Abstract
Genomic information stored in the DNA is transcribed to the mRNA and translated to proteins. The 3′ untranslated regions (3′UTRs) of the mRNA serve pivotal roles in post-transcriptional gene expression, regulating mRNA stability, translation, and localization. Similar to DNA mutations producing aberrant proteins, RNA alterations expand the transcriptome landscape and change the cellular proteome. Recent global analyses reveal that many genes express various forms of altered RNAs, including 3′UTR length variants. Alternative polyadenylation and alternative splicing are involved in diversifying 3′UTRs, which could act as a hidden layer of eukaryotic gene expression control. In this review, we summarize the functions and regulations of 3′UTRs and elaborate on the generation and functional consequences of 3′UTR diversity. Given that dynamic 3′UTR length control contributes to phenotypic complexity, dysregulated 3′UTR diversity might be relevant to disease development, including cancers. Thus, 3′UTR diversity in cancer could open exciting new research areas and provide avenues for novel cancer theragnostics.
Keywords: 3′UTR diversity, alternative polyadenylation, alternative splicing, cancer, RNA alterations, transcriptome
INTRODUCTION
In the nucleus, eukaryotic mRNA is synthesized from a gene in a 5′ to 3′ direction and processed into mature transcripts by 5′-capping, splicing, and 3′-end formation. Genomic information is translated to proteins once mature mRNAs are exported to the cytoplasm. 5′ and 3′ sequences flanking the coding regions are not translated, thus named as untranslated regions (UTRs). What are the roles of UTRs in regulating gene expression? 5′UTRs are the leading sites for ribosome assembly for mRNA translation. In contrast, 3′UTRs play various roles in the post-transcriptional control of gene expressions, including stability, translation, and subcellular localization of mRNAs (Mayr, 2019). We will begin by reviewing why 3′UTR diversity is important, especially in human mRNAs. This review focuses only on the length of 3′UTRs, but not on their sequences and modification, as 3′UTR diversity for simplicity.
Evolutional features of human mRNA 3′UTRs
In general, the noncoding part of the genome has increased in size and complexity during evolution (Pesole et al., 2002). The average length of 5′UTRs is similar in metazoans; whereas, the 3′UTR is significantly longer in humans compared to other species (Mayr, 2016; Pesole et al., 2002; Sood et al., 2006; Wang et al., 2019) (Fig. 1). 3′-end sequencing also revealed that at least half of human genes could generate alternative 3′UTR isoforms from the same genes, demonstrating high diversity in 3′UTR in the human transcriptome (Derti et al., 2012; Hoque et al., 2013).
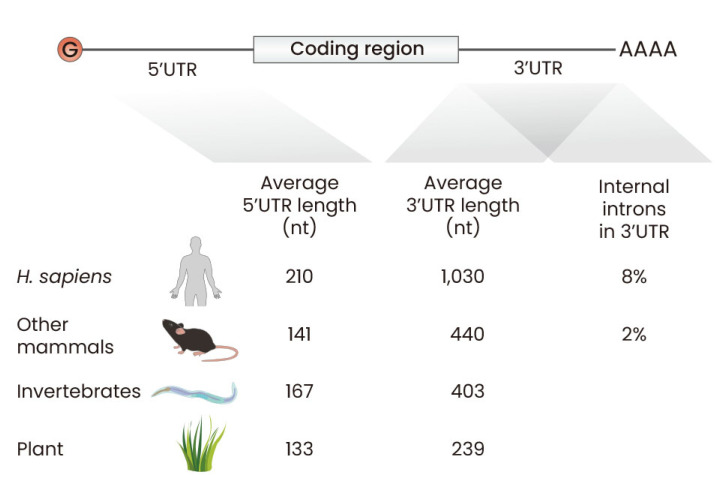
Fig. 1. Length and internal introns in untranslated region (UTR) during evolution.
The average length of the 5′UTR and 3′UTR of mRNA among different organisms. The 5′UTR is typically shorter compared to the 3′UTR. In Homo sapiens, the average length of the 3′UTR is about 1030 nucleotides (nt), which is longer than the average length of the 3′UTR in other species. Percentage of genes containing internal introns is also shown in H. sapiens (8%) and other mammals (2%).
3′UTRs contain cis-regulatory elements recognized by trans-acting factors. Thus, changes in 3′UTR length could alter a number of regulatory elements (Ji et al 2009). In fact, many genes have alternative 3′UTR or contain internal introns which can switch 3′UTR length. Therefore, 3′UTR length changes could affect gene expression by fine-tuning and reprogramming the mRNA regulatory landscape in human cells (Navarro et al., 2021).
FUNCTIONS OF 3′UTRs
3′UTRs contain cis-regulatory elements recognized by trans-acting factors. 3′UTR-mediated mRNA metabolism controls at the steps of the stability, translation, and localization of mRNA (Fig. 2). 3′UTRs form ribonucleoprotein (RNP) with many RNA binding proteins (RBPs). So we will describe representative mRNA 3′UTRs, corresponding main RBPs, and their relevant functions as follows (Table 1).
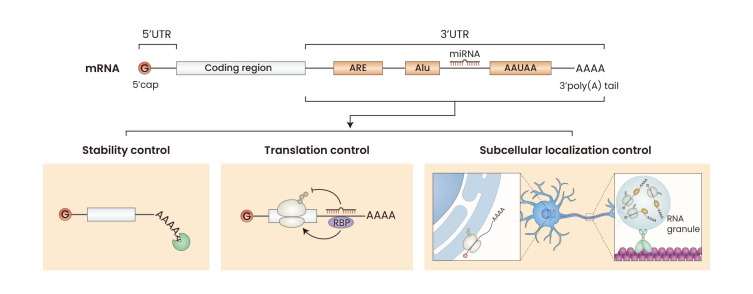
Fig. 2. Roles of 3′ untranslated region (3′UTR) in post-transcriptional regulation..
The diagram illustrates a typical mRNA structure consisting of 5′UTR, coding region, and 3′UTR. The 3′UTR is located following the stop codon and before the poly(A) tail. It includes cisregulatory elements like AU-rich elements (AREs), Alu elements and microRNA (miRNA) targets, and polyadenylation sites. The 3′UTRs play important roles in post-transcriptional gene regulation of mRNA stability, translation, and localization.
Table 1.
List of representative mRNAs with 3’UTR-mediated control
| Transcript | RNA-binding proteins | Control | Function | Reference |
|---|---|---|---|---|
| Hunchback | Pumilio Nanos Brat |
Translation repression | Embryonic axis formation in Drosophila | (Kuersten and Goodwin, 2003) |
| UCP1 | CPEB2 | Translation activation | Maintain body temperature | (Chen et al., 2018) |
| CaMK2A | Mub | Localization translation | Control learning and memory in neuron | (Broix et al., 2021; Chen et al., 2022) |
| ACTB | IGF2BP1 PAT1 |
Stabilization local-translation | Constituent of the cytoskeleton for structural support and movement in neuron | (Wu et al., 2020) |
| Oskar | Bruno Hrp48 |
RNP granule formation Translation |
Embryonic patterning and germline formation in Drosophila | (Bose et al., 2022; Jambor et al., 2014) |
| DMPK | CUG-BP2 | mRNA export Translation |
Maintenance of the myelin sheath | (Amack and Mahadevan, 2001; Taneja et al., 1995) |
|
n-myc
c-fos |
ELAV like RNA binding protein family | ARE-mediated mRNA stabilization | Cellular growth in neuroblastoma | (Chagnovich and Cohn, 1996; Chagnovich et al., 1996) |
| P21 WAF1 | SRSF3 | Translation repression | Regulator of cell cycle and senescence | (Kim et al., 2022) |
| COX-2 | HuR b-catenin |
mRNA stabilization Translation activation |
Cellular proliferation and tumorigenesis | (Kim et al., 2012; Lee and Jeong, 2006) |
Stability control
Most mRNAs undergo stability regulation through specific sequences and structures in 3′UTRs, such as AU-rich elements (AREs), GU-rich elements, CA-rich elements, and miRNA-binding sequences. Some proteins bind to AREs and recruit the degradation machinery to ARE-containing mRNAs. For example, tristetraprolin (TTP) binds AREs within the 3′UTRs of mRNAs, such as tumor necrosis factor (TNF), prostaglandin-endoperoxide synthase 2 (PTGS2/cyclooxygenase-2, [COX-2]), vascular endothelial growth factor A (VEGFA), interleukin 10 (IL10), and immediate early response 3 (IER3), and promote mRNA degradation (Essafi-Benkhadir et al., 2007; Lai et al., 1999; 2006; Sawaoka et al., 2003).
In contrast, Hu-antigen R (HuR) protein binds AREs and stabilizes numerous mRNAs, increasing the corresponding protein level (Brennan and Steitz, 2001). HuR stabilizes mRNAs promoting tumor growth and cell survival, such as VEGFA, COX-2, TNF, CTNNB1//β-catenin, and interferon regulatory factor (IRF9) (Dean et al., 2001; Rothamel et al., 2021; Sengupta et al., 2003; Steinman, 2007; Thiele et al., 2006). It also stabilizes mRNAs involved in cell cycle regulation, such as cyclin dependent kinase inhibitor 1A (CDKN1A/p21) and cyclin (A2, B1, D1, and E1) (Giles et al., 2003; Guo and Hartley, 2006; Wang et al., 2000). In addition, mRNAs encoding prosurvival proteins such as FOS (c-fos) and B-cell CLL/lymphoma 2 (Bcl-2) are stabilized by HuR (Ishimaru et al., 2009; Wang et al., 2001). These data suggest that cis-regulatory elements in 3′UTR participate in the context- and condition-dependent mRNA stability control by binding to the combination of different sets of RBPs.
Translation control
3′UTRs also contain cis-regulatory elements related to translation control. During the development of Drosophila embryo, Hunchback mRNA 3′UTR forms an RNP complex with Pumilio, Nanos, and Brat protein, leading to translation repression for proper development in the posterior region of the embryo (Kuersten and Goodwin, 2003). Likewise, many developmental genes are expressed by the translation regulation mediated by 3′UTR-RNP complexes.
Different lengths of 3′UTR contribute to the translation efficiency control due to the presence or the absense of cis-regulatory elements. In the case of Uncoupling protein 1 (UCP1), the long 3′UTR isoform is predominant and contains the binding sites for the translation regulator, cytoplasmic polyadenylation element-binding protein 2 (CPEB2) (Chen et al., 2018). CPEB2 is required for associating the long 3′UTR isoform for the low-level translation of UCP1 mRNA in steady-state conditions, and translation upregulation in response to cold exposure or adrenaline (Chen et al., 2018). These data suggest the role of 3′UTRs for translation regulation by binding to many RBPs forming RNP complexes. 3′UTR-RNP–mediated translational regulation is also linked to mRNA localization, as described below.
Localization control
The subcellular localization of mRNA is investigated mostly in neurons (Andreassi and Riccio, 2009). Localized mRNA is translated in specific subcellular locations and allows the precise production of proteins in spatiotemporal patterns in response to neural activity. For example, calcium/calmodulin-dependent protein kinase II alpha (CaMK2A) regulates calcium signaling in the nervous system as a critical player in activity-dependent behavioral plasticity (Bae and Miura, 2020). In Drosophila, the 3′UTR of CaMK2A mRNA contains regulatory information necessary for local translation in presynaptic terminals (Broix et al., 2021). In mammals, the resting concentration of CaMK2A should be maintained extremely high, reaching up to 2% of the total protein in the hippocampus. This enrichment is achieved through the enhanced axoplasmic translation of CaMK2A mRNA, which is mediated by the RBP Mub and a 23-base Mub-recognition element in 3′UTR (Chen et al., 2022).
Actin beta (ACTB/β-actin) mRNA is another example of 3′UTR-mediated mRNA transport. 3′UTR of ACTB mRNA associated with the zipcode-binding protein 1 (ZBP1), also known as insulin-like growth factor 2 mRNA-binding protein (IGF2BP) (Wu et al., 2020). This association facilitates the transport of ACTB mRNAs to the specific locations in neurons, such as dendrites, in response to brain-derived neurotrophic factor (BDNF) (Wu et al., 2020). Overall, the 3′UTR of the ACTB mRNA is crucially involved in its transport and localization in neurons, which is essential for the proper development and function of the nervous system.
The control of mRNA localization is also investigated in Drosophila embryo. In the case of oskar mRNA, the 67-nt stem-loop structure in 3′UTRs is essential for transporting the mRNA from the nurse cells into the oocyte. This transport depends on the action of dynein and kinesin motor proteins, which use the oocyte entry signal to guide the mRNA to its proper location within the embryo (Bose et al., 2022; Jambor et al., 2014). Therefore, 3′UTRs of an mRNA molecule are critically involved in determining its subcellular localization and manipulating its translation. In this way, 3′UTRs can regulate the spatiotemporal expression of a gene and control when and where a protein is produced.
Multi-step control
During the lifecycle of mRNAs, some RBPs bind to 3′UTRs and affect multiple steps of post-transcriptional regulation (Moore, 2005). Such multifunctional RBPs associate with 3′UTRs of many mRNAs involved in critical downstream events. Quaking (QK) is an RBP with a STAR domain involved in compact myelin formation (Vernet and Artzt, 1997). QK binds to a specific sequence in the 3′UTRs of several mRNAs that encode proteins related to myelin formation. The QK response element (QRE) is “ACUAAY” in 3′UTRs of mRNAs, such as microtubule associated protein 1B (MAP1B), cyclin dependent kinase inhibitor 1B (CDKN1B/p27kip), and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) mRNAs (Larocque et al., 2005; Li et al., 2000; Zearfoss et al., 2011; Zhao et al., 2006).
Some proteins are newly found RBPs implicated in multiple 3′UTR-mediated control steps. For example, β-catenin is a Wnt-activated transcription factor and an adhesion protein. In addition, it also associates with ARE in 3′UTRs of COX-2 mRNAs as an RBP in colon cancer cells. COX-2 protein expression is regulated by mRNA stability and translation control by 3′UTR-binding β-catenin in the cytoplasm (Lee and Jeong, 2006). Furthermore, the interaction between β-catenin and COX-2 mRNA assembles a tertiary complex with the HuR protein, vital for the upregulation of COX-2 protein levels and colon cancer progression (Kim et al., 2012). Considering that COX-2 protein is the enzyme critical for synthesizing prostaglandins, a high level of β-catenin RBP might have significant pathological effects on cancer cells.
Another example is serine/arginine-rich splicing factor 3 (SRSF3). SRSF3 is a member of the serin/arginine-rich (SR) family proteins, acting in AS mainly in the nucleus. In addition, the shuttling activity of SRSF3 is linked to AS regulation, export, and translation in the cytoplasm (Park and Jeong, 2016). SRSF3 interacts with the 3′UTR of p21cip1/waf1 (CDKN1A) mRNA and represses translation in the cytoplasm (Kim et al., 2022).
These multifunctional RBPs are located in both the nucleus and cytoplasm and form 3′UTR-mediated RNP to control the multiple steps of mRNA metabolism. Further research is needed to fully understand the multiple roles of RBPs in 3′UTR-mediated mRNA regulation.
REGULATION OF 3′UTR FUNCTIONS
Considering the important roles of 3′UTRs in post-transcriptional gene expression, 3′UTR-mediated control should be regulated by many factors, including miRNA, RBPs and RNA granules as follows.
By the interplay of miRNA and RBPs
3′UTR functions are regulated by the crosstalk of miRNAs and RBPs, and cis-regulatory elements within 3′UTRs. miRNAs are small noncoding RNA molecules that are crucially involved in regulating gene expression by binding to complementary sequences in the 3′UTRs of the target mRNA. This binding can lead to the destabilization of mRNAs or the inhibition of their translation (Baek et al., 2008; Friedman et al., 2009). For example, VEGFA mRNA 3′UTR contains several regulatory elements, including CA-rich elements and miRNA-binding sequences. In normal conditions, miR-297 and miR-299 bind to the 3′UTR of the VEGFA mRNA leading to reduced expression of VEGFA proteins (Matoulkova et al., 2012). However, under hypoxic conditions, the silencing activity of miR-297 and miR-299 on VEGFA mRNA is inhibited by the binding of hnRNPL to CA-rich elements in 3′UTRs, allowing mRNAs to be continuously translated into proteins (Jafarifar et al., 2011). These data suggest that the interplay of miRNAs and RBPs are important for the condition-dependent regulation of 3′UTR activities.
The control of 3′UTR length also could affect miRNA-binding sites and leads to functional consequences (Nam et al., 2014). Thus, the dynamic regulation of 3′UTR length could play a significant role in development because it contributes to the gene expression patterns observed in different cell types and tissues. Likely, distinct sets of genes are differentially regulated by the interplay of miRNA and RBPs in a context- and condition-dependent manner (Hoffman et al., 2016).
By forming RNA granules
RNA granules in the cytoplasm are emerging as essential players in mRNA localization and translation regulation (Moore, 2005; Tian et al., 2020). RNA granules are specialized structures in assembled RNAs with RNP particles. Several RNA granules include transport RNPs, stress granules, and processing bodies (P-bodies) (Kiebler and Bassell, 2006). This compartmentalization creates cellular asymmetries, may enhance biological reactions, and promote molecular interactions required for cell growth and development (Tian et al., 2020). How RNAs assemble RNA granules? 3′UTRs within mRNAs could be key components in forming RNA granules. For example, the cytoplasmic polyadenylation element (CPE) in 3′UTR and its binding protein CPEB facilitate mRNA transport to dendrites in rat hippocampal neurons (Huang et al., 2003). 3′UTR-RNP granules are formed on CaMK2A, MAP2, and β-catenin mRNAs with CPEB, maskin, and eIF4E, for transporting corresponding mRNAs (Huang et al., 2003; Kundel et al., 2009; Rook et al., 2000).
Technical advances enhance cellular RNA granule visualization. In situ hybridization (ISH) demonstrated the bicoid and oskar RNP granules with Staufen double-strand binding protein in Drosophila oocytes (Huang et al., 2003; Kiebler and Bassell, 2006; St Johnston, 2005). Moreover, the improvement of in situ RNA visualization methods, such as single-molecular fluorescent ISH (smFISH) and RNAscope® made it possible to observe oskar RNP granules with scaffold protein Bruno and Hrp48 (Bae and Miura, 2020; Bose et al., 2022; de Planell-Saguer et al., 2010). Further enhancement of mRNA visualization techniques will be required to elucidate and visualize the 3′UTR-dependent RNA granule localization.
GENERATING 3′UTR LENGTH DIVERSITY
3′UTR diversity can be caused by various ways, such as genetic variations, RNA modification, alternative cleavage, alternative polyadenylation (APA), and alternative splicing (AS). These could diversify the transcriptome and contribute to epigenetic gene regulation. This section focuses on the length-diversifying mechanism, APA, and AS (Fig. 3). Databases with different 3′UTR lengths formed by APA and AS are also summarized in Table 2.
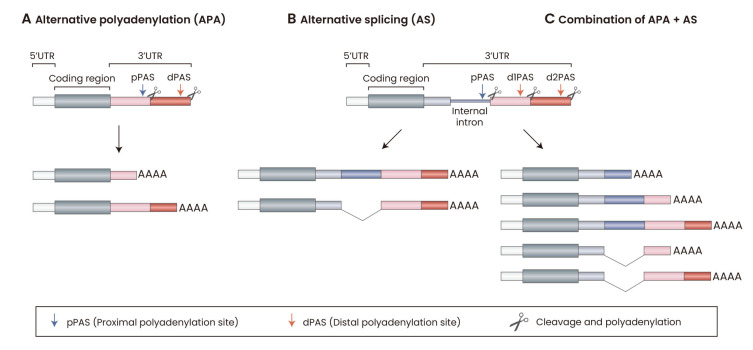
Fig. 3. Regulatory events generating 3′ untranslated region (3′UTR) length diversity.
(A) Alternative polyadenylation (APA) in 3′UTR. The generation of 3′UTR length diversity using different cleavage and polyadenylation sites: proximal polyadenylation site (pPAS) and distal polyadenylation site (dPAS). The selection of pPAS over dPAS, or vice versa, can result in different 3′UTR lengths. If the pPAS is selected, the 3′UTR will be shorter (shown in the pink box); the dPAS is selected, the 3′UTR will be longer (shown in the pink box and red box). (B) Alternative splicing (AS) within 3′UTR. The generation of 3′UTR length diversity using the internal intron in 3′UTR (shown in the blue line). Inclusion or skipping of this intron can affect the length of the 3′UTR. If the intron is included, 3′UTR will contain an intronic sequence (shown in the blue box). (C) Combination of APA and AS events in 3′UTR. More 3'UTR variants can be generated.
Table 2.
List of Database related to 3′UTR divers
| Tool name | Features | URL | Reference |
|---|---|---|---|
| Poly(A) sites databases | |||
| PolyASite 2.0 | APA atlas made from 3′end sequencing data | https://polyasite.unibas.ch | (Herrmann et al., 2020) |
| PolyA_DB 3 | APA atlas built from ~1.2 billion 3′end deep sequencing reads | https://exon.apps.wistar.org/PolyA_DB/v3/ | (Wang et al., 2018) |
| scAPAdb | APA atlas at single-cell resolution from publications and Genomics website | http://www.bmibig.cn/scAPAdb/ | (Zhu et al., 2022) |
| TREND-DB | APA database constructed from bulk RNA-seq data of potential APA regulators | http://shiny.imbei.uni-mainz.de:3838/trend-db | (Marini et al., 2021) |
| 3′UTR alternative splicing databases | |||
| RNA structure databases | 3′UTR splicing events database from RNA-seq data of TCGA and GTEx | https://www.cbrc.kaust.edu.sa/spur/home/ | (Chan et al., 2022b) |
| RNA structure databases | |||
| RNAfold | Prediction tool of secondary structures for single-stranded RNA or DNA | http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi | (Lorenz et al., 2011) |
APA in 3′UTR
Cleavage and polyadenylation is a critical step in the maturation of 3′UTR ends of most eukaryotic mRNAs (Mitschka and Mayr, 2022). The polyadenylation process begins the recognition of specific sequences in the mRNA molecule by the polyadenylation machinery in the nucleus. The polyadenylation machinery consists of several factors, including cleavage factor I, cleavage and polyadenylation specificity factor, cleavage factor II, and cleavage stimulation factor. These complexes bind to specific sequences in mRNA 3′UTRs and facilitate the cleavage at a specific site downstream of the AAUAAA motif. This cleavage is aided by cleavage factor polyribonucleotide kinase subunit 1, followed by adding a polyadenylate tail to the mRNA molecule by the enzyme poly(A) polymerase alpha (Tian and Manley, 2013). A strong polyadenylation signal at the 3′UTR end is important for the efficient cleavage and polyadenylation of mRNAs. However, many genes also have additional polyadenylation signals at their 3′UTRs in eukaryotes, and the usage of these signals can be regulated through APA (Fig. 3A).
APA is a widespread phenomenon in metazoan protein-coding transcripts (70%-79% of mammalian genes). In humans, >70% of genes have more than one polyadenylation site in their 3′UTRs, and approximately 50% have three or more. By contrast, in the mouse liver, >60% of expressed genes had multiple polyadenylation signals in their 3′UTRs (Tian and Manley, 2013). This can result in mRNAs with different 3′UTR ends and different regulatory and functional properties (Mitschka and Mayr, 2022). 3′UTR shortening was proposed to increase mRNA stability by reducing the accessibility of mRNA degradation mechanisms, such as RBP- or miRNA-based degradation. Conversely, 3′UTR lengthening may increase the accessibility of miRNAs, decreasing mRNA stability and translation.
In addition to regulating mRNA stability APA can play a role in mRNA–protein interactions and protein localization via alternative 3′UTRs. For example, the APA of CD47 mRNA 3′UTR can affect the interaction with different protein complexes and thereby direct CD47 localization to either the plasma membrane (long 3′UTRs) or endoplasmic reticulum (short 3′UTRs) in various cancers (Berkovits and Mayr, 2015; Ma and Mayr, 2018). More studies are required to understand this interesting phenomenon by 3′UTR-directed protein localization.
AS in 3′UTRs
AS can also alter the 3′UTR length if 3′UTR contains an internal intron (Fig. 3B). It will increase the length, heterogeneity, and functional diversity of 3′UTRs (Bicknell et al., 2012; Chan et al., 2022b). The combination of APA and AS could further increase mRNA isoform variability resulting in the generation of mRNA transcripts containing various forms (Fig. 3C). The splicing of introns located within 3′UTRs was first described in heterogenous nuclear ribonucleoprotein D (hnRNPD/AUF1) (Wilson et al., 1999). This study reported that splicing of the intron in 3′UTR regulates AUF1 expression, so it may play a significant role in gene expression regulation.
Another study demonstrated that 3′UTRs of integrin subunit alpha 3 (ITGA3) mRNA interacts with muscleblind like splicing regulator 2 (MBNL2) in human lung carcinoma cells (Adereth et al., 2005; Pascual et al., 2006). These splicing factors regulate the AS of ITGA3 3′UTR, in which produces distinct 3′UTR variants with different functions. The authors also showed the ITGA3 mRNA localized to adhesion plaques, specialized structures involved in cell adhesion and signaling. Moreover, in the gene coding for an activity regulated cytoskeleton associated protein (Arc), AS generates isoforms with distinct 3′UTRs. The AS of Arc 3′UTR is induced by long-term potentiation learning and memory (Paolantoni et al., 2018).
The 3′UTR of the β-catenin gene CTNNB1 is also an interesting example. Three 3′UTR variants are expressed following the AS of an intron located within 3′UTRs (Chan et al., 2022b; Thiele et al., 2006). The expression of variant 3′UTRs in β-catenin mRNA was reported in peripheral blood mononuclear leukocytes (PBMCs). This suggests that different isoforms of β-catenin mRNAs may play different roles in regulating gene expression in PBMCs (Thiele et al., 2006). Recently, a pan-cancer analysis revealed pervasive upregulation of 3′UTR splicing driving tumorigenesis with b-catenin as a top-dysregulated 3′UTR (Chan et al., 2022b). Thus, 3′UTR diversity could not only contribute to the overall diversity of the transcriptome and proteome but also drive cancer development and progression.
3′UTR-MEDIATED DISEASES
Transcriptome analyses of human cancer databases revealed that a large percentage of genes in cancers have RNA alterations (PCAWG Transcriptome Core Group et al., 2020). RNA alterations are mostly in protein-coding regions, but 3′UTR diversity also attribute to RNA alterations. Because APA is widespread and alters the regulatory potential of 3′UTRs, APA dysregulation plays a significant role in cancers (Kahles et al., 2018). Global APA within 3′UTRs is involved in the proliferation and metastasis of cancer cells and tumor tissues (Hoque et al., 2013; Mayr and Bartel, 2009). APA dysregulation is exemplified in Cyclin D1 overexpression because of 3′UTR shortening (Mayr and Bartel, 2009; Wiestner et al., 2007). This 3′UTR shortening leads to the loss of binding sites of complementary miRNA to escape repression and potentially contribute to oncogene activation (Yang and Nam, 2020).
Based on genome and transcriptome databases, AS events in cancer tissues are estimated to be 20% higher on average than in normal tissues (Kahles et al., 2018). Recently, a pan-cancer analysis revealed that pervasive upregulation of 3′UTR splicing drives tumorigenesis (Chan et al., 2022b). In hepatocellular carcinoma, aberrant 3′UTR isoforms of β-catenin mRNA escape non-sense-mediated mRNA decay and promote cell proliferation and migration (Chan et al., 2022b). Thus, 3′UTR diversity could not only contribute to the overall complexity of the transcriptome and proteome but also drive cancer development and progression (Chan et al., 2022a).
As in cancer disorders of neuronal plasticity and learning, 3′UTRs appear to be a pathological hotspot (Conne et al., 2000). Because several neuronal mRNAs depend on their 3′UTRs for appropriate subcellular targeting or translational control, perturbations in 3′UTR-mediated functions deserve attention. In addition, alterations in the secondary structure of 3′UTRs are related to the pathology of a certain disease (Reamon-Buettner et al., 2007). Mutations in the 3′UTR of GATA binding protein 4 (GATA4) mRNA alter its secondary structure and may contribute to the development of congenital heart disease (Reamon-Buettner et al., 2007). Therefore, further studies could explain the mechanism of the 3′UTR-mediated disease development (Conne et al., 2000).
CONCLUSIONS AND PERSPECTIVES
As genomic and transcriptomic data accumulated, RNA alterations emerged as important features of cancer development (PCAWG Transcriptome Core Group et al., 2020). Among many RNA alterations, 3′UTR variants received little attention in comparison with those in protein-coding regions (Bicknell et al., 2012). However, a widespread shortening of 3′UTR lengths in cancers and causal UTR variants in human diseases demonstrated the important roles of 3′UTRs in disease development (Griesemer et al., 2021; Mayr and Bartel, 2009). 3′UTR alterations can contribute to the diversification of the transcriptome and protein functions, impacting numerous biological processes.
3′UTR activities are context- and condition-dependent, dynamically affecting gene expression programs by miRNAs, RBPs, and RNA granule formation. Thus, finding rules and factors generating and modulating 3′UTR variants, such as 3′-end processing machinery, 3′UTR-binding proteome and UTR-mediated RNP granules, might be important. As functional alterations in 3′UTRs are found in various diseases, a systemic search for “3′UTR-mediated diseases” will extend the horizons in the diagnosis and treatment of these diseases (Conne et al., 2000). Specifically, functional cis-regulatory elements in 3′UTRs in pathogenic genes, interaction with disease-associated RBPs, and their pathogenic conditions should be understood to elucidate the mechanism and their consequent phenotypic changes in diseases. It could also open exciting new research areas, answering the question of what enables the biological complexity in humans during evolution.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation (NRF), funded by the Ministry of Science & ICT (NRF-2020R1A2C2005358 and NRF-2022M3E5F1016546).
Footnotes
AUTHOR CONTRIBUTIONS
S.J. conceived the idea, wrote the manuscript, and secured funding. D.W. performed experiments and wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Adereth Y., Dammai V., Kose N., Li R., Hsu T. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat. Cell Biol. 2005;7:1240–1247. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amack J.D., Mahadevan M.S. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum. Mol. Genet. 2001;10:1879–1887. doi: 10.1093/hmg/10.18.1879. [DOI] [PubMed] [Google Scholar]
- Andreassi C., Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Bae B., Miura P. Emerging roles for 3' UTRs in neurons. Int. J. Mol. Sci. 2020;21:3413. doi: 10.3390/ijms21103413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits B.D., Mayr C. Alternative 3' UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell A.A., Cenik C., Chua H.N., Roth F.P., Moore M.J. Introns in UTRs: why we should stop ignoring them. Bioessays. 2012;34:1025–1034. doi: 10.1002/bies.201200073. [DOI] [PubMed] [Google Scholar]
- Bose M., Lampe M., Mahamid J., Ephrussi A. Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development. Cell. 2022;185:1308–1324.e23. doi: 10.1016/j.cell.2022.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.M., Steitz J.A. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broix L., Turchetto S., Nguyen L. Coordination between transport and local translation in neurons. Trends Cell Biol. 2021;31:372–386. doi: 10.1016/j.tcb.2021.01.001. [DOI] [PubMed] [Google Scholar]
- Chagnovich D., Cohn S.L. Binding of a 40-kDa protein to the N-myc 3'-untranslated region correlates with enhanced N-myc expression in human neuroblastoma. J. Biol. Chem. 1996;271:33580–33586. doi: 10.1074/jbc.271.52.33580. [DOI] [PubMed] [Google Scholar]
- Chagnovich D., Fayos B.E., Cohn S.L. Differential activity of ELAV-like RNA-binding proteins in human neuroblastoma. J. Biol. Chem. 1996;271:33587–33591. doi: 10.1074/jbc.271.52.33587. [DOI] [PubMed] [Google Scholar]
- Chan J.J., Tabatabaeian H., Tay Y. 3'UTR heterogeneity and cancer progression. Trends Cell Biol. 2022a doi: 10.1016/j.tcb.2022.10.001. 2022 Nov 11 [Epub]. https://doi.org/10.1016/j.tcb.2022.10.001 . [DOI] [PubMed] [Google Scholar]
- Chan J.J., Zhang B., Chew X.H., Salhi A., Kwok Z.H., Lim C.Y., Desi N., Subramaniam N., Siemens A., Kinanti T., et al. Pan-cancer pervasive upregulation of 3' UTR splicing drives tumourigenesis. Nat. Cell Biol. 2022b;24:928–939. doi: 10.1038/s41556-022-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.F., Hsu C.M., Huang Y.S. CPEB2-dependent translation of long 3'-UTR Ucp1 mRNA promotes thermogenesis in brown adipose tissue. EMBO J. 2018;37:e99071. doi: 10.15252/embj.201899071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhang Y., Adel M., Kuklin E.A., Reed M.L., Mardovin J.D., Bakthavachalu B., VijayRaghavan K., Ramaswami M., Griffith L.C. Local translation provides the asymmetric distribution of CaMKII required for associative memory formation. Curr. Biol. 2022;32:2730–2738.e5. doi: 10.1016/j.cub.2022.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conne B., Stutz A., Vassalli J.D. The 3' untranslated region of messenger RNA: a molecular 'hotspot' for pathology? Nat. Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- de Planell-Saguer M., Rodicio M.C., Mourelatos Z. Rapid in situ codetection of noncoding RNAs and proteins in cells and formalin-fixed paraffin-embedded tissue sections without protease treatment. Nat. Protoc. 2010;5:1061–1073. doi: 10.1038/nprot.2010.62. [DOI] [PubMed] [Google Scholar]
- Dean J.L.E., Wait R., Mahtani K.R., Sully G., Clark A.R., Saklatvala J. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A., Garrett-Engele P., Macisaac K.D., Stevens R.C., Sriram S., Chen R., Rohl C.A., Johnson J.M., Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essafi-Benkhadir K., Onesto C., Stebe E., Moroni C., Pages G. Tristetraprolin inhibits Ras-dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Mol. Biol. Cell. 2007;18:4648–4658. doi: 10.1091/mbc.e07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles K.M., Daly J.M., Beveridge D.J., Thomson A.M., Voon D.C., Furneaux H.M., Jazayeri J.A., Leedman P.J. The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J. Biol. Chem. 2003;278:2937–2946. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- Griesemer D., Xue J.R., Reilly S.K., Ulirsch J.C., Kukreja K., Davis J.R., Kanai M., Yang D.K., Butts J.C., Guney M.H., et al. Genome-wide functional screen of 3′UTR variants uncovers causal variants for human disease and evolution. Cell. 2021;184:5247–5260.e19. doi: 10.1016/j.cell.2021.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PCAWG Transcriptome Core Group , Calabrese C., Davidson N.R., Demircioglu D., Fonseca N.A., He Y., Kahles A., Lehmann K.V., Liu F., Shiraishi Y., et al. Genomic basis for RNA alterations in cancer. Nature. 2020;578:129–136. doi: 10.1038/s41586-020-1970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Hartley R.S. HuR contributes to cyclin E1 deregulation in MCF-7 breast cancer cells. Cancer Res. 2006;66:7948–7956. doi: 10.1158/0008-5472.CAN-05-4362. [DOI] [PubMed] [Google Scholar]
- Herrmann C.J., Schmidt R., Kanitz A., Artimo P., Gruber A.J., Zavolan M. PolyASite 2.0: a consolidated atlas of polyadenylation sites from 3' end sequencing. Nucleic Acids Res. 2020;48(D1):D174–D179. doi: 10.1093/nar/gkz918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman Y., Bublik D.R., Ugalde A.P., Elkon R., Biniashvili T., Agami R., Oren M., Pilpel Y. 3'UTR shortening potentiates microRNA-based repression of pro-differentiation genes in proliferating human cells. PLoS Genet. 2016;12:e1005879. doi: 10.1371/journal.pgen.1005879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., Park J.Y., Yehia G., Tian B. Analysis of alternative cleavage and polyadenylation by 3' region extraction and deep sequencing. Nat. Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.S., Carson J.H., Barbarese E., Richter J.D. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru D., Ramalingam S., Sengupta T.K., Bandyopadhyay S., Dellis S., Tholanikunnel B.G., Fernandes D.J., Spicer E.K. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol. Cancer Res. 2009;7:1354–1366. doi: 10.1158/1541-7786.MCR-08-0476. [DOI] [PubMed] [Google Scholar]
- Jafarifar F., Yao P., Eswarappa S.M., Fox P.L. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30:1324–1334. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H., Mueller S., Bullock S.L., Ephrussi A. A stem-loop structure directs oskar mRNA to microtubule minus ends. RNA. 2014;20:429–439. doi: 10.1261/rna.041566.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Lee J.Y., Pan Z., Jiang B., Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahles A., Lehmann K.V., Toussaint N.C., Huser M., Stark S.G., Sachsenberg T., Stegle O., Kohlbacher O., Sander C., Cancer Genome Atlas Research Network, et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell. 2018;34:211–224.e6. doi: 10.1016/j.ccell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M.A., Bassell G.J. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim I., Kwak H., Lee H.K., Hyun S., Jeong S. beta-Catenin recognizes a specific RNA motif in the cyclooxygenase-2 mRNA 3'-UTR and interacts with HuR in colon cancer cells. Nucleic Acids Res. 2012;40:6863–6872. doi: 10.1093/nar/gks331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Park R.Y., Kee Y., Jeong S., Ohn T. Splicing factor SRSF3 represses translation of p21(cip1/waf1) mRNA. Cell Death Dis. 2022;13:933. doi: 10.1038/s41419-022-05371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S., Goodwin E.B. The power of the 3' UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Kundel M., Jones K.J., Shin C.Y., Wells D.G. Cytoplasmic polyadenylation element-binding protein regulates neurotrophin-3-dependent beta-catenin mRNA translation in developing hippocampal neurons. J. Neurosci. 2009;29:13630–13639. doi: 10.1523/JNEUROSCI.2910-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.S., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/MCB.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.S., Parker J.S., Grissom S.F., Stumpo D.J., Blackshear P.J. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocque D., Galarneau A., Liu H.N., Scott M., Almazan G., Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat. Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Jeong S. Beta-Catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3'-UTR. Nucleic Acids Res. 2006;34:5705–5714. doi: 10.1093/nar/gkl698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Li D., Feng Y. Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J. Neurosci. 2000;20:4944–4953. doi: 10.1523/JNEUROSCI.20-13-04944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Mayr C. A membraneless organelle associated with the endoplasmic reticulum enables 3'UTR-mediated protein-protein interactions. Cell. 2018;175:1492–1506.e19. doi: 10.1016/j.cell.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F., Scherzinger D., Danckwardt S. TREND-DB-a transcriptome-wide atlas of the dynamic landscape of alternative polyadenylation. Nucleic Acids Res. 2021;49(D1):563–576. doi: 10.1093/nar/gkaa722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoulkova E., Michalova E., Vojtesek B., Hrstka R. The role of the 3' untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:D243–D253. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- Mayr C. Evolution and biological roles of alternative 3'UTRs. Trends Cell Biol. 2016;26:227–237. doi: 10.1016/j.tcb.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. What are 3' UTRs doing? Cold Spring Harb. Perspect. Biol. 2019;11:a034728. doi: 10.1101/cshperspect.a034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Bartel D.P. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschka S., Mayr C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 2022;23:779–796. doi: 10.1038/s41580-022-00507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nam J.W., Rissland O.S., Koppstein D., Abreu-Goodger C., Jan C.H., Agarwal V., Yildirim M.A., Rodriguez A., Bartel D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell. 2014;53:1031–1043. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro E., Mallen A., Hueso M. Dynamic variations of 3'UTR length reprogram the mRNA regulatory landscape. Biomedicines. 2021;9:1560. doi: 10.3390/biomedicines9111560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolantoni C., Ricciardi S., De Paolis V., Okenwa C., Catalanotto C., Ciotti M.T., Cattaneo A., Cogoni C., Giorgi C. Arc 3' UTR splicing leads to dual and antagonistic effects in fine-tuning Arc expression upon BDNF signaling. Front. Mol. Neurosci. 2018;11:145. doi: 10.3389/fnmol.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.K., Jeong S. SRSF3 represses the expression of PDCD4 protein by coordinated regulation of alternative splicing, export and translation. Biochem. Biophys. Res. Commun. 2016;470:431–438. doi: 10.1016/j.bbrc.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Pascual M., Vicente M., Monferrer L., Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Pesole G., Liuni S., Grillo G., Licciulli F., Mignone F., Gissi C., Saccone C. UTRdb and UTRsite: specialized databases of sequences and functional elements of 5' and 3' untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res. 2002;30:335–340. doi: 10.1093/nar/30.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner S.M., Cho S.H., Borlak J. Mutations in the 3'-untranslated region of GATA4 as molecular hotspots for congenital heart disease (CHD) BMC Med. Genet. 2007;8:38. doi: 10.1186/1471-2350-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook M.S., Lu M., Kosik K.S. CaMKIIalpha 3' untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J. Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothamel K., Arcos S., Kim B., Reasoner C., Lisy S., Mukherjee N., Ascano M. ELAVL1 primarily couples mRNA stability with the 3' UTRs of interferon-stimulated genes. Cell Rep. 2021;35:109178. doi: 10.1016/j.celrep.2021.109178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaoka H., Dixon D.A., Oates J.A., Boutaud O. Tristetraprolin binds to the 3'-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J. Biol. Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- Sengupta S., Jang B.C., Wu M.T., Paik J.H., Furneaux H., Hla T. The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J. Biol. Chem. 2003;278:25227–25233. doi: 10.1074/jbc.M301813200. [DOI] [PubMed] [Google Scholar]
- Sood P., Krek A., Zavolan M., Macino G., Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Steinman R.A. mRNA stability control: a clandestine force in normal and malignant hematopoiesis. Leukemia. 2007;21:1158–1171. doi: 10.1038/sj.leu.2404656. [DOI] [PubMed] [Google Scholar]
- Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele A., Nagamine Y., Hauschildt S., Clevers H. AU-rich elements and alternative splicing in the beta-catenin 3'UTR can influence the human beta-catenin mRNA stability. Exp. Cell Res. 2006;312:2367–2378. doi: 10.1016/j.yexcr.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Tian B., Manley J.L. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Curnutte H.A., Trcek T. RNA granules: a view from the RNA perspective. Molecules. 2020;25:3130. doi: 10.3390/molecules25143130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet C., Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/S0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- Wang R., Nambiar R., Zheng D., Tian B. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res. 2018;46(D1):D315–D319. doi: 10.1093/nar/gkx1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Caldwell M.C., Lin S., Furneaux H., Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Fang D.H., Gan J., Shi Y., Tang H., Wang H., Fu M.Z., Yi J. Evolutionary and functional implications of 3' untranslated region length of mRNAs by comprehensive investigation among four taxonomically diverse metazoan species. Genes Genomics. 2019;41:747–755. doi: 10.1007/s13258-019-00808-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang X., Cristofalo V.J., Holbrook N.J., Gorospe M. Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol. Cell. Biol. 2001;21:5889–5898. doi: 10.1128/MCB.21.17.5889-5898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiestner A., Tehrani M., Chiorazzi M., Wright G., Gibellini F., Nakayama K., Liu H., Rosenwald A., Muller-Hermelink H.K., Ott G., et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599–4606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.M., Sun Y., Sellers J., Lu H., Penkar N., Dillard G., Brewer G. Regulation of AUF1 expression via conserved alternatively spliced elements in the 3' untranslated region. Mol. Cell. Biol. 1999;19:4056–4064. doi: 10.1128/MCB.19.6.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zhou J., Zhu T., Cohen I., Dictenberg J. A kinesin adapter directly mediates dendritic mRNA localization during neural development in mice. J. Biol. Chem. 2020;295:6605–6628. doi: 10.1074/jbc.RA118.005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.D., Nam S.W. Pathogenic diversity of RNA variants and RNA variation-associated factors in cancer development. Exp. Mol. Med. 2020;52:582–593. doi: 10.1038/s12276-020-0429-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zearfoss N.R., Clingman C.C., Farley B.M., McCoig L.M., Ryder S.P. Quaking regulates Hnrnpa1 expression through its 3' UTR in oligodendrocyte precursor cells. PLoS Genet. 2011;7:e1001269. doi: 10.1371/journal.pgen.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ku L., Chen Y., Xia M., LoPresti P., Feng Y. QKI binds MAP1B mRNA and enhances MAP1B expression during oligodendrocyte development. Mol. Biol. Cell. 2006;17:4179–4186. doi: 10.1091/mbc.e06-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Lian Q., Ye W., Qin W., Wu Z., Ji G., Wu X. scAPAdb: a comprehensive database of alternative polyadenylation at single-cell resolution. Nucleic Acids Res. 2022;50(D1):D365–D370. doi: 10.1093/nar/gkab795. [DOI] [PMC free article] [PubMed] [Google Scholar]