Abstract
Background
This study aimed at investigating the characteristics and correlation between oral (tongue coating) and fecal microbiota in patients with diarrheal irritable bowel syndrome (IBS-D).
Methods
Fifty-two IBS-D patients were chosen, with ten healthy volunteers serving as the normal control group. Tongue coating samples and fecal samples of subjects were sequenced for the 16S rRNA gene (V4–V5). Bioinformatics analysis was done on the test data to investigate oral and fecal microbiota composition characteristics in IBS-D patients.
Results
The microbial richness of tongue coating in IBS-D group was lower than that in the normal control group (P < 0.05). The beta diversity of tongue coating microbiota and fecal microbiota was significantly different in the IBS-D group compared to the normal control group (P < 0.05). Pseudomonadales (Pseudomonadaceae and Pseudomonas), Moraxellaceae, Parvimonas, Peptostreptococcus, and Alloprevotella were considerably high in number the tongue coating samples of the IBS-D group in comparison to the normal control group. Similarly, the fecal samples from the IBS-D group were significantly enriched in Alphaproteobacteria, Pseudomonadales (Pseudomonadaceae and Pseudomonas), Acidaminococcaceae, Phascolarctobacterium, Alloprevotella, and Escherichia compared to the normal control group.
Conclusions
The oral and fecal microbiotas of IBS-D patients differ from those of the control group; hence studying IBS-D from the perspective of the oral-gut microbiome axis is an interesting research avenue.
Keywords: Fecal microbiota, Oral microbiota, Irritable bowel syndrome, Diarrhea
1. Introduction
Irritable bowel syndrome (IBS) is a functional gastrointestinal disorder characterized by persistent or intermittent abdominal pain and changes in stool form and frequency, but without morphological and biochemical abnormalities that could explain the symptoms. According to epidemiological data [1], the global prevalence of IBS is around 10%. Still, there are significant differences between regions and populations, with about 7% in Southeast Asia and the Middle East, 11.8–14.0% in North America and Northern Europe, and 15.0–21.0% in southern Europe and South America. In China, the prevalence of IBS in adults ranges from 1.4% to 15.8%, with students accounting for 33.3% [2]. IBS can be classified into four types, i.e., diarrhea-dominant type (IBS-D), constipation-dominant type (IBS–C), mixed type, and unclassified type, with IBS-D being the predominant subtype in China.
IBS pathogenesis is not yet completely understood. However, in recent years, the extensive research on IBS pathophysiology has revealed some IBS etiology [3,4], such as bile acid metabolism disorders, abnormal serotonin metabolism, brain-gut axis dysfunction, and altered intestinal permeability caused by immunological activation, have been closely related to gut microbiota. The role of gut microbiota dysbiosis in IBS-D pathogenesis receives increasing attention. De Palma G et al. [5] reported that germ-free mice with fecal microbiota of IBS-D patients displayed changes in innate immune activation, quicker gastrointestinal transit, intestinal hypersensitivity, and intestinal barrier failure. Most research in recent years has indicated that the gut microbiota of IBS-D patients differs significantly from that of the general population. The gut microbiota has the potential as a biomarker for IBS-D, although there is no conclusion on the characteristics of IBS-D gut microbiota [1,6].
In addition to the gut microbiota, oral microbiota has been found to be closely related to gastrointestinal diseases in recent years. For example, the pathogenesis of colorectal cancer [7], gastric cancer [8], and inflammatory bowel disease [9] may all involve oral microbiota. The relationship between oral microflora and IBS-D is also attracting attention [10]. Therefore, both oral and gut microbiota may be closely related to IBS-D. To explore the characteristics and correlation of oral and fecal microbiota in patients with IBS-D, 16S rRNA gene sequencing technology was used to compare and analyze the differences of oral and fecal microbiota between IBS-D patients and normal population in this study.
2. Methods
2.1. Participants
In this study, 52 patients who met the Roman IV diagnostic criteria for IBS-D were recruited between July and October 2018 from the outpatients and inpatients department of gastroenterology at Hubei Provincial Hospital of Traditional Chinese Medicine, while a group of ten healthy volunteers served as the normal control group. All those patients were excluded from the study who had used antibiotics or probiotics within the past four weeks, were aged below 20 and above 60, had a history of smoking and drinking, and had other digestive system disorders, such as peptic ulcers and inflammatory bowel disease, as well as circulatory, respiratory, urinary, endocrine, blood, and neuropsychiatric system problems; women who are pregnant, menstruating, or breastfeeding; incomplete clinical data or failure to give tongue coating and fecal samples. All participants were Han Chinese. The mean duration of disease of IBS patients was (4.92 + 2.51) years. Table 1 shows the main features of IBS-D and normal control groups. The study was approved by the Hubei provincial hospital of TCM (HBZY2021-c65-02), and all participants signed informed consent forms.
Table 1.
The general characteristics of the IBS-D group and normal control group.
| N | Age (years) | Gender (male/female) | BMI | |
|---|---|---|---|---|
| IBS-D group | 52 | 36.46 ± 10.95 | 30/22 | 21.89 ± 1.99 |
| Control group | 10 | 35.90 ± 9.42 | 6/4 | 21.31 ± 2.13 |
| P-value | – | 0.88 | 0.89 | 0.39 |
2.2. Sample collection
Fresh fecal samples (1–3 g) were collected by the subjects themselves in sterile medical stool collection containers, returned within 60 min, and stored at −80 °C. If conditions do not allow it, the fecal sample can be stored at −20 °C and returned within 48 h. Tongue coating samples were collected as a source of microbiota by scraping from the tongue coating surface of participants from back to front with sterile cotton swabs on site. Cotton swabs with tongue coating samples were placed in a special sterile collection tube and immediately stored at −80 °C.
2.3. Library preparation
The genomic DNA was extracted from fecal and tongue coating samples using QIAamp DNA Stool Mini Kit and the protease K lysis technique, and the V4–V5 region of the 16S rRNA gene was amplified (forward, 515F:5′-GTGCCAGCMGCCG.
CGGTAA-3'; reverse, 926R:5′-CCGTCAATTCMTTTGAGTTT-3′). PCR products were electrophorized on a 2% agarose gel, purified using AxyPrep DNA Gel recovery kit (AXYGEN), and quantified using the FTC-3000TM real-time PCR. After preparing the DNA library, quantitative insights into microbial ecology (QIIME) were utilized for high-throughput sequencing. The raw sequencing reads were deposited into the NCBI Sequence Read Archive (SRA) database (PRJNA873889).
2.4. Bioinformatics processing and statistical analysis
Effective sequence quality control, splicing, and sequence optimization were carried out for the original sequence data. The processed data were clustered at 97% similarity using UPARSE software to obtain the representative sequence of operational taxonomic units (OTUs). Then, using RDP classifier, OTUs representative sequence was compared to the Silva128 database for species notes, with the confidence coefficient set to 0.6. The rarefaction curve was used to assess the sequencing depth of samples. The flat curve indicated that the sequencing data was adequate. The adequacy of the sample size was evaluated by the cumulative species curve. If the curve tended to be flat with the increase of the sample size, it indicated that the sample was sufficient and data analysis could be carried out.
Mothur v1.30.1 and SPSS 22.0 were used for statistical analysis of data. Chao index, ACE index, Shannon index, and Simpson index were used to evaluate the Alpha diversity of data. Beta diversity was assessed using Bray-Curtis and unweighted UniFrac to calculate the distances between samples and visualized by principal coordinate analysis (PCoA). Community structure and community heat map were analyzed at phylum, class, order, family, and genus levels. When the data follow normal distribution and homogeneity of variance, the student's t-test was used for measuring data comparison and Chi-square test was used for counting data comparison. Otherwise, the non-parametric test was applied. Wilcoxon rank sum test was used for Alpha diversity comparison between the two groups, and LEfSe analysis (LDA score >2, P < 0.05) was used for comparison of community differences. P < 0.05 was considered to demonstrate statistically significant differences.
3. Results
3.1. Sequencing quality analysis
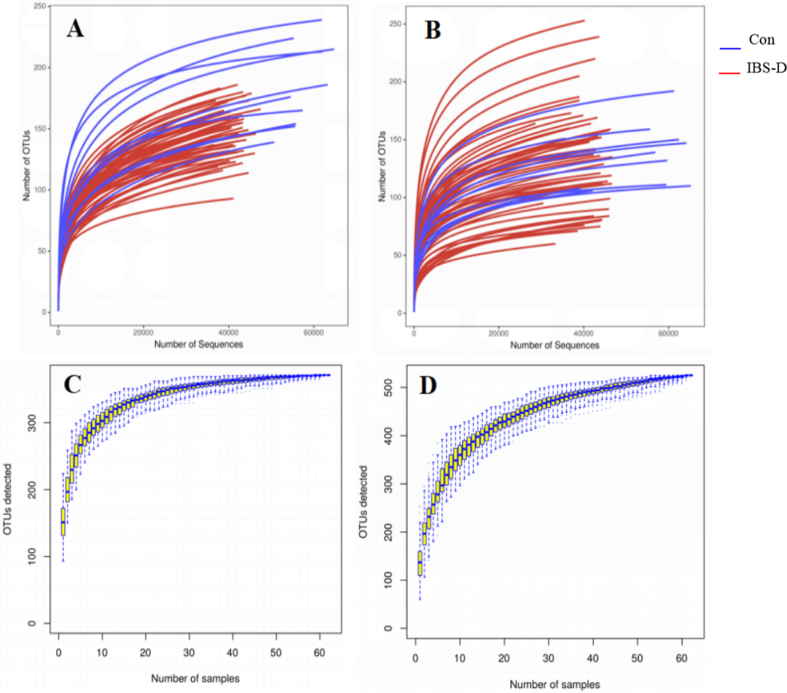
A total of 62 tongue coating and fecal samples were sequenced. Tongue coating samples yielded 3147661 effective sequences and 2729527 optimized sequences. The average length of optimized sequences was 371bp, while a total of 3185812 effective sequences and 2732592 optimized sequences were obtained for fecal samples, where the optimized sequences had an average length of 371bp. As shown in Fig. 1, both the rarefaction curve and the species accumulation curve indicated that the sequencing had reached a plateau, indicating that the sampling and sequencing depth was sufficient for subsequent analysis.
Fig. 1.
Rarefaction curves and species accumulation curves of tongue coating samples and fecal samples. (A) Rarefaction curves of tongue coating samples; (B) rarefaction curves of fecal samples; (C) species accumulation curve of tongue coating samples; (D) species accumulation curve of faecal samples. Con, normal control group.
3.2. OTU-based diversity analysis
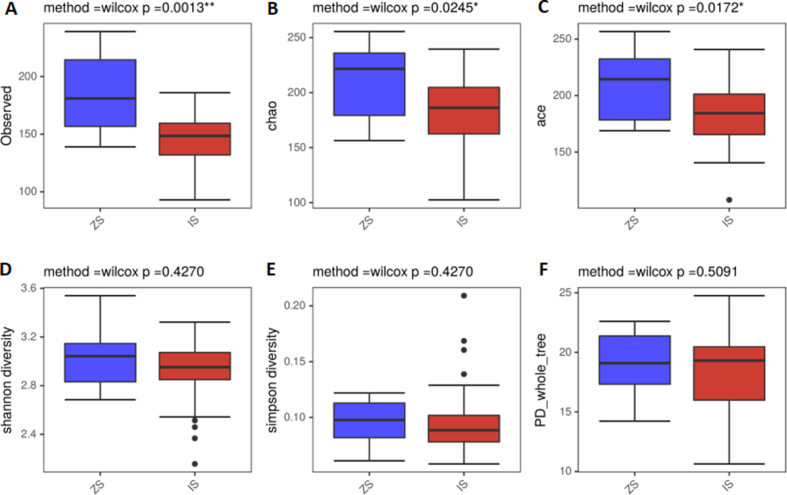
A total of 371 OTUs were obtained from tongue coating samples, 46 of which were unique to the IBS-D group and 28 to the normal control group. Similarly, 525 OTUs were obtained from fecal samples, 194 of which were unique to the IBS-D group and 15 unique to the normal control group. The alpha diversity reflected bacterial abundance and diversity, and the observed index (P = 0.0013), Chao index (P = 0.0245), and ACE index (P = 0.0172) of tongue coating microbiota in IBS-D group were significantly lower than those in the normal control group. As shown in Fig. 2 and Table 2, the Shannon and Simpson indexes revealed no significant difference between the two groups (P > 0.05). Table 2 shows that PD whole tree index of fecal microbiota was significantly higher in IBS-D group than in the normal control group (P = 0.00033). However, there were no significant differences in Chao index, ACE index, or Shannon index between the two groups (P > 0.05).
Fig. 2.
Comparison of alpha diversity of tongue coating microbiota in IBS-D and normal control groups. (A) Observed species index; (B) Chao index; (C) Ace index; (D) Shannon index; (E) Simpson index; (F) PD_whole_tree index. ZS, tongue coating samples from normal control group; IS, tongue coating samples from IBS-D group.
Table 2.
Alpha diversity analysis of IBS-D and normal control groups.
|
tongue coating samples |
observed | chao | ace | shannon | simpson | PD_whole_tree |
|---|---|---|---|---|---|---|
| IBS-D group | 147.40 ± 20.12 | 184.04 ± 29.12 | 182.81 ± 25.52 | 2.93 ± 0.23 | 0.09 ± 0.03 | 18.32 ± 3.22 |
| normal control group | 186.30 ± 34.56 | 211.63 ± 36.09 | 210.39 ± 32.51 | 3.03 ± 0.25 | 0.10 ± 0.02 | 19.19 ± 2.73 |
| P values | 0.0013 | 0.0245 | 0.0172 | 0.4270 | 0.4270 | 0.5091 |
| Fecal samples | ||||||
| IBS-D group | 134.48 ± 42.73 | 162.45 ± 49.06 | 158.34 ± 45.94 | 2.53 ± 0.61 | 0.19 ± 0.12 | 17.99 ± 5.16 |
| normal control group | 135.40 ± 27.87 | 153.32 ± 28.24 | 151.07 ± 29.28 | 2.46 ± 0.38 | 0.19 ± 0.08 | 11.68 ± 2.17 |
| P values | 0.89340 | 0.54658 | 0.68071 | 0.69480 | 0.59866 | 0.00033 |
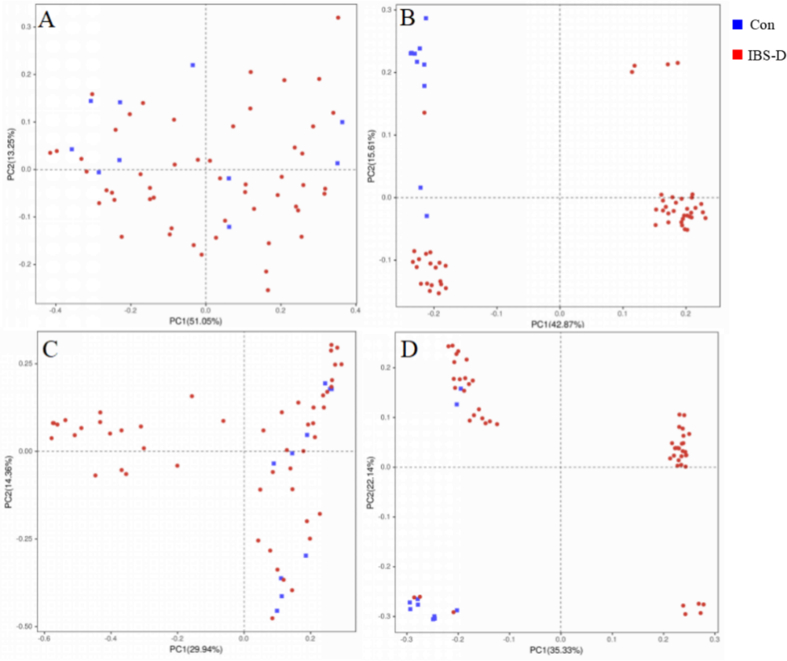
The differences in bacterial community structure are reflected in beta diversity. PCoA plots based on the bray-Curtis method and the unweighted UniFrac method were performed on tongue coating and fecal samples to assess differences between the two groups. As shown in Fig. 3, the closer the distance, the more similar the bacterial structure of the two samples. The Anosim test revealed that the beta diversity of tongue coating microbiota was significantly different in IBS-D group compared to the normal control group (Bray-Curtis: R = 0.170, P = 0.013; UniFrac: R = 0.170, P = 0.013). The beta diversity of fecal microbiota differed from the control group (Bray Curtis: R = 0.0008, P = 0.477; UniFrac: R = 0.529, P = 0.001).
Fig. 3.
Comparison of beta diversity of tongue coating microbiota and fecal microbiota in IBS-D and normal control groups. (A) Bray-Curtis analysis of tongue coating microbiota; (B) Unweighted UniFrac analysis of tongue coating microbiota; (C) Bray-Curtis analysis of fecal microbiota; (D) Unweighted UniFrac analysis of fecal microbiota. The horizontal and vertical coordinates respectively represented a principal component, and the percentage represented the contribution of the principal component to sample differences. Each dot in the figure represented a sample, and samples from the same group were represented in the same color. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Analysis of species composition
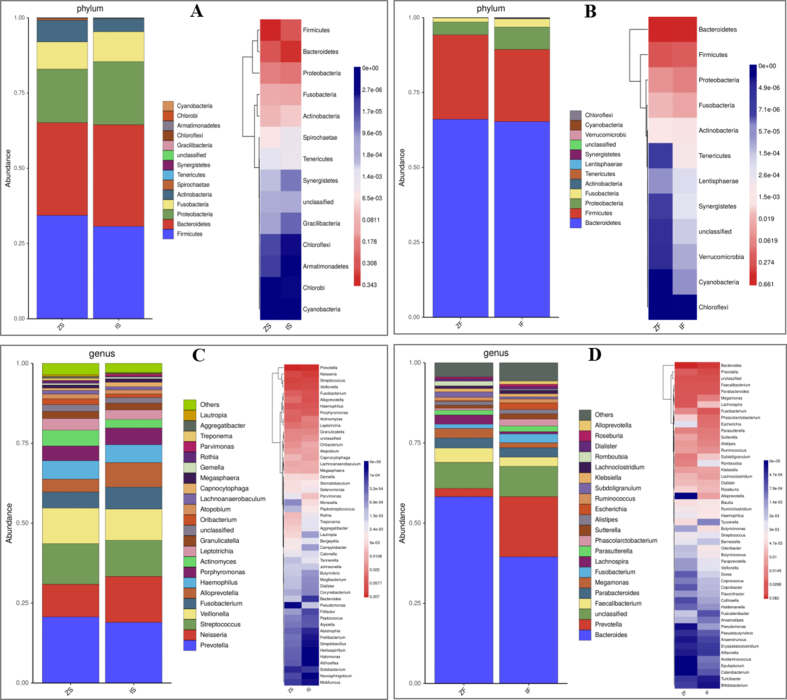
The microbiota can be annotated using 16S rRNA sequencing at six different species classification levels (phylum, class, order, family, genus, and species). This paper primarily explained microbiota species composition of phylum and genus classification levels. At the phylum level, the top four tongue coating microbiota of richness in the IBS-D group compared with normal control group were Bacteroidetes (33.8% vs. 30.8%), Firmicutes (30.7% vs. 34.3%), Proteobacteria (21.0% vs. 17.8%), and Fusobacteria (9.9% vs. 9.0%), while the fecal microbiota were Bacteroidetes (65.3% vs 66.1%), Firmicutes (24.1% vs. 28.2%), Proteobacteria (7.5% vs. 4.3%) and Fusobacteria (2.8% vs. 1.3%). Fig. 4A and B shows the composition of tongue coating and fecal microbiota in two groups at the phylum level. At the genus level, the top four tongue coating microbiota of richness in the IBS-D group compared with normal control group were Prevotella (19.0% vs 20.7%), Neisseria (14.4% vs 10.2%), Streptococcus (11.3% vs. 12.7%), and Veillonella (11.3% vs. 12.7%) (9.7% vs. 11.1%). The fecal microbiota of IBS-D group was dominated by Bacteroides (39.5%) and Prevotella (18.8%), while Bacteroides (58.2%) dominated the normal control group. Fig. 4C and D shows the genus-level composition of tongue coating and fecal microbiota.
Fig. 4.
The composition of tongue coating and fecal microbiota in two groups at phylum and genus level. (A) Species distribution and heatmap of tongue coating microbiota at phylum level; (B) species distribution and heatmap of fecal microbiota at phylum level; (C) species distribution and heatmap of tongue coating microbiota at genus level; (D) species distribution and heatmap of fecal microbiota at genus level. The shades of color in the heatmap indicate the richness of microbiota, and the colors from blue to red indicate the richness from low to high. ZS, tongue coating samples of normal control group; IS, tongue coating samples of IBS-D group; ZF, fecal samples of normal control group; IF, Fecal samples of IBS-D group. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Analysis of differential microbiota
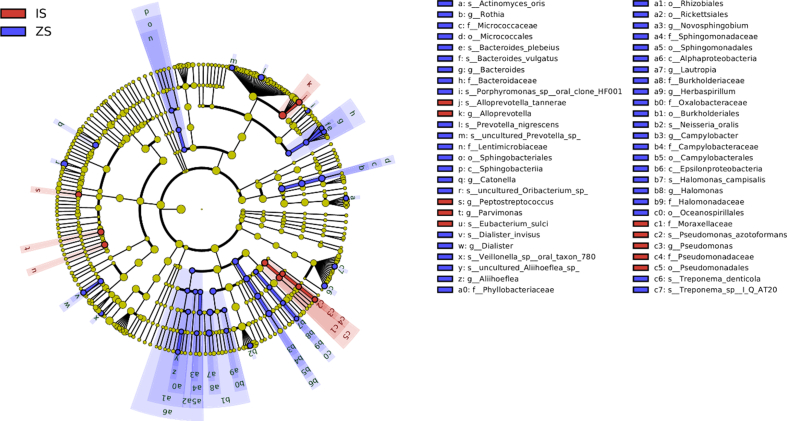
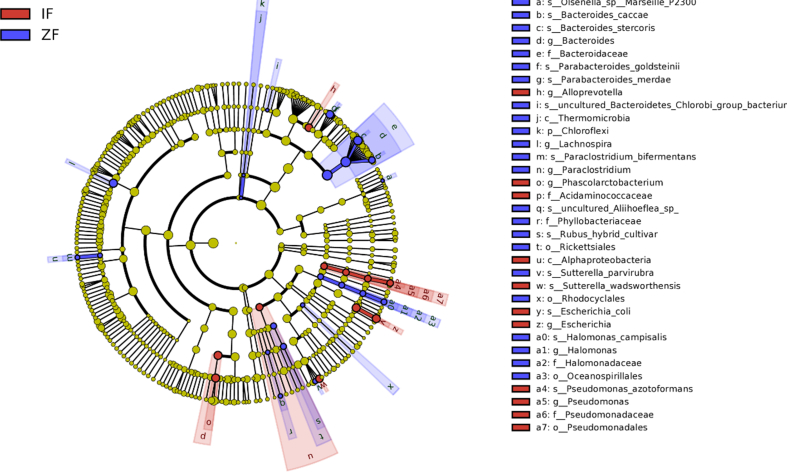
LEfSe analysis results revealed significant differences in tongue coating and fecal microbiota between control and IBS-D groups, As shown in Figs. 5 and 6. Pseudomonadales, Pseudomonadaceae, Pseudomonas, Moraxellaceae, Peptostreptococcus, Parvimonas, and Alloprevotella were relatively enriched in the IBS-D group, while Alphaproteobacteria, Sphingobacteria, Micrococcales, and Lentimicrobiaceae in the normal control group. Similarly, the fecal samples of IBS-D group were relatively enriched in Pseudomonadales, Pseudomonadaceae, Pseudomonas, Alphaproteobacteria, Acidaminococcaceae, Phascolarctobacterium, Alloprevotella, and Escherichia, while in Chloroflexi, Thermomicrobia, Rickettsiales, and Phyllobacteriaceae in the normal control group.
Fig. 5.
LEfSe analysis of tongue coating microbiota of IBS-D and normal control groups. The circles radiating from the inside out represent tvhe classification levels of phylum, class, order, family, genus, and species. Each small circle on a different circle layer represents a classification at that level, and the diameter of the small circle is proportional to richness. Color principle: red is the dominant species of IBS-D group, green is the dominant species of normal control group, yellow is the species with no significant difference between the two groups. ZS, tongue coating samples of normal control group; IS, tongue coating samples of IBS-D group. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
LEfSe analysis of fecal microbiota of IBS-D and normal control groups. ZF, fecal samples of normal control group; IF, fecal samples of IBS-D group.
4. Discussion
Many studies have proved that gut microbiota disorder is closely related to IBS-D [[11], [12], [13]]. The gut microbiota may be involved in IBS-D occurrence and progression in various ways, including neural, immune, and endocrine [14]. The Human Microbiome Project revealed the microbial taxa complexity in the human gut, and also highlighted the highly individualised microbiome composition due to inheritance, diet, and environmental factors. There is no unified conclusion on the characteristics of IBS-D gut microbiota. Some research results are inconsistent, but some studies confirm that the fecal microbiota of IBS-D patients is significantly different from that of the normal population, primarily manifested as a decrease in probiotics, an increase in opportunistic pathogens, and a decrease in Firmicutes/Bacteroides ratio [11,15].
Furthermore, most current studies are based on fecal and mucosal-associated microbiota, with only a few studies on oral (tongue coating) microbiota. The oral cavity and intestinal tract are two important organs of the human digestive system, and their microbiota is closely related. Studies have shown that some resident bacteria and pathogenic immune cells in the oral cavity can migrate to the lower digestive tract, causing gut microbiota imbalance and inflammation [16]. The relationship between oral (tongue coating) microbiota and gastrointestinal diseases is relatively a new research dimension.
Chao and ACE indexes of tongue coating microbiota were significantly lower in IBS-D patients than in the normal population in this study. In contrast, Shannon and Simpson indexes revealed no significant difference, indicating that the richness of IBS-D oral (tongue coating) microbiota was reduced, while the diversity of oral microbiota was not changed significantly. The richness and diversity of fecal microbiota did not decrease significantly in IBS-D patients. Previous research on the alteration of Alpha diversity in IBS-D fecal microbiota is still debatable. Some studies have indicated that the richness or diversity of gut microbiota in IBS-D patients is lower than in the normal population [17,18]. However, several studies have also reported no significant difference in the Alpha diversity of gut microbiota between IBS-D patients and the normal population [19,20]. Pittayanon et al. [11] conducted a meta-analysis and discovered that the decrease in microbial richness in IBS-D patients was not common, which is consistent with the findings of our study. The decrease in Alpha diversity cannot be considered a characteristic manifestation of IBS-D patients. However, this study demonstrated that a decrease in oral (tongue coating) microbial richness might be more directly connected to IBS-D. The relationship between oral microbial richness and IBS-D urges additional investigation.
The gut microbiota composition in IBS-D patients differed significantly from that in the normal population [21], which was also verified in this work. The two most important gut microbiota communities are Bacteroidetes and Firmicutes. This study also discovered that Bacteroidetes and Firmicutes accounted for more than 85% of the gut microbiota in both IBS-D patients and the normal population. Bacteroidetes and Firmicutes are associated with carbohydrate breakdown, which can help the body absorb and store energy. An imbalance between the two may cause obesity and inflammatory bowel disease [22]. It has been reported that IBS-D patients have a lower Firmicutes/Bacteroidetes ratio than the normal population [23]. Although this phenomenon was not observed in the fecal samples collected for this study, Firmicutes/Bacteroidetes < 1 were found in tongue samples, which differed from the normal population. The findings suggest that, compared to fecal microbiota, the imbalance of Firmicutes/Bacteroidetes ratio in oral microbiota of IBS-D patients is more worthy of our concern.
This study discovered that Pseudomonadales, Pseudomonadaceae, Pseudomonas, and Alloprevotella were enriched in the tongue coating and fecal microbiota of IBS-D patients using species difference analysis. Concurrently, Rickettsiales, Oceanospirillales, Bacteroidaceae, Bacteroides, Phyllobacteriaceae, Halomonadaceae, and Halomonas were scarce. Many opportunistic pathogens, such as Pseudomonas aeruginosa, are found in Pseudomonas [24], which might result in a microecological imbalance in the intestine. Alloprevotella can produce diarrhea by promoting the formation of short-chain fatty acids and inflammation [25,26], whereas prolonged low-grade inflammation and excessive short-chain fatty acids can cause diarrhea. The typical microbiota of IBS-D may contain Pseudomonas and Alloprevotella. Rickettsiales also have many opportunistic pathogens [27], and its association with IBS-D is still unknown. According to a previous report [28], the decrease in Oceanospirillales may be associated with an inflammatory reaction. The reduction of Oceanospirillales could be one of the mechanisms encouraging an intestinal low-grade inflammatory response in IBS-D patients. Bacteroides are involved in many intestinal metabolic processes such as glucose metabolism, nitrogenous material utilization, and bile acid biotransformation. However, the different effects of Bacteroides richness on the body are still controversial [29,30]. Brown et al. [31] hypothesized that Bacteroides-derived sphingolipids play an important role in maintaining intestinal homeostasis and symbiosis. Decreased abundance of Bacteroides may lead to IBS-D by altering intestinal metabolic function and compromising homeostasis. Phyllobacteriaceae participates in the nitrate metabolic pathway [32], and nitrate metabolites are associated with intestinal mucosal integrity and inflammation [33,34]. Halomonadaceae and Halomonas are associated with glutamate metabolism [35], and glutamate is an excitatory neurotransmitter in the Microbiota-Gut-Brain axis [36], which plays a vital role in the pathogenesis of IBS-D. It is also worth noting that the vast majority of research to date on the gut-brain axis has been conducted on animal models and much more human research is needed. It remains unclear whether these altered serotonin levels in the gut trigger a cascade of molecular events, which in turn affect brain activity, and whether similar events take place in humans, too. There are still existing gaps in knowledge regarding the interaction between the microbiome and the host in vivo - and the pathway of its metabolites - and how their metabolites influence the microenvironment.
This study also discovered that the abundance of Escherichia and Escherichia coli in fecal samples from IBS-D patients was greater than in the normal population. Our work is in line with past research. A meta-analysis by Wang et al. [37] discovered that the richness of Escherichia coli in the fecal microbiota of IBS patients tended to rise. Therefore, the increase of opportunistic pathogens represented by Escherichia coli is one of the fecal microbiota characteristics of IBS-D. It has been reported [38] that the richness of Lactobacillus and Bifidobacterium decreased in IBS-D patients; however, this result was not in line with the current study, where the results indicated that the decrease of probiotics such as Lactobacillus and Bifidobacterium in IBS-D patients was not common. Ford et al. [39] also considered that the degree of evidence for probiotic treatment of IBS-D is low and that its effectiveness has to be confirmed further. The abundance of Phascolarctobacterium in fecal samples from IBS-D patients was elevated in this study. Phascolarctobacterium is a Gram-negative anaerobic bacterium with a high colonization rate in China that can increase the formation of short-chain fatty acids like acetate and propionate [40]. However, it has been observed [41,42] that Phascolarctobacterium can prevent Clostridium difficile infection and lower intestine succinic levels in treating inflammatory bowel disease. The connection between Phascolarctobacterium and IBS-D is currently unknown. Prevotella is the most common genus of oral (tongue coating) bacteria. There was no significant difference in Prevotella abundance between patients with IBS-D and the normal population in this study; however, the richness of Parvimonas and Peptostreptococcus increased in the oral microbiota of IBS-D patients. Some study has found that oral Parvimonas and Peptostreptococcus are correlated with gastrointestinal diseases in recent years [43], but their relationship with IBS-D has not been established.
Furthermore, this study discovered some similarities between the oral microbiota and fecal microbiome of IBS-D patients. There may be crosstalk between oral and gut microbiota (Oral-Gut Microbiota Axis) which can synergistically participate in the pathogenesis of gastrointestinal diseases [44]. It is a worthwhile research direction to investigate the pathogenesis of IBS-D from the standpoint of the Oral-Gut Microbiota axis. There were some limitations to our study. Firstly, the sample size of our study was small, especially the small number of the normal control group. Thus, large-sample, multicenter studies are still needed to confirm our results. Secondly, the fecal samples or tongue coating samples used in this study do not reflect the microbial environment of the entire gut or oral cavity. Thirdly, the 16S rRNA gene sequencing method does not provide more in-depth and comprehensive species analysis. Therefore, metagenomic sequencing is necessary.
5. Conclusions
Our results showed that there are some differences in the composition of oral and fecal microbiota between IBS-D patients and normal population. Pseudomonadales, Pseudomonadaceae, Pseudomonas, and Alloprevotella were enriched in the tongue coating and fecal microbiota of IBS-D patients. Compared to fecal microbiota, the imbalance of Firmicutes/Bacteroidetes ratio in oral microbiota of IBS-D patients is more worthy of our concern. In addition, a decrease in oral microbial richness also might be more directly connected to IBS-D. Hence studying IBS-D from the perspective of the oral-gut microbiome axis is an interesting research avenue.
Author contribution statement
Binbin tang: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yunlian Hu: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Jianhui Chen; Chengxia Su: Performed the experiments; Analyzed and interpreted the data.
Qian Zhang; Chaoqun Huang: Analyzed and interpreted the data.
Funding statement
This study was supported by Wuhan Applied Foundational Frontier Project (2020020601012244), Special Research Project of Hubei Key Laboratory (2022-7) and excellent discipline team of Hubei University of Chinese Medicine (5431100702020811).
Data availability statement
Data associated with this study has been deposited at “NCBI Sequence Read Archive (SRA) database” under the accession number “PRJNA873889”.
Declaration of interest's statement
The authors declare no competing interests.
Contributor Information
Jianhui Chen, Email: cjh2012hbwh@163.com.
Chengxia Su, Email: 995981764@qq.com.
References
- 1.Black C.J., Ford A.C. Global burden of irritable bowel syndrome: trends, predictions and risk factors. Nat. Rev. Gastroenterol. Hepatol. 2020;17:473–486. doi: 10.1038/s41575-020-0286-8. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y.L., Liu J.S. Irritable bowel syndrome in China: a review on the epidemiology, diagnosis, and management. Chin. Med. J. (Engl) 2021;134:1396–1401. doi: 10.1097/CM9.0000000000001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holtmann G.J., Ford A.C., Talley N.J. Pathophysiology of irritable bowel syndrome. Lancet Gastroenterol. Hepatol. 2016;1:133–146. doi: 10.1016/S2468-1253(16)30023-1. [DOI] [PubMed] [Google Scholar]
- 4.Ng Q.X., Soh A., Loke W., Lim D.Y., Yeo W.S. The role of inflammation in irritable bowel syndrome (IBS) J. Inflamm. Res. 2018;11:345–349. doi: 10.2147/JIR.S174982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Palma G., Lynch M.D., Lu J., Dang V.T., Deng Y., Jury J., Umeh G., Miranda P.M., Pigrau P.M., Sidani S., Pinto-Sanchez M.I., Philip V., McLean P.G., Hagelsieb M.G., Surette M.G., Bergonzelli G.E., Verdu E.F., Britz-McKibbin P., Neufeld J.D., Collins S.M., Bercik P. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaf6397. [DOI] [PubMed] [Google Scholar]
- 6.Das A., O'Herlihy E., Shanahan F., O'Toole P.W., Jeffery I.B. The fecal mycobiome in patients with Irritable Bowel Syndrome. Sci. Rep. 2021;11:124. doi: 10.1038/s41598-020-79478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flemer B., Warren R.D., Barrett M.P., Cisek K., Das A., Jeffery I.B., Hurley E., O'Riordain M., Shanahan F., O'Toole P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67:1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erawijantari P.P., Mizutani S., Shiroma H., Shiba S., Nakajima T., Sakamoto T., Saito Y., Fukuda S., Yachida S., Yamada T. Influence of gastrectomy for gastric cancer treatment on faecal microbiome and metabolome profiles. Gut. 2020;69:1404–1415. doi: 10.1136/gutjnl-2019-319188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas L.R., Grande B.M., Galvez A., Perez P.R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: a state of the science review. APMIS. 2017;125:3–10. doi: 10.1111/apm.12609. [DOI] [PubMed] [Google Scholar]
- 10.Fourie N.H., Wang D., Abey S.K., Sherwin L.B., Joseph P.V., Rahim-Williams B., Ferguson E.G., Henderson W.A. The microbiome of the oral mucosa in irritable bowel syndrome. Gut Microb. 2016;7:286–301. doi: 10.1080/19490976.2016.1162363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pittayanon R., Lau J.T., Yuan Y., Leontiadis G.I., Tse F., Surette M., Moayyedi P. Gut microbiota in patients with irritable bowel syndrome-A systematic review. Gastroenterology. 2019;157:97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 12.Jabbar K.S., Dolan B., Eklund L., Wising C., Ermund A., Johansson A., Tornblom H., Simren M., Hansson G.C. Association between Brachyspira and irritable bowel syndrome with diarrhoea. Gut. 2021;70:1117–1129. doi: 10.1136/gutjnl-2020-321466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J., Xiong T., Grabauskas G., Owyang C. Mucosal serotonin reuptake transporter expression in irritable bowel syndrome is modulated by gut microbiota via mast cell-prostaglandin E2. Gastroenterology. 2022;162:1962–1974. doi: 10.1053/j.gastro.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillestad E., van der Meeren A., Nagaraja B.H., Bjorsvik B.R., Haleem N., Benitez-Paez A., Sanz Y., Hausken T., Lied G.A., Lundervold A., Berentsen B. Gut bless you: the microbiota-gut-brain axis in irritable bowel syndrome. World J. Gastroenterol. 2022;28:412–431. doi: 10.3748/wjg.v28.i4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei L., Zhou J., Su Y., Mao K., Wu J., Zhu C., He L., Cui Y. Gut microbiota composition and functional prediction in diarrhea-predominant irritable bowel syndrome. BMC Gastroenterol. 2021;21:105. doi: 10.1186/s12876-021-01693-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitamoto S., Kamada N. Periodontal connection with intestinal inflammation: microbiological and immunological mechanisms. PERIODONTOL. 2000 2022;89:142–153. doi: 10.1111/prd.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan R., Zhu S., Wang B., Duan L. Alterations of gut microbiota in patients with irritable bowel syndrome based on 16S rRNA-targeted sequencing: a systematic review. Clin. Transl. Gastroenterol. 2019;10:e12. doi: 10.14309/ctg.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maharshak N., Ringel Y., Katibian D., Lundqvist A., Sartor R.B., Carroll I.M., Ringel-Kulka T. Fecal and mucosa-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Dig. Dis. Sci. 2018;63:1890–1899. doi: 10.1007/s10620-018-5086-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z., Xu C.M., Liu Y.X., Wang X.Q., Zhang L., Li M., Zhu S.W., Xie Z.J., Wang P.H., Duan L.P., Zhu H.Q. Characteristic dysbiosis of gut microbiota of Chinese patients with diarrhea-predominant irritable bowel syndrome by an insight into the pan-microbiome. Chin. Med. J. (Engl) 2019;132:889–904. doi: 10.1097/CM9.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S.M., Kim N., Yoon H., Kim Y.S., Choi S.I., Park J.H., Lee D.H. Compositional and functional changes in the gut microbiota in irritable bowel syndrome patients. GUT LIVER. 2021;15:253–261. doi: 10.5009/gnl19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ford A.C., Sperber A.D., Corsetti M., Camilleri M. Irritable bowel syndrome. Lancet (N. Am. Ed.) 2020;396:1675–1688. doi: 10.1016/S0140-6736(20)31548-8. [DOI] [PubMed] [Google Scholar]
- 22.Stojanov S., Berlec A., Strukelj B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. 2020;8 doi: 10.3390/microorganisms8111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhuang X., Tian Z., Li L., Zeng Z., Chen M., Xiong L. Fecal microbiota alterations associated with diarrhea-predominant irritable bowel syndrome. Front. Microbiol. 2018;9:1600. doi: 10.3389/fmicb.2018.01600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Smet J., Hendrix H., Blasdel B.G., Danis-Wlodarczyk K., Lavigne R. Pseudomonas predators: understanding and exploiting phage-host interactions. Nat. Rev. Microbiol. 2017;15:517–530. doi: 10.1038/nrmicro.2017.61. [DOI] [PubMed] [Google Scholar]
- 25.Wang J., Wang P., Li D., Hu X., Chen F. Beneficial effects of ginger on prevention of obesity through modulation of gut microbiota in mice. Eur. J. Nutr. 2020;59:699–718. doi: 10.1007/s00394-019-01938-1. [DOI] [PubMed] [Google Scholar]
- 26.Yang X., He Z., Hu R., Yan J., Zhang Q., Li B., Yuan X., Zhang H., He J., Wu S. Dietary beta-carotene on postpartum uterine recovery in mice: crosstalk between gut microbiota and inflammation. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.744425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salje J. Cells within cells: Rickettsiales and the obligate intracellular bacterial lifestyle. Nat. Rev. Microbiol. 2021;19:375–390. doi: 10.1038/s41579-020-00507-2. [DOI] [PubMed] [Google Scholar]
- 28.Li K.J., Chen Z.L., Huang Y., Zhang R., Luan X.Q., Lei T.T., Chen L. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir. Res. 2019;20:272. doi: 10.1186/s12931-019-1246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zafar H., Saier M.J. Gut Bacteroides species in health and disease. Gut Microb. 2021;13:1–20. doi: 10.1080/19490976.2020.1848158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C., Zhao J., Zhang H., Lee Y.K., Zhai Q., Chen W. Roles of intestinal bacteroides in human health and diseases. Crit. Rev. Food Sci. Nutr. 2021;61:3518–3536. doi: 10.1080/10408398.2020.1802695. [DOI] [PubMed] [Google Scholar]
- 31.Brown E.M., Ke X., Hitchcock D., Jeanfavre S., Avila-Pacheco J., Nakata T., Arthur T.D., Fornelos N., Heim C., Franzosa E.A., Watson N., Huttenhower C., Haiser H.J., Dillow G., Graham D.B., Finlay B.B., Kostic A.D., Porter J.A., Vlamakis H., Clish C.B., Xavier R.J. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe. 2019;25:668–680. doi: 10.1016/j.chom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mantelin S., Desbrosses G., Larcher M., Tranbarger T.J., Cleyet-Marel J.C., Touraine B. Nitrate-dependent control of root architecture and N nutrition are altered by a plant growth-promoting Phyllobacterium sp. Planta. 2006;223:591–603. doi: 10.1007/s00425-005-0106-y. [DOI] [PubMed] [Google Scholar]
- 33.Lundberg J.O., Weitzberg E. Biology of nitrogen oxides in the gastrointestinal tract. Gut. 2013;62:616–629. doi: 10.1136/gutjnl-2011-301649. [DOI] [PubMed] [Google Scholar]
- 34.Rocha B.S., Laranjinha J. Nitrate from diet might fuel gut microbiota metabolism: minding the gap between redox signaling and inter-kingdom communication. Free Radic. Biol. Med. 2020;149:37–43. doi: 10.1016/j.freeradbiomed.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q., Ye J., Fang D., Lv L., Wu W., Shi D., Li Y., Yang L., Bian X., Wu J., Jiang X., Wang K., Wang Q., Hodson M.P., Thibaut L.M., Ho J., Giannoulatou E., Li L. Multi-omic profiling reveals associations between the gut mucosal microbiome, the metabolome, and host DNA methylation associated gene expression in patients with colorectal cancer. BMC Microbiol. 2020;20:83. doi: 10.1186/s12866-020-01762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moroz L.L., Nikitin M.A., Policar P.G., Kohn A.B., Romanova D.Y. Evolution of glutamatergic signaling and synapses. Neuropharmacology. 2021;199 doi: 10.1016/j.neuropharm.2021.108740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Alammar N., Singh R., Nanavati J., Song Y., Chaudhary R., Mullin G.E. Gut microbial dysbiosis in the irritable bowel syndrome: a systematic review and meta-analysis of case-control studies. J. Acad. Nutr. Diet. 2020;120:565–586. doi: 10.1016/j.jand.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 38.Liu H.N., Wu H., Chen Y.Z., Chen Y.J., Shen X.Z., Liu T.T. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: a systematic review and meta-analysis. Dig. Liver Dis. 2017;49:331–337. doi: 10.1016/j.dld.2017.01.142. [DOI] [PubMed] [Google Scholar]
- 39.Ford A.C., Lacy B.E., Talley N.J. Irritable bowel syndrome. N. Engl. J. Med. 2017;376:2566–2578. doi: 10.1056/NEJMra1607547. [DOI] [PubMed] [Google Scholar]
- 40.Wu F., Guo X., Zhang J., Zhang M., Ou Z., Peng Y. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp. Ther. Med. 2017;14:3122–3126. doi: 10.3892/etm.2017.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fremder M., Kim S.W., Khamaysi A., Shimshilashvili L., Eini-Rider H., Park I.S., Hadad U., Cheon J.H., Ohana E. A transepithelial pathway delivers succinate to macrophages, thus perpetuating their pro-inflammatory metabolic state. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109521. [DOI] [PubMed] [Google Scholar]
- 42.Nagao-Kitamoto H., Leslie J.L., Kitamoto S., Jin C., Thomsson K.A., Gillilland M.R., Kuffa P., Goto Y., Jenq R.R., Ishii C., Hirayama A., Seekatz A.M., Martens E.C., Eaton K.A., Kao J.Y., Fukuda S., Higgins P., Karlsson N.G., Young V.B., Kamada N. Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med. 2020;26:608–617. doi: 10.1038/s41591-020-0764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sung J., Coker O.O., Chu E., Szeto C.H., Luk S., Lau H., Yu J. Gastric microbes associated with gastric inflammation, atrophy and intestinal metaplasia 1 year after Helicobacter pylori eradication. Gut. 2020;69:1572–1580. doi: 10.1136/gutjnl-2019-319826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park S.Y., Hwang B.O., Lim M., Ok S.H., Lee S.K., Chun K.S., Park K.K., Hu Y., Chung W.Y., Song N.Y. Oral-gut microbiome Axis in gastrointestinal disease and cancer. Cancers. 2021;13:2124. doi: 10.3390/cancers13092124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at “NCBI Sequence Read Archive (SRA) database” under the accession number “PRJNA873889”.