2 standard sample also provides the similar trend of increase in peak currents by increasing the concentration of target samples. Moreover, the regression calibration plot (Fig. 5b) was drawn and LOD values were calculated to be 2.068 × 10−20 M (R2 = 0.98) for S-probes. These results clearly indicate a good performance from S-probe to detect the SARS-CoV-2 standard solutions.
Fig. 5.
a, b DPV and calibration plots for the detection of SARS-CoV-2 standard using S-probe. c, d DPV and calibration plots for the detection of SARS-CoV-2 standard solutions using ORF 1b-probe in a solution containing 1.0 M KCl and 0.2 M K3[Fe(CN)6]
Moreover, the SARS-CoV-2 standard solutions were also detected using the ORF 1b-probes. For ORF 1b-probe (Fig. 5c, d), the results followed the trend from 1 to 20 copies, which has superior sensitivity as RT-PCR detection method using the TaqPath™ COVID-19 Combo Kit.
Additionally, the performance of the DNA biosensor for the detection of SARS-CoV-2 was compared with previously published studies. According to Table 2, the proposed ZnS/graphene modified GCE achieved a lower detection limit which has 5 orders of magnitude less than the other reports.
Table 2.
Comparison of the designed biosensors for the detection of SARS-CoV-2
| Platform | Detection Method | LOD | Reference |
|---|---|---|---|
| Pt/Ti/glass wafer | Impedance | 1 × 10−8 M | [43] |
| Co-TNTs | Amperometry | 7 × 10−10 M | [44] |
| Graphene | SWV | 2.6 × 10−7 M | [45] |
| Au IDE | EIS | 3.9 × 10−16 M | [46] |
| CSPE/CNF-AuNP | EIS | 7 × 10−13 M | [47] |
| rGO-Au | EIS | 13 × 10−15 M | [48] |
| PANI/GCE | DPV | 3.5 × 10−15 M | [31] |
| MIP/Au-TFE | DPV | 15 × 10−15 M | [49] |
| AuNPs/FTO | DPV | 10 × 10−15 M | [50] |
| ZnS/graphene/GCE | DPV | 2.1 × 10−20 M | This work |
Co-TNTs cobalt functionalized TiO2 nanotubes, IDE interdigitated electrode, CSPE carbon-based screen-printed electrode, CNF carbon nanofibers, MIP molecular imprinted polymer, TFE thin film electrode, FTO fluorine doped tin oxide electrode
Investigating the detection of synthetic DNA samples by one-step hybridization
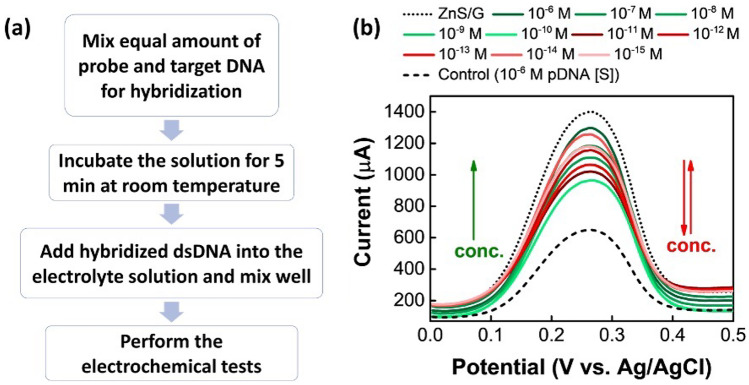
Besides investigating the detection of DNA samples following the multiple-step procedure, the one-step hybridization technique was also applied in this study. The major steps are displayed in Fig. 6a, where instead of drop coating the DNA samples onto the surface of ZnS/G-modified GCE, the probe and target DNA samples were directly mixed into the electrolyte solution. Subsequently, the DPV tests were performed using the ZnS/graphene/GCE as the working electrode. From the results displayed in Fig. 6b, at high concentration range from 1 × 10−6 M to 1 × 10−10 M of cDNA, the results showed the similar trend as before. However, the samples with low concentrations from 1 × 10−11 M to 1 × 10−15 M, the results are inconsistent. These results can be improved by handling the samples more carefully. Moreover, this one-step hybridization can save a lot of sample-preparation time comparing to the multiple-step procedure. Further optimizations, specifically for incubation time and temperature are still required to establish a stable detection method using one-step hybridization.
Fig. 6.
a Detection steps of synthetic DNA samples via one-step hybridization method. b DPV plots to detect S-gene by one-step method [note: probe and complementary DNA samples are for S-gene]
Conclusions
In conclusion, we report the construction of novel electrochemical sensing nanohybrid of ZnS/graphene for detecting SARS-CoV-2 nucleic acids by a one-pot and ultrafast microwave-controlled synthesis approach. The ZnS/graphene modified GCE exhibited promising electrochemical property and faster electron transfer kinetic which improved the detection performance. When the pDNA was immobilized on the surface of ZnS/graphene/GCE, the peak current decreased due to the obstruction of electron transfer process. Hybridization between the probe and target DNA caused the regeneration of the self-signal of ZnS/graphene. The peak current showed linear trend with decreasing the concentration of target DNA. Among different pDNA sequences, S-gene provides a better detection result when hybridized with target DNA or standard SARS-CoV-2 controls where the limits of detection are 4.453 × 10−20 M and 2.068 × 10−20 M, respectively. In addition, we presented a one-step hybridization to save the multiple-step procedure. A similar result was obtained only at high concentrations from 1 × 10−6 M to 1 × 10−10 M. The investigation of the one-step hybridization could be further studied. Overall, the microwave-synthesized composite of ZnS/graphene coating on the GCE electrode showed superior sensitivity and low-concentration detection limit. Our strategy and assays developed here could be useful for rapid virus infection screening, public health surveillance, and in the settings with limited laboratory resources and utilized by users with limited training. Our platform will enable robust, low-cost, multiplex, and quantitative DNA/RNA analysis tools for field-deployable detection devices.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
All authors contributed to the study conception and design. Etinosa Iyayi and Haseena Polk contributed to sample preparation. Material preparation, data collection, and analysis were performed by Shatila Sarwar and Carolina Amezaga. Zhen Wei performed the SEM, XPS, and XRD characterization. Both Shatila Sarwar and Mao-Chia Lin aided in interpreting the results and worked on the manuscript. Ruigang Wang, Honghe Wang, and Xinyu Zhang supervised the project. All authors read and approved the final manuscript.
Funding
This work is supported by the National Science Foundation Award (2039369) and Tuskegee University-Thermo Fisher Scientific COVID-19 testing initiative (Just Project).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Honghe Wang, Email: hwang@tuskegee.edu.
Xinyu Zhang, Email: xzz0004@auburn.edu.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaidai O, Xing Y (2022) A novel multi regional reliability method for COVID-19 death forecast. Eng Sci 1–16. 10.30919/es8d799
- 4.Gaidai O, Yihan Y (2022) A novel bio-system reliability approach for multi-state COVID-19 epidemic forecast. Eng Sci 1–18. 10.30919/es8d797
- 5.Chaudhary AP, Sonar NS, TR J, Banerjee M, Yadav S (2021) Impact of the COVID-19 pandemic on the mental health of college students in India: cross-sectional web-based study. JMIRx Med 2:e28158. 10.2196/28158 [DOI] [PMC free article] [PubMed]
- 6.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen K-Y, Chen R, Tang C, Wang T, Chen P, Xiang J, Li S, Wang J, Liang Z, Peng Y, Wei L, Liu Y, Hu Y, Peng P, Wang J, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Sun J, Lyu S, Lu Z, Li T, Yang Y, Li B, Han H, Wu B, Sun H, Li D, Huang J, Sun D. Poly(lactic acid)/artificially cultured diatom frustules nanofibrous membranes with fast and controllable degradation rates for air filtration. Adv Compos Hybrid Mater. 2022;5:1221–1232. doi: 10.1007/s42114-022-00474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rai P, Kumar BK, Deekshit VK, Karunasagar I, Karunasagar I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl Microbiol Biotechnol. 2021;105:441–455. doi: 10.1007/s00253-020-11061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu A, Wang K, Weng S, Lei Y, Lin L, Chen W, Lin X, Chen Y. Development of electrochemical DNA biosensors. TrAC Trends Anal Chem. 2012;37:101–111. doi: 10.1016/j.trac.2012.03.008. [DOI] [Google Scholar]
- 11.Cagnin S, Caraballo M, Guiducci C, Martini P, Ross M, SantaAna M, Danley D, West T, Lanfranchi G. Overview of electrochemical DNA biosensors: new approaches to detect the expression of life. Sensors. 2009;9:3122–3148. doi: 10.3390/s90403122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerman K, Kobayashi M, Tamiya E. Recent trends in electrochemical DNA biosensor technology. Meas Sci Technol. 2004;15:R1–R11. doi: 10.1088/0957-0233/15/2/R01. [DOI] [Google Scholar]
- 13.Odenthal KJ, Gooding JJ. An introduction to electrochemical DNA biosensors. Analyst. 2007;132:603–610. doi: 10.1039/b701816a. [DOI] [PubMed] [Google Scholar]
- 14.Sassolas A, Leca-Bouvier BD, Blum LJ. DNA biosensors and microarrays. Chem Rev. 2008;108:109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 15.Lv M, Liu Y, Geng J, Kou X, Xin Z, Yang D. Engineering nanomaterials-based biosensors for food safety detection. Biosens Bioelectron. 2018;106:122–128. doi: 10.1016/j.bios.2018.01.049. [DOI] [PubMed] [Google Scholar]
- 16.Mishra G, Barfidokht A, Tehrani F, Mishra R. Food safety analysis using electrochemical biosensors. Foods. 2018;7:141. doi: 10.3390/foods7090141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sin ML, Mach KE, Wong PK, Liao JC. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn. 2014;14:225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Justino CIL, Duarte AC, Rocha-Santos TAP. Recent progress in biosensors for environmental monitoring: a review. Sensors (Basel) 2017;17:2918. doi: 10.3390/s17122918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aydin EB, Aydin M, Sezginturk MK. Biosensors in drug discovery and drug analysis. Curr Anal Chem. 2019;15:467–484. doi: 10.2174/1573411014666180912131811. [DOI] [Google Scholar]
- 20.Hossain SMZ, Mansour N. Biosensors for on-line water quality monitoring–a review. Arab J Basic Appl Sci. 2019;26:502–518. doi: 10.1080/25765299.2019.1691434. [DOI] [Google Scholar]
- 21.Izyumskaya N, Tahira A, Ibupoto ZH, Lewinski N, Avrutin V, Özgür Ü, Topsakal E, Willander M, Morkoç H. Review—electrochemical biosensors based on ZnO nanostructures. ECS J Solid State Sci Technol. 2017;6:Q84–Q100. doi: 10.1149/2.0291708jss. [DOI] [Google Scholar]
- 22.Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 23.Rashid JIA, Yusof NA. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: a review. Sens Bio-Sensing Res. 2017;16:19–31. doi: 10.1016/j.sbsr.2017.09.001. [DOI] [Google Scholar]
- 24.Wang Y-H, He L-L, Huang K-J, Chen Y-X, Wang S-Y, Liu Z-H, Li D. Recent advances in nanomaterial-based electrochemical and optical sensing platforms for microRNA assays. Analyst. 2019;144:2849–2866. doi: 10.1039/c9an00081j. [DOI] [PubMed] [Google Scholar]
- 25.Shakoori Z, Salimian S, Kharrazi S, Adabi M, Saber R. Electrochemical DNA biosensor based on gold nanorods for detecting hepatitis B virus. Anal Bioanal Chem. 2015;407:455–461. doi: 10.1007/s00216-014-8303-9. [DOI] [PubMed] [Google Scholar]
- 26.Chen M, Hou C, Huo D, Yang M, Fa H. An ultrasensitive electrochemical DNA biosensor based on a copper oxide nanowires/single-walled carbon nanotubes nanocomposite. Appl Surf Sci. 2016;364:703–709. doi: 10.1016/j.apsusc.2015.12.203. [DOI] [Google Scholar]
- 27.Wu X, Mu F, Wang Y, Zhao H. Graphene and graphene-based nanomaterials for DNA detection: a review. Molecules. 2018;23:2050. doi: 10.3390/molecules23082050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonanni A, Pumera M. Graphene platform for hairpin-DNA-based impedimetric genosensing. ACS Nano. 2011;5:2356–2361. doi: 10.1021/nn200091p. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Qin Z, Fu H, Li T, Peng R, Li Z, Rini JM, Liu X (2021) Enhancing the performance of paper-based electrochemical impedance spectroscopy nanobiosensors: an experimental approach. Biosens Bioelectron 177:112672. 10.1016/j.bios.2020.112672 [DOI] [PMC free article] [PubMed]
- 30.Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, Lee C-S, Jun S, Park D, Kim HG, Kim S-J, Lee J-O, Kim BT, Park EC, Il KS. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 31.Song Z, Ma Y, Chen M, Ambrosi A, Ding C, Luo X. Electrochemical biosensor with enhanced antifouling capability for COVID-19 nucleic acid detection in complex biological media. Anal Chem. 2021;93:5963–5971. doi: 10.1021/acs.analchem.1c00724. [DOI] [PubMed] [Google Scholar]
- 32.Fang X, Zhai T, Gautam UK, Li L, Wu L, Bando Y, Golberg D. ZnS nanostructures: from synthesis to applications. Prog Mater Sci. 2011;56:175–287. doi: 10.1016/j.pmatsci.2010.10.001. [DOI] [Google Scholar]
- 33.Wang X, Huang H, Liang B, Liu Z, Chen D, Shen G. ZnS nanostructures: synthesis, properties, and applications. Crit Rev Solid State Mater Sci. 2013;38:57–90. doi: 10.1080/10408436.2012.736887. [DOI] [Google Scholar]
- 34.Huang C-P, Li Y-K, Chen T-M. A highly sensitive system for urea detection by using CdSe/ZnS core-shell quantum dots. Biosens Bioelectron. 2007;22:1835–1838. doi: 10.1016/j.bios.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Rajesh, Das BK, Srinives S, Mulchandani A (2011) ZnS nanocrystals decorated single-walled carbon nanotube based chemiresistive label-free DNA sensor. Appl Phys Lett 98:013701. 10.1063/1.3529951 [DOI] [PMC free article] [PubMed]
- 36.Bai Y, Xu T, Zhang X. Graphene-based biosensors for detection of biomarkers. Micromachines. 2020;11:60. doi: 10.3390/mi11010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan K, Gu J. Ordered alignment of liquid crystalline graphene fluoride for significantly enhancing thermal conductivities of liquid crystalline polyimide composite films. Macromolecules. 2022;55:4134–4145. doi: 10.1021/acs.macromol.2c00491. [DOI] [Google Scholar]
- 38.Yang X, Fan S, Li Y, Guo Y, Li Y, Ruan K, Zhang S, Zhang J, Kong J, Gu J (2020) Synchronously improved electromagnetic interference shielding and thermal conductivity for epoxy nanocomposites by constructing 3D copper nanowires/thermally annealed graphene aerogel framework. Compos Part A Appl Sci Manuf 128:105670. 10.1016/j.compositesa.2019.105670
- 39.Wang S, Feng D, Guan H, Guo Y, Liu X, Yan C, Zhang L, Gu J (2022) Highly efficient thermal conductivity of polydimethylsiloxane composites via introducing “Line-Plane”-like hetero-structured fillers. Compos Part A Appl Sci Manuf 157:106911. 10.1016/j.compositesa.2022.106911
- 40.Zhang X, Liu Z. Recent advances in microwave initiated synthesis of nanocarbon materials. Nanoscale. 2012;4:707–714. doi: 10.1039/C2NR11603K. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z, Zhang L, Poyraz S, Smith J, Kushvaha V, Tippur H, Zhang X. An ultrafast microwave approach towards multi-component and multi-dimensional nanomaterials. RSC Adv. 2014;4:9308. doi: 10.1039/c3ra47086e. [DOI] [Google Scholar]
- 42.Meng F, Wang H, Huang F, Guo Y, Wang Z, Hui D, Zhou Z. Graphene-based microwave absorbing composites: a review and prospective. Compos Part B Eng. 2018;137:260–277. doi: 10.1016/j.compositesb.2017.11.023. [DOI] [Google Scholar]
- 43.Hwang C, Park N, Kim ES, Kim M, Kim SD, Park S, Kim NY, Kim JH (2021) Ultra-fast and recyclable DNA biosensor for point-of-care detection of SARS-CoV-2 (COVID-19). Biosens Bioelectron 185:113177. 10.1016/j.bios.2021.113177 [DOI] [PMC free article] [PubMed]
- 44.Vadlamani BS, Uppal T, Verma SC, Misra M. Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors (Basel) 2020;20:5871. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mojsoska B, Larsen S, Olsen DA, Madsen JS, Brandslund I, Alatraktchi FA. Rapid SARS-CoV-2 detection using electrochemical immunosensor. Sensors. 2021;21:390. doi: 10.3390/s21020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramanathan S, Gopinath SCB, Ismail ZH, Md Arshad MK, Poopalan P (2022) Aptasensing nucleocapsid protein on nanodiamond assembled gold interdigitated electrodes for impedimetric SARS-CoV-2 infectious disease assessment. Biosens Bioelectron 197:113735. 10.1016/j.bios.2021.113735 [DOI] [PMC free article] [PubMed]
- 47.Amouzadeh Tabrizi M, Acedo P. An electrochemical impedance spectroscopy-based aptasensor for the determination of SARS-CoV-2-RBD using a carbon nanofiber–gold nanocomposite modified screen-printed electrode. Biosensors. 2022;12:142. doi: 10.3390/bios12030142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali MA, Hu C, Zhang F, Jahan S, Yuan B, Saleh MS, Gao S-J, Panat R. N protein-based ultrasensitive SARS-CoV-2 antibody detection in seconds via 3D nanoprinted, microarchitected array electrodes. J Med Virol. 2022;94:2067–2078. doi: 10.1002/jmv.27591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raziq A, Kidakova A, Boroznjak R, Reut J, Öpik A, Syritski V (2021) Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens Bioelectron 178:113029. 10.1016/j.bios.2021.113029 [DOI] [PMC free article] [PubMed]
- 50.Mahari S, Roberts A, Shahdeo D, Gandhi S (2020) eCovSens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv 2020.04.24.059204. 10.1101/2020.04.24.059204
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.