Abstract
Emergence of various circulating SARS‐CoV‐2 variants of concern (VOCs) promotes the identification of pan‐sarbecovirus vaccines and broadly neutralizing antibodies (bNAbs). Here, to characterize monoclonal antibodies cross‐reactive against both SARS‐CoV‐1 and SARS‐CoV‐2 and to search the criterion for bNAbs against all emerging SARS‐CoV‐2, we isolated several SARS‐CoV‐1‐cross‐reactive monoclonal antibodies (mAbs) from a wildtype SARS‐CoV‐2 convalescent donor. These antibodies showed broad binding capacity and cross‐neutralizing potency against various SARS‐CoV‐2 VOCs, including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta), but failed to efficiently neutralize Omicron variant and its sublineages. Structural analysis revealed how Omicron sublineages, but not other VOCs, efficiently evade an antibody family cross‐reactive against SARS‐CoV‐1 through their escape mutations. Further evaluation of a series of SARS‐CoV‐1/2‐cross‐reactive bNAbs showed a negative correlation between the neutralizing activities against SARS‐CoV‐1 and SARS‐CoV‐2 Omicron variant. Together, these results suggest the necessity of using cross‐neutralization against SARS‐CoV‐1 and SARS‐CoV‐2 Omicron as criteria for rational design and development of potent pan‐sarbecovirus vaccines and bNAbs.
Keywords: broadly neutralizing antibody (bNAb), cross‐reactivity, Omicron, SARS‐CoV‐1, SARS‐CoV‐2
1. INTRODUCTION
The pandemic of coronavirus disease of 2019 (COVID‐19) has already lasted for 3 years. However, the SARS‐CoV‐2 infections are still escalating globally, largely due to the emergence and circulation of several SARS‐CoV‐2 variants of concern (VOCs). The B.1.1.7 lineage (labeled as “Alpha” by World Health Organization, WHO) was first documented in the United Kingdom; the B.1.351 lineage (labeled as “Beta”) was first identified in South Africa; the P.1 lineage (labeled as “Gamma”) was originated from Brazil; the B.1.617.2 lineage (labeled as “Delta”) was earliest documented in India; while the B.1.1.529 lineage (labeled as “Omicron”) was reported from South Africa. 1 Most recently, several Omicron sublineages, including BA.1, BA.2, BA.2.12.1, BA.3, BA.4, and BA.5, have taken over and spread across the globe. 2 , 3 The Omicron variant and its sublineages are associated with increased transmissibility, elevated infectivity, reduced pathogenicity, and decreased effectiveness of vaccines and antibody therapeutics. 2 , 4 , 5 , 6 For instance, D614G substitution in viral spike (S) protein increased the viral infectivity and stability of virions 7 ; N501Y enhanced the binding affinity between the viral S protein and the human receptor ACE2 8 ; E484K/E484A, K417N and several other mutations largely escaped the neutralizing activity of the monoclonal antibodies (mAbs) isolated from convalescent and vaccinated individuals. 6 , 9 These emerging VOCs and variants of interest raised concerns about the effectiveness of current COVID‐19 vaccines and antibody therapeutics. 10
Recently, new SARS‐CoV‐2 variants keep emerging. Among the new variants are XBB and BQ.1.1, which are all rising in numbers and have surpassed some other recent variants. 11 Newly emerging SARS‐CoV‐2 variants promote researchers to identify broadly neutralizing antibodies (bNAbs) that could be of potential clinical benefit. One strategy for identifying bNAbs is to test antibodies one by one against all reported variants. Although this strategy is straightforward, it is time‐consuming and labor‐intensive. Here, we hypothesize another strategy for identifying bNAbs against SARS‐CoV‐2 VOCs by evaluating antibody cross‐reactivity against SARS‐CoV‐1 as an indicator of broad reactivity.
Similar to SARS‐CoV‐2, SARS‐CoV‐1 also utilizes its S protein and host ACE2 receptor for cell entry. The sequence alignment showed high similarities of RNA genome (~79% identity), S protein (~76% identity) and receptor‐binding domain (RBD) (~73% identity) between SARS‐CoV‐1 (Urbani strain) and SARS‐CoV‐2 (Wuhan Hu‐1 strain). Moreover, the vaccination strategy using a combination of SARS‐CoV‐1 and SARS‐CoV‐2 wildtype S proteins elicited cross‐reactive immune responses against SARS‐CoV‐2 VOCs. 12 , 13 , 14 Therefore, we speculate that the antibodies against both SARS‐CoV‐1 and wildtype SARS‐CoV‐2 are more likely to be cross‐reactive against the circulating SARS‐CoV‐2 VOCs.
To investigate whether the cross‐reactivity against both SARS‐CoV‐1 and wildtype SARS‐CoV‐2 could be used as criteria for identifying bNAbs against SARS‐CoV‐2 VOCs, we utilized SARS‐CoV‐1 S protein as the bait protein to perform single B cell sorting from a wildtype SARS‐CoV‐2 convalescent individual, and isolated several antibodies cross‐reactive against both SARS‐CoV‐1 and wildtype SARS‐CoV‐2. These antibodies, recognizing distinct nonoverlapping epitopes, potently bound and even efficiently neutralized SARS‐CoV‐2 VOCs, including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta). However, these antibodies dramatically lost their neutralization against B.1.1.529 (Omicron) and its sublineages, including BA.1, BA.2, BA.2.12.1, BA.3, BA.4, and BA.5.
These findings are consistent with that from a previous serological study, in which SARS‐CoV‐1 convalescent individuals who have been immunized with SARS‐CoV‐2 vaccine could develop potent cross‐clade pan‐sarbecovirus neutralizing antibodies, 15 while their serum neutralization was apparently evaded by Omicron and its sublineages. 16 Based on its serological profiles, Omicron was even proposed to be called as SARS‐CoV‐3. 16
Finally, we examined a series of anti‐SARS‐CoV‐2 bNAbs and showed that their neutralization capacity against SARS‐CoV‐1 negatively correlated with their neutralization capacity against SARS‐CoV‐2 Omicron variant. Taken together, these results confirm the unique serological characteristics of Omicron and its sublineages and suggest that pan‐sarbecovirus therapeutic bNAbs must have cross‐neutralizing activity against: (1) SARS‐CoV‐1, (2) SARS‐CoV‐2 and its VOCs of Alpha, Beta, Gamma, and Delta, and (3) SARS‐CoV‐2 Omicron and its sublineages, and that a pan‐sarbecovirus vaccine should contain the epitopes that could elicit the above bNAbs.
2. MATERIALS AND METHODS
2.1. Single cell sorting of memory B cells binding SARS‐CoV‐1 S1 protein
We performed the single cell sorting from isolated human peripheral blood mononuclear cells (PBMCs) as previously reported, 17 with minor modifications such as using Avi‐tagged SARS‐CoV‐1 S1 recombinant protein 18 as the bait protein during the single cell sorting. Commercially purchased Avi‐tagged SARS‐CoV‐1 S1 protein (Kactus Biosystems) was biotinylated using BirA Biotin‐protein Ligase kit (Avidity) as manufacturer's instructions. Every time before single cell sorting, biotinylated S1 protein was freshly prepared and incubated with streptavidin‐PE (eBioscience) or streptavidin‐APC (BD Biosciences) overnight to generate bait protein‐PE and bait protein‐APC. PBMCs from the selected donor were quickly thawed and incubated with CD19 MicroBeads (Miltenyi Biotech) for positive selection of B lymphocytes. Sequential incubations with human Fc block (BD Biosciences), anti‐CD20‐PECy7 (BD Biosciences), and bait protein‐PE/APC (freshly prepared) were performed to enrich the human memory B cells recognizing SARS‐CoV‐1 S1 protein. PECy7+ PE+ APC+ single memory B cells were sorted into each well of 96‐well plates using a FACSAriaII (Becton Dickinson), and were stored at −80°C for single cell‐based antibody cloning.
2.2. Single cell‐based antibody cloning and sequencing
Single cell‐based antibody sequencing and cloning were performed as previously reported. 19 Briefly, reverse transcription of the sorted single B cells was performed using random primers. Afterwards, the 1st/2nd round of nested PCR reactions were performed. The amplified bands were subjected for Sanger sequencing, and the sequencing results were analyzed by IMGT/V‐QUEST 20 and/or IgBlast 21 to determine the sequences of both heavy and kappa/lambda light chains. Moreover, the V(D)J gene segment and CDR3 sequences of each antibody were identified.
2.3. Antibody cloning and production
The nucleotide sequences of both heavy and light chains of the selected mAbs were inserted into expression vectors for in vitro production. The nucleotide fragments of unmutated common ancestor (UCA) mAbs, including XG051, XG052, XG069, XG070, and XG014, were synthesized (Genscript) and also cloned into the expression vectors. Five UCA antibodies were constructed for each of the five mAb family members, XG051, XG052, XG069, XG070, and XG014, with the corresponding germline IGHV/IGLV genes used and the original CDR3 sequences unchanged (Table S2). Site‐directed mutagenesis of XG014 (Q100H, G102R, G102L, N104D, Y105F, Y109H, and T97A in XG014 heavy chain; S26R, N31Y, and S97A in XG014 lambda light chain) were constructed and performed by using KOD‐PLUS‐Mutagenesis kit (Toyobo) according to the manuscript's instructions. For antibody expression, HEK293F cells were transiently cotransfected with equal amount of plasmid encoding human heavy chain or light chain by using EZ Trans transfection reagents (VIGOROUS). Seven days after transfection, supernatants were harvested, filtered and purified with Protein G‐coupled Sepharose beads.
2.4. Enzyme‐linked immunosorbent assay (ELISA)
The in vitro expressed and purified human recombinant immunoglobulin G1 (IgG1) mAbs were used in ELISA assays, and their binding activity against SARS‐CoV‐2 proteins (S‐ECD, S1, RBD, and N‐terminal domain [NTD]), SARS‐CoV‐1 proteins (S‐ECD and S1), and SARS‐CoV‐2 variant proteins (S‐ECDs of B.1.1.7, B.1.351, B.1.351*, P.1, and B.1.617.2) were measured. A total of 96‐well plates were coated with 50 µl per well of antigen (10 µg/ml) and then blocked with 2% bovine serum albumin (BSA) in phosphate buffered saline (PBS). The primary antibody (10 µg/ml) was threefold serially diluted for eight dilutions and incubated (50 µl per well) for 1 h at room temperature. After incubation, the horseradish peroxidase (HRP)‐conjugated goat anti‐human IgG secondary antibody (Thermo Fisher Scientific) was added for detection. To evaluate the antigen‐binding capacity of mAbs, we calculated the area under the curve (AUC) by using PRISM analysis.
2.5. Competition ELISA
Competition ELISAs were performed as described previously. 22 Briefly, 96‐well plates were coated with SARS‐CoV‐2 S‐ECD protein (2 µg/ml in PBS) overnight at 4°C, and then blocked (2% BSA in PBS) for 2 h at room temperature. Incubation with the 1st unbiotinylated antibodies (15 µg/ml) for 2 h at room temperature was followed by directly adding 2nd biotinylated antibodies (final concentration, 0.25 µg/ml for all mAbs, except 8 µg/ml for CR3022) and incubating for 30 min at room temperature. The much higher concentration used for CR3022 is due to its much weaker ELISA signal generated. Detection was performed with Streptavidin‐HRP (BD Biosciences), and the generated signals were normalized by using the reference wells without the 1st antibody blocking. 22
2.6. In vitro neutralization assay using pseudoviruses
We used Huh‐7 cell lines and performed in vitro neutralization assays against pseudoviruses of SARS‐CoV‐1, SARS‐CoV‐2 and its variant strains as previously reported. 23 Briefly, Huh‐7 cells were seeded in 96‐well plates (1 × 104/well) and incubated with the generated pseudoviruses for 30 min. In vitro expressed and purified mAbs were threefold serially diluted in Dulbecco's modified eagle medium (DMEM) medium for nine dilutions in total (maximum concentration: 10 µg/ml), and were also added into Huh‐7 cells for incubation. After 60 h incubation in DMEM medium containing 10% fetal bovine serum, the cultured supernatants were removed and the cells were lysed for luciferase signal detection by using a Firefly Luciferase Assay Kit (Promega). The reference wells without the addition of any mAb were used for normalization.
2.7. Structural remodeling and analysis
The structure model of the XG014 with mutations Q100H, G102R, N104D, Y105F, Y109H, S26R, N31Y, and S97A were generated by swiss‐model based on the atomic model of SARS‐CoV‐2 S‐XG014 (PDB ID 7V2A). 24 The model was superimposed onto the SARS‐CoV‐2 S‐XG014 structure to analyze the interaction between the mutants and SARS‐CoV‐2 S using UCSF Chimera. 25
2.8. Statistical analysis
The detailed information of statistical analysis could be found in the Results and Figure Legends. The ELISA AUC values and the neutralization titer shown as IC50 values were calculated in PRISM software. Statistical significance was calculated by Wilcoxon rank sum test, while the correlation was evaluated by Spearman's rank correlation method.
3. RESULTS
3.1. Single‐cell sorting using SARS‐CoV‐1 S1 protein as the bait protein
We have previously identified a SARS‐CoV‐2 convalescent individual, whose infection was confirmed by polymerase chain reaction (PCR) and donor's serum had an exceptional IgG neutralizing activity against SARS‐CoV‐2. 17 To identify antibodies cross‐reactive with SARS‐CoV‐1 from this donor, we performed single‐cell sorting from the donor's PBMCs by using Avi‐tagged biotinylated SARS‐CoV‐1 S1 recombinant protein as the bait protein and adopting a dual‐dye labeling strategy 26 (Figure S1). Compared with the uninfected unvaccinated naïve control donor, the convalescent donor (#16) showed a higher percentage of bait protein‐stained B cells (S1‐PE+ S1‐APC+), about 5 times higher than that of naïve control staining (Figure 1A and Figure S1). These gated cells were then single‐cell sorted and processed by nested PCR for amplifying the variable regions of immunoglobulin heavy (IGH) and light (IGL or IGK) chain genes as previously reported. 19 Finally, 20 antibodies with paired heavy and light chains were cloned (Figure S2A) and subjected for antibody expression and further characterization. Since 48 antibodies, XG001 to XG048, have previously been isolated from the same donor, 17 these 20 antibodies were designated as XG051 to XG070.
Figure 1.
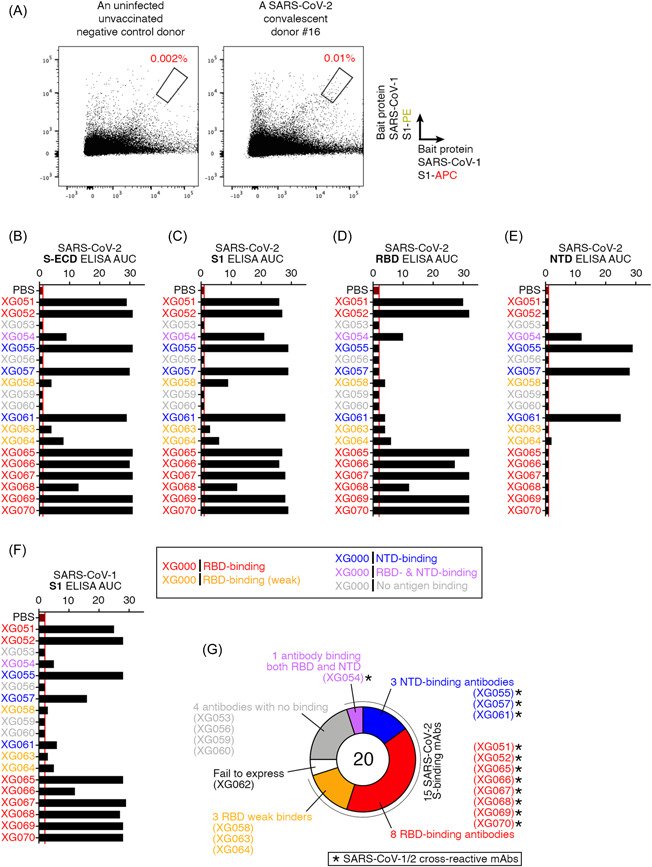
Cloning of cross‐reactive antibodies recognizing both SARS‐CoV‐1 and SARS‐CoV‐2. (A) Flow cytometry plots for single memory B cell sorting by using SARS‐CoV‐1 S1 protein as the bait protein. Red percentage numbers indicate the frequency of B cells recognizing SARS‐CoV‐1 S1 protein (dual S1‐APC+ S1‐PE+ B cells) for an uninfected unvaccinated negative control donor (left panel) and a selected SARS‐CoV‐2 convalescent donor #16, who had a high level of serum neutralizing activity 17 (right panel). (B–F) Area under the curve (AUC) values for ELISA assays using S‐ECD (B), S1 (C), RBD (D), and NTD (E) of SARS‐CoV‐2, and SARS‐CoV‐1 S1 protein (F). Based on the ELISA results, antibody names are color‐coded: red, RBD‐binding antibodies; orange, weak RBD‐binding antibodies; blue, NTD‐binding antibodies; purple, antibodies binding both RBD and NTD; and gray, no binding on the tested antigens. Phosphate‐buffered saline (PBS) was used as a negative control in ELISA assays. Data are representative of at least two independent experiments. (G) Summary of the 20 cloned mAbs. Based on ELISA results, 20 mAbs were categorized into six types. Twelve mAbs with cross‐binding activity against both SARS‐CoV‐1 and ‐2 are labeled with asterisk. ELISA, enzyme‐linked immunosorbent assay; mAb, monoclonal antibodie; NTD, N‐terminal domain.
To analyze these antibody sequences, we determined their V(D)J gene segments and CDR3 sequences (Figure S2A). Sequence analysis and comparison revealed that four mAbs (XG051, XG052, XG069, and XG070), together with XG014, 17 , 22 were encoded by the same Ig variable and joining gene segments with closely related CDR3 sequences, suggesting that these five antibody family members were derived from an expanded B cell clone (Figure S2B and S2C).
3.2. Epitope characterization for cross‐reactive antibodies
XG062 failed to express in vitro. The remaining 19 antibodies were tested by ELISA assays against various SARS‐CoV‐1/2 antigens (Figure 1B–F). Fifteen antibodies showed ELISA binding against the SARS‐CoV‐2 S‐ECD protein and also the SARS‐CoV‐2 S1 protein (Figure 1B,C). Eight of them were RBD‐binding antibodies (red names); three were NTD‐binding antibodies (blue names); three weak binders (orange names); one RBD/NTD‐binding antibody (purple names) and four nonbinders (gray names) (Figure 1D,E). ELISA against SARS‐CoV‐1 S1 proteins showed that 12 antibodies had different levels of cross‐reactivity against SARS‐CoV‐1 (Figure 1F). Together, among the 15 SARS‐CoV‐2 S protein‐binding antibodies, 12 antibodies showed cross‐binding affinity towards SARS‐CoV‐1 S1 protein (Figure 1G).
To determine whether these antibodies bind to nonoverlapping epitopes, we performed competition ELISA assays by coating the plates with SARS‐CoV‐2 S‐ECD proteins. Antibodies with relative weak binding capacities against SARS‐CoV‐2 S protein (XG054, XG058, XG063, XG064, XG068) were excluded. As expected, all the nonbiotinylated 1st antibodies efficiently blocked the binding of the 2nd biotinylated antibody of their own (yellow rectangles, Figure 2A). Four mutually exclusive epitopes (red rectangles, Figure 2A) were identified, with two nonoverlapping epitopes on RBD and two nonoverlapping epitopes on NTD region. Previously, we have identified four nonoverlapping epitope groups on RBD, Group‐I, ‐II, ‐III, and ‐IV. 17 Here, using competition ELISA, we found that the RBD‐binding mAbs, XG051, XG052, XG069, and XG070, blocked the binding of an RBD Group‐IV antibody, XG014 (Figure 2B). Consequently, these five antibody family members (Figure S2B and S2C) shared a similar RBD epitope. The other three RBD‐binding mAbs, XG065‐XG067, had an RBD epitope overlapping with those of RBD Group‐I (XG011) and/or RBD Group‐II mAbs (XG041), but not with RBD Group‐III (XG038) and RBD Group‐IV mAbs (XG014) (Figure 2B). Furthermore, XG065‐XG067 also efficiently blocked the binding of CR3022, a well‐characterized mAb isolated from an antibody‐phage library derived from a convalescent SAR‐CoV‐1 patient 27 and cross‐neutralizing both SARS‐CoV‐1 and ‐2 28 , 29 , 30 (Figure 2C). For the NTD‐binding antibodies, XG055, XG057, and XG061, none of them shared the epitope with FC05, an NTD‐directed neutralizing antibody 31 (Figure 2D).
Figure 2.
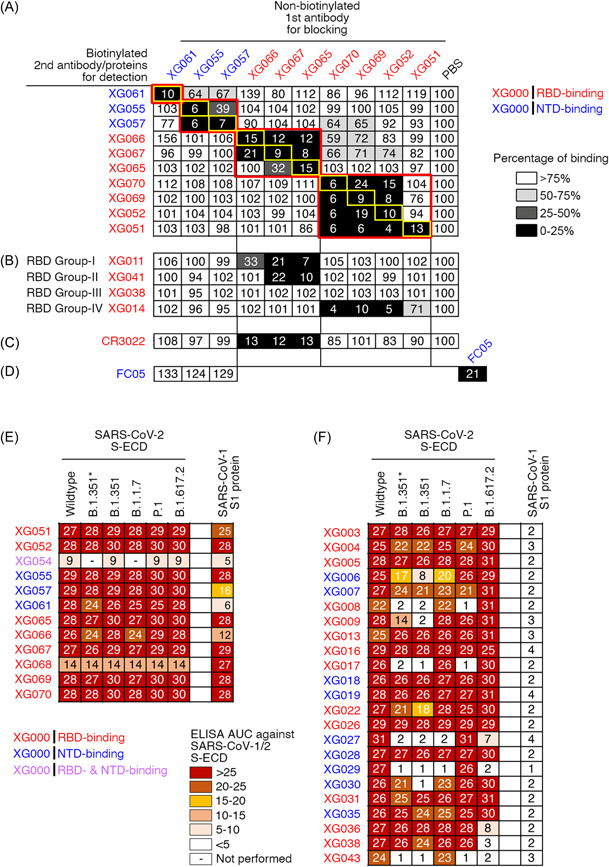
Epitope characterization and cross‐binding activity. (A–D) Competition ELISA assays for the cloned cross‐reactive mAbs. The nonbiotinylated 1st mAbs were added respectively in each well to block the coated SARS‐CoV‐2 S‐ECD recombinant proteins, while the 2nd biotinylated mAbs were used for detection. Results of competition ELISA results were normalized to the signals in the reference wells, in which no 1st mAbs were added. The binding percentages by the 2nd biotinylated antibodies were illustrated in the following colors: black, 0%–25%; dark gray, 26%–50%; light gray, 51%–75%; white, >76%. Representative of two experiments. (A) All tested mAbs efficiently blocked the binding of their own biotinylated versions (yellow rectangles), and four nonoverlapping epitope groups (red rectangles), with two on NTD and two on RBD, were identified. (B) Four RBD mAbs, XG011, XG041, XG038, and XG014 recognized four nonoverlapping RBD epitopes. (C) CR3022 mAb bound an RBD epitope shared by both SARS‐CoV‐1 and ‐2, 31 thus serving as an antibody control. (D) FC05 mAb bound an NTD epitope with potent neutralizing activity. 30 (E–F) ELISA results against various recombinant S‐ECD proteins for mAbs cross‐reactive against SARS‐CoV‐1 (E) and mAbs with no SARS‐CoV‐1 cross‐reactivity (F). The area under the curve (AUC) values were calculated by PRISM software. Experiments were performed at least twice. ELISA, enzyme‐linked immunosorbent assay; mAb, monoclonal antibodie; NTD, N‐terminal domain.
Interestingly, XG065 could compete for XG066, but not vice versa (Figure 2A), suggesting a stronger antibody binding affinity for XG065. Similarly, XG070, XG069, and XG052 could compete for XG051, while XG051 failed to compete for XG070, XG069, and XG052 (Figure 2A), suggesting a weaker binding affinity for XG051. These results suggested that, even for the antibodies in the same binding group, the antibody with stronger binding affinity could compete for the antibodies with weaker binding affinity.
Together, these results suggested that these isolated cross‐reactive antibodies recognized multiple nonoverlapping epitopes on both RBD and NTD of S protein.
3.3. Cross‐reactivity against VOC
To test our hypothesis that the antibodies cross‐reactive with both SARS‐CoV‐1 and ‐2 S proteins could broadly recognize various SARS‐CoV‐2 VOCs, we performed a series of ELISA assays using the identified 12 cross‐reactive mAbs. All of them maintained their binding capacities against SARS‐CoV‐2 RBD or NTD recombinant proteins of B.1.1.7 (Alpha), B.1.351 (Beta), B.1.351* (Beta‐L242H), and P.1 (Gamma) (Figure S3A, and Table S1). ELISA assays using S‐ECD recombinant proteins of several circulating SARS‐CoV‐2 variants, including B.1.1.7 (Alpha), B.1.351 (Beta), B.1.351*, P.1 (Gamma), and B.1.617.2 (Delta), showed that all 12 SARS‐CoV‐1‐cross‐reactive mAbs efficiently bound to the tested VOC S‐ECD proteins, with no reduction on binding capacity (Figure 2E; Figure S3B and S3C). On the contrary, for XG antibodies with no cross‐reactivity against SARS‐CoV‐1, half of them showed reduced binding activity against S‐ECD proteins of various VOCs (Figure 2F; Figure S3D and S3E). Therefore, we concluded that the isolated SARS‐CoV‐1/2‐cross‐reactive antibodies in this study exhibited broad binding capacity against SARS‐CoV‐2 VOCs, including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta).
3.4. In vitro neutralizing activity
To determine whether these mAbs described above neutralize SARS‐CoV‐1 and SARS‐CoV‐2 pseudoviruses in vitro, we performed neutralization assays in Huh‐7 cells as previously described. 17 Seven of these antibodies showed neutralizing activity against SARS‐CoV‐2 pseudovirus, with IC50 values ranging from 0.005 µg/ml to approximately 10 µg/ml (Figure 3A,B). Six antibodies neutralized SARS‐CoV‐1 pseudovirus with IC50 values ranging from 0.021 µg/ml to 8 µg/ml (Figure 3C,D).
Figure 3.
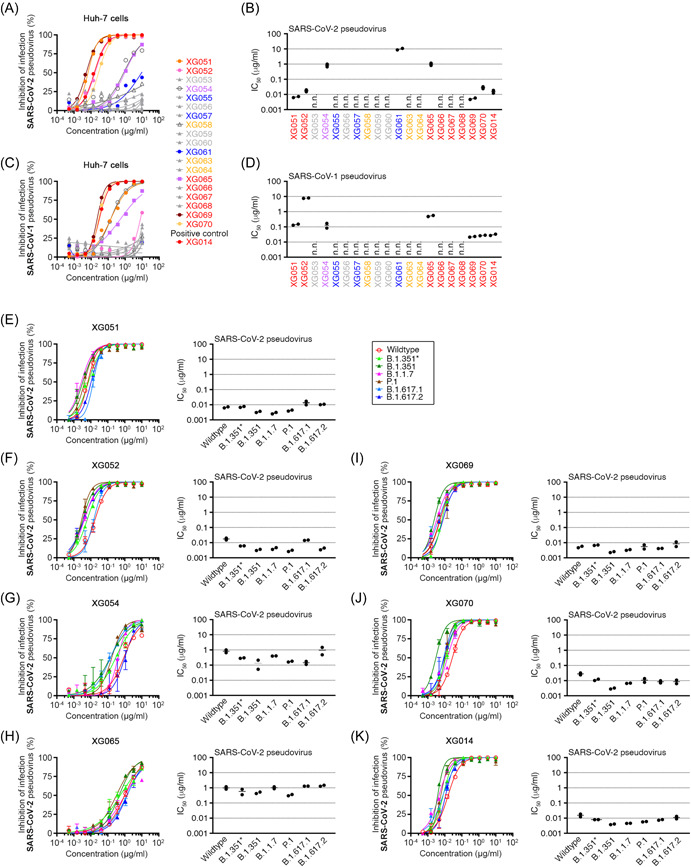
Cross‐neutralizing activity against SARS‐CoV‐1/2 and major VOCs. (A–D) In vitro neutralization assays against SARS‐CoV‐2 (A and B) and SARS‐CoV‐1 (C and D) pseudoviruses in Huh‐7 cells. (A and C) Percent inhibition of infection was determined in the presence of various concentrations of mAbs and was normalized to the control with only pseudovirus. (B and D) IC50 values against wildtype pseudovirus. n.n., not neutralizing in our assays. (E–K) In vitro neutralization assays (left) using cross‐reactive mAb XG051 (E), XG052 (F), XG054 (G), XG065 (H), XG069 (I), XG070 (J), and XG014 (K), and the calculated IC50 values (right). Pseudoviruses included wildtype SARS‐CoV‐2 and its related variants, B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), B.1.617.1 (Kappa), and B.1.351*. Percent inhibition of infection was normalized to no‐antibody‐control, and the data are shown as mean ± SEM. All neutralization experiments were performed at least two times.
To investigate whether these antibodies could neutralize various SARS‐CoV‐2 variants, we generated six pseudoviruses with their S protein containing various mutations, including B.1.1.7 (Alpha), B.1.351* (Beta‐L242H), B.1.351 (Beta), P.1 (Gamma), B.1.617.1 (Kappa), and B.1.617.2 (Delta). Neutralization assays using pseudoviruses of SARS‐CoV‐2 variants revealed that the IC50 values ranged from 0.002 to 0.017 µg/ml for XG051 (Figure 3E), 0.003–0.020 µg/ml for XG052 (Figure 3F), 0.053‐1.457 µg/ml for XG054 (Figure 3G), 0.297–1.478 µg/ml for XG065 (Figure 3H), 0.002–0.011 µg/ml for XG069 (Figure 3I), 0.003–0.032 µg/ml for XG070 (Figure 3J), and 0.004–0.018 µg/ml for XG014 (Figure 3K). All these antibodies exhibited broad neutralizing activity against the tested six variants, with no obvious reduction on their neutralizing capacity. These neutralization results were consistent with the broad antigen‐binding capacity shown by the ELISA assays (Figure 2E). Therefore, we concluded that the mAbs isolated from the SARS‐CoV‐2 convalescent donor by using SARS‐CoV‐1 bait protein, not only cross‐neutralized both SARS‐CoV‐1 and SARS‐CoV‐2 pseudoviruses, but also exhibited potent and broad neutralizing activity against the circulating SARS‐CoV‐2 VOCs, from Alpha to Delta.
3.5. Antibody affinity maturation is essential for neutralizing potency and breadth
Four mAbs (XG051, XG052, XG069, and XG070) derived from the same expanded B cell clone as XG014 (Figure S2), an ultra‐potent cross‐neutralizing antibody we identified from the same donor 23 (Figure 4A). Interestingly, these five antibodies derived from the same B cell ancestor exhibited dramatically different levels of neutralization (Figure 3A–D). Compared with XG014, XG070 showed comparable neutralizing activity; XG069 was a more potent neutralizer against both SARS‐CoV‐1 and SARS‐CoV‐2 pseudoviruses; while XG051 and XG052 showed much less potency against SARS‐CoV‐1 (Figure 4A).
Figure 4.
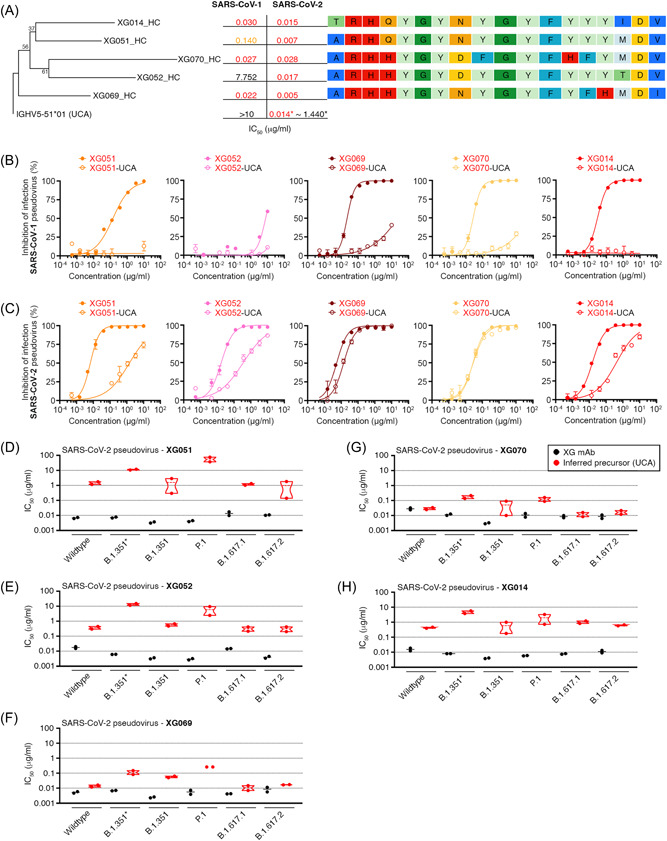
Antibody affinity maturation for neutralization potency and breadth. (A) A phylogenetic tree of IGHV nucleotide sequences of five antibody family members (XG051, XG052, XG069, XG070, and XG014) isolated from an expanded B cell clone. The tree was rooted on the nucleotide sequence of IGHV5‐51*01, which is the unmutated common ancestor (UCA) of these five antibody family members. For each antibody family member, the IC50 values for neutralizing SARS‐CoV‐1 and SARS‐CoV‐2 were labeled, and the corresponding CDR3 amino acid sequences were also aligned. (B–C) In vitro neutralization assay of five antibody family members and their corresponding unmutated common ancestor (UCA) antibodies against SARS‐CoV‐1 (B) and SARS‐CoV‐2 (C). Five UCA antibodies were constructed for each of the five mAb family members, XG051, XG052, XG069, XG070, and XG014, with the corresponding germline IGHV/IGLV genes used and their original CDR3 sequences unchanged (Table S2). Percent inhibition of infection was normalized to the luciferase signals in the control samples, which had no antibody added. The data are shown as mean ± SEM. (D–H) IC50 values, determined by in vitro neutralization assays, of five antibody family members and their corresponding unmutated common ancestor (UCA) antibodies against various SARS‐COV‐2 variants, including B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), B.1.617.1 (Kappa), and B.1.351*.
To study whether antibody affinity maturation was essential for their neutralizing activity, we performed in vitro pseudovirus neutralization assays using the corresponding inferred UCA antibodies (Figure S2B and S2C). Five UCA antibodies were constructed for each of the five mAb family members, XG051, XG052, XG069, XG070, and XG014, with the corresponding germline IGHV/IGLV genes used and their original CDR3 sequences unchanged (Table S2). As illustrated, the inferred UCA antibodies showed reduced neutralizing activity against both SARS‐CoV‐1 and SARS‐CoV‐2 (Figure 4B,C). It is worth mentioning that, for XG070, its UCA version exhibited similar level of neutralization potency against SARS‐CoV‐2 but dramatic reduction in the SARS‐CoV‐1 neutralization (Figure 4B,C). Moreover, the neutralizing activities against various SARS‐CoV‐2 variants by these UCA antibodies also drastically declined (Figure 4D–H; Figure S4). Together, these results suggested that somatic mutations are essential for the potent and broad neutralizing activity of this antibody family against SARS‐CoV‐1 and several SARS‐CoV‐2 variants.
3.6. Amino acid residues essential for neutralization breadth and potency
The cryo‐electron microscopy (cryo‐EM) structure of the SARS‐CoV‐2 S trimer complexed with XG014 (PDB ID: 7V2A) revealed that XG014 recognized an RBD epitope outside the ACE2‐binding site and locked three RBDs in the nonfunctional “closed” (“down”) conformation (Figure S5A), and also explained why XG014 exhibited strong resistance against escape mutations, such as N501Y in B.1.1.7, K417N/E484K in B.1.351 and P.1, and L452R in B.1.617.2 (Figure S5B). 23
Although XG051, XG052, XG069, and XG070 shared a high sequence similarity with XG014, with only a few amino acid residues altered in both immunoglobulin heavy and light chains (Figure S2B and S2C), their neutralization potency and breadth against SARS‐CoV‐1 and SARS‐CoV‐2 variants were very distinct from each other (Figure 3). Therefore, we determined the key amino acid residues responsible for neutralization potency and breadth. To do this, we generated ten XG014 antibody variants, each with only a single amino acid substitution (Figure 5A). Seven of these substitutions were on the XG014 heavy chain and three on its light chain. These mutated residues were all located in the antigen‐antibody binding interface region based on the cryo‐EM structure (Figure 5B,C), and were substituted by the corresponding amino acid residues in the four newly cloned family members (Figure S2B and S2C). Neutralization assays against various pseudoviruses (Figure S6) showed that some amino acids, such as Q100H in heavy chain (Mut‐1) and S97A in light chain (Mut‐9), barely altered neutralizing activities; while several amino acids, G102R (Mut‐2), G102L (Mut‐3), and Y109H (Mut‐6) in heavy chain, and S26R (Mut‐7) in light chain, significantly reduced the neutralization breadth against SARS‐CoV‐1 (Figure 5D). Conversely, N31Y substitution in light chain (Mut‐8) significantly improved the neutralization potency against SARS‐CoV‐1 and the tested SARS‐CoV‐2 variants, but without alteration of neutralizing titer against wildtype SARS‐CoV‐2 (Figure 5D). Structural remodeling showed that N31Y substitution in XG014 light chain enhanced its interaction with N501Y mutation in S protein RBD, thus explaining the enhanced breadth and potency of neutralization (Figure 5C). Therefore, these results identified the key amino acid residues on XG014 essential for neutralizing breadth and potency, strengthening the significance of somatic hypermutation during antibody evolution for combating variant emergence.
Figure 5.
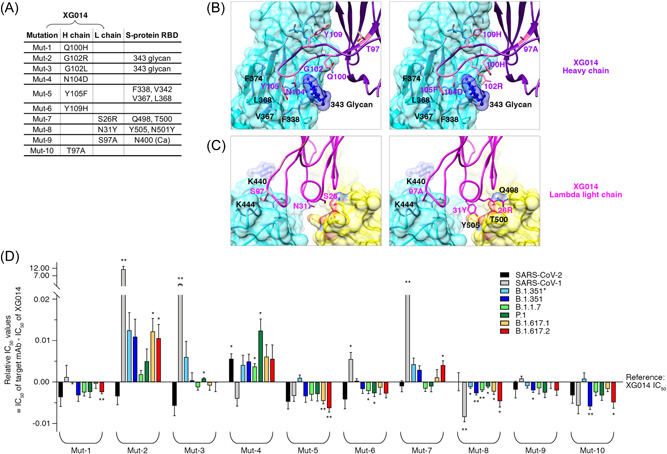
Key amino acid residues for XG014 neutralization breadth and potency. (A) Ten XG014 antibody variants, named from Mut‐1 to Mut‐10, with only a single amino acid substitution on either XG014 heavy or lambda light chains. The interacting amino acid residues on the RBD of SARS‐CoV‐2 S protein were indicated. (B–C) Key interactions between XG014 heavy/lambda light chain and SARS‐CoV‐2 RBD before (left) and after (right) amino acid substitutions. For model building, the atomic model of SARS‐CoV‐2 S‐XG014 (PDB ID 7V2A) was used. (D) The relative IC50 values of the ten XG014 antibody variants against SARS‐CoV‐1, SARS‐CoV‐2 and its variants. Statistical analysis was performed using the Wilcoxon rank‐sum test, and the p values are indicated by stars, *p < 0.05; **p < 0.01.
3.7. Striking neutralization evasion by SARS‐CoV‐2 Omicron sublineages
The recently emerged B.1.1.529 lineage (labeled as “Omicron”) variant and its sublineages, such as BA.1, BA.2, BA.2.12.1, BA.3, BA.4, and BA.5, had more than 30 amino acid substitutions in their S protein, with around half of these substitutions located in the RBD region. Numerous studies have shown that Omicron sublineages caused significant humoral immune evasion, thus posing a remarkable challenge for the effectiveness of SARS‐CoV‐2 vaccines and anti‐SARS‐CoV‐2 mAbs. 6 , 33 , 34 , 35
To further evaluate the neutralizing activity of our cloned cross‐neutralizing mAbs, we constructed several more types of pseudoviruses, including B.1.1.529 (Omicron), BA.1, BA.2, BA.2.12.1, BA.3, BA.4/5, and performed in vitro neutralization assays (Figure 6A–G; Figure S7). For the XG014 family members (XG051, XG052, XG069, XG070, and XG014), all mAbs lost or drastically reduced their neutralizing activities against most of the tested Omicron subvariants, B.1.1.529, BA.1, BA.2, BA.2.12.1, BA.4/5, but maintained their potency against BA.3 (Figure 6A,B and 6E–G). Although XG014‐N31Y (Mut‐8) mAb showed significantly improved neutralization potency against SARS‐CoV‐2 VOCs (Figure 5D), no enhanced neutralization was observed against Omicron subvariants for XG014‐N31Y (Mut‐8) (Figure 6H). Since amino acid residues, G339, S371, S373, and N440 on the RBD were involved in the major interactions with the XG014 family members (Figure S8A), the potent neutralizing activity against BA.3 might be explained by the lack of several escape mutations, such as G339D, S371L/F, S373P, and N440K, on the RBD of BA.3S protein (Figure S8B). For XG069, XG070, and XG014, their neutralizing activity against BA.1 remained detected (Figure 6E–G), possibly due to the lack of escape mutation N440K on the BA.1S protein (Figure S8A and S8B). Moreover, for XG054, although its neutralizing activities against BA.1 and BA.2 were improved, other SARS‐CoV‐2 Omicron sublineages strongly evaded its neutralization (Figure 6C). Further, for cross‐neutralizing mAb XG065, the neutralizing activity against Omicron variant and its sublineages generally declined, compared with its neutralizing activity against wildtype SARS‐CoV‐2 and SARS‐CoV‐1 (Figure 6D). To sum up, the neutralizing activities of our cloned SARS‐CoV‐1‐cross‐neutralizing mAbs decreased dramatically against the newly emerged SARS‐CoV‐2 Omicron and its sublineages.
Figure 6.
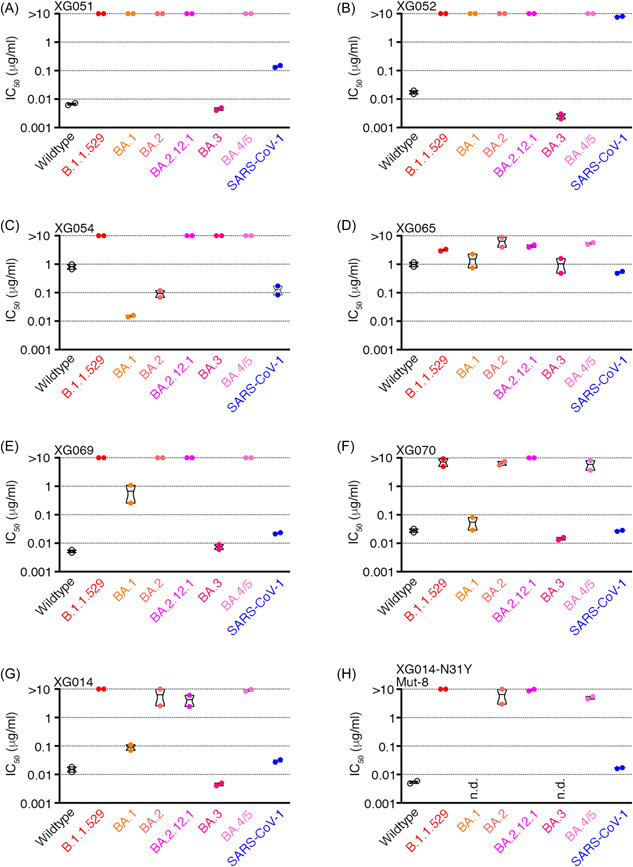
Dramatically reduced neutralizing activity against SARS‐CoV‐2 Omicron and its sublineages for SARS‐CoV‐1‐cross reactive mAbs. (A–G) IC50 values of seven cross‐neutralizing mAbs, XG051 (A), XG052 (B), XG054 (C), XG065 (D), XG069 (E), XG070 (F), XG014 (G), and XG014‐N31Y (Mut‐8) (H), against SARS‐CoV‐1, SARS‐CoV‐2 and its variants. n.d., not determined.
3.8. Negative correlation between SARS‐CoV‐1 and Omicron neutralization
In our study, the seven mAbs with cross‐neutralizing activity against SARS‐CoV‐1 could broadly neutralize SARS‐CoV‐2 VOCs, including Alpha, Beta, Gamma, and Delta, but failed to neutralize SARS‐CoV‐2 Omicron. Thus, these results led us to further investigate whether other reported SARS‐CoV‐1‐cross‐neutralizing mAbs also exhibited reduced neutralizing activity against Omicron variant. To do this, we collected a series of mAbs neutralizing both SARS‐CoV‐1 and SARS‐CoV‐2 from previous literatures (Figure 7A). These mAbs showed broadly neutralizing activity against SARS‐CoV‐1 (IC50 values lower than 0.1 µg/ml) and SARS‐CoV‐2 VOCs, but significantly reduced neutralizing potency against Omicron (Figure 7A,B).
Figure 7.
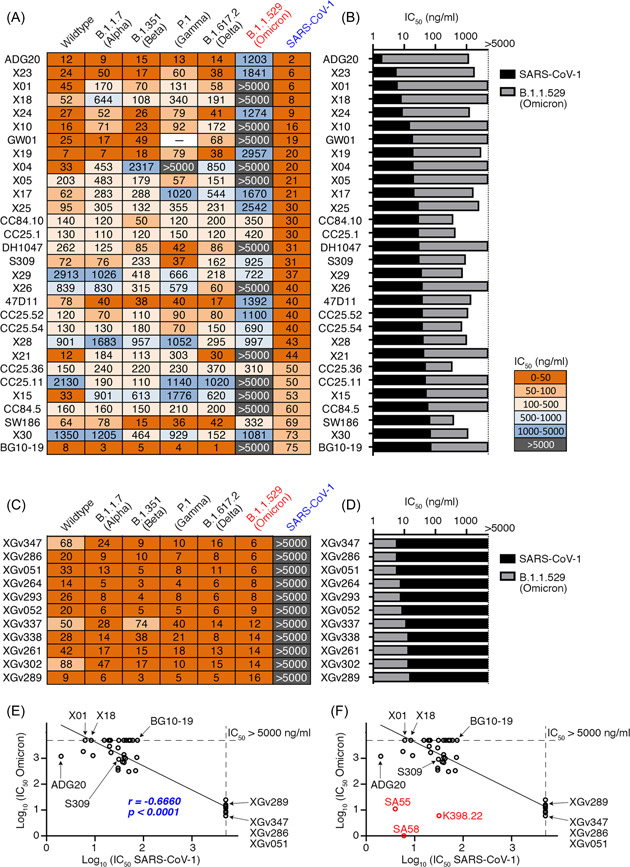
Negative correlation between neutralizing capacity against SARS‐CoV‐1 and Omicron. (A) IC50 values of a series of reported anti‐SARS‐CoV‐2 bNAbs with cross‐neutralizing activity against SARS‐CoV‐1. Most of these IC50 values were collected from previous literatures: ADG20, S309, and DH1047 36 ; X mAbs 37 ; CC mAbs 38 ; GW01 39 ; and SW186. 40 The IC50 values for 47D11 41 and BG10‐19 42 were evaluated in this study. (B) IC50 values of mAbs listed in (A) against SARS‐CoV‐1 and B.1.1.529 (Omicron) are shown side by side in columns. (C) IC50 values of a series of XGv anti‐SARS‐CoV‐2 bNAbs we cloned from three‐dose vaccinees. 43 These XGv mAbs exhibited potent neutralizing activity against all known SARS‐CoV‐2 VOCs, including Omicron, with their IC50 values against SARS‐CoV‐2 and its VOCs collected from a previous literature and their IC50 values against SARS‐CoV‐1 measured in this study. (D) IC50 values of mAbs listed in (C) against SARS‐CoV‐1 and B.1.1.529 (Omicron) are shown side by side in columns. (E) Dot plot showing the negative correlation between neutralizing activity against SARS‐CoV‐1 (x‐axis) and Omicron (y‐axis). Spearman's rank correlation coefficient (r) and significance value (p). (F) Three mAbs with potent neutralizing activities against both SARS‐CoV‐1 and Omicron variant. Neutralization IC50 values against SARS‐CoV‐1 (x‐axis) and Omicron (y‐axis). Compared with Figure 7E, RED dots representing mAbs K398.22, SA55, and SA58 44 , 45 were added.
For example, S309, an antibody isolated from a SARS‐CoV‐1 recovered individual, 46 efficiently neutralized SARS‐CoV‐1 with IC50 of 31 ng/ml, while much weaker neutralization against Omicron (IC50 of 925 ng/ml) was observed. Similarly, mAb 47D11 targeting a comparable epitope of S309 also neutralized SARS‐CoV‐2 variants, 41 with IC50 values against SARS‐CoV‐1 and Omicron of 40 and 1392 ng/ml, respectively. BG10‐19, a mAb recently cloned from a SARS‐CoV‐2 convalescent donor, potently neutralized SARS‐CoV‐2 VOCs B.1.1.7 (Alpha), B.1.351 (Beta), and SARS‐CoV‐1 (IC50 of 75 ng/ml) by locking the S‐trimer in a closed conformation, 42 but failed to neutralize Omicron in our assay (IC50 >5000 ng/ml). Together, most anti‐SARS‐CoV‐2 mAbs with potent SARS‐CoV‐1 cross‐neutralizing activity showed significantly reduced neutralization against SARS‐CoV‐2 Omicron variant.
XGv mAbs were previously isolated from 3‐dose vaccinees (CoronaVac, a β‐propiolactone‐inactivated SARS‐CoV‐2 vaccine against COVID‐19), and some of these XGv mAbs efficiently neutralized wildtype SARS‐CoV‐2 and all its related VOCs, including B.1.1.529 (Omicron). 43 , 47 To test whether potent Omicron neutralizers exhibited neutralizing activity against SARS‐CoV‐1, we selected 11 XGv antibodies with potent neutralizing activity against Omicron, and then tested their neutralization against SARS‐CoV‐1. None of them neutralized SARS‐CoV‐1 in our in vitro pseudovirus assays (Figure 7C,D). Further, correlation analysis showed that anti‐SARS‐CoV‐2 bNAbs exhibited a significant negative correlation between the IC50 values against SARS‐CoV‐1 and Omicron variant (Figure 7E).
Importantly, the potent bNAbs against SARS‐CoV‐1, SARS‐CoV‐2 VOCs, and SARS‐COV‐2 Omicron and its sublineages were indeed identified. For example, antibody K398.22 48 potently neutralized both SARS‐CoV‐1 and Omicron, with its IC50 values of 32 and 6 ng/ml, respectively (Figure S9). Moreover, by screening more than 1400 mAbs, broad‐spectrum neutralizing mAbs, SA58, and SA55, recognizing two nonoverlapping super‐conserved epitopes, were recently identified, with potent neutralizing activities against both SARS‐CoV‐1 (IC50 values: 4–6 ng/ml) and Omicron and its sublineages (IC50 values: 1–11 ng/ml) 44 (Figure 7F). Structural comparison showed that the key resides involved in K398.22, SA58, and SA55 are mostly conserved, explaining their broad neutralization activity (Figure S10). Especially, SA55 showed very potent and broadly neutralizing activity against the very newly emerging subvariants, such as BA.2.75, BJ.1, BU.1, and XBB, which showed the most significant level of immune evasion so far. 45
Taken together, we conclude that anti‐SARS‐CoV‐1/2 cross‐neutralizing mAbs could broadly neutralize SARS‐CoV‐2 VOCs, such as B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta) variants, but their neutralizing potency against Omicron and its sublineages dramatically declined. Therefore, our data emphasized the significance of cross‐neutralizing activity against SARS‐CoV‐1 and SARS‐CoV‐2 Omicron for the development of pan‐sarbecovirus vaccines and the selection of super‐antibodies with potent broad‐spectrum neutralizing activity.
4. DISCUSSION
The newly emerging SARS‐CoV‐2 variants with their numerous escape mutations have successfully evaded many mAbs. Here, in our study, we investigated the possibility of using the cross‐neutralizing activity against SARS‐CoV‐1 to identify bNAbs against SARS‐CoV‐2 and its variants. To do this, we used SARS‐CoV‐1 S1 protein as the bait protein for single B cell sorting, and identified 15 SARS‐CoV‐2 S protein‐binding antibodies. Among them, 12 antibodies showed cross‐binding affinity towards SARS‐CoV‐1 S protein (Figure 1G), thus suggesting our strategy to isolate cross‐reactive mAbs was very successful.
In our analysis, we have successfully identified an antibody family, with their complementarity‐determining region 3 (CDR3) sequences being highly similar with each other, suggesting that these five SARS‐CoV‐1‐cross‐reactive antibodies were derived from an expanded B cell clone. With highly similar amino acid sequences, but slightly distinct somatic mutations, these antibody family members exhibited different levels of neutralization potency and breadth. Our data suggested that the alteration of only one crucial amino acid residue in the Fab region could significantly reduce or increase neutralization potency or breadth. Since antibody evolution occurs by somatic mutation and selection in germinal centers, our results of this specific antibody family showed that the continued antibody clonal evolution boosts the protection against circulating SARS‐CoV‐2 variants.
SARS‐CoV‐2 Omicron variant showed powerful immune escape capacity, 49 , 50 even was proposed to be renamed as SARS‐CoV‐3 based on its serological profiles. 16 , 51 Our results by scrutinizing a series of anti‐SARS‐CoV‐2 bNAbs showed a negative correlation between SARS‐CoV‐1 and Omicron neutralization for most majority of anti‐SARS‐CoV‐2 bNAbs, further demonstrating the unique serological profile of SARS‐CoV‐2 Omicron variant and its sublineages. In other words, for SARS‐CoV‐1‐cross‐neutralizing mAbs, it seems that, structurally, their epitopes were more remarkably evaded by escape mutations in SARS‐CoV‐2 Omicron. On the other hand, for Omicron‐cross‐neutralizing mAbs, their binding capacities against SARS‐CoV‐1 S proteins were strikingly evaded by SARS‐CoV‐1 mutations, though their antigenic epitopes were highly conserved among SARS‐CoV‐2 VOCs, such as Alpha, Beta, Gamma, and Delta. This negative correlation of SARS‐CoV‐1/Omicron neutralization implied that the shared antigenic epitopes between SARS‐CoV‐1 and Omicron S protein were very rare, suggesting the extreme rarity of bNAbs with potent neutralization against both SARS‐CoV‐1 and SARS‐CoV‐2 Omicron variant.
In summary, our cloned SARS‐CoV‐1/2‐cross‐neutralizing mAbs showed broad neutralizing activity against SARS‐CoV‐2 VOCs of Alpha, Beta, Gamma and Delta, but failed to efficiently neutralize SARS‐CoV‐2 Omicron variant and its sublineages. Cryo‐EM data explained how escape mutations, such as G339D, S371L/F, S373P, and N440K, 52 in Omicron and its sublineages, but not in other VOCs, effectively evaded these cloned mAbs. Therefore, to simply use SARS‐CoV‐1 cross‐neutralization as a criterion is not suitable for identifying bNAbs with neutralizing activity against SARS‐CoV‐2 Omicron and its sublineages. In the future, to quickly and efficiently identify pan‐sarbecovirus therapeutic bNAbs and develop pan‐sarbecovirus vaccines, we should evaluate the bNAbs with cross‐neutralizing activity against SARS‐CoV‐1 and SARS‐CoV‐2 Omicron variant as well as its sublineages, and rationally design vaccine antigens containing epitopes with capacity to elicit the above bNAbs.
AUTHOR CONTRIBUTIONS
Qiao Wang, Rong Xia, Lei Sun, Shibo Jiang, Zhenguo Chen, Junqing Wang, and Yunjiao Zhou: designed the experiments. Shibo Li, Jianbo Wu, Weiyu Jiang, Haiyan He, Wei Wu, Yidan Gao, Yunjiao Zhou, Minxiang Xie, Anqi Xia, Jiaying He, Qianqian Zhang, Yuru Han, Nan Wang, and Kang Wang: performed experiments. Shibo Li, Haiyan He, Guangqi Zhu, Qiao Wang, Zheen Zhang, and Rong Xia: recruited volunteers for blood donation. Zhenguo Chen and Lei Sun: conducted Cryo‐EM analysis. Qiao Wang, Rong Xia, Lei Sun, Zhenguo Chen, Yunjiao Zhou, Jianbo Wu, Xiangxi Wang and Yidan Gao: analyzed the data. Qiao Wang: wrote the manuscript. Christian T. Mayer, Jianbo Wu, Shibo Jiang, and Lei Sun: participated in revision.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
We thank members of the Core Facility of Microbiology and Parasitology (SHMC) at Shanghai Medical College of Fudan University. This work was supported by National Key Research and Development Program (2021YFA1301400 to Q.W. and 2019YFA0904402 to S.J.), the National Natural Science Foundation of China (32070947 and 31872730 to Q.W. and 92169112 to S.J.), and the Ministry of Science and Technology of China (2021YFC2302500 to L.S.). Project was supported by Shanghai Municipal Science and Technology Major Project (ZD2021CY001 to Q.W. and S.J.) and by Guangzhou Laboratory (SRPG22‐003 to L.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies or individuals.
Li S, Wu J, Jiang W, et al. Characterization of cross‐reactive monoclonal antibodies against SARS‐CoV‐1 and SARS‐CoV‐2: implication for rational design and development of pan‐sarbecovirus vaccines and neutralizing antibodies. J Med Virol. 2023;95:e28440. 10.1002/jmv.28440
Shibo Li, Jianbo Wu, Weiyu Jiang, Haiyan He, Yunjiao Zhou, Wei Wu, Yidan Gao, and Minxiang Xie contributed equally.
Contributor Information
Yunjiao Zhou, Email: centrallab5th@163.com.
Xiangxi Wang, Email: Xiangxi@ibp.ac.cn.
Junqing Wang, Email: wangjunqingmd@hotmail.com.
Zhenguo Chen, Email: zhenguochen@fudan.edu.cn.
Shibo Jiang, Email: shibojiang@fudan.edu.cn.
Lei Sun, Email: llsun@fudan.edu.cn.
Rong Xia, Email: xiarongcn@126.com.
Qiao Wang, Email: wangqiao@fudan.edu.cn.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Gao SJ, Guo H, Luo G. Omicron variant (B.1.1.529) of SARS‐CoV‐2, a global urgent public health alert! J Med Virol. 2022;94:1255‐1256. 10.1002/jmv.27491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou Y, Zhi H, Teng Y. The outbreak of SARS‐CoV‐2 Omicron lineages, immune escape, and vaccine effectivity. J Med Virol. 2022:95. 10.1002/jmv.28138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desingu PA, Nagarajan K. The emergence of Omicron lineages BA.4 and BA.5, and the global spreading trend. J Med Virol. 2022;94:5077‐5079. 10.1002/jmv.27967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Araf Y, Akter F, Tang Y, et al. Omicron variant of SARS‐CoV‐2: genomics, transmissibility, and responses to current COVID‐19 vaccines. J Med Virol. 2022;94:1825‐1832. 10.1002/jmv.27588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xia S, Wang L, Zhu Y, Lu L, Jiang S. Origin, virological features, immune evasion and intervention of SARS‐CoV‐2 Omicron sublineages. Signal Transduct Target Ther. 2022;7:241. 10.1038/s41392-022-01105-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS‐CoV‐2 neutralizing antibodies. Nature. 2022;602:657‐663. 10.1038/s41586-021-04385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell. 2020;182:812‐827. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Komurcu SZM, Artik Y, Cesur NP, et al. The evaluation of potential global impact of the N501Y mutation in SARS‐COV‐2 positive patients. J Med Virol. 2022;94:1009‐1019. 10.1002/jmv.27413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y, Zhao X, Zhou H, Zhu H, Jiang S, Wang P. Broadly neutralizing antibodies to SARS‐CoV‐2 and other human coronaviruses. Nat Rev Immunol. 2022. 10.1038/s41577-022-00784-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu Y, Yang X, Xun J, et al. Neutralization of five SARS‐CoV‐2 variants of concern by convalescent and BBIBP‐CorV vaccinee serum. Virologica Sinica. 2022;37:831‐841. 10.1016/j.virs.2022.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao Y, Jian F, Wang J, et al. Imprinted SARS‐CoV‐2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2022. 10.1038/s41586-022-05644-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martinez DR, Schäfer A, Leist SR, et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991‐998. 10.1126/science.abi4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen AA, Gnanapragasam PNP, Lee YE, et al. Mosaic nanoparticles elicit cross‐reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735‐741. 10.1126/science.abf6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walls AC, Miranda MC, Pham MN, et al. Elicitation of broadly protective sarbecovirus immunity by receptor‐binding domain nanoparticle vaccines. bioRxiv. 2021. 10.1101/2021.03.15.435528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan CW, Chia WN, Young BE, et al. Pan‐Sarbecovirus neutralizing antibodies in BNT162b2‐Immunized SARS‐CoV‐1 survivors. N Engl J Med. 2021;385:1401‐1406. 10.1056/NEJMoa2108453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vogel G. New subvariants are masters of immune evasion. Science. 2022;376:679‐680. 10.1126/science.adc9448 [DOI] [PubMed] [Google Scholar]
- 17. Zhou Y, Liu Z, Li S, et al. Enhancement versus neutralization by SARS‐CoV‐2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep. 2021;34:108699. 10.1016/j.celrep.2021.108699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, Wang L, Cao H, Liu C. SARS‐CoV‐2 S1 is superior to the RBD as a COVID‐19 subunit vaccine antigen. J Med Virol. 2021;93:892‐898. 10.1002/jmv.26320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou Y, Liu Z, Wang Z, et al. Single‐cell sorting of HBsAg‐Binding memory B cells from human peripheral blood mononuclear cells and antibody cloning. STAR Protocols. 2020;1:100129. 10.1016/j.xpro.2020.100129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brochet X, Lefranc MP, Giudicelli V. IMGT/V‐QUEST: the highly customized and integrated system for IG and TR standardized V‐J and V‐D‐J sequence analysis. Nucleic Acids Res. 2008;36:W503‐W508. 10.1093/nar/gkn316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34‐W40. 10.1093/nar/gkt382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang L, Fu W, Bao L, et al. Selection and structural bases of potent broadly neutralizing antibodies from 3‐dose vaccinees that are highly effective against diverse SARS‐CoV‐2 variants, including Omicron sublineages. Cell Res. 2022;32:691‐694. 10.1038/s41422-022-00677-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Z, Xu W, Chen Z, et al. An ultrapotent pan‐β‐coronavirus lineage B (β‐CoV‐B) neutralizing antibody locks the receptor‐binding domain in closed conformation by targeting its conserved epitope. Protein Cell. 2021;13:655‐675. 10.1007/s13238-021-00871-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Waterhouse A, Bertoni M, Bienert S, et al. SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296‐W303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605‐1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 26. Wang Q, Michailidis E, Yu Y, et al. A combination of human broadly neutralizing antibodies against hepatitis B virus HBsAg with distinct epitopes suppresses escape mutations. Cell Host Microbe. 2020;28:335‐349. 10.1016/j.chom.2020.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. ter Meulen J, van den Brink EN, Poon LLM, et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. 10.1371/journal.pmed.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huo J, Zhao Y, Ren J, et al. Neutralization of SARS‐CoV‐2 by destruction of the prefusion spike. Cell Host Microbe. 2020;28:445‐454. 10.1016/j.chom.2020.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yuan M, Huang D, Lee CCD, et al. Structural and functional ramifications of antigenic drift in recent SARS‐CoV‐2 variants. Science. 2021;373:818‐823. 10.1126/science.abh1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS‐CoV‐2 and SARS‐CoV. Science. 2020;368:630‐633. 10.1126/science.abb7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang N, Sun Y, Feng R, et al. Structure‐based development of human antibody cocktails against SARS‐CoV‐2. Cell Res. 2021;31:101‐103. 10.1038/s41422-020-00446-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382‐385. 10.1080/22221751.2020.1729069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS‐CoV‐2. Nature. 2022;602:676‐681. 10.1038/s41586-021-04388-0 [DOI] [PubMed] [Google Scholar]
- 34. Iketani S, Liu L, Guo Y, et al. Antibody evasion properties of SARS‐CoV‐2 Omicron sublineages. Nature. 2022;604:553‐556. 10.1038/s41586-022-04594-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS‐CoV‐2 Omicron to antibody neutralization. Nature. 2022;602:671‐675. 10.1038/s41586-021-04389-z [DOI] [PubMed] [Google Scholar]
- 36. Yuan M, Zhu X, He W, et al. A broad and potent neutralization epitope in SARS‐related coronaviruses. PNAS. 2022;119:e2205784119. 10.1073/pnas.2205784119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiong H, Sun H, Wang S, et al. The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS‐CoV and SARS‐CoV‐2. Proceedings of the National Academy of Sciences. 2022;119:e2204256119. 10.1073/pnas.2204256119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He W, Musharrafieh R, Song G, et al. Targeted isolation of diverse human protective broadly neutralizing antibodies against SARS‐like viruses. Nature Immunol. 2022;23:960‐970. 10.1038/s41590-022-01222-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y, Liu M, Shen Y, et al. Novel sarbecovirus bispecific neutralizing antibodies with exceptional breadth and potency against currently circulating SARS‐CoV‐2 variants and sarbecoviruses. Cell Discov. 2022;8:36. 10.1038/s41421-022-00401-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fang Y, Sun P, Xie X, et al. An antibody that neutralizes SARS‐CoV‐1 and SARS‐CoV‐2 by binding to a conserved spike epitope outside the receptor binding motif. Science Immunology. 2022;7:eabp9962. 10.1126/sciimmunol.abp9962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fedry J, Hurdiss DL, Wang C, et al. Structural insights into the cross‐neutralization of SARS‐CoV and SARS‐CoV‐2 by the human monoclonal antibody 47D11. Sci Adv. 2021:7. 10.1126/sciadv.abf5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scheid JF, Barnes CO, Eraslan B, et al. B cell genomics behind cross‐neutralization of SARS‐CoV‐2 variants and SARS‐CoV. Cell. 2021;184:3205‐3221. 10.1016/j.cell.2021.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang K, Jia Z, Bao L, et al. Memory B cell repertoire from triple vaccinees against diverse SARS‐CoV‐2 variants. Nature. 2022;603:919‐925. 10.1038/s41586-022-04466-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cao Y, Jian F, Zhang Z, et al. Rational identification of potent and broad sarbecovirus‐neutralizing antibody cocktails from SARS convalescents. Cell Rep. 2022;41:111845. 10.1016/j.celrep.2022.111845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cao Y, et al. Imprinted SARS‐CoV‐2 humoral immunity induces convergent Omicron RBD evolution. bioRxiv. 2022. 2022.2009.2015.507787. 10.1101/2022.09.15.507787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pinto D, Park YJ, Beltramello M, et al. Cross‐neutralization of SARS‐CoV‐2 by a human monoclonal SARS‐CoV antibody. Nature. 2020;583:290‐295. 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 47. Wang L, Fu W, Bao L, et al. Selection and structural bases of potent broadly neutralizing antibodies from 3‐dose vaccinees that are highly effective against diverse SARS‐CoV‐2 variants, including Omicron sublineages. Cell Res. 2022;32:691‐694. 10.1038/s41422-022-00677-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He W, Yuan M, Callaghan S, et al. Broadly neutralizing antibodies to SARS‐related viruses can be readily induced in rhesus macaques. Sci Transl Med. 2022;14:eabl9605. 10.1126/scitranslmed.abl9605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tian D, Sun Y, Xu H, Ye Q. The emergence and epidemic characteristics of the highly mutated SARS‐CoV‐2 Omicron variant. J Med Virol. 2022;94:2376‐2383. 10.1002/jmv.27643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeng G, Wang X. Ending the COVID‐19 pandemic: we still have a long way to go. J Med Virol. 2022;94:5075‐5076. 10.1002/jmv.27980 [DOI] [PubMed] [Google Scholar]
- 51. Qi Z, Wu J, Jiang S. Renaming the SARS‐CoV‐2 Omicron sublineages as SARS‐CoV‐3 is contrary to nomenclature standards based on evolutionary and serological evidence. Clin Transl Med. 2022;12:e924. 10.1002/ctm2.924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of SARS‐CoV‐2 variants and their mutational immune escape. J Med Virol. 2022;94:847‐857. 10.1002/jmv.27376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lv Z, Deng YQ, Ye Q, et al. Structural basis for neutralization of SARS‐CoV‐2 and SARS‐CoV by a potent therapeutic antibody. Science. 2020;369:1505‐1509. 10.1126/science.abc5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article The data that support the findings of this study are available from the corresponding author upon reasonable request.


