Summary
Background
Mobile plasmids play a key role in spurring the global dissemination of multidrug-resistant (MDR) K. pneumoniae, while plasmid curing has been recognized as a promising strategy to combat antimicrobial resistance. Here we exploited a K. pneumoniae native CRISPR system to cure the high-risk IncFII plasmids.
Methods
We examined matched protospacers in 725 completely sequenced IncFII plasmids from K. pneumoniae genomes. Then, we re-engineered a native CRISPR-Cas3 system and deliver the CRISPR-Cas3 system via conjugation. Plasmid killing efficiency and G. mellonella infection model were applied to evaluate the CRISPR-Cas3 immunity in vitro and in vivo.
Findings
Genomic analysis revealed that most IncFII plasmids could be targeted by the native CRISPR-Cas3 system with multiple matched protospacers, and the targeting regions were highly conserved across different IncFII plasmids. This conjugative endogenous CRISPR-Cas3 system demonstrated high plasmid curing efficiency in vitro (8-log decrease) and in vivo (∼100% curing) in a Galleria mellonella infection model, as well as provided immunization against the invasion of IncFII plasmids once the system entering a susceptible bacterial host.
Interpretation
Overall, our work demonstrated the applicability of using native CRISPR-mediated plasmid curing to re-sensitize drug-resistant K. pneumoniae to multiple antibiotics. This work provided strong support for the idea of utilizing native CRISPR-Cas systems to tackle AMR in K. pneumoniae.
Funding
This work was supported by research grants National Natural Science Foundation of China [grant numbers 81871692, 82172315, 82102439, and 82202564], the Shanghai Science and Technology Commission [grant number 19JC1413002], and Shanghai Sailing Program [grant number 22YF1437500].
Keywords: IncFII plasmid, CRISPR-Cas system, K. pneumoniae, Antimicrobial resistance
Research in context.
Evidence before this study
The rapid spread of multidrug-resistant (MDR) Klebsiella pneumoniae poses a major threat to modern medicine, jeopardizing our ability to treat life-threatening infections. Mobile plasmids play a key role in spurring the global dissemination of MDR K. pneumoniae, while plasmid curing has been recognized as a promising strategy to combat antimicrobial resistance. CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) systems have been repurposed as a novel approach for MDR plasmid curing. Cas9 nuclease has been extensively reprogrammed for genome editing in a broad range of organisms, especially eukaryotes. However, for most prokaryotes, the heterologous CRISPR-Cas9 machineries are difficult to exploit due to their large size and severe toxicity to host cells. Hence, repurposing the broadly distributed endogenous CRISPR-Cas systems is emerging as a new CRISPR-based genome-editing strategy in prokaryotes. We previously revealed a significant inverse correlation between the presence of native I-E CRISPR-Cas3 systems and the presence of MDR plasmids in clinical K. pneumoniae isolates. We, therefore, searched PubMed and Google Scholar for studies published between Jan 1, 1990, and Sep 1, 2022, containing the terms “K. pneumoniae”, “CRISPR-Cas”, and “plasmid curing”. We identified 4 studies, including two studies from us, that showed the relationship between native type I-E CRISPR-Cas system and antibiotic-resistance in K. pneumoniae. Notably, we found no evidence for the application of native type I-E CRISPR-Cas system to control the acquisition of MDR plasmids.
Added value of this study
Our study is a step toward the development of K. pneumoniae endogenous CRISPR-Cas3 system to combat the antibiotic resistance in vitro and in vivo. In this study, we re-engineered a native CRISPR-Cas3 system from a clinical K. pneumoniae strain and integrated it into a high-copy-transferable plasmid. This conjugative endogenous CRISPR-Cas3 system demonstrated high plasmid curing efficiency in vitro and in vivo in a Galleria mellonella infection model, as well as immunization against the invasion of MDR plasmids in a susceptible bacterial host. Our work provided strong support for the idea of utilizing native CRISPR-Cas3 systems to tackle antimicrobial resistance (AMR) in K. pneumoniae. Besides the native CRISPR-Cas systems in K. pneumoniae, other endogenously encoded CRISPR-Cas systems have also been increasingly considered as a promising genetic manipulation strategy in prokaryotes. The application of native CRISPR-Cas systems for clinical intervention should be further explored.
Implications of all the available evidence
MDR K. pneumoniae has emerged as a global problem hindering the treatment of bacterial infections. Effective strategies to combat MDR K. pneumoniae are urgently needed, and MDR plasmid curing could be a promising approach to reduce AMR prevalence, and sensitize bacteria to antibiotics. We developed an endogenous CRISPR-Cas3 mediated platform for the curing of high-risk resistant plasmids in K. pneumoniae, which is highly efficient in eliminating plasmids by targeting multiple sites of the conservative plasmid backbone, thereby re-sensitizing MDR strains to antibiotics. Further, we are currently optimizing this endogenous system through customized CRISPR array assembly to improve its applications and coverages. The current study confirmed the concept of using endogenous CRISPR-Cas3-mediated plasmid curing to re-sensitize resistant strains to antibiotics.
Introduction
Antimicrobial resistance (AMR) has emerged as an urgent threat to global public health.1 Klebsiella pneumoniae is one of the most commonly detected multidrug-resistant (MDR) pathogens in clinical settings, frequently associated with various resistance determinants, such as the extended-spectrum β-lactamase and carbapenemase genes.2 Plasmids served as important vectors that promoted the spread of AMR in K. pneumoniae and other Gran-negative pathogens.3 In K. pneumoniae, IncFII plasmids (including IncFIIK, IncFII, IncFIIY, and IncFIIS) is the most predominant plasmid incompatibility type, and plays a critical role in worldwide dissemination of AMR in K. pneumoniae.4, 5, 6, 7, 8, 9, 10, 11, 12, 13
Novel strategies to combat AMR are urgently needed. CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) system has been repurposed as a novel approach for AMR plasmid curing, which can cause single (class 1, Cas3) or double-stranded (class 2, Cas9) breaks in the AMR plasmid and leads to plasmid degradation.3 Cas9 nuclease has been extensively reprogrammed for genome editing in a broad range of organisms, especially eukaryotes.14 However, their applications in prokaryotes are rather limited, in part because the expression of heterologous Cas9 is frequently hindered by the exceptionally diverse and frequently poor DNA homeostasis in microbial cells.15, 16, 17, 18 For most prokaryotes, the exogenous CRISPR-Cas9 machineries are difficult to exploit due to their large size and severe toxicity to host cells.19 Consequently, the application of heterologous Cas9-mediated genome editing in some prokaryotes remain to be challenging.15 Hence, repurposing the broadly distributed endogenous CRISPR-Cas systems is emerging as a new CRISPR-based genome-editing strategy in prokaryotes.20,21
We previously revealed a significant inverse correlation between the presence of native I-E CRISPR systems and the presence of IncFII-MDR plasmids in clinical K. pneumoniae isolates.22,23 Interestingly, I-E CRISPR-Cas matched protospacers were commonly found in IncFII-MDR plasmids.22,23 This suggested that the native type I-E CRISPR-Cas system can be reprogrammed to control the acquisition of IncFII plasmids, thereby reducing the IncFII plasmid-associated AMR. Notably, this type I-E CRISPR-Cas system, similar to the Cascade–Cas3 system, displayed great potential for large genetic element deletion, including gene clusters, genomic islands, prophages and plasmids, a task that Class 2 systems (e.g. Cas9) were less efficient.24 An important distinction between the two systems is that CRISPR-Cas3 system cleaves and degrade DNA through the action of a 3′-to-5′ exonuclease, whereas Cas9 only cleaves DNA.25 The DNA degradation effect of CRISPR-Cas3 system could further improve the cleavage potency by interfering DNA repair.25
In this study, we explored the application of an endogenous type I-E CRISPR-Cas system to cure the epidemic multi-drug-resistant IncFII plasmids. We initially extracted the spacer sequences from 207 sequenced K. pneumoniae genomes from NCBI database. We then examined the matched protospacers in 725 completely sequenced IncFII plasmids from 932 K. pneumoniae genomes from the public domain. An efficient conjugative system was developed to deliver the native CRISPR-Cas3 nuclease to cure high-risk IncFII-MDR plasmids in clinical isolates. Our results also further demonstrated that the presence of type I-E CRISPR-Cas system could provide immunity to the invasion of IncFII resistant plasmids. Overall, this study is the first step toward the development of K. pneumoniae endogenous CRISPR-Cas3 system to combat the antibiotic resistance.
Methods
Bacterial strains
The bacterial strains and plasmids used or generated in this work are listed in Dataset and Table S1. Two clinical K. pneumoniae strains, JS187 (PRJNA422509) and HD5914 (PRJNA904520), were used as the conjugative recipients for the endogenous CRISPR-Cas3 system. Escherichia coli S17-1 (TpR SmR recA, thi, pro, hsdR-M + RP4: 2-Tc: Mu: KmR Tn7 λpir) was used for plasmid cloning and as the conjugative donor.26 All the bacteria were grown in Luria–Bertani (LB) broth or on LB agar plates at 37 °C. When necessary, appropriate antibiotics were added at the following final concentrations: apramycin (Apr, 100 mg/L), ampicillin (Amp, 100 mg/L), and meropenem (MEM, 1 mg/L for JS187 and 16 mg/L for HD5914).
Characterization of K. pneumoniae plasmids
A total of 932 completely sequenced K. pneumoniae genomes, including 3117 complete plasmids, were retrieved from the NCBI genome database (http://ftp.ncbi.nih.gov/genomes/, dated as 9/1/2021). The detailed replicons of these plasmids were further determined by the PlasmidFinder tool (≥90% identities and ≥90% coverage).27 Antibiotic resistance genes were annotated using Mega BLAST with the default parameters against the CRAD 2020 and ResFinder database (https://cge.cbs.dtu.dk/services/ResFinder/). MDR was defined as resistan to three or more antimicrobial classes based on the in-silico resistance gene mining results.27 Virulence genes were identified by BLAST against the VFDB database (http://www.mgc.ac.cn/VFs/main.htm), and the rmpA/rmpA2, aerobactin (iuc) and salmochelin (iro) alleles were classified using Kleborate v0.3.0 (https://github.com/katholt/Kleborate/). The complete conjugal modules in the plasmid sequences were characterized using oriTfinder,28 which detects the origin of transfer site (oriT), relaxase genes, type IV coupling protein (T4CP) genes, and the type IV secretion system gene clusters (T4SS).
Analysis of the protospacers on IncFII plasmids
In our previous study, we identified 71 CRISPR loci from 207 sequenced K. pneumoniae genomes, which contains 34 different layouts of CRISPR array and 415 distinct spacers.22 We then used blastn to query the presence of the protospacers on the 725 IncFII plasmids (characterized as above), with a minimum of 90% identities (29/32 nt).22 The IncFIIK plasmid p187-2 (GenBank accession no. CP025468.1) from our previous studies was used as a reference plasmid for comparison.23,29 The CGView (http://cgview.ca/) was used to visualize the IncFII plasmids to exemplify the conservative backbone sequences targeted by the CRISPR-Cas system (E-value, ≤0.1).
Construction of CRISPR-Cas conjugative delivery system
Plasmids were constructed using the in-fusion cloning method through the NEBuilder HiFi DNA assembly kit (New England BioLabs, Beijing). The list of primers is provided in Table S2. The pEmpty (empty plasmid) and pCRISPR (with CRISPR-Cas3 system) plasmids were constructed by polymerase chain reaction (PCR) amplification of fragments with 25–60 bp homology overlaps. The native type I-E CRISPR-Cas3 system was amplified from the K. pneumoniae KP8 (CP025636.1).23 The oriTRP4 fragment was amplified from pJTOOL-3, while the pBR322 replicon and apramycin-resistant gene aac(3)-IV were amplified from a modified pUC-19(AprR).23 The fragments were then assembled using the NEBuilder HiFi DNA assembly kit to generate the pEmpty (oriTRP4, pBR322 replicon and aac(3)-IV, without CRISPR-Cas3) or pCRISPR (oriTRP4, pBR322 replicon, aac(3)-IV and CRISPR-Cas3) plasmids, respectively. The pEmpty and pCRISPR plasmid vectors are schematically shown in Fig. 3aii. The pEmpty and pCRISPR were transferred into E. coli S17 cells, which contained the broad-host RP4 transfer machinery, allowing the conjugative transfer of pEmpty and pCRISPR into other strains.
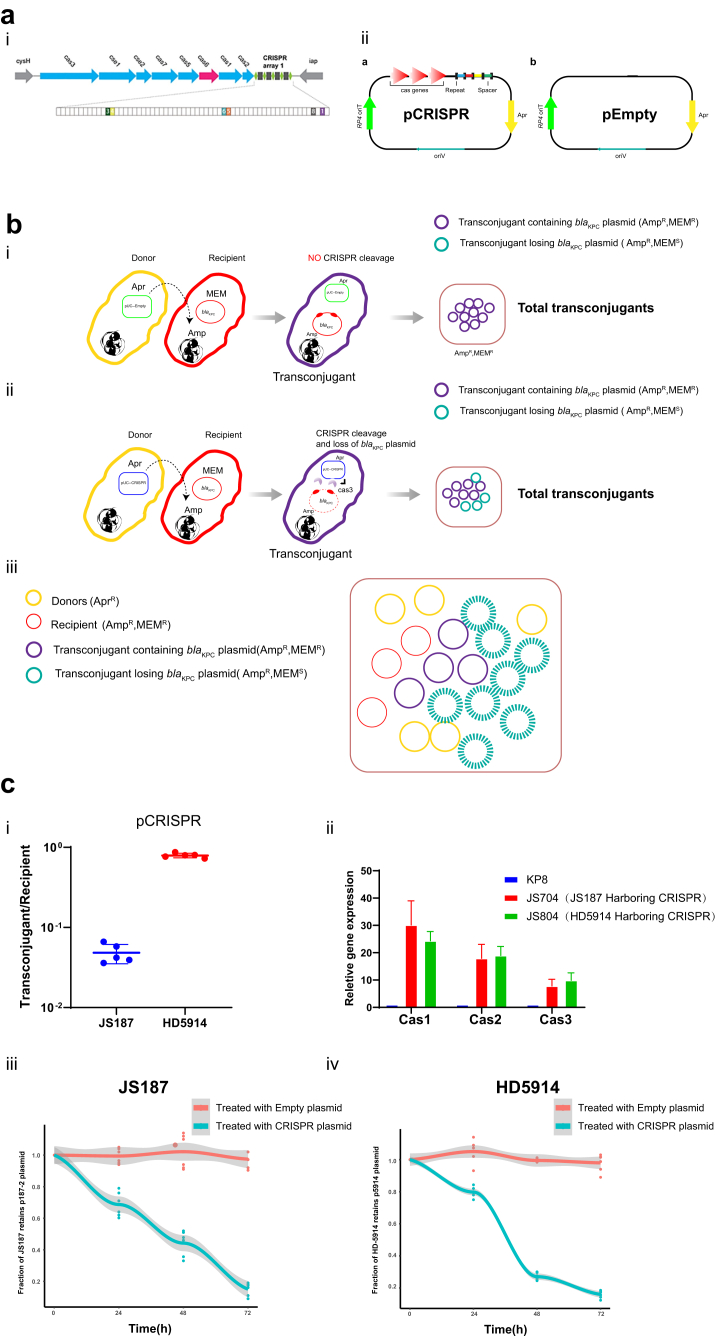
Fig. 3.
Conjugative delivery of endogenous CRISPR-Cas3 platform (a) Schematic of the KP8-CRISPR(i) and pEmpty, pCRISPR plasmid (ii). i. Genes are depicted as arrows in different colours and the IncFII plasmid-matched spacers are shown as colour boxes. ii. The high-copy pBR322 origin of replication was responsible for Cas-operon overexpression, and the oriTRP4 of the IncP RK2 plasmid contributed to the mobilization. (b) Cartoon depicting the delivery of pEmpty (i) or pCRISPR (ii) to the recipient K. pneumoniae cells. The ampicillin is used as the antibiotic to select total transconjugants and recipients (K. pneumoniae), since nearly all K. pneumoniae strains are naturally resistant to ampicillin. Similarly, meropenem was applied to screen the presence of blaKPC plasmids. The purple circle indicated the pCRISPR transconjugants containing blaKPC plasmids, while the green circle represented the blaKPC plasmid cured pCRISPR transconjugants. The total transconjugants include both blaKPC plasmid positive (“escaper”) and negative (cured) strains. (c) (i) Conjugation frequency of pCRISPR to K. pneumoniae recipients. Conjugation frequency was reported as the number of total transconjugants (AprR, AmpR) per total recipient cells (AmpR). The results of the conjugation assay were presented as means ± SD from five independent experiments. (ii) The expression of native CRISPR in K. pneumoniae. Cas1, Cas2 (Adaptation) and Cas3 (DNA degradation) are three core Cas operon genes in type I-E CRISPR. The plasmid killing efficiency of JS187 (iii) and HD5914 (iv) mediated by the CRISPR-Cas3 platform. Markers: Apr, Apramycin; Amp, Ampicillin; MEM, Meropenem.
Plate mating conjugation assay to deliver the CRISPR-Cas3 nuclease
The donors and recipients were cultured to the logarithmic phase, mixed in 10:1 ratio, and then resuspended in 20 μl MgSO4 (10 mM). The 20 μl resuspension was spotted on the Luria Bertani (LB) plate and incubated at 37 °C overnight.23 The cultures were then scraped from the plate and serially diluted, followed by plating on plates with appropriate antibiotics: apramycin and ampicillin for the selection of total transconjugants (K. pneumoniae is naturally-resistant to ampicillin), while apramycin and meropenem for transconjugants retaining IncFII plasmids. The conjugation frequency was calculated as the number of transconjugants per recipient.
Quantitation of mRNA expressions of Cas operon in K. pneumoniae
To examine whether the transferred CRISPR-Cas3 system can successfully express in the new hosts, we applied real-time PCR to assess the transcription of the core Cas operons (Cas1, Cas2, and Cas3). K. pneumoniae strains harboring the engineered plasmid (pEmpty or pCRISPR plasmid) were grown overnight under antibiotic selection. Overnight cultures were diluted 1:100 in LB with 100 mg/L apramycin, and grown to logarithmic phase (OD600 = 0.4). RNA of each strain was isolated using MiniBEST Universal RNA extraction kit (TaKaRa), following the manufacturer's instructions. RNA samples for real-time PCR were pre-treated with DNase I (TaKaRa). Real-time PCR was conducted on a 7500 system (Applied Biosystems, Foster City, CA, USA) using SYBR Premix ExTag (Takara). All assays were performed in triplicate with three independent RNA preparations.
Plasmid killing efficiency in vitro
Donors (E. coli S17 with pEmpty or pCRISPR) and recipients (JS187 and HD5914) were grown overnight. We chose two blaKPC-IncFII plasmid-harboring K. pneumoniae strains with different clonal backgrounds, JS187 (ST11) and HD5914 (ST751). A total of 180 μL donors and 20 μL recipients were added in 5 mL LB. Bead-supplemented conjugations were conducted similar to a previously published method,30 with the addition of 1 mL soda lime glass beads (0.5 mm diameter). Plasmid elimination was proceeded by incubating the culture at 37 °C with 60 RPM agitation for 72 h. Cultures were homogenized by vortexing, serially diluted and spot-plated on plates containing appropriate antibiotics for the selection for K. pneumoniae with or without blaKPC-IncFII plasmid. Plates were incubated at 37 °C for 16–20 h. Colonies were counted manually. Killing efficiency was calculated by the ratio of cells on MEM (Meropenem)-selective to non-selective (Amp-selective) plates.
Plasmid eliminating efficiency by cumulative CRISPR interference
We have previously demonstrated that the plasmid elimination frequency correlated with cumulative CRISPR-Cas interference during the proliferation.23 Here, we evaluated the plasmid curing efficiency in putative “escaper” isolates, in which the plasmid survived from the CRISPR-Cas3 killing in vitro in the first few generations of growth. K. pneumoniae strains JS187 and HD5914, harbouring blaKPC positive IncFII plasmids, were used as the test strains to examine the curing efficiency of IncFII plasmids.29 pEmpty or pCRISPR plasmid was conjugatively delivered into K. pneumoniae strains JS187 and HD5914, followed by selection on agar plates supplemented with apramycin (100 mg/L) and meropenem (1 mg/L or 16 mg/L), as well as PCR confirmation. These obtained transconjugants were named JS701 (JS187 carried pEmpty plasmid), JS801 (HD5914 carried pEmpty plasmid), JS704 (JS187 carried pCRISPR plasmid) and JS804 (HD5914 carried pCRISPR plasmid), respectively. Overnight culture of these strains were diluted 1000-fold in 3 ml LB broth with no antibiotics. The cultures were incubated for 24 h (OD600 = 1.0) at 37 °C with shaking at ∼200 rpm. The cultures were then diluted and plated simultaneously on nonselective LB agar plates and selective plates (MEM) to determine the number of colonies forming units (cfu, colony-forming units). Killing efficiency is determined by the ratio of cells on selective to nonselective plates. The experiments were conducted using four randomly picked JS704/JS804 colonies. Moreover, blaKPC real-time PCR was conducted to confirm the colony counting results. After the plasmid killing, we obtained the IncFII plasmid cured JS187, and we applied Pulsed-field gel electrophoresis (PFGE) assay to further confirm the plasmid deletion.
Plasmid immunization of the delivered CRISPR-Cas system
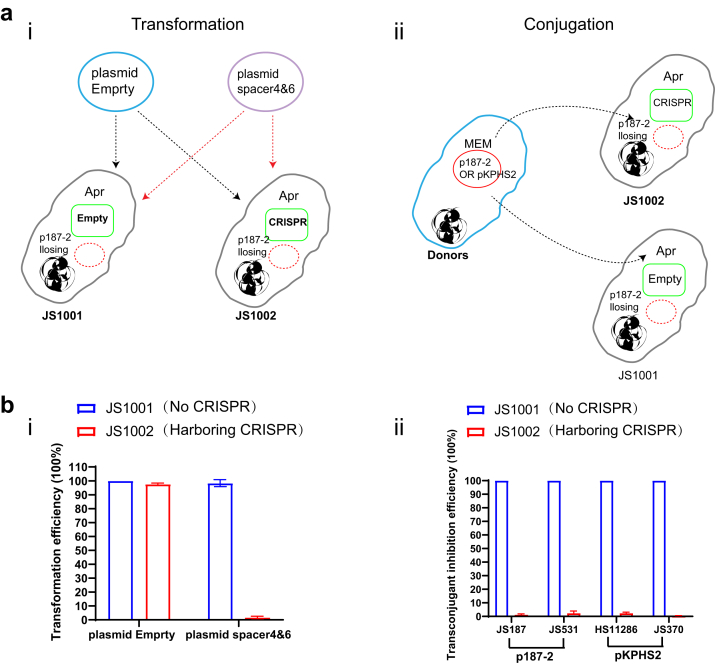
We applied both electro transformation and conjugation assays to examine whether the acquired CRISPR-Cas system will prevent the invasion of target plasmids. The protospacer 4 and protospacer 6 were synthesized by annealing single-stranded, complementary oligonucleotides and then cloned into a Bsa I site in the modified pACYC-184 plasmid (chloramphenicol-resistant) (pACYC-184 spacer)23 to perform the transformation experiment, and the pACYC-184 empty plasmid was used as no-protospacer control (pACYC-184 control). Notably, pACYC-184 is a plasmid cloning vector containing the p15A origin of replication, which allows pACYC-184 to coexist in cells with plasmids of the ColE 1 compatibility group (pBR322, pUC19). The IncFII-p187-2 plasmid cured JS187 strain was introduced the pEmpty or pCRISPR plasmid, to generate the recipient strains JS1001 (harboring pEmpty) and JS1002 (harboring pCRISPR), respectively. 1 μg of plasmid DNA was electroporated to 100 μl electrocompetent cells using a 2-mm electroporation cuvette. A pulse of 2.5 kV, 25 μF, and 200 Ω for 50 ms was used for electroporation. Cells were then resuspended in 1 ml of non-selective LB medium and incubated 1 h at 37 °C (for thermosensitive plasmids) for recovery before plating on selective media (chloramphenicol-positive). The transformation efficiencies of pACYC-184control and pACYC-184spacer were compared in JS1001 (Empty) and JS1002 (CRISPR). The transformation efficiency of pACYC-184control in JS1001 was set as 100%, and the transformation efficiency was calculated by dividing the number of transformants for the pACYC-184spacer by the number of transformants for pACYC-184control.
In the conjugation assay, we selected JS187 (K. pneumoniae harboring IncFII-p187-2 plasmid), JS531 (E. coli Top10 harboring p187-2 plasmid), HS11286 (K. pneumoniae harboring IncFII-pKPHS2 plasmid) and JS370 (E. coli Top10 harboring p187-2 plasmid) as the donor strains, and JS1001 and JS1002 as the recipient strains. The conjugation assay and conjugation frequencies were performed and calculated as above.
G. mellonella infection model to evaluate the CRISPR-Cas3 immunity in vivo
G. mellonella larvae were stored at 4 °C prior to use. Larvae with the weight of 150–200 mg were used. For the CRISPR-Cas3 treated groups, larvae were inoculated with 10 μL of JS902 (E. coli S17 with pCRISPR) at a concentration of ∼4 × 108 cfu/mL prepared in sterilized saline, and after 2 h the same amount of K. pneumoniae JS187 was injected. The empty plasmid treated group (E. coli S17 with pEmpty) and normal saline group were used as the control groups. A minimum of 30 larvae were used in each treatment group, and they were kept in three Petri dishes at 37 °C and inspected daily for 3 days. Survival rates were recorded for each day. To evaluate the hemolymph burdens in different G. mellonella groups, we also establish a similar experiment to examine the cfu counts of bacteria at 24 h, 48 h, and 72 h post infection (hpi).
Statistical analysis
Statistical significance was assessed using a Mann–Whitney non-parametric test and A log-rank (Mantel– 638 Cox) test by the GraphPad Prism 9 software. p < 0.05 was considered statistically significant.
Ethics
The research protocol was approved by the Ethics Committee of Shanghai Pulmonary Hospital (K21-371Y).
Role of funders
The funders played no role in the study design, data collection, data analyses, interpretation, or writing of the report.
Results
IncFII plasmids are the dominant multidrug-resistance plasmid types in Klebsiella pneumoniae
Previous studies demonstrated that IncFII was a major plasmid incompatibility group carrying AMR genes, and played a critical role in the dissemination of antibiotic resistance in Enterobacteriaceae. In this study, we analysed 3117 plasmids from 932 completely sequenced K. pneumoniae from the public domain, and obtained 1439 antimicrobial-resistant plasmids (at least harbouring one resistance gene), with 38.5% (554/1439) being characterized as IncFII plasmids. By contrast, other epidemic plasmids, for example, the IncX type only accounted for 4.7% (68/1439). In particular, 63.1% (147/233) of blaKPC plasmids belonged to IncFII plasmids (Dataset).
We identified a total of 3327 antibiotic resistance genes (ARGs) from 77.52% (562/725) of the IncFII plasmids, covering 172 non-redundant genes that encode resistance to a wide spectrum of antibiotics (Fig. S1 and Dataset). These ARGs mediated resistance to almost all the common clinical antibiotics (Figs. S1 and S2). Notably, most IncFII plasmids were predicted to be conjugative, and a ∼35-kb conjugation module4 was found in >90% IncF plasmids, suggesting the ARGs on IncFII plasmids may be readily transferred to other bacterial hosts through conjugation.
IncFII plasmids are well targeted by K. pneumoniae native CRISPR -Cas3
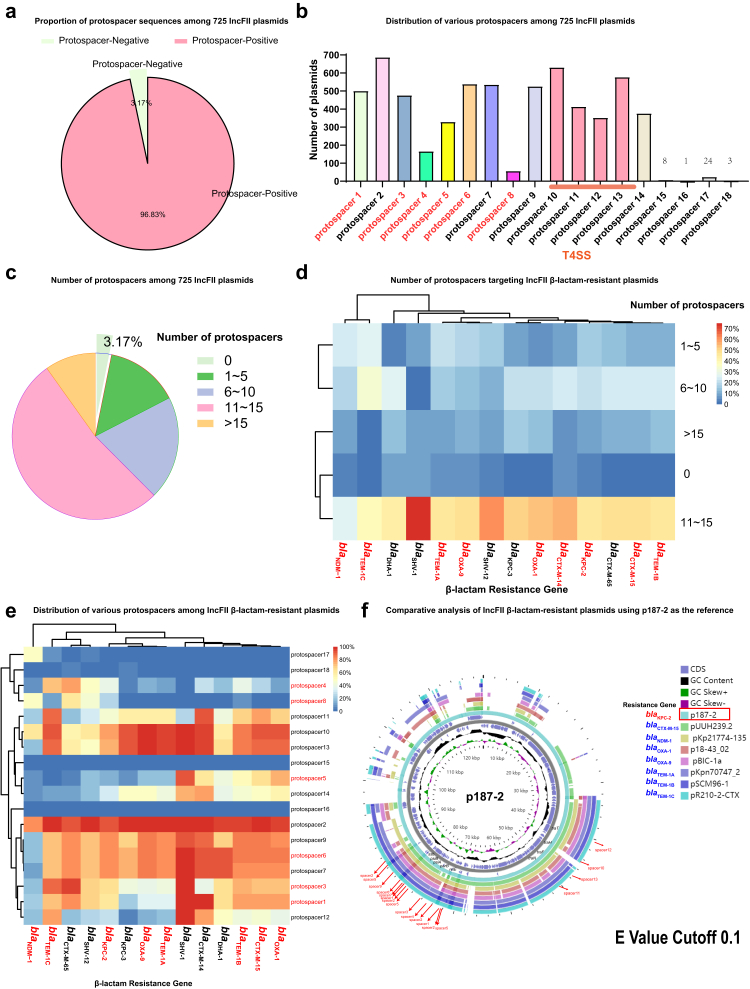
To harness the endogenous system for IncFII plasmid curing, we firstly assessed whether the IncFII plasmids could be good targets for native CRISPR-Cas3 systems. To this end, we examined the presence of matched protospacers on the 725 K. pneumoniae IncFII plasmids against a set of 415 spacer sequences we identified previously from 207 K. pneumoniae genomes.22 We found the presence of 18 matched protospacers among these IncFII plasmids, and 96.83% (702/725) plasmids harbours at least one protospacer (Fig. 1a). Importantly, most of the plasmids could be targeted by multiple spacers, and more than half of the plasmids (52.4%, 380/725) contained 11–15 protospacers (Dataset, Fig. 1c and d). Among them, protospacer 1, 2, 3, 5, 6, 7, and 9 were the most abundant (Fig. 1b and e, and Fig. S3), and a combination of the five protospacer could cover> 70% of IncFII plasmids.
Fig. 1.
Characteristics of protospacers on IncFII plasmids. (a) Proportion of protospacer sequences among 725 IncFII plasmids. (b) Distribution of various protospacers among 725 IncFII plasmids. (c) Number of protospacers among 725 IncFII plasmids. (d) Number of protospacers targeting IncFII β-lactam-resistant plasmids. (e) Distribution of various protospacers among IncFII β-lactam-resistant plasmids. (f) Comparative analysis of IncFII β-lactam-resistant plasmids using p187-2 as the reference. The CRISPR targeted regions were illustrated with red arrows. The detailed information of these plasmids were listed in the Dataset.
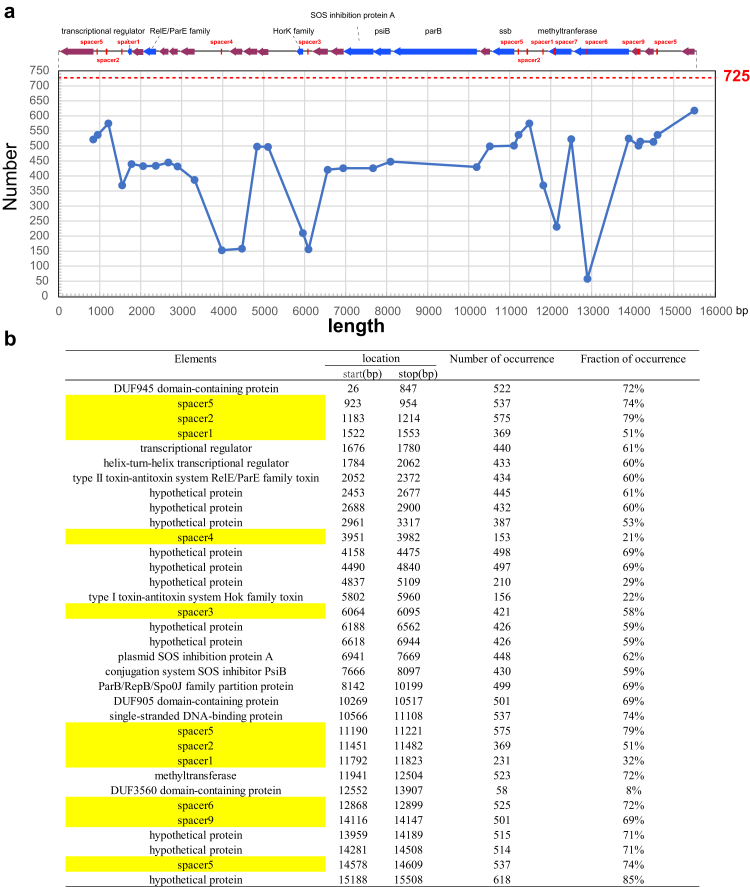
We then examined the spacer target regions on the IncFII plasmids. Interestingly, we found most protospacers corresponded to the plasmid stability regions, such as ssb or the region adjacent to ssb, parB, ardA, psiA and psiB (Figs. 1f and 2 and Fig. S4). Inactivation of these plasmid regions will likely interfere the plasmid stability, leading to the curing of a plasmid from its bacterial host. The above results suggested that the native CRISPR-Cas3 carried by K. pneumoniae may be repurposed to combat the IncFII plasmids.
Fig. 2.
Protospacers targeting the IncFII plasmids. (a) p187-2 (GenBank accession no. CP025468.1) was used as the reference plasmid for illustration. Its CRISPR targeting regions (involving plasmid stability) are shown on the top. The blue line in the chart below indicates how frequent the corresponding protospacers found in the 725 IncFII plasmids. The horizontal axis indicates the coordinates of p187-2. The red line highlights the total numbers of analysed plasmids (725 plasmids). (b) Detailed information of the spacer targeting regions in p187-2.
In vitro IncFII plasmids curing by high-efficiency conjugative delivery of K. pneumoniae native CRISPR-Cas3
Based on the protospacer analysis of IncFII plasmids, we choose K. pneumoniae KP8 (CP025636.1) carried type I-E CRISPR(CRISPR-Cas3) system for plasmid curing (Fig. 3a), which harboured 6 matched spacers.23 pCRISPR was derivate from the synthetic pUC-RP4 (pEmpty) plasmid, and carry the pBR322 origin of replication (high-copy number) and the oriTRP4 of the IncP RK2 plasmid (moveable) as well as the CRISPR-Cas3 system from KP8 (Fig. 3aii). pCRISPR can be mobilized with the help from the chromosomally integrated RP4 plasmid from the E. coli S17-1 donor cells.
To determine whether our pCRISPR system can efficiently deliver CRISPR-Cas3 nucleases to K. pneumoniae, we assessed the plasmid conjugation frequencies in two K. pneumoniae strains, JS187 (ST11) and HD5914 (ST751), which originated from two different genetic backgrounds, and harboured two different blaKPC-2 plasmids (IncFIIK type in JS187 and IncFII (pHN7A8) type in HD5914), with different matched spacers. As shown in Fig. 3ci, conjugation frequency for pCRISPR in plate mating condition was high (∼10−1, ranged from 3.6 × 10−2 to 8.68 × 10−1) in both K. pneumoniae strains. Notably, the cas3 system was in a conjugative plasmid, so it can self-replicate and transfer. Further real-time PCR analysis confirmed that the type I-E CRISPR-Cas3 system was successfully expressed in the new K. pneumoniae hosts (JS187 and HD5914) (Fig. 3cii).
Previous studies suggested the in vitro plasmid curing efficiency may be affected by different conjugation conditions.30 We therefore tested the curing efficiency of pCRISPR in various conjugation conditions, including different incubating temperatures (37 °C or 25 °C), incubation time lengths (24 h, 48 h, and 72 h), donor to recipient ratios (10:1 or 50:1), agitation speeds (0 rpm or 60 rpm) and liquid culture mixtures (with or without 0.5 mm glass beads).30 The results showed that with or without 0.5 mm glass beads liquid culture, and 0 or 60 rpm resulted in similar killing frequencies (Fig. S5), but 10:1 donor to recipient ratio and 37 °C outperformed 50:1 ratio and 25 °C culture conditions. We then use 10:1 donor to recipient ratio, 37 °C, 60 rpm agitation and without glass beads as the test condition for plasmid curing.
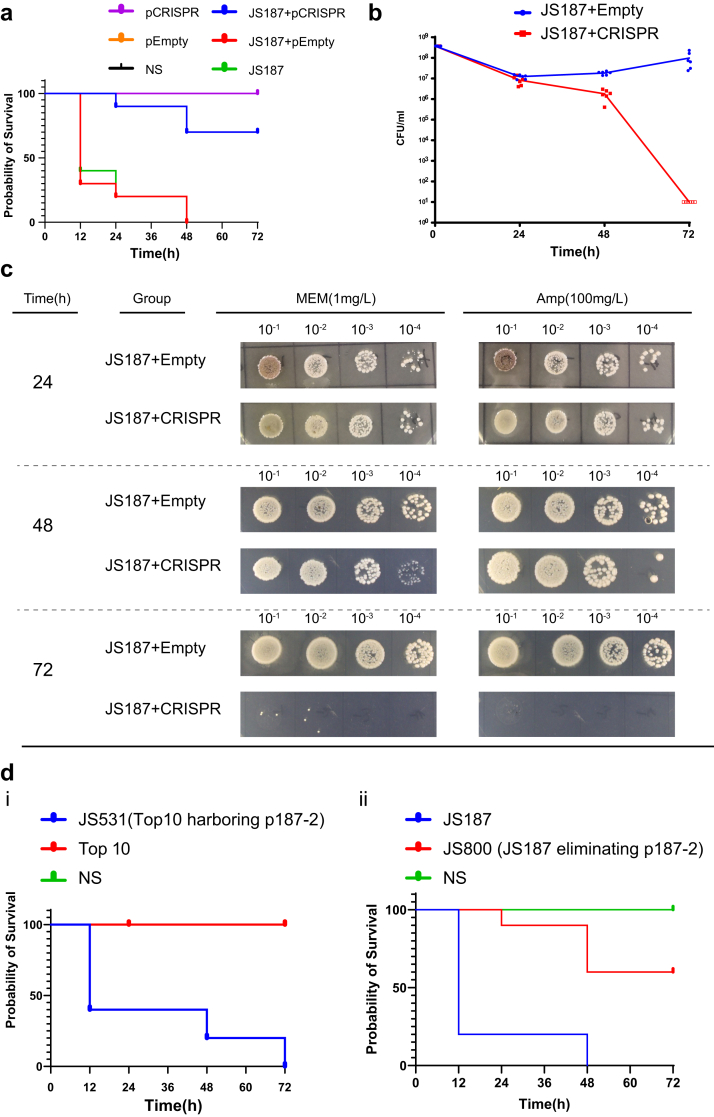
The curing efficiency displayed a time dependent increase, and 72 h incubation yielded the largest number reduction of MEM-resistant transconjugants (Fig. 3ciii and civ, Figs. S5 and S6), which is similar to the results from a previous study.30 Notably, we only observed apparent curing among the pCRISPR transconjugants, yet the transconjugants with empty plasmid were ineffective at reducing antibiotic resistance (Fig. 3ciii and civ). After 72 h incubation, the MEM-resistant colonies (both JS187 and HD5914) have decreased almost 90% (Fig. S6). For K. pneumoniae HD5914, the decrease efficiency reached ∼95% (Fig. 3ciii and civ). These results demonstrated that our engineered CRISPR-Cas3 platform can be successfully delivered through conjugation in vitro, and mediated a remarkable strand break of the target IncFII plasmids, leading to the elimination of the resistant plasmids.
IncFII plasmid curing in escapers by endogenous CRISPR-Cas3
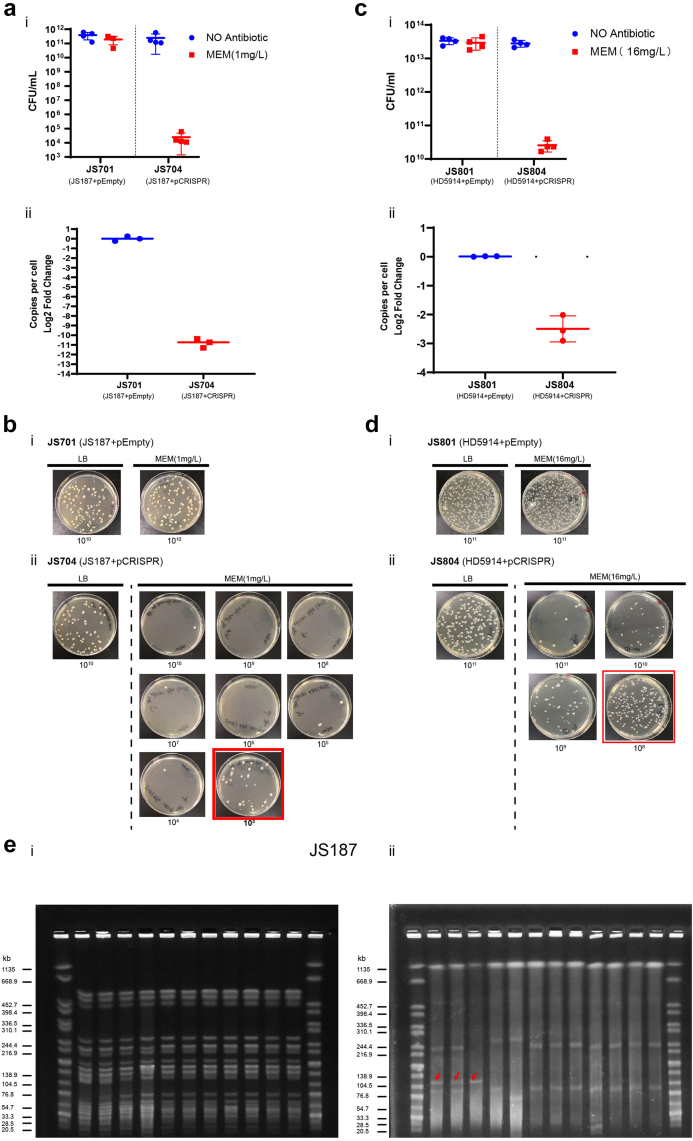
Although our above in vitro conjugation plasmid curing experiment showed significant elimination of IncFII plasmids, there still were some “escapers” from the CRISPR cleavage process. Previous studies demonstrated that both original and acquired type I-E CRISPR-Cas system could contribute to restraining the retention of target plasmids in K. pneumoniae, and complete plasmid elimination required cumulative CRISPR interference.23 Here, we further explored whether the engineered CRISPR-Cas3 platform could effectively eliminate target plasmids from putative “escapers” during growth. The K. pneumoniae JS704 (JS187 carried pCRISPR, blaKPC positive) and JS804 (HD5914 carried pCRISPR, blaKPC positive) were used to conduct plasmid killing, and their corresponding isolates carrying CRISPR-empty plasmids were used as the negative controls. The proportion of the IncFII plasmid retaining cells in the population was measured as the ratio of MEM-resistant cfu to total cfu. Our results showed the strains with CRISPR-Cas3 system (pCRISPR) had 108-fold and 103-fold reduction of the population with blaKPC IncFII plasmids in the JS704 (JS187 carried pCRISPR) and JS804 (HD5914 carried pCRISPR) (Fig. 4) respectively in 6 h growth. The relative copy numbers of blaKPC-IncFII plasmids determined by quantitative PCR in both JS704 and JS804 decreased 1011-fold and 103-fold compared with the empty plasmid-harbouring JS701 and JS801, which were consistent with plasmid curing results (Fig. 4).
Fig. 4.
Killing efficiency of IncFII plasmids by native CRISPR. Plasmid curing of JS187 (blaKPC, MEM-resistant) (ai, b) and HD5914 (blaKPC, MEM-resistant) (ci, d) with pEmpty (negative control) or pCRISPR. The plasmid killing efficiency was calculated by the reduced MEM-resistant colonies (LB with MEM) in total cell counts (LB with no antibiotic) (n = 4). CFU, colony-forming units. p187-2 plasmid (IncFII-blaKPC plasmid) copy number (aii) and pHD5914 plasmid (IncFII-blaKPC plasmid) copy number (cii) determined by quantitative PCR in either JS701/JS801(JS187/HD5914 harbouring pEmpty plasmid) or JS704/JS804 (JS187/HD5914 harbouring pCRISPR plasmid). (e) XbaI (i)and S1 (ii) PFGE of genomic DNA of JS187 and its derivatives. Marker: XbaI-digested DNA of Salmonella Braenderup H9812. The red arrows in S1-PFGE (ii) represent the p187-2 plasmid (129,684 bp, CP025468.1). JS704: JS187 harbouring pCRISPR; JS531: E. coli Top 10 harbouring p187-2 plasmid. JS708, JS709, JS800, and JS805-JS900: p187-2 cured JS187. Two examples of potential recombination events associated with p187-2 curing are found in JS708 and JS709 and a larger size of p187-1 plasmid (246,557 bp, CP025467.1) and p187-4 plasmid (106,402 bp, CP025470.1).
To further confirm whether the CRISPR-Cas3-mediated curing resulted in the complete loss of the IncFII plasmids, we selected 9 IncFII plasmid-cured colonies from JS187 and conducted XbaI-PFGE and S1-PFGE. All the colonies had the same XbaI-PFGE pattern (Fig. 4ei) as the parent JS187, suggesting they were isogenic strains, and the S1-PFGE results showed that all nine curated colonies lost the plasmid of p187-2 (129.684 kb), confirming that the native CRISPR-Cas3 system could successfully eliminate the target IncFII plasmids in K. pneumoniae (Fig. 4eii).
Prevent the invasion of IncFII-resistant plasmids by CRISPR-Cas3
Our above results showed that the delivered CRISPR-Cas3 system could effectively eliminate the IncFII plasmids. We then explored whether the K. pneumoniae harboring the CRISPR-Cas3 platform could prevent the invasion of IncFII plasmids, and we assessed the Cas3-immunity using in vitro electro-transformation and conjugation experiments.
For the electro-transformation experiment, two plasmids (chloramphenicol-resistant) with or without matched proto-spacers (spacer4&6) were used as the donor plasmids (pACYC-184spacer and pACYC-184control), and the JS800 (JS187 losing the IncFII-p187-2 plasmid) with CRISPR (JS1002) or without CRISPR (JS1001) was used as the recipient. The results showed the pre-existed CRISPR-Cas3 system in JS1002 significantly inhibit the pACYCY-184spacer plasmid transfer compared with the CRISPR-Cas3 negative strain (JS1001) (p < 0.001, Mann–Whitney non-parametric test), while the pACYC-184control plasmid showed similar transform frequency between JS1001 and JS1002 (Fig. 5bi).
Fig. 5.
Plasmid immunization of CRISPR-Cas3 platform in clinical isolates. (a) Cartoon depicting the transformation (i) and conjugation (ii) processes. In the transformation process, two plasmids (chloramphenicol-resistant) were constructed with or without matched proto-spacers (spacer4&6) (pUC-19control and pUC-19spacer) and were used as the donor plasmids, while the JS800 (the IncFII-p187-2 plasmid cured JS187) with or without CRISPR were used as the recipient (JS1001 and JS1002). In the conjugation assay, JS187 (K. pneumoniae harbouring IncFII-p187-2 plasmid, CP025468.1), JS531 (E. coli Top10 harboring p187-2 plasmid), HS11286 (K. pneumoniae harbouring IncFII-pKPHS2 plasmid, CP003224.1) and JS370 (E. coli Top10 harbouring p187-2 plasmid) were used as the donors and the recipients were the same as transformation process. (b) The transformation and conjugation efficiencies of different plasmids and donors. (i) The transformation efficiency of Empty plasmid was set as 100% and those of plasmid spacer4&6 were calculated verse the empty plasmid. (ii) Conjugation inhibition efficacy of p187-2 and pKPHS2 in JS800 strains with or without KP8 CRISPR. The conjugation efficiency of plasmids in JS1001 was set as 100%. Data are presented as means ± SD from three independent experiments.
To examine if the pre-existed CRISPR-Cas3 system can reduce the conjugation efficiency of the target plasmids, we used four isolates harbouring two different IncFII plasmids29,31 as the donors, including JS187 (K. pneumoniae harbouring IncFII-p187-2 plasmid, CP025468.1), JS531 (E. coli Top10 harbouring p187-2 plasmid), HS1128631 (K. pneumoniae harbouring IncFII-pKPHS2 plasmid, CP003224.1) and JS370 (E. coli Top10 harbouring pKPHS2 plasmid), while JS1001 and JS1002 were used as the recipient strains as above. We observed that the presence of the type I-E CRISPR-Cas3 significantly decreased the conjugation frequencies in JS1002 by approximately 100-fold for all four donors, in comparison to the CRISPR-Cas3 negative JS1001 (Fig. 5bii) (p < 0.001, Mann–Whitney non-parametric test). All these results indicated the acquired CRISPR-Cas3 platform could provide the clinical isolate the ability to resist the invasion of IncFII plasmids.
IncFII plasmid elimination by CRISPR-Cas3 platform in vivo
In order to probe whether the native CRISPR-Cas3 system can be potentially implemented for in vivo plasmid curing, we applied the G. mellonella infection model to evaluate the effects of the native CRISPR-Cas3 on IncFII plasmid eliminating in vivo. We firstly injected the JS902 (E. coli S17-1 harboring CRISPR-Cas3 system) into G. mellonella, aiming to establish an immune barrier in advance, and 2 h later we injected the targeting clinical K. pneumoniae stain JS187. Our results showed the pre-injection of JS902 not only eliminate the p187-2 plasmid, but also improved the survival rates (70% survival rate in 72 h), compared with the JS187 infection controls with pre-treated normal saline or Empty plasmid (100% mortality) (Fig. 6a). In addition, we also examined the G. mellonella hemolymph bacterial burdens at 24, 48, and 72 hpi, respectively. We observed a rapid reduction of JS187 burden from 48 h to 72 h, when pre-treated with the CRISPR-Cas3 plasmid in comparison to the empty plasmid (Fig. 6bc). The above results suggested that CRISPR-Cas3 plasmid conjugation treatment demonstrated high plasmid curing efficiency (∼100% curing), and reduced the relative virulence (mortality and hemolymph burdens) in comparison with empty plasmid or saline controls, which is unexpected. In contrast, the pEmpty plasmid group didn't showed apparent impact on the survival (Fig. 6a). One plausible explanation is that CRISPR-Cas3-mediated p187-2 (blaKPC-IncFII plasmid in JS187) curing reduced the overall pathogenicity, and the plasmid p187-2 may harbour virulence factors contributing to overall pathogenicity of JS187.
Fig. 6.
IncFII plasmid curing by CRISPR-Cas3 in vivo. (a) The survival of JS187 infecting G. mellonella treated with CRISPR plasmid and Empty plasmid. (b, c) The burden of the JS187 in the G. mellonella model. (d) (i) Survival rates of G. mellonella infected with E. coli Top 10 with our without p187-2 plasmid; (ii) the survival rates of G. mellonella infected with JS187 and p187-2 cured JS187 (JS800).
We then transformed the p187-2 plasmid into E. coli Top10 (JS531), and compared the survival rate in the same G. mellonella infection model with that of E. coli Top10 strain (without plasmid). In addition, we also compared the survival rate between the parental JS187 strain and its p187-2 plasmid cured isogenic strain JS800 (Fig. 4). The results showed that in both bacterial hosts (E. coli Top10 and K. pneumoniae JS187) the presence of p187-2 significantly increased the mortalities in comparison to their p187-2 negative isogenic counterparts (Fig. 6d) (p < 0.001, log-rank (Mantel– 638 Cox) test), which further confirmed that p187-2 increased the pathogenicity in G. mellonella.
Discussion
MDR K. pneumoniae has emerged as a global problem hindering the treatment of bacterial infections.32 Of particular concern is that most of these resistance determinants were harboured by mobile genetic elements, like conjugative plasmids, facilitating the horizontal transfer of AMR between different bacterial hosts. Effective strategies to combat MDR K. pneumoniae are urgently needed, and plasmid curing could be a promising approach to reduce AMR prevalence, and sensitize bacteria to antibiotics.3 Our genomic data mining results showed that IncFII plasmids are the most common vectors underlying the rapid spread of various ARGs, including the genes encoding carbapenemase and ESBL, in clinical K. pneumoniae strains. Besides ARGs, IncFII plasmids were also found to promote the dissemination of hypervirulent genes (Fig. S1).13,33 Previous genomic studies suggested that hybrid virulence plasmids were likely originated from the recombination of IncFII and the virulence IncHIB-FIB plasmids.13 Moreover, multiple studies have demonstrated such hybrid plasmids could be transferred by conjugation to different types of Klebsiella strains and augmented the virulence levels of the host strain.13,34,35
Above findings further highlighted the important role of high-risk of IncFII plasmids, and emphasized an urgent need to control plasmid-mediated resistance in clinical K. pneumoniae strains. In this study, we engineered an endogenous CRISPR-Cas3 based platform to eliminate the high-risk IncFII plasmids, providing an alternative to tackle AMR in K. pneumoniae. Due to the simplicity and high efficiency,21 repurposing widespread, endogenously encoded CRISPR-Cas systems for ‘built-in’ genome editing is emerging as a promising genetic manipulation strategy in prokaryotes. Our previous studies showed a significantly inverse correlation between the presence of CRISPR-Cas3 and IncFII plasmids in K. pneumoniae, indicating the native CRISPR immunity may influence the dissemination of this common plasmid type.22,23 This finding prompts us to consider the possibility of repurposing endogenous CRISPR-Cas3 system for IncFII plasmid curing. We firstly analysed the distribution of protospacers on IncFII plasmids and found 18 matched protospacers on 96.83% IncFII plasmids, targeting multiple sites in the same plasmid. Among these protospacers, protospacer 1, 2, 3, 5 and 6 were both abundant on IncFII plasmids and on the CRISPR loci identified in K. pneumoniae. These photospacers located in the regions adjacent to ssb, parB, adrA, psiA and psiB, which are involved with plasmid stability and propagation, and served as good targets for plasmid curing.23
In this study, the native CRISPR obtained from K. pneumoniae KP8 isolate was selected for IncFII plasmid curing, since it carried several common spacers (spacer 1,3,4,5,6,8). We then used conjugation to deliver CRISPR-Cas3 nucleases, which has been recognized a better route over transformable plasmids or phages as the delivery system.30 We constructed a high copy number pCRISPR plasmid, which contains the CRISPR-Cas3 system from KP8. Unlike other Cas9 mediated plasmid curing platforms,36, 37, 38 the Cas9 was usually controlled by an inducible promoter, e.g. pBAD or pTet, our CRISPR-Cas3 system used the native promoter in K. pneumoniae, without the needs of external inducers. In addition, the high plasmid copy number also help to maintain higher-level expression of CRISPR-Cas3 modules. We observed over 20-fold Cas expressions in pCRISPR transformants in comparison to parental KP8 strain (Fig 3cii).
This system displayed a high conjugation frequency (∼10−1) in the selected K. pneumoniae strains. The conjugative transfer of the native CRISPR-Cas3 system into recipient K. pneumoniae cells resulted in a significant reduction of meropenem resistant population (blaKPC IncFII plasmids) (Fig. 3c). This indicated our conjugative CRISPR-Cas3 system can effectively reduce the resistance burden in a population. Although we still found a small percentage of “escapers” from the CRISPR-Cas3 conjugation mediated cleavage process, these “escapers” can be effectively eliminated during propagation, as the constitutively expressed CRISPR-Cas3 system could continually cleavage the plasmids through the accumulative CRISPR immunity. Unlike the Cas9 system of which the escaper primarily originated from protospacer or cas9 mutations, resulting in the loss of the cleavage activities, the “escapers” in Cas3 system were mainly due to the alterations to the expression of CRISPR RNAs,39 and the system remains the cleavage activities. In addition, CRISPR-Cas3 creates a single-strand nick at the DNA sequence, followed by processive exonucleolytic degradation of the targeted strand; while CRISPR-Cas9 only cause double-strand DNA break without additional degradation.25 Consequently, previous study40 observed CRISPR-Cas9 system failed to completely cure AMR genes harboured by high copy number plasmids. In this case, the CRISPR-Cas3 could be more efficient, which deserves further comparison. Furthermore, the multiple native cleavage sites that existed in our native CRISPR-Cas3 system also improved the cleavage efficiency in clinically complex MDR plasmids.25 In addition, our results showed that the native type I-E CRISPR could inhibit the invasion of target plasmids in K. pneumoniae. As such, once a strain obtains this CRISPR-Cas3 system, it could actively prevent the acquisition of other IncFII plasmid DNA.
We also evaluated the activity of the native CRISPR-Cas3 nuclease in removing the IncFII plasmids in vivo using a G. mellonella infection model. Similar to the in vitro results, at 72 hpi, the conjugative delivery of exogenous CRISPR-Cas3 significantly reduced the MEM resistant population in comparison with the empty plasmid treated group, confirming that the CRISPR-Cas3 system could effectively cure the IncFII plasmids in vivo. Intriguingly, our results indicated that along with the curing of MEM resistant IncFII p178-2 plasmids, the overall virulence was significantly reduced in the G. mellonella infection model, suggesting that plasmid borne genes contributed to the overall virulence in JS187. Our results demonstrated the native CRISPR-platform may not only reduce the AMR burden but the plasmid-mediated virulence. As the IncFII-hypervirulent plasmids are increasingly described,12,13,41 future work is needed to evaluate how effective our system act against these IncFII-hypervirulent plasmids.
The study has some limitations. Firstly, a native CRISPR array from a clinical strain (KP8) was used in this study, and the spacers primarily target the IncFII plasmids, which limits its application against other AMR genes or plasmids. We are currently optimizing this endogenous system through customized CRISPR array assembly to make this platform for other targets curing. Secondly, although the Galleria mellonella model showed good efficacy to test plasmid curing efficiency in vivo (∼100% curing), a more relevant animal model or human gut microbiome editing experiment are needed to assess the in vivo resistance gene curing efficacy, and the animal model is currently under development in our lab. Lastly, only two different IncFII plasmids [IncFII (pHN7A8) and IncFIIK] from distinct K. pneumoniae backgrounds [JS187 (ST11, CG258) and HD5914 (ST751)] were tested in our study. The IncFII (pHN7A8) and IncFIIK type plasmids were the two most common IncFII plasmid identified in K. pneumoniae. Although our results showed great efficacy in curing the two IncFII plasmids in JS187 and HD5914, additional strains and plasmids should be included in future studies to define the target curing efficacy in different K. pneumoniae bacterial hosts.
Taken together, we developed an endogenous CRISPR-Cas3 mediated platform for the curing of high-risk IncFII resistant plasmids in K. pneumoniae. This platform is highly efficient in eliminating IncFII plasmids by targeting multiple sites of the conservative plasmid backbone, thereby re-sensitizing MDR strains to antibiotics. Interestingly, in vivo G. mellonella model demonstrated that the platform not only re-sensitize carbapenem susceptibility, but also reduce the relative virulence. The current study confirmed the proof of concept of using endogenous CRISPR-Cas3-mediated plasmid curing to resensitize resistant strains to antibiotics, and its application for clinical intervention should be further evaluated.
Contributors
XJ, LC, and FY contributed to conceptualization, funding acquisition, supervision, and designed the study. YZ, YY, and XL performed data analysis, including methodology, software, and interpretation. YZ, YY, XL, XJ, LC, and FY verified the underlying data. YZ and LC wrote the original draft. DT, WA, WW, BW, and BK contributed to the interpretation of data. All the authors contributed to the manuscript drafting. All authors read and approved the final version of the manuscript.
Data sharing statement
The authors declare that the data supporting the findings of this study are available from the corresponding author upon request.
Declaration of interests
None to declare.
Acknowledgment
This work was supported by research grants National Natural Science Foundation of China [grant numbers 81871692, 82172315, 82102439, and 82202564], the Shanghai Science and Technology Commission [grant number 19JC1413002], and Shanghai Sailing Program [grant number 22YF1437500].
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104445.
Contributor Information
Fangyou Yu, Email: wzjxyfy@163.com.
Liang Chen, Email: Liang.Chen@hmh-cdi.org.
Xiaofei Jiang, Email: Jiangxi2154@sina.com.
Appendix A. Supplementary data
References
- 1.Spellberg B., Guidos R., Gilbert D., et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46(2):155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 2.Zohra T., Numan M., Ikram A., et al. Cracking the challenge of antimicrobial drug resistance with crispr/cas9, nanotechnology and other strategies in eskape pathogens. Microorganisms. 2021;9(5):954. doi: 10.3390/microorganisms9050954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckner M.M.C., Ciusa M.L., Piddock L.J.V. Strategies to combat antimicrobial resistance: anti-plasmid and plasmid curing. FEMS Microbiol Rev. 2018;42(6):781–804. doi: 10.1093/femsre/fuy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi D., Zheng J., Li J., et al. In silico typing and comparative genomic analysis of incfiik plasmids and insights into the evolution of replicons, plasmid backbones, and resistance determinant profiles. Antimicrob Agents Chemother. 2018;62(10):e00764–e00818. doi: 10.1128/AAC.00764-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villa L., García-Fernández A., Fortini D., Carattoli A. Replicon sequence typing of incf plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65(12):2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 6.Fu P., Tang Y., Li G., Yu L., Wang Y., Jiang X. Pandemic spread of bla((kpc-2)) among klebsiella pneumoniae st11 in China is associated with horizontal transfer mediated by incfii-like plasmids. Int J Antimicrob Agents. 2019;54(2):117–124. doi: 10.1016/j.ijantimicag.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Löhr I.H., Hülter N., Bernhoff E., Johnsen P.J., Sundsfjord A., Naseer U. Persistence of a pkpn3-like ctx-m-15-encoding incfiik plasmid in a klebsiella pneumonia st17 host during two years of intestinal colonization. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0116516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolejska M., Brhelova E., Dobiasova H., et al. Dissemination of incfii(k)-type plasmids in multiresistant ctx-m-15-producing enterobacteriaceae isolates from children in hospital paediatric oncology wards. Int J Antimicrob Agents. 2012;40(6):510–515. doi: 10.1016/j.ijantimicag.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Leavitt A., Chmelnitsky I., Carmeli Y., Navon-Venezia S. Complete nucleotide sequence of kpc-3-encoding plasmid pkpqil in the epidemic klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother. 2010;54(10):4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y., Yu D., Wei Z., Shen P., Zhou Z., Yu Y. Complete nucleotide sequence of klebsiella pneumoniae multidrug resistance plasmid pkp048, carrying blakpc-2, bladha-1, qnrb4, and arma. Antimicrob Agents Chemother. 2010;54(9):3967–3969. doi: 10.1128/AAC.00137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carattoli A. Resistance plasmid families in enterobacteriaceae. Antimicrob Agents Chemother. 2009;53(6):2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y., Zhang J., Wang M., et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent klebsiella pneumoniae. Genome Med. 2021;13(1):119. doi: 10.1186/s13073-021-00936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X., Dong N., Chan E.W., Zhang R., Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in klebsiella pneumoniae. Trends Microbiol. 2021;29(1):65–83. doi: 10.1016/j.tim.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Fagerlund R.D., Staals R.H., Fineran P.C. The cpf1 crispr-cas protein expands genome-editing tools. Genome Biol. 2015;16:251. doi: 10.1186/s13059-015-0824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z., Li M., Li Y., et al. Native crispr-cas-mediated genome editing enables dissecting and sensitizing clinical multidrug-resistant p. aeruginosa. Cell Rep. 2019;29(6):1707–1717. doi: 10.1016/j.celrep.2019.10.006. e3. [DOI] [PubMed] [Google Scholar]
- 16.Pyne M.E., Bruder M.R., Moo-Young M., Chung D.A., Chou C.P. Harnessing heterologous and endogenous crispr-cas machineries for efficient markerless genome editing in clostridium. Sci Rep. 2016;6 doi: 10.1038/srep25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker P.L., Orf G.S., Kevershan K., et al. Using the endogenous crispr-cas system of heliobacterium modesticaldum to delete the photochemical reaction center core subunit gene. Appl Environ Microbiol. 2019;85(23):e01644–e01719. doi: 10.1128/AEM.01644-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Zong W., Hong W., Zhang Z.T., Wang Y. Exploiting endogenous crispr-cas system for multiplex genome editing in clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab Eng. 2018;47:49–59. doi: 10.1016/j.ymben.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y., Han J., Wang B., et al. Characterization and repurposing of the endogenous type i-f crispr–cas system of zymomonas mobilis for genome engineering. Nucleic Acids Res. 2019;47(21):11461–11475. doi: 10.1093/nar/gkz940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hidalgo-Cantabrana C., Goh Y.J., Pan M., Sanozky-Dawes R., Barrangou R. Genome editing using the endogenous type i crispr-cas system inlactobacillus crispatus. Proc Natl Acad Sci U S A. 2019;116(32):15774–15783. doi: 10.1073/pnas.1905421116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z., Li Y., Li M., Xiang H., Yan A. Harnessing the type icrispr-cas systems for genome editing in prokaryotes. Environ Microbiol. 2021;23(2):542–558. doi: 10.1111/1462-2920.15116. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y., Fu P., Zhou Y., et al. Absence of the type i-e crispr-cas system in klebsiella pneumoniae clonal complex 258 is associated with dissemination of incf epidemic resistance plasmids in this clonal complex. J Antimicrob Chemother. 2020;75(4):890–895. doi: 10.1093/jac/dkz538. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y., Tang Y., Fu P., et al. The type i-e crispr-cas system influences the acquisition of bla kpc-incf plasmid in klebsiella pneumonia. Emerg Microbes Infect. 2020;9(1):1011–1022. doi: 10.1080/22221751.2020.1763209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csörgő B., León L.M., Chau-Ly I.J., et al. A compact cascade–cas3 system for targeted genome engineering. Nat Methods. 2020;17(12):1183–1190. doi: 10.1038/s41592-020-00980-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y., Battalapalli D., Hakeem M.J., et al. Engineered crispr-cas systems for the detection and control of antibiotic-resistant infections. J Nanobiotechnology. 2021;19(1):401. doi: 10.1186/s12951-021-01132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Aartsen J.J., Rajakumar K. An optimized method for suicide vector-based allelic exchange in klebsiella pneumoniae. J Microbiol Methods. 2011;86(3):313–319. doi: 10.1016/j.mimet.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Clausen P., Aarestrup F.M., Lund O. Rapid and precise alignment of raw reads against redundant databases with kma. Bmc Bioinformatics. 2018;19(1):307. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Xie Y., Liu M., et al. Oritfinder: a web-based tool for the identification of origin of transfers in dna sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018;46(W1):W229–W234. doi: 10.1093/nar/gky352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Y., Zhou Y., Meng C., Huang Y., Jiang X. Co-occurrence of a novel vim-1 and fosa3-encoding multidrug-resistant plasmid and a kpc-2-encoding pkp048-like plasmid in a clinical isolate of klebsiella pneumoniae sequence type 11. Infect Genet Evol. 2020;85 doi: 10.1016/j.meegid.2020.104479. [DOI] [PubMed] [Google Scholar]
- 30.Hamilton T.A., Pellegrino G.M., Therrien J.A., et al. Efficient inter-species conjugative transfer of a crispr nuclease for targeted bacterial killing. Nat Commun. 2019;10(1):4544. doi: 10.1038/s41467-019-12448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi D., Jiang X., Sheng Z.K., et al. Mapping the resistance-associated mobilome of a carbapenem-resistant klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a ‘resistance-disarmed’ model organism. J Antimicrob Chemother. 2015;70(10):2770–2774. doi: 10.1093/jac/dkv204. [DOI] [PubMed] [Google Scholar]
- 32.Chen L., Mathema B., Chavda K.D., Deleo F.R., Bonomo R.A., Kreiswirth B.N. Carbapenemase-producing klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian D., Wang M., Zhou Y., Hu D., Ou H., Jiang X. Genetic diversity and evolution of the virulence plasmids encoding aerobactin and salmochelin in klebsiella pneumoniae. Virulence. 2021;12(1):1323–1333. doi: 10.1080/21505594.2021.1924019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam M.M.C., Wyres K.L., Wick R.R., et al. Convergence of virulence and mdr in a single plasmid vector in mdr klebsiella pneumoniae st15. J Antimicrob Chemother. 2019;74(5):1218–1222. doi: 10.1093/jac/dkz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y., Chou S., Liang S., et al. Emergence of an xdr and carbapenemase-producing hypervirulent klebsiella pneumoniae strain in taiwan. J Antimicrob Chemother. 2018;73(8):2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 36.Kim J.S., Cho D.H., Park M., et al. Crispr/cas9-mediated re-sensitization of antibiotic-resistant escherichia coli harboring extended-spectrum β-lactamases. J Microbiol Biotechnol. 2016;26(2):394–401. doi: 10.4014/jmb.1508.08080. [DOI] [PubMed] [Google Scholar]
- 37.Wang P., He D., Li B., et al. Eliminating mcr-1-harbouring plasmids in clinical isolates using the crispr/cas9 system. J Antimicrob Chemother. 2019;74(9):2559–2565. doi: 10.1093/jac/dkz246. [DOI] [PubMed] [Google Scholar]
- 38.Hao M., He Y., Zhang H., et al. Crispr-cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistantenterobacteriaceae. Antimicrob Agents Chemother. 2020;64(9):e00843–e00920. doi: 10.1128/AAC.00843-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomaa A.A., Klumpe H.E., Luo M.L., Selle K., Barrangou R., Beisel C.L. Programmable removal of bacterial strains by use of genome-targeting crispr-cas systems. mBio. 2014;5(1):e00928–e001013. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tagliaferri T.L., Guimarães N.R., Pereira M., et al. Exploring the potential of crispr-cas9 under challenging conditions: facing high-copy plasmids and counteracting beta-lactam resistance in clinical strains of enterobacteriaceae. Front Microbiol. 2020;11:578. doi: 10.3389/fmicb.2020.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie M., Chen K., Ye L., et al. Conjugation of virulence plasmid in clinical klebsiella pneumoniae strains through formation of a fusion plasmid. Adv Biosyst. 2020;4(4) doi: 10.1002/adbi.201900239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.