Abstract
Fusarium graminearum is the causal agent of Fusarium Head Blight, a serious disease affecting grain crops worldwide. Biological control involves the use of microorganisms to combat plant pathogens such as F. graminearum. Strains of Bacillus velezensis are common biological control candidates for use against F. graminearum and other plant pathogens, as they can secrete antifungal secondary metabolites. Here we study the interaction between B. velezensis E68 and F. graminearum DAOMC 180378 by employing a dual RNA-seq approach to assess the transcriptional changes in both organisms. In dual culture, B. velezensis up-regulated genes related to sporulation and phosphate stress and down-regulated genes related to secondary metabolism, biofilm formation and the tricarboxylic acid cycle. F. graminearum up-regulated genes encoding for killer protein 4-like proteins and genes relating to heavy metal tolerance, and down-regulated genes relating to trichothecene biosynthesis and phenol metabolism. This study provides insight into the molecular mechanisms involved in the interaction between a biocontrol bacterium and a phytopathogenic fungus.
Introduction
Fusarium graminearum is the causal agent of Fusarium Head Blight (FHB) in wheat and barley [1]. It is considered a major global threat that impacted great economic losses on the cereal industry because of reduced grain yield and grain quality. FHB can also cause contamination of crops and grains with diverse mycotoxins, including deoxynivalenol (DON) and zearalenone, which are harmful for humans and animals [2, 3]. FHB has severe impacts on grain yield, with reported losses of up to 80% [4]. It has been estimated that in 2015, FHB costed the US wheat and barley industries $1.469 billion in value of yield forgone [5]. Recent data for Canada is not available, though it has been estimated that $520 million were lost by wheat producers in Ontario, Quebec and Manitoba during the 1990s [6]. Grain quality is affected as well, as kernels become discoloured, damaged and light in weight. Additionally, Fusarium may secrete mycotoxins into the crops. Contaminated grain can have adverse health effects when consumed, whether by humans or livestock. Contaminated grain can also lead to reduced seed germination and vigour [7]. Due to these factors, market price for affected grain becomes severely discounted.
Control of FHB requires a combination of multiple management strategies, including a well-thought out crop rotation strategy and the use of resistant crop cultivars. Another management strategy for FHB is the use of chemical fungicides. Triazole fungicides inhibit the cytochrome P450 sterol 14α-demethylase, and consequently, ergosterol biosynthesis. Triazoles could reduce disease index (diseased spikelets per spike) by 52%, and reduce DON concentration by 45% [8]. However, it has been shown that fungicides become much more effective when used on cultivars with at least moderate resistance to FHB [9]. Triazole residues have been detected in many locations, from wastewater, to soil, to human hair and urine samples. Evidence shows that triazoles can lead to endocrine disruption, hepatotoxicity, cytotoxicity and liver carcinogenesis [10]. In addition, chemical fungicides are quite expensive and may lead to farmers choosing not to produce FHB-susceptible crops [5]. Concerns have emerged as numerous strains of F. graminearum have been identified with resistance to triazoles [11–13]. As a result, there is a need for an alternative to these fungicides.
A common tactic for phytopathogen management is the use of biological control methods. This implies the use of living organisms (viruses, bacteria, fungi, insects, etc.) to suppress or eliminate the growth of plant pests. The use of microorganisms is most common, whether employing them live, or extracting their antimicrobial metabolites. Microorganisms offer a distinct advantage in that their mass production and application is very convenient. Many different mechanisms of action are possible. The microorganisms may induce systemic resistance in the plant without direct interaction with the pathogen, compete for nutrients and space, parasitize or antagonize the pathogen or secrete antimicrobial compounds which inhibit the pathogen (antibiosis). Alternatively, the microorganisms may deal specifically with mycotoxins, and sequester or degrade them in some fashion [14].
Biological control methods tend to offer less disadvantages than chemical control, as there are many less health risks to the consumer, and no risks of environmental contamination. Additionally, biological control is usually narrower in scope, avoiding non-specific effects to other non-target organisms. Isolates of Trichoderma harzianum Rifai reduced F. graminearum perithecial formation on wheat straw residues in the field and might have the potential to reduce inoculum development [15]. Compared to the registered fungicide Folicur, a strain of the fungus Clonostachys rosea reduced infected spikelets by 58%, and reduced Fusarium damaged kernels by 49% and DON by 21%. These effects were significant but of lesser magnitude than those achieved by the tebuconazole fungicide [16]. Other effective microorganisms include species of Pseudomonas, B acillus, Streptomyces and yeasts [17, 18]. These originated from various sources such as sourdough bread, peanut shells, soil and wheat itself. These biocontrol agents have been tested both in vitro and in greenhouse/field experiments. Biocontrol in FHB specifically has been mediated mostly through 2 mechanisms: either secretion of antifungal compounds or competition for nutrients and space, particularly iron [17].
Bacillus velezensis is a bacterial species, member of the Bacillus subtilis species complex. Bacteria from this group can be found in several different environments, with some living as plant endophytes [19, 20]. B. velezensis and its related species can produce and secrete secondary metabolites known as cyclic lipopeptides. These molecules are biosurfactants which have demonstrated antimicrobial ability against fungal phytopathogens in both field studies and in vitro [21]. B. velezensis and molecules derived from it have shown to inhibit growth of F. graminearum in vitro and limit FHB disease progression in planta when applied as biocontrol agents [22–24]. The bacterial strains from these studies and similar ones are typically endophytes isolated directly from plants, ensuring their ability to colonize the hosts. However, it is important to assess the abilities of bacterial strains from other environments as they may have differing effects as biocontrol candidates as compared to endophytes.
Bacillus velezensis strain E68 was previously isolated from crude oil samples from the Albertan oil sands [25, 26]. Oil-dwelling bacteria must be adapted to live in the harsh conditions: high temperature, high pressure and low carbon [27, 28]. These bacteria have adapted to the particular organic carbon sources present in these environments, including hydrocarbons. Biosurfactants secreted by these bacteria are used to degrade hydrocarbons for use as a carbon source [29]. Much research has gone into studying the potential of these biosurfactants for bioremediation of hydrocarbon pollutants [30]. However, oil-dwelling bacteria are unexplored when it comes to biocontrol. In preliminary experiments, B. velezensis E68 was found to inhibit the growth of various fungal phytopathogens such as Rhizoctonia solani and Fusarium graminearum (S1 Fig). Also, the cell-free supernatant of strain E68 exhibited biosurfactant activity using drop collapse and agar spreading assays [26]. These results suggest strain E68 has a wide inhibition spectrum against plant fungal pathogens.
The main task of the present research is to dissect the undergoing changes in the transcriptome profiles of both partners following dual culture of B. velezensis E68 and F. graminearum DAOMC 180378 using RNA-seq. This could provide valuable insights into the main factors that determine its outcome. Using the RNA-seq method, we observed the transcriptional activity of both organisms in response to the presence of one another.
Methods
Biological material and culture conditions
Cultures of Fusarium graminearum strain DAOMC 180378 were provided by the Canadian Collection of Fungal Cultures (DAOMC; Agriculture and Agri-Food Canada). Bacillus velezensis strain E68 was previously isolated from an oil well in Alberta [25, 26]. Microbial strains were retrieved from 20% glycerol stock stored at -80°C. F. graminearum DAOMC 180378 was grown on potato dextrose agar (PDA; Difco Laboratories) at 25°C in the dark. B. velezensis E68 was grown on Luria-Bertani agar (LBA) at 37°C.
Inhibitory effects of Bacillus velezensis E68 on Fusarium graminearum DAOMC 180378 mycelial growth
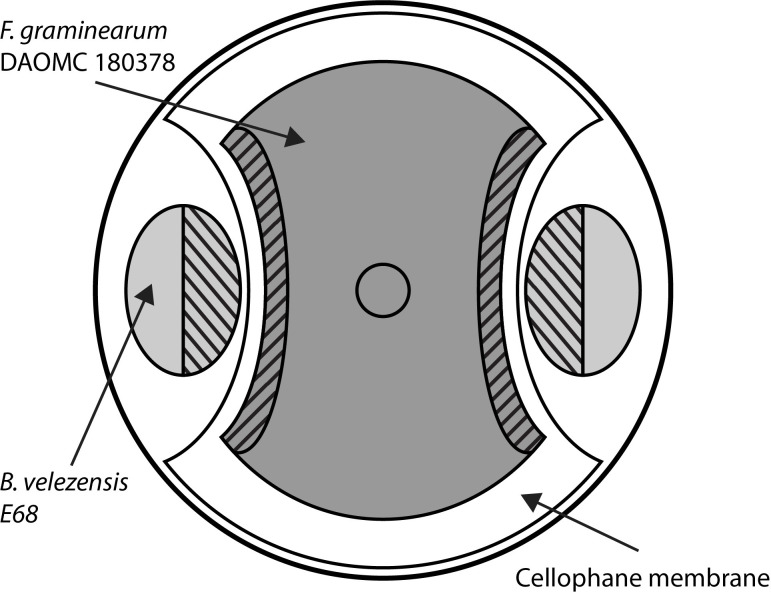
In order to test the inhibitory effect of Bacillus velezensis E68 on Fusarium graminearum DAOMC 180378 mycelial growth, the two organisms were grown in dual culture. Sterilized circular cellophane membranes (UCB, North Augusta, USA) were overlaid on 9 cm diameter petri plates containing 20 mL of PDA. Two half-moon sections were cut out from opposite sides of the membrane up to 2 cm from its center in order for B. velezensis to grow directly on the culture media (Fig 1). An agar plug (5 mm diameter) was taken from 7-day old cultures of actively growing F. graminearum, and were placed at the center of each plate overlaid with cellophane. A 10 μL aliquot of Bacillus velezensis (OD600 of 1.0) from an overnight LB broth was spotted on 2 opposite edges of the plate where the cellophane membrane was cut out, 2.5 cm from the center of the plate. A 0.5 cm agar plug of F. graminearum placed at the center of a PDA plate overlaid with a cellophane membrane served as control (Fig 1). Three 10 μL aliquots of Bacillus velezensis (OD600 of 1.0) spotted apart on a PDA plate served as control (Fig 1). Dual culture and control plates were incubated at 24°C for 3 days before RNA extraction.
Fig 1. Dual-culture interaction plate setup.
Fusarium graminearum DAOMC 180378 hyphae (dark grey) was grown from an agar plug placed at the centre of the plate on a cellulose membrane. Colonies of Bacillus velezensis E68 (light grey) were grown from aliquots of cell suspension spotted 2.5 cm from the centre of the plate. Striped areas were collected with a sterile loop or spatula for RNA extraction. Diagram is not to scale.
Dual culture and control plates were set up for microscopy as described above. A strip of cellophane membrane (0.5 cm x 2 cm) containing edges of the fungal hyphae was carefully excised from both a dual culture and a control treatment plate. The strips were placed on a microscopy slide, stained with bromophenol blue and mounted for microscopy on an EVOS XL Core Imaging System (Invitrogen).
Sample preparation and RNA extraction
Bacterial colonies were collected with a sterile loop by removing the half of each colony that was facing the center of the plate (Fig 1). In total, 6 halves of bacterial colonies were pooled into one RNA sample (i.e. 3 dual-culture plates pooled into 1 sample and 1 control plate pooled into 1 sample). The bacterial cells was placed into 500 μL of sterile water per sample for immediate extraction. Strips of fungal hyphae (5 x 50 mm) closest to the bacterial spots were collected with a sterile spatula (Fig 1). Six hyphal strips were pooled into one RNA sample. For the control treatment, an equivalent amount of hyphae was taken from the edges of a fungal colony and used as one sample. The fungal material was immediately ground with a mortar and pestle in liquid nitrogen and stored at -80°C before RNA extraction.
High quality total RNA from B. velezensis and F. graminearum was extracted using the Nucleospin RNA kit (Takara). RNA extraction from B. velezensis followed manufacturer’s instructions for Gram-positive bacteria, with modifications: the cell pellet was resuspended in 100 μL TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) with 3 mg/mL lysozyme, then incubated for 15 minutes at 37°C. RNA extraction from F. graminearum followed the instructions for cultured cells and tissues. For both organisms, 600 μL of buffer RA1, 6 μL of β-mercaptoethanol and 600 μL of 70% ethanol were used instead of 350 μL, 3.5 μL and 350 μL respectively. In all cases, elution was done by eluting twice in 60 μL of RNase-free water. RNA samples integrity and concentration were assessed by 1% bleach agarose gel and Nanodrop ND-1000. Samples were stored at -80°C for downstream applications.
RNA sequencing
Five RNA biological replicates for each treatment (B. velezensis control and dual-culture, F. graminearum control and dual-culture) were sent for library preparation and sequencing at IDSeq Inc, (Sacramento, California). The B. velezensis libraries were prepared with a TruSeq RNA library prep kit, with a Ribo-Zero Plus rRNA depletion kit. The F. graminearum libraries were prepared with the NEBNext Ultra II RNA library prep kit with poly-A selection. Samples were sequenced on an Illumina HiSeq 4000.
RNA-seq analysis
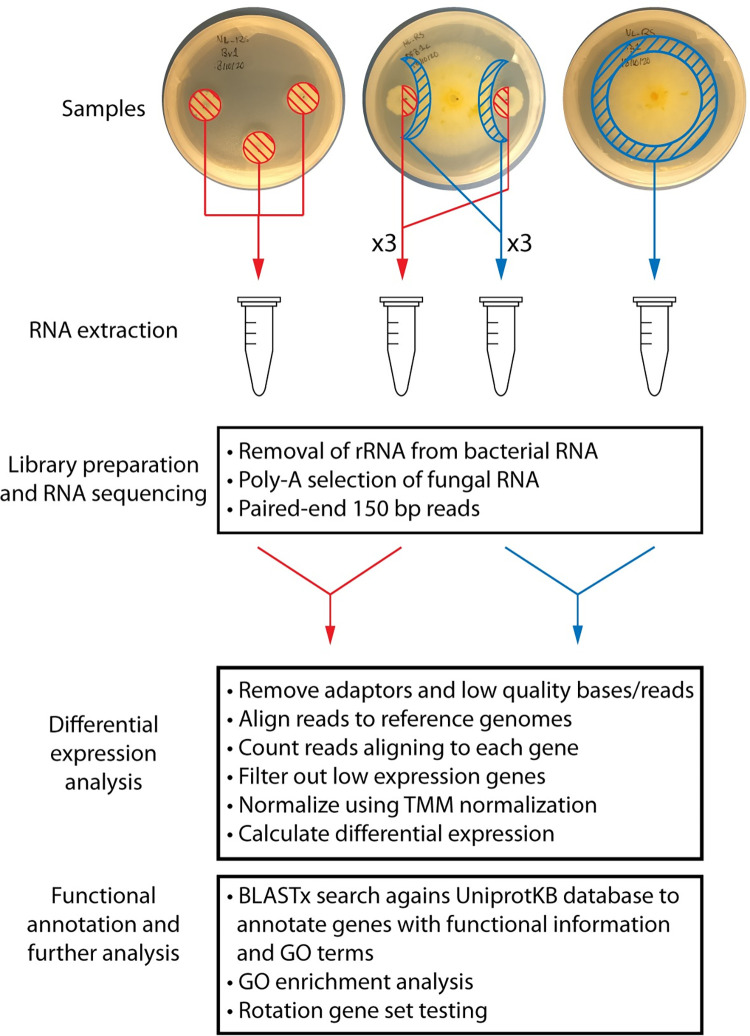
Fig 2 depicts the pipeline followed from sample preparation to RNA-seq data analysis. Sequencing reads were checked for quality and adaptor contamination before and after trimming with FastQC 0.11.9 [31]. Low quality bases and adaptor sequences were trimmed from the raw sequencing reads with Trim Galore 0.6.5 with stringency set to 3 [32]. The genome assembly for F. graminearum DAOMC 180378 is available on GenBank, however no reference annotation is available. The genome annotation version 5 [33] from Fusarium graminearum PH-1 was transferred to the genome assembly of DAOMC 180378 using Liftoff 1.6.1 [34]. The resulting annotation was converted from GFF3 format to GTF format with gffread 0.12.3 [35]. Trimmed reads were mapped to the reference genomes for F. graminearum DAOMC180378 and Bacillus velezensis E68 using STAR 2.7.5a on default settings [25, 36, 37]. The GeneCounts function from STAR was used to obtain read counts. Differential gene expression was assessed using edgeR 3.32.1 [38]. Genes were filtered by CPM expression using edgeR default settings. The edgeR GLM framework was used. Genes with a log2 fold change under 1.5 were deemed non-significant using the glmTreat function. Genes with a p-value under 0.05 after correction for multiple testing were deemed significant and differentially expressed. edgeR was also used to perform rotation gene set testing (ROAST) [39]. Genes were functionally annotated with gene ontology data by using Trinotate 3.2.2 to facilitate BLASTx searches of the gene coding sequences against the UniProtKB protein database [40]. Gene ontology enrichment analysis was carried out with GOSeq 1.44.0 to detect enriched gene ontology terms among up and down-regulated genes in both organisms, using a false discovery rate < 0.05 [41]. Biosynthetic gene clusters in B. velezensis E68 were identified with antiSMASH in previous work [25].
Fig 2. Schematic of experimental workflow of RNA-seq analysis of Bacillus velezensis and Fusarium graminearum.
Cultures of B. velezensis E68 and F. graminearum DAOMC 180378 were grown in single and dual culture. The striped areas were collected with a sterile spatula before RNA extraction. Total RNA was sent for library preparation and RNA sequencing. Bacterial RNA was depleted for ribosomal RNA, while fungal RNA was enriched with poly-A selection. Samples were sequenced on an Illumina HiSeq using a paired-end 150 bp protocol. Raw sequencing reads were pre-processed by removing adaptors and poor quality bases and reads. Filtered reads were aligned to their respective genomes and counted for each gene. Gene counts were normalized by the TMM method, low expression genes were removed from analysis and differential expression was calculated. In order to functionally annotate the genes, each gene sequence was used in a BLASTx search to compare to the UniprotKB protein database, annotating each gene with a description and gene ontology terms. The resulting GO terms were used in GO enrichment analysis. Known biosynthetic gene clusters were used in rotation gene set testing to determine their overall regulation pattern.
Validation of RNA-seq by qRT-PCR
In order to confirm the differential expression results obtained by RNA-seq, 8 genes for each organism were selected for validation by qRT-PCR (Table 1). Three RNA samples from each condition were used to validate the RNA-seq. The RNA samples were transcribed into cDNA using the Agilent AffinityScript qPCR cDNA Synthesis Kit. Specific primers for the selected genes were designed using Geneious 8.1.9 and the OligoAnalyzer tool from IDT. Primers are listed in Table 1. Specificity was assessed with PCR and agarose gel. recA (GYA98_RS07520) and secA (GYA98_RS15380) were used as housekeeping genes for B. velezensis [42]. GzUBH, ubiquitin thiolesterase (FGRAMPH1_01G03049) and TUB2, tubulin beta chain (FGRAMPH1_01G26865) were used as housekeeping genes for F. graminearum [43, 44]. Dilutions of pooled cDNA were used to create standard curves for each primer to calculate reaction efficiency and determine the optimal cDNA concentration for final usage. The geometric mean of the housekeeping genes was used as a normalization factor. The reaction efficiency and normalization factor were used to adjust the ΔCq values of the genes of interest as described in Taylor et al. [45]. Log transformed normalized expression values were compared between single and dual culture using a t-test. Three technical replicates were run per sample.
Table 1. List of primers used in this study.
| Target locus ID | Gene product, Gene symbol | Primer ID | Sequence (5’– 3’) | Source |
|---|---|---|---|---|
| Primers for use with Bacillus velezensis E68 | ||||
| GYA98_RS07520$ | Multifunctional protein RecA | recA-F | AAAAAACAAAGTCGCTCCTCCG | [42] |
| recA | recA-R | CGATATCCAGTTCAGTTCCAAG | ||
| GYA98_RS15380$ | Protein translocase subunit SecA | oNL25_secA_f | CGTTTAAAGTGCAGCTGATGG | This paper |
| secA | oNL26_secA_r | GGCTCGCCAGATATTCGTTG | ||
| GYA98_RS15140 | Heme response regulator HssR | oNL45_hssR_F | TATGAAGGTGACGAGCGTAC | This paper |
| hssR | oNL46_hssR_R | TGGATGGCAAACGGATAGTC | ||
| GYA98_RS00430 | Alkaline phosphatase D | oNL47_phoD_F | GCGTTTGAAGTGAATGCCG | This paper |
| phoD | oNL48_phoD_R | CAGAGAATGACGCTGTCAGAC | ||
| GYA98_RS07565 | Spore coat protein E | oNL49_cotE_F | ACCAACACGATCTCACCTGG | This paper |
| cotE | oNL50_cotE_R | TCGGCGTAAGAGTACCAGAC | ||
| GYA98_RS00125 | ECF RNA polymerase sigma factor SigW | oNL53_sigW_F | GCCTGACAAGTACAGAACGG | This paper |
| sigW | oNL54_sigW_R | CTGCCTCTGTGTATCCTTGTC | ||
| GYA98_RS03850 | ECF RNA polymerase sigma factor SigM | oNL67_sigM_F | GACCACAGCAAAGTAAAGCC | This paper |
| sigM | oNL68_sigM_R | ATACCCTGTCAGCACTTCC | ||
| GYA98_RS15180 | ATP phosphoribosyltransferase | oNL69_hisG_F | AGTTATTTCAGAGAGCAGGGC | This paper |
| hisG | oNL70_hisG_R | CTACGATTCTGTCAGCAAGCC | ||
| GYA98_RS03165 | Catechol-2,3-dioxygenase | oNL71_catE_F | GTCATCTCGCAGACAGATCG | This paper |
| catE | oNL72_catE_R | GGTCAGGCAGAAGTATCGC | ||
| GYA98_RS12240 | Citrate synthase 2 | oNL73_citZ_F | TGAGATGTCAATCCGTGTCG | This paper |
| citZ | oNL74_citZ_R | GATCGGCGTGAACAGATCG | ||
| Primers for use with Fusarium graminearum DAOMC 180378 | ||||
| FGRAMPH1_01G03049# | Ubiquitin thiolesterase | oNL29_GzUBH-F | CTCGAGGCCAGCAAAAAGTCA | [43] |
| UBH | oNL30_GzUBH-R | ATCGCCGTTAGGGGTGTCTG | ||
| FGRAMPH1_01G26865# | Tubulin beta chain | oNL43_TUB2-F | GTTGATCTCCAAGATCCGTG | [44] |
| TUB2 | oNL44_TUB2-R | CATGCAAATGTCGTAGAGGG | ||
| FGRAMPH1_01G01687 | Pleiotropic ABC efflux transporter of multiple drugs YBT1 | oNL55_ybt1_F | GCTACGCCTGCAAATGAACC | This paper |
| YBT1 | oNL56_ybt1_R | CTTAGGTCGAGCTACAATGGC | ||
| FGRAMPH1_01G15627 | ZEB2-regulated ABC transporter 1 | oNL57_zra1_F | GCTTCCAGGACATGAACGTC | This paper |
| ZRA1 | oNL58_zra1_R | GTCAATTCTGCGCTTGTTGG | ||
| FGRAMPH1_01G00143 | Nonribosomal peptide synthetase GRA1 | oNL59_gra1_F | ACTTGGAAGCTTTCTACCTGG | This paper |
| GRA1 | oNL60_gra1_R | CGTTGATCTATCACCAGCGAG | ||
| FGRAMPH1_01G05661 | Acetylesterase | oNL61_aes1_F | TGGTCAAGTCGATCAACCTC | This paper |
| AES1 | oNL62_aes1_R | CAGTGGGATTCTTGATTGCC | ||
| FGRAMPH1_01G12231 | D-malate dehydrogenase | oNL63_dmlA_F | GGTAATGACGAACCTGAGCC | This paper |
| DmlA | oNL64_dmlA_R | CGATGGTGACTGCATGAAGG | ||
| FGRAMPH1_01G05741 | Mitochondrial aldehyde dehydrogenase | oNL65_aldh_F | GGAGGAAATCTTTGGACCGG | This paper |
| ALDH2B4 | oNL66_aldh_R | GAGTTGAGCTTCTGAGTGACC | ||
| FGRAMPH1_01G19889 | RuvB-like helicase 2 | oNL75_RVB2_F | TTCGCAACTTGTCTCTGCC | This paper |
| RVB2 | oNL76_RVB2_R | GGCTACAAACTTGATGCTGC | ||
| FGRAMPH1_01G06539 | DNA repair protein | oNL77_RAD5_F | AGTTCACATCTTTCCTCAGCC | This paper |
| RAD5 | oNL78_RAD5_R | CGTTAAGTACAGCGGCTCG | ||
$Housekeeping genes for Bacillus velezensis E68.
#Housekeeping genes for F. graminearum DAOMC 180378
Results
Dual RNA-seq analysis of B. velezensis and F. graminearum
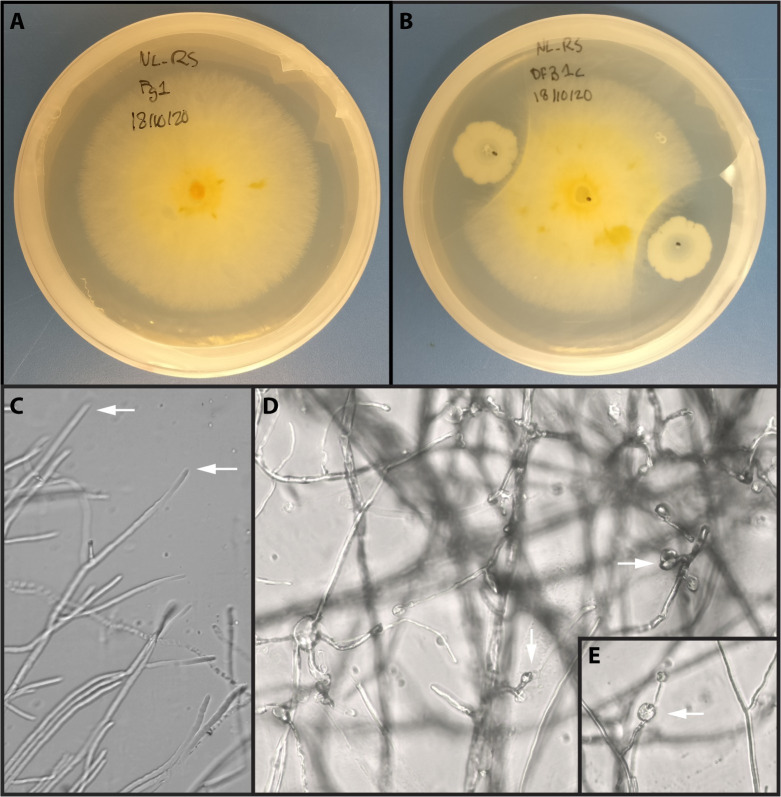
Cultures of Bacillus velezensis E68 and Fusarium graminearum DAOMC 180378 were grown together on potato dextrose agar as described in the Methods. Following 3 days of incubation, a clear inhibition of fungal growth was observed (Fig 3B). The morphology of F. graminearum appeared healthy in monoculture (Fig 3C). When grown in dual culture with B. velezensis, the morphology of F. graminearum appeared collapsed, with hyphal tips disrupted and developing circular spheres (Fig 3D and 3E). To evaluate the changes in total gene expression in both organisms, RNA was separately extracted from both organisms growing alone in single and together in dual culture. RNA from both organisms from both conditions was sequenced. After adaptor trimming and filtering the sequencing reads for quality, between 2.9 and 10.4 million sequencing reads were obtained for B. velezensis samples and between 16.4 and 24.5 million reads were obtained for F. graminearum samples (Table 2). The remaining reads were aligned to the reference genomes and reads aligning to each gene were quantified. The R package edgeR was used to filter out lowly expressed genes, normalize the libraries and calculate differential expression for each gene between single and dual culture. A total of 168 genes from B. velezensis and 4207 genes from F. graminearum were filtered out due to low expression. After calculation of differential expression, genes with a log2 fold change under 1.5 between conditions were not considered significant, as well as genes with a false discovery rate above 0.05. With these requirements, 1155 genes (32.4% of all expressed genes) were differentially expressed in B. velezensis between single and dual culture. In F. graminearum, 1503 genes (15.3% of all expressed genes) were differentially expressed (Table 3). Of the differentially expressed genes (DEGs), 47.2% were down-regulated in B. velezensis, whereas 66.1% of DEGs were down-regulated in F. graminearum. Genes were functionally annotated by BLASTx searches to the UniprotKB database, which also annotated many genes with gene ontology (GO) terms. In B. velezensis, 2394 genes were functionally annotated with GO terms (67.2% of all expressed genes), while 5464 genes were functionally annotated with GO terms in F. graminearum (55.6% of all expressed genes).
Fig 3. B. velezensis E68 inhibits the growth of F. graminearum DAOMC 180378 in dual culture.
(A) F. graminearum DAOMC 180378 grown in single culture on PDA, (B) Dual culture condition for B. velezensis E68 and F. graminearum DAOMC 180378 on PDA. (C) Hyphal morphology of F. graminearum in single culture. (D) and (E) Hyphal morphology of F. graminearum in dual culture with B. velezensis. Microscopy performed at 40x magnification.
Table 2. Number of sequencing reads from each sample after trimming.
| Sample ID | Organism | Condition | Number of sequencing reads |
|---|---|---|---|
| DB1 | Bacillus velezensis E68 | Dual culture | 6,895,885 |
| DB2 | Bacillus velezensis E68 | Dual culture | 5,930,991 |
| DB3 | Bacillus velezensis E68 | Dual culture | 2,902,440 |
| DB5 | Bacillus velezensis E68 | Dual culture | 4,826,251 |
| DB6 | Bacillus velezensis E68 | Dual culture | 3,998,583 |
| Bv1 | Bacillus velezensis E68 | Single culture | 7,051,088 |
| Bv2 | Bacillus velezensis E68 | Single culture | 6,818,309 |
| Bv3 | Bacillus velezensis E68 | Single culture | 5,535,266 |
| Bv5 | Bacillus velezensis E68 | Single culture | 10,013,420 |
| Bv6 | Bacillus velezensis E68 | Single culture | 10,482,205 |
| DF1 | Fusarium graminearum DAOMC 180378 | Dual culture | 23,178,176 |
| DF2 | Fusarium graminearum DAOMC 180378 | Dual culture | 24,528,876 |
| DF3 | Fusarium graminearum DAOMC 180378 | Dual culture | 20,797,041 |
| DF5 | Fusarium graminearum DAOMC 180378 | Dual culture | 21,440,526 |
| DF6 | Fusarium graminearum DAOMC 180378 | Dual culture | 23,730,823 |
| Fg1 | Fusarium graminearum DAOMC 180378 | Single culture | 18,261,933 |
| Fg2 | Fusarium graminearum DAOMC 180378 | Single culture | 18,966,692 |
| Fg3 | Fusarium graminearum DAOMC 180378 | Single culture | 19,694,534 |
| Fg5 | Fusarium graminearum DAOMC 180378 | Single culture | 19,549,899 |
| Fg6 | Fusarium graminearum DAOMC 180378 | Single culture | 16,435,517 |
Table 3. Genes of B. velezensis E68 and F. graminearum DAOMC 180378.
| Not significantly differentially expressed genes (% of all expressed genes) | Significantly down-regulated genes (% of all DEGs) | Significantly up-regulated genes (% of all DEGs) | Total expressed genes (% of all expressed genes) | |
|---|---|---|---|---|
| B. velezensis E68 | ||||
| Hypothetical | 102 (2.86) | 31 (2.68) | 41 (3.55) | 174 (4.88) |
| Annotated | 2308 (64.7) | 514 (44.5) | 569 (49.2) | 3391 (95.1) |
| Without GO terms | 852 (23.9) | 153 (13.2) | 166 (14.4) | 1171 (32.8) |
| Annotated with GO terms | 1558 (43.7) | 392 (33.9) | 444 (38.4) | 2394 (67.2) |
| Total | 2410 (67.6) | 545 (47.2) | 610 (52.8) | 3565 (100) |
| F. graminearum DAOMC 180378 | ||||
| Hypothetical | 2490 (25.3) | 337 (22.4) | 161 (10.7) | 2988 (30.4) |
| Annotated | 5838 (59.3) | 656 (43.6) | 349 (23.2) | 6843 (69.6) |
| Without GO terms | 3637 (37.0) | 484 (32.2) | 246 (16.4) | 4367 (44.4) |
| Annotated with GO terms | 4691 (47.7) | 509 (33.9) | 264 (17.6) | 5464 (55.6) |
| Total | 8328 (84.7) | 993 (66.1) | 510 (33.9) | 9831 (100) |
Genes were annotated with functional information and gene ontology (GO) terms based on BLASTx searches against the Swiss-Prot database. Genes with minimal expression as determined by edgeR were not included in this table.
Transcriptomic changes of B. velezensis in dual culture with F. graminearum
Following gene ontology enrichment analysis, 16 GO terms were enriched among significantly up-regulated genes in B. velezensis, whereas 7 GO terms were enriched among significantly down-regulated genes (Table 4). Genes associated to sporulation and related GO terms such as the spore wall and spore germination were up-regulated. Many genes were annotated as spore coat proteins as well as many different genes from sporulation stages II, III, IV and V. These included spoIIE, spoIIGA, spoIIR, and sigE from stage II, spoIID, spoIIP, spoIIQ, spoIIIAH, FisB, sigG and sigK from stage III, spoIVB, ctpB, spoIVA, sleB and cwlJ from stages IV and V (S2 Table). The master sporulation regulator gene spo0A which initiates sporulation (locus tag GYA98_RS10380) was not significantly differentially regulated (S1 Table). The phosphotransferase spo0B (GYA98_RS11640) was also not regulated, but the phosphotransferase spo0F (GYA98_RS16270) was down-regulated. The GO term for oxidoreductase activity, acting on CH or CH2 groups, was enriched in up-regulated genes. Genes associated with this term included 2 subunits of ribonucleoside-diphosphate reductase: nrdE (GYA98_RS07750) and nrdF (GYA98_RS07755) (S1 Table). This enzyme contributes to DNA synthesis by converting ribonucleotides to deoxyribonucleotides. The 4 other up-regulated genes associated to this term were subunits of xanthine dehydrogenase (GYA98_RS17550, 17555, 17560, 17570) (S1 Table). Another subunit GYA98_RS17565 was also up-regulated, though not annotated with the GO term. Xanthine dehydrogenase participates in purine degradation by degrading xanthine into uric acid. Two terms for teichuronic acid biosynthesis were enriched in up-regulated genes (Table 4). All genes associated to these terms were part of the tua operon. These genes synthesize teichuronic acid, which replaces teichoic acid in the cell wall in phosphate starvation conditions. In addition, all 5 genes annotated with the ATPase-coupled phosphate ion transmembrane transporter activity GO term were up-regulated. These genes are all part of the PstSABC complex, responsible for phosphate import (GYA98_RS10730, 10735, 10740, 10745, 10750) [46] (S2 Table). Both the tua and PstSABC operons are controlled by the Pho regulon. Another member of this regulon, glpQ (GYA98_RS00270), was also up-regulated in dual culture [47] (S2 Table).
Table 4. Significantly over-represented Gene Ontology (GO) terms following enrichment analysis on up- and down-regulated genes from B. velezensis E68 and F. graminearum DAOMC 180378 in dual culture.
| Enrichment group | GO term ID | Term | Ontology | False Discovery Rate | DEGs in category | Expressed genes in category |
|---|---|---|---|---|---|---|
| Bacillus velezensis E68 up-regulated genes | GO:0032502 | developmental process | BP | 9.85E-31 | 122 | 238 |
| GO:0048869 | cellular developmental process | BP | 3.96E-29 | 114 | 222 | |
| GO:0030154 | cell differentiation | BP | 3.96E-29 | 113 | 219 | |
| GO:0030435 | sporulation resulting in formation of a cellular spore | BP | 3.96E-29 | 113 | 219 | |
| GO:0048646 | anatomical structure formation involved in morphogenesis | BP | 3.96E-29 | 113 | 219 | |
| GO:0043934 | sporulation | BP | 1.03E-28 | 113 | 221 | |
| GO:0031160 | spore wall | CC | 5.77E-08 | 19 | 24 | |
| GO:0005618 | cell wall | CC | 9.34E-05 | 23 | 44 | |
| GO:0030312 | external encapsulating structure | CC | 9.34E-05 | 23 | 44 | |
| GO:0050845 | teichuronic acid biosynthetic process | BP | 1.58E-03 | 7 | 7 | |
| GO:0050846 | teichuronic acid metabolic process | BP | 1.58E-03 | 7 | 7 | |
| GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups | MF | 8.02E-03 | 6 | 6 | |
| GO:0016020 | membrane | CC | 9.06E-03 | 176 | 758 | |
| GO:0009847 | spore germination | BP | 1.29E-02 | 11 | 18 | |
| GO:0045229 | external encapsulating structure organization | BP | 3.27E-02 | 26 | 71 | |
| GO:0015415 | ATPase-coupled phosphate ion transmembrane transporter activity | MF | 3.27E-02 | 5 | 5 | |
| Bacillus velezensis E68 down-regulated genes | GO:0016999 | antibiotic metabolic process | BP | 1.21E-05 | 44 | 59 |
| GO:0017144 | drug metabolic process | BP | 1.21E-05 | 44 | 59 | |
| GO:0017000 | antibiotic biosynthetic process | BP | 5.11E-05 | 41 | 54 | |
| GO:0006099 | tricarboxylic acid cycle | BP | 4.60E-03 | 9 | 11 | |
| GO:0006101 | citrate metabolic process | BP | 6.31E-03 | 10 | 14 | |
| GO:0072350 | tricarboxylic acid metabolic process | BP | 6.31E-03 | 10 | 14 | |
| GO:0000271 | polysaccharide biosynthetic process | BP | 3.93E-02 | 15 | 34 | |
| Fusarium graminearum DAOMC 180378 up-regulated genes *no GO terms were significantly enriched | GO:0036246 | phytochelatin 2 import into vacuole | BP | 1.69E-01 | 3 | 3 |
| GO:0071993 | phytochelatin transport | BP | 1.69E-01 | 3 | 3 | |
| GO:0071994 | phytochelatin transmembrane transport | BP | 1.69E-01 | 3 | 3 | |
| GO:0071995 | phytochelatin import into vacuole | BP | 1.69E-01 | 3 | 3 | |
| GO:0071996 | glutathione transmembrane import into vacuole | BP | 1.69E-01 | 3 | 3 | |
| Fusarium graminearum DAOMC 180378 down-regulated genes | GO:0016491 | oxidoreductase activity | MF | 2.39E-09 | 112 | 614 |
| GO:0003824 | catalytic activity | MF | 2.95E-07 | 326 | 2802 | |
| GO:0009056 | catabolic process | BP | 9.19E-05 | 108 | 724 | |
| GO:0009712 | catechol-containing compound metabolic process | BP | 7.04E-04 | 9 | 13 | |
| GO:1901575 | organic substance catabolic process | BP | 7.04E-04 | 97 | 665 | |
| GO:0044282 | small molecule catabolic process | BP | 1.16E-03 | 38 | 180 | |
| GO:0018958 | phenol-containing compound metabolic process | BP | 3.28E-03 | 10 | 19 | |
| GO:0042537 | benzene-containing compound metabolic process | BP | 4.69E-03 | 11 | 24 | |
| GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | MF | 6.32E-03 | 29 | 127 | |
| GO:0032787 | monocarboxylic acid metabolic process | BP | 9.29E-03 | 39 | 209 | |
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF | 9.29E-03 | 34 | 169 | |
| GO:0046278 | 3,4-dihydroxybenzoate metabolic process | BP | 3.46E-02 | 5 | 6 | |
| GO:0016054 | organic acid catabolic process | BP | 3.46E-02 | 26 | 126 | |
| GO:0046395 | carboxylic acid catabolic process | BP | 3.46E-02 | 26 | 126 | |
| GO:0016829 | lyase activity | MF | 3.53E-02 | 33 | 176 | |
| GO:0044248 | cellular catabolic process | BP | 3.87E-02 | 77 | 550 |
Secondary metabolites in B. velezensis are produced through enzyme complexes encoded by biosynthetic gene clusters. The transcription of these gene clusters was assessed by treating each cluster as a gene set and using rotation gene set testing (ROAST). The genome of B. velezensis E68 contains biosynthetic gene clusters which may synthesize 9 known secondary metabolites. Rotation gene set testing showed that all gene clusters were significantly down-regulated, with the exception of surfactin, which was not significant (Table 5). This was also reflected in gene ontology, as GO terms related to antibiotic biosynthesis and metabolism were enriched among down-regulated genes in dual culture (Table 4). Despite being down-regulated, genes responsible for synthesis of secondary metabolites remained fairly highly expressed in dual culture. The median transcripts per million (TPM) of all genes across all dual culture B. velezensis samples was 73.207. In comparison, the expression of genes annotated as core biosynthetic genes by antiSMASH remained high in down-regulated clusters encoding secondary metabolites (Table 6).
Table 5. Rotation gene set testing of biosynthetic gene clusters encoding secondary metabolites in B. velezensis E68.
| Compound | Number of genes | Direction of regulation | False discovery rate |
|---|---|---|---|
| Bacilysin | 7 | Down | 6.79E-08 |
| Macrolactin H | 10 | Down | 2.98E-07 |
| Difficidin | 19 | Down | 3.72E-06 |
| Bacillaene | 14 | Down | 3.72E-06 |
| Bacillibactin | 13 | Down | 6.13E-05 |
| Bacillomycin D | 10 | Down | 4.60E-04 |
| Amylocyclicin | 6 | Down | 1.52E-03 |
| Fengycin | 17 | Down | 0.026 |
| Surfactin | 25 | Down | 0.35 |
Table 6. Normalized expression of core biosynthetic genes from biosynthetic gene clusters encoding secondary metabolites in B. velezensis E68.
| Compound | Core biosynthetic gene locus ID | TPM$ in single culture | TPM in dual culture |
|---|---|---|---|
| Surfactin | GYA98_RS00790 | 41.62444 | 54.49393 |
| GYA98_RS00795 | 39.72677 | 36.2239 | |
| GYA98_RS00800 | 10.51762 | 14.11884 | |
| GYA98_RS00810 | 49.16629 | 56.89863 | |
| GYA98_RS00815 | 55.97234 | 52.11947 | |
| GYA98_RS00820 | 77.41527 | 64.22825 | |
| Macrolactin H | GYA98_RS06300 | 237.6562 | 90.06079 |
| GYA98_RS06305 | 521.1573 | 100.0418 | |
| GYA98_RS06310 | 527.2992 | 96.8625 | |
| GYA98_RS06315 | 622.4262 | 113.8966 | |
| GYA98_RS06320 | 783.2996 | 181.6299 | |
| GYA98_RS06325 | 440.9877 | 118.3611 | |
| GYA98_RS06330 | 744.2543 | 223.779 | |
| Bacillaene | GYA98_RS07605 | 410.8624 | 488.6164 |
| GYA98_RS07610 | 420.911 | 373.5302 | |
| GYA98_RS07615 | 558.4976 | 478.7145 | |
| GYA98_RS07625 | 660.5088 | 360.3584 | |
| GYA98_RS07640 | 1183.568 | 361.8864 | |
| GYA98_RS07645 | 1198.974 | 260.8726 | |
| GYA98_RS07650 | 792.9114 | 160.4905 | |
| GYA98_RS07655 | 943.0376 | 233.3925 | |
| GYA98_RS07660 | 1074.126 | 305.5386 | |
| Bacillomycin D | GYA98_RS08205 | 345.918 | 221.1193 |
| GYA98_RS08210 | 604.1638 | 437.0655 | |
| GYA98_RS08215 | 565.6975 | 475.9636 | |
| GYA98_RS08220 | 277.014 | 215.3259 | |
| Fengycin | GYA98_RS08300 | 3.060367 | 33.54516 |
| GYA98_RS08315 | 2.122462 | 36.92852 | |
| GYA98_RS08335 | 571.6845 | 177.9275 | |
| GYA98_RS08340 | 543.9846 | 173.68 | |
| GYA98_RS08345 | 693.3596 | 238.2508 | |
| GYA98_RS08350 | 463.5909 | 187.7093 | |
| GYA98_RS08355 | 580.417 | 294.5991 | |
| GYA98_RS08365 | 281.5708 | 150.397 | |
| Difficidin | GYA98_RS10060 | 1267.013 | 857.4785 |
| GYA98_RS10065 | 1026.037 | 719.9703 | |
| GYA98_RS10070 | 899.1106 | 563.8603 | |
| GYA98_RS10075 | 1083.195 | 696.7137 | |
| GYA98_RS10080 | 901.9802 | 558.2665 | |
| GYA98_RS10085 | 912.1075 | 526.2694 | |
| GYA98_RS10090 | 1367.896 | 835.8435 | |
| GYA98_RS10095 | 1072.906 | 647.6522 | |
| GYA98_RS10100 | 1126.655 | 623.7472 | |
| GYA98_RS10105 | 510.1765 | 295.7482 | |
| GYA98_RS10110 | 953.4498 | 515.7316 | |
| GYA98_RS10135 | 573.8718 | 259.0871 | |
| Bacillibactin | GYA98_RS13635 | 159.2003 | 49.03839 |
| GYA98_RS13645 | 259.1896 | 116.4149 | |
| Amylocyclicin | GYA98_RS13745 | 25433.92 | 14390.07 |
| Bacilysin | GYA98_RS16580 | 1691.767 | 989.5184 |
Genes were identified as core biosynthetic genes by antiSMASH.
$transcripts per million (TPM)
GO terms related to the tricarboxylic acid (TCA) cycle were also enriched among down-regulated genes. Significantly down-regulated genes associated to these terms included the enzymes of the TCA cycle: 2 citrate synthases (GYA98_RS03820, 12240), aconitase (GYA98_RS08030), isocitrate dehydrogenase (GYA98_RS12235), 2-oxoglutarate dehydrogenase (GYA98_RS08685, 08690), succinate dehydrogenase (GYA98_RS11900, 11905), succinyl-CoA ligase (GYA98_RS07090, 07095) and fumarate dehydrogenase (GYA98_RS14210), as well as a citrate/malate transporter (GYA98_RS17095) (S2 Table). Malate dehydrogenase (GYA98_RS12230), which was not annotated with the TCA cycle GO terms, was also significantly down-regulated. The GO term for polysaccharide biosynthetic process was also enriched among the down-regulated genes. Of the significantly down-regulated genes associated to this term, 14 of the 15 genes of the eps operon (epsB-O, GYA98_RS14870-14935) were included (S2 Table). The epsA homolog in B. velezensis E68 (GYA98_RS14940) was not significantly differentially expressed. The eps operon encodes enzymes responsible for synthesis of exopolysaccharide, a component of the extracellular biofilm matrix [48, 49]. In addition, tasA (GYA98_RS10575), the major protein component of the extracellular matrix, was also down-regulated in dual culture, though it wasn’t annotated with GO terms.
Transcriptomic changes of F. graminearum in dual culture with B. velezensis
In F. graminearum, no GO terms were significantly enriched among up-regulated genes in dual culture. Despite not being significantly enriched, 5 GO terms had all 3 of their associated genes be up-regulated in F. graminearum (Table 4). FGRAMPH1_01G11895 and FGRAMPH1_01G06603, both annotated as heavy metal tolerance proteins which may transport phytochelatin, and FGRAMPH1_01G26673, an ATP-binding cassette transporter, were all up-regulated in dual culture conditions and were the only genes associated with the 5 GO terms for phytochelatin transport/import and glutathione import into the vacuole (S3 Table). The relative lack of total genes associated to the terms prevented these terms from being significantly enriched after correction for multiple testing. The genome of F. graminearum DAOMC 180378 was annotated with 4 genes described as killer protein 4-like. All 4 genes were up-regulated in dual culture (S4 Table).
In the 16 GO terms enriched among down-regulated genes in F. graminearum, many were related to the broad categories of the catabolic process term and the oxidoreductase activity term. More specifically, many of the terms related to the metabolism of aromatic compounds, notably the terms for phenol containing, benzene-containing and catechol-containing compound metabolic processes, as well as the term for 3,4-dihydroxybenzoate (also known as protocatechuate) metabolic process (Table 4). Significantly down-regulated genes associated to these compounds included genes upstream of protocatechuate/catechol: 2 genes annotated as quinate dehydrogenases (FGRAMPH1_01G13935 and FGRAMPH1_01G18555), one gene annotated either as a quinate or shikimate dehydrogenase (FGRAMPH1_01G13945), 2,3-dihydroxybenzoate decarboxylase (FGRAMPH1_01G28015) and a 3-dehydroshikimate dehydratase (FGRAMPH1_01G08441) (S3 Table). In addition, genes degrading catechol and protocatechuate were down regulated: a catechol 1,2-dioxygenase (FGRAMPH1_01G13423) and another gene annotated as a catechol 1,2-dioxygenase, but a BLAST search against the DAOMC 180378 genome showed that protocatechuate-3,4-dioxygenase from Aspergillus nidulans was homologous to this gene (FGRAMPH1_01G12667) [50]. Also part of the significantly down-regulated genes associated to benzene compounds were 3 genes from the homogentisate pathway, which utilizes phenylacetate and phenylalanine as carbon sources. Homogentisate 1,2-dioxygenase (FGRAMPH1_01G05639), 3-hydroxyphenylacetate 6-hydroxylase (FGRAMPH1_01G22299) and fumarylacetoacetase (FGRAMPH1_01G11459) were all down-regulated (S3 Table). In addition, 3 genes from the kynurenine pathway were down-regulated: 2 kynureninases (FGRAMPH1_01G16409 and FGRAMPH1_01G14561), as well as an indoleamine 2,3-dioxygenase (FGRAMPH1_01G16407) (S3 Table). This pathway degrades tryptophan into NAD+.
In dual culture, F. graminearum hyphae was distorted and swelling occurred, based on microscopy (Fig 3). However, the differential gene expression data from RNA-seq did not show concerted differential regulation of genes relating to this phenotype. This included genes related to ergosterol synthesis, glucan or chitin synthases and genes related to autophagy. Similarly, F. graminearum also did not show large scale differential regulation of genes relating to oxidative stress, specifically genes encoding for superoxide dismutases and catalases (S4 Table). Genes annotated as glutathione S-transferases were also mostly not differentially expressed, though some were significantly down-regulated in dual culture (S4 Table).
F. graminearum DAOMC 180378 is known to produce deoxynivalenol, a trichothecene mycotoxin [37]. To assess the effect of dual culture with B. velezensis on trichothecene mycotoxin expression, genes from the core trichothecene gene cluster, as well as other genes from the trichothecene pathway were evaluated for differential expression (Table 7) [51]. Biosynthetic enzymes TRI3, TRI8 and TRI11 did not show significant differential expression. The biosynthetic enzymes TRI1, TRI5, TRI13 and TRI101 were significantly down-regulated, while TRI4 was down-regulated, only approaching significance. The accessory proteins TRI6, TRI9, TRI10, TRI11 and TRI14 did not show significant differential expression. The transcription factor TRI15 showed significant up-regulation in dual culture. TRI15 is likely a negative regulator of trichothecene expression [52].
Table 7. Fold change of expression of trichothecene pathway genes in F. graminearum DAOMC 180378 in dual culture with B. velezensis E68.
| Locus ID | Gene | Log2 fold change | False discovery rate | Function |
|---|---|---|---|---|
| FGRAMPH1_01G13947 | TRI15 | 3.03529 | 3.72E-06 | Transcription factor, negative regulator |
| FGRAMPH1_01G00223 | TRI1 | -1.06495 | 0.014269 | Biosynthetic enzyme |
| FGRAMPH1_01G13121 | TRI13 | -2.08402 | 0.021416 | Biosynthetic enzyme |
| FGRAMPH1_01G25923 | TRI101 | 0.982818 | 0.027902 | Biosynthetic enzyme |
| FGRAMPH1_01G13111 | TRI5 | -1.19642 | 0.028048 | Biosynthetic enzyme |
| FGRAMPH1_01G13107 | TRI4 | -1.03885 | 0.053723 | Biosynthetic enzyme |
| FGRAMPH1_01G13101 | TRI8 | -0.9157 | 0.176208 | Biosynthetic enzyme |
| FGRAMPH1_01G13123 | TRI14 | -0.84547 | 0.185704 | Virulence factor, unknown function |
| FGRAMPH1_01G13105 | TRI3 | 0.889523 | 0.345196 | Biosynthetic enzyme |
| FGRAMPH1_01G13115 | TRI9 | -0.54126 | 0.751079 | Unknown function |
| FGRAMPH1_01G13109 | TRI6 | -0.52321 | 0.778677 | Transcription factor |
| FGRAMPH1_01G13113 | TRI10 | 0.359458 | 1 | Transcription factor |
| FGRAMPH1_01G13117 | TRI11 | -0.36532 | 1 | Biosynthetic enzyme |
| FGRAMPH1_01G05631 | TRI12 | -0.15582 | 1 | Transporter |
Functions were assigned to genes based on [51].
Validation of RNA-seq via quantitative PCR
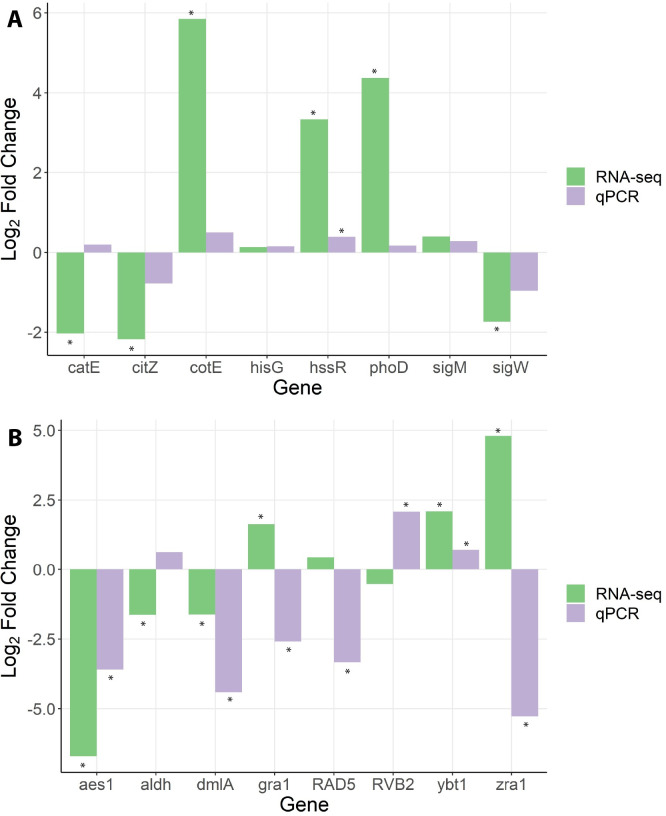
In order to validate the results of RNA-seq, 8 genes were selected per organism to evaluate their differential expression through qRT-PCR (Table 1). Three RNA samples per condition were used for analysis (Bv1, Bv2, Bv5, DB1, DB2, DB5, Fg1, Fg2, Fg5, DF1, DF2, DF5). The housekeeping genes were selected based on use in previous studies as well as lack of differential expression in the RNA-seq dataset. In B. velezensis E68, the 2 selected non-differentially expressed genes (hisG and sigM) saw identical results in qRT-PCR (Fig 4A). The gene hssR showed significant up-regulation in both RNA-seq and qRT-PCR. Four significantly differentially expressed genes (phoD, sigW, cotE, citZ) had similar trends between RNA-seq and qRT-PCR, though the qRT-PCR fold-changes were not significant after t-test. One gene, catE showed opposite trends in qPCR, though the result was not significant. In F. graminearum DAOMC 180378, the 2 selected non-differentially expressed genes RVB2 and RAD5 showed significant differential expression using qRT-PCR (Fig 4B). The 3 genes YBT1, AES1 and DMLA showed significant differential expression in the same direction in both RNA-seq and qRT-PCR. The genes ZRA1 and GRA1 showed significant regulation in opposite directions in RNA-seq and qRT-PCR. The ALDH gene also showed opposite regulation by RNA-seq and qRT-PCR, though the qRT-PCR result was not significant.
Fig 4.
Log2 fold change of selected genes between single and dual culture based on RNA-seq and qPCR for (A) B. velezensis E68 and (B) F. graminearum DAOMC 180378. Asterisks (*) indicate significantly differentially expressed genes for RNA-seq and indicate p < 0.05 for qPCR.
Discussion
A dual RNA-seq approach was developed to compare the transcriptional profiles of two organisms, Bacillus velezensis E68 and Fusarium graminearum DAOMC 180378 in single and dual culture. Previously, dual RNA-seq was employed to investigate the interaction between the rhizobacteria Lysobacter capsici and the oomycete Phytophthora infestans [53]. In addition, a dual microarray approach was used to assess the transcriptional changes from both sides of a Bacillus subtilis and Aspergillus niger interaction [54]. However, to the best of our knowledge, this is the first time a dual RNA-seq approach has been used to study the transcriptional changes between a biocontrol candidate bacteria and a filamentous fungus. Strains of B. velezensis and the closely related B. subtilis were previously shown to inhibit growth of F. graminearum on solid media, demonstrating the potential of these strains as biological control candidates [23, 24, 55]. In our study, B. velezensis E68 showed very similar inhibition of F. graminearum DAOMC 180378. Microscopic observations of F. graminearum exposed to B. velezensis showed distorted hyphal tips and circular swellings, most likely due to secretion of antifungal lipopeptides by B. velezensis. This phenotype was also observed in other studies exposing F. graminearum to both B. velezensis cultures and also purified lipopeptide extracts from B. velezensis and B. subtilis [23, 56–59]. Lipopeptides secreted from B. velezensis and similar species are known to cause damage to the plasma membrane and cell wall of various fungal phytopathogens, including F. graminearum [23, 58, 60, 61].
When confronted with F. graminearum, B. velezensis showed clear up-regulation of many genes related to sporulation. Strains of Bacillus tend to sporulate under conditions of limited nutrient availability, though the specific signals by which this is determined are unknown [62]. In our study, genes related to stages 0 and I of sporulation did not show significant differential expression in B. velezensis. This includes the master regulator of sporulation spo0A, though since its activity is reliant on its phosphorylation state, its transcriptional status may not be of importance [63]. However, multiple genes from stages II through V were up-regulated. SigF, the first sigma factor related to sporulation is produced prior to stage II. The gene product of spoIIE is responsible for activating this sigma factor in the forespore [64]. Consistent with this idea, in our study spoIIE was up-regulated in B. velezensis E68 in dual culture. In addition, spoIIGA and spoIIR, the activators of sigE, as well as sigE itself, were all up-regulated in dual culture, triggering stage III: engulfment [65]. Genes required for engulfment and membrane fission were up-regulated in dual culture: spoIID, spoIIP, spoIIQ, spoIIIAH and FisB [66, 67]. The forespore-specific sigma factor sigG and the mother cell specific sigma factor sigK are both activated at the end of engulfment, these are both up-regulated in dual culture. In addition the sigK activators spoIVB and ctpB were up-regulated [68–70]. Many spore cortex coat proteins related to stages IV and V were up-regulated, including spoIVA, which forms the base layer of the spore coat [68]. Furthermore, spore germination genes were also up-regulated in dual culture. sleB and cwlJ are enzymes which lyse peptidoglycan during germination and were both up-regulated. However, they lie dormant in the spore in a mature form [71]. Taken together, these results suggest that in the presence of F. graminearum, B. velezensis cells sporulate, at stages II through V of sporulation on day 3 during our dual culture assay. Previous research showed B. subtilis up-regulated late sporulation genes when grown on Aspergillus niger hyphae, though no significant difference in spore content was found [54].
In dual culture, B. velezensis E68 down-regulated key components of the extracellular biofilm matrix: the eps operon and the tasA gene [72]. The decision to begin biofilm formation and the decision to sporulate are both governed by the Spo0A protein [73]. Spo0A is a transcriptional regulator whose activity depends on its phosphorylation state. High levels of phosphorylated Spo0A promote entry into sporulation through repression of sinI expression [63]. Research also shows that sporulation may require biofilm formation to occur [73]. As a result, the down-regulation of biofilm components in B. velezensis coincides with the up-regulation of sporulation genes.
The genes encoding for the tricarboxylic acid cycle enzymes were down-regulated in B. velezensis in dual culture. This is consistent with previous research studying the transcriptomics of the life cycle of Bacillus pumilus [74]. B. pumilus showed decreased expression of TCA cycle genes in the stationary growth phase as compared to the exponential growth phase, in addition to an increase in expression of sporulation-related genes, much the same as B. velezensis in our experiment. Furthermore, TCA cycle genes were down-regulated and sporulation genes were up-regulated in B. subtilis which were attached to Aspergillus niger hyphae as compared to planktonic cells [54].
Secondary metabolism in B. velezensis E68 was evidently altered, with all known biosynthetic gene clusters (with the exception of surfactin) showing reduced expression in dual culture with F. graminearum. Despite the down-regulation, these metabolites remained expressed in dual culture and likely secreted into the media, as reflected by the changes in fungal hyphae appearance in dual culture (Fig 3). This down-regulation is unexpected, as multiple studies of lipopeptide-secreting Bacillus report up-regulation of lipopeptide related genes or increased lipopeptide concentrations in confrontation with fungi [75–79]. However, surfactin was found to be down-regulated in B. subtilis cells attached to hyphae of A. niger [54]. Our experiment was performed at a single time-point: 3 days after establishment of interaction. It may be possible that different regulation patterns could be expressed for secondary metabolism genes in earlier or later stages of the interaction. Using proteomic methods for the detection of lipopeptides and other secondary metabolites could also show variations from the gene expression data obtained here.
In dual culture, B. velezensis also showed signs of phosphate stress. In times of phosphate starvation, Bacillus strains induce genes of the Pho regulon, controlled by the PhoP sensor and the PhoR regulator, though these were not differentially expressed in our study [47]. When confronted with F. graminearum, B. velezensis up-regulated members of the Pho regulon, such as the tua operon. This operon synthesizes teichuronic acid, a component of the cell wall. In phosphate starvation conditions, teichuronic acid replaces teichoic acid in the cell wall [80]. Phosphate can then be collected from the teichoic acid through the action of phoD, which was also up-regulated in dual culture [81]. The PstSABC operon is another part of the Pho regulon which was up-regulated in dual culture. The PstSABC complex is a phosphate importer which may also play a role in sensing extracellular phosphate concentrations through interaction with PhoR [46, 82]. Furthermore, B. velezensis up-regulated glpQ, also part of the Pho regulon. This gene encodes a glycerophosphoryl diester phosphodiesterase, which hydrolyzes deacetylated phospholipids [83].
Taken together, these results indicate that in dual culture with F. graminearum, B. velezensis E68 entered sporulation, as shown by the up-regulation of sporulation and biofilm formation genes. This could be due to different stresses, including phosphate starvation. Consistent with its entry into sporulation, B. velezensis down-regulated genes related to carbohydrate metabolism and secondary metabolite biosynthesis.
In our study, there were no GO terms enriched among up-regulated genes in F. graminearum. This can be partially attributed to the low number of expressed genes annotated with GO terms (55.6% of expressed genes). Additionally, only 510 genes were significantly up-regulated, as compared to 993 down-regulated genes. As a result, global up-regulation of particular molecular functions or biological processes was not found. When the fungal phytopathogen Sclerotinia sclerotiorum was confronted to B. velezensis fermentation broth, the fungus was found to induce genes related to cell wall and ergosterol synthesis, fatty acid synthesis, antioxidants and autophagy [84]. In this study, genes related to cell wall synthesis via either glucan synthase or chitin synthase were mostly not differentially expressed, with chitin synthase 6 (FGRAMPH1_01G22563) being the only significantly up-regulated chitin synthase and chitin synthase 5 (FGRAMPH1_01G10489) being down-regulated. Distinct up-regulation of ergosterol pathway genes as defined through annotation or previous evidence was also not found. It may be that fungal cells most affected by the bacterial lipopeptides were lysed and their RNA could not be sequenced. Genes related to autophagy were also not differentially expressed in dual culture [85]. In addition to damage to the cell wall and membranes, Bacillus lipopeptides can also induce accumulation of reactive oxygen species [58, 86]. However, our data signalled that F. graminearum did not show concerted up-regulation of genes relating to oxidative stress. Five genes previously identified as superoxide dismutases did not show differential expression [87]. From a list of 7 previously identified catalases in F. graminearum, 6 showed no differential expression, while 1 was significantly down-regulated [88]. One catalase which was separately identified using its annotation was up-regulated in dual culture (FGRAMPH1_01G05343). In addition, there were no genes annotated as glutathione S-transferase that were up-regulated in dual culture, though several were significantly down-regulated. F. graminearum did show up-regulation of 3 genes related to heavy metal tolerance through phytochelatin or glutathione transport into the vacuole. Phytochelatin is a polymer of glutathione which is used to sequester cadmium. Upon transport into the vacuole via HMT1, an ABC transporter, the phytochelatin-cadmium complex can integrate sulfide to achieve higher cadmium binding and stability. Phytochelatin may also confer tolerance to copper, mercury, silver and arsenate ions [89].
While not associated to particular GO terms, all 4 genes previously identified as killer protein 4-like proteins were up-regulated in dual culture. These had previously been shown to be induced by F. graminearum in stressful conditions such as high salt concentrations or osmotic pressures, and it is believed they play a role in virulence, though it is uncertain which specific function they perform in the stress response [90].
F. graminearum showed a clear down-regulation of genes associated to metabolism of phenolic compounds in dual culture with B. velezensis. In nature, F. graminearum overwinters on infested crop residues, adopting a saprotrophic lifestyle [91]. Fungi living on plant litter must decompose the lignin-related aromatic compounds to use as a carbon source [92]. Fungi employ various pathways to degrade and recycle aromatic compounds, usually through one of 5 intermediates: protocatechuate, catechol, homogentisate, gentisate or hydroquinone [50]. Potato, a major component of the culture media, is known to contain various phenolic compounds such as phenolic acids and lignin [93, 94]. F. graminearum showed down-regulation of genes in the protocatechuate, catechol and homogentisate pathways. It also down-regulated genes of the kynurenine pathway, responsible for degrading the aromatic amino acid tryptophan into NAD+. It is possible that in dual culture, B. velezensis is metabolizing some of the aromatics in the media, lessening the requirements for aromatic metabolism in the fungus. Phenolic compounds such as 2,3-dihydroxybenzoate are required for the synthesis of bacillibactin, a secondary metabolite produced by B. velezensis [95]. This notion merits further investigation.
In dual culture, F. graminearum showed significant down-regulation of 3 biosynthetic enzymes: TRI1, TRI5 and TRI13, with TRI4 approaching significance. TRI1, TRI4 and TRI5 are all key components of the trichothecene synthesis pathway [51]. TRI4 and TRI5 carry out the first 5 steps of the biosynthetic pathway, while TRI1 performs a later step. TRI101 was significantly up-regulated in dual culture, however, which is unexpected given its position between TRI4 and TRI1 in the biosynthetic pathway. TRI13 has no active functionality in deoxynivalenol-producing chemotypes of F. graminearum, which include strain DAOMC 180378 [51]. This is reflected by its low expression relative to the other genes of the TRI locus. TRI15 is a negative regulator of trichothecene biosynthesis which was up-regulated in dual culture [52]. Its specific gene targets are unknown, though exogenously added trichothecenes induced its expression in F. sporotrichoides, a trichothecene producer. Studies of trichothecene gene expression have not analyzed TRI15 expression, though we speculate its up-regulation in dual culture caused down-regulation of trichothecene biosynthetic genes. Down-regulation of trichothecene biosynthetic genes or reduction in trichothecene production through dual culture with Bacillus has been observed before [22–24]. The mechanism through which this gene inhibition occurs remains to be elucidated.
In summary, this work investigated the transcriptomic response of both a biocontrol candidate bacteria and its fungal phytopathogenic target in a dual culture assay. B. velezensis E68 was found to up-regulate genes related to sporulation and phosphate stress response and down-regulate genes related to secondary metabolism, biofilm formation and the TCA cycle. F. graminearum DAOMC 180378 saw up-regulation of heavy metal tolerance genes and killer protein-4 like proteins. It down-regulated trichothecene biosynthesis and phenol metabolism.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Fungal plugs from 7–10 day old cultures on PDA were placed at the centre of an LBA (A-H) or PDA (I-P) plate. On interaction plates (left) B. velezensis strain E68 was grown overnight in LB media at 37°C and diluted to OD600 of 1.0. 10 μL of cell suspension was dropped 2 cm from the fungal agar plug. Fungal cultures were grown alone on control plates (right). Plates were incubated at 25°C for 4 days and then imaged. (A-B) Rhizoctonia solani AG-4, (C-D) Rhizoctonia solani AG-1-1A, (E-F) Fusarium solani, (G-H) Fusarium oxysporum, (I-J) Fusarium graminearum, (K-L) Rhizoctonia solani AG-3, (M-N) Fusarium proliferatum, (O-P) Chaetomium globosum.
(TIF)
Acknowledgments
We thank Calcul Québec and the Digital Research Alliance of Canada for allowing us use of their high performance computing resources.
Data Availability
All relevant data are within the article and its Supporting Information files.
Funding Statement
This research was funded through an NSERC Discovery Grant (RGPIN-2016-04805) to SJ. Natural Sciences and Engineering Research Council of Canada: https://www.nserc-crsng.gc.ca/index_eng.asp. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Guenther JC, Trail F. The development and differentiation of Gibberella zeae (anamorph: Fusarium graminearum) during colonization of wheat. Mycologia. 2005;97: 229–237. doi: 10.3852/mycologia.97.1.229 [DOI] [PubMed] [Google Scholar]
- 2.Langevin F, Eudes F, Comeau A. Effect of Trichothecenes Produced by Fusarium graminearum during Fusarium Head Blight Development in Six Cereal Species. Eur J Plant Pathol. 2004;110: 735–746. doi: 10.1023/B:EJPP.0000041568.31778.ad [DOI] [Google Scholar]
- 3.Pestka J. Toxicological mechanisms and potential health effects of deoxynivalenol and nivalenol. World Mycotoxin J. 2010;3: 323–347. doi: 10.3920/WMJ2010.1247 [DOI] [Google Scholar]
- 4.McMullen M, Bergstrom G, De Wolf E, Dill-Macky R, Hershman D, Shaner G, et al. A Unified Effort to Fight an Enemy of Wheat and Barley: Fusarium Head Blight. Plant Dis. 2012;96: 1712–1728. doi: 10.1094/PDIS-03-12-0291-FE [DOI] [PubMed] [Google Scholar]
- 5.Wilson W, Dahl B, Nganje W. Economic costs of Fusarium Head Blight, scab and deoxynivalenol. World Mycotoxin J. 2018;11: 291–302. doi: 10.3920/WMJ2017.2204 [DOI] [Google Scholar]
- 6.Windels CE. Economic and Social Impacts of Fusarium Head Blight: Changing Farms and Rural Communities in the Northern Great Plains. Phytopathology. 2000;90: 17–21. doi: 10.1094/PHYTO.2000.90.1.17 [DOI] [PubMed] [Google Scholar]
- 7.Parry DW, Jenkinson P, McLEOD L. Fusarium ear blight (scab) in small grain cereals—a review. Plant Pathol. 1995;44: 207–238. doi: 10.1111/j.1365-3059.1995.tb02773.x [DOI] [Google Scholar]
- 8.Paul PA, Lipps PE, Hershman DE, McMullen MP, Draper MA, Madden LV. Efficacy of Triazole-Based Fungicides for Fusarium Head Blight and Deoxynivalenol Control in Wheat: A Multivariate Meta-Analysis. Phytopathology. 2008;98: 999–1011. doi: 10.1094/PHYTO-98-9-0999 [DOI] [PubMed] [Google Scholar]
- 9.Mesterházy Á, Bartók T, Lamper C. Influence of Wheat Cultivar, Species of Fusarium, and Isolate Aggressiveness on the Efficacy of Fungicides for Control of Fusarium Head Blight. Plant Dis. 2003;87: 1107–1115. doi: 10.1094/PDIS.2003.87.9.1107 [DOI] [PubMed] [Google Scholar]
- 10.Lv X, Pan L, Wang J, Lu L, Yan W, Zhu Y, et al. Effects of triazole fungicides on androgenic disruption and CYP3A4 enzyme activity. Environ Pollut. 2017;222: 504–512. doi: 10.1016/j.envpol.2016.11.051 [DOI] [PubMed] [Google Scholar]
- 11.Spolti P, Del Ponte EM, Dong Y, Cummings JA, Bergstrom GC. Triazole Sensitivity in a Contemporary Population of Fusarium graminearum from New York Wheat and Competitiveness of a Tebuconazole-Resistant Isolate. Plant Dis. 2014;98: 607–613. doi: 10.1094/PDIS-10-13-1051-RE [DOI] [PubMed] [Google Scholar]
- 12.Yin Y, Liu X, Li B, Ma Z. Characterization of Sterol Demethylation Inhibitor-Resistant Isolates of Fusarium asiaticum and F. graminearum Collected from Wheat in China. Phytopathology. 2009;99: 487–497. doi: 10.1094/PHYTO-99-5-0487 [DOI] [PubMed] [Google Scholar]
- 13.Klix MB, Verreet J-A, Beyer M. Comparison of the declining triazole sensitivity of Gibberella zeae and increased sensitivity achieved by advances in triazole fungicide development. Crop Prot. 2007;26: 683–690. doi: 10.1016/j.cropro.2006.06.006 [DOI] [Google Scholar]
- 14.Köhl J, Kolnaar R, Ravensberg WJ. Mode of Action of Microbial Biological Control Agents Against Plant Diseases: Relevance Beyond Efficacy. Front Plant Sci. 2019;10: 845. doi: 10.3389/fpls.2019.00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inch S, Gilbert J. Effect of Trichoderma harzianum on perithecial production of Gibberella zeae on wheat straw. Biocontrol Sci Technol. 2007;17: 635–646. doi: 10.1080/09583150701408865 [DOI] [Google Scholar]
- 16.Xue AG, Voldeng HD, Savard ME, Fedak G, Tian X, Hsiang T. Biological control of fusarium head blight of wheat with Clonostachys rosea strain ACM941. Canadian Journal of Plant Pathology. 2009;31: 169–179. doi: 10.1080/07060660909507590 [DOI] [Google Scholar]
- 17.Legrand F, Picot A, Cobo-Díaz JF, Chen W, Le Floch G. Challenges facing the biological control strategies for the management of Fusarium Head Blight of cereals caused by F. graminearum. Biol Control. 2017;113: 26–38. doi: 10.1016/j.biocontrol.2017.06.011 [DOI] [Google Scholar]
- 18.Palazzini JM, Ramirez ML, Torres AM, Chulze SN. Potential biocontrol agents for Fusarium head blight and deoxynivalenol production in wheat. Crop Prot. 2007;26: 1702–1710. doi: 10.1016/j.cropro.2007.03.004 [DOI] [Google Scholar]
- 19.Reva ON, Swanevelder DZH, Mwita LA, Mwakilili AD, Muzondiwa D, Joubert M, et al. Genetic, Epigenetic and Phenotypic Diversity of Four Bacillus velezensis Strains Used for Plant Protection or as Probiotics. Front Microbiol. 2019;10. doi: 10.3389/fmicb.2019.02610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinke K, Mohite OS, Weber T, Kovács ÁT. Phylogenetic Distribution of Secondary Metabolites in the Bacillus subtilis Species Complex. mSystems. 2021;6: e00057–21. doi: 10.1128/mSystems.00057-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16: 115–125. doi: 10.1016/j.tim.2007.12.009 [DOI] [PubMed] [Google Scholar]
- 22.Palazzini J, Roncallo P, Cantoro R, Chiotta M, Yerkovich N, Palacios S, et al. Biocontrol of Fusarium graminearum sensu stricto, Reduction of Deoxynivalenol Accumulation and Phytohormone Induction by Two Selected Antagonists. Toxins. 2018;10: 88. doi: 10.3390/toxins10020088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanif A, Zhang F, Li P, Li C, Xu Y, Zubair M, et al. Fengycin Produced by Bacillus amyloliquefaciens FZB42 Inhibits Fusarium graminearum Growth and Mycotoxins Biosynthesis. Toxins. 2019;11: 295. doi: 10.3390/toxins11050295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W, Zhang L, Goodwin PH, Xia M, Zhang J, Wang Q, et al. Isolation, Identification, and Complete Genome Assembly of an Endophytic Bacillus velezensis YB-130, Potential Biocontrol Agent Against Fusarium graminearum. Front Microbiol. 2020;11: 3151. doi: 10.3389/fmicb.2020.598285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang N, Jabaji S. Draft Genome Sequence of Bacillus velezensis Strain E68, Isolated from an Oil Battery. Microbiol Resour Announc. 2020;9. doi: 10.1128/MRA.00332-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rani M, Weadge JT, Jabaji S. Isolation and Characterization of Biosurfactant-Producing Bacteria From Oil Well Batteries With Antimicrobial Activities Against Food-Borne and Plant Pathogens. Front Microbiol. 2020;11. doi: 10.3389/fmicb.2020.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos-Padrón E, Bordenave S, Lin S, Bhaskar IM, Dong X, Sensen CW, et al. Carbon and Sulfur Cycling by Microbial Communities in a Gypsum-Treated Oil Sands Tailings Pond. Environ Sci Technol. 2011;45: 439–446. doi: 10.1021/es1028487 [DOI] [PubMed] [Google Scholar]
- 28.Varjani SJ, Gnansounou E. Microbial dynamics in petroleum oilfields and their relationship with physiological properties of petroleum oil reservoirs. Bioresour Technol. 2017;245: 1258–1265. doi: 10.1016/j.biortech.2017.08.028 [DOI] [PubMed] [Google Scholar]
- 29.Varjani SJ, Upasani VN. Biodegradation of petroleum hydrocarbons by oleophilic strain of Pseudomonas aeruginosa NCIM 5514. Bioresour Technol. 2016;222: 195–201. doi: 10.1016/j.biortech.2016.10.006 [DOI] [PubMed] [Google Scholar]
- 30.Pacwa-Płociniczak M, Płaza GA, Piotrowska-Seget Z, Cameotra SS. Environmental Applications of Biosurfactants: Recent Advances. Int J Mol Sci. 2011;12: 633–654. doi: 10.3390/ijms12010633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data. 2010. [cited 1 Sep 2022]. Available: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 32.Krueger F. Trim Galore!: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. 2015. [cited 1 Sep 2022]. Available: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ [Google Scholar]
- 33.King R, Urban M, Hammond-Kosack KE. Annotation of Fusarium graminearum (PH-1) Version 5.0. Genome Announc. 2017;5: 2. doi: 10.1128/genomeA.01479-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shumate A, Salzberg SL. Liftoff: accurate mapping of gene annotations. Bioinformatics. 2020;37: 1639–1643. doi: 10.1093/bioinformatics/btaa1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pertea G, Pertea M. GFF Utilities: GffRead and GffCompare. F1000Res. 2020;9: 304. doi: 10.12688/f1000research.23297.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29: 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walkowiak S, Rowland O, Rodrigue N, Subramaniam R. Whole genome sequencing and comparative genomics of closely related Fusarium Head Blight fungi: Fusarium graminearum, F. meridionale and F. asiaticum. BMC Genom. 2016;17: 1–15. doi: 10.1186/s12864-016-3371-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26: 139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26: 2176–2182. doi: 10.1093/bioinformatics/btq401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017;18: 762–776. doi: 10.1016/j.celrep.2016.12.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11: R14. doi: 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan J, Zhang N, Huang Q, Raza W, Li R, Vivanco JM, et al. Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6. Sci Rep. 2015;5: 13438. doi: 10.1038/srep13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim H-K, Yun S-H. Evaluation of Potential Reference Genes for Quantitative RT-PCR Analysis in Fusarium graminearum under Different Culture Conditions. Plant Pathol J. 2011;27: 301–309. doi: 10.5423/PPJ.2011.27.4.301 [DOI] [Google Scholar]
- 44.Puri KD, Yan C, Leng Y, Zhong S. RNA-Seq Revealed Differences in Transcriptomes between 3ADON and 15ADON Populations of Fusarium graminearum In Vitro and In Planta. PLOS ONE. 2016;11: e0163803. doi: 10.1371/journal.pone.0163803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor SC, Nadeau K, Abbasi M, Lachance C, Nguyen M, Fenrich J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019;37: 761–774. doi: 10.1016/j.tibtech.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 46.Vuppada RK, Hansen CR, Strickland KAP, Kelly KM, McCleary WR. Phosphate signaling through alternate conformations of the PstSCAB phosphate transporter. BMC Microbiol. 2018;18: 8. doi: 10.1186/s12866-017-1126-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prágai Z, Harwood CR 2002. Regulatory interactions between the Pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology. 2002;148: 1593–1602. doi: 10.1099/00221287-148-5-1593 [DOI] [PubMed] [Google Scholar]
- 48.Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes Involved in Formation of Structured Multicellular Communities by Bacillus subtilis. J Bacteriol. 2004;186: 3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elsholz AKW, Wacker SA, Losick R. Self-regulation of exopolysaccharide production in Bacillus subtilis by a tyrosine kinase. Genes Dev. 2014;28: 1710–1720. doi: 10.1101/gad.246397.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martins TM, Hartmann DO, Planchon S, Martins I, Renaut J, Silva Pereira C. The old 3-oxoadipate pathway revisited: New insights in the catabolism of aromatics in the saprophytic fungus Aspergillus nidulans. Fungal Genet Biol. 2015;74: 32–44. doi: 10.1016/j.fgb.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 51.Foroud NA, Baines D, Gagkaeva TY, Thakor N, Badea A, Steiner B, et al. Trichothecenes in Cereal Grains–An Update. Toxins. 2019;11: 634. doi: 10.3390/toxins11110634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexander NJ, McCormick SP, Larson TM, Jurgenson JE. Expression of Tri15 in Fusarium sporotrichioides. Curr Genet. 2004;45: 157–162. doi: 10.1007/s00294-003-0467-3 [DOI] [PubMed] [Google Scholar]
- 53.Tomada S, Sonego P, Moretto M, Engelen K, Pertot I, Perazzolli M, et al. Dual RNA-Seq of Lysobacter capsici AZ78 –Phytophthora infestans interaction shows the implementation of attack strategies by the bacterium and unsuccessful oomycete defense responses. Environ Microbiol. 2017;19: 4113–4125. doi: 10.1111/1462-2920.13861 [DOI] [PubMed] [Google Scholar]
- 54.Benoit I, Esker MH van den, Patyshakuliyeva A, Mattern DJ, Blei F, Zhou M, et al. Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ Microbiol. 2015;17: 2099–2113. doi: 10.1111/1462-2920.12564 [DOI] [PubMed] [Google Scholar]
- 55.Kiesewalter HT, Lozano-Andrade CN, Wibowo M, Strube ML, Maróti G, Snyder D, et al. Genomic and Chemical Diversity of Bacillus subtilis Secondary Metabolites against Plant Pathogenic Fungi. mSystems. 2021;6: e00770–20. doi: 10.1128/mSystems.00770-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y, Selvaraj JN, Xing F, Zhou L, Wang Y, Song H, et al. Antagonistic Action of Bacillus subtilis Strain SG6 on Fusarium graminearum. PLOS ONE. 2014;9: e92486. doi: 10.1371/journal.pone.0092486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong A-D, Li H-P, Yuan Q-S, Song X-S, Yao W, He W-J, et al. Antagonistic Mechanism of Iturin A and Plipastatin A from Bacillus amyloliquefaciens S76-3 from Wheat Spikes against Fusarium graminearum. PLOS ONE. 2015;10: e0116871. doi: 10.1371/journal.pone.0116871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu Q, Yang Y, Yuan Q, Shi G, Wu L, Lou Z, et al. Bacillomycin D Produced by Bacillus amyloliquefaciens Is Involved in the Antagonistic Interaction with the Plant-Pathogenic Fungus Fusarium graminearum. Appl Environ Microbiol. 2017;83: e01075–17. doi: 10.1128/AEM.01075-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L, Heng J, Qin S, Bian K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLOS ONE. 2018;13: e0198560. doi: 10.1371/journal.pone.0198560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi G, Zhu F, Du P, Yang X, Qiu D, Yu Z, et al. Lipopeptide induces apoptosis in fungal cells by a mitochondria-dependent pathway. Peptides. 2010;31: 1978–1986. doi: 10.1016/j.peptides.2010.08.003 [DOI] [PubMed] [Google Scholar]
- 61.Liu J, Zhou T, He D, Li X, Wu H, Liu W, et al. Functions of Lipopeptides Bacillomycin D and Fengycin in Antagonism of Bacillus amyloliquefaciens C06 towards Monilinia fructicola. MIP. 2011;20: 43–52. doi: 10.1159/000323501 [DOI] [PubMed] [Google Scholar]
- 62.Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6: 212–225. doi: 10.1111/1758-2229.12130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chai Y, Kolter R, Losick R. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol. 2010;78: 218–229. doi: 10.1111/j.1365-2958.2010.07335.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Activation of Cell-Specific Transcription by a Serine Phosphatase at the Site of Asymmetric Division. Science. 1995;270: 641–644. doi: 10.1126/science.270.5236.641 [DOI] [PubMed] [Google Scholar]
- 65.Diez V, Schujman GE, Gueiros-Filho FJ, de Mendoza D. Vectorial signalling mechanism required for cell–cell communication during sporulation in Bacillus subtilis. Mol Microbiol. 2012;83: 261–274. doi: 10.1111/j.1365-2958.2011.07929.x [DOI] [PubMed] [Google Scholar]
- 66.Broder DH, Pogliano K. Forespore Engulfment Mediated by a Ratchet-Like Mechanism. Cell. 2006;126: 917–928. doi: 10.1016/j.cell.2006.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doan T, Coleman J, Marquis KA, Meeske AJ, Burton BM, Karatekin E, et al. FisB mediates membrane fission during sporulation in Bacillus subtilis. Genes Dev. 2013;27: 322–334. doi: 10.1101/gad.209049.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Resnekov O, Losick R. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. PNAS. 1998;95: 3162–3167. doi: 10.1073/pnas.95.6.3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campo N, Rudner DZ. A Branched Pathway Governing the Activation of a Developmental Transcription Factor by Regulated Intramembrane Proteolysis. Molecular Cell. 2006;23: 25–35. doi: 10.1016/j.molcel.2006.05.019 [DOI] [PubMed] [Google Scholar]
- 70.Regan G, Itaya M, Piggot PJ. Coupling of σG Activation to Completion of Engulfment during Sporulation of Bacillus subtilis Survives Large Perturbations to DNA Translocation and Replication. J Bacteriol. 2012;194: 6264–6271. doi: 10.1128/JB.01470-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christie G, Setlow P. Bacillus spore germination: Knowns, unknowns and what we need to learn. Cell Signal. 2020;74: 109729. doi: 10.1016/j.cellsig.2020.109729 [DOI] [PubMed] [Google Scholar]
- 72.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59: 1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x [DOI] [PubMed] [Google Scholar]
- 73.Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD Is a Checkpoint Protein Linking Spore Formation to Extracellular-Matrix Production in Bacillus subtilis Biofilms. mBio. 2010;1: e00035–10. doi: 10.1128/mBio.00035-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han L-L, Shao H-H, Liu Y-C, Liu G, Xie C-Y, Cheng X-J, et al. Transcriptome profiling analysis reveals metabolic changes across various growth phases in Bacillus pumilus BA06. BMC Microbiol. 2017;17: 156. doi: 10.1186/s12866-017-1066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li B, Li Q, Xu Z, Zhang N, Shen Q, Zhang R. Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production. Front Microbiol. 2014;5: 636. doi: 10.3389/fmicb.2014.00636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cawoy H, Debois D, Franzil L, Pauw ED, Thonart P, Ongena M. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microb Biotechnol. 2015;8: 281–295. doi: 10.1111/1751-7915.12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zihalirwa Kulimushi P, Argüelles Arias A, Franzil L, Steels S, Ongena M. Stimulation of Fengycin-Type Antifungal Lipopeptides in Bacillus amyloliquefaciens in the Presence of the Maize Fungal Pathogen Rhizomucor variabilis. Front Microbiol. 2017;8: 850. doi: 10.3389/fmicb.2017.00850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao Y, Pi H, Chandrangsu P, Li Y, Wang Y, Zhou H, et al. Antagonism of Two Plant-Growth Promoting Bacillus velezensis Isolates Against Ralstonia solanacearum and Fusarium oxysporum. Sci Rep. 2018;8: 1–14. doi: 10.1038/s41598-018-22782-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Farzand A, Moosa A, Zubair M, Khan AR, Ayaz M, Massawe VC, et al. Transcriptional profiling of diffusible lipopeptides and fungal virulence genes during Bacillus amyloliquefaciens EZ1509 mediated suppression of Sclerotinia sclerotiorum. Phytopathology. 2019;110: 317–326. doi: 10.1094/PHYTO-05-19-0156-R [DOI] [PubMed] [Google Scholar]
- 80.Lahooti M., Harwood CR. Y 1999. Transcriptional analysis of the Bacillus subtilis teichuronic acid operon. Microbiology. 1999;145: 3409–3417. doi: 10.1099/00221287-145-12-3409 [DOI] [PubMed] [Google Scholar]
- 81.Rodriguez F, Lillington J, Johnson S, Timmel CR, Lea SM, Berks BC. Crystal Structure of the Bacillus subtilis Phosphodiesterase PhoD Reveals an Iron and Calcium-containing Active Site. J Biol Chem. 2014;289: 30889–30899. doi: 10.1074/jbc.M114.604892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qi Y, Kobayashi Y, Hulett FM. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J Bacteriol. 1997;179: 2534–2539. doi: 10.1128/jb.179.8.2534-2539.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Antelmann H, Scharf C, Hecker M. Phosphate Starvation-Inducible Proteins of Bacillus subtilis: Proteomics and Transcriptional Analysis. J Bacteriol. 2000;182: 4478–4490. doi: 10.1128/JB.182.16.4478-4490.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yun Y, Yin D, Dawood DH, Liu X, Chen Y, Ma Z. Functional characterization of FgERG3 and FgERG5 associated with ergosterol biosynthesis, vegetative differentiation and virulence of Fusarium graminearum. Fungal Genet Biol. 2014;68: 60–70. doi: 10.1016/j.fgb.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 85.Lv W, Wang C, Yang N, Que Y, Talbot NJ, Wang Z. Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci Rep. 2017;7: 11062. doi: 10.1038/s41598-017-11640-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang L, Sun C. Fengycins, Cyclic Lipopeptides from Marine Bacillus subtilis Strains, Kill the Plant-Pathogenic Fungus Magnaporthe grisea by Inducing Reactive Oxygen Species Production and Chromatin Condensation. Appl Environ Microbiol. 2018;84: e00445–18. doi: 10.1128/AEM.00445-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yao S-H, Guo Y, Wang Y-Z, Zhang D, Xu L, Tang W-H. A cytoplasmic Cu-Zn superoxide dismutase SOD1 contributes to hyphal growth and virulence of Fusarium graminearum. Fungal Genet Biol. 2016;91: 32–42. doi: 10.1016/j.fgb.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 88.Guo Y, Yao S, Yuan T, Wang Y, Zhang D, Tang W. The spatiotemporal control of KatG2 catalase-peroxidase contributes to the invasiveness of Fusarium graminearum in host plants. Mol Plant Pathol. 2019;20: 685–700. doi: 10.1111/mpp.12785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clemens S, Simm C. Schizosaccharomyces pombe as a model for metal homeostasis in plant cells: the phytochelatin-dependent pathway is the main cadmium detoxification mechanism. New Phytol. 2003;159: 323–330. doi: 10.1046/j.1469-8137.2003.00811.x [DOI] [PubMed] [Google Scholar]
- 90.Lu S, Faris JD. Fusarium graminearum KP4-like proteins possess root growth-inhibiting activity against wheat and potentially contribute to fungal virulence in seedling rot. Fungal Genet Biol. 2019;123: 1–13. doi: 10.1016/j.fgb.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 91.Leplat J, Heraud C, Gautheron E, Mangin P, Falchetto L, Steinberg C. Colonization dynamic of various crop residues by Fusarium graminearum monitored through real-time PCR measurements. J Appl Microbiol. 2016;121: 1394–1405. doi: 10.1111/jam.13259 [DOI] [PubMed] [Google Scholar]
- 92.Lubbers RJM, Dilokpimol A, Visser J, Mäkelä MR, Hildén KS, de Vries RP. A comparison between the homocyclic aromatic metabolic pathways from plant-derived compounds by bacteria and fungi. Biotechnol Adv. 2019;37: 107396. doi: 10.1016/j.biotechadv.2019.05.002 [DOI] [PubMed] [Google Scholar]
- 93.Akyol H, Riciputi Y, Capanoglu E, Caboni MF, Verardo V. Phenolic Compounds in the Potato and Its Byproducts: An Overview. Int J Mol Sci. 2016;17: 835. doi: 10.3390/ijms17060835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheng L, Hu X, Gu Z, Hong Y, Li Z, Li C. Characterization of physicochemical properties of cellulose from potato pulp and their effects on enzymatic hydrolysis by cellulase. Int J Biol Macromol. 2019;131: 564–571. doi: 10.1016/j.ijbiomac.2019.02.164 [DOI] [PubMed] [Google Scholar]
- 95.May JJ, Wendrich TM, Marahiel MA. The dhb Operon of Bacillus subtilisEncodes the Biosynthetic Template for the Catecholic Siderophore 2,3-Dihydroxybenzoate-Glycine-Threonine Trimeric Ester Bacillibactin. J Biol Chem. 2001;276: 7209–7217. doi: 10.1074/jbc.M009140200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Fungal plugs from 7–10 day old cultures on PDA were placed at the centre of an LBA (A-H) or PDA (I-P) plate. On interaction plates (left) B. velezensis strain E68 was grown overnight in LB media at 37°C and diluted to OD600 of 1.0. 10 μL of cell suspension was dropped 2 cm from the fungal agar plug. Fungal cultures were grown alone on control plates (right). Plates were incubated at 25°C for 4 days and then imaged. (A-B) Rhizoctonia solani AG-4, (C-D) Rhizoctonia solani AG-1-1A, (E-F) Fusarium solani, (G-H) Fusarium oxysporum, (I-J) Fusarium graminearum, (K-L) Rhizoctonia solani AG-3, (M-N) Fusarium proliferatum, (O-P) Chaetomium globosum.
(TIF)
Data Availability Statement
All relevant data are within the article and its Supporting Information files.