Abstract
Antisense oligonucleotide (ASO) technology has become an attractive therapeutic modality for various diseases, including Mendelian disorders. ASOs can modulate the expression of a target gene by promoting mRNA degradation or changing pre-mRNA splicing, nonsense-mediated mRNA decay, or translation. Advances in medicinal chemistry and a deeper understanding of post-transcriptional mechanisms have led to the approval of several ASO drugs for diseases that had long lacked therapeutic options. For instance, an ASO drug called nusinersen became the first approved drug for spinal muscular atrophy, improving survival and the overall disease course. Mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene cause cystic fibrosis (CF). Although Trikafta and other CFTR-modulation therapies benefit most CF patients, there is a significant unmet therapeutic need for a subset of CF patients. In this review, we introduce ASO therapies and their mechanisms of action, describe the opportunities and challenges for ASO therapeutics for CF, and discuss the current state and prospects of ASO therapies for CF.
Keywords: antisense oligonucleotide, cystic fibrosis, cystic fibrosis transmembrane conductance regulator, nonsense-mediated mRNA decay, RNA therapeutics, splicing
INTRODUCTION
Antisense oligonucleotide (ASO) technology has emerged as an attractive therapeutic modality for various diseases, thanks to important advances in synthetic nucleic-acid chemical modifications and ligand conjugation that resulted in improved potency, delivery, biodistribution, and stability. ASOs can modulate the expression of target genes by promoting mRNA degradation or otherwise changing post-transcriptional processing or translation. The recent approval of ASO drugs for various diseases that previously lacked adequate treatment options demonstrated their efficacy and safety as precision medicines that could fill the gap in unmet therapeutic needs (Table 1). This review introduces ASO therapies and their mechanisms of action, describes opportunities and challenges of ASO therapeutics for cystic fibrosis (CF), and discusses the current state of such ASO therapies.
Table 1.
Clinically approved ASO therapies
| Drug | Chemistry (mechanism) | Target (organ) | Indication | Delivery route | Key observation | Trial info/source |
|---|---|---|---|---|---|---|
| Mipomersen | 2’-MOE/PS (gapmer) | ApoB-100 (liver) | Familial hypercholesterolemia | Subcutaneous | Reduced apoB-100, LDL-C and VLDL | (Raal et al., 2010) |
| Inotersen | 2’-MOE/PS (gapmer) | TTR (liver) | Hereditary transthyretin amyloidosis | Subcutaneous | Slower progression of neuropathy | (Benson et al., 2018) |
| Volanesorsen | 2’-MOE/PS (gapmer) | ApoC-III (liver) | Familial chylomicronaemia syndrome | Subcutaneous | Reduction of triglycerides and reduced pancreatitis | (Witztum et al., 2019) |
| Eteplirsen | PMO (exon 51 skipping) | Dystrophin exon 51 (muscle) | Duchenne muscular dystrophy | Intravenous | Increased dystrophin production in skeletal muscle | (Pascual-Morena et al., 2020) |
| Golodirsen | PMO (exon 53 skipping) | Dystrophin exon 53 (muscle) | Duchenne muscular dystrophy | Intravenous | Increased dystrophin production in skeletal muscle | (Aartsma-Rus and Corey, 2020) |
| Viltolarsen | PMO (exon 53 skipping) | Dystrophin exon 53 (muscle) | Duchenne muscular dystrophy | Intravenous | Increased dystrophin production in skeletal muscle | (Dhillon, 2020) |
| Casimersen | PMO (exon 45 skipping) | Dystrophin exon 45 (muscle) | Duchenne muscular dystrophy | Intravenous | Increased dystrophin production in skeletal muscle | (Shirley, 2021) |
| Nusinersen | 2’-MOE/PS (exon 7 inclusion) | SMN2 intron 7 (CNS) | Spinal muscular atrophy | Intrathecal | Increased SMN2 exon 7 inclusion, slows or improves the disease course | (Darras et al., 2019; Finkel et al., 2017) |
ASO, antisense oligonucleotide; 2’-MOE, 2’-O-methoxyethyl; PS, phosphorothioate; PMO, phosphorodiamidate morpholino oligomers; CNS, central nervous system.
General properties and chemical modifications of ASOs
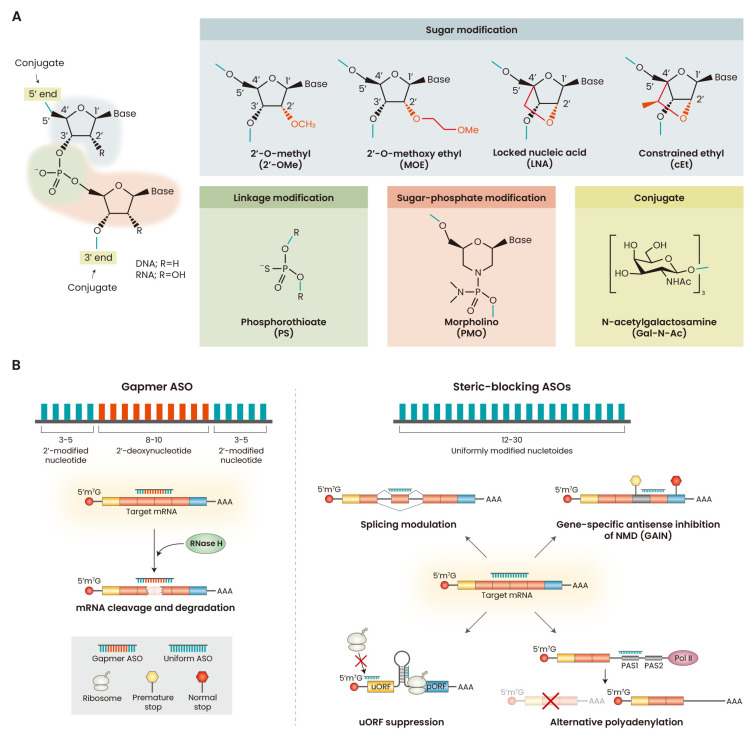
ASOs are synthetic polymers―generally 12-30 nucleotides long―that structurally or functionally mimic DNA/RNA and bind to target RNA via Watson-Crick base paring (Bennett, 2019). ASOs used for biomedical applications have modifications in their sugar-phosphate backbone and nucleobases to confer nuclease resistance and higher binding affinity (Fig. 1A).
Fig. 1. Overview of ASO properties and mechanisms.
(A) ASO chemical modifications. ASOs can be modified at the phosphate inter-nucleotide linkage, and at the 2’ position in the sugar. Replacement of phosphate linkages with PS linkages improves nuclease resistance and improves binding to plasma proteins. Modification of the 2’ position of the sugar (e.g., 2’-OMe, 2’-MOE, LNA, and cEt) enhances nuclease stability and increases affinity for target RNAs. PMO modification replaces the sugar with the morpholino group and the phosphate linkages with neutral phosphorodiamidate linkages. Various molecules can be conjugated to the 3’ or 5’ termini of ASOs (Winkler, 2013). The most popular and successful conjugate is the N-acetylgalactosamine (GalNAc) moiety, which binds to the asialoglycoprotein receptor and enhances delivery of ASOs to hepatocytes. (B) General mechanisms of gene-expression modulation by ASO. Gapmer ASOs downregulate gene expression by inducing RNase H-mediated degradation of the target RNA. Steric-blocking ASOs are uniformly modified ASOs that sterically hinder the binding of proteins or RNPs that regulate post-transcriptional RNA processing (e.g., splicing, NMD, or polyadenylation) or translation. ASO, antisense oligonucleotide; PMO, phosphorodiamidate morpholino oligomers; NMD, nonsense-mediated mRNA decay; uORF, upstream open reading frame; pORF, primary open reading frame; PAS, polyadenylation signal.
Sugar modifications
Various chemical modifications of the ribose have been developed to increase the affinity of ASO for its target sequence (Wan and Seth, 2016). 2’-O-methyl (2’-OMe) and 2’-O-methoxyethyl (2’-MOE) modifications improve the binding affinity and nuclease resistance, prevent cleavage of the RNA strand in the ASO:RNA duplex by RNase H, and reduce pro-inflammatory properties, compared to unmodified nucleotides (Bennett, 2019). Locked nucleic acid (LNA) or constrained ethyl (cEt) nucleotides are nuclease resistant and have enhanced affinity and stability, attributable to the constrained sugar pucker; cEt was reported to have a better toxicity profile than LNA (Wan and Seth, 2016). The increased affinity imparted by LNA/cEt nucleotides necessitates using mixed modified and unmodified nucleotides, resulting in more complex ASO design and screening (Wan and Seth, 2016).
Phosphate backbone modifications
A phosphorothioate (PS) modification that replaces the natural phosphodiester backbone improves plasma protein binding, nuclease resistance, and overall pharmacokinetics (Bennett, 2019). Phosphorodiamidate morpholino oligomers (PMO), which have an uncharged, non-sugar backbone and the standard nucleobases, have also been widely developed and investigated for therapeutic purposes (Shimo et al., 2018). PMOs are neutral, resistant to nucleases, and do not activate RNase H-mediated RNA cleavage; they are typically used for splicing modulation or to inhibit translation (Crooke et al., 2021).
Clinically approved ASOs
The mechanisms of ASO drugs can be categorized into targeted degradation of RNA and occupancy-mediated steric hindrance (Fig. 1B). ‘Gapmer’ ASOs are composed of a window of >5 contiguous DNA nucleotides, flanked by modified RNA-like nucleotides, and they promote RNase H-mediated cleavage of the target RNA, which becomes destabilized (Bennett, 2019; Crooke et al., 2021). In contrast, steric-blocking ASOs are designed to prevent the binding of proteins or RNPs (ribonucleoprotein complexes) that are important for RNA metabolism and processing (Crooke et al., 2021). These ASOs are composed of uniformly modified nucleotides or a combination of modified and DNA nucleotides (Khvorova and Watts, 2017). Steric-blocking ASOs are particularly useful for modulating splicing or other post-transcriptional gene-expression steps. Since the first approval of the ASO drug fomiversen―for cytomegalovirus-associated retinitis, but later withdrawn―three other gapmer ASOs and five splice-switching ASOs have been approved (Table 1). These ASO therapies have significantly impacted the clinical course of patients who lacked therapeutic options for a long time.
Overview of CF and CF transmembrane conductance regulator (CFTR)-targeted therapies
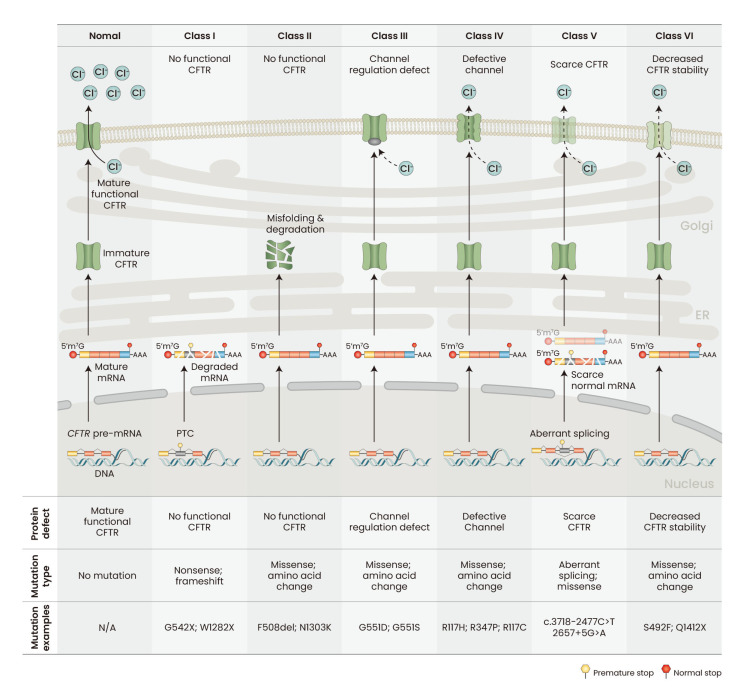
CF is caused by loss-of-function mutations in the CFTR gene, inherited in an autosomal recessive fashion. CFTR encodes the CFTR protein, which regulates chloride transport across the plasma membrane, thus playing a critical role in the homeostasis of water and electrolytes in the airway and various epithelial-layer surfaces. Respiratory failure is the most common cause of CF morbidity and mortality, but CF chronically and progressively affects multiple other organ systems (Shteinberg et al., 2021). CFTR mutations are categorized into six classes (I-VI) (Fig. 2) (De Boeck and Amaral, 2016). Class I mutations result in the lack of CFTR protein production (e.g., G542X, W1282X). Class II mutations are missense mutations that lead to improper post-translational processing of CFTR protein; a representative example is the F508del mutation, present in most CF patients globally (CFTR2, 2022; Cystic Fibrosis Foundation, 2021). Class III mutations (e.g., G551D) are defective in channel-gating regulation. Class IV mutations (e.g., R117H) produce CFTR proteins with channel defects. Class V mutations lead to reduced CFTR protein synthesis, and many of them affect pre-mRNA splicing (e.g., c.3718-2477C>T). Finally, class VI mutations lead to decreased CFTR stability, due to missense mutations or truncation of the C-terminus (e.g., S492F or Q1412X).
Fig. 2. Classification of CFTR mutations.
CFTR, cystic fibrosis transmembrane conductance regulator; ER, endoplasmic reticulum; PTC, premature termination codon; N/A, not applicable. Mutation classification based on De Boeck and Amaral (2016).
The development of CFTR modulators, which are small-molecule drugs that modify the function or post-translational processing of the protein, resulted in significant improvements in the lung function and life expectancy of CF patients. CFTR modulators are categorized into potentiators, correctors, and amplifiers. Several potentiators and correctors have been approved for the treatment of CF, and have been extensively reviewed elsewhere (Zaher et al., 2021). Potentiators increase CFTR function by increasing the channel-opening probability (Ramsey et al., 2011; Van Goor et al., 2009), whereas correctors improve the trafficking of mutant CFTR, such as F508del, from the endoplasmic reticulum to the plasma membrane (Fiedorczuk and Chen, 2022; Southern et al., 2020). Amplifiers enhance CFTR function by increasing mRNA or CFTR protein stability, but they are not yet approved for therapy (Dukovski et al., 2020; Giuliano et al., 2018). The most recently approved drug, Trikafta, is a cocktail of a potentiator (VX-770) and two correctors (VX-445 and VX-661) that significantly enhances lung function and quality of life, and reduces severe pulmonary infection (Heijerman et al., 2019; Keating et al., 2018; Middleton et al., 2019). Trikafta is approved for use in patients with at least one F508del allele, i.e., homozygous or compound heterozygous with another CF-causing mutation, thus benefiting about 85% of CF patients (Voelker, 2019). The remaining patients are not eligible for Trikafta, so there is still a significant unmet need for new CFTR-enhancing drugs. CFTR modulators are helpful when there is sufficient CFTR protein expression for them to act on. Various mutations, including nonsense mutations (e.g., G542X and W1282X) and splicing mutations (e.g., c.3718-2477C>T and 2657+5G>A) result in very low levels of CFTR expression, precluding the effective use of modulators (Voelker, 2019).
ASOs UNDER DEVELOPMENT FOR CF
ASOs targeting CFTR nonsense mutations
Nonsense-mediated mRNA decay (NMD) is a cellular mechanism that prevents the accumulation of potentially harmful truncated proteins translated from PTC (premature termination codon)-containing mRNAs (Kurosaki et al., 2019). Nonsense mutations account for ~8% of CF-causing CFTR mutations (CFTR1, 2022), and CFTR mRNAs with nonsense mutations in the penultimate exon or farther upstream are degraded through NMD, leading to very low CFTR expression and function (Clarke et al., 2019). NMD inhibition can potentially increase the expression of some truncated CFTR proteins with residual function. For instance, W1282X protein retains 1281 out of 1480 amino acids in the full-length CFTR protein, and its function can be improved with CFTR modulators if its expression is sufficiently increased (Haggie et al., 2017; Mutyam et al., 2017; Sanderlin et al., 2022; Wang et al., 2016). A relatively small increase in CFTR function, to 10%-30% of normal CFTR function, is expected to be clinically beneficial (Kerem, 2004; Van Goor et al., 2009).
A class of drugs called read-through compounds (RTCs) increases the level of full-length protein by reducing the fidelity of the ribosome at the PTC (Lentini et al., 2014). Studies showed that NMD is an important therapeutic target for CF caused by class I mutations; combining newer-generation RTCs currently under pre-clinical and clinical investigation with NMD inhibition could be useful for CF (Clancy et al., 2007; Huang et al., 2018; Keeling et al., 2013; Keenan et al., 2019; Kerem, 2020; Linde et al., 2007; Sharma et al., 2021; Valley et al., 2019; Zainal Abidin et al., 2017).
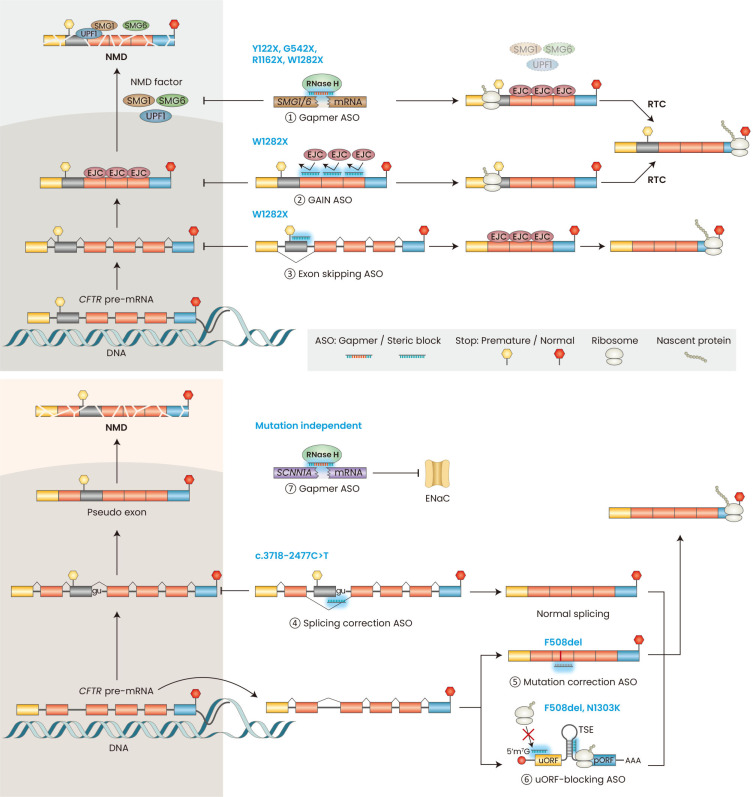
SMG1 is a kinase that phosphorylates the RNA helicase UPF1—an essential step in NMD that leads to the recruitment of the endonuclease SMG6, which cleaves the target mRNA (Kurosaki et al., 2019). cEt/PS-modified gapmer ASOs that reduce SMG1 or SMG6 expression inhibit NMD and increase the level of PTC-containing CFTR mRNA close to that of wild-type CFTR mRNA in human bronchial epithelial cell lines (CFF16HBEge) harboring homozygous Y122X, G542X, R1162X, or W1282X mutations (Fig. 3, Table 2) (Keenan et al., 2019; Sanderlin et al., 2022). CFF16HBEge cells were generated by introducing homozygous mutations into the endogenous CFTR loci of the parental human bronchial epithelial cell line (16HBE14o-) using CRISPR-Cas9 technology (Valley et al., 2019). In W1282X-mutant CFF16HBEge cells, SMG1 or SMG6 gapmer ASO increased CFTR protein levels; combining the gapmer ASO with VX-770 increased CFTR function; and adding the RTC G418 further increased CFTR function in these cells (Keenan et al., 2019; Sanderlin et al., 2022). In contrast, CFTR function significantly increased in Y122X, G542X, or R1162X mutant CFF16HBEge cells only when the gapmer ASO, VX-770, and G418 were combined (Sanderlin et al., 2022). Thus, the residual CFTR function of each truncating mutation affects the response to NMD inhibition and RTC. The SMG1 and SMG6 gapmers plus VX-770 plus G418 treatments increased CFTR function to ~2% to 4% of wild-type levels (Sanderlin et al., 2022). The addition of correctors, such as VX-445/661, may further increase CFTR function.
Fig. 3. Mechanisms of ASOs for CF.
Overview of the available mechanisms for class I mutations (top panel) and other mutations (bottom panel). Gapmer ASOs can trigger RNase H-mediated downregulation of NMD-factor mRNAs (①) in the cytoplasm and nucleus. Uniformly modified ASOs that block the binding of EJCs downstream of a PTC can specifically stabilize nonsense-mutant CFTR mRNA (②), which is translated to make truncated CFTR protein or, in the presence of RTC, to make full-length CFTR. Uniformly modified ASOs targeting the splice sites or ESEs (exonic splicing enhancers) of an in-frame exon containing a PTC can induce exon skipping, and the resulting mRNA can be translated to produce partially functional CFTR (③). Uniformly modified ASOs can correct aberrant splicing mutations (④). Fully modified ASOs can also insert missing bases in the mutant CFTR mRNA through an unknown RNA-repair mechanism (⑤). Enhance translation in the pORF of CFTR by suppressing translation in the uORF by binding to the uORF start codon or TSE in the 5’ UTR (⑥). Gapmer ASOs can trigger RNase H-mediated downregulation of CF modifier genes, such as SCNN1A, to improve mucociliary clearance (⑦). ASO, antisense oligonucleotide; CF, cystic fibrosis; NMD, nonsense-mediated mRNA decay; EJCs, exon-junction complex(es); PTC, premature termination codon; CFTR, cystic fibrosis transmembrane conductance regulator; RTC, read-through compound; TSE, translational-suppression elements; uORF, upstream open reading frame; pORF, primary open reading frame.
Table 2.
ASOs under preclinical development for CF therapy
| Mutation | Target gene | Chemistry | Mechanism | Key observation | Reference |
|---|---|---|---|---|---|
| Y122X G542X R1162X W1282X |
SMG1
SMG6 |
cEt/PS gapmer | RNase H-mediated knockdown | SMG1/6 knockdown increases CFTR expression. The ASOs combined with read-through compound increases CFTR function in HBE cells. | (Keenan et al., 2019; Sanderlin et al., 2022) |
| W1282X | CFTR | Uniform 2’-MOE/PS |
GAIN | Gene-specific NMD inhibition of CFTR-W1282X mRNA increases CFTR expression and CFTR function in HBE cells. | (Kim et al., 2022a) |
| W1282X | CFTR | PMO Uniform 2’-MOE/PS |
Exon skipping | Skipping of exon 23 containing W1282X mutation increases expression of CFTR-Δex23 isoform and increases CFTR function in HBE and HNE cells. | (Kim et al., 2022b; Michaels et al., 2022; Oren et al., 2022) |
| c.3718-2477C>T | CFTR | PMO PPMO Uniform 2’-MOE/PS 2’-OMe/PS |
Exon skipping | Splice correction ASOs suppress inclusion of pseudo-exon, restores normal splicing, and increase CFTR function in HBE and HNE cells. | (Dang et al., 2021; Michaels et al., 2020; Oren et al., 2021) |
| F508del N1303K |
CFTR | Uniform 2’-MOE/PS |
uORF suppression | Suppressing uORF translation increases CFTR expression and function in HBE cells. | (Sasaki et al., 2019) |
ASO, antisense oligonucleotide; CF, cystic fibrosis; cEt, constrained ethyl; PS, phosphorothioate; CFTR, cystic fibrosis transmembrane conductance regulator; HBE, human bronchial epithelial; 2’-MOE, 2’-O-methoxyethyl; GAIN, gene-specific antisense inhibition of NMD (nonsense-mediated mRNA decay); PMO, phosphorodiamidate morpholino oligomers; HNE, human nasal epithelial; PPMO, peptide-conjugated PMO; 2’-OMe, 2’-O-methyl; uORF, upstream open reading frame.
Global inhibition of NMD by gapmer ASOs may disrupt mRNA homeostasis in a broad range of tissues, considering that the NMD machinery post-transcriptionally regulates gene expression of a subset of normal and physiologically functional mRNA isoforms (Kurosaki et al., 2019). Thus, inhibiting NMD of CFTR mRNA specifically may be therapeutically advantageous. CFTR-specific NMD inhibition can be achieved by using a technique dubbed gene-specific antisense inhibition of NMD (GAIN), in which ASOs are used to disrupt the binding of exon-junction complex(es) (EJC) downstream of a PTC (Fig. 3, Table 2) (Kim et al., 2022a; Nomakuchi et al., 2016). GAIN ASO cocktails composed of three 2’-MOE/PS-modified ASOs that disrupt EJC binding at three sites downstream of the W1282X mutation specifically increase CFTR mRNA without suppressing global NMD in CFF16HBEge-W1282X cells (Kim et al., 2022a). The increase in the CFTR mRNA expression was close to the levels reported for wild-type CFTR mRNA (Sanderlin et al., 2022). The GAIN ASO cocktail plus VX-770/809 or VX-770/445/661 increased CFTR-mediated chloride current in the mutant human bronchial epithelial (HBE) cells up to 30% of wild-type CFTR (Kim et al., 2022a). The GAIN ASO cocktail also enhanced the effect of G418 on CFTR function. The GAIN ASOs caused some exon-skipping events downstream of the PTC on CFTR-W1282X mRNA, thus not affecting the resulting amino-acid sequence. However, because of these splicing changes upstream of the normal termination codon, the GAIN strategy may only be suitable for homozygous W1282X mutations or compound heterozygous mutations with W1282X and a severe CFTR allele not amenable to currently approved therapies.
Alternatively, exon-skipping ASOs can restore CFTR function in HBE cells harboring the W1282X mutation by inducing the skipping of the exon containing the PTC. The resulting CFTR-Δ23 mRNA isoform, which lacks the in-frame exon 23 with the W1282X mutation, is not degraded by NMD (Kim et al., 2022b; Michaels et al., 2022; Oren et al., 2022). CFTR-Δ23 protein lack residues 1240 through 1291, but retains functional domains important for CFTR function that are missing in CFTR-W1282X protein (Kim et al., 2022b). Exon-skipping ASOs with uniform 2’-MOE/PS (Kim et al., 2022b; Oren et al., 2022) or PMO (Michaels et al., 2022) modification targeting the splice sites, exonic splicing enhancer (ESE) elements, or both, induced efficient exon 23 skipping in W1282X mutant CFF16HBEge cells (Fig. 3, Table 2). Exon 23 skipping ASOs combined with VX-770/809 or VX-770/445/661 led to higher CFTR protein levels and CFTR function in the W1282X mutant CFF16HBEge- cells, patient-derived HBE cells (heterozygous W1282X/F508del), and patient-derived human nasal epithelial (HNE) cells (homozygous W1282X/W1282X) (Kim et al., 2022b; Michaels et al., 2022; Oren et al., 2022). Oren et al. (2022) estimated that one of their lead exon-skipping ASOs combined with VX-770/445/661 increased CFTR function to up to 20% of wild-type CFTR. The exon-skipping approach cannot be combined with RTC, due to the lack of a PTC on CFTR-Δ23 mRNA, but it may be an effective approach if combined with a CFTR modulator therapy.
ASOs targeting CFTR splicing mutations
Splicing mutations account for approximately ~10% of CF-causing CFTR mutations (CFTR1, 2022). CFTR c.3718-2477C>T (also known as 3849+10 kb C>T) is the 10th most common CFTR mutation (CFTR2, 2022). The C>T mutation generates a new 5’ splice site (5’ss) far downstream of the canonical 5’ss in intron 22, resulting in splicing of an 84-nucleotide pseudo-exon with a PTC; to the resulting mRNA undergoes NMD, with the consequent reduction in functional CFTR (Chiba-Falek et al., 1998). 2’-OMe/PS splice switching ASOs targeting the 3’ splice site (3’ss) or 5’ss of the pseudo-exon restored the normal splicing pattern of a CFTR minigene with the c.3718-2477C>T mutation (Friedman et al., 1999). Since then, three other studies showed that splice-switching ASOs targeting the splice sites or ESEs suppress pseudo-exon inclusion and increase the normal CFTR mRNA isoform (Fig. 3, Table 2) (Dang et al., 2021; Michaels et al., 2020; Oren et al., 2021). Michaels et al. (2022) used uniformly PMO-modified ASOs; Dang et al. (2021) used peptide-conjugated PMO (PPMO) ASOs combined with oligonucleotide-enhancing compounds (OEC, dubbed UNC7938 and UNC2383) that promote endosomal escape of ASOs and aid their delivery to the nucleus; Oren et al. (2021) used 2’-MOE/PS ASOs based on the prototype ASOs with 2’-OMe/PS modification. These ASOs significantly reduced the inclusion of the pseudo-exon in patient-derived HBE or HNE cells. Even though these ASOs do not completely suppress pseudo-exon inclusion, the increase in the normal CFTR mRNA led to a substantial increase in normal CFTR protein and CFTR-mediated chloride current. In patient-derived HNE and HBE cells harboring compound c.3718-2477C>T/F508del mutations, the estimated CFTR function with 2’-MOE/PS ASO plus VX-770 treatment was ~43% of wild-type CFTR function. In patient-derived HBE cells with homozygous c.3718-2477C>T mutation, the PMO ASO plus a potentiator (C-18) increased CFTR by 3-fold, and PPMO plus OEC plus Trikafta increased CFTR activity by 6- to 10-fold. Currently, SpliSense is pursuing development of SPL84-23-1, one of the lead ASOs described by Oren et al. (2021) (Table 3) (Cystic Fibrosis Foundation, 2022). All groups showed that splice-switching ASOs plus CFTR modulators increased CFTR function more than CFTR modulators alone. Even though Trikafta is approved for patients with the c.3718-2477C>T/F508del mutations (Zaher et al., 2021), splice-switching ASOs combined with CFTR modulators may be a useful therapeutic strategy for CF patients with the homozygous or other compound heterozygous c.3718-2477C>T mutation.
Table 3.
Commercially developed ASOs for CF therapy
| Drug | Chemistry | Does route | Development Phase | Key observation | Trial info/source |
|---|---|---|---|---|---|
| IONIS-ENaC-2.5-Rx | cEt/PS gapmer | Inhaled aerosolized ASO | Phase-1/2a (discontinued) | Decrease in SCNN1A mRNA levels in lung gavage samples Well tolerated |
NCT03647228 |
| Eluforsen | 2’O-Me/PS | Inhaled aerosolized ASO | Phase-1b (discontinued) | Improved CF quality of life in patient survey Well tolerated |
NCT02532764 |
| SPL84-23-1 | 2’MOE/PS | N/A | Preclinical | Corrects aberrant splicing caused by c.3718-2477C>T | (Oren et al., 2021) |
| SPL23 | 2’MOE/PS | N/A | Preclinical | Skips exon 23 containing the nonsense W1282X mutation | (Oren et al., 2022) |
ASO, antisense oligonucleotide; CF, cystic fibrosis; cEt, constrained ethyl; PS, phosphorothioate; 2’-OMe, 2’-O-methyl; 2’-MOE, 2’-O-methoxyethyl; N/A, not applicable.
ASOs targeting CFTR mRNA translation and stability
ASOs can be designed to upregulate translation. For example, ASOs complementary to upstream open reading frames (uORFs) and translation-inhibitory elements can increase gene expression in vivo (Crooke et al., 2021). The 5’ untranslated region (UTR) of CFTR mRNA comprises an upstream AUG (uAUG) translational initiation site and translational-suppression elements (TSE) that can be blocked with uniformly 2’-MOE/PS-modified ASOs (Fig. 3, Table 2) (Sasaki et al., 2019). The uORF-blocking ASOs suppressed translation initiation at the uORF and significantly increased the CFTR protein localized to the membrane and the CFTR-mediated chloride current in 16HBE14o- cells. uORF-blocking ASOs increased the expression and function of F508del and N1303K mutant CFF16HBEge cells, but not W1282X mutant CFF16HBEge cells. In patient-derived primary HBE cells harboring at least one F508del mutation and another CFTR mutation that retains partial function, combining the uORF-blocking ASOs with VX-770/661 increased CFTR function to 15% to 45% of wild-type CFTR, depending on the mutations. One challenge of this approach is that dose-dependent effects of the uAUG and TSE-blocking ASOs were not evident in primary HBE cells. Further investigating the interplay among the ASO’s uORF-suppression mechanism, dose-dependent toxicity, and ASO delivery method may facilitate development of this approach.
PERSPECTIVES FOR THE FUTURE DEVELOPMENT OF ASO DRUGS FOR CF
Delivery of ASOs for CF to the lung
Chronic pulmonary inflammation and mucus plugging are hallmarks of CF (Shteinberg et al., 2021). Thus, efficient delivery of ASOs to ciliated airway epithelial cells in the upper airway remains an important challenge for therapeutic development. Aerosolized ASO, rather than systemic delivery, is a clinically relevant method of intratracheal delivery to the airways (Templin et al., 2000). Studies of aerosolized ASOs in mice and monkeys demonstrated that aerosolized ASOs have relatively long half-lives in the lungs (4 and 7 days, respectively), low toxicity, favorable lung-tissue accumulation, and superior bioavailability compared to intravenously administered ASOs (Crooke, 2007; Crosby et al., 2017; Templin et al., 2000). Systemic exposure is minimal; kidney and liver exposure of inhaled ASOs has been reported, but the level of exposure is significantly less than obtained by systemic administration of ASOs, as the amount of drug is much lower (Crooke, 2007; Moschos et al., 2017).
In clinical trials, several aerosolized ASOs developed for CF therapy had favorable safety profiles and showed exposure in the intended lung tissues. IONIS-ENaC-2.5-Rx, developed by Ionis Pharmaceuticals, is a cEt/PS gapmer ASO that downregulates SCNNA1 to prevent and reverse lung pathology in CF (Table 3, Fig. 3). In a phase-1/2a clinical trial (NCT03647228), the ASO showed significant in vivo activity without significant safety concerns, but its development was recently discontinued due to long-term toxicity in the preclinical model (Pinto et al., 2021). Eluforsen (also known as QR-010), developed by ProQR Therapeutics, is a uniform 2’-OMe/PS ASO that inserts the three missing bases in the mutant CFTR-F508del mRNA through an unknown mechanism, converting it into wild-type CFTR mRNA (Table 3, Fig. 3) (Drevinek et al., 2020). In a phase-1b clinical trial, eluforsen improved patients’ quality of life but did not significantly improve lung function or sweat chloride, compared to the placebo (Drevinek et al., 2020). Even though a subgroup analysis showed some improvement in pulmonary function compared to the placebo, its development was discontinued (Cystic Fibrosis Foundation, 2022).
Future challenges and considerations
ASO-based therapeutics provide a unique opportunity to address the unmet needs of CF, especially in combination with CFTR modulator therapies. Despite favorable characteristics, significant challenges for ASO drug development remain. A major challenge is maximizing the efficacy and minimizing the toxicity of ASO drugs. The potency and toxicity of ASOs can be affected by various factors, such as chemical modifications, off-target effects, and delivery methods. Safety profiles are well-established for various ASO chemical modifications, and each chemical class of ASO has unique safety profiles (Crooke et al., 2021). Optimizing the chemical modification, sequence, and delivery methods of ASOs could maximize on-target effects and reduce off-target effects (Scharner et al., 2020).
Localized delivery of aerosolized ASO for CF depends in part on the properties of the diseased airway but can be improved by optimizing the properties of aerosolized ASO droplets (Labiris and Dolovich, 2003). Delivery of ASOs to the desired cell types for CF therapy may be improved by using conjugated ASOs. GalNAc-conjugated ASO is widely used to enhance delivery to hepatocytes, and other conjugates such as lipids, carbohydrates, peptides, or aptamers may enhance delivery to other target tissues (Fig. 1) (Crooke et al., 2021; Winkler, 2013). CFTR is highly expressed in ciliated cells and serous cells of the submucosal glands, but ciliated-cell-specific expression of CFTR in Cftr-null mice does not rescue the ion-transport defects (Ostrowski et al., 2007). Hence, the target cell type of ASO therapy for CF is not entirely clear. Recently, single-cell RNA sequencing experiments revealed a cell type dubbed ‘pulmonary ionocyte,’ with high expression of Cftr in mice and CFTR in humans, which could play an important role in CF pathology (Harris, 2021). Future investigation may reveal which cell type(s) should be targeted for effective CF ASO therapies.
There is much room to improve ASO therapeutics for CF. Ongoing research and improvements in ASO technology, together with a better understanding of RNA regulatory mechanisms, may provide new therapeutic modalities for CF patients with unmet therapeutic needs.
ACKNOWLEDGMENTS
A.R.K. acknowledges support from NIH grant R37GM042699.
Footnotes
AUTHOR CONTRIBUTIONS
Y.J.K. and A.R.K. wrote the article.
CONFLICT OF INTEREST
The authors declare the following competing interests: A.R.K. is an inventor in issued patent US20160194630A1, “Reducing nonsense-mediated mRNA decay”, assigned to Cold Spring Harbor Laboratory. A.R.K. is a co-founder, Director, and Chair of the SAB of Stoke Therapeutics. The other author has no potential conflicts of interest to disclose.
REFERENCES
- Aartsma-Rus A., Corey D.R. The 10th oligonucleotide therapy approved: golodirsen for Duchenne muscular dystrophy. Nucleic Acid Ther. 2020;30:67–70. doi: 10.1089/nat.2020.0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.F. Therapeutic antisense oligonucleotides are coming of age. Annu. Rev. Med. 2019;70:307–321. doi: 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- Benson M.D., Waddington-Cruz M., Berk J.L., Polydefkis M., Dyck P.J., Wang A.K., Planté-Bordeneuve V., Barroso F.A., Merlini G., Obici L., et al. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:22–31. doi: 10.1056/NEJMoa1716793. [DOI] [PubMed] [Google Scholar]
- CFTR1 (Cystic Fibrosis Mutation Database), author CFMDB Statistics. 2022. [Retrieved October 22, 2022]. from http://www.genet.sickkids.on.ca/StatisticsPage.html.
- CFTR2 (Clinical and Functional TRanslation of CFTR), author CFTR2 Variant List History. 2022. [Retrieved April 29, 2022]. from https://cftr2.org/sites/default/files/CFTR2_29April2022.xlsx .
- Chiba-Falek O., Kerem E., Shoshani T., Aviram M., Augarten A., Bentur L., Tal A., Tullis E., Rahat A., Kerem B. The molecular basis of disease variability among cystic fibrosis patients carrying the 3849+10 kb C→T mutation. Genomics. 1998;53:276–283. doi: 10.1006/geno.1998.5517. [DOI] [PubMed] [Google Scholar]
- Clancy J.P., Rowe S.M., Bebok Z., Aitken M.L., Gibson R., Zeitlin P., Berclaz P., Moss R., Knowles M.R., Oster R.A., et al. No detectable improvements in cystic fibrosis transmembrane conductance regulator by nasal aminoglycosides in patients with cystic fibrosis with stop mutations. Am. J. Respir. Cell Mol. Biol. 2007;37:57–66. doi: 10.1165/rcmb.2006-0173OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L.A., Awatade N.T., Felicio V.M., Silva I.A., Calucho M., Pereira L., Azevedo P., Cavaco J., Barreto C., Bertuzzo C., et al. The effect of premature termination codon mutations on CFTR mRNA abundance in human nasal epithelium and intestinal organoids: a basis for read-through therapies in cystic fibrosis. Hum. Mutat. 2019;40:326–334. doi: 10.1002/humu.23692. [DOI] [PubMed] [Google Scholar]
- Crooke S.T. Antisense Drug Technology: Principles, Strategies, and Applications. (2nd Edition) CRC Press; Boca Raton: 2007. [DOI] [Google Scholar]
- Crooke S.T., Baker B.F., Crooke R.M., Liang X.H. Antisense technology: an overview and prospectus. Nat. Rev. Drug Discov. 2021;20:427–453. doi: 10.1038/s41573-021-00162-z. [DOI] [PubMed] [Google Scholar]
- Crosby J.R., Zhao C., Jiang C., Bai D., Katz M., Greenlee S., Kawabe H., McCaleb M., Rotin D., Guo S., et al. Inhaled ENaC antisense oligonucleotide ameliorates cystic fibrosis-like lung disease in mice. J. Cyst. Fibros. 2017;16:671–680. doi: 10.1016/j.jcf.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Cystic Fibrosis Foundation, author. 2020 Patient Registry Annual Data Report. 2021. [Retrieved September 15, 2021]. from https://www.cff.org/sites/default/files/2021-10/2019-Patient-Registry-Annual-Data-Report.pdf .
- Cystic Fibrosis Foundation, author. Drug Development Pipeline. 2022. [Retrieved October 23, 2022]. from https://apps.cff.org/trials/pipeline/
- Dang Y., van Heusden C., Nickerson V., Chung F., Wang Y., Quinney N.L., Gentzsch M., Randell S.H., Moulton H.M., Kole R., et al. Enhanced delivery of peptide-morpholino oligonucleotides with a small molecule to correct splicing defects in the lung. Nucleic Acids Res. 2021;49:6100–6113. doi: 10.1093/nar/gkab488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras B.T., Farrar M.A., Mercuri E., Finkel R.S., Foster R., Hughes S.G., Bhan I., Farwell W., Gheuens S. An integrated safety analysis of infants and children with symptomatic spinal muscular atrophy (SMA) treated with nusinersen in seven clinical trials. CNS Drugs. 2019;33:919–932. doi: 10.1007/s40263-019-00656-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boeck K., Amaral M.D. Progress in therapies for cystic fibrosis. Lancet Respir. Med. 2016;4:662–674. doi: 10.1016/S2213-2600(16)00023-0. [DOI] [PubMed] [Google Scholar]
- Dhillon S. Viltolarsen: first approval. Drugs. 2020;80:1027–1031. doi: 10.1007/s40265-020-01339-3. [DOI] [PubMed] [Google Scholar]
- Drevinek P., Pressler T., Cipolli M., De Boeck K., Schwarz C., Bouisset F., Boff M., Henig N., Paquette-Lamontagne N., Montgomery S., et al. Antisense oligonucleotide eluforsen is safe and improves respiratory symptoms in F508DEL cystic fibrosis. J. Cyst. Fibros. 2020;19:99–107. doi: 10.1016/j.jcf.2019.05.014. [DOI] [PubMed] [Google Scholar]
- Dukovski D., Villella A., Bastos C., King R., Finley D., Kelly J.W., Morimoto R.I., Hartl F.U., Munoz B., Lee P.S., et al. Amplifiers co-translationally enhance CFTR biosynthesis via PCBP1-mediated regulation of CFTR mRNA. J. Cyst. Fibros. 2020;19:733–741. doi: 10.1016/j.jcf.2020.02.006. [DOI] [PubMed] [Google Scholar]
- Fiedorczuk K., Chen J. Mechanism of CFTR correction by type I folding correctors. Cell. 2022;185:158–168.e11. doi: 10.1016/j.cell.2021.12.009. [DOI] [PubMed] [Google Scholar]
- Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- Friedman K.J., Kole J., Cohn J.A., Knowles M.R., Silverman L.M., Kole R. Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J. Biol. Chem. 1999;274:36193–36199. doi: 10.1074/jbc.274.51.36193. [DOI] [PubMed] [Google Scholar]
- Giuliano K.A., Wachi S., Drew L., Dukovski D., Green O., Bastos C., Cullen M.D., Hauck S., Tait B.D., Munoz B., et al. Use of a high-throughput phenotypic screening strategy to identify amplifiers, a novel pharmacological class of small molecules that exhibit functional synergy with potentiators and correctors. SLAS Discov. 2018;23:111–121. doi: 10.1177/2472555217729790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggie P.M., Phuan P.W., Tan J.A., Xu H., Avramescu R.G., Perdomo D., Zlock L., Nielson D.W., Finkbeiner W.E., Lukacs G.L., et al. Correctors and potentiators rescue function of the truncated W1282X-cystic fibrosis transmembrane regulator (CFTR) translation product. J. Biol. Chem. 2017;292:771–785. doi: 10.1074/jbc.M116.764720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. Human Molecular Genetics and the long road to treating cystic fibrosis. Hum. Mol. Genet. 2021;30:R264–R273. doi: 10.1093/hmg/ddab191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijerman H.G.M., McKone E.F., Downey D.G., Van Braeckel E., Rowe S.M., Tullis E., Mall M.A., Welter J.J., Ramsey B.W., McKee C.M., et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet. 2019;394:1940–1948. doi: 10.1016/S0140-6736(19)32597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Low A., Damle S.S., Keenan M.M., Kuntz S., Murray S.F., Monia B.P., Guo S. Antisense suppression of the nonsense mediated decay factor Upf3b as a potential treatment for diseases caused by nonsense mutations. Genome Biol. 2018;19:4. doi: 10.1186/s13059-017-1386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating D., Marigowda G., Burr L., Daines C., Mall M.A., McKone E.F., Ramsey B.W., Rowe S.M., Sass L.A., Tullis E., et al. VX-445-tezacaftor-ivacaftor in patients with cystic fibrosis and one or two Phe508del alleles. N. Engl. J. Med. 2018;379:1612–1620. doi: 10.1056/NEJMoa1807120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling K.M., Wang D., Dai Y., Murugesan S., Chenna B., Clark J., Belakhov V., Kandasamy J., Velu S.E., Baasov T., et al. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One. 2013;8:e60478. doi: 10.1371/journal.pone.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan M.M., Huang L., Jordan N.J., Wong E., Cheng Y., Valley H.C., Mahiou J., Liang F., Bihler H., Mense M., et al. Nonsense-mediated RNA decay pathway inhibition restores expression and function of W1282X CFTR. Am. J. Respir. Cell Mol. Biol. 2019;61:290–300. doi: 10.1165/rcmb.2018-0316OC. [DOI] [PubMed] [Google Scholar]
- Kerem E. Pharmacologic therapy for stop mutations: how much CFTR activity is enough? Curr. Opin. Pulm. Med. 2004;10:547–552. doi: 10.1097/01.mcp.0000141247.22078.46. [DOI] [PubMed] [Google Scholar]
- Kerem E. ELX-02: an investigational read-through agent for the treatment of nonsense mutation-related genetic disease. Expert Opin. Investig. Drugs. 2020;29:1347–1354. doi: 10.1080/13543784.2020.1828862. [DOI] [PubMed] [Google Scholar]
- Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Nomakuchi T., Papaleonidopoulou F., Yang L., Zhang Q., Krainer A.R. Gene-specific nonsense-mediated mRNA decay targeting for cystic fibrosis therapy. Nat. Commun. 2022a;13:2978. doi: 10.1038/s41467-022-30668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Sivetz N., Layne J., Voss D.M., Yang L., Zhang Q., Krainer A.R. Exon-skipping antisense oligonucleotides for cystic fibrosis therapy. Proc. Natl. Acad. Sci. U. S. A. 2022b;119:e2114858118. doi: 10.1073/pnas.2114858118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Popp M.W., Maquat L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019;20:406–420. doi: 10.1038/s41580-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labiris N.R., Dolovich M.B. Pulmonary drug delivery. Part I: physiological factors affecting therapeutic effectiveness of aerosolized medications. Br. J. Clin. Pharmacol. 2003;56:588–599. doi: 10.1046/j.1365-2125.2003.01892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini L., Melfi R., Di Leonardo A., Spinello A., Barone G., Pace A., Palumbo Piccionello A., Pibiri I. Toward a rationale for the PTC124 (Ataluren) promoted readthrough of premature stop codons: a computational approach and GFP-reporter cell-based assay. Mol. Pharm. 2014;11:653–664. doi: 10.1021/mp400230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde L., Boelz S., Nissim-Rafinia M., Oren Y.S., Wilschanski M., Yaacov Y., Virgilis D., Neu-Yilik G., Kulozik A.E., Kerem E., et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J. Clin. Invest. 2007;117:683–692. doi: 10.1172/JCI28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels W.E., Bridges R.J., Hastings M.L. Antisense oligonucleotide-mediated correction of CFTR splicing improves chloride secretion in cystic fibrosis patient-derived bronchial epithelial cells. Nucleic Acids Res. 2020;48:7454–7467. doi: 10.1093/nar/gkaa490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels W.E., Pena-Rasgado C., Kotaria R., Bridges R.J., Hastings M.L. Open reading frame correction using splice-switching antisense oligonucleotides for the treatment of cystic fibrosis. Proc. Natl. Acad. Sci. U. S. A. 2022;119:e2114886119. doi: 10.1073/pnas.2114886119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton P.G., Mall M.A., Drevinek P., Lands L.C., McKone E.F., Polineni D., Ramsey B.W., Taylor-Cousar J.L., Tullis E., Vermeulen F., et al. Elexacaftor-tezacaftor-ivacaftor for cystic fibrosis with a single Phe508del allele. N. Engl. J. Med. 2019;381:1809–1819. doi: 10.1056/NEJMoa1908639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschos S.A., Usher L., Lindsay M.A. Clinical potential of oligonucleotide-based therapeutics in the respiratory system. Pharmacol. Ther. 2017;169:83–103. doi: 10.1016/j.pharmthera.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Mutyam V., Libby E.F., Peng N., Hadjiliadis D., Bonk M., Solomon G.M., Rowe S.M. Therapeutic benefit observed with the CFTR potentiator, ivacaftor, in a CF patient homozygous for the W1282X CFTR nonsense mutation. J. Cyst. Fibros. 2017;16:24–29. doi: 10.1016/j.jcf.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomakuchi T.T., Rigo F., Aznarez I., Krainer A.R. Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat. Biotechnol. 2016;34:164–166. doi: 10.1038/nbt.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren Y.S., Avizur-Barchad O., Ozeri-Galai E., Elgrabli R., Schirelman M.R., Blinder T., Stampfer C.D., Ordan M., Laselva O., Cohen-Cymberknoh M., et al. Antisense oligonucleotide splicing modulation as a novel Cystic Fibrosis therapeutic approach for the W1282X nonsense mutation. J. Cyst. Fibros. 2022;21:630–636. doi: 10.1016/j.jcf.2021.12.012. [DOI] [PubMed] [Google Scholar]
- Oren Y.S., Irony-Tur Sinai M., Golec A., Barchad-Avitzur O., Mutyam V., Li Y., Hong J., Ozeri-Galai E., Hatton A., Leibson C., et al. Antisense oligonucleotide-based drug development for Cystic Fibrosis patients carrying the 3849+10 kb C-to-T splicing mutation. J. Cyst. Fibros. 2021;20:865–875. doi: 10.1016/j.jcf.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski L.E., Yin W., Diggs P.S., Rogers T.D., O'Neal W.K., Grubb B.R. Expression of CFTR from a ciliated cell-specific promoter is ineffective at correcting nasal potential difference in CF mice. Gene Ther. 2007;14:1492–1501. doi: 10.1038/sj.gt.3302994. [DOI] [PubMed] [Google Scholar]
- Pascual-Morena C., Cavero-Redondo I., Alvarez-Bueno C., Mesas A.E., Pozuelo-Carrascosa D., Martinez-Vizcaino V. Restorative treatments of dystrophin expression in Duchenne muscular dystrophy: a systematic review. Ann. Clin. Transl. Neurol. 2020;7:1738–1752. doi: 10.1002/acn3.51149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M.C., Silva I.A., Figueira M.F., Amaral M.D., Lopes-Pacheco M. Pharmacological modulation of ion channels for the treatment of cystic fibrosis. J. Exp. Pharmacol. 2021;13:693–723. doi: 10.2147/JEP.S255377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raal F.J., Santos R.D., Blom D.J., Marais A.D., Charng M.J., Cromwell W.C., Lachmann R.H., Gaudet D., Tan J.L., Chasan-Taber S., et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- Ramsey B.W., Davies J., McElvaney N.G., Tullis E., Bell S.C., Drevinek P., Griese M., McKone E.F., Wainwright C.E., Konstan M.W., et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N. Engl. J. Med. 2011;365:1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderlin E.J., Keenan M.M., Mense M., Revenko A.S., Monia B.P., Guo S., Huang L. CFTR mRNAs with nonsense codons are degraded by the SMG6-mediated endonucleolytic decay pathway. Nat. Commun. 2022;13:2344. doi: 10.1038/s41467-022-29935-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S., Sun R., Bui H.H., Crosby J.R., Monia B.P., Guo S. Steric inhibition of 5′ UTR regulatory elements results in upregulation of human CFTR. Mol. Ther. 2019;27:1749–1757. doi: 10.1016/j.ymthe.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharner J., Ma W.K., Zhang Q., Lin K.T., Rigo F., Bennett C.F., Krainer A.R. Hybridization-mediated off-target effects of splice-switching antisense oligonucleotides. Nucleic Acids Res. 2020;48:802–816. doi: 10.1093/nar/gkz1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J., Du M., Wong E., Mutyam V., Li Y., Chen J., Wangen J., Thrasher K., Fu L., Peng N., et al. A small molecule that induces translational readthrough of CFTR nonsense mutations by eRF1 depletion. Nat. Commun. 2021;12:4358. doi: 10.1038/s41467-021-24575-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimo T., Maruyama R., Yokota T. Designing effective antisense oligonucleotides for exon skipping. Methods Mol. Biol. 2018;1687:143–155. doi: 10.1007/978-1-4939-7374-3_10. [DOI] [PubMed] [Google Scholar]
- Shirley M. Casimersen: first approval. Drugs. 2021;81:875–879. doi: 10.1007/s40265-021-01512-2. [DOI] [PubMed] [Google Scholar]
- Shteinberg M., Haq I.J., Polineni D., Davies J.C. Cystic fibrosis. Lancet. 2021;397:2195–2211. doi: 10.1016/S0140-6736(20)32542-3. [DOI] [PubMed] [Google Scholar]
- Southern K.W., Murphy J., Sinha I.P., Nevitt S.J. Corrector therapies (with or without potentiators) for people with cystic fibrosis with class II CFTR gene variants (most commonly F508del) Cochrane Database Syst. Rev. 2020;12:CD010966. doi: 10.1002/14651858.CD010966.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templin M.V., Levin A.A., Graham M.J., Aberg P.M., Axelsson B.I., Butler M., Geary R.S., Bennett C.F. Pharmacokinetic and toxicity profile of a phosphorothioate oligonucleotide following inhalation delivery to lung in mice. Antisense Nucleic Acid Drug Dev. 2000;10:359–368. doi: 10.1089/oli.1.2000.10.359. [DOI] [PubMed] [Google Scholar]
- Valley H.C., Bukis K.M., Bell A., Cheng Y., Wong E., Jordan N.J., Allaire N.E., Sivachenko A., Liang F., Bihler H., et al. Isogenic cell models of cystic fibrosis-causing variants in natively expressing pulmonary epithelial cells. J. Cyst. Fibros. 2019;18:476–483. doi: 10.1016/j.jcf.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Van Goor F., Hadida S., Grootenhuis P.D., Burton B., Cao D., Neuberger T., Turnbull A., Singh A., Joubran J., Hazlewood A., et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R. Patients with cystic fibrosis have new triple-drug combination. JAMA. 2019;322:2068. doi: 10.1001/jama.2019.19351. [DOI] [PubMed] [Google Scholar]
- Wan W.B., Seth P.P. The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 2016;59:9645–9667. doi: 10.1021/acs.jmedchem.6b00551. [DOI] [PubMed] [Google Scholar]
- Wang W., Hong J.S., Rab A., Sorscher E.J., Kirk K.L. Robust stimulation of W1282X-CFTR channel activity by a combination of allosteric modulators. PLoS One. 2016;11:e0152232. doi: 10.1371/journal.pone.0152232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J. Oligonucleotide conjugates for therapeutic applications. Ther. Deliv. 2013;4:791–809. doi: 10.4155/tde.13.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witztum J.L., Gaudet D., Freedman S.D., Alexander V.J., Digenio A., Williams K.R., Yang Q., Hughes S.G., Geary R.S., Arca M., et al. Volanesorsen and triglyceride levels in familial chylomicronemia syndrome. N. Engl. J. Med. 2019;381:531–542. doi: 10.1056/NEJMoa1715944. [DOI] [PubMed] [Google Scholar]
- Zaher A., ElSaygh J., Elsori D., ElSaygh H., Sanni A. A review of Trikafta: triple cystic fibrosis transmembrane conductance regulator (CFTR) modulator therapy. Cureus. 2021;13:e16144. doi: 10.7759/cureus.16144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zainal Abidin N., Haq I.J., Gardner A.I., Brodlie M. Ataluren in cystic fibrosis: development, clinical studies and where are we now? Expert Opin. Pharmacother. 2017;18:1363–1371. doi: 10.1080/14656566.2017.1359255. [DOI] [PubMed] [Google Scholar]