Abstract
Aging and age-related diseases have been linked to microbial dysbiosis with changes in blood bacterial DNA concentration. This condition may promote chronic low-grade inflammation, which can be further aggravated by antioxidant nutrient deficiency. Low plasma carotenoids are associated with an increased risk of inflammation and cellular damage and predict mortality. However, no evidence is yet available on the relationship between antioxidants and the blood bacterial DNA (BB-DNA). Therefore, this study aimed to compare BB-DNA from (a) GO (nonagenarian offspring), (b) age-matched controls (Randomly recruited Age-Stratified Individuals from the General population [RASIG]), and (c) spouses of GO (SGO) recruited in the MARK-AGE project, as well as to investigate the association between BB-DNA, behavior habits, Charlson Comorbidity Index (CCI), leucocyte subsets, and the circulating levels of some antioxidants and oxidative stress markers. BB-DNA was higher in RASIG than GO and SGO, whereas GO and SGO participants showed similar values. BB-DNA increased in smokers and males with CCI ≥ 2 compared with those with CCI ≤ 1 within RASIG. Moreover, BB-DNA was positively associated with lymphocyte, neutrophil, and monocyte counts, but not with self-reported dietary habits. Higher quartiles of BB-DNA were associated with low lutein and zeaxanthin and elevated malondialdehyde plasma concentrations in RASIG. BB-DNA was also positively correlated with nitric oxide levels. Herein, we provide evidence of a reduced BB-DNA in individuals from long-living families and their spouses, suggesting a decreased microbial dysbiosis and bacterial systemic translocation. BB-DNA was also associated with smoking, CCI, leukocyte subsets, and some redox biomarkers in older participants.
Keywords: Aging, Blood bacterial DNA, Dysbiosis, Inflammation, Longevity
The gastrointestinal tract and other epithelial surfaces such as the skin, oral cavity, and lung mucosa host a complex ecosystem of bacteria (1,2). Although the colonization of specific body sites in contact with the external environment by microorganisms is well described and known (3), the existence of microbial populations in other sterile compartments, such as the blood, is a relatively new concept. Previous studies suggest that the presence of bacteria in the blood is a consequence of translocation from other body sites, particularly the gastrointestinal tract, the oral cavity, and the respiratory tract (4–6). With regards to the precise location of microorganisms inside human blood, current evidence suggests that microorganisms can survive inside erythrocytes and leukocytes (7), and in healthy individuals, most bacterial DNA (94%) has been found to be localized within the buffy coat (5).
Recent studies show age-related differences in microbial DNA profiles present in the blood of healthy humans and changes in microbial community composition with aging (8) and with the onset of age-related diseases (5,9,10).
Gut barrier function alterations can occur as an effect of microbial dysbiosis and immune activation, causing bacterial translocation in the bloodstream with possible increment of total blood bacterial DNA (BB-DNA), amplifying the immune-inflammatory responses and promoting the pathophysiology of noninfectious disorders such as obesity, type 2 diabetes, depression, irritable bowel syndrome, cardiovascular, and chronic kidney diseases (11–14).
Although there is evidence that microbial dysbiosis can promote age-associated inflammation (9,15–17), there are no studies investigating BB-DNA in longevity models. Deficiencies in antioxidant nutritional factors may accentuate chronic inflammation (18). For example, low plasma levels of carotenoids have been associated with a higher risk of inflammation and cellular damage (19) with altered intestinal barrier function (20) and may predict mortality in the older adult population (19).
Furthermore, despite the proven antimicrobial activity of some antioxidants such as carotenoids (21), tocopherols (22), and vitamin C (23), no study has investigated a possible relationship between concentrations of circulating antioxidants and BB-DNA.
Therefore, the objective of this study is to compare BB-DNA in GO (offspring of nonagenarians), community-dwelling older adults (Randomly recruited Age-Stratified Individuals from the General population [RASIG]), and spouses of GO (SGO) recruited in the MARK-AGE project, as well as to investigate the association among BB-DNA, behavior habits (diet, smoking, alcohol consumption, and physical activity), Charlson Comorbidity Index (CCI), and oxidant and antioxidant biomarkers.
Materials and Methods
Study Population, Recruitment, Data, and Blood Collection
In the present study, we measured BB-DNA in peripheral blood from 1 130 participants recruited during MARK-AGE cross-sectional study (24,25). Participants were in the age range of 54–75 years and came from 8 different European countries. The population studied consisted of 799 RASIG (mean age 64.1 ± 6.1), 212 GO (GEHA offspring; mean age 64.6 ± 5.3), 119 SGO (spouses of GO; mean age 65.3 ± 5.1). GO comprised participants born from a long-living parent belonging to a family with long-living sibling(s), such as the “90+ sib-pairs” recruited within the framework of the EU Integrated Project GEHA, and designated GEHA offspring (GO).
Seropositivity for HIV, HBV (except seropositivity by vaccination), and HCV represented exclusion criteria. Details of the recruitment procedures and the collection of anthropometric, clinical, and demographic data and behavior habits, as well as details of laboratory parameter assays have already been reported (26–28). Comorbidities were evaluated using the CCI (29).
Anticoagulated whole blood, obtained by phlebotomy after overnight fasting, was collected. Prepared samples of plasma, peripheral blood mononuclear cells, and whole blood from the various recruitment centers were shipped to the MARK-AGE Biobank located at the University of Hohenheim, Stuttgart, Germany, on dry ice. From the Biobank, coded samples were subsequently sent to the different laboratories on dry ice, where they were stored at −80°C until use (27).
16S rRNA Quantification by Real-Time qPCR
DNA was extracted from 300 µL of whole blood biobanked samples using QIAamp DNA Blood mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions.
Details in the methodology of 16S rRNA quantification are previously reported (30). Briefly, highly sensitive and specific universal primers targeting the V3–V4 hypervariable region of the bacterial 16S rDNA were used in real-time qPCR reactions to quantify the 16S rRNA gene copy number in DNA samples (31). The PCR mixture (20 µL) consisted of 20 ng of DNA, SensiFAST SYBR Hi-ROX Mix 1× (Bioline, London, UK), and 0.4 µM of the following primers: For 5′-TCCTACGGGAGGCAGCAGT-3′ and Rev 5′-GGACTACCAGGGTATCTAATCCTGTT-3′.
The thermal profile used for the reaction included a heat activation of the enzyme at 95°C for 2 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 60 seconds, followed by melt analysis ramping at 60–95°C. All measurements were taken in the log phase amplification. Standard curves obtained using a 10-fold dilution series of bacterial DNA standards (Femto bacterial DNA quantification kit, Zymo research) ranging from 0.0002 to 2 pg were routinely run with each sample set and compared with previous standard curves to check for consistency between runs. Amplicon quality was ascertained by melting curves. Amplifications of samples and standard dilutions were performed in triplicate on the StepOne Real-Time PCR System (Applied Biosystems by Life Technologies, Waltham, MA). BB-DNA levels were expressed as nanogram per milliliter of whole blood and were calculated by normalizing the absolute quantities of BB-DNA of each sample to their dilution factors and to the volume of starting blood used for the extraction.
A series of controls both in silico and in vitro was performed to exclude artifacts from sample manipulation, reagent contamination, and nonspecific amplifications. The primers were checked for possible cross-hybridization with genes from eukaryotic and mitochondrial genomes using the database similarity search program BLAST. The BLAST search results showed no hits, thus confirming the specificity of primers for the bacterial 16S rRNA as previously reported (31). Separate working areas for real-time PCR mix preparation, template addition, and performing the PCRs were used, and all experimental procedures were performed under a laminar flow hood by using dedicated pipettes, filter-sealed tips, and plasticware guaranteed to be DNA free. Negative controls, in which ultrapure water instead of DNA was added, were also run in each plate.
Determination of Total Glutathione and Total Free Cysteine in Whole Blood
Total glutathione and total free cysteine in whole blood were measured by RP-HPLC with UV-Vis detection after reduction and modifications as previously reported (32).
Determination of Ascorbic Acid, Nitric Oxide, and Uric Acid in Plasma
Plasma ascorbic acid and uric acid were analyzed by RP-HPLC and UV detection after reduction with tris-(2-carboxyethyl)-phosphine. The details of the method have been previously reported (32). The concentration of the oxidized metabolites of NO was determined with Nitric Oxide Assay Kit (Abcam, ab 65328), which provides an accurate measure of total nitrate/nitrite reflecting nitric oxide amount in samples. The detection limit of the assay is approximately 1 nmol nitrite/well.
Determination of Carotenoids, Tocopherols, and Retinol in Plasma
The carotenoids (α-carotene, β-carotene, β-cryptoxanthin, lutein, zeaxanthin, and lycopene), α-tocopherol, ϒ-tocopherol, and retinol were analyzed in plasma and determined simultaneously by RP-HPLC with UV and fluorescence detection as previously described (33).
Malondialdehyde and Protein Carbonyl Analysis in Plasma
Plasma malondialdehyde was determined by RP-HPLC coupled with fluorescence detection after derivatization with thiobarbituric acid as previously described (32). The determination of plasma protein carbonyls was performed by a sensitive ELISA method as previously reported (34).
Plasma Isoprostanes
The levels of plasma isoprostanes were detected by a Human 8-epi-prostaglandin F2 alpha (8-iso-PGF2α) time-resolved fluorescence immunoassay on an AutoDELFIA (PerkinElmer).
Statistical Analysis
Participant characteristics were reported as mean ± SEM or percentages for continuous and categorical variables, respectively. For continuous variables, the normal distribution was verified by the 1-sample Kolmogorov–Smirnov test. All the variables not normally distributed were log-transformed. Differences between groups were checked by the 1-way analysis of variance (ANOVA) for continuous variables and Pearson’s χ 2 test for categorical variables. ANOVA (after correction for age, sex, countries, smoke habit, and CCI) was used to evaluate differences in BB-DNA levels among GO, SGO, and RASIG or differences in redox parameters among bacterial DNA quartiles. Generalized linear models were also performed to analyze the effect of dietary and lifestyle habits on BB-DNA in RASIG. Multivariate linear regression analysis, with enter method, was performed to evaluate the association between leucocyte subsets and BB-DNA in RASIG participants after sex stratification and adjusting for age and country. A linear regression analysis, using the stepwise method, was also carried out to explore the main predictors of bacterial DNA. The variables included were as follows: age, sex, country, body mass index, oxidative stress markers, and antioxidant nutrients.
The level of statistical significance was set at α ≤ .05.
All the analyses were performed using the SPSS/Win program (version 22.0; SPSS Inc., Chicago, IL).
Results
Characteristics of the MARK-AGE Population
Table 1 shows the main characteristics of the RASIG, GO, and SGO populations. All participants had similar age, whereas SGO had a lower percentage of females than males compared with GO and RASIG. Moreover, no significant differences were found in the examined laboratory parameters such as red blood cell number, white blood cell count, number of neutrophils, lymphocytes, monocytes and platelets, albumin, CRP, lipid serum levels, fasting glucose, hemoglobin A1c, and creatinine serum levels. No significant differences were observed in the frequency of cardiovascular diseases, diabetes, depressive symptoms, chronic obstructive pulmonary disease, osteoporosis, and hypothyroidism among groups.
Table 1.
Characteristics of Participants Selected From the Whole MARK-AGE Population for Circulating Bacterial DNA Analysis
| RASIG | GO | SGO | p-Value | |
|---|---|---|---|---|
| n = 799 | n = 212 | n = 119 | ||
| Age (y) | 64.06 ± 0.22 | 64.64 ± 0.36 | 65.34 ± 0.47 | NS |
| Females (n, %) | 413 (51.7) | 126 (59.4) | 53 (44.5) | .026 |
| RBC (×106/μL) | 4.71 ± 0.02 | 4.71 ± 0.03 | 4.76 ± 0.04 | NS |
| WBC (×103/μL) | 5.97 ± 0.06 | 6.14 ± 0.14 | 6.41 ± 0.21 | NS |
| Neutrophils (×103/μL) | 3.32 ± 0.04 | 3.33 ± 0.10 | 3.49 ± 0.15 | NS |
| Lymphocytes (×103/μL) | 1.94 ± 0.26 | 2.06 ± 0.62 | 2.11 ± 0.91 | NS |
| Monocytes (×103/μL) | 0.48 ± 0.01 | 0.49 ± 0.01 | 0.50 ± 0.02 | NS |
| Platelets (×103/μL) | 230.0 ± 2.0 | 232 ± 5.0 | 221 ± 7.3 | NS |
| Albumin (g/dL) | 4.04 ± 0.01 | 4.02 ± 0.03 | 3.99 ± 0.04 | NS |
| CRP (μg/L) | 2.14 ± 0.12 | 2.75 ± 0.30 | 2.98 ± 0.45 | NS |
| TC (mmol/L) | 5.73 ± 0.04 | 5.59 ± 0.09 | 5.71 ± 0.14 | NS |
| HDL (mmol/L) | 1.53 ± 0.01 | 1.50 ± 0.04 | 1.43 ± 0.05 | NS |
| LDL (mmol/L) | 3.36 ± 0.03 | 3.52 ± 0.08 | 3.28 ± 0.12 | NS |
| TG (mmol/L) | 1.30 ± 0.03 | 1.27 ± 0.07 | 1.63 ± 0.10 | NS |
| FG (mmol/L) | 5.40 ± 0.04 | 5.21 ± 0.10 | 5.14 ± 0.15 | NS |
| HbA1c (%) | 6.07 ± 0.02 | 6.05 ± 0.06 | 6.06 ± 0.08 | NS |
| Creatinine (μmol/L) | 74.2 ± 0.5 | 75.3 ± 1.2 | 75.2 ± 1.8 | NS |
| Cardiovascular diseases | 90 (11.3 %) | 19 (9.0%) | 10 (8.4%) | NS |
| Hypertension | 260 (32.5%) | 50 (23.6%)* | 38 (31.9%) | .041 |
| Diabetes | 75 (9.4%) | 16 (7.5%) | 10 (8.4%) | NS |
| Depressive symptoms | 106 (13.3%) | 25 (11.8%) | 9 (7.6%) | NS |
| COPD | 57(7.1%) | 15 (7.1%) | 4 (3.4%) | NS |
| Osteoporosis | 84 (10.5%) | 16 (7.5%) | 6 (5.0%) | NS |
| Hypothyroidism | 72 (9%) | 14 (6.6%) | 7 (5.9%) | NS |
Notes: COPD = chronic obstructive pulmonary disease; CRP = C-reactive protein; FG = fasting glucose; GO = nonagenarian offspring; HbA1c = hemoglobin A1c; HDL = high-density lipoprotein cholesterol; NS = nonsignificant; LDL = low-density lipoprotein cholesterol; RASIG = Randomly recruited Age-Stratified Individuals from the General population; RBC = red blood cells; SGO = spouses of GO; TG = triglycerides; TC = total cholesterol; WBC = white blood cells. Data are reported as mean ± SEM.
*p < .05 versus RASIG.
The laboratory parameter analysis was adjusted for age, sex, countries.
GO participants showed a lower prevalence of hypertension when compared with RASIG. Dietary and lifestyle habits in RASIG, GO, and SGO population are reported in Supplementary Table S1. No significant differences were observed on smoking habit, alcohol consumption, and physical activity among groups. Differences in the consumption of vegetables, meat, dairy products, eggs, and brown and white bread were found among RASIG, GO, and SGO participants.
BB-DNA Levels in GO, SGO, and RASIG Participants and Their Relation With CCI, Dietary, and Lifestyle Habits
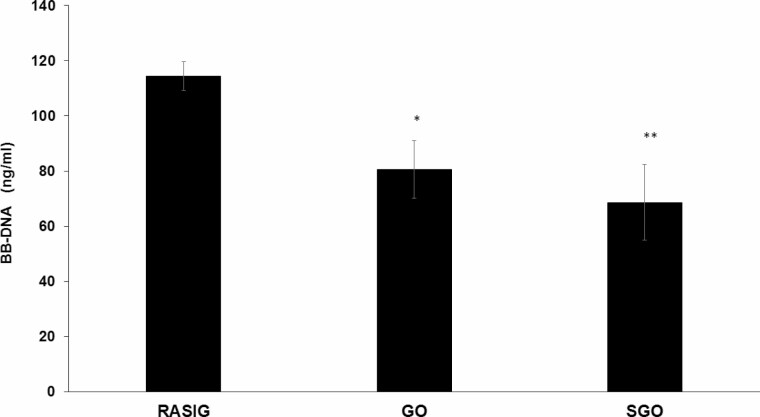
BB-DNA levels were significantly higher in RASIG compared with GO and SGO participants (Figure 1, p < .001), whereas GO and SGO participants showed similar values. Analyzing differences within-participant groups subdivided by sex, RASIG maintained a higher BB-DNA levels only with respect to GO in males and females (Supplementary Table S2).
Figure 1.
Blood bacterial DNA in peripheral blood from GO, SGO, and RASIG participants. RASIG showed significantly higher BB-DNA levels when compared with the other study groups. GO and SGO participants displayed similar BB-DNA levels. ANCOVA correcting for age, countries, sex, smoke habit, and CCI was applied; data are reported from the model adjusted mean ± SEM; *p < .05 when compared with GO; **p < .01 when compared with SGO. BB-DNA = blood bacterial DNA; CCI = Charlson Comorbity Index; GO = nonagenarian offspring; RASIG = Randomly recruited Age-Stratified Individuals from the General population; SGO = spouses of GO.
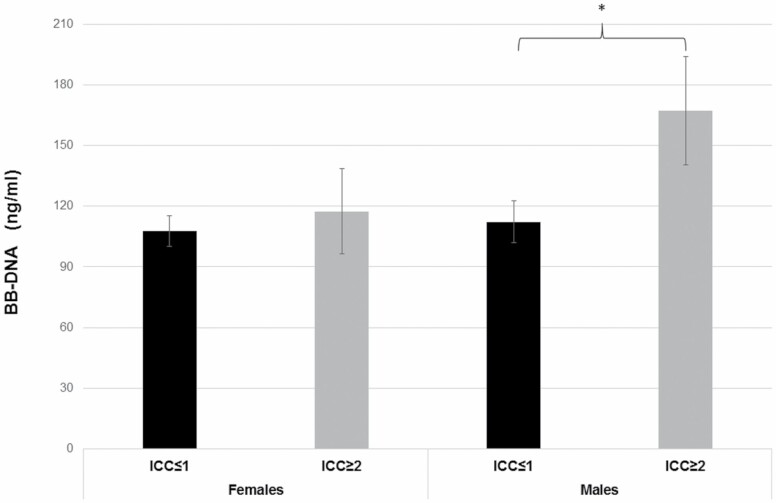
BB-DNA levels were also evaluated in relation to CCI in RASIG population stratified by sex. Characteristics of RASIG population in relation to CCI are reported in Supplementary Table S3. BB-DNA levels increased in males with CCI ≥ 2 compared with those with CCI ≤ 1 (Figure 2, p < .05), whereas no significant differences were observed in females. We have also evaluated the influence of dietary and lifestyle habits on BB-DNA in RASIG population. Generalized linear models evidenced that current smokers had higher levels of BB-DNA than former or never smokers in males and only versus never smokers in females (Supplementary Table S4 and Supplementary Figure S1). A lack of association was observed between BB-DNA and self-reported dietary habits, alcohol consumption, or physical activity.
Figure 2.
BB-DNA levels in relation to CCI in RASIG population stratified by gender. Males with CCI ≥ 2 showed increased BB-DNA levels compared with males with CCI ≤ 1. *p < .05. ANCOVA stratifying by gender and correcting for age, smoke habit, and countries was applied. BB-DNA = blood bacterial DNA; CCI = Charlson Comorbity Index.
Multivariate Stepwise Linear Regression Analysis for Antioxidant and Oxidant Parameters Independently Associated With BB-DNA Levels in the RASIG Sample
Independent biomarkers associated with BB-DNA levels (dependent variable) were identified by a linear regression analysis using a stepwise method (Table 2). BB-DNA levels were positively associated with nitric oxide levels (β coefficient = 0.105; p < .01) and negatively associated with zeaxanthin plasma levels (β coefficient = −0.082; p < .05). No association of BB-DNA was found with age, sex, country, ascorbic acid, total glutathione, α-carotene, β-carotene, β-cryptoxanthin, lutein, and lycopene, α-tocopherol, ϒ-tocopherol and retinol, malondialdehyde, protein carbonyls, total free cysteine, uric acid, and isoprostanes.
Table 2.
Multivariate Stepwise Linear Regression Analysis for Antioxidant and Oxidant Parameters Independently Associated With BB-DNA Levels in the RASIG Sample
| Model | Unstandardized Coefficients | Standardized Coefficients | p-Value | ||
|---|---|---|---|---|---|
| B | SE | β | |||
| 1 | NO | 0.228 | 0.080 | 0.102 | .005 |
| 2 | NO | 0.235 | 0.080 | 0.105 | .004 |
| Zeaxanthin | −0.141 | 0.062 | -0.082 | .024 |
Notes: BB-DNA = blood bacterial DNA; NO = nitric oxide; RASIG = Randomly recruited Age-Stratified Individuals from the General population.
Association Between BB-DNA, Lymphocyte, Monocyte, and Neutrophil Counts After Sex Stratification in RASIG Population
Multivariate linear regression analysis was performed to evaluate the association between leucocyte subsets and BB-DNA in RASIG participants after sex stratification (Supplementary Figure S2). BB-DNA was positively associated with lymphocyte (male β coefficient = 0.114, p < .05; female β coefficient = 0.180, p < .01) and neutrophil counts (male β coefficient = 0.224, p < .001; female β coefficient = 0.153, p < .01) in both sexes. Monocyte count was significantly associated with BB-DNA only in males (male β coefficient = 0.126, p < .05; female β coefficient = 0.076, p = .136).
Quartiles of BB-DNA Levels in Relation to Oxidant and Antioxidant Parameters in the RASIG Participants
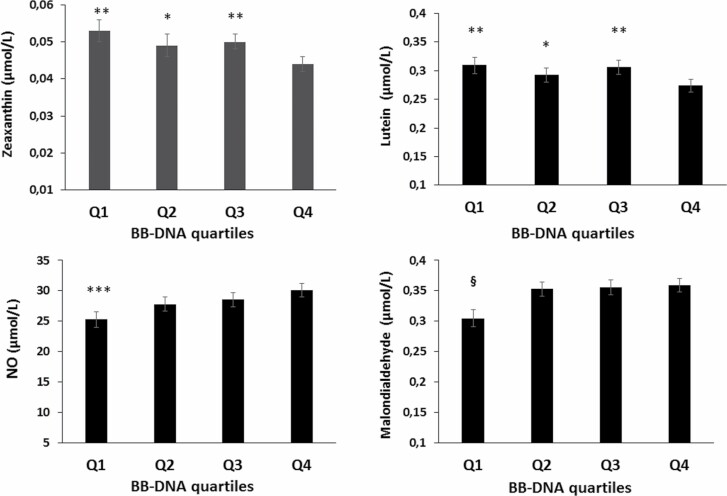
RASIG sample was divided into quartiles of BB-DNA levels, and the association with oxidant and antioxidant parameters was evaluated. The fourth quartile (Q4) of BB-DNA levels was associated with lower plasma levels of lutein and zeaxanthin when compared with the other quartiles (Figure 3, p < .05). The first quartile (Q1) of circulating BB-DNA showed lower levels of plasma malondialdehyde when compared with the fourth quartile (Figure 3, p < .05). Moreover, the first quartile of BB-DNA displayed lower NO values with respect to Q3 and Q4 quartiles (Figure 3, p < .05). No significant association was found with the other oxidant and antioxidant parameters (Supplementary Table S5).
Figure 3.
Relationship between BB-DNA quartiles and oxidant and antioxidant parameters in RASIG population. The fourth quartile (Q4) of BB-DNA levels showed the lowest plasma levels of zeaxanthin and lutein when compared with the other quartiles. Nitric oxide was diminished in the first quartile compared with Q3 and Q4 quartiles. The first quartile (Q1) of BB-DNA showed lower levels of malondialdehyde with respect to the fourth quartile. ANCOVA correcting for age, countries, sex, smoke habit, and CCI was applied. §p < .05 as compared with Q3 and Q4. ***p < .05 when compared with Q3 and Q4. **p < .01 when compared with Q4. *p < .05 when compared with Q4. BB-DNA = blood bacterial DNA; CCI = Charlson Comorbity Index; RASIG = Randomly recruited Age-Stratified Individuals from the General population.
Discussion
The presence of the circulating bacterial DNA in blood has been associated with pathological conditions such as diabetes, cardiovascular diseases, chronic kidney disease, obesity, and microbial dysbiosis (8,11–13). Gut dysbiosis is increased in aged people, and it is characterized by microbial composition changes (35), with a reduction of commensals such as Bacteroides, Bifidobacterium, and Firmicutes and an expansion of opportunistic bacteria (34). These proliferating microorganisms can induce chronic low-grade inflammation and promote the onset of age-related degenerative diseases (36,37).
We previously measured BB-DNA levels in RASIG participants. Although we did not find a difference in circulating BB-DNA levels with age (range 35–75 years), we found an association with free fatty acids, insulin, and glucose levels (30), suggesting that persistent high levels of BB-DNA in the bloodstream might predispose to metabolic syndrome or diabetes. Although a causal relationship between increased BB-DNA and the onset of metabolic syndrome or diabetes has not been fully demonstrated, there is evidence showing that insulin resistance has been implicated in impairment of intestinal barrier integrity and increased gut permeability (38,39).
To the best of our knowledge, this is the first study that analyses the differences in BB-DNA levels present in blood of older participants, nonagenarian offspring, and their spouses, as well as the association of DNA amount with oxidative and antioxidant biomarkers.
GO participants had lower bacterial DNAemia than RASIG, suggesting an improved intestinal barrier function and reduced inflammaging (40). It has been shown that persistent antigenic stimulation by an increased pathogen load may be responsible for immunosenescence, leading to increased morbidity and mortality in the older adults (41).
In centenarian offspring, the immune system is more preserved and their proinflammatory status more reduced when compared with older adults without centenarian parents (42). Therefore, the lower DNAemia in GO than RASIG is consistent with this evidence. However, we found no higher anti-inflammatory markers in GO than in RASIG, but only reduced adiponectin levels in SGO females compared with the other 2 participant classes (Supplementary Table S6).
Interestingly, GO and SGO showed similar values of BB-DNA levels probably due to sharing a common environment and lifestyle factors, as also suggested in previous results from MARK-AGE project (43–45).
Herein, we analyzed the differences in certain lifestyle factors such as smoking, alcohol consumption, physical activity, and dietary habits among the participant groups and assessed whether these factors were associated with BB-DNA levels.
Differences were only found in self-reported dietary habits, but not in lifestyle in RASIG compared with GO and SGO, who had similar behavior habits.
Surprisingly, no significant association between self-reported dietary habits and BB-DNA levels was found, although diet affects gut microbiome composition and intestinal barrier integrity (46,47). This fact could be due to the limitation of using a self-reported dietary questionnaire that represents an underaccurate estimation of nutrient consumption compared with measures of nutrient intake, which avoid systematic bias.
Among lifestyle factors, only smoking affects BB-DNA levels and current smokers showed increased bacterial DNAemia when compared with nonsmokers. This result was expected, as the effects of smoking on gut dysbiosis and intestinal permeability changes have already been described (48).
Previous evidence demonstrates that host genetic background influences gut microbiome composition (43,49), whereas the environmental factors such as smoking, exercise, and diet only partially explain changes in the microbiome (43,49).
Many studies conducted in long-lived individuals and their offspring have demonstrated an increased abundance of bacterial species which are considered as potentially beneficial bacteria and linked to body mass index, immunomodulation, and healthier metabolic profile and healthy homeostasis (35,49,50). On the other hand, many commensal bacteria produce useful metabolites that preserve gut barrier integrity (51). It has been demonstrated that cohabitation, especially in spouse pairs, entails a sharing of microbiome composition at the species level (43). On this basis, we can hypothesize that GO have a favorable microbiome composition and preserved intestinal permeability, resulting in reduced BB-DNA compared with RASIG. In addition, GO participants, sharing the same environment and having similar behavior habits (diet, smoking, physical activity) as shown in our data, can pass on to their spouses their own microbiome, likely associated with improved intestinal integrity, and thus explaining the similar BB-DNA levels between GO and SGO.
Of note is also the finding that BB-DNA increases in males with more comorbidities (CCI ≥ 2). As shown in Supplementary Table S3, male participants with CCI ≥ 2 are characterized by a cardiovascular risk profile due to the presence of dyslipidaemia, hyperglycemia, overweight, and subclinical systemic inflammation as suggested by the higher Cu/Zn ratio levels (Supplementary Table S3) (44) and by the increased proinflammatory cytokines observed in a subgroup of male RASIG participants (Supplementary Table S7). These results are consistent with previous evidence that demonstrated as bacterial translocation in the circulation, likely due to altered intestinal barrier, can affect the inflammatory state, contributing to the onset and progression of cardiovascular diseases (11). There are many causes that can affect intestinal barrier permeability including hyperglycemia, increased alcohol consumption, chronic liver disease, alterations in epithelial stem cell turn over, mucus layer thickness, and tight junctions disruption (52). Moreover, innate or adaptive immune dysfunction that occurs with aging can also contribute to the translocation of microbes. The commensal microbiota play a crucial role in reinforcing the gut barrier by preventing colonization by pathogens and producing useful metabolites such as short-chain fatty acids, which promote epithelial health and integrity. Therefore, overgrowth and alterations in the diversity of the intestinal bacterial populations (dysbiosis) can lead to intestinal inflammation and gut barrier alteration (51). It has also been observed a positive association was also observed between BB-DNA and neutrophil, lymphocyte, and monocyte counts, which was partly described in our previous study (30) and which is consistent with recent findings on the implication of gut microbiota in immune cell modulation (53).
Our findings regarding the relationship between BB-DNA levels and NO are concordant with previous evidence showing a positive association between NO blood levels and BB-DNA abundance in cirrhotic patients (54). Circulating BB-DNA triggers an immune response inducing secretion of proinflammatory cytokines that activate the inducible form of NO synthase (iNOS) and release NO with the induction of hemodynamic disturbances (55). NO overproduction by iNOS may lead to decreased endothelial viability and may contribute to tissue damage through both direct cytotoxic effects and the interaction with reactive oxygen intermediates with detrimental effects on the gut barrier, increasing the intensity of bacterial translocation (56,57). Therefore, elevated circulating BB-DNA could induce NO release, and, at the same time, NO overproduction could promote bacterial translocation. Accordingly, GO had lower plasma NO levels than RASIG that in part could explain the reduced presence of BB-DNA levels in the bloodstream (Supplementary Table S8). However, further investigations are needed to better understand these phenomena in aging.
In this study, higher quartiles of BB-DNA were associated with lower levels of certain carotenoids such as lutein and zeaxanthin and higher malondialdehyde plasma concentrations in RASIG, suggesting that an increased bacterial translocation could also depend on an altered redox homeostasis in these participants. In fact, previous evidence shows higher levels of oxidative stress biomarkers in RASIG when compared with GO and different plasma concentrations of some antioxidants (32). Becasue carotenoids have antioxidant and anti-inflammatory properties, improve gut immune function (58), and influence gut microbiome diversity and composition (46), their higher intake and bioavailability may protect intestinal barrier function and prevent gut dysbiosis (47).
Our study has some limitations. First, the 16S microbiome analysis does not discriminate between the presence of just microbial DNA fragments and viable microbes. Second, our study lacked the measurement of markers associated with gut permeability such as zonulin and a characterization of the blood microbiome composition. Third, this study does not clarify whether there are causal relationships between redox changes and increased BB-DNA levels; thus, future studies are needed to explain these aspects.
Conclusion
In summary, our study provides the first evidence of a reduced BB-DNA presence in the bloodstream of individuals from long-living families and also in their spouses, advocating a decreased microbial dysbiosis and bacterial systemic translocation. Bacterial DNAemia increases in smokers and in males with 2 or more comorbidities, and it is also associated with leukocyte subset counts and some redox biomarkers in older participants, suggesting a role for redox imbalance in promoting bacterial translocation. Our study provides basic evidence for further investigations that can elucidate blood microbiome implications in healthy longevity and age-related diseases.
Supplementary Material
Acknowledgments
The authors thank the participants who were enrolled in MARK-AGE study; a special thanks to Oliver Toussaint who dedicated his life to research activity with great energy, creativity, and enthusiasm. We remember him with great admiration and affection.
Contributor Information
Robertina Giacconi, Advanced Technology Center for Aging Research, IRCCS INRCA, Ancona, Italy.
Patrizia D’Aquila, Department of Biology, Ecology and Earth Sciences (DIBEST), University of Calabria, Rende, Italy.
Marco Malavolta, Advanced Technology Center for Aging Research, IRCCS INRCA, Ancona, Italy.
Francesco Piacenza, Advanced Technology Center for Aging Research, IRCCS INRCA, Ancona, Italy.
Alexander Bürkle, Molecular Toxicology Group, Department of Biology, University of Konstanz, Konstanz, Germany.
María Moreno Villanueva, Molecular Toxicology Group, Department of Biology, University of Konstanz, Konstanz, Germany; Human Performance Research Centre, Department of Sport Science, University of Konstanz, Konstanz, Germany.
Martijn E T Dollé, Centre for Health Protection, National Institute for Public Health and the Environment, Bilthoven, The Netherlands.
Eugène Jansen, Centre for Health Protection, National Institute for Public Health and the Environment, Bilthoven, The Netherlands.
Tilman Grune, Department of Molecular Toxicology, German Institute of Human Nutrition Potsdam-Rehbruecke (DIfE), Nuthetal, Germany; University of Potsdam, Institute of Nutritional Science, Nuthetal, Germany; Department of Physiological Chemistry, Faculty of Chemistry, University of Vienna, Vienna, Austria.
Efstathios S Gonos, National Hellenic Research Foundation, Institute of Biology, Medicinal Chemistry and Biotechnology, Athens, Greece.
Claudio Franceschi, Department of Experimental, Diagnostic and Specialty Medicine, Alma Mater Studiorum, University of Bologna, Bologna, Italy; Institute of Information Technologies, Mathematics and Mechanics, Lobachevsky University, Nizhniy Novgorod, Russia.
Miriam Capri, Department of Experimental, Diagnostic and Specialty Medicine, Alma Mater Studiorum, University of Bologna, Bologna, Italy; Interdepartmental Center—Alma Mater Research Institute on Global Challenges and Climate Change, University of Bologna, Bologna, Italy.
Daniela Gradinaru, Ana Aslan National Institute of Gerontology and Geriatrics, Bucharest, Romania; Faculty of Pharmacy, Department of Biochemistry, Carol Davila University of Medicine and Pharmacy, Bucharest, Romania.
Beatrix Grubeck-Loebenstein, Research Institute for Biomedical Aging Research, University of Innsbruck, Innsbruck, Austria.
Ewa Sikora, Laboratory of the Molecular Bases of Ageing, Nencki Institute of Experimental Biology, Polish Academy of Sciences, Warsaw, Poland.
Wolfgang Stuetz, Institute of Nutritional Sciences, Department of Food Biofunctionality, University of Hohenheim, Stuttgart, Germany.
Daniela Weber, Department of Molecular Toxicology, German Institute of Human Nutrition Potsdam-Rehbruecke (DIfE), Nuthetal, Germany; University of Potsdam, Institute of Nutritional Science, Nuthetal, Germany.
Olivier Toussaint, URBC-NARILIS, University of Namur, Namur, Belgium.
Florence Debacq-Chainiaux, URBC-NARILIS, University of Namur, Namur, Belgium.
Antti Hervonen, The Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland.
Mikko Hurme, The Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland.
P Eline Slagboom, Department of Molecular Epidemiology, Leiden University Medical Centre, Leiden, The Netherlands.
Christiane Schön, BioTeSys GmbH, Esslingen, Germany.
Jürgen Bernhardt, BioTeSys GmbH, Esslingen, Germany.
Nicolle Breusing, Department of Applied Nutritional Science/Dietetics, Institute of Nutritional Medicine, University of Hohenheim, Stuttgart, Germany.
Talbot Duncan, Unilever Science and Technology, Bedford, UK.
Giuseppe Passarino, Department of Biology, Ecology and Earth Sciences (DIBEST), University of Calabria, Rende, Italy.
Dina Bellizzi, Department of Biology, Ecology and Earth Sciences (DIBEST), University of Calabria, Rende, Italy.
Mauro Provinciali, Advanced Technology Center for Aging Research, IRCCS INRCA, Ancona, Italy.
Funding
This work was supported by the European Commission (Project Full Name: European Study to Establish Biomarkers of Human Ageing; Project Acronym: MARK-AGE; Project No. 200880) and in part by the Project: PROMISING: “aPpROccio alla MedIcina di preciSIoNe in Geriatria: dalle basi biomolecolari dell’ invecchiamento e delle malattie età correlate ai modelli clinico/assistenziali” RCR-2021-23671216—Cat.B-PRV-RETE AGING—by Italian Health Ministry. We thank all study participants for their willingness to participate. The work has been made possible by the collaboration with the nursing homes of SADEL S.p.A (San Teodoro, San Raffaele, Villa del Rosario, A.G.I srl, SAVELLI HOSPITAL., Casa di Cura Madonna dello Scoglio) in the frame of the agreement “SOLUZIONI INNOVATIVE PER L’INNALZAMENTO DELLA SALUTE E DELLA SICUREZZA DELLA POPOLAZIONE” with the University of Calabria.
Conflict of Interest
None declared.
Author Contributions
Conceptualization: R.G., D.B., M.P.; Methodology: P.D., R.G., F.P.; Software: R.G., M.M., M.M.V.; BioBank: T.G, N.B.; recruitment of participants: B.G.-L. (Austrian cohort), O.T. and F.D.-C. (Belgian cohort) C.S. and J.B. (German cohort), E.S.G. (Greek cohort), C.F. and M.C. (Italian cohort), E.S. (Polish cohort), and A.H. (Finnish cohort); Validation: P.D., R.G.; Formal analysis: D.B., M.P., G.P., A.B., M.M.V., D.G., W.S., D.W., T.D., M.E.T.D.; Investigation: R.G., P.D.; Resources: A.B, D.B.; Data curation: R.G., M.M., P.D., E.J., M.E.T.D., E.S.; D.G., D.W., T.D., M.H., E.S.; Writing—original draft preparation: R.G.; Writing—review and editing: P.D., R.G., D.B., M.P., A.B., M.M.V., M.M., F.P.; Supervision: D.B., M.P., G.P; Project administration: A.B.; Funding acquisition: A.B. All authors have read and agreed to the published version of the manuscript.
Ethics Approval and Consent to Participate
This study was approved by the local ethics committees of each Recruitment Centre. The written informed consent for each participant was obtained before study enrollment.
Data Availability
The data sets used and analyzed during the current study are available from the authors upon reasonable request and with permission of the MARK-AGE Consortium.
References
- 1. Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Segal LN, Blaser MJ. A brave new world: the lung microbiota in an era of change. Ann Am Thorac Soc. 2014;11:S21–7. doi: 10.1513/AnnalsATS.201306-189MG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Opazo MC, Ortega-Rocha EM, Coronado-Arrázola I, et al. Intestinal microbiota influences non-intestinal related autoimmune diseases. Front Microbiol. 2018;9(March):432. doi: 10.3389/fmicb.2018.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Whittle E, Leonard MO, Harrison R, Gant TW, Tonge DP. Multi-method characterization of the human circulating microbiome. Front Microbiol. 2018;9(January):3266. doi: 10.3389/fmicb.2018.03266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Païssé S, Valle C, Servant F, et al. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56(5):1138–1147. doi: 10.1111/trf.13477 [DOI] [PubMed] [Google Scholar]
- 6. Horliana ACRT, Chambrone L, Foz AM, et al. Dissemination of periodontal pathogens in the bloodstream after periodontal procedures: a systematic review. PLoS One. 2014;9(5):e98271. doi: 10.1371/journal.pone.0098271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Damgaard C, Magnussen K, Enevold C, et al. Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS One. 2015;10(3):e0120826. doi: 10.1371/journal.pone.0120826m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gargari G, Mantegazza G, Taverniti V, et al. Bacterial DNAemia is associated with serum zonulin levels in older subjects. Sci Rep. 2021;11(1):11054. doi: 10.1038/s41598-021-90476-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buford TW, Carter CS, VanDerPol WJ, et al. Composition and richness of the serum microbiome differ by age and link to systemic inflammation. GeroScience. 2018;40(3):257–268. doi: 10.1007/s11357-018-0026-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kowarsky M, Camunas-Soler J, Kertesz M, et al. Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. Proc Natl Acad Sci USA. 2017;114(36):9623–9628. doi: 10.1073/pnas.1707009114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dinakaran V, Rathinavel A, Pushpanathan M, Sivakumar R, Gunasekaran P, Rajendhran J. Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PLoS One. 2014;9(8): 10.1371/journal.pone.0105221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qiu J, Zhou H, Jing Y, Dong C. Association between blood microbiome and type 2 diabetes mellitus: a nested case–control study. J Clin Lab Anal. 2019;33(4):e22842. doi: 10.1002/jcla.22842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shah NB, Allegretti AS, Nigwekar SU, et al. Blood microbiome profile in CKD: a pilot study. Clin J Am Soc Nephrol. 2019;14(5):692–701. doi: 10.2215/CJN.12161018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Slyepchenko A, Maes M, Jacka FN, et al. Gut microbiota, bacterial translocation, and interactions with diet: pathophysiological links between major depressive disorder and non-communicable medical comorbidities. Psychother Psychosom. 2016;86(1):31–46. doi: 10.1159/000448957 [DOI] [PubMed] [Google Scholar]
- 15. Thevaranjan N, Puchta A, Schulz C, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21(4):455–466.e4. doi: 10.1016/j.chom.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biagi E, Candela M, Fairweather-Tait S, Franceschi C, Brigidi P. Ageing of the human metaorganism: the microbial counterpart. Age (Omaha). 2012;34(1):247–267. doi: 10.1007/s11357-011-9217-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kavanagh K, Hsu FC, Davis AT, Kritchevsky SB, Rejeski WJ, Kim S. Biomarkers of leaky gut are related to inflammation and reduced physical function in older adults with cardiometabolic disease and mobility limitations. GeroScience. 2019;41(6):923–933. doi: 10.1007/s11357-019-00112-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reed S, Neuman H, Moscovich S, Glahn RP, Koren O, Tako E. Chronic zinc deficiency alters chick gut microbiota composition and function. Nutrients. 2015;7(12):9768–9784. doi: 10.3390/nu7125497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lauretani F, Semba RD, Dayhoff-Brannigan M, et al. Low total plasma carotenoids are independent predictors of mortality among older persons: The InCHIANTI study. Eur J Nutr. 2008;47(6):335–340. doi: 10.1007/s00394-008-0732-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vieira MM, Paik J, Blaner WS, et al. Carotenoids, retinol, and intestinal barrier function in children from Northeastern Brazil. J Pediatr Gastroenterol Nutr. 2008;47(5):652–659. doi: 10.1097/MPG.0b013e31816bf4bf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cucco M, Guasco B, Malacarne G, Ottonelli R. Effects of β-carotene on adult immune condition and antibacterial activity in the eggs of the grey partridge, Perdix perdix. Comp Biochem Physiol A Mol Integr Physiol. 2007;147(4):1038–1046. doi: 10.1016/j.cbpa.2007.03.014 [DOI] [PubMed] [Google Scholar]
- 22. Hartmann MS, Mousavi S, Bereswill S, Heimesaat MM. Vitamin E as promising adjunct treatment option in the combat of infectious diseases caused by bacterial including multi-drug resistant pathogens—results from a comprehensive literature survey. Eur J Microbiol Immunol. 2020;10(4):193–201. doi: 10.1556/1886.2020.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mumtaz S, Mumtaz S, Ali S, et al. Evaluation of antibacterial activity of vitamin C against human bacterial pathogens. Braz J Biol. 2021;83:e247165. doi: 10.1590/1519-6984.247165 [DOI] [PubMed] [Google Scholar]
- 24. Bürkle A, Moreno-Villanueva M, Bernhard J, et al. MARK-AGE biomarkers of ageing. Mech Ageing Dev. 2015;151:2–12. doi: 10.1016/j.mad.2015.03.006 [DOI] [PubMed] [Google Scholar]
- 25. Capri M, Moreno-Villanueva M, Cevenini E, et al. MARK-AGE population: from the human model to new insights. Mech Ageing Dev. 2015;151:13–17. doi: 10.1016/j.mad.2015.03.010 [DOI] [PubMed] [Google Scholar]
- 26. Moreno-Villanueva M, Kötter T, Sindlinger T, et al. The MARK-AGE phenotypic database: structure and strategy. Mech Ageing Dev. 2015;151:26–30. doi: 10.1016/j.mad.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 27. Moreno-Villanueva M, Capri M, Breusing N, et al. MARK-AGE standard operating procedures (SOPs): a successful effort. Mech Ageing Dev. 2015;151:18–25. doi: 10.1016/j.mad.2015.03.007 [DOI] [PubMed] [Google Scholar]
- 28. Jansen E, Beekhof P, Cremers J, et al. Quality control data of physiological and immunological biomarkers measured in serum and plasma. Mech Ageing Dev. 2015;151:54–59. doi: 10.1016/j.mad.2015.06.004 [DOI] [PubMed] [Google Scholar]
- 29. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- 30. D’Aquila P, Giacconi R, Malavolta M, et al. Microbiome in blood samples from the general population recruited in the MARK-AGE project: a pilot study. Front Microbiol. 2021;12:707515. doi: 10.3389/fmicb.2021.707515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(1):257–266. doi: 10.1099/00221287-148-1-257 [DOI] [PubMed] [Google Scholar]
- 32. Weber D, Stuetz W, Toussaint O, et al. Associations between specific redox biomarkers and age in a large European cohort: the MARK-AGE project. Oxid Med Cell Longev. 2017;2017:1–12. doi: 10.1155/2017/1401452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stuetz W, Weber D, Dollé MET, et al. Plasma carotenoids, tocopherols, and retinol in the age-stratified (35–74 years) general population: a cross-sectional study in six European countries. Nutrients. 2016;8(10):614. doi: 10.3390/nu8100614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rampelli S, Candela M, Turroni S, et al. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY). 2013;5(12):902–912. doi: 10.18632/aging.100623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Biagi E, Franceschi C, Rampelli S, et al. Gut microbiota and extreme longevity. Curr Biol. 2016;26(11):1480–1485. doi: 10.1016/j.cub.2016.04.016 [DOI] [PubMed] [Google Scholar]
- 36. Dumitrescu L, Marta D, Dănău A, et al. Serum and fecal markers of intestinal inflammation and intestinal barrier permeability are elevated in Parkinson’s disease. Front Neurosci. 2021;15:689723. doi: 10.3389/fnins.2021.689723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun Y, Wu D, Zeng W, et al. The role of intestinal dysbacteriosis induced arachidonic acid metabolism disorder in inflammaging in atherosclerosis. Front Cell Infect Microbiol. 2021;11:618265. doi: 10.3389/fcimb.2021.618265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gueddouri D, Caüzac M, Fauveau V, et al. Insulin resistance per se drives early and reversible dysbiosis-mediated gut barrier impairment and bactericidal dysfunction. Mol Metab. 2022;57:101438. doi: 10.1016/j.molmet.2022.101438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mkumbuzi L, Mfengu MMO, Engwa GA, Sewani-Rusike CR. Insulin resistance is associated with gut permeability without the direct influence of obesity in young adults. Diabetes Metab Syndr Obes. 2020;13:2997–3008. doi: 10.2147/DMSO.S256864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Derhovanessian E, Maier AB, Beck R, et al. Hallmark features of immunosenescence are absent in familial longevity. J Immunol. 2010;185(8):4618–4624. doi: 10.4049/jimmunol.1001629 [DOI] [PubMed] [Google Scholar]
- 41. Ng TP, Lu Y, Tan CTY, et al. Pathogenic load and frailty in older adults: Singapore Longitudinal Ageing Study. Aging (Albany NY). 2020;12(21):22139–22151. doi: 10.18632/aging.104076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rubino G, Bulati M, Aiello A, et al. Sicilian centenarian offspring are more resistant to immune ageing. Aging Clin Exp Res. 2019;31(1):125–133. doi: 10.1007/s40520-018-0936-7 [DOI] [PubMed] [Google Scholar]
- 43. Finnicum CT, Beck JJ, Dolan CV, et al. Cohabitation is associated with a greater resemblance in gut microbiota which can impact cardiometabolic and inflammatory risk. BMC Microbiol. 2019;19(1):230. doi: 10.1186/s12866-019-1602-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Piacenza F, Giacconi R, Costarelli L, et al. Age, sex, and BMI influence on copper, zinc, and their major serum carrier proteins in a large European population including nonagenarian offspring from MARK-AGE study. J Gerontol A Biol Sci Med Sci. 2021;76:20907–2106. doi: 10.1093/gerona/glab134 [DOI] [PubMed] [Google Scholar]
- 45. Ciccarone F, Malavolta M, Calabrese R, et al. Age-dependent expression of DNMT1 and DNMT3B in PBMCs from a large European population enrolled in the MARK-AGE study. Aging Cell. 2016;15(4):755–765. doi: 10.1111/acel.12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Frankenfeld CL, Hullar MAJ, Maskarinec G, et al. The gut microbiome is associated with circulating dietary biomarkers of fruit and vegetable intake in a multiethnic cohort. J Acad Nutr Diet. 2021;122:78–98. doi: 10.1016/j.jand.2021.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grar H, Dib W, Gourine H, et al. β-Carotene improves intestinal barrier function by modulating proinflammatory cytokines and improving antioxidant capacity in β-lactoglobulin-sensitized mice. J Biol Regul Homeost Agents. 2020;34(5):1689–1697. doi: 10.23812/20-24-A [DOI] [PubMed] [Google Scholar]
- 48. Gui X, Yang Z, Li MD. Effect of cigarette smoke on gut microbiota: state of knowledge. Front Physiol. 2021;12. doi: 10.3389/fphys.2021.673341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Santoro A, Ostan R, Candela M, et al. Gut microbiota changes in the extreme decades of human life: a focus on centenarians. Cell Mol Life Sci. 2018;75(1):129–148. doi: 10.1007/s00018-017-2674-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ren M, Li H, Fu Z, Li Q. Succession analysis of gut microbiota structure of participants from long-lived families in Hechi, Guangxi, China. Microorganisms. 2021;9(12):2524. doi: 10.3390/microorganisms9122524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chopyk DM, Grakoui A. Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology. 2020;159(3):849–863. doi: 10.1053/j.gastro.2020.04.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thaiss CA, Levy M, Grosheva I, et al. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science. 2018;359(6382):1376–1383. doi: 10.1126/science.aar3318 [DOI] [PubMed] [Google Scholar]
- 53. Schluter J, Peled JU, Taylor BP, et al. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588(7837):303–307. doi: 10.1038/s41586-020-2971-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Caro E, Francés R, Zapater P, Pascual S, Bellot P, Such J. Grade of soluble inflammatory response is mainly affected by circulating bacterial DNA concentrations in cirrhosis. Liver Int. 2016;36(10):1473–1480. doi: 10.1111/liv.13118 [DOI] [PubMed] [Google Scholar]
- 55. Francés R, Muñoz C, Zapater P, et al. Bacterial DNA activates cell mediated immune response and nitric oxide overproduction in peritoneal macrophages from patients with cirrhosis and ascites. Gut. 2004;53(6):860–864. doi: 10.1136/gut.2003.027425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Quirino IEP, Cardoso VN, Dos Santos RDGC, et al. The role of l-arginine and inducible nitric oxide synthase in intestinal permeability and bacterial translocation. J Parenter Enter Nutr. 2013;37(3):392–400. doi: 10.1177/0148607112458325 [DOI] [PubMed] [Google Scholar]
- 57. Grishin A, Bowling J, Bell B, Wang J, Ford HR. Roles of nitric oxide and intestinal microbiota in the pathogenesis of necrotizing enterocolitis. J Pediatr Surg. 2016;51(1):13–17. doi: 10.1016/j.jpedsurg.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lyu Y, Wu L, Wang F, Shen X, Lin D. Carotenoid supplementation and retinoic acid in immunoglobulin A regulation of the gut microbiota dysbiosis. Exp Biol Med. 2018;243(7):613–620. doi: 10.1177/1535370218763760 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and analyzed during the current study are available from the authors upon reasonable request and with permission of the MARK-AGE Consortium.