SUMMARY
The thalamus is the principal information hub of the vertebrate brain, with essential roles in sensory and motor information processing, attention, and memory. The complex array of thalamic nuclei develops from a restricted pool of neural progenitors. We apply longitudinal single-cell RNA sequencing and regional abrogation of Sonic hedgehog (Shh) to map the developmental trajectories of thalamic progenitors, intermediate progenitors, and post-mitotic neurons as they coalesce into distinct thalamic nuclei. These data reveal that the complex architecture of the thalamus is established early during embryonic brain development through the coordinated action of four cell differentiation lineages derived from Shh-dependent and -independent progenitors. We systematically characterize the gene expression programs that define these thalamic lineages across time and demonstrate how their disruption upon Shh depletion causes pronounced locomotor impairment resembling infantile Parkinson’s disease. These results reveal key principles of thalamic development and provide mechanistic insights into neurodevelopmental disorders resulting from thalamic dysfunction.
In brief
Govek et al. describe the developmental trajectory of thalamic nuclei from Shh-dependent and -independent neural progenitors. They show that molecular signatures of thalamic neuronal subtypes can be distinguished prior to their aggregation into histologically distinct thalamic nuclei and that alterations in Shh cause neurodevelopmental disorders attributed to thalamic dysfunction.
Graphical Abstract

INTRODUCTION
The thalamus develops in the posterior region of the diencephalon, between the mesencephalon and telencephalon. This location is important for unique aspects of thalamic function, to process and relay sensory and motor information to and from the cerebral cortex, and to regulate sleep, alertness, and consciousness.1 How the thalamus comes to reside within this region of the central nervous system (CNS) has been the subject of much investigation.2,3 Extracellular signals secreted from key locations both extrinsic and intrinsic to the thalamic primordium have been shown to play important roles in the growth, regionalization, and specification of thalamic progenitors.4–16 One factor in particular, the secreted morphogen Sonic hedgehog (Shh), has been implicated in spatiotemporal and threshold models of thalamic development that differ from other areas of the CNS. This impact of Shh is due, in large part, to its expression within two signaling centers, the basal plate and the zona limitans intrathalamica (ZLI), a dorsally projecting spike that separates the thalamus from the prethalamic territory.17,18 Shh signaling from these sources exhibits both unique and overlapping functions in the control of thalamic progenitor identity and specification of thalamic nuclei.11–13
The adult thalamus is subdivided into 44 distinct nuclei that are categorized on the basis of their positioning (anterior, medial, lateral, ventral, posterior, and intralaminar groups), cytoarchitectural properties, types of input (sensory, motor, or limbic), and hierarchical connectivity patterns to and from the cerebral cortex and subcortical areas.1,19–25 The spatial arrangement of these nuclei is important for generating the precise topographical relationship needed to fulfill its role as a relay and information processing center. Despite advances in our understanding of the early events regulating thalamic growth and regionalization, there remain major gaps in knowledge of the mechanisms by which heterogeneous clusters of mostly excitatory relay neurons are specified and aggregate into distinct thalamic nuclei. One particular challenge has been to decipher the full complement of thalamic progenitor identities and to elucidate their contribution to specific thalamic nuclei.11,12,26–30 Thus far, only two distinct thalamic progenitor domains have been defined by gene expression and fate mapping studies. The caudal population of thalamic progenitors, cTh.Pro, gives rise to all glutamatergic thalamic nuclei that extend axonal projections to the neocortex.28 The rostral population of thalamic progenitors, rTh.Pro, comprises a narrow band of cells sandwiched between cTh.Pro and the ZLI. Thalamic neurons derived from rTh.Pro are GABAergic and contribute to the ventrolateral geniculate nucleus (VLG) and the intergeniculate leaflet (IGL).1,12,28,31–33
Multiple studies have shown that thalamic nuclei exhibit extensive heterogeneity at the level of gene expression.2,25,34–38 Nevertheless, we still lack a clear understanding of the molecular logic and developmental trajectories by which thalamic nuclei acquire their distinct identities. Here, we make use of highly parallelized single-cell RNA sequencing (RNA-seq) to molecularly and anatomically characterize thalamic progenitor subtypes, intermediate progenitors, and post-mitotic neurons across multiple stages of mouse embryonic development. Our approach overcomes limitations of conventional single-cell transcriptomic atlases, which often lack mechanistic detail, by investigating how regional abrogation of Shh expression alters thalamic lineage progression at single-cell scale. Our findings unify models of thalamic development and provide a detailed understanding of a neurodevelopmental disorder resulting from alterations in thalamic architecture.
RESULTS
Deletion of SBE1 and SBE5 abrogates Shh expression in the ZLI
Shh expression in the ZLI and basal plate of the caudal diencephalon is dependent on two Shh brain enhancers: SBE1 and SBE5.39 Mouse embryos homozygous for targeted deletions of SBE1 and SBE5 (ShhΔSBE1ΔSBE5/ΔSBE1ΔSBE5, herein referred to as ΔSBE1/5) fail to activate Shh transcription in the ZLI and basal plate after E10.0, compared with control littermates (ShhΔSBE1ΔSBE5/+) (Figure 1A).39 Consequently, Shh signaling activity in thalamic and prethalamic territories is compromised in ΔSBE1/5 embryos, as indicated by the loss of Gli1 expression (Figure 1A). Despite the absence of Shh expression and Shh signaling activity, a GFP reporter transgene driven by SBE1 continued to be expressed in the ZLI of ΔSBE1/5 embryos (Figure 1A). Moreover, genes coding for transcription factors that are normally expressed in the ZLI, such as Pitx2 and Foxa1, maintained much of their expression in ΔSBE1/5 mutants, albeit in a partially reduced area (Figure S1A). These results indicate that the cellular integrity of the ZLI remains intact in ΔSBE1/5 embryos and highlight the utility of this mouse model for studying the implications of Shh signaling in mammalian thalamic development.
Figure 1. A high-resolution single-cell transcriptomic atlas of the developing caudal diencephalon.
(A) Deletion of the SBE1 and SBE5 enhancers leads to a loss of Shh expression and Shh signaling activity in the caudal diencephalon. Whole-mount RNA in situ hybridization for Shh (left) and the Shh-responsive gene Gli1 (right) in bisected heads from control (top) and ΔSBE1/5 embryos (bottom) at E12.5 shows reduced expression in the caudal diencephalon of mutant embryos (n = 3). Scale bar, 500 μm. A Shh bacterial artificial chromosome (BAC) transgene (Shh-GFP) expressing eGFP in place of Shh shows persistent reporter activity in the ZLI of ΔSBE1/5 and control embryos (center).
(B) Schematic of the experimental study design. The caudal diencephalon of control and ΔSBE1/5 embryos was micro-dissected at E12.5, E14.5, E16.5, and E18.5 in three replicates per time point and genotype, fixed in methanol, and profiled with single-cell RNA-seq.
(C) 3D UMAP representation of the consolidated single-cell gene expression space across all samples, colored by the genotype of the cells. Cells from control and ΔSBE1/5 embryos substantially overlap in the representation.
(D) 3D UMAP representation of the single-cell transcriptomic atlas colored by the 23 cell populations identified in the clustering analyses.
(E) Fraction of cells in each cell population for each of the four time points. Only cells from control mice were considered in this analysis. The observed expansion of glial cells starting between E14.5 and E16.5 is consistent with the transition between neurogenesis and gliogenesis at this stage of embryonic development. Cell populations are listed in the legend in the same order as they appear in the band plot. cTh.N, caudal thalamic neurons; rTh.N, rostral thalamic neurons; cTh.Pro, caudal thalamic progenitors; preTh.N, prethalamic neurons; MB, midbrain; ZI, zona incerta; preThE, prethalamic eminence; OPCs, oligodendrocyte precursor cells.
A high-resolution single-cell transcriptomic atlas of the developing caudal diencephalon in control and ΔSBE1/5 embryos
To uncover the molecular logic driving the specification of distinct thalamic nuclei, we generated a high-resolution single-cell RNA-seq atlas of the developing caudal diencephalon in control and ΔSBE1/5 embryos. The thalamic primordia were micro-dissected from three embryos per genotype according to anatomical landmarks (STAR Methods) at four developmental stages (E12.5, E14.5, E16.5, and E18.5) coinciding with peak periods of thalamic proliferation, neurogenesis, and differentiation (Figure 1B; n = 24 embryos in total). Overall, we captured the transcriptome of 249,071 cells (121,878 cells from control and 127,193 cells from ΔSBE1/5 embryos) with a median of 2,466 genes and 4,891 unique molecular identifiers (UMIs) detected per cell. We consolidated the single-cell gene expression space of all the samples and clustered the cells in this space (Figures 1C and 1D).
Our analysis identified 23 distinct cell populations, comprising most of the known cell types in the caudal diencephalon (Figures 1D and 1E). Cells from three independent biological replicates overlapped in the consolidated representation and were similarly delineated across all cell populations (Figure S1B), indicating the absence of large batch effects. Most of the identified cell populations were part of continuous developmental trajectories, such as the differentiation of thalamic progenitors (Th.Pro) into rostral GABAergic and caudal glutamatergic thalamic neurons (denoted as rTh.N and cTh.N, respectively), or the transition from neurogenesis into gliogenesis at E14.5–16.5 (Figures 1D, 1E, S1C, and S1D).
Of note, we found that these data correctly recapitulated the effect of SBE1/5 deletions on the expression of Shh and Shh target genes. Expression of the basic-helix-loop-helix (bHLH) transcription factor Olig3 distinguishes thalamic progenitors from other diencephalic cell types.28 As expected, differential gene expression analysis in the Olig3+ progenitor cell subpopulation showed a downregulation of Shh-responsive genes (Gli1, Ptch1, Nkx2–2, and Olig2) in ΔSBE1/5 compared with control cells (Figures S1E, S1F, and S1G; Table S1). In addition, a detailed examination of the larger cluster of progenitor cells identified a subgroup with an expression profile consistent with the ZLI (Shh, Foxa1, Pitx2, Sim2)26,40 (Figure S1H). Differential expression analysis confirmed the strong depletion of Shh expression from this subgroup in ΔSBE1/5 compared with control cells (Figure S1H; Table S2). The persistence of other ZLI markers in mutant cells indicates that the ZLI is not dependent on Shh for the bulk of its formation after E10.0. Altogether, these data represent a unique resource for studies of the developing caudal diencephalon and a substantial increase in cell type resolution with respect to previous single-cell RNA-seq datasets of this brain region.37,38,41–43
Most thalamic nuclei have well-defined molecular identities at E18.5
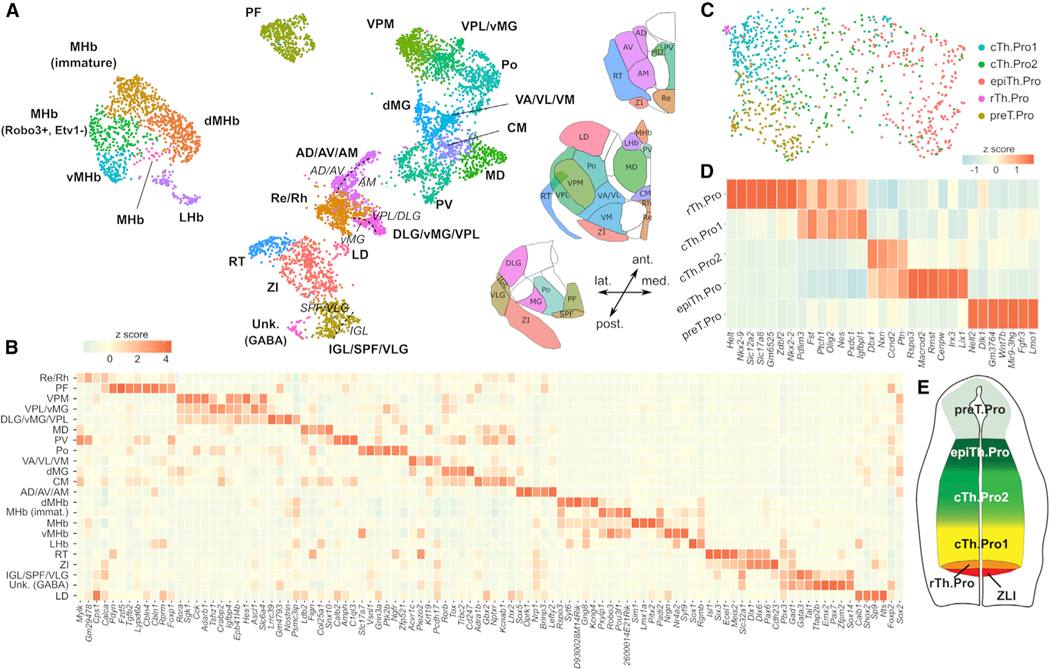
Gene expression signatures of distinct thalamic nuclei have been described in adult mice or at single stages of embryonic development, but not in a coordinated manner across developmental time.2,25,36–38,42 We sought to systematically characterize the cell populations that define thalamic nuclei throughout embryonic development. For this purpose, we performed a separate clustering and differential gene expression analysis of E18.5 control cells from the cTh.N, rTh.N, zona incerta (ZI), habenula, and reticular complex (RT) post-mitotic cell populations. This analysis identified 23 cell subpopulations with distinct transcriptomic profiles (Figures 2A and 2B; Table S3). By comparing differentially expressed genes across subpopulations with RNA in situ hybridization data from the Allen Developing Mouse Brain Atlas,44 we were able to assign one or more thalamic nuclei to each of these subpopulations (Figures 2A, S2A, and S2B). Most nuclei in the thalamic, habenular, and reticular complexes were localized to distinct regions of the Uniform Manifold Approximation and Projection (UMAP) representation (Figure 2A). However, in some cases, closely related thalamic nuclei, such as the anterodorsal (AD), anteroventral (AV), and anteromedial (AM), were assigned to the same transcriptomic cell subpopulation. We used a spectral graph method45 to dissect the transcriptional heterogeneity within the cell populations identified in our clustering analysis and further resolved the transcriptomic signatures of some of these closely related thalamic nuclei (Figures 2A, S2C, and S2D). We were able to distinguish the transcriptomic profile of the AD/AV thalamic nuclei from that of the AM nucleus, and the profile of the IGL nucleus from that of the VLG and subparafascicular (SPF) nuclei. The identities of other nuclei, such as the distinction between the ventral anterior (VA), ventral lateral (VL), and ventral medial (VM) nuclei, were only partially resolved at this stage of development (Figures S2C and S2D). In other cases, such as for the ventral posterolateral (VPL) and the medial geniculate (MG) thalamic nuclei, we observed multiple transcriptomic subpopulations associated with the same nucleus, suggesting the existence of several cell populations within these nuclei. For example, we identified three distinct transcriptomic populations contributing to the MG nucleus. All of these populations expressed Gbx2, Lhx2, Rorα, and Rorβ, and demonstrate co-expression of Sox2 and Foxp2 in the same cells. However, two of these transcriptomic populations had high expression of ventromedial geniculate (vMG) markers (Slc6a4, Adarb1, Tshz1, Dlk1), whereas the other one expressed a dorsomedial geniculate (dMG) marker (Prox1).25 In addition, the two populations with high expression of vMG markers differed in the expression of multiple genes (Figures 2B and S2A; Table S3), such as Sp9, Synp2, Nostrin, and Psmc3ip.
Figure 2. Thalamic nuclei emerge from a diverse pool of progenitor populations and acquire unique transcriptional identities during embryogenesis.
(A) UMAP representation of the mRNA expression data of E18.5 control cells from the post-mitotic cTh.N, rTh.N, ZI, habenula, and reticular complex cell populations. In total, this analysis identified 23 cell subpopulations with distinct transcriptomic profiles (bold labels). Using a spectral graph method, we split some of these populations into smaller transcriptional identities (italic labels).
(B) Heatmap depicting the expression of the top differentially expressed genes in the cell populations from (A).
(C) UMAP representation of the mRNA expression data of E12.5 control cells from the Olig3+ thalamic progenitor population. In total, three thalamic (rTh.Pro, cTh.Pro1, and cTh.Pro2), one epithalamic (epiTh.Pro), and one pretectal (preT.Pro) progenitor populations were identified with distinct transcriptomic profiles.
(D) Heatmap depicting the expression of the top differentially expressed genes in the cell populations identified in (C).
(E) Schematic showing the rostro-caudal organization of the identified progenitor cell populations in the developing thalamus as inferred from RNA in situ hybridization data.
Sensory inputs in the brain follow thalamocortical loops, where first-order thalamic nuclei project peripheral sensory inputs onto the primary sensory cortex, and higher-order thalamic nuclei receive their input from the primary sensory cortex and project it back to a secondary cortex.19,46,47 The hierarchical position that each nucleus occupies in these circuits has been shown to be the primary determinant of the postnatal transcriptional identity of somatosensory, visual, and auditory thalamic nuclei in postnatal mice.25 To assess whether the shared transcriptional programs between same-order nuclei are established earlier during embryogenesis, we projected the postnatal day 3 (P3) differential gene expression signatures of first- and higher-order nuclei from a previous study25 onto our single-cell gene expression data of E18.5 thalamic nuclei (Methods S1). This analysis revealed that the postnatal gene expression signatures are already present at E18.5 (Figure S3). Taken together, these data unveil unique molecular signatures that distinguish most thalamic nuclei prior to birth and further suggest that much of the complex structure of the caudal diencephalon is encoded by genetic programs that are active during embryonic development.
Thalamic progenitors are a heterogeneous cell population
We next characterized the cellular heterogeneity of the Olig3+ progenitor subpopulation in E12.5 control embryos. Since cell cycle is tightly coupled to cell differentiation during early neurogenesis,48 an unsupervised analysis of the progenitor cell population using the most variable genes failed to reveal distinct progenitor types. Regressing out the expression of cell cycle genes did not rectify this issue. To circumvent this problem in the analysis, we adopted a semi-supervised approach in which we compared the transcriptional profile of progenitor cells based on the expression of genes that were highly correlated or anti-correlated with a set of pre-specified markers, including Nkx2–2, Olig2, Dbx1, and Rspo3. These genes were previously shown to mark distinct and partially overlapping progenitor domains distributed along the rostral to caudal axis of the thalamus.28,37
Clustering and differential expression analysis based on this approach identified five transcriptionally distinct populations of progenitors (Figures 2C and 2D; Table S4). We named these progenitor populations as rTh.Pro (rostral thalamic progenitor), cTh.Pro1 (caudal thalamic progenitor 1), cTh.Pro2 (caudal thalamic progenitor 2), epiTh.Pro (epithalamic progenitor), and preT.Pro (pretectal progenitor) based on their rostro-caudal order in the neural tube (Figure 2E). The two most rostral thalamic progenitor populations (rTh.Pro and cTh.Pro1) were characterized by the expression of Shh-responsive genes, such as Nkx2–2, Olig2, and Ptch1 (Figure 2D), consistent with previous findings.12 High levels of expression of Shh-responsive genes were excluded from cTh.Pro2, epiTh.Pro, and preT.Pro domains, which were defined by distinct sets of differentially expressed transcripts (Figure 2D). EpiTh.Pro progenitors were characterized by the expression of markers, such as Macrod2 and Rspo3, that continued to be expressed in neurons with an epithalamic identity across all differentiation stages, including neurons of the habenula (Figure S4). These markers were upregulated in distinct but partially overlapping subpopulations of the epiTh.Pro domain, suggesting the presence of additional heterogeneity within these progenitors. EpiTh.Pro progenitors had lower but non-vanishing expression of Olig3 compared with other thalamic progenitors (Figure S4A). This result is consistent with a gradual transition between thalamic progenitor domains and Olig3− habenular and pretectal progenitors. The most caudal progenitor subpopulation (preT.Pro) was characterized by the expression of pretectal markers (Pax3, Meis1, Lmo1) (Table S4). On the basis of these results, we hypothesize that different cell lineages derived from distinct thalamic progenitor populations give rise to the diversity of thalamic nuclei. Our results also suggest the presence of soft boundaries between progenitor domains, where some cells co-express markers from multiple progenitor domains.
Glutamatergic thalamic nuclei emerge sequentially through the coordinated action of three distinct cell lineages
To uncover the transcriptomic lineages that relate thalamic progenitors at E12.5 to post-mitotic thalamic neurons at E18.5, we devised a semi-supervised approach for transferring the E12.5 and E18.5 cell annotations across all time points and identified cells with a similar gene expression profile (STAR Methods). We disaggregated the progenitor and post-mitotic populations into smaller clusters in the full single-cell RNA-seq representation and used the E12.5 and E18.5 cell annotations to label each cluster. We then constructed separate single-cell gene expression representations for glutamatergic cell lineages (Olig3+ cTh.Pro, cTh.N early post-mitotic, and cTh.N clusters) and computed the RNA velocity vector field49,50 in these representations (Figure 3A).
Figure 3. Glutamatergic thalamic cell lineages regionalize the thalamus into distinct molecular domains.

(A) UMAP representation and RNA velocity field of the single-cell RNA-seq data of the glutamatergic cell lineage in control embryos at each developmental stage. For reference, the same UMAP is also colored by the inferred cell differentiation pseudo-time and the gene expression levels of Sox2, Foxp2, and Cadm1. The expression of Sox2 and Foxp2 genes separates differentiation trajectories into Sox2+ Foxp2− (cTh.N1) and Sox2− Foxp2+ (cTh.N2) populations.
(B) Proportion of cells belonging to different glutamatergic thalamic nuclei at each developmental stage. Distinct thalamic subpopulations emerge at different time points. Late- and intermediate-emerging cell subpopulations are indicated by double asterisk (**) and single asterisk (*), respectively.
(C) Whole-mount RNA in situ hybridization for Sox2, Foxp2, and Cadm1 in sagittal sections of the E13.5 diencephalon. The expression of these markers regionalizes the developing thalamus into distinct domains. Image credit: Allen Institute.
(D) Glutamatergic thalamic IPCs are a heterogeneous population of cells with contributions from both cTh.Pro1 and cTh.Pro2 progenitors. The UMAP representation of the single-cell RNA-seq data corresponding to the glutamatergic thalamic IPC population from E14.5 control embryos is colored by the inferred cell differentiation pseudo-time and the gene expression levels of Olig2, Dbx1, Foxp2, and Sox2. CPT, counts per thousand.
Each embryonic day was analyzed independently, since neurodevelopmental lineages may change across time as a consequence of shifts in the progenitor identities. As expected, the inferred differentiation trajectories connected the progenitors with the post-mitotic neurons in each developmental stage (Figure 3A). The differentiation trajectories included a population of intermediate progenitor cells (IPCs), characterized by the co-expression of neurogenic determinants (Neurog1, Neurog2) and mitotic markers (Top2a, Mki67).51 The number of Olig3+ progenitors and IPCs rapidly diminished after E14.5 due to the transition into gliogenesis, while post-mitotic cells continued to differentiate well beyond E14.5.
Our analysis revealed the sequential emergence of glutamatergic thalamic nuclei, where lateral and intermediate nuclei such as the AD/AV/AM, dorsolateral geniculate (DLG), MG, VPL, parafascicular (PF), and posterior (Po) nuclei differentiate first, and medial nuclei such as the centromedian (CM), MD, and paraventricular (PV) differentiate later (Figures 3A and 3B). Analysis of gene expression along the differentiation trajectories showed that the expression of Sox2 and Foxp2 was anti-correlated with each other (Spearman’s correlation coefficient r = − 0.2, p < 2 × 10−16) and separated glutamatergic cell differentiation trajectories into Sox2+ Foxp2− and Sox2− Foxp2+ trajectories at all developmental stages (Figure 3A). These results suggest the presence of two major cell differentiation lineages of glutamatergic thalamic neurons, which we denote as cTh.N1 (caudal thalamic neuron 1) and cTh.N2 (caudal thalamic neuron 2). Differential gene expression analysis at E12.5 revealed 148 differentially expressed genes between cTh.N1 and cTh.N2 cell populations with a fold change >2 (Table S5, adjusted p value <0.05).
To understand the relationship between these putative cell differentiation lineages and the observed diversity of thalamic nuclei and progenitors, we examined the expression of Sox2 and Foxp2 in these populations. The expression of Sox2 was ubiquitous in the progenitors and was preserved throughout the cTh.N1 population, including in the differentiated ventroposterior medial (VPM), AD/AV, AM, and VA/VL/VM nuclei (Figures 3A and S5A). By contrast, the expression of Foxp2 was initiated in a subset of IPCs (Figure 3D) and preserved throughout the cTh.N2 population, including the PF, Po, PV, MD, and CM nuclei (Figures 3A and S5A). Importantly, these results establish a basis for the distinction between Sox2− and Foxp2-expressing thalamic nuclei observed in E18.5 cells (Figure 2B, far right columns).
Our analysis also revealed a separation of the cTh.N1 differentiation trajectory into Cadm1+ and Cadm1− subpopulations (Figure 3A). The expression of Cadm1 was initiated in a subset of early cTh.N1 post-mitotic cells and preserved in the thalamic nuclei that derive from this population, including the AD/AV, AM, VA/VL, and VM nuclei (Figures 3A and S5A). RNA in situ hybridization confirmed the regionalization of the thalamus into three distinct domains based on the expression of Sox2, Foxp2, and Cadm1, with Sox2 being expressed rostrally and Foxp2 caudally (Figure 3C). Thus, our analysis of differentiation trajectories in the single-cell data is consistent with the emergence of glutamatergic thalamic nuclei from distinct transcriptomic lineages marked by the expression of Sox2 and Foxp2. The rostro-caudal organization of these transcriptomic populations together with the organization of thalamic progenitors in the neural tube suggests that these lineages originate from distinct subpopulations of glutamatergic progenitors. A similar analysis of the GABAergic cell populations showed that GABAergic neurons of the thalamus derive from Tal1+ neural progenitors and are characterized by the expression of Shh-responsive genes (Methods S2 and Figure S6).
Glutamatergic thalamic IPCs are a heterogeneous population of cells with contributions from both cTh.Pro1 and cTh.Pro2 progenitors
Despite our ability to transfer annotations across time points, a gene expression bottleneck within IPCs prevented us from relating the aforementioned transcriptomic cell trajectories to individual thalamic progenitor identities. Most of the genes expressed in progenitor cells ceased to be expressed after mitotic arrest, except for Hs3st1, which marked both the cTh.Pro1 and cTh.N1 populations (Figure S5B). However, a closer look at the IPC population and its RNA velocity field revealed the presence of two subsets of early IPCs, characterized by the expression of the cTh.Pro1 marker Olig2, and the cTh.Pro2 marker Dbx1, as well as two subsets of late IPCs, respectively, characterized by the presence and absence of Foxp2 expression (Figure 3D). The Foxp2+ subpopulation of IPCs appeared contiguous in the gene expression space to the Dbx1+ subpopulation, whereas the Foxp2− subpopulation appeared contiguous to the Olig2+ subpopulation. We did not observe the expression of epiTh.Pro markers in the IPC cluster, indicating that the formation of habenular nuclei does not involve the generation of IPCs from these progenitors. These results suggest that the cTh.N1 and cTh.N2 cell populations are respectively derived from cTh.Pro1 and cTh.Pro2 progenitors, and that both lineages expand through the generation of distinct IPCs.
Shh signaling is required for rTh.Pro and cTh.Pro1 progenitor specification and expansion
The inference of developmental trajectories from unperturbed single-cell transcriptomic data has proved to be misleading in some situations.52 Therefore, we studied the effect of the ΔSBE1/5 deletions on the transcriptomic cell populations to further test our model of thalamic development. To determine the effect of Shh signaling on thalamic progenitor specification, we examined the differences between Olig3+ progenitors in control and ΔSBE1/5 embryos at E12.5. Our analysis of the single-cell RNA-seq data identified a strong depletion of rTh.Pro and cTh.Pro1 progenitors in ΔSBE1/5 embryos (Figure 4A, odds ratio = 13.5, Fisher’s exact test p < 10−10), consistent with previous studies demonstrating that the specification of these two progenitor identities is dependent on Shh signaling.12,13 RNA in situ hybridization for Nkx2–2 and Olig2 confirmed the strong depletion of rTh.Pro and cTh.Pro1 progenitors, respectively, in ΔSBE1/5 embryos (Figure 4B). Our analysis of the single-cell RNA-seq data also identified a moderate expansion of the cTh.Pro2 progenitor pool in mutant embryos (Figure 4A, odds ratio = 0.23, Fisher’s exact test p < 10−10). However, we were unable to confirm this expansion with in situ data (Figure 4B). Differential gene expression analysis between control and ΔSBE1/5 Olig3+ glutamatergic progenitor cells revealed the downregulation of cell cycle genes in mutant cells (Figures 4C and S7A; Table S1). This effect was particularly prominent in the IPC population (Figure 4C). We confirmed the observed downregulation of cell cycle in ΔSBE1/5 cTh.Pro1 progenitors by means of a 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay (Figures 4D, 4E, and S7B). These results suggest that the specification and expansion of cTh.Pro1 progenitors is dependent on Shh. Moreover, consistent with our hypothesis that cTh.N1 cells derive from cTh.Pro1 progenitors, we observed that the reduction in cTh.Pro1 progenitors in ΔSBE1/5 mice was accompanied by a substantial reduction in the number of Sox2+ post-mitotic cells in the glutamatergic cTh.N1 transcriptomic lineage in our single-cell RNA-seq data and a higher proportion of Foxp2 expressing cells in this lineage (Figures 4F and S8A). These results were confirmed by immunofluorescence staining for Sox2 and Foxp2 in control and ΔSBE1/5 embryos at E14.5 (Figures 4G and 4H). We conclude that Shh signaling is required for the specification and expansion of rTh.Pro- and cTh.Pro1-derived thalamic cell populations.
Figure 4. Shh signaling is required for the specification and expansion of rTh.Pro and cTh.Pro1 thalamic progenitors.
(A) UMAP representation of the mRNA expression data of E12.5 control and ΔSBE1/5 cells from the Olig3+ thalamic progenitor population (left). The bar plot shows the depletion or enrichment of cells between control and ΔSBE1/5 embryos in each progenitor cluster, indicating a strong depletion of rTh.Pro and cTh.Pro1 cells in ΔSBE1/5 embryos (****p < 0.0001, two-sided Fisher’s exact test, n = 1,789 cells).
(B) Whole-mount RNA in situ hybridization for Nkx2–2, Olig2, Dbx1, and Rspo3 in E12.5 control and ΔSBE1/5 embryos, confirming the strong depletion of rTh.Pro and cTh.Pro1 cells in mutant embryos (n = 3). Scale bar, 500 μm.
(C) UMAP representation of the glutamatergic thalamic cell lineage at E14.5 colored by the difference in the G2/M cell cycle gene expression score between control and ΔSBE1/5 cells. The analysis shows G2/M cell cycle genes are downregulated in thalamic progenitors and IPCs in ΔSBE1/5 embryos.
(D and E) EdU incorporation and immunofluorescence staining for Olig2 on coronal sections of the thalamus in control and ΔSBE1/5 E13.5 embryos, showing a depletion of Olig2-expressing cells (cTh.Pro1 cells) and a reduction of cell cycle in IPCs (**p < 0.01, two-sided t test, n = 4 mice of each condition; error bars represent standard deviation). Scale bar, 100 μm.
(F) The total number of Sox2-expressing cells in the post-mitotic cTh.N1 and cTh.N2 transcriptomic cell lineages is largely depleted in ΔSBE1/5 compared with control embryos at E14.5. For reference, the location of the post-mitotic cTh.N1 and cTh.N2 clusters in the UMAP representation of the glutamatergic thalamic cell lineage is shown on the left (**p < 0.01, two-sided t test, n = 3 mice; error bars represent standard deviation).
(G and H) Immunofluorescence staining for Sox2 and Foxp2 in anterior, medial, and posterior sections of E14.5 control and ΔSBE1/5 embryos. A depletion of Sox2+ cells and an expansion of Foxp2+ cells is observed in mutant embryos (*p < 0.05, **p < 0.01, ***p < 0.001, two-sided t test, n = 3 mice of each condition; error bars represent standard deviation). Scale bar, 100 μm.
See Figure S7.
Nkx2–2 and Sox2 expressing thalamic nuclei are reduced in ΔSBE1/5 mice
Based on our model of thalamic development, the observed depletion of cTh.Pro1 progenitors in ΔSBE1/5 embryos should lead to a failure in the development of Sox2-expressing cTh.N1 thalamic nuclei. Our analysis of single-cell RNA-seq data from post-mitotic glutamatergic cells in control and ΔSBE1/5 embryos at E18.5 confirmed a large depletion of cells in thalamic nuclei expressing high levels of Sox2, including the AD/AV/AM, VA/VL/VM, DLG/MG/VPL, and VPM nuclei (Figure 5A). Consistent with these results, RNA in situ hybridization for some of the markers identified in our differential gene expression analysis of these nuclei (Figure 2B) showed a large reduction in staining in ΔSBE1/5 newborn (P0) mice (Figures 5B and S8B). In particular, markers of the AD/AV/AM and VM nuclei were completely absent in mutant mice, whereas markers of the VA/VL, DLG/MG/VPL, and VPM were largely reduced (Figures 5B and S8B). Additionally, Nkx2–2 immunostaining was absent in the IGL/VLG nuclei in ΔSBE1/5 mice at P0 (Figure S8B), consistent with the depletion of GABAergic rTh.Pro progenitors (Figure 4A). Taken together, the analysis of single-cell RNA-seq and in situ expression data is consistent with a model of thalamic development where glutamatergic Sox2-expressing nuclei (cTh.N1 cells) are derived from Shh-dependent cTh.Pro1 progenitors, glutamatergic Foxp2-expressing nuclei (cTh.N2 cells) are derived from Shh-independent cTh.Pro2 progenitors, and GABAergic Nkx2–2-expressing nuclei (rTh.N cells) are derived from Shh-dependent rTh.Pro progenitors.
Figure 5. Shh-dependent thalamic nuclei fail to develop in ΔSBE1/5 mice.
(A) UMAP representation of the single-cell RNA-seq expression data from control and ΔSBE1/5 embryos at E18.5 corresponding to post-mitotic cTh.N, rTh.N, ZI, habenula, and reticular complex cell populations (left). The UMAP is colored by the amount of depletion or enrichment in the number of cells of each progenitor subpopulation between control and ΔSBE1/5 embryos (right).
(B) RNA in situ hybridization for Neurog2, Cbln1, and Rorα on coronal sections through the thalamus of control and ΔSBE1/5 mice at P0 showing reduced expression in AV/AM/AD, VA/VL/VM, DLG, and VPM/VPL thalamic nuclei, consistent with the results of the single-cell RNA-seq analysis (**p < 0.01, ***p < 0.001, ****p < 0.0001, two-sided t test; n = 3 mice of each condition for Cbln1 and Rorα; n = 4 mice of each condition for Neurog2; error bars represent standard error of the mean).
(C) Locomotor behavior is compromised in adult ΔSBE1/5 mice as determined by force plate actometer (top four graphs) and rotarod (bottom) assays (*p < 0.05, **p < 0.01, ****p < 0.0001, two-sided t test; n = 8–9 mice of each condition for force plate; n = 10 mice of each condition for rotarod; error bars represent standard error of the mean).
See Figure S8.
Locomotor deficits in ΔSBE1/5 mice
We next assessed the consequences that abnormal thalamic development might have on animal behavior in ΔSBE1/5 mutant mice. ΔSBE1/5 mice are viable and exhibit reduced locomotor activity and rearing in the open field, as well as a significantly impaired initial coordination and subsequent motor learning on the accelerating rotarod test (Figure 5C). These early-onset motor deficits resemble several cardinal features of infantile Parkinson’s disease, including bradykinesia, abnormal gait, and poor coordination (Figure 5C).53
The nigrostriatal pathway is a critical component of the basal ganglia motor circuit that degenerates in individuals with Parkinson’s disease. However, unlike other mouse models of the infantile form of this condition that result from tyrosine hydroxylase deficiency,53 the rate-limiting enzyme in dopamine synthesis, we observed no defects in the development or maintenance of dopaminergic neurons in the substantia nigra pars compacta or their projections to the striatum in ΔSBE1/5 mice (Figures S8C and S8D). This result is particularly relevant since SBE1 and SBE5 also regulate Shh expression in the ventral midbrain (Figure 1A), which is the source of midbrain dopaminergic neurons. However, since these neurons develop properly in ΔSBE1/5 mice, likely due to their dependency on an earlier source of Shh, we suspect that another component of the basal ganglia motor circuit is compromised in ΔSBE1/5 mice.
As described above, the failure to specify sufficient numbers of cTh.Pro1 progenitors in the absence of Shh results in a reduction in the cTh.N1 lineage, which consequently disrupts the formation of motor thalamic nuclei (VA/VL, VM). The deficits in motor thalamic nuclei observed in ΔSBE1/5 embryos and newborn pups extends into adulthood (Figure S8E). Lesions to VA/VL are known to cause severe motor dysfunction in a variety of animal models.54–57 Therefore, the most parsimonious explanation for the motor impairment phenotype in ΔSBE1/5 mice is the neurodevelopmental defect in motor thalamic nuclei formation, although secondary effects in other motor circuits cannot be ruled out.
DISCUSSION
Single-cell trajectories support the “outside-in” model of thalamic neurogenesis
Our study provides a comprehensive transcriptome-wide analysis of thalamic progenitors and their trajectories into thalamic nuclei during embryonic stages of brain development. We demonstrate that molecular signatures of thalamic neuronal subtypes can be readily distinguished as early as E12.5, prior to their aggregation into histologically distinct thalamic nuclei.26 These data lend support to the outside-in model of thalamic neurogenesis, whereby early-born neurons contribute to lateral thalamic nuclei and later-born neurons contribute to medial thalamic nuclei.36,58 Our work further extends this model with molecular insights into the mechanisms by which diverse thalamic nuclei acquire their identities. By following single-cell trajectories over developmental time, our data are consistent with a model in which thalamic nuclei arise from three distinct cellular lineages. In this model, Shh-responsive cTh.Pro1 progenitors give rise to glutamatergic neurons (cTh.N1) within sensory (DLG, VPM/VPL, vMG), motor (VA/VL, VM), and anterior (AD, AV, AM) nuclei that occupy predominantly ventrolateral regions of the thalamus. Shh-responsive rTh.Pro progenitors give rise to GABAergic neurons (rTh.N) in VLG and IGL thalamic nuclei, in agreement with previous findings.12,13 In contrast, Shh-independent cTh.Pro2 progenitors give rise to neurons (cTh.N2) within nuclei (e.g., PV, MD, CM) that form at more dorsomedial positions of the thalamus.
The bifurcation of transcriptomic trajectories according to thalamic progenitor identity is in general agreement with results from in vivo clonal analyses.36,59 These studies revealed that individual thalamic progenitors give rise to many neurons that populate multiple thalamic nuclei. They also demonstrated that the clonal relationship between thalamic nuclei is determined primarily by the rostro-caudal and dorsoventral positions of thalamic progenitors. These findings are consistent with our observations that thalamic nuclei originate from a small number of spatially and temporally segregated neural progenitors located along the primary axes of the developing thalamus (Figure 6A).
Figure 6. Model of thalamic development.
(A) Thalamic progenitors are organized rostro-caudally in the neural tube and acquire distinct identities based on their exposure to the Shh morphogen gradient originating from the ZLI and basal plate (top). In ΔSBE1/5 mutants, the pool of rTh.Pro and cTh.Pro1 progenitors are not specified and fail to expand due to the lack of Shh signaling (bottom).
(B) rTh.Pro, cTh.Pro1, and cTh.Pro2 give rise to different cell lineages, which are characterized, respectively, by the expression of Tal1, Sox2, and Foxp2 at all post-mitotic stages of cell differentiation. The Sox2+ lineage is divided into two sub-lineages soon after post-mitotic arrest of the progenitors. These sub-lineages can be distinguished by the expression of Cadm1. Different cell lineages contribute to different thalamic nuclei in a sequential temporal manner, where lateral nuclei are formed first and medial nuclei are formed later. An exception to this model is the MG/VPL/DLG, which receives contributions from both cTh.Pro1 and cTh.Pro2 progenitors.
(C) Thalamic nuclei derived from Shh-dependent (rTh.Pro and cTh.Pro1) progenitors (labeled in orange and yellow, respectively) are either absent, or significantly reduced, in ΔSBE1/5 mice, whereas nuclei derived from cTh.Pro2 progenitors (labeled in green) are partially expanded in the absence of Shh.
IPCs mediate the transition between thalamic progenitors and neurons
Previous work described the presence of IPCs in the thalamus but not their specific lineage relationships with thalamic progenitors and neurons.37,51 Our data are consistent with Olig2+ Sox2+ IPCs giving rise to ventrolateral thalamic nuclei and Foxp2+ IPCs giving rise to dorsomedial thalamic nuclei (Figures 6B and 6C). Intermixing of Sox2+ and Foxp2+ populations was observed in a subset of sensory nuclei (Figures 6B and 6C). The Sox2+ transcriptomic trajectory is further partitioned by Cadm1 expression. In addition to marking distinct and overlapping thalamic IPCs and neurons, Sox2 and Foxp2 are required to regulate functional properties of their respective thalamic lineages.60,61 In particular, the cell-autonomous loss of VP (VPM/VPL), PF, and Po nuclei in Foxp2 mutants at E14.560 suggests that Foxp2 may be required for the transition of IPCs to a subset of cTh.N2 thalamic neurons. It is worth noting that Foxp2 has a similar role in the cerebral cortex, where it regulates the formation of IPCs and their transition to cortical neurons.62 Less is known about the role of Sox2 in thalamic neurogenesis. However, the conditional knockout of Sox2 in post-mitotic thalamic neurons results in a reduction in the size and connectivity of sensory nuclei.61 Thus, the developmental segregation of cTh.Pro, IPC, and cTh.N subtypes by lineage-determining transcription factors likely plays an important role in the formation of distinct thalamic nuclei.
Impaired locomotor activity in ΔSBE1/5 mutant mice
Deciphering the pathogenic mechanisms of thalamic dysfunction in ΔSBE1/5 mutants provided novel insight into the etiology of a neurodevelopmental movement disorder with a similar phenotype to infantile Parkinson’s disease. We have shown that the spatiotemporal regulation of Shh expression in the ZLI and basal plate of the caudal diencephalon is critical for the elaboration of thalamic progenitor identities that populate multiple thalamic nuclei, including the motor thalamus (VA/VL, VM), principal sensory nuclei (DLG, VPM/VPL, vMG), and anterior thalamic nuclei (AD, AV, AM) (Figures 5A and 5B). It would therefore appear that the timing of Shh depletion may explain many of the unique features of our mouse model compared with other conditional Shh mutants in this brain region.11,12
We propose that the motor impairment in ΔSBE1/5 mutant mice is attributed to loss of the Shh-dependent cTh.Pro1 subtype of thalamic progenitors, resulting in a reduced number of Olig2+ Sox2+ IPCs and, subsequently, fewer cTh.N1 thalamic neurons populating motor thalamic nuclei (VA/VL, VM) during embryonic development. Future studies will address the consequences that alterations of other thalamic nuclei have for sensory, motor, and other behaviors in ΔSBE1/5 mutant mice. These experiments have the potential to further improve our basic understanding of thalamic development, which has consistently lagged behind other brain regions, and may also provide novel insights into the etiology of other circuit-level endophenotypes associated with abnormal motor and sensory information processing that occur in a variety of neurodevelopmental disorders.63–66
Limitations of the study
Our study maps the transcriptomic trajectories and Shh dependencies of thalamic progenitors into specific thalamic nuclei. However, since not all thalamic nuclei were readily discerned by our single-cell methods at E18.5, it is likely that their differentiation continues postnatally, beyond the scope of our analysis. Complementary lineage tracing experiments will be useful to confirm and potentially refine some of the sub-lineages presented in this paper. Moreover, elucidating the cell-intrinsic mechanisms by which thalamic nuclei acquire their unique transcriptional identities will require follow-up experiments integrating functional and multi-omics approaches (e.g., assay for transposase-accessible chromatin with high-throughput sequencing [ATAC-seq], chromatin immunoprecipitation sequencing [ChIP-seq]). Finally, despite our determination that Shh instructs cTh.Pro1 and rTh.Pro progenitors to adopt specific trajectories, the extrinsic signals responsible for patterning cTh.Pro2 and epiTh.Pro remain to be confirmed.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Pablo G. Camara (pcamara@pennmedicine.upenn.edu).
Materials availability
This study has generated mouse lines, which are listed in the key resource table. Mouse lines are available upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Chicken anti-GFP (1:200) | Aves Labs | Cat# GFP-1020; RRID:AB_10000240 |
| Mouse anti-Nkx2.2 (1:300) | DSHB | Cat# 74.5A; RRID:AB_2314951 |
| Rabbit anti-Olig2 (1:300) | Millipore | Cat# AB9610; RRID:AB_570666 |
| Rabbit anti-Foxp2 (1:200) | Abcam | Cat# ab16046; RRID:AB_2107107 |
| Mouse anti-Sox2 (1:100) | R&D Systems | Cat# MAB2018; RRID:AB_358009 |
| Rabbit anti-Sox2 (1:300) | Millipore | Cat# AB5603; RRID:AB_2286686 |
| Rabbit anti-Tyrosine Hydroxylase (1:1000) | Pel-Freez Biologicals | Cat# P40101–0; RRID:AB_461064 |
| Goat anti-Mouse Alexa 488 (1:400) | Thermo Fisher Scientific | Cat# A28175; RRID:AB_2536161 |
| Goat anti-Rabbit Alexa 488 (1:400) | Thermo Fisher Scientific | Cat# A-11008; RRID:AB_143165 |
| Goat anti-Rabbit Alexa 594 (1:400) | Thermo Fisher Scientific | Cat# A-11037; RRID:AB_2534095 |
| Goat anti-Chicken Alexa 488 (1:350) | Thermo Fisher Scientific | Cat# A-11039; RRID:AB_2534096 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Click-iT® EdU Imaging Kit | Molecular Probes | C10339 |
| Papain Dissociation System | Worthington Biochemical Corporation | LK003153 |
| Chromium Single Cell 3′ library and Gel Bead Kit v3 Deposited data |
10x Genomics | PN-1000075 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and processed single-cell RNA-seq data | This paper | GEO: GSE211701 |
| Interactive UMAP visualizations | This paper | CELLxGENE: https://cellxgene.cziscience.com/collections/d5cad3f0–56b6–4fbe-8f2b-be92a8c7820f |
| 3D UMAP visualization | This paper | Mendeley Data: https://doi.org/10.17632/kjpj9n66jg (https://data.mendeley.com/datasets/kjpj9n66jg) |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| ShhΔSBE1ΔSBE5/ΔSBE1ΔSBE5 (ΔSBE1/5) | Yao et al., (2016)39 | N/A |
| Shh-GFP (429M20eGFP) | Jeong et al., (2006)67 | N/A |
| Cbln2-mVenus | Seigneur and Sudhof (2017)68 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Cell Ranger (v2.1) | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| R (v3.4.1) | The R Foundation | https://cran.r-project.org/ |
| Seurat (v3.0.2) | Stuart et al., 201969 | https://satijalab.org/seurat/index.html |
| Harmony | Korsunsky et al., 201970 | https://portals.broadinstitute.org/harmony/ |
| edgeR (v3.20.1) | Robinson et al., 201071 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| RayleighSelection | Govek et al., 201945 | https://github.com/CamaraLab/RayleighSelection |
| velocyto command line tool (v0.17.13) | La Manno et al., 201849 | http://velocyto.org/ |
| Python (v3.9.10) | Python Software Foundation | https://www.python.org/ |
| scvelo (v0.2.4) | Bergen et al., 202050 | https://scvelo.readthedocs.io/ |
|
| ||
| Other | ||
|
| ||
| Leica TCS SP8 MP system | Leica | https://www.leica-microsystems.com/products/confocal-microscopes/p/leica-tcs-sp8-mp/downloads/ |
| Leica DM 5500 upright microscope system | Leica | https://www.leica-microsystems.com/products/light-microscopes/p/leica-dm5500-b/ |
Data and code availability
Single-cell RNA-seq data have been deposited at Short Read Archive (SRA), Gene Omnibus Archive (GEO) (GSE211701), Mendeley Data, and cellxgene and are publicly available as the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All mouse experiments were performed in accordance with the ethical guidelines of the National Institutes of Health and with the approval of the Institutional Animal Care and Use Committee of the University of Pennsylvania. Mice were housed in Thoren caging units under a constant 12-h light/dark cycle. Mice carrying targeted deletions of SBE1 and SBE5 were described previously.13,39 To generate ShhΔSBE1ΔSBE5/ΔSBE1ΔSBE5 double homozygous mutant embryos, the ShhΔSBE5/+ line was first crossed with ShhΔSBE1/ΔSBE1 mutants. ShhΔSBE1/+; ΔSBE5/+ males, carrying the SBE1 and SBE5 deletions in trans, were then bred to wild type CD1 females. The progeny from this cross were screened for recombination events that placed the SBE1 and SBE5 deletions in cis (1/600 offspring). The ShhΔSBE1ΔSBE5/+ double heterozygous animals were then intercrossed to generate ShhΔSBE1ΔSBE5/ΔSBE1ΔSBE5 double homozygous mice and embryos. The 429M20eGFP BAC transgenic mouse reporter line (referred to herein as Shh-GFP) expresses eGFP in the ZLI and basal plate of the caudal diencephalon under the transcriptional control of SBE1, as described previously.67 Cbln2-mVenus mice were described previously.68 Both male and female mice (3–5 months of age) were used in behavioral studies. No sex specific differences were observed between genotypes.
METHOD DETAILS
In situ hybridization
Embryonic or neonatal (P0) brains were collected from timed pregnant females (vaginal plug = E0.5). For whole-mount RNA in situ hybridization, heads were fixed in 4% paraformaldehyde at 4°C for overnight, bisected along the mid-sagittal plane and hybridized with digoxygenin-UTP-labeled riboprobes as previously described.67 For RNA in situ hybridization on sections, heads were dissected and fixed for 2 h in 4% paraformaldehyde at 4°C, then washed in PBS. Samples were cryoprotected overnight in 30% sucrose/PBS then snap frozen in OCT embedding compound (Sakura Finetek Torrence, CA). Samples were serially sectioned along the coronal plane at 16 μm (for E12.5 and E13.5 embryos), 18 μm (for E14.5 embryos) or 20 μm (for E18.5 embryos) thickness using a cryostat (Leica Biosystems, CM3050 S). Sections were hybridized with digoxigenin- UTP-labeled riboprobes as previously described.72
Immunohistochemistry
Brains were processed for immunohistochemistry in the same fashion as for in situ hybridization on sections. Brain sections were stained with DAPI and incubated with the following primary antibodies: rabbit anti-Foxp2 (1:200, Abcam, ab16046), mouse anti-Nkx2.2 (1:300, DSHB, 74.5A), rabbit anti-Olig2 (1:300, Millipore, AB9610), mouse anti-Sox2 (1:100, R&D Systems, MAB2018), rabbit anti-Sox2 (1:300, Millipore, Cat#AB5603), and rabbit anti-Tyrosine Hydroxylase (1:1000, Pel-Freez, P40101–0). Detection of primary antibodies was achieved using secondary antibodies conjugated to goat anti-mouse Alexa 488 (1:400, Thermo Fisher, Cat#A28175), goat anti-rabbit Alexa 488(1:400, Thermo Fisher, Cat#A-11008), and goat anti-rabbit Alexa 594 (1:400, Thermo Fisher, Cat#A-11037). Specimens were imaged on a Leica TCS SP8 MP system.
EdU incorporation
EdU was dissolved in sterile water and administered to pregnant dams via intraperitoneal injection at a concentration of 50 μg/g of body weight, 2 h prior to embryo harvest (Molecular Probes). Embryos were fixed in 4% paraformaldehyde at 4°C for 2 h, then were processed in the same fashion as for in situ hybridization on sections. EdU incorporation was detected at room temperature with the Click-iT® EdU Imaging Kit (Molecular Probes C10339). The staining protocol was optimized for frozen sections using the following modifications: 2 × 10′ PBS-Tween wash, 2 × 10′ 3% BSA incubation, 30′ incubation in the dark with Click-iT® reaction cocktail assembled in the recommended order immediately prior to application, 3% BSA wash, 2x PBS wash.
Isolation of embryonic thalamus and single-cell dissociation
The thalamus was manually dissected from control (ShhΔSBE1;ΔSBE5/+) and mutant (ShhΔSBE1;ΔSBE5/ΔSBE1;ΔSBE5) brains at four embryonic stages (E12.5, E14.5, E16.5 and E18.5) in ice-cold PBS. The caudal and rostral boundaries of dissection coincided with the cephalic flexure and the mammillary body, respectively. Each thalamus was cut into small pieces and dissociated into a single cell suspension using the Papain Dissociation System (Worthington, LK003153) according to the manufacturer’s protocol. Samples were dissociated in Papain-EBSS solution for 40 min at 37°C. Papain was inactivated with ovomucoid protease inhibitor, and the digested tissue was resuspended in PBS (calcium and magnesium free) containing 0.04% weight/volume BSA (400 μg/mL). Cell suspensions were stained with Trypan Blue to determine the ratio of viable to damaged cells and counted using a haemocytometer (Thermo Fisher Scientific). Single cell suspensions containing more than 90% viable cells were fixed in methanol for 1 week. Samples were rehydrated at a concentration of 700–1200 cells/μL in ice-cold PBS containing 0.04% weight/volume BSA (400 μg/mL) immediately prior to the generation of single-cell RNA sequencing libraries.
Single-cell RNA library preparation and sequencing
Single-cell RNA-seq libraries were generated using the 10X Genomics platform (Chromium Single Cell 3′ library and Gel Bead Kit v3, PN-1000075) and sequenced on a NovaSeq 6000 system (Illumina) at the Center for Applied Genomics (Children’s Hospital of Philadelphia). Three independent libraries were generated for each time point and genotype (n = 24 libraries).
Processing of RNA sequencing data
Fastq files were aligned to the mm10 mouse reference genome and count matrices were generated using the CellRanger (v2.1) pipeline. Except where otherwise specified, we processed and visualized the scRNA-seq counts with the following Seurat-based pipeline, using Seurat v3.0.2 70. We filtered out cells with less than 2,000 UMIs based on the inflection point of the log-transformed barcode rank plot of each sample, or more than 15% of the UMIs coming from mitochondrially encoded genes. In total, 249,071 cells passed these filters, with a median of 4,891 UMIs/cell, (interquartile range: 3,795–6,372 UMIs/cell), 2,466 genes/cell (interquartile range: 2,092–2,943 genes/cell), and 2.7% of UMIs in each cell coming from mitochondrially encoded genes (interquartile range: 2.1–3.5%). We next scaled and centered the UMI counts and used the default vst method to identify the top 2,000 variable genes. We removed all genes from the X and Y chromosomes to reduce the effect of unequal male and female mouse replicates between conditions. To correct for non-biological batch effects between conditions and time points, we used the Harmony algorithm70 with its Seurat integration, run on the top principal components (PCs) of the variable genes. Harmony outputs a batch-corrected representation of the scRNA-seq data of same dimensionality as the input PCs. We ran Louvain clustering and UMAP on the output of Harmony to visualize this consolidated gene expression space. The full dataset was visualized using 20 PCs and 3 UMAP dimensions, while the rest of the subset analyses used 20 PCs and 2 UMAP dimensions. To compute differentially expressed genes (DEGs) in the clusters, we used edgeR’s generalized linear model likelihood ratio test (glmLRT)71 to compare the gene expression in cells from control mice in a single cluster versus all other clusters in the representation. We also computed DEGs between cells from control and ΔSBE1/5 mice in each cluster using the same method. To reduce the running time of edgeR, we subsampled large clusters to 2,000 cells when computing DEGs.
Annotation of cell populations in the full atlas
We followed the steps outlined above (Processing of RNA sequencing data) to visualize and cluster all cells in our scRNA-seq dataset. We used the DEGs in each cluster to annotate it based on cell type, differentiation stage, or area of the brain according to published literature and ISH images from the Allen Developing Mouse Brain Atlas. We did not observe the presence of abundant doublets based on the co-expression of markers.
Identification of thalamic nuclei at E18.5
We selected all E18.5 cells from the clusters that we annotated as cTh.N, rTh.N, RT, ZI, and habenula and followed the steps outlined above (Processing of RNA sequencing data) to visualize and cluster differentiated cells from thalamic nuclei. We compared DEGs from each cluster to known markers of thalamic nuclei and E18.5 mice ISH data from the Allen Developing Mouse Brain Atlas. We used RayleighSelection45 to identify significantly localized genes marking nuclei that could not be disaggregated by unsupervised clustering. Enrichment of each annotated thalamic nucleus in control or ΔSBE1/5 mice compared to the rest of the E18.5 thalamic clusters was computed using a Fisher’s exact test.
Identification of progenitor populations at E12.5
We selected all E12.5 cells from the progenitor cell clusters with Olig3 expression: cTh.Pro, GABAergic progenitors, and astroglia. We used the same approach described above (processing of RNA sequencing data) to produce higher resolution clusters of just these cells and selected those clusters that had high expression of Olig3 and Vim but did not yet express neuronal differentiation markers (Neurod1, Stmn2). We created a UMAP visualization of the resulting 1,885 cells using the top 10 genes that were correlated and the top 10 genes that were anti-correlated with Nkx2–2, Olig2, Dbx1, and Rspo3 (62 genes in total). We associated each progenitor subpopulation (rTh.Pro, cTh.Pro1, cTh.Pro2, epiTh.Pro, cTh.IPC, or preT.Pro) with a subset of these 62 genes based on known markers and a correlation-based hierarchical clustering of the genes. We then assigned each cell to a progenitor subpopulation based on the total counts for each set of genes. Enrichment of each subpopulation in control or ΔSBE1/5 mice compared to the rest of the E12.5 progenitor clusters was computed using a Fisher’s exact test.
Transferring annotations across time points
We transferred thalamic nuclei and progenitor identities across time points by building representations from the clusters cTh.N, rTh.N, RT, ZI, and habenula (for thalamic nuclei), and cTh.Pro, GABAergic progenitors, and astroglia (for progenitors). We processed and clustered these representations following the same approach as in Processing of RNA sequencing data. We annotated each cluster, containing cells from all time points, based on the proportion of annotated E18.5 or E12.5 cells, as long as they represented at least 2% of the cells in the cluster. To identify Shh-responsive clusters, we calculated the GSEA score of Shh-responsive genes (Gli1, Ptch1, Olig2, Nkx2–2, Pdlim3, Fst, Zdbf2, Hs3st1, and Slc38a11) in the list of all genes ordered by fold change in expression between control and ΔSBE1/5 mice.
Thalamic lineages across time
We reconstructed the GABAergic, glutamatergic thalamic, and habenula lineages separately for each time point. For GABAergic lineages, we selected cells from the GABAergic progenitors, rTh.N (early post-mitotic), rTh.N, preTh.N (early post-mitotic), RT, and ZI clusters, as well as the cells labeled as rTh.Pro at E12.5 (Identification of progenitor populations at E12.5). For glutamatergic thalamic lineages, we selected cells from the cTh.N and cTh.N (early post-mitotic) clusters, as well as progenitors from all time points in Olig3+ progenitor clusters. For habenula lineages, we selected cells from the same Olig3+ progenitor clusters, plus cTh.N (early post-mitotic), habenula, and pretectum clusters. We prepared UMAP representations of these differentiation lineages as described in processing of RNA Sequencing data, with the addition of Seurat’s cell cycle regression to balance out the clear cell cycle effect differences between progenitor and differentiated cells. Cell cycle scores were computed using Seurat’s CellCycleScoring function. We used the velocyto command line interface49 and scVelo50 to infer and visualize RNA velocity streamlines and pseudotime on the UMAP representations. We identified gene patterns associated with the differentiation trajectory from the top DEG lists, then further identified correlated and anti-correlated genes of interest. The significance of a change in Seurat’s G2/M score between control and ΔSBE1/5 cells in each cluster was calculated using a Wilcoxon rank-sum test.
Locomotor behavior
Locomotor activity was assayed using the force plate actometer as previously described.73 Briefly, mice (10–14 weeks of age) were acclimated to the room for 15 min prior to the start of each experiment. Individual mice were placed on an open field plate (28 cm × 28 cm) with four force transducers and sampled at 200 scans/second for 60 min. Each session was digitally recorded. The force plate was wiped down with 70% ethanol after each session. The activity of the mouse was tracked with high accuracy, including total distance traveled, rearing events, low mobility bouts, and time spent in the center 25% of the open field. Data acquisition and calibration procedures were followed as previously described.74
Rotarod
Balance and coordination were assessed on a five-station Rotarod treadmill (IITC Life Science Inc.). Each mouse was tested three times per day for two consecutive days. All trials lasted for five minutes, the time when maximum speed was reached at a constant rate of acceleration from 4–40 rpm. A trial was terminated when a mouse fell off, made one complete backward revolution while hanging on, or after five minutes. The mice were acclimated to the room for 30 min on each testing day. The machine was wiped down with 70% ethanol in between each trial. Mice (10–14 weeks of age) were tested in four separate cohorts comprising five mice per cohort (n = 10 control and n = 10 ΔSBE1/5 mutant littermates from 4 separate litters).
QUANTIFICATION AND STATISTICAL ANALYSIS
All cell counts in imaging data were performed using the cell counter function in ImageJ (NIH) on tissue sections from at least three control and mutant embryos. In cases where double labeling was examined the tissue was imaged at a single Z-plane. Each channel (green for marker 1, red for marker 2) was first examined independently, assigning a positive count for a given marker to the DAPI stained nucleus most closely associated with the staining. A cell was only counted as double labeled if a single nucleus marked by DAPI had been assigned to the cell labeled by marker1 and marker 2. Statistical analysis of all cell counts was performed in GraphPad Prism using a two-side t test. For a given in situ probe, expression area was measured from at least three control and mutant embryos using ImageJ software. Quantification of the spatial distribution of genes expressed in the zli was normalized to head size. Statistical analysis of all area and length measurements was performed in GraphPad Prism using a two-sided t test. Differential gene expression analyses of single-cell RNA-seq data were performed using edgeR’s generalized linear model likelihood ratio test (glmLRT).71 Control/mutant cell enrichments in single-cell RNA-seq clusters were assessed using a two-sided Fisher’s exact test. Differences in G2/M score between control and ΔSBE1/5 cells in each single-cell RNA-seq cluster were evaluated using a two-side Wilcoxon rank-sum test. Locomotor, balance, and coordination data was compared between control and ΔSBE1/5 mice using two-sided t-tests. For t-tests, normality of the data was not tested due to the small sample size. p values and sample sizes for each statistical test are described in the respective figure legend.
Supplementary Material
Highlights.
Molecular signatures distinguish most thalamic nuclei prior to birth
Thalamic nuclei emerge from four distinct cell lineages
Defects in motor thalamic nuclei formation cause infantile Parkinson’s disease
ACKNOWLEDGMENTS
The authors are grateful to Drs. Stephen Liebhaber, Zhaolan Zhou, and Hao Wu for their constructive comments on the manuscript. They also thank the Center for Applied Genomics of the Children’s Hospital of Philadelphia for excellent technical assistance with the preparation and sequencing of single-cell RNA-seq libraries. This work was supported by NIH grant R01 NS039421 (D.J.E.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111768.
REFERENCES
- 1.Jones EG (2007). The Thalamus (Cambridge University Press; ). [Google Scholar]
- 2.Gezelius H, Moreno-Juan V, Mezzera C, Thakurela S, Rodríguez-Malmierca LM, Pistolic J, Benes V, Tiwari VK, and López-Bendito G.(2017). Genetic labeling of nuclei-specific thalamocortical neurons reveals putative sensory-modality specific genes. Cereb. Cortex 27, 5054–5069. 10.1093/cercor/bhw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakagawa Y.(2019). Development of the thalamus: from early patterning to regulation of cortical functions. Wiley Interdiscip. Rev. Dev. Biol 8, e345. 10.1002/wdev.345. [DOI] [PubMed] [Google Scholar]
- 4.Ishibashi M, and McMahon AP (2002). A sonic hedgehog-dependent signaling relay regulates growth of diencephalic and mesencephalic primordia in the early mouse embryo. Development 129, 4807–4819. [DOI] [PubMed] [Google Scholar]
- 5.Braun MM, Etheridge A, Bernard A, Robertson CP, and Roelink H(2003). Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development 130, 5579–5587. 10.1242/dev.00685. [DOI] [PubMed] [Google Scholar]
- 6.Zhou CJ, Pinson KI, and Pleasure SJ (2004). Severe defects in dorsal thalamic development in low-density lipoprotein receptor-related protein-6 mutants. J. Neurosci 24, 7632–7639. 10.1523/JNEUROSCI.2123-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyake A, Nakayama Y, Konishi M, and Itoh N.(2005). Fgf19 regulated by Hh signaling is required for zebrafish forebrain development. Dev. Biol 288, 259–275. 10.1016/j.ydbio.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 8.Kataoka A, and Shimogori T.(2008). Fgf8 controls regional identity in the developing thalamus. Development 135, 2873–2881. 10.1242/dev.021618. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Ferre A, and Martinez S.(2009). The development of the thalamic motor learning area is regulated by Fgf8 expression. J. Neurosci 29, 13389–13400. 10.1523/JNEUROSCI.2625-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bluske KK, Kawakami Y, Koyano-Nakagawa N, and Nakagawa Y.(2009). Differential activity of Wnt/beta-catenin signaling in the embryonic mouse thalamus. Dev. Dyn 238, 3297–3309. 10.1002/dvdy.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabó NE, Zhao T, Zhou X, and Alvarez-Bolado G.(2009). The role of Sonic hedgehog of neural origin in thalamic differentiation in the mouse. J. Neurosci 29, 2453–2466. 10.1523/JNEUROSCI.4524-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vue TY, Bluske K, Alishahi A, Yang LL, Koyano-Nakagawa N, Novitch B, and Nakagawa Y.(2009). Sonic hedgehog signaling controls thalamic progenitor identity and nuclei specification in mice. J. Neurosci 29, 4484–4497. 10.1523/JNEUROSCI.0656-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong Y, Dolson DK, Waclaw RR, Matise MP, Sussel L, Campbell K, Kaestner KH, and Epstein DJ (2011). Spatial and temporal requirements for sonic hedgehog in the regulation of thalamic interneuron identity. Development 138, 531–541. 10.1242/dev.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peukert D, Weber S, Lumsden A, and Scholpp S.(2011). Lhx2 andLhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PLoS Biol. 9, e1001218. 10.1371/journal.pbio.1001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bluske KK, Vue TY, Kawakami Y, Taketo MM, Yoshikawa K, Johnson JE, and Nakagawa Y.(2012). beta-Catenin signaling specifies progenitor cell identity in parallel with Shh signaling in the developing mammalian thalamus. Development 139, 2692–2702. 10.1242/dev.072314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee M, Yoon J, Song H, Lee B, Lam DT, Yoon J, Baek K, Clevers H, and Jeong Y.(2017). Tcf7l2 plays crucial roles in forebrain development through regulation of thalamic and habenular neuron identity and connectivity. Dev. Biol 424, 62–76. 10.1016/j.ydbio.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Scholpp S, and Lumsden A.(2010). Building a bridal chamber: development of the thalamus. Trends Neurosci. 33, 373–380. 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein DJ (2012). Regulation of thalamic development by sonic hedgehog. Front. Neurosci 6, 57. 10.3389/fnins.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herkenham M.(1980). Laminar organization of thalamic projections to the rat neocortex. Science 207, 532–535. 10.1126/science.7352263. [DOI] [PubMed] [Google Scholar]
- 20.Theyel BB, Llano DA, and Sherman SM (2010). The corticothalamo-cortical circuit drives higher-order cortex in the mouse. Nat. Neurosci 13, 84–88. 10.1038/nn.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris JA, Mihalas S, Hirokawa KE, Whitesell JD, Choi H, Bernard A, Bohn P, Caldejon S, Casal L, Cho A, et al. (2019). Hierarchical organization of cortical and thalamic connectivity. Nature 575, 195–202. 10.1038/s41586-019-1716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puelles L, Martinez-de-la-Torre M, Ferran J-L, and Watson C.(2012). Diencephalon. In The Mouse Nervous System (Elsevier; ), pp. 313–336. [Google Scholar]
- 23.Sherman SM (2012). Thalamocortical interactions. Curr. Opin. Neurobiol 22, 575–579. 10.1016/j.conb.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clascá F, Rubio-Garrido P, and Jabaudon D.(2012). Unveiling the diversity of thalamocortical neuron subtypes. Eur. J. Neurosci 35, 1524–1532. 10.1111/j.1460-9568.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- 25.Frangeul L, Pouchelon G, Telley L, Lefort S, Luscher C, and Jabaudon D.(2016). A cross-modal genetic framework for the development and plasticity of sensory pathways. Nature 538, 96–98. 10.1038/nature19770. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa Y, and O’Leary DD (2001). Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J. Neurosci 21, 2711–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones EG, and Rubenstein JLR (2004). Expression of regulatory genes during differentiation of thalamic nuclei in mouse and monkey. J. Comp. Neurol 477, 55–80. 10.1002/cne.20234. [DOI] [PubMed] [Google Scholar]
- 28.Vue TY, Aaker J, Taniguchi A, Kazemzadeh C, Skidmore JM, Martin DM, Martin JF, Treier M, and Nakagawa Y.(2007). Characterization of progenitor domains in the developing mouse thalamus. J. Comp. Neurol 505, 73–91. 10.1002/cne.21467. [DOI] [PubMed] [Google Scholar]
- 29.Yuge K, Kataoka A, Yoshida AC, Itoh D, Aggarwal M, Mori S,Blackshaw S, and Shimogori T.(2011). Region-specific gene expression in early postnatal mouse thalamus. J. Comp. Neurol 519, 544–561. 10.1002/cne.22532. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki-Hirano A, Ogawa M, Kataoka A, Yoshida AC, Itoh D, Ueno M, Blackshaw S, and Shimogori T.(2011). Dynamic spatiotemporal gene expression in embryonic mouse thalamus. J. Comp. Neurol 519, 528–543. 10.1002/cne.22531. [DOI] [PubMed] [Google Scholar]
- 31.Horowitz SS, Blanchard JH, and Morin LP (2004). Intergeniculate leaflet and ventral lateral geniculate nucleus afferent connections: an anatomical substrate for functional input from the vestibulo-visuomotor system. J. Comp. Neurol 474, 227–245. 10.1002/cne.20125. [DOI] [PubMed] [Google Scholar]
- 32.Morin LP, and Blanchard JH (2005). Descending projections of the hamster intergeniculate leaflet: relationship to the sleep/arousal and visuomotor systems. J. Comp. Neurol 487, 204–216. 10.1002/cne.20546. [DOI] [PubMed] [Google Scholar]
- 33.Delogu A, Sellers K, Zagoraiou L, Bocianowska-Zbrog A, Mandal S, Guimera J, Rubenstein JLR, Sugden D, Jessell T, and Lumsden A.(2012). Subcortical visual shell nuclei targeted by ipRGCs develop from a Sox14+−GABAergic progenitor and require Sox14 to regulate daily activity rhythms. Neuron 75, 648–662. 10.1016/j.neuron.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Hunnicutt BJ, Jongbloets BC, Birdsong WT, Gertz KJ, Zhong H, and Mao T.(2016). A comprehensive excitatory input map of the striatum reveals novel functional organization. Elife 5, e19103. 10.7554/eLife.19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagalski A, Puelles L, Dabrowski M, Wegierski T, Kuznicki J, and Wisniewska MB (2016). Molecular anatomy of the thalamic complex and the underlying transcription factors. Brain Struct. Funct 221, 2493–2510. 10.1007/s00429-015-1052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong SZH, Scott EP, Mu W, Guo X, Borgenheimer E, Freeman M, Ming GL, Wu QF, Song H, and Nakagawa Y.(2018). In vivo clonal analysis reveals spatiotemporal regulation of thalamic nucleogenesis. PLoS Biol. 16, e2005211. 10.1371/journal.pbio.2005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Q, and Li JYH (2019). Defining developmental diversification ofdiencephalon neurons through single cell gene expression profiling. Development 146, dev174284. 10.1242/dev.174284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips JW, Schulmann A, Hara E, Winnubst J, Liu C, Valakh V, Wang L, Shields BC, Korff W, Chandrashekar J, et al. (2019). A repeated molecular architecture across thalamic pathways. Nat. Neurosci 22, 1925–1935. 10.1038/s41593-019-0483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao Y, Minor PJ, Zhao YT, Jeong Y, Pani AM, King AN, Symmons O, Gan L, Cardoso WV, Spitz F, et al. (2016). Cis-regulatory architecture of a brain signaling center predates the origin of chordates. Nat. Genet 48, 575–580. 10.1038/ng.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitamura K, Miura H, Yanazawa M, Miyashita T, and Kato K.(1997). Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech. Dev 67, 83–96. 10.1016/s0925-4773(97)00110-x. [DOI] [PubMed] [Google Scholar]
- 41.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16. 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalish BT, Cheadle L, Hrvatin S, Nagy MA, Rivera S, Crow M, Gillis J, Kirchner R, and Greenberg ME (2018). Single-cell transcriptomics of the developing lateral geniculate nucleus reveals insights into circuit assembly and refinement. Proc. Natl. Acad. Sci. USA 115, E1051–E1060. 10.1073/pnas.1717871115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hashikawa Y, Hashikawa K, Rossi MA, Basiri ML, Liu Y, Johnston NL, Ahmad OR, and Stuber GD (2020). Transcriptional and spatial resolution of cell types in the mammalian habenula. Neuron 106, 743–758.e5. 10.1016/j.neuron.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, Hawrylycz M, and Dang C.(2013). Allen Brain Atlas: an integrated spatiotemporal portal for exploring the central nervous system. Nucleic Acids Res. 41, D996–D1008. 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Govek KW, Yamajala VS, and Camara PG (2019). Clustering-independent analysis of genomic data using spectral simplicial theory. PLoS Comput. Biol 15, e1007509. 10.1371/journal.pcbi.1007509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman SM, and Guillery RW (1998). On the actions that one nerve cell can have on another: distinguishing “drivers” from “modulators. Proc. Natl. Acad. Sci. USA 95, 7121–7126. 10.1073/pnas.95.12.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman SM, and Guillery RW (2006). Exploring the Thalamus and its Role in Cortical Function (MIT press). [Google Scholar]
- 48.Ohnuma S, Philpott A, and Harris WA (2001). Cell cycle and cell fate inthe nervous system. Curr. Opin. Neurobiol 11, 66–73. 10.1016/s0959-4388(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 49.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Bluske KK, Dickel LK, and Nakagawa Y.(2011). Basal progenitor cells in the embryonic mouse thalamus - their molecular characterization and the role of neurogenins and Pax6. Neural Dev. 6, 35. 10.1186/1749-8104-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E, Dueck H, Stefanik D, Tan K, Trapnell C, Kim J, et al. (2019). A line-age-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science 365, eaax1971. 10.1126/science.aax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korner G, Noain D, Ying M, Hole M, Flydal MI, Scherer T, Allegri G, Rassi A, Fingerhut R, Becu-Villalobos D, et al. (2015). Brain catecholamine depletion and motor impairment in a Th knock-in mouse with type B tyrosine hydroxylase deficiency. Brain 138, 2948–2963. 10.1093/brain/awv224. [DOI] [PubMed] [Google Scholar]
- 54.Canavan AG, Nixon PD, and Passingham RE (1989). Motor learning in monkeys (Macaca fascicularis) with lesions in motor thalamus. Exp. Brain Res 77, 113–126. 10.1007/BF00250573. [DOI] [PubMed] [Google Scholar]
- 55.van Donkelaar P, Stein JF, Passingham RE, and Miall RC (2000). Temporary inactivation in the primate motor thalamus during visually triggered and internally generated limb movements. J. Neurophysiol 83, 2780–2790. 10.1152/jn.2000.83.5.2780. [DOI] [PubMed] [Google Scholar]
- 56.Jeljeli M, Strazielle C, Caston J, and Lalonde R.(2003). Effects of ventrolateral-ventromedial thalamic lesions on motor coordination and spatial orientation in rats. Neurosci. Res 47, 309–316. 10.1016/s0168-0102(03)00224-4. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka YH, Tanaka YR, Kondo M, Terada SI, Kawaguchi Y, and Matsuzaki M.(2018). Thalamocortical axonal activity in motor cortex exhibits layer-specific dynamics during motor learning. Neuron 100, 244–258.e12. 10.1016/j.neuron.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 58.Angevine JB Jr. (1970). Time of neuron origin in the diencephalon of the mouse. An autoradiographic study. J. Comp. Neurol 139, 129–187. 10.1002/cne.901390202. [DOI] [PubMed] [Google Scholar]
- 59.Shi W, Xianyu A, Han Z, Tang X, Li Z, Zhong H, Mao T, Huang K, and Shi SH (2017). Ontogenetic establishment of order-specific nuclear organization in the mammalian thalamus. Nat. Neurosci 20, 516–528. 10.1038/nn.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ebisu H, Iwai-Takekoshi L, Fujita-Jimbo E, Momoi T, and Kawasaki H.(2017). Foxp2 regulates identities and projection patterns of thalamic nuclei during development. Cereb. Cortex 27, 3648–3659. 10.1093/cercor/bhw187. [DOI] [PubMed] [Google Scholar]
- 61.Mercurio S, Serra L, Motta A, Gesuita L, Sanchez-Arrones L, Inverardi F, Foglio B, Barone C, Kaimakis P, Martynoga B, et al. (2019). Sox2 acts in thalamic neurons to control the development of retina-thalamus-cortex connectivity. iScience 15, 257–273. 10.1016/j.isci.2019.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsui D, Vessey JP, Tomita H, Kaplan DR, and Miller FD (2013). FoxP2 regulates neurogenesis during embryonic cortical development. J. Neurosci 33, 244–258. 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chun S, Westmoreland JJ, Bayazitov IT, Eddins D, Pani AK, Smeyne RJ, Yu J, Blundon JA, and Zakharenko SS (2014). Specific disruption of thalamic inputs to the auditory cortex in schizophrenia models. Science 344, 1178–1182. 10.1126/science.1253895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wells MF, Wimmer RD, Schmitt LI, Feng G, and Halassa MM (2016). Thalamic reticular impairment underlies attention deficit in Ptchd1(Y/-) mice. Nature 532, 58–63. 10.1038/nature17427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krol A, Wimmer RD, Halassa MM, and Feng G.(2018). Thalamic reticular dysfunction as a circuit endophenotype in neurodevelopmental disorders. Neuron 98, 282–295. 10.1016/j.neuron.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steullet P.(2020). Thalamus-related anomalies as candidate mechanism-based biomarkers for psychosis. Schizophr. Res 226, 147–157. 10.1016/j.schres.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 67.Jeong Y, El-Jaick K, Roessler E, Muenke M, and Epstein DJ (2006). A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133, 761–772. 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 68.Seigneur E, and Südhof TC (2017). Cerebellins are differentially expressed in selective subsets of neurons throughout the brain. J. Comp. Neurol 525, 3286–3311. 10.1002/cne.24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R.(2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, and Raychaudhuri S.(2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nissim S, Allard P, Bandyopadhyay A, Harfe BD, and Tabin CJ (2007). Characterization of a novel ectodermal signaling center regulating Tbx2 and Shh in the vertebrate limb. Dev. Biol 304, 9–21. 10.1016/j.ydbio.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tischfield DJ, Saraswat DK, Furash A, Fowler SC, Fuccillo MV, and Anderson SA (2017). Loss of the neurodevelopmental gene Zswim6 alters striatal morphology and motor regulation. Neurobiol. Dis 103, 174–183. 10.1016/j.nbd.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fowler SC, Birkestrand BR, Chen R, Moss SJ, Vorontsova E, Wang G, and Zarcone TJ (2001). A force-plate actometer for quantitating rodent behaviors: illustrative data on locomotion, rotation, spatial patterning, stereotypies, and tremor. J. Neurosci. Methods 107, 107–124. 10.1016/s0165-0270(01)00359-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at Short Read Archive (SRA), Gene Omnibus Archive (GEO) (GSE211701), Mendeley Data, and cellxgene and are publicly available as the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.