Abstract
RNAs are versatile molecules that are primarily involved in gene regulation and can thus be widely used to advance the fields of therapeutics and diagnostics. In particular, circular RNAs which are highly stable, have emerged as strong candidates for use on next-generation therapeutic platforms. Endogenous circular RNAs control gene regulatory networks by interacting with other biomolecules or through translation into polypeptides. Circular RNAs exhibit cell-type specific expression patterns, which can be altered in tissues and body fluids depending on pathophysiological conditions. Circular RNAs that are aberrantly expressed in diseases can function as biomarkers or therapeutic targets. Moreover, exogenous circular RNAs synthesized in vitro can be introduced into cells as therapeutic molecules to modulate gene expression networks in vivo. Depending on the purpose, synthetic circular RNA sequences can either be identical to endogenous circular RNA sequences or artificially designed. In this review, we introduce the life cycle and known functions of intracellular circular RNAs. The current stage of endogenous circular RNAs as biomarkers and therapeutic targets is also described. Finally, approaches and considerations that are important for applying the available knowledge on endogenous circular RNAs to design exogenous circular RNAs for therapeutic purposes are presented.
Keywords: biomarkers, circular RNAs, therapeutic molecules, therapeutic targets
INTRODUCTION
RNAs regulate diverse biological processes in addition to participating in translation. For several decades, novel regulatory RNAs and their biological roles have been identified (Morris and Mattick, 2014), and the new functions of previously known RNAs, such as fragmented tRNAs, have been revealed (Gebetsberger and Polacek, 2013; Kumar et al., 2014; Morris and Mattick, 2014). Identifying novel regulatory RNAs and understanding their mechanisms of action have expanded our RNA metabolism-related knowledge regardless of the RNAs’ coding potentials. RNAs have been investigated from multiple perspectives; for example, their use as potential therapeutic molecules has been explored, and they have been targeted for their potential treatment benefits. Antisense oligonucleotide (ASO), which modulates RNA splicing and stability, is an exemplary therapeutic molecule that regulates endogenous RNAs (Stephenson and Zamecnik, 1978). Introducing small interfering RNAs (siRNAs) synthesized outside of cells is another approach to suppressing the expression of endogenous genes. Exogenous siRNAs cleave target RNAs inside cells and control gene expression. Exogenous mRNAs can also be introduced to increase the levels of endogenous proteins or to express foreign proteins in cells.
RNA therapeutics have recently undergone a historical transition since the U.S. Food and Drug Administration urgently approved mRNA vaccines, enabling a quick response to the coronavirus disease 2019 (COVID-19) pandemic (Hodgson, 2020; Jung and Shin, 2021; Lee and Oh, 2021; Park et al., 2021). The knowledge accumulated in the field of RNA biology was applied when developing COVID-19 mRNA vaccines (Pardi et al., 2018). N1-methylpseudouridine (m1Ψ) was incorporated into mRNAs to reduce the likelihood of undesirable immune responses (Nelson et al., 2020), and the 5′ and 3′ untranslated region sequences were modified to increase the mRNA stability and translational efficiency (Nelson et al., 2020; Orlandini von Niessen et al., 2019). Although the results of these experiments were successful, some aspects remain to be improved. Because a decrease in the number of neutralizing antibodies over time is unstoppable in RNA-based vaccine systems, studies to find new platforms that can produce higher amounts of proteins and maintain the levels of RNAs introduced into the body are underway (Qu et al., 2022). Circular RNAs have been attracting attention as a next-generation vaccine platform to improve both the immunological and economical efficiencies of vaccines.
Circular RNAs are closed forms of single-stranded RNAs in which the 5′ and 3′ ends are connected with covalent bonds. The absence of both ends protects circular RNAs from exonucleases and results in increased RNA stability. The average half-life of circular RNAs is longer than that of linear mRNA isoforms in mammalian cells (Jeck and Sharpless, 2014). Thus, using a highly stable circular RNA is beneficial for its application as a biomarker or therapeutic molecule. In this review, we discuss the current therapeutic approaches of circular RNAs in two main categories—the utilization of endogenous circular RNAs and the application of exogenous circular RNAs synthesized in vitro.
INTRACELLULAR CIRCULAR RNAs AS BIOMARKERS AND THERAPEUTIC TARGETS
Most endogenous circular RNAs are produced through pre-mRNA back-splicing, whereas some categories of circular RNAs are produced from exon-containing lariats and intronic lariats that escape debranching (Salzman et al., 2012; Zhang et al., 2013) (Fig. 1A). Back-splicing junctions (BSJs) have been characterized as a key feature in distinguishing circular RNAs from linear RNAs. Besides the identification of BSJs, the development of RNA sequencing and computational analysis technologies has enabled the discovery of huge numbers of novel circular RNAs (Chen et al., 2021). Circular RNAs show cell-type specific expression patterns that change with variations in pathological conditions (Bachmayr-Heyda et al., 2015; Zhang et al., 2018c). Such changes in circular RNA expression patterns highlight the existence of several other ways to develop unconventional biomarkers to diagnose various disease states. Currently, circular RNA-based therapies are actively being developed; for instance, methods to modulate the circular RNA expression levels may be invented to reverse disease states.
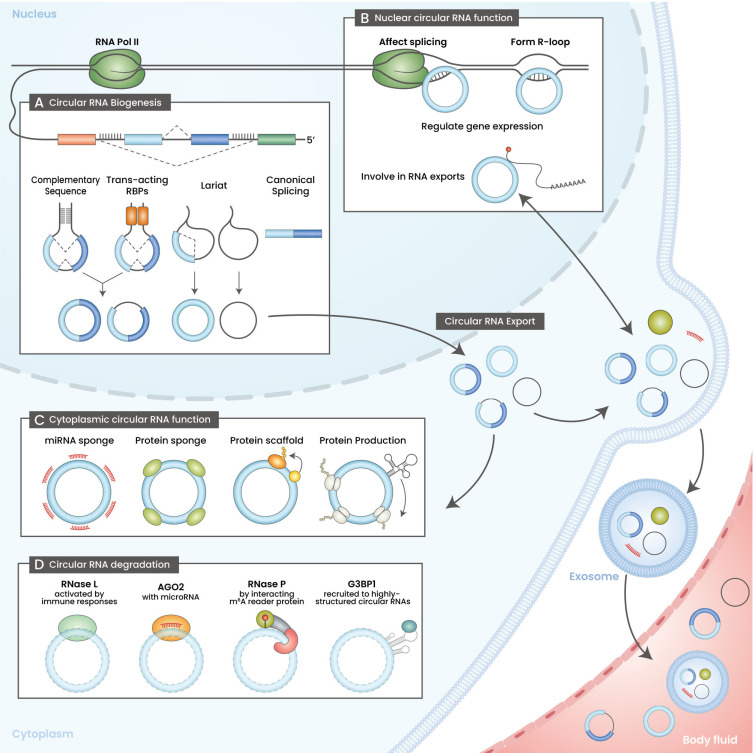
Fig. 1. The life cycle of circular RNA.
(A) Circular RNA biogenesis. Circular RNAs are produced through pre-mRNA back-splicing or by-products of splicing lariats. Back-splicing is facilitated by complementary sequences present in the flanking introns or trans-acting RBPs (RNA binding proteins). (B) Biological roles of nuclear circular RNAs. Circular RNAs can remain in the nucleus and function as gene expression regulators. Nuclear circular RNAs affect splicing by interacting with RNA polymerase II or forming R-loops with genomic DNA. Moreover, nuclear circular RNAs can modulate RNA export. (C) Biological roles of cytoplasmic circular RNAs. Most of the well-known functions of circular RNAs are cytoplasmic (e.g., miRNA sponge, protein sponge, protein scaffold, and template for protein production). Moreover, circular RNAs can be packaged into exosomes and circulated throughout the body as a body fluid. (D) Circular RNA degradation. The known mechanisms accelerating circular RNA degradation have been illustrated.
Biogenesis of circular RNAs
Back-splicing can occur when the upstream splicing acceptor site and downstream splicing donor site in a pre-mRNA become physically close. The proximal distancing of the back-splicing sites is regulated by cis- and trans-acting elements; intronic complementary sequences (ICSs) pairing on flanking introns, and RNA binding proteins (RBPs), respectively. ICSs, which are mainly composed of Alu repeats (Jeck et al., 2013), and RBPs (e.g., MBL, QK1, and FUS), facilitate the back-splicing process (Ashwal-Fluss et al., 2014; Conn et al., 2015; Errichelli et al., 2017; Jeck et al., 2013). However, other RBPs, such as ADAR and DHX9, impede circular RNA production by destabilizing ICSs (Aktas et al., 2017; Ivanov et al., 2015).
The factors determining the initiation of non-canonical back-splicing rather than canonical linear splicing remain largely unknown. Yet, clues as to which factors promote the production of circular RNAs were obtained by analyzing the structural characteristics of the host genes from which highly expressed circular RNAs originated; genes containing long introns between exons and actively transcribed genes were found to frequently generate circular RNAs (Cocquerelle et al., 1992; Kristensen et al., 2018). Moreover, although the causality between circular RNA expression levels and cellular stress is unknown, it has been reported that the process of circular RNA biogenesis increases when cells are under stress caused by DNA damage, oxidation, and heat shock (Lee et al., 2022). Accordingly, elucidating the molecular mechanisms that increase circular RNA levels under stress conditions should enhance our understanding of the link between circular RNAs and diseases.
Biological roles of circular RNAs
While most circular RNAs are localized in the cytoplasm, subsets of circular RNAs are localized in the nucleus (Zhang et al., 2019). Nuclear circular RNAs interact with RNA polymerase II and certain transcription factors to regulate gene expression at the transcriptional level (Guarnerio et al., 2019; Li et al., 2015; Zhang et al., 2013), whereas some other nuclear circular RNAs affect RNA export and splicing by binding RBPs or forming R-loops with the host gene (Conn et al., 2017; Mao et al., 2021; Xu et al., 2020) (Fig. 1B).
The first identified biological role of circular RNAs was that of a microRNA sponge (Hansen et al., 2013; Memczak et al., 2013). A circular RNA localized in the cytoplasm can sequester a specific microRNA and relieve target mRNAs from the microRNA. Currently, additional roles of cytoplasmic circular RNAs, such as protein sponges (Abdelmohsen et al., 2017; Huang et al., 2020) and scaffolds (Zeng et al., 2017), which suppress and facilitate protein functions, respectively, have been reported. Furthermore, in some cases, circular RNAs function as translational templates for producing polypeptides (Legnini et al., 2017; Lei et al., 2020). Polypeptides translated from circular RNAs modulate the biological roles of other intact proteins (Liang et al., 2019; Yang et al., 2018; Zhang et al., 2018a; 2018b; Zheng et al., 2019) (Fig. 1C).
Applications as biomarkers
Circular RNAs are great biomarker candidates. All tissues in the human body express circular RNAs; the expression profiles change depending on the context. During pathogenesis, altered circular RNA expression patterns reflect disease states. The lesions of patients with various types of cancers, neurological diseases, retinal diseases, and diabetes all have been shown to have significant changes in circular RNA expression (Hanineva et al., 2022; Lei et al., 2020; Li et al., 2021; Sakshi et al., 2021; Sun et al., 2020). For example, circ_0044234, a circular RNA that was found to be significantly downregulated in triple-negative breast cancer (TNBC) compared to other breast cancer types was suggested to be a promising biomarker for the diagnosis of TNBC (Darbeheshti et al., 2021).
Circular RNAs can also be detected in various body fluids such as cerebrospinal fluid (CSF), blood, urine, and saliva (Lai et al., 2022; Zhang et al., 2021b). Abundant levels of circular RNAs are expressed in the CSF, although these are less diverse than those in the blood (Wang et al., 2022); by contrast, much more diverse and higher levels of circular RNAs are expressed in the blood than in urine or saliva (Bahn et al., 2015; Hutchins et al., 2021; Koh et al., 2014). Circular RNAs may also be secreted into body fluids through exosomes—one of the extracellular vesicles (Oh et al., 2022; Wang et al., 2019). Changes in the type or composition of circular RNAs in exosomes can be representative of the pathophysiological states of some organs.
Liquid biopsies for circular RNA detection have enormous value as diagnostic and prognostic tools as they are more accessible than tissue biopsies (Lener et al., 2015). A recent study described a group of altered circular RNAs in serum from patients with gastric cancer, supporting the reliability of circular RNAs in body fluids as biomarkers (Roy et al., 2022); the expression of eight circular RNAs was increased in patients, but the expression of seven of the eight circular RNAs returned to normal after surgery. Using circular RNAs from liquid biopsies as biomarkers may be very useful, particularly for the diagnosis and prognosis of central nervous system diseases. Blood, rather than CSF, may be used for diagnosing brain diseases because a large percentage of blood circular RNAs overlap those present in the cerebral cortex; moreover, blood biopsies are less invasive than CSF biopsies (Memczak et al., 2015).
Efforts have been made to find human disease-associated circular RNAs that can be used as biomarkers. The disease-associated circular RNAs identified and analyzed thus far have been uploaded to several databases (Chen et al., 2021; Fan et al., 2021). By accessing the information available in open databases, we can hope for the development of early diagnostic methods and effective treatments for human diseases.
Approaches to regulating circular RNAs as therapeutic targets
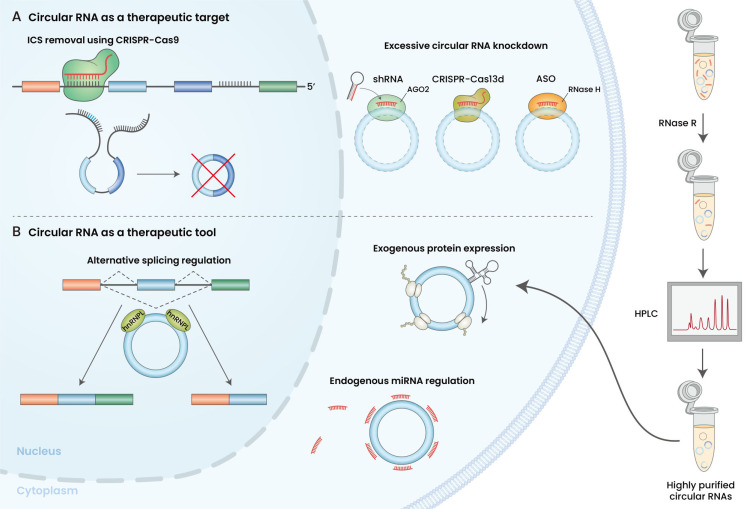
Circular RNAs differentially expressed in certain diseases may be used to modulate and reverse pathologic conditions. Many techniques have been developed to adjust the expression of disease-associated circular RNAs. Clustered regularly interspaced short palindromic repeats (CRISPR) and siRNA systems have been applied to reduce the circular RNAs expression levels (Fig. 2A). The initial step of circular RNA biogenesis can be blocked through CRISPR-Cas9-mediated ICS removal from the genome (Zhang et al., 2016). Alternatively, circular RNAs can be degraded through RNA cleavage using siRNAs (including small hairpin RNAs or shRNAs) or CRISPR-Cas13d (Jiang et al., 2020; Zhang et al., 2021a). Sequences targeted by siRNA or CRISPR-Cas13d should span the BSJs to exclude off-target linear RNA cleavage. siRNAs are practically advantageous for target cleavage as they employ endogenous protein machinery. To cut target RNAs using the CRISPR-Cas13d system, Cas13d in the form of either protein or mRNA has to be delivered along with guide RNAs. Despite these delivery issues, CRISPR-Cas13d is advantageous compared with siRNA, owing to its reduced off-target effects on a partial complementary sequence (Zhang et al., 2021a). Besides the RNA interference and CRISPR systems, ASOs such as GapmeR have been suggested as a method to deplete nuclear circular RNA levels (Santer et al., 2019). By contrast, to increase the expression levels of downregulated circular RNAs in cells, vectors expressing circular RNAs (Hu et al., 2021; Wang et al., 2018) or synthetic circular RNAs can be delivered into cells. If the molecular mechanisms underlying circular RNA biogenesis are elucidated in further detail, CRISPR systems and ASOs may also be utilized to increase endogenous circular RNA expression levels.
Fig. 2. Therapeutic approaches using circular RNA.
(A) Circular RNAs as therapeutic targets. ICS removal using the CRISPR-Cas9 system at the biogenesis stage and circular RNA knockdown using shRNA, CRISPR-Cas13, and ASO were examined for the downregulation of circular RNAs. (B) Circular RNAs as a therapeutic tool. Highly purified in vitro synthetic circular RNAs can be delivered to cells to function as sponges and play roles in alternative splicing regulation, exogenous protein expression, and microRNA suppression.
SYNTHETIC CIRCULAR RNAs AS THERAPEUTIC MOLECULES
Circular RNAs can be used as gene-delivery platforms for disease prevention or treatment. Circular RNAs, which lack ends, have great advantages in terms of stability and can help resolve critical issues in the developmental processes of RNA-based therapeutic molecules. When circular RNAs synthesized in vitro are introduced into cells, compared with circular RNAs expressed through DNA plasmids, synthetic circular RNAs may not be sustained at high levels in cells. However, compared with plasmid DNAs expressing circular RNAs, it is safer to deliver synthetic circular RNAs into cells because DNA may be integrated into the host genome. Moreover, synthetic circular RNAs can be used to express exogenous proteins or regulate endogenous gene networks (Fig. 2B).
Synthesis of circular RNA in vitro
Circularization is the most important step of in vitro circular RNA synthesis. Three different methods have been developed to connect both ends of linear RNAs. First, by adding condensation reagents, a phosphodiester bond can be formed between the 5′-phosphate and 3′-hydroxyl groups of a linear RNA (Dolinnaya et al., 1991). However, the production of by-products, i.e., 2′, 5′- phosphodiester linkages from the same condensation reaction, is unavoidable. Next, enzymatic ligation using DNA/RNA ligases can circularize linear RNAs. Oligonucleotides with complementary sequences spanning the 5′ and 3′ ends of linear RNAs work as splints to physically bring each end of a linear RNA into proximity; ligases can covalently connect both ends (Abe et al., 2018). A caveat of this approach is the possibility of concatemer generation, reducing the overall circular RNA yield. Finally, ribozymes can be conveniently used to generate circular RNAs. Linear RNAs can circularize and form circular RNAs if the sequences required for self-splicing are introduced to the linear forms. However, to produce intact circular RNAs with the desired sequences but without any partial ribozyme sequences originating from other organisms, sequences in a linear RNA need to be used as exonic ribozyme sequences. Through linear RNA sequence permutations, nucleotide sequences functioning as ribozyme exons in circularization can be identified (Rausch et al., 2021).
Protein expression
Circular RNA-based protein expression has been attempted to extend the retention time of protein-expressing RNAs in the body. Protein expression using circular RNAs has been examined in vaccine development, and the results have been remarkable. The same amounts of circular RNAs and linear mRNAs were assessed, and circular RNAs were found to produce more antigens than linear mRNAs (Wesselhoeft et al., 2018). Circular RNAs showed immunogenicity comparable to that of m1Ψ-modified linear RNAs, which have a higher protein translatability than unmodified linear RNAs (Qu et al., 2022). The results indicated that modified nucleotides are unnecessary in circular RNAs and that lesser amounts of circular RNAs than linear RNAs are needed to achieve the same level of protein expression. Furthermore, circular RNAs were found to be durable at room temperature, making the supply of circular RNA-based therapeutic molecules convenient and easy to handle.
To generate proteins, circular RNAs lacking a 5′ cap structure need to recruit translational machinery using other systems. The internal ribosome entry site (IRES) is the most representative structure that can be used for cap-independent translation. A recent study that performed extensive IRES screening identified the most effective functional IRES in circular RNAs (Chen et al., 2022); briefly, human rhinovirus-B IRES was reported to yield the highest level of translation products from circular RNAs. Besides facilitating protein production over a long period, the incorporation of aptamers or chemical modifications to circular RNAs can promote the recruitment of translation initiation factors to IRES (Chen et al., 2022; Tusup et al., 2018).
Noncoding elements regulating cellular gene expression networks
Circular RNAs with engineered sequences have been used to regulate microRNAs and proteins (Jost et al., 2018; Lavenniah et al., 2020; Schreiner et al., 2020) (Fig. 2B). Heterogeneous nuclear ribonucleoprotein L (hnRNPL), which is involved in splicing, can be sequestered by artificial circular RNAs (Schreiner et al., 2020). The splicing patterns were altered when artificial circular RNAs, which function as a hnRNPL sponge, were introduced into cells. Apart from regulating proteins, injecting circular RNAs engineered to function as microRNA sponges for miR-132 and miR-212 into a mouse model of transverse aortic constriction, as well as injecting circular RNAs having twelve miR-29b binding sites into a mouse model of muscle atrophy, resulted in pathological symptom improvement (Lavenniah et al., 2020; Li et al., 2022).
Circular RNAs synthesized in vitro can be introduced into cells to modulate the endogenous circular RNA-centric gene regulatory network. In conditions where the equilibrium between circular RNAs and other interacting molecules is disrupted, circular RNAs mimicking endogenous circular RNAs can be delivered into cells. Nevertheless, further investigations are needed to identify the cellular functions and intermolecular interactions of endogenous circular RNAs.
Considerations
To achieve long-lasting effects of circular RNA-based therapeutics, features that may accelerate circular RNA decay must be avoided during circular RNAs design (Fig. 1D). Complementary sequences that perfectly match those of microRNAs recruit AGO2; such sequences should be excluded to prevent circular RNA cleavages. Because highly structured RNAs preferentially bind G3BP1, which contains endonuclease activity, highly structured sequences in circular RNAs are not conducive to circular RNA stability. The association between RNAs and G3BP1 consequently degrades structured RNAs (Fischer et al., 2020). Moreover, whether the m6A modification helps increase the circular RNA stability remains unclear. The m6A modification helps exogenous circular RNAs be recognized as “self,” reducing the chances of RNA degradation caused by immune responses (Chen et al., 2019). By contrast, another report showed that the m6A modification facilitates circular RNA degradation by recruiting RNase P through YTHDF2, an m6A reader protein (Park et al., 2019). Given these opposing arguments, a substantial amount of attention is needed to progress the field of circular RNA biology.
Another important point concerning persistent synthetic circular RNA expression is purity (Fig. 2). By-products of circular RNAs could stimulate innate immune responses, thereby activating RNase L, which can induce global circular RNA decay. RNase R, a 3′-5′ exoribonuclease, has been used for circular RNA purification. However, only isolating circular RNAs was incomplete despite efforts to maximize the enzymatic activities of RNase R (Xiao and Wilusz, 2019). Nowadays, size-exclusion high-performance liquid chromatography together with RNase R treatment is being used to obtain high-purity circular RNAs. High-purity circular RNAs leading to less innate immune responses provide additional benefits to the uses of circular RNAs encoding proteins, thereby guaranteeing higher translation efficiency (Wesselhoeft et al., 2018).
CONCLUSION AND FUTURE PROSPECTS
Circular RNAs in and out of cells are multifunctional molecules that can be used as biomarkers, therapeutic targets, and therapeutic molecules. Although efforts have been made to understand how circular RNAs are generated and act upon production, the majority of reports are observations, and studies to understand the detailed mechanism are lacking. To use circular RNAs for clinical purposes, additional fundamental knowledge on the molecular mechanisms of circular RNA biogenesis and decay, cell-type specific circular RNA expression profiles in a given condition, and subcellular localization of circular RNAs need to be revealed in further detail.
Regarding circular RNA biogenesis, different angles of basic research may reveal currently unknown cis- and trans-acting elements and factors, inducing back-splicing of the preferred host genes. Moreover, different types of cellular stress, other than previously discovered types of damage, may be unveiled. From another perspective, besides acting as a protein and microRNA sponge, pathological conditions altering the expression levels of novel circular RNAs in diverse common or rare diseases could open a new field of study, thus advancing the diagnosis and treatment of hard-to-treat and incurable human diseases.
ACKNOWLEDGMENTS
This work was supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science and ICT (NRF-2022M3E5F1016556). D.K. was supported by the Fostering the Next Generation of Researchers Program of the NRF of Korea (NRF-2022R1A6A3A13073152).
Footnotes
AUTHOR CONTRIBUTIONS
M.J. and J.H. conceived and designed the study. M.J. and D.K. performed the literature search and reviewed the previous publications on specific topics. M.J. and D.K. wrote the initial draft and designed figures. M.J., D.K., G.S., and J.H. wrote and revised the manuscript. J.H. supervised the paper and provided feedback.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe N., Kodama A., Abe H. Preparation of circular RNA in vitro. Methods Mol. Biol. 2018;1724:181–192. doi: 10.1007/978-1-4939-7562-4_15. [DOI] [PubMed] [Google Scholar]
- Aktas T., Avsar Ilik I., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Bachmayr-Heyda A., Reiner A.T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., Mesteri I., Grunt T.W., Zeillinger R., Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J.H., Zhang Q., Li F., Chan T.M., Lin X., Kim Y., Wong D.T., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang C., Sun H., Wang J., Liang Y., Wang Y., Wong G. The bioinformatics toolbox for circRNA discovery and analysis. Brief. Bioinform. 2021;22:1706–1728. doi: 10.1093/bib/bbaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Wang S.K., Belk J.A., Amaya L., Li Z., Cardenas A., Abe B.T., Chen C.K., Wender P.A., Chang H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2022 Jul 18; doi: 10.1038/s41587-022-01393-0. [Epub]. https://doi.org/10.1038/s41587-022-01393-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.G., Chen R., Ahmad S., Verma R., Kasturi S.P., Amaya L., Broughton J.P., Kim J., Cadena C., Pulendran B., et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C., Daubersies P., Majerus M.A., Kerckaert J.P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C., et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- Darbeheshti F., Zokaei E., Mansoori Y., Emadi Allahyari S., Kamaliyan Z., Kadkhoda S., Tavakkoly Bazzaz J., Rezaei N., Shakoori A. Circular RNA hsa_circ_0044234 as distinct molecular signature of triple negative breast cancer: a potential regulator of GATA3. Cancer Cell Int. 2021;21:312. doi: 10.1186/s12935-021-02015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinnaya N.G., Sokolova N.I., Ashirbekova D.T., Shabarova Z.A. The use of BrCN for assembling modified DNA duplexes and DNA-RNA hybrids; comparison with water-soluble carbodiimide. Nucleic Acids Res. 1991;19:3067–3072. doi: 10.1093/nar/19.11.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfo R., Peruzzi G., et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Lei X., Tie J., Zhang Y., Wu F., Pan Y. CircR2Disease v2.0: an updated web server for experimentally validated circRNA-disease associations and its application. Genomics Proteomics Bioinformatics. 2021 Nov 29; doi: 10.1016/j.gpb.2021.10.002. [Epub]. https://doi.org/10.1016/j.gpb.2021.10.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J.W., Busa V.F., Shao Y., Leung A.K.L. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell. 2020;78:70–84.e6. doi: 10.1016/j.molcel.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J., Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnerio J., Zhang Y., Cheloni G., Panella R., Mae Katon J., Simpson M., Matsumoto A., Papa A., Loretelli C., Petri A., et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 2019;29:628–640. doi: 10.1038/s41422-019-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanineva A., Park K.S., Wang J.J., DeAngelis M.M., Farkas M.H., Zhang S.X. Emerging roles of circular RNAs in retinal diseases. Neural Regen. Res. 2022;17:1875–1880. doi: 10.4103/1673-5374.335691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hodgson J. The pandemic pipeline. Nat. Biotechnol. 2020;38:523–532. doi: 10.1038/d41587-020-00005-z. [DOI] [PubMed] [Google Scholar]
- Hu G., Zhai S., Yu S., Huang Z., Gao R. Circular RNA circRHOBTB3 is downregulated in hepatocellular carcinoma and suppresses cell proliferation by inhibiting miR-18a maturation. Infect. Agent. Cancer. 2021;16:48. doi: 10.1186/s13027-021-00384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins E., Reiman R., Winarta J., Beecroft T., Richholt R., De Both M., Shahbander K., Carlson E., Janss A., Siniard A., et al. Extracellular circular RNA profiles in plasma and urine of healthy, male college athletes. Sci. Data. 2021;8:276. doi: 10.1038/s41597-021-01056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Liu C., Li C.P., Xu S.S., Yao M.D., Ge H.M., Sun Y.N., Li X.M., Zhang S.J., Shan K., et al. Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J. Clin. Invest. 2020;130:3833–3847. doi: 10.1172/JCI123353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost I., Shalamova L.A., Gerresheim G.K., Niepmann M., Bindereif A., Rossbach O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018;15:1032–1039. doi: 10.1080/15476286.2018.1435248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M.K., Shin E.C. Phenotypes and functions of SARS-CoV-2-reactive T cells. Mol. Cells. 2021;44:401–407. doi: 10.14348/molcells.2021.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh W., Pan W., Gawad C., Fan H.C., Kerchner G.A., Wyss-Coray T., Blumenfeld Y.J., El-Sayed Y.Y., Quake S.R. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7361–7366. doi: 10.1073/pnas.1405528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen L.S., Okholm T.L.H., Veno M.T., Kjems J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15:280–291. doi: 10.1080/15476286.2017.1409931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Anaya J., Mudunuri S.B., Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H., Li Y., Zhang H., Hu J., Liao J., Su Y., Li Q., Chen B., Li C., Wang Z., et al. exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 2022;50(D1):D118–D128. doi: 10.1093/nar/gkab1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenniah A., Luu T.D.A., Li Y.P., Lim T.B., Jiang J., Ackers-Johnson M., Foo R.S. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. 2020;28:1506–1517. doi: 10.1016/j.ymthe.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Oh J.E. Humoral immunity against SARS-CoV-2 and the impact on COVID-19 pathogenesis. Mol. Cells. 2021;44:392–400. doi: 10.14348/molcells.2021.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.C., Wang W.Y., Lin H.H., Huang Y.R., Lin Y.C., Hsiao K.Y. The functional roles and regulation of circular RNAs during cellular stresses. Noncoding RNA. 2022;8:38. doi: 10.3390/ncrna8030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Zheng G., Ning Q., Zheng J., Dong D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lener T., Gimona M., Aigner L., Borger V., Buzas E., Camussi G., Chaput N., Chatterjee D., Court F.A., Del Portillo H.A., et al. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J. Extracell. Vesicles. 2015;4:30087. doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen R., Zheng Y., Yuan W., Yang T., Zhu X., Yan Y., Jin B., Xu W., Zhang Z., et al. Engineered circular RNA circmiR-29b attenuates muscle atrophy by sponging miR-29b. Adv. Ther. (Weinh.) 2022;5:2200029. doi: 10.1002/adtp.202200029. [DOI] [Google Scholar]
- Li M.L., Wang W., Jin Z.B. Circular RNAs in the central nervous system. Front. Mol. Biosci. 2021;8:629593. doi: 10.3389/fmolb.2021.629593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Liang W.C., Wong C.W., Liang P.P., Shi M., Cao Y., Rao S.T., Tsui S.K., Waye M.M., Zhang Q., Fu W.M., et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S., Zhang W., Yang F., Guo Y., Wang H., Wu Y., Wang R., Maskey N., Zheng Z., Li C., et al. Hsa_circ_0004296 inhibits metastasis of prostate cancer by interacting with EIF4A3 to prevent nuclear export of ETS1 mRNA. J. Exp. Clin. Cancer Res. 2021;40:336. doi: 10.1186/s13046-021-02138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., Khatwani N., Su S.V., Miracco E.J., Issa W.J., et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6:eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C., Koh D., Jeon H.B., Kim K.M. The role of extracellular vesicles in senescence. Mol. Cells. 2022;45:603–609. doi: 10.14348/molcells.2022.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandini, von Niessen A.G., Poleganov M.A., Rechner C., Plaschke A., Kranz L.M., Fesser S., Diken M., Lower M., Vallazza B., Beissert T., et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3' UTRs identified by cellular library screening. Mol. Ther. 2019;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.W., Lagniton P.N.P., Liu Y., Xu R.H. mRNA vaccines for COVID-19: what, why and how. Int. J. Biol. Sci. 2021;17:1446–1460. doi: 10.7150/ijbs.59233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- Qu L., Yi Z., Shen Y., Lin L., Chen F., Xu Y., Wu Z., Tang H., Zhang X., Tian F., et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185:1728–1744.e16. doi: 10.1016/j.cell.2022.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Kanda M., Nomura S., Zhu Z., Toiyama Y., Taketomi A., Goldenring J., Baba H., Kodera Y., Goel A. Diagnostic efficacy of circular RNAs as noninvasive, liquid biopsy biomarkers for early detection of gastric cancer. Mol. Cancer. 2022;21:42. doi: 10.1186/s12943-022-01527-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakshi S., Jayasuriya R., Ganesan K., Xu B., Ramkumar K.M. Role of circRNA-miRNA-mRNA interaction network in diabetes and its associated complications. Mol. Ther. Nucleic Acids. 2021;26:1291–1302. doi: 10.1016/j.omtn.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer L., Bar C., Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 2019;27:1350–1363. doi: 10.1016/j.ymthe.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner S., Didio A., Hung L.H., Bindereif A. Design and application of circular RNAs with protein-sponge function. Nucleic Acids Res. 2020;48:12326–12335. doi: 10.1093/nar/gkaa1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson M.L., Zamecnik P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. U. S. A. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Li B., Shu C., Ma Q., Wang J. Functions and clinical significance of circular RNAs in glioma. Mol. Cancer. 2020;19:34. doi: 10.1186/s12943-019-1121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusup M., Kundig T., Pascolo S. An eIF4G-recruiting aptamer increases the functionality of in vitro transcribed mRNA. EPH - Int. J. Med. Health Sci. 2018;4:29–37. [Google Scholar]
- Wang H., Xiao Y., Wu L., Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int. J. Oncol. 2018;52:743–754. doi: 10.3892/ijo.2018.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu J., Ma J., Sun T., Zhou Q., Wang W., Wang G., Wu P., Wang H., Jiang L., et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yu R., Chen X., Bao H., Cao R., Li A.N., Ou Q., Tu H.Y., Zhou Q., Wu X., et al. Clinical utility of cerebrospinal fluid-derived circular RNAs in lung adenocarcinoma patients with brain metastases. J. Transl. Med. 2022;20:74. doi: 10.1186/s12967-022-03274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M.S., Wilusz J.E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3' ends. Nucleic Acids Res. 2019;47:8755–8769. doi: 10.1093/nar/gkz576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang J., Tian Y., Gao Y., Dong X., Chen W., Yuan X., Yin W., Xu J., Chen K., et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer. 2020;19:128. doi: 10.1186/s12943-020-01246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A., et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang X., Li C., Yue L., Ding N., Riordan T., Yang L., Li Y., Jen C., Lin S., et al. Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol. 2019;16:220–232. doi: 10.1080/15476286.2019.1565284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H., et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018a;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018b;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Nguyen T.M., Zhang X.O., Wang L., Phan T., Clohessy J.G., Pandolfi P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021a;22:41. doi: 10.1186/s13059-021-02263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Su X., Wang P., Lin W. The value of circulating circular RNA in cancer diagnosis, monitoring, prognosis, and guiding treatment. Front. Oncol. 2021b;11:736546. doi: 10.3389/fonc.2021.736546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Yang T., Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018c;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Chen L., Zhou Y., Wang Q., Zheng Z., Xu B., Wu C., Zhou Q., Hu W., Wu C., et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]