Abstract
Germplasm is a long-term resource management mission and investment for civilization. An estimated ∼7.4 million accessions are held in 1750 plant germplasm centres around the world; yet, only 2% of these assets have been utilized as plant genetic resources (PGRs). According to recent studies, the current food yield trajectory will be insufficient to feed the world's population in 2050. Additionally, possible negative effects in terms of crop failure because of climate change are already being experienced across the world. Therefore, it is necessary to reconciliation of research advancement and innovation of practices for further exploration of the potential of crop germplasm especially for the complex traits associated with yield such as water- and nitrogen use efficiency. In this review, we tried to address current challenges, research gaps, physiological and molecular aspects of two broad spectrum complex traits such as water- and nitrogen-use efficiency, and advanced integrated strategies that could provide a platform for combined stress management for climate-smart crop development. Additionally, recent development in technologies that are directly related to germplasm characterization was highlighted for further molecular utilization towards the development of elite varieties.
Keywords: Core set, Climate change, Nitrogen use efficiency, Omic approaches, Plant genetic resources, Water use efficiency
1. Introduction
The 60-year-old global success story of the first green revolution (1960s) was accomplished by introducing the Rht (reduced height). Subsequently, the importances of allelic variation, genetic diversity, as well as conservation of plant genetic resources (PGRs) have been recognized worldwide. The major goals of germplasm exploration and collection are emphasized to limit the risk of essential genes, genetic stocks being lost to genetic erosion and utilization of important trait/gene(s) in crop breeding programs to stabilize crop productivity by reducing yield loss [1]. Germplasm material such as traditional crops and developed varieties, crop wild relatives, trait-specific genotypes, ecotypes, and wild types of species/accessions serve as a natural resource for civilization [2]. To date, an estimated ∼1750 plant germplasm centres globally hold 7.4 million accessions; however, <2% of these assets have been used as plant genetic resources (PGRs) [3]. Recent studies suggested that the existing crop yield trajectory is insufficient to nourish the global population in 2050 [4] and, by 2050, the global food demand will be expected to increase by 70% [5]. Hence, challenges associated with PGRs that hinder their utilization need to be considered for further development programs as well as for the conservation of potential germplasm materials. Additionally, to mitigate climate change imposed challenges and for further sustainability, urgent attention to systematically assess PGRs is essential besides the upgrading of maintenance strategies (field genebank, backup plantation, water harvest strategies, etc.), as well as the implementation of new technologies for the conservation of PGR (ex- and in situ). At present, the major challenge is how to maintain, exploit and capitalize on this abundant genetic resource.
Increased stress, altering agro-ecosystems, invasion of pathogens and pests, as well as an increase in the frequency of extreme weather events are all examples of the possible negative effects (crop failure) of climate change that is already being experienced [6]. The evolution of crops and their complex traits—such as crop yield, rooting ability, high moisture retention capacity, and biotic and abiotic stress tolerance—has received significant attention when examining the effects of climate change on plants. The impact of climate change on physiological processes such as WUE, NUE, and photosynthesis, which are involved in plant growth and survival are recognized [[6], [7], [8]]. Hence, understanding of physiological and molecular responses of complex traits in course of stress acclimation or adaption is a prerequisite to drawing the mechanisms underlying climate stress conditions, especially drought, high temperature, high light, cold, and salinity.
Excellent genetic and germplasm resources provide the foundation for new variety development. Breeding practice demonstrates that significant improvements in breeding depend on finding and utilizing potential germplasm and genetic resources. In this article, we emphasize the current challenges of plant genetic resources as well as physiological and molecular aspects of crop germplasm for further sustainability through advanced integrated methodologies that may serve as a base for future crop development research.
2. Germplasm and current challenges
The overall objective of PGR collection could be achieved through the conservation of maximum genetic diversity of a given gene pool and the maintenance of breeding resources. To achieve sustainable productivity the major activities such as long-term maintenance, pre-breeding program, gene discovery, and integrated breeding approaches were manifested (Fig. 1). Despite the several importance of crop germplasm resources, the utilization rate of PGRs reported to be significantly low and some major challenges associated with potential genetic materials for sustainable development and conservation is as follows-
-
(1)
Accumulation of duplicates: During a long period of collection, and conservation the chances of the introduction of duplicate accession were increased. Hence, the estimation of duplication/uniqueness across the germplasm accessions is considered to be a prime aspect for improving the efficiency of breeding as well as the cost of ex situ conservation could also be reduced [9,10].
-
(2)
Reduction of genetic diversity: Molecular analysis suggests that changes in allele frequencies and subsequent genetic diversity were observed in Pisum sativum L., because of long-term domestication [11]. Evidence also suggests that productivity is also affected by the reduction of genetic diversity with an increase in the number of gene bank accessions [12]. One of the interesting consequences of a long period of crop domestication as well as artificial selection, plants have evolved a series of fundamental mechanisms to acclimate the environment and the genetic basis of adaptation studies suggest co-transcriptional processing, alternative splicing (AS) played a significant role [13]. Hence, the convolution of isoforms associated with trait plasticity creates further challenges to understanding adaptation.
-
(3)
Accumulation of deleterious alleles: Another common consequence is an accumulation of rare deleterious alleles during crop domestication and their mode of action also varied [14,15].
-
(4)
Low recombination frequency: The lower recombination frequency between the wild relatives and subsequent crop productivity has been considered one of the major bottlenecks for crop improvement [16].
-
(5)
Negative impact of climate change: The potential negative impacts are already being experienced, in the form of increasing stress, shifting agro-ecosystem, invasive pathogens/pests, and more frequent extreme weather events. Hence, the maintenance of genetic resources is challenging in the upcoming days [6].
-
(6)
Complication of core set utilization: The core set collections comprise the greatest diversity of total genetic resources and served as a reference population for additional breeding programs [10]. The core set can be used for innovative field experiments and subsequent parental material in the breeding program. However, the characterization of core set germplasm is further complicated by (a) a higher level of whole-genome duplication (WGD) or polyploidization; (b) a higher level of heterozygosity; (c) poor functional annotation of the plant genome; (d) lack of cell-type-specific marker, (e) lack of information of plant-cell-types and sub-cellular compartments across tissues and species; (f) high level of phenotypic plasticity.
-
(7)
Limitation of understanding complex traits: Complex traits are polygenic and highly influenced by environmental factors. Additionally, complex compartmentalization networking and multilevel-gene regulation of genes involved in complex traits further prevent the understanding of complex traits [17]. Remarkably, a large number of published complex trait-based experiments on controlled-environment conditions did not show consistency when it was executed in real field trials [[17], [18]].
-
(8)
Inadequate information on below-ground traits: Insufficient information on below-ground traits (e.g. root architecture, cross-kingdom interaction, etc.) of germplasm resources limiting the improvement ecosystem processes, maintaining the fertility of the soil, maintaining microbial community and improving the stress-tolerant capacity of the plant [19,20].
-
(9)
Limitation of digitalization and transformation of PGR: Transforming PGR into a digital resource system is another important challenge for future crop improvement, because of low-quality reference genome assemblies as in crop wild relatives, challenging assemblage of pangenome, maintenance of accuracy and specificity on base editing, unavailability of specialized expertise especially for machine learning technique [21], in addition with inadequate information of phenomics, transcriptomics, and genomics of large-scale germplasm accessions/genotypes.
-
(10)
Downsize of the genome in polyploid: Importance of polyploid in the breeding program was well established as crop productivity across the major crops. Recent reports suggest that to reduce nitrogen and phosphate costs associated with DNA replication, transcription, and translation, as well as mentioned cell size from carbon fixation and water use, polyploid involved in genome downsize in terms of DNA loss at a rate of 4–7MB/million years and 4–482 bp/generation [22]. Hence, conservation/maintenance of polyploid crops need to be relooked for future ploidy breeding.
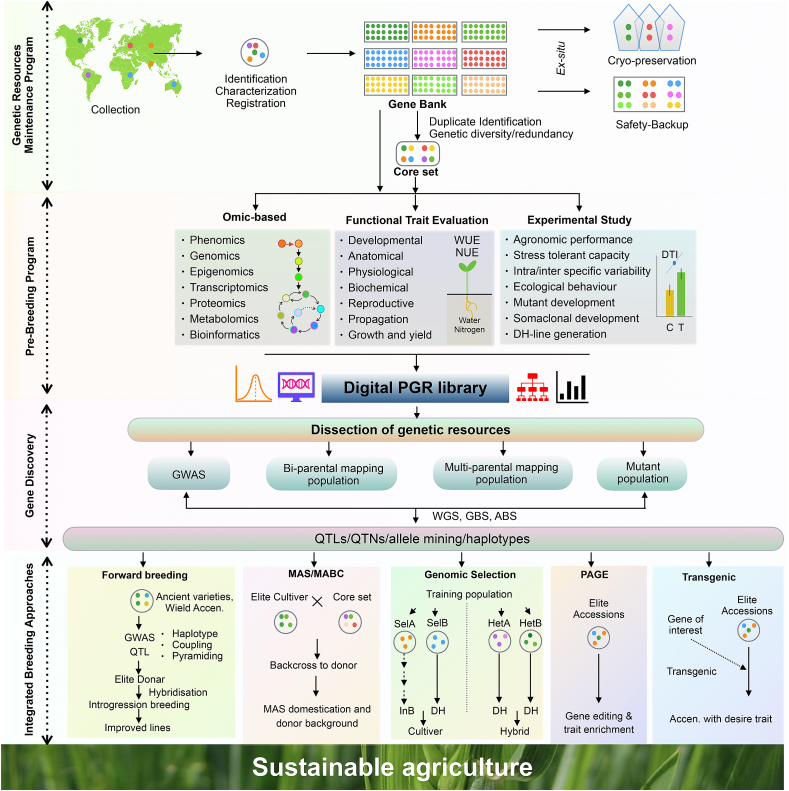
Fig. 1.
A collective model of structure, activity and utility of crop germplasm. The crop improvement program of active germplasm is divided into four major strategies. (A) Genetic Resource Maintenance Program: A combined model of identification, collection, characterization, registration, and ex-situ conservation (e.g. cryopreservation) of breeding resources. Identification of duplicates and development of different populations through integrated taxonomy, cytology, and marker-based approaches have been useful. Additionally, development and maintenance of unique core set germplasm is one of the most promising approaches for the breeding resource. A core set collection represents a reference population of total germplasm with unique accessions. (B) Pre-Breeding Program: An advanced pre-breeding strategy has been emphasized to reveal the potentiality of the core collection. Integrated omic approaches, functional trait evaluation, and different experimental datasets can be generating a database for plant geneticists/breeders. (C) Gene Discovery: For both complex traits and simple inheritance traits, the most challenging job is to harness beneficial gene dissection. GWAS, bi/multi-parental, and mutation population mapping are preferably powerful strategies for establishing genetic trait association. Phenome QTL (phQTL), metabolome QTL (mQTL), proteome QTL (pQTL), and expression QTL (eQTL) have been extensively used to discover genes and functional annotation. (D) Integrated Breeding Approaches: Forward breeding, MAS/MABC, PAGE, and transgenic approach have been employed to illustrate and capitalize on the benefit of simply inherited traits, whereas genomic selection (GS) can be employed for the complex inherited trains. The aim of global sustainability will be fulfilled by this systematic evaluation of genetic resources that will simultaneously save time and budget expenditure. Abbreviation: WUE, water use efficiency; NUE, nitrogen use efficiency; DTI, drought stress index=(T/C)*100; C, control; T, treatment; GWAS, genome-wide association study; QTL, quantitative trait locus; QTN, quantitative trait nucleotide; WGS, whole-genome sequencing; GBS, genotyping by sequencing; ABS, array-based sequencing; MAS, marker-assisted selection; MABC, marker-assisted backcrossing; PAGE, promotion of alleles through genome editing; SelA/B, selfing line A/B; HetA/B, heterozygous line A/B; InB, Inbred; DH, doubled haploid.
Hence, it is understandable that precious ways of maintenance, as well as innovative utilization strategies, are required to capitalize on the benefit of PGRs.
3. Physiological and molecular aspects of germplasm
Agriculture is the world's largest consumer of water and accounts for 70% of all water used worldwide. The ongoing global catastrophe of water is one of the major problems in the present-day climate scenarios and drought is considered the most severe abiotic stress affecting crop productivity at a global level [7]. Besides that, nitrogen is the most essential nutrient for plant growth, development, and yield. Increasing trends of fertilizer application to achieving productivity, resulting in the decreasing the acquisition and subsequent utilization of applied nitrogen in crop plants. Additionally, the surplus amounts of inorganic and organic nitrogen fertilizers often imposed drastic negative impacts on crop plants like delayed flowering and ripening, as well as on the environment [20,23,24]. The NUE for cereal production is ∼33% globally and the unaccountable 67% corresponds to a $15.9 billion loss/year of nitrogen fertilizer [[25], [26],]. Moreover, it is estimated that ∼1% increase in crop NUE could annually save $1.1 billion [17,26,27]. Concerned with current climatic change and improving nutritional quality and quantity, plant breeders mostly target climate resilience crop productivity and functional traits such as WUE and NUE [29]. To achieve greater agricultural sustainability, the development of crop plants with drought-tolerant and more efficient nitrogen usage is, therefore, an important research aspect.
The primary reasons for crop failure include membrane damage, photosynthetic inhibition, oxidative stress, including other impacts of stress on morphological, physiological, and growth changes/damage have been extensively studied across the agriculture, horticulture and other crop systems [8,30,31]. Additional research has been done on the reproductive stage of plants because it is more vulnerable to stress than the vegetative phases as well as negative impacts on reproductive processes that are correlate directly with lower yields [6]. Moreover, stress disturbs the homeostasis of plants, disrupting crucial physiological and molecular processes, sinking energy production, and affecting cellular integrity [6,8]. Upon exposure to stress, plants instantly modify their physiological and molecular responses in order to produce a new state of homeostasis, which is considered acclimatization [32]. Although, plants may change their anatomy, growth, and reproductive methods over extended periods through a process known as adaptation [33]. The acclimation or adaption responses of plants to each single stress state may necessitate a different method since different stressors may impact plants differently. In light of acclimatization and adaptation, an advanced understanding of the relationship between photosynthesis, WUE, NUE, and stress tolerance is a prerequisite to designing climate-smart crop varieties.
Stomata represent a critical target for enhancing yield since they are crucial in controlling plant water use and carbon gain. In addition to that, optimum soil water, as well as coordination between nitrogen uptakes, and transport is essential for plant lifestyle. Recent studies suggest that each type of stress may cause a unique acclimation and/or adaptation response in plants, and these responses may consist of similar or dissimilar mechanization [6]. For example, (a) plants close their stomata during drought to reduce water loss, but they open them during the heat to increase transpiration and cool their leaves [34–36]; (b) stomata of many plants remain closed when co-occurrence of heat and drought stress is present, indicating that during a stress combination, drought-driven control of stomata prevails over heat stress-driven regulation [35,37]; (c) contrarily, in response to combined high light and heat stress, heat stress-induced stomata regulation (stomata opening) overwhelmed high light stress-induced stomata regulation (stomata closure), causing stomata to open under the stress-combination condition [38]. Therefore, depending on the stress condition, the plant may choose one acclimation/adaptation technique over another. Thus, the fundamental issue that plants experience (by the intensity as well as the time of exposure), when two separate and/or combined stressors are present at once is that they may call for diverse, and perhaps conflicting, physiological and molecular processes.
In recent, with the aid of transgenic, genomics, transcriptomics, and performance-based phenotyping, germplasm accessions of Oryza sativa L. [30,39], Triticum aestivum L. [40,41], Glycine max L. [42,43], Malus domestica [44], Sorghum bicolor [45], etc. were studied to understand WUE. Jia et al. [44] overexpressed MdATG8i in M. domestica and exhibited higher WUE under long-term moderate drought conditions. Pignon et al. [45] identified a total of 77 genes involved in WUE, where 24 genes are involved in stomatal opening/closing, 35 genes stomatal/epidermal cell development, 12 genes leaf/vasculature development, 8 genes-chlorophyll metabolism/photosynthesis. Additionally, the association of WUE traits and ABA signaling was reported by Arab et al. [46] in Juglans regia. AtDREB1C overexpressed lines of New Rice for Africa demonstrated improved growth under moderate drought/osmotic stress, notably by reducing water demand together with a reduction in growth time, which may be a consequence of increased WUE and osmotic adjustment [47]. However, an advanced understanding of the relationship between crop yield, drought tolerance, and WUE remains to be understood to design climate-smart crop varieties.
Besides that, higher yield is concurrently associated with N-application, availability in soil, and utilization by plants. Developing varieties with higher NUE is one potential strategy to reduce N fertilizer application. Plants' NUE is significantly impacted by genetic and physiological factors such as nitrogen uptake efficiency, physiological NUE, nitrogen utilization efficiency, and photosynthetic NUE, which in turn are associated with biomass, rapid vegetation, flowering time, germination, RuBisCO activity, leaf senescence, root system architecture traits as well as stress adaptation [[48], [49], [50], [51], [52], [53], [54]]. N-stress response and high NUE are correlated with the activation of transcription, phytohormone signalling, biosynthesis, assimilation and metabolism and/or catabolism of different elements [55]. Several reports were available to understand NUE using O. sativa, Hordeum vulgare, T. aestivum [56], Solanum melongena [57], Solanum tuberosum, Brassica napus [58]. Tiong et al. [56] demonstrated that overexpression of Alanine aminotransferase (HvAlaAT) enhances NUE in different crops. Based on QTL analysis, Zhang et al. [59] identified DULL NITROGEN RESPONSE1 involved NUE by auxin-mediated signaling. Mauceri et al. [57] identified genes like light-harvesting complex, ferredoxin–NADP reductase, catalase, and WRKY33 are associated with high-NUE genotypes. However, a large number of genes are involved in phytohormone signaling, N-uptake and assimilation, amino acid metabolism, fatty acid biosynthesis, photosynthesis, C assimilation, starch biosynthesis and carbohydrate catabolism are associated with N-stress response and high NUE also reported [55]. Zhang et al. [59] reported nitrogen deficiency decreased the activity of nitrate reductase (NR), glutamine synthetase (GS) in the root as compared to shoot. A nitrate transporter gene NRT2.1 and two vacuole nitrate transporter CLC genes were up-regulated by N starvation in the N-efficient genotype in Brassica napus [28]. Hence, the genetic architecture of NUE is modulated by a number of genes, and molecular processes are rather complex. The complex cellular mechanism, as well as physiological responses of WUE and/or NUE are reported to be varied across the crops. For further understanding, advancement of experimental design and system biology approceh may serve the purpose.
4. System biology and improvement of PGRs
The classical paradigm of plant breeding, recurring cycles of crosses, and selections (MAS/MABC) have long been considered a tedious method, however, the implementation of advanced technologies including speed breeding dramatically accelerates crop development by facilitating fast generation cycles [60]. Beside, the targeted genetic recombination approach has been considered a quicker method for the genetic transformation of the gene of interest from superior donors. An array of reports suggested that plant tissue culture technology has shown a considerable impact on the crop enhancement program [61]. However, because of the lower frequency of transformation, in vitro recalcitrance, implementation of different bio-safety rules, and costly evaluation processes are becomes limits to achieve the targets to date [62,63]. As compared to conventional genetic methods the integrated strategy has the following advantages: (a) less tedious because the multiplication of population is not required; (b) large plant sample size, mutants, and stable homozygous overexpressed plants are not required; (c) low-cost of mining of publicly available database resources makes the comparative study of omics data easier; (d) an integrated approach could identify and provide comprehensive interactions, co-expression, and metabolic/biosynthetic grid; and finally (e) integration of advanced artificial intelligence (AI) gives the sufficient multi-dimensional data. Hence, to understand the complex biological phenomena, ongoing trends of omics will be helpful for future crop improvement. Although, in order to improve fundamental understanding and development of new climate-resilient crops, genomic and transcriptomic information is essential, often relevant strategies like transcriptomics and/or genome-wide association studies (GWAS) are powerful.
Currently, the system biology approach is considered a potential move toward the complete understanding of biological systems and provides the information to breeders for mining superior alleles/haplotypes that can serve as the key to selecting precious parental material for breeding [64]. Integrated phenomics and next-generation sequencing (NGS) technologies have been implemented for identifying genetic markers associated with desirable traits in multiple crops, including rice, foxtail millet, pigeon pea, pearl millet, cotton, rapeseed, chickpea, and grape. Moreover, the large-scale germplasm accessions has been re-sequenced in rice (3010), pearl millet (994), and chickpea (429) [65]. Additionally large-scale sequencing generate big data that has storage and computational challenges.
To understand the genetic basis of trait variation, NGS-based genome-wide association studies (GWAS) and genomic prediction (GP)/genomic selection (GS) are emerging powerful tools used in recent years [[66], [67], [68], [69], [70], [71]]. GWAS is an alternate approach for the detection of QTL in a large collection of diverse germplasm due to higher mapping resolution as compared to linkage mapping. GWAS often identifies tens to hundreds of loci containing hundreds of single-nucleotide polymorphisms (SNPs) associated with a trait of interest in natural populations or breeding germplasm [72,73]. This approach was successfully applied to understand the genetic basis of many important traits in crops, including yield-relevant traits [[74], [75], [76], [77]]. However, larger population sizes with high-density markers are required to increase the statistical power of GWAS.
The efficiency of MAS (Marker-assisted selection) in pyramiding favourable alleles is very effective only for major-effect QTL and it is limited when the traits are governed by many QTL with minor effects influenced by the genetic background of the population. Most of the complex traits like yield and stress tolerance in agriculturally important plants are controlled by many minor QTLs. To capture those minor effects QTL genomic prediction (GP)/genomic selection (GS) are widely applied to increase the efficiency and speed of plant breeding [78]. GS predicts the genomic-estimated breeding values (GEBVs) of an individual by computing the effects of both minor and major genes [79]. MAS uses only the selected markers that are linked to a trait of interest, whereas GS estimates a larger proportion of genetic variation of the selected trait using a large set of marker data. With the advancement of prediction models and the development of highly efficient-low cost genotyping, GS is a widely used breeding approach in crop improvement programs [21,80].
Breeding for NUE and WUE is hindered by expensive phenotypic evaluations and trait complexity. Despite the widespread application of GWAS, limited studies have been carried out to dissect the genetic architecture of complex traits like NUE. Allelic variations in key genes regulate differences in NUE among diverse rice germplasm [81]. Ertiro et al. [68] effectively used GWAS to identify markers associated with grain yield and NUE in maize, further incorporating these markers into the prediction model will improve NUE in maize. GWAS on a set of diverse rice populations identified an elite haplotype of nitrate transporter OsNPF6.1HapB which enhanced the nitrate uptake and confers high NUE by increasing yield under low nitrogen supply [82]. And also it was reported that due to the increased application of fertilizer the elite nitrate transporter has been lost in 90.3% of rice varieties. Integrated GWAS and transcriptome-wide association study (TWAS) identified putative candidate genes to investigate the genetic basis of WUE in a C4 model grass for bio-energy, food, and forage production [83]. Similarly, Bhaskara et al. [84] use GWAS with integrated WUE measured by gravimetric and δ13C analyses in Arabidopsis thaliana accessions identified 25 genes affecting WUE. All these identified genes may be manipulated to increase WUE and make significant contributions to the sustainable use of water in agriculture.
Recently, Wei et al. [85] determined that rice exhibits a considerable expression of the DREB family TF under low-N conditions. DREB1C controls many important growth-related genes including the nitrate transporters, nitrate reductase, the flowering regulator, and RuBisCO. Overexpressed DREB1C showed early seedling vigor, an increased amount of photosynthetic pigments, RuBisCO content and higher rates of photosynthesis, N intake, and NUE. Due to the modification of the flowering gene, the DREB1C-OE plants demonstrated a greater harvest index, increased remobilization of N and C to sink organs, and a much shorter blooming time and crop duration. Besides that, Ishizaki et al. [47] reported that AtDREB1C overexpressed rice transgenic lines confer moderate drought tolerance by increasing WUE. Recent reports suggest that OsDREB1C/1E can also be a suitable target for the drought tolerance mechanism in rice [86]. Additionally, under drought and low-N, there are a few common cellular and molecular processes like (1) osmotic adjustment by increasing free sugar [87]; (2) phytohormone signalling e.g. ABA, auxin, salicylic acid [[88], [89], [90]]; (3) gene expression e.g. light-harvesting complex, RuBisCO, DREB, and WRKY [85,86,91] were reported (Fig. 2A–B). However, further research will require a deeper understanding and establishment of molecular mechanisms in direction of the complexity of polygenic regulation, coordination of nuclear to organelle proteome, compartmentalized metabolic networks, and associated multilevel gene regulation.
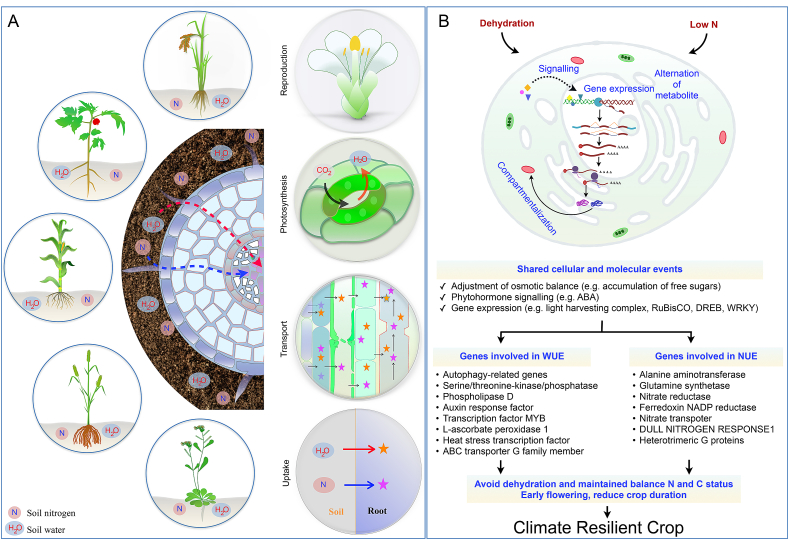
Fig. 2.
Diagrammatic presentation illustrated physiological and molecular coordination of two complex traits like water- and nitrogen use efficiency in different crop/non-crop systems. (A) Water- and nitrogen use efficiency played a central role in growth, development, and response to different stress conditions such as high temperature, light, drought, low-N-condition, etc. Both traits are concurrently associated with below-ground traits. N-uptake and/or transport efficiency, gas exchange through stomata, photosynthesis, as well as reproduction are directly connected with water- and nitrogen-use efficiency. (B) Under drought and low-N, there are several common cellular and molecular processes like (1) osmotic adjustment; (2) phytohormone signalling; (3) gene expression are common. Additionally, a wide range of genes is expressed in response to specific stress conditions also reported. For example, autophagy-related genes are specifically associated with WUE, whereas nitrate reductase is expressed under N-limited conditions. However, to maintain the balance between C, N, and water under drought and low-N conditions, modulation of RuBisCO activity, flowering time, and activation of different genes like transcription factors DREB1 might be a precise coordination strategy.
5. Utilization and prospect of PGRs
Germplasm is the platform to dissect potential genes associated with desirable traits. Once the traits are correlated with specific biosynthetic/metabolic pathways and desired alleles have been identified, researchers can take up a broader understanding of crop biology to anticipate parental and allelic combinations that will discover superior agronomic traits [21]. Although analyzing the genome sequences or genome-wide association study will become a null hypothesis, until it has been complemented by a functional study like promoter structure analysis, the impact of alternative splicing, miRNA mediated post-transcriptional silencing, and post-translation modification. To date, the identification of candidate genes through transcriptomics approaches and/or mapping is restricted in most crops [92]. Furthermore, it is important to understand in-depth molecular mechanisms of their possible agronomic values. In addition to structural genomics, information on gene expression, epigenome maps, proteome maps, and metabolome maps are a prerequisite in crop species [93]. For example, identification and characterization of grain yield by KERNEL NUMBER PER ROW6 in maize [94]. Recent advancements in cloning strategies, genetic transformation methods, gene-editing tools, co-immunoprecipitation, two-hybrid systems, mass spectrometry, and data-mining bioinformatics have shown a global transformation in agriculture. Moreover, the information on the genetic architecture of important agronomical traits, their associated alleles, and the underlying molecular mechanism remains to be elucidated.
At present, automated robotics phenomic platforms have been used to quantify genotypes based on functional traits such as growth form, woodiness, leaf economic spectrum, WUE, NUE, as well as stress tolerance capacity [95]. To explore the diverse gene bank resources for developing climate resilience crops, the evaluation of agronomic performance and stress-tolerant capacity under different climatic conditions along with crop behaviour, intra/inter-specific variability, and mutant performance evaluation is necessary. Therefore, to quantify the potentiality of genotypes, next-generation trait-based evaluation should be performed on field conditions. This will help us identify superior genotypes/accessions that are stable, abundant, and environmentally less sensitive [96]. For identifying the desired genes and associated traits, the recent trend of the functional phenomic approach will be helpful [97]. The availability of these resources will be accelerated by increasing systems biology approaches to understand the molecular mechanism of complex traits such as WUE and/or NUE (Fig. 3).
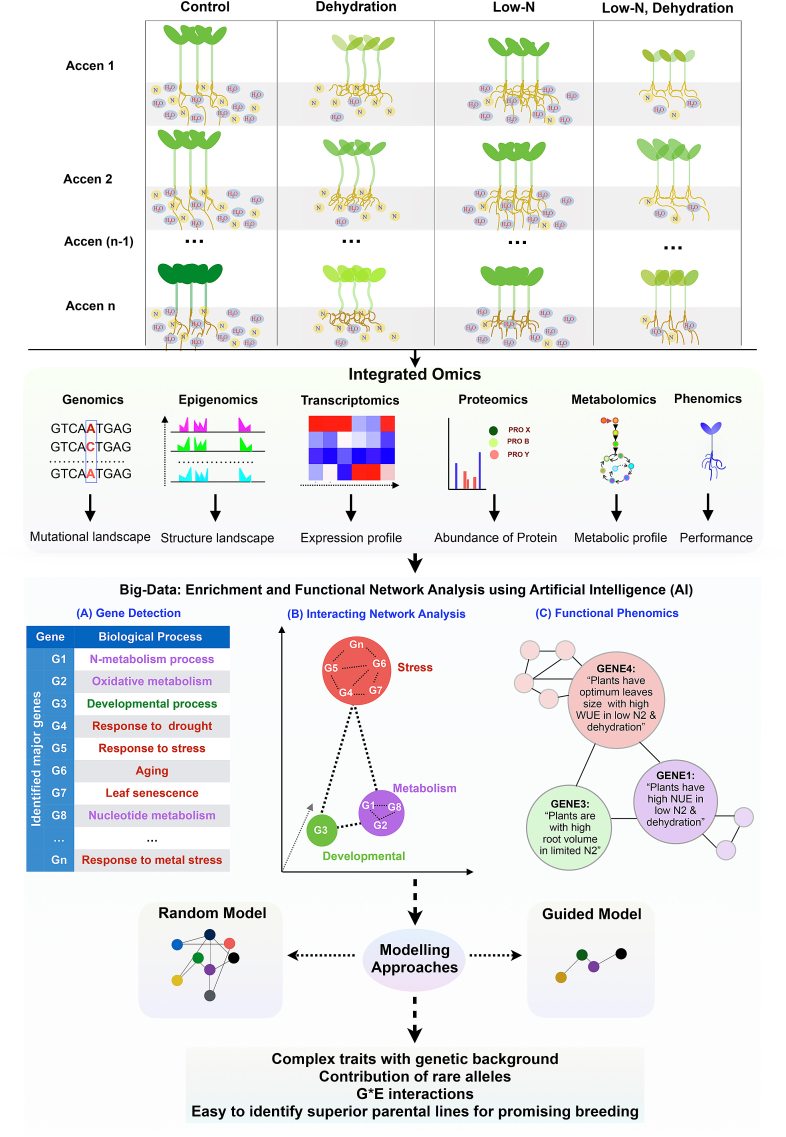
Fig. 3.
A comprehensive gene discovery strategy of water- and nitrogen-use efficiency. Synthesized high-throughput multi-omic integration data can confirm the phenotypic behavior, mutational landmark (SNP), expression pattern, an abundance of protein, and metabolic signature with associated treatment. Different transcript, metabolic, and protein-protein interaction libraries of different genotypes are useful for enrichment and functional network analysis. NGS-based approaches and in silico data-mining bioinformatics-based analysis can be helpful to identify abundant genes, followed by multidimensional clustering and gene network analysis, proteomic study (2D gel electrophoresis and co-immunoprecipitation, etc.), are useful to complement functional phenomics that will help to predict as well as discriminate the desired gene with the associated trait. The identification of a gene that is strongly associated with a particular phenotype, e.g., GENE 1 strongly associated with those plants that have high NUE. Big-data enrichment and functional network analysis by artificial intelligence (AI) will provide the opportunity to develop different types of models such as random models and guided models. Based on the accuracy and requirement, the model can be developed. An intelligently guided model can help in the understanding of complex traits with genetic background, G*E interaction patterns, developing ideotype breeding, especially for underground traits, identifying superior parental lines for promising breeding, and improving smart farming.
In order to increase production under stressful conditions, several molecular techniques and methodologies, including omics, molecular breeding, and transgenic, were used. Furthermore, the following examples of molecular exploitation of germplasm resources have been emphasized in order to maximize the utilization of potential genetic resources:
-
(1)
Most of the germplasm comprises a wide range of trait-specific collections like thermo-, drought-, salt-, cold-tolerance, high NUE, high WUE, etc. Hence, the utilization of trait-specific accession/genotypes for development of climate-smart crops can be a logistic approach for further sustainability.
-
(2)
Crop wild relatives (CWRs) are accredited as a potential source of genetic diversity with a wide range of adaptability against various natural stress conditions, and they provided useful genetic resources that complement the complexity associated with crop domestication and breeding, as well as the development of elite varieties.
-
(3)
Germplasm is the source of novel genes and the discovery of novel genes associated with stress signalling/mitigation can be a promising approach. Additionally, genetically diverse accessions/genotypes can be a potential resource for molecular screening and robust evaluation processes. However, single reference genomes do not depict all variation at the species level, hence the construction of pangenomes or genus-wide pangenomes is another sophisticated method to create structural genetic diversity of prospective genetic resources.
-
(4)
The breeding technique can be further sped up (in just one generation) by developing homozygous/pure-breeding lines using haploid induction (HI) with the manipulation of the centromere-specific histone H3 (CENH3) variant.
-
(5)
The molecular information of genetic resources especially at the level of gene-isoforms, impact of alternating splicing, correlation between genome size and transcriptome and/or proteome are the precondition for further application of breeding.
-
(6)
Germplasm encompasses a wide range of ploidy collection; therefore screening/evaluation of parental material for ploidy breeding is a logistic utilization strategy.
-
(7)
To enhance resilience or improve capacity to adapt and grow, germplasm accessions/genotypes can be exposed under different climate scenarios that include the simultaneous exposure of crops to combined stresses (biotic and abiotic) and other climate-related risks. That can help to better understand physiological, biochemical, and molecular responses under combined stresses.
-
(8)
Special attention is required to create and introduce novel resistance and acclimation strategies through synthetic biology, artificial intelligence, and nanotechnology.
-
(9)
Finally, a vision has to be made for transforming PGRs into a digital resource system. For the digitalization, detailed molecular information of genes, promoter/cis-element enrichment, co-expression/interaction patterns, the influence of cis/trans-elements, metabolic pathways along with high-throughput phenotype data of PGRs are essential for future molecular utilization.
6. Conclusion
Post-Green revolutionary events and the negative impact of climate conditions imposed an urgent action to reconciliation of research challenges, gaps, and advancement/innovation of practices to harness the benefit of ∼7.4 million gene bank germplasm materials. In this connection, we have emphasized (1) the key elements of germplasm, and their functions; (2) major challenges associated with PGRs, (3) the physiological and molecular perspective of two broad spectrum complex traits such as water-and nitrogen-use efficiency to mitigate challenges and sustainable development, (4) proposed integrated approaches that are essential to find out useful candidate genes involved in water-and nitrogen-use efficiency using large collections of PGRs, and (5) molecular utilization of PGRs. Despite tremendous advancements in marker-assisted selection (MAS) over the past few decades, several obstacles still exist in the characterization of germplasm. Therefore, the advancement of sequence-based study in next-generation breeding high-throughput bioinformatics platforms, and passionate scientists is required. More emphasis on the selection of promising genetic resources, as well as the consistent association between loci and traits through integrated approaches, should be considered for promising outcomes. The precise genetic engineering technology will allow the rapid transfer of advantageous traits between crops and their wild relatives in both furrowed and reverse directions. Furthermore, for a high-resolution understanding of complex genetic mechanisms, an integrated approach, and genome editing toolkits should be taken into consideration. Our present discussion will provide the advancement of germplasm assets that will meet the required sustainability in the future.
Author contribution statement
RM conceived the idea, executed research and wrote the manuscript with the help of AK and BNG.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data associated with this study has been deposited at One basic version of this manuscript available in Preprint (https://doi.org/10.20944/preprints202107.0359.v1).
Declaration of interest’s statement
The authors declare no conflict of interest.
Contributor Information
Raju Mondal, Email: rmcrijaf@yahoo.in, mondal.raju2202@gmail.com.
Amit Kumar, Email: amit_bio80@yahoo.com.
Belaghihalli N. Gnanesh, Email: gnaneshbn@gmail.com.
References
- 1.Singh D.P., Singh A.K., Singh A. Academic Press; 2021. Plant Breeding and Cultivar Development. [Google Scholar]
- 2.Ferranti P. Food sustainability, security, and effects of global change. Ref. Mod. Food Sci. 2016 doi: 10.1016/B978-0-08-100596-5.03332-1. [DOI] [Google Scholar]
- 3.Janzen G.M., Wang L., Hufford M.B. The extent of adaptive wild introgression in crops. New Phytol. 2019;221(3):1279–1288. doi: 10.1111/nph.15457. [DOI] [PubMed] [Google Scholar]
- 4.Bailey-Serres J., Parker J.E., Ainsworth E.A., Oldroyd G.E., Schroeder J.I. Genetic strategies for improving crop yields. Nature. 2019;575(7781):109–118. doi: 10.1038/s41586-019-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor R., Renata A., Ortigara C., Koncagül E., Uhlenbrook S., Lamizana-Diallo B.M.…Brdjanovic D. The United Nations World Water Development Report; 2017. The United Nations World Water Development Report 2017. Wastewater: the Untapped Resource. [Google Scholar]
- 6.Rivero R.M., Mittler R., Blumwald E., Zandalinas S.I. Developing climate‐resilient crops: improving plant tolerance to stress combination. Plant J. 2022;109(2):373–389. doi: 10.1111/tpj.15483. [DOI] [PubMed] [Google Scholar]
- 7.Gupta A., Rico-Medina A., Caño-Delgado A.I. The physiology of plant responses to drought. Science. 2020;368(6488):266–269. doi: 10.1126/science.aaz7614. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhary S., Devi P., HanumanthaRao B., Jha U.C., Sharma K.D., Prasad P.V.…Nayyar H. Physiological and molecular approaches for developing thermo tolerance in vegetable crops: a growth, yield and sustenance perspective. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.878498. 1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Hintum T.J. Duplication within and between germplasm collections. III. A quantitative model. Genet. Resour. Crop Evol. 2000;47(5):507–513. doi: 10.1023/A:1008703031415. [DOI] [Google Scholar]
- 10.Mascher M., Schreiber M., Scholz U., Graner A., Reif J.C., Stein N. Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat. Genet. 2019;51(7):1076–1081. doi: 10.1038/s41588-019-0443-6. [DOI] [PubMed] [Google Scholar]
- 11.Cieslarová J., Smýkal P., Dočkalová Z., Hanáček P., Procházka S., Hýbl M., Griga M. Molecular evidence of genetic diversity changes in pea (Pisum sativum L.) germplasm after long-term maintenance. Genet. Resour. Crop Evol. 2011;58(3):439–451. doi: 10.1007/s10722-010-9591-3. [DOI] [Google Scholar]
- 12.Jia J., Li H., Zhang X., Li Z., Qiu L. Genomics-based plant germplasm research (GPGR) Crop J. 2017;5(2):166–174. doi: 10.1016/j.cj.2016.10.006. [DOI] [Google Scholar]
- 13.Huang J., Gao Y., Jia H., Liu L., Zhang D., Zhang Z. Comparative transcriptomics uncovers alternative splicing changes and signatures of selection from maize improvement. BMC Genom. 2015;16(1):1–11. doi: 10.5061/dryad.tk2fn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaut B.S., Seymour D.K., Liu Q., Zhou Y. Demography and its effects on genomic variation in crop domestication. Nature Plants. 2018;4(8):512–520. doi: 10.1038/s41477-018-0210-1. [DOI] [PubMed] [Google Scholar]
- 15.Renaut S., Rieseberg L.H. The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other compositae crops. Mol. Biol. Evol. 2015;32(9):2273–2283. doi: 10.1093/molbev/msv106. [DOI] [PubMed] [Google Scholar]
- 16.Langridge P., Waugh R. Harnessing the potential of germplasm collections. Nat. Genet. 2019;51(2):200–201. doi: 10.1038/s41588-018-0340-4. [DOI] [PubMed] [Google Scholar]
- 17.Mondal R., Kumar A., Chattopadhyay S.K. Structural property, molecular regulation and functional diversity of Glutamine Synthetase in higher plants: a data-mining bioinformatics approach. Plant J. 2021;108(6):1565–1584. doi: 10.1111/tpj.15536. https://doi: 10.1111/tpj.15536 [DOI] [PubMed] [Google Scholar]
- 18.Rebetzke G.J., Fischer R.T.A., Van Herwaarden A.F., Bonnett D.G., Chenu K., Rattey A.R., Fettell N.A. Plot size matters: interference from intergenotypic competition in plant phenotyping studies. Funct. Plant Biol. 2013;41(2):107–118. doi: 10.1071/FP13177. [DOI] [PubMed] [Google Scholar]
- 19.Laliberté E. Below‐ground frontiers in trait‐based plant ecology. New Phytol. 2017;213(4):1597–1603. doi: 10.1111/nph.14247. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A., Medhi K., Fagodiya R.K., Subrahmanyam G., Mondal R., Raja P.…Pathak H. Molecular and ecological perspectives of nitrous oxide producing microbial communities in agro-ecosystems. Rev. Environ. Sci. Biotechnol. 2020;19(4):717–750. doi: 10.1007/s11157-020-09554-w. [DOI] [Google Scholar]
- 21.Saad N.S.M., Neik T.X., Thomas W.J., Amas J.C., Cantila A.Y., Craig R.J.…Batley J. Advancing designer crops for climate resilience through an integrated genomics approach. Curr. Opin. Plant Biol. 2022;67 doi: 10.1016/j.pbi.2022.102220. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Morton J.A., Pellicer J., Leitch I.J., Leitch A.R. Genome downsizing after polyploidy: mechanisms, rates and selection pressures. Plant J. 2021;107(4):1003–1015. doi: 10.1111/tpj.15363. [DOI] [PubMed] [Google Scholar]
- 23.Ye T., Li Y., Zhang J., Hou W., Zhou W., Lu J., Xing Y., Li X. Nitrogen, phosphorus, and potassium fertilization affects the flowering time of rice (Oryza sativa L.) Glob. Ecol. Conserv. 2019;20 [Google Scholar]
- 24.Zhang S., Zhang Y., Li K., Yan M., Zhang J., Yu M.…Xu G. Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr. Biol. 2021;31(4):671–683. doi: 10.1016/j.cub.2020.10.095. [DOI] [PubMed] [Google Scholar]
- 25.Raun W.R., Johnson G.V. Improving nitrogen use efficiency for cereal production. Agron. J. 1999;91(3):357–363. doi: 10.2134/agronj1999.00021962009100030001x. [DOI] [Google Scholar]
- 26.Fagodiya R.K., Kumar A., Kumari S., Medhi K., Shabnam A.A. Contaminants in Agriculture. Springer; Cham: 2020. Role of nitrogen and its agricultural management in changing environment; pp. 247–270. [DOI] [Google Scholar]
- 27.Kant S., Bi Y.M., Rothstein S.J. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 2011;62(4):1499–1509. doi: 10.1093/jxb/erq297. [DOI] [PubMed] [Google Scholar]
- 28.Li M., Xu J., Gao Z., Tian H., Gao Y., Kariman K. Genetically modified crops are superior in their nitrogen use efficiency-A meta-analysis of three major cereals. Sci. Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-65684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tracy S.R., Nagel K.A., Postma J.A., Fassbender H., Wasson A., Watt M. Crop improvement from phenotyping roots: highlights reveal expanding opportunities. Trends Plant Sci. 2020;25(1):105–118. doi: 10.1016/j.tplants.2019.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Dharmappa P.M., Doddaraju P., Malagondanahalli M.V., Rangappa R.B., Mallikarjuna N.M., Rajendrareddy S.H.…Sheshshayee S.M. Introgression of root and water use efficiency traits enhances water productivity: an evidence for physiological breeding in rice (Oryza sativa L.) Rice. 2019;12(1):1–14. doi: 10.1186/s12284-019-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zandalinas S.I., Fritschi F.B., Mittler R. Signal transduction networks during stress combination. J. Exp. Bot. 2020;71(5):1734–1741. doi: 10.1093/jxb/erz486. [DOI] [PubMed] [Google Scholar]
- 32.Walters R.G. Towards an understanding of photosynthetic acclimation. J. Exp. Bot. 2005;56(411):435–447. doi: 10.1093/jxb/eri060. [DOI] [PubMed] [Google Scholar]
- 33.Bohnert H.J., Nelson D.E., Jensen R.G. Adaptations to environmental stresses. Plant Cell. 1995;7(7):1099. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu P.K., Dubeaux G., Takahashi Y., Schroeder J.I. Signaling mechanisms in abscisic acid‐mediated stomatal closure. Plant J. 2021;105(2):307–321. doi: 10.1111/tpj.15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zandalinas S.I., Balfagón D., Arbona V., Gómez-Cadenas A., Inupakutika M.A., Mittler R. ABA is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress. J. Exp. Bot. 2016;67(18):5381–5390. doi: 10.1093/jxb/erw299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zandalinas S.I., Fichman Y., Devireddy A.R., Sengupta S., Azad R.K., Mittler R. Systemic signaling during abiotic stress combination in plants. Proc. Natl. Acad. Sci. USA. 2020;117(24):13810–13820. doi: 10.1073/pnas.2005077117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizhsky L., Liang H., Shuman J., Shulaev V., Davletova S., Mittler R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 2004;134(4):1683–1696. doi: 10.1104/pp.103.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balfagón D., Sengupta S., Gómez-Cadenas A., Fritschi F.B., Azad R.K., Mittler R., Zandalinas S.I. Jasmonic acid is required for plant acclimation to a combination of high light and heat stress. Plant Physiol. 2019;181(4):1668–1682. doi: 10.1104/pp.19.00956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raju B.R., Mohankumar M.V., Sumanth K.K., Rajanna M.P., Udayakumar M., Prasad T.G., Sheshshayee M.S. Discovery of QTLs for water mining and water use efficiency traits in rice under water-limited condition through association mapping. Mol. Breed. 2016;36(3):1–16. doi: 10.1007/s11032-016-0457-z. [DOI] [Google Scholar]
- 40.Jamil M., Ali A., Akbar K.F., Ghafoor A., Aziz A.B.D.U.L., Napar S.A., Mujeeb-Kazi A. Relationship among water use efficiency, canopy temperature, chlorophyll content and spot blotch (Cochliobolus sativus) resistance in diverse wheat (Triticum aestivum L.) Germplasm. Pakistan J. Bot. 2016;48(3):993–998. [Google Scholar]
- 41.Meena R.P., Karnam V., Sendhil R., Sharma R.K., Tripathi S.C., Singh G.P. Identification of water use efficient wheat genotypes with high yield for regions of depleting water resources in India. Agric. Water Manag. 2019;223 doi: 10.1016/j.agwat.2019.105709. [DOI] [Google Scholar]
- 42.Fried H.G., Narayanan S., Fallen B. Evaluation of soybean [Glycine max (L.) Merr.] genotypes for yield, water use efficiency, and root traits. PLoS One. 2019;14(2) doi: 10.1371/journal.pone.0212700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez M.A., Xavier A., Rainey K.M. Phenotypic variation and genetic architecture for photosynthesis and water use efficiency in soybean (Glycine max L. Merr) Front. Plant Sci. 2019;10:680. doi: 10.3389/fpls.2019.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia X., Mao K., Wang P., Wang Y., Jia X., Huo L.…Ma F. Overexpression of MdATG8i improves water use efficiency in transgenic apple by modulating photosynthesis, osmotic balance, and autophagic activity under moderate water deficit. Horticult. Res. 2021;8(81) doi: 10.1038/s41438-021-00521-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pignon C.P., Leakey A.D., Long S.P., Kromdijk J. Drivers of natural variation in water-use efficiency under fluctuating light are promising targets for improvement in sorghum. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.627432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arab M.M., Marrano A., Abdollahi-Arpanahi R., Leslie C.A., Cheng H., Neale D.B., Vahdati K. Combining phenotype, genotype, and environment to uncover genetic components underlying water use efficiency in Persian walnut. J. Exp. Bot. 2020;71(3):1107–1127. doi: 10.1093/jxb/erz467. [DOI] [PubMed] [Google Scholar]
- 47.Ishizaki T., Maruyama K., Obara M., Fukutani A., Yamaguchi-Shinozaki K., Ito Y., Kumashiro T. Expression of Arabidopsis DREB1C improves survival, growth, and yield of upland New Rice for Africa (NERICA) under drought. Mol. Breed. 2013;31(2):255–264. doi: 10.1007/s11032-012-9785-9. [DOI] [Google Scholar]
- 48.Devika S., Ravichandran V., Boominathan P. Physiological analyses of nitrogen use efficiency and yield traits of rice genotypes. Indian J. Plant Physiol. 2018;23(1):100–110. doi: 10.1007/s40502-018-0358-8. [DOI] [Google Scholar]
- 49.Sharma N., Sinha V.B., Gupta N., Rajpal S., Kuchi S., Sitaramam V.…Raghuram N. Phenotyping for nitrogen use efficiency: rice genotypes differ in N-responsive germination, oxygen consumption, seed urease activities, root growth, crop duration, and yield at low N. Front. Plant Sci. 2018;9:1452. doi: 10.3389/fpls.2018.01452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen G.N., Maharjan P., Maphosa L., Vakani J., Thoday-Kennedy E., Kant S. A robust automated image-based phenotyping method for rapid vegetative screening of wheat germplasm for nitrogen use efficiency. Front. Plant Sci. 2019;10:1372. doi: 10.3389/fpls.2019.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zakari S.A., Asad M.A.U., Han Z., Zhao Q., Cheng F. Relationship of nitrogen deficiency-induced leaf senescence with ROS generation and ABA concentration in rice flag leaves. J. Plant Growth Regul. 2020;39(4):1503–1517. doi: 10.1007/s00344-020-10128-x. [DOI] [Google Scholar]
- 52.Jia C., Wang F., Yuan J., Zhang Y., Zhao Z., Abulizi B., Wen X., Kang M., Tang F. Screening and comprehensive evaluation of rice (Oryza sativa L. subsp. japonica Kato) germplasm resources for nitrogen efficiency in Xinjiang, China. Plant Genet. Res. 2020;18(3):179–189. doi: 10.1017/S1479262120000118. [DOI] [Google Scholar]
- 53.Ma L., Qing C., Frei U., Shen Y., Lübberstedt T. Association mapping for root system architecture traits under two nitrogen conditions in germplasm enhancement of maize doubled haploid lines. Crop J. 2020;8(2):213–226. doi: 10.1016/j.cj.2019.11.004. [DOI] [Google Scholar]
- 54.Pujarula V., Pusuluri M., Bollam S., Das R.R., Ratnala R., Adapala G.…Gupta R. Genetic variation for nitrogen use efficiency traits in global diversity panel and parents of mapping populations in pearl millet. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.625915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang N., Li S., Wang S., Li Q., Xu F., Shi L.…Ding G. Dynamic transcriptome analysis indicates extensive and discrepant transcriptomic reprogramming of two rapeseed genotypes with contrasting NUE in response to nitrogen deficiency. Plant Soil. 2020;456(1):369–390. doi: 10.1007/s11104-020-04720-z. [DOI] [Google Scholar]
- 56.Tiong J., Sharma N., Sampath R., MacKenzie N., Watanabe S., Metot C.…Okamoto M. Improving nitrogen use efficiency through overexpression of alanine aminotransferase in rice, wheat, and barley. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.628521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mauceri A., Rosa Abenavoli M., Toppino L., Panda S., Mercati F., Miyassa Aci M.…Lupini A. Transcriptomic insights on molecular regulation of Solanum melongena L. N-Use Efficiency. J. Exp. Bot. 2021;72(22):4237–4253. doi: 10.1093/jxb/erab121. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J., Wang Y., Zhao Y., Zhang Y., Zhang J., Ma H., Han Y. Transcriptome analysis reveals Nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (Solanum tuberosum L.) PLoS One. 2020;15(10) doi: 10.1371/journal.pone.0240662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang S., Zhu L., Shen C., Ji Z., Zhang H., Zhang T.…Li S. Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell. 2021;33(3):566–580. doi: 10.1093/plcell/koaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watson A., Ghosh S., Williams M.J., Cuddy W.S., Simmonds J., Rey M.D.…Hickey L.T. Speed breeding is a powerful tool to accelerate crop research and breeding. Nature Plants. 2018;4(1):23–29. doi: 10.1038/s41477-017-0083-8. [DOI] [PubMed] [Google Scholar]
- 61.Soumare A., Diédhiou A.G., Arora N.K., Tawfeeq Al-Ani L.K., Ngom M., Fall S.…Sy M.O. Potential role and utilization of plant growth promoting microbes in plant tissue culture. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.649878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019;70(1):667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 63.Mushtaq M., Ahmad Dar A., Skalicky M., Tyagi A., Bhagat N., Basu U.…El Sabagh A. CRISPR-based genome editing tools: insights into technological breakthroughs and future challenges. Genes. 2021;12(6):797. doi: 10.3390/genes12060797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lavarenne J., Guyomarc’h S., Sallaud C., Gantet P., Lucas M. The spring of systems biology-driven breeding. Trends Plant Sci. 2018;23(8):706–720. doi: 10.1016/j.tplants.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 65.Varshney R.K., Sinha P., Singh V.K., Kumar A., Zhang Q., Bennetzen J.L. 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 2020;56:190–196. doi: 10.1016/j.pbi.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Esvelt Klos K., Huang Y.F., Bekele W.A., Obert D.E., Babiker E., Beattie A.D.…Tinker N.A. Population genomics related to adaptation in elite oat germplasm. Plant Genome. 2016;9(2):10. doi: 10.3835/plantgenome2015.10.0103. plantgenome2015. [DOI] [PubMed] [Google Scholar]
- 67.Crossa J., Pérez-Rodríguez P., Cuevas J., Montesinos-López O., Jarquín D., De Los Campos G., Burgueño J., González-Camacho J.M., Pérez-Elizalde S., Beyene Y., Dreisigacker S. Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 2017;22(11):961–975. doi: 10.1016/j.tplants.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 68.Ertiro B.T., Labuschagne M., Olsen M., Das B., Prasanna B.M., Gowda M. Genetic dissection of nitrogen use efficiency in tropical maize through genome-wide association and genomic prediction. Front. Plant Sci. 2020;11:474. doi: 10.3389/fpls.2020.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Srivastava R.K., Singh R.B., Pujarula V.L., Bollam S., Pusuluri M., Chellapilla T.S.…Gupta R. Genome-wide association studies and genomic selection in Pearl Millet: advances and prospects. Front. Genet. 2020;10:1389. doi: 10.3389/fgene.2019.01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi A., Bhattarai G., Xiong H., Avila C.A., Feng C., Liu B., Joshi V., Stein L., Mou B., du Toit L.J., Correll J.C. Genome-wide association study and genomic prediction of white rust resistance in USDA GRIN spinach germplasm. Horticult. Res. 2022;9 doi: 10.1093/hr/uhac069. uhac069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhat J.A., Adeboye K.A., Ganie S.A., Barmukh R., Hu D., Varshney R.K., Yu D. Genome-wide association study, haplotype analysis, and genomic prediction reveal the genetic basis of yield-related traits in soybean (Glycine max L.) Front. Genet. 2022;13 doi: 10.3389/fgene.2022.953833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y., Hu G., Zhang A., Loladze A., Hu Y., Wang H., Qu J., Zhang X., Olsen M., San Vicente F., Crossa J. Genome-wide association study and genomic prediction of Fusarium ear rot resistance in tropical maize germplasm. Crop J. 2021;9(2):325–341. doi: 10.1016/j.cj.2020.08.008. [DOI] [Google Scholar]
- 73.McMullen M.D., Kresovich S., Villeda H.S., Bradbury P., Li H., Sun Q.…Buckler E.S. Genetic properties of the maize nested association mapping population. Science. 2009;325(5941):737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 74.Wang H., Xu X., Vieira F.G., Xiao Y., Li Z., Wang J.…Chu C. The power of inbreeding: NGS-based GWAS of rice reveals convergent evolution during rice domestication. Mol. Plant. 2016;9(7):975–985. doi: 10.1016/j.molp.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 75.Ta K.N., Khong N.G., Ha T.L., Nguyen D.T., Mai D.C., Hoang T.G.…Jouannic S. A genome-wide association study using a Vietnamese landrace panel of rice (Oryza sativa) reveals new QTLs controlling panicle morphological traits. BMC Plant Biol. 2018;18(1):1–15. doi: 10.1186/s12870-018-1504-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X., Luo Z., Todd J., Sood S., Wang J. Genome‐wide association study of multiple yield traits in a diversity panel of polyploid sugarcane (Saccharum spp.) Plant Genome. 2020;13(1) doi: 10.1002/tpg2.20006. [DOI] [PubMed] [Google Scholar]
- 77.Sehgal D., Mondal S., Crespo-Herrera L., Velu G., Juliana P., Huerta-Espino J., Shrestha S., Poland J., Singh R., Dreisigacker S. Haplotype-based, genome-wide association study reveals stable genomic regions for grain yield in CIMMYT spring bread wheat. Front. Genet. 2020;11:1427. doi: 10.3389/fgene.2020.589490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhat J.A., Yu D., Bohra A., Ganie S.A., Varshney R.K. Features and applications of haplotypes in crop breeding. Commun. Biol. 2021;4(1):1–12. doi: 10.1038/s42003-021-02782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu Z., Chang F., Lv W., Sharmin R.A., Wang Z., Kong J., Bhat J.A., Zhao T. Identification of QTN and candidate gene for seed-flooding tolerance in soybean [Glycine max (L.) Merr.] using genome-wide association study (GWAS) Genes. 2019;10(12):957. doi: 10.3390/genes10120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang A., Chen S., Cui Z., Liu Y., Guan Y., Yang S., Qu J., Nie J., Dang D., Li C., Dong X. Genomic prediction of drought tolerance during seedling stage in maize using low-cost molecular markers. Euphytica. 2022;218(11):1–21. doi: 10.1007/s10681-022-03103-y. [DOI] [Google Scholar]
- 81.Wang F., Yoshida H., Matsuoka M. Making the ‘Green Revolution’truly green: improving crop nitrogen use efficiency. Plant Cell Physiol. 2021;62(6):942–947. doi: 10.1093/pcp/pcab051. [DOI] [PubMed] [Google Scholar]
- 82.Tang W., Ye J., Yao X., Zhao P., Xuan W., Tian Y., Zhang Y., Xu S., An H., Chen G., Yu J. Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 2019;10(1):1–11. doi: 10.1038/s41467-019-13187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pignon C.P., Fernandes S.B., Valluru R., Bandillo N., Lozano R., Buckler E.…Leakey A.D. Phenotyping stomatal closure by thermal imaging for GWAS and TWAS of water use efficiency-related genes. Plant Physiol. 2021;187(4):2544–2562. doi: 10.1093/plphys/kiab395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bhaskara G.B., Lasky J.R., Razzaque S., Zhang L., Haque T., Bonnette J.E.…Juenger T.E. Natural variation identifies new effectors of water-use efficiency in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2022;119(33) doi: 10.1073/pnas.2205305119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei S., Li X., Lu Z., Zhang H., Ye X., Zhou Y., Li J., Yan Y., Pei H., Duan F., Wang D. A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science. 2022;377(6604):eabi8455. doi: 10.1126/science.abi8455. [DOI] [PubMed] [Google Scholar]
- 86.Wang H., Lu S., Guan X., Jiang Y., Wang B., Hua J., Zou B. Dehydration-Responsive element binding protein 1C, 1E, and 1G promote stress tolerance to chilling, heat, drought, and salt in rice. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.851731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kenawy E.R., Hosny A., Saad-Allah K. Reducing nitrogen leaching while enhancing growth, yield performance and physiological traits of rice by the application of controlled-release urea fertilizer. Paddy Water Environ. 2021;19(1):173–188. doi: 10.1007/s10333-020-00828-1. [DOI] [Google Scholar]
- 88.Zhao Y., Li D., Xu J.W., Zhao P., Li T., Ma H., Yu X. Melatonin enhances lipid production in Monoraphidium sp. QLY-1 under nitrogen deficiency conditions via a multi-level mechanism. Bioresour. Technol. 2018;259:46–53. doi: 10.1016/j.biortech.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 89.Hu S., Zhang M., Yang Y., Xuan W., Zou Z., Arkorful E., Chen Y., Ma Q., Jeyaraj A., Chen X., Li X. A novel insight into nitrogen and auxin signaling in lateral root formation in tea plant [Camellia sinensis (L.) O. Kuntze] BMC Plant Biol. 2020;20(1):1–17. doi: 10.1186/s12870-020-02448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iqbal N., Fatma M., Gautam H., Sehar Z., Rasheed F., Khan M.I.R., Sofo A., Khan N.A. Salicylic acid increases photosynthesis of drought grown mustard plants effectively with sufficient-N via regulation of ethylene, abscisic acid, and nitrogen-use efficiency. J. Plant Growth Regul. 2022:1–12. doi: 10.1007/s00344-021-10565-2. [DOI] [Google Scholar]
- 91.Kayum M., Jung H.J., Park J.I., Ahmed N.U., Saha G., Yang T.J., Nou I.S. Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa. Mol. Genet. Genom. 2015;290(1):79–95. doi: 10.1007/s00438-014-0898-1. [DOI] [PubMed] [Google Scholar]
- 92.Li T., Wang Y.H., Liu J.X., Feng K., Xu Z.S., Xiong A.S. Advances in genomic, transcriptomic, proteomic, and metabolomic approaches to study biotic stress in fruit crops. Crit. Rev. Biotechnol. 2019;39(5):680–692. doi: 10.1080/07388551.2019.1608153. [DOI] [PubMed] [Google Scholar]
- 93.Zogli P., Pingault L., Grover S., Louis J. Ento (o) mics: the intersection of ‘omic’approaches to decipher plant defense against sap-sucking insect pests. Curr. Opin. Plant Biol. 2020;56:153–161. doi: 10.1016/j.pbi.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 94.Jia H., Li M., Li W., Liu L., Jian Y., Yang Z.…Zhang Z. A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat. Commun. 2020;11(1):1–11. doi: 10.1038/s41467-020-14746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gonzalez Guzman M., Cellini F., Fotopoulos V., Balestrini R., Arbona V. New approaches to improve crop tolerance to biotic and abiotic stresses. Physiol. Plantarum. 2022;174(1) doi: 10.1111/ppl.13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin A.R., Isaac M.E. Plant functional traits in agroecosystems: a blueprint for research. J. Appl. Ecol. 2015;52(6):1425–1435. doi: 10.1111/1365-2664.12526. [DOI] [Google Scholar]
- 97.Braun I.R., Yanarella C.F., Lawrence-Dill C.J. Computing on phenotypic descriptions for candidate gene discovery and crop improvement. Plant Phen. 2020;2020 doi: 10.34133/2020/1963251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at One basic version of this manuscript available in Preprint (https://doi.org/10.20944/preprints202107.0359.v1).