Abstract
The unprecedented generation of large volumes of biodiversity data is consistently contributing to a wide range of disciplines, including disease ecology. Emerging infectious diseases are usually zoonoses caused by multi-host pathogens. Therefore, their understanding may require the access to biodiversity data related to the ecology and the occurrence of the species involved. Nevertheless, despite several data-mobilization initiatives, the usage of biodiversity data for research into disease dynamics has not yet been fully leveraged.
To explore current contribution, trends, and to identify limitations, we characterized biodiversity data usage in scientific publications related to human health, contrasting patterns of studies citing the Global Biodiversity Information Facility (GBIF) with those obtaining data from other sources.
We found that the studies mainly obtained data from scientific literature and other not aggregated or standardized sources. Most of the studies explored pathogen species and, particularly those with GBIF-mediated data, tended to explore and reuse data of multiple species (>2). Data sources varied according to the taxa and epidemiological roles of the species involved. Biodiversity data repositories were mainly used for species related to hosts, reservoirs, and vectors, and barely used as a source of pathogens data, which was usually obtained from human and animal-health related institutions. While both GBIF- and not GBIF-mediated data studies explored similar diseases and topics, they presented discipline biases and different analytical approaches.
Research on emerging infectious diseases may require the access to geographical and ecological data of multiple species. The One Health challenge requires interdisciplinary collaboration and data sharing, which is facilitated by aggregated repositories and platforms. The contribution of biodiversity data to understand infectious disease dynamics should be acknowledged, strengthened, and promoted.
Keywords: GBIF, Vector-borne diseases, Zoonoses, Bioinformatics, Risk assessment
Abbreviations: GBIF, Global Biodiversity Information Facility
Highlights
-
•
Infectious diseases affecting humans tend to be multi-hosts pathogens.
-
•
Biodiversity data contribute to explore the species involved in disease dynamics.
-
•
Pathogen, host, and vector data is obtained from multiple non-centralized data sources.
-
•
Biodiversity repositories mainly used for hosts and vector, barely for pathogens data.
-
•
Open data principles present disciplinary biases: One Health approach needed.
1. Introduction
The current threat of emerging zoonotic diseases, involving non-human animal species, is motivating research on several wild and domestic species, usually requiring access to primary biodiversity data related to their occurrences and distributions [[1], [2], [3], [4]]. In general, biodiversity data tend to be associated with ecological and environmental disciplines; however, for disease research purposes, multiple disciplines may need to manage, share and integrate knowledge, including those related to human and animal health [2,5,6]. In fact, considering the COVID-19 pandemic, the need for an interdisciplinary and collaborative One Health approach has never been more urgent [7,8].
Unprecedented volumes of biodiversity data—including species' morphology, ecology, occurrences, taxonomy, and molecular sequence—are continuously generated, based on born-digital records or originated from the digitization of specimens maintained by natural history collections, field observations, citizen science, molecular sampling, among others [4,[9], [10], [11]]. Primary biodiversity data related to occurrences —which reports a named organism observed or collected in a given time and place—can be then organized in datasets and published in supplementary materials, and then may become available in data repositories such as DRYAD [12], Zenodo [13], among others. In some cases, these datasets are available in standardized and fit-for-use formats following the FAIR principles (findability, accessibility, interoperability and reusability) [14,15]. For example, the Darwin Core standards offers a stable framework for compiling biodiversity data from variable sources, with structures terms that facilitate data access and reuse, which can be then aggregated into portals, platforms, and facilities [11,16,17].
Among these aggregated platforms, the Global Biodiversity Information Facility (GBIF) was formed in 2001 as an intergovernmental initiative, following the recommendation of the Working Group on Biological Informatics of the Mega-Science Forum of the Organization for Economic Cooperation and Development (OECD) [18], with the purpose of promoting the development of infrastructures for diverse, high quality and integrated biodiversity data access. Currently, GBIF is the world's largest biodiversity data platform mediating over 2 billion species occurrence records, with an annual rate of increase of 250–300 million [1].
Biodiversity data of species´ occurrences are widely used for geospatial analysis in disciplines such as conservation, biogeography, wildlife management, among many others [1,10,14,19], including infectious disease research [1,3]. In this context, documenting the occurrence of pathogens and other organisms involved in disease circulation is fundamental [20], and their value to support research concerning human health and infectious diseases is becoming more apparent [4,21,22]. For example, occurrence data have been used in distribution modelling to predict the spread of pathogens and vectors, incorporating an ecological understanding of disease dynamics [23,24]. Nevertheless, systematic analyses of the patterns of use of biodiversity data for human health has not been carried out, which could give evidence to improve the processes and systems involved. The present study develops an in-depth exploration of human health studies that have made use of biodiversity data, defining biodiversity as all living organisms, including viruses [25]. For this, we characterize and compare studies that obtained data from GBIF with those that use other data sources, identifying those sources used instead of, and together with GBIF. We discuss current challenges and steps that holders and mediators of biodiversity data resources could consider to promote its use for zoonotic disease research.
2. Methods
We generated two lists of scientific studies related to human health that reuse biodiversity data, separated into those with GBIF-mediated data (positive list) and those that used other data sources (negative list). The positive studies were obtained from the scientific literature database tracked and maintained by the GBIF Secretariat since 2015 (details in Appendix A). After exclusion filtering (duplicates, out of the scope), the final positive list was generated by selecting those specifically related to human infectious diseases. The negative list was generated by searching in the Dimensions database (www.dimensions.ai, August 2021), using a keyword string based on terms obtained from the positive list (including ‘zoonoses’, ‘bat borne disease’, ‘rodent borne’; Appendix A). Negative list was generated by randomly selecting studies from these results mirroring the positive list size. We excluded studies with pathogens only related to humans, those not reusing data from other sources, and studies with data without GBIF scope (e.g., only with captive domestic animals). We did not consider pathogen variables based on serology testing, as the presence of antibodies may not necessarily represent pathogens' occurrence.
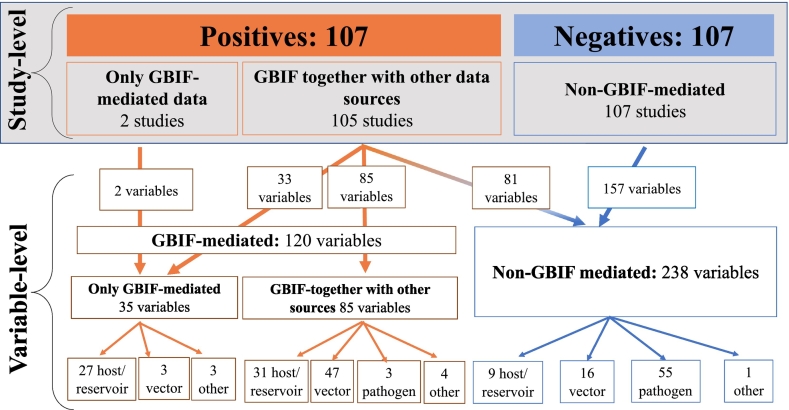
Analyses were developed at study- and variable-levels (Fig. 1). The studies were grouped according to GBIF usage in three categories: (a) only using GBIF-mediated data; (b) studies using GBIF together with other data sources; and (c) studies not using GBIF (from the negative list). As targets for analyses, studies explored single species or taxon or, alternatively, explored multi-species groups, and therefore could be disaggregated in different analytical entities or variables (variable-level). For example, one study may explore mosquitoes and rodents, representing two different variables, which could have different analytical approaches, data sources, and represent different epidemiological levels, i.e., hosts (e.g., rodents), vectors (e.g., mosquitoes), or pathogens (e.g., viruses) (a.k.a. disease compartment [26]). We only included variables related to biodiversity, and occurrence data of pathogens and vector species recorded in domestic animals but not the occurrence of domestic animals themselves.
Fig. 1.
General framework of analyses at study- and variable-levels. In the upper section (study-level, in grey), studies are divided in those that used GBIF-mediated data (positives, in orange) and those that did not (negatives, in blue). Positive studies were group according if GBIF was used as the only data source for all variables (2 studies), or if the variables were based on GBIF together with other data sources (105 studies). In the variable-level section (bottom, white background) the total 358 variables extracted from the positive and negative studies were categorized according to the specific use of GBIF, resulting in five types of variables, four of them extracted from the positive studies. Note that in those studies based on GBIF, the different variables could be based on GBIF alone (33 variables), GBIF together with other sources (85), or specific variables may not be based on GBIF-mediated data at all (81 variables). Finally, each variable was related to different epidemiological roles, resulting in a larger number hosts/reservoirs variables, mostly based on GBIF-mediated data, and a higher presence of pathogen species-variables not using GBIF. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We first developed a bibliometric analysis using the Biblioshine platform (R Bibliometrix package) [27], including parameters such as journals, authors' affiliations, among others. To characterize and compare topics and research areas, we used three approaches, starting with the Bibliometrix theme analysis, which combines performance analysis and science mapping, and identifies conceptual subdomains and thematic structure based on the co-occurrence of key terms [27]. The resulting thematic map consists of a Cartesian representation with clusters distributed into four quadrants organized according to their centrality (degree of interaction with other clusters and citation dynamics), and themes' development or evolution (clusters' internal strength and consistency). Complementarily, we identified and compare the most frequent words extracted from the titles and abstracts, and the research areas of the journals where they were published and authors' affiliations.
For each study we recorded the infectious diseases explored and characterized them according to the causal pathogen (virus, bacteria, parasite, fungus) and transmission mode (mediated by vectors and/or vertebrate animals). At variable-level, we described the species investigated according to their taxa-class (taxonomic groups), and epidemiological level. We identified and characterized the additional data sources used according to their type, disciplines or scope, scale, and governance, and recorded if they were used together or instead of GBIF-mediated data.
3. Results
The GBIF literature tracking system returned 228 studies related to human health citing GBIF-mediated data, reduced to 220 after filtering. Nearly half (113; 51.4%) were not related to infectious diseases, covering topics of medicinal plants (89; 40.5%), venomous species (snakes, scorpions, spiders) (8; 3.6%), pollen and allergies (7; 3.2%), and others (9; 4.1%). Among the 107 studies related to infectious diseases (i.e., positives), 29 (27.1%) included diseases mediated only by vectors, 40 (37.4%) zoonoses only related to vertebrate animals, and 38 (35.5%) included diseases with both transmission modes.
General results of the bibliometric analyses are summarized in Table 1. Both positive and negative studies presented a consistent and similar annual growth rate along the period (18.5% positives, 18.1% negatives), and coincided in four of their most relevant journals, although negatives studies were published in a larger number of journals. We identified 207 authors' affiliations in the positive studies and 236 in the negatives, coinciding in 62 of them. Relevant affiliations (i.e., those participating in >2 studies) related to medical, clinical, or public health disciplines were more frequent in negative studies (43.9%) compared with positives (23.5%). Universities were more frequent in positive studies, while affiliations related to governmental institutions were more frequent in the negatives.
Table 1.
Bibliometric analyses: comparison between positive and negative studies.
| Variable | Positive studies | Negative studies |
|---|---|---|
| Core journals1 (in parenthesis the ones only present in the corresponding group) | 5 journals (BioRxiv) | 7 journals (Int. J. of Health Geographics; J. of Medical Entomology, Scientific Reports; Animals) |
| Number of authors' affiliations | 207 | 236 |
| 1st most frequent affiliation | U. of Kansas, USA* (11 studies; 20.3%) | Tehran U. of Medical Sciences, Iran (9 studies; 8.9%) |
| 2nd most frequent affiliation | National Autonomous U. of Mexico (10 studies; 9.3%) | U. of Oxford, UK** (8 studies; 7.9%) |
| 3rd most frequent affiliation | U. of California, USA* (9 studies; 8.4%) | U. of Florida, USA* (6 studies; 5.9%) |
| N° affiliations related to medical, clinical, and public health+ | 12 (23.5%) | 18 (43.9%) |
| N° affiliations related to universities+ | 39 (76.5%) | 24 (58.5%) |
| N° affiliations related to governmental institutions+ | 6 (11.8%) | 9 (21.9%) |
| N° of countries (authors' affiliations) | 35 countries | 65 countries |
| Most frequent countries (authors' affiliations) | USA*, 44 studies UK**, 16 studies Australia, 14 studies |
USA*, 33 studies UK**, 19 studies Iran, 12 studies |
| Total number of species and average | 2669 total; 32.5 species per study |
1136 total, 12 species per study |
| N° studies with 1 species | 24 (22.4%) | 45 (42.1%) |
| N° studies with 2–50 species | 74 (69.1%) | 59 (55.1%) |
| N° studies with >51 species | 7 (6.5%, max 457 spp.) | 3 (2.8%, max 317 spp.) |
| N° of diseases explored | 42 | 34 |
| Most relevant pathogen taxa class | Virus (49 studies; 47.8%) | Parasites (41 studies; 38.3%) |
1Positive and negative lists presented four core journals in common: Acta Tropica; Parasites & Vectors; PloS Neglected Tropical Diseases; PLoS ONE. *USA: the United States of America; **UK: the United Kingdom; +: only considers affiliations with at least two studies. U.: University.
Topics and diseases: The thematic cartesian map resulted in clusters with similar general topics, presenting differences in terms related to analytical methods and specialized research areas (Appendix B). In both groups, clusters located as relevant themes (i.e., located in Motor and Basic quadrants) presented similar terms such as animals, climate change, and animal distributions, with higher intra-group consistency (establishment) in clusters with terms related to geography and disease vectors. Positive clusters included also analytical terms (theoretical models, risk assessment). In the Niche themes quadrant —specialized topics, low relevance but with high internal consistency—there were no matching terms: topics of negative studies were related to malaria, Macaca, and Culicidae, meanwhile in the positives, were related to yellow fever, Rabies, and cattle.
In general, the most frequent words were similar between groups, both presenting terms such as spatial distributions, geography, climate change, disease reservoirs, and other vector-related terms (Appendix B). Negative studies presented more frequency of words related to tick-borne diseases and leishmaniases, mirroring the trend observed in the thematic map, meanwhile frequent terms in the positive studies were more related to mosquitoes' species and ecological niche models.
We identified 31 research areas in the journals, separated into 16 subcategories, and grouped into six categories named as: Biology, Ecology, Engineer-informatics-mathematics, Medicine, Veterinary sciences, and Others (Fig. 2). Most of these categories presented differences between positives and negatives, with major relevance of biological and ecological areas in the positives, and with medical, public health and veterinarian sciences in negative studies.
Fig. 2.
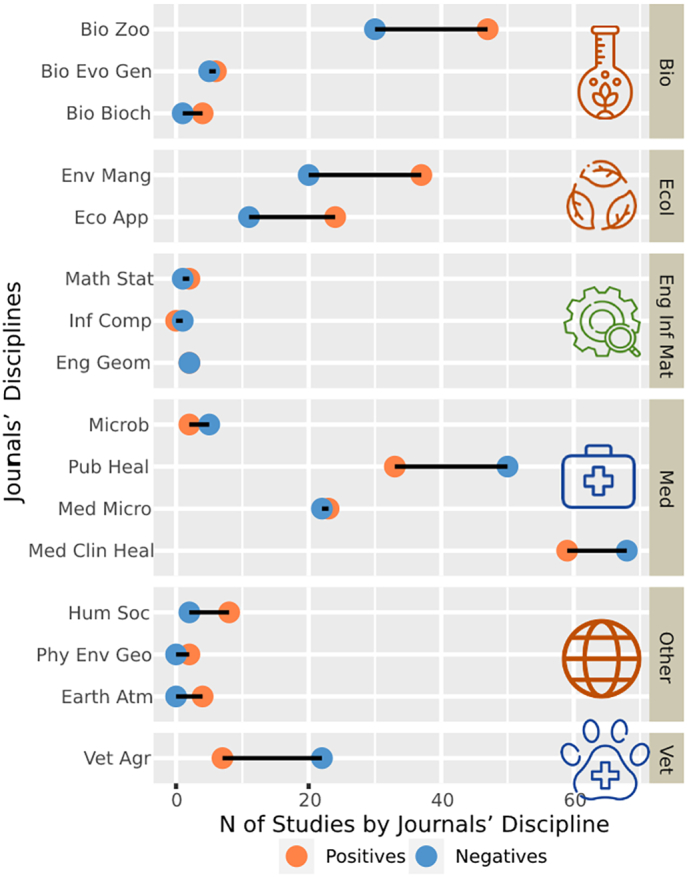
Research areas identified in the studies. Research areas subcategories are represented in the left axis, and general research area groups in the right axis. Orange circles represent the number of positive studies, the blue circles the negatives, and the black lines between them represent the differences in the number of studies, in which larger lines represent larger differences between positive and negatives. Orange icons correspond to research areas with larger number of positive studies, i.e., positive studies were more related to Biology (Bio), Ecology (Ecol) and Other (Hum Soc: Human society; Phy Env Geo: Physical environmental geology; Earth Atm: Earth and atmospheric sciences). Negative studies were more frequent in research areas with blue icons, including Medical (Med) and Veterinary sciences (Vet: Veterinarian and agriculture). In the green icon (Eng Inf Mat: Engineering, informatics, and mathematics) there was no major differences between groups. Subcategories: Bio Zoo: Biology and zoology; Bio Evo Gen: Biology, evolution, and genetics; Bio Bioch: Biology and biochemistry; Env Mang: Environmental management and sciences; Eco App: Ecological applications; Math Stat: Mathematical statistics; Inf Comp: informatics and computing; Eng Geom: Engineer and geometrics; Microb: Microbiology; Pub Heal: Public health; Med Micro: Medical microbiology; Med Clin Heal: Medical clinical and health. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
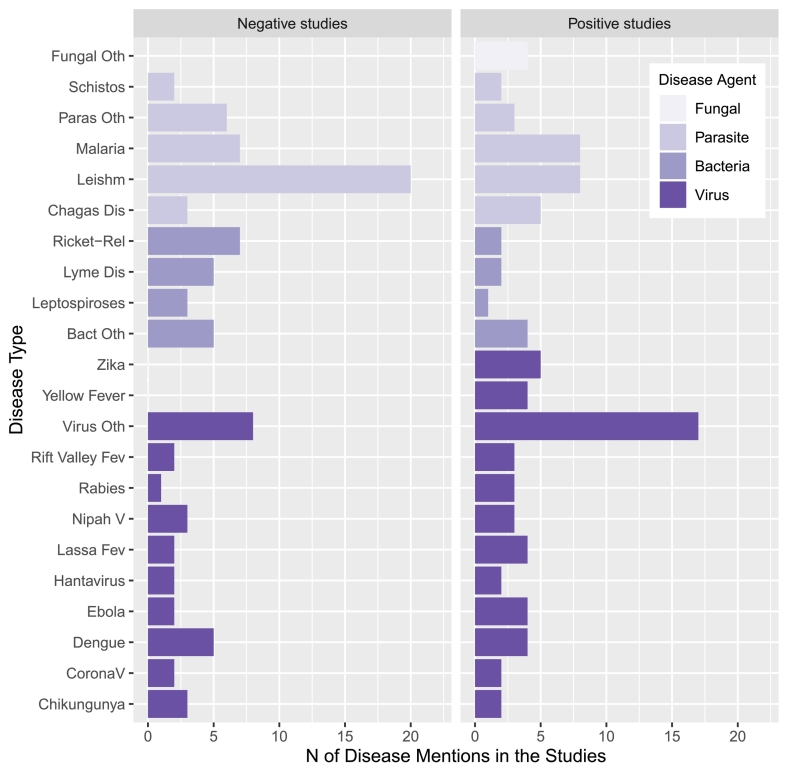
Positive studies explored 42 diseases, and negative studies 34, and in both groups malaria and leishmaniasis were the most frequent diseases (Fig. 3, Table 1, Appendix C). Viral diseases were explored in almost half of the positive studies (54; 50.5%), and 41 (38.3%) of the negative studies were related to parasites, with a remarkable relevance of leishmaniasis.
Fig. 3.
Diseases explored in the studies according to the use of GBIF and the taxa class of the causal pathogen. In the right panel: positive studies (i.e., those studies that used GBIF-mediated data for at least one of the variables explored), negative studies in the left. Bars represent the number of studies exploring each disease, and filling colors represent the corresponding taxa class of the disease agent or causal pathogen (Purple scale, with lighter coloration for fungal diseases, followed by parasites, bacteria, and viruses with the darker purple). Abbreviations: the abbreviation Oth (Fungal Oth, Parasite Oth, Bacteria Oth, Virus Oth) represents a category with multiple species, merged to simplify the figure due to the low number of studies of each disease. Ricket-related: diseases related to Rickettsia species; Paras: parasites; Schistos: Schistosomiases; Leishm: Leishmaniases (both cutaneous and visceral); Dis: disease; Bact: bacteria; Fev: fever; V: virus; CoronaV: Coronavirus. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Variable-level: Studies were disaggregated by 358 variables, from which 157 (43.9%) originated in the negative studies and 201 (56.1%) in the positives (Fig. 1). Most of the 238 non-GBIF variables were extracted from the negative studies. The studies in both groups tended to analyse two or more species (positives: 81; 75.7%; negatives: 61; 57.0%), with a larger number of species in the positive studies (2669 species; average 32.5 per study) compared with the negatives (1136 species; 12 per study) (Table 1). In both groups we found studies not specifying the total number of species explored (23 in positives,13 in negatives).
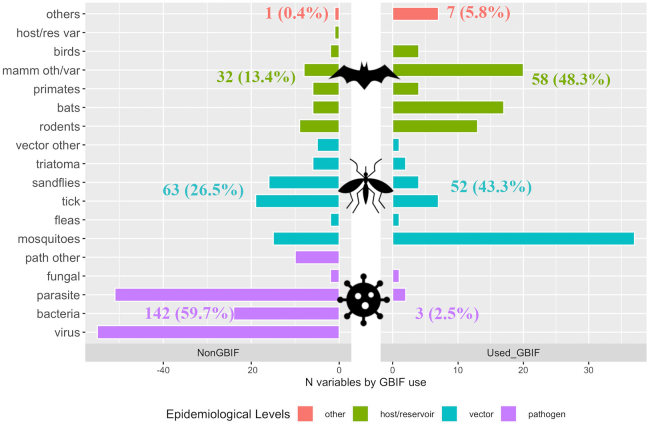
The two groups presented remarkable differences in the epidemiological levels of the variables explored (Fig. 1, Fig. 4, Fig. 5). Overall, a 40.5% of the variables were related to pathogen species (145; 40.5%), with 58 of them (40.0%) obtained from the positive studies; however, GBIF was seldomly used as the data source (only in 3 variables), and in none of these used as an exclusive source (i.e., GBIF was used with other sources). We identified 115 variables related to vectors (32.1%), 68 of them (59.1%) found in positive studies. GBIF was the data source in 52 (85.2%), being particularly relevant for variables related to Aedes spp. and Culicidae mosquitoes. Finally, 90 variables (25.1%) were related to host/reservoirs, mostly obtained from the positive studies (67; 74.4%), with GBIF-mediated data used in 58 of them (86.6%). GBIF was the only source for 27 variables related to host/reservoirs, being particularly relevant for Pteropus bats and murid rodents.
Fig. 4.
Variables according to taxon class (Y- axis) epidemiological level (bar colour) and the use of GBIF-mediated data. Bars represent the number of variables by each taxon class (Y-axis), separated in two panels according to the use of GBIF-mediated data. In the right panel, variables in which GBIF-mediated data was used (Used_GBIF), in the left panel variables in which data was not obtained from GBIF (NonGBIF). Next to the bars, the number of variables by each epidemiological level, and percentage in relation to the total number of variables of each group (Used_GBIF: 120 and NonGBIF: 238). Taxon classes are grouped by taxonomic associations (e.g., birds, primates, ticks, mosquitoes); however, some were merged to simplify the figure. For example, mamm/oth/var includes multiple mammal species which were sparsely mentioned; similarly, hosts/res var, vector other and path other grouped several species participating as hosts/reservoirs, vectors, and pathogens, respectively. Bar colors represent epidemiological levels (pathogens, vectors, hosts/reservoirs), and Other (in sienna) includes species participating as hosts' regulator, predators, among others. GBIF-mediated data was only used in three pathogen variables (purple), representing only 2.5% of the variables in which GBIF-mediated data was used, resulting in a remarkable difference with other sources (NonGBIF), in which pathogens represented a 59.7%. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
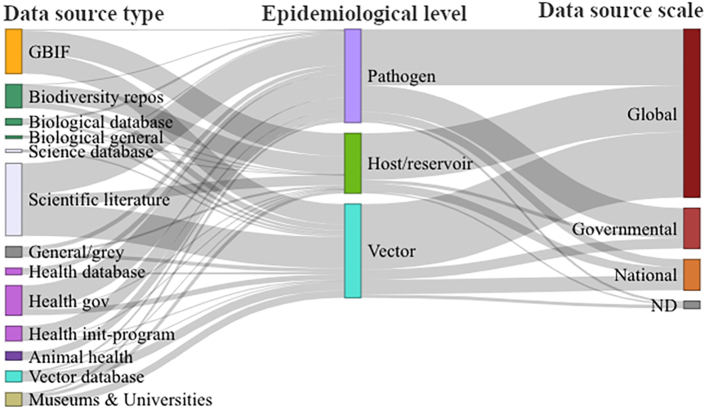
Fig. 5.
Data sources according to epidemiological level and scale. Representation of the data sources (left column) used for each epidemiological level (central column), and the scale of the corresponding data sources (right). Colors of the left column correspond to general data-sources categories; for example, green corresponds to biological/biodiversity data sources (e.g., Biodiversity repositories and biological general source). Health-related sources are represented in purple (Health gov: governmental, init-program: initiative or programs). Using this broad categorization, most of the sources contribute with data related to the three epidemiological levels, although with an unpaired flow. For example, scientific literature has a lower contribution for hosts/reservoirs, and biodiversity-biological sources have a minor contribution for pathogens. Most data sources have a global scale meanwhile governmental sources have a relevant contribution to pathogen data. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Regarding the proportion of the epidemiological levels within each group, in the positive studies 33.4% of the 201 variables were related to vectors, 33.3% to hosts/reservoirs and 28.7% were related to pathogens. The remaining eight variables (4.9%) were related to other epidemiological roles, such as regulator or vector predators, and almost half of the positive studies (52; 48.6%) did not include any pathogen variable. In contrast, near half (86; 54.7%) of the 157 variables from the negative studies were pathogens, followed by vectors (47; 29.9%) and hosts/reservoirs (22; 14.0%). Thirty-six negative studies (33.6%) did not include any pathogen variable.
Overall, 108 (74.5%) of the 145 pathogen variables were recorded in humans, 52 (35.9%) in non-human vertebrates, and 19 in vectors (13.1%). Most of the non-human vertebrate records (33; 70.2%) were obtained from wild species and 22 (46.8%) from domestic animals (e.g., cattle, dogs, pigs). In some variables pathogens were recorded in more than one species (e.g., from humans and rodents), therefore, total percentages do not sum 100.
We identified 172 additional sources used 517 times (Fig. 5, Appendix D). Unidentified sources included those with generic or unspecified descriptions, such as ‘national reports’, or ‘unpublished data’. Data sources were grouped into 27 categories based on type (e.g., scientific literature, repositories), scale (e.g., national, global), topic (e.g., health, biodiversity), among others. For scientific literature we recognized three subcategories: (a) literature search when data was obtained from studies searched in databases (e.g., PubMed, Google Scholar, Scopus) and original studies; (b) if data was obtained from scientific reviews; (c) or from data journals. Scientific search was the most frequent subcategory, used for 142 variables (39.7%), followed by governmental health institutions (83; 23.2%), scientific reviews (37; 10.3%), and global biodiversity/biological platforms (35; 9.8%). Additional sources with global scale were used 314 times (60.7%), and among the 184 times in which national-scale sources were used, 20.3% (105) were related to governmental and 15.3% (79) to private institutions.
We identified 94 additional sources used for pathogen variables (average 1.61 sources per variable), with scientific search used in 67 of them (46.2%), and governmental health institutions in 62 (42.7%) (Fig. 5). For vectors we identified 75 additional sources (1.63 per variable), with scientific literature used in 77 (66.9%) and governmental health institutions in 19 (16.5%) variables. Finally, 36 additional sources were used for host/reservoirs (1.10 per variable), with global biological/biodiversity repositories and platforms used in 28 variables (31.1%), and scientific search in 17 (18.9%). Only 30 additional sources (17.4%) were used for multiple epidemiological levels, and only scientific literature was used for the three levels.
Among the sources used together with GBIF, the most frequent was scientific search (Appendix E). For vectors, GBIF was also complemented with VectorMap and SpeciesLink, and for host/reservoirs used with IUCN and VectorMap. In general, GBIF was infrequently used together with health-related sources.
4. Discussion
Our results give evidence of the growing contribution of biodiversity data for emerging infectious diseases research, which are frequently zoonotic and transmitted by vectors, influenced by the ecology and interactions of multiple species [1,3,4,21,[28], [29], [30]]. Our results address this complexity, while most of the studies explored (especially those using GBIF-mediated data) include the analyses of multiple variables, species, and epidemiological levels. Here, we characterize usage trends of biodiversity data for infectious diseases research and identify other data sources used for similar proposes. Our findings reveal usage-gaps and current limitations, summarized in Appendix F together with potential actions to be taken.
Data sources uses and gaps: In general, biological and biodiversity repositories (including GBIF) were relevant for facilitating data related to hosts/reservoirs and vector species (Fig. 4, Fig. 5), with GBIF frequently used as the data source for mosquitoes and mammals such as bats and rodents. Mosquitoes are considered the most lethal species for humans, responsible for at least 700,000 deaths per year [31], and bats and rodents are well-known hosts and reservoirs of several zoonotic pathogens [32]. Therefore, GBIF contributes with aggregated and standardized data to the understanding of these species' distribution and ecology, which may give important clues about disease potential distribution and risk. This contribution is observed independently of the distribution (proportion) of species' occurrences in the GBIF database.
In positive studies (i.e., those using GBIF), GBIF presented an unpaired contribution depending on the epidemiological level. Even though the similar proportion of variables for the three epidemiological levels in positive studies (33.4%, 33.2%, 28.7% for vectors, hosts/reservoirs and pathogens, respectively), GBIF had a remarkable lower use for pathogens (2.5%; Fig. 4, Fig. 5). In fact, GBIF was not used as the only source in any study exploring multiple variables, revealing a selective use in which even though authors were familiar with GBIF, they tended to exclude it for some variables, especially when related to pathogens.
However, this unpaired use was not exclusive of GBIF. Most of the data sources were used for specific species groups related to only one or two epidemiological levels, i.e., not used to obtain data related to pathogens, hosts/reservoir, and vector species. However, GBIF and scientific literature represented the only data source that, exceptionally, were used to obtain data of species related to the three epidemiological levels. Therefore, even with its limited use for pathogens, GBIF represented the only individual data source with a wider usage.
The selective use of data sources according to the species and their epidemiological levels may be influenced by the scope of the data provided. For example, biodiversity data repositories tend to exhibit taxonomic biases, in general towards social preferences and charismatic species (e.g., birds, mammals) [33,34]. GBIF, for example, presents a median of 317 occurrences per bird species, in contrast to only three for arachnids [33]. In addition, biodiversity data tend to be biased towards free-living organisms, as opposed to mutualistic species, parasites and pathogens [35,36]. In fact, besides a few exceptions (e.g., Biological Abstracts, GenBank), most of the additional biodiversity/ biological data sources were seldomly used to obtain pathogen data (Fig. 5, Appendix D). This bias contrasts with the following definition of biodiversity: ‘the sum of all plants, animals, fungi, and microorganisms on Earth, their genotypic and phenotypic variation, and the communities and ecosystems of which they are a part’ [37]. For example, GBIF offers occurrence data of only three viruses known to infect humans (Zika, West Nile, and Hepatitis B; GBIF.org, February 2022).
Nevertheless, despite this limited availability of virus data, we found that GBIF supported 49 studies related to viral diseases (Fig. 3), in which viral occurrences were not always required. In fact, near half of the positive studies and one-third of the negatives did not include any pathogen species data, revealing that disease research, modelling and risk assessments may be developed using vector or host species' occurrences. For example, the major threat suddenly imposed by SARS-CoV-2 motivated the access to ecological data concerning potential reservoirs of coronavirus and others with zoonotic potential, not necessarily requiring virus occurrences [20,32,38]. Similarly, Estrada-Peña et al. (2019) generated maps of potential tick-borne pathogen distribution based on cooccurrences data of ticks and hosts, the latter obtained from GBIF [39]. Thus, the low availability of pathogen data in repositories such as GBIF, does not necessarily hamper their contribution to disease research.
However, biodiversity repositories could consider expanding their taxa coverage and scope, potentially strengthening the representation of non-free-living organisms such as pathogens. The Museums and Emerging Pathogens ECHO Program may represent a good example of an initiative that aims to integrate wildlife biorepositories with data related to emerging infectious diseases [40]. On the other hand, taxa coverage may consider including domestic animal records. Near one-third of the pathogen records were obtained from non-human vertebrates, from which half were domestic. In this context, pathogen occurrences detected in domestic animals fall into a grey area of biodiversity repositories scope. Furthermore, domestic animals are widely distributed and participate in several zoonotic diseases, such as cattle in Rift Valley fever [41], and domestic dogs in multiple helminthiases [42], and their contact with humans and wild animals may facilitate pathogen transmission. In this study, we excluded domestic animal occurrences, following the general scope of GBIF and other biodiversity repositories, mainly focused on natural and wild-based records [43]. However, domestic animal occurrences in natural and unsupervised settings may be considered as relevant for ecological monitoring [44]. In fact, GBIF offers data of domestic animals' occurrences, usually individuals under natural settings, living under free-ranging conditions with minor influence of humans (e.g., [45]).
Even though being a species out of GBIF scope, humans were in fact the most frequent host of pathogen records. Coinciding with previous studies [24,46], governmental health institutions and other related human-health initiatives were a relevant source for human records of pathogens, meanwhile pathogens from animals were mainly obtained from other specific animal-health and veterinary institutions (Appendix D). In fact, besides some global initiatives such as ProMed, HealthMap, and GIDEON, only a few sources were used to obtain pathogens data from both humans and animals, revealing the lack of centralized repositories for multi-species pathogens (Fig. 5). This could explain in part the use of multiple and scattered sources across observed in our results, contradicting with data related to host/reservoirs, in which biodiversity/biological repositories are frequently used.
The type of sources from which pathogens, vectors and hosts/reservoirs also presented differences. We found that, besides scientific literature, data sources for pathogens tended to be more related to governmental institutions and health-related initiatives, usually managing health primary data, with no integration of multiple data generators, and with no standardized formats. Instead, for hosts and reservoirs, data repositories and aggregators were more usually used. This could represent a general disciplinary bias, with medical and clinical disciplines tending to present a lower and unpaired commitment with some data sharing and reusing practices when compared to ecology and environmental sciences [[47], [48], [49], [50], [51]]. We also found this discipline bias when comparing research areas of the journals in which studies were published, even though positive and negative studies explored similar general topics, terms, and diseases (Fig. 2, Fig. 3, Appendix B). Considering the threat of multi-species pathogens, we highlight the need to follow One Health principles, and improve the publication of data related to pathogens, its mobilization to aggregated repositories, and the cross-linking between data managers from ecological, veterinarian, biomedical and human health related fields [4,20,52].
Compiled data (i.e., not raw data), or with non-standardized nor interoperable formats were frequently observed in the data sources identified in our results (Appendix D). Instead, data was mostly available as texts, tables, pdf, among others (e.g., Governmental Institutions [53,54]), requiring manual management. In other cases, additional data sources have options to export raw data in manageable formats (e.g., .cvs, .xls), such as FAO for pathogens [55]. However, even raw data is mostly presented with different information, different terms, or taxonomic mismatches, i.e., not following FAIR principles. Standardized data were exceptions, such as GenBank [56], Mammals Networked Information System (MaNIS [57]), Atlas of Living Australia (ALA [58]), and GBIF, most of them related to biodiversity data management. Standardisation of data facilitates sharing and reuse, and it has been adopted for several data types (a list of standards can be found in FAIRSharing resources, [59]). Scientific journals, publishers and other open data initiatives have make great advances in the open data policies [15,[60], [61], [62], [63]]. However, most of the repositories of supplementary materials and data journals do not request standardized protocols, such DRYAD [12] and Zenodo [13]. This reflects an issue across scientific domains whereby simply placing data online is seen as sufficient to meet requirements for ‘open data’; however, data only becomes truly open when it succeeds to conform to the FAIR principles, with standardized, indexed and interoperable data [17,50,59,61,64].
Data provided following FAIR principles may also facilitate more complex analyses, which may serve to understand, predict and quantify human risk for pathogen transmission [23,24,65]. We found that studies citing GBIF-mediated data tended to include more variables and more species (23.2 species per study in positives, 8.3 in negatives), with topics and terms related to more complex analyses, including theoretical models and ecological niche modelling (Table 1, Appendix B), which often require robust data quality [23]. This findings support that the access to standardized and interoperable data provided by integrated repositories may facilitate complex and broader scale analyses [3,4,22].
The frequent use of scientific literature could be also explained by the vast amount of information not always propagated to data repositories, and the dynamic publication of results delivered at a faster rate compared to aggregated biorepositories [14,66]. Data sharing and flow from scientific publications to aggregators and other repositories may be promoted by incentives from academic institutions, universities, and other affiliations. Scientists should be aware about the potential benefits of data sharing such as professional networking and increased citation rates, facilitated by the DOI citation systems [47,67] and the opportunities in data journals—used here as a source in six studies—.
The contribution of biodiversity data repositories for infectious disease research can be also strengthened by means of incorporating systems in their searching platforms that consider specific requirements of disease-ecology research. For example, beyond offering single species record, searching engines may consider interactions or associated occurrences expressed as host-pathogen, host-host, predator-host, vector-pathogen, among others, reflecting the observation of an individual host together with the pathogen occurrence [3,68]. These systems have been already implemented by platforms such as the Global Biotic Interactions database (GloBI; [69]) and ARCTOS [70], both of which enable the search of ‘preys on’ and ‘host of’. In the Darwin Core standard these associations can be informed in the terms AssociatedOccurrences and a class of terms ResourceRelationship; however, they have not been used extensively [17]. A recent publication proposes a framework for applying the Darwin Core data standards to diseases, based in a hierarchical structure whereby each ‘parent’ occurrence of the host (term parentOccurrenceID) may be associated with multiple ‘child’ occurrences or material samples (basisOfRecord) [3]. This metadata harmonization at record-level —which corresponds to a publisher responsibility— could be complemented with an identification at species-level, tagged for example as ‘host’ or ‘reservoir’ (Appendix F). This latter task, however, requires that data managers define a particular role of a species in disease dynamics, which may be a complex process, vary according to the specific epidemiological interactions, and not always under scientific consensus [26,71].
5. Conclusions
This study gives evidence and confirms the contribution of biodiversity data to human health research, identifying the usage of biodiversity data repositories as trusted, well positioned, and valuable infrastructures to obtain information related to the multiple species involved in infectious disease dynamics. In addition, biodiversity data infrastructures have an opportunity to provide and promote data integration, interoperability, and access at global scale.
Here, we identify relevant gaps and limitations that may be considered in the future for specific improvements of biodiversity data repositories, summarized in Appendix F. Further actions could include the creation of new international communication pipelines that better unite public and animal health institutions, governmental agencies, scientific community, diverse disciplines, and local communities, so that they together may better coordinate data sharing and global disease surveillance. In these emerging challenges, clinical and health databases must not be on the side-lines. In addition, an improvement to the contribution of biodiversity repositories for infectious disease research may require a discussion in relation to the specific institutional scopes and taxonomic extents of the data provided.
We advocate for the formal incorporation and recognition of biodiversity aggregated databases as critical infrastructure for multi-host disease understanding, predictions, and general human and animal health research, and invite researchers to share and reuse data, incorporating interdisciplinarity and cross-domain approaches in data-intensive research. The evidence of the contribution for infectious disease research justifies current and future actions to support biodiversity data mobilization, from generation, aggregation, accessibility, and final reuse.
Funding
Francisca Astorga was financed by the Global Biodiversity Information Facility (GBIF) as a consultant. Quentin Groom is supported by the BiCIKL project, which receives funding from the European Union's Horizon 2020 Research and Innovation action under grant agreement No 101007492.
Declaration of Competing Interest
Francisca Astorga was financed by the Global Biodiversity Information Facility (GBIF) as a consultant. Daniel Noesgaard, Tim Hirsch and Dmitry Schigel have institutional affiliations related to GBIF
Acknowledgements
We thank Theeraphap Chareonviriyaphap, Florence Fouque, Luna Kamau, and Carlos Zambrana-Torrelio for their support as members of the GBIF Task-group to Enhance GBIF Enabled Research on Species Linked to Human Diseases. We also thank Roderic Page and Arturo Ariño for stimulating discussions that inspired planning of this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2023.100484.
Appendix A. Supplementary data
Appendix A to F Supplementary material
Data availability
No data was used for the research described in the article.
References
- 1.Heberling J.M., Miller J.T., Noesgaard D., Weingart S.B., Schigel D. Data integration enables global biodiversity synthesis. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2018093118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doherty J.-F., Chai X., Cope L.E., de Angeli Dutra D., Milotic M., Ni S., Park E., Filion A. The rise of big data in disease ecology. Trends Parasitol. 2021;37:1034–1037. doi: 10.1016/j.pt.2021.09.003. [DOI] [PubMed] [Google Scholar]
- 3.ENETWILD consortium, F, Jaroszynska G. Body, Pamerlon S., Archambeau A. Applying the Darwin Core data standard to wildlife disease – advancements toward a new data model. EFS3. 2022;19 doi: 10.2903/sp.efsa.2022.EN-7667. [DOI] [Google Scholar]
- 4.Zaspel J.M., Allen J.M., Tyrrell C.D., Lemoine N., Jacobus L.M., Klem C., Goodwin J., Bates J.M. Human health, interagency coordination, and the need for biodiversity data. BioScience. 2020;70:527. doi: 10.1093/biosci/biaa065. [DOI] [Google Scholar]
- 5.SciColl . Scientific Collections International; Washington, D.C.: 2015. Scientific Collections and Emerging Infectious Diseases: Report of an Interdisciplinary Workshop. [Google Scholar]
- 6.Bartlow A.W., Machalaba C., Karesh W.B., Fair J.M. Biodiversity and global health: intersection of health, security, and the environment. Health Secur. 2021;19:214–222. doi: 10.1089/hs.2020.0112. [DOI] [PubMed] [Google Scholar]
- 7.Glidden C.K., Nova N., Kain M.P., Lagerstrom K.M., Skinner E.B., Mandle L., Sokolow S.H., Plowright R.K., Dirzo R., De Leo G.A., Mordecai E.A. Human-mediated impacts on biodiversity and the consequences for zoonotic disease spillover. Curr. Biol. 2021;31:1342–1361. doi: 10.1016/j.cub.2021.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.FAO . FAO/OIE/WHO; Rome, Italy: 2019. Taking a Multisectoral One Health Approach: A Tripartite Guide to Addressing Zoonotic Diseases in Countries. [Google Scholar]
- 9.Chapman A.D. 2005. Principles of Data Quality, Version 1.0, Report for the Global Biodiversity Information Facility. Copenhagen. [Google Scholar]
- 10.Secretariat G.B.I.F. Global Biodiversity Information Facility; Copenhagen: 2021. Introduction to GBIF. Version 981bc3d. [Google Scholar]
- 11.Nelson G., Ellis S. The history and impact of digitization and digital data mobilization on biodiversity research. Philos. Trans. R. Soc. B. 2019;374:20170391. doi: 10.1098/rstb.2017.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DRYAD Dryad Digital Repository. 2022. https://datadryad.org/stash
- 13.European Organization For Nuclear Research OpenAIRE. Zenodo. 2013 doi: 10.25495/7GXK-RD71. [DOI] [Google Scholar]
- 14.Hugo W., Hobern D., Kõljalg U., Tuama É.Ó., Saarenmaa H. In: The GEO Handbook on Biodiversity Observation Networks. Walters M., Scholes R.J., editors. Springer International Publishing; Cham: 2017. Global infrastructures for biodiversity data and services; pp. 259–291. [DOI] [Google Scholar]
- 15.Wilkinson M.D., Dumontier M., Ij J., Aalbersberg G., Appleton M., Axton A., Baak N., Blomberg J.-W., Boiten L.B., Silva Santos P.E., Bourne J., Bouwman A.J., Brookes T., Clark M., Crosas I., Dillo O., Dumon S., Edmunds C.T., Evelo R., Finkers A., Gonzalez-Beltran A.J.G., Gray P., Groth C., Goble J.S., Grethe J., Heringa P.A.C., Hoen R. Hooft, Kuhn T., Kok R., Kok J., Lusher S.J., Martone M.E., Mons A., Packer A.L., Persson B., Rocca-Serra P., Roos M., van Schaik R., Sansone S.-A., Schultes E., Sengstag T., Slater T., Strawn G., Swertz M.A., Thompson M., van der Lei J., van Mulligen E., Velterop J., Waagmeester A., Wittenburg P., Wolstencroft K., Zhao J., Mons B. The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016;3 doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman A.D., Belbin L., Zermoglio P., Wieczorek J., Morris P., Nicholls M., Rees E.R., Veiga A., Thompson A., Saraiva A., James S., Gendreau C., Benson A., Schigel D. Developing standards for improved data quality and for selecting fit for use biodiversity data. BISS. 2020;4 doi: 10.3897/biss.4.50889. [DOI] [Google Scholar]
- 17.TDWG Darwin Core Quick Reference Guide, Darwin Core Standards. 2021. https://dwc.tdwg.org/terms/#dwc:associatedOccurrences
- 18.OECD . Organization for Economic Cooperation and Development; 1999. Final Report of the OECD Megascience Forum. [Google Scholar]
- 19.Stein B.R., Wieczorek J.R. Mammals of the world: MaNIS as an example of data integration in a distributed network environment. Biodivers. Inform. 2004;1 doi: 10.17161/bi.v1i0.7. [DOI] [Google Scholar]
- 20.Brooks D.R., Hoberg E.P., Boeger W.A., Gardner S.L., Araujo S.B.L., Bajer K., Botero-Cañola S., Byrd B.D., Földvári G., Cook J.A., Dunnum J.L., Dursahinhan A.T., Garamszegi L.Z., Herczeg D., Jakab F., Juarrero A., Kemenesi G., Kurucz K., León-Règagnon V., Mejia-Madrid H.H., Molnár O., Nisbett R.A., Preiser W., Stuart M., Szathmáry E., Trivellone V. Before the pandemic ends: making sure this never happens again. World Complex. Sci. Acad. J. 2020;1:1–10. [Google Scholar]
- 21.Cook J.A., Arai S., Armién B., Bates J., Bonilla C.A.C., Cortez M.B.S., Dunnum J.L., Ferguson A.W., Johnson K.M., Khan F.A.A., Paul D.L., Reeder D.M., Revelez M.A., Simmons N.B., Thiers B.M., Thompson C.W., Upham N.S., Vanhove M.P.M., Webala P.W., Weksler M., Yanagihara R., Soltis P.S. Integrating biodiversity infrastructure into pathogen discovery and mitigation of emerging infectious diseases. BioScience. 2020;70:531–534. doi: 10.1093/biosci/biaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunnum J.L., Yanagihara R., Johnson K.M., Armien B., Batsaikhan N., Morgan L., Cook J.A. Biospecimen repositories and integrated databases as critical infrastructure for pathogen discovery and pathobiology research. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson A. Ecologic niche modeling and spatial patterns of disease transmission. Emerg. Infect. Dis. 2006;12:1822–1826. doi: 10.3201/eid1212.060373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purse B.V., Golding N. Tracking the distribution and impacts of diseases with biological records and distribution modelling. Biol. J. Linn. Soc. Lond. 2015;115:664–677. doi: 10.1111/bij.12567. [DOI] [Google Scholar]
- 25.Pearson H. “Virophage” suggests viruses are alive. Nature. 2008;454:677. doi: 10.1038/454677a. [DOI] [PubMed] [Google Scholar]
- 26.Johnson E.E., Escobar L.E., Zambrana-Torrelio C. An ecological framework for modeling the geography of disease transmission. Trends Ecol. Evol. 2019;34:655–668. doi: 10.1016/j.tree.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aria M., Cuccurullo C. Bibliometrix: an R-tool for comprehensive science mapping analysis. J. Inform. 2017;11:959–975. doi: 10.1016/j.joi.2017.08.007. [DOI] [Google Scholar]
- 28.Jones B.A., Grace D., Kock R., Alonso S., Rushton J., Said M.Y., McKeever D., Mutua F., Young J., McDermott J., Pfeiffer D.U. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8399–8404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor L.H., Latham S.M., Woolhouse M.E.J. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doherty J.-F., Chai X., Cope L.E., de Angeli Dutra D., Milotic M., Ni S., Park E., Filion A. The rise of big data in disease ecology. Trends Parasitol. 2021;37:1034–1037. doi: 10.1016/j.pt.2021.09.003. [DOI] [PubMed] [Google Scholar]
- 31.WHO . World Health Organization; 2020. Vector-borne diseases.https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases [Google Scholar]
- 32.Grange Z.L., Goldstein T., Johnson C.K., Anthony S., Gilardi K., Daszak P., Olival K.J., O’Rourke T., Murray S., Olson S.H., Togami E., Vidal G., Expert Panel, PREDICT Consortium, Mazet J.A.K. Ranking the risk of animal-to-human spillover for newly discovered viruses. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2002324118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troudet J., Grandcolas P., Blin A., Vignes-Lebbe R., Legendre F. Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 2017;7:9132. doi: 10.1038/s41598-017-09084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Troia M.J., McManamay R.A. Filling in the GAPS: evaluating completeness and coverage of open-access biodiversity databases in the United States. Ecol. Evol. 2016;6:4654–4669. doi: 10.1002/ece3.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlson C.J., Burgio K.R., Dougherty E.R., Phillips A.J., Bueno V.M., Clements C.F., Castaldo G., Dallas T.A., Cizauskas C.A., Cumming G.S., Doña J., Harris N.C., Jovani R., Mironov S., Muellerklein O.C., Proctor H.C., Getz W.M. Parasite biodiversity faces extinction and redistribution in a changing climate. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1602422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velasco D., García-Llorente M., Alonso B., Dolera A., Palomo I., Iniesta-Arandia I., Martín-López B. Biodiversity conservation research challenges in the 21st century: a review of publishing trends in 2000 and 2011. Environ. Sci. Pol. 2015;54:90–96. doi: 10.1016/j.envsci.2015.06.008. [DOI] [Google Scholar]
- 37.Dirzo R., Raven P.H. Global state of biodiversity and loss. Annu. Rev. Environ. Resour. 2003;28:137–167. doi: 10.1146/annurev.energy.28.050302.105532. [DOI] [Google Scholar]
- 38.Upham N., Agosti D., Poelen J., Penev L., Paul D., Reeder D., Simmons N.B., Csorba G., Groom Q., Dimitrova M., Miller J. Liberating biodiversity data from COVID-19 lockdown: toward a knowledge hub for mammal host-virus information. BISS. 2020;4 doi: 10.3897/biss.4.59199. [DOI] [Google Scholar]
- 39.Estrada-Peña A., Adkin A., Bertolini S., Cook C., Crescio M.I., Grosbois V., Horigan V., Ip S., Leger A., Mastrantonio G., Maurella C., de Nardi M., Ru G., Simons R., Snary E., Staerk K., Taylor R., Smith G.C. Evaluating a mixed abiotic–biotic model for the distribution and host contact rates of an arthropod vector of pathogens: an example with Ixodes ricinus (Ixodidae) Microbial Risk Anal. 2019;13 doi: 10.1016/j.mran.2018.12.001. [DOI] [Google Scholar]
- 40.UNM . University of New Mexico; 2022. Museums and Emerging Pathogens ECHO Program.https://hsc.unm.edu/echo/partner-portal/programs/global/mepa/ Health Sciences. [Google Scholar]
- 41.Redding D.W., Tiedt S., Lo Iacono G., Bett B., Jones K.E. Spatial, seasonal and climatic predictive models of Rift Valley fever disease across Africa. Philos. Trans. R. Soc. B. 2017;372:20160165. doi: 10.1098/rstb.2016.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tull A., Valdmann H., Tammeleht E., Kaasiku T., Rannap R., Saarma U. High overlap of zoonotic helminths between wild mammalian predators and rural dogs – an emerging one health concern? Parasitology. 2022;149:1565–1574. doi: 10.1017/S0031182022001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnaud E., Castañeda-Álvarez N.P., Cossi J.G., Endresen D., Jahanshiri E., Vigouroux Y., Schigel D. 2016. Final Report of the Task Group on GBIF Data Fitness for Use in Agrobiodiversity. [Google Scholar]
- 44.Groom Q., Adriaens T., Bertolino S., Phelps K., Poelen J., Reeder D., Richardson D., Simmons N., Upham N. Holistic understanding of contemporary ecosystems requires integration of data on domesticated, captive and cultivated organisms. BDJ. 2021;9 doi: 10.3897/BDJ.9.e65371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwin Scholes P.D., III . 2017. Macaulay Library Audio and Video Collection. [DOI] [Google Scholar]
- 46.Reznik O.N., Kuzmin D.O., Reznik A.O. Biobanks as the basis for developing biomedicine: problems and prospects. Mol. Biol. 2017;51:666–673. doi: 10.1134/S0026893317050156. [DOI] [PubMed] [Google Scholar]
- 47.Enke N., Thessen A., Bach K., Bendix J., Seeger B., Gemeinholzer B. The user’s view on biodiversity data sharing — investigating facts of acceptance and requirements to realize a sustainable use of research data —. Ecol. Inform. 2012;11:25–33. doi: 10.1016/j.ecoinf.2012.03.004. [DOI] [Google Scholar]
- 48.Oushy M.H., Palacios R., Holden A.E.C., Ramirez A.G., Gallion K.J., O’Connell M.A. To share or not to share? A survey of biomedical researchers in the U.S. Southwest, an ethnically diverse region. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tedersoo L., Küngas R., Oras E., Köster K., Eenmaa H., Leijen Ä., Pedaste M., Raju M., Astapova A., Lukner H., Kogermann K., Sepp T. Data sharing practices and data availability upon request differ across scientific disciplines. Sci. Data. 2021;8:192. doi: 10.1038/s41597-021-00981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenopir C., Allard S., Douglass K., Aydinoglu A.U., Wu L., Read E., Manoff M., Frame M. Data sharing by scientists: practices and perceptions. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wicherts J.M., Bakker M., Molenaar D. Willingness to share research data is related to the strength of the evidence and the quality of reporting of statistical results. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson C.W., Phelps K.L., Allard M.W., Cook J.A., Dunnum J.L., Ferguson A.W., Gelang M., Khan F.A.A., Paul D.L., Reeder D.M., Simmons N.B., Vanhove M.P.M., Webala P.W., Weksler M., Kilpatrick C.W. Preserve a voucher specimen! The critical need for integrating natural history collections in infectious disease studies. MBio. 2021;12 doi: 10.1128/mBio.02698-20. e02698-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gobierno de Mexico Datos Abiertos del Gobierno de Mexico. 2022. https://datos.gob.mx/busca/dataset
- 54.ISP Sistema Interactivo de Resultados de Vigilancia del Instituto de Salud Publica. 2022. https://www.ispch.gob.cl/andid/sistema-interactivo-de-resultados-de-vigilancia/ Chile.
- 55.EMPRES-i FAO . Food and Agriculture Organization of the United Nations; 2022. EMPRES-i - Global Animal Disease Information System Disease Events from FAO Global Animal Disease Information System.https://empres-i.apps.fao.org/ [Google Scholar]
- 56.GenBank GenBank: National Library Medicine, International Nucleotide Sequence Database Collaboration. 2022. https://www.ncbi.nlm.nih.gov/genbank/
- 57.MaNIS . 2022. Mammal Networked Information System Project. [Google Scholar]
- 58.ALA, Atlas of Living Australia 2022. http://www.ala.org.au/
- 59.FairSharing FairSharing: Tandars, Databases, Policies. 2022. https://fairsharing.org/
- 60.BMC Data Standardization, Sharing and Publication. 2022. https://www.biomedcentral.com/collections/datasharing
- 61.Hrynaszkiewicz I. A call for BMC research notes contributions promoting best practice in data standardization, sharing and publication. BMC Res. Notes. 2010;3(235) doi: 10.1186/1756-0500-3-235. 1756-0500-3–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ODI, The Open Data Institute 2022. https://theodi.org/
- 63.OBK, Open Knowledge Foundation A Fair, Free and Open Future. 2022. https://okfn.org/
- 64.Costello M.J., Michener W.K., Gahegan M., Zhang Z.-Q., Bourne P.E. Biodiversity data should be published, cited, and peer reviewed. Trends Ecol. Evol. 2013;28:454–461. doi: 10.1016/j.tree.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 65.Colella J.P., Bates J., Burneo S.F., Camacho M.A., Carrion Bonilla C., Constable I., D’Elía G., Dunnum J.L., Greiman S., Hoberg E.P., Lessa E., Liphardt S.W., Londoño-Gaviria M., Losos E., Lutz H.L., Ordóñez Garza N., Peterson A.T., Martin M.L., Ribas C.C., Struminger B., Torres-Pérez F., Thompson C.W., Weksler M., Cook J.A. Leveraging natural history biorepositories as a global, decentralized, pathogen surveillance network. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gadelha L.M.R., Siracusa P.C., Dalcin E.C., Silva L.A.E., Augusto D.A., Krempser E., Affe H.M., Costa R.L., Mondelli M.L., Meirelles P.M., Thompson F., Chame M., Ziviani A., Siqueira M.F. A survey of biodiversity informatics: concepts, practices, and challenges. WIREs Data Mining Knowl. Discov. 2021;11 doi: 10.1002/widm.1394. [DOI] [Google Scholar]
- 67.Piwowar H.A. A method to track dataset reuse in biomedicine: filtered GEO accession numbers in PubMed central: a method to track dataset reuse in biomedicine: filtered GEO accession numbers in PubMed central. Proc. Am. Soc. Info. Sci. Tech. 2010;47:1–2. doi: 10.1002/meet.14504701450. [DOI] [Google Scholar]
- 68.Dallas T. helminthR: an R interface to the London natural history museum’s host-parasite database. Ecography. 2016;39:391–393. doi: 10.1111/ecog.02131. [DOI] [Google Scholar]
- 69.Plazi-Zenodo-GloBI integration GloBI. 2020. https://www.globalbioticinteractions.org/plazi-zenodo/
- 70.ARCTOS, ARCTOS Collaborative Collection Management Solution. 2022. https://arctosdb.org/about/
- 71.Haydon D.T., Cleaveland S., Taylor H.T., Laurenson M.K. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg. Infect. Dis. 2002;8:1468–1473. doi: 10.3201/eid0812.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A to F Supplementary material
Data Availability Statement
No data was used for the research described in the article.