Abstract
Coronavirus disease 2019 (COVID‐19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which led to the current pandemic. Many factors, including age and comorbidities, influence the severity and mortality of COVID‐19. SARS‐CoV‐2 infection can cause pulmonary vascular dysfunction. The COVID‐19 case‐fatality rate in patients with pulmonary arterial hypertension (PAH) is higher in comparison with the general population. In this study, we aimed to identify pathobiological processes common to COVID‐19 and PAH by utilizing the human protein–protein interactome and whole‐genome transcription data from peripheral blood mononuclear cells (PBMCs) and from lung tissue. We found that there are significantly more interactions between SARS‐CoV‐2 targets and PAH disease proteins than expected by chance, suggesting that the PAH disease module is in the neighborhood of SARS‐CoV‐2 targets in the human interactome. In addition, SARS‐CoV‐2 infection‐induced changes in gene expression significantly overlap with PAH‐induced gene expression changes in both tissues, indicating SARS‐CoV‐2 and PAH may share common transcriptional regulators. We identified many upregulated genes and downregulated genes common to COVID‐19 and PAH. Interestingly, we observed different co‐regulation patterns and dysfunctional signaling pathways in PBMCs versus lung tissue. Endophenotype enrichment analysis revealed that genes regulating fibrosis, inflammation, hypoxia, oxidative stress, immune response, and thromboembolism are significantly enriched in the COVID‐19‐PAH co‐expression modules. We examined the network proximity of the targets of repositioned drugs for COVID‐19 to the co‐expression modules in PBMCs and lung tissue, and identified 42 drugs that can be potentially used for COVID‐19 patients with PAH as a comorbidity. The uncovered common pathobiological pathways are crucial for discovering therapeutic targets and designing tailored treatments for COVID‐19 patients who also have PAH.
Keywords: COVID‐19, networks, Omics data, pathobiological processes, pulmonary arterial hypertension
INTRODUCTION
Coronavirus disease 2019 (COVID‐19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) that has caused the current global pandemic. The spectrum of COVID‐19 disease severity ranges from asymptomatic to critical. Data show that comorbidities such as hypertension, diabetes, asthma, and other pulmonary diseases in COVID‐19 patients are associated with higher risk of severe illness and increasing mortality. 1 , 2 , 3
Pulmonary arterial hypertension (PAH) is a progressive disorder characterized by pulmonary vascular remodeling leading to increased pulmonary vascular resistance and pulmonary arterial pressure. SARS‐CoV‐2 can infect vascular endothelium and cause endothelial dysfunction, 4 , 5 , 6 which impairs vascular function. Although some studies based on small numbers of patients show that PAH patients may be at a lower risk for severe COVID‐19, 7 the case‐fatality rate related to COVID‐19 was found to be significantly higher in patients with PAH compared to the general population. 8 , 9 In addition, SARS‐CoV‐2 infection can cause long‐term pulmonary complications, such as pulmonary fibrosis 10 and, importantly, PAH. 11
Several clinical studies have addressed the comorbidities underlying COVID‐19 and its cardiovascular and pulmonary complications 3 , 12 , 13 , 14 ; however, the common molecular‐level mechanisms accounting for these comorbidities and complications have not been fully explored. In this study, we attempt to comprehend the molecular basis of the comorbidity of COVID‐19 and PAH and post‐COVID‐19 pulmonary complications by an integrated omics analysis that includes whole‐genome transcription data and protein‐protein interactions as depicted by the human interactome. We evaluated the network proximity between SARS‐COV‐2 targets and the PAH disease module in the human interactome as well as gene co‐expression induced by COVID‐19 and PAH. We found common endophenotypes underlying these two diseases by performing enrichment analysis in COVID‐19‐PAH co‐expression modules. We also identified potential drugs for COVID‐19 patients with PAH as a comorbidity by assessing the network proximity of drug targets to the co‐expression modules. The identified common molecular alterations and pathways could potentially accelerate drug development and shed light on the design of tailored treatment for COVID‐19 patients with PAH as a comorbidity.
METHODS
Consolidated human protein–protein interactome
We used the comprehensive human protein–protein interactome consolidated in our previous study. 15 This human interactome contains physical, macromolecular interaction data from different human sources, including protein–protein interactions, protein complexes, kinase‐substrate interactions, and signaling pathways. High‐quality protein–protein interactions are identified from several high‐throughput yeast‐two‐hybrid studies, mass spectrometry, as well as the literature. 16 , 17 , 18 , 19 The latest large‐scale binary protein‐protein interactions were retrieved from HuRI. 20 In addition, experimental signaling interactions and kinase‐substrate interactions, as well as high‐quality literature‐based signaling interactions, were also incorporated. 21 , 22 , 23 , 24 This version of the consolidated human interactome has 16,470 proteins and 233,957 interactions.
Compiling SARS‐CoV‐2 targets and PAH disease genes
We retrieved the 332 human protein targets of SARS‐CoV‐2 from a previous study. 25 We mapped these SARS‐CoV‐2 targets to our human interactome, and 326 remained for further analysis. We first retrieved 276 PAH disease genes from our previous work, 15 which were compiled from the literature, Phenopedia in the HuGE Navigator, 26 the Human Gene Mutation Database, Online Mendelian Inheritance in Man, and Human Phenotype Ontology databases. We also obtained PAH‐related genes from DisGeNET. 27 Altogether, 357 were obtained by integrating these resources. We next mapped these genes (gene products) to the human interactome and denoted the subnetwork formed by the PAH disease proteins as a PAH disease module.
Gene expression data of COVID‐19 and PAH
We retrieved an RNA‐seq dataset of the human peripheral blood mononuclear cells (PBMCs) in a group of 16 COVID‐19 patients (four with moderate symptoms and 12 with severe symptoms) and 17 healthy controls from GSE152418. 28 We also retrieved the RNA‐seq whole transcriptomes of lung biopsies from 31 deceased COVID‐19 patients and 10 healthy individuals (GSE183533). 29 An R package, EdgeR, was used to identify differentially expressed genes in COVID‐19. 30 A gene is said to be significantly differentially expressed if the corresponding false discovery rate (FDR) is less than 0.05 and the fold change is greater than 2. Differentially expressed genes in PBMCs of PAH patients were compiled from a meta‐analysis of blood genome‐wide expression profiling studies in PAH. 31 In addition, we used the microarray data of lung tissue from 58 PAH patients and 25 healthy controls from GSE117261 32 to identify differentially expressed genes in PAH using the R package, Limma. 33 A gene is said to be significantly differentially expressed if the adjusted p‐value is less than 0.05.
Network proximity for drug identification
To determine which drugs that have been experimentally demonstrated to have effects on COVID‐19 can be potentially used for COVID‐19 patients with PAH, we characterized the proximity of the targets of drugs to the COVID‐19‐PAH co‐expression modules using a network proximity measure. 15 This parameter is defined as the average minimum shortest path length in the interactome from the targets of a drug T to the co‐expression module S:
where d(t, s) is the shortest path length in the human interactome from drug target t to gene s in a COVID‐19‐PAH co‐expression module. The significance of the network proximity between the targets of a drug and a COVID‐19‐PAH co‐expression module was evaluated by creating 1000 random modules of the same size and comparing the observed proximity value with the null model (random control) through fitting normal distributions. All p‐values were adjusted by the Benjamini‐Hochberg procedure when applicable.
RESULTS
Construction of COVID‐19 and PAH bipartite network
We first collected 332 SARS‐CoV‐2 human protein targets (i.e., host proteins that bind to 26 SARS‐CoV‐2 proteins) from a previous study, 25 326 of which can be mapped to the human protein interactome to form a distinct COVID‐19 disease module. We also compiled PAH disease genes from different databases (Section 2) among which 357 were mapped to the human interactome to create a PAH disease module. We found 8 overlapping proteins between SARS‐CoV‐2 targets and PAH disease genes, which is not statistically significant based on a hypergeometric test. This observation is consistent with our finding in a previous study that SARS‐CoV‐2 targets do not significantly overlap with any disease module (the proteins associated with a particular disease). 34 We next constructed a bipartite network of SARS‐CoV‐2 targets and PAH disease proteins (Figure 1). Although the COVID‐19 disease module and PAH disease module have few overlapping proteins (nodes), there are significantly more interactions (edges) (409, Figure 2a) between the two modules than random expectation (p = 7.8e‐16). The largest connected component (LCC) of the bipartite network is also significantly larger than the LCC of a bipartite network induced by two random protein sets of the same sizes (p = 1.6e‐5, Figure 2b). These results indicate that the COVID‐19 and PAH modules are adjacent in the human interactome.
Figure 1.
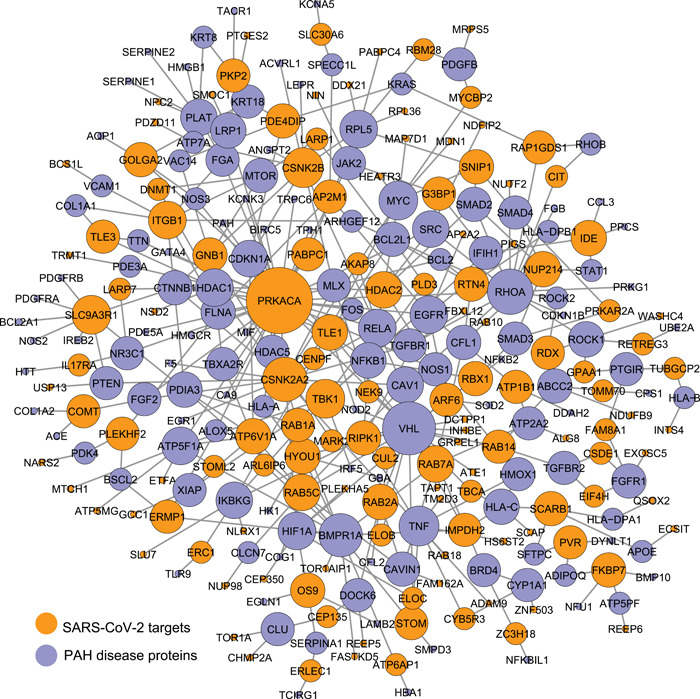
The bipartite network of SARS‐CoV‐2 targets and PAH disease proteins. The size of a node is proportional to its degree (i.e. number of interactors). PAH, pulmonary arterial hypertension; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Figure 2.
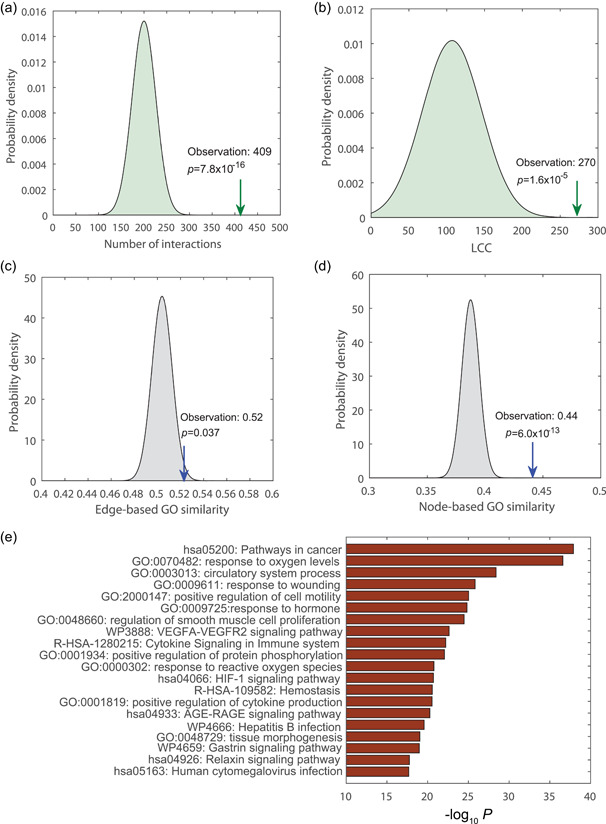
PAH module is in the neighborhood of SARS‐CoV‐2 targets. (a) There are significantly more interactions between SARS‐CoV‐2 targets and PAH disease proteins than random expectation. (b) GO terms and functional pathways enriched in the COVID‐19‐PAH bipartite network. (c) Edge‐based GO similarity between SARS‐CoV‐2 targets and PAH disease proteins. (d) Node‐based GO similarity between SARS‐CoV‐2 targets and PAH disease proteins. COVID‐19, coronavirus disease 2019; PAH, pulmonary arterial hypertension; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
To evaluate whether or not SARS‐CoV‐2 targets and PAH disease proteins have similar functions, we used GS2 (GO‐based similarity of gene sets) 35 to quantify the semantic GO similarity of SARS‐CoV‐2 target and PAH disease protein pairs that themselves interact (i.e., edge‐based GO similarity) and the semantic GO similarity of all SARS‐CoV‐2 target and PAH disease protein pairs (i.e., node‐based GO similarity). From Figure 2c, we can see that compared to randomly selected interacting protein pairs, SARS‐CoV‐2 target and PAH disease protein pairs that interact have significantly higher GO similarity. In fact, the semantic GO similarity of any SARS‐CoV‐2 target and PAH disease protein pair (no matter whether they interact or not) is also significantly higher than that of randomly selected protein pairs (Figure 2d). To determine which pathways are commonly enriched in this bipartite network, we performed a pathway enrichment analysis using Metascape. 36 We found that many pathways are enriched in this network (Figure 2d), such as “response to oxygen levels”, “response to oxidative stress”, and “the HIF‐1 signaling pathway”. Indeed, consistent with this observation, a few studies have shown that COVID‐19 patients have severely increased levels of oxidative stress and oxidant damage. 37 , 38
Gene expression induced by COVID‐19 and PAH
Inspired by the proximity of SARS‐CoV‐2 targets to the PAH disease module, we hypothesized that there may be some common transcription factors regulating downstream gene expression induced by COVID‐19 and PAH. We first identified differentially expressed genes in COVID‐19 from a RNA‐seq dataset of PBMCs 28 (Figure S1). After mapping them to the human interactome, there are 4717 genes differentially expressed in COVID‐19 patients vs healthy controls, among which 2394 are upregulated protein‐coding genes and 2323 are downregulated protein‐coding genes. We compared these genes with the 1085 differentially expressed genes (427 upregulated genes and 658 downregulated genes) in PBMCs of PAH patients from a meta‐analysis study 31 and found that there are a significant number of overlapping differentially expressed genes (451) in COVID‐19 and PAH (Table 1). In addition, there are 175 upregulated genes and 195 downregulated genes common to both COVID‐19 and PAH, which are statistically significant (p = 8.8e‐42 and 3.8e‐26, respectively) (Figure 3a).
Table 1.
Overlapping analysis between differentially expressed genes in COVID‐19 and PAH.
| PAH disease genes (357) | PAH‐PBMC (1085) | PAH‐Lung (2863) | |
|---|---|---|---|
| SARS‐CoV‐2 targets (326) | 8 | 23 | 63 |
| COVID‐19‐PBMC (4717) | 88 | 451 (p = 1.3e‐21) | 1016 (p = 1.0e‐18) |
| COVID‐19‐Lung (3470) | 122 (p = 4.8e‐09) | 185 | 728 (p = 8.5e‐05) |
Abbreviations: COVID‐19, coronavirus disease 2019; PAH, pulmonary arterial hypertension; PBMC, peripheral blood mononuclear cell; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Figure 3.
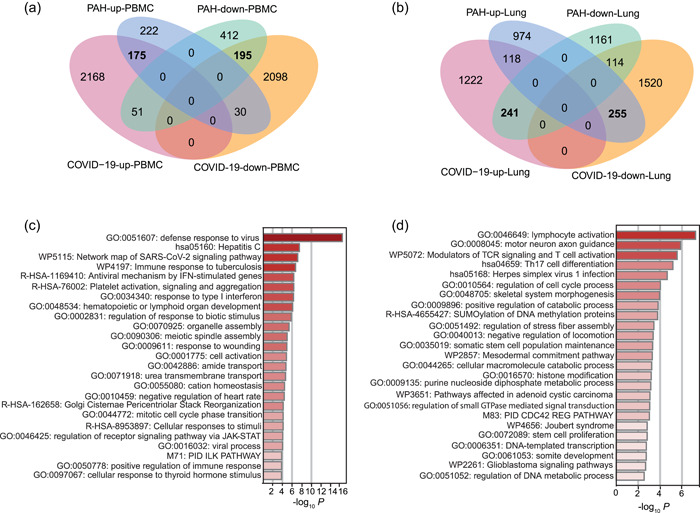
Overlapping genes and pathways in COVID‐19 and PAH. Significant overlaps are highlighted in bold. (a) Venn diagram of overlapping differentially expressed genes in PBMCs of COVID‐19 and PAH. (b) Venn diagram of overlapping differentially expressed genes in lung tissue of COVID‐19 and PAH. (c) Enriched functions and pathways in the co‐expression gene set COVID‐19‐PAH‐up‐PBMC. (d) Enriched functions and pathways in the co‐expression gene set COVID‐19‐PAH‐down‐PBMC. COVID‐19, coronavirus disease 2019; PAH, pulmonary arterial hypertension; PBMC, peripheral blood mononuclear cell; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
In the transcriptomes of lung tissue from COVID‐19 patients, 29 we identified 3470 differentially expressed genes, among which 1581 are upregulated and 1889 are downregulated. We then identified differentially expressed genes in PAH using the microarray data from lung tissues 32 (Figure S2). In this case, we found 2863 protein‐coding genes that are differentially expressed after FDR adjustment (FDR < 0.05). Among these genes, 1347 are upregulated and 1516 are downregulated. We compared the transcription profiles of the two diseases in lung tissue and found that there are a significant number of overlapping differentially expressed genes (728) between COVID‐19 and PAH (Table 1). We identified 118 upregulated genes and 114 downregulated genes common to both COVID‐19 and PAH. There are 241 genes upregulated in COVID‐19 and downregulated in PAH and 255 genes downregulated in COVID‐19 and upregulated in PAH, which are significant (p = 2.7e‐16 and 3.9e‐17, respectively) (Figure 3b). The co‐regulated genes in COVID‐19 and PAH are provided in File S2.
We next focused on four significant co‐expression gene sets affected by COVID‐19 and PAH: COVID‐19‐PAH‐up‐PBMC, COVID‐19‐PAH‐down‐PBMC, COVID‐19‐up‐PAH‐down‐Lung, COVID‐19‐down‐PAH‐up‐Lung. We then performed GO function and pathway enrichment analysis using Metascape for each of the four co‐expression gene sets and found different pathways enriched in the gene sets (Figure 3c). Specifically, in COVID‐19‐PAH‐up‐PBMC we found a few significant GO terms related to virus response, such as “defense response to virus,” “regulation of response to type I interferon,” “response to biotic stimulus,” and “positive regulation of immune response.” SARS‐CoV‐2, indeed, can inhibit various steps in type I interferon production and response and can impair type I interferon activity. 39 In addition, “regulation of receptor signaling pathway via JAK‐STAT” is significant. SARS‐CoV‐2 infection triggers inflammation via the JAK‐STAT pathway, and, therefore, JAK‐STAT signaling may be a potential therapeutic target for developing therapies for COVID‐19. 40 , 41 The JAK‐STAT signaling pathway is also overactivated in the pulmonary arteries of patients as chronic inflammation contributes to pulmonary artery remodeling. 42 The COVID‐19‐PAH‐down‐PBMC gene set is significantly enriched with “lymphocyte activation.” Indeed, SARS‐CoV‐2 can induce lymphocyte subset changes. 43 In addition, some immune‐related pathways and T cell pathways are significant (Figure 3d). A recent study based on single cell omics data confirmed that T cell populations rather than B cell populations are significantly reduced in critically ill COVID‐19 patients. 44 , 45
In the COVID‐19‐up‐PAH‐down‐Lung gene set (File S2 and Figure S3a), many metabolic processes are significant, such as fatty acid metabolism, tryptophan metabolism, and glyoxylate and dicarboxylate metabolism. In both COVID‐19 and PAH, metabolic perturbation and reprogramming are intimately connected to the mechanisms of disease pathogenesis and pathophysiology. 46 , 47 In addition, we found “Neutrophil degranulation” is significantly enriched in COVID19‐up‐PAH‐down‐Lung genes, which is consistent with the recent finding that neutrophils play an important role in the pathobiology of COVID‐19, particularly in those with severe disease courses. 48 In the COVID‐19‐down‐PAH‐up‐Lung gene set (Figure S3b), some relevant GO terms are significant, as well, such as “cellular response to cytokine stimulus,” “positive regulation of protein phosphorylation,” and “SMAD protein signal transduction.”
Biological networks induced by COVID‐19 and PAH
We then mapped the differentially expressed gene sets COVID‐19‐PAH‐up‐PBMC and COVID‐19‐PAH‐down‐PBMC to the human interactome and obtained a network of 128 proteins and 126 interactions. The LCC of the network is shown in Figure 4a. This protein–protein interaction network represents the dysfunctional network module underlying both COVID‐19 and PAH in PBMCs. We also mapped gene sets for COVID‐19‐up‐PAH‐down‐Lung and COVID‐19‐down‐PAH‐up‐Lung to the human interactome and obtained a network of 209 proteins and 197 interactions. The LCC of the network is shown in Figure 4b, representing the dysfunctional network module underlying both COVID‐19 and PAH in lung tissue.
Figure 4.
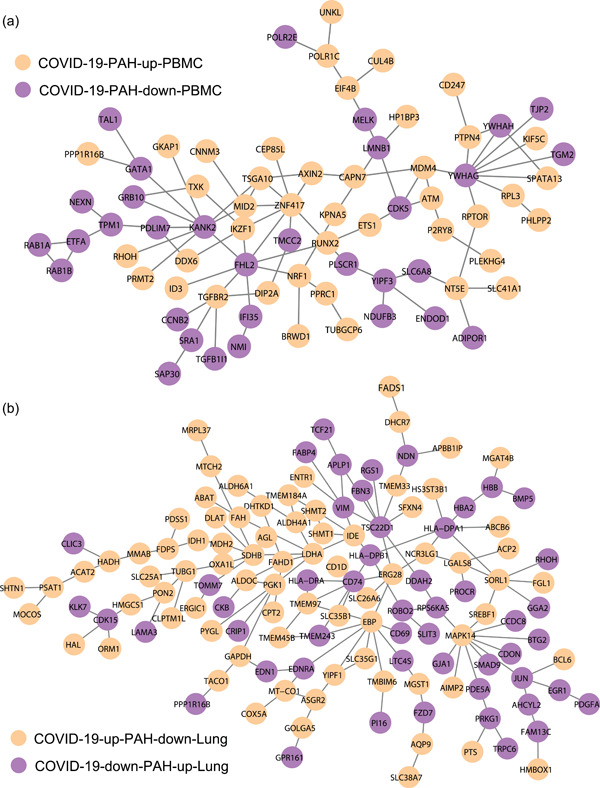
Dysfunctional network modules of COVID‐19‐PAH in PBMCs (a) and lung tissue (b). COVID‐19, coronavirus disease 2019; PAH, pulmonary arterial hypertension; PBMC, peripheral blood mononuclear cell.
Previous studies have shown that PAH is linked with some endophenotypes, such as fibrosis, hypoxia, and thrombosis. 49 To determine whether there are some common endophenotypes enriched in COVID‐19 and PAH, we next collected the genes associated with hypoxia from a microarray gene expression dataset 50 and the genes associated with cell proliferation and immune response from the GO database (geneontology.org). The genes associated with other endophenotypes were compiled from Phenopedia 26 and DisGeNet. 27 Pulmonary thromboembolism may also represent a pathobiological condition seen in both COVID‐19 and PAH. 51 For this reasson, we considered three additional related endophenotypes: pulmonary embolism, thromboembolism and venous thrombosis based on the availablity of associated genes in the databases and their association with complicated COVID‐19 infection. The endophenotype‐associated genes can be found in File S3. We performed a hypergeometric test and found that COVID‐19 and PAH share some common endophenotypes, such as fibrosis, inflammation, hypoxia, oxidative stress, immune response, and thromboembolism (Figure 5a).
Figure 5.
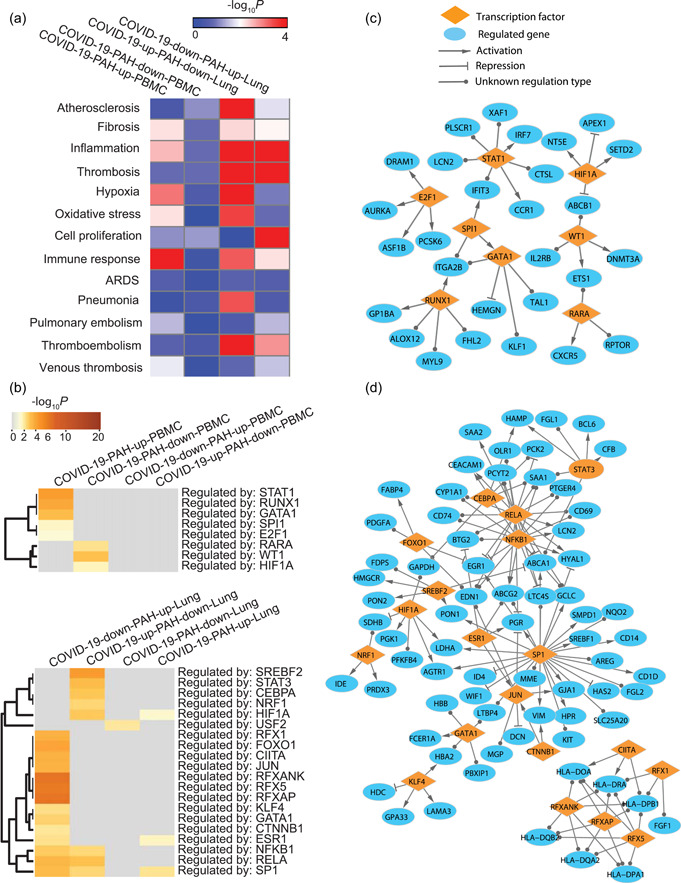
Common endophenotypes and regulators in COVID‐19 and PAH. (a) Endophenotype enrichment in each of the co‐expression gene sets. (b) Transcription factors whose targets are significantly enriched in the co‐expression gene sets. (c) Transcriptional regulatory network for COVID‐19‐PAH in PBMCs. (d) Transcriptional regulatory network for COVID‐19‐PAH in lung tissue. COVID‐19, coronavirus disease 2019; PAH, pulmonary arterial hypertension; PBMC, peripheral blood mononuclear cell.
We also used the manually curated database TRUST to examine the common transcription factors in PAH and COVID‐19, 52 and found some common transcriptional regulators, such as RUNX1, HIF1A, and STAT1 in PBMCs, and HIF1A, JUN, STAT3, RFX5, NFKB1 in lung tissue (Figure 5b). A transcriptional regulatory network was constructed for the regulation of COVID‐19 and PAH in PBMCs (Figure 5c). The transcriptional regulatory network for COVID‐19 and PAH in lung tissue is shown in Figure 5d. These transcriptional regulatory networks represent the common regulatory pathways underlying COVID‐19 and PAH.
Drug selection for COVID‐19 patients with PAH
There have been many studies focusing on drug repositioning for COVID‐19. Given the comorbitity of COVID‐19 and PAH, we next aimed to identify which repositioned drugs for COVID‐19 could be particulary useful for COVID‐19 patients with PAH as a comorbidity. A previous study evaluated 6710 clinical and preclinical drugs by immunocytofluorescence‐based screening to identify SARS‐CoV‐2 infection inhibitors 53 and characterized those with strong effects, weak effects, very weak effects, and no effects on COVID‐19. We collected 631 drugs that have been experimentally demonstrated to have strong, weak, or very weak effects on COVID‐19 from this study and another set of 77 drugs that have experimental strong or weak effects on COVID‐19, 34 and examined which of them could be potentially repurposed for patients with COVID‐19 who also had PAH. Only drugs that have at least one target included in the human protein‐protein interactome were considered, leading to a total of 491 drug candidates under consideration. We used these drugs as candidates and applied a network proximity method 15 to determine whether the targets of these drugs are significantly close to the dysfunctional network modules of COVID‐19 and PAH in the human protein‐protein interactome. After adjusting the p‐value by the Benjamini‐Hochberg correction, in total, we found 30 drugs whose targets are significantly proximate to the COVID‐19‐PAH dysfunctional network modules in PBMCs (Table 2). In lung tissue, the targets of 21 drugs are significantly close the dysfunctional network module of COVID‐19 and PAH (Table S1). There are 9 drugs common to the two tissues (p = 6.1e‐7, hypergeometric test). Altogether, 42 drugs, we suggest, can be potentially used specifically for COVID‐19 patients with comorbid PAH.
Table 2.
A list of drugs that have biologically plausible potential to be used for COVID‐19 patients with PAH as a comorbidity based on data from PBMCs.
| DrugBank/ChEMBL/PubChem ID | Drug name | Description | Network proximity | Adjusted p‐value |
|---|---|---|---|---|
| DB17060 | U‐0126 | MEK1/2 inhibitor | 1.06 | 2.7e‐8 |
| CHEMBL278041 | SB‐202190 | p38 MAPK inhibitor | 1.07 | 9.6e‐07 |
| DB08597 | Dorsomorphin | AMPK inhibitor | 1.23 | 5.7e‐04 |
| DB11794 | Berzosertib | ATR kinase inhibitor | 0.75 | 8.8e‐04 |
| DB05013 | Ingenol mebutate | PKC activator | 1.00 | 4.2e‐03 |
| DB00619 | Imatinib | Tyrosine kinase inhibitor | 1.13 | 4.5e‐03 |
| PubChemCID135398499 | OTS167 | MELK inhibitor | 0.00 | 4.6e‐03 |
| DB04879 | Vatalanib | VEGFR inhibitor | 1.00 | 6.3e‐03 |
| DB04865 | Omacetaxine mepesuccinate | Protein synthesis inhibitor | 0.00 | 6.3e‐03 |
| DB13874 | Enasidenib | Isocitrate dehydrogenase inhibitor | 0.00 | 6.3e‐03 |
| PubChemCID15953870 | KU‐60019 | ATM kinase inhibitor | 0.00 | 6.3E‐03 |
| DB01169 | Arsenic trioxide | Apoptosis stimulant | 1.14 | 6.5e‐03 |
| CHEMBL123292 | Cycloheximide | Protein synthesis inhibitor | 0.00 | 6.5e‐03 |
| DB15408 | Silmitasertib | Bcasein kinase inhibitor | 1.00 | 8.6e‐03 |
| DB12180 | Apitolisib | mTOR/PI3K inhibitor | 1.25 | 0.011 |
| DB17046 | PI‐103 | mTOR/PI3K inhibitor | 1.17 | 0.012 |
| DB12986 | VS‐5584 | mTOR/PI3K inhibitor | 1.2 | 0.030 |
| DB11851 | Bafetinib | Bcr‐Abl kinase inhibitor | 1.00 | 0.035 |
| DB05294 | Vandetanib | EGFR/VEGFR inhibitor | 1.58 | 0.035 |
| DB07662 | PD‐168393 | EGFR inhibitor | 1.00 | 0.035 |
| PubChemCID6852167 | PIK‐93 | PI3K inhibitor | 1.00 | 0.040 |
| DB11973 | Tesevatinib | EGFR/VEGFR inhibitor | 1.20 | 0.040 |
| DB05243 | XL019 | JAK inhibitor | 1.00 | 0.040 |
| DB16828 | MK‐2206 | AKT inhibitor | 1.00 | 0.042 |
| DB12703 | Omipalisib | mTOR/PI3K inhibitor | 1.20 | 0.042 |
| DB00128 | Aspartic acid | Metallic radical formation stimulant | 1.60 | 0.042 |
| DB04813 | Bithionol | Autotaxin inhibitor | 1.00 | 0.042 |
| DB01396 | Digitoxin | ATPase inhibitor | 1.48 | 0.044 |
| DB08889 | Carfilzomib | Proteasome inhibitor | 1.58 | 0.045 |
| DB01078 | Deslanoside | Na/K‐ATPase inhibitor | 1.38 | 0.049 |
Abbreviations: COVID‐19, coronavirus disease 2019; PAH, pulmonary arterial hypertension; PBMC, peripheral blood mononuclear cell; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
In this study, we explored the pathophysiological relationships between COVID‐19 and PAH in the human protein‐protein interactome and integrated transcriptomic data from COVID‐19 and PAH patients to identify common pathobiological processes and transcriptional regulators underlying the two diseases. We found that the PAH disease module is in the neighborhood of SARS‐CoV‐2 targets in the human interactome, and that SARS‐CoV‐2 infection‐induced gene expression overlaps significantly with PAH‐induced gene expression in both PBMCs and lung tissue. Through function and pathway enrichment analyses, we found some common pathways and transcriptional regulators, such as RUNX1, HIF1A, JUN, STAT1, STAT3, and NFKB1. We also identified some significant common endophenotypes in COVID‐19 and PAH, such as fibrosis, inflammation, hypoxia, oxidative stress, immune response, and thromboembolism. Uncovering such common molecular alterations and pathways is crucial for discovering tailored treatment for COVID‐19 patients with comorbid PAH. We examined the network proximity of the targets of effective, repositioned drugs for COVID‐19 to the dysfunctional network modules in COVID‐19‐PAH, and identified 42 drugs that potentially can be used for COVID‐19 patients with PAH.
While this work is a pilot study investigating common molecular mechanisms underlying COVID‐19 and PAH, there are some potential limitations. First, the transcriptomic datasets of COVID‐19 and PAH are from populations of cells and bulk tissue. There are increasingly available single‐cell RNA‐seq datasets for both COVID‐19 and PAH; however, open‐access to single‐cell datasets from consistent cell types for both diseases are currently unavailable. Although we found significant overlap between COVID‐19‐induced gene expression and PAH‐induced gene expression, it would be helpful to investigate the comorbidity of COVID‐19 and PAH using single‐cell data from the same cell types when available in the future. Second, the gene expression analysis was performed on samples obtained at a single time point and at different phases of the patients’ illnesses, which may confound efforts to identify pathways or key drug targets that should be considered in a given individual. In addition, clinical data regarding COVID‐19 and PAH increasingly suggest that right ventricular (RV) failure is more commonly seen in COVID‐19 and PAH than pulmonary vascular remodeling. Comparison of COVID‐19 and PAH in RV tissue would, therefore, be interesting when data ultimately become available.
Pulmonary thromboembolism may represent another pathobiological condition observed in both COVID‐19 and chronic thromboembolic pulmonary hypertension (CTEPH). 8 , 51 , 54 We, therefore, collected CTEPH‐associated genes from DisGeNet 27 and found that there are a significant number of interactions between CTEPH genes and SARS‐CoV‐2 targets, as well (Figure S4). However, the availability of gene expression datasets from RV tissue for CTEPH is very limited. Endophenotype enrichment analysis indicates that thromboembolism is significantly enriched in the co‐expression gene sets of COVID‐19 and PAH in lung tissue. Comparing gene expression in RV tissue from patients with CTEPH versus COVID‐19 with pulmonary embolism is a topic worthy of exploration as datasets become available, as well.
AUTHOR CONTRIBUTIONS
Rui‐Sheng Wang: was responsible for the concept, design, analysis and data interpretation. Joseph Loscalzo: interpreted the results. Rui‐Sheng Wang: drafted the article, and Joseph Loscalzo: revised it critically. Both authors edited and approved the final version to be submitted.
CONFLICTS OF INTEREST STATEMENT
Joseph Loscalzo is scientific co‐founder of Scipher Medicine, Inc., which uses network medicine analyses to identify disease biomarkers and potential therapies. Rui‐Sheng Wang has no conflicts of interest to declare.
ETHICS STATEMENT
N/A.
Supporting information
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
This work is supported in part by National Institutes of Health Grants (HL119145, HL155107, HL155096, HG007690, and GM107618), and by American Heart Association Grants (D700382 and 957729). The authors wish to thank Stephanie Tribuna for expert secretarial assistance.
Wang R‐S, Loscalzo J. Uncovering common pathobiological processes between COVID‐19 and pulmonary arterial hypertension by integrating Omics data. Pulm Circ. 2023;13:e12191. 10.1002/pul2.12191
DATA AVAILABILITY STATEMENT
N/A. Publicly available databases were used.
REFERENCES
- 1. Skevaki C, Karsonova A, Karaulov A, Fomina D, Xie M, Chinthrajah S, Nadeau KC, Renz H. SARS‐CoV‐2 infection and COVID‐19 in asthmatics: a complex relationship. Nat Rev Immunol. 2021;21:202–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ng WH, Tipih T, Makoah NA, Vermeulen JG, Goedhals D, Sempa JB, Burt FJ, Taylor A, Mahalingam S. Comorbidities in SARS‐CoV‐2 patients: a systematic review and meta‐analysis. mBio. 2021;12:e03647–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clerkin KJ, Fried JA, Raikhelkar J, Sayer G, Griffin JM, Masoumi A, Jain SS, Burkhoff D, Kumaraiah D, Rabbani L, Schwartz A, Uriel N. COVID‐19 and cardiovascular disease. Circulation. 2020;141:1648–55. [DOI] [PubMed] [Google Scholar]
- 4. Huertas A, Montani D, Savale L, Pichon J, Tu L, Parent F, Guignabert C, Humbert M. Endothelial cell dysfunction: a major player in SARS‐CoV‐2 infection (COVID‐19)? Eur Respir J. 2020;56:2001634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonaventura A, Vecchié A, Dagna L, Martinod K, Dixon DL, Van Tassell BW, Dentali F, Montecucco F, Massberg S, Levi M, Abbate A. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID‐19. Nat Rev Immunol. 2021;21:319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang R, Mamun A, Dominic A, Le NT. SARS‐CoV‐2 mediated endothelial dysfunction: the potential role of chronic oxidative stress. Front Physiol. 2021;11:605908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horn EM, Chakinala M, Oudiz R, Joseloff E, Rosenzweig EB. Could pulmonary arterial hypertension patients be at a lower risk from severe COVID‐19? Pulm Circ. 2020;10:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belge C, Quarck R, Godinas L, Montani D, Escribano Subias P, Vachiéry JL, Nashat H, Pepke‐Zaba J, Humbert M, Delcroix M. COVID‐19 in pulmonary arterial hypertension and chronic thromboembolic pulmonary hypertension: a reference centre survey. ERJ Open Res. 2020;6:00520–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JD, Burger CD, Delossantos GB, Grinnan D, Ralph DD, Rayner SG, Ryan JJ, Safdar Z, Ventetuolo CE, Zamanian RT, Leary PJ. A survey‐based estimate of COVID‐19 incidence and outcomes among patients with pulmonary arterial hypertension or chronic thromboembolic pulmonary hypertension and impact on the process of care. Ann Am Thorac Soc. 2020;17:1576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McGroder CF, Zhang D, Choudhury MA, Salvatore MM, D'Souza BM, Hoffman EA, Wei Y, Baldwin MR, Garcia CK. Pulmonary fibrosis 4 months after COVID‐19 is associated with severity of illness and blood leucocyte telomere length. Thorax. 2021;76(12):1242–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raval A, Edwards N, Kant R, Verma V. COVID‐19 as a primary cause of pulmonary arterial hypertension. Chest. 2021;160(4):A2198. [Google Scholar]
- 12. Chung MK, Zidar DA, Bristow MR, Cameron SJ, Chan T, Harding CV, Kwon DH, Singh T, Tilton JC, Tsai EJ, Tucker NR, Barnard J, Loscalzo J. COVID‐19 and cardiovascular disease: from bench to bedside. Circ Res. 2021;128:1214–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fraser E. Long term respiratory complications of covid‐19. BMJ. 2020;370:m3001. [DOI] [PubMed] [Google Scholar]
- 14. Halawa S, Pullamsetti SS, Bangham CRM, Stenmark KR, Dorfmüller P, Frid MG, Butrous G, Morrell NW, de Jesus Perez VA, Stuart DI, O'Gallagher K, Shah AM, Aguib Y, Yacoub MH. Potential long‐term effects of SARS‐CoV‐2 infection on the pulmonary vasculature: a global perspective. Nat Rev Cardiol. 2022;19:314–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang RS, Loscalzo J. Network module‐based drug repositioning for pulmonary arterial hypertension. CPT: Pharmacometrics Syst Pharmacol. 2021;10:994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein‐protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–68. [DOI] [PubMed] [Google Scholar]
- 17. Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis AR, Convery‐Zupan D, Corominas R, Coulombe‐Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere JC, Vega K, Walsh J, Cusick ME, Xia Y, Barabási AL, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M. A proteome‐scale map of the human interactome network. Cell. 2014;159:1212–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, Chessman K, Pal S, Cromar G, Papoulas O, Ni Z, Boutz DR, Stoilova S, Havugimana PC, Guo X, Malty RH, Sarov M, Greenblatt J, Babu M, Derry WB, Tillier ER, Wallingford JB, Parkinson J, Marcotte EM, Emili A. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–23. [DOI] [PubMed] [Google Scholar]
- 20. Luck K, Kim DK, Lambourne L, Spirohn K, Begg BE, Bian W, Brignall R, Cafarelli T, Campos‐Laborie FJ, Charloteaux B, Choi D, Coté AG, Daley M, Deimling S, Desbuleux A, Dricot A, Gebbia M, Hardy MF, Kishore N, Knapp JJ, Kovács IA, Lemmens I, Mee MW, Mellor JC, Pollis C, Pons C, Richardson AD, Schlabach S, Teeking B, Yadav A, Babor M, Balcha D, Basha O, Bowman‐Colin C, Chin SF, Choi SG, Colabella C, Coppin G, D'Amata C, De Ridder D, De Rouck S, Duran‐Frigola M, Ennajdaoui H, Goebels F, Goehring L, Gopal A, Haddad G, Hatchi E, Helmy M, Jacob Y, Kassa Y, Landini S, Li R, van Lieshout N, MacWilliams A, Markey D, Paulson JN, Rangarajan S, Rasla J, Rayhan A, Rolland T, San‐Miguel A, Shen Y, Sheykhkarimli D, Sheynkman GM, Simonovsky E, Taşan M, Tejeda A, Tropepe V, Twizere JC, Wang Y, Weatheritt RJ, Weile J, Xia Y, Yang X, Yeger‐Lotem E, Zhong Q, Aloy P, Bader GD, De Las Rivas J, Gaudet S, Hao T, Rak J, Tavernier J, Hill DE, Vidal M, Roth FP, Calderwood MA. A reference map of the human binary protein interactome. Nature. 2020;580:402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vinayagam A, Stelzl U, Foulle R, Plassmann S, Zenkner M, Timm J, Assmus HE, Andrade‐Navarro MA, Wanker EE. A directed protein interaction network for investigating intracellular signal transduction. Sci Signaling. 2011;4(189):rs8. [DOI] [PubMed] [Google Scholar]
- 22. Türei D, Korcsmáros T, Saez‐Rodriguez J. OmniPath: guidelines and gateway for literature‐curated signaling pathway resources. Nature Methods. 2016;13:966–7. [DOI] [PubMed] [Google Scholar]
- 23. Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newman RH, Hu J, Rho H‐S, Xie Z, Woodard C, Neiswinger J, Cooper C, Shirley M, Clark HM, Hu S, Hwang W, Seop Jeong J, Wu G, Lin J, Gao X, Ni Q, Goel R, Xia S, Ji H, Dalby KN, Birnbaum MJ, Cole PA, Knapp S, Ryazanov AG, Zack DJ, Blackshaw S, Pawson T, Gingras AC, Desiderio S, Pandey A, Turk BE, Zhang J, Zhu H, Qian J. Construction of human activity‐based phosphorylation networks. Mol Syst Biol. 2013;9:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio‐Hernandez I, Memon D, Hernandez‐Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy‐Lugo J, Lin Y, Huang XP, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Train QD, Shengjuler D, Fletcher SJ, O'Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu‐Ozturk D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d'Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, García‐Sastre A, Shokat KM, Shoichet BK, Krogan NJ. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and Genopedia: disease‐centered and gene‐centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26:145–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piñero J, Bravo À, Queralt‐Rosinach N, Gutiérrez‐Sacristán A, Deu‐Pons J, Centeno E, García‐García J, Sanz F, Furlong LI. DisGeNET: a comprehensive platform integrating information on human disease‐associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak‐Yin Tsang O, Wagh D, Coller J, Pellegrini KL, Kazmin D, Alaaeddine G, Leung WS, Chan JMC, Chik TSH, Choi CYC, Huerta C, Paine McCullough M, Lv H, Anderson E, Edupuganti S, Upadhyay AA, Bosinger SE, Maecker HT, Khatri P, Rouphael N, Peiris M, Pulendran B. Systems biological assessment of immunity to mild versus severe COVID‐19 infection in humans. Science. 2020;369:1210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Budhraja A, Basu A, Gheware A, Abhilash D, Rajagopala S, Pakala S, Sumit M, Ray A, Subramaniam A, Mathur P, Nambirajan A, Kumar S, Gupta R, Wig N, Trikha A, Guleria R, Sarkar C, Gupta I, Jain D. Molecular signature of postmortem lung tissue from COVID‐19 patients suggests distinct trajectories driving mortality. Dis Models & Mech. 2022;15(5):dmm049572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elinoff JM, Mazer AJ, Cai R, Lu M, Graninger G, Harper B, Ferreyra GA, Sun J, Solomon MA, Danner RL. Meta‐analysis of blood genome‐wide expression profiling studies in pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2020;318(1):L98–L111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stearman RS, Bui QM, Speyer G, Handen A, Cornelius AR, Graham BB, Kim S, Mickler EA, Tuder RM, Chan SY, Geraci MW. Systems analysis of the human pulmonary arterial hypertension lung transcriptome. Am J Respir Cell Mol Biol. 2019;60:637–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morselli Gysi D, do Valle Í, Zitnik M, Ameli A, Gan X, Varol O, Ghiassian SD, Patten JJ, Davey RA, Loscalzo J, Barabási AL. Network medicine framework for identifying drug‐repurposing opportunities for COVID‐19. Proc Natl Acad Sci USA. 2021;118:e2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruths T, Ruths D, Nakhleh L. GS2: an efficiently computable measure of GO‐based similarity of gene sets. Bioinformatics. 2009;25:1178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist‐oriented resource for the analysis of systems‐level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laforge M, Elbim C, Frère C, Hémadi M, Massaad C, Nuss P, Benoliel JJ, Becker C. Tissue damage from neutrophil‐induced oxidative stress in COVID‐19. Nat Rev Immunol. 2020;20:515–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paul BD, Lemle MD, Komaroff AL, Snyder SH. Redox imbalance links COVID‐19 and myalgic encephalomyelitis/chronic fatigue syndrome. Proc Natl Acad Sci USA. 2021;118:e2024358118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier‐Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pène F, Marin N, Roche N, Szwebel TA, Merkling SH, Treluyer JM, Veyer D, Mouthon L, Blanc C, Tharaux PL, Rozenberg F, Fischer A, Duffy D, Rieux‐Laucat F, Kernéis S, Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID‐19 patients. Science. 2020;369(6504):718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Satarker S, Tom AA, Shaji RA, Alosious A, Luvis M, Nampoothiri M. JAK‐STAT pathway inhibition and their implications in COVID‐19 therapy. Postgrad Med. 2021;133(5):489–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo W, Li YX, Jiang LJ, Chen Q, Wang T, Ye DW. Targeting JAK‐STAT signaling to control cytokine release syndrome in COVID‐19. Trends Pharmacol Sci. 2020;41(8):531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yerabolu D, Weiss A, Kojonazarov B, Boehm M, Schlueter BC, Ruppert C, Günther A, Jonigk D, Grimminger F, Ghofrani HA, Seeger W, Weissmann N, Schermuly RT. Targeting Jak‐Stat signaling in experimental pulmonary hypertension. Am J Respir Cell Mol Biol. 2021;64(1):100–14. [DOI] [PubMed] [Google Scholar]
- 43. Yua X, Yang R. Changes of peripheral lymphocyte subset in patients with SARS‐CoV‐2 infection during the whole course of disease. Expert Rev Respir Med. 2021;15(4):553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fava VM, Bourgey M, Nawarathna PM, Orlova M, Cassart P, Vinh DC, Cheng MP, Bourque G, Schurr E, Langlais D. A systems biology approach identifies candidate drugs to reduce mortality in severely ill patients with COVID‐19. Sci Adv. 2022;8:eabm2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moss P. The T cell immune response against SARS‐CoV‐2. Nature Immunol. 2022;23(2):186–93. [DOI] [PubMed] [Google Scholar]
- 46. Ayres JS. A metabolic handbook for the COVID‐19 pandemic. Nature Metabolism. 2020;2:572–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu W, Janocha AJ, Erzurum SC. Metabolism in pulmonary hypertension. Annu Rev Physiol. 2021;83:551–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reusch N, De Domenico E, Bonaguro L, Schulte‐Schrepping J, Baßler K, Schultze JL, Aschenbrenner AC. Neutrophils in COVID‐19. Front Immunol. 2021;12:652470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leopold JA, Maron BA, Loscalzo J. The application of big data to cardiovascular disease: paths to precision medicine. J Clin Invest. 2020;130:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang RS, Oldham WM, Loscalzo J. Network‐based association of hypoxia‐responsive genes with cardiovascular diseases. New J Phys. 2014;16:105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malas MB, Naazie IN, Elsayed N, Mathlouthi A, Marmor R, Clary B. Thromboembolism risk of COVID‐19 is high and associated with a higher risk of mortality: a systematic review and meta‐analysis. EClinicalMedicine. 2020;29:100639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Patten JJ, Keiser PT, Morselli‐Gysi D, Menichetti G, Mori H, Donahue CJ, Gan X, Valle I, Geoghegan‐Barek K, Anantpadma M, Boytz R, Berrigan JL, Stubbs SH, Ayazika T, O'Leary C, Jalloh S, Wagner F, Ayehunie S, Elledge SJ, Anderson D, Loscalzo J, Zitnik M, Gummuluru S, Namchuk MN, Barabási AL, Davey RA. Identification of potent inhibitors of SARS‐CoV‐2 infection by combined pharmacological evaluation and cellular network prioritization. iScience. 2022;25(9):104925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Newman J, Boubriak I, Jenkins D, Ng C, Ruggiero A, Screaton N, Cannon J, Toshner M. Rising COVID‐19 related acute pulmonary emboli but falling national chronic thromboembolic pulmonary hypertension referrals from a large national dataset. ERJ Open Res. 2021;7:00431‐2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
N/A. Publicly available databases were used.


