Abstract
The immune response is crucial for coronavirus disease 19 (COVID‐19) progression, with the participation of proinflammatory cells and cytokines, inducing lung injury and loss of respiratory function. CLEC5A expression on monocytes can be triggered by viral and bacterial infections, leading to poor outcomes. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is able to induce neutrophil activation by CLEC5A and Toll‐like receptor 2, leading to an aggressive inflammatory cascade, but little is known about the molecular interactions between CLEC5A and SARS‐CoV‐2 proteins. Here, we aimed to explore how CLEC5A expression could be affected by SARS‐CoV‐2 infection using immunological tools with in vitro, in vivo, and in silico assays. The findings revealed that high levels of CLEC5A expression were found in monocytes from severe COVID‐19 patients in comparison with mild COVID‐19 and unexposed subjects, but not in vaccinated subjects who developed mild COVID‐19. In hamsters, we detected CLEC5A gene expression during 3–15 days of Omicron strain viral challenge. Our results also showed that CLEC5A can interact with SARS‐CoV‐2, promoting inflammatory cytokine production, probably through an interaction with the receptor‐binding domain in the N‐acetylglucosamine binding site (NAG‐601). The high expression of CLEC5A and high levels of proinflammatory cytokine production were reduced in vitro by a human CLEC5A monoclonal antibody. Finally, CLEC5A was triggered by spike glycoprotein, suggesting its involvement in COVID‐19 progression; therapy with a monoclonal antibody could be a good strategy for COVID‐19 treatment, but vaccines are still the best option to avoid hospitalization/deaths.
Keywords: CLEC5A, COVID‐19, immunotherapy, monocytes, spike protein
1. INTRODUCTION
C‐type lectins play an important role during microbial infection, as it mediates inflammatory responses and can contribute to poor outcomes, as already described for flaviviruses and bacterial infections. 1 , 2 , 3 , 4 CLEC5A is expressed by mononuclear cells, such as monocytes, macrophages, neutrophils, and dendritic cells, and can interact with virions and bacterial walls. It is known that CLEC5A is a potential target for therapeutic purposes to attenuate inflammatory reactions because this pattern recognition receptor can form multivalent heterocomplexes with DC‐SIGN and other ligands, promoting cascades of inflammatory cytokines by Syk activation. 3 , 4 , 5
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is responsible for the pandemic situation that has occurred worldwide since March 2020, causing COVID‐19 with a remarkable number of hospitalization and deaths. 6 , 7 Licensed vaccines have helped to reduce the poor outcomes; however, special populations, such as pregnant women, immunocompromised subjects, and elderly people still need caution regarding the frequency of new variant emergence. 8 , 9 , 10 , 11 , 12 Additionally, products for COVID‐19 treatment are still under investigation, showing positive results for the use of monoclonal antibodies (mAbs). 13 , 14 , 15 , 16
It is known that a major part of COVID‐19 severity is mediated by the immune response, with the involvement of activated monocytes, neutrophils, and natural killer cells producing proinflammatory cytokines, inducing lung injury and compromising respiratory function. 17 , 18 , 19 , 20 However, the expression of C‐type lectins, such as CLEC5A on mononuclear cells, during COVID‐19 or induced by SARS‐CoV‐2 remains unexplored.
Regarding this gap in the knowledge of how C‐type lectins are affected by SARS‐CoV‐2 infection, we aimed to explore (1) if CLEC5A is expressed by monocytes in COVID‐19 patients and vaccinated subjects; (2) if the CLEC5A gene has a kinetic of expression during SARS‐CoV‐2 infection using in vivo murine model; (3) whether the spike glycoprotein is able to induce CLEC5A expression on monocytes from COVID‐19 patients; and (4) if there is a molecular interaction between the CLEC5A receptor and spike protein using in silico investigations.
2. MATERIALS AND METHODS
2.1. Human blood samples
Blood samples were obtained from 70 volunteers, divided into five different groups with descriptive clinical characteristics displayed in Supporting Information: Table 1. Blood collection was performed at different times: February to March 2020 from subjects unexposed to SARS‐CoV‐2 (n = 18); July to November 2020 from mild (n = 17) and severe (n = 10) COVID‐19 patients; April to October 2021 from fully vaccinated and unexposed subjects (n = 12); and January to February 2022 from fully vaccinated subjects with recent SARS‐CoV‐2 infection (n = 13). Unexposed subjects were monitored biweekly by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) on naso‐oropharyngeal swabs for SARS‐CoV‐2 detection, as described by Matos et al. 21 The signs and symptoms reported by all subjects who had confirmed COVID‐19 were: fever, headache, myalgia, cough, and loss of smell/taste; each volunteer reported at least three of those symptoms, and severity was defined as hospitalized patients. The symptoms lasted for up to 2 weeks. Peripheral blood mononuclear cells (PBMC) were obtained by Ficoll Histopaque™ density gradient medium centrifugation (30 min at 400g at 18°C). 22 Cells were frozen using Cryostor cell preservation media (Sigma‐Aldrich) and kept in liquid nitrogen until use. Plasma samples separated by centrifugation were used to detect SARS‐CoV‐2 specific immunoglobulin G (IgG) antibodies targeting spike protein using commercial kits (SARS‐CoV‐2 IgG II Quant Architect; Abbott).
2.2. Syrian golden hamster blood samples
Blood samples were obtained at days 3, 5, 10, and 15 through exsanguination by cardiac puncture from a 36 Syrian golden hamster (Mesocricetus auratus) at 1 year of age and 150 ± 1.4 g infected intranasally with SARS‐CoV‐2 strains Delta (1.0 × 106 PFU/ml) and Omicron (1.0 × 106 PFU/ml). 23 , 24 The SARS‐CoV‐2 detection after infection was performed using RT‐qPCR by oropharyngeal swabs from the same time points for blood collection.
2.3. Immunophenotyping by flow cytometry
PBMC were thawed at 37°C and suspended in Roswell Park Memorial Institute (RPMI) 1640 medium, centrifuged at 10 min at 400g at 18°C, and washed in fluorescence‐activated cell sorting (FACS) buffer solution (2% fetal bovine serum in phosphate buffer solution, pH 7.4). The cells were centrifuged at 400g for 10 min, and the cell pellet was homogenized and stained with live/dead cell viability dye (Thermo Fisher Scientific; L23105) according to the manufacturer's protocol. PBMC were washed in FACS buffer and stained with surface antibodies: CD3‐APC‐Cy7, clone: SK7; CD8‐Brilliant Violet 605, clone: SK1; CD14‐Brilliant Blue 700, clone: MOP9; CD16‐FITC, clone: 3G8; CD56‐Brilliant Violet 605, clone: B159; CLEC5A‐PE, clone: FAB2384P. All antibodies were purchased from BD Biosciences, except for anti‐CLEC5A, which was purchased from R&D Systems. After surface staining, cells were washed with FACS buffer and fixed (Cytofix; BD Biosciences). All the steps were performed at 4°C unless otherwise specified by the manufacturer. Compensation beads (UltraCompBeads; Invitrogen™; cat #01‐2222‐42 and ArC™ Armine Reactive Compensation Bead Kit; Invitrogen™; cat#A10346) were used for compensation set‐up. The acquisition was performed using an LSR Fortessa flow cytometer (Becton Dickinson) and data were analyzed using FlowJo™ software (Becton Dickinson).
2.4. Detection of CLEC5A gene expressed in blood from hamsters by RT‐qPCR
To evaluate the CLEC5A gene expression after the viral challenge, blood samples were treated with TRIzol (Invitrogen) to obtain total RNA extracted for RNA detection. RNA was quantified in a spectrophotometer (Nanodrop Technologies), followed by a complementary DNA (cDNA) synthesis from 250 ng of total RNA using the High‐Capacity cDNA Reverse‐Transcription Kit (Thermo Fisher Scientific), both procedures according to manufacturer's instructions. Analysis of gene expression was performed using Fast SYBR™ Green Master Mix (Applied Biosystems). Reference genes PPIA and GAPDH were chosen as constitutive control. The primer set for the CLEC5A gene is as follows: TTTTTCTCGTGTATTTCCCACA (forward primer); ACGAAGCCATCATTACTTTTGC (reverse primer). For the qualitative qRT‐PCR reaction, 4 μl of diluted cDNA (1:2) was added to the reaction using Syber green (Applied Biosystems). Quantistudio (Applied Biosystem) instrument was used to calculate the C t values during the qRT‐PCR assay and the protocol involved: polymerase activation at 95°C for 20 s; followed by up to 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min.
2.5. In vitro assay for CLEC5a binding with spike glycoprotein
Available PBMC samples from mild and unexposed volunteers were selected for in vitro assays (n = 20; 10 unexposed and 10 mild COVID‐19) to evaluate CLEC5a binding with spike glycoprotein. Cells were thawed at 37°C and suspended in supplemented RPMI 1640 medium (R10), centrifuged at 10 min at 400g, and 1 × 106 cells/well were cultured in a 96‐well round bottom plate for 18 h at 37°C in a humidified chamber at 5% CO2 before stimulation. After 18 h of resting, cells were cultured with spike glycoprotein (1 μg/ml, JPT peptides, PepMix™ SARS‐CoV‐2, Spike Glycoprotein, PM‐WCPV‐S), and the mix of human cytokines interleukin‐2 (IL‐2), IL‐6, IL‐8, tumor necrosis factor‐α(TNF‐α) and interferon γ (IFN‐γ) (50 ng/ml each, all from R&D Systems) as a positive control. Negative control was set up with RPMI 1640 media with dimethyl sulfoxide (diluted 1:1000). In a separate plate, cells were cultured with 50 μg/ml of human anti‐CLEC5A for 2 h at 4°C, according to that described by others. 4 After incubation at 4°C, the plate was centrifuged (10 min at 400g), the supernatant was discarded, and spike glycoprotein was added to an equal volume at 1 μg/ml; in separate wells, the human cytokines IL‐2, IL‐6, IL‐8, TNF‐α, and IFN‐γ at 50 ng/ml each were also added to the cell cultivation for 24 h. After that, the PBMC was submitted to immunophenotyping staining as described previously for the flow cytometry assay, including the monocyte activation marker CD69, 25 , 26 purchased from BD Biosciences (CD69‐Brilliant Violet 421, clone: FN50). The unstimulated condition was used to subtract any background staining from the analysis. Positive spike protein or cytokine stimulation was determined by the detection of at least 10 antigen‐specific CD69+ cells and a frequency of antigen‐specific cells of at least twice the corresponding unstimulated signal.
2.6. Cytokine detection
Quantification of the cytokines IL‐2, IFN‐γ, IL‐6, and IL‐1β was performed using an in‐house multiplex liquid microarray test. Briefly, 106 xMAP® microspheres (Luminex Corporation) were coupled with anti‐human mouse purified mAbs with the following concentrations: 10 μg/ml of anti‐IL‐2, 500 μg/ml of anti‐IFN‐γ, 100 μg/ml of anti‐IL‐6, and 50 μg/ml of anti‐IL1β (all from Abcam Plc). Coupling reactions were performed using an Amine Coupling Kit (Bio‐Rad) following the manufacturer's instructions. For the quantification assay, the Bio‐Plex Pro™ Human Cytokine Standard 27‐plex, Group I (Bio‐Rad) was used as the standard curve following the manufacturer's instructions. Cell supernatant samples (diluted 1:2) and standards were incubated in duplicate with coupled microspheres for 30 min at 37°C under rotation of 600 rpm. The microspheres were washed three times with wash solution (Phosphate‐buffered saline pH 7.4 + 1% bovine serum albumin + 0.02% Tween 20 + 0.005% sodium azide) and incubated with 0.1 μg/ml of anti‐human goat polyclonal biotinylated antibodies against all cytokines analyzed (R&D Systems) for 30 min at 37°C at 600 rpm. Then, the microspheres were washed three times, incubated with 1X streptavidin‐phycoerythrin (BD Biosciences) for 10 min at 37°C at 600 rpm, and resuspended in the wash solution. The median fluorescence intensity (MFI) of each reaction was quantified using the Luminex MagPix® system (Luminex Corporation). Cytokine concentrations in pg/ml were calculated by interpolating samples on the MFI standard curve by four‐parameter analysis (Softmax v5.4; Molecular Devices). The mix of human cytokines added in the in vitro assay, as a positive control, had the initial value subtracted from the final quantification.
2.7. Molecular docking for binding affinity and residue interaction prediction
The C‐lectin receptors on myeloid cells are extremely important for innate immunity response concerning dysregulation induced by several viruses, as described previously, 27 , 28 but the absence of information about the interactions with SARS‐CoV‐2 proteins led us to investigate their participation in COVID‐19. Molecular docking experiments were performed to generate a complex between the CLEC5A receptor (PBD ID: 2YHF) and spike protein with the human angiotensin‐converting enzyme 2 (hACE2) receptor (PBD ID: 6LZG). ClusPro 2.0 server (https://cluspro.bu.edu/login.php) 29 was used to dock the designed extracellular monocyte receptor to spike protein from SARS‐CoV‐2. ClusPro is a fully automated protein–protein docking server that evaluates the docked complexes and retains a limited number of poses. The retained poses were then clustered, and the top poses were assessed. After the docking prediction by Cluspro, the interactions between the CLEC5A receptor and spike protein were visualized using PyMOL software (http://www.pymol.org/pymol), 30 which provided the binding residue prediction, as well as the root‐mean‐square deviation (RMSD). The RMSD was calculated by superimposition between the ligands atoms in the model provided by ClusPro top poses.
2.8. Statistical analysis
The normality assumption of the data was initially evaluated by the Kolmogorov–Smirnov test or Shapiro–Wilk test. Fisher's exact test was used to evaluate any associations among studied variables. Differences between groups were assessed using the Mann–Whitney t test, considering the groups of investigation. Bar graphs show the mean ± standard error. GraphPad Prism for Macintosh, version 8.4.2 was used to perform the statistical analysis. The significance for all statistical analyses was defined as p < 0.05.
In the graphs, the y‐axis displays the difference between stimulated cells and nonstimulated cells (mock) percentages, with the information provided in the legend. Cell frequencies above 1% were considered for the final analysis.
3. RESULTS
3.1. High expression of CLEC5A on monocytes is associated with severe COVID‐19
Immunophenotyping of PBMCs from the five studied groups (1: unexposed, 2: mild COVID‐19, 3: severe COVID‐19, 4: vaccinated for COVID‐19, and 5: participants with SARS‐CoV‐2 infection after full immunization [IAFV]) revealed that people who were hospitalized with severe COVID‐19 presented a significantly lower percentage of classical monocytes (CD14++CD16−) and higher percentages of intermediate (CD14+CD16+) and nonclassical monocytes (CD14+CD16++) in comparison with subjects who did not have any exposure to SARS‐CoV‐2 (Figure 1A,E). Meanwhile, CLEC5A was expressed on intermediate monocytes (CD14+CD16+) in COVID‐19 patients, independent of vaccination status. Surprisingly, nonclassical monocytes (CD14+CD16++) expressed significantly higher percentages of CLEC5A in the severe COVID‐19 group, distinct from the mild COVID‐19 and IAFV groups (Figure 1B–F). The expression of CLEC5A on nonclassical monocytes was also significantly higher in mild COVID‐19 patients without vaccination than in the IAFV group (Figure 1D,F). Considering these findings, a receiver operating characteristic curve was performed to evaluate a cut‐off for CLEC5A expression on nonclassical monocytes in our study population; when the percentage was above 20.9% (area under the curve = 0.79, p = 0.007), it had a strong association with severe COVID‐19 (odds ratio = 58.50, 95% confidence interval = 5.75–594.53, p = 0.0006).
Figure 1.
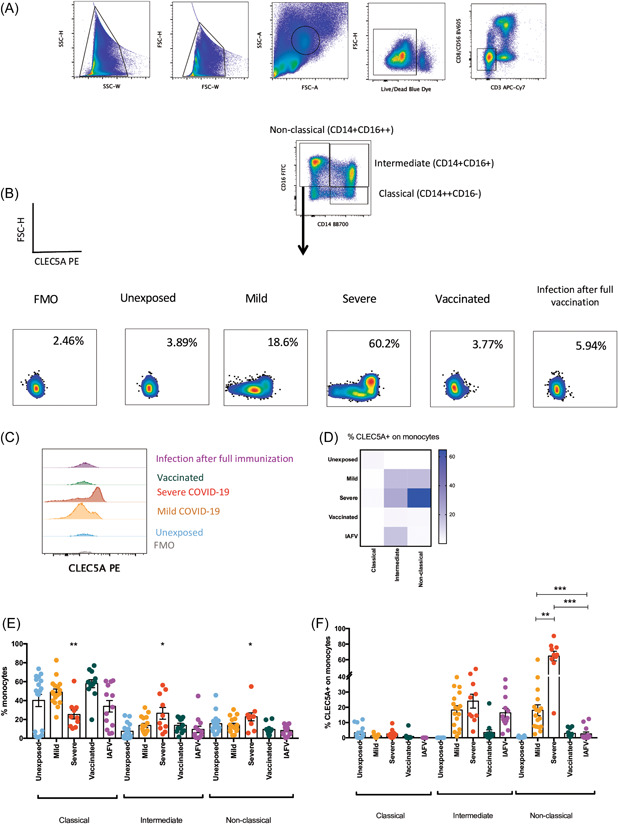
Immunophenotyping of CLEC5A expression by monocytes subpopulations. (A) Gate strategy to get monocytes subpopulations, (B) percentage of CLEC5A expression on monocytes using PBMC samples from different groups evaluated (left to right): Fluorescence minus one (FMO), unexposed, mild COVID‐19, severe COVID‐19, vaccinated for COVID‐19 (with two or three doses), Infected after full vaccination (IAFV, subjects who had mild COVID‐19). (C) Histogram of CLEC5A expression on nonclassical monocytes. (D) Mean of CLEC5A expression on the three subpopulations of monocytes according to the five groups evaluated. (E) Percentage of monocyte subpopulations divided by studied groups. (F) Percentage of CLEC5A expressed by monocytes subpopulations by studied groups. FSC‐A,forward scatter area;FSC‐H,forward scatter height;FSC‐W, forward light scatter width; PBMC, peripheral blood mononuclear cell; SSC‐A, side scatter area; SSC‐H,side scatter height; SSC‐W, side scatter width. *p < 0.05; **p < 0.01; ***p < 0.001.
3.2. CLEC5A gene expression is detected in hamsters infected with different strains of SARS‐CoV‐2
To explore whether and when the CLEC5A gene expression will be detected in blood cells from animal models infected by SARS‐CoV‐2, we followed 36 hamsters for 15 days with four‐time points: 3, 5, 10, and 15 days after the viral challenge. Twelve hamsters were noninfected (control group), 12 animals received the SARS‐CoV‐2 Delta strain, and the other 12 animals received the SARS‐CoV‐2 Omicron strain. After RT‐qPCR assays, it was possible to detect CLEC5A gene expression in 3/12 hamsters after 10 days of Delta strain challenge (Figure 2). In the other group, one animal expressed the CLEC5A gene on the third day, and 6/9 animals expressed the CLEC5A gene during 5–15 days of the Omicron strain challenge (Figure 2). The SARS‐CoV‐2 viral load was detected in oropharyngeal swabs of hamsters infected with Delta ([mean of viral particles/ml] − 3 days: 5.07 × 106; 5 days: 5.09 × 106; 10 days: 1.19 × 106; 15 days: 1.59 × 106) and Omicron strain ([mean of viral particles/ml] − 3 days: 5.26 × 106; 5 days: 5.12 × 106; 10 days: 3.8 × 105; 15 days: not detected). The CLEC5A gene expression and/or SARS‐CoV‐2 viral load were not detected in noninfected (control group) (Figure 2).
Figure 2.
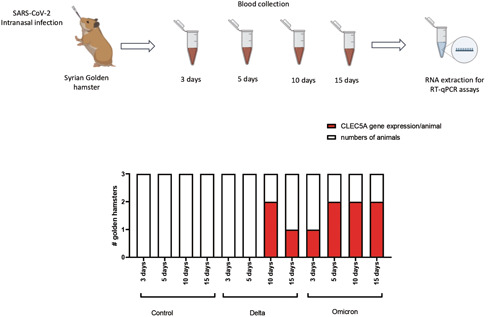
In vivo assay to monitor the kinetic of CLEC5A gene expression in hamster blood samples. The graph represents the detection of CLEC5A gene expression in the hamster blood samples collected between 3 and 15 days in noninfected animals (control), followed by SARS‐CoV‐2 infection using Delta variant (B.1.617.2) and Omicron variant (B.1.1.529). RT‐qPCR, reverse transcription‐quantitative polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.3. CLEC5A expression is induced by spike protein in vitro stimulation
We investigated the potential binding of spike protein with CLEC5A to induce expression of its receptor on nonclassical monocytes, and to determine if a human CLEC5A mAb (hCLEC5A mAb) could interfere in this interaction using the in vitro cultivation of PBMCs from mild COVID‐19 and unexposed subjects stimulated for 24 h under different conditions, including with and without hCLEC5A mAb (Figure 3A–D). The data show that spike protein elevated CLEC5A expression on nonclassical monocytes in mild COVID‐19 and in unexposed subjects (Figure 3C); the hCLEC5A mAb was able to inhibit the interaction between CLEC5A and spike protein in this monocyte subpopulation, similar to that observed for the mix of the cytokines (IFN‐γ, TNF‐α, IL‐2, IL‐6, and IL‐8), used as a positive control in the in vitro experiments, independent of COVID‐19 status (Figure 3D). Cytokine measurements in the supernatant after stimulation conditions showed that IFN‐γ was significantly elevated in unexposed subjects after cytokine stimulation or the hCLEC5 mAb with cytokines. For mild COVID‐19 subjects, the mix of cytokines and spike protein was able to induce IL‐2, IFN‐γ, IL‐6, and IL‐1β production. The hCLEC5A mAb reduced the production of IL‐2 and IL‐1β, even with the mix of cytokines or spike protein stimulation (Table 1).
Figure 3.
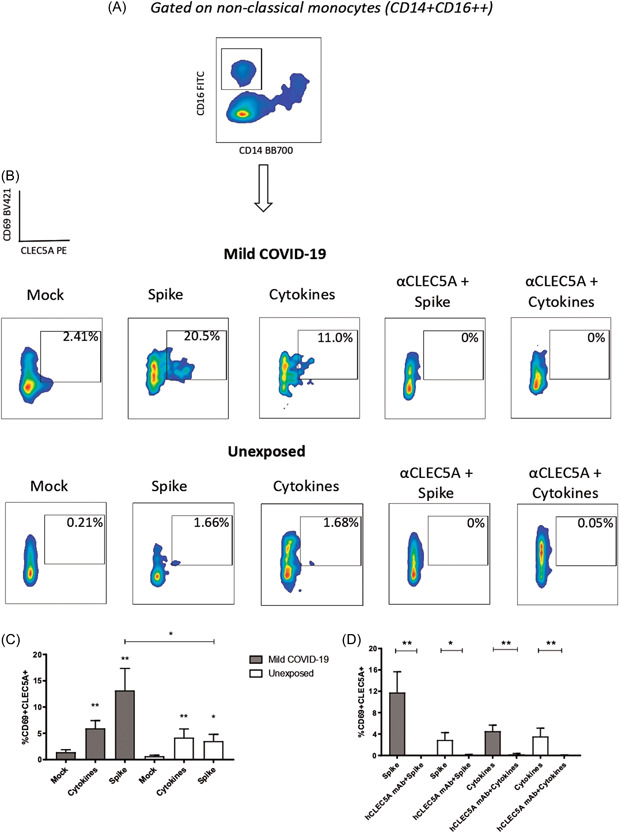
In vitro interaction between CLEC5A and spike protein (SARS‐CoV‐2). (A) Nonclassical monocytes were gated to access the information of activation (CD69+) and CLEC5A expression induced by stimulatory ligands, as (B) spike protein, proinflammatory cytokines, and after treatment with a monoclonal antibody, anti‐CLEC5A using PBMC samples from mild COVID‐19 patients and unexposed subjects. (C) Graphical representation of spike protein and the mix of cytokines stimulation comparing with unstimulated conditions (mock). (D) The values of the stimulated conditions were subtracted from unstimulated conditions showing the percentage of CD69 and CLEC5A coexpression on nonclassical monocytes in the groups of cell cultivation. PBMC, peripheral blood mononuclear cell; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2. *p < 0.05; **p < 0.01; ***p < 0.001.
Table 1.
Cytokines were measured after in vitro stimulation with cytokines, spike protein, and human CLEC5A monoclonal antibody
| Unexposed | |||||
|---|---|---|---|---|---|
| IL‐2 | IFN‐γ | IL‐6 | IL‐1β | ||
| In vitro conditions | Mock | 151.70 ± 39.27 | 55.74 ± 41.97 | 1092 ± 554.50 | 151.7 ± 39.27 |
| Cytokines | 155.80 ± 25.36 | 1200 ± 350.40 | 1953 ± 915.80 | 155.8 ± 25.36 | |
| aCLEC5A + cytokines | 83.70 ± 59.00 | 3788 ± 1028 | 1230 ± 445.88 | 83.70 ± 59.00 | |
| Spike | 177.80 ± 83.41 | 25.03 ± 5.15 | 585.3 ± 383.80 | 186.8 ± 83.41 | |
| aCLEC5A + spike | 172.10 ± 84.21 | 102.80 ± 23.89 | 439.00 ± 237.61 | 172.1 ± 84.21 | |
| p Value | Mock versus cytokines | 0.29 | 0.02 | 0.34 | 0.88 |
| Mock versus aCLEC5A + cytokines | 0.12 | 0.02 | 0.20 | 0.20 | |
| Mock versus spike | 0.11 | 0.77 | 0.06 | 0.89 | |
| Mock versus aCLEC5A + spike | 0.15 | 0.40 | 0.06 | 0.89 | |
| Cytokines versus aCLEC5A + cytokines | 0.34 | 0.06 | 0.90 | 0.12 | |
| Spike versus aCLEC5A + spike | 0.49 | 0.06 | 0.88 | 1.00 | |
| Mild COVID‐19 | |||||
|---|---|---|---|---|---|
| IL‐2 | IFN‐γ | IL‐6 | IL‐1β | ||
| In vitro conditions | Mock | 170.10 ± 46.69 | 10.97 ± 3.19 | 1075 ± 461.8 | 170.1 ± 46.69 |
| Cytokines | 320.70 ± 50.41 | 7147 ± 2470 | 3067 ± 108.61 | 395.7 ± 48.74 | |
| aCLEC5A + cytokines | 109.70 ± 24.30 | 4457 ± 1287 | 2578 ± 236.4 | 109.7 ± 12.15 | |
| Spike | 307.90 ± 87.03 | 29.16 ± 7.33 | 5395 ± 709.10 | 682.90 ± 186.81 | |
| aCLEC5A + spike | 153.55 ± 11.65 | 24.40 ± 12.01 | 3452 ± 2101 | 153.51 ± 11.65 | |
| p Value | Mock versus cytokines | 0.02 | 0.02 | 0.02 | 0.02 |
| Mock versus aCLEC5A + cytokines | 0.12 | 0.04 | 0.02 | 0.34 | |
| Mock versus spike | 0.02 | 0.03 | 0.02 | 0.02 | |
| Mock versus aCLEC5A + spike | 0.56 | 0.07 | 0.20 | 0.88 | |
| Cytokines versus aCLEC5A + cytokines | 0.02 | 0.62 | 0.34 | 0.02 | |
| Spike versus aCLEC5A + spike | 0.04 | 0.69 | 0.11 | 0.02 | |
Note: Cytokine levels are expressed in pg/ml.
Abbreviations: aCLEC5A, human CLEC5A monoclonal antibody; IFN‐γ,interferon‐γ; IL‐6, interleukin 6.
3.4. Molecular docking shows binding sites between CLEC5A and spike protein
Docking of the CLEC5A and spike proteins was investigated using the ClusPro 2.0 docking program, which resulted in the generation of 30 docked models, each assigned with a weightage score representing the lowest binding energy. Based on these scores, the model from Cluster‐0 was chosen as a putative binding mode between the monocyte receptor CLEC5A (PDB ID: 2YHF) and spike protein (receptor‐binding domain, RBD) (PBD ID: 6LZG). There were nine copies of CLEC5A within the asymmetric unit, representing a range of structural conformers, thus providing valuable insight into the range of possible conformations that this receptor may adopt in vivo.
The binding energy between nine copies of CLEC5A and the RBD of spike protein was similar to the binding energy for hACE2 and spike protein (Table 2, Figure 4A,B). Moreover, the interactions between the nine copies of CLEC5A and dengue (serotype 2) (PDB ID: 1OAN) were stronger than those of CLEC5A and spike protein RBD (Table 2, Figure 4C). Six different copies of CLEC5A seemed to tightly interact with spike protein RBD (Figure 4D,E), but no interactions were observed between CLEC5A and hACE2 when spike protein was bound to hACE2 (Figures 4B,D).
Table 2.
A detailed comparison of the top docking model of protein–protein docking complex by ClusPro 2.0 method
| Weighted score (lowest energy) | |||
|---|---|---|---|
| ClusPro 2.0 models | CLEC5A—dengue virus envelop | Spike S1—human ACE2 | Spike S1—CLEC5A |
| Balanced | −999.4 | −752.7 | −793.5 |
| Electrostatic favored | −1009.8 | −785.5 | −826.2 |
| Hydrophobic favored | −1100.6 | −932.8 | −1048.7 |
| Van de Waals + electrostatic | −261.2 | −236.2 | −303.2 |
| RMSDa | 1.817 | 0.952 | 1.137 |
Note: Values are described in kcal/mol.
Abbreviation: ACE2, angiotensin converting enzyme 2.
RMSD: Root‐mean‐square deviation calculated by superimposition between the ligands atoms in the model measured in Angstroms (Å) by PyMOL 2.5 software.
Figure 4.
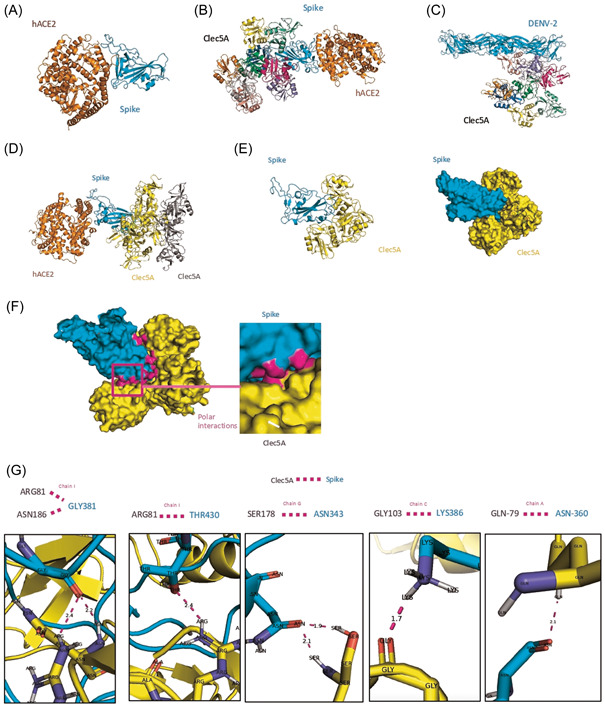
In silico interaction between CLEC5A and spike protein (SARS‐CoV‐2). Docking was performed using the software ClusPro 2.0 to show the binding energy among (A) human angiotensin‐converting enzyme 2 (hACE2) and spike protein (Spike S1 RBD) from SARS‐CoV‐2, (B) binding energy among the nine copies of CLEC5A structure and spike protein with hACE2, and (C) binding energy among the nine copies of CLEC5A and dengue virus envelope (serotype 2). (D) Definition of chains that may interact to explore the interactions between CLEC5A and spike protein, which regions in color cyan for spike and yellow for CLEC5A chains were chosen. (E) The three forms among nine copies of CLEC5A structure with a capacity of polar contacts with spike residues. (F) Binding site showing polar interactions between CLEC5A and spike protein (magenta). (G) Interaction representation of the residues from CLEC5A and residues from spike protein capable of polar contacts (magenta) with the distance measurement in Angstroms. Created using PyMOL 2.5 software (https://pymol.org). RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
CLEC5A structures are organized as homodimers at the cell surface, and the docking analysis showed that four different forms (Chains A, C, G, and I) were able to bind with spike protein through polar interactions using PyMOL 2.5 software (Figure 4F,G, Supporting Information: Video 1) with a good RMSD score (<2 Å) (Table 2). Interestingly, the N‐acetylglucosamine ligand site (NAG‐601) in spike protein RBD bound to an asparagine residue (Asn‐343), indicating interchain interactions with serine (Ser‐178) in one of CLEC5A forms (Chain G) through polar contacts (Figure 4G, Supporting Information: Video 2).
4. DISCUSSION
Monocytes have an important role in the innate immune response against COVID‐19, as these cells are directly affected by SARS‐CoV‐2 replication 19 , 31 , 32 , 33 , 34 and involved in the activation of the inflammatory cascade, that is, the phenomenon known as the cytokine storm, which is related to severity and lethality. 31 , 32 , 35 , 36 Although the function of monocytes is very well described by literature, there is a gap in the information about C‐type lectin receptors expressed by these cells during SARS‐CoV‐2 infection. It has been shown that the influenza virus activates CLEC5A to enhance inflammatory reactions and induce severe lung damage in animal models. Proinflammatory cytokine production and neutrophil extracellular trap (NET) formation can be driven by the CLEC5A receptor in myeloid cells during microbial infection. 1 , 2 , 3 , 4 Sung et al., 37 using unexposed blood samples and a murine model, demonstrated that CLEC5A and Toll‐like receptor 2 have critical roles in neutrophil activation during SARS‐CoV‐2 infection, inducing NET formation and lung inflammation. Nevertheless, the expression of CLEC5A in PBMCs from COVID‐19 patients is still unexplored.
In our study population, we detected high percentages of CLEC5A on intermediate monocytes and nonclassical monocytes in severe COVID‐19 cases. Notably, CLEC5A on nonclassical monocytes was extremely elevated, and significantly different compared to mild COVID‐19 cases and has a significant association with severity. Inflammatory monocytes are commonly related to the pathogenesis of COVID‐19, 32 and we previously demonstrated that nonclassical monocytes can be activated by SARS‐CoV‐2 using in vitro assays. 19 As expected, vaccinated people had low expression of CLEC5A on monocytes, which could be explained by mild infection with minor signs and symptoms without hospitalization or death after full vaccination (two or three doses of a COVID‐19 vaccine) (Supporting Information: Table 1), as mentioned by others. 38 , 39 In our findings with hamster samples, the animals were challenged with SARS‐CoV‐2 to monitor the kinetic of CLEC5A gene expression 15 days after infection, and the data showed that after 3 days of viral infection CLEC5A gene is expressed in blood samples. Similar results were seen by other authors, in which CLEC5A markers were detected in murine samples during pulmonary mycobacterial infection and other viral infections, but it was not detected in healthy samples. 32 , 37 , 38 , 39 , 42 , 43 In addition, some studies mentioned that inflammatory cells are detected in the lung at 2–10 days postinfection with SARS‐CoV‐2 particles, being part of the disease's pathology in hamster models. 42 , 43
Cytokines and viral proteins have interactions with several receptors on monocytes, including CLEC5A, evoking inflammatory processes during infections. 1 , 2 , 4 , 11 , 32 , 38 , 39 Considering this, our data provided by in vitro assays showed that CLEC5A expression can be activated by spike glycoprotein in samples from nonhospitalized COVID‐19 patients (mild infection) and unexposed individuals. Hu and colleagues 4 , 36 , 44 mentioned that IL‐2, IL‐6, IL‐8, IFN‐γ, and TNF‐α were cytokines related to cytokine storm during coronaviruses infection, and little can explain which cytokine is responsible for CLEC5A activation during a cytokine storm. Given that CLEC5A activation can be regulated by oxidative stress caused by inflammation, here, in the experimental assay, it was used a mix of cytokines to mimic a cytokine storm event using in vitro conditions, which was successfully able to induce CLEC5A expression on nonclassical monocytes.
In general, our in vitro data was relevant to demonstrate that spike protein can induce higher activation of nonclassical monocytes expressing CLEC5A in people who had COVID‐19 in comparison with unexposed subjects. Therefore, it seems that activation is harmless in unexposed subjects, even more, when this group was vaccinated. This effect is not surprising, considering that, after vaccination, the adaptive immune response cross‐talks with innate immunity and collaborate rapidly to reduce the negative aspects of viral infection during COVID‐19. 40 , 41 , 42
mAbs or gene silencing for the CLEC5A receptor have been used as a good therapeutic approach to reduce proinflammatory cytokine production and the lethality of flaviviruses and influenza viruses following challenges in mouse models. 1 , 2 , 3 , 4 Here, we also demonstrated that a human mAb for CLEC5A (hCLEC5A mAb) is an option for further therapeutic purposes since our findings show that hCLEC5A mAb inhibited CLEC5A expression in monocytes upon spike protein stimulation and induced a significant decrease in proinflammatory cytokines, such as IL‐2 and IL‐1β, in mild COVID‐19 samples. Chen et al. 4 showed that proinflammatory cytokines such as CXCL8 and TNF‐α were reduced after treatment with a monoclonal CLEC5A antibody in a murine model challenged with Dengue virus, but this did not affect IFN‐α production. Here, we noted that proinflammatory cytokines were also reduced by hCLEC5A mAb treatment using in vitro cell cultivation, but this did not affect IFN‐γ production in PBMC from mild COVID‐19 subjects.
Molecular docking provides important in silico information to preclinical studies based on protein–protein binding interactions. Several studies using bioinformatics to study drugs and biopharmaceutical products have been applied to explore a way to reduce the risk of COVID‐19 effects in severe acute respiratory syndrome poor outcomes. 27 , 28 There are nine copies of CLEC5A within the asymmetric unit, representing a range of structural conformers and so providing insight into the range of possibilities in the interactions that this receptor may adopt with different ligands in vivo. Dynamic simulations of how the transmembrane portions can arrange on the cellular surface were also diverse, 5 which means that our docking findings indicate a number of interactions between this host cell receptor and SARS‐CoV‐2. Watson et al. 5 mentioned that regions of high glutamine/asparagine content on CLEC5A often act as ligand binding sites, as seen by our docking results showing that asparagine was bound to glycine in the spike RBD structure. 5 Furthermore, Chen et al. 4 and Watson et al. 5 demonstrated that interactions between CLEC5A and Dengue virions could be inhibited in vitro by fucose and mannan, and Golay et al. 45 mentioned that low levels of fucose were linked to disease severity in SARS‐CoV‐2 and Dengue virus infection, mainly during antibody production. Despite the lack of information here about how fucose and mannan could interact in the CLEC5A and spike binding, the literature suggests that the presence/absence of those carbohydrate content could be relevant during SARS‐CoV‐2 infection, inhibiting or facilitating cell host entry, 4 , 5 , 45 and it should be further explored in the CLEC5A and spike interactions by experimental assays.
NAG binding on asparagine (Asn‐343) is a binding site of the spike RBD structure and is unlikely to be part of the connection with hACE2. Pokhrel et al. 46 mentioned that the only asparagine conserved in several variants and mutations found in spike RBD structure N‐glycosylation sites is Asn‐343. Rajendaran et al. 30 suggested that this ligand site is a good key to developing therapies against COVID‐19. CLEC5A structures do not have any special ligand site but can interact with the structures of several viruses, and its expression is related to lethality. 1 , 3 , 4 It is important to note that there is no information about the CLEC5A interaction with the binding site of the spike protein‐ligand, and our data suggest that SARS‐CoV‐2 may lead to stimulation as well as signal transduction through these observed interactions, increasing CLEC5A expression by inflammatory monocytes in vitro and as noted in severe cases (Figure 5).
Figure 5.
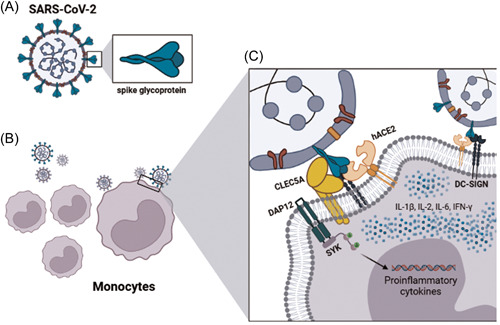
Mechanism suggestion of spike protein interaction with CLEC5A. Spike glycoprotein structure from a SARS‐CoV‐2 virion (A) and the probable contact among monocyte surface and viral particles (B) with the details of molecular interactions between viral protein (spike) and cellular receptors in the monocytes surface (CLEC5A, DC‐SIGN, and ACE2) showing the intracellular signaling activation cascade (cytokines activation and production) induced by the protein–protein binding. Created with biorender.com. hACE2, human angiotensin‐converting enzyme 2; IFN‐γ,interferon‐γ; IL‐6, interleukin 6; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
This study has limitations, such as the limited available samples, the unknown diversity of CLEC5A molecular interactions with SARS‐CoV‐2 proteins, as well as the impact of viral protein mutations occurring all the time. Despite these, our data bring important knowledge to medical science and may influence the immunotherapeutic approach to respiratory infections such as COVID‐19 and other diseases without an available vaccine. Our results also reinforce the importance of virological surveillance for new variants of concern, which could lead to immune escape. Moreover, viral proteins can use molecular machinery to promote inflammatory damage, triggering poor outcomes. The search for new therapies is a part of the constant effort of the scientific community to provide solutions to the pandemic.
AUTHOR CONTRIBUTIONS
Thiago L. Machado performed the fluorescence‐activated cell sorting experiments, molecular assays, and wrote and revised the first draft of the manuscript. Thiago L. Machado, Alanna C. Santos, Luciana N. Tubarão, and Livia M. Villar performed the human blood collection, peripheral blood mononuclear cells preparation, and patient information organization. Tamiris Azamor, Andrea M. V. da Silva, and Vanessa R. Pimenta performed serological and cytokines detection assays. Alexandre dos Santos da Silva, Daniela Del Rosário Flores Rodrigues, Rodrigo Müller, and Marcelo A. Pinto performed the in vivo study, hamster samples collection, and preparation. Ana P. A. Bom edited and revised the final version of the manuscript. Juliana G. Melgaço worked on the study conceptualization, performed the statistical analysis, and in vitro and in silico experiments, and wrote and revised the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The study protocol was approved by our Institutional Review Board (CAAE 34728920.4.0000.5262, CAAE 30468620.5.0000.5248, CAAE 37079320.4.0000.5258), in which all human participants provided signed informed consent according to the Helsinki Declaration for ethical procedures. For animal experiments, the study protocol was also approved by Fundação Oswaldo Cruz (LW‐9/20), and SARS‐CoV‐2 infection, as well as blood collection, were carried out in biosafety level III laboratory specific for animal procedures.
Supporting information
Supplementary information.
Supplementary information.
Supplementary information.
ACKNOWLEDGMENTS
The authors would like to thank the volunteers for blood donation and the BSL‐3 facility for the animal study. They would also like to thank the financial support, as Fundação Oswaldo Cruz (VPPCB‐05‐FIO‐20‐2‐52) to Livia Melo Villar, (VPPCB‐005‐FIO‐20‐2‐38) to Marcelo Alves Pinto, and (VPPCB‐005‐FIO‐20‐2‐11) to Ana Paula Dinis Ano Bom, as well as Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ—SEI‐260003/002688/2020) to Livia Melo Villar, (FAPERJ‐E‐26/210.189/2020; E‐26/210.242/2020; E‐26/202.158/2020; E‐26/202.161/2020) to Marcelo Alves Pinto, and (FAPERJ‐ARC 010.002596/2019) to Juliana Gil Melgaço.
Machado TL, Santos AC, Azamor T, et al. CLEC5A expression can be triggered by spike glycoprotein and may be a potential target for COVID‐19 therapy. J Med Virol. 2022;95:e28427. 10.1002/jmv.28427
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Teng O, Chen ST, Hsu TL, et al. CLEC5A‐mediated enhancement of the inflammatory response in myeloid cells contributes to influenza virus pathogenicity in vivo. J Virol. 2017;91(1):e01813‐e01816. 10.1128/JVI.01813-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sung PS, Hsieh SL. CLEC2 and CLEC5A: pathogenic host factors in acute viral infections. Front Immunol. 2019;10:2867. 10.3389/fimmu.2019.02867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen ST, Liu RS, Wu MF, et al. CLEC5A regulates Japanese encephalitis virus‐induced neuroinflammation and lethality. PLoS Pathog. 2012;8(4):e1002655. 10.1371/journal.ppat.1002655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen ST, Lin YL, Huang MT, et al. CLEC5A is critical for dengue‐virus‐induced lethal disease. Nature. 2008;453(7195):672‐676. 10.1038/nature07013 [DOI] [PubMed] [Google Scholar]
- 5. Watson AA, Lebedev AA, Hall BA, et al. Structural flexibility of the macrophage dengue virus receptor CLEC5A. J Biol Chem. 2011;286(27):24208‐24218. 10.1074/jbc.M111.226142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. To KK, Sridhar S, Chiu KH, et al. Lessons learned 1 year after SARS‐CoV‐2 emergence leading to COVID‐19 pandemic. Emerg Microbes Infect. 2021;10(1):507‐535. 10.1080/22221751.2021.1898291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khan M, Adil SF, Alkhathlan HZ, et al. COVID‐19: A global challenge with old history, epidemiology and progress so far. Molecules. 2020;26(1):39. 10.3390/molecules26010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vitiello A, Ferrara F, Troiano V, La Porta R. COVID‐19 vaccines and decreased transmission of SARS‐CoV‐2. Inflammopharmacology. 2021;29(5):1357‐1360. 10.1007/s10787-021-00847-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sadarangani M, Marchant A, Kollmann TR. Immunological mechanisms of vaccine‐induced protection against COVID‐19 in humans. Nat Rev Immunol. 2021;21(8):475‐484. 10.1038/s41577-021-00578-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soiza RL, Scicluna C, Thomson EC. Efficacy and safety of COVID‐19 vaccines in older people. Age Ageing. 2021;50(2):279‐283. 10.1093/ageing/afaa274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dagan N, Barda N, Biron‐Shental T, et al. Effectiveness of the BNT162b2 mRNA COVID‐19 vaccine in pregnancy. Nature Med. 2021;27(10):1693‐1695. 10.1038/s41591-021-01490-8 [DOI] [PubMed] [Google Scholar]
- 12. Lv G, Yuan J, Xiong X, Li M. Mortality rate and characteristics of deaths following COVID‐19 vaccination. Front Med. 2021;8:670370. 10.3389/fmed.2021.670370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rezagholizadeh A, Khiali S, Sarbakhsh P, Entezari‐Maleki T. Remdesivir for treatment of COVID‐19; an updated systematic review and meta‐analysis. Eur J Pharmacol. 2021;897:173926. 10.1016/j.ejphar.2021.173926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jahanshahlu L, Rezaei N. Monoclonal antibody as a potential anti‐COVID‐19. Biomed Pharmacother. 2020;129:110337. 10.1016/j.biopha.2020.110337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deb P, Molla MMA, Saif‐Ur‐Rahman KM. An update to monoclonal antibody as therapeutic option against COVID‐19. Biosaf Health. 2021;3(2):87‐91. 10.1016/j.bsheal.2021.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loganathan S, Athalye SN, Joshi SR. Itolizumab, an anti‐CD6 monoclonal antibody, as a potential treatment for COVID‐19 complications. Expert Opin Biol Ther. 2020;20(9):1025‐1031. 10.1080/14712598.2020.1798399 [DOI] [PubMed] [Google Scholar]
- 17. Triggle CR, Bansal D, Ding H, et al. A comprehensive review of viral characteristics, transmission, pathophysiology, immune response, and management of SARS‐CoV‐2 and COVID‐19 as a basis for controlling the pandemic. Front Immunol. 2021;12:631139. 10.3389/fimmu.2021.631139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID‐19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1‐9. 10.12932/AP-200220-0772 [DOI] [PubMed] [Google Scholar]
- 19. Melgaço JG, Azamor T, Silva AMV, et al. Two‐step in vitro model to evaluate the cellular immune response to SARS‐CoV‐2. Cells. 2021;10(9):2206. 10.3390/cells10092206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Melgaço JG, Brito e Cunha D, Azamor T, et al. Cellular and molecular immunology approaches for the development of immunotherapies against the new coronavirus (SARS‐CoV‐2): challenges to near‐future breakthroughs. J Immunol Res. 2020;2020:8827670. 10.1155/2020/8827670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matos AR, Motta FC, Caetano BC, et al. Identification of SARS‐CoV‐2 and additional respiratory pathogens cases under the investigation of COVID‐19 initial phase in a Brazilian reference laboratory. Mem Inst Oswaldo Cruz. 2020;115:e200232. 10.1590/0074-02760200232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Azamor T, da Silva AMV, Melgaço JG, et al. Activation of an effective immune response after yellow fever vaccination is associated with the genetic background and early response of IFN‐γ and CLEC5A. Viruses. 2021;13(1):96. 10.3390/v13010096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dejnirattisai W, Huo J, Zhou D, et al. SARS‐CoV‐2 Omicron‐B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467‐484.e15. 10.1016/j.cell.2021.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arantes I, Gomes Naveca F, Gräf T, et al. Emergence and spread of the SARS‐CoV‐2 variant of concern Delta across different Brazilian regions. Microbiol Spectrum. 2022;10:e0264121. 10.1128/spectrum.02641-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Maria R, Cifone MG, Trotta R, et al. Triggering of human monocyte activation through CD69, a member of the natural killer cell gene complex family of signal transducing receptors. J Exp Med. 1994;180(5):1999‐2004. 10.1084/jem.180.5.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kusdra L, Rempel H, Yaffe K, Pulliam L. Elevation of CD69+ monocyte/macrophages in patients with alzheimer's disease. Immunobiology. 2000;202(1):26‐33. 10.1016/S0171-2985(00)80049-2 [DOI] [PubMed] [Google Scholar]
- 27. Mohammadi S, Heidarizadeh M, Entesari M, et al. In silico investigation on the inhibiting role of Nicotine/Caffeine by blocking the S protein of SARS‐CoV‐2 versus ACE2 receptor. Microorganisms. 2020;8(10):1600. 10.3390/microorganisms8101600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marhaeny HD, Widyawaruyanti A, Widiandani T, Fuad Hafid A, Wahyuni TS. Phyllanthin and hypophyllanthin, the isolated compounds of Phyllanthus niruri inhibit protein receptor of corona virus (COVID‐19) through in silico approach. J Basic Clin Physiol Pharmacol. 2021;32(4):809‐815. 10.1515/jbcpp-2020-0473 [DOI] [PubMed] [Google Scholar]
- 29. Kozakov D, Hall DR, Xia B, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12(2):255‐278. 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rajendaran S, Jothi A, Anbazhagan V. Targeting the glycan of receptor binding domain with jacalin as a novel approach to develop a treatment against COVID‐19. R Soc Open Sci. 2020;7(9):200844. 10.1098/rsos.200844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fintelman‐Rodrigues N, Sacramento CQ, Lima CR, et al. Atazanavir, alone or in combination with ritonavir, inhibits SARS‐CoV‐2 replication and proinflammatory cytokine production. Antimicrob Agents Chemother. 2020;64:e00825. 10.1101/2020.04.04.020925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harrison AG, Lin T, Wang P. Mechanisms of SARS‐CoV‐2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100‐1115. 10.1016/j.it.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferreira AC, Soares VC, de Azevedo‐Quintanilha IG, et al. SARS‐CoV‐2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021;7(1):43. 10.1038/s41420-021-00428-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu Q, Liu J, Zhao S, et al. SARS‐CoV‐2 exacerbates proinflammatory responses in myeloid cells through C‐type lectin receptors and Tweety family member 2. Immunity. 2021;54(6):1304‐1319.e9. 10.1016/j.immuni.2021.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J, Jiang M, Chen X, Montaner LJ. Cytokine storm and leukocyte changes in mild versus severe SARS‐CoV‐2 infection: review of 3939 COVID‐19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. 2020;108(1):17‐41. 10.1002/JLB.3COVR0520-272R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu B, Huang S, Yin L. The cytokine storm and COVID‐19. J Med Virol. 2021;93(1):250‐256. 10.1002/jmv.26232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sung PS, Yang SP, Peng YC, Sun CP, Tao MH, Hsieh SL. CLEC5A and TLR2 are critical in SARS‐CoV‐2‐induced NET formation and lung inflammation. J Biomed Sci. 2022;29(1):52. 10.1186/s12929-022-00832-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Williams SV, Vusirikala A, Ladhani SN, et al. An outbreak caused by the SARS‐CoV‐2 Delta (B.1.617.2) variant in a care home after partial vaccination with a single dose of the COVID‐19 vaccine Vaxzevria, London, England, April 2021. Euro Surveill. 2021;26(27):2100626. 10.2807/1560-7917.ES.2021.26.27.2100626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brunner‐Ziegler S, Spath T, Kornek G, et al. Postvaccination infections among staff of a tertiary care hospital after vaccination with severe acute respiratory syndrome coronavirus 2 vector and mRNA‐based vaccines. Clin Microbiol Infect. 2022;28(4):596‐601. 10.1016/j.cmi.2021.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aoki N, Kimura Y, Kimura S, et al. Expression and functional role of MDL‐1 (CLEC5A) in mouse myeloid lineage cells. J Leukoc Biol. 2009;85(3):508‐517. 10.1189/jlb.0508329 [DOI] [PubMed] [Google Scholar]
- 41. Cheng YL, Lin YS, Chen CL, et al. Activation of Nrf2 by the dengue virus causes an increase in CLEC5A, which enhances TNF‐α production by mononuclear phagocytes. Sci Rep. 2016;6:32000. 10.1038/srep32000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Imai M, Iwatsuki‐Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS‐CoV‐2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2022;117:16587‐16595. 10.1073/pnas.2009799117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sia SF, Yan LM, Chin AWH, et al. Pathogenesis and transmission of SARS‐CoV‐2 in golden hamsters. Nature. 2020;583(7818):834‐838. 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Joyce‐Shaikh B, Bigler ME, Chao CC, et al. Myeloid DAP12‐associating lectin (MDL)‐1 regulates synovial inflammation and bone erosion associated with autoimmune arthritis. J Exp Med. 2010;207(3):579‐589. 10.1084/jem.20090516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Golay J, Andrea AE, Cattaneo I. Role of Fc core fucosylation in the effector function of IgG1 antibodies. Front Immunol. 2022;13:929895. 10.3389/fimmu.2022.929895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pokhrel S, Kraemer BR, Burkholz S, Mochly‐Rosen D. Natural variants in SARS‐CoV‐2 Spike protein pinpoint structural and functional hotspots with implications for prophylaxis and therapeutic strategies. Sci Rep. 2021;11(1):13120. 10.1038/s41598-021-92641-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Supplementary information.
Supplementary information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


