DNA methylation provides a mechanism by which additional information is imparted to DNA, and such epigenetic information can alter the timing and targeting of cellular events (47). DNA methylation occurs throughout the living world, including bacteria, plants, and mammals. Until recently, methylated DNA sequences were not detected in the fruit fly, in brewer's yeast, or in the nematode. However, analysis by Lyko and colleagues showed that Drosophila melanogaster does contain methylated DNA (42, 43), and thus it is possible that yeast and worms may also have it. In this review, we focus our attention on the roles of DNA methylation in regulating bacterial gene expression and virulence. Although some background information about DNA methylation is presented, we refer the reader to excellent reviews on the subject (5, 15, 28, 47, 64).
DNA methylation occurs at the C-5 or N-4 positions of cytosine and at the N-6 position of adenine and is catalyzed by enzymes known as DNA methyltransferases (MTases) (57, 59). All MTases use S-adenosyl methionine as a methyl donor. DNA methylation has historically been associated with DNA restriction-modification systems thought to be important in protecting cells from foreign DNAs such as transposons and viral DNAs (35, 50, 69). Restriction-modification systems contain a DNA methylase that protects host DNA sequences from restriction with their cognate restriction enzymes which digest unmodified foreign DNAs. Certain MTases, including DNA cytosine MTase (Dcm), which methylates the C-5 position of cytosine in CC(A/T)GG sequences, DNA adenine methylase (Dam), which methylates N-6 of adenine in GATC sequences, and cell cycle-regulated methylase (CcrM), which methylates the N-6 adenine of GAnTC, do not have cognate restriction enzymes associated with them (64). These methylases participate in cellular regulatory events, including those that control bacterial virulence, which are the primary focus of this review.
DAM FAMILY
Background.
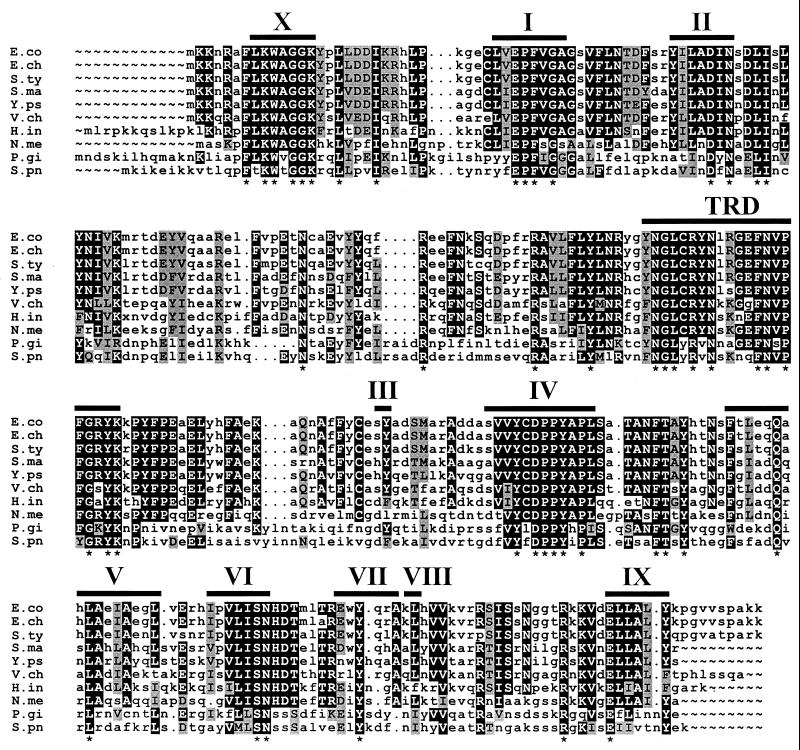
Based on the organization of 10 amino acid domains present in MTases, Dam is classified in the α group (Fig. 1) (46). Dam homologues are widespread among enteric bacteria, including Escherichia coli, Salmonella spp., Serratia marcescens, Yersinia spp., and Vibrio cholerae, cholerae, but are also present in disparate genera, including Neisseria among others (Table 1 and Fig. 1). Dam methylation is not essential for viability of E. coli (3); however, recent data indicate that Dam is an essential gene in Vibrio cholerae and Yersinia pseudotuberculosis (31, 45), similar to results showing that the CcrM methylase is essential in Caulobacter crescentus, Brucella abortus, Rhizobium meliloti, and Agrobacterium tumefaciens (32, 70, 73, 93). Certain α-group methylases, including the DpnII methylase, share significant sequence identity with Dam (32% for DpnII) and methylate GATC sites like Dam but are part of restriction-modification systems (Fig. 1).
FIG. 1.
Sequence alignment of selected E. coli Dam homologues. Residues that are identical in all 10 proteins are noted with asterisks. Residues that are identical or similar in 70% of the sequences are boxed in black and gray, respectively. Lowercase letters indicate residues that occur in less than 70% of the listed sequences. Motifs implicated in the binding of S-adenosyl methionine and methyl transfer are labeled with Roman numerals according to the nomenclature of Tran et al. (80; also see reference 46). The putative GATC site recognition domain of DpnM, an α family DNA adenine methylase (80), is indicated by TRD. The alignment was created using the PILEUP program of the Genetics Computer Group sequencing package (17). E. chrysanthemi sequences were provided by C.-H. Yang and Noel T. Keen (unpublished data). E.co, Escherichia coli (P00475) (SWISSPROT or GenPept database accession numbers are listed in parentheses); E.ch, Erwinia chrysanthemi; S.ty, Salmonella serovar Typhimurium (P55893); S.ma, Serratia marcescens (P45454); Y.ps, Yersinia pseudotuberculosis YPIII (AF274318); V.ch, Vibrio cholerae O395 (AF274317); H.in, Haemophilus influenzae (P44431); N.me, Neisseria meningitidis (AAD34292); P.gi, Porphyromonas gingivalis (S34414); and S.pn, Streptococcus pneumoniae (P04043).
TABLE 1.
Roles of Dam in bacterial virulence
| Bacterial pathogena | Methylase family | Target sequence | % identity to E. coli Damb | Role for methylase
in:
|
Reference | |
|---|---|---|---|---|---|---|
| Growth | Virulence | |||||
| Escherichia coli | Dam (α) | GATC | 100 | No | Yes | 36 |
| Erwinia chrysanthemi | Dam (α) | GATC | 99 | No | Yes | C.-H. Yang and N. Keenc |
| Salmonella enterica serovar Typhimurium | Dam (α) | GATC | 92 | No | Yes | 21, 25, 26 |
| Yersinia pseudotuberculosis | Dam (α) | GATC | 71 | Yes | Yes | 31 |
| Vibrio cholerae | Dam (α) | GATC | 64 | Yes | Yes | 31 |
| Neisseria meningitidis | Dam (α) | GATC | 47 | No | Yes | 14 |
| Brucella abortus | CerM (β) | GAnTC | NA | Yes | Yes | 70 |
The SWISSPROT or GenPept database accession numbers are listed in parentheses for the following strains: E. coli (P00475), Salmonella serovar Typhimurium (P55893), Yersinia pseudotuberculosis YPIII (AF274318), Vibrio cholerae O395 (AF274317), Haemophilus influenzae (P44431), and Neisseria meningitidis (AAD34292).
Percent identities among the selected E. coli Dam homologs were calculated using the GAP program of the Genetics Computer Group sequencing package (17). NA, not applicable.
Yang and Keen, unpublished data.
Functions.
Adenine methylation can alter the interactions of regulatory proteins with DNA, either by a direct steric effect or by an indirect effect on DNA structure (18, 61, 62). Initial studies with dam mutants showed that Dam regulates the expression of certain genes in E. coli including trpR (60), Tn10 transposase (68), and dnaA (13) as well as phage genes including mom of Mu (24). Methylation of a GATC site(s) within the consensus RNA polymerase binding site inhibits (trpR and Tn10 transposase) or enhances (dnaA) transcription, by altering the interaction with the transcription apparatus.
As discussed above, methylation can alter the affinity of regulatory proteins for DNA. Conversely, DNA binding proteins have been shown to inhibit methylation of specific DNA sequences. For example, the SeqA protein involved in the timing of DNA replication binds specifically to hemimethylated DNA sequences near the origin of replication (oriC), thereby sequestering oriC from Dam methylation for a part of the cell cycle and maintaining it in a hemimethylated state (41). Other regulatory proteins bind nonmethylated DNAs with highest affinity, protect specific DNA sequences from methylation, and form DNA methylation patterns (DMPs), which are present in certain eukaryotes as well (see Fig. 2) (28). DMPs are formed when regulatory factors bind to DNA target sites that overlap or are near methylation sites and inhibit their methylation (12).
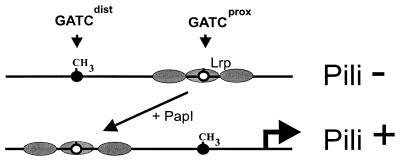
FIG. 2.
Regulation of Pap pilus expression by DNA methylation patterns. Cooperative binding of Lrp to sites proximal to the papBA pilin promoter blocks transcription, inhibits methylation of GATCprox, and forms the phase-OFF DNA methylation pattern (Pili− state) (90). According to the Pap phase variation model (see the text) (36, 84), transition to the Pili+ state occurs following DNA replication under conditions which induce PapI expression. PapI binds specifically to Lrp, increases the affinity of Lrp for distal pap DNA binding sites, and forms the phase-ON DNA methylation pattern (33, 55, 56).
Regulation by hemimethylation.
Work carried out with E. coli Dam indicates that it acts as an efficient de novo methylase, methylating both nonmethylated and hemimethylated GATC sites with similar efficiency (82). Dam plays an important role in regulating the timing and targeting (51) of a number of cellular functions including DNA replication (9, 34, 41, 72), segregation of chromosomal DNA (52, 58), mismatch repair (29, 48, 71), and transposition (19, 66, 68, 77, 89). In all of these events, hemimethylated GATC sites, present immediately following DNA replication, control the binding of proteins to specific DNA target sites. For example, DNA replication is controlled in part by SeqA, which binds specifically to hemimethylated GATC sites near the origin of replication and delays their methylation (34, 41). Segregation of newly replicated DNA may occur by binding of hemimethylated DNA to membrane-bound factors. In methyl-directed mismatch repair, MutH binds to hemimethylated DNA and cleaves the nonmethylated strand (1). Certain transposases, including Tn10 transposase, bind with highest affinity to hemimethylated binding sites, limiting transposition to a time immediately following DNA replication (68).
Regulation by DNA methylation patterns.
Dam also plays pivotal roles in controlling gene expression by the formation of DMPs. DMPs have long been known to be present in eukaryotes and appear to regulate gene expression (5, 92). The first reported DMPs which regulate gene expression in prokaryotes are within the pyelonephritis-associated pilus (pap) operon of uropathogenic E. coli (7).
Most nonmethylated GATC sites are found within noncoding regions that likely have a regulatory function. For example, the GutR repressor binds in the upstream regulatory region of the glucitol (gut) operon, blocking methylation of a GATC site designated GATC −44.5 within the GutR binding domain and forming a specific DMP (85). In the presence of glucitol, GutR no longer blocks DNA methylation, indicating that it no longer binds gut regulatory DNA. This is an example of environmental control of a DMP. Notably, although catabolite gene activator protein (CAP) also binds to a site overlapping GATC −44.5, it does not protect this site from methylation.
It appears that most, if not all, DMPs are formed by the binding of regulatory proteins such as GutR (85), Lrp (12), histone-like nucleoid-structuring protein (H-NS) (91), and OxyR (22) to upstream regulatory DNA sequences. In these examples, purified proteins have been shown to block methylation of GATC sites that are contained in or near the DNA recognition sequence in vitro. Inhibition of methylation could occur by direct steric occlusion of Dam binding or by alterations in DNA conformation which change the configuration of the Dam target (GATC) site (61).
Analysis of the E. coli chromosome has shown that there are at least 50 GATC sites that are stably undermethylated (67, 75). The locations of these sites may vary depending upon environmental conditions (23), which can alter the expression and/or binding of regulatory factors that bind to DNA and specifically block DNA methylation. Analysis of the Salmonella chromosome by pulsed-field gel electrophoresis has shown that, similar to E. coli, specific DNA methylation patterns are present (26). Although many nonmethylated GATC sites have been identified in E. coli, methylation in only a subset has been shown to control the expression of linked genes. For example, GutR blocks methylation of GATC −44.5 in the gut operon, but methylation of GATC −44.5 does not alter the binding of GutR to gut DNA in vitro nor does it alter gut expression in vivo (85). The CarP regulatory protein and integration host factor (IHF) protect a GATC site in the carAB operon, but it is not clear if methylation controls CarAB expression (16). In contrast, methylation of regulatory GATC sites in the pap and agn operons directly controls expression of Pap pili and the Ag43 outer membrane protein, respectively (12, 22, 27). This occurs by reduction of the affinities of the Lrp and OxyR proteins for pap and agn regulatory DNAs, respectively. In pap and related methylation-controlled operons, methylation of two GATC sites spaced 102 bp apart regulates Lrp binding, whereas in agn, methylation of three closely spaced GATC sites inhibits OxyR binding (22). In these instances, there is a mutual competition between the methylase and the DNA binding protein, forming a DMP which heritably controls gene expression and provides a form of cellular memory.
ROLES OF DAM IN VIRULENCE
Alterations in the levels of Dam attenuate the virulence of a number of pathogens, including Salmonella spp. (21, 25, 26), Y. pseudotuberculosis, and V. cholerae (31). Because Dam plays multiple roles in cell physiology (see above), it is possible that pleiotropic effects not related to alterations in gene expression may be responsible for the virulence defects of dam mutants. However, the growth rates of dam mutant and Dam-overproducer Salmonella enterica serovar Typhimurium were similar to that of wild-type Salmonella (25). In addition, levels of overproduction of Dam in Y. pseudotuberculosis and V. cholerae that inhibited virulence had no significant effect on in vitro growth rates (31). These data strongly indicate that the virulence defect of dam mutants is directly the result of alterations in gene expression and not due to a nonspecific growth defect. Dam has been reported to control the expression of a number of virulence genes (12, 21, 25, 26, 31, 44, 45). Deletion of dam erases DNA methylation patterns, which could alter the binding of regulatory proteins to a number of regions on the bacterial chromosome. In the absence of Dam, overexpression of genes could occur if GATC methylation blocked binding of an activator or enhanced the binding of a repressor. Conversely, underexpression of a gene would occur in the absence of Dam if GATC methylation blocked binding of a repressor or enhanced binding of an activator.
E. coli.
Dam regulates the expression of a large group of pilus operons that play important roles in virulence in urinary tract infections (e.g., Pap, Prf, and S pili) and diarrheal diseases (e.g., Afa, CS31a, and K88 pili) (30, 36, 49, 53, 87). All of these pilus-adhesin operons share common regulatory features including control by Lrp, the presence of a promoter proximal GATC site (GATCprox) located within one set of Lrp binding sites and a promoter distal site (GATCdist) within a second set of Lrp binding sites, inter-GATC spacing of 102 or 103 bp, and a homologue to the PapI regulatory protein that binds to Lrp, increasing its affinity for the promoter distal Lrp sites which then helps activate transcription (7, 8, 11, 12, 33, 55, 56, 86) (Fig. 2).
In the pap operon, DNA methylation directly regulates the switch between pilus expression (phase ON) and nonexpression (phase OFF) by dictating the binding of Lrp (Fig. 2). At low PapI levels, Lrp binds with high affinity to promoter proximal sites, blocks transcription from the papBA promoter, and inhibits methylation of GATCprox (90). However, the promoter distal GATCdist site is not bound by Lrp and thus becomes methylated. Methylation of GATCdist inhibits movement of Lrp to the distal set of sites and thus is presumed to lock cells in the phase-OFF state until DNA replication generates a hemimethylated GATCdist site which binds Lrp with a higher affinity (36). Methylation of the promoter proximal GATCprox site is required for the expression of Pap pili (12). Mutations in the Lrp binding sites near GATCprox result in a phase-locked ON transcription phenotype that is Dam and PapI independent (55). These results indicate that methylation of GATCprox may help displace Lrp from its promoter proximal DNA binding sites that overlap GATCprox, with the aid of PapI (33). Binding of Lrp-PapI at the promoter distal GATCdist site blocks its methylation, forming a DNA methylation pattern that is characteristic of cells expressing pili. The cell environment controls the pap DNA methylation pattern since in poor carbon sources the cyclic AMP level is high and stimulates PapI expression via a cyclic AMP-CAP binding site in the pap regulatory region (2). PapI facilitates movement of Lrp to the GATCdist site, and Lrp blocks methylation of GATCdist and helps activate pap transcription (84). Other members of the Pap family, including sfa (87), daa (87), fae (30), and clp (49) and pef in Salmonella serovar Typhimurium (53), have been shown to be regulated by DNA methylation patterns as well (36).
Salmonella.
Torreblanca and Casadesus first identified genes regulated by Dam in Salmonella serovar Typhimurium using a genetic approach (78). One of these genes mapped to the pSLT virulence plasmid and was later shown to be finP, which expresses an antisense RNA controlling the F-type pili required for conjugative plasmid transfer (79). The result is that under conditions of low levels of Dam, transfer of the Salmonella pSLT plasmid is elevated. The physiologic connection between Dam-controlled pilus expression and mating is not yet clear but could function to coordinate mating with virulence plasmid replication or to enable environmental control of mating (79). Dam also regulates the expression of plasmid-encoded fimbriae (Pef) encoded by pSLT by a mechanism that shares features with pap (53). Work from Heffron's laboratory indicates that Pef may play a role in Salmonella virulence (83). Like pap, expression of Pef fimbriae is turned off in the absence of Dam since methylation of a promoter-proximal GATC site of pef is essential for transcription.
Recently, Dam was shown to be essential for the virulence of Salmonella serovar Typhimurium in a murine model of typhoid fever (21, 25, 26). Dam− Salmonella shows reduced M-cell cytotoxicity and invasion of enterocytes but appears to grow normally within cells (21). In the absence of Dam, serovar Typhimurium is avirulent when given orally and intraperitoneally and fails to kill mice at 10,000 times the lethal dose required to kill half of the animals (LD50). Dam-deficient Salmonella colonizes Peyer's patches in a manner similar to that of wild-type bacteria but attains only very low numbers in systemic tissues and is totally cleared from mice after about 4 weeks. The failure of dam mutants to cause disease is not the result of defects in mismatch repair since mutS and mutL serovar Typhimurium is fully virulent (21, 26).
Why is Dam− Salmonella avirulent? We hypothesize that dam mutant Salmonella is markedly attenuated as a result of dysregulation of gene expression. Dam-deficient Salmonella serovar Typhimurium up-regulates the expression of over 35 genes that are induced during infection (26), including spvB, a cytotoxin which causes apoptosis of macrophages (39). In contrast, Dam positively regulates the secretion of the SipA, SipB, and SipC proteins coded for by the Salmonella pathogenicity island type 1 virulence locus (21). Thus, in the absence of Dam, virulence factors such as SpvB are predicted to be overexpressed (the SpvB protein was recently shown to be ectopically expressed at very high levels in the absence of Dam [D Guiney, unpublished results]), whereas other factors such as SipABC are underexpressed. We hypothesize that this combination of overexpression and underexpression of virulence proteins inhibits virulence (25, 45). If this is correct, then overexpression of Dam might also block virulence since negatively regulated factors such as SpvB would be underexpressed and positively regulated factors such as SipABC would be overexpressed. In fact, overexpression of Dam reduces the virulence of Salmonella serovar Typhimurium 10,000-fold (25). As predicted, the protein profiles from Dam− and Dam-overproducing Salmonella strains are different from each other and from wild-type Salmonella (25). In addition, Dam− Salmonella releases a high level of outer membrane vesicles, suggesting an instability defect in the outer membrane. Since OmpA is a highly immunogenic protein, vesicle release by dam mutants might contribute to their efficacy as live attenuated vaccines (J. Casadesus, personal communication). These data support the hypothesis that Dam is a global regulator of virulence genes in Salmonella and that Dam levels regulate virulence.
Both Dam-deficient and Dam-overproducing Salmonella serovar Typhimurium strains are highly effective vaccines against salmonellosis. As few as 90 Dam− bacteria administered intraperitoneally provide significant protection (21) and oral vaccination of mice with 109 Dam− Salmonella bacteria completely protects them from challenge with 109 wild-type Salmonella bacteria (10,000 times the oral LD50) (26). Vaccination with Dam− Salmonella protects mice against challenge with other S. enterica serovars including Enteritidis and Dublin (25). Additionally, Dam− Salmonella conferred cross-protective immunity in chickens (20a). This cross-protection could occur as a result of aberrant expression of a number of virulence determinants of serovar Typhimurium, some of which might be shared with other Salmonella serovars. The dysregulation of expression of Salmonella virulence determinants not only could disrupt the normal pathogenic cycle but also may enable the host immune system to mount an effective response. This response could be elicited to Dam-controlled bacterial antigens which are normally under temporal and spatial control and not easily detected. Consistent with this hypothesis, S. enterica serovar Typhimurium overproducing Dam conferred significant protection against homologous Salmonella (25) although not to the same extent as Dam-deficient Salmonella. This discrepancy could be due, in part, to the differences in antigen expression observed between these two vaccines (see above). Dam is also essential for full virulence of S. enterica serovar Enteritidis (25), which can invade the yolk sac and contaminate eggs. Because Dam− S. enterica serovars Typhimurium and Enteritidis are highly attenuated, it seems likely that Dam might also be essential for virulence of S. enterica serovar Typhi, the causative agent of typhoid fever.
Other pathogens
Although Dam from E. coli and Salmonella spp. is not essential for growth, dam is an essential gene in V. cholerae and Y. pseudotuberculosis (31) (Table 1). Since V. cholerae has two chromosomes (81), it is possible that Dam plays the same roles as those in E. coli and Salmonella, but additionally, Dam may coordinate the timing and segregation of the two chromosomes. Recent data indicate that the virulence of Dam− Erwinia chrysanthemi is greatly reduced for African violets and lettuce, two of its hosts (C.-H. Yang and N. Keen, unpublished data). Thus, Dam is important for the virulence of both animal and plant pathogens.
Overproduction of Dam inhibits the colonization of V. cholerae based on a suckling mouse model (31). Moreover, overexpression of Dam in Y. pseudotuberculosis greatly attenuates virulence in a murine model (>6,000-fold) and alters the protein expression profile. Oral immunization of mice with Dam-overproducing Y. pseudotuberculosis protects against challenge with at least 1,000 times the LD50. Thus, it appears that the Dam-based vaccination strategy developed using S. enterica serovar Typhimurium can be extended to other enteric pathogens.
In contrast to the essentiality of Dam for virulence and/or growth in enteric bacteria, all pathogenic isolates of group B Neisseria meningitidis have a mutation within dam (dam gene replacement, or drg) and thus do not express Dam (14). These meningococci are deficient in methyl-directed mismatch repair, as expected, and show high rates of phase variation of a neuraminic acid capsule controlled by the polysialyltransferase (siaD) gene. Frameshift mutations within a poly-deoxycytosine repeat in the siaD gene coding sequence regulate the on-off expression of capsule. A high rate of capsular phase variation was observed only in dam mutant meningococci. Thus, selection for Dam-deficient meningococci may occur as a result of increased phase variation rates.
We are only beginning to understand the roles that Dam plays in regulating the interactions between bacterial pathogens and their hosts that contribute to virulence. Many bacterial pathogens, including S. enterica serovar Typhi, Shigella spp. (dysentery), pathogenic E. coli including O157:H7 (hemolytic-uremic syndrome), Haemophilus influenzae (pneumonia and otitis media), and Legionella pneumophila (pneumonia), have been reported to contain dam homologues and/or Dam activity (25, 38, 45) (Table 1 and Fig. 1). It seems likely that Dam is important for the pathogenesis of a number of diseases. An important next step will be to determine the mechanism by which Dam controls virulence and whether there are any unifying concepts that can be gleaned from the analysis of genes regulated by Dam in a number of pathogens.
Alteration in DMPs caused by fluctuations in the binding of regulatory proteins such as Lrp or OxyR to DNA target sites can be viewed as a novel mechanism for global regulation. The relevance of DMPs to pathogenesis could be determined by identification of all Dam-regulated virulence genes in a pathogen by microarray analysis and matching them to the nonmethylated GATC sites in the chromosome (75, 88). The first part of this analysis has recently been carried out in E. coli, with the data indicating that the mRNA levels of about 50 to 60 genes are significantly altered in the absence of Dam (N. Bourquard, G. W. Hatfield, and D. A. Low, unpublished data).
CCRM FAMILY
Background.
In contrast to Dam, CcrM is classified in the β group of DNA methylases (Table 1). CcrM was originally discovered as a cell cycle-regulated MTase in Caulobacter (95). CcrM is 49% identical to the HinfI MTase from H. influenzae, which shares the same recognition sequence but is part of a retriction-modification system. CcrM homologues are found in the α-proteobacteria, including the plant pathogen A. tumefaciens and symbiont Rhizobium meliloti and B. abortus (64), which causes brucellosis in cattle and humans (70). All CcrM homologues tested appear to be essential for cell viability, similar to the essential roles of MTases in mammalian cells and Dam in V. cholerae and Y. pseudotuberculosis. Moreover, based on complementation analysis of Caulobacter and Rhizobium, the CcrM proteins are functionally interchangeable (64).
Functions.
The CcrM methylase from C. crescentus plays an essential role in the cell cycle of this developmentally programmed bacterium. CcrM, in contrast to Dam, appears to be a “maintenance” methylase with preference for hemimethylated DNA over nonmethylated DNA (4). CcrM methylase is essential for the viability of α-proteobacteria including C. crescentus, B. abortus, R. meliloti, and A. tumefaciens (32, 70, 73, 93).
Initial evidence that CcrM regulates gene expression came from analysis of the ccrM gene itself. Methylation of GAnTC sites within the ccrM promoter inhibits transcription. Mutation of these CcrM target sites prevents shutdown of methylase gene transcription after cell division (74). Analysis of the complete genome sequence of C. crescentus showed that CcrM target sites were less abundant than predicted from random occurrence and had a bias to intergenic regions (54). These data support the hypothesis that CcrM is a global regulator of gene expression. Indeed, recent microarray analysis by Lucy Shapiro and colleagues indicates that approximately 100 genes in C. crescentus are affected by depletion of CcrM (L. Shapiro, unpublished data).
Roles in virulence.
CcrM may play an important role in the virulence of B. abortus based on the analysis of bacteria overexpressing the methylase (70). Overexpression of CcrM on recombinant plasmids inhibited growth of Brucella within murine peritoneal macrophages. This attenuation did not appear to be due to alterations in bacterial growth rates or alterations in cell morphology or DNA replication initiation at lower CcrM expression levels. It thus appears that the defect in intracellular replication is due to some other effect of CcrM on cellular function such as regulation of a gene(s) required for adapting to the intracellular environment (64).
REGULATION OF DNA METHYLATION
Dam and CcrM methylase activities are under complex regulatory control, as expected for global regulators. CcrM transcription is activated by the cell cycle transcription regulator (CtrA) in late S phase in predivisional cells, resulting in fully methylated chromosomes which initiate replication. By the time cell division occurs, the CcrM level is greatly reduced via cleavage with Lon protease (65). Thus, both the level of CcrM and its cellular location in the Caulobacter morphogenic pathway are strictly regulated. In addition, ccrM may be under autoregulatory control since methylation of two CcrM target sites in the ccrM promoter may play a role in inhibiting ccrM transcription (74).
There are only about 130 molecules of Dam in rapidly growing cells, a number sufficient to methylate all of the available GATC sites within a single DNA replication cycle (10). dam methylase from E. coli contains five promoters, with the major promoter (P2) located about 3.5 kbp upstream of the dam AUG translation start site (40). The dam P2 promoter is controlled by growth rate, with high levels of dam transcription present in cells with high growth rates (63). The functions of the other dam promoters are unknown but may be responsive to in vivo growth conditions. Precedent for this possibility comes from the analysis of Helicobacter pylori, the causative agent of chronic gastritis (6). H. pylori contains a number of putative methylases without cognate restriction enzymes, and thus it has been hypothesized that these methylases may control cellular functions by analogy with CcrM and Dam (76). One Helicobacter gene, hpyIM (GenBank accession number AAC45818), codes for an adenine methylase which recognizes the sequence CATG (20, 94). HpyIM appears to be a member of the α group of methylases (46), sharing 61 and 34% identity with NlaIII methylase (Swiss Prot accession number P24582) and a Campylobacter methylase (EMBL accession number CAB72691), respectively. Notably, hpyIM expression appears to be induced following attachment of H. pylori to gastric epithelial cells, suggesting that induction of HpyIM in the host may play a role in the regulation of virulence (37).
CONCLUSIONS
Clearly, DNA methylation plays important roles in the virulence of a growing list of bacterial pathogens (Table 1). DNA methylation provides an additional level of regulatory control since the binding of many different regulatory factors to target DNA sequences can potentially be affected in a heritable fashion. Moreover, alteration of methylase levels in response to environmental stimuli could control the temporal expression of specific gene subgroups depending on the effect of methylation on the affinity of each regulatory protein for target DNA. Important questions for future research include the following: What is the spectrum of pathogens in which DNA methylation plays a role(s) in virulence? What types of virulence genes are regulated by DNA methylation? What mechanisms are involved in controlling and coordinating virulence gene expression by DNA methylation? How are DNA methylase levels altered in response to environmental stimuli? Does methylation provide a memory system to help bacterial pathogens time and coordinate the expression of virulence determinants?
ACKNOWLEDGMENTS
We are thoroughly indebted to Josep (Pepe) Casadesus, Lucy Shapiro, and Marjan van der Woude for their helpful comments and unpublished data presented here. We also thank Bruce Braaten, Douglas Heithoff, and Robert Sinsheimer for helpful comments.
This work was supported by National Institutes of Health grant AI23348 (to D.A.L.), by private donations from Jim and Deanna Dehlsen, University of California Biotech Program, the Santa Barbara Cottage Hospital Research Program, Santa Barbara, Calif., and by USDA grant 2000 to 02539 (to M.J.M).
REFERENCES
- 1.Au K G, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 2.Baga M, Goransson M, Normark S, Uhlin B E. Transcriptional activation of a pap pilus virulence operon from uropathogenic Escherichia coli. EMBO J. 1985;4:3887–3893. doi: 10.1002/j.1460-2075.1985.tb04162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bale A, d'Alarcao M, Marinus M G. Characterization of DNA adenine methylation mutants of Escherichia coliK12. Mutat Res. 1979;59:157–165. doi: 10.1016/0027-5107(79)90153-2. [DOI] [PubMed] [Google Scholar]
- 4.Berdis A J, Lee I, Coward J K, Stephens C, Wright R, Shapiro L, Benkovic S J. A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentusprocessively methylates GANTC sites on hemimethylated DNA. Proc Natl Acad Sci USA. 1998;95:2874–2879. doi: 10.1073/pnas.95.6.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird A P, Wolffe A P. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 6.Blaser M J, Kirschner D. Dynamics of Helicobacter pyloricolonization in relation to the host response. Proc Natl Acad Sci USA. 1999;96:8359–8364. doi: 10.1073/pnas.96.15.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blyn L B, Braaten B A, Low D A. Regulation of pap pilin phase variation by a mechanism involving differential dammethylation states. EMBO J. 1990;9:4045–4054. doi: 10.1002/j.1460-2075.1990.tb07626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blyn L B, Braaten B A, White-Ziegler C A, Rolfson D H, Low D A. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 1989;8:613–620. doi: 10.1002/j.1460-2075.1989.tb03416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogan J A, Helmstetter C E. DNA sequestration and transcription in the oriC region of Escherichia coli. Mol Microbiol. 1997;26:889–896. doi: 10.1046/j.1365-2958.1997.6221989.x. [DOI] [PubMed] [Google Scholar]
- 10.Boye E, Marinus M G, Lobner-Olesen A. Quantitation of Dam methyltransferase in Escherichia coli. J Bacteriol. 1992;174:1682–1685. doi: 10.1128/jb.174.5.1682-1685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braaten B A, Blyn L B, Skinner B S, Low D A. Evidence for a methylation-blocking factor (mbf) locus involved in pap pilus expression and phase variation in Escherichia coli. J Bacteriol. 1991;173:1789–1800. doi: 10.1128/jb.173.5.1789-1800.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braaten B A, Nou X, Kaltenbach L S, Low D A. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell. 1994;76:577–588. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 13.Braun R E, Wright A. DNA methylation differentially enhances the expression of one of the two E. coli dnaApromoters in vivo and in vitro. Mol Gen Genet. 1986;202:246–250. doi: 10.1007/BF00331644. [DOI] [PubMed] [Google Scholar]
- 14.Bucci C, Lavitola A, Salvatore P, Del Giudice L, Massardo D R, Bruni C B, Alifano P. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol Cell. 1999;3:435–445. doi: 10.1016/s1097-2765(00)80471-2. [DOI] [PubMed] [Google Scholar]
- 15.Casadesus J, Torreblanca J. Methylation-related epigenetic signals in bacterial DNA. In: Russo V E A, Martienssen R A, Riggs A D, editors. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 141–153. [Google Scholar]
- 16.Charlier D, Gigot D, Huysveld N, Roovers M, Pierard A, Glansdorff N. Pyrimidine regulation of the Escherichia coli and Salmonella typhimurium carABoperons: CarP and integration host factor (IHF) modulate the methylation status of a GATC site present in the control region. J Mol Biol. 1995;250:383–391. doi: 10.1006/jmbi.1995.0384. [DOI] [PubMed] [Google Scholar]
- 17.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diekmann S. DNA methylation can enhance or induce DNA curvature. EMBO J. 1987;6:4213–4217. doi: 10.1002/j.1460-2075.1987.tb02769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodson K W, Berg D E. Factors affecting transposition activity of IS50 and Tn5ends. Gene. 1989;76:207–213. doi: 10.1016/0378-1119(89)90161-3. [DOI] [PubMed] [Google Scholar]
- 20a.Dueger E L, House J K, Heithoff D M, Mahan M J. SalmonellaDNA adenine methylase mutants elicit protective immune responses to homologous and heterologous serotypes in chickens. Infect Immun. 2001;69:7950–7954. doi: 10.1128/IAI.69.12.7950-7954.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donahue J P, Peek R M, Van Doorn L J, Thompson S A, Xu Q, Blaser M J, Miller G G. Analysis of iceA1 transcription in Helicobacter pylori. Helicobacter. 2000;5:1–12. doi: 10.1046/j.1523-5378.2000.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-F. Del Portillo, Pucciarelli M G, Casadesus J. DNA adenine methylase mutants of Salmonella typhimuriumshow defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc Natl Acad Sci USA. 1999;96:11578–11583. doi: 10.1073/pnas.96.20.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haagmans W, van der Woude M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol. 2000;35:877–887. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- 23.Hale W B, van der Woude M W, Low D A. Analysis of nonmethylated GATC sites in the Escherichia colichromosome and identification of sites that are differentially methylated in response to environmental stimuli. J Bacteriol. 1994;176:3438–3441. doi: 10.1128/jb.176.11.3438-3441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hattman S. DNA methyltransferase-dependent transcription of the phage Mu momgene. Proc Natl Acad Sci USA. 1982;79:5518–5521. doi: 10.1073/pnas.79.18.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heithoff D M, Enioutina E Y, Daynes R A, Sinsheimer R L, Low D A, Mahan M J. SalmonellaDNA adenine methylase mutants confer cross-protective immunity. Infect Immun. 2001;69:6725–6730. doi: 10.1128/IAI.69.11.6725-6730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence [see comments] Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 27.Henderson I R, Owen P. The major phase-variable outer membrane protein of Escherichia colistructurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J Bacteriol. 1999;181:2132–2141. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrich B, Bird A. Mammalian methyltransferases and methyl-CpG-binding domains: proteins involved in DNA methylation. Curr Top Microbiol Immunol. 2000;249:55–74. doi: 10.1007/978-3-642-59696-4_4. [DOI] [PubMed] [Google Scholar]
- 29.Herman G E, Modrich P. Escherichia coli K-12 clones that overproduce dam methylase are hypermutable. J Bacteriol. 1981;145:644–646. doi: 10.1128/jb.145.1.644-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huisman T T, de Graaf F K. Negative control of fae (K88) expression by the ‘global’ regulator Lrp is modulated by the ‘local’ regulator FaeA and affected by DNA methylation. Mol Microbiol. 1995;16:943–953. doi: 10.1111/j.1365-2958.1995.tb02320.x. [DOI] [PubMed] [Google Scholar]
- 31.Julio S M, Heithoff D M, Provenzano D, Klose K K, Sinsheimer R L, Low D A, Mahan M J. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect Immun. 2001;69:7610–7615. doi: 10.1128/IAI.69.12.7610-7615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahng L S, Shapiro L. The CcrM DNA methyltransferase of Agrobacterium tumefaciensis essential, and its activity is cell cycle regulated. J Bacteriol. 2001;183:3065–3075. doi: 10.1128/JB.183.10.3065-3075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaltenbach L S, Braaten B A, Low D A. Specific binding of PapI to Lrp-papDNA complexes. J Bacteriol. 1995;177:6449–6455. doi: 10.1128/jb.177.22.6449-6455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang S, Lee H, Han J S, Hwang D S. Interaction of SeqA and Dam methylase on the hemimethylated origin of Escherichia colichromosomal DNA replication. J Biol Chem. 1999;274:11463–11468. doi: 10.1074/jbc.274.17.11463. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome–restriction-modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9:649–656. doi: 10.1016/s0959-437x(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 36.Krabbe M, Weyand N, Low D. Environmental control of pilus gene expression. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 305–321. [Google Scholar]
- 37.Lee A, Mégraud F. Helicobacter pylori: techniques for clinical diagnosis and basic research. W. B. Philadelphia, Pa: Saunders; 1996. [Google Scholar]
- 38.Lema M W, Brown A. Dam-like methylation in legionellae. Curr Microbiol. 1996;32:64–66. doi: 10.1007/s002849900011. [DOI] [PubMed] [Google Scholar]
- 39.Libby S J, Adams L G, Ficht T A, Allen C, Whitford H A, Buchmeier N A, Bossie S, Guiney D G. The spv genes on the Salmonella dublinvirulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect Immun. 1997;65:1786–1792. doi: 10.1128/iai.65.5.1786-1792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobner-Olesen A, Boye E, Marinus M G. Expression of the Escherichia coli damgene. Mol Microbiol. 1992;6:1841–1851. doi: 10.1111/j.1365-2958.1992.tb01356.x. [DOI] [PubMed] [Google Scholar]
- 41.Lu M, Campbell J L, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 42.Lyko F. DNA methylation learns to fly. Trends Genet. 2001;17:169–172. doi: 10.1016/s0168-9525(01)02234-x. [DOI] [PubMed] [Google Scholar]
- 43.Lyko F, Ramsahoye B H, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408:538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 44.Mahan M J, Heithoff D M, Sinsheimer R L, Low D A. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu Rev Genet. 2000;34:139–164. doi: 10.1146/annurev.genet.34.1.139. [DOI] [PubMed] [Google Scholar]
- 45.Mahan M J, Low D A. DNA methylation regulates bacterial gene expression and virulence. ASM News. 2001;67:356–361. [Google Scholar]
- 46.Malone T, Blumenthal R M, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 47.Marinus M G. Methylation of DNA. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 782–791. [Google Scholar]
- 48.Marinus M G. Recombination is essential for viability of an Escherichia coli dam(DNA adenine methyltransferase) mutant. J Bacteriol. 2000;182:463–468. doi: 10.1128/jb.182.2.463-468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin C. The clp (CS31A) operon is negatively controlled by Lrp, ClpB, and L-alanine at the transcriptional level. Mol Microbiol. 1996;21:281–292. doi: 10.1046/j.1365-2958.1996.00651.x. [DOI] [PubMed] [Google Scholar]
- 50.Meselson M, Yuan R, Heywood J. Restriction and modification of DNA. Annu Rev Biochem. 1972;41:447–466. doi: 10.1146/annurev.bi.41.070172.002311. [DOI] [PubMed] [Google Scholar]
- 51.Messer W, Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988;54:735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- 52.Meury J, Bahloul A, Kohiyama M. Importance of the replication origin sequestration in cell division of Escherichia coli. Biochimie. 1995;77:875–879. doi: 10.1016/0300-9084(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 53.Nicholson B, Low D. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol Microbiol. 2000;35:728–742. doi: 10.1046/j.1365-2958.2000.01743.x. [DOI] [PubMed] [Google Scholar]
- 54.Nierman W C, Feldblyum T V, Laub M T, Paulsen I T, Nelson K E, Eisen J, Heidelberg J F, Alley M R, Ohta N, Maddock J R, Potocka I, Nelson W C, Newton A, Stephens C, Phadke N D, Ely B, DeBoy R T, Dodson R J, Durkin A S, Gwinn M L, Haft D H, Kolonay J F, Smit J, Craven M B, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg S L, Venter J C, Shapiro L, Fraser C M. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nou X, Braaten B, Kaltenbach L, Low D A. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 1995;14:5785–5797. doi: 10.1002/j.1460-2075.1995.tb00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nou X, Skinner B, Braaten B, Blyn L, Hirsch D, Low D. Regulation of pyelonephritis-associated pili phase-variation in Escherichia coli: binding of the PapI and the Lrp regulatory proteins is controlled by DNA methylation. Mol Microbiol. 1993;7:545–553. doi: 10.1111/j.1365-2958.1993.tb01145.x. [DOI] [PubMed] [Google Scholar]
- 57.Noyer-Weidner M, Trautner T A. Methylation of DNA in prokaryotes. In: Jost J P, Saluz H P, editors. DNA methylation: molecular biology & biological significance. Basel, Switzerland: Birkhauser; 1992. pp. 39–108. [Google Scholar]
- 58.Ogden G B, Pratt M J, Schaechter M. The replicative origin of the E. colichromosome binds to cell membranes only when hemimethylated. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- 59.Palmer B R, Marinus M G. The dam and dcm strains of Escherichia coli—a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 60.Peterson K R, Wertman K F, Mount D W, Marinus M G. Viability of Escherichia coliK-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol Gen Genet. 1985;201:14–19. doi: 10.1007/BF00397979. [DOI] [PubMed] [Google Scholar]
- 61.Polaczek P, Kwan K, Campbell J L. GATC motifs may alter the conformation of DNA depending on sequence context and N6-adenine methylation status: possible implications for DNA-protein recognition. Mol Gen Genet. 1998;258:488–493. doi: 10.1007/s004380050759. [DOI] [PubMed] [Google Scholar]
- 62.Polaczek P, Kwan K, Liberies D A, Campbell J L. Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol Microbiol. 1997;26:261–275. doi: 10.1046/j.1365-2958.1997.5701931.x. [DOI] [PubMed] [Google Scholar]
- 63.Rasmussen L J, Lobner-Olesen A, Marinus M G. Growth-rate-dependent transcription initiation from the dam P2 promoter. Gene. 1995;157:213–215. doi: 10.1016/0378-1119(94)00619-4. [DOI] [PubMed] [Google Scholar]
- 64.Reisenauer A, Kahng L S, McCollum S, Shapiro L. Bacterial DNA methylation: a cell cycle regulator? J Bacteriol. 1999;181:5135–5139. doi: 10.1128/jb.181.17.5135-5139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reisenauer A, Quon K, Shapiro L. The CtrA response regulator mediates temporal control of gene expression during the Caulobactercell cycle. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reznikoff W S. The Tn5transposon. Annu Rev Microbiol. 1993;47:945–963. doi: 10.1146/annurev.mi.47.100193.004501. [DOI] [PubMed] [Google Scholar]
- 67.Ringquist S, Smith C L. The Escherichia coli chromosome contains specific, unmethylated dam and dcmsites. Proc Natl Acad Sci USA. 1992;89:4539–4543. doi: 10.1073/pnas.89.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roberts D, Hoopes B C, McClure W R, Kleckner N. IS10transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- 69.Roberts R J, Macelis D. REBASE— restriction enzymes and methylases. Nucleic Acids Res. 2000;28:306–307. doi: 10.1093/nar/28.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robertson G T, Reisenauer A, Wright R, Jensen R B, Jensen A, Shapiro L, Roop R M., 2nd The Brucella abortusCcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J Bacteriol. 2000;182:3482–3489. doi: 10.1128/jb.182.12.3482-3489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schlagman S L, Hattman S, Marinus M G. Direct role of the Escherichia coliDam DNA methyltransferase in methylation-directed mismatch repair. J Bacteriol. 1986;165:896–900. doi: 10.1128/jb.165.3.896-900.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stancheva I, Koller T, Sogo J M. Asymmetry of Dam remethylation on the leading and lagging arms of plasmid replicative intermediates. EMBO J. 1999;18:6542–6551. doi: 10.1093/emboj/18.22.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stephens C M, Zweiger G, Shapiro L. Coordinate cell cycle control of a CaulobacterDNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol. 1995;177:1662–1669. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tavazoie S, Church G M. Quantitative whole-genome analysis of DNA-protein interactions by in vivo methylase protection in E. coli. Nat Biotechnol. 1998;16:566–571. doi: 10.1038/nbt0698-566. [DOI] [PubMed] [Google Scholar]
- 76.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D J C, Venter, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori[see comments] Nature. 1997;388:539–547. doi: 10.1038/41483. . (Erratum 389:412.) [DOI] [PubMed] [Google Scholar]
- 77.Tomcsanyi T, Berg D E. Transposition effect of adenine (Dam) methylation on activity of O end mutants of IS50. J Mol Biol. 1989;209:191–193. doi: 10.1016/0022-2836(89)90271-4. [DOI] [PubMed] [Google Scholar]
- 78.Torreblanca J, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium and a novel dam-regulated locus. Genetics. 1996;144:15–26. doi: 10.1093/genetics/144.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Torreblanca J, Marques S, Casadesus J. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics. 1999;152:31–45. doi: 10.1093/genetics/152.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tran P H, Korszun Z R, Cerritelli S, Springhorn S S, Lacks S A. Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of Streptococcus pneumoniaebound to S-adenosylmethionine. Structure. 1998;6:1563–1575. doi: 10.1016/s0969-2126(98)00154-3. [DOI] [PubMed] [Google Scholar]
- 81.Trucksis M, Michalski J, Deng Y K, Kaper J B. The Vibrio choleraegenome contains two unique circular chromosomes. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Urieli-Shoval S, Gruenbaum Y, Razin A. Sequence and substrate specificity of isolated DNA methylases from Escherichia coliC. J Bacteriol. 1983;153:274–280. doi: 10.1128/jb.153.1.274-280.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van der Velden A W, Baumler A J, Tsolis R M, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimuriumin mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van der Woude M, Braaten B, Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 1996;4:5–9. doi: 10.1016/0966-842x(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 85.van der Woude M, Hale W B, Low D A. Formation of DNA methylation patterns: nonmethylated GATC sequences in gut and papoperons. J Bacteriol. 1998;180:5913–5920. doi: 10.1128/jb.180.22.5913-5920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van der Woude M W, Kaltenbach L S, Low D A. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli papfimbrial operon. Mol Microbiol. 1995;17:303–312. doi: 10.1111/j.1365-2958.1995.mmi_17020303.x. [DOI] [PubMed] [Google Scholar]
- 87.van der Woude M W, Low D A. Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the sfa and daa pili operons in Escherichia coli. Mol Microbiol. 1994;11:605–618. doi: 10.1111/j.1365-2958.1994.tb00340.x. [DOI] [PubMed] [Google Scholar]
- 88.Wang M X, Church G M. A whole genome approach to in vivo DNA-protein interactions in E. coli. Nature. 1992;360:606–610. doi: 10.1038/360606a0. [DOI] [PubMed] [Google Scholar]
- 89.Weinreich M D, Reznikoff W S. Fis plays a role in Tn5 and IS50transposition. J Bacteriol. 1992;174:4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weyand N J, Low D A. Regulation of Pap phase variation. Lrp is sufficient for the establishment of the phase off pap DNA methylation pattern and repression of pap transcription in vitro. J Biol Chem. 2000;275:3192–3200. doi: 10.1074/jbc.275.5.3192. [DOI] [PubMed] [Google Scholar]
- 91.White-Ziegler C A, Angus Hill M L, Braaten B A M, van der Woude W, Low D A. Thermoregulation of Escherichia coli paptranscription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol Microbiol. 1998;28:1121–1137. doi: 10.1046/j.1365-2958.1998.00872.x. [DOI] [PubMed] [Google Scholar]
- 92.Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- 93.Wright R, Stephens C, Shapiro L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol. 1997;179:5869–5877. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu Q, Peek R M, Jr, Miller G G, Blaser M J. The Helicobacter pylori genome is modified at CATG by the product of hpyIM. J Bacteriol. 1997;179:6807–6815. doi: 10.1128/jb.179.21.6807-6815.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zweiger G, Marczynski G, Shapiro L. A CaulobacterDNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]